Abstract
Recent studies have shown that microRNAs and long noncoding RNAs (lncRNAs) regulate the expression of drug metabolizing enzymes (DMEs) in human hepatic cells and that a set of DMEs, including UDP glucuronosyltransferase (UGT) 2B15, is down-regulated dramatically in liver cells by toxic acetaminophen (APAP) concentrations. In this study we analyzed mRNA, microRNA, and lncRNA expression profiles in APAP-treated HepaRG cells to explore noncoding RNA-dependent regulation of DME expression. The expression of UGT2B15 and lncRNA LINC00574 was decreased in APAP-treated HepaRG cells. UGT2B15 levels were diminished by LINC00574 suppression using antisense oligonucleotides or small interfering RNA. Furthermore, we found that hsa-miR-129-5p suppressed LINC00574 and decreased UGT2B15 expression via LINC00574 in HepaRG cells. In conclusion, our results indicate that LINC00574 acts as an important regulator of UGT2B15 expression in human hepatic cells, providing experimental evidence and new clues to understand the role of cross-talk between noncoding RNAs.
SIGNIFICANCE STATEMENT
We showed a molecular network that displays the cross-talk and consequences among mRNA, micro RNA, long noncoding RNA, and proteins in acetaminophen (APAP)-treated HepaRG cells. APAP treatment increased the level of hsa-miR-129-5p and decreased that of LINC00574, ultimately decreasing the production of UDP glucuronosyltransferase (UGT) 2B15. The proposed regulatory network suppresses UGT2B15 expression through interaction of hsa-miR-129-5p and LINC00574, which may be achieved potentially by recruiting RNA binding proteins.
Introduction
Drug metabolizing enzymes (DMEs) play crucial roles in drug metabolism and toxicity in the liver. For example, cytochrome P450 (P450) enzymes in hepatic cells are essential in acetaminophen (APAP) activation, whereas UDP glucuronosyltransferases (UGTs) and sulfotransferases are key DMEs for APAP elimination (McGill and Jaeschke, 2013). The expression levels of DMEs are influenced by both genetic polymorphism and epigenetic factors (Sheweita, 2000). For instance, decreased APAP glucuronide formation and increased protein adduct levels were found in individuals with the UGT2B15 *2/*2 genotype compared with those with *1/*2 or *1/*1 genotypes (Court et al., 2017). Conversely, Court et al. (2013) reported enhanced APAP glucuronidation in individuals bearing the UGT1A1 c.2042C>G (rs8330) allele and decreased risk for unintentional APAP-induced acute liver failure.
Increasing evidence has illuminated the importance of epigenetic factors, including DNA methylation, histone modification, and noncoding RNAs, in regulating DME expression (Zhong and Leeder, 2013; Peng and Zhong, 2015; Li et al., 2019c). MicroRNAs (miRNAs) are small noncoding RNAs (∼22 nucleotides long) that are important epigenetic regulators affecting posttranscriptional gene expression. Many DMEs, such as CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, multidrug resistance-associated protein 1, sulfotransferase (SULT) 1A1, SULT2A1, and aldehyde dehydrogenase 5 family, member A1, have been identified as the targets of miRNAs in hepatic cells (Yu et al., 2015a,b,c; Jin et al., 2016; Chen et al., 2017; Wang et al., 2017; Zeng et al., 2017; Li et al., 2019a). In addition to direct modulation of DME expression by miRNAs, our previous work demonstrated that miRNAs can regulate DME expression indirectly by targeting the expression of nuclear receptors (NRs), including NR1I2 (also known as pregnane X receptor), hepatocyte nuclear factor (HNF) 1A, and HNF4A, in an APAP-induced hepatotoxicity model (Yu et al., 2018). In recent years the regulatory roles of miRNAs and long noncoding RNAs (lncRNAs) have been recognized in the modulation of DME expression at the transcriptional and translational levels (Li et al., 2019b). For example, lncRNA HNF1α-AS1 was reported to regulate constitutive androstane receptor and pregnane X receptor directly and P450 enzymes indirectly (Wang et al., 2019b). However, no regulatory function of lncRNAs in coordination of miRNAs to modulate DME expression have been reported yet.
Although only a small proportion of lncRNAs have been functionally annotated, accumulating evidence suggests that interactions between lncRNAs and RNA binding protein (RBPs) (RNA-protein interactions) and interactions between lncRNAs and miRNA molecules (RNA-RNA interactions) form complicated networks that achieve fine levels of control over gene expression. Most lncRNAs interact with one or more RBPs to accomplish their regulatory functions, and these lncRNA and RBP interactions are important topics of research (Ferrè et al., 2016). In addition, lncRNAs may bind to miRNAs via complementary base pairing and function as decoys to sequester miRNAs sacrificially, blocking miRNA-dependent posttranscriptional degradation of other mRNA transcripts (Yamamura et al., 2018).
Our previous study reported expression profiles for protein-coding genes and miRNAs in HepaRG cells exposed to APAP and identified 2758 genes and 47 miRNAs significantly deregulated upon APAP exposure (Yu et al., 2018). Intriguingly, nine genes encoding pivotal enzymes involved in APAP metabolism were significantly down-regulated after the treatment of APAP at a toxic concentration of 10 mM. The down-regulated genes included UGT1A1 and UGT2B15 that encode UDP glucuronosyltransferases, SULT1A1 and SULT2A1 that encode sulfotransferases, CYP2A6, CYP2E1, and CYP3A4 that encode cytochrome P450 enzymes, and glutathione S-transferase (GST) M1 and GSTT1 that encode glutathione S-transferases. This phenomenon suggested that DME expression in liver cells could be “shut down” upon exposure to a high concentration of APAP (Thorgeirsson et al., 1976; Snawder et al., 1994). However, this “shut-down” phenomenon also suppresses DMEs important for APAP detoxification, including UGT2B15 and SULT2A1, making the balance between self-protection and toxicity more complicated (Yu et al., 2018).
In this study we analyzed the lncRNA expression profile in HepaRG cells treated with APAP using RNA sequencing (RNA-seq) data described in our former report to identify lncRNA candidates that are potentially associated with DME modulation. We found that LINC00574, a liver-enriched lncRNA with a stable secondary structure, was significantly down-regulated in APAP-treated HepaRG cells. More importantly, LINC00574 levels exhibited a significant positive correlation with UGT2B15 expression liver tissues, suggesting its potential regulatory role in APAP metabolism. We then further investigated its biologic significance in DME regulation through in vitro and in vivo analyses. We demonstrated that LINC00574 regulated the expression of UGT2B15 in HepaRG cells; in silico evidence suggested that this regulation may involve interactions of LINC00574 with potential mRNA splicing regulators. We also discovered that the level of the miRNA hsa-miR-129-5p increased in HepaRG cells after APAP exposure and that hsa-miR-129-5p suppressed UGT2B15 expression via LINC00574.
Materials and Methods
Cell Culture.
The human hepatoma cell line HepG2, obtained from the American Type Culture Collection (Manassas, VA), was used in luciferase reporter gene assays because of its higher transfection efficiency compared with HepaRG cells. HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS.
HepaRG cells were purchased from Life Technologies (Carlsbad, CA) and cultured according to the manufacturer’s instructions. Briefly, HepaRG cells were thawed, seeded in Williams’ E medium supplemented with the Thaw, Plate, and General Purpose Medium Supplement (Life Technologies) for 24 hours, and then maintained in Williams’ E medium supplemented with the Maintenance/Metabolism Medium Supplement (Life Technologies) for 7 days prior to experiments.
lncRNA Profiling.
RNA samples were extracted from APAP-treated or control HepaRG cells using miRNeasy Mini kits (Qiagen, Valencia, CA). Total RNAs were subjected to high-throughput RNA-seq using an Illumina HiSeq1500 system, and then the paired-end sequencing reads were mapped to Gencode (version 19) that was employed as the reference. To focus on lncRNA molecules we selected as lncRNA candidates in the current study any RNA species with a term of “LINCxxxxx” that were differentially expressed in cells treated with 5 or 10 mM APAP. In the volcano plots that display the expression of differentially expressed lncRNAs, thresholds were set at absolute log2-fold changes >1.0 or < −1.0 and P values <0.05.
RNA Extraction and Quantitative Real-Time PCR.
RNeasy Mini kits (Qiagen) were used to extract total RNAs from cell lines. The syntheses of the first-strand cDNAs from mRNAs and the lncRNA were achieved using a QuantiTect Reverse Transcription kit (Qiagen), whereas the cDNA for miRNA was reverse transcribed using an NCode microRNA First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Tewksbury, MA). The expression levels of candidate mRNAs and lncRNAs were determined as relative levels to the RNA expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), whereas the miRNA levels were calculated relative to the expression levels of U6 small nuclear RNA using a QuantiFast SYBR Green RT-PCR kit (Qiagen). All primers or oligonucleotides (sequences available in Table 1) were synthesized by Integrated DNA Technologies (Coraville, IA).
TABLE 1.
Sequences of oligonucleotides and primers
| Name | Sequence (5′—3′) |
|---|---|
| For quantitative real-time PCR | |
| LINC00574 forward | CGG AGA AGC TCC TCT TGT GA |
| LINC00574 reverse | GCC CAG TAC TCT CCT GGA TG |
| UGT2B15 forward | GAA AAT TCT CGA TAG ATG GAT ATA TGG TG |
| UGT2B15 reverse | AAC TGC ATC TTT ACA GAG CTT GTT ACT G |
| GAPDH forward | GAA ATC CCA TCA CCA TCT TCC AGG |
| GAPDH reverse | GAG CCC CAG CCT TCT CCA TG |
| hsa-miR-129-5p forward | CUU UUU GCG GUC UGG GCU UGC |
| miRNA Universal reverse | AAC GCT TCA CGA ATT TGC GT |
| U6 forward | CGC TTC GGC AGC ACA TAT AC |
| U6 reverse | AAA ATA TGG AAC GCT TCA CGA |
| For LINC00574 knockdown | |
| Negative control ASO | CGA CUA TAC GCG CAA UAU GG |
| LINC00574 ASO | GUG ACA GCA AAC TAC CGA AC |
| LINC00574 siRNA forward | GUC CCU CCU GCC UCA GUU UCU CAG A |
| LINC00574 siRNA reverse | UCU GAG AAA CUG AGG CAG GAG GGA CUG |
| For luciferase gene reporter assay | |
| LINC00574 PCR forward | GGG AAA GUT CTC CCA GCC TAG CCC TAC T |
| LINC00574 PCR reverse | GGA GAC AUA AGG GCG AGT AGC AAG AAC A |
| For FREMSA | |
| LINC00574 oligo | GGC AGA GCU CGG GGU UGC UGC GGA GC |
| hsa-miR-129-5p oligo | CUU UUU GCG GUC UGG GCU UGC |
| miRNA negative control oligo | AAC GCT TCA CGA ATT TGC GT |
Subcellular Fractionation.
Cytoplasmic and nuclear fractions of RNA molecules were extracted from HepaRG cells using SurePrep Nuclear and Cytoplasmic RNA Purification kits (Thermo Fisher Scientific), respectively. The subcellular localization of LINC00574 was detected by quantitative real-time polymerase chain reaction (PCR).
Transfection Assays in HepaRG Cells.
The small interfering RNA (siRNA) and antisense oligonucleotide (ASO), targeting LINC00574 located in the cytoplasmic and nuclear, respectively, were designed and synthesized by Integrated DNA Technologies, and transfected into HepaRG cells (both final concentration: 50 nM) using Lipofectamine 2000 (Life Technologies) to down-regulate LINC00574 production.
To test the regulatory role of hsa-miR-129-5p on the endogenous expression of UGT2B15, miRNA negative control, hsa-miR-129-5p mimic, miRNA inhibitor control, and hsa-miR-129-5p inhibitor (all final concentration: 50 nM) were transiently transfected separately into HepaRG cells using Lipofectamine 2000 reagent.
Cells were harvested 48 hours after transfection or chemical treatments to extract total RNA or protein for further experiments. Each assay was performed at least three times.
Luciferase Reporter Gene Assays.
The pGL3-CU vector that expresses firefly luciferase, modified based on pGL3-Control vector (Promega, Madison, WI), has been described in our previous reports (Yu et al., 2015a; Wang et al., 2017). The core RNA sequences of LINC00574 harboring the putative targeting sites for hsa-miR-129-5p were PCR amplified and ligated into linearized pGL3-CU following the Universal USER Cassette protocol (New England Biolabs, Beverly, MA). The resultant construct was sequenced to confirm its authenticity.
HepG2 cells were seeded into 96-well plates at a density of 1 × 105 cells per well, cultured for 24 hours to reach approximately 80% confluence, and then transfected with the constructed plasmid (100 ng/well) together with the pRL-SV40 Renilla plasmid (1 ng/well; Promega) and the hsa-miR-129-5p mimic or inhibitor (final concentration: 50 nM) using Lipofectamine 2000 reagent. A Dual Luciferase Reporter Assay kit (Promega) was used to measure the firefly and Renilla luciferase signals 24 hours after transfection. Three independent experiments were conducted in triplicate.
Fluorescence-Based RNA Electrophoretic Mobility Shift Assay.
The oligonucleotides for hsa-miR-129-5p were synthesized and 5′-modified using IRDye800 dye, whereas the cognate LINC00574 oligonucleotides were 5′-labeled using cy5.5 dye. Cold probes, i.e., unlabeled oligonucleotides for negative control and hsa-miR-129-5p, were used in the competition assays. To detect the proteins involved in the miRNA-lncRNA interaction, NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo Fisher Scientific) were used to extract cytoplasmic proteins from HepaRG cells.
The detailed protocol for fluorescence-based RNA electrophoretic mobility shift assay (FREMSA) has been introduced in Molecular Toxicology Protocols (Yu et al., 2020). Briefly, 200 fmol hsa-miR-129-5p and cognate LINC00574 oligonucleotides were added to the reaction buffer containing 10 mM HEPES buffer (pH 7.3), 0.5% glycerol, 20 mM KCl, and 10 mM MgCl2. The reaction solution was incubated for 20 minutes at room temperature, separated by 10% native PAGE at 4°C, and then mobility shifts were detected with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Fifty-fold molar excesses of cold oligonucleotides were used in competition assays. Antibodies against argonaute RNA-induced silencing complex catalytic component (Ago) 1, Ago2, Ago3, and Ago4 were obtained from Abcam (Cambridge, MA) and used in supershift assays.
Western Blotting.
Total proteins were extracted from cell lines using detergent lysis buffer (radioimmunoprecipitation assay buffer; Thermo Fisher Scientific). Antibodies against UGT2B15 and GAPDH were obtained from Abcam. Quantitative analyses based on the Odyssey CLx Infrared Imaging System for Western blotting were conducted.
In silico and Statistical Analyses.
RNAfold (Lorenz et al., 2011) (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) was used to predict secondary structures for LINC00574. starBase version 3.0 (Li et al., 2014) (http://starbase.sysu.edu.cn/) was used to analyze the heterogeneous nuclear ribonucleoproteins (hnRNPs) interacting with LINC00574. RNAhybrid (Krüger and Rehmsmeier, 2006) (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid) was used to measure the free energy of miRNA-mRNA duplexes. Pearson correlation analysis was used to calculate the correlation between LINC00574 and UGT2B15 RNA levels in liver tissue samples deposited in the data base of The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/). The differences between subgroups for luciferase signals, protein, or RNA levels in this study were tested using one-way ANOVA on ranks test, and a P value <0.05 was considered statistically significant.
Accession Codes.
RNA-seq and miRNA sequencing data have been deposited at the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) with the accession number SRP094716.
Results
Differentially Expressed lncRNAs and DMEs in HepaRG Cells Exposed to APAP.
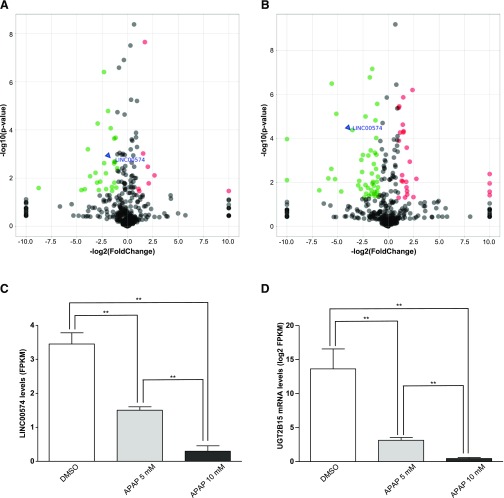
As reported previously (Yu et al., 2018), hepatic cell treatments with 5 or 10 mM APAP were considered moderately or severely toxic, respectively. Both concentrations were selected to screen the deregulated expression of lncRNAs. Using LINCxxxxx as a term to “define” lncRNAs in the study, a total of 558 lncRNAs were identified. The expression levels of lncRNAs are displayed in volcano plots. In the 5 mM APAP-treated cells, 8 lncRNAs were significantly up-regulated and 26 lncRNAs were significantly down-regulated (Fig. 1A), whereas 26 lncRNAs were significantly up-regulated and 47 lncRNAs were significantly down-regulated in cells treated with 10 mM APAP (Fig. 1B). LINC00574 was significantly down-regulated in both experiments (green node, labeled in blue). Specifically, LINC00574 levels were reduced by more than 3-fold with 5 mM APAP treatment and by more than 10-fold with 10 mM treatment (Fig. 1C). Similarly, together with other DMEs such as P450 enzymes and GSTs (Yu et al., 2018), the expression of UGT2B15 was down-regulated significantly in HepaRG cells treated with either 5 or 10 mM APAP (Fig. 1D). Thus, we selected LINC00574 for further investigation based on the magnitude of its down-regulation in response to APAP and because a positive correlation between the levels of a lncRNA species and its targeted mRNAs could be consistent with a novel lncRNA-dependent regulatory mechanism.
Fig. 1.
Deregulated lncRNAs in HepaRG cells exposed to APAP. (A and B) show the volcano plots in HepaRG cells treated with 5 and 10 mM APAP for 24 hours, respectively. The x-axis represents the log2-fold change, and the y-axis represents the P value (−log10 based). A P value <0.05 and absolute log2-fold change >1 were used as thresholds to differentiate significantly up- and down- expressed lncRNAs. Significantly up- and down- expressed LincRNAs are labeled in red and green colors, respectively. (C) displays the specific fold changes of LINC00574 in cells treated with 5 or 10 mM APAP. (D) shows the changes of the expression of UGT2B15 upon exposure to 5 or 10 mM APAP. **P < 0.01. FPKM, fragments per kilobase of transcript per million mapped reads.
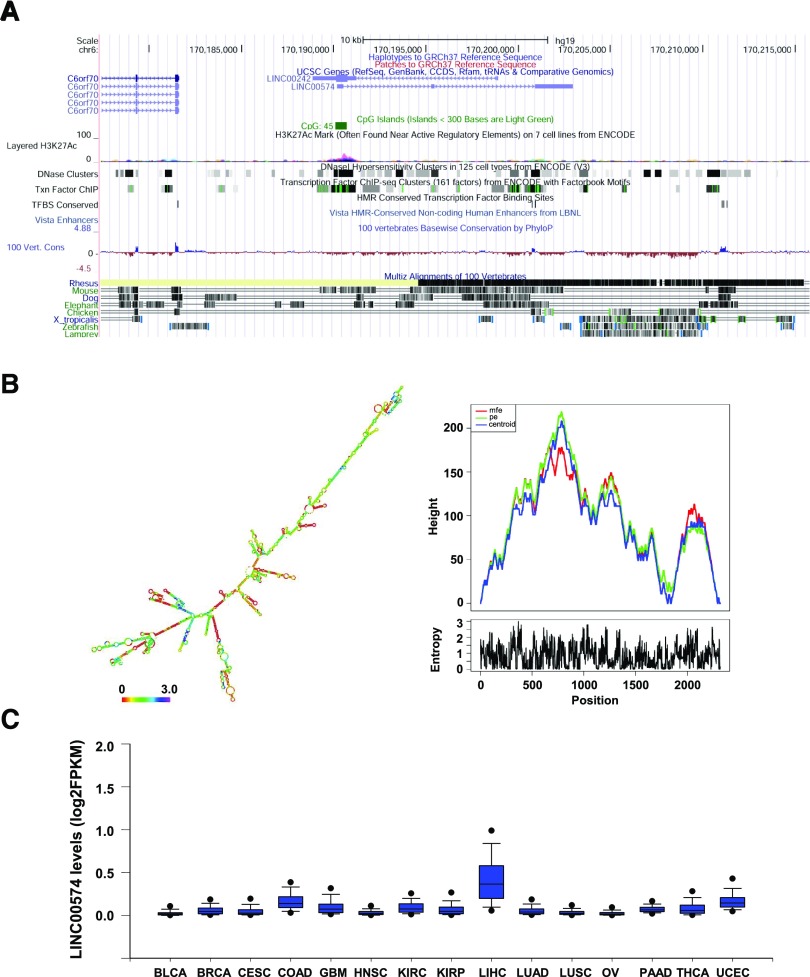
LINC00574 Is a Liver-Enriched lncRNA.
LINC00574 is 2566 nucleotides in length and is transcribed from an intergenic region on chromosome 6. Two CpG islands were observed around the exon 1 of LINC00574. A histone mark H3K27Ac with a peak signal adjacent to active regulatory elements was also identified in the gene promoter region, suggesting that the transcription of LINC00574 could be influenced by environmental stimuli. LINC00574 exhibited sequence conservation only in human and rhesus monkey, indicating its characteristics as a nonconserved lncRNA that may have obtained survival advantages from ancestors in the process of environmental adaptation (Fig. 2A). Structure prediction results showed that LINC00574 should adopt a stable secondary structure with a minimal free energy (MFE) of –1060.10 kcal/mol at 37°C, far above the stability threshold of RNA structures (MFE < –80 kcal/mol) (Mohammadin et al., 2015) (Fig. 2B, left panel). The centroid structure, MFE structure, and partition function structure of LINC00574 were in close agreement in the mountain plot (Fig. 2B, right panel), supporting the notion that the secondary structure of LINC00574 is very stable. The tissue distribution of LINC00574 was analyzed using the RNA-seq data from tumor and adjacent nontumor samples of 15 different types of tissues in TCGA data base, and the liver was identified as the most enriched organ for LINC00574 production (Fig. 2C). These data indicate that LINC00574 is a liver-enriched lncRNA with a stable secondary structure; as such, we speculated that it might have a functional role involved regulating metabolism in response to environmental challenges.
Fig. 2.
LINC00574 is a liver-specific lncRNA with a stable secondary structure. (A) Schematic representation of LINC00574 on chromosome 6 based on University of California Santa Cruz Genome Browser tracks showing CpG island information, peak signal of H3K27Ac, mammalian conservation. (B) Secondary structure (color indicates positional entropy) and mountain plot of LINC00574. A secondary structure of LINC00574 was predicted based on minimal free energy (–1060.10 kcal/mol, left panel). Right panel shows a mountain plot of minimum free energy structure, centroid structures, and partition function structure. The closely related status for these curves indicated the stable secondary structure of RNA molecule. The height (y-axis) of the mountain plot indicates the number of base pairs enclosing a sequence position indicated in the x-axis. pe indicates partition function structure; Centroid indicates centroid structure. (C) RNA levels (log2 fragments per kilobase of transcript per million mapped reads) of LINC00574 in 15 different types of tissues (tumors and adjacent normal tissues) from TCGA data base. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma.
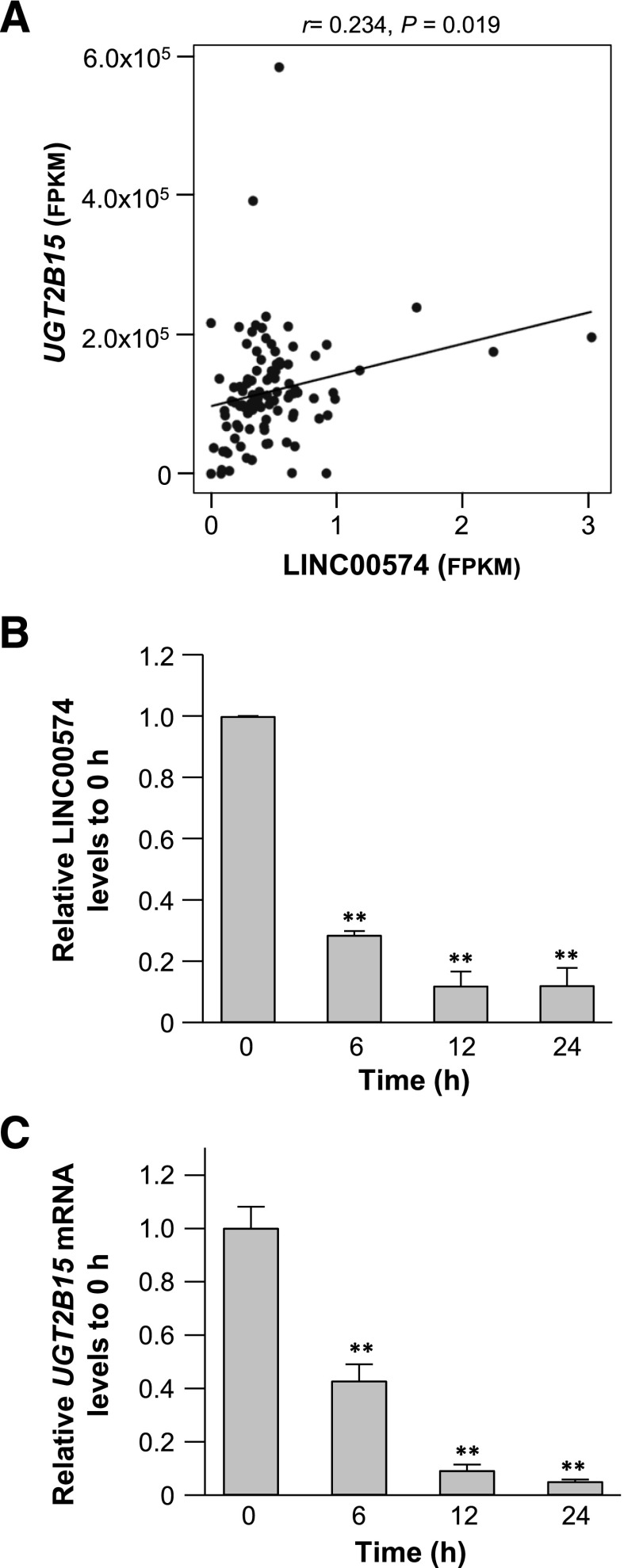
LINC00574 Levels in Liver Correlated with UGT2B15 mRNA Levels.
To investigate whether LINC00574 could be potentially involved in the cell responses to APAP exposure, we evaluated the intrinsic correlations between the expression of LINC00574 and the expression of nine key DMEs that are involved in the metabolism of APAP (Yu et al., 2018) based on their RNA levels in 49 pairs of hepatocellular carcinoma and adjacent normal tissues. As shown in Fig. 3A, the expression of LINC00574 exhibited a significantly positive correlation with UGT2B15 RNA levels (r = 0.234, P = 0.019). No correlations between LINC00574 and the other APAP metabolizing enzymes was observed (data not shown). Since we reported a time-dependent reduction of UGT2B15 after APAP exposure in our previous study (Yu et al., 2018), we further investigated the time course for the alteration of LINC00574 upon APAP treatment. Similar decreasing trends of LINC00574 and UGT2B15 levels were observed in HepaRG cells that were harvested at multiple time points after APAP treatment (Fig. 3, B and C). These results implicated a potential role for LINC00574 in the regulation of UGT2B15 in APAP metabolism.
Fig. 3.
LINC00574 level correlated with the expression UGT2B15. (A) Significantly positive correlation between the expression of LINC00574 and the expression of UGT2B15 in liver tissues (tumors and adjacent normal tissues) in TCGA data base. (B) and (C) Significant down-regulation of relative LINC00574 and UGT2B15 in HepaRG cells obtained at multiple time points after 10 mM APAP exposure. Each assay was carried out in triplicate. **P < 0.01. FPKM, fragments per kilobase of transcript per million mapped reads.
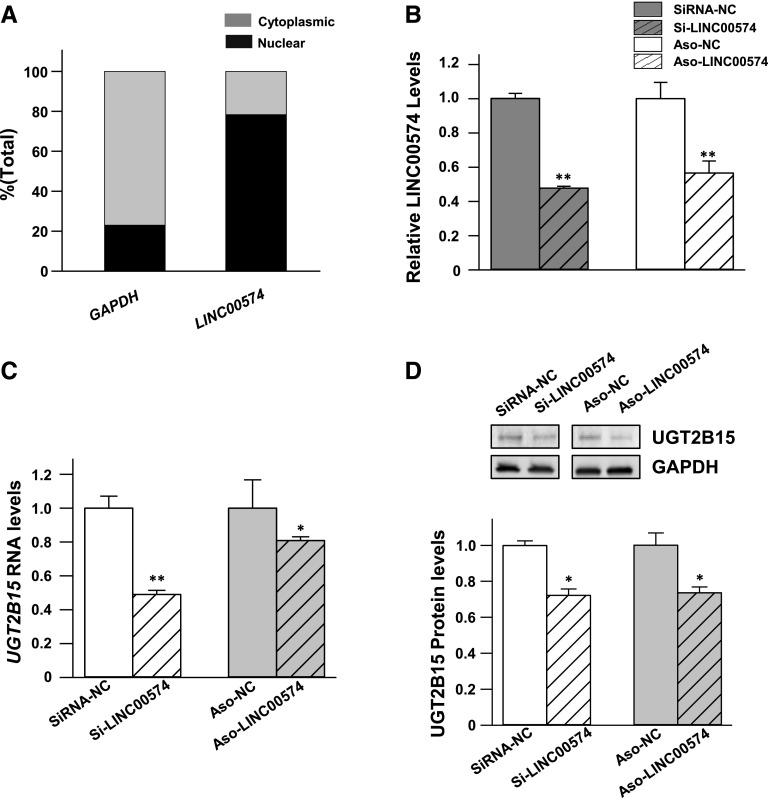
LINC00574 Modulated the Expression of UGT2B15.
Cellular localization analyses of LINC00574 showed that LINC00574 was significantly enriched in the nuclear fraction compared with the cytoplasmic fraction in HepaRG cells (Fig. 4A, 78% and 22%, respectively). Typically, ASOs are 15–25 base pair DNA sequences designed to bind and degrade complementary RNA present in the nuclear fraction, whereas commonly used siRNAs are theoretically more efficient for degrading cytoplasmic RNAs (Wheeler et al., 2012). Both ASO and siRNA species were designed in a sequence-specific manner and applied to knock down LINC00574. As shown in Fig. 4B, the expression levels of LINC00574 were significantly down-regulated by both ASO and siRNA in HepaRG cells (by 43% and 52%, respectively; both P < 0.001) compared with the cognate negative controls. The down-regulation of LINC00574 by ASO and siRNA reduced endogenous RNA and protein levels of UGT2B15 (by 19% and 51% for RNA and 23% and 22% for protein; all P < 0.05) (Fig. 4, C and D). These results indicate that LINC00574 regulates UGT2B15 at both mRNA and protein levels.
Fig. 4.
Knockdown of LINC00574 decreases UGT2B15 expression. (A) Enriched LINC00574 RNAs were observed in the nuclear fraction, compared with the cytoplasmic fraction. (B–D) Differentiated HepaRG cells were transfected with 20 nM siRNA, ASO, or their cognate controls. Both siRNA and ASO, designed to target LINC00574, decreased LINC00574 (B) and the expression of UGT2B15 mRNA (C) and protein (D) in HepaRG cells. Each assay was carried out in triplicate. *P < 0.05; **P < 0.01. ASO-LINC00574, antisense oligonucleotide (ASO) against LINC00574; ASO-NC, negative control ASO; si-LINC00574, small interfering RNA (siRNA) against LINC00574; siRNA-NC, negative control siRNA.
hsa-miR-129-5p Targeted LINC00574 RNA.
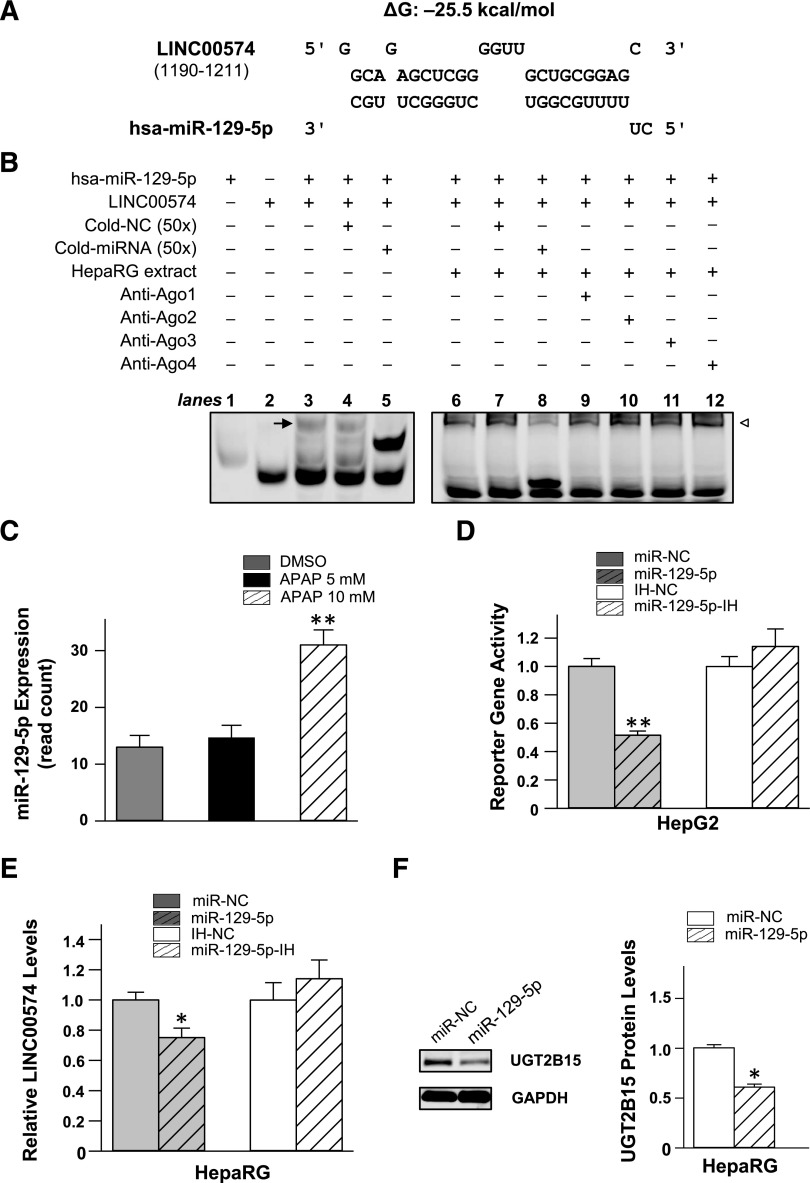
To investigate the potential RNA-RNA interactions between LINC00574 and miRNAs, we used RNAhybrid to analyze whether LINC00574 contains potential miRNA response elements (MREs) for the 47 miRNAs that were found to be up-regulated upon APAP exposure in our previous study (Yu et al., 2018). The miRNA hsa-miR-129-5p was predicted to interact with LINC00574 with an MFE of –25.5 kcal/mol, suggesting a high binding affinity (Fig. 5A). Subsequent FREMSA analyses provided experimental evidence of the binding between hsa-miR-129-5p and LINC00574 in vitro. As shown in Fig. 5B, hsa-miR-129-5p was able to form stable complexes together with the targeted sequences in LINC00574 (lane 3). A competition assay was conducted to evaluate the specificity of the interaction. The intensity of lncRNA-miRNA complexes displayed a significant decrease when excess unlabeled hsa-miR-129-5p was added (lane 5), in comparison with the negative control sample (lane 4), indicating sequence-specific interaction between hsa-miR-129-5p and LINC00574. Although the lncRNA-miRNA complexes also formed protein-RNA complexes when HepaRG cytoplasmic extracts were added (lane 6), no supershift band was observed when antibodies against Ago1, Ago2, Ago3, or Ago4 was added to the reaction system (lanes 9–12), indicating unknown proteins rather than Ago family members were recruited in the interaction between hsa-miR-129-5p and LINC00574. For practical reasons our FREMSA experiments were performed using a 26 nt LINC00574 fragment containing its hsa-miR-129-5p MRE and not the complete 2566 nt LINC00574 sequence. Therefore, supershifts would be unlikely using antibodies against RBPs expected to interact with intact LINC00574.
Fig. 5.
hsa-miR-129-5p modulates the expression of UGT2B15 by interacting with LINC00574. (A) Free energy analysis shows that hsa-miR-129-5p is able to target LINC00574 with an MFE of –25.2 kcal/mol. ΔG indicates Gibbs energy. (B) hsa-miR-129-5p oligonucleotides interact with LINC00574 in vitro. Lanes 1 and 2 indicate the mobility of hsa-miR-129-5p and LINC00574 oligonucleotides, respectively; lane 3 indicates the mobility shift of the miRNA-lncRNA; lanes 4 and 5 indicate the competition assays to diminish the complex formed by hsa-miR-129-5p and LINC00574 oligonucleotides using excess unlabeled nonspecific competitors (cold negative control, cold-NC) or hsa-miR-129-5p (cold-miRNA). Lane 6 indicates the RNA-protein complexes formed by cytoplasmic extracts from HepaRG cells together with hsa-miR-129-5p or LINC00574 oligonucleotides. Lanes 7 and 8 show the competition assays to diminish the RNA-protein complex. Lanes 9–12 indicate the mobility status of RNA-protein complex together with antibodies against Ago1, Ago2, Ago3, and Ago4, respectively. The arrow indicates the oligonucleotide complexes in lane 3. The hollow triangle indicates the RNA-protein complexes in lanes 6–12. (C) Differentially expressed miR129-5p upon treating cells with 5 or 10 mM APAP. (D) The luciferase reporter gene activity in HepG2 cells was suppressed by transfection with hsa-miR-129-5p mimics. The luciferase reporter plasmid (100 ng) containing response elements of hsa-miR-129-5p in LINC00574 was cotransfected with 50 nM of miRNA negative control (miR-NC), hsa-miR-129-5p mimics, miRNA inhibitor negative control (IH-NC), or miR-129-5p-inhibitor (miR-129-5p-IH). (E) Exogeneous hsa-miR-129-5p inhibited the endogenous expression of LINC00574 in HepaRG cells. Differentiated HepaRG cells were transiently transfected with 50 nM of miRNA negative control (miR-NC), hsa-miR-129-5p mimics, miRNA inhibitor negative control (IH-NC), or miR-129-5p-inhibitor (miR-129-5p-IH). A decreased LINC00574 level was observed in cells transfected with hsa-miR-129-5p mimics. (F) Exogeneous hsa-miR-129-5p inhibited the protein expression of UGT2B15 in HepaRG cells detected by Western blot. Each assay was conducted in triplicate. Data are presented as means ± S.D. *P < 0.05; **P < 0.01.
The level of miR-129-5p increased >2-fold in HepaRG cells treated with 10 mM APAP (Fig. 5C). To investigate the regulatory effects dependent on interactions between hsa-miR-129-5p and LINC00574, we constructed a reporter gene plasmid harboring the predicted LINC00574 hsa-miR-129-5p MRE. HepG2 cells were cotransfected with the reporter plasmid and with miRNA negative control, hsa-miR-129-5p mimic, miRNA inhibitor control, or hsa-miR-129-5p inhibitor. Figure 5D shows that the luciferase activity of the reporter plasmid containing the LINC00574 hsa-miR-129-5p MRE was significantly suppressed by hsa-miR-129-5p mimics compared with its negative controls (by 47%; P < 0.001). However, under our experimental conditions no increase of the luciferase activity was observed in hsa-miR-129-5p inhibitor cotransfected cells. Taken together, these results indicated that hsa-miR-129-5p binds to its cognate response element in LINC00574 in a sequence-specific manner and suppresses LINC00574 levels.
hsa-miR-129-5p Suppresses UGT2B15 Expression by Interacting with LINC00574.
We reasoned that the principal role of hsa-miR-129-5p in suppressing UGT2B15 expression in response to toxic levels of APAP would be via direct molecular interactions between hsa-miR-129-5p and LINC00574 to destabilize LINC00574-dependent UGT2B15 expression. First, the obvious absence of a potential hsa-miR-129-5p MRE in the UGT2B15 mRNA transcript is consistent with the requirement of a mediator, such as LINC00574, to influence UGT2B15 expression. Second, our LINC00574 knockdown experiments suppressed UGT2B15 expression in HepaRG cells that contain negligible hsa-miR-129-5p levels, suggesting that LINC00574 acts downstream from hsa-miR-129-5p to suppress UGT2B15. These findings led us to investigate further the effects of exogeneous hsa-miR-129-5p on the suppression of endogenous LINC00574 and UGT2B15. As shown in Fig. 5E, the exogeneous overexpression of hsa-miR-129-5p by transfection significantly decreased LINC00574 RNA levels in HepaRG cells (by 25%; P < 0.05); no change in LINC00574 expression was observed in the cells transfected with the hsa-miR-129-5p inhibitor. The observed lack of a response to the hsa-miR-129-5p inhibitor is consistent with very low basal expression of hsa-miR-129-5p in HepaRG cells. Western blot results showed that the hsa-miR-129-5p mimic also suppressed the protein level of UGT2B15 by a substantial portion (by 36%; P < 0.05) in HepaRG cells (Fig. 5F). These data suggested that hsa-miR-129-5p regulates UGT2B15 by targeting LINC00574.
Discussion
In this study we screened deregulated lncRNAs in HepaRG cells after APAP exposure and identified LINC00574 as a new epigenetic agent that regulates UGT2B15 expression. Subsequent analyses of RNA-RNA interactions revealed that miRNA hsa-miR-129-5p was able to bind to LINC00574 and suppress its regulatory function. Deciphering the complex regulatory network controlling DME expression, with mechanistic interactions involving transcription factors, DMEs, miRNAs, and lncRNAs, should improve our understanding of drug metabolism and drug-induced hepatotoxicity.
We identified 47 lncRNAs that were substantially deregulated after APAP exposure through the reanalysis of RNA-seq data obtained in our former report, which focused on deregulated protein-coding genes (Yu et al., 2018). Our data showed that LINC00574 is enriched in liver cells, and its secondary structure was predicted to be quite stable. Importantly, the level of LINC00574 expression was decreased (by >90%) in HepaRG cells after treatment with a toxic concentration of APAP, suggesting that LINC00574 could be a key response mediator for APAP overexposure in hepatic cells. The expression of LINC00574 exhibited a significant positive correlation with UGT2B15 expression liver tissues by in silico analyses, providing additional supporting evidence of a potential regulatory role for LINC00574. We used the term “LINCxxxxx” to select intergenic lncRNA candidates for the current study. In the future we will investigate other types of lncRNA candidates involved in APAP-induced hepatotoxicity as well.
RBPs play a key role in the regulation of RNA processing and stability, including gene transcription, pre-mRNA splicing and polyadenylation, mRNA transport, translation, and decay (Glisovic et al., 2008). It is well known that lncRNAs often recruit RBPs to perform their biologic functions (Ferrè et al., 2016). In an ongoing study, we employed proteomics and RNA-seq approaches to demonstrate that the nuclear riboproteins heterogeneous nuclear ribonucleoproteins (hnRNP) Q and HNRNPL were recruited directly by lncRNA RP11.116D2.1, resulting in a decreased level of RNA stability and splicing of several NRs and DMEs. In the current study we analyzed the potential interactions between LINC00574 and RBPs in silico based on the individual-nucleotide resolution cross-linking immunoprecipitation (iCLIP), enhanced cross-linking immunoprecipitation (eCLIP), and photoactivatable ribonucleoside-enhanced cross-linking immunoprecipitation (PAR-CLIP) experiments contained in the starBase data base. As shown in Table 2, HNRNPA1, HNRNPC, serine/arginine-rich splicing factor 1 (SRSF1), and Dyskerin Pseudouridine Synthase 1 (DKC1) were identified as RBPs potentially able to bind LINC00574. HNRNPA1, the most abundant isoform of hnRNPs, is involved in RNA splicing, mRNA maturation, and translation processes (Roy et al., 2017), and HNRNPC is also believed to be involved in RNA splicing (Geuens et al., 2016). SRSF1 exhibits versatile roles in RNA splicing, RNA stability, and protein translation (Das and Krainer, 2014), whereas DKC1 could affect rRNA processing and telomere stability (Penzo et al., 2013). Together, the insights from in silico predictions of interactions between RBPs and LINC00574, and the experimental results of RBPs interacting with RP11.116D2.1, suggested a plausible mechanism in which lncRNAs modulate the levels of DMEs and NRs through coupling with RNA processing proteins and may provide a self-protective response network for hepatic cells exposed to toxic xenobiotic agents.
TABLE 2.
RNA binding proteins interacting with LINC00574
| RNA binding proteina | Binding site in LINC00574 | Seq type | Cell line | Accession |
|---|---|---|---|---|
| HNRNPA1 | chr6:170202698–170202865[+] | iCLIP | HEK 293T | GSM2221653 |
| HNRNPA1 | chr6:170202699–170202910[+] | iCLIP | HEK 293T | GSM2221654 |
| HNRNPA1 | chr6:170202899–170202957[+] | iCLIP | HEK 293T | GSM2221653 |
| SRSF1 | chr6:170200944–170200945[+] | eCLIP | HepG2 | ENCSR989VIY |
| SRSF1 | chr6:170202217–170202265[+] | eCLIP | HepG2 | ENCSR989VIY |
| SRSF1 | chr6:170202714–170202845[+] | iCLIP | HEK 293T | GSM2221667 |
| SRSF1 | chr6:170202732–170202871[+] | iCLIP | HEK 293T | GSM2221666 |
| SRSF1 | chr6:170202760–170202870[+] | iCLIP | HEK 293T | GSM2221668 |
| SRSF1 | chr6:170202762–170202853[+] | iCLIP | HEK 293T | GSM2221664 |
| SRSF1 | chr6:170202767–170202861[+] | iCLIP | HEK 293T | GSM2221665 |
| DKC1 | chr6:170190453–170190468[+] | PAR-CLIP | HEK 293 | GSM1067866 |
| DKC1 | chr6:170202508–170202531[+] | PAR-CLIP | HEK 293 | GSM1067866 |
| HNRNPC | chr6:170201725–170201734[+] | eCLIP | HepG2 | ENCSR550DVK |
Data obtained from starBase data base.
To better understand the regulation of UGT2B15 expression by LINC00574, we analyzed the expression profiles of LINC00574 and miRNAs in APAP-treated cells and found that LINC00574 expression was correlated inversely with hsa-miR-129-5p expression. miRNA hsa-miR-129-5p has been reported to be associated with multiple cancers, including breast cancer, prostate cancer, or gastrointestinal cancer (He et al., 2014; Meng et al., 2018; Wang et al., 2019a). We found that hsa-miR-129-5p was significantly elevated in HepaRG cells treated by 10 mM APAP. Additionally, a potential hsa-miR-129-5p MRE in LINC00574 was predicted by in silico analysis, and FREMSA provided evidence for the physical interaction between LINC00574 and hsa-miR-129-5p in vitro. Luciferase reporter gene assays showed decreased activity with constructs containing the LINC00574 hsa-miR-129-5p MRE in HepG2 cells cotransfected with exogenous hsa-miR-129-5p mimics. HepG2 instead of HepaRG cells were used in the luciferase assay due to the much higher transfection efficiency observed in HepG2 cells. Loss-of-function experiments confirmed that LINC00574 down-regulation using siRNA and ASO significantly suppressed the endogenous UGT2B15 RNA and protein levels in HepaRG cells. These results provided evidence that the cross-talk between long noncoding RNA LINC00574 and microRNA-129-5p modulates UGT2B15 in APAP-induced liver toxicity.
Previous studies have shown that 14 microRNAs, including miR-376c-3p, miR-331-5p, miR-455-5p, miR548as-3p, miR624-3p, miR-605-5p, miR-4292, miR-376b-3p, miR-103b, miR-6500-5p, miR3675-3p, miR-4712-5p, miR-770-5p, and miR-3924 may regulate the expression of UGT2B15 (Wijayakumara et al., 2015, 2018; Papageorgiou and Court, 2017). In the current study none of these 14 miRNAs met the selection criteria for the set of 47 differentially expressed miRNA we detected upon APAP exposure. Furthermore, we checked the possibility of the interaction between LINC00574 and the 14 miRNAs above with two in silico prediction tools, LncBase Predicted version 2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted) and miRDB Prediction (http://mirdb.org/). The results confirmed that LINC00574 does not possess potential MREs for these miRNAs. Moreover, our in silico analyses showed that there was no hsa-miR-129-5p targeting site in UGT2B15 mRNA. These data suggest that UGT2B15 expression was regulated by miR-129-5p via LINC00574 in a molecule-specific manner.
In summary, our study demonstrated a distinct scheme of key molecular events, including the cross-talk and consequences among mRNA, miRNA, lncRNA, and proteins, for regulation of UGT2B15 expression in HepaRG cells. Specifically, high concentrations of APAP increased the expression of hsa-miR-129-5p and decreased the expression of LINC00574, which in turn, inhibited the expression or production of a key DME, UGT2B15. The proposed regulatory network involves increased expression of hsa-miR-129-5p, which mediates the suppression of UGT2B15 via LINC00574. RBPs, such as HNRNPA1, HNRNPC, SRSF1, and DKC1, are predicted to participate in regulation of UGT2B15 expression by interacting with LINC00574, possibly through decreasing splicing efficiency or reducing mRNA stability for UGT2B15; however, the role of RBPs interacting with LINC00574 in UGT2B15 regulation requires validation in future studies.
Abbreviations
- Ago
argonaute RNA-induced silencing complex catalytic component
- APAP
acetaminophen
- ASO
antisense oligonucleotide
- DME
drug metabolizing enzyme
- eCLIP
enhanced cross-linking immunoprecipitation
- FREMSA
fluorescence-based RNA electrophoretic mobility shift assay
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- hnRNP
heterogeneous nuclear ribonucleoprotein
- iCLIP
individual-nucleotide resolution cross-linking immunoprecipitation
- lncRNA
long noncoding RNA
- MFE
minimal free energy
- miRNA
microRNA
- MRE
miRNA response element
- NR
nuclear receptor
- P450
cytochrome P450
- PAR-CLIP
photoactivatable ribonucleoside-enhanced cross-linking immunoprecipitation
- PCR
polymerase chain reaction
- RBP
RNA binding protein
- RNA-seq
RNA sequencing
- siRNA
small interfering RNA
- TCGA
The Cancer Genome Atlas
- UGT
UDP glucuronosyltransferase
Authorship Contributions
Participated in study design: Yu, Tong, Ning.
Conducted experiments: Yu, J. Chen, S. Chen, Xu, Luo, Jin, Knox.
Performed data analysis: Yu, Xu, Wu, Li, Jin, Zhao, Wang.
Wrote or contributed to the writing of the manuscript: Yu, J. Chen, S. Chen, Tolleson, Guo, Ning.
Footnotes
This study was partially supported and funded by the National Key Research and Development Program of China [Grant SQ2017YFC16008401 to D.Y.] and the National Natural Science Foundation of China [Grant 91743113 to D.Y.] and partially supported by the National Center for Toxicological Research (NCTR)/US Food and Drug Administration (FDA) Project E0753201. D.Y. and D.L. were sponsored by the Oak Ridge Institute for Science and Education (ORISE) fellowships.
Disclaimer: The views presented in this paper are those of the authors and do not necessarily represent those of the US Food and Drug Administration.
References
- Chen Y, Zeng L, Wang Y, Tolleson WH, Knox B, Chen S, Ren Z, Guo L, Mei N, Qian F, et al. (2017) The expression, induction and pharmacological activity of CYP1A2 are post-transcriptionally regulated by microRNA hsa-miR-132-5p. Biochem Pharmacol 145:178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Freytsis M, Wang X, Peter I, Guillemette C, Hazarika S, Duan SX, Greenblatt DJ, Lee WM, Acute Liver Failure Study Group (2013) The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure. J Pharmacol Exp Ther 345:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Zhu Z, Masse G, Duan SX, James LP, Harmatz JS, Greenblatt DJ.(2017) Race, gender, and genetic polymorphism contribute to variability in acetaminophen pharmacokinetics, metabolism, and protein-adduct concentrations in healthy African-American and European-American volunteers. J Pharmacol Exp Ther 362:431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Krainer AR.(2014) Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 12:1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrè F, Colantoni A, Helmer-Citterich M.(2016) Revealing protein-lncRNA interaction. Brief Bioinform 17:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V.(2016) The hnRNP family: insights into their role in health and disease. Hum Genet 135:851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G.(2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Huang C, Zhang L, Li J.(2014) Epigenetic repression of miR-129-2 in cancer. Liver Int 34:646. [DOI] [PubMed] [Google Scholar]
- Jin Y, Yu D, Tolleson WH, Knox B, Wang Y, Chen S, Ren Z, Deng H, Guo Y, Ning B.(2016) MicroRNA hsa-miR-25-3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochem Pharmacol 113:88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M.(2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Knox B, Chen S, Wu L, Tolleson WH, Liu Z, Yu D, Guo L, Tong W, Ning B.(2019a) MicroRNAs hsa-miR-495-3p and hsa-miR-486-5p suppress basal and rifampicin-induced expression of human sulfotransferase 2A1 (SULT2A1) by facilitating mRNA degradation. Biochem Pharmacol 169:113617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tolleson WH, Yu D, Chen S, Guo L, Xiao W, Tong W, Ning B.(2019b) MicroRNA-dependent gene regulation of the human cytochrome P450,in Pharmacoepigenetics pp 129–138, Elsevier, London. [Google Scholar]
- Li D, Tolleson WH, Yu D, Chen S, Guo L, Xiao W, Tong W, Ning B.(2019c) Regulation of cytochrome P450 expression by microRNAs and long noncoding RNAs: epigenetic mechanisms in environmental toxicology and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 37:180–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JH, Liu S, Zhou H, Qu LH, Yang JH.(2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL.(2011) ViennaRNA package 2.0. Algorithms Mol Biol 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Jaeschke H.(2013) Metabolism and disposition of acetaminophen: recent advances in relation to hepatotoxicity and diagnosis. Pharm Res 30:2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng R, Fang J, Yu Y, Hou LK, Chi JR, Chen AX, Zhao Y, Cao XC.(2018) miR-129-5p suppresses breast cancer proliferation by targeting CBX4. Neoplasma 65:572–578. [DOI] [PubMed] [Google Scholar]
- Mohammadin S, Edger PP, Pires JC, Schranz ME.(2015) Positionally-conserved but sequence-diverged: identification of long non-coding RNAs in the Brassicaceae and Cleomaceae. BMC Plant Biol 15:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou I, Court MH.(2017) Identification and validation of the microRNA response elements in the 3′-untranslated region of the UDP glucuronosyltransferase (UGT) 2B7 and 2B15 genes by a functional genomics approach. Biochem Pharmacol 146:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Zhong X.(2015) Epigenetic regulation of drug metabolism and transport. Acta Pharm Sin B 5:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo M, Casoli L, Ceccarelli C, Treré D, Ludovini V, Crinò L, Montanaro L.(2013) DKC1 gene mutations in human sporadic cancer. Histol Histopathol 28:365–372. [DOI] [PubMed] [Google Scholar]
- Roy R, Huang Y, Seckl MJ, Pardo OE.(2017) Emerging roles of hnRNPA1 in modulating malignant transformation. Wiley Interdiscip Rev RNA 8:e1431. [DOI] [PubMed] [Google Scholar]
- Sheweita SA.(2000) Drug-metabolizing enzymes: mechanisms and functions. Curr Drug Metab 1:107–132. [DOI] [PubMed] [Google Scholar]
- Snawder JE, Roe AL, Benson RW, Roberts DW.(1994) Loss of CYP2E1 and CYP1A2 activity as a function of acetaminophen dose: relation to toxicity. Biochem Biophys Res Commun 203:532–539. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson SS, Sasame HA, Mitchell JR, Jollow DJ, Potter WZ.(1976) Biochemical changes after hepatic injury from toxic doses of acetaminophen or furosemide. Pharmacology 14:205–217. [DOI] [PubMed] [Google Scholar]
- Wang S, Chen Y, Yu X, Lu Y, Wang H, Wu F, Teng L.(2019a) miR-129-5p attenuates cell proliferation and epithelial mesenchymal transition via HMGB1 in gastric cancer. Pathol Res Pract 215:676–682. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yan L, Liu J, Chen S, Liu G, Nie Y, Wang P, Yang W, Chen L, Zhong X, et al. (2019b) The HNF1α-regulated lncRNA HNF1α-AS1 is involved in the regulation of cytochrome P450 expression in human liver tissues and Huh7 cells. J Pharmacol Exp Ther 368:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu D, Tolleson WH, Yu LR, Green B, Zeng L, Chen Y, Chen S, Ren Z, Guo L, et al. (2017) A systematic evaluation of microRNAs in regulating human hepatic CYP2E1. Biochem Pharmacol 138:174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA.(2012) Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488:111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayakumara DD, Hu DG, Meech R, McKinnon RA, Mackenzie PI.(2015) Regulation of human UGT2B15 and UGT2B17 by miR-376c in prostate cancer cell lines. J Pharmacol Exp Ther 354:417–425. [DOI] [PubMed] [Google Scholar]
- Wijayakumara DD, Mackenzie PI, McKinnon RA, Hu DG, Meech R.(2018) Regulation of UDP-glucuronosyltransferase 2B15 by miR-331-5p in prostate cancer cells involves canonical and noncanonical target sites. J Pharmacol Exp Ther 365:48–59. [DOI] [PubMed] [Google Scholar]
- Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R.(2018) Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci 75:467–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, Fuscoe J, Pogribny I, Ning B.(2015a) Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep 5:8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Tolleson WH, Jin Y, Mei N, Guo Y, Deng H, Pogribny I, Ning B.(2015b) MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochem Pharmacol 98:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Tolleson WH, Knox B, Jin Y, Guo L, Guo Y, Kadlubar SA, Ning B.(2015c) Modulation of ALDH5A1 and SLC22A7 by microRNA hsa-miR-29a-3p in human liver cells. Biochem Pharmacol 98:671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Wu L, Gill P, Tolleson WH, Chen S, Sun J, Knox B, Jin Y, Xiao W, Hong H, et al. (2018) Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Arch Toxicol 92:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen S, Li D, Knox B, Guo L, Ning B. (2020) FREMSA: A Method That Provides Direct Evidence of the Interaction between microRNA and mRNA. Methods Mol Biol 2102:557–566, doi: 10.1007/978-1-0716-0223-2_30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Chen Y, Wang Y, Yu LR, Knox B, Chen J, Shi T, Chen S, Ren Z, Guo L, et al. (2017) MicroRNA hsa-miR-370-3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol 140:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XB, Leeder JS.(2013) Epigenetic regulation of ADME-related genes: focus on drug metabolism and transport. Drug Metab Dispos 41:1721–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]