Abstract
We investigated the transcriptomic response and epigenetic changes in the lungs of mice exposed to inhalation of copper(II) oxide nanoparticles (CuO NPs) (8 × 105 NPs/m3) for periods of 3 days, 2 weeks, 6 weeks, and 3 months. A whole genome transcriptome and miRNA analysis was performed using next generation sequencing. Global DNA methylation was assessed by ELISA. The inhalation resulted in the deregulation of mRNA transcripts: we detected 170, 590, 534, and 1551 differentially expressed transcripts after 3 days, 2 weeks, 6 weeks, and 3 months of inhalation, respectively. Biological processes and pathways affected by inhalation, differed between 3 days exposure (collagen formation) and longer treatments (immune response). Periods of two weeks exposure further induced apoptotic processes, 6 weeks of inhalation affected the cell cycle, and 3 months of treatment impacted the processes related to cell adhesion. The expression of miRNA was not affected by 3 days of inhalation. Prolonged exposure periods modified miRNA levels, although the numbers were relatively low (17, 18, and 38 miRNAs, for periods of 2 weeks, 6 weeks, and 3 months, respectively). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis based on miRNA–mRNA interactions, revealed the deregulation of processes implicated in the immune response and carcinogenesis. Global DNA methylation was not significantly affected in any of the exposure periods. In summary, the inhalation of CuO NPs impacted on both mRNA and miRNA expression. A significant transcriptomic response was already observed after 3 days of exposure. The affected biological processes and pathways indicated the negative impacts on the immune system and potential role in carcinogenesis.
Keywords: copper(II) oxide nanoparticles, inhalation, mouse, gene expression, DNA methylation
1. Introduction
Copper(II) oxide nanoparticles (CuO NPs) may enter the body by means of inhalation, dermal or ingestion. In human exposure, the inhalation route is the most significant one, particularly for occupational exposures. In the body, nanoparticles (NPs) may reach the lungs where they are deposited, and additionally other organs including the central nervous system. Their presence usually causes oxidative stress or inflammatory responses.
Copper (Cu) is a biogenic element with many biological functions that include the appropriate growth, development, and maintenance of bones, connective tissues and various organs [1]. It is further involved in the stimulation of the immune system in the protection against infection and in the repair of injuries and healing. It plays an important role as a cofactor of some enzymes (e.g., superoxide dismutase and cytochrome oxidase) and is involved in cell signaling and cell proliferation. In addition, its metabolism is critical for tumor progression [2]. Nanosized Cu possesses antimicrobial and antiviral properties and is therefore used in face masks, wound dressings, and socks [3]. It is applied in conductive films, lubrication, nanofluids, gas sensors, solar cells, lithium batteries, and catalysts. Cu-based nanostructures are also used in various electronic devices, e.g., inkjet printers [4]. In comparison with other metal-based NPs (e.g., Ag or Zn), Cu-based NPs are less commonly used: the global production of CuO NPs was 570 tons in 2014 and it is estimated to increase to 1600 tons in 2025 [5]. The toxicity of Cu-based NPs depends on the solubility of the material that is affected by pH, dissolved organic carbon content, and water hardness [3].
Although the production of CuO NPs is at present relatively low, their properties and behavior in biological systems make them an important subject for toxicological investigation. The theoretical mechanisms of CuO NPs toxicity include: (1) release of Cu ions from the surface of NPs, (2) oxidative damage mediated by Fenton-type reactions [reactive oxygen species (ROS) generation] induced by particles, and (3) cell death associated with autophagy caused by CuO NP exposure. While metal oxide NPs (e.g., Cu, Zn, Ti, Fe, and Si) share similarities in their biological effects often mediated by ROS production, CuO NPs have been shown to have the highest potential to induce cytotoxicity and DNA damage in vitro [6]. This seems to be related to the ability of CuO NPs to better overwhelm the antioxidant defenses of the organism.
The biological effects of the various methods of CuO NPs pulmonary delivery, including intratracheal instillation, oropharyngeal instillation, or whole-body inhalation, were investigated in both rats and mice (reviewed in [1,6]). The majority of the studies reported inflammatory responses in the lungs that included neutrophil and eosinophil infiltration after acute exposures, and neutrophilic, neutrophilic/lymphocytic, eosinophilic/fibrotic/granulomatous, and fibrotic granulomatous inflammation after chronic exposures. Pulmonary administration may even cause systemic effects affecting the liver, kidney, or spleen, as shown by Liu et al. [7] and in our own recent study [8]. Inflammatory processes may induce DNA damage which may cause mutations and tumor formation in the lungs, as demonstrated by Yokohira et al. in a study in rats [9].
Despite the potential serious health effects of human exposure to CuO NPs, only one study that focused on the analysis of whole genome transcriptional changes, following the inhalation of CuO NPs, has been published [10]. Studies of global gene expression changes are valuable as they have the potential to reveal a wide range of processes which are affected by a respective exposure, unable to be detected by analyses focused on a defined, limited number of pathological changes. To address these gaps, we conducted a study in which we exposed ICR mice to CuO NPs by continuous whole-body inhalation. We used four time periods to mimic acute and chronic exposure and analyzed global mRNA expression changes and epigenetic alterations by analysis of miRNA and global DNA methylation changes. Moreover, we identified mRNA targets of miRNAs found to be deregulated in our study and performed an analysis of pathways affected by CuO NPs exposure.
2. Materials and Methods
2.1. CuO NPs Generation, Exposure of Mice, Analysis of Cu and CuO NPs in the Lungs
The generation of CuO NPs, mice exposure and analysis of Cu and CuO NPs in the lungs, were reported in our previous study [8]. Briefly, CuO NPs were continuously generated by using the thermal decomposition of metal organic precursor copper (II) acetylacetonate and a subsequent oxidation at a temperature of 700 °C. CuO NPs were diluted with a U-HEPA filtrated air stream, and then split into two streams at a ratio of 1:1. Prior to entering the inhalation chamber, both streams were further diluted with a stream of purified, humidified air and used for whole-body inhalation experiments. Female ICR mice (8 animals/group) were exposed in a whole-body inhalation chamber [11] to 8 × 105 CuO NP/cm3 (geometric mean diameter 29.4 nm) for 3 days, 2 weeks, 6 weeks, and 3 months; the control animals inhaled clean air. The conditions of inhalation were selected based on our previous study [8]. At the selected time intervals mice were sacrificed by chloroform inhalation and the lungs were collected for further analyses. The animal experiments were approved by the Ethical Board of the Institute of Analytical Chemistry in Brno.
An analysis of Cu content was performed using electrothermal atomic absorption spectrometry, employing AAnalyst 600 (Perkin-Elmer Inc., Wellesley, MA, USA) instrumentation. The presence of CuO NPs in the lungs was assessed using scanning transmission electron microscopy (STEM) and scanning electron microscopy-energy-dispersive X-ray spectroscopy (SEM-EDX). The sections were examined under a Philips EM 208 S Morgagni transmission electron microscope (FEI, Brno, Czech Republic). For the SEM-EDX analysis, a Magellan 400 SEM (FEI) equipped with an Energy Dispersive Spectroscopy Detector Octane Elect Super (EDAX, Baltimore, MD, USA) was used.
The number concentration and the size distribution of CuO NPs in the inhalation chambers were measured continuously in the size range of 7.64–229.6 nm using a Scanning Mobility Particle Sizer Spectrometer (SMPS; TSI, Shoreview, MN, USA) at 5 min intervals.
2.2. DNA, RNA, and miRNA Extraction
Lung tissue was disrupted using a mortar and pestle, and homogenized with a needle and syringe. From tissue homogenates, the total RNA and DNA was extracted using AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN Manchester Ltd; Manchester, UK). The extraction protocol followed the manufacturer’s instructions. The concentration of nucleic acids was determined by Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific; Waltham, MA, USA); RNA integrity number (RIN) was assessed by Agilent Bioanalyzer (Agilent Technologies Inc.; Santa Clara, CA, USA) with RNA 6000 Nano kit (Agilent Technologies Inc.; Santa Clara, CA, USA). RIN values were > 7.5 in all samples.
2.3. Whole Genome Transcriptome Analysis by Next Generation Sequencing
RNA samples (concentration > 260 ng/µl, RNA integrity number > 7.5) were used for RNA libraries preparation using (Poly(A)RNA Selection, Lexogen Sense Total RNA-Seq Library Prep Kit (Lexogen GmbH; Vienna, Austria). For sequencing, NextSeq® 500/550 High Output Kit v2 (75 cycle) and NextSeq 500/550 system (Illumina, Inc.; San Diego, CA, USA) were used. The reactions were performed according to the manufacturers’ recommendations.
2.4. miRNA Expression Analysis
Small RNA libraries were prepared from 150 ng of total RNA using Qiaseq miRNA library kit (QIAGEN Manchester Ltd; Manchester, UK). This kit allows detection of not only miRNAs, but also other small RNAs, including piRNAs. In our data, a vast majority of results consisted of deregulated miRNAs, with a small proportion of piRNAs. Thus, “miRNA expression” reported here denotes expression of both miRNAs and piRNAs. Briefly, adaptors were ligated to the 3′ and 5′ ends of all miRNAs. After adaptor ligation, miRNA was reverse transcribed to cDNA and the library was amplified (95 °C 15 min, (95 °C 15 s, 60 °C 30 s, 72 °C 15 s, 16×), 72 °C 2 min, 4 °C for at least 5 min) on a Mastercycler® Nexus (Eppendorf; Hamburg, Germany). Small RNA libraries were validated on a Fragment Analyzer (Agilent Technologies Inc.; Santa Clara, CA, USA) with a High sensitivity NGS kit. Sequencing of sRNA libraries was performed on MiSeq system (Illumina, Inc.; San Diego, CA, USA) using MiSeq Reagent Kit V3 (Illumina, Inc.; San Diego, CA, USA).
2.5. Global DNA Methylation Analysis
The content of 5-methylcytosine (5-mC) was assessed in DNA samples (100 ng/well) using 5-mC DNA ELISA Kit (Zymo Research; Irvine, CA, USA) following the manufacturer’s instructions. The samples were analyzed in triplicate. The results were expressed as % 5-mC/total cytosine content.
2.6. Data Analysis
An NGI-RNAseq pipeline (https://github.com/SciLifeLab/NGI-RNAseq) was used for RNA sequencing data. It pre-processed raw data from FastQ inputs (FastQC, Trim Galore!), aligned the reads (HiSAT2) using reference genome Mus musculus assembly GRCm38.91, Ensembl annotation version 91, generated gene counts (featureCounts) and performed extensive quality-control on the results (RSeQC, dupRadar, Preseq, edgeR, and MultiQC). DESeq2 with default parameter settings was applied to normalize read counts and to identify differences in gene expression between sample groups. The standard deviations for the numbers of differentially expressed mRNAs were estimated with bootstrapping (25 times repeated sampling with replacement carried out independently in each of the treatment groups). A functional enrichment analysis of differential gene expression data was performed by ToppFun tool, using the features “Biological process” and “Pathway” [12].
Differential miRNA expression count data were generated by QIAseq miRNA Quantification, and the UMI counts were taken to compensate for sequencing bias. The DESeq2-package was used to test for differential expression by the application of negative binomial generalized linear models. A p < 0.02 was used as the cut-off for statistically significant deregulated miRNAs between sample groups.
mRNA-miRNA correlation analysis was performed with the R package miRComb [13]. First, the sets of differentially expressed mRNAs and miRNAs were identified (the detection thresholds were adjusted pval < 0.1, FC > 1.5; the detection method was DESeq). Correlations between all the mRNA and miRNA candidate pairs were then calculated. We used the default parameter settings proposed in miRComb (Pearson method). Only the pairs with a negative correlation were considered. The pairs with a significantly adjusted correlation p-value were reported and used to construct the mRNA-miRNA interaction networks. We extended the interaction networks using information regarding validated and predicted miRNA targets. These targets were taken from the multiMiR package [14] that comprises of 14 different human and mouse miRNA target databases. The predicted and validated targets were used to highlight the edges in mRNA-miRNA interaction correlation networks and also helped to identify miRNAs with the highest number of deregulated targets. The R package edgeR served to report the enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [15].
3. Results
3.1. mRNA Expression Changes Induced by CuO NPs Inhalation
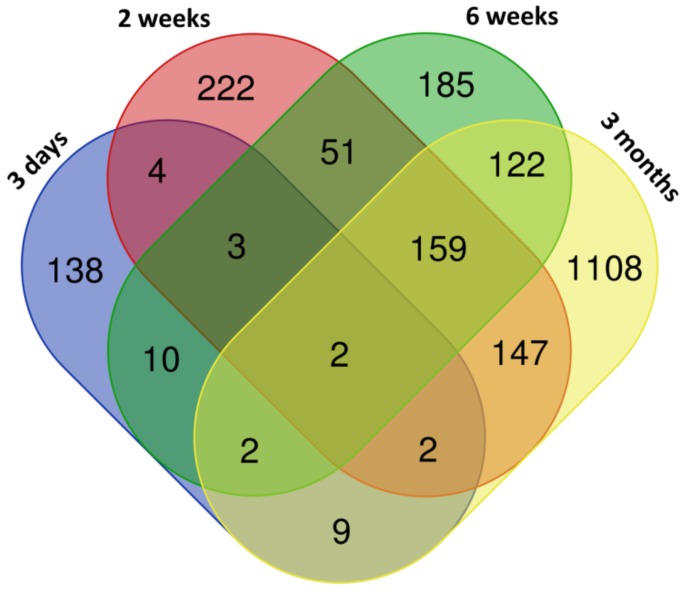
Exposure to CuO NPs was associated with the deregulation of mRNA expression. The numbers of differentially expressed genes increased with the length of exposure, although the change was not consistent and standard deviations for these numbers are large as a consequence of small sample sizes (170 ± 84, 590 ± 331, 534 ± 172, and 1551 ± 387 deregulated genes for 3 days, 2 weeks, 6 weeks, and 3 months of exposure, respectively) (Supplementary Table S1). The number of upregulated genes was generally higher than those that were downregulated; this difference ranged from 1.37-fold to 3.27-fold, depending on the exposure period. The distribution of common and unique deregulated genes is shown in the Venn diagram (Figure 1). Only two transcripts were common for all of the exposure periods (Hmox1, Slc7a11). In contrast, 159 genes were commonly deregulated for 2 weeks, 6 weeks, and 3 months exposure. Interestingly, few common transcripts were found for any comparison in which 3 days exposure was included. This suggests that the effects associated with the shortest (acute) exposure, substantially differed from those induced by longer exposure periods. The list of the common and unique deregulated genes is reported in Supplementary Table S2.
Figure 1.
The distribution of common and unique genes differentially expressed after copper(II) oxide nanoparticles (CuO NPs) inhalation.
In agreement with findings for differentially expressed mRNAs, the functional enrichment analysis revealed differences between biological processes and pathways deregulated after 3 days exposure and longer treatments. The short inhalation period mostly affected collagen metabolism and extracellular matrix organization. Prolonged exposure caused the deregulation of immune response-related processes including cytokine production and cell activation (selected biological processes and pathways are reported in Table 1). We further investigated the biological processes and pathways unique for individual, longer exposure periods. For 2 weeks of treatment, the processes associated with immune response and cell death were deregulated, 6 weeks of inhalation affected cell division and cell cycle regulation and 3 months of treatment involved changes in cell adhesion, migration, and cytokine production (selected unique biological processes are shown in Table 2).
Table 1.
Selected biological processes and pathways deregulated after inhalation of CuO NPs.
| ID | Deregulated Biological Process/Pathway | q-Value | Deregulated Genes (N) | Genes in Pathway (N) |
|---|---|---|---|---|
| 3 Days | ||||
| GO:0030574 | collagen catabolic process | 0.003 | 7 | 67 |
| GO:0043062 | extracellular structure organization | 0.005 | 13 | 355 |
| GO:0030198 | extracellular matrix organization | 0.005 | 13 | 354 |
| GO:0032963 | collagen metabolic process | 0.028 | 7 | 122 |
| GO:0061077 | chaperone-mediated protein folding | 0.028 | 5 | 52 |
| 1269985 | Chondroitin sulfate biosynthesis | 0.009 | 4 | 22 |
| 82982 | Glycosaminoglycan biosynthesis - chondroitin sulfate/dermatan sulfate | 0.009 | 4 | 20 |
| 545276 | chondroitin sulfate biosynthesis (late stages) | 0.009 | 3 | 8 |
| 1270245 | Collagen formation | 0.019 | 6 | 93 |
| 1270247 | Assembly of collagen fibrils and other multimeric structures | 0.019 | 5 | 60 |
| 2 Weeks | ||||
| GO:0006955 | immune response | < 0.001 | 165 | 1572 |
| GO:0002682 | regulation of immune system process | < 0.001 | 140 | 1506 |
| GO:0006952 | defense response | < 0.001 | 142 | 1651 |
| GO:0002684 | positive regulation of immune system process | < 0.001 | 102 | 976 |
| GO:0001775 | cell activation | < 0.001 | 96 | 1001 |
| 1269203 | Innate Immune System | < 0.001 | 101 | 1312 |
| 1470924 | Interleukin-10 signaling | < 0.001 | 21 | 49 |
| 83051 | Cytokine-cytokine receptor interaction | < 0.001 | 41 | 270 |
| 1457780 | Neutrophil degranulation | < 0.001 | 50 | 492 |
| 1269310 | Cytokine Signaling in Immune system | < 0.001 | 64 | 763 |
| 6 Weeks | ||||
| GO:0006955 | immune response | < 0.001 | 122 | 1572 |
| GO:0002682 | regulation of immune system process | < 0.001 | 113 | 1506 |
| GO:0006954 | inflammatory response | < 0.001 | 76 | 711 |
| GO:0006952 | defense response | < 0.001 | 115 | 1651 |
| GO:0001816 | cytokine production | < 0.001 | 70 | 700 |
| 1269203 | Innate Immune System | < 0.001 | 97 | 1312 |
| 1457780 | Neutrophil degranulation | < 0.001 | 56 | 492 |
| 1470924 | Interleukin-10 signaling | < 0.001 | 17 | 49 |
| M5889 | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins | < 0.001 | 70 | 1028 |
| M5885 | Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins, ECM regulators and secreted factors | < 0.001 | 57 | 753 |
| 3 Months | ||||
| GO:0006955 | immune response | < 0.001 | 282 | 1572 |
| GO:0006952 | defense response | < 0.001 | 277 | 1651 |
| GO:0051240 | positive regulation of multicellular organismal process | < 0.001 | 260 | 1610 |
| GO:0002682 | regulation of immune system process | < 0.001 | 249 | 1506 |
| GO:0001816 | cytokine production | < 0.001 | 154 | 700 |
| 1269203 | Innate Immune System | < 0.001 | 210 | 1312 |
| 1457780 | Neutrophil degranulation | < 0.001 | 105 | 492 |
| 1269310 | Cytokine Signaling in Immune system | < 0.001 | 125 | 763 |
| 1470924 | Interleukin-10 signaling | < 0.001 | 25 | 49 |
| 83051 | Cytokine-cytokine receptor interaction | < 0.001 | 62 | 270 |
Table 2.
Selected unique biological processes deregulated after inhalation of CuO NPs.
| ID | Deregulated Biological Process | q-Value | Deregulated Genes (N) | Genes in Pathway (N) |
|---|---|---|---|---|
| 2 Weeks | ||||
| GO:0006955 | immune response | 0.019 | 33 | 1572 |
| GO:0045776 | negative regulation of blood pressure | 0.019 | 6 | 56 |
| GO:0010941 | regulation of cell death | 0.019 | 33 | 1650 |
| GO:0002682 | regulation of immune system process | 0.019 | 31 | 1506 |
| GO:0043067 | regulation of programmed cell death | 0.022 | 31 | 1536 |
| GO:0002683 | negative regulation of immune system process | 0.022 | 14 | 420 |
| GO:0012501 | programmed cell death | 0.022 | 36 | 1952 |
| GO:0001775 | cell activation | 0.025 | 23 | 1001 |
| GO:0042981 | regulation of apoptotic process | 0.025 | 30 | 1519 |
| GO:0006915 | apoptotic process | 0.027 | 35 | 1923 |
| GO:0006955 | immune response | 0.019 | 33 | 1572 |
| 6 Weeks | ||||
| GO:0000280 | nuclear division | < 0.001 | 24 | 599 |
| GO:0048285 | organelle fission | < 0.001 | 24 | 636 |
| GO:1903047 | mitotic cell cycle process | < 0.001 | 26 | 931 |
| GO:0000278 | mitotic cell cycle | < 0.001 | 27 | 1016 |
| GO:0022402 | cell cycle process | < 0.001 | 32 | 1385 |
| GO:0000070 | mitotic sister chromatid segregation | < 0.001 | 10 | 136 |
| GO:0051241 | negative regulation of multicellular organismal process | 0.001 | 27 | 1127 |
| GO:0051301 | cell division | 0.001 | 20 | 668 |
| GO:0007049 | cell cycle | 0.001 | 35 | 1766 |
| GO:0055118 | negative regulation of cardiac muscle contraction | 0.001 | 3 | 4 |
| 3 Months | ||||
| GO:0051240 | positive regulation of multicellular organismal process | < 0.001 | 171 | 1610 |
| GO:0022610 | biological adhesion | < 0.001 | 164 | 1542 |
| GO:0007155 | cell adhesion | < 0.001 | 161 | 1530 |
| GO:0040011 | locomotion | < 0.001 | 170 | 1735 |
| GO:0016477 | cell migration | < 0.001 | 137 | 1300 |
| GO:0001816 | cytokine production | < 0.001 | 90 | 700 |
| GO:0051674 | localization of cell | < 0.001 | 141 | 1428 |
| GO:0048870 | cell motility | < 0.001 | 141 | 1428 |
| GO:0006928 | movement of cell or subcellular component | < 0.001 | 170 | 1882 |
| GO:0009611 | response to wounding | < 0.001 | 106 | 967 |
| GO:0051240 | positive regulation of multicellular organismal process | < 0.001 | 171 | 1610 |
3.2. Changes of miRNA Expression Affected by CuO NPs
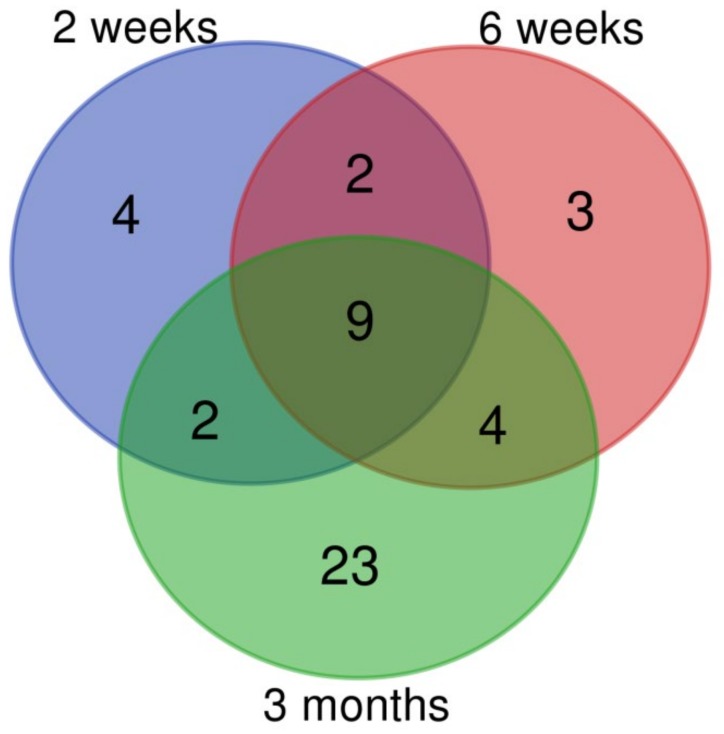
The short, 3-day inhalation period had no effect on miRNA expression, but the longer exposure periods were associated with significant changes in miRNA levels. We observed 17, 18, and 38 deregulated miRNAs for 2 weeks, 6 weeks, and 3 months exposure, respectively (Supplementary Table S3). Similarly to mRNA expression, the number of upregulated miRNAs was higher than those that were downregulated. The greatest difference (4.6-fold) was observed for the 2-week inhalation period, followed by 6 weeks (1.57-fold) and 3 months (1.11-fold) exposure. The numbers of common and unique deregulated miRNAs are shown in Figure 2. There were nine miRNAs common for all three exposure periods. The number of unique deregulated miRNAs differed by the exposure time: the highest proportion of such miRNAs (61% of all deregulated miRNAs) was detected after 3 months of inhalation. The common and unique deregulated miRNAs are shown in Supplementary Table S4.
Figure 2.
The numbers of common and unique miRNAs deregulated after CuO NPs inhalation.
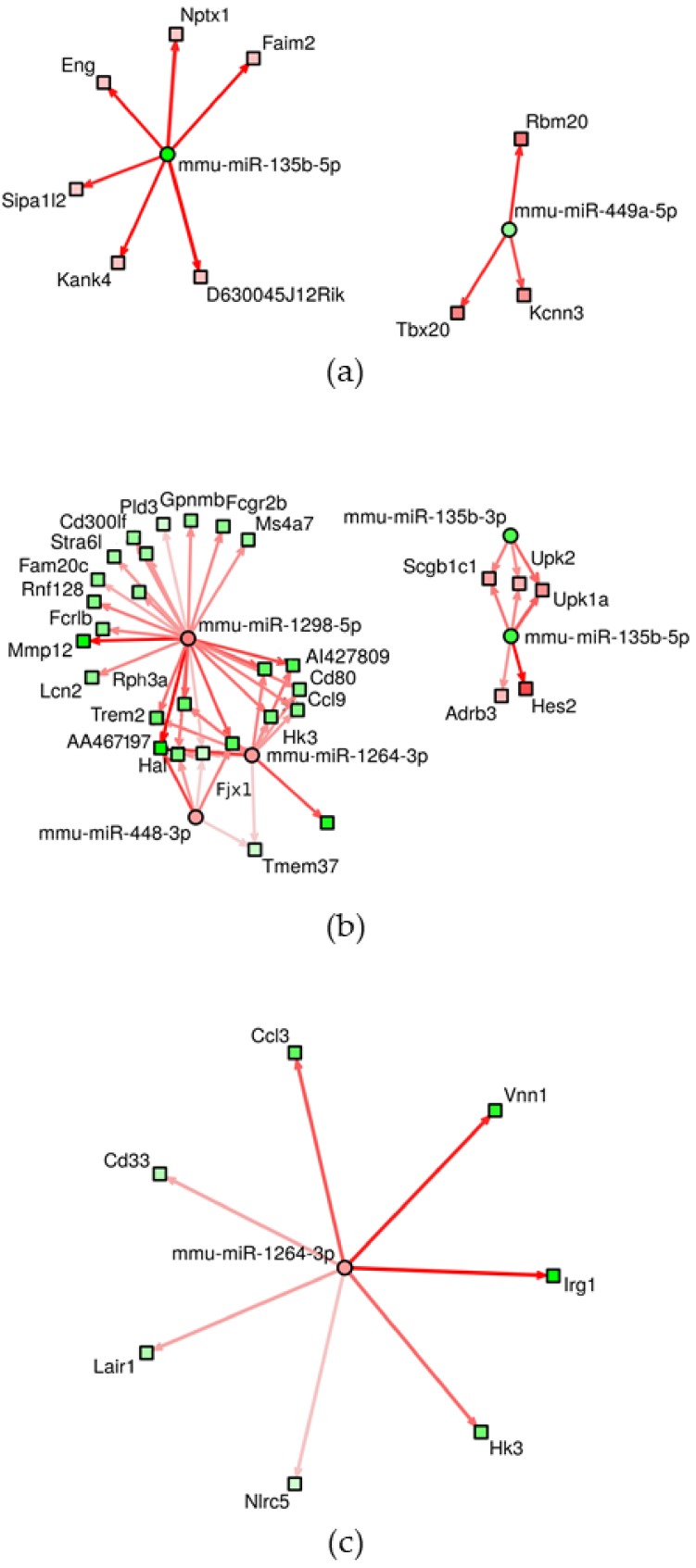
The interactions between experimentally validated, significantly deregulated miRNAs and mRNAs are reported in Supplementary Table S5. The number of significant results increased again with the length of exposure: 148 mRNAs targeted by three miRNAs, 575 mRNAs targeted by 18 miRNAs and 1542 mRNAs targeted by 15 miRNAs were found after 2 weeks, 6 weeks, and 3 months of inhalation, respectively. A summary of miRNAs involved in the interactions and the respective number of target mRNAs are reported in Table 3. The highest number of miRNAs was affected by mmu-miR-449a-5p (2 weeks of inhalation), mmu-miR-448-3p (6 weeks exposure) and mmu-miR-1264-3p, mmu-miR-1298-5p, mmu-miR-144-5p (6 weeks and 3 months of treatment). We further studied miRNA–mRNA interactions using interaction networks. These networks graphically depict targets for individual miRNAs and the potential relationship with targeted mRNAs. The density of the networks and number of visualized interactions depend on the selected, adjusted p-value for interactions and a score and generally increase with a higher p-value and lower score. The score is computed as a measure of impact of miRNA on respective mRNA expression and a higher score means a tighter miRNA–mRNA association [13]. The results for individual miRNAs and their targets for adjusted p-value <0.001 and score ≥ 3 are visualized in Figure 3. Using these conditions, the highest density of interactions was found with 6 weeks of inhalation, this was as expected considering the highest number of miRNAs significantly targeting mRNAs was for this exposure period. In these interactions, five miRNAs were involved (mmu-miR-135b-3p, mmu-miR-135b-5p, mmu-miR-448-3p, mmu-miR-1264-3p, mmu-miR-1298-5p). Interestingly, despite the highest number of targeted mRNAs after 3 months of inhalation, the density of interaction networks was low and only one miRNA was detected (mmu-miR-1264-3p). For 2 weeks of inhalation, mmu-miR-135b-5p and mmu-miR-449a-5p participated in miRNA–mRNA interactions. The details on miRNA–mRNA interactions depicted in Figure 3 are provided in Supplementary Table S6.
Table 3.
miRNAs and numbers of corresponding targeted mRNAs after inhalation of CuO NPs for individual exposure periods.
| miRNA | Target mRNA (N) |
|---|---|
| 2 Weeks | |
| mmu-miR-135b-5p | 50 |
| mmu-miR-147-3p | 11 |
| mmu-miR-449a-5p | 87 |
| 6 Weeks | |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:17:27580702:27580732:Minus | 1 |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:18:60298547:60298577:Plus | 1 |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:3:46956408:46956438:Minus | 2 |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:6:128725729:128725759:Minus | 4 |
| mmu-miR-1264-3p | 134 |
| mmu-miR-1298-5p | 183 |
| mmu-miR-135b-3p | 22 |
| mmu-miR-135b-5p | 68 |
| mmu-miR-146b-5p | 11 |
| mmu-miR-147-3p | 13 |
| mmu-miR-155-5p | 15 |
| mmu-miR-21a-5p | 5 |
| mmu-miR-448-3p | 104 |
| mmu-miR-489-3p | 1 |
| mmu-miR-672-5p | 2 |
| mmu-miR-6972-5p | 4 |
| mmu-miR-703 | 1 |
| mmu-miR-7048-5p | 4 |
| 3 Months | |
| mmu-miR-1264-3p | 515 |
| mmu-miR-1298-5p | 461 |
| mmu-miR-135b-5p | 29 |
| mmu-miR-144-5p | 249 |
| mmu-miR-146a-5p | 23 |
| mmu-miR-146b-5p | 15 |
| mmu-miR-147-3p | 26 |
| mmu-miR-155-5p | 34 |
| mmu-miR-21a-3p | 43 |
| mmu-miR-21a-5p | 11 |
| mmu-miR-21b | 11 |
| mmu-miR-223-3p | 8 |
| mmu-miR-342-3p | 12 |
| mmu-miR-3967 | 14 |
| mmu-miR-451a | 91 |
Figure 3.
Network of miRNA–mRNA interactions after 2 weeks (a), 6 weeks (b), and 3 months (c) of inhalation. The results for individual miRNAs and their targets for adjusted p-value <0.001 and score ≥3 are visualized. Circles represent miRNAs, squares mRNAs. Red fill indicates upregulation, green fill corresponds to downregulation. The lines depict miRNA–mRNA pairs [13].
The KEGG pathway analysis based on miRNA–mRNA interactions revealed 3, 25, 1, and 20 significantly deregulated pathways after 3 days, 2 weeks, 6 weeks, and 3 months of inhalation, respectively (Table 4). Most of these pathways were unique for individual exposure periods. Potentially important results include axon guidance, gap junction, dilated cardiomyopathy, Wnt signaling, calcium signaling (2 weeks), and immune response related pathways (3 months). In addition, olfactory transduction was induced by 2 weeks and 3 months of treatment. The short exposure periods (3 days and 2 weeks) commonly affected the following pathways: ECM receptor interaction, focal adhesion and pathways in cancer. None of the pathways were commonly deregulated in all of the time periods.
Table 4.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways based on miRNA–mRNA interactions deregulated after inhalation of CuO NPs.
| ID | Deregulated Biological Process/Pathway | q-Value | Deregulated Genes (N) | Genes in Pathway (N) |
|---|---|---|---|---|
| 3 Days | ||||
| mmu04510 | Focal adhesion | <0.001 | 19 | 199 |
| mmu04512 | ECM receptor interaction | <0.001 | 11 | 88 |
| mmu05200 | Pathways in cancer | 0.003 | 21 | 539 |
| 2 Weeks | ||||
| mmu04520 | Adherens junction | <0.001 | 27 | 71 |
| mmu04510 | Focal adhesion | <0.001 | 44 | 199 |
| mmu04270 | Vascular smooth muscle contraction | <0.001 | 29 | 140 |
| mmu04360 | Axon guidance | <0.001 | 30 | 180 |
| mmu04916 | Melanogenesis | <0.001 | 24 | 100 |
| mmu05414 | Dilated cardiomyopathy | <0.001 | 23 | 94 |
| mmu05200 | Pathways in cancer | <0.001 | 51 | 539 |
| mmu04330 | Notch signaling pathway | <0.001 | 16 | 54 |
| mmu04540 | Gap junction | <0.001 | 21 | 86 |
| mmu04810 | Regulation of actin cytoskeleton | <0.001 | 36 | 216 |
| mmu04740 | Olfactory transduction | <0.001 | 5 | 1134 |
| mmu04912 | GnRH signaling pathway | <0.001 | 21 | 90 |
| mmu04310 | Wnt signaling pathway | <0.001 | 26 | 162 |
| mmu05217 | Basal cell carcinoma | <0.001 | 14 | 63 |
| mmu04020 | Calcium signaling pathway | 0.001 | 27 | 192 |
| mmu05410 | Hypertrophic cardiomyopathy (HCM) | 0.001 | 17 | 91 |
| mmu04512 | ECM receptor interaction | 0.002 | 16 | 88 |
| mmu05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 0.002 | 15 | 77 |
| mmu05213 | Endometrial cancer | 0.003 | 11 | 58 |
| mmu04144 | Endocytosis | 0.003 | 27 | 269 |
| mmu11190 | Dorso-ventral axis formation | 0.005 | 7 | 22 |
| mmu05211 | Renal cell carcinoma | 0.005 | 13 | 68 |
| mmu05016 | Huntingtons disease | 0.005 | 25 | 199 |
| mmu04720 | Long term potentiation | 0.006 | 13 | 67 |
| mmu04260 | Cardiac muscle contraction | 0.010 | 14 | 86 |
| 6 Weeks | ||||
| mmu04142 | Lysosome | <0.001 | 23 | 124 |
| 3 Months | ||||
| mmu04142 | Lysosome | <0.001 | 43 | 124 |
| mmu04740 | Olfactory transduction | <0.001 | 4 | 1134 |
| mmu04060 | Cytokine-cytokine receptor interaction | <0.001 | 60 | 296 |
| mmu05140 | Leishmania infection | <0.001 | 27 | 69 |
| mmu04062 | Chemokine signaling pathway | <0.001 | 45 | 198 |
| mmu04650 | Natural killer cell mediated cytotoxicity | <0.001 | 34 | 117 |
| mmu04612 | Antigen processing and presentation | <0.001 | 23 | 91 |
| mmu05332 | Graft versus host disease | <0.001 | 15 | 65 |
| mmu04662 | B cell receptor signaling pathway | <0.001 | 22 | 82 |
| mmu04666 | Fc gamma R-mediated phagocytosis | <0.001 | 26 | 90 |
| mmu04620 | Toll like receptor signaling pathway | <0.001 | 25 | 99 |
| mmu05330 | Allograft rejection | <0.001 | 14 | 63 |
| mmu16848 | Epithelial cell signaling in helicobacter pylori infection | <0.001 | 21 | 69 |
| mmu04940 | Type I diabetes mellitus | <0.001 | 15 | 70 |
| mmu04664 | Fc epsilon RI signaling pathway | 0.001 | 20 | 67 |
| mmu05320 | Autoimmune thyroid disease | 0.002 | 14 | 78 |
| mmu05416 | Viral myocarditis | 0.006 | 17 | 88 |
| mmu04660 | T cell receptor signaling pathway | 0.006 | 23 | 103 |
| mmu04640 | Hematopoietic cell lineage | 0.008 | 18 | 95 |
| mmu04672 | Intestinal immune network for IgA production | 0.009 | 12 | 42 |
3.3. Global DNA Methylation after CuO NPs Inhalation
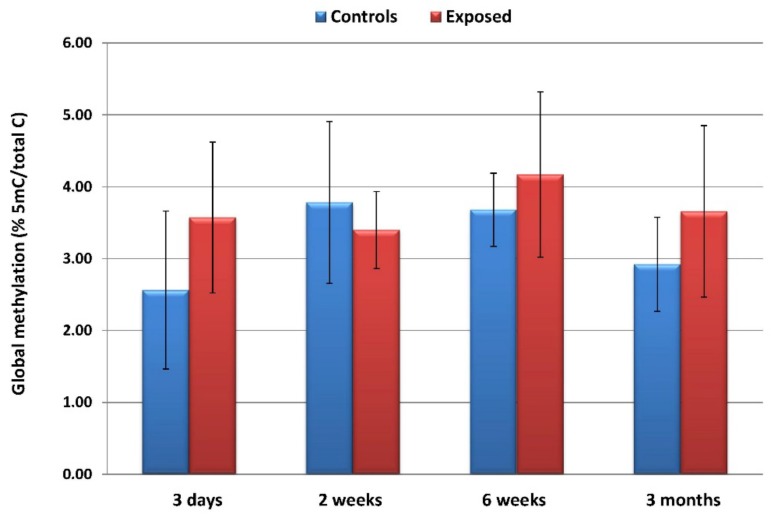
The inhalation of CuO NPs had no significant effect on global DNA methylation in any of the exposure periods. Although a higher proportion of 5-methylcytosine/total cytosine in the exposed than in the controls could be seen after 3 days, 6 weeks, and 3 months of inhalation, the differences did not reach statistical significance. Furthermore, no significant trends in global DNA methylation changes relating to the exposure time were observed (Figure 4).
Figure 4.
The average (±SD) global DNA methylation levels reported as a proportion of 5-methylcytosine/total cytosine in the exposed and control samples.
4. Discussion
In this study, we aimed to investigate the effects of whole-body CuO NPs inhalation on the expression of mRNA and epigenetic markers (expression of miRNA and global DNA methylation) in mice. The strategy of our experiment allowed us to assess the impact of both short-term acute inhalation and long-term chronic exposure.
The toxicity of CuO NPs is relatively high when compared with other metal oxide NPs. The negative effects are mostly mediated by ROS production followed by oxidative stress, destruction of mitochondrial membranes and the induction of downstream processes, resulting in autophagy and/or apoptosis [16], including e.g., activation of p53 which increases Bax/Bcl2 ratio. CuO NPs also cause cell cycle arrest and DNA damage manifested by γ-H2AX formation. They further induce interleukin (IL)-8 production, which consequently causes the activation of NF-κB pathway [17]. CuO NPs have been shown to bind to proteins via hydrogen bonds [18] causing changes of protein phosphorylation and ubiquitination, which plays an important role in cell signaling. They upregulate the expression of DNA repair proteins and directly interact with the structural elements of the cell (e.g., cytoskeleton). This may disrupt the mitotic spindle and cause aberrant cell division [16]. CuO NPs further directly interact with DNA, causing a replication arrest possibly leading to genome instability. Finally, copper has been shown to affect gene expression through epigenetic mechanisms, specifically by histone modification [19].
Despite many biological effects associated with CuO NPs exposure, studies on gene expression and epigenetic changes induced by these NPs are very rare. To date, only one study that focused on the analysis of whole genome transcriptional changes following the inhalation of CuO NPs [10] has been published. The authors exposed rats for 6 hours to two doses (3.3 and 13.2 mg/m3) of CuO NPs and analyzed the transcriptomics response 1 day post-exposure and after a 22 day recovery period. The exposure resulted in the deregulation of approximately 1000 genes in the high-dose group and approximately 200 genes in the low-dose group. After the recovery period, the number of deregulated genes dropped to around 20. The main processes affected by the exposure included cell proliferation/survival and inflammation. Interestingly, no oxidative stress-related pathways were affected. The authors observed proliferation of alveolar epithelial cells and detected the upregulation of CCl2 (monocyte chemoattractant protein 1), both on the level of mRNA and protein and the upregulation of Epithelial Cell Transforming 2 ECT2 oncoprotein. They concluded that short-term exposure to CuO NPs affected the processes associated with acute inflammation in the lungs and tumorigenesis. They pointed out that long-term exposure studies would be of interest.
Recently, we have shown that the inhalation of CuO NPs, affects the systemic immune response of the exposed mice [8]. Changes to the proportion of eosinophils, neutrophils, macrophages, and antigen-presenting cells (innate immunity), along with the modulation of proliferative and secretory activity of T cells were observed. The effects were dependent on the time of exposure: the production of cytokines increased after 3 days of inhalation, decreased after 2 weeks and then returned to the control levels at later time intervals. The proportion of cells of innate immunity decreased on day 3, increased after 2 weeks, and again decreased after 3 months of exposure.
In this study, we extended our investigation to gene expression and the epigenetic effects in the lungs of CuO NPs-exposed mice. In contrast to the study by Costa et al. [10], we analyzed whole genome mRNA expression changes, using next generation sequencing of the transcriptome, which allowed us to detect a higher number of deregulated mRNAs with greater sensitivity. We further assessed miRNA expression and focused on the detection of miRNA–mRNA interactions, using experimentally validated deregulated molecules. Although the number of differentially expressed mRNAs generally increased with the exposure time, the data suggested substantial differences between the effects induced by 3 days of inhalation and longer exposure periods. There was very little overlap between mRNAs commonly affected by all of the inhalation periods: only two genes (heme oxygenase 1, involved in oxidative stress response, and solute carrier family 7, encoding cystine/glutamate transporter) were deregulated, while for 2 weeks, 6 weeks, and 3 months exposures, 159 common transcripts were detected. Among these transcripts, immune response-related genes were prevalent [e.g., interleukin 1 beta, tumor necrosis factor, CD14, CD33, and CD80 antigens, chemokine (C–C motif) ligands, or toll-like receptor]. For the short exposure period, the greatest proportion of unique mRNA transcripts was found (138 unique of 170 total deregulated transcripts, 81%), followed by 3 months of exposure (1108 unique of 1551 total transcripts, 71%), while for 2 weeks and 6 weeks of treatments, the corresponding percentage was 38% and 35%, respectively. Genes deregulated after 3 days exposure included e.g., several collagen-encoding genes, zinc finger proteins, or chondroitin polymerizing factor. Two-week exposure resulted in changes of expression of apoptosis-relates genes (BCL2-associated athanogene 3, cell death inducing Trp53 target 1, and programmed cell death 1), as well as cytochrome P450 1B1 and superoxide dismutase 2 genes. After 6 weeks of inhalation, expression of genes involved in cell cycle was detected (cyclin-dependent kinase 1, aurora kinase B, spindle and kinetochore associated complex subunit 3, regulator of cell cycle, and cyclin A2). Three months of exposure was associated with deregulation of e.g., WNT1 inducible signaling pathway protein 2, jun B proto-oncogene, B cell leukemia/lymphoma 2 related protein A1a, mitogen-activated protein kinase 4, and B cell leukemia/lymphoma 3 that are all potentially implicated in carcinogenesis.
To further investigate the biological impacts of CuO NPs inhalation, we assessed the biological processes and pathways deregulated as a result of exposure. Again, a distinct response was found for the short inhalation period. It was generally associated with collagen metabolism and extracellular matrix organization. Collagen, the major part of extracellular matrix, is fundamental to maintain the structure and function of the lungs. In a recent study, collagen accumulation, along with the expression of the progressive fibrosis marker α-SMA in the lungs of C57BL/6 mice exposed to CuO NPs by intranasal delivery, was detected [20] suggesting the possible induction of pulmonary fibrosis. Similarly, we also found the upregulation of α-SMA expression. Our results therefore indicate that even a short, 3-day inhalation of CuO NPs, may negatively affect lung tissues. Longer inhalation periods were commonly characterized by changes of immune response-related biological processes and pathways. This observation is in agreement with previous studies [1,6,8], indicating the induction of local and systemic inflammatory responses after chronic CuO NPs exposures. We further focused on the identification of biological processes and pathways unique for 2 weeks, 6 weeks, and 3 months of inhalation, respectively. A two-week inhalation period affected the processes associated with apoptosis and programmed cell death. This result is comparable with the observations of Lai et al., who found time- and dose-dependence increased the intensity of the TUNEL-positive cells in the lung sections of mice, after the intranasal delivery of CuO NPs [20]. A six-week exposure period impacted on cell cycle-related processes. Although no similar results in mice have been reported, it has been shown that CuO NP may cause a cell cycle arrest in human keratinocytes, mouse embryonic fibroblasts [21], and in human umbilical vein endothelial cells (HUVECs) [22]. In a model of poorly differentiated hepatocellular carcinoma cells, the cell cycle arrest was possibly caused by histone H2AX phosphorylation, resulting from DNA damage [17]. The longest 3-month inhalation period caused changes in cell adhesion, cell migration, and related processes. These processes depend on extracellular matrix organization and their misregulation is implicated in tumorigenesis [23]. Although no relevant in vivo animal studies have been published, in vitro experiments in model cell lines suggest the inhibition of cell migration after exposure to CuO NPs [21,22].
To the best of our knowledge, our study is the first one to investigate miRNA expression changes after CuO NPs inhalation in mice. We found relatively low numbers of differentially expressed miRNAs that increased with the length of inhalation. For the short (3 day) exposure period no significant results were observed. Although no reports on miRNA expression after CuO NPs inhalation have been published, some of the miRNAs we detected were implicated after exposure to other NPs. Specifically, the expression of mmu-miR-21a-5p, mmu-miR-147-3p, mmu-miR-449a-5p, and mmu-miR-703 was deregulated in the lungs of mice exposed to multi-walled carbon nanotubes [24] and mmu-miR-342-3p in mice exposed to nano-TiO2 particles [25]. The deregulation of the miRNAs observed in our study was further found in the lungs of mice after cigarette smoke exposure (mmu-miR-21a-5p, mmu-miR-135b-5p, mmu-miR-146a-5p, mmu-miR-146b-5p, mmu-miR-155-5p, mmu-miR-342-3p, mmu-miR-449a-5p, mmu-miR-489-3p, and mmu-miR-672-5p) [26], in mice with cigarette smoke induced atherosclerosis (mmu-miR-21a-5p and mmu-miR-155-5p) [27], in the lungs of mice after radon inhalation (mmu-miR-21a-3p, mmu-miR-135b-5p, mmu-miR-146b-5p, mmu-miR-1298-5p, mmu-miR-6972-5p, and mmu-miR-703) [28], in mice with lung tumors (mmu-miR-135b-5p, mmu-miR-146b-5p, mmu-miR-223-3p, and mmu-miR-1298-5p) [29,30,31], in an ovalbumin-induced asthma mouse model (mmu-miR-21a-5p, mmu-miR-135b-5p, mmu-miR-146a-5p, mmu-miR-155-5p, mmu-miR-223-3p, and mmu-miR-451a) [32], and in mice with pneumonia and influenza infections (mmu-miR-21a-5p, mmu-miR-144-5p, mmu-miR-155-5p, and mmu-miR-223-3p) [33,34,35]. This overall suggests that the inhalation of CuO NPs induces a similar response as the above-mentioned adverse conditions and therefore likely has negative effects on the organism.
To further study the biological impacts of CuO NPs, we searched for miRNA–mRNA interactions by correlating the miRNA expression with mRNAs significantly deregulated by the inhalation. In contrast to the number of deregulated mRNAs that was lower after 6 weeks than 2 weeks of exposure, the number of interactions increased with the exposure time. In addition, the number of miRNAs targeting mRNAs was highest after 6 weeks of inhalation. Although it is difficult to explain these discrepancies, we may speculate that involvement of piRNAs detected in these interactions after 6 weeks of inhalation plays a role. piRNAs have been shown to be expressed in a tissue-specific manner and modulate various signaling pathways on both transcriptional and post-transcriptional manner [36]. Unlike miRNAs, they preferentially target transposons. Their abnormal expression in various cancers has been reported [36].
The results of miRNA–mRNA interactions analysis became a basis for the assessment of deregulated KEGG pathways. These pathways, specific for the response to CuO NPs exposure, partly overlapped with the deregulated biological processes and pathways identified for mRNA expression. Such pathways included e.g., ECM receptor interaction and focal adhesion (3-day exposure) or immune response related pathways (3-month inhalation). In addition, some unique pathways have been identified. Deregulation of Axon guidance (2 weeks) suggests changes in cell migration, which are potentially linked to cancer development [37]. Changes in Gap junction pathway (2 weeks) may affect intercellular communications which are frequently dysregulated in cancer [38]. Cell–cell communication deregulation is further related to Wnt signaling pathway (2 weeks) [39]. Calcium signaling, a key mechanism for the intercellular communication that is altered in lung cancer [40] also shows potential adverse effects of CuO NPs inhalation. The association of CuO NPs inhalation with cancer is further supported by the fact that pathways in cancer were deregulated after 3 days and 2 weeks of exposure. The deregulation of dilated cardiomyopathy pathway is most likely linked to changes in the immune response in the exposed mice [41]. The induction of olfactory transduction pathway after 2 weeks and 3 months of treatment is unexpected, considering the fact that non-olfactory tissues were investigated. However, it has been shown that mouse pulmonary macrophages can express olfactory receptors as a result of their activation by interferon-γ [42]. Thus, the deregulation of this pathway may again reflect changes in immune response following the inhalation. Finally, we should mention the deregulation of lysosome pathway, detected after 6 weeks and 3 months of exposure. This pathway is related to synthesis of lysosomal enzymes that help to degrade foreign structures in the cells. In a recent study, SiO2 NPs were found in the lysosomes of macrophages of exposed mice [43]. We can therefore expect that CuO NPs inhalation affected the lysosome pathway in our study.
We performed global DNA methylation analysis to identify the possible effects of CuO NPs inhalation on the epigenome but found no differences between the exposed and control animals in any of the time intervals. Global DNA methylation has recently been investigated by Lu et al. in mice exposed to CuO NPs by intratracheal instillation [44]. In contrast to our results, the authors found increased levels of 5-hydroxymethylcytosine in the lungs after 24-h exposure. In another study, CuO NPs had no effect on DNA methylation of selected inflammation-related genes [10]. Thus, CuO NPs have the potential to modify DNA methylation levels, but the response depends on the mode of administration and length of exposure. It should also be noted that the results may depend on the method used to analyze DNA methylation: a gene-specific approach would most likely yield a different outcome than the global DNA methylation analysis.
5. Conclusions
CuO NPs are used in many industrial products and as antimicrobial agents. Although their production is lower than other metal-based NPs, their toxicity is high. They have been shown to induce oxidative stress, apoptosis, and cell cycle arrest in vitro. In laboratory animals exposed to CuO, NPs inflammation was most commonly detected, but neurotoxicity was also noted. Our study focused on transcriptomic and epigenetic changes in mice following CuO NPs inhalation and revealed distinct effects depending on the length of exposure. We observed a deregulation of extracellular matrix organization, impacts on immune response, cell cycle regulation, cell adhesion and apoptosis, as well as pathways potentially implicated in carcinogenesis. Due to the potential negative health effects of exposure in humans, more studies are needed to better characterize the mechanisms of CuO NPs toxicity in more detail.
Acknowledgments
Michal Kolar and Jan Kubovciak, Core facility of the Institute of Molecular Genetics of the Czech Academy of Sciences, Prague, Czech Republic are acknowledged for providing mRNA and miRNA sequencing services.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-4991/10/3/550/s1. Supplementary Table S1. Lists of differentially expressed genes after 3 days, 2 weeks, 6 weeks, and 3 months of exposure to CuO NPs. The numbers of deregulated genes after individual exposure periods are also shown. Supplementary Table S2. Identification of common and unique deregulated genes after exposure to CuO NPs. The numbers of genes are shown in the Venn diagram (Figure 1). Supplementary Table S3. Lists of deregulated miRNAs after 2 weeks, 6 weeks, and 3 months of exposure to CuO NPs. The numbers of deregulated miRNAs after individual exposure periods are also shown. Supplementary Table S4. Identification of common and unique deregulated miRNAs after exposure to CuO NPs. The numbers of miRNAs are shown in the Venn diagram (Figure 2). Supplementary Table S5. The interactions between experimentally validated, significantly deregulated miRNAs and mRNAs after individual exposure periods. The numbers of interactions are also shown. miRNAs involved in the interaction and numbers of target mRNAs are reported in Table 3. Supplementary Table S6. The details on interactions between miRNAs and mRNAs identified by the interaction networks shown in Figure 3.
Author Contributions
Conceptualization: P.R., A.R., V.H., M.M. and J.T.; Data curation: J.K.; Formal analysis: J.K.; Funding acquisition: P.R.; Investigation: K.V., T.Z., A.M., Z.V., P.M., P.C., L.C., K.K. and B.D.; Methodology: Z.V. and P.M.; Project administration: P.R.; Software: J.K.; Supervision: P.R.; Writing—original draft: P.R.; Writing—review & editing: K.V., A.R., T.Z., A.M., J.K., Z.V., P.M., P.C., L.C., K.K., B.D., V.H., M.M. and J.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by project LO1508 from the Ministry of Education, Youth and Sports of the Czech Republic and by project P503/12/G147 from Czech Science Foundation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Magaye R., Zhao J., Bowman L., Ding M. Genotoxicity and carcinogenicity of cobalt-, nickel- and copper-based nanoparticles. Exp. Ther. Med. 2012;4:551–561. doi: 10.3892/etm.2012.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sengupta J., Ghosh S., Datta P., Gomes A., Gomes A. Physiologically important metal nanoparticles and their toxicity. J. Nanosci. Nanotechnol. 2014;14:990–1006. doi: 10.1166/jnn.2014.9078. [DOI] [PubMed] [Google Scholar]
- 3.Bondarenko O., Juganson K., Ivask A., Kasemets K., Mortimer M., Kahru A. Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: A critical review. Arch. Toxicol. 2013;87:1181–1200. doi: 10.1007/s00204-013-1079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubilar O., Rai M., Tortella G., Diez M.C., Seabra A.B., Durán N. Biogenic nanoparticles: Copper, copper oxides, copper sulphides, complex copper nanostructures and their applications. Biotechnol. Lett. 2013;35:1365–1375. doi: 10.1007/s10529-013-1239-x. [DOI] [PubMed] [Google Scholar]
- 5.Su Y., Zheng X., Chen Y., Li M., Liu K. Alteration of intracellular protein expressions as a key mechanism of the deterioration of bacterial denitrification caused by copper oxide nanoparticles. Sci. Rep. 2015;5:15824. doi: 10.1038/srep15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahamed M., Akhtar M.J., Alhadlaq H.A., Alrokayan S.A. Assessment of the lung toxicity of copper oxide nanoparticles: Current status. Nanomedicine. 2015;10:2365–2377. doi: 10.2217/nnm.15.72. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Gao Y., Zhang L., Wang T., Wang J., Jiao F., Li W., Liu Y., Li Y., Li B., et al. Potential health impact on mice after nasal instillation of nano-sized copper particles and their translocation in mice. J. Nanosci. Nanotechnol. 2009;9:6335–6343. doi: 10.1166/jnn.2009.1320. [DOI] [PubMed] [Google Scholar]
- 8.Holan V., Javorkova E., Vrbova K., Vecera Z., Mikuska P., Coufalik P., Kulich P., Skoupy R., Machala M., Zajicova A., et al. A murine model of the effects of inhaled CuO nanoparticles on cells of innate and adaptive immunity—a kinetic study of a continuous three-month exposure. Nanotoxicology. 2019;13:952–963. doi: 10.1080/17435390.2019.1602679. [DOI] [PubMed] [Google Scholar]
- 9.Yokohira M., Hashimoto N., Yamakawa K., Suzuki S., Saoo K., Kuno T., Imaida K. Lung Carcinogenic Bioassay of CuO and TiO2 Nanoparticles with Intratracheal Instillation Using F344 Male Rats. J. Toxicol. Pathol. 2009;22:71–78. doi: 10.1293/tox.22.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa P.M., Gosens I., Williams A., Farcal L., Pantano D., Brown D.M., Stone V., Cassee F.R., Halappanavar S., Fadeel B. Transcriptional profiling reveals gene expression changes associated with inflammation and cell proliferation following short-term inhalation exposure to copper oxide nanoparticles. J. Appl. Toxicol. 2018;38:385–397. doi: 10.1002/jat.3548. [DOI] [PubMed] [Google Scholar]
- 11.Vecera Z., Mikuska P., Moravec P., Smolik J. Unique Exposure System for the Whole Body Inhalation Experiments with Small Animals. Tanger Ltd; Brno, Czech Republic: 2011. [Google Scholar]
- 12.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vila-Casadesús M., Gironella M., Lozano J.J. MiRComb: An R Package to Analyse miRNA–mRNA Interactions. Examples across Five Digestive Cancers. PLoS ONE. 2016;11:e0151127. doi: 10.1371/journal.pone.0151127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ru Y., Kechris K.J., Tabakoff B., Hoffman P., Radcliffe R.A., Bowler R., Mahaffey S., Rossi S., Calin G.A., Bemis L., et al. The multiMiR R package and database: Integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42:e133. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson M.D., McCarthy D.J., Smyth G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chibber S., Shanker R. Can CuO nanoparticles lead to epigenetic regulation of antioxidant enzyme system? CuO NPs can result in epigenetic regulation: A hypothetical view. J. Appl. Toxicol. 2017;37:84–91. doi: 10.1002/jat.3392. [DOI] [PubMed] [Google Scholar]
- 17.Kung M.-L., Hsieh S.-L., Wu C.-C., Chu T.-H., Lin Y.-C., Yeh B.-W., Hsieh S. Enhanced reactive oxygen species overexpression by CuO nanoparticles in poorly differentiated hepatocellular carcinoma cells. Nanoscale. 2015;7:1820–1829. doi: 10.1039/C4NR05843G. [DOI] [PubMed] [Google Scholar]
- 18.Rabbani G., Khan M.J., Ahmad A., Maskat M.Y., Khan R.H. Effect of copper oxide nanoparticles on the conformation and activity of β-galactosidase. Colloids Surf. B Biointerfaces. 2014;123:96–105. doi: 10.1016/j.colsurfb.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Cheng T.F., Choudhuri S., Muldoon-Jacobs K. Epigenetic targets of some toxicologically relevant metals: A review of the literature. J. Appl. Toxicol. 2012;32:643–653. doi: 10.1002/jat.2717. [DOI] [PubMed] [Google Scholar]
- 20.Lai X., Zhao H., Zhang Y., Guo K., Xu Y., Chen S., Zhang J. Intranasal Delivery of Copper Oxide Nanoparticles Induces Pulmonary Toxicity and Fibrosis in C57BL/6 mice. Sci. Rep. 2018;8:4499. doi: 10.1038/s41598-018-22556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo C., Li Y., Yang L., Long J., Zheng Y., Liu J., Xiao S., Jia J. Activation of Erk and p53 regulates copper oxide nanoparticle-induced cytotoxicity in keratinocytes and fibroblasts. Int. J. Nanomed. 2014;9:4763–4772. doi: 10.2147/IJN.S67688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song H., Wang W., Zhao P., Qi Z., Zhao S. Cuprous oxide nanoparticles inhibit angiogenesis via down regulation of VEGFR2 expression. Nanoscale. 2014;6:3206. doi: 10.1039/c3nr04363k. [DOI] [PubMed] [Google Scholar]
- 23.Andl C.D. The Misregulation of Cell Adhesion Components during Tumorigenesis: Overview and Commentary. J. Oncol. 2010;2010:1–9. doi: 10.1155/2010/174715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dymacek J., Snyder-Talkington B.N., Porter D.W., Mercer R.R., Wolfarth M.G., Castranova V., Qian Y., Guo N.L. mRNA and miRNA Regulatory Networks Reflective of Multi-Walled Carbon Nanotube-Induced Lung Inflammatory and Fibrotic Pathologies in Mice. Toxicol. Sci. 2015;144:51–64. doi: 10.1093/toxsci/kfu262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halappanavar S., Jackson P., Williams A., Jensen K.A., Hougaard K.S., Vogel U., Yauk C.L., Wallin H. Pulmonary response to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in microRNAs: A toxicogenomic study. Environ. Mol. Mutagen. 2011;52:425–439. doi: 10.1002/em.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conickx G., Avila Cobos F., Van den Berge M., Faiz A., Timens W., Hiemstra P.S., Joos G.F., Brusselle G.G., Mestdagh P., Bracke K.R. microRNA profiling in lung tissue and bronchoalveolar lavage of cigarette smoke-exposed mice and in COPD patients: A translational approach. Sci. Rep. 2017;7:12871. doi: 10.1038/s41598-017-13265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama Y., Mise N., Suzuki Y., Tada-Oikawa S., Izuoka K., Zhang L., Zong C., Takai A., Yamada Y., Ichihara S. MicroRNAs as Potential Mediators for Cigarette Smoking Induced Atherosclerosis. Int. J. Mol. Sci. 2018;19:1097. doi: 10.3390/ijms19041097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei W., Tao L., Zhang L.W., Zhang S., Cao J., Jiao Y., Tong J., Nie J. Circular RNA profiles in mouse lung tissue induced by radon. Environ. Health Prev. Med. 2017;22:36. doi: 10.1186/s12199-017-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luettich K., Xiang Y., Iskandar A., Sewer A., Martin F., Talikka M., Vanscheeuwijck P., Berges A., Veljkovic E., Gonzalez-Suarez I., et al. Systems toxicology approaches enable mechanistic comparison of spontaneous and cigarette smoke-related lung tumor development in the A/J mouse model. Interdiscip. Toxicol. 2014;7:73–84. doi: 10.2478/intox-2014-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y., Zhang Q., Du M., Xiong D., Wang Y., Mohammed A., Lubet R., Wang L., You M. Exosomal miRNAs as Novel Pharmacodynamic Biomarkers for Cancer Chemopreventive Agent Early Stage Treatments in Chemically Induced Mouse Model of Lung Squamous Cell Carcinoma. Cancers. 2019;11:477. doi: 10.3390/cancers11040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong D., Pan J., Zhang Q., Szabo E., Miller M.S., Lubet R.A., You M., Wang Y. Bronchial airway gene expression signatures in mouse lung squamous cell carcinoma and their modulation by cancer chemopreventive agents. Oncotarget. 2017;8:18885–18900. doi: 10.18632/oncotarget.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plank M.W., Maltby S., Tay H.L., Stewart J., Eyers F., Hansbro P.M., Foster P.S. MicroRNA Expression Is Altered in an Ovalbumin-Induced Asthma Model and Targeting miR-155 with Antagomirs Reveals Cellular Specificity. PLoS ONE. 2015;10:e0144810. doi: 10.1371/journal.pone.0144810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan K., Choi H., Jiang X., Yin L., Seet J., Patzel V., Engelward B.P., Chow V.T. Micro-RNAs in regenerating lungs: An integrative systems biology analysis of murine influenza pneumonia. BMC Genom. 2014;15:587. doi: 10.1186/1471-2164-15-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez J.C., Dang H., Kanke M., Hagan R.S., Mock J.R., Kelada S.N.P., Sethupathy P., Doerschuk C.M. Predicted effects of observed changes in the mRNA and microRNA transcriptome of lung neutrophils during S. pneumoniae pneumonia in mice. Sci. Rep. 2017;7:11258. doi: 10.1038/s41598-017-11638-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi E.-J., Kim H.B., Baek Y.H., Kim E.-H., Pascua P.N.Q., Park S.-J., Kwon H., Lim G.-J., Kim S., Kim Y.-I., et al. Differential microRNA expression following infection with a mouse-adapted, highly virulent avian H5N2 virus. BMC Microbiol. 2014;14:252. doi: 10.1186/s12866-014-0252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Dou M., Song X., Dong Y., Liu S., Liu H., Tao J., Li W., Yin X., Xu W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer. 2019;18:123. doi: 10.1186/s12943-019-1052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasarre P., Potiron V., Drabkin H., Roche J. Guidance molecules in lung cancer. Cell Adhes. Migr. 2010;4:130–145. doi: 10.4161/cam.4.1.10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aasen T., Sansano I., Montero M.Á., Romagosa C., Temprana-Salvador J., Martínez-Marti A., Moliné T., Hernández-Losa J.Y., Cajal S.R. Insight into the Role and Regulation of Gap Junction Genes in Lung Cancer and Identification of Nuclear Cx43 as a Putative Biomarker of Poor Prognosis. Cancers. 2019;11:320. doi: 10.3390/cancers11030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan T., Rindtorff N., Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H., Zhang Q., He J., Lu W. Regulation of calcium signaling in lung cancer. J. Thorac. Dis. 2010;2:52–56. [PMC free article] [PubMed] [Google Scholar]
- 41.Sanghera C., Wong L.M., Panahi M., Sintou A., Hasham M., Sattler S. Cardiac phenotype in mouse models of systemic autoimmunity. Dis. Model. Mech. 2019;12:036947. doi: 10.1242/dmm.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J.J., Tay H.L., Plank M., Essilfie A.-T., Hansbro P.M., Foster P.S., Yang M. Activation of Olfactory Receptors on Mouse Pulmonary Macrophages Promotes Monocyte Chemotactic Protein-1 Production. PLoS ONE. 2013;8:e80148. doi: 10.1371/journal.pone.0080148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsugita M., Morimoto N., Nakayama M. SiO2 and TiO2 nanoparticles synergistically trigger macrophage inflammatory responses. Part. Fibre Toxicol. 2017;14:11. doi: 10.1186/s12989-017-0192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu X., Miousse I.R., Pirela S.V., Moore J.K., Melnyk S., Koturbash I., Demokritou P. In vivo epigenetic effects induced by engineered nanomaterials: A case study of copper oxide and laser printer-emitted engineered nanoparticles. Nanotoxicology. 2016;10:629–639. doi: 10.3109/17435390.2015.1108473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.