Abstract
In spite of great developments in the agricultural field and plant productivity in the last decades, the concern about the control of agricultural pests is still continuous. However, pest management is expected to have more effects from nanomaterials by providing innovative solutions. The current study confirms the biotransformation of copper nanoparticles (CuNPs) using a cell-free culture extract of metal copper-resistant bacteria Pseudomonas fluorescens MAL2, which was isolated from heavy metal-contaminated soils collected from Sharqia Governorate, Egypt. The local screened bacterial isolate, Pseudomonas fluorescens MAL2, is similar to Pseudomonas fluorescens DSM 12442T DSM. After optimization of growth conditions, F-Base medium was found to be the best medium and pH 7, temperature 35 °C, concentration of CuSO4·5H2O 300 ppm, 10 mL supernatant: 40 mL CuSO4·5H2O (300 ppm), and reaction time 90 min were recorded as the best growth conditions to the fabrication of CuNPs. The formed CuNPs were characterized using initially visual observation of the change in the color of the reaction mixture from blue color to the dark green as an indication of CuNPs biotransformation. Then, UV–Vis spectroscopy showed a maximum absorption at 610 nm under the optimum conditions performed. Transmission Electron Microscopy (TEM) revealed the formation of spherical aspect with size ranges from 10:70 nm; moreover, Energy Dispersive X-ray spectroscopy (EDX) indicated the presence of CuNPs and other elements. In addition, the presence of alcohols, phenols, alkenes, and amines is confirmed by Fourier-Transform Infrared spectroscopy (FTIR) spectroscopy analysis. Dynamic Light Scattering (DLS) supported that the Zeta-average size of nanoparticle was 48.07 with 0.227 PdI value. The Zeta potential showed −26.00mV with a single peak. The biosynthesized CuNPs (Bio CuNPs) showed toxicity against the stored grain pest (Tribolium castaneum), where LC50 value was 37 ppm after 5 days of treatment. However, the negligible effect was observed with chemical synthesis of CuNPs (Ch CuNPs) at the same concentration. The results suggest that Bio CuNPs could be used not only as a biocontrol agent, but also as an ecofriendly and inexpensive approach for controlling the stored grain pests.
Keywords: copper nanoparticles, Pseudomonas fluorescens, insecticidal, Tribolium castaneum
1. Introduction
Over the past decade, many exploratory experiments have been conducted to demonstrate the effectiveness of the powerful impact of nanotechnology and its applications [1,2]. The size of solid nanoparticles is ranging from 1 to 100 nm [3,4,5]. Copper nanoparticles (CuNPs) have been used in full applications such as agriculture, medicine, environment, industrial engineering, and various technological fields. Recently, studies in the agriculture field emphasized the influence of some simple elements related to the economics of plants [4,5,6,7]. The combination of biological principles with different chemical and physical methods is called bio-nanotechnology producing nanoparticles with specific functions [8,9]. The chemical synthesis of nanoparticles is a very costly method, and it is considered toxic with low productivity. Subsequently, the biological synthesis methods from plants or microbes have attractive application because they are costless, safe, rapid, and ecofriendly [8,9,10].
The biotransformation of NPs using microbes has attracted much researcher interest because of its intrinsic qualities and potential applications in different fields. Each microorganism has a specific mechanism to form nanoparticles [11]. Target ions from the environment were grasped on the surface or inside the microbial cells, and then the cell enzymes can reduce the metal ions size to nanoparticles by the electrostatic interaction [12]. CuO NPs can be synthesized from Gram-negative bacterium, which belongs to the genus Serratia [13]. Non-pathogenic Pseudomonas stutzeri can produce spherical CuNPs with size ranges from 8 to 15 nm; however, it can synthesize cubical CuNPs sized 50 to 150 nm from electroplating wastewater [14]. Furthermore, Escherichia coli can synthesize CuO NPs with variable sizes and shapes [15]. Currently, there is an increasing interest in using nanoscience in agriculture for several purposes such as enhancing nutrient absorption and delivery, detecting diseases, and improving the pesticides deliveries to the target sites; thereby, it can enhance our understanding of the crop’s biology [16,17].
The essential role of nanoparticles for the control of agricultural arthropods and medical-veterinary pests was reported [18]. The arthropods represent the most abundant phylum of the animal kingdom with fundamental economic importance. Arthropods which include phytophagous insects and mite species can destroy not only the grown crops but also the stored agricultural products [19]. The red flour beetle, Tribolium castaneum Herbst (Coleoptera: Tenebrionidae), is a global stored product pest particularly stored in grains and foodstuffs [20], damaging their quality and quantity [21]. This beetle causes a significant lack in the weight and nutritive contents of grains, reducing the commercial value and reducing the germination ratio of grains [22]. T. castaneum has been globally used as a model organism in pesticide and ecotoxicology researches [23]. Copper nanoparticles (CuNPs) exhibited fungicidal and insecticidal activity against the pests of crop plants. They can be used as CuNP-based nano-pesticides, nano-herbicides, and nano-fertilizers [24].
Therefore, the current investigation describes the biotransformation of CuNPs using metal copper-resistant Pseudomonas fluorescens under optimized growth conditions, and it sheds lights on the characterization of the produced CuNPs by available advanced techniques, as well as investigating the CuNPs bioresources and their potential pesticidal activity against the storage pest (T. castaneum).
2. Materials and Methods
Metal contaminated soil samples were collected from different sites next to some metal factories in Sharqia Governorate, Egypt. They were mixed and considered to be source material for bacterial isolation. The isolation of metal (Cu)-resistant bacteria was done by Replica plate technique using nutrient agar (NA) plates supplemented with 100 ppm of filtered and sterilized CuSO4·5H2O solution incubated at 37 °C for 48 h, and they were observed for the presence of bacterial growth [25], then the recovered cultured isolates were considered as copper-resistant bacteria. Only three bacterial isolates were recovered because of their good aspects and distribution on the plates. The bacterial isolates (MAL1, MAL2, and MAL3) were subjected to a screening process, which was done to choose the best isolate that biosynthesizes copper nanoparticles.
2.1. Screening for Copper Nanoparticles Synthesis
All the metal (Cu)-resistant bacterial isolates were further studied qualitatively for copper nanoparticle synthesis [25,26]. The cultures were cultivated separately at pH 7 and 37 °C for 24 h in Luria–Bertani broth (LB). Bacterial cells were separated by centrifugation at 20,000 rpm for 10 min. Cell-free filtrate material was separated out and used for extracellular synthesis of nanoparticles. For nanoparticle synthesis, approximately 10 mL of supernatant from each cell-free culture was mixed individually with 40 mL of 100 ppm of filtered and sterilized CuSO4·5H2O solutions (1:4 ratio) in 250 mL conical flasks containing 100 mL of the reaction mixture and placed in a 120 rpm incubatory shaker (LAB—Line R ORBIT environ shaker, AK, USA) at 37 °C up to 24 h. Qualitative screening for the presence of copper nanoparticles was done by periodic visual observation to check the color change, where the color changed from blue to heavy green. The appearance of heavy or dark green color in the flask reveals the biotransformation of CuNPs in the reaction mixture [27], also a set of flasks without CuSO4·5H2O were maintained as control.
2.2. Bacterial Isolates Identification
The chosen isolate was identified as per their morphological, biochemical, and physiological characteristics through tests designated in Manual of Systematic Bacteriology by Bergey’s et al. [28]. It was then identified at molecular level by matrix-assisted laser desorption ionization-time of flight (MALDI TOF, VITEK MS RUO; bioMérieux, Marcy l’Etoile, France) mass spectrometry [29], to confirm the previous identity of isolated bacteria.
2.3. Biosynthesis of CuNPs by Pseudomonas fluorescens
The isolated bacteria, Pseudomonas fluorescens MAL2, was grown in F-Base medium flasks (Kings B medium), which contained peptone, 20 g/L; K2HPO4, 1.5 g/L; MgSO4, 1.5 g/L; Agar, 15 g/L and glycerol, 10 mL pH 7.2, where the flasks were incubated at 30 °C and 150 rpm for incubation (Lab-line® Incubator-Shaker-ORBIT-USA, Suite D Annapolis Junction, MD 20701, USA). Just after 48 h, the culture was subjected to centrifugation at 10,000 rpm for 20 min at 30 °C for cell pellet and cell-free extract collection [27].
One gram of cell pellet was suspended separately in 20 mL of (100, 300, 500, 700 and 900 ppm) CuSO4·5H2O solutions. Some altered volumes (10, 20, 10, 40, 40 and 50mL) of cell-free supernatant were also added to several concentrations (100, 300, 500, 700, and 900 ppm) of CuSO4·5H2O solutions in the prepared conical flasks. Then, all flasks were set in shaker incubator for 24 to 48 h at 35 °C and 150 rpm, and then they were investigated for CuNPs biotransformation by using UV–Vis analysis (400–800 nm “model Laxco™, Alpha-1502 Alpha Series Spectrophotometer (Mettler-Toledo, LLC 1900 Polaris Parkway Columbus, OH 43240S)”, 200–1000 nm [30].
2.4. Optimization of CuNPs Biotransformation
Six different parameters, i.e., media type, CuSO4·5H2O concentration, different volumes of cell-free supernatant, pH values, reaction time, and temperature degrees were tested to attain the maximum growth conditions for the biosynthesis of CuNPs, where only one factor was variable, while the other factors were kept constant. Pseudomonas fluorescens MAL2 inoculum (2.5 × 106 CFU/ mL) was prepared and inoculated in three different media flasks): Nutrient broth (NB), F-Base media (Kings B media), and Luria–Bertani broth (LBB), and then incubated at 30 °C and 150 rpm for 48 h. Different concentrations of CuSO4·5H2O (100, 300, 500, 700, and 900 ppm) were added to prepared cell-free supernatants. After this, the optimum concentration recorded from (CuSO4·5H2O) solution was added to the cell-free supernatants at different pH values (4, 5, 6, 7, 8, and 9), and then incubated at 30 °C. The recorded values from the previously testes parameters were used to determine the mixing ratios of cell-free supernatant to CuSO4·5H2O (10:50, 20:40, 10:40, 40:10, 40:20, and 50:10). Finally, the reaction time (30, 60, 90, 150, and 180 min) was also studied using the optimized different parameters. The obtained mixture reactions samples were analyzed at the appropriate times using UV–Vis spectral analysis (400–800 nm model “Laxco™, Alpha-1502 Alpha Series (Mettler-Toledo, LLC 1900 Polaris Parkway Columbus, OH 43240)” Spectrophotometer, 200–1000 nm [30]. After optimization of the measured parameters, the recorded values were applied collectively in an experiment to yield CuNPs, separation and purification processes.
2.5. Characterization of Copper Nanoparticles
Initially, CuNPs formation was recorded by visual observation of color change by the naked eye. Then, the formed CuNPs were again characterized by UV–Vis spectroscopy, prepared mixture reaction was monitored using Laxco™ dual-beam spectrophotometer (Mettler-Toledo, LLC 1900 Polaris Parkway Columbus, OH 43240), model Laxco™, Alpha-1502 Alpha Series Spectrophotometer (Mettler-Toledo, OH 43240), 200–1000 nm) [30] for detecting surface plasmon resonance (SPR) of the obtained absorbance peak for CuNPs. Fourier Transform Infrared Spectroscopy (FTIR) analysis was performed as per [31] (Bruker Tensor 37, Kaller, Germany). Moreover, the suspension of CuNPs was dried [32] and exposed to EDX using a JEOL MODEL JSM-IT200 Series (Peabody, MA 01960, USA) [33]. The morphology and size of CuNPs were determined by TEM. To this end, an aliquot of an aqueous suspension of CuNPs was transferred onto an amorphous carbon-coated copper grid, left for drying, and investigated using TEM JEOL 1010, Japan, as per [34]. A Zetasizer analyzer (Nano “Z2 Malven, Malvern Hills, UK”), for the analysis of Zeta potential, was employed as per the work in [35].
2.6. Chemical Synthesis of CuNPs
The copper nanoparticles were synthesized by using a chemical reduction method described in [36].
2.7. Insect Rearing
The red flour beetles, T. castaneum, were reared on ground wheat grains mixed with 5% dried yeast under laboratory conditions 32 ± 2 °C and 68 ± 2% rh., and 12:12 h (Light: Dark). Beetle adults used in the bioassay experiment were 7–14 days old.
2.8. Insect Bioassay
Efficacy of chemical and biosynthesis of CuNPs against T. castaneum was tested at six different concentrations (300, 250, 200, 150, 100, and 50 μg mL−1), which were prepared by dilutions with distilled water. Distilled water was used as a negative control treatment. Procedures materials used in the biosynthesis of copper nanoparticles (Pseudomonas supernatant and CuSO4·5H2O with 300 ppm concentration) were evaluated as a positive control. Two milliliters of each concentration were added carefully to 20 g of wheat grains and shaken to uniformly distribute the used concentration on wheat grains, then left to air dry. Twenty adults were exposed to each concentration separately. Three replicates were performed in all experiments. The mortality rate of adults was registered after 24 h till the fifth day.
3. Statistical Analysis
Probit analysis, according to the work in [37], using IBM SPSS software (Armonk, NY 10504-1722, USA, version 20) was employed in analyzing the concentration-mortality response. LC50 values for each exposure period and their fiducial limits were estimated. Control mortality was zero, and no corrections were necessary.
4. Results and Discussion
4.1. Isolation of Metal Resistant Bacteria from Soil
From the isolation process, it was observed that among the cultured colonies, only three isolates were recovered on the basis of their good growth and aspect on nutrient agar plates supplemented with 100 ppm concentration of CuSO4·5H2O. Therefore, they were considered as copper-resistant bacterial isolates, and they have the ability to biosynthesis the CuNPs. Findings on the existence of copper-resistant bacteria in copper-contaminated soils are in accordance with Altimira et al. [38], as they have isolated the silver-resistant bacteria from heavy metal contaminated areas. The copper-resistant soil isolates, showing the growth in the presence of CuSO4·5H2O, were stained with Gram’s stain. The results confirmed the presence of similar morphological forms such as short bacilli with a variety of arrangements and three species of Gram-negative bacilli were observed and slanted.
4.2. Screening for Copper Nanoparticles Synthesis
Cell-free supernatants of all copper-resistant bacterial isolates were separately treated with 100 ppm of filtered and sterilized CuSO4·5H2O solutions. After the incubation time, the reaction mixtures in the growing flasks were observed for color change from blue to dark green (Figure 1). Only one isolate out of three clearly showed the synthesis of CuNPs. The second isolate showed the synthesis of CuNPs quickly within one hour, whereas the first and the third did not have the ability to synthesize CuNPs. These findings are in agreement with the experimental findings of Zaki et al. [39]. The aforementioned authors reported that the color change from blue to dark green is due to the biosynthesis of CuNPs. From the results of the qualitative screening of CuNPs producer, it was also sensible that the character to have copper resistant traits among the strains must not be taken as the only final decision or indicator for the biotransformation of CuNPs. The qualitative screened isolate was nominated as Pseudomonas fluorescens MAL2 as CuNPs producer and subjected to molecular identification.
Figure 1.
Observation of the color change in the reaction mixture of Pseudomonas fluorescens MAL2 during the biosynthesis of CuNPs: (A) P. fluorescens MAL2 in F-Base medium. (B) Cell-free extract supernatant. (C) CuSO4·5H2O solution. (D) One gram of cell pellet added to CuSO4·5H2O. (E) Color change in the reaction mixture from blue to dark green indicating the formation of CuNPs.
4.3. Identification of the Screened Bacteria
The screened microbe was Gram-negative, motile, short rod, and non-spore-forming under a light microscope, and aerobic in its oxygen requirements. Regarding the morphological results and biological tests that were conducted on the selected microbe as per protocols recommended in Bergey’s Manual [40] and comparison process, which was held in this respect, it can be inferred that the local isolate was Pseudomonas fluorescens, and it was denoted as Pseudomonas fluorescens MAL2. Then, it was further exposed again to fast and precise identification by MALDI TOF Mass spectrometry (VITEK MS RUO; bioMérieux, Marcy l’Etoile, France). The registered results observed its maximum similarity of 98% to several Pseudomonas spp. predominantly Pseudomonas fluorescens DSM 12442T DSM [38]. Thus, the local screened bacterial isolate (Pseudomonas fluorescens MAL2) is similar to Pseudomonas fluorescens DSM 12442T DSM.
4.4. Optimization Factors
4.4.1. Medium Type
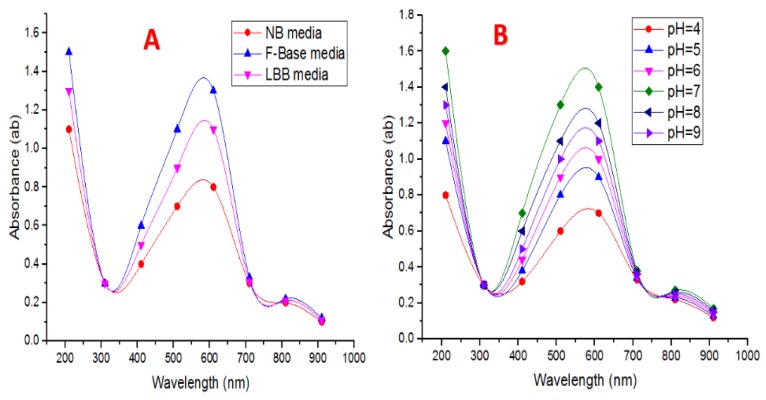
Three different media—Nutrient broth (NB), F-Base broth (Kings B medium), and Luria–Bertani broth (LBB)—were utilized to explore the best medium type in this experiment. UV–Vis spectra of CuNPs produced by the tested bacteria after incubation and growth in three different media are presented in Figure 2A. The absorbance in the case of NB was 0.81 nm, whereas it was 1.43 nm in the case of F-Base medium and this absorbance in LBB was 1.19 nm at 650 nm. The F-Base medium was found to be the best medium for the fabrication of CuNPs using the tested Pseudomonas fluorescens MAL2 bacteria. As for medium type, F-Base medium was found to be the best medium for the fabrication of CuNPs by Pseudomonas fluorescens MTCC 103 [41].
Figure 2.
(A) UV–Vis spectra of medium type. (B) UV–Vis spectra of different pH values utilized throughout CuNPs biotransformation by P. fluorescens MAL2.
4.4.2. pH Levels
It is well-known that the change in the pH values can control the shape and size of NPs [42]. Peaks recorded at acidic pH of four, five, and six in the media revealed that low pH decreased the biotransformation of CuNPs. Low pH did not favor CuNPs biotransformation (Figure 2B). Alkaline pH presented a sharp peak at 561 nm, indicating the presence of larger-sized particles or aggregation. Increase or decrease of alkalinity can lead to aggregation or alteration of nanoparticles which was likewise found in E. coli. In this experiment, at pH 7, a characteristic peak observed at 610 nm proves CuNP fabrication. Figure 2B reveals that pH 7 is the best for the NPs fabrication. Gold nanoparticles are biotransformed at pH 7 using Rhodopseudomonas capsulate, as stated earlier by He et al. [43]. Shantkriti and Rani [27] reported that pH seven was found to be the optimum pH for the production of CuNPs by Pseudomonas fluorescens MTCC 103.
4.4.3. Temperature Degrees
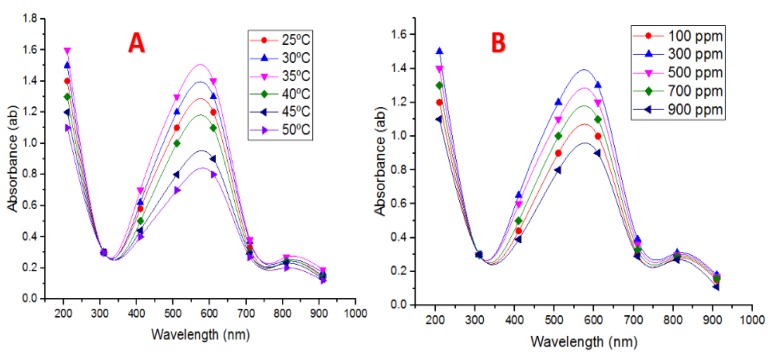
Six different temperature degrees were performed for studying this factor, i.e., 25, 30, 35, 40, 45, and 50 °C. It was easy to see that 35 °C was recorded to be the optimum temperature for the biotransformation of CuNPs by the tested Pseudomonas fluorescens MAL2 bacteria (Figure 3A). The obtained results proved that the increase in temperature reduced the production of CuNPs, and this reduction could have occurred because of the inactivation or degradation of biocompounds in the reaction mixture responsible for the bioreduction process. As the reaction temperature increased in the reaction mixture, both synthesis rate and conversion to copper nanoparticles in the medium increased [44].
Figure 3.
(A) UV–Vis spectra of different temperatures utilized. (B) UV–Vis spectra of CuSO4·5H2O concentration used during CuNPs biotransformation by P. fluorescens MAL2.
4.4.4. CuSO4·5H2O Concentrations and Supernatant on CuNPs Biotransformation
To study the influence of CuSO4·5H2O concentration, five concentrations of CuSO4·5H2O (100, 300, 500, 700, and 900 ppm Cu) were utilized as revealed in Figure 3B. Control samples did not show any distinct peak in the range of 550 to 650 nm and indicated no formation of CuNPs. As the cell-free extract concentration was increased in the reaction mixture to 700 and 900 ppm Cu, clear absorption peaks were perceived in the region above 700 nm. When 10 mL of cell-free extract was added to 300 ppm CuSO4·5H2O solution, a specific peak was detected; suggesting 10 mL supernatant was sufficient to reduce copper ions in mixture reaction to form CuNPs. A further increase in the volume of cell-free extract (40 mL) to 900 ppm copper solution leads to peak sharpening with the aggregation of CuNPs. This result is in accordance with Shantkriti and Rani [27], as they found that when 10 mL of a cell-free extract of Pseudomonas fluorescens was added to 318.4 ppm copper sulfate solution, a characteristic peak was recorded, suggesting that 10 mL cell-free extract was adequate to decrease Cu+2 ions in solution to form CuNPs.
4.4.5. Mixing Ratio of Cell-Free Extract and CuSO4·5H2O
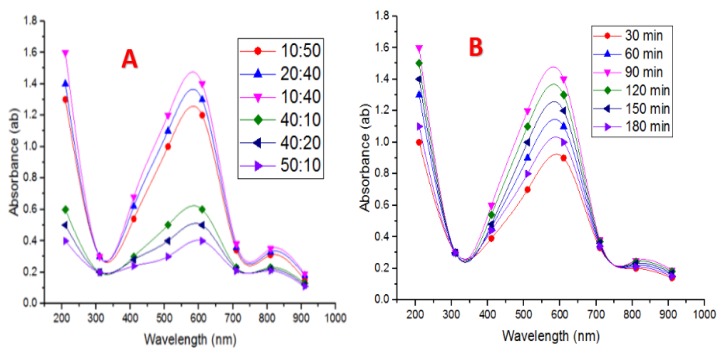
Six different volumes from 300 ppm CuSO4·5H2O as stock solution were mixed with six different volumes of a cell-free extract of Pseudomonas fluorescens MAL2 (10:50, 20:40, 10:40, 40:10, 40:20, and 50:10, respectively) and incubated for 24 h at 35 °C. UV–Vis spectra of CuNPs were recorded, and are given in (Figure 4A). The results showed that volumes of 10 mL of 300 ppm CuSO4·5H2O, which were added with 40 mL of cell-free extract at 1: 4 ratios, were deemed to be the best ratio. As it provided final absorbance at 610 nm, the increment in cell-free extracts volumes caused a reduction in the absorbance. This decline proved a reduction in NPs size. This indicates that by increasing the amount of cell-free extract in the reaction mixture, the amount and size of NPs become smaller [45].
Figure 4.
(A) UV–Vis spectra of mixing relation of culture supernatant and CuSO4·5H2O. (B) UV–Vis spectra of incubation times used during CuNPs biofabrication by P. fluorescens MAL2.
4.4.6. Reaction Time on Nanoparticle Production
Time is an important feature that enhances NPs biotransformation and stability. From the reaction time experiment performed (30, 60, 90, 120, 150, and 180 min), it was seen that the absorbance of the reaction mixtures at 610 nm increased gradually up to 90 min, and then it started to decrease (Figure 4B). This trend suggests that CuNP biotransformation occurs, with an increase in reaction time; size reduction takes place, and CuNP biotransformation was performed under optimum conditions. To clarify the reaction time process, the UV–Vis absorption spectra were registered every 10 min [46].
4.5. Characterization of Copper Nanoparticles
4.5.1. UV–Vis Analysis of CuNPs Biosynthesis
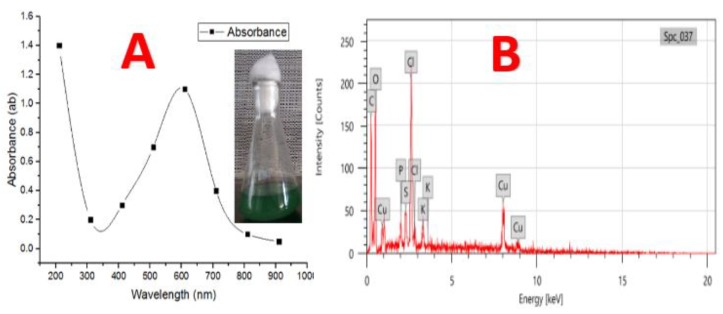
When a cell-free extract of Pseudomonas fluorescens MAL2 was added to CuSO4·5H2O solution in the growing flasks and left for incubation for 48 h, the reaction mixtures color changed from blue to heavy green (Figure 5A), proving the biotransformation of CuNPs. The analogous appearance of a dark green color solution due to the addition of 5 mM CuSO4 to a flask comprising Morganella sp., indicated CuNPs biotransformation [47,48]. When different concentrations of CuSO4·5H2O solutions were added to the pellet, no color change occurred (Figure 5B). The UV–Vis absorption spectrum of this reaction mixture exhibited a specific absorption peak in the range of 550 to 650 nm (Figure 6A), which can be attributed to CuNPs [49]. The exact position of Surface Plasmon Resonance (SPR) band may shift, which depends on the properties of individual nanoparticles, including size, shape, and capping agents [50].
Figure 5.
(A) Color change in the in the intensity of the reaction of CuNPs formation. (B) No color change occurs when 1 g of cell pellet is added to the CuSO4·5H2O solution.
Figure 6.
(A) UV–Vis spectrum of CuNPs. (B) Energy-dispersive X-ray spectroscopy (EDX) analyzer image of biotransformation of CuNPs by P. fluorescens MAL2.
4.5.2. EDX
EDX analysis proved that CuNPs were designed as aggregates and had mutable morphology (Figure 6B). The EDX analysis illustrated that 13.21% of copper and 22.55% of oxygen and another element were present in nanoparticles. The appearance of other elements may be due to a mixture of reaction components or other biomolecules secreted by the tested bacteria. Another element appears with the nanoparticles as per the medium components used in bacterial growth, which was also reported by Shantkriti and Rani [27]. The EDX results showed the presence of oxygen, carbon, phosphorus, chlorine, potassium, and sulfur elements in the purified bioflocculant. The presence of oxygen, copper, phosphorus, magnesium, calcium, chlorine, and sulfur elements in the as-synthesized nanoparticles represented the passivation of the copper nanoparticles with the bioflocculant [51]. The EDX results confirmed the presence of Cu in a sample which was an indication that the bioflocculant could be used as a capping agent in CuNPs biotransformation.
4.5.3. Transmission Electron Microscope (TEM)
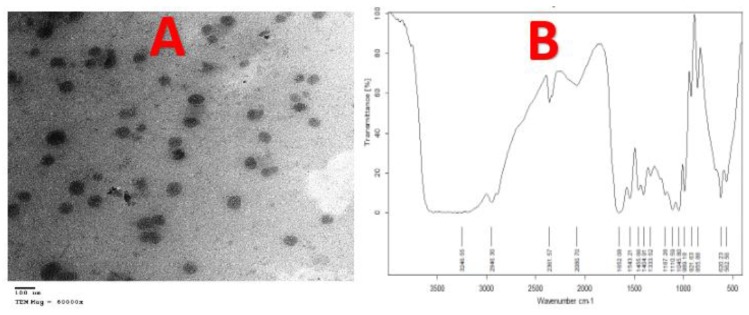
TEM was performed to analyze the shape and size of CuNPs [52]. Figure 7A revealed TEM images of CuNPs attained after 48 h of the reaction of cell-free extract (P. fluorescens MAL2) with CuSO4·5H2O; the CuNPs size was in range of 10 to 70 nm with the presence of spherical aspect. The size of CuNPs produced by Pseudomonas fluorescens ranged between 20 and 80 nm with the presence of spherical and hexagonal NPs. These results are in line with those reported by Shantkriti and Rani [27]. From the study of Dlamini et al. [51], the obtained TEM image showed that as-synthesized copper nanoparticles, using a bioflocculant extracted from Alcaligenes faecalis HCB2, were close to the spherical shape. The copper nanoparticles appeared to be close to spherical in shape and aggregation [51].
Figure 7.
(A) TEM electron micrograph of the CuNPs. (B) FTIR analysis of the biotransformation CuNPs by P. fluorescens MAL2.
4.5.4. Fourier Transform Infrared (FTIR) Analysis
This analysis gave data of the functional groups presented in compounds in the bacterial supernatant that interacted with metal ions. The FTIR spectrum of CuNPs synthesized using Pseudomonas fluorescens MAL2 supernatant is shown in Figure 7B. The presence of seventeen bands at 3246.56, 2946.30, 2361.57, 2080.70, 1652.08, 1543.21, 1455.66, 1404.91, 1333.52, 1187.28, 1110.59, 1045.80, 989.18, 921.62, 855.88, 620.23, and 562.56 cm−1 were recorded. These bands represented different functional groups such as primary and secondary amines, amides, alcohols, phenols, and strong and broad peak with the bonds of O–H stretching and N–H stretching at 3246.56 cm−1, whereas at 2946.30, 2361.57, and 2080.70 cm−1 gave strong and weak peaks, having functional groups of aromatics and alkynes, respectively. The bands appearing at 1652.08, 1455.66, 1404.91, and 1333.52 cm−1 were assigned for –C≡C– stretching and C–C stretching (in-ringing) of alkenes and aromatics, respectively. Moreover, a strong peak at 1543.21 cm−1 corresponded to N–O asymmetric stretching vibrations of nitro molecules. The crest of absorbance at 1187.28, 1110.59, and 1045.80 cm−1 shows C–O stretching of phenol and alcoholic compounds. The peaks at 989.18, 921.62, and 855.88 cm−1 are due to aromatic C–H bending. The peak at 620.23 cm−1 is as per aromatic C–H bending. The peak at 562.56 cm−1 is as per metal–carbon stretch. The recorded FTIR analysis suggests that the CuNPs might be surrounded by organic molecules such as polyphenols, alkaloids, and terpenoids, as already reported [53]. The carbonyl groups of the amino acids residues and some peptides have a strong ability to couple to the NPs [54]. Many scholars reported that the proteins could bind to NPs either through free amine or cysteine groups in proteins. These proteins that prevail over the NPs surface might act as a capping agent for stabilization [55].
4.6. Dynamic Light Scattering (DLS) Analysis
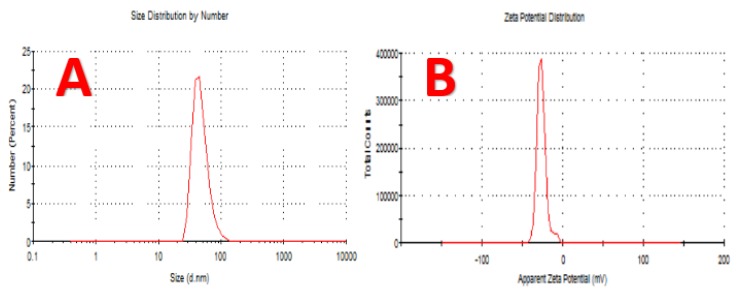
The average size of the CuNPs, size distribution, and polydispersity index (PDI) of the biotransformation CuNPs were estimated by Dynamic Light Scattering (DLS), and the results are given in Figure 8A. The results show that the average CuNPs size was 48.07 nm and the polydispersity index was 0.227. The average NPs size and PDI showed that the biotransformed CuNPs were monodispersed.
Figure 8.
(A) The particle size distribution of biotransformation CuNPs. (B) Zeta potential of the biofabrication CuNPs by P. fluorescens MAL2.
4.7. Zeta Potential Analysis
To observe the surface charges acquired by CuNPs, Zeta potential analysis was conducted, which can be performed to have clear prevision into the stability of the formed colloidal CuNPs. The magnitude of zeta potential gives an insight into inherent stability of colloid. It should be observed that the particles of CuNPs, with zeta potential values which were more positive than +30 mV or more negative than −30 mV, are deemed to be stable [56]. The zeta potential of the CuNPs in this study was found to be −26.00 mV (Figure 8B), and it can be inferred that the biotransformed CuNPs are very stable [57]. The electrostatic repulsive force between the NPs depends on the charge that resides on the surface of nanoparticles. When the charge of the nanoparticles is negative, it does not cause the aggregation of these nanoparticles, causing long-term stability.
4.8. Toxic Effect of Nano-Copper on Tribolium castaneum
Table 1 shows the toxic effect of Bio CuNPs against T. castaneum in comparison with Ch CuNPs. The mortality rate of T. castaneum adults increased with the increase in Bio CuNPs concentration concerning the exposure time. More specifically, the LC50 of Bio CuNPs after 24 h exposure of T. castaneum adults was 693.7 ppm. This value decreased dramatically to 130.5 and 36.89 ppm after three and five days, respectively. Besides, T. castaneum was suppressed at the higher concentration of Bio CuNPs (300 ppm) after 5 days of exposure. This effect may be attributed to the ability of nanoparticles to pass through the epithelial and endothelial cells by transcytosis [58]. Furthermore, nanoparticles can proceed easily along the dendrites, axons, blood, and lymphatic vessels, causing oxidative stress and other effects [59]. On the contrary, any mortality of T. castaneum adults was observed with Ch CuNPs at all used concentrations. Also, the control samples did not show any mortality. Therefore, it was obvious that the same concentration of Bio CuNPs and Ch CuNPs did not provide the same effect on T. castaneum. Two possible reasons could explain the negative effect of Ch CuNPs: First, the concentrations of Ch CuNPs were not adequate to kill T. castaneum and should be increased. This hypothesis was further supported by Shaker [60] who reported a mortality rate of 95% when cotton leafworm Spodoptera littoralis was exposed to chemically synthesized CuO-NPs at 1000 ppm. Secondly, Gudikandula [61] demonstrated that a stabilizer agent should be added to enhance the effect of Ch CuNPs and prevent agglomeration.
Table 1.
Probit analysis for the data on adult mortality of Tribolium castaneum treated with CuNPs at 95% confidence limits.
| Treatments | CuNPs Concentration (μg/mL) | 300 | 250 | 200 | 150 | 100 | 50 | LC50 (μg/mL) | Chi-square Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Period | %Mortality | ||||||||||||
| Bio CuNPs | First day | 30 | 20 | 17 | 10 | 7 | 3 | 693.7 | 0.589 | ||||
| Third day | 67 | 60 | 50 | 47 | 43 | 43 | 130.5 | 1.67 | |||||
| Fifth day | 100 | 93 | 83 | 73 | 70 | 67 | 36.89 | 6.63 | |||||
| Ch CuNPs | - | - | - | - | - | - | - | - | - | ||||
| Negative Control (distilled H2O) |
- | - | - | - | - | - | |||||||
| Positive Control | Pseudomonas supernatant | - | - | - | - | - | - | ||||||
| CuSO4·5H2O | - | - | - | - | - | - | |||||||
LC50: lethal concentration that kills 50% of the exposed adults; Bio CuNPs: biological of Copper nanoparticles; Ch CuNPs: chemical synthesis of Copper nanoparticles.
On the other hand, the positive effect of the different chemical under nanosize as biocontrol against different pests has been reported in several investigations. For instance, the populations of two-spotted spider mites, the most critical bean pests, were significantly decreased with the addition of nano-capsule of CuO [62]. Selvan [63] found larvicidal activity against the transmit vector Aedes aegypti when treating with CuO-NPs using greenly synthesis using leaf extract of Tridax procumbens. Besides, it was proposed that the application of nanotechnology in agrochemical formulations can reduce the petrochemicals used in pesticides [64]. Recent studies demonstrated that the combination of AgNPs with a lesser quantity of Malathion produced high insecticidal efficacy for T. castaneum control [65]. Taking into consideration the positive effects of nanoparticles as biocontrol agents that have been confirmed by aforementioned studies together with the fact that the concentration of nanoparticles could be reduced with an effective mortality rate when they were produced biologically, it can be concluded that the biological production of nanoparticles could be considered an ecofriendly approach. This conclusion is also supported by Selvan and Gunalan, who demonstrated that both biogenic and green synthesis of Cu-NPs emphasized their insecticidal efficacy as an ecofriendly and inexpensive method [63,66]. Note that even if the current investigation demonstrated that nanoparticles, i.e., Cu-NPs, have the insecticidal potential against the stored product pests, with particular reference to T. castaneum, the same conclusion should not be assumed in other pests and each pest should be evaluated separately. Accurately, it was reported that larvae of T. confusum were more susceptible to the exposure of nano-Diatomaceous earth than T. castaneum larvae [67]. In addition, further investigations are still required for the utilization of high concentration of such nanoparticles. For instance, despite CuO-NPs showing the potential to eliminate alachlor and phenanthrene in aqueous solutions [68], the high concentration of CuO-NPs in aqueous solutions led to intense accumulation in the larval body of the freshwater shredder Allogamus ligonifer; where LC50 value was 569 mg L−1 after 96 h of exposure, resulting in negative effects on their growth and feeding behavior [69].
5. Conclusions
Green manufacturing of copper nanoparticles is a safe, cost-effective, and ecofriendly process against the most critical economic insects such as Tribolium castaneum, which cause significant losses in crops that enter human and animal food chains. The Pseudomonas fluorescens strain that was isolated in the present study showed competency in copper nanoparticle synthesis in sizes ranging from 10 to 70 nm. It could be concluded that the biologically synthesized of copper nanoparticles have an excellent insecticidal activity at low concentrations against the stored product pest T. castaneum. The effect of biologically synthesized of copper nanoparticles excelled that of chemically synthesized of copper nanoparticles.
Abbreviations
FTIR: Fourier-transform infrared spectroscopy; LC50: lethal concentration 50; PDI: polydispersity index; Ch CuNPs: Chemical copper nanoparticles; DSM: Deutsche Sammlung von Mikroorganismen; CFU: Colony forming unit; Rh: Relative humidity; Cu+2: Copper ion.
Author Contributions
Conceptualization and methodology, M.T.E.-S. and N.E.; M.E.A.E.-H., A.E.T., and M.M.G.F.; data curation, J.S.A.; writing—original draft preparation, M.E.A.E.-H. and A.E.T.; writing—review and editing, S.N.M. and A.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Dean of Scientific Research, King Saud University, for funding the work through the research group project number RGP-VPP-240.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Rajak A. Nanotechnology and Its Application. J. Nanomed. Nanotechnol. 2018;9:502. doi: 10.4172/2157-7439.1000502. [DOI] [Google Scholar]
- 2.Bahru T.B., Ajebe E.G. A Review on Nanotechnology: Anal. Tech.Use and Appl. IRJPAC. 2019;19:1–10. doi: 10.9734/irjpac/2019/v19i430117. [DOI] [Google Scholar]
- 3.Khan I., Saeed K. Idrees Khan Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2017;12:908–931. doi: 10.1016/j.arabjc.2017.05.011. [DOI] [Google Scholar]
- 4.Gahlawat G., Choudhury A.R. A review on the biosynthesis of metal and metal salt nanoparticles by microbes. RSC Adv. 2019;9:12944–12967. doi: 10.1039/C8RA10483B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gour A., Jain N. Advances in green synthesis of nanoparticles. Artif. Cells Nanomed. Biotechnol. 2019;47:844–851. doi: 10.1080/21691401.2019.1577878. [DOI] [PubMed] [Google Scholar]
- 6.Chung I.M., Abdul Rahuman A., Marimuthu S., Kirthi A.V., Anbarasan K., Padmini P., Rajakumar G. Green synthesis of copper nanoparticles using Eclipta prostrata leaves extract and their antioxidant and cytotoxic activities. Exp. Ther. Med. 2017;14:18–24. doi: 10.3892/etm.2017.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yusof H.M., Mohamad R., Zaidan U.H. Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: A review. J. Anim. Sci. Biotechnol. 2019;10:57. doi: 10.1186/s40104-019-0368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shobha G., Vinutha M., Ananda S. Biological synthesis of copper nanoparticles and its impact—A Review. Int. J. Pharma. Sci. Invent. 2014;3:6–38. [Google Scholar]
- 9.Thakur S., Sharma S., Thakur S., Rai R. Green synthesis of copper nano-particles using Asparagus adscendens Roxb root and leaf extract and their antimicrobial activities. Int. J. Curr. Microbiol. Appl. Sci. 2018;7:683–694. doi: 10.20546/ijcmas.2018.704.077. [DOI] [Google Scholar]
- 10.Vasudev D.K., Pramod S.K. Green synthesis of copper nanoparticles using Ocimum sanctum Leaf Extract. Int. J. Chem. Stud. 2013;1:1–4. [Google Scholar]
- 11.Lee H.J., Lee G., Jang N.R., Yun J.H., Song J.Y., Kim B.S. Biological synthesis of copper nanoparticles using plant extract. Nanotechnology. 2011;1:361–764. [Google Scholar]
- 12.Li X., Xu H., Chen Z.S., Chen G. Biosynthesis of nanoparticles by microorgan-isms and their applications. J. Nanomater. 2011;2011:16. doi: 10.1155/2011/270974. [DOI] [Google Scholar]
- 13.Hasan S.S., Singh R.Y., Parikh M.S., Dharne M.S., Patole B.L., Prasad P., Shouche Y.S. Bacterial synthesis of copper/copper oxide nanoparticles. J. Nanosci. Nanotechnol. 2018;8:3191–3196. doi: 10.1166/jnn.2008.095. [DOI] [PubMed] [Google Scholar]
- 14.Varshney R., Bhadauria S., Gaur M.S., Pasricha R. Characterization of copper nanoparticles synthesized by a novel microbiological method. JOM. 2010;62:100–102. doi: 10.1007/s11837-010-0171-y. [DOI] [Google Scholar]
- 15.Singh V., Patil R., Ananda A., Milani P., Gade W. Biological synthesis of copper oxide nanoparticles using Escherichia coli. Curr. Nanosci. 2010;6:365–369. doi: 10.2174/157341310791659062. [DOI] [Google Scholar]
- 16.Kole C., Kole P., Randunu K.M., Choudhary P., Podila R., Ke P.C., Rao A.M., Marcus R.K. Nanobiotechnology can boost crop production and quality: First evidence from increased plant biomass, fruit yield and phytomedicine content in bitter melon (Momordica charantia) BMC Biotechnol. 2013;13:1–10. doi: 10.1186/1472-6750-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqui M.H., Al-Whaibi M.H. Role of nano-SiO2 in germination of tomato (Lycopersicum esculentum seeds Mill.) Saudi J. Biol. Sci. 2014;21:13–17. doi: 10.1016/j.sjbs.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benelli G. Plant-mediated biosynthesis of nanoparticles as an emerging tool against mosquitoes of medical and veterinary importance: A review. Parasitol. Res. 2017;115:23–34. doi: 10.1007/s00436-015-4800-9. [DOI] [PubMed] [Google Scholar]
- 19.Stevenson P.C., Arnold S.E.J., Belmain S.R. Advances in Plant Biopesticides. Springer; New Delhi, India: 2014. Pesticidal plants for stored product pests on small-holder farms in Africa. [Google Scholar]
- 20.Opit G.P., Phillips T.W., Aikins M.J., Hasan M.M. Phosphine resistance in Tribolium castaneum and Rhyzopertha dominica from stored wheat in Oklahoma. J. Econ. Entomol. 2012;105:1107–1114. doi: 10.1603/EC12064. [DOI] [PubMed] [Google Scholar]
- 21.Arthur F.H., Hagstrum D.W., Flinn P.W., Reed C.R., Phillips T.W. Insect populations in grain residues associated with commercial Kansas grain elevators. J. Stored Prod. Res. 2006;42:226–239. doi: 10.1016/j.jspr.2005.02.003. [DOI] [Google Scholar]
- 22.Tefera T., Kanampiu F., De Groote H., Hellin J., Mugo S., Kimenju S., Beyene Y., Boddupalli P.M., Shiferaw B., Banziger M. The metal silo: An effective grain storage technology for reducing post-harvest insect and pathogen losses in maize while improving smallholder farmers’ food security in developing countries. Crop Prot. 2011;30:240–245. doi: 10.1016/j.cropro.2010.11.015. [DOI] [Google Scholar]
- 23.Silver K., Jiang H., Fu J., Phillips T.W., Beeman R.W., Park Y. The Tribolium castaneum cell line TcA: A new tool kit for cell biology. Sci. Rep. 2014;4:6840. doi: 10.1038/srep06840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rai M., Ingle A.P., Pandit R., Paralikar P., Shende S., Gupta I., Biswas J.K., Silvério da Silva S. Copper and copper nanoparticles: Role in management of insect-pests and pathogenic microbes. Nanotechnol. Rev. 2018;7:303–315. doi: 10.1515/ntrev-2018-0031. [DOI] [Google Scholar]
- 25.Lakshmi D.V., Thomas R., Varghese R.T., Soniya E.V., Mathew J., Radhakrishnan E.K. Extracellular synthesis of silver nanoparticles by the Bacillus strain CS 11 isolated from industrialized area. 3 Biotech. 2014;4:121–126. doi: 10.1007/s13205-013-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannan N., Subbalaxmi S., Ramachandra Murty V. Microbial production of silver nanoparticles. Nanomater. Biostructure. 2010;5:135–140. [Google Scholar]
- 27.Shantkriti S., Rani P. Biological synthesis of Copper nanoparticles using Pseudomonas fluorescens. Int. J. Curr. Microbiol. Appl. Sci. 2014;3:374–383. [Google Scholar]
- 28.Brenner J., Kreig R., Stanley T. The Probacteria, Part A. Introductory Essay. Springer; New York, NY, USA: 2005. Bergey’s Manual of Systematic Bacteriology. [Google Scholar]
- 29.Bille E., Dauphin B., Leto J., Bougnoux M.E., Beretti J.L., Lotz A., Suarez S., Meyer J., Join Lambert O., Descamps P., et al. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin. Microbiol. Infect. 2012;18:1117–1125. doi: 10.1111/j.1469-0691.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- 30.Forough M., Farhadi K. Biological and green synthesis of silver nanoparticles. Turk. J. Eng. Environ. Sci. 2010;34:281–287. [Google Scholar]
- 31.Aguilar M.M.A., Martinez E.S.M., Arroyo L.O., Portillo G.C., Espindola E.S. Synthesis and characterization of silver nanoparticles: Effect on Phytopathogen Colletotrichum gloeosporioides. J. Nanoparticle Res. 2011;13:2525–2532. doi: 10.1007/s11051-010-0145-6. [DOI] [Google Scholar]
- 32.Duran N., Marcato P.D., Alves O.L., DeSouza G.I.H., Esposito E. Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J. Nanobiotechnol. 2005;3:8. doi: 10.1186/1477-3155-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavani K.V., Nandigam S., Guntur P., Tandale S. Synthesis of copper nanoparticles by Aspergillus sp. Lett. Appl. Nanobiosci. 2013;2:110–113. [Google Scholar]
- 34.Ganachari S.V., Bhat R., Deshpande R., Venkataraman A. Extracellular biosynthesis of silver nanoparticles using fungi Penicillium diversum and their antimicrobial activity studies. Biol. Nanosci. 2012;2:316–321. doi: 10.1007/s12668-012-0046-5. [DOI] [Google Scholar]
- 35.Dash S.K., Ghosh T., Roy S., Chattopadhyay S., Das D. Zinc sulfide nanoparticles selectively induce cytotoxic and genotoxic effects on leukemic cells: Involvement of reactive oxygen species and tumor necrosis factor alpha. J. Appl. Toxicol. 2014;34:1130–1144. doi: 10.1002/jat.2976. [DOI] [PubMed] [Google Scholar]
- 36.Khalid H., Shamaila S., Zafar N. Synthesis of copper nanoparticles by chemical reduction method. Sci. Int. 2015;27:3085–3088. [Google Scholar]
- 37.Finney D.J. Probit Analysis. Cambridge University; London, UK: 1971. pp. 68–78. [Google Scholar]
- 38.Altimira F., Yáñez C., Bravo G. Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol. 2012;12:1–12. doi: 10.1186/1471-2180-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaki S., Kady M.F., Abd-EL-Haleem D. Biosynthesis and Structural Characterization of silver nanoparticles from bacterial isolates. Mater. Res. Bull. 2011;46:1571–1576. doi: 10.1016/j.materresbull.2011.06.025. [DOI] [Google Scholar]
- 40.Logan N.A., Devos P., Genus I. Bacillus Cohn 1872, 174AL. In: De Vos P., Garrity G.M., Jones D., Krieg N.R., Ludwig W., Rainey F.A., Schleifer K.H., Whitman W.B., editors. Bergey’s Man. of System. Bacter. 2nd ed. Vol. 3. Springer; New York, NY, USA: 2009. pp. 21–128. [Google Scholar]
- 41.Krasny L., Hyneka R., Hochela I. Identification of bacteria using mass spectrometry techniques. Int. J. Mass Spectrom. 2013;353:67–79. doi: 10.1016/j.ijms.2013.04.016. [DOI] [Google Scholar]
- 42.Gurunathan S., Lee K.J., Kalishwaralal K., Sheikpranbabu S., Vaidyanathan R., Eom S.H. Antiangiogenic properties of silver nanoparticles. Biomaterials. 2009;30:6341–6350. doi: 10.1016/j.biomaterials.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 43.He S., Guo Z., Zhang Y., Zhang S., Wang J., Gu N. Biosynthesis of gold nanoparticles using the bacteria Rhodopseudomonas capsulata. Mater. Lett. 2007;61:3984–3987. doi: 10.1016/j.matlet.2007.01.018. [DOI] [Google Scholar]
- 44.Lee S.H., Jun B.H. Silver Nanoparticles: Synthesis and application for nanomedicine. Int. J. Mol. Sci. 2019;20:865. doi: 10.3390/ijms20040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safekordi A.A., Attar H., Ghorbani H. Optimization of silver nanoparticles production by E. coli and the study of reaction kinetics. Int. Conf. Chem. Ecol. Environ. Sci. 2011;33:5111–5118. [Google Scholar]
- 46.Caroling G., Vinodhini E., Mercy Ranjitham A., Shanthi P. Biosynthesis of copper nanoparticles using aqueous Phyllanthus Embilica (Gooseberry) Extract- characterisation and study of antimicrobial effects. Int. J. Nano. Chem. 2015;1:53–63. [Google Scholar]
- 47.Yu E.K., Piao L., Kim S.H. Sintering behavior of copper nanoparticles. Bull. Korean Chem. Soc. 2011;32:4099–4102. doi: 10.5012/bkcs.2011.32.11.4099. [DOI] [Google Scholar]
- 48.Ramanathan R., Mullane A.P.O’. Bacterial kinetics-controlled shape directed biosynthesis of silver nanoplates using Morganella psychrotolerans. Langmuir. 2011;27:714–719. doi: 10.1021/la1036162. [DOI] [PubMed] [Google Scholar]
- 49.Dhas N.A., Raj C.P., Gedanken A. Synthesis, characterization, and properties of metallic copper nanoparticles. Chem. Mater. 1998;10:1446–1452. doi: 10.1021/cm9708269. [DOI] [Google Scholar]
- 50.Mott D., Galkowski J., Wang L., Luo J., Zhong C.J. Synthesis of size-controlled and shaped copper nanoparticles. Langmuir. 2007;23:5740–5745. doi: 10.1021/la0635092. [DOI] [PubMed] [Google Scholar]
- 51.Dlamini N.G., Basson A.K., Pullabhotla V.S.R.R. Biosynthesis and characterization of copper nanoparticles using a bioflocculant extracted from Alcaligenes faecalis HCB2. Adv. Sci. Eng. Med. 2019;11:1–7. doi: 10.1166/asem.2019.2448. [DOI] [Google Scholar]
- 52.Aswathy Aromal S., Philip D. Green synthesis of gold nanoparticles using Trigonella foenumgraecum and its size-dependent catalytic activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012;97:1–5. doi: 10.1016/j.saa.2012.05.083. [DOI] [PubMed] [Google Scholar]
- 53.Kalainila P., Subha V., Ernest ravindran R.S., Sahadevan R. Synthesis and characterization of silver nanoparticle from Erythrina indic. Asian J. Pharm. Clin. Res. 2014;7:39–43. [Google Scholar]
- 54.Balaji D.S., Basavaraja S., Deshpande R., Mahesh D.B., Prabhakar B.K., Venkataraman A. Extracellular biosynthesis of functionalized silver nanoparticles by strains of Cladosporium cladosporioides fungus. Colloids Surf. B. 2009;68:88–92. doi: 10.1016/j.colsurfb.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 55.Mandal S., Phadtare S., Sastry M. Interfacing Biology with Nanoparticles. Curr. Appl. Phys. 2005;5:118–127. doi: 10.1016/j.cap.2004.06.006. [DOI] [Google Scholar]
- 56.Meléndrez M.F., Cárdenas G., Arbiol J. Synthesis and characterization of gallium colloidal nanoparticles. J. Colloid Interface Sci. 2010;346:279–287. doi: 10.1016/j.jcis.2009.11.069. [DOI] [PubMed] [Google Scholar]
- 57.Netala V.R., Kotakadi V.S., Domdi L., Gaddam S.A., Bobbu P., Venkata S.K., Ghosh S.B., Tartte V. Biogenic silver nanoparticles: Efficient and effective antifungal agents. Appl. Nanosci. 2016;6:475–484. doi: 10.1007/s13204-015-0463-1. [DOI] [Google Scholar]
- 58.Yamanaka Y.J., Leong K.W. Engineering strategies to enhance nanoparticle-mediated oral delivery. J. Biomater. Sci. Polym. Ed. 2008;19:1549–1570. doi: 10.1163/156856208786440479. [DOI] [PubMed] [Google Scholar]
- 59.Oberdörster G., Oberdörster E., Oberdörster J. Nanotoxicology: Discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 2005;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaker A.M., Zaki Elham A.H., Abdel-Rahim F., Khedr M.H. Novel CuO nanoparticles for pest management and pesticides photodegradation. Adv. Environ. Biol. 2016;10:274–283. [Google Scholar]
- 61.Gudikandula K., Maringanti S.C. Synthesis of silver nanoparticles by chemical and biological methods and their antimicrobial properties. J. Exp. Nanosci. 2016;11:9. doi: 10.1080/17458080.2016.1139196. [DOI] [Google Scholar]
- 62.Dorri H.R., Khaghani S.h., Moghadam A., Ghanbari D., Bihamta M.R. The Effect of copper nano-capsules on the control of two spotted spider mite (Tetranychus urticae) J. Nanostruct. 2018;8:316–324. [Google Scholar]
- 63.Selvan S.M., Anand K.V., Govindaraju K., Tamilselvan S., Kumar V.G., Subramanian K.S., Kannan M., Raja K. Green synthesis of copper oxide nanoparticles and mosquito larvicidal activity against dengue, zika and chikungunya causing vector Aedes aegypti. IET Nanobiotechnol. 2018;12:1042–1046. doi: 10.1049/iet-nbt.2018.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castro M.L., Ojeda C., Cirelli A. Advances in surfactants for agrochemicals. Environ. Chem. Lett. 2014;12:85–95. doi: 10.1007/s10311-013-0432-4. [DOI] [Google Scholar]
- 65.Alif Alisha A.S., Thangapandiyan S. Comparative bioassay of silver nanoparticles and malathion on infestation of red flour beetle Tribolium castaneum. JoBAZ. 2019;80:1–10. [Google Scholar]
- 66.Gunalan S., Sivaraj R., Venckatesh R. Aloe barbadensis miller mediated green synthesis of mono-disperse copper oxide nanoparticles: Optical properties, Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012;97:1140–1144. doi: 10.1016/j.saa.2012.07.096. [DOI] [PubMed] [Google Scholar]
- 67.Sabbour M.M., El-Aziz S.E. Efficacy of Nano-diatomaceous earth against red flour beetle, Tribolium castaneum and confused Flour beetle, Tribolium confusum (Coleoptera: Tenebrionidae) under laboratory and storage conditions. Bull. Environ. Pharmacol. Life Sci. 2015;4:54–59. [Google Scholar]
- 68.Ben-Moshe T., Dror I., Berkowitz B. Oxidation of organic pollutants in aqueous solutions by nanosized copper oxide catalysts. Appl. Catal. B Environ. 2009;85:207–211. doi: 10.1016/j.apcatb.2008.07.020. [DOI] [Google Scholar]
- 69.Pradhan A., Seena S., Pascoal C., Cássio F. Copper oxide nanoparticles can induce toxicity to the freshwater shredder Allogamus ligonifer. Chemosphere. 2012;89:1142–1150. doi: 10.1016/j.chemosphere.2012.06.001. [DOI] [PubMed] [Google Scholar]