Abstract
The findings that ALS patients almost universally display pathological mislocalization of the RNA-binding protein TDP-43 and that mutations in its gene cause familial ALS have nominated altered RNA metabolism as a disease mechanism. However, the RNAs regulated by TDP-43 in motor neurons and their connection to neuropathy remain to be identified. Here, we report transcripts whose abundance in human motor neurons are sensitive to TDP-43 depletion. Notably, expression of STMN2, which encodes a microtubule regulator, declined after TDP-43 knockdown and TDP-43 mislocalization as well as in patient-specific motor neurons and postmortem patient spinal cord. STMN2 loss upon reduced TDP-43 function was due to altered splicing, which is functionally important, as we show STMN2 is necessary for normal axonal outgrowth and regeneration. Importantly, post-translational stabilization of STMN2 rescued neurite outgrowth and axon regeneration deficits induced by TDP-43 depletion. We propose restoring STMN2 expression warrants examination as a therapeutic strategy for ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by the loss of motor neurons1. Patients with ALS experience progressive paralysis, usually succumbing to disease 1–5 years after diagnosis2. Aside from two FDA approved drugs, which only modestly slow disease, treatment for ALS is limited to supportive care3. ALS is recognized to be on the same pathological spectrum as frontotemporal dementia (FTD), the most common cause of pre-senile dementia4. Although the causes of most ALS and FTD cases remains unknown, pathological findings and family-based linkage studies have demonstrated overlap in molecular pathways involved in both diseases1. In a landmark finding, TDP-43 was discovered to be a major constituent of inclusions in many sporadic cases of ALS and FTD5. TDP-43 is a predominantly nuclear DNA/RNA-binding protein with functional roles in transcriptional regulation, splicing, pre-miRNA processing, stress granule formation, and mRNA transport and stability6,7. Subsequently, autosomal-dominant mutations in its gene TARDBP were identified in both ALS and FTD families, linking genetics and pathology with neurodegeneration7,8. Thus, elucidating the role that perturbations to TDP-43 play in disease is essential to understanding both sporadic and familial ALS.
Whether neurodegeneration associated with TDP-43 pathology is the result of loss-of-function mechanisms, toxic gain-of-function mechanisms, or both, remains unclear7. Early studies showed that overexpression of wildtype and mutant TDP-43 led to its aggregation and nuclear loss7. While these studies along with the autosomal dominant inheritance pattern of TARDBP mutations support a gain-of-function view, the loss of nuclear TDP-43 and its aggregation suggests its normal functions might also become impaired. Subsequent findings revealed that TDP-43 depletion in the developing embryo or motor neurons have profound consequences9.
Given the myriad roles TDP-43 plays, a key question is: what RNA substrates are perturbed upon alterations of TDP-43 localization or by TDP-43 mutation, and how do they contribute to neurodegeneration? Initial efforts to answer this question utilized cross-linking and immunoprecipitation with RNA sequencing of whole brain homogenates10,11. These studies led to an understanding that many transcripts are regulated by TDP-43 with preference towards RNAs containing UG repeats and long introns; however, the prominence of glial RNAs in the samples limited insights into the neuronal targets of TDP-43. Consequently, few clear connections between the TDP-43 target RNAs and neurodegenerative mechanisms have been forged.
To identify substrates potentially contributing to neurodegeneration, we sought to identify RNAs regulated by TDP-43 in human motor neurons. Because the vulnerable neurons in living ALS patients are fundamentally inaccessible for isolation, we and others have developed approaches for directing the differentiation of human pluripotent stem cells into motor neurons (hMNs)12,13,14. Here, we performed RNA-seq after TDP-43 knockdown in hMNs and identified TDP-43-regulated transcripts in this cellular context. In total, we found 885 transcripts for which TDP-43 was needed to maintain normal RNA levels. Although any number of these targets may play subtle roles in neurodegeneration, we noted that one of the most abundant transcripts in hMNs encoding STMN2, also known as SCG10 a regulator of microtubule stability, was particularly sensitive to a decline in TDP-43. Additionally, we determined that STMN2 levels were also decreased following pharmacological induction of TDP-43 relocalization from the nucleus and in iPS cell-derived hMNs generated from patients with TDP-43 mutations15. We further show that STMN2 encodes a protein necessary for normal hMN outgrowth and repair. Importantly, we established that post-translational stabilization of STMN2 through inhibition of c-Jun N-terminal kinases (JNK) can rescue deficits in motor neurite outgrowth and axon regeneration induced by TDP-43 knock-down. Finally, STMN2 depletion resulting from loss of TDP-43 activity is likely to be relevant to people with ALS as we found that STMN2 expression is decreased in the spinal cord of ALS patients postmortem.
Results
Motor neuron RNAs regulated by TDP-43
In order to produce hMNs for studying RNAs regulated by TDP-43, we differentiated the human embryonic stem cell line HUES3 Hb9::GFP12 into GFP+ hMNs under adherent culture conditions. On day 14 of differentiation, we routinely obtained cultures comprised of ~18–20% GFP+ cells. Subsequent analysis of these GFP+ hMNs following purification by flow cytometry demonstrated that they possessed expected functional and molecular properties (Supplementary Fig.1a–i).
The reduced levels of nuclear TDP-43 observed in ALS has been hypothesized to contribute to downstream neurodegenerative events through alterations in the metabolism of the RNAs it binds7. We, therefore, sought to identify the RNAs regulated by TDP-43 in purified hMNs through a combination of knock-down and RNA-Seq approaches. Using a fluorescently conjugated siRNA (siRED), we validated transfection conditions to achieve high-levels of siRNA delivery (~94%). We then used qRT-PCR to validate that TDP-43 mRNA levels had been depleted in hMNs treated with siTDP43s, but not in those exposed to a scrambled siRNAs. We further confirmed TDP-43 depletion at the protein level by immunoblot, with siTDP43-treated hMNs showing a 54–65% reduction in TDP-43 levels (Supplementary Fig. 2a–e).
To measure global alterations in transcript abundance following partial loss of TDP-43, we prepared RNA-Seq libraries from siRNA-treated hMNs (Fig. 1a). After next-generation sequencing, we obtained expression data for each gene annotated as transcripts per million (TPMs). After initial clustering (Supplementary Fig. 3a), principle components analyses focused on the 500 most differentially abundant transcripts segregated the samples based on siTDP-43 treatment (PC1), indicating that reduction of TDP-43 levels accounted for the greatest differences in transcript abundance, followed by the particular batch of hMNs produced (PC2) (Fig. 1b). Inspection of TPM values for TDP-43 expression confirmed that its abundance was significantly reduced in siTDP43-treated hMNs (Supplementary Fig. 3b).
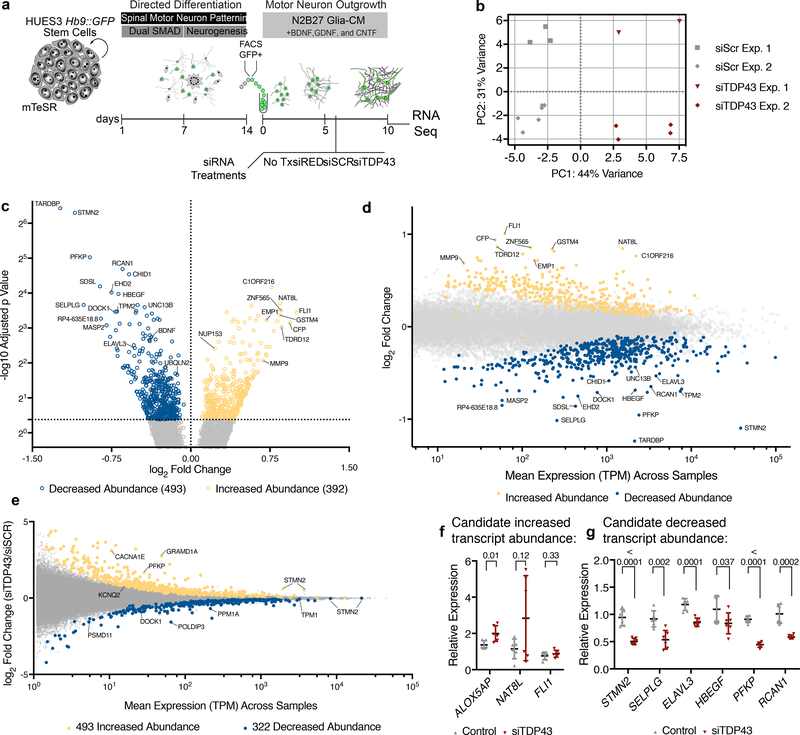
Figure 1. RNA-Seq following TDP-43 knockdown in hMNs.
(a) hMN differentiation, purification, and RNAi strategy for TDP-43 knockdown in cultured hMNs. (b) Multidimensional scaling analysis for RNA-Seq data sets obtained from n=2 independent MN differentiation and siRNA transfection experiments based on 500 most differentially expressed transcripts. (c) Volcano plot showing transcripts with significantly altered abundance in hMNs treated with siTDP43 relative to those with scrambled controls. We tested for significant differences between control (n=9) and TDP43 knockdown (n=6) samples, which are highlighted, using the Wald test and a cutoff of 0.05 for Benjamini-Hochberg adjusted p-values with no log2 fold-change ratio cutoff. (d) Differential transcript abundance scatter plot comparing TPM values for all transcripts expressed in hMNs treated with control siRNAs versus the fold change in expression for those transcripts in cells treated with siTDP43. (e) Differential exon usage scatter plot comparing TPM values for individual exons expressed in hMNs treated with siTDP43 siRNAs versus the fold change in expression for those exons in cells treated with control siRNAs. (f-g) A subset of 9 transcripts initially identified as ‘hits’ (significantly increased abundance (f) or decreased abundance (g)) in the TDP43 knockdown experiment were selected for validation by qRT-PCR. Data are displayed as mean with SD of technical replicates from n=2 independent experiments (Unpaired t-test, two-sided, P value < 0.05).
We next performed differential gene expression analysis, which at a false discovery rate (FDR) of 5%, identified 885 significantly differentially expressed transcripts after TDP-43 knockdown (Fig. 1c–d). Expression was significantly higher for 392 transcripts (‘increased abundance’) and significantly lower for 493 transcripts (‘decreased abundance’) in TDP-43-depleted hMNs (Fig. 1c–d). In addition to altering transcript levels of hundreds of genes10, reduced levels of TDP-43 can also influence RNA splicing7,10. We therefore analyzed differential exon usage, which at an FDR of 10%, identified 815 statistically differentially expressed exons in hMNs after TDP-43 knockdown (Fig. 1e). In these cells, values were significantly higher for 493 exons (‘increased abundance’), and lower for 322 exons (‘decreased abundance’) (Fig. 1e). Reassuringly, we detected the POLDIP3 as top candidates for altered splicing (Fig. 1e), which has previously been associated with deficits in TDP-43 function16,17. Additionally, we detected splicing changes in 50 of the 885 transcripts differentially expressed following TDP-43 knockdown including the abundant transcripts STMN2 and PFKP (Fig. 1c–e).
Of the 885 altered transcripts, we selected a subset for further validation. We considered transcripts with enriched neuronal expression (STMN218,19, ELAVL320), those associated with neurodevelopment/neurological disorders (RCAN121, NAT8L22) and those with reasonable expression levels or large fold changes (SELPLG, NAT8L). To this end, we obtained RNA from independent biological MN replicates after TDP-43 knockdown and determined by qRT-PCR the relative abundance of 9 candidate transcripts. Notably, we confirmed differential expression for 7/9 RNAs after treatment with siTDP43, validating the quality and reproducibility of our RNA-Seq analyses (Fig. 1f–g).
STMN2 levels downregulated in hMNs expressing mutant TDP-43
We proceeded to ask if any RNAs with altered abundances after TDP-43 depletion were perturbed by expression of mutant TDP-43. Directed differentiation approaches tend to yield heterogeneous cultures making quantitative, comparative analyses between individuals iPS cell lines challenging23. Residual, mitotic progenitor cells are especially troublesome as they can overtake cultures. To overcome this, we performed an unbiased FACS-based immunoprofiling analysis on HUES3 Hb9::GFP+ hMNs to identify antigenic signatures enriched on both postmitotic neurons and cycling progenitors (Supplementary Fig. 4a). These studies revealed that by isolating NCAM+/EpCAM− cells, we could obtain neuronal cultures with reproducible properties (Supplementary Fig. 4h–i).
Using this method, we isolated neurons from 5 control iPSC lines and 4 iPSC lines with distinct TDP-43 mutations (Fig. 2a and Supplementary Fig. 5a). As anticipated, each cell line exhibited its own differentiation propensity into hMNs (Supplementary Fig. 4f–g), but after sorting we could readily obtain homogenous neuronal cultures from each line (Fig. 2b). We used these hMNs to explore how mutations in TARDBP influence TDP-43 localization, solubility and ability to regulate select RNA targets. The majority of mutations in TDP-43 are dominant missense mutations in the C-terminal glycine-rich domain, but how mutations trigger disease or if all mutations are equivalent remains unclear7. Previous studies have reported that iPSC-derived neuronal cultures expressing mutant TDP-43 recapitulate some aspects of TDP-43 pathology including its accumulation in both soluble and insoluble cell protein extracts, as well as some cytoplasmic mislocalization24–26. We first tested whether expression of mutant TDP-43 was sufficient to induce cytoplasmic mislocalization using immunofluorescence (Fig. 2b and Supplementary Fig. 5b). However, we did not find this to be the case. In both control and mutant neurons, we observed primarily nuclear TDP-43 staining using two different metrics. Pearson’s coefficient analysis revealed a strong correlation between TDP-43 immunostaining and the DNA counterstain for both groups (Fig 2c). We also calculated the nuclear to cytoplasmic ratio of TDP-43 staining with no significant difference observed between groups (Supplementary Fig. 5c). Moreover, we did not detect differences in the abundance of TDP-43 protein by immunoblot analysis in either the soluble or insoluble fractions (Supplementary Fig. 5d–e). These results are consistent with some TDP-43 iPS disease modeling studies26, yet inconsistent with others24, and to us suggested that additional perturbations could be required to induce TDP-43 mislocalization27.
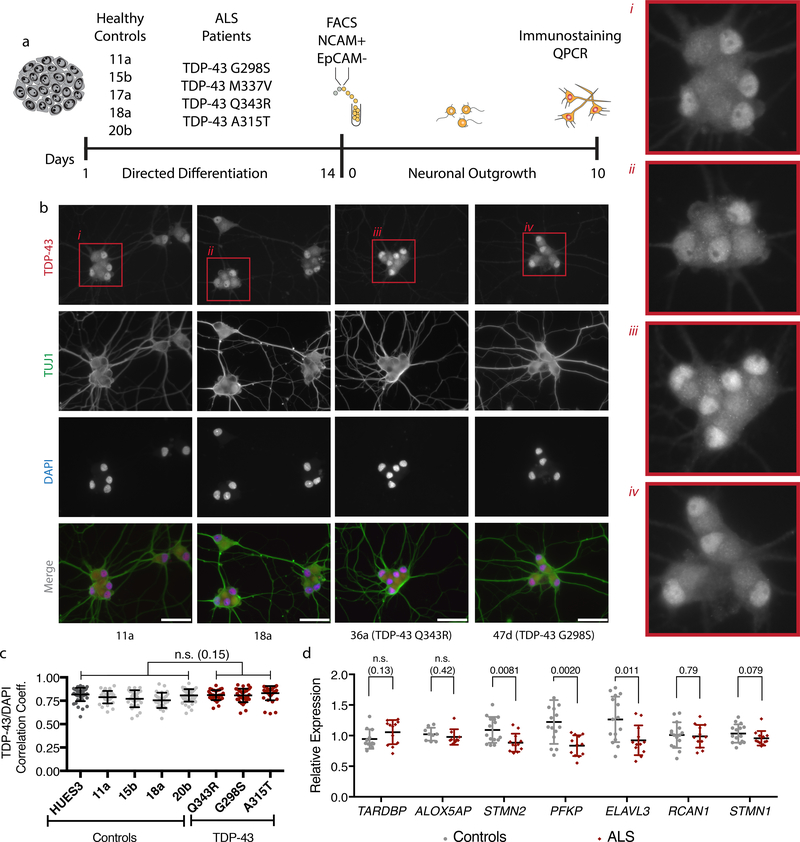
Figure 2. A subset of transcripts with altered abundance after TDP-43 depletion also displayed altered abundance in hMNs expressing mutant TDP-43.
(a) Strategy for assessing candidate TDP-43 target transcripts in ALS patient iPS cell-derived hMNs expressing mutant TDP-43. (b) Representative micrographs of control and patient neurons immunostained for TDP-43 (red), β-III tubulin (green) and counterstained with DAPI (blue). Scale bar, 100 μm. n=4 control and 3 patient lines with similar results in two independent experiments (c) Pearson’s correlation analysis for TDP-43 immunostaining and DAPI fluorescence comparing control neurons to those with TDP-43 mutations. Dots represent individual cells and are displayed as mean with SD for 60 cells from n=4 control and 3 patient lines (Unpaired t-test, two-sided, P value < 0.05). Two independent experiments were performed. (d) qRT-PCR analysis of the transcripts with altered abundance after TDP-43 knockdown in neurons differentiated from controls or TDP-43 patients. Data are displayed as mean with SD from two independent experiments with n=4 control and 3 patient lines in experiment 1 and n=5 control and 4 patient lines in experiment 2 (Unpaired t-test, two-sided, P value < 0.05).
Although we did not detect gross changes to TDP-43, we hypothesized that TARDBP mutations might impact the metabolism of a subset of TDP-43 target RNAs. Thus, we collected RNA from hMNs and performed qRT-PCR to investigate levels of the transcripts most reproducibly impacted by TDP-43 depletion (ALOX5AP, STMN2, ELAVL3, PFKP, and RCAN1). For three of the genes (STMN2, PFKP, and ELAVL3), we observed a significant decrease in abundance (Fig. 2d). Consistent with the TDP-43 depletion experiments, we did not observe significant changes to the abundance of the closely related STMN1 RNA suggesting a specific relationship between TDP-43 and STMN2 (Fig. 2d and Supplementary Fig. 6e). Additionally, we did not observe significant differences in TARDBP transcript levels between mutant and control neurons (Fig. 2d).
TDP-43 regulates STMN2 levels
Due to their broad expression pattern, previous studies have detected links between both PFKP as well as ELAVL328,29 and TDP-43. We, therefore, focused our efforts on exploring how STMN2, which shows a more neuronally restricted expression pattern, might be regulated by TDP-43. STMN2 is one of four proteins belonging to the Stathmin family of microtubule-binding proteins, with functional roles in neuronal cytoskeletal regulation and axonal regeneration pathways18,19,30. In humans, STMN1 and STMN3 transcripts exhibit ubiquitous expression, whereas STMN2 and STMN4 are enriched in CNS tissues31. Considering the growing evidence for cytoskeletal defects in ALS32–34 and the importance of cytoskeletal function in projection neurons, characterizing a putative relationship between STMN2 and TDP-43 seemed to be of potential relevance to ALS.
To this end, we first examined whether the decline in the STMN2 transcript abundance after TDP-43 knockdown was reflected at the protein level. Following independent RNAi experiments, we performed qRT-PCR with two different sets of primer pairs and found STMN2 (~50–60%) transcripts abundance significantly reduced in hMNs relative to controls (Fig. 3a). Immunoblot assays on protein lysates from these same hMN cultures demonstrated STMN2 protein levels were also significantly reduced following siTDP-43-treatment (Fig. 3b).
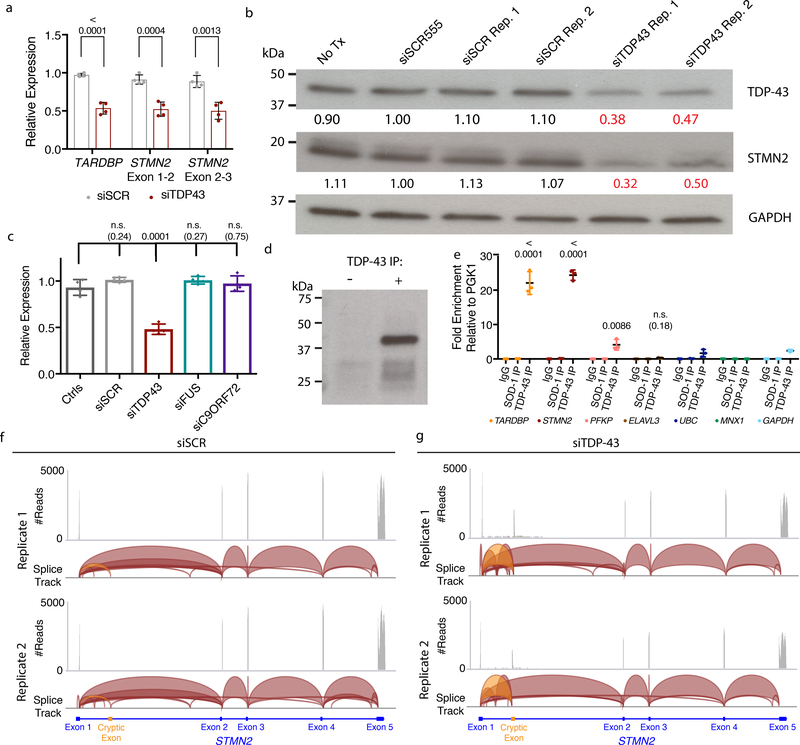
Figure 3. TDP-43 regulates STMN2 RNA and protein levels through direct interactions and suppression of a cryptic exon.
(a) qRT-PCR analysis for the STMN2 transcript using two different sets of exon spanning primer pairs. Data are displayed as mean with SD of two technical replicates from n=2 independent experiments (Unpaired t-test, two-sided, P value < 0.05). (b) Immunoblot analysis for TDP-43 and STMN2 protein levels following partial depletion of TDP-43 by siRNA knockdown. Protein levels were normalized to GAPDH and are expressed relative to the levels in hMNs treated with the siRED control. Similar results were obtained in n=2 independent experiments. (c) qRT-PCR analysis for STMN2 transcript analysis in Hb9::GFP+ hMNs treated with siRNAs targeting three ALS-linked genes (TARDBP, FUS, and C9ORF72). Data are displayed as mean with SD of replicates from n=2 independent experiments (Dunnett’s multiple comparison test, Alpha value < 0.05). Similar results were obtained in n=3 independent experiments. (d-e) Formaldehyde RNA immunoprecipitation was used to identify transcripts bound to TDP-43. After TDP-43 immunoprecipitation (d), qRT-PCR analysis was used to test for enrichment of TARDBP transcripts, STMN2, PFKP, and ELAVL3 (e) normalized to the sample input then relative to IgG and the house keeping pulled down by TDP-43. Data are displayed as mean with SD of replicates for one of n=2 independent experiments (Unpaired t-test, two-sided, P value < 0.05 compared to the house keeping genes MNX1, GAPDH, and UBC). (f-g) Visualization of RNA-seq reads mapping to STMN2 from neurons treated with scrambled siRNAs (f) or siTDP43 (g). Read coverage and splice junctions are shown for alignment of the samples to the human hg19 genome. Splice ribbons from exon 1 to the cryptic exon are highlighted in orange.
To probe how selective the changes in STMN2 levels were to TDP-43 knockdown, we knocked down two additional ALS-linked genes, FUS and C9ORF721,4. FUS protein is structurally similar to TDP-43 and is also involved in RNA metabolism. The function of C9ORF72 is under investigation, but a large intronic GGGGCC repeat expansion is responsible for a substantial number ALS and FTD cases through three proposed mechanisms: the creation of long repetitive RNAs, the translation of these repeats into toxic, repetitive dipeptides, or haploinsufficiency. Following induction of RNAi targeting TDP-43, FUS, or C9ORF72, we found significant downregulation of the respective siRNA-targeted genes by qRT-PCR (Supplementary Fig. 6a–c). Although knockdown of TARDBP reduced levels of STMN2, STMN2 levels were unaltered following knockdown of FUS and C9ORF72 (Fig. 3c). These results confirm that STMN2 downregulation was not a trivial consequence of RNAi induction. Furthermore, these data suggest that reduced function of FUS and C9ORF72 were not sufficient to induce a reduction in STMN2 abundance.
TDP-43 can bind to and regulate several aspects of RNA metabolism. To determine whether TDP-43 associated directly with STMN2 RNA, which contains many TDP-43 binding motifs (Supplementary Fig. 6f–g), we developed conditions for TDP-43 immunoprecipitation and subsequent formaldehyde RNA immunoprecipitation (fRIP) (Fig. 3d). After reversal of cross-linking, we performed quantitative RT-PCR to detect RNA molecules bound to TDP-43. We first examined whether transcripts encoding TDP-43 itself were enriched as this autoregulation is well-established10. Indeed, we observed enrichment of TARDBP RNA following TDP-43 pulldown, but not in an IgG control or when we pulled down a distinct ALS-associated protein, SOD-1 (Fig. 3e). We next asked whether additional transcripts influenced by TDP-43 knockdown, housekeeping transcripts (PGK1, UBC, GAPDH), or a MN-specific transcript (MNX1) were enriched. Of these, we observed strong and significant enrichment for STMN2 transcripts (Fig. 3e).
TDP-43 suppresses cryptic exon inclusion
Emerging evidence highlights the importance of nuclear TDP-43 in suppressing non-conserved or cryptic exons to maintain splicing integrity35. When cryptic exons are included in RNA transcripts, their inclusion can affect transcript abundance by disrupting translation and promoting nonsense-mediated decay35. Interestingly, little or no overlap in the genes regulated by TDP-43 through cryptic exon suppression has been observed between mouse and man35. We, therefore, examined our MN sequencing data for evidence of cryptic exons in the 95 human genes reported to contain TDP-43-regulated cryptic exons36. We found reads mapping to cryptic exons in 9 of these genes including PFKP (Supplementary Fig. 7a). Encouraged, we utilized our data to search for neuronal transcripts that might be regulated by this mechanism. We found strong evidence for the presence of novel cryptic exons in both ELAVL3 and STMN2 (Supplementary Fig. 7b–c). In the case of STMN2, we observed splicing tracks emanating from exon 1 and entering a cryptic exon, but no splice ribbons from the cryptic exon to either exon 2 or other exons downstream (Fig. 3f–g). Moreover, the inclusion of this cryptic exon would lead to a very early stop codon (Supplementary Fig. 10h). As a result, STMN2 transcripts that include this cryptic exon cannot yield functional STMN2 protein.
STMN2 levels tuned by TDP-43 levels and localization
TARDBP mutations and overexpression have been proposed to perturb autoregulation of its transcript leading to altered TDP-43 protein levels37. Thus, we asked whether expression of additional WT or mutant TDP-43 could act dominantly to influence expression of STMN2. To answer this question in a controlled manner, we knocked-in a single copy of either wildtype TDP-43 or mutant TDP-43 M337V into the AAVS1 safe harbor locus of the HUES3 Hb9::GFP cell line (Supplementary Fig. 8a). Genotyping of the AAVS1 locus confirmed single copy integration of each construct (Supplementary Fig. 8b). Following directed differentiation, GFP+ hMN purification, and RNA isolation, we found that additional expression of either WT or mutant TDP-43 significantly depressed the abundance of STMN2 (Supplementary Fig. 8c). These data together with our siRNA data indicate that both depleting and increasing TDP-43 levels can alter STMN2 expression.
Aggregation and loss of nuclear TDP-43 are pathological hallmarks of ALS. To investigate a potential connection between these postmortem findings and STMN2 abundance, we explored whether MG-132-mediated proteasome inhibition could influence TDP-43 localization in hMNs and serve as a potential model of acute TDP-43 dysfunction. After establishing the range and timing of tolerable proteasome inhibition (Supplementary Fig. 9a–c), we performed a pulse-chase experiment to determine the consequences of proteasome inhibition on TDP-43 localization (Supplementary Fig. 10a). Strikingly, using the Pearson’s coefficient analysis and nuclear to cytoplasmic ratio, we observed that TDP-43 staining in the nucleus was greatly diminished after 24 hours (Supplementary Fig. 10b–d). Following MG-132 washout, we found that TDP-43 staining became indistinguishable from that in unchallenged neurons after 4 days (Supplementary Fig. 10b–d). Thus, proteasome inhibition in hMNs induced reversible TDP-43 mislocalization.
In ALS patients, TDP-43 mislocalization is associated with accumulation of insoluble TDP-43 protein. After proteasome inhibition in hMNs, we examined TDP-43 levels by immunoblot analysis in both the detergent-soluble and detergent-insoluble fractions. In lysates from neurons treated with MG-132 (Supplementary Fig. 10a), we observed a significant decline in soluble TDP-43 (Supplementary Fig. 10e), which could be readily explained by an increase of insoluble TDP-43 found after urea exposure (Supplementary Fig. 10e). After establishing that proteasome inhibition induced TDP-43 mislocalization and insolubility, we asked if this change in TDP-43 also influenced STMN2 transcript levels and again found a significant decline in its abundance (Supplementary Fig. 10f). We next used RT-PCR with primers spanning into the cryptic exon observed after TDP-43 depletion to asked if proteasome inhibition also induced its inclusion in hMNs (Supplementary Fig. 10g). Although hMNs growing under normal conditions showed no detectable evidence for this cryptic event, following proteasome inhibition, it could be readily observed (Supplementary Fig. 10g) Sequencing of the PCR product confirmed the splice junction and premature stop codon within the cryptic splice-form predicted by our RNA-seq analysis (Supplementary Fig. 10h). These data connect impaired protein homeostasis to TDP-43 localization and solubility as well as STMN2 levels. We found it notable that an identical cryptic splicing event was induced in STMN2 following either RNAi-mediated TDP-43 depletion or proteasome inhibition induced TDP-43 insolubility.
STMN2 localization in human neurons
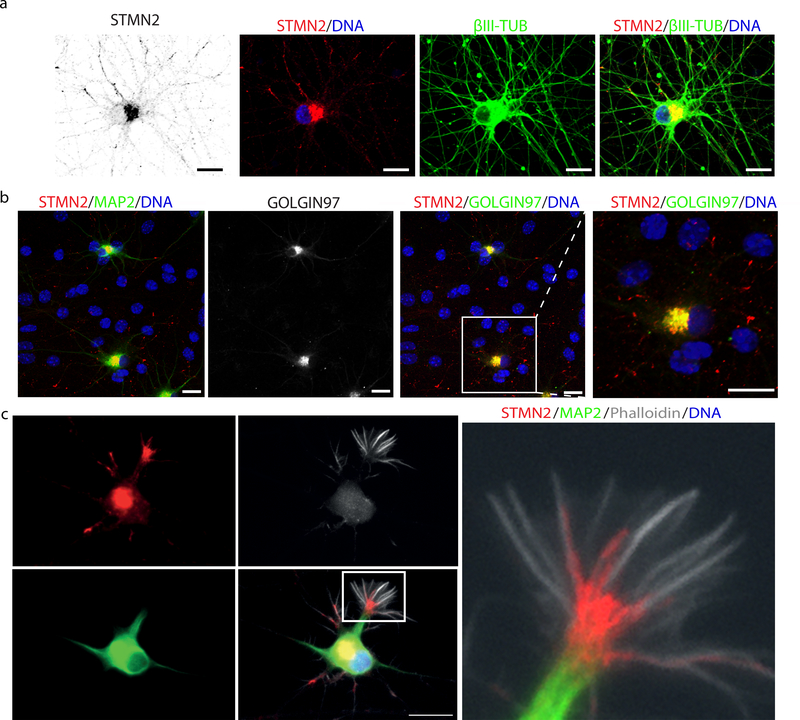
Given the importance of TDP-43 in ALS and its tight regulation of STMN2, we wondered if alterations in STMN2 might influence motor axon biology. Supporting previous expression studies30, we found that STMN2 protein was selectively expressed in differentiated neurons and could not be as readily detected in stem cells or neuronal progenitors (Supplementary Fig. 6d). We used immunocytochemistry to probe the subcellular localization of STMN2 and found it both at neurite tips and in peri-nuclear regions of the cell bodies (Fig. 4a). Further co-staining for the Golgi-associated protein GOLGIN97, indicated STMN2 was localizing to this sub-cellular compartment (Fig. 4b). Based on rodent studies18, STMN2 would be predicted to function at the human growth cone during neurite extension and repair. When we stained hMNs after differentiation and sorting, we observed strong staining of STMN2 at the interface between microtubules and F-actin bundles; cytoskeletal components defining the growth cone (Fig. 4c).
Figure 4. STMN2 localizes to the Golgi apparatus and growth cone in hMNs.
(a) Micrographs of Hb9::GFP+ hMNs immunostained for STMN2 (red), β-III tubulin (green) and counterstained with DAPI (blue). (b) Micrographs of Hb9::GFP+ hMNs co-cultured on glia immunostained for STMN2 (red) and MAP2 (green) and GOLGIN97 (green). (c) Micrograph of Hb9::GFP+ hMNs day 3 after sorting immunostained for STMN2 (red), MAP2 (green) and counterstained with F-actin-binding protein phalloidin (white). Scale bars, 5 μm. Similar results were obtained in n=3 independent experiments.
TDP-43 depletion inhibits neurite and axon growth
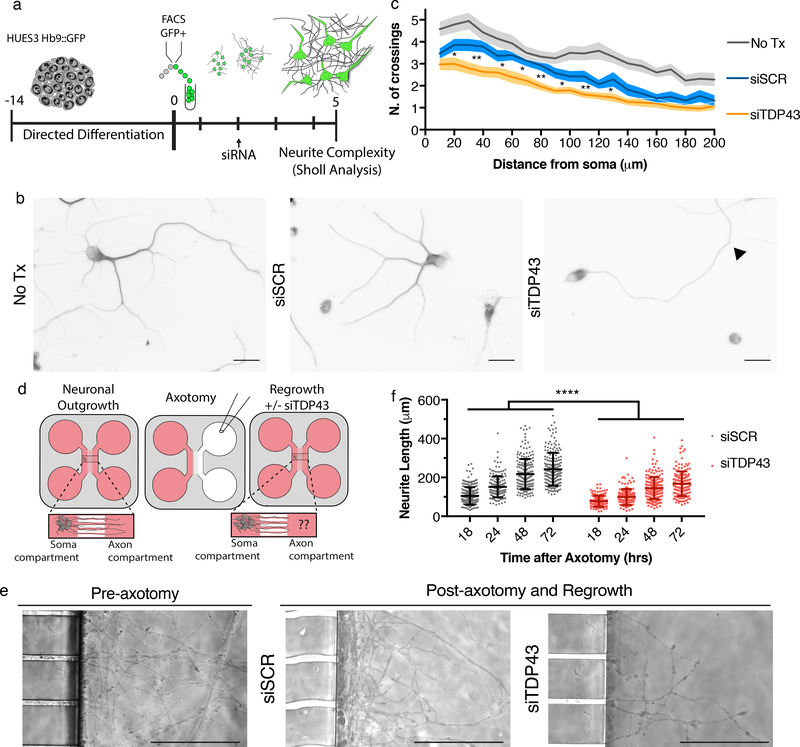
Based on predicted STMN2 function, we investigated whether TDP-43 knockdown compromised neurite outgrowth and axonal regeneration in hMNs. Three days after siTDP43 transfection, hMNs were fixed and stained for β-III-tubulin to label neuronal processes. Sholl analysis, which quantifies the number of neurite branches at a given interval from the center of the soma, revealed significantly reduced neurite complexity in neurons treated with siTDP-43 (Fig. 5a–c).
Figure 5. TDP-43 depletion leads to neurite outgrowth and axonal regrowth defects.
(a) Experimental strategy used to assess the cellular effect of TDP-43 depletion on hMN neurites. (b) Representative micrographs of hMNs treated with indicated siRNAs and immunostained for β-III tubulin to perform Sholl analysis. Arrow head indicates an example branch point. Scale bar, 50 μm. (c) Sholl analysis of hMNs after siRNA treatment. Lines represent sample means and shading represent the SEM with unpaired t-test between siTDP43 and siSCR, two-sided, P value < 0.05 with all values in Supplementary Table 1. Similar results were obtained in n=2 independent experiments. (d) Experimental strategy used to assess the cellular effect of TDP-43 depletion in hMNs after axonal injury. (e) Representative micrographs of hMNs in the microfluidics device prior to and after axotomy. Scale bars, 150 μm. (f) Measurements of axonal regeneration after axotomy. Individual neurites are displayed as dots along with the mean and SD (Unpaired t-test, two-sided, P value < 0.05 18hrs=<0.0001, 24hrs=<0.0001, 48hrs=<0.0001, and 72hrs=<0.0001). Similar results were obtained in n=4 devices from 2 independent experiments.
To test if reduced TDP-43 levels inhibited axonal regrowth after injury, we plated hMNs in microfluidic devices that permitted axon growth into a chamber distinct from the neuronal cell bodies. Neurons cultured for 7 days in the soma compartment of the device extended axons through the microchannels into the axon chamber. We severed axons without disturbing cell bodies in the soma compartment and then measured axon extension from the microchannel across a time course to assess regrowth after injury. Our analysis revealed significantly reduced regrowth in the siTDP-43-treated neurons for all time points measured (Fig. 5d–f).
STMN2 is necessary for normal neurite growth
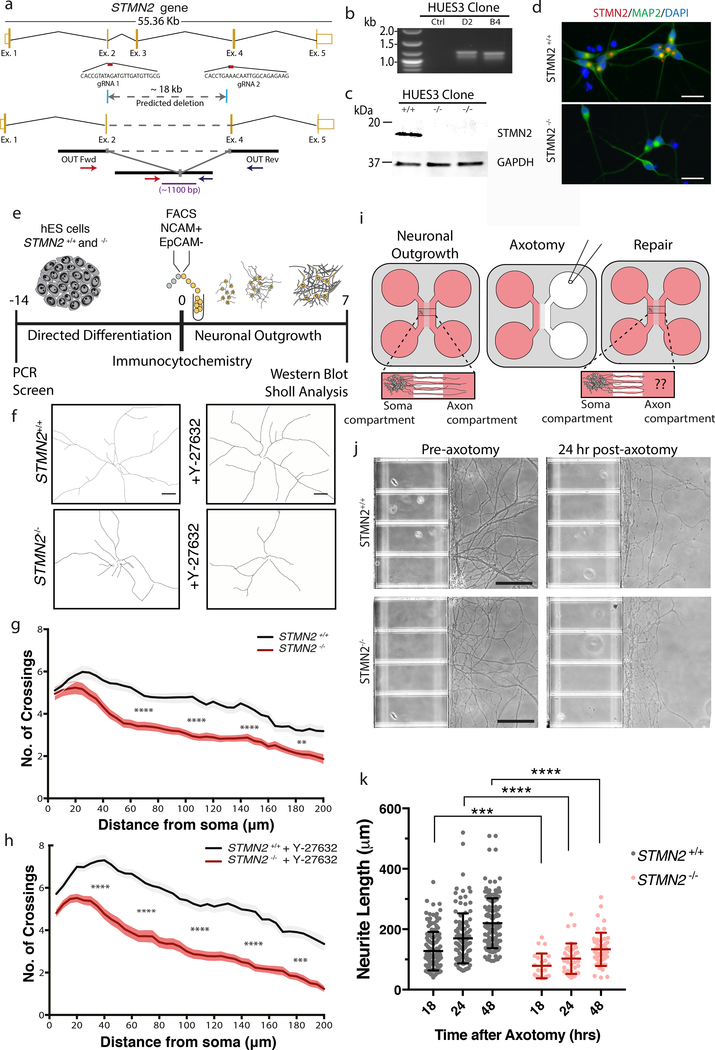
Because TDP-43 regulates many transcripts, we confirmed that loss of STMN2 was sufficient to induce phenotypes similar those observed after TDP-43 knockdown. We used CRISPR/Cas9 to produce a large deletion within STMN2 in two independent hES cell lines (WA01 and HUES3 Hb9::GFP). We confirmed loss of protein expression in differentiated hMNs by both immunoblotting and immunocytochemistry (Fig. 6a–d). We then characterized neurite outgrowth in STMN2−/− mutant hMNs (Fig. 6e). After 7 days in culture, Sholl analysis revealed significantly reduced neurite extension in STMN2−/− mutant neurons relative to controls (Fig. 6f–g). Separately, we cultured neurons in the presence of a ROCK inhibitor, Y-27632, which has been shown to increase neurite extension. The difference in neurite outgrowth was even more striking in these experiments with the molecule enhancing the outgrowth of the STMN+/+ line but not the STMN−/− line (Fig. 6h). Inactivation of STMN2 in the WA01 background resulted in hMNs with similar outgrowth phenotypes (Supplementary Fig. 11). We also queried whether STMN2 functions in neuronal repair after injury (Fig. 6i). To this end, hMNs were cultured for 7 days in microfluidic devices before performing axotomy. Our experiments revealed significantly reduced regrowth in the STMN2−/− mutant hMNs when compared to STMN2+/+ controls for all time points measured (Fig. 6j–k and Supplementary Fig. 11).
Figure 6. STMN2 mutant neurons have neurite outgrowth and regrowth deficits similar to neurons treated with siTDP43.
(a) Knockout strategy targeting two constitutive exons of the human STMN2 gene. (b-d) STMN2 elimination was confirmed in the HUES3 Hb9::GFP line by RT-PCR analysis of genomic DNA (b), by immunoblot analysis (c), and by immunofluorescence (d). Similar results were obtained from n=2 biologically independent experiments. (e) Experimental strategy used to assess the cellular effect of STMN2 elimination in hMNs. (f-h) Sholl analysis of hMNs with and without STMN2 and in the absence (g) or presence (h) of an ROCK inhibitor (Y-27632, 10 μM). Lines represent sample means and shading represent the SEM with unpaired t-test between siTDP43 and siSCR, two-sided, P value < 0.05 with all values in Supplementary Table 1. Similar results were obtained in n=2 biologically independent experiments. (i) Experimental strategy used to assess the cellular effect of STMN2 elimination in hMNs after axonal injury. (j-k) Axonal regrowth after injury. Representative micrographs of hMNs in the microfluidics device prior to and after axotomy (j). Measurements of axonal regeneration after axotomy. Individual neurites are displayed as dots along with the mean and SD (Unpaired t-test, two-sided, P value < 0.05 18hrs=0.0005, 24hrs=0.0001, 48hrs=<0.0001) (k). Similar results were obtained in n=4 devices from 2 independent experiments. Similar results were obtained in n=2 biologically independent experiments.
STMN2 is altered in postmortem ALS spinal cord
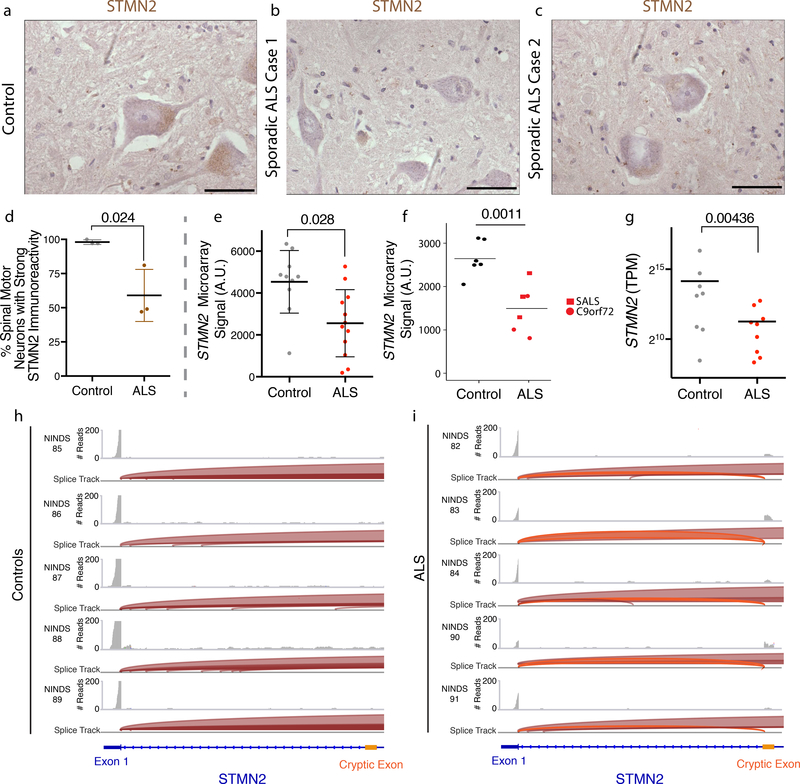
We next asked if our findings were relevant to the motor neurons of ALS patients in vivo. To this end, we used immunohistochemistry to investigate STMN2 expression in control and ALS patient post-mortem spinal cord. Similar to our observations in stem cell-derived hMNs, strong STMN2 immunoreactivity was present in the cytoplasmic region of lumbar spinal MNs, but absent in the surrounding glial cells (Fig. 7a–c). We determined the percentage of MNs exhibiting STMN2 immunoreactivity in lumbar spinal cord tissue sections in 3 controls and in 3 ALS cases. Consistent with our hypothesis, we found that the percentage of lumbar MNs with immunoreactivity to STMN2-specific antibody was significantly reduced in sporadic ALS cases (Fig. 7d). Our results are further supported by several independent expression analyses of ALS postmortem spinal cord samples38–40. We independently interrogated these data and observed decreased STMN2 transcript levels for the ALS patient samples relative to control samples in each instance (Fig. 7e–g). For the single study that performed RNA-Seq40, we also examined the sequencing reads mapping to STMN2 and identified the presence of the aforementioned cryptic exon in 5 of the 6 ALS samples and no control samples (Fig. 7h–i). These results provide an important in vivo validation of our disease modeling results.
Figure 7. ALS patient spinal cord motor neurons have decreased expression of STMN2 and express transcripts containing the cryptic exon.
(a-c) Histologic analysis of human adult lumbar spinal cord from post-mortem samples collected from a subject with no evidence of spinal cord disease (control) (a) or two patients diagnosed with sporadic ALS (b-c). The experiment was performed with n= 3 controls and 3 ALS cases. STMN2 immunoreactivity in lumbar spinal motor neurons from control and ALS cases was scored as ‘strong’ or as ‘absent’. Scale bars, 50 μm. (d) The percentage of lumbar spinal motor neurons with strong STMN2 immunoreactivity was significantly lower in ALS tissue samples. Data are displayed as the mean with SD for n= 3 controls and 3 ALS cases; approximately 40 MNs were scored for each subject (Unpaired t-test, two-sided, P value < 0.05). (e-g) Meta-analysis of STMN2 transcript abundance in previously published data sets for laser captured lumbar motor neurons analyzed by microarray n= 10 controls and 12 ALS cases displayed (Rabin et al 2009, e), laser captured lower motor neurons analyzed by microarray n= 6 controls and 6 ALS cases (Highley et al 2014, f), individuals are displayed as dots with mean and SD (moderated t-test, P value < 0.05, (e-f)), and spinal cord ventral horns analyzed by RNA-Seq for n= 8 controls and 9 ALS cases, individuals are displayed as dots with mean (D’Erchia et al 2017, Wald test and a cutoff of 0.1 for Benjamini-Hochberg adjusted p values with no log2 fold-change ratio cutoff, (g)). (h-i) Visualization of the cryptic exon for STMN2 from the NINDS datasets (g) for the ventral horns of controls (h) and sporadic ALS patient (i) spinal cords. Read coverage and splice junctions are shown for alignment of the samples to the human hg19 genome. Splice ribbons from exon 1 to the cryptic exon are highlighted in orange.
JNK inhibition rescues STMN2 and neurite defects
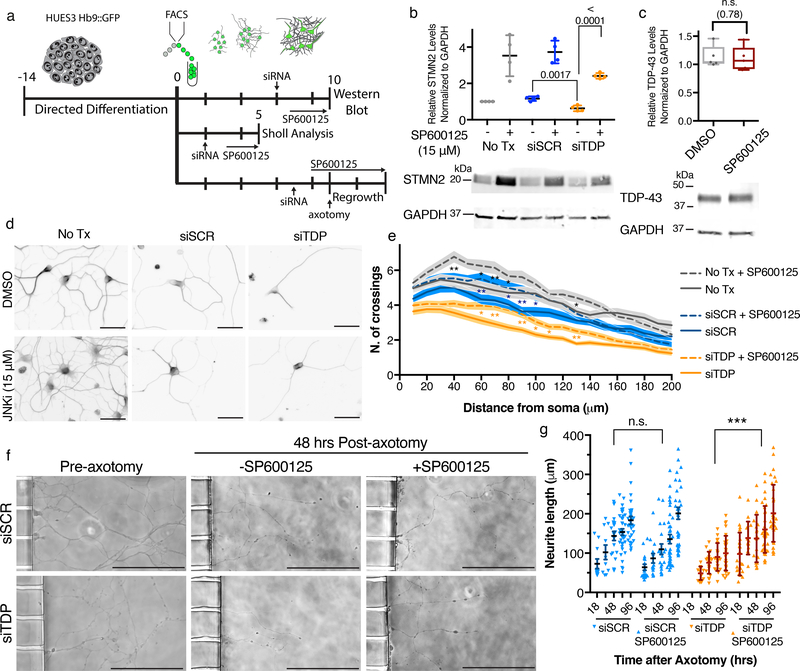
Finally, we wondered if STMN2 levels might be pharmacologically elevated through posttranslational means to rescue TDP-43 depletion induced neuropathy. Previous studies indicated that JNK1 can bind to and phosphorylate STMN2 and that phosphorylation of STMN2 promotes its degradation41,42. We therefore tested whether JNK inhibition could rescue STMN2 levels after TDP-43 knockdown (Fig. 8a). hMNs were treated with siRNAs then cultured in the presence or absence of SP600125, an established JNK inhibitor. Subsequent immunoblot analysis showed STMN2 protein levels were significantly increased after JNK inhibition (Fig. 8b) with all uncropped blots available for this and all other experiments in supplementary materials (Supplementary Fig. 12). Notably, the reduced levels of STMN2 observed after siTDP43 treatment could be boosted to levels greater than those observed in control hMNs (Fig. 8b) and, importantly, did not affect the levels of TDP-43 (Fig. 8c).
Figure 8. STMN2 protein and outgrowth deficits following TDP-43 depletion can be rescued by JNK inhibition.
(a) Experimental strategy used to assess the effect of JNK inhibitor SP600125, 15 μM, on STMN2 protein levels and neurite outgrowth after TDP-43 depletion. (b) Immunoblot analysis for STMN2 protein levels following partial depletion of TDP-43 by siRNA knockdown and then treatment with SP600125 for 3 days. Protein levels were normalized to GAPDH and are expressed relative to the levels in hMNs not treated with the siRNAs. Data are displayed as mean with SD of technical replicates from n=2 independent experiments (Unpaired t-test, two-sided, P value < 0.05). (c) Immunoblot analysis for TDP-43 protein levels in hMNs treated with SP600125 for 3 days. Protein levels were normalized to GAPDH and are expressed relative to the levels in hMNs treated with DMSO. Data are displayed as mean with SD of technical replicates from n=2 independent experiments (Unpaired t-test, two-sided, P value < 0.05). (d) Representative micrographs of HUES3 hMNs treated with indicated siRNAs with and without SP600125 for 3 days before being immunostained for β-III tubulin to perform Sholl analysis. Scale bar, 100 μm. (e) Sholl analysis of hMNs after siRNA treatment with and without SP600125. Lines represent sample means and shading represent the SEM. Data represent n= 60 cells (Unpaired t-test between samples with and without SP600125, two-sided, P value < 0.05). Similar results were obtained in n=2 independent experiments. (f-g) Axonal regrowth after injury. Representative micrographs of hMNs in the microfluidics device prior to and after axotomy with and without SP600125 (f). Measurements of axonal regeneration after axotomy. Individual neurites are displayed as dots along with the mean and SD (Unpaired t-test, two-sided, P value < 0.05 (siTDP43 vs siTDP43 SP600125; 24hr=<0.0001, 48hrs=0.0002, 72rs=<0.0001, 96hrs=<0.0001)(siSCR vs siSCR SP600125; 18hr=0.52, 24hr=0.48, 48hrs=0.062, 72rs=0.19, 96hrs=0.39). Similar results were obtained in n=2 independent experiments.
After rescuing STMN2 protein levels, we investigated whether SP600125 could rescue the TDP-43 depletion induced neuronal phenotypes. Sholl analysis reconfirmed decreased outgrowth following TDP-43 depletion and demonstrated that JNK inhibition significantly rescued outgrowth (Fig. 8d–e). Lastly, we determined whether SP600125 could also rescue defects in axonal regrowth after injury (Fig. 8a). Replicating our previous findings, we observed decreased regeneration following injury in siTDP43-treaed neurons (Fig. 8f–g). SP600125 did not alter axonal repair in neurons treated with scrambled siRNAs (Fig. 8g). However, JNK inhibition did significantly rescue axon regrowth following TDP-43 depletion (Fig. 8f–g).
Discussion
An outstanding question has been: what are the mechanistic consequences of TDP-43 mutations and how do their effects relate to the events that occur when TDP-43 becomes pathologically re-localized in patients? Identifying transcripts regulated by TDP-43 provided us the opportunity to explore the potential impact of different types of manipulations of TDP-43 relevant to both familial and sporadic disease. First, we addressed whether a subset of the target RNAs identified after TDP-43 depletion change in hMNs produced from patients with TDP-43 mutations. Interestingly, we found modest but significant changes in the expression of a number of interesting genes43 including the microtubule regulator STMN2. Thus, cells expressing patient mutations displayed partial loss-of-function. But if so, how can their dominant nature be explained?
Upon over-expression, mutant TDP-43 is prone to aggregation7, and some studies suggest mutant TDP-43 is prone to aggregation when expressed at native levels in patient specific motor neurons24,26,27. To determine whether aggregation or loss of nuclear mutant TDP-43 could be contributing to decreased expression, we carefully monitored TDP-43 in these patient hMNs, but could not detect such a defect. We are, however, confident that if larger scale aggregation, or nuclear loss of mutant TDP-43 were occurring in iPS hMNs that we would have detected it, as we found that proteasome inhibition induced dramatic nuclear loss of TDP-43 along with its insoluble accumulation. These results are akin to our previous results with mutant SOD-1 being selectively insoluble after proteasome inhibition44. We also found these data captivating because disrupted proteostasis induced by any number of ALS implicated mutations or events could be upstream of the most common histopathological finding in sALS, TDP-43 pathology, and our finding that TDP-43 targets are depleted following proteasome inhibition is consistent with that model. This model is attractive because it reconciles how overexpression of an additional transgenic copy of TDP-43 might lead to a decrease in STMN2 expression. Two recent publications describing mice with endogenous TDP-43 mutations also provide insight into the potential gain and loss of TDP-43 functions. Both studies suggest splicing abnormalities are a major consequence of TDP-43 mutations37,45. Our study also spotlights splicing defects downstream of TDP-43 perturbations, and, because of the lack of cryptic exon conservation amongst species35, showcases the advantage of human cellular disease models in identifying STMN2 RNA as a TDP-43 target.
Interestingly, partial depletion of FUS and C9ORF72 did not alter STMN2 abundance. It is well known that patients with C9ORF72 display TDP-43 pathology and consistent with this we found reduced STMN2 levels in post-mortem material from such patients. These findings suggest that either the gain of function effects of the C9ORF72 mutation or further events subsequent to knockdown are required to induce TDP-43 pathology and reduced STMN2 levels. In the instance of patients with mutations in FUS, which do not display TDP-43 pathology, it seems plausible that distinct defects in RNA metabolism underlie neuropathy. While they are beyond the scope of this study, further studies with iPS cell-derived neurons from FUS and C9ORF72 patients could shed light on these hypotheses.
The Stathmin family of proteins are regulators of microtubule stability30. Using gene editing we demonstrated STMN2 has important functions in both motor axon outgrowth and regeneration, phenocopying results in neurons with diminished TDP-43 levels. Although hMNs generated in vitro share many molecular and functional properties with bona fide MNs13, the in vivo validation of stem cell-based ALS model discoveries is a critical test of their relevance46. Using spinal cord tissues and published expression studies, we provide in vivo evidence corroborating the finding that STMN2 levels are altered in ALS. Furthermore, we found that the likely mechanism for this decline, as in our cell models, was through splicing into a cryptic exon. Finally, we demonstrate JNK inhibition increases STMN2 levels, which was sufficient to rescue axonal phenotypes induced by TDP-43 depletion. While we suggest that restoring STMN2 levels in ALS patients to normal levels is an attractive target, it is important this be done with care as overexpression of Stathmins can cause Golgi fragmentation as shown in the transgenic SOD1 mouse model of ALS47.
Methods
Life Sciences Reporting Summary
A Life Sciences Reporting Summary is available online
Cell culture and Differentiation of hESCs and hiPSCs into MNs
Pluripotent stem cells were grown with mTeSR1 medium (Stem Cell Technologies) on tissue culture dishes coated with Matrigel™ (BD Biosciences), and maintained in 5% CO2 incubators at 37°C. Stem cells were passaged as small aggregates of cells after 1 mM EDTA treatment. 10 μM rock inhibitor (Sigma, Y-27632) was added to the cultures for 16–24 hours after dissociation to prevent cell death. The HUES3 Hb9::GFP cell line has been previously described12,48. MN differentiation was achieved using a modified 14-day strategy. This approach relies on neural induction through small molecule inhibition of SMAD signaling, accelerated neural differentiation through FGF and NOTCH signaling inhibition, and MN patterning through the activation of retinoic acid (RA) and Sonic Hedgehog signaling pathways. In brief, ES cells were dissociated to single cells using accutase™ (Stem Cell Technologies) and plated at a density of 80,000 cells/cm2 on Matrigel-coated culture plates with mTeSR1 medium (Stem Cell Technologies) supplemented with rock inhibitor (10μM Y-27632, Sigma). When cells reached 100% confluency, medium was changed to differentiation medium (1/2 Neurobasal (Life Technologies™) 1/2 DMEM-F12 (Life Technologies™) supplemented with 1x B-27 supplement (Gibco®), 1x N-2 supplement (Gibco®), 1x Gibco® GlutaMAX™ (Life Technologies™) and 100μM non-essential amino-acids (neaa)). This time point was defined as day 0 (d0) of motor neuron differentiation. Treatment with small molecules was carried out as follows: 10μM SB431542 (Custom Synthesis), 100nM LDN-193189 (Custom Synthesis), 1μM retinoic acid (Sigma) and 1μM Smoothend agonist (Custom Synthesis) on d0-d5; 5μM DAPT (Custom Synthesis), 4μM SU-5402 (Custom Synthesis), 1μM retinoic acid (Sigma) and 1μM Smoothend agonist (Custom Synthesis) on d6-d14.
Cell acquisition and authentication
The hES cell and iPS cells used in this study were previously approved by the institutional review boards (IRBs) of Harvard University, Massachusetts General Hospital, and Columbia University. Specific point mutations were confirmed by PCR amplification followed by Sanger sequencing. Weekly, our lab checks for mycoplasma contamination using the MycoAlert kit (Lonza) with no cell lines used in this study testing positive. The use of these cells at Harvard was further approved and determined not to constitute Human Subjects Research by the Committee on the Use of Human Subjects in Research at Harvard University.
Fluorescent Activated Cell Sorting (facs) of hMNs
After 14 days, differentiated cultures were dissociated to single cells using accutase™ treatment for 1 hour inside a 5% CO2 / 37°C incubator. Repeated (10–20 times) but gentle pipetting with a 1000μL Pipetman® was used to achieve a single cell preparation. Cells were spun down, washed 1x with PBS and resuspended in sorting buffer (1x cation-free PBS 15mM HEPES at pH 7 (Gibco®), 1% BSA (Gibco®), 1x penicillin-streptomycin (Gibco®), 1 mM EDTA, and DAPI (1μg/mL). Cells were passed through a 45μm filter immediately before FACS analysis and purification. The BD FACS Aria II cell sorter was routinely used to purify Hb9::GFP+ cells into collection tubes containing MN medium (Neurobasal (Life TechnologiesTM), 1x N-2 supplement (Gibco®), B-27 supplement (Gibco®), GlutaMax and neaa) with 10μM rock inhibitor (Sigma, Y-27632) and 10ng/mL of neurotrophic factors gdnf, bdnf and cntf (R&D). Forward and side scatter was used to resolve cells from debris with doublet discrimination by forward scatter area vs forward scatter width. DAPI signal was then used to determine cell viability, and differentiated cells not exposed to MN patterning molecules (ra and sag) were used as negative controls to gate for green fluorescence.
For cell lines not containing the Hb9::GFP reporter, the BD Lyoplate was used to identify a cell surface signature49. After 14 days of differentiation, HUES3 Hb9::GFP cultures were dissociated to single cells using Accutase supplemented with DNAse I, washed by gentle pipetting and 500000 cells were aliquoted across three 96-well plates and stained for 242 monoclonal antibodies (BD Lyoplate, BD Bioscience 560747). Cells were washed, stained with secondary antibody conjugated to Alexa Fluor 647, washed, fixed and analyzed using the high throughput sampler module of BD LSR II. To sort cells based on the BD Lyoplate findings, single cell suspensions were incubated with antibodies against NCAM (BD Bioscience, BDB557919, 1:200) and EpCAM (BD Bioscience, BDB347198, 1:50) for 25 minutes in sorting buffer, then washed once with PBS 1x and resuspended in sorting buffer. The population that was negative for EpCAM but positive for NCAM was sorted. For RNA-Seq experiments, 200,000 GFP+ cells per well were plated in 24-well tissue culture dishes pre-coated with Matrigel. MN medium supplemented with 10 ng/mL of each gdnf, bdnf and cntf (R&D Systems) was used to feed and mature the purified hMNs. RNA-Seq experiments and most downstream assays were carried with d10 purified hMNs (10 days in culture after FACS) grown plates coated with 0,1 mg/ml poly-D-lysine (Invitrogen) and 5 μg/ml laminin (Sigma-Aldrich) at a concentration of around 130000 cells/cm2.
RNAi
RNAi in cultures of purified GFP+ MNs was induced with Silencer® Select siRNAs (Life Technologies™) targeting the TDP-43 mRNA or with a non-targeting siRNA control with scrambled sequence that is not predicted to bind to any human transcripts. Lyophilized siRNAs were resuspended in nuclease-free water and stored at −20°C as 20μM stocks until ready to use. For transfection, siRNAs were diluted in Optimem (Gibco®) and mixed with RNAimax (Invitrogen) according to manufacturer’s instructions. After 30 min incubation, the mix was added drop-wise to the MN cultures, so that the final siRNA concentration in each well was 60nM in 1:1 Optimem:MN medium (Neurobasal (Life Technologies™, N2 supplement (Gibco®), B-27 supplement (Gibco®), GlutaMax and neaa) and 10ng/mL of each gdnf, bdnf and cntf (R&D). 12–16 hours post-transfection media was changed. RNA-Seq experiments and validation assays were carried out with material collected 4 days after transfection.
Immunocytochemistry
For immunofluorescence, cells were fixed with ice-cold 4% PFA for 15 minutes at 4°C, permeabilized with 0.2% Triton-X in 1x PBS for 45 minutes and blocked with 10% donkey serum in 1x PBS-T (0.1% Tween-20) for 1 hour. Cells were then incubated overnight at 4°C with primary antibody (diluted in blocking solution). At least 4 washes (5 min incubation each) with 1xPBS-T were carried out, before incubating the cells with secondary antibodies for 1 hour at room temperature (diluted in blocking solution). Nuclei were stained with DAPI. The following antibodies were used in this study: Hb9 (1:100, DSHB, MNR2 81.5C10-c), TUJ1 (1:1000, R&D Systems MAB NL493), MAP2 (1:10000, Abcam ab5392), GFP (1:500, Life Technologies™, A10262), Islet1 (1:500, Abcam ab20670), TDP-43 (1:500, ProteinTech Group), and STMN2 (1:4000, Novus). Secondary antibodies used (488, 555, 594, and 647) were AlexaFluor™ (1:1000, Life Technologies™). Images were acquired using a Nikon Eclipse Ti microscope and analyzed using NIS-Elements (Nikon).
Immunoblot Assays
For analysis of TDP-43 and STMN2 protein expression levels, d10 MNs were lysed in RIPA buffer (150mM Sodium Chloride; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% SDS; 50 mM Tris pH 8.0) containing protease and phosphatase inhibitors (Roche) for 20 min on ice, and centrifuged at high speed. 200μL of RIPA buffer per well of 24-well culture were routinely used, which yielded ~20μg of total protein as determined by BCA (Thermo Scientific). After two washes with RIPA buffer, insoluble pellets were resuspended in 200 μl of urea buffer (Bio-Rad). For immunoblot assays 2–3μg of total protein were separated by SDS-PAGE (BioRad), transferred to PDVF membranes (BioRad) and probed with antibodies against TDP-43 (1:1000, ProteinTech Group), GAPDH (1:1000, Millipore) and STMN2 (1:3000, Novus). Insoluble pellets were loaded based on protein concentration of correspondent RIPA-soluble counterparts. The same PDVF membrane was immunoassayed 2–3 times using Restore™ PLUS Western Blot Stripping Buffer (Thermo Scientific). GAPDH levels were used to normalized each sample, and LiCor software (Image Studios) was used to quantitate protein band signal. Full size blots have been provided (Supplementary Fig. 13)
RNA preparation, qRT-PCR and RNA sequencing
Total RNA was isolated from d10 MNs for RNA-Seq experiments and validation assays using Trizol LS (Invitrogen) according to manufacturer’s instructions. 500μL were added per well of the 24-well cultures. A total of 300–1000ng of total RNA was used to synthesize cDNA by reverse transcription according to the iscript kit (Bio-rad). Quantitative RT-PCR (qRT-PCR) was then performed using sybr green (Bio-Rad) and the iCycler system (Bio-rad). Quantitative levels for all genes assayed were normalized using gapdh expression. Normalized expression was displayed relative to the relevant control sample. For comparison between patient lines, normalized expression was displayed relative to the average of pooled data points. The qPCR experiment comparing control and ALS cell lines was performed with 5 control lines and 4 patient lines with 3 technical replicates. All primer sequences are available upon request. For next-generation RNA sequencing (RNA-Seq), two independent experiments were performed (Exp 1 and Exp 2) with multiple unique siRNAs used for the two groups control (siRED, siSCR1, and siSCR2) vs siTDP43 (siTDP1 and siTDP2) with a total of n=15 samples. After RNA extraction, samples with RNA integrity numbers (RIN) above 7.5, determined by a bioAnalyzer, were used for library preparation. In brief, RNA sequencing libraries were generated from ~250ng of total RNA using the illumina TruSeq RNA kit v2, according to the manufacturer’s directions. Libraries were sequenced at the Harvard Bauer Core Sequencing facility on a HiSeq 2000 platform. All FASTQ files were analyzed using the bcbioRNASeq workflow and toolchain50. The FASTQ files were aligned to the GRCh37/hg19 reference genome. Differential expression testing was performed using DESeq2 suite of bioinformatics tools51. The DEXSeq module of Bioconductor was used to identify differential splicing52. We used salmon to generate the counts and tximport to load them at gene level53,54. All p-values are then corrected for multiple comparisons using the method of Benjamini and Hochberg55. We used an adjusted p-value cutoff of 0.05 with no log fold-change ratio cutoff for differential gene expression analysis and a 0.1 cutoff with no log fold-change ratio cutoff for differential exon usage.
TDP-43 localization analysis
For analysis of TDP-43 nuclear localization analysis, hMNs were stained for TDP-43 (ProteinTech), β-III tubulin (R&D Systems) and counterstained with DAPI. Images were acquired using a Nikon Eclipse Ti microscope. β-III tubulin staining was used to determine the cell body as the region of interest and then the Pearson’s correlation coefficient was calculated for TDP-43 and DAPI staining using NIS-Elements (Nikon) with a minimum of 30 neurons analyzed. NIS-Elements was used to segment cells into the nuclear (DAPI positive) and cytoplasmic (β-III tubulin positive DAPI negative) regions. The mean TDP-43 intensity in these two regions were used to calculate the nuclear to cytoplasmic ratio for TDP-43 immunofluorescence.
Electrophysiology recordings
GFP+ MNs were plated at a density of 5,000 cells/cm2 on poly-D-lysine/laminin-coated coverslips and cultured for 10 days in MN medium, conditioned for 2–3 days by mouse glial cells and supplemented with 10ng/mL of each gdnf, bdnf and cntf (R&D Systems). Electrophysiology recordings were carried out as previously reported44,56. Briefly, whole-cell voltage-clamp or current-clamp recordings were made using a Multiclamp 700B (Molecular Devices) at room temperature (21–23C). Data were digitized with a Digidata 1440A A/D interface and recorded using pCLAMP 10 software (Molecular Devices). Data were sampled at 20 kHz and low-pass filtered at 2 kHz. Patch pipettes were pulled from borosilicate glass capillaries on a Sutter Instruments P-97 puller and had resistances of 2–4 MW. The pipette capacitance was reduced by wrapping the shank with Parafilm and compensated for using the amplifier circuitry. Series resistance was typically 5–10 MW, always less than 15 MW, and compensated by at least 80%. Linear leakage currents were digitally subtracted using a P/4 protocol. Voltages were elicited from a holding potential of −80 mV to test potentials ranging from −80 mV to 30 mV in 10 mV increments. The intracellular solution was a potassium-based solution and contained KCl, 150; MgCl2, 2; HEPES, 10; Mg ATP, 4; EGTA, 1 (pH 7.4 with KOH). The extracellular was sodium-based and contained NaCl, 135; KCl, 5; CaCl2, 2; MgCl2, 1; glucose, 10; HEPES, 10, pH 7.4 with NaOH). Kainate was purchased from Sigma. For MEA analysis, hMNs were plated onto the multielectrode array (MEA) plate and co-cultured with mouse p0–2 astrocytes (Axion Biosystems M768-KAP-96). The neuronal activities including spike and bursting number for individual wells were measured and analyzed by Axion NeuroMetric Tool software.
Formaldehyde RNA Immunoprecipitation
1 well of a 6 well plate of hMNs (2 million cells) were crosslinked and processed according to the MagnaRIP instructions (Millipore). The following antibodies were used in this study: SOD1 (1:50, Cell Signaling Technologies), TDP-43 (1:50 FL9, gift of D. Cleveland), and mouse IgG, (1:50, Cell Signaling Technology). We normalized each RIP RNA fractions’ Ct value to the Input RNA fraction Ct value for the same qPCR Assay to account for RNA sample preparation differences. To calculate the dCt [normalized RIP], we determined Ct[RIP] – (Ct[Input] – log2 (Input Dilution Factor)), where the dilution factor was 100 or 1%. To determine the fold enrichment, we calculated the ddCt by dCt [normalized RIP] – dCt[normalized IgG] then fold enrichment = 2^-ddCt.
STMN2 knockout generation
STMN2 guide RNAs were designed using the following web resources: CHOPCHOP (https://chopchop.rc.fas.harvard.edu) from the Schier Lab57. Guides were cloned into a vector containing the human U6 promotor (custom synthesis Broad Institute, Cambridge) followed by the cloning site available by cleavage with BbsI, as well as ampicillin resistance. To perform the cloning, all the gRNAs were modified before ordering. The following modifications were used in order to generate overhangs compatible with a BbsI sticky end: if the 5’ nucleotide of the sense strand was not a G, this nucleotide was removed and substituted with a G; for the reverse complement strand, the most 3’ nucleotide was removed and substituted with a C, while AAAC was added to the 5’ end. The resulting modified STMN2 gRNA sequences were used for Cas9 nuclease genome editing: guide 1: 5’ CACCGTATAGATGTTGATGTTGCG 3’ (Exon 2), guide 2: 5’ CACCTGAAACAATTGGCAGAGAAG 3’ (Exon 3), guide 3: 5’ CACCAGTCCTTCAGAAGGCTTTGG 3’ (Exon 4). Cloning was performed by first annealing and phosphorylating both the gRNAs in PCR tubes. 1 μL of both the strands at a concentration of 100 μM was added to 1 μL of T4 PNK (New England Biolabs), 1 μL of T4 ligation buffer and 6 μL of H2O. The tubes were placed in the thermocycler and incubated at 37°C for 30 mins, followed by 5 mins at 95°C and a slow ramp down to 25°C at a rate of 5°C/minute. The annealed oligos were subsequently diluted 1:100 and 2 μL was added to the ligation reaction containing 2 μL of the 100 μM pUC6 vector, 2 μL of NEB buffer 2.1, 1 μL of 10mM DTT, 1 μL of 10mM ATP, 1 μL of BbsI (New England Biolabs), 0.5 μL of T7 ligase (New England Biolabs) and 10.5 μL of H2O. This solution was incubated in a thermocycler with the following cycle, 37°C for 5 minutes followed by 21°C for 5 minutes, repeated a total of 6 times. The vectors were subsequently cloned in OneShot Top10 (ThermoFisher Scientific) cells and plated on LB-ampicilin agar plates and incubated overnight on 37°C. The vectors were isolated using the Qiagen MIDI-prep kit (Qiagen) and measured DNA concentration using the nanodrop. Proper cloning was verified by sequencing the vectors by Genewiz using the M13F(−21) primer.
Stem cell transfection was performed using the Neon Transfection System (ThermoFisher Scientific) with the 100 μL kit (ThermoFisher Scientific). Prior to the transfection, stem cells were incubated in mTeSR1 containing 10μM Rock inhibitor for 1 hour. Cells were subsequently dissociated by adding accutase and incubating for 5 min at 37°C. Cells were counted using the Countess and resuspended in R medium at a concentration of 2,5*106 cells/mL. The cell solution was then added to a tube containing 1 μg of each vector containing the guide and 1.5 μg of the pSpCas9n(BB)-2A-Puro (PX462) V2.0, a gift from Feng Zhang (Addgene). The electroporated cells were immediately released in pre-incubated 37°C mTeSR medium containing 10μM of Rock inhibitor in a 10-cm dish when transfected with the puromycin resistant vector. 24 hours after transfection with the Puromycin resistant vector, selection was started. Medium was aspirated and replaced with mTESR1 medium containing different concentrations of Puromycin: 1μg/μL, 2μg/μL and 4 μg/μL. After an additional 24 hours, the medium was aspirated and replaced with mTeSR1 medium. Cells were cultured for 10 days before colony picking the cells into a 24-well plate for expansion.
Genomic DNA was extracted from puromycin-selected colonies using the Qiagen DNeasy Blood and Tissue kit (Qiagen) and PCR screened to confirm the presence of the intended deletion in the STMN2 gene. PCR products were analyzed after electrophoresis on a 1% Agarose Gel. In brief, the targeted sequence was PCR amplified by a pair of primers external to the deletion, designed to produce a 1100 bp deletion-band in order to detect deleted clones. Sequences of the primers used are as follows: OUT_FWD, 5’ GCAAAGGAGTCTACCTGGCA 3’ and OUT_REV, 5’ GGAAGGGTGACTGACTGCTC 3’. Knockout lines were further confirmed using immunoblot analysis.
Neurite outgrowth assay
Individual Tuj1-positive neurons used for Sholl analyses58 were randomly selected and imaged using a Nikon Eclipse TE300 with a 40× objective. The neurites were traced using the ImageJ (NIH) plugin NeuronJ58, and Sholl analysis was performed using the Sholl tool of Fiji59, quantifying the number of intersections at 10-μm intervals from the cell body. The Sholl analysis was performed on a minimum of n= 50 neurons. Statistical analysis was performed by comparing the number of intersections of KO clones with the parental WT line for each 10-μm interval using Prism 7 (Graph Pad, La Jolla, CA, USA). Significance was assessed by a standard Student’s t-test, with a p value of p<0.05 considered as significant.
Axotomy
Sorted motor neurons were cultured in standard neuron microfluidic devices60 (SND150, XONA Microfluidics) mounted on glass coverslips coated with with 0.1 mg/ml poly-L-ornithine (Sigma-Aldrich) diluted in 50 mM Borate buffer, pH=8.5 and 5 μg/ml laminin (Invitrogen) at a concentration of around 250,000 neurons/device. Axotomy was performed at day 7 of culture by repeated vacuum aspiration and reperfusion with 1X PBS in the axon chamber until axons were cut effectively without disturbing cell bodies in the soma compartment. For HUES3 experiments, 2 independent replicates were performed with the first have 1 device and 20–30 neurites measured and the second experiment 3 devices were used with 70–90 neurites measured. For the WA01 cell lines, 1 replicate was performed with 2 devices and 50–60 neurites measured.
TDP-43 and STMN2 immunohistochemical analyses
Post-mortem samples from 3 sporadic ALS cases (1M, 2F) and 3 controls (2M, 1F) (no evidence of spinal cord disease) were gathered from the Massachusetts Alzheimer’s Disease Research Center (ADRC) in accordance with Partners and Harvard IRB protocols. Histologic analysis of TDP-43 immunoreactivity (rabbit polyclonal, ProteinTech Group) was performed to confirm the diagnosis. For STMN2 analyses, sections of formalin fixed lumbar spinal cord were stained using standard immunohistochemical procedure with the exception that citrate buffer antigen retrieval was performed before blocking. Briefly, samples were rehydrated, rinsed with water, blocked in 3% hydrogen peroxide then normal serum, incubated with primary STMN2 rabbit-derived antibody (1:100 dilution, Novus), followed by incubation with the appropriate secondary antibody (anti-rabbit IgG conjugated to horseradish peroxidase 1:200), and exposure to ABC Vectastain kit and DAB peroxidase substrate, and briefly counterstained with hematoxylin before mounting. Multiple levels were examined for each sample.
STMN2 splicing analysis
Total RNA was isolated from neurons using RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. A total of 300–1000ng of total RNA was used to synthesize cDNA by reverse transcription according to the iscript kit (Bio-rad). RT-PCR was then performed using one cryptic exon-specific primer and then analyzed using the Agilent 2200 Tapestation.
Data presentation and Statistical analysis
In the figure elements, bars and lines represent the median with error bars representing standard deviation. The box and whisker plots display the minimum to maximum. Data distribution was assumed to be normal but this was not formally tested. Unless stated otherwise, the statistical analyses were performed using a 2-tail unpaired Student’s t-test, with a p value of *p<0.05 considered as significant using Prism 7 (Graph Pad, La Jolla, CA, USA).
Data Availability
The authors will make all data available to readers upon reasonable request. The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus61 and are accessible through GEO Series accession number GSE121569 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121569). The patient spinal cord RNA-Seq is available through dbGAP (phs000747.v2.p1).
Supplementary Material
Acknowledgements
This research was supported by HHMI, Project ALS, HSCI, Target ALS and the NINDS grant NIH5R01NS089742 to K. Eggan. J. Klim is the Project ALS Tom Kirchhoff Family Postdoctoral Fellow. B. Davis-Dusenbery was supported by the Milton Safenowitz postdoctoral fellowship from the ALS Association. A. Burberry was supported by the U.S. National Institutes of Health (1K99AG057808-01A1). D. Mordes was funded by the MGH training grant (5T32CA009216) and is grateful for the assistance of the Massachusetts ADRC neuropathology core in preparing tissue samples. B. Wainger is a New York Stem Cell Foundation – Robertson Investigator. We thank D. Cleveland for the generous gift of TDP-43 (FL9) antibody.
Footnotes
Competing Financial Interests
The authors have no competing financial interests.
Accession Codes
GEO Series accession number for the hMN RNA-seq data is GSE121569.
References
- 1.Taylor JP, Brown RH & Cleveland DW Decoding ALS: from genes to mechanism. 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravits J et al. Deciphering amyotrophic lateral sclerosis: What phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotroph Lateral Scler Frontotemporal Degener 14, 5–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller RG, Mitchell JD & Moore DH Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst Rev 3, CD001447 (2012). [Google Scholar]
- 4.Ling S-C, Polymenidou M & Cleveland DW Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann M et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Alami NH et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 81, 536–543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee EB, Lee VM-Y & Trojanowski JQ Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. 13, 38–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sreedharan J et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraemer BC et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol 119, 409–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polymenidou M et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14, 459–468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tollervey JR et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14, 452–458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Giorgio FP, Boulting GL, Bobrowicz S & Eggan KC Human Embryonic Stem Cell-Derived Motor Neurons Are Sensitive to the Toxic Effect of Glial Cells Carrying an ALS-Causing Mutation. Stem Cell 3, 637–648 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Davis-Dusenbery BN, Williams LA, Klim JR & Eggan K How to make spinal motor neurons. Development 141, 491–501 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Han SSW, Williams LA & Eggan KC Constructing and deconstructing stem cell models of neurological disease. Neuron 70, 626–644 (2011). [DOI] [PubMed] [Google Scholar]
- 15.van Eersel J et al. Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS ONE 6, e22850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiga A et al. Alteration of POLDIP3 Splicing Associated with Loss of Function of TDP-43 in Tissues Affected with ALS. PLoS ONE 7, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A 111, E1121–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grenningloh G, Soehrman S, Bondallaz P, Ruchti E & Cadas H Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J. Neurobiol 58, 60–69 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Shin JE, Geisler S & DiAntonio A Dynamic regulation of SCG10 in regenerating axons after injury. Exp Neurol 252, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasashima K, Sakashita E, Saito K & Sakamoto H Complex formation of the neuron-specific ELAV-like hu RNA-binding proteins. Nucleic Acids Res 30, 4519–4526 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin KR et al. Over-expression of RCAN1 causes Down syndrome-like hippocampal deficits that alter learning and memory. Human Molecular Genetics 21, 3025–3041 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Ariyannur PS et al. Methamphetamine-induced neuronal protein NAT8L is the NAA biosynthetic enzyme: implications for specialized acetyl coenzyme A metabolism in the CNS. Brain Research 1335, 1–13 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Boulting GL et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol 29, 279–286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egawa N et al. Drug Screening for ALS Using Patient-Specific Induced Pluripotent Stem Cells. Sci Transl Med 4, 145ra104–145ra104 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Serio A et al. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc. Natl. Acad. Sci. U.S.A 110, 4697–4702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bilican B et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc. Natl. Acad. Sci. U.S.A 109, 5803–5808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z et al. Downregulation of MicroRNA-9 in iPSC-Derived Neurons of FTD/ALS Patients with TDP-43 Mutations. PLoS ONE 8, e76055 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park Y-Y et al. TARDBP regulates glycolysis in hepatocellular carcinoma by regulating PFKP through miR-520. Hepatology (Baltimore, Md.) 58, 182–191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colombrita C et al. From transcriptomic to protein level changes in TDP-43 and FUS loss-of-function cell models. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1849, 1398–1410 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Chauvin S & Sobel A Neuronal stathmins: A family of phosphoproteins cooperating for neuronal development, plasticity and regeneration. Progress in Neurobiology 126, 1–18 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Bieche I et al. Expression of stathmin family genes in human tissues: non-neural-restricted expression for SCLIP. Genomics 81, 400–410 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Smith BN et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 84, 324–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C-H et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488, 499–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolas A et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97, 1268–1283.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling JP, Pletnikova O, Troncoso JC & Wong PC TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349, 650–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey J, Emmett W, Fratta P, Isaacs AM & Plagnol V Quantitative analysis of cryptic splicing associated with TDP-43 depletion. BMC Med Genomics 10, 38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White MA et al. TDP-43 gains function due to perturbed autoregulation in a Tardbp knock-in mouse model of ALS-FTD. Nat Neurosci 1–20 (2018). doi: 10.1038/s41593-018-0113-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabin SJ et al. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell–matrix adhesion biology. Human Molecular Genetics 19, 313–328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Highley JR et al. Loss of nuclear TDP‐43 in amyotrophic lateral sclerosis (ALS) causes altered expression of splicing machinery and widespread dysregulation of RNA splicing in motor neurones. Neuropathology and Applied Neurobiology 40, 670–685 (2014). [DOI] [PubMed] [Google Scholar]
- 40.D’Erchia AM et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci Rep 7, 10046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tararuk T et al. JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J Cell Biol 173, 265–277 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin JE et al. SCG10 is a JNK target in the axonal degeneration pathway. P Natl Acad Sci Usa 109, E3696–E3705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan A et al. Neuronal Matrix Metalloproteinase-9 Is a Determinant of Selective Neurodegeneration. Neuron 81, 333–348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiskinis E et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 14, 781–795 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fratta Pietro et al. Mice with endogenous TDP‐43 mutations exhibit gain of splicing function and characteristics of amyotrophic lateral sclerosis. EMBO J 37, e98684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Boer AS et al. Genetic validation of a therapeutic target in a mouse model of ALS. Sci Transl Med 6, 248ra104–248ra104 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Bellouze S et al. Stathmin 1/2-triggered microtubule loss mediates Golgi fragmentation in mutant SOD1 motor neurons. Mol Neurodegeneration 11, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 48.Amoroso MW et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J Neurosci 33, 574–586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan SH et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS ONE 6, e17540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinbaugh MJ et al. bcbioRNASeq: R package for bcbio RNA-seq analysis. F1000Research 6, 1976 (2017). [Google Scholar]
- 51.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Reyes A & Huber W Detecting differential usage of exons from RNA-seq data. Genome Research 22, 2008–2017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soneson C, Love MI & Robinson MD Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. http://www.jstor.org/stable/2346101 1, 289–300 (1995).
- 56.Son EY et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Labun K, Montague TG, Gagnon JA, Thyme SB & Valen E CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44, W272–W276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meijering E et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry Part A 58A, 167–176 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Ferreira TA et al. Neuronal morphometry directly from bitmap images. Nat Methods 11, 982–984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor AM et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2, 599–605 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors will make all data available to readers upon reasonable request. The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus61 and are accessible through GEO Series accession number GSE121569 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121569). The patient spinal cord RNA-Seq is available through dbGAP (phs000747.v2.p1).