Abstract
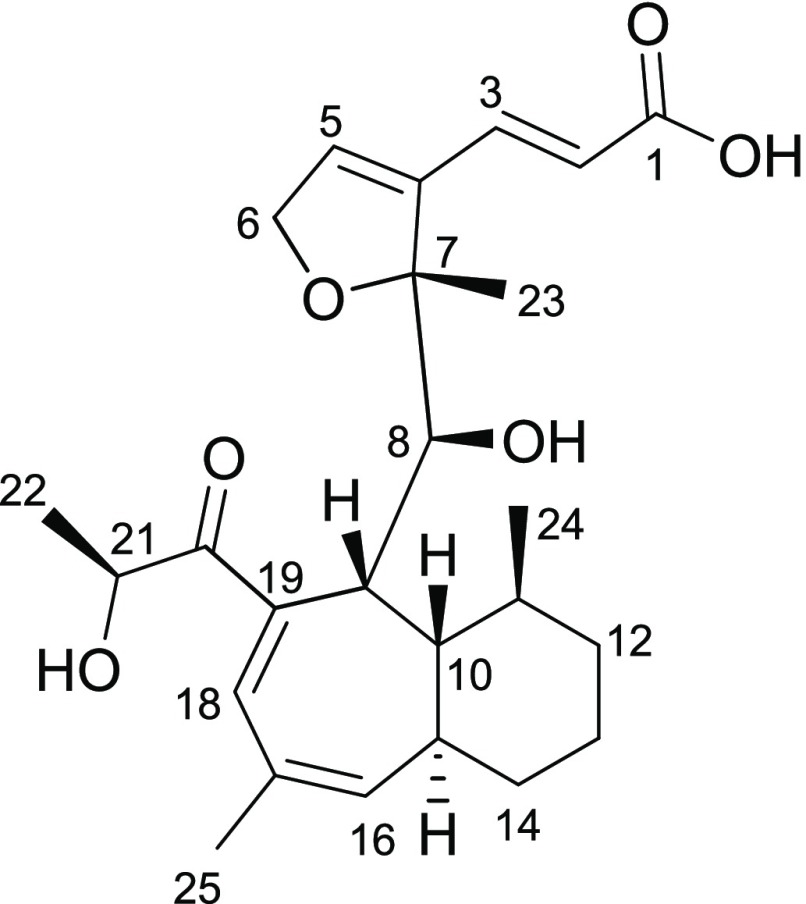
Wheldone (1) was isolated and elucidated from a coculture of Aspergillus fischeri (NRRL 181) and Xylaria flabelliformis (G536), where secondary metabolite biosynthesis was stimulated by antagonism between these fungi. First observed via in situ analysis between these competing fungal cultures, the conditions were scaled to reproducibly generate 1, whose novel structure was elucidated by one- and two-dimensional NMR and mass spectrometry. Compound 1 displayed cytotoxic activity against breast, ovarian, and melanoma cancer cell lines.
Fungi have been explored for new compounds for nearly 100 years, and that has led to the discovery of unique chemical diversity possessing promising biological activities, ranging from antibiotic, to immunosuppressant, to cholesterol-lowering properties.1−5 In nature, fungi grow in competition for resources, and as such, they have evolved the ability to adapt to changes in their environment. One of the ways they stave off rival organisms is through the activation of biosynthetic gene clusters, thereby stocking their arsenal for chemical warfare.6−10 Under standard lab conditions, fungi have been shown to produce only a fraction of their potential secondary metabolites.11,12 As such, coculturing fungi, forcing them to compete for limited resources, may present a pragmatic strategy to stimulate the biosynthesis of novel chemical diversity.11,13−16
To test this, fungi with antagonistic properties were chosen to participate in coculture experiments. The draft genome for Xylaria flabelliformis (strain G536; previously named Xylaria cubensis) was reported recently,17 and this strain biosynthesizes griseofulvin, which is an FDA-approved fungistatic compound that is known to interact with a broad range of fungi.18−20 Fungistatic denotes that it inhibits fungal growth, rather than killing competing fungi.21 We hypothesized that griseofulvin (and cobiosynthesized analogues) would impart stress on the competing fungal culture, especially because we observed that X. flabelliformis exudes these compounds into its surroundings.18,22
Aspergillus fischeri (strain NRRL 181) was chosen as the challenger due to its genetic tractability23 and the biosynthesis of metabolite weaponry in the form of mycotoxins.22−25 Indeed, bioinformatic analysis of the genomes of both organisms predicted the presence of as many as 48 biosynthetic gene clusters for A. fischeri(23) and 86 biosynthetic gene clusters for X. flabelliformis,17 yet only a relatively narrow range of secondary metabolites has been reported from either fungus. Our hypothesis was that the stress caused by the chemical warfare between these organisms would activate “silent” biosynthetic gene clusters and generate unprecedented chemical diversity.13,26−29
In a previous study, we reported the biosynthesis of several compounds that were found only in the coculture, including one putative new structure.22 As reported herein, the isolation and characterization of a secondary metabolite (1) (Figure 1) with a novel chemical scaffold supported our postulate that coculturing could generate new chemical diversity. This experiment was repeated several times in Petri dishes and in Erlenmeyer flasks (i.e., scaled up five times), demonstrating both a reproducible and scalable way to generate new fungal metabolites.
Figure 1.
Wheldone (1) was isolated from the coculture of Aspergillus fischeri and Xylaria flabelliformis.
To initiate this experiment, monocultures of X. flabelliformis and A. fischeri were examined first in situ (Petri plates) by the droplet probe30 to generate baseline profiles of the secondary metabolites. X. flabelliformis concentrates its fungistatic metabolites toward the colony edge (i.e., the youngest part of the fungal culture).18 Alternatively, A. fischeri had an even distribution of secondary metabolites across its mycelium (i.e., the colony edge and the colony center had similar metabolites and relative abundances).22
Next, cocultures of X. flabelliformis and A. fischeri were examined by droplet probe once a clear “junction” was formed (Figure 2 and larger version in Figure S8), which is the dividing area that is essentially the “battlefield” between the two competing fungi. The profile of compounds in the X. flabelliformis side of the coculture was interesting, as secondary metabolites were primarily observed in the junction. In contrast, A. fischeri was able to upregulate the biosynthesis of mycotoxins, suggesting that it was responding to the fungistatic properties imparted by the other fungus.22
Figure 2.
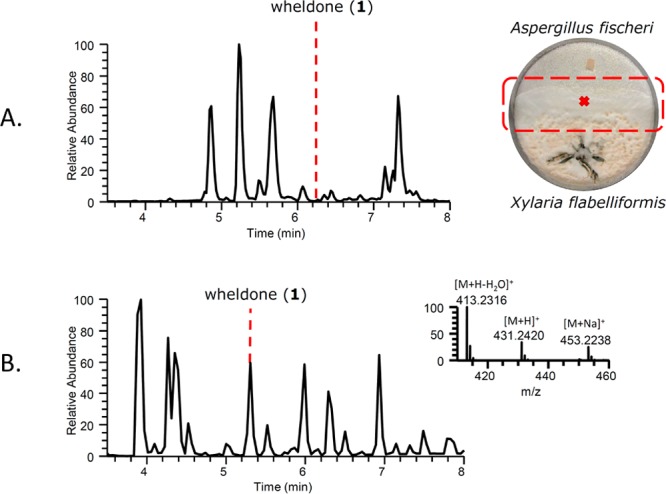
(A) Wheldone (1) was first noted as a minor component in the base peak chromatogram during in situ analysis22 of the junction that developed between A. fischeri and X. flabelliformis (shown in the box in the coculture Petri dish at the right). (B) Base peak chromatogram of the scaled-up coculture experiment (250 mL Erlenmeyer flasks), with the inset showing the mass spectrum of 1 and its adducts. Note that the chromatographic conditions were different between panels A (in situ analysis) and B (UPLC–MS), which is why the retention time of 1 varies.
There were several known metabolites identified in the junction that were not observed in the monoculture, as reported recently.23,25,26 However, there was one minor peak that did not match with any metabolites in an in-house database of over 525 fungal metabolites,31 that could not be characterized via mass defect filtering,32 and whose molecular formula and spectroscopic data did not correlate to any organic compounds in the literature. Thus this metabolite was targeted for isolation and characterization. Collectively, these data suggested that biosynthetic gene clusters, which were previously silent, could be activated via coculturing experiments to generate new chemical diversity.
To isolate and characterize the compound observed in situ in the coculture experiments, solid-phase cocultures of A. fischeri and X. flabelliformis were grown on oatmeal. (See the Supporting Information.) The organic extract (CHCl3–MeOH (1:1)) of the fermentation product underwent purification using normal-phase flash chromatography to afford six fractions. Upon further purification via C18 preparative HPLC, fraction 2 yielded compound 1 (5.48 mg) (Figure 1). The purity (99%) of 1 was assessed via UPLC–MS (Figure S1). This process was repeated five times to isolate larger quantities of 1 (>15 mg), showing the reproducibility and scalability of coculturing experiments.
Compound 1 was obtained as a white amorphous powder with a molecular formula of C25H34O6, as determined via HRESIMS along with 1H, 13C, and edited-HSQC NMR data (Table 1 and Figures S2 and S3), demonstrating an index of hydrogen deficiency of 9. The 13C NMR data (Table 1) indicated the presence of 25 carbons, inclusive of 2 carbonyl, 8 vinylic, 4 oxygenated, and 11 aliphatic carbons. The 1H and edited-HSQC NMR data (Table 1) indicated four methyls, five olefinic protons, four methines, and three methylenes. The HMBC correlations from H-3 to C-1 and C-2 and from H-2 to C-1 as well as the COSY cross correlations between H-3 and H-2 indicated a trans (JH-3/H-2 = 15.97 Hz) α,β-unsaturated carboxylic acid (Figures S4 and S5). The COSY correlation between H-5 and H2-6, the HMBC correlations from H2-6 to C-5, H-5 to C-7 and C-4, and H3-23 to C-7 and C-5, along with the oxygenated carbons at C-6 (δC 74.8) and C-7 (δC 95.2) established the methylated 2,5-dihydrofuran ring. HMBC correlations from H-2 to C-4 and H-3 to C-5 and C-4 formed the connection between the furan ring and the α,β-unsaturated carboxylic acid.
Table 1. 1H (700 MHz), 13C (175 MHz), and HMBC NMR Data for 1 in CD3OD.
| pos | δC, type | δH (J, Hz) | HMBC (H → C) |
|---|---|---|---|
| 1 | 172.2, C | ||
| 2 | 122.6, CH | 5.71 (d, 15.97) | 4, 1 |
| 3 | 137.4, CH | 7.40 (d, 16.00) | 6, 2, 4, 5, 1 |
| 4 | 137.1, C | ||
| 5 | 142.8, CH | 6.46 (t, 1.96) | 6, 7, 3, 4 |
| 6 | 74.8, CH2 | 4.75 (dd, 12.01, 1.30) | 4, 5 |
| 4.81 (dd, 12.01, 1.03) | |||
| 7 | 95.2, C | ||
| 8 | 76.1, CH | 3.54 (d, 10.41) | 23, 9, 7, 19 |
| 9 | 43.0, CH | 3.84 (dd, 10.45, 2.12) | 15, 10, 8, 7, 17, 19 |
| 10 | 44.0, CH | 1.96 (dt, 10.78, 3.49) | 16 |
| 11 | 31.6, CH | 1.19 (m) | |
| 12 | 36.4, CH2 | 0.94 (dq, 12.82, 2.63) | |
| 1.56 (m) | |||
| 13 | 23.5, CH2 | 1.23 (m) | |
| 1.48 (m) | |||
| 14 | 32.8, CH2 | 1.50 (m) | |
| 1.74 (m) | |||
| 15 | 34.6, CH | 2.88 (br s) | |
| 16 | 142.9, CH | 5.81 (br s) | 25, 14, 10, 19 |
| 17 | 133.6, C | ||
| 18 | 121.9, CH | 6.57 (s) | 9, 8, 17, 19, 20 |
| 19 | 157.3, C | ||
| 20 | 208.7, C | ||
| 21 | 74.2, CH | 4.41 (q, 7.04) | 22, 20 |
| 22 | 20.8, CH3 | 1.35 (d, 7.04) | 21, 20 |
| 23 | 19.3, CH3 | 1.42 (s) | 8, 7, 5 |
| 24 | 20.5, CH3 | 0.78 (d, 6.47) | 11, 12, 10 |
| 25 | 20.0, CH3 | 1.91 (d, 1.11) | 17, 16, 19 |
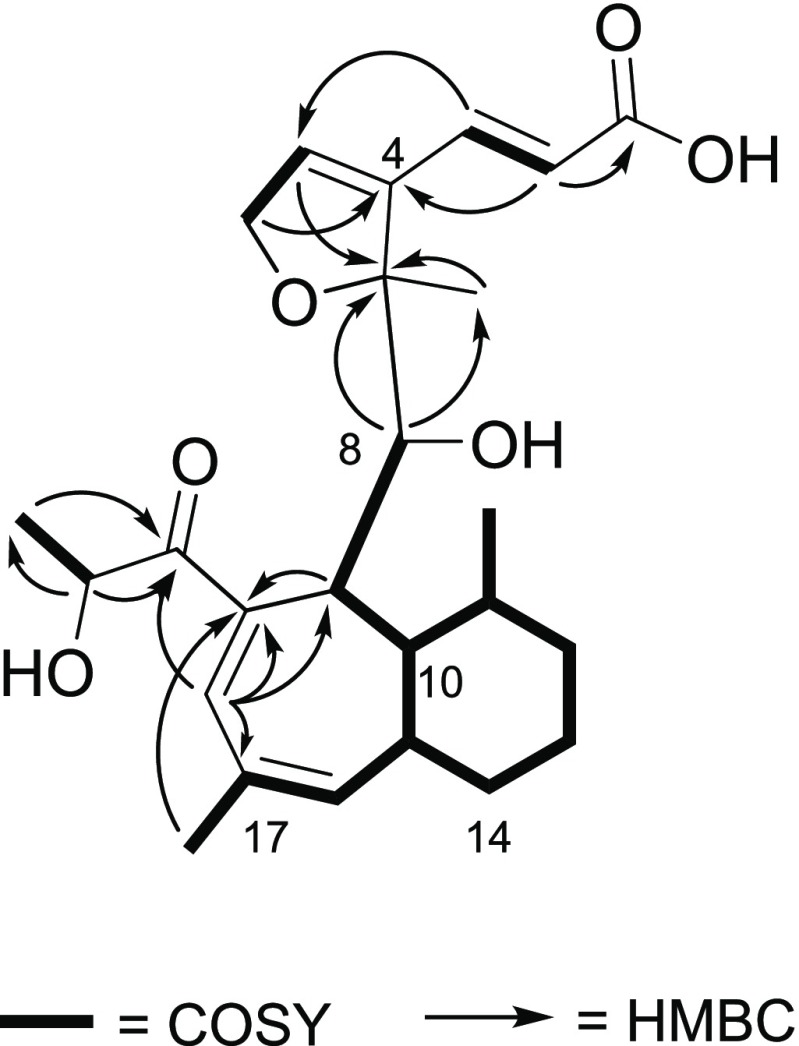
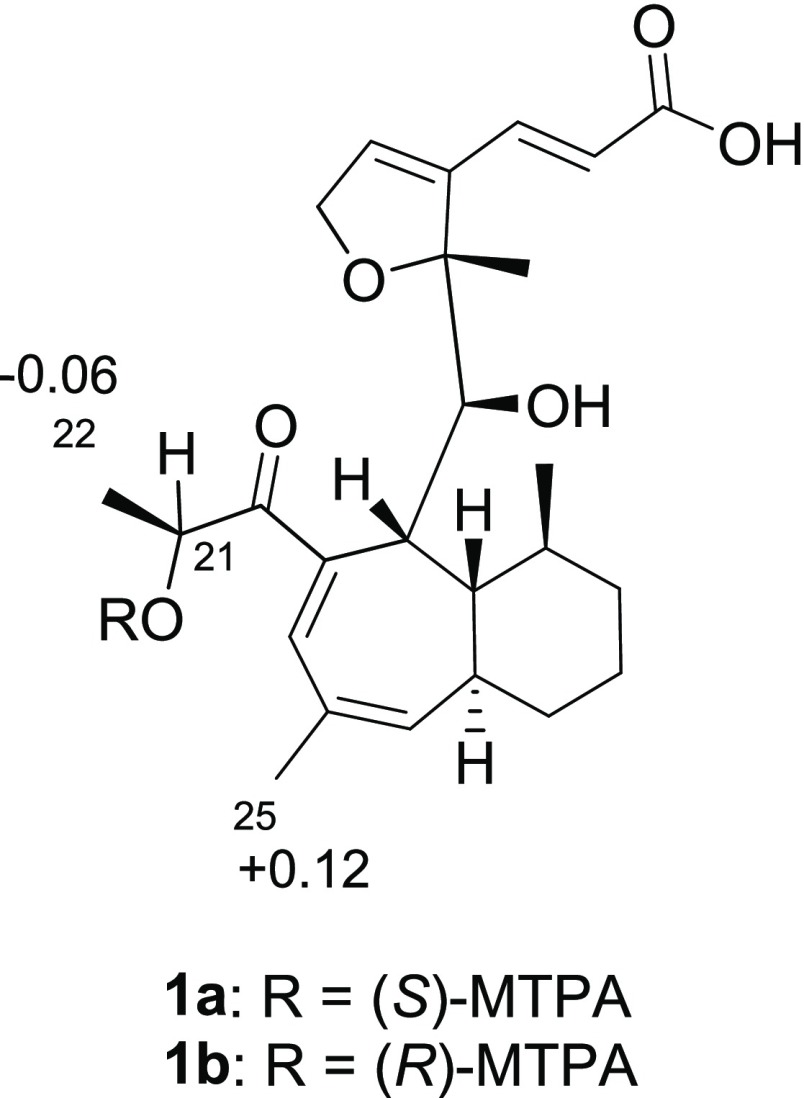
The COSY NMR spectrum of 1 displayed an 11-proton spin system (H-8/H-9/H-10/H-11/H3-24/H2-12/H213/H2-14/H-15/H-16/H3-25), which served to frame the bicyclic system (Figure 3). The seven membered-ring was discerned via HMBC correlations from H3-25 to C-19, H-9 to C-19 and C-17, and H-18 to C-9, C-17, and C-19. The α-hydroxy-1-propanone side chain was elucidated through the COSY correlations of H3-22 (δH/δC 1.35/20.8) and H-21 (δH/δC 4.41/74.2) and the HMBC correlations to C-20 from both H3-22 and H-21; this side chain was connected to the seven-member ring via an HMBC correlation between H-18 and C-20. The bicyclic system was connected to the furan ring system via HMBC correlations from H-8 to C-7 and C-23 and H3-23 to C-8. The absolute configuration of 1 was assigned via the Mosher ester method33 and NOESY correlations (Figures S6 and S7), establishing the configuration as (7R,8S,9R,10R,11S,15R,21S) (Figure 4).
Figure 3.
Key COSY and HMBC correlations for 1.
Figure 4.
ΔδH values (Δδ = δS – δR) obtained for (S)- and (R)-MTPA esters of wheldone (1) (1a and 1b, respectively) in pyridine-d5.
Compound 1 was tested against a panel of tumor cell lines (Table 2), MDA-MB-231 (triple-negative human breast cancer), OVCAR-3 (human ovarian cancer), and MDA-MB-435 (human melanoma cancer), using methods described previously.34 (See the Cytotoxicity Assay section in the Supporting Information.) Although taxol was more potent, these data demonstrated that the cell lines responded to both compounds in the same rank (i.e., taxol and 1 display the highest and lowest activities in the same cell lines), with the highest response seen in MDA-MB-435, followed by OVCAR3 and then MDA-MB-231.
Table 2. Activity of 1 against Three Tumor Cell Lines.
| IC50 (μM)a |
|||
|---|---|---|---|
| compd | MDA-MB-231 | OVCAR-3 | MDA-MB-435 |
| 1 | 7.6 | 3.8 | 2.4 |
| taxol | 0.17 | 0.0051 | 0.00043 |
IC50 values were determined as the concentration required to inhibit growth to 50% of control with a 72 h incubation.
Using the coculturing of A. fischeri and X. flabelliformis, one novel compound (1) with cytotoxic activity was isolated and characterized. Our strategy employed the droplet probe30 to first pilot the coculture conditions in a Petri dish, essentially scouting for changes in the secondary metabolite profile at the intersection of the fungal cultures.22 Then, the cocultures were scaled to reproducibly generate 1 on the milligram scale, biosynthesizing enough material for further chemical and biological evaluation. Importantly, the scaled growths imparted a much higher concentration of 1 as compared with the Petri plates, likely because the antagonistic fungi were in close contact throughout the Erlenmeyer flask, as opposed to when they grow into each other in a Petri dish, which gives a visual indication of the battlefield (Figure 2 and Figure S8) but is likely less efficient than constant interaction.
During the peer review of this manuscript, we started coculturing X. flabelliformis with another ascomycete fungus (strain MSX79272). Natural product studies on strain MSX79272 will be reported in more detail in the future. However, on the basis of DNA barcoding,35 we know that this strain belongs to the order Hypocreales; Aspergillus spp. are in the order Eurotiales. We were encouraged when we observed a peak in the extract from a coculture experiment of these fungi that aligned with the retention time and HRMS data for 1 (Figure S9). The peak was isolated via HPLC, and a comparison between the 1H NMR data for 1 from both coculture experiments showed that they were in concordance (Figure S9). Because 1 was generated when X. flabelliformis was used in coculture experiments with two different fungal strains, we hypothesize biosynthesis by this organism. Of the limited fungal–fungal coculture experiments in the literature (∼40),36 this is the first example of using an alternate fungus to narrow down the biosynthetic source for the new chemical entity. Given that the genomes of both fungal strains used in this study have been sequenced and putative biosynthetic gene clusters have been predicted,17,23 future studies will take advantage of the development of improved heterologous gene expression platforms for the targeted production of fungal secondary metabolites37 and to identify the biosynthetic gene cluster responsible for the biosynthesis of wheldone (1).
Acknowledgments
This Letter is dedicated to the late Daniel (Dan) Clive Wheldon (06/22/1978–10/16/2011), who was a two-time winner of the Indianapolis 500 (2005 and 2011). This research was supported in part by the National Institutes of Health via the National Cancer Institute (P01 CA125066) and the National Center for Complementary and Integrative Health (T32 AT008938 and F31 AT010558). This work was performed in part at the Joint School of Nanoscience and Nanoengineering, a member of the Southeastern Nanotechnology Infrastructure Corridor (SENIC) and National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (grant ECCS-1542174).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.orglett.0c00219.
Experimental procedures, HRESIMS, UPLC chromatograms, 1D and 2D NMR spectra, and key NOESY correlations of compound 1 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Bills G. F.; Gloer J. B. Biologically Active Secondary Metabolites from the Fungi. Microbiol. Spectrum 2016, 4, 1–32. 10.1128/microbiolspec.FUNK-0009-2016. [DOI] [PubMed] [Google Scholar]
- El-Elimat T.; Raja H. A.; Ayers S.; Kurina S. J.; Burdette J. E.; Mattes Z.; Sabatelle R.; Bacon J. W.; Colby A. H.; Grinstaff M. W.; Pearce C. J.; Oberlies N. H. Meroterpenoids from Neosetophoma sp.: A Dioxa[4.3.3]propellane Ring System, Potent Cytotoxicity, and Prolific Expression. Org. Lett. 2019, 21, 529–534. 10.1021/acs.orglett.8b03769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-S.; Ding Y.; Yang B.-J.; Miklossy G.; Yin H.-Q.; Walker L. A.; Turkson J.; Cao S. A new metabolite with a unique 4-Pyranone– γ-Lactam–1, 4-Thiazine moiety from a hawaiian-plant associated fungus. Org. Lett. 2015, 17, 3556–3559. 10.1021/acs.orglett.5b01650. [DOI] [PubMed] [Google Scholar]
- Vinale F.; Sivasithamparam K.; Ghisalberti E.; Marra R.; Barbetti M.; Li H.; Woo S.; Lorito M. A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 2008, 72, 80–86. 10.1016/j.pmpp.2008.05.005. [DOI] [Google Scholar]
- Li K.; Wang Y.-F.; Li X.-M.; Wang W.-J.; Ai X.-Z.; Li X.; Yang S.-Q.; Gloer J. B.; Wang B.-G.; Xu T. Isolation, Synthesis, and Radical-Scavenging Activity of Rhodomelin A, a Ureidobromophenol from the Marine Red Alga Rhodomela confervoides. Org. Lett. 2018, 20, 417–420. 10.1021/acs.orglett.7b03716. [DOI] [PubMed] [Google Scholar]
- Chatterjee S.; Kuang Y.; Splivallo R.; Chatterjee P.; Karlovsky P. Interactions among filamentous fungi Aspergillus niger, Fusarium verticillioides and Clonostachys rosea: fungal biomass, diversity of secreted metabolites and fumonisin production. BMC Microbiol. 2016, 16, 83. 10.1186/s12866-016-0698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver J. H.; Slot J. C.; Rokas A. The evolution of fungal metabolic pathways. PLoS Genet. 2014, 10, e1004816 10.1371/journal.pgen.1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada L.; Ajayi O.; Frisvad J. C.; Yu J.; Nierman W. C. Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med. Mycol. 2009, 47, S88–S96. 10.1080/13693780802409542. [DOI] [PubMed] [Google Scholar]
- Keller N. P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167. 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A.; Mead M. E.; Steenwyk J. L.; Raja H. A.; Oberlies N. H. Biosynthetic Gene Clusters and the Evolution of Fungal Chemodiversity. Nat. Prod. Rep. 2020, 10.1039/C9NP00045C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherlach K.; Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- Nützmann H.-W.; Schroeckh V.; Brakhage A. A. Regulatory Cross Talk And Microbial Induction Of Fungal Secondary Metabolite Gene Clusters. Methods Enzymol. 2012, 517, 325–341. 10.1016/B978-0-12-404634-4.00016-4. [DOI] [PubMed] [Google Scholar]
- Stierle A. A.; Stierle D. B.; Decato D.; Priestley N. D.; Alverson J. B.; Hoody J.; McGrath K.; Klepacki D. The berkeleylactones, antibiotic macrolides from fungal coculture. J. Nat. Prod. 2017, 80, 1150–1160. 10.1021/acs.jnatprod.7b00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.-Y.; Shen X.-T.; Yuan X.-J.; Zhou Y.-M.; Fan H.; Zhu L.-P.; Du F.-Y.; Sadilek M.; Yang J.; Qiao B.; et al. Metabolomics investigation of an association of induced features and corresponding fungus during the co-culture of Trametes versicolor and Ganoderma applanatum. Front. Microbiol. 2018, 8, 2647. 10.3389/fmicb.2017.02647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnani N.; Chevrette M. G.; Adibhatla S. N.; Zhang F.; Yu Q.; Braun D. R.; Nelson J.; Simpkins S. W.; McDonald B. R.; Myers C. L. Coculture of marine invertebrate-associated Bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem. Biol. 2017, 12, 3093–3102. 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S.; Bohni N.; Schnee S.; Schumpp O.; Gindro K.; Wolfender J.-L. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Mead M. E.; Raja H. A.; Steenwyk J. L.; Knowles S. L.; Oberlies N. H.; Rokas A. Draft Genome Sequence of the Griseofulvin-Producing Fungus Xylaria flabelliformis Strain G536. Microbiol. Resour. Announc. 2019, 8, e00890. 10.1128/MRA.00890-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica V. P.; Rees E. R.; Tchegnon E.; Bardsley R. H.; Raja H. A.; Oberlies N. H. Spatial and temporal profiling of griseofulvin production in Xylaria cubensis using mass spectrometry mapping. Front. Microbiol. 2016, 7, 544. 10.3389/fmicb.2016.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraballo-Rodríguez A. M.; Mayor C. A.; Chagas F. O.; Pupo M. T. Amphotericin B as an inducer of griseofulvin-containing guttate in the endophytic fungus Xylaria cubensis FLe9. Chemoecology 2017, 27, 177–185. 10.1007/s00049-017-0243-3. [DOI] [Google Scholar]
- Paguigan N. D.; Al-Huniti M. H.; Raja H. A.; Czarnecki A.; Burdette J. E.; González-Medina M.; Medina-Franco J. L.; Polyak S. J.; Pearce C. J.; Croatt M. P.; Oberlies N. H. Chemoselective fluorination and chemoinformatic analysis of griseofulvin: Natural vs fluorinated fungal metabolites. Bioorg. Med. Chem. 2017, 25, 5238–5246. 10.1016/j.bmc.2017.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill J.; Burgess D.; Hardin T. Key issues concerning fungistatic versus fungicidal drugs. Eur. J. Clin. Microbiol. Infect. Dis. 1997, 16, 42–50. 10.1007/BF01575120. [DOI] [PubMed] [Google Scholar]
- Knowles S. L.; Raja H. A.; Wright A. J.; Lee A. M. L.; Caesar L. K.; Cech N. B.; Mead M. E.; Steenwyk J. L.; Ries L.; Goldman G. H.; Rokas A.; Oberlies N. H. Mapping the Fungal Battlefield: Using in situ Chemistry and Deletion Mutants to Monitor Interspecific Chemical Interactions Between Fungi. Front. Microbiol. 2019, 10, 285. 10.3389/fmicb.2019.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead M. E.; Knowles S. L.; Raja H. A.; Beattie S. R.; Kowalski C. H.; Steenwyk J. L.; Silva L. P.; Chiaratto J.; Ries L. N.; Goldman G. H.; et al. Characterizing the pathogenic, genomic, and chemical traits of Aspergillus fischeri, a close relative of the major human fungal pathogen Aspergillus fumigatus. mSphere 2019, 4, e00018–19. 10.1128/mSphere.00018-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverberi M.; Ricelli A.; Zjalic S.; Fabbri A. A.; Fanelli C. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 2010, 87, 899–911. 10.1007/s00253-010-2657-5. [DOI] [PubMed] [Google Scholar]
- Ehrlich K. C.; Chang P.-K.; Bhatnagar D. Mycotoxins. eLS 2011, 1–14. 10.1002/9780470015902.a0000373.pub2. [DOI] [Google Scholar]
- Huang S.; Ding W.; Li C.; Cox D. G. Two new cyclopeptides from the co-culture broth of two marine mangrove fungi and their antifungal activity. Pharmacogn. Mag. 2014, 10, 410–414. 10.4103/0973-1296.141781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhar F.; Castiglia V.; Levin L. Enhancement of laccase production and malachite green decolorization by co-culturing Ganoderma lucidum and Trametes versicolor in solid-state fermentation. Int. Biodeterior. Biodegrad. 2015, 104, 238–243. 10.1016/j.ibiod.2015.06.017. [DOI] [Google Scholar]
- Kettering M.; Sterner O.; Anke T. Antibiotics in the Chemical Communication of Fungi. Z. Naturforsch., C: J. Biosci. 2004, 59, 816–823. 10.1515/znc-2004-11-1209. [DOI] [PubMed] [Google Scholar]
- Schroeckh V.; Scherlach K.; Nützmann H.-W.; Shelest E.; Schmidt-Heck W.; Schuemann J.; Martin K.; Hertweck C.; Brakhage A. A. Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 14558–14563. 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlies N. H.; Knowles S. L.; Amrine C. S. M.; Kao D.; Kertesz V.; Raja H. A. Droplet probe: coupling chromatography to the in situ evaluation of the chemistry of nature. Nat. Prod. Rep. 2019, 36, 944–959. 10.1039/C9NP00019D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T.; Figueroa M.; Ehrmann B. M.; Cech N. B.; Pearce C. J.; Oberlies N. H. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 2013, 76, 1709–1716. 10.1021/np4004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paguigan N. D.; El-Elimat T.; Kao D.; Raja H. A.; Pearce C. J.; Oberlies N. H. Enhanced dereplication of fungal cultures via use of mass defect filtering. J. Antibiot. 2017, 70, 553. 10.1038/ja.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoye T. R.; Jeffrey C. S.; Shao F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nat. Protoc. 2007, 2, 2451. 10.1038/nprot.2007.354. [DOI] [PubMed] [Google Scholar]
- Chen W.-L.; Ren Y.; Ren J.; Erxleben C.; Johnson M. E.; Gentile S.; Kinghorn A. D.; Swanson S. M.; Burdette J. E. (+)-Strebloside-induced cytotoxicity in ovarian cancer cells is mediated through cardiac glycoside signaling networks. J. Nat. Prod. 2017, 80, 659–669. 10.1021/acs.jnatprod.6b01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja H. A.; Miller A. N.; Pearce C. J.; Oberlies N. H. Fungal identification using molecular tools: a primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. 10.1021/acs.jnatprod.6b01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora D.; Gupta P.; Jaglan S.; Roullier C.; Grovel O.; Bertrand S. Expanding the chemical diversity through microorganisms co-culture: Current status and outlook. Biotechnol. Adv. 2020, 107521. 10.1016/j.biotechadv.2020.107521. [DOI] [PubMed] [Google Scholar]
- Greco C.; Keller N. P.; Rokas A. Unearthing Fungal Chemodiversity and Prospects for Drug Discovery. Curr. Opin. Microbiol. 2019, 51, 22–29. 10.1016/j.mib.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.