Summary
Regeneration involves regulating tissue proportionality across considerable size ranges through unknown mechanisms. In planarians, which scale reversibly over 40x through regeneration, we identify the Striatin-interacting phosphatase and kinase (STRIPAK) complex as a potent negative regulator of axis length. Inhibition of two proteins in the STRIPAK complex, mob-4 and striatin, dramatically increased posterior length, through expansion of a posterior wnt1+ signaling center within midline muscle cells. wnt1 was required for tail expansion after mob4 inhibition and dynamically reestablishes proportionality after amputation in normal animals, indicating STRIPAK represses Wnt signaling for scaling. Regulation of wnt1 expansion was stem-cell dependent, demonstrating that control of signaling center production through stem cell differentiation underlies proportional growth in adult regenerative tissue.
One Sentence Summary:
Organ size in regeneration is controlled by STRIPAK-mediated differentiation of WNT producing cells.
Introduction
Proportional tissue scaling is fundamental to organismal form. Within species, the relative sizes of appendages or organs scale in stereotyped ways with respect to body size, for example arm span versus height in humans. Across species, modifications to organ proportionality can underlie morphological evolution [1]. Morphogens that enable communication across cell populations have been implicated in both size attainment and proportional scaling through growth. Examples include the Dpp gradient within Drosophila imaginal discs, the Bicoid gradient within the Drosophila syncytial blastoderm, or the dorsoventral BMP gradient of Xenopus and Zebrafish embryos, all of which function over the range of ~10s to 100s of microns [2–4]. However, it is unclear what mechanisms ensure proportional growth across the much larger scale of adult growth, which can occur from millimeters to meters. Organisms capable of adult regeneration offer an intriguing view into this phenomenon, because the regeneration of new tissue involves restoration of relative tissue proportions without transit through embryogenesis and also across these larger length scales.
Freshwater planarians are well known for their ability to regenerate after essentially any injury using neoblast adult pluripotent stem cells [5], but they also undergo robust regulation of proportional growth (Figure 1A). The planarian Schmidtea mediterranea grows after feeding and “degrows” after starvation by scaling reversibly over a 40x range in body size from 0.5 to 2 centimeters in length, largely by regulating total cell numbers from ~1-20 million cells [6–8]. Regeneration itself in these animals involves the restoration of proportionality and not initially a restoration of absolute size. After severe body amputations, planarians regenerate missing tissues through a combination of blastema outgrowth to produce new tissue and also the re-scaling of pre-existing tissue regions, which together produce a smaller but well-proportioned animal over several weeks. The scaling of the planarian head-to-tail anteroposterior (AP) is easily accessible to measurement and manipulation for surgery, and regenerated planarians of different sizes attain stereotyped proportions of body features with respect to size (Figure 1B). This biology, combined with genome resources and tools for molecular and functional analysis has made planarians an emerging model for studying the biology of adult tissue growth and proportionality at the millimeter-to-centimeter scale.
Figure 1. mob4 regulates the ability of planarians to proportionally scale body regions.
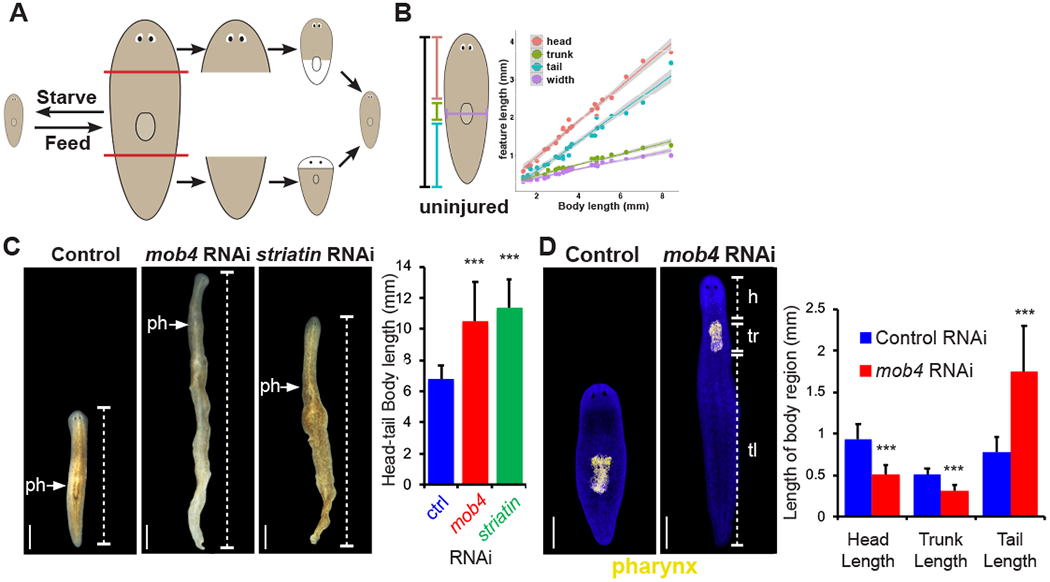
(A) Planarians scale tissue across a 40x size range during homeostasis, and during regeneration rescale existing tissue with newly produced tissue.
(B) The length of different regions of the body (head, trunk, tail, and width) scale proportionally relative to overall body length during homeostasis. Trendlines are linear regressions with 95% confidence intervals calculated in R.
(C) Live images showing long term homeostatic mob4(RNAi) and striatin(RNAi) animals have disproportionate body tissue, and an anteriorly placed pharynx (arrows) Brackets indicate tail (tl). Graph indicates total length of animals of each RNAi condition. N=>10 animals, error bars are standard deviations and asterisks, p<0.0005 by 2-tailed t-test.
(D) Fluorescent in situ hybridization (FISH) of pharynx progenitor ASXL_059179 to compare the anterior-posterior position of the pharynx of mob4(RNAi) animals to controls. Brackets indicate tail (tl), trunk (tr) length, and head (h) length. Right- graph shows average head, trunk, and tail length. N=15 animals, error bars are standard deviations and asterisks, p<0.0005 by 2-tailed t-test. In panels A, B, C and D, bars are 300 microns.
See also Figure S1.
Planarians use a system of nested posterior Wnts and anterior Wnt inhibitors to control tissue identity along the anteroposterior (AP) axis running from head to tail, which in principle could also regulate AP axis proportionality. Wnt expression, transcriptional interdependence, and functional hierarchy suggest a particular importance of signaling from the axis termini [9–13]. notum and wnt1 are expressed at the anterior and posterior poles and their expression is induced early near amputation sites to control a head-versus-tail decision of the outgrowing blastema through control of canonical Wnt signaling [9–12]. Inhibition of canonical Wnt signaling results in ectopic head regeneration and over-activation results in ectopic tail regeneration [9–18]. However, the distinct contributions of wnt1 to regeneration through its expression at the posterior pole versus near injury sites, if any, have not yet been resolved. Two other constitutive signaling systems use alternate Wnts and associated factors in order to restrict anterior regionalization along the AP axis: inhibition of wnt11-6/wntA, fzd5/8-4, or NDK results in posterior expansion of brain and posterior duplication of eyes, notum or prep inhibition results in anterior duplication of eyes, and wntP-2/wnt11-5, fzd1/2/7, ptk7 or ndl-3 inhibition results in posterior duplication of the pharynx [19–21]. Although inhibition of Wnts and associated factors modify AP pattern, it is unclear whether the same positional information system underlies proportional axis scaling in general. If so, tissue proportionality could be the result of regulation at the levels of production, propagation, or reception of regionalized signals.
Results
To resolve possible models for planarian proportional scaling, we used RNAi to seek specific phenotypes of aberrant relative body size. We found that long term inhibition (34 days) of a kinase activator, mob4 (phocein), caused significant changes to proportionality of the planarian body plan resulting in a doubling of the size of the head-to-tail axis, and we verified RNAi knockdown of mob4 transcript using qPCR (Figures 1C, S1A–D). MOB4 factors are a conserved subfamily of Mob-domain kinase activators that act within the striatin-interacting phosphatase and kinase complex (STRIPAK) to regulate a variety of targets and cellular behaviors [22–24] (Figure S1E). By contrast, other distinct Mob-domain containing subfamilies include Mob1 factors Mst1/2 that activate NDR/LATS kinases. To determine if mob4 likely functioned within a planarian STRIPAK complex, we inhibited striatin, the core scaffolding protein of the STRIPAK complex [25]. After 34 days of inhibition, striatin(RNAi) animals displayed changes to body size similar to mob4(RNAi) (Figure 1C). In addition to overall changes in body size, mob4(RNAi) animals had a visibly altered anterior-posterior axis. Staining for pharynx markers revealed that mob4(RNAi) animals underwent a dramatic enlargement of tail tissue, reduction of head and trunk (defined as the length of the region containing the pharynx) regions, and an increased length: width ratio (Figures 1D, S1D, F). Compared with controls, mob4(RNAi) animals had reduced numbers of cintillo+ brain neurons that typically scale to body size [7, 8, 26], indicating a decrease in numbers of anterior cells (Figure S1G). These results suggest that mob4 could be acting to exclusively restrict posterior tissue, or alternatively could both restrict posterior tissues and promote anterior tissue. To test these possibilities, we compared total cintillo+ cell number in untreated animals at day 0 versus animals fed either control dsRNA or mob4 dsRNA for 34 days (Figure S1H). We found that compared to untreated animals, control animals increased their cintillo+ cell number due to growth after the 34 days of feeding. However, mob4(RNAi) animals had no change in total cintillo+ cell numbers compared to untreated animals (Figure S1H). Therefore, tail enlargement in mob4(RNAi) is accompanied by a failure in anterior growth. Additionally, the ratio of head length to trunk length was unchanged in mob4(RNAi) compared to controls (Figure S1I), suggesting that relative proportionality between these tissues are unchanged with respect to each other when mob4 is inhibited. Finally, mob4(RNAi) animals had broadly normal composition of non-regionalized cell types such as neoblasts, epidermal progenitors, muscle, and intestine (Figure S2A). Together these findings indicate that mob4 specifically restricts relative and absolute posterior identity and is important for anterior growth.
Positional control genes (PCGs) are signaling molecules expressed in body-wide gradients within body-wall muscle cells that provide cues for body regionalization in planarians [27]. Given the role of mob4 in regulating body proportionality, we tested the hypothesis that mob4 inhibition would shift PCG gradients to reflect the new body proportions after mob4 inhibition. We examined posteriorly expressed Wnt signaling genes wnt1, wnt11-2, wnt11-1, fz4, and wntP-2 and found each domain shifted anteriorly with respect to body length (Figure 2A–C). By contrast, trunk-expressed ptk7 and anteriorly expressed ndl-5 and sfrp-1 were only slightly restricted compared to total body length, with no statistically significant changes to ndl-3 or ndk (Figure 2A–B). However, mob4 inhibition also had the effect of increasing both the absolute and relative size of the tail, so we further examined whether any posterior PCG expression domains expanded when normalized to tail length. Of these, wnt1 prominently increased ~10-fold in size into a domain extending beyond the tail, with other posterior PCGs showing smaller increases with respect to tail length (Figure 2C). Furthermore, because wnt1 is expressed on the dorsal midline, we examined for possible changes to PCGs marking the medial-lateral and dorsal-ventral axes, but these appeared broadly normal after mob4 inhibition (Figure S2B). Therefore, these analyses pointed to wnt1 as a primary candidate PCG strongly misregulated in mob4 RNAi that we hypothesized could mediate some aspects of this phenotype. Indeed, tail expansion phenotypes from inhibition of both mob4 or striatin each corresponded with a dramatic expansion of wnt1 along the midline (Figures 2A, S2C). wnt1 encodes the most posteriorly expressed Wnt factor and is required for posterior regeneration, but it is unknown whether it is sufficient to drive tail growth [9].
Figure 2. mob4 regulates the anteroposterior axis.
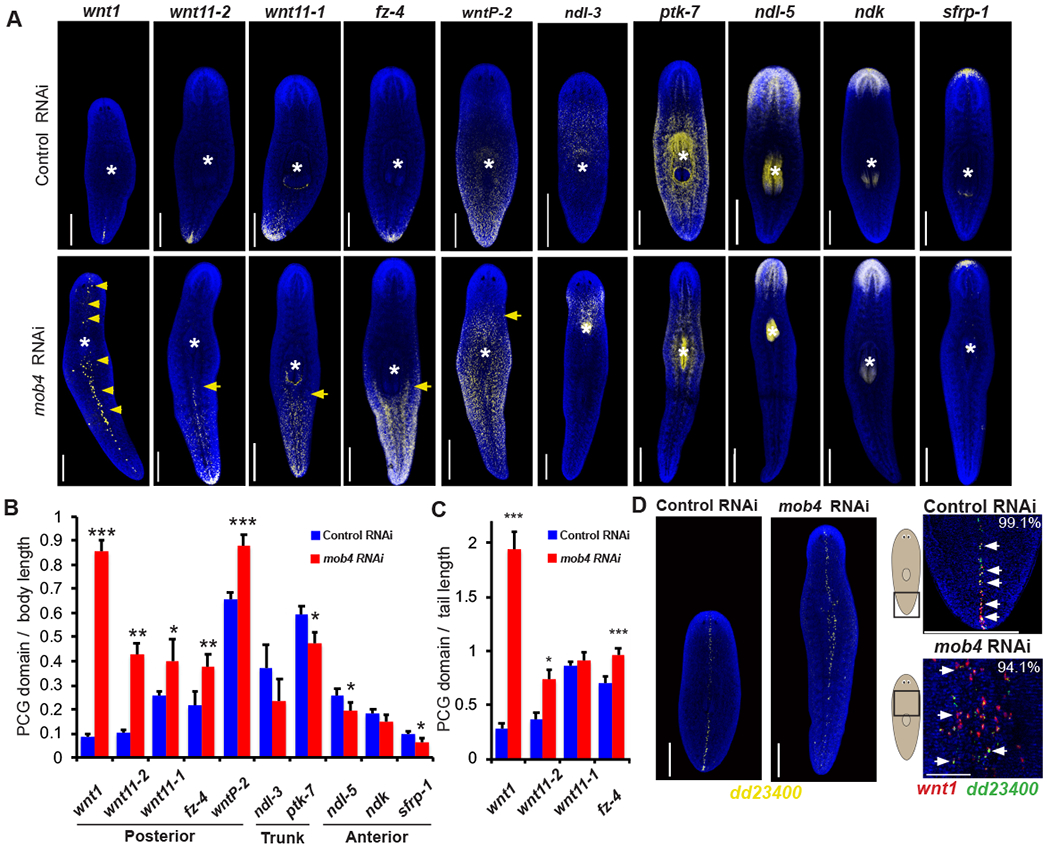
(A) Animals were fixed after 34 days of mob4 or control RNAi and stained by FISH as indicated for PCGs marking the anteroposterior axis. mob4 RNAi caused anterior shift to posterior markers wnt-1, wnt11-2, wnt11-1, fzd4, and wntP-2 and had proportionally slightly reduced expression of trunk markers ndl-3 and ptk7 and anterior markers sfrp-1, ndl-5, ndk. Yellow arrows indicate expanded expression of posterior markers in mob4 RNAi animals. White asterisks indicate the pharynx location. n>4 animals.
(B) Graph showing the length of indicated PCG domain divided by total length in mob4(RNAi) animals compared to controls. mob4 inhibition had the greatest effect on wnt1 expression. Error bars are standard deviations, ***=p<0.0005, **=p<0.005, and *=p<0.05 by 2-tailed t-test. n>4 animals.
(C) Graph showing the length of indicated PCG domain divided by tail length domain in mob4(RNAi) animals compared to controls. wnt1 is exceptional in the extent of ectopic expression present after mob4 inhibition. Error bars are standard deviations, ***=p<0.0005 and *=p<0.05 by 2-tailed t-test n=>4 animals.
(D) Left 2 images- FISH showing dd23400 expression along dorsal midline in controls and mob4(RNAi). Right- (Upper panel) Co-labeling of wnt1 (red) and dd23400 (green) showing wnt1 is exclusively expressed in a population of dorsal midline muscle cells at the posterior pole. (Lower panel) wnt1 expansion in mob4(RNAi) animals occurs in dd_23400+ cells. In panel A and B and left two images of D, bars are 300 microns. In the right two images of D, bars are 150 microns.
See also Figure S2.
We noted that ectopic wnt1 expression in mob4(RNAi) animals was primarily found within dorsal cells along the midline, similar to normal animals, so we sought to identify markers of putative midline cell types that could undergo regulation of wnt1 expression. Prior single-cell RNA-seq studies revealed a muscle cell type present along the dorsal midline marked by expression of dd_23400 [28] (Figure 2D). Using double-FISH, we found that wnt1 is exclusively expressed in this subpopulation of muscle cells in untreated animals (112/113 cells counted) and that ectopic wnt1+ cells in mob4(RNAi) animals also express dd_23400 (190/202) cells counted) (Figure 2D). We next investigated whether mob4(RNAi) animals have increased dd_23400+ cells. mob4(RNAi) animals had normal numbers of dd_23400+ cells in the head domain relative to head length as well as normal numbers of total dd_23400 relative to total body length (Figure S2D). We additionally assessed the expression of notum, a Wnt antagonist expressed in the anterior pole and required for regeneration of anterior tissue [11]. However, mob4 inhibition did not alter the expression of notum at the anterior pole, indicating mob4 likely does not limit wnt1 at the expense of notum+ tissue (Figure S2E). Consistent with this, notum+ pole cells did not express dd_23400. Together, these results suggest that mob4 and striatin specifically inhibit wnt1 expression within midline muscle cells outside of the posterior pole, normally limiting the domain of the wnt1+ midline posterior pole.
In principle, mob4 might directly restrict posterior identity and indirectly modify wnt1 expression, or alternatively mob4 could restrict wnt1 expression to in turn control tail regionalization. To distinguish these possibilities, we first asked whether wnt1 expression changes preceded changes to overall body proportionality after mob4 inhibition. During short-term inhibition of mob4 (three dsRNA feedings totaling 10 days or RNAi), wnt1 expression expansion preceded tail enlargement (Figure 3A). We further tested the interaction of the two genes using double RNAi experiments. After 3 weeks of inhibition, mob4(RNAi) animals had expanded tails, while simultaneous inhibition of wnt1 and mob4 suppressed the mob4 tail expansion phenotype (Figure 3B). We confirmed that mob4 expression was knocked down in mob4(RNAi);wnt1(RNAi) animals (Figure 3C). To determine the specificity of the mob4-wnt1 relationship, we tested whether inhibition of another posterior PCG, wntP-2, could suppress the mob4 RNAi phenotype. mob4(RNAi);wntP-2(RNAi) animals formed elongated tails similar to mob4(RNAi) animals, suggesting mob4 does not act through wntP-2 (Figure S3A). Using qPCR, we confirmed that mob4 expression was successfully knocked down in mob4(RNAi):wntP-2(RNAi) animals compared to controls (Figure S3B). Posterior Wnts have been reported to undergo cross-regulation for a variety of outputs in the posterior [10, 13, 29], but these results indicate that mob4 regulation of wnt1 expression is of particular importance to tail size. In addition, long-term homeostatic inhibition of wnt1 produced a phenotype effectively opposite to mob4 RNAi in which tail length was reduced (along with formation of a posterior pharynx), and did not coincide with ectopic posterior expression of trunk (ndl-3, ptk-7) or anterior (ndl-5, sfrp-1) markers. (Figure S3C, S3D). We conclude that wnt1 activity is required for mob4 control of body proportion, and wnt1 can act as a regulator of posterior size.
Figure 3. wnt1 mediates mob4 control of body proportionality.
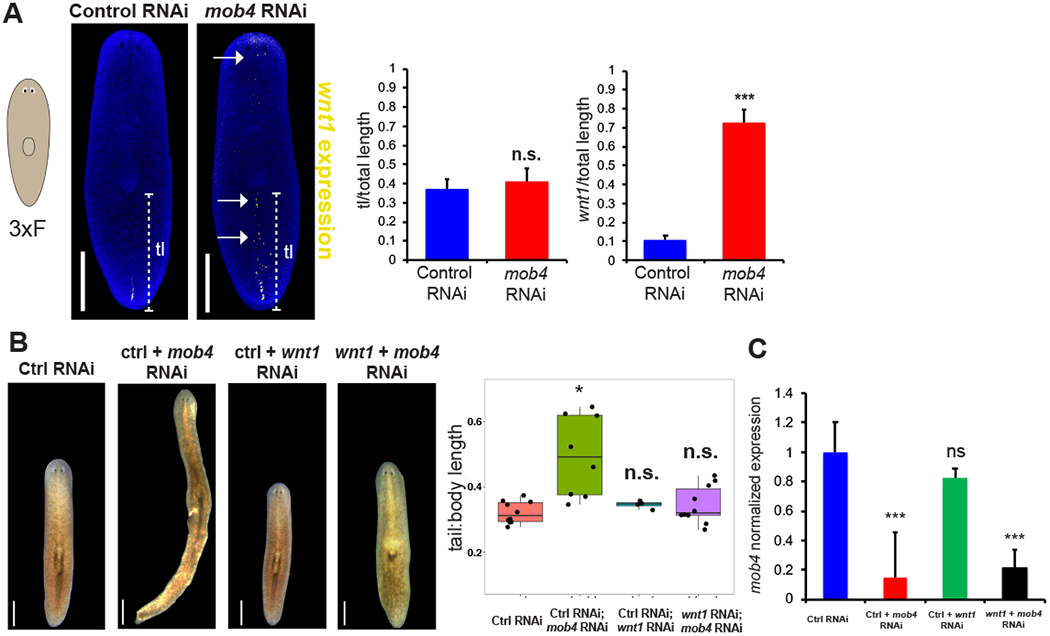
(A) FISH to detect wnt1 showing expanded expression in mob4(RNAi) animals (right image) compared to controls (left image) after only 3 dsRNA feedings totaling 10 days of RNAi. Middle-graph showing tail length divided by total body length. Right- graph showing length of wnt1 expression divided by total length. N=10 animals, error bars are standard deviations and ns= not significant, p>0.05 by 2-tailed t-test.
(B) Live images showing body morphology of double-RNAi experiments. Compared to controls, mob4(RNAi) animals show characteristic body patterning changes, while wnt1(RNAi) and mob4(RNAi);wnt1(RNAi) animals are unchanged. Right- Graph showing tail length of each RNAi condition. N=10 animals, error bars are standard deviations and asterisks, p<0.05 by 2-tailed t-test.
(C) mob4 mRNA expression measured by qPCR from RNA collected after 17 days of RNAi as indicated. Simultaneous treatment with wnt1 and mob4 dsRNA resulted in mob4 transcript reduction similar to mob4 dsRNA treatment alone. Error bars are standard deviations of 4 biological replicates. Asterisks denote p<0.0005 by 2-tailed t-test. Bars are 300 microns.
See also Figure S3.
Planarians not only actively maintain tissue proportionality during homeostatic maintenance but also undergo substantial rescaling of pre-existing tissues in regeneration after amputation [7, 8], also referred to as morphallaxis or tissue remodeling. Decapitated tail fragments exemplify an extreme case of these transformations as they must reduce the size of the pre-existing posterior, form a new trunk within the old posterior, and grow a head from the amputation site. mob4(RNAi) tail fragments failed to undergo posterior reduction and positioned trunk tissue too far anteriorly, indicating the importance of mob4 for regeneration-induced tissue rescaling (Figure 4A). In addition, mob4(RNAi) head fragments failed to form a pharynx, and instead wnt1 expression extended far into the anterior (Figure S4A). Therefore, mob4 is required for properly defining new posterior territory through the rescaling that occurs during whole-body regeneration.
Figure 4. Restriction of wnt1+ midline cells controls rescaling during regeneration.
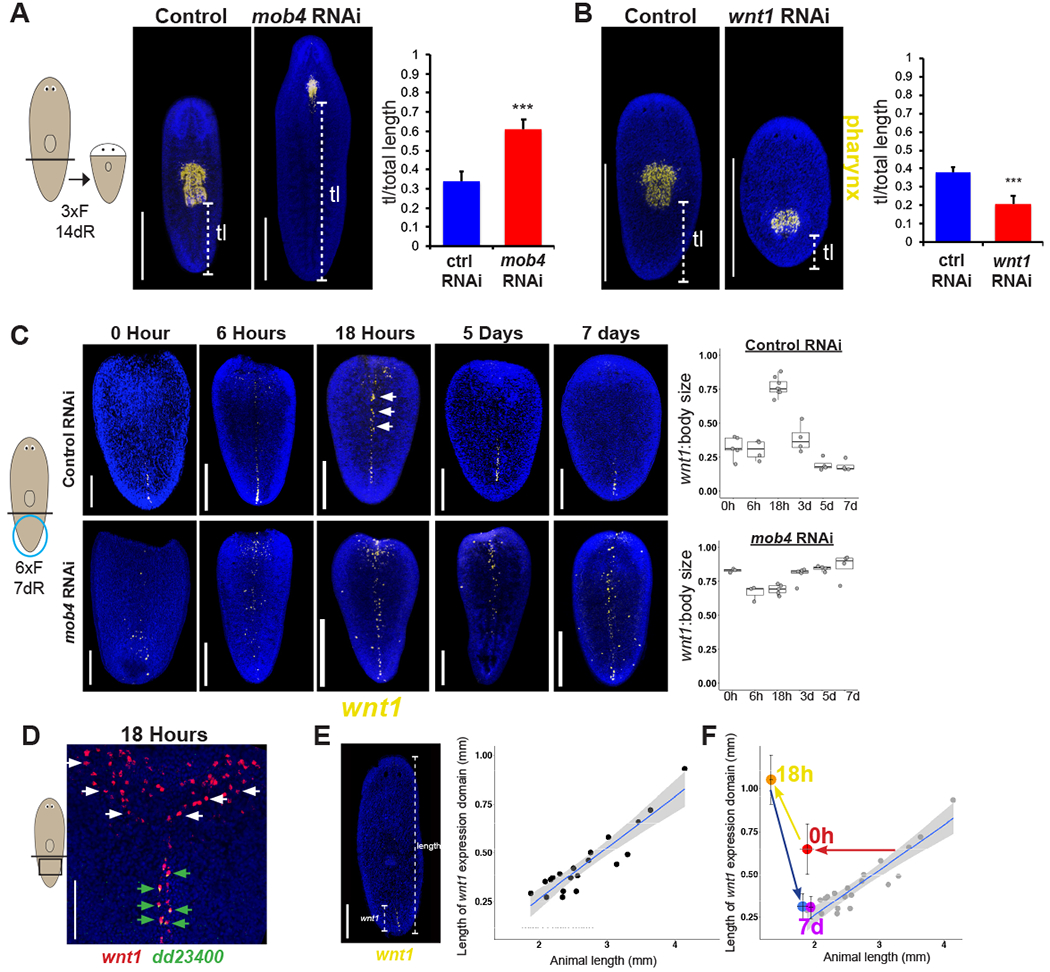
(A) FISH to detect ASXL_059179 in regenerating tail fragments of controls and mob4(RNAi) animals. Animals were fed dsRNA 3 times and regenerated for 14 days. Right- graph showing tail length (brackets) divided by total length of the tail fragment. N>10 animals, error bars are standard deviations and asterisks, p<0.0005 by 2-tailed t-test.
(B) FISH detecting ASXL_059179 to measure tail length in regenerating tail fragments in wnt1(RNAi) animals compared to controls. Right- graph showing tail length divided by total body length. N=10 animals, error bars are standard deviations and asterisks, p<0.0005 by 2-tailed t-test.
(C) Top panels- FISH showing wnt1 expression expansion in controls at 18 hours after injury along the dorsal midline in tail fragments, followed by a restriction to the posterior tip after 7 days. Arrows indicate expanded wnt1 expression along the midline at 18hr. Bottom panels- FISH showing wnt1 expression in mob4(RNAi) animals persists at the dorsal midline throughout regeneration. Animals were fed dsRNA 6 times and regenerated for 7 days. Right- graph showing the quantification of the most anterior midline wnt1+ cell outside the region proximal to the wound site divided by total length of the tail fragment in a time series after injury. N>3 animals for each time point, error bars are standard deviations.
(D) Double FISH showing wnt1 (red) is expressed in dd23400+ cells (green) in tail fragments 18 hours after injury along the dorsal midline, but wnt1 expression at the wound site is not expressed in dd23400+ cells.
(E) FISH to determine relationship between length of the wnt1 expression domain versus total length of individual animals (dots). Linear regression line with 95% confidence interval was calculated in R/ggplot2, R^2=0.85.
(F) Average wnt1:body length of regenerating animals from Fig. 4C mapped onto uninjured animals from Fig. 4E. Bars in panels A, B, E are 300 microns, 150 microns in panels C and D.
See also Figure S4.
We further examined the relationship between wnt1 and mob4 in regeneration-induced rescaling. Because wnt1 is necessary for formation of an entirely new posterior end after tail amputation, we examined this relationship in tail fragments that undergo a reduction in posterior territory. Consistent with opposite action with mob4 in homeostatic maintenance, wnt1(RNAi) tail fragments formed their pharynx too far posteriorly indicating a loss of tail tissue (Figure 4B). In addition to undergoing early wound-site proximal expression of wnt1, we confirmed prior observations that regenerating tail fragments also undergo anterior expansion of wnt1’s midline expression (Figure 4C) [9, 12]. Regeneration-induced midline wnt1 was expressed within dd_23400+ muscle cells, in contrast to wound-proximal wnt1 expression, and it peaked at 18 hours, followed by gradual restriction to the new posterior over time (Figure 4C–D). We hypothesized that the rescaling process requires restriction of this domain to the new posterior pole to allow for formation of trunk and head domains, because the wnt1 expression domain ultimately scales directly with animal length in animals that have completed regeneration (Figure 4E). To test this, we utilized the mob4(RNAi) phenotype of wnt1 pole expansion. In tail fragments following 6 feedings of mob4 dsRNA, wnt1 expression was present at the time of wounding (0 hour), was maintained over all times over the course of regeneration (Figure 4C). Under these dsRNA feeding conditions, 8/12 mob4(RNAi) animals regenerate dramatically smaller brains as measured by cintillo+ cells and significantly longer tails, while 4/12 completely lacked regeneration of any anterior or trunk tissues (Figure S4B). By contrast, control animals underwent wnt1 midline expansion followed by restriction to allow for proper proportion restoration through regeneration (Figure 4F). Together, these results suggest wnt1+ expression at the pole is necessary for posterior proportionality, and mob4 normally restricts wnt1+ pole cells to allow for appropriate anterior and trunk fates.
We next considered the possible mechanisms accounting for mob4’s negative regulatory effect on wnt1 expression. wnt1 expression could be directly regulated by mob4 in a signaling event occurring within pre-existing muscle cells, or alternatively by controlling the production of the cells that express wnt1. To examine these models, we first considered the expression of mob4 with respect to midline muscle cells. FISH showed that both mob4 and striatin mRNA were expressed broadly and were not regionally restricted (Figure S5A–B), with mob4 mRNA enriched in the parenchyma. Prior single-cell RNA-seq experiments showed broad and low expression of both genes, with the caveat that low abundance transcripts may be underrepresented in such datasets (Figure S5A–B). Puncta observed from staining with a mob4 riboprobe were eliminated in mob4(RNAi) animals, consistent with low expression of this gene. Because mob4 expression appeared stronger in the parenchyma, we hypothesized that mob4 could be expressed in neoblasts or their differentiating progeny and tested this by examining expression in animals treated with lethal doses of X-ray irradiation. This treatment almost completely eliminated expression of mob4 by 7 days, suggesting that mob4 expression depends on stem cells or early progenitor cells (Figure 5A). We then used double-fluorescent in situ hybridizations to determine co-expression of mob4 and smedwi-1, a marker of neoblasts. In confocal slices, we found that mob4 expression could be detected at low levels in in many but not all neoblasts (Figure S5D). Because neoblasts are 5-10 μm, we used 10 μm maximum projections to quantify coexpression of mob4 and smedwi-1 and found that 59.7% of neoblasts had clear mob4 expression (Figure 5B, S5D). Using similar approaches, we could not detect mob4 mRNA in either wnt1+ posterior pole cells (Figure 5C), or further anterior midline muscle cells (dd_23400+) that do not express wnt1 (Figure 5D).
Figure 5. mob4 limits differentiation of stem cells into wnt1+ midline cells.
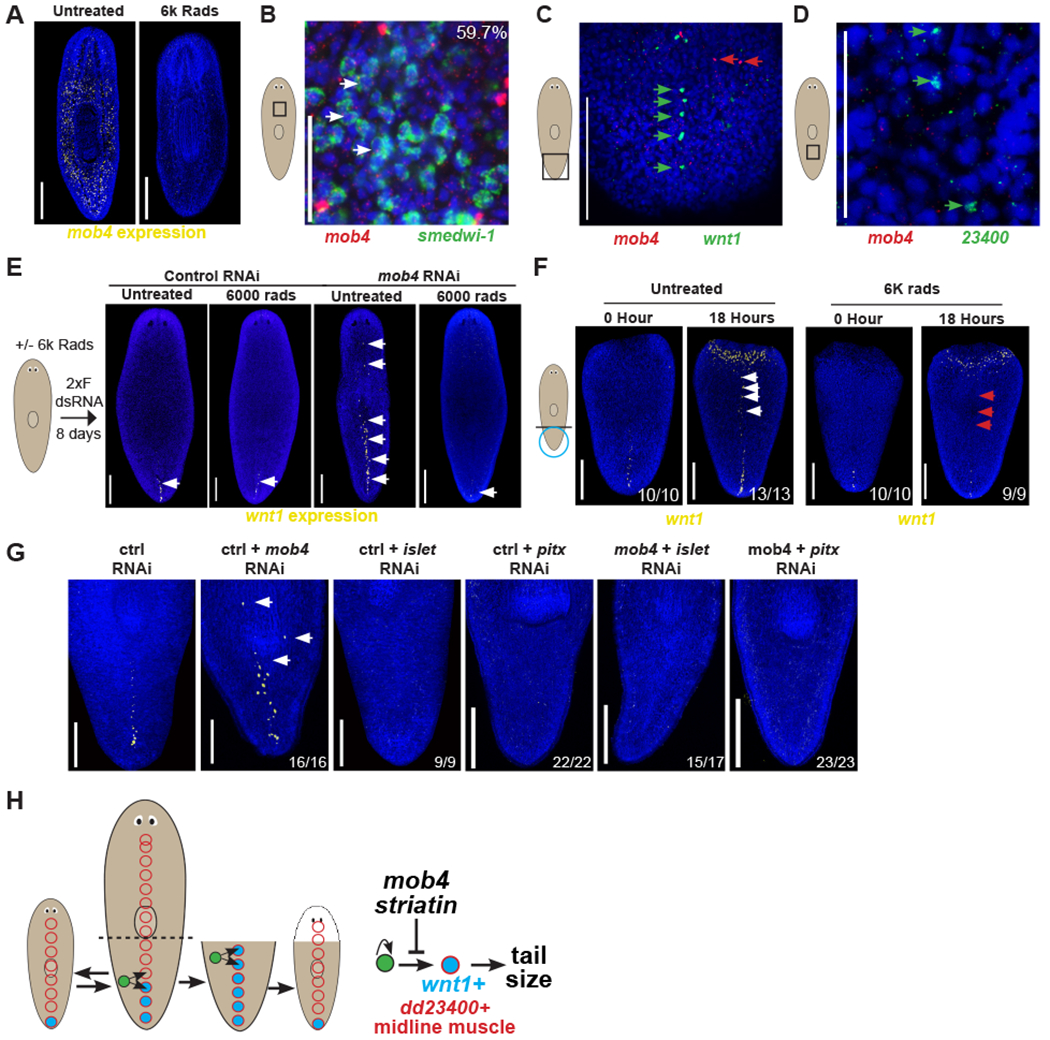
(A) FISH to detect mob4 showing reduced expression in irradiated animals depleted for stem cells (right) compared to controls (left).
(B) Double FISH of mob4 (red) and smedwi-1 (green) showing mob4 is expressed in 59.7% (242/405) smedwi-1+ stem cells.
(C) Double FISH of mob4 (red) and wnt1 (green) showing mob4 is not expressed in wnt1+ posterior pole cells.
(D) Double FISH of mob4 (red) and dd_23400 (green) showing mob4 is not expressed in dd_23400+ midline muscle cells.
(E) Left 2 images- FISH showing wnt1 expression in untreated vs. neoblast depleted irradiated animals after control RNAi feeding. Right 2 images- FISH showing wnt1 expression in untreated vs. neoblast depleted irradiated animals after mob4 inhibition.
(F) FISH showing expanded wnt1 expression along the dorsal midline in tail fragments at 0 hours or 18 hours after injury in untreated animals (left panels) or treated with 6K rads of X-ray irradiation 2 days prior to surgery. N>5 animals for each time point.
(G) FISH to detect wnt1 showing expanded expression in mob4(RNAi) animals but not in islet(RNAi) or pitx(RNAi) animals. Double RNAi of mob4 with either islet or pitx resulted in the absence of wnt expansion. Bars are 300 microns.
(H) Model showing stem cells (green) giving rise to either wnt1-/dd23400+ (red circle) or wnt1+/dd23400+ pole cells (blue with red border) in the far posterior. mob4/striatin inhibits the differentiation of wnt1+/dd23400+ pole cells. Bars in panels A, E, F are 300 microns and in B-D are 150 microns.
See also Figure S5.
These results suggested that mob4 controls wnt1 expression and body proportionality through a stem-cell mediated process rather than cell-autonomously within differentiated muscle cells. However, because of the broadness of mob4 expression, it remained possible that stem cell or progenitor expression of mob4 was unrelated to control of wnt1 expression. To examine this possibility, we determined whether wnt1 expansion after mob4 RNAi could still occur in animals depleted of neoblasts. Some PCG expression in planarians, including wound-proximal injury-induced wnt1 expression, can occur in irradiated animals lacking neoblasts, likely because of signaling events occurring only within differentiated cells. We identified the earliest mob4 RNAi dosing conditions resulting in wnt1 expression expansion (2 feedings encompassing 8 days) and compared this cohort to animals irradiated prior to the same RNAi treatment. Irradiated animals that were fed mob4 dsRNA failed to undergo wnt1 expression expansion (Figure 5E). Therefore, neoblasts are required for mob4’s effects on wnt1 expression. Consistent with these results, rescaling-induced wnt1 expressed along the midline of regenerating tail fragments, but not wound-proximal wnt1 expression in dd_23400− cells, was also eliminated by lethal irradiation (Figure 5F). Based on these results, we reasoned that mob4 either controls neoblasts in the process of differentiating wnt1+ cells or instead might influence wnt1 expression in pre-existing muscle cells in some process indirectly involving neoblasts. To test these models, we made use of prior work which identified the transcription factors islet and pitx as required for the production of wnt1+ pole cells from neoblasts [30–32]. Inhibition of either islet or pitx suppressed the mob4 RNAi phenotype on wnt1 expression, indicating the effects of mob4 inhibition require wnt1+ cell differentiation. These results suggest that mob4 primarily acts to inhibit the production of wnt1+ cells rather than to repress wnt1 expression in preexisting muscle cells (Figure 5G). Using qPCR, we confirmed that mob4 expression was successfully knocked down in both mob4(RNAi);islet(RNAi) and mob4(RNAi):pitx(RNAi) animals compared to controls (Figure S5E). Taken together, these results support a model in which wnt1+ pole cells determine tail proportionality along the A-P axis, with mob4 controlling wnt1+ pole cell numbers through limiting stem-cell dependent production (Figure 5G).
Discussion
Together, our results indicate that STRIPAK suppression of Wnt signaling is a critical pathway regulating whole-body proportional scaling in planarians. STRIPAK complexes regulate a variety of processes including cell growth, differentiation, signaling, proliferation, apoptosis and tumorigenesis, using a variety of inputs [24]. In planarians, the activity of a separate striatin gene was possibly needed for the functioning of neoblast progeny [33]. Recently, multiple studies have identified STRIPAK as a key negative regulator of Hippo signaling [34, 35], although planarian Hippo pathway components do not obviously function in A-P tissue proportionality. Hippo RNAi results in modifications to proliferation, decreased cell death, and formation of undifferentiated lesions, while yorkie(RNAi) animals have elevated injury-induced wnt1, as well as regeneration failure and excretory cell dysfunction [36–38]. Additionally, yorkie(RNAi) animals have both ectopic wnt1 and notum cells during homeostasis, but without obvious changes to position of tissues along the anterior-posterior axis. We hypothesize that the pleiotropic roles of yorkie may mask specific contributions to tail size via wnt1 expression regulation. Our results raise the possibility that STRIPAK could exert context-specific regulation of Yorkie functions.
wnt1 is a multifunctional factor in planarian regeneration. Inhibition of wound-induced follistatin results in excess wound-proximal, injury-induced wnt1 expression leading to the failure of head regeneration [39]. In addition, RNAi of wnt1 only after injury in head fragments caused ectopic head regeneration, indicating its effects on axis polarization are injury-induced [10]. By contrast, in regenerating animals, pole expression of wnt1 is impaired in irradiated animals or after inhibition of several transcription factors Djislet, junli, and pitx [10, 30–32, 40]. How wnt1 may act upstream or in parallel to other posteriorly expressed factors for tail specification and sizing has not yet been resolved. However, the mob4(RNAi) phenotype does not involve an inversion of axis polarization, indicating the independence between tissue polarity and body regionalization used for tissue proportionality in regeneration.
Planarians scale all body regions with respect to each other and across all body axes. Factors responsible for overall animal growth could include pathways responsive to nutrient status and link to generalized control of neoblast differentiation rates. By contrast, proportional scaling involves controlling the relative sizes of local regions with respect to each other. Here we implicate mob4 and the STRIPAK complex in scaling tail size with respect to body size via regulation of wnt1. Homeostatic wnt1-dependent tail enlargement in mob4(RNAi) coincided with a lack of anterior growth, suggesting that at least some aspects of the planarian tissue scaling system could be mediated through individual control of discrete body regions. These findings raise the possibility that planarian positional control genes expressed from muscle might in general function both in regional identity determination and also in regional proportional scaling. Because these genes are constitutively expressed in the absence of injury, the re-attainment of normal proportions across the body through regeneration might then involve a process that achieving a stable equilibrium of these local signaling processes across all body regions.
In principle, the determinants of axis scale could be regulated through signaling across fields of existing cells, either by ensembles of cells controlled locally through junctional signals or directed by morphogens acting at a distance. Our results indicate the importance of generating the cells that produce a secreted factor, wnt1, at an axis boundary in order to control the regional proportionality along that body axis. Signaling through planarian stem cells is necessary for production of the notum+ anterior pole and both to produce [41–44] and limit the production of the wnt1+ posterior pole. Therefore, regulating the production of signaling center cells through stem cell differentiation could be a critical component of establishing proportional scaling in regeneration.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Contact, Dr. Christian Petersen, christian-p-petersen@northwestern.edu. All unique/stable reagents generated in this study are available from the lead contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Asexual Schmidtea mediterranea animals (CIW4 strain) were maintained in 1x Montjuic salts (1.6 mmol/l NaCl, 1.0 mmol/l CaCl2, 1.0 mmol/l MgSO4, 0.1 mmol/l MgCl2, 0.1 mmol/l KCl and 1.2 mmol/l NaHCO3 in Milli-Q water, pH 6.9-8.1) between 18-20°C. Animals were fed a puree of calf liver (Western Veal Liver Non-Formula Feed product #2180 from Golden Gate Meat Co., 550 Seventh Street, San Francisco CA 94103), homogenized using a Juiceman JM400 Electric Juicer after removal of the casing and stored at −80 in aliquots. Animals were starved for at least 5 days before experiments.
METHOD DETAILS
Fluorescent in situ hybridization (FISH)
Animals were killed in 5% NAC in PBS, fixed in 4% formaldehyde and bleached in 6% H2O2 in either Methanol or PBS as described previously [45, 46]. Riboprobes (digoxigenin- or fluorescein-labeled) were synthesized by in vitro transcription [45, 46] using PCR templates amplified to contain T7 RNA binding sites for antisense transcription. Anti-Digoxigenin-POD or Anti-Fluorescein-POD Antibodies were used in TNTx/10% horse serum/10% Western Blocking Reagent (Roche, Basel Switzerland) at a concentration of 1:2000 for anti-DIG-POD (Roche, Basel Switzerland), and 1:2000 for anti-FL-POD (Roche). For multiplex FISH, peroxidase conjugated enzyme activity was quenched between tyramide reactions by sodium azide treatment (100 mM in 1xPBSTx) for 45 min at room temperature. Nuclear counterstaining was performed using Hoechst 33342 (Invitrogen, 1:1000 in 1xPBSTx). Primers for probe design are indicated in Table S1.
RNAi administration
dsRNA was generated by T7 in vitro transcription of PCR templates to generate sense and antisense transcripts, Ethanol precipitated, annealed at 72 degrees, and verified by gel electrophoresis. RNAi feeding was performed from in vitro transcribed dsRNA mixed with 80% liver paste and 5% red food dye [47]. In all RNAi experiments animals were fed RNAi food every 2–3 days for the length of experiment indicated. Figure 1C–D, S1D, S1F–I, 2D, S2A–E, 3B and S3A–C are homeostatic animals fed 12 times or for 34 days of RNAi. Figure 2A–C are homeostatic animals fed 9 times or a total of 26 days RNAi. Figure S3D are homeostatic animals fed 10 times or a total of 28 days RNAi. Figure 5G, S5F are homeostatic animals were fed 6 times for a total of 19 days of RNAi. Figure S5C are homeostatic animals fed 4 times or a total of 12 days of RNAi. Figures 4A–B, S4A are regenerating animals fed 3 times, while figures 4C and S4B are regenerating animals fed 6 times. Primers for RNAi are indicated in Table S1.
Image Acquisition and Quantification
Live images of animals were performed with a Leica M210F dissecting microscope scope with a Leica DFC295 camera. FISH images were performed a Leica DM5500B compound microscope with optical sectioning by Optigrid structured illumination or a Leica SP5 or Leica TCS SPE confocal compound microscopes. Fluorescent images collected by compound microscopy are maximum projections of a z-stack and adjusted for brightness and contrast using Metamorph, Adobe Photoshop, or Fiji/ImageJ.
Body Length Measurements
Total animal and regional lengths were measured with Metamorph as visualized with Hoechst or the pharynx progenitor marker asxl_059179. Changes to tissue domain lengths during homeostasis were assessed following at least 34 days of RNAi feeding. Changes to tissue domain lengths during regeneration were assessed following at least 10 days of RNAi feeding. Domain lengths were averaged and significant differences determined by two-tailed Student’s t-tests. Quantification in Figure 4C is the length of the most anterior midline wnt1+ cell outside the region 200 microns proximal to the wound site divided by the total body length. Significant differences determined by two-tailed Student’s t-tests.
Cell Counting
In Figure S1D, cto+ cells were imaged using a Leica DM5500B compound microscope with optical sectioning equipped with an Optigrid structured illumination device. After a maximum projection of z-stacks, cto+ cells were counted manually in Metamorph. cto+ cell numbers were averaged and significance was determined by two-tailed t-tests. In Figure 5B, and S5C using a Leica TCS SPE confocal microscope, we took a 10uM projection of mob4 and smedwi-1 expression to ensure that mob4 expression was in the same plane as neoblasts, which are 5-10 μm in size. Double positive cells were counted manually from 5 separate worms Leica LAS software.
DATA CODE AND AVAILABILITY
This study did not generate any unique datasets or code.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed in Microsoft Excel unless indicated otherwise. Statistical tests, significance, data points, error bars, replicates, and other information relevant to figures are described and explained in corresponding legends. Experiments were conducted with at least 5 animals per condition unless otherwise noted. No specific strategy was employed for randomization, blinding, sample-size estimation, or testing normality. No subjects or data were excluded from analysis.
Supplementary Material
Acknowledgements
We thank members of the Petersen lab and Dr. Eric Hill for critical comments. Funding: CP acknowledges funding from NIGMS R01GM129339, R01GM130835, and pilot project funding from the NSF-Simons Center for Quantitative Biology at Northwestern University, an NSF (1764421)-Simons/SFARI (597491-RWC) MathBioSys Research Center.
Footnotes
Declaration of Interests: There are no competing interests.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Thompson D.a.W. (1942). On Growth and Form, (Cambridge, England: Cambridge University Press; ). [Google Scholar]
- 2.Hariharan IK (2015). Organ Size Control: Lessons from Drosophila. Dev Cell 34, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kicheva A, and Briscoe J (2015). Developmental Pattern Formation in Phases. Trends Cell Biol 25, 579–591. [DOI] [PubMed] [Google Scholar]
- 4.Schwank G, and Basler K (2010). Regulation of organ growth by morphogen gradients. Cold Spring Harb Perspect Biol 2, a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner DE, Wang IE, and Reddien PW (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baguñà J, Romero R, Saló E, Collet J, Carme A, Ribas M, Riutort M, García-Fernàndez J, Burgaya F, and Bueno D (1990). Growth, degrowth and regeneration as developmental phenoma in adult freshwater planarians In Experimental Embryology in Aquatic Plants and Animals. , Volume 195, Marthy HJ, ed. (New York: Springer; ), pp. 129–162. [Google Scholar]
- 7.Hill EM, and Petersen CP (2015). Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain. Development 142, 4217–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oviedo NJ, Newmark PA, and Sanchez Alvarado A (2003). Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Developmental dynamics : an official publication of the American Association of Anatomists 226, 326–333. [DOI] [PubMed] [Google Scholar]
- 9.Petersen CP, and Reddien PW (2008). Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327–330. [DOI] [PubMed] [Google Scholar]
- 10.Petersen CP, and Reddien PW (2009). A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci U S A 106, 17061–17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen CP, and Reddien PW (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, and Sanchez Alvarado A (2010). Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347, 24–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuckemann T, Cleland JP, Werner S, Thi-Kim Vu H, Bayersdorf R, Liu SY, Friedrich B, Julicher F, and Rink JC (2017). Antagonistic Self-Organizing Patterning Systems Control Maintenance and Regeneration of the Anteroposterior Axis in Planarians. Dev Cell 40, 248–263 e244. [DOI] [PubMed] [Google Scholar]
- 14.Gurley KA, Rink JC, and Sanchez Alvarado A (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iglesias M, Gomez-Skarmeta JL, Salo E, and Adell T (2008). Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development 135, 1215–1221. [DOI] [PubMed] [Google Scholar]
- 16.Owen JH, Wagner DE, Chen CC, Petersen CP, and Reddien PW (2015). teashirt is required for head-versus-tail regeneration polarity in planarians. Development 142, 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reuter H, Marz M, Vogg MC, Eccles D, Grifol-Boldu L, Wehner D, Owlarn S, Adell T, Weidinger G, and Bartscherer K (2015). Beta-catenin-dependent control of positional information along the AP body axis in planarians involves a teashirt family member. Cell Rep 10, 253–265. [DOI] [PubMed] [Google Scholar]
- 18.Beane WS, Morokuma J, Lemire JM, and Levin M (2013). Bioelectric signaling regulates head and organ size during planarian regeneration. Development 140, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felix DA, and Aboobaker AA (2010). The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet 6, e1000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander R, and Petersen CP (2016). Wnt, Ptk7, and FGFRL expression gradients control trunk positional identity in planarian regeneration. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scimone ML, Cote LE, Rogers T, and Reddien PW (2016). Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baillat G, Moqrich A, Castets F, Baude A, Bailly Y, Benmerah A, and Monneron A (2001). Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol Biol Cell 12, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno CS, Lane WS, and Pallas DC (2001). A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J Biol Chem 276, 24253–24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Z, Jiao S, and Zhou Z (2016). STRIPAK complexes in cell signaling and cancer. Oncogene 35, 4549–4557. [DOI] [PubMed] [Google Scholar]
- 25.Moreno CS, Park S, Nelson K, Ashby D, Hubalek F, Lane WS, and Pallas DC (2000). WD40 repeat proteins striatin and S/G(2) nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J Biol Chem 275, 5257–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeda H, Nishimura K, and Agata K (2009). Planarians maintain a constant ratio of different cell types during changes in body size by using the stem cell system. Zoolog Sci 26, 805–813. [DOI] [PubMed] [Google Scholar]
- 27.Witchley JN, Mayer M, Wagner DE, Owen JH, and Reddien PW (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep 4, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, and Reddien PW (2018). Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sureda-Gomez M, Pascual-Carreras E, and Adell T (2015). Posterior Wnts Have Distinct Roles in Specification and Patterning of the Planarian Posterior Region. Int J Mol Sci 16, 26543–26554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Currie KW, and Pearson BJ (2013). Transcription factors lhx1/5-1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development 140, 3577–3588. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi T, Motoishi M, Yazawa S, Itomi K, Tanegashima C, Nishimura O, Agata K, and Tarui H (2011). A LIM-homeobox gene is required for differentiation of Wnt-expressing cells at the posterior end of the planarian body. Development 138, 3679–3688. [DOI] [PubMed] [Google Scholar]
- 32.Marz M, Seebeck F, and Bartscherer K (2013). A Pitx transcription factor controls the establishment and maintenance of the serotonergic lineage in planarians. Development 140, 4499–4509. [DOI] [PubMed] [Google Scholar]
- 33.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, and Sanchez Alvarado A (2005). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen M, Zhang H, Shi Z, Li Y, Zhang X, Gao Z, Zhou L, Ma J, Xu Q, Guan J, et al. (2018). The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer. J Biol Chem 293, 14455–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Y, Liu B, Wang L, Lei H, Pulgar Prieto KD, and Pan D (2017). Homeostatic Control of Hpo/MST Kinase Activity through Autophosphorylation-Dependent Recruitment of the STRIPAK PP2A Phosphatase Complex. Cell Rep 21, 3612–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Sousa N, Rodriguez-Esteban G, Rojo-Laguna JI, Salo E, and Adell T (2018). Hippo signaling controls cell cycle and restricts cell plasticity in planarians. PLoS Biol 16, e2002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin AY, and Pearson BJ (2014). Planarian yorkie/YAP functions to integrate adult stem cell proliferation, organ homeostasis and maintenance of axial patterning. Development 141, 1197–1208. [DOI] [PubMed] [Google Scholar]
- 38.Lin AYT, and Pearson BJ (2017). Yorkie is required to restrict the injury responses in planarians. PLoS Genet 13, e1006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tewari AG, Stern SR, Oderberg IM, and Reddien PW (2018). Cellular and Molecular Responses Unique to Major Injury Are Dispensable for Planarian Regeneration. Cell Rep 25, 2577–2590 e2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tejada-Romero B, Carter JM, Mihaylova Y, Neumann B, and Aboobaker AA (2015). JNK signalling is necessary for a Wnt- and stem cell-dependent regeneration programme. Development 142, 2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CC, Wang IE, and Reddien PW (2013). pbx is required for pole and eye regeneration in planarians. Development 140, 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scimone ML, Lapan SW, and Reddien PW (2014). A forkhead transcription factor is wound-induced at the planarian midline and required for anterior pole regeneration. PLoS Genet 10, e1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasquez-Doorman C, and Petersen CP (2014). zic-1 Expression in Planarian neoblasts after injury controls anterior pole regeneration. PLoS Genet 10, e1004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogg MC, Owlarn S, Perez Rico YA, Xie J, Suzuki Y, Gentile L, Wu W, and Bartscherer K (2014). Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Dev Biol 390, 136–148. [DOI] [PubMed] [Google Scholar]
- 45.Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, and Sanchez Alvarado A (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn 238, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King RS, and Newmark PA (2013). In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouhana L, Weiss JA, Forsthoefel DJ, Lee H, King RS, Inoue T, Shibata N, Agata K, and Newmark PA (2013). RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: methodology and dynamics. Dev Dyn 242, 718–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


