Abstract
Multidrug efflux is a major contributor to antibiotic resistance in Gram-negative bacterial pathogens. Inhibition of multidrug efflux pumps is a promising approach for reviving the efficacy of existing antibiotics. Previously, inhibitors targeting both the efflux transporter AcrB and the membrane fusion protein AcrA in the Escherichia coli AcrAB-TolC efflux pump were identified. Here we use existing physicochemical property guidelines to generate a filtered library of compounds for computational docking. We then experimentally test the top candidate coumpounds using in vitro binding assays and in vivo potentiation assays in bacterial strains with controllable permeability barriers. We thus identify a new class of inhibitors of E. coli AcrAB-TolC. Six molecules with a shared scaffold were found to potentiate the antimicrobial activity of erythromycin and novobiocin in hyperporinated E. coli cells. Importantly, these six molecules were also active in wild-type strains of both Acinetobacter baumannii and Klebsiella pneumoniae, potentiating the activity of erythromycin and novobiocin up to 8-fold.
Keywords: Gram-negative, permeability, resistance nodulation division, substrate, docking, minimum inhibitory concentration
Introduction
Multidrug antibiotic resistance is a global public health crisis that leads to the deaths of ~35,000 people every year in the United States alone (https://www.cdc.gov/drugresistance/about.html). Infections caused by Gram-negative bacteria, such as multidrug-resistant Enterobacteriaceae and Acinetobacter baumannii are particularly challenging to treat.1 Significant efforts are currently focused on discovery of new antibiotics that will address this growing public health threat. One of the major hurdles in the discovery of new antibiotics effective against Gram-negative pathogens is the antibiotic permeation barrier due to the synergistic relationship between low intrinsic permeability of the Gram-negative outer membrane (OM) and multidrug efflux.2, 3
The OM consists of an asymmetric bilayer comprising a phospholipid inner leaflet and a lipopolysacchararide outer leaflet in which polyanionic lipids are bridged by divalent cations.4, 5 Porins in the OM, such as OmpF and OmpC in Escherichia coli provide a selective barrier that restricts the entry of large >600 Da molecules into the cell.6 Furthermore, multidrug efflux pumps expel a wide range of antibiotics from the periplasm into the extracellular medium.7 Among the most well studied efflux pumps is AcrAB-TolC from E. coli. Cryo-EM structures have been determined for the assembled AcrAB-TolC pump, which consists of three main components, AcrB, AcrA and TolC (Figure 1A). AcrB is a homotrimeric protein that consists of an α-helical integral membrane domain, a periplasmic porter domain that binds and extrudes substrates, and a docking domain that interacts with AcrA.8, 9 The periplasmic porter domain of AcrB contains a narrow cleft within the substrate binding pocket that acts as a hydrophobic trap.10 AcrA is a membrane fusion protein that consists of four domains: α-hairpin, lipoyl, β-barrel, and membrane-proximal (Figure 1B). TolC is a trimeric protein that consists of a β-barrel domain embedded in the OM and a periplasmic α-helical coiled-coil domain.11 AcrB, AcrA, and TolC assemble in a 3:6:3 stoichiometry12 to form a complex that spans the entire Gram-negative cell envelope.13
Figure 1.

(A) Cryo-EM structure of the AcrAB-TolC complex (PDB Entry 5NG5) shown in cartoon and surface representations. Individual subunits of TolC (purple), AcrA (dark green), and AcrB (blue) are shown. (B) Sites I-IV on AcrA used for docking are color coded by domain:α-hairpin (light green), lipoyl (orange), β-barrel (yellow) and membrane-proximal (red). Residues used to define the center of each site are shown as spheres and colored by domain.
Many antibiotics are substrates of one or more efflux transporters, making efflux a major limiting factor in antibiotic activity.14, 15 One approach to overcoming this barrier is the use of efflux pump inhibitors (EPIs): small molecules that potentiate the activity of existing antibiotics. The first EPI to be discovered, MC-207,110 (phenylalanyl-β-naphtylamide, PAβN), potentiates the activity of fluoroquinolones in E. coli and Pseudomonas aeruginosa.16 This compound also disrupts the structural integrity of the OM in efflux-deficient cells and increases the accumulation of levofloxacin.17 MC-207,110 and other known EPIs targeting AcrB are thought to be non-competitive inhibitors that bind in the hydrophobic trap.18 Surface plasmon resonance (SPR) experiments have shown that MC-207,110 binds both AcrB and AcrA with μM affinity.19 The pyranopyridine compound MBX2319 and its derivatives potentiate the activity of levofloxacin and piperacillin in E. coli.20, 21 MBX2319 binds tightly to the hydrophobic trap22 but does not have any detectable binding affinity for AcrA.19
Recently, EPIs targeting AcrA have also been developed in an effort to disrupt assembly of the complex, thereby preventing or limiting efflux. By combining computational docking with in vivo potentiation and binding assays, four compounds were identified as EPIs that bind to AcrA, inhibit the efflux of fluorescent probes, and potentiate the activities of novobiocin (NOV) and erythromycin (ERY) in E. coli and other Gram-negative bacteria.19 On the basis of in vivo proteolysis experiments, it was proposed that one of these EPIs, NSC60339, functions by disrupting pump assembly. Second-generation analogs of NSC60339 were subsequently synthesized that penetrate the OM, exhibit improved efflux inhibition compared to the parent compound, and potentiate the activity of NOV and ERY in E. coli.23 A combination of computational docking, site-directed mutagenesis, tryptophan fluorescence quenching, and antibiotic susceptibility assays indicated that NSC60339 binds at a site located between the lipoyl and β-barrel domains of AcrA (Figure 1B).24
Antibacterial drugs tend to occupy different regions of physicochemical property space relative to conventional small molecule drugs. In general, antibiotics tend to be more polar and charged and have higher molecular weights.25,26 The OM in Gram-negative bacteria acts as an additional barrier to entry that many Gram-positive antibiotics are unable to overcome. Therefore, it has been suggested that Gram-positive and Gram-negative antibiotics may have divergent patterns in physicochemical properties that trend with activity. For example, Gram-negative antibiotics were found to contain on average four additional hydrogen bond donors and acceptors and have log D values more than four log units lower on average compared to the Comprehensive Medicinal Chemistry (CMC) data set (https://www.3dsbiovia.com/). However, analysis of the ChEMBL database suggests that both types of antibiotics overlap in size, polarity, and flexibility.25 Therefore, selecting compounds with optimal molecular properties is not guaranteed to be successful because it is challenging to identify compounds that simultaneously permeate the OM, evade efflux, and have the desired activity. Simply occupying this subset of physicochemical space is not sufficient to meet these criteria.27
Recently, physicochemical properties have been identified for compounds that permeate the OM in E. coli.15, 28, 29 Liquid chromatography with tandem mass spectroscopy (LC–MS/MS) analysis revealed that small molecule compounds containing amine functional groups were most likely to accumulate in E. coli cells, with primary amines having the highest accumulation.28 Incorporation of a primary amine into the Gram-positive antibiotic deoxynybomycin (6DNM) resulted in a new antibiotic (6DNM-NH3) that exhibited broad-spectrum activity against a panel of multidrug-resistant Gram-negative bacteria. In addition to containing an amine, antibiotics that tended to be successful at bypassing the OM permeability barrier were polar, amphiphilic, relatively rigid, and had low globularity.
The use of engineered bacterial strains with controllable permeability barriers, i.e. hyperporinated OM and efflux deletion, has provided key insight into factors that limit antibiotic and EPI activity.14, 15, 19 Separation of these two barriers revealed that, for many existing antibiotics and EPIs, molecules that are permeable also tend to be efflux substrates. In addition, there is overlap in the physicochemical properties that trend with efflux and OM permeability. Unsurprisingly, though, in the cases of E. coli and P. aeruginosa, several of the properties appear to be barrier and strain specific. Although these two species have similar OM lipid compositions, their porins differ.5, 30 In addition, whereas efflux in E. coli can be inactivated by deleting a single gene (tolC),31, 32 multiple pumps must be inactivated to abolish efflux in P. aeruginosa.33 Although the physicochemical guidelines for permeability and efflux may differ between species as for E. coli and P. aeruginosa, antibiotic activity and potentiation is not always species specific. For example, although NSC60339 appears to be specific for E. coli, NSC33353 also potentiated NOV and ERY activity in Acinetobacter baumannii, Enterobacter cloacae, and Klebsiella pneumoniae.19 Therefore, EPIs identified for one species (i.e., E. coli with only a single efflux pump to target) may also potentiate antibiotic activity in other Gram-negative bacteria with different efflux pumps and OM compositions.
Here, we used existing physicochemical rules for OM permeability and efflux in E. coli in combination with in vitro binding assays and in vivo potentiation assays in bacterial strains with controllable permeability barriers to identify a new class of EPIs with activity against several Gram-negative bacteria. By searching the ZINC15 database34 for compounds with physicochemical properties that fit selected guidelines for Gram-negative antibiotics, we greatly reduced the size of the molecular search space, from ~13M to 1,427. We then computationally docked the resulting 1,427 compounds to an ensemble of conformations of E. coli AcrA generated from molecular dynamics (MD) simulation at four sites: the proposed binding site of NSC60339 and three other sites that have been shown previously to disrupt efflux when residues within those sites were mutated.24, 35 Thirty-four compounds were selected from among the top 500 of all docking poses at each site and SPR was used to measure their ability to bind to both AcrA and AcrB. The ability of these compounds to restore the antibiotic activity of NOV and ERY, two antibiotics that are substrates of AcrAB-TolC, was assessed by determining minimal potentiating concentrations (MPCs) in E. coli and several other Gram-negative bacteria. We identified six molecules with a shared scaffold that potentiate the antibiotic activity of ERY and NOV in hyperporinated E. coli cells and in wild-type strains of both A. baumannii and K. pneumoniae.
Materials and Methods
Generation of a small molecule library.
We used the Tranche Browser to search a subset of the ZINC 15 database34 for 3D representations of in-stock compounds with “standard” reactivity, molecular charge in the range of −2 to 2 at pH “ref” or “mid”, log P between 2.5 and 3, and no molecular weight cutoff, resulting in ~1.8M small molecules (as of November 26, 2018). Alternate protonation and tautomeric states at pH 7.4 were included for each molecule. Coordinates and charges were downloaded and used as provided by ZINC15. We then used a custom Python script to select compounds containing a primary amine using the SMILES tag [NH3+], resulting in 22,842 compounds. From this list we selected molecules that are relatively rigid (number of rotatable bonds ≤ 4), polar (dipole moment ≥ 5.5 D), amphiphilic (amphiphilic moment ≥ 4.0 Å2), and have low globularity (≤ 0.14).15, 28 The globularity, rigidity, and amphiphilic moment thresholds were chosen on the basis of previous work28. The threshold of 5.5 D for the dipole moment was the average cutoff value for antibiotics that trended with MIC and MIC ratios in E. coli and P. aeruginosa.15 Antibiotics with a dipole moment greater than 5.5 D exhibited activity in P. aeruginosa and were therefore able to permeate the OM. Globularity is defined as the deviation of the shape of the compound from a perfect sphere.28 The dipole moment quantifies the degree of charge separation and was estimated based on molecular topology using the partial equalization of orbital electronegativity (PEOE) method.15 The amphiphilic moment quantifies the degree of separation between hydrophobic and hydrophilic regions of a molecule. Molecular properties were calculated with MOE 2016.36
Ensemble docking to AcrA.
Previously, we generated a full-length model of AcrA from E. coli, performed a 50-ns molecular dynamics simulation of the model, and extracted 29 representative conformations using RMSD clustering.24 In the present work, ensemble docking37 was performed using these 29 conformations24 with VinaMPI,38 which is a massively parallel version of AutoDock Vina.39 The following residues were selected as the centers of the 25 Å × 25 Å × 25 Å docking search spaces: E67 (site I), K241 (site II), I343 and I252 (site III), and F81 and F254 (site IV) (Figure 1B). The exhaustiveness was set to 10 and the top 500 poses for each site were selected for further analysis based on the docking scores. We then selected compounds based on docking scores at any site. Of the resulting ~50 top compounds, commercially available compounds were purchased from ChemBridge (San Diego, CA). A summary of the physicochemical properties and docking scores is provided in Table S1.
Microbiological and biochemical
Wild-type (WT) and ΔtolC strains of E. coli and their hyperporinated (i.e.,WT-Pore) variants used in this study are derivatives of BW 25113 and GD102, respectively.14 The E. coli BL21(DE3) strain was used for overexpression and purification of AcrA40 and AG100AX strain was used for overexpression and purification of AcrB.41
MICs were analyzed using a two-fold broth dilution method.42 For the checkerboard assay, an antibiotic and a test compound were serially diluted into 96-well plates as described previously.16 Minimal potentiating concentration (MPC) was defined as a concentration of a compound that descreases the MIC of an antibiotic by four (MPC4) or more fold. SPR experiments were carried out with purified AcrA and AcrB immobilized onto a CM5 chip (Biacore).43 AcrA and AcrB were immobilized at densities of 7167 and 7072 response units (RU), respectively, and compounds were injected in HEPES-NaCl buffer supplemented with 5% DMSO at compound concentrations 12.5, 25, 50, 100, 200, and 400 μM. The HT uptake assay was performed in a temperature-controlled micro-plate reader (Tecan Spark 10M) equipped with a sample injector, in fluorescence mode. Data were fitted to extract initial rates as described previously.44
Results and Discussion
Selection of candidate compounds.
Recently, properties of molecules that are favorable for OM permeability in E. coli have been identified and it was shown that compounds that permeate the OM are often efflux pump substrates.15, 28 An important feature of OM-permeable compounds is the presence of a cationic amine, with primary amines being the most permeable. However, primary amines are relatively rare in chemical databases. For example, only ~0.1% of the ChemBridge Microformat Set contains this functional group.28 Of the limited number of amines, antibacterials should also be relatively rigid, polar, amphiphilic, and have low globularity. Interestingly, many existing EPIs have cationic amines and many of these other properties.
To generate a focused library of molecules that could potentially serve as efflux pump inhibitors, we searched a subset of the ZINC database and performed additional filtering for compounds with appropriate properties. We then docked the resulting ~1,400 compounds to an ensemble of conformations of monomeric AcrA at four different potential binding sites that were identified previously.24 In ensemble docking,37 small molecules are computationally docked to a set of representative conformations of a protein to mimic conformational selection. We then selected compounds based on docking scores at any of the four sites. Of the resulting ~50 top predicted binders, 34 commercially available compounds were purchased and tested experimentally (Table 1 and Table S2).
Table 1.
Antibiotic Susceptibilities of Top Hits
| MIC (μM) | ||||
|---|---|---|---|---|
| compound | WT | WT-Pore | ΔTolC | ΔTolC-Pore |
| 23380367 | ||||
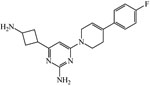 |
≥200 | 200 | 200 | 200 |
| 24123034 | ||||
 |
≥200 | ≥200 | ≥200 | 50 |
| 36287038 | ||||
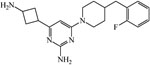 |
≥200 | ≥200 | ≥200 | 200 |
| 36579393 | ||||
 |
≥200 | 200 | 200 | 200 |
| 56871955 | ||||
 |
≥200 | ≥200 | ≥200 | 200 |
| 58997260 | ||||
 |
≥200 | ≥200 | ≥200 | 100 |
| 65071797 | ||||
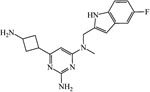 |
≥200 | ≥200 | ≥200 | 200 |
| 98577577 | ||||
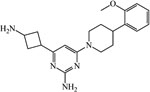 |
≥200 | ≥200 | ≥200 | 200 |
Compounds with intrinsic antibiotic activity in WT-Pore cells.
To test for any intrinsic antibacterial activity of the 34 selected compounds toward E. coli and determine the contributions of permeation and efflux that limit that activity, we measured the minimum inhibitory concentrations (MICs) in WT, WT-Pore, ΔTolC and ΔTolC-Pore cells. WT-Pore and ΔTolC-Pore cells contain ~2.4 nm diameter pores in the OM that allow small molecules to overcome the permeability barrier without affecting efflux.14 None of the compounds had any antibiotic activity in WT cells (Table 1 and Table S2). However, 23380367 and 36579393 showed weak antibacterial activity (MIC = 200μM) in WT-Pore cells, indicating that the lack of OM permeability in the WT prevents activity. Twelve compounds inhibited growth of the efflux-deficient and hyperporinated ΔTolC-Pore cells, suggesting that these compounds possess weak antibacterial activities and are efflux substrates.
Compounds that potentiate the activity of novobiocin and erythromycin in WT-Pore cells and bind AcrA
To qualify as EPIs, compounds must satisfy at least three criteria: (i) they must enhance the activities of antibiotics that are effluxed in strains containing functioning pumps, (ii) they must not significantly potentiate the activities of antibiotics in strains that lack efflux pumps, and (iii) must interact with AcrA or AcrB.16, 19 Therefore, we next measured the ability of each compound to potentiate the activity of NOV and ERY, which have MICs of 32–64 μg/mL and 16–32 μg/mL in E. coli WT-Pore cells, respectively. These two antibiotics have different mechanisms of antibacterial action and are well-characterized substrates of the AcrAB-TolC pump. We performed standard checkerboard assays in which the MICs of antibiotics were determined in the presence of different concentrations of hits (Table 1 and Table S2) and assays were carried out in WT-Pore cells to enable unrestricted access of antibiotics and inhibitors to the transporter. Of the compounds tested, eight potentiated the activities of NOV and ERY in WT-Pore cells by at least fourfold. Six of the compounds contained the same structural core and were analyzed in further detail.
The six compounds potentiated the activity of NOV and ERY in WT cells when present at concentrations 100–200 μM, but were more effective in WT-Pore cells and potentiated the activities of antibiotics at concentrations of 12.5–50 μM. Therefore, these six compounds potentiated antibiotic activity in the presence of efflux pumps but did not readily permeate the OM. While 24123034 itself was devoid of significant antibacterial activity (MICs of 50 and >200 μM for the efflux-deficient ΔTolC and WT-Pore strains, respectively), it increased the susceptibilities only of the efflux-proficient WT-Pore to ERY and NOV by 16- and 8-fold, respectively (Tables 2–3). Compound 58997260 also potentiated ERY activity by at least 16-fold, but possessed only modest activity against NOV in both WT-Pore and in ΔTolC-Pore cells (MPC4 = 50 μM), indicating that the potentiation by this compound may be due not only to interaction with AcrAB-TolC. None of the other compounds potentiated antibiotic activities in ΔTolC-Pore cells, indicating that the activities of the remaining five compounds are related to efflux inhibition.
Table 2.
Potentiation of Erythromycin Activity by Top Hits in Efflux-proficient (WT-Pore) and Efflux-deficient (ΔTolC) Cells
| ERY MIC (mg/L) in the presence of compounds at a given concentration (μM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT-Pore | ΔTolC | |||||||||
| compound | 0 | 6.25 | 12.5 | 25 | 50 | 0 | 6.25 | 12.5 | 25 | 50 |
| 24123034 | 16 | 8 | 4 | 1 | 1 | 2 | 2 | 2 | 2 | 2 |
| 36287038 | 16 | 16 | 16 | 4 | 1–2 | 2 | 2 | 2 | 2 | 1 |
| 56871955 | 16 | 16 | 16 | 8 | 4 | 2 | 2 | 2 | 2 | 2 |
| 58997260 | 16 | 8 | 4 | 2 | 1 | 2 | 2 | 2 | 2 | 1 |
| 65071797 | 16 | 16 | 8 | 2–4 | 1 | 2 | 2 | 2 | 2 | 2 |
| 98577577 | 16 | 16 | 16 | 8 | 2–4 | 2 | 2 | 2 | 2 | 2 |
Table 3.
Potentiation of Novobiocin Activity by Top Hits in Efflux-proficient (WT-Pore) and –deficient (ΔTolC) Cells
| NOV MIC (mg/L) in the presence of compounds at a given concentration (μM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT-Pore | ΔTolC | |||||||||
| compound | 0 | 6.25 | 12.5 | 25 | 50 | 0 | 6.25 | 12.5 | 25 | 50 |
| 24123034 | 32 | 32 | 16 | 8 | 4 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 |
| 36287038 | 32 | 32 | 32 | 32 | 16 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 |
| 56871955 | 32 | 32 | 32 | 16 | 8 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 |
| 58997260 | 32 | 32 | 32 | 16 | 8 | 0.5 | 0.5 | 0.25 | 0.25 | 0.125 |
| 65071797 | 32 | 32 | 32 | 4 | 4 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| 98577577 | 32 | 32 | 32 | 32 | 4 | 0.5 | 0.5 | 0.5 | 0.25 | 0.5 |
We next used SPR to measure the binding of the compounds to immobilized AcrA. Eleven compounds were found to bind AcrA, in an unusually high hit rate of 32%, among which are the six hits that potentiate the activities of NOV and ERY (Figure 2 and Table S3). Hence, these six compounds satisfy the requirements for EPIs. The collected concentration-dependent sensorgrams of these compounds were fit to different kinetic models to extract the equilibrium dissociation constants (Kd) and on- (ka) and off (kd) rates of compound binding to AcrA (Table S3). The best fits for all six compounds were obtained using a two-state kinetic model that postulates a conformational change after the initial binding step. The Kd values derived from fitting the sensorgram were in the mid-micromolar to low millimolar range with relatively slow on- and off- rates. The relatively low affinity of compounds to AcrA could be one of the factors defining the MPCs of these compounds in the growth inhibition assays.
Figure 2.
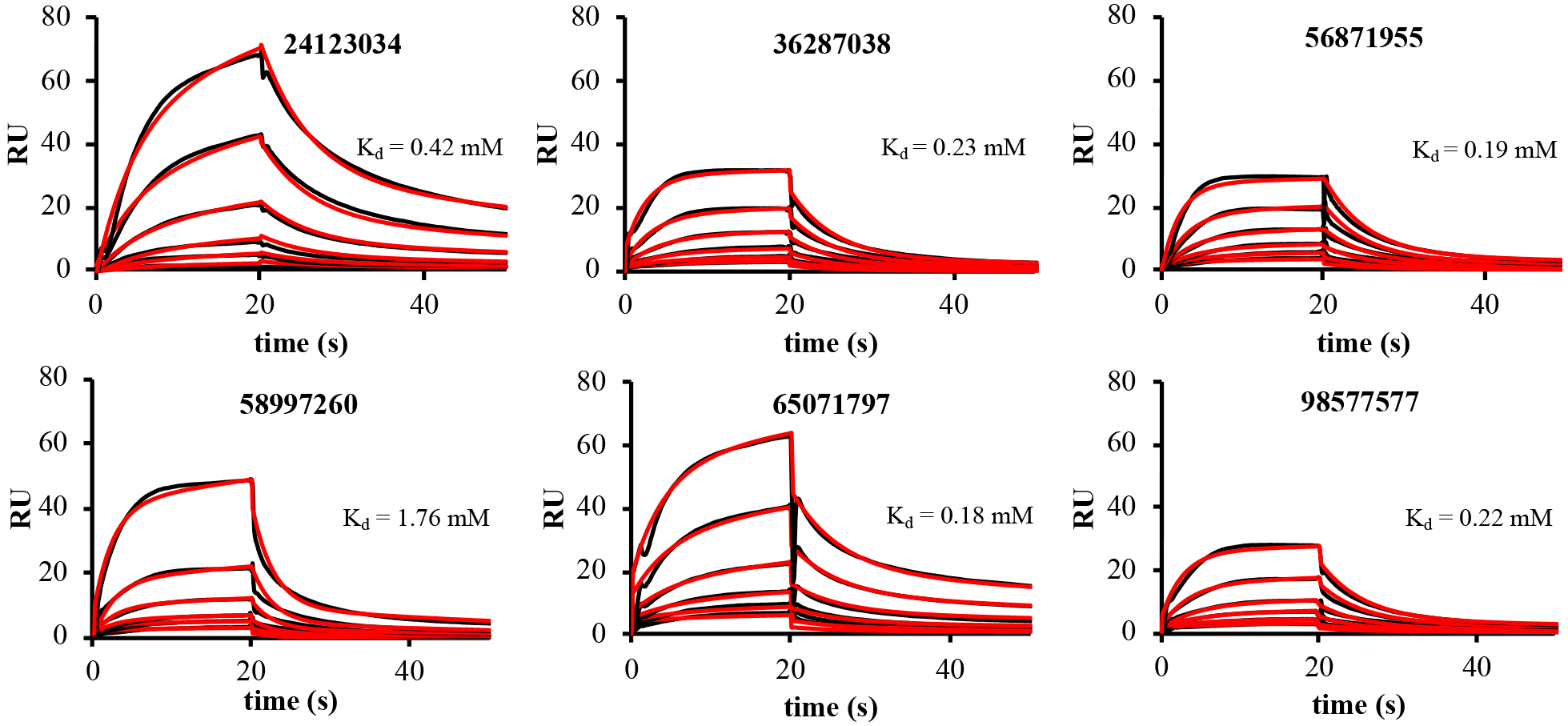
SPR sensorgrams (black lines) for six compounds that bind AcrA in vitro. Sensorgrams were fitted to different kinetic models and the best fits obtained with Two-States kinetic model are shown as red lines. Kd values are labeled.
Taken together, the binding of the six compounds to AcrA and their potentiation of NOV and ERY in WT-Pore cells suggest that they inhibit the efflux activity of AcrAB-TolC. Interestingly, the six compounds that potentiate antibiotics and bind AcrA all are substituted 4(3-aminocyclobutyl)pyrimidin-2-amine compounds. However, not all compounds with this scaffold potentiated antibiotics and bound to AcrA (Table S2). Each of these six compounds has low globularity (0.05–0.12), few rotatable bonds (2–4), relatively high dipole moment (5–10 D) and relatively high amphiphilic moment (~4.5–7) (Table S4). Based on the docking scores, the compounds were predicted in some cases to bind at multiple sites. For example, the most favorable docking score for each compound (Table S4) corresponds to binding at either site II or site III, which flank the β-barrel domain of AcrA (Figure 3).The AutoDock Vina scoring function has a standard error of ~2.9 kcal/mol.39 Many of the docking scores for each ligand at the different binding sites are within this range, so we do not consider the scores for each site to be significantly different. The experimentally measured binding affinities for these compounds from SPR are in the millimolar range. Given these weak binding affinities, it is possible that the compounds do indeed bind weakly at multiple sites.
Figure 3.

Top: Highest-scoring docked pose for the six compounds that bind to AcrA and potentiate antibiotic activity. Individual snapshots of AcrA were aligned to chain A (colored by domain) in the cryo -EM structure of the AcrAB-TolC complex (PDB entry 5NG5), which is shown in cartoon and surface representations (Green = AcrA, Blue = AcrB). Bottom: Individual docking poses for each of the six compounds highlighting interactions with residues in AcrA colored by domain.
Efflux inhibitory activity of substituted 4(3-aminocyclobutyl)pyrimidin-2-amine compounds is antibiotic-specific.
The inhibition of efflux of fluorescent probes is an assay that is broadly used to analyze the mechanism of action of EPIs.19, 45 Depending on the interactions with AcrA, AcrB, or both, the EPIs could potentially inhibit efflux of all substrates by disrupting assembly of the AcrAB-TolC complex19 or by trapping AcrB in a specific conformation.20 Alternatively, EPIs could potentiate the activities of only specific antibiotics by binding the AcrB transporter in overlapping substrate binding sites, as proposed for MC-207,110.45 Furthermore, some derivatives of MC-207,110 are not only substrates of AcrAB-TolC and competitive inhibitors but also are able to stimulate efflux of other substrates by lowering Km of the transporter to antibiotics.45, 46 We next tested whether the substituted 4(3-aminocyclobutyl)pyrimidin-2-amines are able to inhibit efflux of the fluorescent probe Hoechst H33342,20 which fluoresces upon intercalation with DNA inside the cell and is also an excellent AcrAB-TolC substrate.20 The addition of increasing concentrations of an EPI should therefore lead to an increase in fluorescence.
No increase in fluorescence was observed upon addition of any of the six compounds, indicating that efflux of H33342 was not inhibited (Figure 4). Furthermore, increasing concentrations of compounds such as 65071797 and 98577577 decreased the rate of accumulation of H33342 in a EPI concentration-dependent manner, indicating that these compounds promote efflux of this probe, as has been reported for some AcrAB-TolC substrates.45, 46 Taken together with the more efficient potentiation of ERY in the growth inhibition assays (Tables 2–3), these results suggest that these six substituted 4(3-aminocyclobutyl)pyrimidin-2-amines inhibit efflux at the substrate binding site, i.e. hydrophobic trap, of AcrB. Therefore, we also measured the binding of these six compounds to AcrB using SPR. We found that all six active hits bind the purified, immobilized AcrB transporter in the SPR assays (Figure 5). As with AcrA, the best fits for the AcrB binding sensorgrams were obtained using the two-state kinetic model and the KD and kinetic parameters values for the binding of compounds were in the same range for the two proteins (Table S5). Thus, these compounds are not only inhibitors but also are likely to be substrates of AcrB transporter.
Figure 4.

Kinetics of uptake of the fluorescent probe Hoechst H33342 (final concentration 4 μM) into WT-Pore cells in the presence of increasing concentrations of the indicated substituted 4(3-aminocyclobutyl)pyrimidin-2-amines.
Figure 5.

SPR sensorgrams (black lines) for six compounds that bind AcrB in vitro. Sensorgrams were fitted to different kinetic models and the best fits are shown as red lines. Kd values are labeled.
Potentiation in other Gram-negative pathogens
We also tested the six top hits for their ability to potentiate the activity of NOV and ERY in four other Gram-negative pathogens. We first considered WT and hyperporinated strains of A. baumannii and P. aeruginosa. The substituted 4(3-aminocyclobutyl)pyrimidin-2-amines potentiated activities of both antibiotics in A. baumannii either with native or hyperporinated OM at similar concentrations (Table 4). This result suggests that these compounds have broad-spectrum activity and permeate the OM of A. baumannii better than the OM of E. coli. In contrast, we found no potentiation of antibiotic activities in P. aeruginosa (Table S6). We also measured potentiation of antibiotic activity in K. pneumoniae and E. cloacae. None of the compounds potentiated antibiotic activity in E. cloacae. However, compounds potentiated both antibiotics in Klebsiella pneumoniae (Table 4), albeit at the same concentrations of 100–200 μM as in E. coli WT cells. Thus, some of these compounds increased the efficacy of novobiocin and erythromycin as measured by MPC4 in WT cells of both A. baumannii (up to 8-fold) and K. pneumoniae (up to 2-fold) (Table 4).
Table 4.
MIC and Potentiation Data for Top Hits in A. baumannii and K. pneumoniae.
| Aclnetobacter baumannii | Klebsiella pneumoniae | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | WT-Pore | WT | |||||||
| MIC (μM) | MPC4 (μM) | MIC (μM) | MPC4 (μM) | MIC (μM) | MPC4 (μM) | ||||
| compound | NOV | ERY | NOV | ERY | NOV | ERY | |||
| 24123034 | 200 | 100 | 100 | >200 | 100 | 200 | >200 | 200 | 100 |
| 36287038 | >200 | 25 | 200 | >200 | 50 | 200 | >200 | 100 | 100 |
| 56871955 | >200 | 50 | 200 | >200 | 50 | 100 | >200 | 200 | 200 |
| 58997260 | >200 | 50 | 50 | >200 | 100 | 100 | >200 | 200 | 100 |
| 65071797 | >200 | 50 | 100 | >200 | 100 | 50 | >200 | 200 | 100 |
| 98577577 | >200 | 100 | 200 | >200 | 200 | 100 | >200 | 100 | 200 |
Conclusions
By combining physicochemical property filtering, ensemble docking, in vitro binding studies, and in vivo potentiation assays in bacterial strains with controllable permeability barriers, we have identified a new class of EPIs with activity against several Gram-negative bacteria.Six molecules with a shared scaffold were found to potentiate the antibiotic activity of ERY and NOV in hyperporinated E. coli cells and in wild-type strains of both A. baumannii and K. pneumoniae. The physicochemical property space covered by our compound search was relatively small and not necessarily optimal. Therefore, refinement of properties and expansion of the number of molecules searched is expected to lead to the identification of more potentent EPIs.
Supplementary Material
Generated a focused library of compounds with specific physicochemical properties
Performed ensemble docking of compounds to multiple sites on AcrA
Measured in vivo activity of top compounds from docking
Identified six molecules with a shared scaffold that potentiate antibiotic activity
Acknowledgements
This work was supported by National Institutes of Health Grant AI052293 to HIZ. C.J.C. was supported by a National Science Foundation Graduate Research Fellowship under Grant No. 2017219379. SPR experiments were carried out using a Biacore T200 instrument at the Oklahoma Medical Research Foundation Biacore Facility, which is funded by Shared Instrumentation Grant S10 OD025014. This work used resources of the Compute and Data Environment for Science (CADES) at Oak Ridge National Laboratory, which is managed by UT-Battelle for the U.S. Department of Energy under contract no. DE-AC05-00OR22725. We thank Illia S. Afanasiev for help with measuring MICs and MPCs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Lewis K, Platforms for antibiotic discovery. Nat Rev Drug Discov 2013, 12 (5), 371–87. [DOI] [PubMed] [Google Scholar]
- 2.Zgurskaya HI; Lopez CA; Gnanakaran S, Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis 2015, 1 (11), 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zgurskaya HI; Rybenkov VV, Permeability barriers of Gram-negative pathogens. Ann N Y Acad Sci 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raetz CR; Whitfield C, Lipopolysaccharide endotoxins. Annu Rev Biochem 2002, 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H, Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 2003, 67 (4), 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pages JM; James CE; Winterhalter M, The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 2008, 6 (12), 893–903. [DOI] [PubMed] [Google Scholar]
- 7.Levy SB; Marshall B, Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med 2004, 10, S122–S129. [DOI] [PubMed] [Google Scholar]
- 8.Murakami S; Nakashima R; Yamashita E; Matsumoto T; Yamaguchi A, Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 2006, 443 (7108), 173–179. [DOI] [PubMed] [Google Scholar]
- 9.Murakami S; Nakashima R; Yamashita E; Yamaguchi A, Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419 (6907), 587–593. [DOI] [PubMed] [Google Scholar]
- 10.Nakashima R; Sakurai K; Yamasaki S; Hayashi K; Nagata C; Hoshino K; Onodera Y; Nishino K; Yamaguchi A, Structural basis for the inhibition of bacterial multidrug exporters. Nature 2013, 500 (7460), 102–106. [DOI] [PubMed] [Google Scholar]
- 11.Koronakis V; Sharff A; Koronakis E; Luisi B; Hughes C, Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 2000, 405 (6789), 914–919. [DOI] [PubMed] [Google Scholar]
- 12.Du D; Wang Z; James NR; Voss JE; Klimont E; Ohene-Agyei T; Venter H; Chiu W; Luisi BF, Structure of the AcrAB-TolC multidrug efflux pump. Nature 2014, 509 (7501), 512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zgurskaya HI; Nikaido H, Bypassing the periplasm: Reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA 1999, 96 (13), 7190–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnamoorthy G; Wolloscheck D; Weeks JW; Croft C; Rybenkov VV; Zgurskaya HI, Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob Agents Chemother 2016, 60 (12), 7372–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper SJ; Krishnamoorthy G; Wolloscheck D; Walker JK; Rybenkov VV; Parks JM; Zgurskaya HI, Molecular properties that define the activities of antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect Dis 2018, 4 (8), 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomovskaya O; Warren MS; Lee A; Galazzo J; Fronko R; Lee M; Blais J; Cho D; Chamberland S; Renau T; Leger R; Hecker S; Watkins W; Hoshino K; Ishida H; Lee VJ, Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob Agents Chemother 2001, 45 (1), 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto Y; Hayama K; Sakakihara S; Nishino K; Noji H; Iino R; Yamaguchi A, Evaluation of multidrug efflux pump inhibitors by a new method using microfluidic channels. PLoS One 2011, 6 (4), e18547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sjuts H; Vargiu AV; Kwasny SM; Nguyen ST; Kim HS; Ding X; Ornik AR; Ruggerone P; Bowlin TL; Nikaido H; Pos KM; Opperman TJ, Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc Natl Acad Sci U S A 2016, 113 (13), 3509–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdali N; Parks JM; Haynes KM; Chaney JL; Green AT; Wolloscheck D; Walker JK; Rybenkov VV; Baudry J; Smith JC; Zgurskaya HI, Reviving antibiotics: Efflux pump inhibitors that interact with AcrA, a membrane fusion protein of the AcrAB-TolC multidrug efflux pump. ACS infectious diseases 2017, 3 (1), 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opperman TJ; Kwasny SM; Kim HS; Nguyen ST; Houseweart C; D’Souza S; Walker GC; Peet NP; Nikaido H; Bowlin TL, Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother 2014, 58 (2), 722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen ST; Kwasny SM; Ding X; Cardinale SC; McCarthy CT; Kim HS; Nikaido H; Peet NP; Williams JD; Bowlin TL; Opperman TJ, Structure-activity relationships of a novel pyranopyridine series of Gram-negative bacterial efflux pump inhibitors. Bioorg Med Chem 2015, 23 (9), 2024–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vargiu AV; Ruggerone P; Opperman TJ; Nguyen ST; Nikaido H, Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother 2014, 58 (10), 6224–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes KM; Abdali N; Jhawar V; Zgurskaya HI; Parks JM; Green AT; Baudry J; Rybenkov VV; Smith JC; Walker JK, Identification and structure-activity relationships of novel compounds that potentiate the activities of antibiotics in Escherichia coli. J Med Chem 2017, 60 (14), 6205–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darzynkiewicz ZM; Green AT; Abdali N; Hazel A; Fulton RL; Kimball J; Gryczynski Z; Gumbart JC; Parks JM; Smith JC; Zgurskaya HI, Identification of binding sites for efflux pump inhibitors of the AcrAB-TolC component AcrA. Biophys J 2019, 116 (4), 648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebejer J-P; Charlton MH; Finn PW, Are the physicochemical properties of antibacterial compounds really different from other drugs? J Cheminform 2016, 8, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Shea R; Moser HE, Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med Chem 2008, 51 (10), 2871–2878. [DOI] [PubMed] [Google Scholar]
- 27.Silver LL, A Gestalt approach to Gram-negative entry. Bioorg Med Chem 2016, 24 (24), 6379–6389. [DOI] [PubMed] [Google Scholar]
- 28.Richter MF; Drown BS; Riley AP; Garcia A; Shirai T; Svec RL; Hergenrother PJ, Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 2017, 545 (7654), 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acosta-Gutierrez S; Ferrara L; Pathania M; Masi M; Wang J; Bodrenko I; Zahn M; Winterhalter M; Stavenger RA; Pages JM; Naismith JH; van den Berg B; Page MGP; Ceccarelli M, Getting drugs into Gram-negative bacteria: Rational rules for permeation through general porins. ACS infectious diseases 2018, 4 (10), 1487–1498. [DOI] [PubMed] [Google Scholar]
- 30.Tamber S; Hancock RE, The outer membranes of Pseudomonads In Pseudomonas, Ramos J-L, Ed. Kluwer Academic/Plenum Publishers: New York, 2004; Vol. 1, pp 575–601. [Google Scholar]
- 31.Fralick JA, Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriology 1996, 178 (19), 5803–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zgurskaya HI; Krishnamoorthy G; Ntreh A; Lu S, Mechanism and function of the outer membrane channel TolC in multidrug resistance and physiology of Enterobacteria. Front Microbiol 2011, 2, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole K, Multidrug resistance in Gram-negative bacteria. Curr Opin Microbiol 2001, 4 (5), 500–508. [DOI] [PubMed] [Google Scholar]
- 34.Sterling T; Irwin JJ, ZINC 15 - Ligand discovery for everyone. J Chem Inf Model 2015, 55 (11), 2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazel AJ; Abdali N; Leus IV; Parks JM; Smith JC; Zgurskaya HI; Gumbart JC, Conformational dynamics of AcrA govern multidrug efflux pump assembly. ACS infectious diseases 2019, 5 (11), 1926–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molecular Operating Environment (MOE), 2012.10. Chemical Computing Group Inc; 1010 Sherbooke St. West, Suite 910, Montreal, QC, Canada, H3A 2R7, 2015. [Google Scholar]
- 37.Ellingson SR; Miao Y; Baudry J; Smith JC, Multi-conformer ensemble docking to difficult protein targets. J Phys Chem B 2015, 119 (3), 1026–1034. [DOI] [PubMed] [Google Scholar]
- 38.Ellingson SR; Smith JC; Baudry J, VinaMPI: Facilitating multiple receptor high-throughput virtual docking on high-performance computers. J Comput Chem 2013, 34 (25), 2212–2221. [DOI] [PubMed] [Google Scholar]
- 39.Trott O; Olson AJ, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010, 31 (2), 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zgurskaya HI; Nikaido H, AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol 1999, 285 (1), 409–420. [DOI] [PubMed] [Google Scholar]
- 41.Tikhonova EB; Yamada Y; Zgurskaya HI, Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chemistry & Biology 2011, 18 (4), 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tikhonova EB; Wang Q; Zgurskaya HI, Chimeric analysis of the multicomponent multidrug efflux transporters from Gram-negative bacteria. J Bacteriol 2002, 184 (23), 6499–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tikhonova EB; Dastidar V; Rybenkov VV; Zgurskaya HI, Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci USA 2009, 106 (38), 16416–16421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westfall DA; Krishnamoorthy G; Wolloscheck D; Sarkar R; Zgurskaya HI; Rybenkov VV, Bifurcation kinetics of drug uptake by Gram-negative bacteria. PLoS One 2017, 12 (9), e0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinana AD; Vargiu AV; May T; Nikaido H, Aminoacyl beta-naphthylamides as substrates and modulators of AcrB multidrug efflux pump. Proc Natl Acad Sci USA 2016, 113 (5), 1405–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinana AD; Vargiu AV; Nikaido H, Some ligands enhance the efflux of other ligands by the Escherichia coli multidrug pump AcrB. Biochemistry 2013, 52 (46), 8342–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


