Mutations affecting spliceosomal proteins are frequently found in hematological malignancies. DDX41/Abstrakt is a metazoan-specific spliceosomal DEAD-box RNA helicase found to be recurrently mutated in relapsing cases of acute myeloid leukemia, as well as in inherited myelodysplastic syndromes...
Keywords: SACY-1/DDX-41, spliceosome, myelodysplastic disorders, C. elegans
Abstract
Mutations affecting spliceosomal proteins are frequently found in hematological malignancies, including myelodysplastic syndromes and acute myeloid leukemia (AML). DDX41/Abstrakt is a metazoan-specific spliceosomal DEAD-box RNA helicase that is recurrently mutated in inherited myelodysplastic syndromes and in relapsing cases of AML. The genetic properties and genomic impacts of disease-causing missense mutations in DDX41 and other spliceosomal proteins have been uncertain. Here, we conduct a comprehensive analysis of the Caenorhabditis elegans DDX41 ortholog, SACY-1. Biochemical analyses defined SACY-1 as a component of the C. elegans spliceosome, and genetic analyses revealed synthetic lethal interactions with spliceosomal components. We used the auxin-inducible degradation system to analyze the consequence of SACY-1 depletion on the transcriptome using RNA sequencing. SACY-1 depletion impacts the transcriptome through splicing-dependent and splicing-independent mechanisms. Altered 3′ splice site usage represents the predominant splicing defect observed upon SACY-1 depletion, consistent with a role for SACY-1 in the second step of splicing. Missplicing events appear more prevalent in the soma than the germline, suggesting that surveillance mechanisms protect the germline from aberrant splicing. The transcriptome changes observed after SACY-1 depletion suggest that disruption of the spliceosome induces a stress response, which could contribute to the cellular phenotypes conferred by sacy-1 mutant alleles. Multiple sacy-1/ddx41 missense mutations, including the R525H human oncogenic variant, confer antimorphic activity, suggesting that their incorporation into the spliceosome is detrimental. Antagonistic variants that perturb the function of the spliceosome may be relevant to the disease-causing mutations, including DDX41, affecting highly conserved components of the spliceosome in humans.
MUTATIONS affecting components of the spliceosome are frequently found in hematological malignancies, including myelodysplastic syndromes (MDS; Yoshida et al. 2011; reviewed by Yoshida and Ogawa 2014; Coltri et al. 2019), which comprise a heterogeneous set of myeloid neoplasms characterized by anemia and cytopenia that progress to acute myeloid leukemia (AML) to varying degrees (Tefferi and Vardiman 2009). The genetic properties and genomic impacts of disease-causing missense mutations in spliceosomal proteins have been uncertain. Nonetheless, mutations affecting spliceosomal components are predictive of poor clinical outcomes in AML patients (Papaemmanuil et al. 2016). Exactly how mutations in spliceosomal components contribute to malignancy is uncertain. An attractive model, but one that has not been firmly established, is that aberrant splicing might interfere with tumor suppressor activity. This model suggests that malignancy-associated spliceosomal mutations are either loss-of-function or confer antimorphic (i.e., antagonistic) activity. Importantly, genome sequencing data in patients is currently being used in the clinic to generate personalized prognoses, with the idea of optimally targeting existing therapies and generating new treatment strategies (Grinfeld et al. 2018). One potential therapeutic approach under development is the discovery of splicing inhibitors (Effenberger et al. 2017; Kim and Abdel-Wahab 2017; DeNicola and Tang 2019). Although mutations affecting several spliceosomal proteins appear to be beneficial to tumor cells, excessive splicing abnormalities are likely to be lethal to all cells. Splicing inhibitors have been demonstrated to target tumor cells with splicing mutations by inducing excessive splicing abnormalities, but cells with intact splicing machinery appear to be resistant to these agents (Seiler et al. 2018). In fact, several new splicing inhibitors are currently in clinical trials.
The spliceosomal components frequently affected in MDS, occurring in ∼60–70% of patients, include the biochemically well-defined factors SF3B1, SRSF2, and U2AF1 (Yoshida et al. 2011; reviewed by Yoshida and Ogawa 2014). More recent studies have implicated DDX41, a DEAD-box RNA helicase highly conserved in metazoans, whose precise biochemical function in the spliceosome is less well understood (Ding et al. 2012; Polprasert et al. 2015; Cardoso et al. 2016; Lewinsohn et al. 2016; Li et al. 2016; Diness et al. 2018; reviewed by Maciejewski et al. 2017). DDX41 appears to be specifically recruited to the catalytically active C complex (Jurica et al. 2002; Bessonov et al. 2008), which performs the second step of splicing, in which the 5′ and 3′ exons are ligated and an intronic lariat is released. DDX41 is one of many spliceosomal proteins specific to metazoans and not found in budding yeast (Bessonov et al. 2008).
Whole genome sequencing studies suggest that DDX41 mutations are associated with hematological malignancies that are considered to be different clinical entities. For example, examination of clonal evolution of relapsed AML cases identified DDX41 as one of several genes found to be mutated in secondary, but not primary, tumors, suggesting that de novo DDX41 mutations might contribute to disease progression (Ding et al. 2012). By contrast, studies of familial AML syndromes suggest that preexisting germline DDX41 mutations in trans to newly arising somatic mutations cause the development of hematological malignancies (Polprasert et al. 2015; Cardoso et al. 2016; Lewinsohn et al. 2016; Li et al. 2016). Germline biallelic DDX41 missense mutations were recently reported in two siblings that exhibited intellectual disability, psychomotor delays, and facial and skeletal dysmorphologies, with one sibling presenting with childhood leukemia (Diness et al. 2018). Other work suggests that DDX41 might be a multifunctional protein; in addition to its nuclear function in RNA splicing, it has been suggested to function as a cytoplasmic DNA sensor in a signaling pathway that detects infecting double-stranded DNA and initiates an antiviral interferon response (Zhang et al. 2011; Parvatiyar et al. 2012; Stavrou et al. 2015, 2018; reviewed by Jiang et al. 2017). However, more recent work suggests that cyclic GMP-AMP synthase (cGAS) functions as the major DNA sensor and is several orders of magnitude more effective in inducing interferon beta synthesis than DDX41 (Sun et al. 2013). Two studies, one of DDX41 and another of its Drosophila ortholog, Abstrakt, suggested a role in regulating translation of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 (Peters et al. 2017) and the Inscuteable protein (Irion et al. 2004), respectively, though the exact mechanism of these activities has not been elucidated, and indirect effects acting at the level of splicing were not addressed in these studies.
To better understand the highly conserved functions of DDX41, we undertook a comprehensive molecular genetic analysis of its ortholog, sacy-1, in the nematode Caenorhabditis elegans. The cGAS and STING-mediated cytoplasmic DNA sensing pathway is absent in C. elegans (Wu et al. 2014), suggesting that this model may be useful for addressing spliceosomal functions of DDX41/SACY-1. Our prior work identified the DEAD-box helicase SACY-1 as a negative regulator of oocyte meiotic maturation functioning in the germline upstream of the TIS11 CCCH zinc-finger RNA-binding proteins OMA-1 and OMA-2 (Kim et al. 2012). Genetic analysis also established roles for SACY-1 in regulating the hermaphrodite sperm-to-oocyte switch, and in preventing necrotic cell death of gametes. Genetic experiments further suggested an essential role for sacy-1(+) in early embryos and larvae that appeared to be maternally rescued. At the time of our original study, searchable databases of the scientific literature had not yet annotated DDX41 (or its Drosophila ortholog, Abstrakt) as spliceosomal components identified by proteomics. We therefore did not recognize that SACY-1 was likely involved in splicing.
In this study, we undertook a comprehensive molecular genetic analysis of the functions of SACY-1 in C. elegans. Our results demonstrate that SACY-1 is a component of the C. elegans spliceosome that exhibits genetic interactions with other spliceosomal components. Depletion of SACY-1 in the germline or soma was found to have major impacts on the transcriptome through splicing-dependent and splicing-independent mechanisms. Alterations in 3′ splice site selection represent the most prevalent changes in splicing patterns observed following SACY-1 depletion, consistent with its function as a component of the spliceosomal C complex, which carries out the second step in splicing. Missplicing events are more prevalent upon SACY-1 depletion in the soma than in the germline, leading us to suggest that surveillance mechanisms protect the germline from aberrant splicing events. The gene expression changes observed after SACY-1 depletion suggest that perturbations of spliceosomal function might induce a stress response, which could be relevant to the cellular phenotypes conferred by sacy-1 mutant alleles. Further, our genetic results reveal that multiple sacy-1 missense mutations confer a dosage-sensitive antimorphic activity, most consistent with the possibility that they compromise the function of the spliceosome by perturbing the action of other spliceosomal proteins. Moreover, the human oncogenic DDX41 R525H mutation was introduced into the C. elegans genome using CRISPR-Cas9 genome editing and found to exhibit weak antagonistic activity. Antagonistic activities that perturb the function of the spliceosome, as defined by genetic analysis in C. elegans, may be relevant to the disease-causing mutations affecting highly conserved components of the spliceosome in humans, including DDX41.
Materials and Methods
C. elegans strains and genetic analysis
The genotypes of strains used in this study are reported in Supplemental Material, Table S1. Genes and mutations are described in WormBase (www.wormbase.org; Harris et al. 2014) or in the indicated references. Culture and genetic manipulations were conducted at 20° unless specified otherwise. The following mutations were used: LGI– fog-1(q253ts), dpy-5(e61), gld-1(tn1478), unc-13(e51), unc-13(e1091), lin-41(n2914), lin-41(tn1541[gfp::tev::s-tag::lin-41]), sacy-1(tm5503), sacy-1(tn1385), sacy-1(tn1479), sacy-1(tn1480), sacy-1(tn1481Mog), sacy-1(tn1482), sacy-1(tn1602), sacy-1(tn1603), sacy-1(tn1604), sacy-1(tn1605), sacy-1(tn1606), sacy-1(tn1607), sacy-1(tn1608), sacy-1(tn1609), sacy-1(tn1610), sacy-1(tn1611), sacy-1(tn1612), sacy-1(tn1615), sacy-1(tn1616), sacy-1(tn1617), sacy-1(tn1632[3xFLAG::PreScission protease site::gfp::tev::s-tag::sacy-1]), sacy-1(tn1880[aid::gfp::tev::myc::sacy-1]), and sacy-1(tn1887); LGII–tra-2(e2020), ieSi57[eft-3p::TIR1::mRuby::unc-54 3′UTR + Cb unc-119(+)], ieSi64[gld-1p::TIR1::mRuby::gld-1 3′UTR + Cb unc-119(+)]; LGIII–unc-119(ed3); LGIV–unc-24(e138), fem-3(e1996), and dpy-20(e1282); LGV–acy-4(ok1806), her-1(hv1y101), emb-4(sa44), unc-51(e369), and fog-2(oz40). The following rearrangements were used: hT2[bli-4(e937) let-?(q782) qIs48] (I;III), tmC18[dpy-5(tmIs1236) + pmyo-2::mCherry] I (Dejima et al. 2018), mIn1[dpy-10(e128) mIs14] II, and tmC12[egl-9(tmIs1194) + pmyo-2::Venus] V (Dejima et al. 2018). The following transgenes were used: tnEx37[acy-4(+) + sur-5::gfp], tnEx159[gfp::sacy-1 + pDPMM0016B(unc-119(+))] and tnIs102[sacy-1p::gfp::tev::s-tag::sacy-1 + Cb unc-119(+)] III.
For the analysis of genetic interactions between sacy-1(tn1481) and fem-3(e1996), non-Unc non-Dpy non-GFP animals from sacy-1(tn1481)/hT2[bli-4(e937) let-?(q782) qIs48]; fem-3(e1996)/unc-24(e138) dpy-20(e1282) were individually cultured and scored for germline phenotypes. Following scoring, the fem-3 genotype of each animal was scored by conducting PCR with primers fem-3 F2 and fem-3 R2 and sequencing the products.
To map the cold-sensitive (15°) and temperature-sensitive (25°) phenotypes of sacy-1(tn1480), 34 Unc non-Dpy recombinants were obtained from sacy-1(tn1480)/dpy-5(e61) unc-13(e1091) heterozygotes. The recombinant chromosomes were bred to homozygosity and scored for the presence or absence of the sacy-1(tn1480) mutation by conducting PCR with primers H27M09.1F1 and H27M09.1R4, and sequencing purified PCR products with primer H27M09.1F2. We found that 7 of the 34 recombinants contained sacy-1(tn1480) and were cold-sensitive and temperature-sensitive. By contrast, 27 recombinants were sacy-1(+) and grew at 15 and 25°. These data indicate that sacy-1(tn1480) mutation is inseparable from the cold-sensitive and temperature-sensitive phenotypes (e.g., within ∼0.06 map units). In addition, 32 Dpy non-Unc recombinants were selected. Interestingly, all the homozygous recombinants were fertile at both 15 and 25°, including the 22 recombinants that contained the sacy-1(tn1480) mutation. Although these dpy-5(e61) sacy-1(tn1480) recombinants grew at 15 and 25°, they produced appreciable numbers of dead embryos and grew more slowly than their sacy-1(+) counterparts. This result suggests that the dpy-5(e61) mutation suppresses the cold-sensitive and temperature-sensitive phenotypes of sacy-1(tn1480). Previous work has shown that mutant alleles of collagen genes can suppress temperature-sensitive mutations in other gene products, possibly by triggering a stress response (Levy et al. 1993; Maine and Kimble 1993; Nishiwaki and Miwa 1998). That dpy-5(e61) suppresses sacy-1(tn1480) was further shown by constructing dpy-5(e61) sacy-1(tn1480)/sacy-1(tn1480) unc-13(e1091) heterozygotes (n = 30), of which 20 exhibited the sacy-1(tn1480) sperm-defective phenotype at 25° and 10 were fertile. Thus, dpy-5(e61) exhibits semidominance for its effects on body morphology and for suppression of sacy-1(tn1480). To examine the dominant high incidence of males (Him) phenotype of sacy-1(tn1480) and its interaction with sacy-1(tn1887), we compared the percentage of males produced at 25° by dpy-5(e61)/sacy-1(tn1480) unc-13(e1091) and dpy-5(e61) sacy-1(tn1887)/sacy-1(tn1480) unc-13(e1091) heterozygotes.
RNA interference
Genome-wide RNA interference (RNAi) screening employed the Ahringer feeding library (Kamath et al. 2003) using the RNAi culture media described by Govindan et al. (2006) at 22°. The empty vector L4440 was used as a control. The identity of RNAi clones was verified by DNA sequencing. Gene-specific RNAi was performed by placing wild-type or sacy-1(tn1385rf) gravid hermaphrodites on RNAi medium seeded with double-stranded RNA (dsRNA)-expressing Escherichia coli (Timmons and Fire 1998). The gravid hermaphrodites were immediately treated with 20% bleach to release the F1 embryos. Phenotypes were assessed 3–4 days later. In scoring RNAi clones for enhanced defects specific to the sacy-1(tn1385rf) mutant background, we could not reliably score clones that produced high levels of embryonic lethality in the wild-type genetic background. For quantification of phenotypes, sterility and gamete degeneration were scored in the F1 generation, and embryonic lethality was scored in the F2 generation produced by the RNAi-treated F1 animals.
Immunofluorescence, fluorescent labeling, and microscopy
Dissected gonads were fixed in 3% paraformaldehyde as described (Rose et al. 1997). Fixed gonads were stained with rabbit anti-RME-2 antibody (Grant and Hirsh 1999; kindly provided by B. Grant, Rutgers University, 1:50), a mixture of two purified mouse monoclonal anti-MSP antibodies (Kosinski et al. 2005, each at 1:300), rabbit anti-phospho-histone H3 (Ser10) antibody (1:400; Millipore). Secondary antibodies were Alexa 488-conjugated donkey anti-rabbit antibodies (1:500; Jackson ImmunoResearch) and Cy3-conjugated goat anti-mouse antibodies (1:500; Jackson ImmunoResearch). 4′,6-diamidino-2-phenylindole (DAPI) was used to detect DNA. DIC and fluorescent images were acquired on a Zeiss motorized Axioplan 2 microscope with either a 40× Plan-Neofluar (numerical aperture 1.3) or a 63× Plan-Apochromat (numerical aperture 1.4) objective lens using a AxioCam MRm camera and AxioVision software (Zeiss). DIC and GFP fluorescent images of sacy-1(tm5503); tnEx159 and sacy-1(tn1632) adults were acquired on a Nikon A1R resonant scanning confocal microscope using a Plan Fluor 40× Oil DIC (numerical aperture 1.3) objective lens. Image acquisition utilized the large image function of NIS-Elements AR (v. 5.11.00) with an image overlap setting of 15%.
Genome editing and generation of transgenics
CRISPR-Cas9 genome editing used pRB1017 to express single guide RNA (sgRNA) under control of the C. elegans U6 promoter (Arribere et al. 2014). The sequences of all oligonucleotides used are listed in Table S2. To generate sgRNA clones, annealed oligonucleotides were ligated to BsaI-digested pRB1017 plasmid vector, and the resulting plasmids were verified by Sanger sequencing. pDD162 served as the source of Cas9 expressed under control of the eef-1A.1/eft-3 promoter (Dickinson et al. 2013). Indels were targeted to exon 2 of sacy-1 using sacy-1 sgRNA7 (pCS520). The injection mix contained pCS520 (25 ng/μl), pDD162 (50 ng/μl), and Pmyo-2::tdTomato (4 ng/μl). sacy-1(tn1602–tn1612) were recovered from injections into DG3913 lin-41(tn1541[gfp::tev::s-tag::lin-41]) and sacy-1(tn1615–tn1617) were recovered from injections into the wild type (strain N2).
An N-terminal gfp fusion to endogenous sacy-1, sacy-1(tn1632[3xflag::PreScission protease site::gfp::tev::s-tag::sacy-1]), was constructed using sacy-1 sgRNA1 (pCS486) and a repair template generated by conducting the PCR with oligonucleotide primers sacy-1 5HAF and sacy-1 3HAR, using a gfp::tev::s-tag::sacy-1 recombineered fosmid (SK212; Kim et al. 2012) as template. Genome editing employed the dpy-10 coconversion method (Arribere et al. 2014). The injection mix contained pJA58 (7.5 ng/μl), AF-ZF-827 (500 nM), pCS486 (50 ng/μl), repair template (50 ng/μl), and pDD162 (50 ng/μl) and was injected into wild-type worms. Correct targeting was verified by conducting PCR with primer pairs GFP_7215 and H27M09.1_R5 and GFP_1094R and H27M09.1_seqF1 followed by DNA sequencing. DG3768 was constructed using microparticle bombardment with SK212 as described (Praitis et al. 2001).
An N-terminal auxin-inducible degron (aid) fusion to sacy-1, sacy-1(tn1880[aid::gfp::myc::sacy-1]), was constructed using sacy-1 sgRNA1 and a repair template generated by conducting the PCR with oligonucleotide primers sacy-1 AID5F and sacy-1 AID3R using a wee-1.3::aid::gfp::myc clone (pCS575, C.A. Spike, unpublished results) as template. The injection mix, prepared as described above, was injected into CA1352 worms. sacy-1(tn1880[aid::gfp::myc::sacy-1]) was identified by screening the progeny of 414 F1 Roller animals for GFP fluorescence. Correct targeting was verified by conducting PCR with primer pairs GFP_R1 and H27M09.1_F5 and GFP_F1 and H27M09.1_R5 followed by DNA sequencing.
The R525H mutation in DDX41 was imported into C. elegans (e.g., SACY-1[R534H]) using genome editing (Paix et al. 2014) with sacy-1 sgRNA11 and sacy-1 sgRNA12 and a single-stranded repair oligonucleotide (sacy-1 GM1), which introduces the R534H mutation and two synonymous changes to alter the protospacer adjacent motif and to facilitate screening using an introduced AvaI restriction site. The injection mix contained pJA58 (7.5 ng/μl), AF-ZF-827 (500 nM), sacy-1 sgRNA11 (25 ng/μl), sacy-1 sgRNA12 (25 ng/μl), sacy-1 GM1 (500 nM), and pDD162 (50 ng/μl) and was injected into wild-type worms. Edited loci were verified by PCR and DNA sequencing using primers sacy-1 seq F1 and sacy-1 seq R1.
Antibody production, purification, and western blotting
sacy-1 cDNA sequences were cloned into the E. coli expression vector pMal-c2 to create an inducible fusion protein wherein maltose binding protein was fused to amino acids 411–578 of SACY-1 (MBP::SACY-1(411–578)). MBP::SACY-1(411–578) was column- and gel-purified and used to immunize rabbits. Immunizations and sera collection were performed using standard protocols (Cocalico Biologicals, Reamstown, PA). Rabbit antibody (R217) was affinity purified, and was suitable for use in western blots with partially purified SACY-1 preparations. Hybridoma cell lines producing anti-GFP monoclonal antibodies 12A6 and 4C9 (Sanchez et al. 2014) were obtained from the Developmental Studies Hybridoma Bank and prepared as described (Tsukamoto et al. 2017). Proteins were separated using NuPAGE 4–12% Bis-Tris gels (Invitrogen, Carlsbad, CA) and visualized after western blotting. Blots were blocked with 5% nonfat dried milk. Primary antibodies used to detect proteins were affinity-purified rabbit anti-SACY-1(411–578) R217 antibody (100 ng/ml) and rabbit anti-GFP NB600-308 antibody (250 ng/ml; Novus Biologicals). The secondary antibody used for western blots was peroxidase-conjugated donkey anti-rabbit antibody (1:30,000; Jackson ImmunoResearch). Detection was performed using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
Identification of SACY-1-associated proteins
Tandem affinity purification of SACY-1 was conducted using strains DG4068 and DG4070 using modifications of a previously described protocol (Tsukamoto et al. 2017). Immunopurified proteins were precipitated with 16.7% trichloroacetic acid (TCA), washed with acetone at −20°, and briefly separated on a 12% NuPAGE Bis-Tris gel, stained with Colloidal Blue Staining Kit (Invitrogen). Lanes were subdivided into eight gel slices and mass spectrometry was performed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School) using an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific). Protein identification used the Sequest software program (Thermo Fisher Scientific) to match the fragmentation pattern of tryptic peptides to the C. elegans proteome. The data were filtered to a 1–2% peptide false discovery rate. File S1 reports the mass spectrometry results and the additional filtering criteria for identifying nonspecific interactions.
Single-step immunopurifications were also conducted on a smaller scale using 80 mg of protein lysate from DG3768 sacy-1(tm5503) I; unc-119(ed3) tnIs102[sacy-1p::gfp::tev::s-tag::sacy-1 + Cb unc-119(+)] III. GFP::TEV::S-tag::SACY-1 and associated proteins were isolated using monoclonal anti-GFP antibody 12A6 and TEV protease cleavage as described (Tsukamoto et al. 2017). In addition to our standard conditions (300 mM KCl) higher stringency washes were conducted using 1 M KCl or 300 mM KCl with 5 μg/ml RNase A.
RNA sequencing
The auxin-inducible degradation system (Zhang et al. 2015) was used to deplete SACY-1 using strain backgrounds in which TIR1 was expressed in the germline (CA1352) or soma (CA1200). Experimental (DG4700 and DG4703) and control strains (CA1352 and CA1200) were grown on peptone-enriched nematode growth medium with NA22 as a food source. Embryos were isolated by alkaline hypochlorite treatment (20% bleach and 0.5 N NaOH), washed in M9 buffer and allowed to hatch overnight in the absence of food. For each of three biological replicates, 60,000 L1-stage larvae were cultured on two 150 × 15 mm Petri dishes containing peptone-enriched medium with OP50. The worms were grown to the young adult stage and harvested by washing off the plates with M9, then placed on fresh plates containing peptone-enriched medium and 2 mM auxin seeded with OP50. Plates were cultured in the dark at 20° for 24 hr. The worms were then harvested and washed with M9 repeatedly to reduce the presence of E. coli. Total RNA was isolated using TRIzol LS Reagent (Invitrogen) and the RNeasy Micro Kit (Qiagen, Valencia, CA). Poly(A)+ RNA was selected from 1 μg of total RNA using the NEBNext Ultra Kit (New England Biolabs, Ipswich, MA). Libraries were prepared and sequenced by Genewiz (South Plainfield, NJ). Paired-end reads of 150 bp were obtained on an Illumina HiSeq 4000 instrument with an average depth >31 million reads per sample.
Bioinformatics
After trimming adapters with Trim Galore (v0.6.0) and cutadapt (v1.18), reads were assessed for quality with FastQC (v0.11.8), mapped to the WBcel235/ce11 genome with STAR (v2.7.2a) guided by gene annotations defined in Ensembl (release 97) and sorted and indexed with samtools (v1.7). Gene-level abundance was estimated for Ensembl defined annotations using the featureCounts function in the Bioconductor package Rsubread (v1.28.1). An average of 28 million high-quality (MAPQ > 55) reads mapped to annotated genes within each sample. Principal component analysis and inspection of 5′ vs. 3′ read coverage indicated that one soma control sample (CA1200-2) contained degraded RNA and was excluded from further analysis. Differential gene expression of Ensembl defined genes was determined using DESeq2 (v1.26.0). P values were adjusted for multiple test correction using the Benjamini–Hochberg procedure. The fold change, adjusted P values, the mean number of counts across samples, and the number of complementary DNA fragments per kilobase of transcript per million mapped reads (FPKM) were used to define differentially expressed genes. Gene ontology (GO) data were obtained from WormBase release WS273, and analyzed taking length bias into account using the Goseq (v1.38.0) package. Novel transcripts in each of the high-quality samples and in the previously published GSE57109 (Ortiz et al. 2014) dataset were identified using StringTie (v2.0.4) and merged together with the Ensembl annotations to generate a comprehensive annotation set. These annotations were used with RMATS (v4.0.2 turbo) to determine statistically significant differences, expressed as false discovery rates (FDRs), for splicing events between conditions. Coverage data were visualized with Gviz (v 1.30.0). A custom R script with details for the analysis and figure generation is available at https://github.com/micahgearhart/sacy1.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains and reagents are available upon request. RNA sequencing data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through accession number GSE144003. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11825673.
Results
SACY-1 is a component of the C. elegans spliceosome
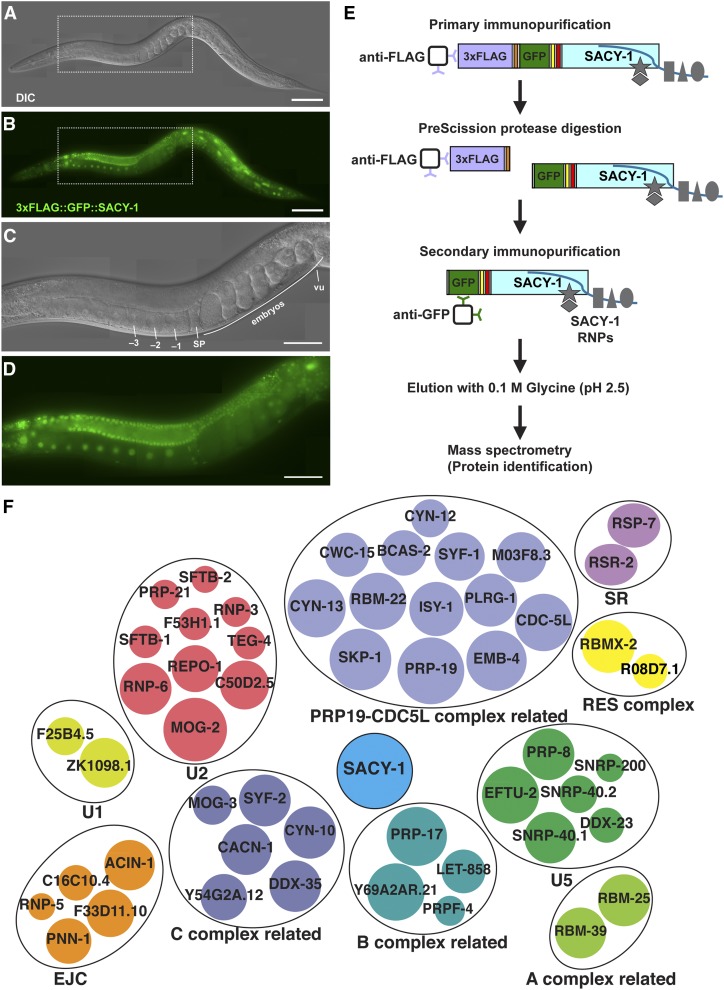
To characterize SACY-1-associated proteins, we conducted tandem affinity purifications using strains in which we used CRISPR-Cas9 genome editing to insert 3xFLAG and eGFP tags at the SACY-1 N-terminus, separated by a PreScission protease recognition sequence (Figure 1 and Figure S1). The resulting sacy-1(tn1632[3x flag::PreScission::gfp::tev::s-tag::sacy-1]) strain was viable and fertile and exhibited no apparent germline or somatic defects. Although 3xFLAG::GFP::SACY-1 is expressed in all cells except sperm, it appears most abundant in the female germline (Figure 1, A–D). Thus, we conducted purifications from protein lysates prepared from adult animals in which the germline was feminized (experiment I) and also from adult hermaphrodites (experiment II). This approach was taken to utilize the most abundant source of SACY-1, and also to potentially gain insights of how it might function as a negative regulator of meiotic maturation in the absence of sperm, though our data were not informative from this standpoint. In both experiments, we found that 3xFLAG::GFP::SACY-1 associated with 52 proteins defined as spliceosomal proteins in other systems (Figure 1F and Table S3; Jurica et al. 2002; Bessonov et al. 2008). We also detected nine additional spliceosomal proteins in the purification from the female, but not the hermaphrodite, genetic background (Table S4), but this might be a consequence of the fact that more protein extract was used in that experiment. We also detected 29 other proteins in our tandem affinity purifications (Table S5). Homologs of several of these factors have been implicated in the regulation of RNA splicing, including NRDE-2 (Jiao et al. 2019), CIR-1 (Maita et al. 2005; Kasturi et al. 2010), and CDK-12 (Rodrigues et al. 2012). These biochemical studies, taken together with the results from the genome-wide RNAi screen described below, are consistent with the possibility that both specific and pleiotropic defects conferred by sacy-1 mutant alleles result from spliceosomal defects.
Figure 1.
Tandem affinity purification of SACY-1 to identify associated proteins. DIC (A and C) and fluorescence (B and D) photomicrographs of an adult hermaphrodite generated by CRISPR-Cas9 genome editing to express an epitope-tagged version of SACY-1 suitable for tandem affinity purification (strain DG4070 sacy-1(tn1632[3xflag::PreScission protease site::gfp::tev::s-tag::sacy-1]) I). The regions within the dotted rectangles are magnified in (C and D). Oocytes (−1, −2, −3), the spermatheca (sp), and the vulva (vu) are indicated. Bars, 100 μm in (A and B); 50 μm in (C and D). (E) Strategy for tandem affinity purification. Cleavage with PreScission protease releases GFP::SACY-1 and associated proteins from the affinity matrix, whereas the fragment with the 3xFLAG epitope and nonspecifically bound proteins are retained on the matrix. The second purification step utilized an anti-GFP affinity resin and a low pH elution. Western blot analysis of fractions from the tandem affinity purification procedure analyzed with anti-GFP or R217 anti-SACY-1(411–578) antibodies are shown in Figure S1. (F) Spliceosomal proteins reproducibly associated with SACY-1 by tandem affinity purification. Spliceosomal proteins are organized into spliceosomal subcomplexes according to Bessonov et al. (2008) or the supplemental references in Table S3. The primary data on which figure is based are presented in File S1 and Table S3. The area of each circle represents the maximal percentage coverage of each protein in a single gel slice from the purification from DG4068 (see Table S3). For example, the percent coverage of SACY-1 was 78.9% and the percent coverage of MOG-2 was 56.1%. The names of the human orthologs of the C. elegans proteins are listed in Table S3.
Biochemical studies established that DDX41/Abstrakt is a component of the C complex, which requires the presence of an RNA splicing substrate to form, and is sufficient to catalyze exon ligation in the absence of additional factors (Jurica et al. 2002; Bessonov et al. 2008). Treatment of isolated C complexes with high salt was observed to remove DDX41/Abstrakt and dozens of other spliceosomal proteins from a C complex RNP core (Bessonov et al. 2008). To begin to identify spliceosomal proteins that might associate with SACY-1 at high stringency, we conducted purifications in the presence of 1 M KCl or RNase A. We identified eight spliceosomal proteins that maintained high levels of association with SACY-1 (recovery of at least 50% of peptides) under both stringent conditions (RBM-39, RNP-6, DDX-35, ACIN-1, CYN-13, PRPF-4, CYN-12, and C16C10.4; Table S6). Interestingly, DDX35/PPWD1 is a C complex protein, which is removed from the C complex RNP together with DDX41/Abstrakt after high salt treatment (Bessonov et al. 2008). Additional studies will be needed to determine whether SACY-1 and DDX-35 (or the other spliceosomal proteins) might interact directly.
Genetic interactions between sacy-1 and several genes encoding spliceosomal proteins
To better understand the function of sacy-1, we conducted a genome-wide RNAi screen for loci that resulted in more severe phenotypes when knocked down by RNAi in a sacy-1 reduction-of-function (rf) genetic background [sacy-1(tn1385rf)] than in the wild type. We screened 18,101 RNAi clones from the Ahringer RNAi library, and identified five clones that produced increased levels of sterility, gamete degeneration, or embryonic lethality when knocked down in the sacy-1(tn1385rf) genetic background (Table 1). Because these three phenotypes are observed in a variety of sacy-1 mutant alleles (Kim et al. 2012 and as described below), we consider these genetic interactions to reflect an enhancement of sacy-1 mutant phenotypes or to be synthetic lethal interactions with the rf allele. The five RNAi clones target the transcripts of three genes (Table 1): mog-2 (one clone), Y111B2A.25 (one clone), and emb-4 (three clones). To test whether sacy-1 expression and/or localization is affected by RNAi of mog-2, Y111B2A.25, or emb-4, we conducted RNAi of these genes in sacy-1(tm5503) mutant animals expressing the rescuing gfp::sacy-1 transgene (tnEx159). In no case did we observe that an RNAi treatment altered the expression or localization of the GFP::SACY-1 transgene; the expression level and predominant nuclear localization of GFP::SACY-1 after RNAi was similar to that of the control animals (S. Kim and D. Greenstein, unpublished results). This result suggests that the defects caused by the RNAi treatments in the sacy-1(tn1385rf) mutant background are not caused by changes in the expression or localization of SACY-1.
Table 1. RNAi of sacy-1 enhancer loci increases the penetrance of germline or lethal phenotypes in sacy-1(tn1385) reduction-of-function mutants.
| RNAia | Genotype | Sterileb (%) | Gamete degenerationb (%) | Embryonic lethalc (%) |
|---|---|---|---|---|
| L4440 (control) | Wild type | 0 (n = 338) | 0 (n = 338) | 1 (n = 674) |
| sacy-1(tn1385) | 0 (n = 256) | 0 (n = 256) | 1 (n = 502) | |
| mog-2 (II-3D16) | Wild type | 4 (n = 172) | 0 (n = 54) | 7 (n = 574) |
| sacy-1(tn1385) | 43 (n = 254) | 47 (n = 72) | 95 (n = 426) | |
| Y111B2A.25 (III-6G22) | Wild type | 22 (n = 230) | 1 (n = 94) | 84 (n = 463) |
| sacy-1(tn1385) | 95 (n = 272) | 16 (n = 110) | 93 (n = 42) | |
| cacn-1d (II-9E09) | Wild type | 100 (n = 164) | 35 (n = 40) | ND |
| sacy-1(tn1385) | 100 (n = 224) | 81 (n = 52) | ND | |
| emb-4 (V-12E12) | Wild type | 2 (n = 184) | 0 (n = 110) | 12 (n = 337) |
| sacy-1(tn1385) | 89 (n = 176) | 42 (n = 82) | ND | |
| emb-4 (V-12E14) | Wild type | 4 (n = 288) | 0 (n = 96) | 13 (n = 421) |
| sacy-1(tn1385) | 91 (n = 202) | 80 (n = 120) | ND | |
| emb-4 (V-12E16) | Wild type | 4 (n = 210) | 0 (n = 96) | 9 (n = 433) |
| sacy-1(tn1385) | 91 (n = 326) | 87 (n = 140) | ND |
ND, not determined.
RNAi clones showing genetic interactions with sacy-1 are listed with the target gene name in italics and the location of the clone in the RNAi library in parentheses. The identity of clones was verified by DNA sequencing.
Sterility and gamete degeneration were scored by DIC microscopy on the first day of adulthood 24 hr post-L4 at 22°. Gonad arms were scored as sterile if they did not produce embryos and exhibited defects in gametogenesis. Number of gonad arms scored is reported.
Embryonic lethality was measured by conducting daily egg lays over the reproductive lifespan and determining the number of embryos that failed to hatch by 48 hr after egg laying. The number of embryos scored is reported.
cacn-1 was not initially identified as an enhancer of sacy-1 during the genome-wide RNAi screen because cacn-1(RNAi) results in complete sterility in both sacy-1(tn1385) and wild-type animals. However, we determined that RNAi to the Y111B2A.25 pseudogene likely targets cacn-1 (see text for details).
mog-2(RNAi) induces a higher penetrance of sterility, gamete degeneration, and embryonic lethality in the sacy-1(tn1385) mutant genetic background in comparison to the wild type (Figure S2 and Table 1). mog-2 encodes the U2 small nuclear ribonucleoprotein (snRNP) A’ (Zanetti et al. 2011), which is a constitutive component of the spliceosome (Jurica et al. 2002; Bessonov et al. 2008, 2010; Herold et al. 2009).
Similarly, Y111B2A.25(RNAi) specifically enhances the penetrance of multiple sacy-1 mutant phenotypes, including sterility, gamete degeneration, and embryonic lethality (Figure S2 and Table 1). Y111B2A.25 is annotated as a pseudogene (Agarwal et al. 2010; www.wormbase.org). Y111B2A.25 is part of an operon, and the expressed sequence tag (EST) data show that the Y111B2A.25 locus is transcribed, but the transcript lacks protein-coding ability. In C. elegans, ∼40 bp of sequence identity is sufficient to induce off-target RNAi effects (Rual et al. 2007). Use of the Basic Local Alignment Search Tool (BLAST) indicates that Y111B2A.25(RNAi) might target the cacn-1 locus, which encodes a spliceosomal protein and shares ∼200 bp of sequence identity with Y111B2A.25. To test whether the enhanced sterility induced by Y111B2A.25(RNAi) in the sacy-1(tn1385) genetic background might be explained by an off-target effect to cacn-1, we conducted cacn-1(RNAi) and found that the cacn-1(RNAi) induces complete sterility in both the sacy-1(tn1385) and wild-type animals (Figure S2 and Table 1). Interestingly, under the cacn-1(RNAi) condition, the sacy-1(tn1385) animals show additional phenotypes, such as high penetrance of a protruding vulva (Pvl) phenotype, and slow growth compared to the wild type, suggesting a genetic interaction between cacn-1 and sacy-1 that might be partially masked by the strong phenotypes induced by cacn-1(RNAi). Thus, we reasoned that the short identity shared between Y111B2A.25 and cacn-1 dsRNA might induce strong sterility in the sacy-1(tn1385rf) genetic background, but not in the wild type, through weaker interference with cacn-1. To test this possibility, we systematically reduced the efficacy of the cacn-1(RNAi) response by serially diluting the cacn-1(RNAi)-inducing bacteria with bacteria containing the empty vector control (L4440). Consistent with the possibility that Y111B2A.25(RNAi) targets cacn-1, limiting the efficacy of the cacn-1(RNAi) response revealed specific enhancement of sterility, gamete degeneration, and embryonic lethality in the sacy-1(tn1385) genetic background (Table S7). Notably, the response to limited cacn-1(RNAi) exhibited by sacy-1(tn1385) animals was remarkably similar to their response to Y111B2A.25(RNAi) (Table S7). The human and Drosophila orthologs of CACN-1 have been identified as components of spliceosomal C complexes (Jurica et al. 2002; Bessonov et al. 2008, 2010; Herold et al. 2009; Fica et al. 2019). Like DDX41/Abstrakt, Cactin is recruited to the C complex of the spliceosome.
In addition to mog-2 and Y111B2A.25, we identified three different RNAi clones targeting the emb-4 locus as strong enhancers of the sacy-1(tn1385) sterility and gamete degeneration phenotypes (Figure S2 and Table 1). emb-4 encodes a nuclear protein orthologous to human Aquarius/IBP160/KIAA0560/fSAP164, an intron-binding spliceosomal protein with a helicase-like domain (Sam et al. 1998; Jurica et al. 2002; Bessonov et al. 2008; Herold et al. 2009; De et al. 2015; Bertram et al. 2017; Haselbach et al. 2018). To extend these RNAi results, we examined genetic interactions between sacy-1 and emb-4, employing the emb-4(sa44) reduction-of-function allele. When combined with the sacy-1(tm5503) null allele, we observed enhancement of lethal vulval rupture and protruding vulva (Pvl) phenotypes in sacy-1(tm5503); emb-4(sa44) double mutants (Table 2). We also observed enhancement of these phenotypes in sacy-1(tn1385); emb-4(sa44) double mutants, which were derived from sacy-1(tn1385)/+; emb-4(sa44) parents (Table 2). Interestingly, the F1 progeny of fertile sacy-1(tn1385); emb-4(sa44) homozygous adults exclusively produced dead embryos or arrested L1-stage larvae, unlike each of the single mutants, which were viable and fertile (Table 3).
Table 2. Genetic interactions between sacy-1 and emb-4: enhancement of germline and somatic sacy-1 mutant defects.
| Genotype | Vulval rupture (%) | Sterile and Pvla (%) | Sterilea (%) | Fertile (%) |
|---|---|---|---|---|
| sacy-1(tm5503) (n = 284) | 3 | 1 | 96 | 0 |
| sacy-1(tm5503)/+; emb-4(sa44)b (n = 205) | 0 | 0 | 0 | 100 |
| sacy-1(tm5503); emb-4(sa44)b (n = 242) | 83 | 16 | 1 | 0 |
| sacy-1(tn1385) (n = 278) | 0 | 0 | 0 | 100 |
| sacy-1(tn1385)/+; emb-4(sa44)b (n = 201) | 0 | 0 | 0 | 100 |
| sacy-1(tn1385); emb-4(sa44)b,c (n = 144) | 36 | 4 | 3 | 57d |
Sterile animals exhibit the sacy-1(lf) gamete degeneration phenotype.
The hT2(qIs48) balancer chromosome, which is dominantly marked with GFP, was used to differentiate between sacy-1(tm5503) heterozygotes and homozygotes.
The progeny of sacy-1(tn1385)/hT2(qIs48); emb-4(sa44) hermaphrodites; the balancer chromosome provides maternal sacy-1(+) function. The fertile F1 progeny of these animals are maternal-effect lethal, see Table 3.
These adult hermaphrodites produce a majority of embryos that fail to hatch, see Table 3.
Table 3. Genetic interactions between sacy-1 and emb-4: enhancement of embryonic lethality.
| Genotypea | Embryonic lethal (%) | L1 lethal (%) | Viable (%) |
|---|---|---|---|
| sacy-1(tn1385) (n = 949) | 1 | 0 | 99 |
| emb-4(sa44) (n = 1348) | 4 | 0 | 96 |
| sacy-1(tn1385); emb-4(sa44)b (n = 620) | 97 | 3 | 0 |
The number of embryos examined.
The F1 progeny of fertile sacy-1(tn1385); emb-4(sa44) parents derived from the sacy-1(tn1385)/+; emb-4(sa44) heterozygotes analyzed in Table 2.
The genetic interactions between sacy-1 and the spliceosomal proteins identified in our genome-wide RNAi screen for sacy-1 enhancers (MOG-2, EMB-4, and CACN-1) likely reflect biochemical interactions because these three proteins were very well represented in our tandem affinity purifications of SACY-1-associated proteins (∼35–56% peptide coverage; Table S3). Taken together, these genetic and biochemical interactions are consistent with the possibility that the multiple sacy-1 mutant phenotypes result from compromised functions of the spliceosome.
Impact of sacy-1 on the transcriptome
Depletion of SACY-1 using the auxin-inducible degradation system:
RNA sequencing studies using human patient samples suggested a role for DDX41 in splice site selection for a small number of human genes (Polprasert et al. 2015). Thus, we sought to address the impact of SACY-1 on the transcriptome by exploiting the power of the C. elegans system for transcriptomics under well-controlled conditions. We chose to use the auxin-inducible degradation system (Zhang et al. 2015) to acutely deplete SACY-1 in the adult stage, and, thus, avoid indirect impacts on the transcriptome arising as a developmental consequence of strong loss-of-function phenotypes (e.g., germline degeneration and cell fate changes). Because SACY-1 is expressed in the germline and soma, we used strains bearing germline (CA1352 ieSi64) or somatically expressed (CA1200 ieSi57) TIR1 F-box proteins to deplete AID::GFP::SACY-1 in each tissue individually (Figure S3 and Figure S4). Depletion of AID::GFP::SACY-1 in the germline starting at approximately the L3 stage phenocopied the gamete degeneration phenotype observed in sacy-1 null mutants (see below) in a small proportion of the animals (3 of 270; Figure S3). This result is consistent with genetic mosaic analysis showing that the gamete degeneration phenotype is cell autonomous (Kim et al. 2012), and it also highlights the difficulty of recapitulating null phenotypes through auxin-inducible degradation. When AID::GFP::SACY-1 is depleted in the germline starting at the L4 stage, many F1 progeny arrest as embryos or larvae, consistent with the idea that maternally contributed sacy-1(+) activity is essential. Animals that escape the lethality and progress to adulthood often display the germline degeneration phenotype (40%; n = 139). When AID::GFP::SACY-1 is depleted in the soma beginning at the L4 stage, the resulting F1 progeny grew very slowly, taking ∼4–6 days to reach adulthood (instead of 2.5 days) and were sterile (Figure S3). Taken together, depletion of SACY-1 using the auxin-inducible degradation system resulted in a rf condition less severe than the null phenotype, but more severe than conferred by rf missense alleles (see below).
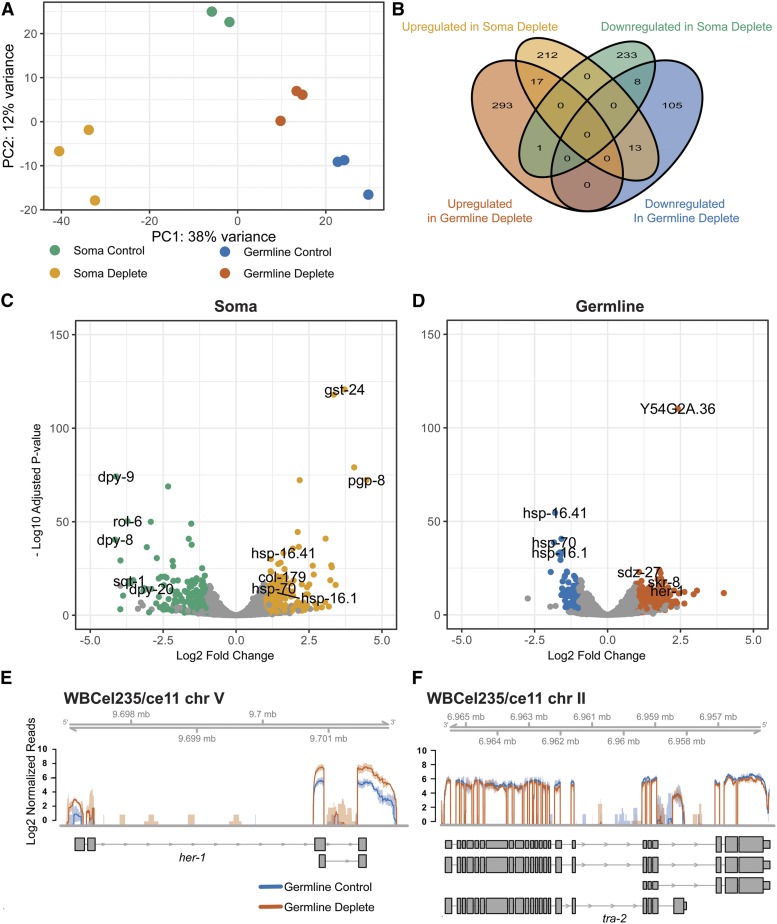
For analysis of transcriptomes, we exposed young adult hermaphrodites to auxin for 24 hr before preparing total RNA for sequencing. Examination of the worms showed that AID::GFP::SACY-1 was depleted from the targeted tissues (Figure S4). Total RNA was prepared from each of three biological replicates and their respective controls, which were the parent strains expressing TIR1 in the germline (ieSi64) or soma (ieSi57) also treated with auxin. Poly(A)+ mRNA was sequenced using 150 bp paired-end reads, and the sequencing reads were aligned to the WB235/ce11 genome. Principal component analysis (PCA) revealed that the biological replicates clustered together (Figure 2A), which is indicative of experimental reproducibility. However, PCA indicated that the control strains for the germline (CA1352) and soma (CA1200) depleted samples did not cluster together, which indicates that, under these conditions, the two strain backgrounds exhibit marked differences in their transcriptomes (Figure 2A)—a finding that was further confirmed with a more granular assessment of mRNA expression level differences of individual genes (Figure S5A). Thus, in our analysis, we compared the germline- and soma-depleted SACY-1 transcriptomes only to their respective controls.
Figure 2.
Transcriptome changes upon SACY-1 depletion. (A) PCA comparison of RNA-seq data of control strains and the experimental samples in which SACY-1 was depleted in the germline or soma, as indicated. Three biological replicates were analyzed for each sample; however, one of the control samples for the soma depletion exhibited evidence of RNA degradation and was excluded from the analysis. (B) A Venn diagram showing the limited overlap of upregulated genes (twofold; adjusted P < 0.05, FPKM deplete > 2.5 and mean counts > 25) and downregulated (twofold; adjusted P < 0.05, FPKM control > 2.5 and mean counts > 25) genes in the RNA-seq datasets. (C and D) Volcano plots showing the log2 fold change in expression vs. the −log10 of the adjusted P value of genes following SACY-1 depletion in the soma (C) or germline (D). (E and F) The normalized coverage of sequencing reads across her-1 (E) and tra-2 (F) following depletion of SACY-1 in the germline. The solid lines represent the mean of the biological replicates and shaded regions represent the corresponding confidence interval. Note, the pattern of tra-2 splicing is not affected.
Changes in transcript abundance following SACY-1 depletion:
We observed two classes of transcriptome alterations upon depletion of SACY-1 in the germline or soma: changes in transcript abundance and alterations in splicing patterns. In terms of transcript abundance, we observed 242 down-regulated genes (twofold down-regulation, adjusted P < 0.05, FPKM in soma control ≥ 2.5, mean counts across samples > 25) in the RNA samples depleted for somatic SACY-1 (Figure 2, B and C and File S2). Notably these down-regulated genes included many cuticle collagen genes and genes affecting cuticular morphology and body size (col-17, col-41, col-46, col-47, col-90, col-128, col-149, dpy-3, dpy-4, dpy-5, dpy-6, dpy-8, dpy-9, dpy-13, dpy-20, lon-3, mlt-7, qua-1, rol-6, rol-8, sqt-1, and sqt-2). Consistent with this observation, the top enriched GO term for transcripts reduced in abundance in the SACY-1 soma-depleted samples was “cuticle development involved in collagen and cuticulin-based cuticle molting cycle” (Figure S5B). We also observed 242 up-regulated genes (twofold up-regulation, adjusted P < 0.05, FPKM in SACY-1 soma-deplete ≥ 2.5, mean counts across samples > 25) in the SACY-1 soma-depleted samples (Figure 2, B and C and File S2). The top enriched GO term for transcripts with increased abundance in the SACY-1 soma-depleted samples related to cellular responses to heat stress, the unfolded protein response, and innate immune responses (Figure S5B), suggesting that the organism might perceive the reduction of sacy-1(+) function as a stressor, and then mount a response that then alters the transcriptome.
In the SACY-1 germline-depleted samples, we observed 126 down-regulated genes (twofold down-regulation, adjusted P < 0.05, FPKM in germline control ≥ 2.5, mean counts across samples > 25; Figure 2, B and D and File S2). The top enriched GO terms for transcripts with decreased abundance in the SACY-1 germline-depleted samples included the response to heat stress and the unfolded protein response (Figure S5B), suggesting the response to sacy-1(+) depletion differs between the soma and germline. We observed 311 transcripts to exhibit increased abundance in the SACY-1 germline-depleted sample (twofold up-regulation, adjusted P < 0.05, FPKM in SACY-1 germline-deplete ≥ 2.5, mean counts across samples > 25; Figure 2, B and D and File S2). The top enriched GO terms for transcripts with increased abundance in the SACY-1-germline depleted samples included SCF-dependent proteasomal ubiquitin-dependent protein catabolic processes (Figure S5B). As discussed below, this observation may be relevant to the role of SACY-1 in germline sex determination.
Alteration of splicing patterns following SACY-1 depletion:
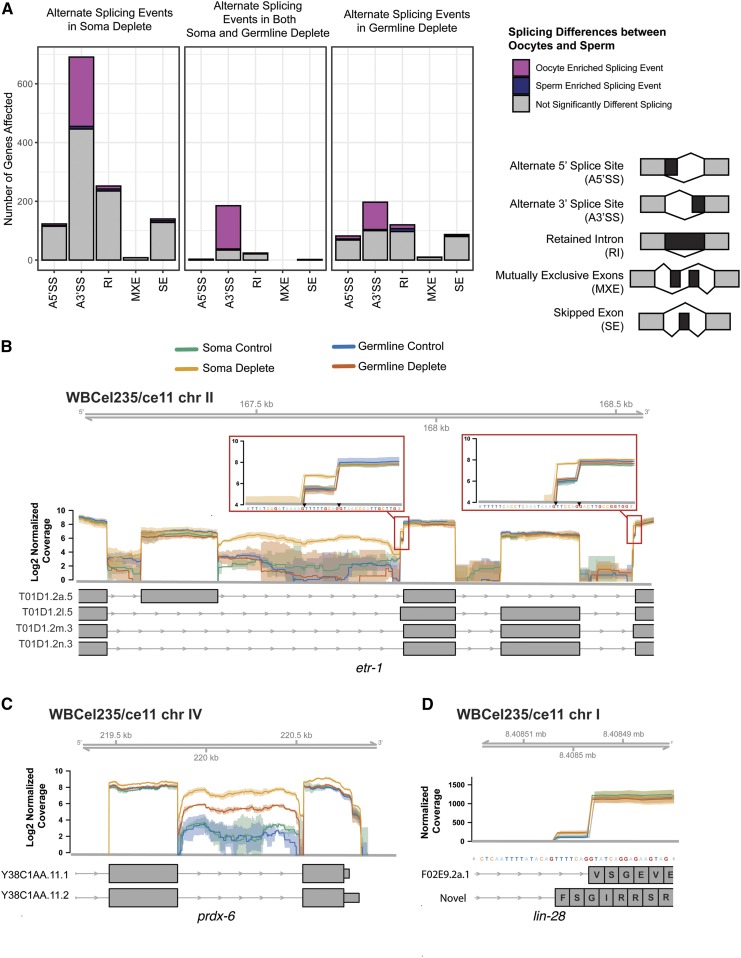
In the SACY-1 soma-depleted samples, we observed significant (FDR < 0.05) alterations in splicing patterns for 1606 transcripts (Figure 3A). These splicing alterations fell into several broad classes: the use of alternative 5′ splice sites, the use of alternative 3′splice sites, abnormal splicing within an exon, skipped exons, and retained introns. The largest class of splicing changes was the use of alternative 3′ splice sites, consistent with the fact that DDX41 was shown to be recruited to the C complex, which mediates the second step in splicing (Bessonov et al. 2008). Multiple splicing defects were sometimes observed within a single gene. For example, in the case of the RNA-binding protein ETR-1, which has multiple isoforms and is expressed in the soma and germline (Boateng et al. 2017), depletion of SACY-1 results in intron retention and multiple alterations in 3′-splice-site usage (Figure 3B). In the SACY-1 germline-depleted samples we observed significant (FDR < 0.05) alterations in splicing for 796 transcripts (Figure 3A). Thus, splicing defects appeared less prevalent in the SACY-1 germline-depleted samples than the SACY-1 soma-depleted samples. One possibility is that nonsense mediated decay or other surveillance pathways actively clear misspliced mRNAs from the germline. Some alternative splicing events were observed in both the RNA preparations depleted for SACY-1 in the germline and the soma (Figure 3A). For example, we observed retention of an intron in prdx-6 mRNA in both experiments (Figure 3C). Likewise, we observed alternative 3′ splice site selection for the heterochronic gene lin-28 in both SACY-1-depleted samples, which results in a change in reading frame (Figure 3D).
Figure 3.
Quantification of altered splicing patterns upon SACY-1 depletion. (A) Bar graphs showing the number of genes with statistically significant (FDR < 0.05) changes in splicing patterns. The legend at the right depicts the nature of the observed splicing changes: A5′SS, Alternate 5′ Splice Site; A3′SS, Alternate 3′ Splice Site; RI, Retained Intron; MXE, Mutually Exclusive Exons; SE, Skipped Exon (B–D) Examples of alterations in splicing patterns following SACY-1 depletion in the germline or soma as indicated. The etr-1 gene shows pronounced intron retention and two alternatively spliced 3′ sites in the SACY-1 soma depleted (gold) sample (B). A subset of etr-1 transcript annotations are shown. The prdx-6 gene has a retained intron in the SACY-1 soma depleted (gold) and germline depleted (red) samples (C). The soma and germline depleted samples have an increase in the usage of alternate splice acceptor in the lin-28 gene that results in an altered reading frame (D). The solid lines represent the mean of the biological replicates and shaded regions represent the corresponding confidence interval.
Germline sex-specific splicing patterns and the involvement of SACY-1:
Alternative splicing events enriched in oogenic or spermatogenic germlines were previously identified (Ortiz et al. 2014). Consequently, we reanalyzed their dataset in order to directly compare with the alternative splicing events observed upon SACY-1 depletion. We observed 1600 genes for which there was a significant (FDR < 0.05) germline sex-specific splicing pattern (Figure S6 and File S3). We noted that upon SACY-1 depletion in either the soma or germline, oocyte-enriched splicing events were favored (Figure 3A and Figure S6). This result suggests that SACY-1 plays a role in the selection of 3′ splice sites for many genes, and raises the possibility that the appropriate balance of splice variants may play a role in cellular differentiation.
sacy-1 reduction-of-function mutation enhances germline tumor formation
Prior work showed that mutational or RNAi treatments affecting the function of multiple spliceosomal components enhance weak gain-of-function (gf) mutations in glp-1/Notch, resulting in the ectopic proliferation of undifferentiated germ cells in the proximal gonad arm (Mantina et al. 2009; Kerins et al. 2010; Wang et al. 2012). This phenotype, which is referred to as a proximal proliferation or Tumorous phenotype, can also be enhanced by mutations in genes that function in GLP-1/Notch signaling (reviewed by Hubbard and Schedl 2019). Since the preceding analysis suggested that SACY-1 is a functional component of the spliceosome, we predicted that the sacy-1(tn1385rf) mutation would enhance the Tumorous phenotype conferred by the glp-1(ar202gf) mutation (Pepper et al. 2003). Thus, we constructed the sacy-1(tn1385rf); glp-1(ar202gf) double mutant and analyzed its phenotype at the permissive temperature of 15° at which glp-1(ar202gf) mutants exhibit a low penetrance of the Tumorous phenotype (Table 4; Pepper et al. 2003). Under these conditions (40 hr post-L4 at 15°), very few glp-1(ar202gf) young adult hermaphrodites were observed to exhibit a proximal proliferation phenotype with undifferentiated germ cells in the proximal gonad arm (∼0.8%, Table 4). By contrast, many sacy-1(tn1385rf); glp-1(ar202gf) adults exhibited a Tumorous phenotype (∼50%, Table 4). This phenotype was not observed in sacy-1(tn1385rf) single mutants (Table 4). To examine this phenotype further, we stained dissected and fixed gonads with the phosphohistone H3(Ser10) M-phase marker. Of 25 sacy-1(tn1385); glp-1(ar202) gonads scored, 16 (64%) contained phosphohistone H3-positive undifferentiated germ cells in the proximal gonad arm at 15°. The average number of proximal phosphohistone H3-positive germ cells in the Tumorous gonads was 22 ± 12. None of the sacy-1(tn1385) (n = 18) or glp-1(ar202) (n = 31) dissected gonads examined contained phosphohistone H3-positive undifferentiated germ cells in the proximal gonad arm at 15°. These results are consistent with the idea that the sacy-1(tn1385rf) mutation, though homozygous viable and fertile (brood size ∼350; Kim et al. 2012), compromises the function of the spliceosome, as assessed in a sensitized genetic background.
Table 4. sacy-1(tn1385rf) enhances the glp-1(ar202) Tumorous phenotype.
| Strain | Gonad arms containing mitotic undifferentiated germ cells in the proximal gonad arma |
|---|---|
| sacy-1(tn1385[G533R]) | 0 (n = 256) |
| glp-1(ar202) | 0.8 (n = 364) |
| sacy-1(tn1385[G533R]); glp-1(ar202) | 49.6 (n = 415) |
| sacy-1(tn1887[R534H]); glp-1(ar202) | 0 (n = 62) |
The percentage of young adult hermaphrodites were examined by DIC microscopy ∼40 hr post-L4 at 15°. The number of gonad arms scored is listed.
sacy-1(tm5503) and sacy-1(tn1615) define the null phenotype
The strong sacy-1 mutant allele, sacy-1(tm5503), deletes exons 2 and 3 and a portion of exon 4, and is homozygous sterile, displaying the gamete degeneration phenotype (Figure 4A; Kim et al. 2012). Consistent with the idea that sacy-1(tm5503) defines the null phenotype, an antibody specific to a portion of the DEAD-box domain downstream of the tm5503 deletion (residues 411–578) fails to detect a protein product in extracts from sacy-1(tm5503) adults (Figure S1C). To further define the sacy-1 null phenotype, we used CRISPR/Cas9 genome editing to generate indels upstream of the DEAD-box-encoding regions by targeting Cas9 double-strand DNA breaks to exon 2 with an efficient sgRNA. We generated sacy-1 indels in both wild-type as well as lin-41(tn1541[gfp::lin-41]) hermaphrodites, the latter serving to provide a marker for oocyte development (Spike et al. 2014a,b). In these experiments, we generated 14 new sacy-1 alleles (tn1602–tn1612 and tn1615–tn1617). Of these, 13 displayed the gamete degeneration phenotype, again consistent with this representing the null phenotype. Not surprisingly, GFP::LIN-41 levels declined and the protein became undetectable as oocytes degenerated (D. Greenstein, unpublished results). One new CRISPR-Cas9-induced allele, sacy-1(tn1615), was sequenced, and found to result from a 10-bp deletion at the end of exon 2, which is predicted to introduce a stop codon prior to the DEAD-box domain (Figure 4). sacy-1(tn1615) and sacy-1(tm5503) exhibited indistinguishable phenotypes (Table 5), consistent with the possibility that they define the null phenotype. Among the CRISPR-Cas9-induced alleles, sacy-1(tn1617) was exceptional in that it was homozygous viable and fertile, though slow growing, despite the fact that the deletion removes the initiation codon of sacy-1 (Figure 4A). This exceptional allele might utilize an alternative start codon just prior to the DEAD-box domain, although this possibility was not explored. Since null mutations in sacy-1 result in hermaphrodite sterility, there is the possibility that maternal sacy-1(+) activity contributes to the development of the germline and soma. Indeed, when the gamete degeneration phenotype is delayed through germline feminization, the mating of sacy-1(tm5503) null females to wild-type males produces embryos that arrest prior to morphogenesis (Kim et al. 2012).
Figure 4.
(A) The structure of sacy-1. Newly isolated mutations reported in this study are displayed in red font beneath the exons. The mutations in blue font shown above the exons were reported previously (Kim et al. 2012). The extent of two deletions, tm5503 and tn1615, that result in sacy-1 null mutations are shown with black bars. A third deletion, tn1617, which is a reduction-of-function mutation, is shown with a gray bar. (B) A protein sequence alignment of SACY-1 (NP_491962.1), Drosophila Abstrakt (NP_524220.1), and human DDX41 (NP_057306.2). Mutations isolated in C. elegans are shown above that sequence, whereas the human mutations associated with myelodysplastic syndromes are shown beneath the human sequence. Conserved domains [DEAD-box domain (DEADc), helicase domain (HELICc), and zinc finger domain (ZnF)] and motifs (Q, I Ia, Ib, II, III, IV, V, and VI) are indicated as described by Henn et al. (2012). (C) The locations of SACY-1 missense mutations are shown on structures of the DDX41 DEADc (Omura et al. 2016) and HELICc (Schütz et al. 2010) domains. The side chains of the amino acids in the human structure are labeled with amino acid numbering that corresponds to the SACY-1 missense mutations in this study.
Table 5. sacy-1 alleles relevant to this study.
| Allele | Alteration | Phenotypes | Inferred activity |
|---|---|---|---|
| Loss-of-function alleles | |||
| sacy-1(tm5503)a | 619 bp deletion | Sterile, gamete degeneration | Likely null |
| sacy-1(tn1615)b | 10 bp deletion | Sterile, gamete degeneration | Likely null |
| sacy-1(tn1385)a | G533R | Viable and fertile, suppresses acy-4 sterility, suppresses fog-2 sterility | Reduction of function |
| sacy-1(tn1391)a | G473R | Viable and fertile, suppresses acy-4 sterility, suppresses fog-2 sterility | Reduction of function |
| sacy-1(tn1440)a | G331R | Viable and fertile, suppresses acy-4 sterility, suppresses fog-2 sterility | Reduction of function |
| sacy-1(tn1482)b | D506N | Viable and fertile, suppresses fog-2 sterilityc | Reduction of function |
| sacy-1(tn1483)b | G269E | Viable and fertile, suppresses fog-2 sterilityc,d | Reduction of function |
| Alleles with antagonistic activity | |||
| sacy-1(tn1479)b | G504E | Sterile, adult lethal (rupture) or gamete degeneration | Strong loss of function with antimorphic activity |
| sacy-1(tn1480)b | H527Y | Viable and fertile at 20° and suppresses fog-2 sterility.c Embryonic lethal or larval arrest at 15°. Sterile and spermatogenesis-defective at 25° | Loss of function with antimorphic activity, dominant Him |
| sacy-1(tn1481)b | P222L | Sterile, masculinization of germline | Recessive gain-of-function |
| sacy-1(tn1887)b | R534H | Viable and fertile | Weak antagonistic activity; enhances the dominant Him phenotype of sacy-1(tn1480) |
This work.
Suppression of acy-4 sterility was not tested.
sacy-1(tn1483) adult hermaphrodites have smaller gonad arms, suggesting effects on germline proliferation (T. Tsukamoto and D. Greenstein, unpublished results).
Reduction-of-function sacy-1 mutations in C. elegans affect highly conserved residues in the DEAD-box and helicase domains
To better understand the functions and activities of the highly conserved SACY-1/DDX41 protein (Figure 4B), we conducted forward genetic screens for new sacy-1 mutations, taking advantage of the fact that reductions of sacy-1 function by mutation or RNAi can suppress the self-sterility of fog-2 null mutations (Kim et al. 2012), which is caused by a failure to produce sperm (Schedl and Kimble 1988). Thus, we conducted a noncomplementation screen for new mutations that enable fertility in trans to the sacy-1(tn1385) rf allele in the fog-2(oz40) genetic background (Figure S7). In a screen of 15,577 haploid genomes, we isolated five new sacy-1 missense alleles (tn1479–tn1483; Figure 4; see Table 5 for a list of all sacy-1 alleles central to this work and their properties). Phenotypic analyses, which will be described in detail below (see Table 5 and Table 6), indicate that three of these missense alleles (tn1479, tn1480, and tn1481) appear to confer antagonistic activities. All the sacy-1 missense alleles thus far isolated alter highly conserved amino acids, and several of these mutations are nearby or in subdomains of the DEAD-box affected by DDX41 mutations found in human neoplasms (Figure 4B). These missense alleles were modeled onto the crystal structures of DEADc and the HELICc domains of DDX41 (Schütz et al. 2010; Omura et al. 2016) and found likely to be surface accessible, suggesting that some of these mutant alleles could interfere with the function of other protein components of the spliceosome.
Table 6. sacy-1(tn1479), sacy-1(tn1480), and sacy-1(tn1481) confer antagonistic activity.
| Allele | Class | Gamete degenerationa | Vulval rupturea | Moga,b | T (°) |
|---|---|---|---|---|---|
| sacy-1(tm5503) (n = 143) | Strong loss-of-function | 5.6 | 94.4 | 0 | 15° |
| sacy-1(tm5503) (n = 96) | Strong loss-of-function | 96.9 | 3.1 | 0 | 20° |
| sacy-1(tm5503) (n = 106) | Strong loss-of-function (from tm5503/+ parents) | 98.0 | 2.0 | 0 | 25° |
| sacy-1(tm5503)c (n = 231) | Strong loss-of-function (from tm5503/tn1480 parents) | 14.7 | 85.3 | 0 | 25° |
| sacy-1(tn1615) (n = 92) | Strong loss-of-function | 97.8 | 2.2 | 0 | 20° |
| sacy-1(tn1479) (n = 170) | Strong loss-of-function with antimorphic activity | 11.2 | 88.8 | 0 | 20° |
| sacy-1(tn1481) (n = 125) | Recessive gain-of-function | 0 | 0 | 100 | 20° |
The percentage of adult hermaphrodites exhibiting the reported phenotype is shown. Adults were scored 24 hr post-L4 at 20 or 25° or 48 hr post-L4 at 15°.
sacy-1(tn1481) adult hermaphrodites produce large numbers of sperm but no oocytes and are sterile.
The sacy-1(tm5503) progeny of sacy-1(tm5503)/unc-13(e1091) sacy-1(tn1480) hermaphrodites grown at 25°.
Novel sacy-1 mutant alleles appear to antagonize essential functions of the spliceosome
A strong loss-of-function sacy-1 mutation is more severe than a null allele:
The strongest newly isolated allele, sacy-1(tn1479[G504E]), confers a more severe phenotype than sacy-1 null alleles—the majority of sacy-1(tn1479) adult hermaphrodites die by vulval rupture at 20° (Figure S8 and Table 6). This phenotype is observed only in a small minority of sacy-1(tm5503) hermaphrodites at 20° (Table 6). This observation suggests that sacy-1(tn1479) is a strong loss-of-function allele that confers antagonistic activity. One possibility is that the SACY-1[G504E] product is nonfunctional but incorporates into the spliceosome and antagonizes its function.
A recessive gain-of-function sacy-1 mutation masculinizes the hermaphrodite germline:
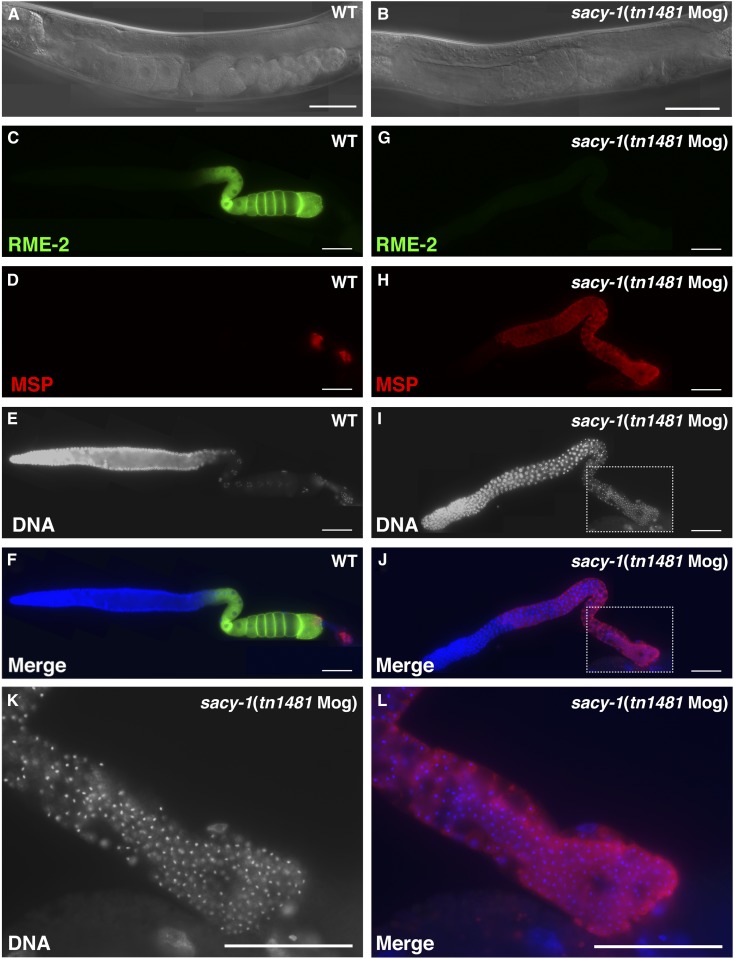
A novel mutation isolated in the noncomplementation screen was tn1481, which confers a masculinization of germline (Mog) phenotype (Figure 5 and Table 6). All sacy-1(tn1481) homozygous hermaphrodites produce excess numbers of sperm but no oocytes (n = 125). Staining of dissected gonads from adults showed that whereas all wild-type hermaphrodite gonad arms examined (n = 64) expressed both the major sperm protein (MSP) and the RME-2 oocyte yolk receptor, all sacy-1(tn1481) gonad arms (n = 178) expressed only MSP but not RME-2 (Figure 5). In our noncomplementation screen, we also isolated a gld-1 Mog allele, tn1478, as a dominant suppressor of fog-2(oz40). gld-1(tn1478) results from the same G248R amino acid substitution reported for the gld-1(q93) Mog allele (Francis et al. 1995a,b; Jones and Schedl 1995). Thus it was important to determine whether the sacy-1(tn1481[P222L]) mutation was the cause of the Mog phenotype. This was ascertained by crossing a GFP::SACY-1 extrachromosomal array (tnEx159) into the sacy-1(tn1481) genetic background. The tnEx159 extrachromosomal array expresses GFP::SACY-1 in the germline and soma at wild-type levels and rescues the sacy-1(tm5503) null allele (Figure S9). We found that all sacy-1(tn1481); tnEx159[gfp::sacy-1 + unc-119(+)] hermaphrodites (n = 30) produced oocytes and sperm and were self-fertile. This result established that the P222L mutation in SACY-1 causes the Mog phenotype.
Figure 5.
sacy-1(tn1481) adult hermaphrodites exhibit a masculinization of germline (Mog) sterile phenotype. DIC images of (A) wild-type and (B) sacy-1(tn1481) adult hermaphrodites. The wild-type animal contains oocytes and sperm and produces embryos but the sacy-1(tn1481) animal only produces sperm. (C–L) Dissected gonads stained for the RME-2 yolk receptor (C and G), the major sperm protein (D and H), or DNA (E, I, and K). Merged images are also shown (F, J, and L). The sacy-1(tn1481) mutant overproduces sperm to the exclusion of oocytes and is sterile. This phenotype is completely penetrant. Bar, 50 μm.
The suppression of fog-2 sterility by rf sacy-1 mutations is consistent with sacy-1(+) possessing a function that promotes the oocyte fate; this function is nonessential, however, because the strongest loss-of-function sacy-1 alleles are able to produce oocytes, which nevertheless undergo necrotic degeneration. Thus, the sacy-1(tn1481) Mog phenotype suggests this mutant allele, although recessive, possesses an activity antagonistic to this oocyte-promoting function in the sperm-to-oocyte switch. To genetically characterize sacy-1(tn1481) further, we analyzed the phenotype of sacy-1(tn1481)/sacy-1(tm5503 null) heterozygotes. Whereas all sacy-1(tn1481) homozygotes (n = 50) displayed a Mog phenotype, all sacy-1(tn1481)/sacy-1(tm5503) heterozygotes (n = 48) produced both oocytes and sperm and were self-fertile. This result suggests that the sacy-1(tn1481) Mog phenotype is dosage sensitive—two copies of the SACY-1(P222L) protein produce a Mog phenotype, whereas one copy does not. Thus, the SACY-1(P222L) protein possesses an activity that a null allele cannot provide. If this activity resulted from a loss of a sacy-1 function, the expectation would be that the sacy-1(tn1481)/sacy-1(tm5503 null) heterozygotes should exhibit a Mog phenotype, which is not the case. Thus, the sacy-1(tn1481Mog) allele appears to be a recessive gain-of-function mutation that might antagonize proteins that normally function with SACY-1. Given that many spliceosomal proteins functioning at different stages of the splicing process are needed for the sperm-to-oocyte switch (Kerins et al. 2010; reviewed by Zanetti and Puoti 2013), we suggest that incorporation of SACY-1(P222L) into the spliceosome is detrimental to its roles in germline sex determination. Although the SACY-1(P222L) protein confers an antagonistic activity, it likely possesses reduced activity compared to the wild-type protein because it was isolated by virtue of failing to complement sacy-1(tn1385rf).
Genetic interactions between sacy-1 and regulators of germline sex determination:
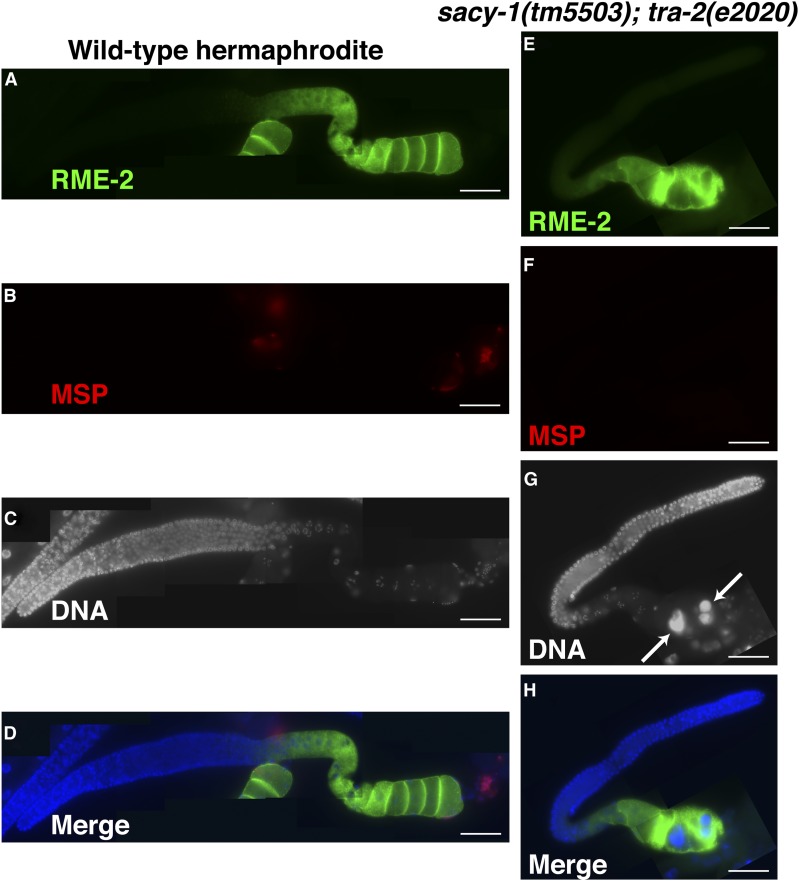
In C. elegans, a genetic hierarchy controls germline sex determination (Figure 6). The failure of sacy-1(RNAi) to suppress the sterility of the dominant strongly feminizing tra-2(e2020) mutation, which deletes GLD-1 binding sites within the tra-2 3′-UTR, was interpreted in the context of a model in which sacy-1(+) promotes the oocyte fate in opposition to fog-2 and gld-1 at the level of tra-2 (Figure 6; Kim et al. 2012). Because the evaluation of potential interactions between sacy-1 and tra-2 relied on sacy-1(RNAi), there was the concern that this treatment reduced, but did not eliminate, the function of sacy-1. Thus, we reevaluated the interaction between tra-2 and sacy-1 genetically. In the first approach, we combined the sacy-1(tm5503) null allele with tra-2(e2020). We analyzed the sexual fate of the germline by staining dissected gonads from adult animals with oocyte (RME-2) and sperm (MSP) markers. Whereas all wild-type gonad arms examined (n = 30) expressed RME-2 and MSP, all gonad arms of sacy-1(tm5503); tra-2(e2020) animals (n = 26) expressed only RME-2 and not MSP (Figure 7). This result is consistent with a model in which sacy-1 promotes the oocyte fate by promoting the function of tra-2 (Figure 6). This possibility is consistent with the finding that sacy-1(rf) alleles and sacy-1(RNAi) suppress the sterility of fog-2 mutants (Kim et al. 2012; this work), and the observation that a decrease in tra-2 dosage can also partially suppress fog-2 sterility (Schedl and Kimble 1988). Although the germlines of sacy-1(tm5503); tra-2(e2020) adults were feminized, oocytes underwent meiotic maturation constitutively, consistent with the finding that sacy-1 is a negative regulator of meiotic maturation (Kim et al. 2012). sacy-1(tm5503); tra-2(e2020) animals did, however, exhibit a highly penetrant ovulation defect, which caused endomitotic oocytes to accumulate in the gonad arm (Emo phenotype; Figure 7).
Figure 6.
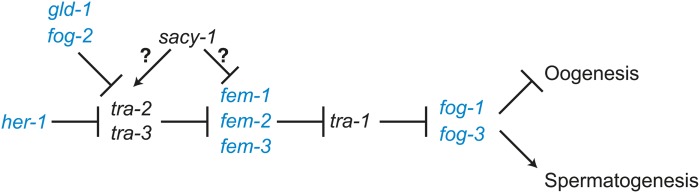
The C. elegans germline sex determination pathway. Genes promoting the male and female fate are shown in blue and black, respectively. sacy-1 promotes the oocyte fate antagonistically to fog-2, which promotes spermatogenesis.
Figure 7.
Genetic interactions between sacy-1 and tra-2 in germline sex determination were analyzed by combining the strong germline-feminizing dominant tra-2(e2020) mutation with the sacy-1(tm5503) null mutation. Dissected gonads of wild-type hermaphrodites (A–D) and sacy-1(tm5503); tra-2(e2020) females were analyzed by staining for the oocyte RME-2 yolk receptor (A and E) and the major sperm protein (B and F). DNA was detected with DAPI (C and G). Merged images are also shown (D and H). Note gonads from sacy-1(tm5503); tra-2(e2020) females do not express MSP and frequently contain endomitotic oocytes (arrows). Bar, 50 μm.
To extend these observations, we examined germline sexual fates in dissected gonads from sacy-1(tn1481Mog); tra-2(e2020) adults. Whereas all wild-type gonad arms examined (n = 21) expressed MSP and contained sperm, none of the sacy-1(tn1481Mog); tra-2(e2020) gonad arms (n = 37) expressed MSP or contained sperm. We noted that the Emo phenotype was less penetrant in sacy-1(tn1481Mog); tra-2(e2020) gonad arms (46% penetrance). These results are consistent with the possibility that sacy-1(+) promotes the oocyte fate by promoting the function of tra-2 in the germline and suggests that sacy-1(tn1481Mog) may interfere with this function.
Interestingly, recessive loss-of-function mutations in six genes, mog-1–mog-6, cause a Mog phenotype and encode spliceosomal components (Graham and Kimble 1993; Graham et al. 1993; Puoti and Kimble 1999, 2000; Belfiore et al. 2004; Zanetti et al. 2011). Mutation and RNAi depletion of many splicing factors have been observed to result in a Mog phenotype, suggesting that the germline sex determination process is particularly sensitive to disruptions in RNA splicing (Konishi et al. 2008; Mantina et al. 2009; Kerins et al. 2010; Wang et al. 2012; Novak et al. 2015). Prior studies focusing on the C. elegans soma were interpreted in the context of a model in which mog-1–mog-6 might function at the level of fem-3 through 3′UTR-dependent translational regulation (Gallegos et al. 1998); however, the experiments in that study did not address the regulation of fem-3 in the germline. We previously showed that sacy-1(tm5503); fem-3(e1996) adult XX animals had feminized germlines (Kim et al. 2012). To examine the genetic relationship between sacy-1 and fem-3 further, we generated sacy-1(tn1481Mog); fem-3(e1996) double mutants. We observed that 92% (n = 23) of sacy-1(tn1481Mog); fem-3(e1996) animals were feminized. Mating of sacy-1(tn1481Mog); fem-3(e1996) females (n = 29) to wild-type males resulted in the production of embryos that failed to hatch (n = 4140, 99.8%) or arrested as larvae (n = 7, 0.2%). This result indicates that two copies of sacy-1(tn1481) in the maternal germline, but not one [e.g., sacy-1(tn1481)/sacy-1(tm5503) heterozygotes are fertile] are incompatible with embryonic development. We found that 8% of sacy-1(tn1481Mog); fem-3(e1996) animals (n = 25) produced oocytes and sperm and a few dead embryos (one of these animals produced sperm in one gonad arm but not the other). The production of sperm in a small percentage of sacy-1(tn1481Mog); fem-3(e1996) hermaphrodites might depend on maternal fem-3(+) activity. We did observe that a reduction in fem-3 dosage could suppress the sacy-1(tn1481) Mog phenotype (n = 60). Specifically, whereas 65% (n = 39) of sacy-1(tn1481); fem-3(e1996)/+ animals were Mog, exclusively producing sperm, 33% (n = 20) produced sperm and oocytes, and one animal (2%) was feminized. Taken together, our results seem most consistent with the possibility that sacy-1 functions at the level of tra-2 in the genetic epistasis pathway (Figure 6; Zanetti and Puoti 2013). However, since this analysis of genetic interactions involves the use of dominant and recessive gain-of-function mutations [e.g., tra-2(e2020) and sacy-1(tn1481)], we cannot rule out the possibility that sacy-1 affects other steps, including the regulation of the fem-1–fem-3 genes (Figure 6).
We noted that among the 311 transcripts increased in abundance in the SACY-1 germline-depleted sample was her-1 (Figure 2, B, D, and E and File S2). This might be due to an increase in X chromosome nondisjunction in embryos located in the uterus following germline depletion of sacy-1(+), but this possibility was not investigated. Because her-1 likely encodes an inhibitory ligand for the TRA-2 receptor in the sex-determination pathway (Perry et al. 1993; Figure 6), we tested whether the her-1(hv1y101) null mutation could suppress the Mog phenotype of the recessive gain-of-function sacy-1(tn1481) mutation, but this proved not to be the case (n = 53 gonad arms). Since the genetic epistasis results suggest that SACY-1 might promote the expression of TRA-2 (Figure 6), we examined the effect of SACY-1 depletion in the germline on the levels of tra-2 mRNA and the fidelity of its splicing. We observed no statistically significant change in tra-2 mRNA levels or its splicing patterns (Figure 2F). We did not examine the expression of TRA-2 protein after SACY-1 depletion because a recent study suggested that TRA-2 protein expression in the wild-type germline is below the detection limit of immunofluorescence (Hu et al. 2019). Since SACY-1 genetically and biochemically interacts with components of the spliceosome, we suggest that the sacy-1(tn1481) mutation antagonizes functions of the spliceosome needed for germline sex determination and proper oogenesis. Interestingly, the top enriched GO terms for transcripts with increased abundance in the SACY-1 germline-depleted samples included SCF-dependent proteasomal ubiquitin-dependent protein catabolic processes (Figure S5B). This may be relevant to the role of SACY-1 in germline sex determination because the FEM-1–3 proteins are components of a CUL-2 E3 ubiquitin ligase that controls sex-determination in the germline and soma (Starostina et al. 2007).
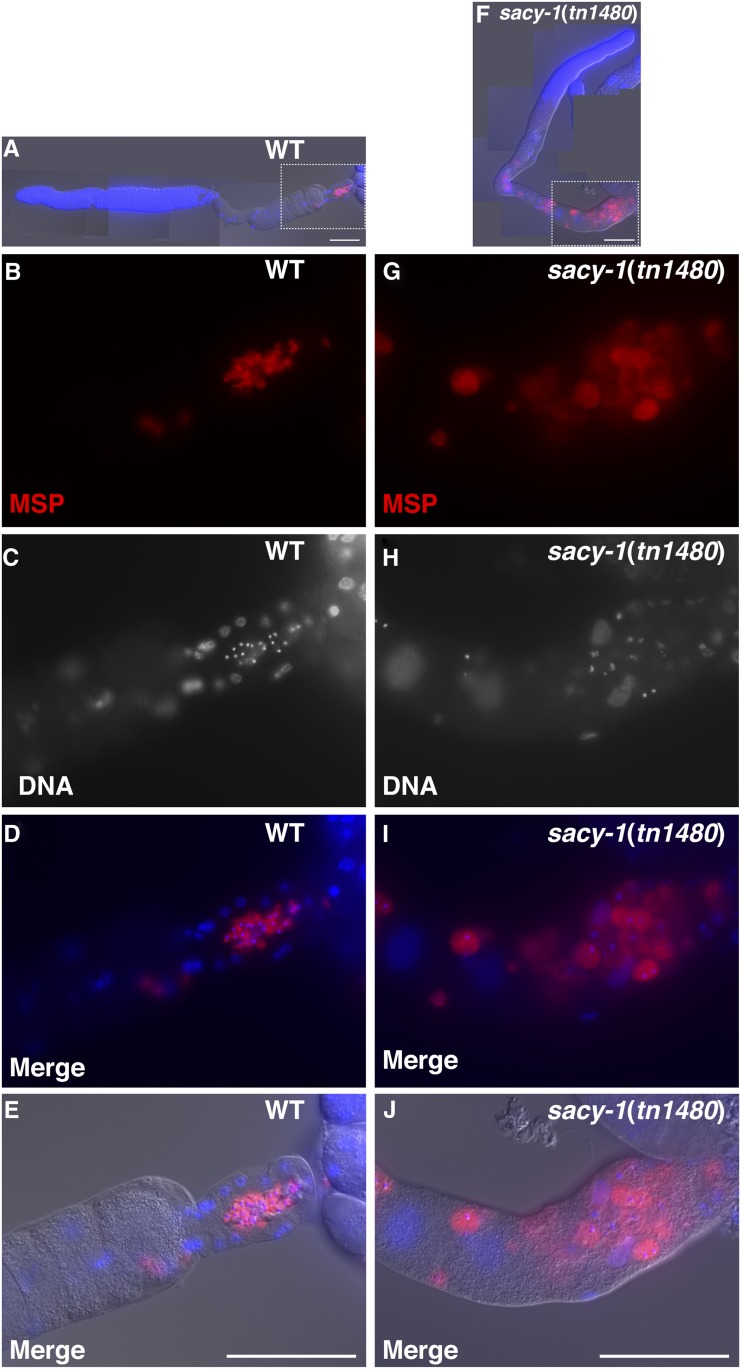
The recessive gain-of-function sacy-1(tn1480) allele confers multiple pleiotropic phenotypes in a temperature-dependent manner:
Interestingly, sacy-1(tn1480) displayed both cold-sensitive (15°) and temperature-sensitive (25°) defects. At 15°, sacy-1(tn1480) homozygotes (n = 55), which were the offspring of heterozygous parents grown at 15°, laid dead eggs and produced arrested larvae (91%) or produced very few progeny (9%). At 25°, sacy-1(tn1480) hermaphrodites were sterile and produced abnormal and fertilization-defective sperm (Figure 8). sacy-1(tn1480) hermaphrodite sterility is rescued by mating with wild-type males at 25°. In addition to producing oocytes, some sacy-1(tn1480) adult hermaphrodites continued to produce sperm, as swollen germ cells specified in the male fate were detected distal to the loop region in 42% of gonad arms examined (Figure 8; n = 12). Thus, in addition to the other phenotypes it confers, the sacy-1(tn1480) mutation perturbs the sperm-to-oocyte switch at 25°. We also observed that sacy-1(tn1480) conferred a dominant high-incidence of males (Him) phenotype; sacy-1(tn1480)/+ heterozygous hermaphrodites produced 1.6% male progeny (n = 3759), as compared to 0.1% for the wild-type control (n = 6044; P < 0.01, Fisher’s exact test). Genetic mapping showed that the temperature-dependent pleiotropic defects were inseparable from the sacy-1(tn1480) mutation (see Materials and Methods). Like sacy-1(tn1481), the defects conferred by sacy-1(tn1480) were dosage sensitive. At 25°, nearly all sacy-1(tn1480)/sacy-1(tm5503) heterozygotes (99%, n = 88) were fertile (the average brood size was 132 ± 64, n = 34); however, the majority of their sacy-1(tm5503) homozygous progeny (85.3%, n = 231) undergo vulval rupture and burst as adults (Table 6). This is in contrast to sacy-1(tm5503) homozygotes derived from sacy-1(tm5503)/+ heterozygous parents, only 2% of which undergo vulval rupture (n = 106; Table 6). This result suggests that maternal sacy-1(tn1480) activity can antagonize the spliceosome in the absence of zygotic sacy-1(+) function. By these genetic criteria, sacy-1(tn1480) exhibits recessive and weakly dominant gain-of-function properties, depending on the phenotype examined.
Figure 8.
Sperm-defective phenotype of sacy-1(tn1480) at 25°. Dissected gonads of wild-type (A–E) and sacy-1(tn1480) (F–J) stained with anti-MSP antibodies (B and G) and DAPI to detect DNA (C and H). Merged images are also shown (A, D, E, F, I, and J). At 25°, sacy-1(tn1480) hermaphrodites produce swollen and abnormal sperm, which are incapable of fertilization. Note that defective sperm are also found near the bend region in sacy-1(tn1480) adults (F) indicating that there is a defect in the sperm-to-oocyte switch. Bar, 50 μm.
The human oncogenic DDX41 R525H mutation confers weak antagonistic activity in C. elegans
The R525H mutation in DDX41 has been reported in myeloid leukemias both as newly arising somatic mutations specific to the neoplastic cells, as well as inherited germline mutations (Polprasert et al. 2015; Lewinsohn et al. 2016; Sébert et al. 2019). Thus, it was of interest to examine the impact of the analogous mutation (R534H) on sacy-1 function. Interestingly, a substitution at the adjacent amino acid (G533R) results in the sacy-1(tn1385) reduction-of-function mutation (Figure 4). Thus, we used genome editing to introduce the R534H mutation in the C. elegans genome (see Materials and Methods). By several criteria, sacy-1(tn1887[R534H]) homozygotes were indistinguishable from the wild type. Neither did sacy-1(tn1887) suppress acy-4 sterility (n = 140; sacy-1(tn1887); acy-4(ok1806) brood size was 1.5 ± 2.3) nor did it suppress fog-2 sterility (n = 48). All unmated sacy-1(tn1887); fog-2(oz40) gonad arms examined (n = 96) exhibited stacked oocytes, which indicates that the sacy-1(tn1887) mutation does not derepress meiotic maturation in the absence of sperm, like strong reduction-of-function mutations do. Further, the sacy-1(tn1887[R534H]) did not enhance the Tumorous phenotype of a gain-of-function glp-1/Notch allele (Table 4). The brood size of sacy-1(tn1887) (204 ± 37; n = 20) at 25° was similar to that of the wild type (202 ± 62, n = 30; P > 0.8, two-sample Z-test). Also, the incidence of males (0.2%, n = 4087) was similar to that observed in the wild type (0.1%, n = 6044). When placed in trans to the sacy-1(tm5503) null mutation, sacy-1(tn1887)/sacy-1(tm5503) heterozygotes were found to be fertile at all temperatures examined (15, 20, and 25°; n > 100). However, we did observe that sacy-1(tn1887) significantly enhanced the dominant Him phenotype of sacy-1(tn1480) (P < 0.05, Fisher’s exact test); sacy-1(tn1887)/sacy-1(tn1480) heterozygotes produced 5.7% males at 25° (n = 1483) as compared to 1.6% for the +/sacy-1(tn1480) control (n = 3759). The brood size of sacy-1(tn1887)/sacy-1(tn1480) heterozygotes at 25° (74 ± 65, n = 20) was also significantly lower than that of the +/sacy-1(tn1480) control (188 ± 68, n = 20; P < 0.001, two-sample Z-test). Because sacy-1(tn1480)/sacy-1(tm5503) heterozygotes do not exhibit an enhanced Him phenotype at 25° (0.7% males, n = 4493), the sacy-1(tn1887[R534H]) mutation appears to possess a weak antagonistic activity.
Discussion
In this study, we report the results of a molecular genetic and biochemical analysis of SACY-1/DDX41 in C. elegans, conducted to gain potential insights into how DDX41 mutations might contribute to MDS and AML in humans. DDX41 is a component of the spliceosomal C complex (Jurica et al. 2002; Bessonov et al. 2008), which carries out the second step in splicing. Pre-mRNA splicing is an essential process in eukaryotes (reviewed by Wahl et al. 2009). Thus, the finding that mutations in highly conserved genes encoding spliceosomal components are frequently found in hematological malignancies was unexpected (reviewed by Yoshida and Ogawa 2014). Unfortunately, the genetic properties of disease-causing mutations in spliceosomal proteins have been difficult to assess. Nonetheless, the observation that many of these missense mutations map to highly conserved amino acids, within biochemically defined structural and functional domains, might suggest that they reduce but not eliminate gene function. An alternative possibility is that some of the oncogenic missense mutations might confer antagonistic gain-of-function properties. Indeed, oncogenic mutations in several splicing factors (e.g., SF3B1, U2AF1, SRSDF2, and SRSR2) present in the heterozygous condition (Yoshida and Ogawa 2014); however, it is probable that additional newly arising mutations in hematopoetic lineages contribute to neoplastic development.
In this regard, the DDX41 mutations inherited and arising in familial cases of AML are particularly informative (Polprasert et al. 2015). Affected individuals often inherit one copy containing a mutation, which is likely to be a null mutation (e.g., a frameshift mutation, pD140fs). In addition to this DDX41 germline mutation, the neoplasms in these patients invariably contain a second copy with a somatic mutation that arose in the hematopoietic lineage. Several specific DDX41 somatic mutations (e.g., R525H) arise independently with high prevalence, suggesting that they markedly contribute to neoplastic development. These somatic DDX41 mutations are invariably missense mutations affecting conserved amino acids but have never been found to include candidate null alleles (e.g., frameshift mutations or premature termination codons). Thus, these somatically arising mutations are unlikely to eliminate DDX41 function. The presumption is that biallelic mutations disrupting DDX41 function might be lethal. In line with this view, a recent report found that two related patients with biallelic DDX41 missense mutations exhibited a more severe syndrome characterized by dysmorphic skeletal and facial features, psychomotor delays, intellectual disability, and early onset leukemia (Diness et al. 2018).
Here, we build upon our prior work (Kim et al. 2012) to conduct a comprehensive molecular genetic analysis of SACY-1/DDX41 function in C. elegans. Recent work of others has used the C. elegans system to gain information on oncogenic mutations in the SF3B1 spliceosomal protein (Serrat et al. 2019). Our results reveal that sacy-1 mutations confer a range of phenotypes, from highly pleiotropic defects affecting the germline and soma to very specific defects affecting cell differentiation and cell cycle regulation. Reduction-of-function sacy-1 alleles are homozygous viable and fertile, yet affect germline sex determination and the regulation of oocyte meiotic maturation (Kim et al. 2012; this work). Animals homozygous for sacy-1 null mutations grow to adulthood but exhibit a gamete degeneration phenotype and are sterile. Feminization of the germline delays oocyte degeneration, which enables sacy-1 null mutant females to be mated and produce embryos. However, these embryos invariably die, revealing an essential maternal requirement for development. Maternally provided sacy-1(+) must in turn be sufficient for homozygous sacy-1 null mutant animals (produced from heterozygous parents) to grow to adulthood.
Several sacy-1 mutant alleles exhibit genetic properties that suggest they can counteract sacy-1(+) function, potentially by compromising the function of the spliceosome. Most notable among these mutations is sacy-1(tn1481), which confers a masculinization of the germline phenotype resulting from the overproduction of sperm to the exclusion of the oocyte fate. Since multiple reduction-of-function sacy-1 alleles suppress the feminization of the germline phenotype caused by null mutations in fog-2 (Kim et al. 2012; this work), sacy-1(+) must possess a function to promote the oocyte fate. This oocyte-promoting function of sacy-1(+) might not be essential because sacy-1 null mutants produce oocytes, which nonetheless undergo necrotic degeneration. The caveat here is that sacy-1 null mutants only develop to adulthood because of maternally provided sacy-1(+), which might be sufficient to promote oogenesis. In any case, the sacy-1(tn1481) mutation might disrupt an oocyte-promoting function, either by interfering with maternally provided SACY-1 activity or the proteins with which it associates.
Several observations are consistent with the possibility that sacy-1(tn1481) interferes with, or compromises, the function of the spliceosome in germline sex determination in a dosage-sensitive manner. This view is also supported by our tandem affinity purification results showing that SACY-1 is a component of the C. elegans spliceosome, and interacts genetically with spliceosomal components. Significantly, multiple C. elegans spliceosomal components, functioning at different steps of the splicing reaction, can mutate to a masculinization of germline phenotype (reviewed by Zanetti and Puoti 2013). Interestingly, when placed in trans to a sacy-1 null mutation (e.g., tm5503), the resulting sacy-1(tn1481)/sacy-1(tm5503) heterozygous animals are viable and fertile, and can be maintained indefinitely as a heterozygous strain. This highly informative genetic result suggests that a single dose of sacy-1(tn1481) can mediate all its essential functions, but that two doses of the mutant protein interferes with the normal mechanisms of germline sex determination [i.e., sacy-1(tn1481) is a recessive gain-of-function antimorphic mutation]. Although sacy-1(tn1481)/sacy-1(tm5503) heterozygous adult hermaphrodites are fertile, when sacy-1(tn1481); fem-3(e1996) females are mated to wild-type males, they produce embryos that invariably fail to hatch. Similar results are obtained with the six other mog genes (Graham and Kimble 1993; Graham et al. 1993), which encode spliceosomal proteins (Puoti and Kimble 1999, 2000; Belfiore et al. 2004; Kasturi et al. 2010; Zanetti et al. 2011). Interestingly, the P222L amino acid substitution found in sacy-1(tn1481) is adjacent to the Q motif, which participates in nucleotide binding and hydrolysis (Schütz et al. 2010). It is tempting to speculate that SACY-1/DDX41 might promote remodeling of spliceosomes during the splicing reaction, and that two doses of SACY-1 P222L might disrupt these rearrangements. In a similar vein, sacy-1(tn1479[G504E]) exhibits a phenotype more severe than a null allele, suggesting that this mutation in the helicase domain too might possess a dosage-sensitive antimorphic activity. Whether missense alleles in human DDX41 also possess antimorphic activity will require additional work, including biochemical analyses.
We imported the human oncogenic DDX41[R525H] mutation into C. elegans using CRISPR-Cas9 genome editing (R534H in SACY-1). Our analysis revealed that, in C. elegans, this mutation possesses very weak antagonistic activity. While it is possible that the genetic properties of this mutation might differ between C. elegans and mammalian systems, it is worthwhile noting that the human oncogenic variant must support the high levels of proliferation characteristic of neoplastic cells. While all the sacy-1 mutations we isolated in forward genetic screens occur at conserved amino acids, none of them match the human oncogenic mutations thus far isolated (Polprasert et al. 2015; Cardoso et al. 2016; Lewinsohn et al. 2016; Li et al. 2016; Diness et al. 2018; Sébert et al. 2019). This observation is consistent with the idea that the human oncogenic mutations are, at most, weakly antagonizing or weak rf mutations. Our forward genetic screens in C. elegans may have required a substantial reduction of function and thus might have missed very weak mutations like SACY-1(R534H) or other human oncogenic variants.
An attractive idea is that human oncogenic mutations affecting the spliceosome contribute to neoplastic development through effects on gene expression occurring through alterations in RNA splicing, as well as effects on the transcriptional machinery or RNA stability (Yoshimi et al. 2019). The very specific defects observed in individual sacy-1/ddx41 mutant alleles in C. elegans could be due to a failure to properly splice key genes, as has been documented for several splicing-related mutations in Drosophila melanogaster (Van Buskirk and Schüpbach 2002; Roignant and Treisman 2010). In this study, we examined the effects of SACY-1 depletion at the adult stage on the transcriptome. We observed many instances of missplicing following SACY-1 depletion, though missplicing events were more prevalent when SACY-1 was depleted from the soma as compared with the germline, suggesting the possibility that surveillance mechanisms might serve to protect the germline by removing the products of aberrant splicing events. Although we observed missplicing events in the germline and soma upon SACY-1 depletion, no clear candidates emerged that might account for its role in germline sex determination. Genetic evidence suggests that sacy-1 promotes the activity of tra-2, but this appears to be independent of splicing because the expression level and splicing pattern of tra-2 mRNA was unchanged following depletion of sacy-1 in the germline. Whether sacy-1 mutations might affect the localization of tra-2 mRNA or its translation remains to be determined. In both the soma and the germline, we observed splicing-independent impacts on the abundance of many transcripts. The gene expression changes observed suggest that depletion of SACY-1 might elicit a stress response. One possibility is that defective proteins produced via missplicing contribute to the induction of stress responses. Alternatively, there might be surveillance pathways that respond to dysfunctional splicing by evoking homeostatic mechanisms. If analogous processes occur after perturbations of the spliceosome in humans, such stress responses might contribute to the vitality of tumor cells and could represent therapeutic targets. The extent to which gene expression changes and splicing alterations contribute to the various sacy-1 mutant phenotypes will require further study but will likely provide insights relevant to spliceosomal perturbations in humans.
Acknowledgments
This paper is dedicated to the memory of Michael A. Miller, our colleague and friend whose scientific contributions will not be forgotten. We thank Donna Coetzee for technical assistance. We are grateful to Joshua Arribere, Daniel Dickinson, Barth Grant, Tim Schedl, and Jordan Ward for providing strains or reagents. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by grant P40OD010440 from the National Institutes of Health (NIH) Office of Research Infrastructure Programs. We thank Gabriela Huelgas-Morales, Zohar Sachs, and Todd Starich for their helpful suggestions for the manuscript. This work was supported by NIH grant GM57173 to D.G.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11825673.
Communicating editor: E. Tran
Literature Cited
- Agarwal A., Koppstein D., Rozowsky J., Sboner A., Habegger L. et al. , 2010. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 11: 383 10.1186/1471-2164-11-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S. et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore M., Pugnale P., Saudan Z., and Puoti A., 2004. Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development 131: 2935–2945. 10.1242/dev.01154 [DOI] [PubMed] [Google Scholar]
- Bertram K., Agafonov D. E., Liu W.-T., Dybkov O., Will C. L. et al. , 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. 10.1038/nature21079 [DOI] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Will C. L., Urlaub H., and Lührmann R., 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850. 10.1038/nature06842 [DOI] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Krasauskas A., Golas M. M., Sander B. et al. , 2010. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16: 2384–2403. 10.1261/rna.2456210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng R., Nguyen K. C. Q., Hall D. H., Golden A., and Allen A. K., 2017. Novel functions for the RNA-binding protein ETR-1 in Caenorhabditis elegans reproduction and engulfment of germline apoptotic corpses. Dev. Biol. 429: 306–320. 10.1016/j.ydbio.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso S. R., Ryan G., Walne A. J., Ellison A., Lowe R. et al. , 2016. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia 30: 2083–2086. 10.1038/leu.2016.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltri P. P., dos Santos M. G. P., and da Silva G. H. G., 2019. Splicing and cancer: challenges and opportunities. WIREs RNA 10: e1527 10.1002/wrna.1527 [DOI] [PubMed] [Google Scholar]
- De I., Bessonov S., Hofele R., dos Santos K., Will C. L. et al. , 2015. The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat. Struct. Mol. Biol. 22: 138–144. 10.1038/nsmb.2951 [DOI] [PubMed] [Google Scholar]
- Dejima K., Hori S., Iwata S., Suehiro Y., Yoshina S. et al. , 2018. An aneuploidy-free and structurally defined balancer chromosome toolkit for Caenorhabditis elegans. Cell Rep. 22: 232–241. 10.1016/j.celrep.2017.12.024 [DOI] [PubMed] [Google Scholar]
- DeNicola A. B., and Tang Y., 2019. Therapeutic approaches to treat human spliceosomal diseases. Curr. Opin. Biotechnol. 60: 72–81. 10.1016/j.copbio.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., and Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diness B. R., Risom L., Frandsen T. L., Hansen B., Andersen M. K. et al. , 2018. Putative new childhood leukemia cancer predisposition syndrome caused by germline bi-allelic missense mutations in DDX41. Genes Chromosomes Cancer 57: 670–674. 10.1002/gcc.22680 [DOI] [PubMed] [Google Scholar]
- Ding L., Ley T. J., Larson D. E., Miller C. A., Koboldt D. C. et al. , 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effenberger K. A., Urabe V. K., and Jurica M. S., 2017. Modulating splicing with small molecular inhibitors of the spliceosome. WIREs RNA 8: e1381 10.1002/wrna.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica S. M., Oubridge C., Wilkinson M. E., Newman A. J., and Nagai K., 2019. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science 363: 710–714. 10.1126/science.aaw5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine E., and Schedl T., 1995a Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., and Schedl T., 1995b gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos M., Ahringer J., Crittenden S., and Kimble J., 1998. Repression by the 3′ UTR of fem-3, a sex-determining gene, relies on a ubiquitous mog-dependent control in Caenorhabditis elegans. EMBO J. 17: 6337–6347. 10.1093/emboj/17.21.6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan J. A., Cheng H., Harris J. E., and Greenstein D., 2006. Gαo/i and Gαs signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol. 16: 1257–1268. 10.1016/j.cub.2006.05.020 [DOI] [PubMed] [Google Scholar]
- Graham P. L., and Kimble J., 1993. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics 133: 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P. L., Schedl T., and Kimble J., 1993. More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev. Genet. 14: 471–484. 10.1002/dvg.1020140608 [DOI] [PubMed] [Google Scholar]
- Grant B., and Hirsh D., 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311–4326. 10.1091/mbc.10.12.4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinfeld J., Nangalia J., Baxter E. J., Wedge D. C., Angelopoulos N. et al. , 2018. Classification and personalized prognosis in myeloproliferative neoplasms. N. Engl. J. Med. 379: 1416–1430. 10.1056/NEJMoa1716614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. W., Baran J., Bieri T., Cabunoc A., Chan J. et al. , 2014. WormBase 2014: new views of curated biology. Nucleic Acids Res. 42: D789–D793. 10.1093/nar/gkt1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbach D., Komarov I., Agafonov D. E., Hartmuth K., Graf B. et al. , 2018. Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172: 454–464.e11. 10.1016/j.cell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Henn A., Bradley M. J., and De La Cruz E. M., 2012. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu. Rev. Biophys. 41: 247–267. 10.1146/annurev-biophys-050511-102243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N., Wil C. L., Wolf E., Kastner B., Urlaub H. et al. , 2009. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 29: 281–301. 10.1128/MCB.01415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Skelly L. E., Kaymak E., Freeberg L., Lo T.-W. et al. , 2019. Multi-modal regulation of C. elegans hermaphrodite spermatogenesis by the GLD-1-FOG-2 complex. Dev. Biol. 446: 193–205. 10.1016/j.ydbio.2018.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J. A., and Schedl T., 2019. Biology of the Caenorhabditis elegans germline stem cell system. Genetics 213: 1145–1188. 10.1534/genetics.119.300238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U., Leptin M., Siller K., Fuerstenberg S., Cai Y. et al. , 2004. Abstrakt, a DEAD box protein, regulates Insc levels and asymmetric division of neural and mesodermal progenitors. Curr. Biol. 14: 138–144. 10.1016/j.cub.2004.01.002 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Zhu Y., Liu Z.-J., and Ouyang S., 2017. The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell 8: 83–89. 10.1007/s13238-016-0303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao A. L., Perales R., Umbreit N. T., Haswell J. R., Piper M. E. et al. , 2019. Human nuclear RNAi-defective 2 (NRDE2) is an essential RNA splicing factor. RNA 25: 352–363. 10.1261/rna.069773.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. R., and Schedl T., 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affects a conserved domain also found in Src-associated protein Sam68. Genes Dev. 9: 1491–1504. 10.1101/gad.9.12.1491 [DOI] [PubMed] [Google Scholar]
- Jurica M. S., Licklider L. J., Gygi S. P., Grigorieff N., and Moore M. J., 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. 10.1017/S1355838202021088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R. et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kasturi P., Zanetti S., Passannante M., Saudan Z., Müller F. et al. , 2010. The C. elegans sex determination protein MOG-3 functions in meiosis and binds to the CSL co-repressor CIR-1. Dev. Biol. 344: 593–602. 10.1016/j.ydbio.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Kerins J. A., Hanazawa M., Dorsett M., and Schedl T., 2010. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation vs. meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev. Dyn. 239: 1555–1572. 10.1002/dvdy.22274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Govindan J. A., Tu Z. J., and Greenstein D., 2012. SACY-1 DEAD-Box helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in Caenorhabditis elegans. Genetics 192: 905–928. 10.1534/genetics.112.143271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., and Abdel-Wahab O., 2017. Therapeutic targeting of RNA splicing in myelodysplasia. Semin. Hematol. 54: 167–173. 10.1053/j.seminhematol.2017.06.007 [DOI] [PubMed] [Google Scholar]
- Konishi T., Uodome N., and Sugimoto A., 2008. The Caenorhabditis elegans DDX-23, a homolog of yeast splicing factor PRP28, is required for the sperm-oocyte switch and differentiation of various cell types. Dev. Dyn. 237: 2367–2377. 10.1002/dvdy.21649 [DOI] [PubMed] [Google Scholar]
- Kosinski M., McDonald K., Schwartz J., Yamamoto I., and Greenstein D., 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132: 3357–3369. 10.1242/dev.01916 [DOI] [PubMed] [Google Scholar]
- Levy A. D., Yang J., and Kramer J. M., 1993. Molecular and genetic analyses of the Caenorhabditis elegans dpy-2 and dpy-10 collagen genes: a variety of molecular alterations affect organismal morphology. Mol. Biol. Cell 4: 803–817. 10.1091/mbc.4.8.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn M., Brown A. L., Weinel L. W., Phung C., Rafidi G. et al. , 2016. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood 127: 1017–1023. 10.1182/blood-2015-10-676098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Sobreira N., Witmer P. D., Pratz K. W., and Braunstein E. M., 2016. Two novel germline DDX41 mutations in a family with inherited myelodysplasia/acute myeloid leukemia. Hematologica 101: e228–e231. 10.3324/haematol.2015.139790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewski J. P., Padgett R. A., Brown A. L., and Müller-Tidow C., 2017. DDX41-related myeloid neoplasia. Semin. Hematol. 54: 94–97. 10.1053/j.seminhematol.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. M., and Kimble J., 1993. Suppressors of glp-1, a gene required for cell communication during development in Caenorhabditis elegans, define a set of interacting genes. Genetics 135: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maita H., Kitaura H., Ariga H., and Iguchi-Ariga S. M., 2005. CIR, a corepressor of CBF1, binds to PAP-1 and effects alternative splicing. Exp. Cell Res. 303: 375–387. 10.1016/j.yexcr.2004.10.012 [DOI] [PubMed] [Google Scholar]
- Mantina P., McDonald L., Kulaga A., Zhao L., and Hansen D., 2009. A mutation in teg-4, which encodes a protein homologous to the SAP130 pre-mRNA splicing factor, disrupts the balance between proliferation and differentiation in the C. elegans germ line. Mech. Dev. 126: 417–429. 10.1016/j.mod.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Nishiwaki K., and Miwa J., 1998. Mutations in genes encoding extracellular matrix proteins suppress the emb-5 gastrulation defect in Caenorhabditis elegans. Mol. Gen. Genet. 259: 2–12. 10.1007/s004380050782 [DOI] [PubMed] [Google Scholar]
- Novak P., Wang X., Ellenbecker M., Feilzer S., and Voronina E., 2015. Splicing machinery facilitates post-transcriptional regulation by FBFs and other RNA-binding proteins in the Caenorhabditis elegans germline. G3 (Bethesda) 5: 2051–2059. 10.1534/g3.115.019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura H., Oikawa D., Nakane T., Kato M., Ishii R. et al. , 2016. Structural and functional analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide. Sci. Rep. 6: 34756 10.1038/srep34756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Noble D., Sorokin E. P., and Kimble J., 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4: 1765–1772. 10.1534/g3.114.012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C. Y., Calidas D. et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198: 1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V. I., Paschka P. et al. , 2016. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374: 2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K., Zhang Z., Teles R. M., Ouyang S., Jiang Y. et al. , 2012. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13: 1155–1161. 10.1038/ni.2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A. S.-R., Killian D. J., and Hubbard E. J. A., 2003. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163: 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. D., Li W., Trent C., Robertson B., Fire A. et al. , 1993. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes Dev. 7: 216–228. 10.1101/gad.7.2.216 [DOI] [PubMed] [Google Scholar]
- Peters D., Radine C., Reese A., Budach W., Sohn D. et al. , 2017. The DEAD-box RNA helicase DDX41 is a novel repressor of p21WAF1/CIP1 mRNA translation. J. Biol. Chem. 292: 8331–8341. 10.1074/jbc.M116.772327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polprasert C., Schulze I., Sekeres M. A., Makishima H., Przchodzen B. et al. , 2015. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell 27: 658–670. 10.1016/j.ccell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., and Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A., and Kimble J., 1999. The Caenorhabditis elegans sex determination gene mog-1 encodes a member of the DEAH-Box protein family. Mol. Cell. Biol. 19: 2189–2197. 10.1128/MCB.19.3.2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A., and Kimble J., 2000. The hermaphrodite sperm/oocyte switch requires the Caenorhabditis elegans homologs of PRP2 and PRP22. Proc. Natl. Acad. Sci. USA 97: 3276–3281. 10.1073/pnas.97.7.3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues F., Thuma L., and Klämbt C., 2012. The regulation of glial-specific splicing of Neurexin IV requires HOW and Cdk12 activity. Development 139: 1765–1776. 10.1242/dev.074070 [DOI] [PubMed] [Google Scholar]
- Roignant J.-Y., and Treisman J. E., 2010. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochomatic gene. Cell 143: 238–250. 10.1016/j.cell.2010.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K. L., Winfrey V. P., Hoffman L. H., Hall D. H., Furuta T. et al. , 1997. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 192: 59–77. 10.1006/dbio.1997.8728 [DOI] [PubMed] [Google Scholar]
- Rual J.-F., Klitgord N., and Achaz G., 2007. Novel insights into RNAi off-target effects using C. elegans paralogs. BMC Genomics 8: 106 10.1186/1471-2164-8-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam M., Wurst W., Klüppel M., Jin O., Heng H. et al. , 1998. Aquarius, a novel gene isolated by gene trapping with an RNA-dependent RNA polymerase motif. Dev. Dyn. 212: 304–317. https://anatomypubs.onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291097-0177%28199806%29212%3A2%3C304%3A%3AAID-AJA15%3E3.0.CO%3B2-3 [DOI] [PubMed] [Google Scholar]
- Sanchez P., Daniels K. J., Park Y.-N., and Soll D. R., 2014. Generating a battery of monoclonal antibodies against native green fluorescent protein for immunostaining, FACS, IP, and ChIP using a unique adjuvant. Monoclon. Antib. Immunodiagn. Immunother. 33: 80–88. 10.1089/mab.2013.0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., and Kimble J., 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119: 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz P., Karlberg T., van den Berg S., Collins R., Lehtiö L. et al. , 2010. Comparative structural analysis of human DEAD-box RNA helicases. PLoS One 5: e12791 10.1371/journal.pone.0012791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sébert M., Passet M., Raimbault A., Rahmé R., Raffoux E. et al. , 2019. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood 134: 1441–1444. 10.1182/blood.2019000909 [DOI] [PubMed] [Google Scholar]
- Seiler M., Yoshimi A., Darman R., Chan B., Keaney G. et al. , 2018. H3B–8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 24: 497–504. 10.1038/nm.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrat X., Kukhtar D., Cornes E., Esteve-Codina A., Benlloch H. et al. , 2019. CRISPR editing of sftb-1/SF3B1 in Caenorhabditis elegans allows the identification of synthetic interactions with cancer-related mutations and the chemical inhibition of splicing. PLoS Genet. 15: e1008464 10.1371/journal.pgen.1008464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Eichten C., Wang X., Hansen D. et al. , 2014a The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics 198: 1535–1558. 10.1534/genetics.114.168831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Nishi Y., Guven-Ozkan T., Oldenbroek M. et al. , 2014b Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics 198: 1513–1533. 10.1534/genetics.114.168823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina N. G., Lim J., Schvarzstein M., Wells L., Spence A. M. et al. , 2007. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell 13: 127–139. 10.1016/j.devcel.2007.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou S., Blouch K., Kotla S., Bass A., and Ross S. R., 2015. Nucleic acid recognition orchestrates the anti-viral response to retroviruses. Cell Host Microbe 17: 478–488. 10.1016/j.chom.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou S., Aguilera A. N., Blouch K., and Ross S. R., 2018. DDX41 recognizes RNA/DNA retroviral reverse transcripts and is critical for in vivo control of murine leukemia virus infection. MBio 9: pii: e00923-18. 10.1128/mBio.00923-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z. J., 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A., and Vardiman J. W., 2009. Myelodysplastic syndromes. N. Engl. J. Med. 361: 1872–1885. 10.1056/NEJMra0902908 [DOI] [PubMed] [Google Scholar]
- Timmons L., and Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Gearhart M. D., Spike C. A., Huelgas-Morales G., Mews M. et al. , 2017. LIN-41 and OMA ribonucleoprotein complexes mediate a translational repression-to-activation switch controlling oocyte meiotic maturation and the oocyte-to-embryo transition in Caenorhabditis elegans. Genetics 206: 2007–2039. 10.1534/genetics.117.203174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C., and Schüpbach T., 2002. half pint regulates alternative splice site selection in Drosophila. Dev. Cell 2: 343–353. 10.1016/S1534-5807(02)00128-4 [DOI] [PubMed] [Google Scholar]
- Wahl M. C., Will C. L., and Lührmann R., 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wang C., Wilson-Berry L., Schedl T., and Hansen D., 2012. TEG-1 CD2BP2 regulates stem cell proliferation and sex determination in the C. elegans germ line and physically interacts with the UAF-1 U2AF65 splicing factor. Dev. Dyn. 241: 505–521. 10.1002/dvdy.23735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Wu F.-H., Wang X., Wang L., Siedow J. N. et al. , 2014. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 42: 8243–8257. 10.1093/nar/gku569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., and Ogawa S., 2014. Splicing factor mutations and cancer. WIREs RNA 5: 445–459. 10.1002/wrna.1222 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y. et al. , 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. 10.1038/nature10496 [DOI] [PubMed] [Google Scholar]
- Yoshimi A., Lin K.-T., Wiseman D. H., Rahman M. A., Pastore A. et al. , 2019. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574: 273–277. 10.1038/s41586-019-1618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti S., and Puoti A., 2013. Sex determination in the Caenorhabditis elegans germline. Adv. Exp. Med. Biol. 757: 41–69. 10.1007/978-1-4614-4015-4_3 [DOI] [PubMed] [Google Scholar]
- Zanetti S., Meola M., Bochud A., and Puoti A., 2011. Role of the C. elegans U2 snRNP protein MOG-2 in sex determination, meiosis, and splice site selection. Dev. Biol. 354: 232–241. 10.1016/j.ydbio.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Zhang L., Ward J. D., Cheng Z., and Dernburg A. F., 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142: 4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yuan B., Bao M., Lu N., Kim T. et al. , 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12: 959–965 [corrigenda: Nat. Immunol. 13: 196 (2012)]. 10.1038/ni.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains and reagents are available upon request. RNA sequencing data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through accession number GSE144003. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11825673.