Abstract
Genetic variance of a phenotypic trait can originate from direct genetic effects, or from indirect effects, i.e., through genetic effects on other traits, affecting the trait of interest. This distinction is often of great importance, for example, when trying to improve crop yield and simultaneously control plant height. As suggested by Sewall Wright, assessing contributions of direct and indirect effects requires knowledge of (1) the presence or absence of direct genetic effects on each trait, and (2) the functional relationships between the traits. Because experimental validation of such relationships is often unfeasible, it is increasingly common to reconstruct them using causal inference methods. However, most current methods require all genetic variance to be explained by a small number of quantitative trait loci (QTL) with fixed effects. Only a few authors have considered the “missing heritability” case, where contributions of many undetectable QTL are modeled with random effects. Usually, these are treated as nuisance terms that need to be eliminated by taking residuals from a multi-trait mixed model (MTM). But fitting such an MTM is challenging, and it is impossible to infer the presence of direct genetic effects. Here, we propose an alternative strategy, where genetic effects are formally included in the graph. This has important advantages: (1) genetic effects can be directly incorporated in causal inference, implemented via our PCgen algorithm, which can analyze many more traits; and (2) we can test the existence of direct genetic effects, and improve the orientation of edges between traits. Finally, we show that reconstruction is much more accurate if individual plant or plot data are used, instead of genotypic means. We have implemented the PCgen-algorithm in the R-package pcgen.
Keywords: Structural equation models, multivariate mixed models, causal inference
TO attain higher genetic gains, modern plant and animal breeders increasingly scale up their programs via the implementation of genomic prediction technologies (Cooper et al. 2014). Most genomic prediction applications are based on linear mixed- or Bayesian models that predict the phenotype for the target trait (yield) as a function of a multivariate distribution for single nucleotide polymorphism (SNP) effects. In these models, the physiological mechanisms and traits that modulate the genotypic response to the environment over time are modeled implicitly via the SNP effects directly affecting the target trait (Calus and Veerkamp 2011; Zhou and Stephens 2014). The availability of high throughput phenotyping technologies has enabled breeders to characterize additional traits, and to monitor growth and development during the season. This opens new opportunities in breeding strategies, in which better-adapted genotypes result from combining loci that regulate complementary physiological mechanisms. This kind of breeding strategy is called physiological breeding (Reynolds and Langridge 2016).
In physiological breeding, prediction accuracy for the target trait potentially benefits from a joint model for all underlying traits. This is partly because of the physiological knowledge that can be incorporated, but also because the use of genetically correlated traits with sufficiently large heritability increases accuracy (Thompson and Meyer 1986; van Eeuwijk et al. 2019). Often, however, a realistic model should account for at least some of the causal relations between traits, which is difficult with the regression models that are used in most genomic prediction literature. Structural equation models (SEMs), proposed by Wright (1921), and extended with random genetic effects in Gianola and Sorensen (2004), are a promising approach to deal with this problem (Rosa et al. 2011). In SEMs, each trait is modeled explicitly as a function of the other traits and a noise term. Therefore, SEMs are a useful tool to identify which are the key traits that could be selection targets, or be incorporated in multi-trait genomic prediction models to improve the prediction accuracy for the target trait.
A first advantage of SEMs, compared to regression models, is that one can predict the behavior of the system when one or more of the structural equations are modified by some kind of intervention. Figure 1 shows an illustration of an intervention. For example, a question that could be of interest to plant breeders is: which would be the contribution of a trait (say, radiation use efficiency) to yield, if flowering time is fixed for all genotypes at a particular value?
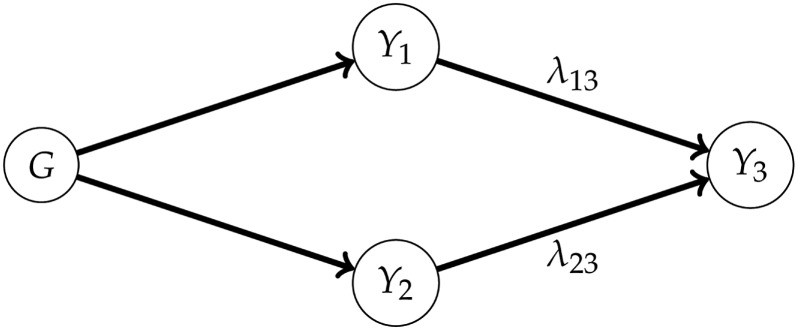
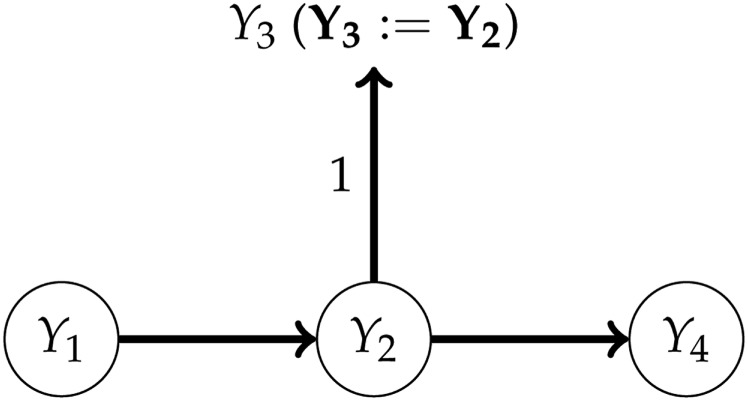
Figure 1.
An example of a linear SEM. The SEM can be represented by a graph (middle), which is defined by the nonzero elements of Λ, the matrix containing the path coefficients (top). The total effect of on can be obtained by summing the contributions of the directed paths and , where each contribution is the product of the corresponding path coefficients. After the intervention (bottom), the effect changes from to .
Second, SEMs make possible to distinguish between direct and indirect effects of one trait on another, and, similarly, between direct and indirect genetic effects. For example, let plant height (trait ) be modeled as , i.e., as the sum of a random genetic and residual effect, where all terms are vectors, containing the values for a population of n individuals. Suppose plant height has a linear effect on yield (), with additional random effects and :
where λ is the path (or structural) coefficient associated with the effect of on . On the one hand, we have the direct genetic effects and ; on the other, we have the total (or marginal) genetic effects and , the indirect effects being and . Similarly, we can distinguish between the matrices , containing the (co) variances of and , and , with the (co) variances of and . The latter is the matrix of genetic (co) variances, appearing in the usual MTM (multi-trait mixed model) for and ; here, it is a function of and λ.
Knowledge of the direct genetic effects is often of great interest to breeders (Valente et al. 2013, 2015). However, routine use of these models is currently difficult for two reasons. First, for a given SEM, not all parameters may be identifiable, i.e., because of overparameterization, different sets of parameter values can lead to the same model, making estimation infeasible. Gianola and Sorensen (2004) provided criteria for identifiability, and suggested putting constraints on some parameters, although automatic generation of interpretable and meaningful constraints remains difficult, especially in high-dimensional settings.
A second (and more fundamental) obstacle for the use of SEMs with genetic effects is that the underlying structure is often unknown. In such cases, causal inference methods (Spirtes et al. 2001; Pearl 2009; Maathuis and Nandy 2016) can be used, which reconstruct causal models that are, in some sense, most compatible with the observed data. Most causal inference methods, however, require independent samples, and cannot account for genetic relatedness. For this reason, genotypic differences are most often modeled using a small number of quantitative trait loci (QTL) with fixed effects (Chaibub Neto et al. 2008, 2013; Scutari et al. 2014), but when part of the genetic variance is not explained by QTL (missing heritability), the use of random genetic effects seems inevitable. Only a few works have studied reconstruction in the presence of such effects. Valente et al. (2010) and Töpner et al. (2017) proposed to perform causal inference after subtracting genomic predictions obtained from an MTM. Similarly, Gao and Cui (2015) applied the PC algorithm (Spirtes et al. 2001) to the residuals of multi-single nucleotide polymorphism (SNP) models. The difficulty with these approaches is that the MTM is limited to small numbers of traits, and that the existence of direct genetic effects cannot be tested. For example, if the causal graph among three traits is , and there are direct genetic effects on and , then the absence of a direct genetic effect on cannot be inferred from MTM residuals.
Inspired by these problems, we define a framework in which direct genetic effects are part of the causal graph, and a single node G represents all direct genetic effects. For each trait an arrow is present if and only if the direct genetic effect on is nonzero, i.e., if the jth diagonal element of is positive. See Figure 3 below for an example. Although our causal interpretation of genetic effects is not new (Stephens 2013; Valente et al. 2013, 2015), this work appears to be the first to formalize it. In particular, we show that the Markov property holds for the graph extended with genetic effects (Theorem 1 below). Informally speaking, this means that there is a one-to-one correspondence between edges in the causal graph and conditional dependencies in the distribution of the traits and genetic effects. This means that edges (either between two traits, or between a trait and G) can be inferred from multi-trait data. Consequently, while some of the covariances between direct genetic effects (contained in ) may still be unidentifiable, we can at least identify which rows and columns in are zero.
Figure 3.
An example of a graph representing a genetic structural equation model (GSEM), with path-coefficients and . There is no direct genetic effect on and therefore no edge
Building on the Markov-property, we propose the PCgen algorithm. PCgen stands for PC with genetic effects, and is an adaptation of the general PC-algorithm (named after its inventors Peter Spirtes and Clark Glymour). Briefly, PCgen assesses the existence of a direct genetic effect on a given trait by testing whether its genetic variance is zero, conditional on various sets of other traits. For the existence of an edge between traits and we test whether, in a bivariate MTM, the residual covariance between and is zero, again conditional on sets of other traits. Under the usual assumptions of independent errors, recursiveness, and faithfulness, we show that PCgen can recover the underlying partially directed graph (Theorem 2). Because fitting an MTM for all traits simultaneously is no longer necessary, PCgen can handle a considerably larger number of traits.
While our approach is generally applicable to any species and relatedness matrix, our implementation of PCgen is currently limited to the specific (but important) case of populations where observations on genetically identical replicates are available, assuming independent genetic effects (i.e., as in the classical estimation of broad-sense heritability). This is partly for pragmatic reasons (e.g., lower computational requirements), and partly for statistical reasons. In particular, successful reconstruction requires sufficient power in the tests for direct genetic effects and those for the between traits relations . Given the availability of replicates, this power is likely to be highest when the original observations are used, instead of genotypic means and a marker-based genetic relatedness matrix (GRM), modeling additive effects (Kruijer et al. 2015). Although mixed models with both replicates and a GRM may further increase power, the increase is often modest, and statistical inference can become biased under model misspecification (e.g., when the GRM models additive effects, and the true architecture is partly epistatic; see Kruijer 2016). By contrast, using only replicates, unbiased estimation of broad-sense heritability is always possible, regardless of the population structure and genetic architecture. The downside is that the contributions of different types of genetic effects cannot be distinguished. On the positive side, PCgen appears to be the first algorithm that can infer the presence of direct genetic effects based on phenotypic data alone.
Our approach is related to that of Stephens (2013), who inferred the sets of traits being affected directly and indirectly by a given locus, assuming unrelated individuals and using only summary statistics. Here, we instead consider sums of individual locus effects, for possibly related individuals. Moreover, PCgen also aims to reconstruct structural relations among the traits themselves, and can deal with larger numbers of traits.
The paper is organized as follows. After introducing SEMs with genetic effects, we define their graphical structure, and, from this perspective, review existing approaches. We then describe the general form of the PCgen-algorithm for estimation of the graphical structure, followed by various proposals for the required conditional independence tests. Next, we test PCgen performance in data simulated with both statistical and crop-growth models, and analyze a maize and a rice dataset. Finally, we state several results regarding PCgen’s statistical properties. Supplemental Material, Table S1 provides an overview of the notation, and Appendix A.1 contains the necessary graph-theoretic definitions. Figure 2 provides a graphical summary of our theory and methodology.
Figure 2.
Graphical summary of the theory and methodology. The Markov property on the right (green; for the residuals) is well known from the literature, while the Markov property on the left (blue, for the conditional distributions of ) is established in Theorem 1. Table S1 contains an overview of the notation, and Appendix A.1 provides the necessary graph-theoretic definitions.
Materials and Methods
Structural equation models
To introduce structural models, we first consider a simple linear SEM without genetic effects:
| (1) |
where is a vector of phenotypic values for p traits measured on the ith individual, is a vector of random errors, and is a matrix of structural coefficients. The matrix contains intercepts and trait-specific fixed effects of (exogenous) covariates, whose values are contained in the vector .
To write Equation 1 in matrix-form, we define the design matrix with rows . Similarly, we define matrices and , with rows and , and columns and . We can then write
| (2) |
has zeros on the diagonal, and defines a directed graph over the traits , containing the edge if and only if the th entry of is nonzero. The columns in Equation 2 correspond to p linear structural equations, one for each trait. These are determined by the path coefficients, the nonzero elements in . For example, in Figure 1, if is the vector of ones and , the third trait has values . The equality sign here should be understood as an assignment, i.e., is determined by the values of and (its parents in the graph ) and an error. If the directed graph does not contain any cycle (i.e., a directed path from a trait to itself), it is a directed acyclic graph (DAG), and the SEM is said to be recursive. In the notation, we will distinguish between the nodes in the graph (normal type), and the random vectors that these nodes represent (bold face).
As mentioned above, SEMs can be used to predict the effects of interventions, which mathematically correspond to changes in the structural equations. For example, suppose that, in Figure 1, , and are the expression levels of three genes, and is plant height. Then, after forcing to be zero (e.g., by knocking out the gene), the total effect of on changes from to (File S7.3 and File S7.4 provide other examples, involving genomic prediction). More generally, the new joint distribution of after an intervention can be obtained from the manipulation or truncated factorization theorem (Pearl 2009), without observations from the new distribution. For the consequences for genomic prediction, see Valente et al. (2013) and the Discussion section below.
GSEM: structural equation models with genetic effects
Gianola and Sorensen (2004) extended model (1) with random genetic effects : for individuals , it is then assumed that
| (3) |
where the vectors, contain the direct genetic effects for individuals . We will refer to model (3) as a linear genetic structural equation model (GSEM). While the genetic effects introduce relatedness between individuals, there is no form of social interaction [as in Moore et al. (1997) and Bijma (2014)]. Each follows a distribution, where is a matrix of genetic variances and covariances. The vectors are independent of the ’s, but not independent among themselves. Defining a matrix with rows and columns , we can extend Equation 2 as follows:
| (4) |
Each vector contains the direct genetic effects on the jth trait. We make the following assumptions about the GSEM defined by (4), which are summarized in Figure 2:
All traits are measured in the same experiment: the rows of may be either observations at plot or plant level or genotypic means across plots or plants, but the observations should always come from the same experiment. In addition, the residual errors originate from biological variation, i.e., measurement errors are negligible [this in contrast to related work on Mendelian randomization (Hemani et al. 2017)].
Recursiveness: the graph defined by is a DAG. Consequently, there should be no feedback loops.
Causal sufficiency: the covariance matrix of the error vectors is diagonal, i.e., there are no latent variables. This means that all nonzero (nongenetic) correlations between traits must be the consequence of causal relations between the traits. We also assume the diagonal elements of to be strictly positive.
Genetic relatedness among individuals: is independent of , and has a matrix-variate normal distribution with row-covariance K and column covariance , where K is a relatedness matrix, which we describe in more detail below (see the section Genetic relatedness). Equivalent to this, the vector is multivariate normal with covariance , where denotes the operation of creating a column vector by stacking the columns of a matrix. Consequently, each is multivariate normal with covariance , where the variances form the diagonal of . Using the same notation, we can write that is matrix-variate normal with row-covariance and column covariance , and that .
No collinear genetic effects: the diagonal elements of do not need to be strictly positive, but, for all nonzero elements, the corresponding correlation should not be 1 or .
Assumptions 1–4 were also made in related work on structural models with random genetic effects (Valente et al. 2010; Töpner et al. 2017), and 1–3 are commonly made for structural models without such effects. Assumption 1 is implicit in the GSEM model (4) itself, as it is assumed that the structural equations propagate all errors to traits further down in the graph. Network reconstruction with traits from different experiments would rely completely on the genetic effects, requiring to be diagonal, which is a rather unrealistic assumption (see the Discussion, section Data from different experiments). A small amount of measurement error does not seem to pose problems for our PCgen algorithm. Larger amounts of measurement error will decrease power, which can, however, be avoided by increasing the number of genotypes or replicates (see Table S4, discussed below). Assumption 1 does not require traits to be measured at the same time. In particular, it is possible to include the same trait measured at different time-points, which, of course, puts constraints on causality. Such constraints can, in principle, be incorporated in our model, just as other biological constraints (see e.g., Peters et al. 2017), although we will not explore this here. What is also implicit in Equation 4 is that all causal relations between traits are linear. Our PCgen algorithm relies on this rather heavily, and we discuss the consequences of nonlinearity in the Results below (specifically, in the APSIM simulations, and the example just before the Discussion). In specific cases, it may be possible to obtain linearity by certain transformations of the data, but this requires prior knowledge that is typically unavailable. In the Discussion, we suggest various directions of future work to deal with nonlinear relationships, as well as non-Gaussian errors. In any case, as long as the other assumptions hold, the core of our framework (the graphical representation of genetic effects with a single node G, and the Markov property in Theorem 1 below) is still valid for nonlinear GSEMs.
Assumption 2 (no cycles) is essential given the type of data considered here, as the reconstruction of feedback loops requires time-course data (Peters et al. 2017), typically with high-resolution. Without such data (or only a few time-points), it is impossible to verify this assumption, but Maathuis et al. (2010) provide examples of interventions in yeast data, where cycles are likely to exist, but structural models still outperform nonstructural models.
Assumption 3 (no latent variables) is important for orientation of the edges, and has been studied in detail by many authors. In particular, Spirtes et al. (2001) and Colombo et al. (2012) proposed the FCI and RFCI algorithms, which are extensions of the PC-algorithm, and allow for latent variables. These algorithms could be extended with genetic effects, as we do here for the PC-algorithm (see the Discussion). Apart from nonlinear trait-to-trait relations, the APSIM simulations below also contain latent variables.
As in related work (Valente et al. 2010; Töpner et al. 2017), as well as in much of the literature on multi-trait genomic prediction and genome-wide association studies (GWAS), the relatedness matrix K is the same for all traits (Assumption 4). This may not hold if traits have very different genetic architectures, but seems a good approximation if most of the underlying QTL are small. Large QTL may be added as additional fixed effects.
Assumption 5 implies that, for each pair of traits with direct genetic effects, these effects should not be the result of exactly the same set of QTL, with exactly the same effect sizes. This seems a reasonable assumption whenever the underlying biological structures or processes are really different; see the section Dealing with derived traits in the Discussion. Of course, reconstruction of direct genetic effects will be more difficult under strong correlations, similar to, for example, the reduced power in GWAS when two causal loci are in strong LD.
Finally, there are a few additional assumptions which are required for the PCgen-algorithm, and are not essential for the definition of GSEM; see the overview in Figure 2 and the section Statistical properties of PCgen in the Results. In particular, we require the faithfulness assumptions defined by expressions 9 and 10 below, and assumptions about the conditional distributions. Appendices A.5 and A.6 provide additional examples and results regarding faithfulness.
Graphical representation of GSEM: extending with genetic effects
In contrast to previous work, we will explicitly take into account the possibility that there are no direct genetic effects on some of the traits. In this case, the corresponding rows and columns in are zero. Following the notation of Stephens (2013), we use to denote the index set of traits with direct genetic effects, and write for the submatrix with rows and columns restricted to D. From Assumption 5 above, it follows that is nonsingular, i.e., there can be no perfect correlations between direct genetic effects.
We graphically represent model (4) by a graph with nodes and a node G, which represent, respectively, and the matrix . contains an edge if the th entry of is nonzero, and an edge if is nonzero with probability 1, i.e., if . See Figure 3 for an example. In words, is defined as the original graph over the traits, extended with the node G and arrows for traits with a direct genetic effect, i.e., for all . Consequently, our main objective of reconstructing trait-to-trait relationships and direct genetic effects translates as reconstructing .
As for the ’s, we distinguish between the node G in the graph (normal type) and the random matrix it represents (bold face). is represented by a single node G, instead of multiple nodes . This choice is related to our assumption that K is the same for all traits; see File S7.1 for a motivating example. The orientation of any edge between G and is restricted to , because the opposite orientation would be biologically nonsensical. Because of our assumption that is a DAG, it follows that is a DAG as well, as a cycle would require at least one edge pointing into G.
We emphasize that is just a mathematical object and not a complete visualization of all model terms and their distribution, as is common in the SEM literature. In particular, does not contain nodes for the residual errors, path coefficients, or information about the off-diagonal elements of . While in general, is not entirely identifiable (Gianola and Sorensen 2004), we will see that is identifiable in terms of its skeleton (the undirected graph obtained when removing the arrowheads) and some of the orientations. The skeleton is generally not equal to the conditional independence graph, which is the undirected graph associated with the inverse covariance or precision matrix (Spirtes et al. 2001; Kalisch and Bühlmann 2007). See File S6.2 for an example.
Direct and indirect genetic effects
As pointed out by various authors (Gianola and Sorensen 2004; Valente et al. 2010, 2013; Töpner et al. 2017), the genetic variance of a trait is driven not only by its direct genetic effect , but also by direct genetic effects on traits affecting it, i.e., its parents in the graph . Assuming that the inverse exists, it follows from Equation 3 that
| (5) |
where the vector contains the total genetic effects for the ith individual. The vector contains the total genetic effects for the jth trait, where is defined as the jth column of . The vector of indirect genetic effects is the difference . In Figure 3 for example, and .
Likewise, we can distinguish between the contribution of direct and indirect genetic effects to the genetic covariance. The th element of in Equation 5 is the total genetic covariance between and . This is what is usually meant with genetic covariance. Most often, this is different from the covariance between the direct genetic effects and , given by . Indeed, affects the total genetic covariance, but the latter is also driven by causal relationships between traits, as defined by . If no such relationship exist, then contains only zeros, and . In general, however, these matrices are different, and, depending on the structure of the graph and the path coefficients, the correlation may be much larger than , or vice versa. For example, given direct effects and with equal variance and correlation 0.9, and an effect of size , the total genetic correlation is . Regarding the diagonal of , we note that traits without a direct genetic effect may still have positive genetic variance.
Genetic relatedness:
The genetic relatedness matrix K introduced in Assumption 4 determines the covariance between the rows of . In principle, our approach allows for any type of GRM, but, for simplicity, we focus on the following types. In all cases, K has dimension .
1. , Z being the incidence matrix assigning plants (or plots) to m genotypes, in a balanced design with r replicates for each genotype. This K is obtained when each genotype has an independent effect, as in the classical estimation of broad-sense heritability (or repeatability). Since no marker information is included, the model cannot be used directly for genomic prediction, but we will see that, for the reconstruction of (using the training genotypes), it has considerable computational and statistical advantages.
-
2. Given only a single individual per genotype (or genotypic means), we assume A being a GRM estimated from a dense set of markers, assuming additive infinitesimal effects.
3. Given both r replicates of m genotypes and a GRM A of dimension , we assume that . In absence of nonadditive effects, this covariance structure uses all available information. However, for computational reasons it is usually easier to work with either the replicates or with genotypic means and the GRM A. We further explore this issue in the simulations below and in the Discussion.
The balance required when is necessary in Theorems 5 and 6 below, but is not a general requirement for our models, nor for the PCgen algorithm.
The joint distribution implied by the GSEM:
The sum does not, in general, have a matrix-variate normal distribution, but from our Assumption 4, it still follows that is multivariate normal with covariance . We can rewrite Equation 4 as
| (6) |
where is the matrix of total genetic effects, with columns . Equation 5 now generalizes to
| (7) |
where is the matrix of fixed effects transformed by . This is a common model for multi-trait GWAS and genomic prediction [see, among others, Korte et al. (2012), Stephens (2013), and Zhou and Stephens (2014)]. In those works, however, and are arbitrary covariance matrices, whereas here they are modeled as functions of , and .
Under Assumption 3 ( diagonal), , and together have, at most, parameters, as many as and together. This suggests that , and might be identifiable from the distribution in Equation 7. In Appendix A.2 we show how , and can be obtained from and . Apart from Assumption 3, this requires knowledge of the graph, and the faithfulness Assumptions (9)–(10) given below. Equations 17 and 18 in Appendix A.2 can be used to derive estimates , and from estimates and , although the development of good estimators of , and is beyond the scope of this work. Such estimators should account for the structure of the GSEM, as defined by and , and might rely on alternative restrictions on the parameters (instead of diagonal ); see Gianola and Sorensen (2004).
Using the results of Spirtes et al. (2001) (p. 371), it turns out that can be written directly in terms of sums of products of path coefficients (see Appendix A.3). Consequently, there is no need to invert , although it still holds that , provided the inverse exists. Recalling that is the jth column of , we can express the jth trait as
| (8) |
which is Equation 6 restricted to the jth column. Similarly, for any nonempty index-set , the matrix of traits in S (i.e., with columns restricted to S) equals , where is the matrix with columns . Equation (26) (Appendix A.7) provides an expression for the covariance of . For the corresponding nodes in the graph, we write .
Causal inference without genetic effects:
So far, we have assumed that is known, in which case estimation of , and is usually of interest. In this work, however, we focus on the reconstruction of an unknown , based on observations from a GSEM given in Equation 4. We will do this with the PCgen algorithm introduced below, but will first review the necessary concepts, as well as existing methods. Appendix A.1 contains a more detailed introduction.
Suppose for the moment we have observations generated by an acyclic SEM without latent variables, and without genetic effects. From the pioneering work of Judea Pearl and others in the 1980s, it is known that we can recover the skeleton of the DAG and some of the orientations, i.e., those given by the v-structures. A v-structure is any triple of nodes such that , without an edge between and . All DAGs with the same skeleton and v-structures form an equivalence class, which can be represented by a completed partially directed acyclic graph (CPDAG). DAGs from the same equivalence class cannot be distinguished using observational data, at least not under the assumptions we make here. For the reconstruction of the CPDAG, constraint-based and score-based methods have been developed (for an overview, see Peters et al. 2017).
Here, we focus on constraint-based methods, which rely on the equivalence of conditional independence (a property of the distribution) and directed separation (d-separation; a property of the graph). An important result is that an edge is missing in the skeleton of the DAG if and only if and are d-separated by at least one (possibly empty) set of nodes, . Such is called a separating set for and . Given the equivalence of d-separation and conditional independence, this means that we can infer the presence of the edge in the skeleton by testing for all . The PC- and related algorithms therefore start with a fully connected undirected graph, and remove the edge whenever and are found to be conditionally independent for some . While the first constraint-based algorithms such as IC (Pearl et al. 1991) exhaustively tested all possible subsets, the PC-algorithm (Spirtes et al. 2001) can often greatly reduce the number of subsets to be considered. Although this is not essential for the equivalence of d-separation and conditional independence, most constraint-based algorithms assume that observations be indendently and identically distributed, and structural equations with additional random effects are usually not considered.
Existing approaches for estimating , given genetic effects:
To deal with the dependence introduced by the genetic effects, Valente et al. (2010) and Töpner et al. (2017) proposed to predict the total genetic effects (i.e., the term in Equation 6), and perform causal inference on the residuals. These methods are flexible, in the sense that any genomic prediction method can be used, and combined with any causal inference method. A disadvantage, however, is that the presence of direct genetic effects cannot be tested. Suppose, for example, that , and we subtract the total genetic effects. Then, given only the residuals, we can never know if part of the genetic variance of was due to a direct effect . Another disadvantage is that fewer of the between-trait edges can be oriented. Technically, this is because, in the CPDAG (showing which orientations can be recovered from data), typically more edges are undirected; see Appendix A.1 for more details. In the preceding example, the CPDAG associated with is , i.e., all orientations can be recovered. By contrast, the CPDAG associated with is , and we only know that the orientation is not (see Figure 2).
To use the causal information associated with genetic effects, Töpner et al. (2017) estimated “genomic networks”, based on the predictions themselves. These, however, seem to require additional assumptions, which are not required for the residual networks (in particular, diagonal ). Moreover, it seems difficult to relate edges in such a network to direct genetic effects (see the section Data from different experiments in the Discussion, and File S7.2). In summary, residual and genomic networks only estimate the (CPDAG associated with the) subgraph of trait-to-trait relations, instead of the complete graph .
Another disadvantage is that, without specific models putting restrictions on and , the MTM (7) can only be fitted for a handful of traits (Zhou and Stephens 2014) for statistical as well as computational reasons. For example, Zwiernik et al. (2017) showed that, for general Gaussian covariance models, (residual) ML-estimation behaves like a convex optimization problem only when . Similar problems are likely to occur for Bayesian approaches. The problem with fitting the MTM to data from the GSEM model (Equation 4) is that one cannot exploit the possible sparseness of . Even for sparse graphs with few direct genetic effects, the matrices and may still be dense, requiring a total of parameters. To overcome these limitations, we explicitly consider the presence or absence of direct genetic effects to be part of the causal structure, and develop PCgen, a causal inference approach directly on .
The PCgen algorithm:
The main idea behind PCgen is that the PC-algorithm is applicable to any system in which d-separation and conditional independence are equivalent, and where conditional independence can be tested. We first describe the algorithm and propose the various independence tests; the equivalence is addressed in Theorems 1 and 2 below. If we define and temporarily rename the node G as , PCgen is essentially the PC-algorithm applied to :
Skeleton-stage. Start with the fully connected undirected graph over and an empty list of separation sets. Then, test the conditional independence between all pairs and given subsets of other variables . Whenever a p-value is larger than the prespecified significance threshold α, update the skeleton by removing the edge and add to the list of separation sets for and . This is done for conditioning sets of increasing size, starting with the empty set (; marginal independence between and ). Only consider S that, in the current skeleton, are adjacent to or .
Orientation-stage. Apply the orientation rules given in File S1 (R1–R3 in Algorithm 1) to the skeleton and separating sets found in the skeleton-stage. For example, if the skeleton is and is not a separating set for and , the skeleton is oriented ; otherwise, neither of the two edges can be oriented.
In order to obtain PCgen, we need to make a few refinements to these steps. First, in the skeleton stage, we need to specify how to test conditional independence statements. Clearly, independence between two traits requires a different test than independence between a trait and (i.e., ), in particular because the latter is not directly observed. Second, we need to modify the orientation rules, in order to avoid edges pointing into G. The usual rules give the correct orientations when given perfect conditional independence information, but statistical errors in the tests may lead to edges of the form . Third, statistical errors can also make the output of PC(gen) order-dependent, i.e., putting the columns (traits) in a different order may lead to a different reconstruction. We therefore adopt the PC-stable algorithm of Colombo and Maathuis (2014), who proposed to perform all operations in the skeleton- and orientation-stage list-wise (details given in File S1). Apart from eliminating the order-dependence, this has the advantage that all conditional independence tests of a given size can be performed in parallel.
In summary, PCgen is the PC-stable algorithm with: (1) specific conditional independence tests (described below); and (2) modified orientation rules, in order to avoid edges pointing into G (File S1.2). As in the original PC-algorithm, the number of type-I and type-II errors occurring in the tests is determined by the choice of the significance threshold α, which is discussed in section Assessing uncertainty below and in the Discussion.
Skeleton stage: conditional independence tests.
We can distinguish between the following types of conditional independence statements in the skeleton stage:
| (A) |
| (B) |
| (C) |
where and [or, in statement (A), In words, (A) means that the trait is independent of all genetic effects , conditional on the traits . If S is the empty set, this is understood as marginal independence of and . Similarly, (B) and (C) express conditional independence of traits and given and or given alone.
We now propose statistical tests for statements (A) and (B), which rely on the linearity of our GSEM, as well as some additional assumptions, which we discuss in more detail below (section Statistical properties of PCgen, and Figure 2). Statement (C) can be tested using standard partial correlations and Fisher’s z-transform. However, as we show in File S6, this test is redundant, since for any set that d-separates and the set will also d-separate them. We therefore skip any test for and instead test the corresponding statement including , i.e., .
Testing :
Our test for statement (A) is based on the intuition that is independent of given whenever there is no direct genetic effect on (i.e., ), and all directed paths from G to are blocked by the set . In particular, if S is the empty set, there should not be any directed path from G to Because directed paths from G to will generally introduce some genetic variance in the idea is to test whether there is significant genetic variance in the conditional distribution of given This is done as follows:
When , we use the classical F-test in a one-way ANOVA, with and as covariates. Technically, this is an ANCOVA (analysis of covariance), where the treatment factor genotype is tested conditional on the covariates being in the model.
For other K, one can use a likelihood ratio test (LRT). The asymptotic distribution under the null-hypothesis is a mixture of a point mass at zero and a chi square.
In both cases, it is assumed that the conditional distribution of given is that of a single-trait mixed model, the mean being a linear regression over the conditioning traits. This assumption is made mathematically precise below in Equations 12 and 14.
Testing :
For statement (B), we mostly use the residual covariance (RC) test, which is based on the conditional distribution of and given the observed It is assumed that this distribution is that of a bivariate MTM, again with the mean being a linear regression over the conditioning traits; see Equations 13 and 14 below. Assuming the bivariate MTM, we test whether the residual covariance is zero, using the LRT described in File S1.3. The underlying idea is that a nonzero residual covariance must be the consequence of an edge or , because of the assumed normality and causal sufficiency. On the other hand, a nonzero genetic covariance may also be due to covariance between direct genetic effects on these variables, or due to a genetic effect on a common ancestor. The RC-test therefore compares the full bivariate mixed model with the submodel with diagonal residual covariance, while accounting for all genetic (co)variances. The RC-test is not to be confused with a test for zero genetic covariance. The latter is often useful for data exploration, but has no role in PCgen (although in File S1.4 we describe a LRT test that is implemented in our software).
An alternative to the RC-test is the RG-test [Residuals of GBLUP, i.e., the best linear unbiased prediction of the genetic effects]. Fitting the MTM (Equation 7), we obtain the BLUP of the total genetic effects and the BLUE of the fixed effects. We then test the significance of partial correlations among residuals, i.e., the columns of . When is close enough to , it follows from Equations 6 and 7 that the covariance of is approximately , i.e., that of independent samples, without any genetic relatedness. This approach is very similar to the work of Valente et al. (2010) and Töpner et al. (2017), who instead took a fully Bayesian approach to predict . In either case, the performance of the RG-test critically depends on the prediction error . As mentioned before, fitting an MTM is usually challenging for >5–10 traits; we therefore also consider residuals of single-trait GBLUP, as an approximation.
PCres: reconstructing only trait-to-trait relationships:
Testing only conditional independencies of the form (B), one can reconstruct the graph of trait-to-trait relations (see the green boxes in Figure 2). Moreover, if this is done with the RG-test, the algorithm is very similar to the residual approaches of Valente et al. (2010) and Töpner et al. (2017). Staying within the context of the PC-algorithm, and using residuals from GBLUP, we will call this PCres. As for the RG-test in PCgen, PCres can be based on residuals of either single or multi-trait mixed models.
Software:
In our R-package pcgen, we implemented PCgen for the case PCres is implemented for as well as . Moreover, PCres can be based on either residuals of the full MTM (Equation 7) (only for small numbers of traits), or from univariate models (the default). Tables 1 and 2 in File S2 provide a complete overview of the options, with the required R-commands. The package is freely available at https://cran.r-project.org/web/packages/pcgen/index.html. pcgen is built on the pcalg package (Hauser and Bühlmann 2012; Kalisch et al. 2012), in which we modified the orientation rules and the default conditional independence test.
Assessing uncertainty:
The PC-algorithm is asymptotically correct, in the sense that the underlying CPDAG is recovered if conditional independence can be tested without error (Spirtes et al. 2001). In Theorem 2 below, we provide a similar consistency result for PCgen. In practice however, type-I or type-II errors are likely to occur, leading to incorrect edges in the graph. Depending on the significance level α used in each test, there may be more type-I errors (large α) or rather more type-II errors (small α). Reliable control of the (expected) false positive rate, or total number of false positives, remains challenging; see the Discussion (Assessing uncertainty). We will therefore just consider the P-values as they are, and analyze the real datasets for different significance thresholds. Following Kalisch and Bühlmann (2007) and Kalisch et al. (2012), we report, for each remaining edge, the largest P-value found across all conditioning sets for which the edge was tested.
Extensions of PCgen:
File S3 describes several extensions of PCgen, which are partly implemented in our software. Among others, the causal graph and PCgen could be extended with fixed effect QTL, and PCgen can be sped up by starting with a skeleton obtained from PCres (“prior screening”). As in the pcalg-package, it is possible to restrict the maximum size of the conditioning sets, also to improve computation time.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The maize and rice data used below can be accessed at https://doi.org/10.15454/IASSTN and https://doi.org/10.6084/m9.figshare.7964357.v1, respectively. Supplemental materials are available at figshare: https://doi.org/10.6084/m9.figshare.11635392.
Results
Simulations with randomly drawn graphs
To compare the different algorithms, we simulated random GSEMs by randomly sampling the sets D (defining the traits with direct genetic effects) and the covariance matrices combined with randomly drawn DAGs over the traits Traits were simulated for an existing population of 256 maize hybrids (Millet et al. 2016). Two replicates of each genotype were simulated. Given the additive relatedness matrix A based on 50k SNPs, genetic effects were simulated such that (i.e., the vector of genetic effects for all the replicates, and all traits). File S4.1 provides further details, such as the magnitude of genetic (co) variances. We focus here on the comparison of:
PCgen based on the replicates, assuming (i.e., ignoring A). By default, we apply the prior-screening with PCres.
PCres (replicates): PCres based on residuals from univariate GBLUP, again using only the replicates.
PCres (means): PCres based on residuals from multivariate GBLUP, using genotypic means and the relatedness matrix A that was used to simulate the data.
Table S2 provides results for variations on these algorithms, including PCgen without prior screening. In all simulations, the significance threshold was . The effect of sample size, and the trade-off between power and false positives as function of α, was already investigated by Kalisch and Bühlmann (2007) for the standard PC-algorithm, and is likely to be similar for PCgen.
We separately evaluated the reconstruction of and the edges as the latter is only possible with PCgen. To assess the difference between estimated and true skeleton of we considered the true positive rate (TPR), the true discovery rate (TDR), and the false positive rate (FPR). Additionally, we used the Structural Hamming Distance (SHD), which also takes into account the orientation of the edges. File S4.2 provides definitions of these criteria. Reconstruction of is assessed only in terms of TPR, TDR, and FPR, as these edges can have only one orientation.
Simulation results:
We first performed simulations with traits (scenario 1), with each potential edge between traits occurring in the true graph with probability Hence, for any given trait, the expected number of adjacent traits was The edges were included in the true graph with probability In a related set of simulations (scenario 2), was increased to 0.5, giving denser graphs. In both scenarios, PCgen reconstructed the edges with little error, the average TPR being ∼0.97 and FPR ∼0.03 (Table 1). In the first scenario, about one-third of the actual edges between traits was not detected with PCgen (TPR i.e., the proportion of true edges that was discovered). At the same time, the number of false edges in was very low, which is also reflected in high TDR values (the proportion of edges in the reconstruction that is true). In scenario 2, the TPR, FPR, and TDR all increased. Hence, for denser graphs, more of the true edges were found, at the expense of a somewhat higher number of false edges.
Table 1. Performance of PCgen and residuals-based approaches, averaged over 500 simulated datasets per scenario.
| TPR | FPR | TDR | SHD | TPR | FPR | TDR | |
|---|---|---|---|---|---|---|---|
| Scenario 1 | |||||||
| PCgen | 0.647 | 0.006 | 0.981 | 0.442 | 0.982 | 0.026 | 0.995 |
| PCres (replicates) | 0.650 | 0.039 | 0.908 | 1.410 | |||
| PCres (means) | 0.521 | 0.390 | 0.438 | 3.174 | |||
| Scenario 2 | |||||||
| PCgen | 0.804 | 0.033 | 0.986 | 1.246 | 0.976 | 0.031 | 0.995 |
| PCres (replicates) | 0.819 | 0.073 | 0.939 | 2.320 | |||
| PCres (means) | 0.672 | 0.364 | 0.659 | 3.628 | |||
| Scenario 3 | |||||||
| PCgen | 0.895 | 0.002 | 0.985 | 6.806 | 0.969 | 0.018 | 0.991 |
| PCres (replicates) | 0.911 | 0.004 | 0.961 | 9.874 | |||
| Scenario 4 | |||||||
| PCgen | 0.959 | 0.001 | 0.942 | 27.288 | 0.976 | 0.022 | 0.943 |
| PCres (replicates) | 0.962 | 0.001 | 0.940 | 38.410 | |||
SE for the TPR, FPR, and TDR were between 0.001 and 0.015. SE for the SHD were ∼0.06 (scenarios 1 and 2), 0.18 (scenario 3), and 0.28 (scenario 4). For the performance of other variants of PCgen and PCres in scenarios 1 and 2, see Table S2. In scenario 4, we used PCgen with the RG-test (PCgen-RG-uni); in the other scenarios we used the RC-test, with prior screening (PCgen-RC-screening). All acronyms are explained in Table 1 in File S2. PCres (replicates) and PCres (means) refer to PCres-uni-R and PC-multi-A.
PCres (replicates) outperformed PCres (means), in spite of the use of univariate GBLUP, and ignoring the actual relatedness matrix. Hence, the information contained in the replicates appears much more important than the precise form of the relatedness matrix, or unbiased estimation of genetic correlations. The performance of PCres strongly depends on the prediction error of the GBLUP, and, in line with the results of Kruijer et al. (2015), this error appeared lowest when using the replicates. The use of both the replicates and the marker-based GRM (i.e., assuming as the data were generated), further improved performance, but only slightly (Table S1, PCres-uni-RA). Unsurprisingly, the MTM required for PCres (means) was computationally more demanding, and could often not be obtained for more than four traits. Motivated by this computational advantage, and the statistical advantages mentioned in the Discussion, all analyses in the remainder will consider only PCgen and PCres based on replicates.
For trait-to-trait relations, PCgen and PCres (replicates) had very similar performance in terms of TPR, TDR, and FPR. However, PCgen substantially improved the orientation of these edges, as shown by the reduced SHD. This is a consequence of the additional edges in the underlying graph: because of the fixed orientation of these edges, this generally increases the number of v-structures, and, hence, the number of between-trait edges that can be oriented. See again the example in Figure 2.
To assess performance in higher dimensions, we simulated data sets with traits, and (scenario 3), and with , and (scenario 4). Both scenarios consider sparse graphs; denser graphs can be analyzed as well, but, for p >20–30, require several hours, or even days, unless the size of the conditioning sets is restricted, or our implementation of PCgen would be parallelized. Here, we limited the size of conditioning sets to three (scenario 3) and two (scenario 4). As in the first two scenarios, PCgen achieved a strong reduction in SHD, and reliable reconstruction of the direct genetic effects (Table 1).
To assess the effect of thresholding the size of conditioning sets, we simulated 200 datasets with traits and a relatively dense graph ( and ), and used PCgen with various thresholds (Table S3). The restricted maximum size means that a certain number of conditional independence tests is skipped, which may lead to extra false positives. However, the thresholding is only done in PCgen itself and not in the prior screening with PCres (which is much faster, and already removes most false edges). Consequently, thresholding had very little effect on the reconstruction of trait-to-trait relations but did lead to a higher FPR in the reconstruction of the direct genetic effects (0.07 without thresholding, 0.08 with , and 0.48 with ). Also, the accuracy in the orientations of slightly decreased (SHD increasing from 15.9 to 16.2).
In another set of simulations, we explored the effect of measurement error. As expected, increasing amounts of measurement error decreased the power to detect between-trait edges as well as direct genetic effects (Table S4). However, the loss in power could largely be compensated by increasing the number of replicates, or the number of genotypes. The latter was most effective for between-trait edges, while increased replication gave the highest power for the edges
In our final set of simulations (Table S5), we explored the effect of strong correlations in i.e., when Assumption 5 is close to being violated. We simulated an example with two traits whose direct genetic effects had unit variance, and increasing covariance (0, 0.5, and 0.95). The corresponding TPR values for the genetic effects were respectively 0.94, 0.85, and 0.60. Consequently, even in the presence of strong correlations, PCgen still had some power to detect direct genetic effects.
Simulations using a crop-growth model
We also simulated data using the popular crop growth model APSIM-wheat (Keating et al. 2003; Holzworth et al. 2014). Compared to the preceding simulations, this represents a more challenging scenario, as several of the underlying assumptions are violated. In particular, the data-generating process introduces nonlinearities and latent variables. We simulated 12 traits for an existing wheat population of 199 genotypes, with three replicates each. The traits included seven primary traits, four secondary traits, and yield (Y). File S5 provides further details, and trait acronyms are given in Table S6. Traits were simulated by running a discrete dynamic model from the beginning to the end of the growing season. Motivated by the fact that some trait measurements are destructive, observations are taken only at Figure S1A shows the summary graph, defining the causal effects from one time-step to the next (Peters et al. 2017). We note that the summary graph does not directly describe the distribution of the traits at (obtained by marginalizing over previous time points), which can be represented by an ancestral graph (Richardson and Spirtes, 2002). As such graphs are outside the scope of this work, we investigate the extent to which we can reconstruct the summary graph, given observations taken at There are direct genetic effects on all of the primary traits, which have heritability 0.9. The genetic effects originate from 300 trait-specific QTL, with randomly drawn effect sizes. There are no direct genetic effects on secondary traits and yield.
Compared to the simulations above, it turned out to be much harder to detect the absence of direct genetic effects: in the PCgen reconstruction, all 12 traits had such effects (Figure S1B; highest P-value: ). These false positives seemed to be a consequence of the nonlinearities in the data-generating distribution, which are not accounted for in our tests. The reconstructed trait-to-trait relations were mostly correct, except for the missing edge and one incorrect orientation PCres made the same errors (Figure S1C), with an additional false arrow The standard PC-stable algorithm applied to all traits and QTL led to many more errors (Figure S1D), such as the false edge between and , the missing edge and some incorrect orientations. These errors occurred because, for various traits many QTL-effects were removed from the graph, i.e., for some set of traits , the conditional independence was mistakenly accepted. This, in turn, led to problems in the remaining tests, where part of the genetic variance was not taken into account. We emphasize that all 300 QTL were available to the PC-algorithm, and no other markers were provided. Hence, the poor performance in this case is really a consequence of the small effects, rather than the difficulty of QTL detection.
Two case-studies
We now use PCgen to analyze real data from four field trials and one experiment in a phenotyping platform. In all network reconstructions, we used a significance threshold of . Reconstructions with are shown in Figures S2 and S4. Figure S5 and Table S9 contain P-values for the remaining edges. In all datasets, we removed traits that were derived as sums or ratios of other traits, rather than being directly measured. In particular, the maize data do not contain grain number, which was defined as the ratio of yield over grain weight. We return to this issue in the Discussion.
Maize:
First, we analyze the field trials described by Millet et al. (2016, 2019), with phenotypic data for 254 hybrids of maize (Zea mays). We consider a subset of four trials, representing four (out of a total of five) different environmental scenarios described in Millet et al. (2016). See Table S8 for an overview. The scenarios were derived from physiological knowledge, crop-growth models, and environmental sensors in the fields. Scenarios were defined as a combination of well-watered or water-deficient conditions (WW vs. WD) and temperature. The latter was classified as “Cool” (average maximum and night temperature below respectively 33 and 20°), “Hot” (above 33 and 20°) or “Hot (days)” (maximum temperature above 33, night temperature below 20). Most trials included seven traits:
three height traits, i.e., tassel height , ear height , and plant height the latter is missing in the Ner12R trial.
two flowering time traits: anthesis (A) and silking , which are male and female flowering, respectively.
two yield-related traits: grain weight and yield (Y).
Table S7 provides an overview of trait acronyms. Each trial was laid out as an alpha-lattice design, with either two or three replicates. Spatial trends and (in)complete block effects were estimated using the mixed model of Rodríguez-Álvarez et al. (2018) (R-package SpATS), and subtracted from the original data; PCgen was then applied to the detrended data, assuming a completely randomized design. Residuals from SpATS appeared approximately Gaussian, and no further transformation was applied.
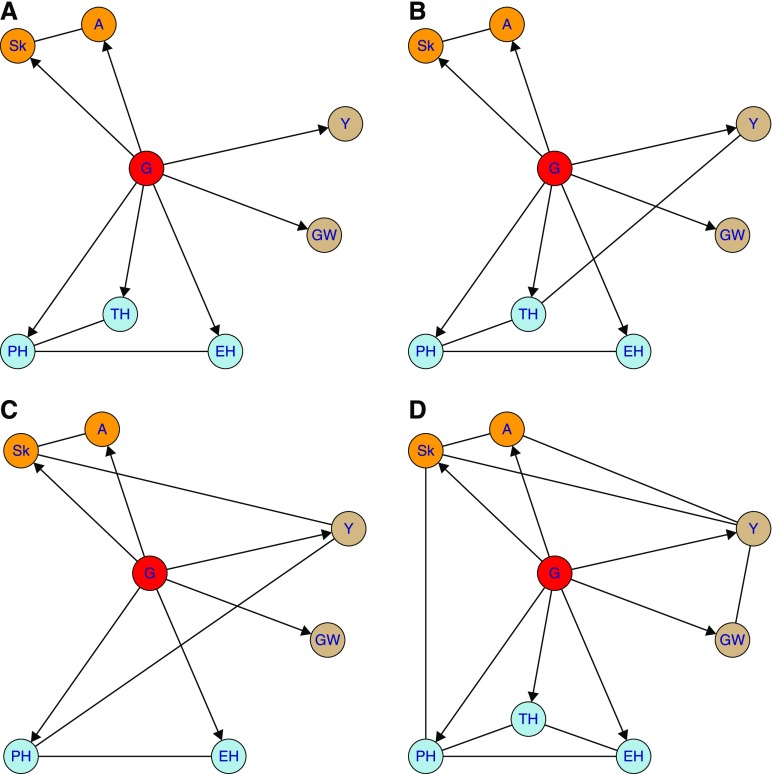
In the PCgen reconstruction, all traits have direct genetic effects, and traits mostly cluster according to their biological category (height, flowering, and yield-related), especially in the WW scenarios (Figure 4, A and B). In the Ner13W and Ner12R trials (Figure 4, B and C), there are edges between yield and respectively tassel- and plant-height, but these (conditional) dependencies are weak and disappear in the reconstruction with (Figure S2). Much stronger are the edges between yield and the flowering traits in the water-deficit trials (Figure 4, C and D); the corresponding conditional independence tests gave highly significant P-values for all of the considered conditioning sets (Table S9). By contrast, in the trial without heat or drought stress (Kar12W), the and edges were already removed in the test conditioning only on the genetic effects; Figure S3 provides an illustration. The relation between yield and delay in silking in maize is well known [see e.g., Borrás et al. (2007) and Araus et al. (2012)]. In the most stressed environment (Bol12R), there is an additional edge between plant height and silking. This may relate to the fact that the timing of anthesis determines the number of phytomeres (number of internodes and leaves) that a plant will generate, which, in turn, affects plant height (McMaster et al. 2005). The strong correlation between anthesis (A) and silking may explain the presence of the edge (rather than ).
Figure 4.
Estimated networks with for four of the DROPS field trials. Trait categories are flowering (orange), height (blue), and yield (brown). Each trial represents a different environmental scenario, arising from well-watered (WW) or water-deficit (WD) conditions, and different temperatures (see text). (A) Kar12W, (WW, Cool). (B) Ner13W (WW, Hot). (C) Ner12R (WD, Hot (days)). (D) Bol12R (WD, Hot). Trait acronyms are given in Table S7.
Finally, apart from the Bol12R trial, there is never an edge between Y and which seems due to the choice of the genetic material (giving little variation in grain weight) and the design of the trials (targeting stress around flowering time, rather than the grain filling period). See Millet et al. (2016) for further details. For all trials, the structure of the graphs is such that none of the between-trait edges can be oriented (technically, this is due to a lack of v-structures). However, for some of these edges, physiological knowledge clearly suggests a certain orientation, in particular for and
The trials also illustrate the difference between the total genetic covariance and the covariance among direct genetic effects, as defined by For most pairs of traits, the total genetic correlation was between 0.3 and 0.9 (Table S10). The (total) genetic correlation between yield and silking was strongly negative in both WD trials ( and ), and, in the Bol12R trial, also for yield and anthesis In all trials, genetic correlation with was negative for most traits, but not always significant. In the Kar12W trial for example, we found for and and for and (silking). In both cases, the two traits are d-separated in the graph (conditioning on ), but only for is the genetic covariance significant .
As we have seen in the examples following Equation (5), the existence of an edge between two traits in the graph does not necessarily imply a strong genetic correlation. In other words, having a shared genetic basis is not the same thing as the presence of a causal effect, found after conditioning on the genetic effects and other traits. In the Ner12R trial, for example, there is no edge between yield and grain weight, but a significant genetic correlation, whereas in the Bol12R trial it is the other way round.
Rice:
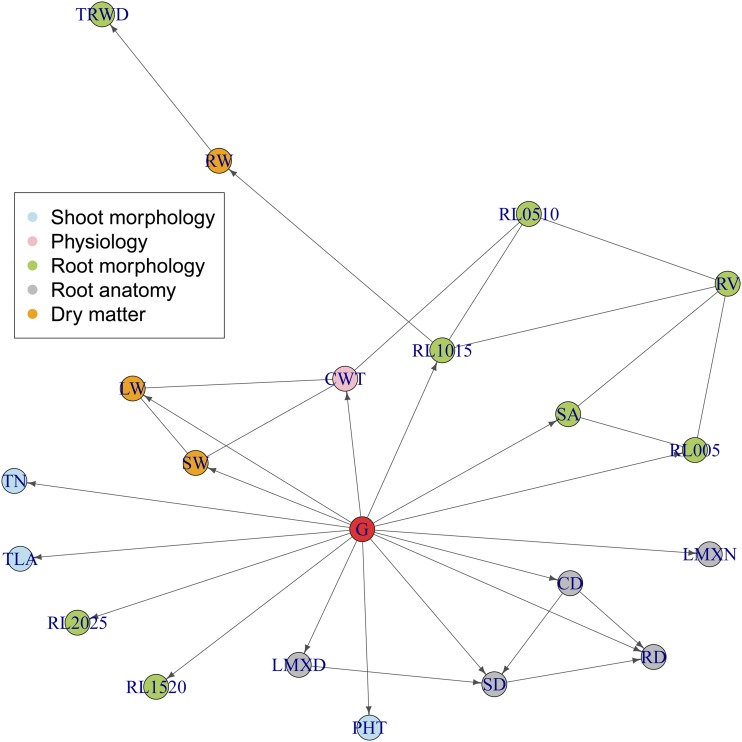
Next we analyze 25 traits measured on 274 indica genotypes of rice (Oryza sativa) under water deficit, reported by Kadam et al. (2017). Three replicates of each genotype were phenotyped in a randomized complete block design, and block was included as a covariate in all conditional independence tests. Tests were restricted to conditioning sets of, at most, four traits. A first run of PCgen produced several inconsistencies in the genetic effects, i.e., traits with significantly positive heritability but without a partially directed path coming from the node G. We therefore applied the correction described in File S3, adding edges for all traits with this inconsistency, and then re-ran PCgen. The final reconstruction is given in Figure 5, where traits are grouped into 3 shoot morphological traits (blue), 1 physiological trait (rose), 13 root morphological traits (green), 5 root anatomical traits (gray), and 3 dry matter traits (orange).
Figure 5.
PCgen-reconstruction for the rice data from Kadam et al. (2017), with Five traits (MRL, ART, RL2530, RL3035, and RL35) are not shown, as they were completely isolated in the graph, without any connections to other traits or G. Trait acronyms are given in Table S11.
After correcting the inconsistencies, there were nine traits without a direct genetic effect. Five of these (MRL, ART, RL2530, RL3035, and RL35) were completely isolated in the graph, without edges connecting to any other trait. All of these traits are related to either root length, or to the length of thicker roots, which contribute to drought adaptation under field conditions (Uga et al. 2013). However, as the experiment was done in pots, roots were constrained in their exploration range and therefore genotypic differences in root length would not translate into differential access to water and biomass (Poorter et al. 2012). Four other traits (TRWD, RW, RL0510, and RV) had at least one adjacent trait in the graph, but no direct genetic effect. At a lower significance level (, Figure S4), direct genetic effects disappeared also for cumulative water transpiration (CWT), and for three root anatomical traits (RD, CD, and SD). For RV (root volume), a direct genetic effect was only present with , which was an artifact of the way the initial consistencies were resolved.
Traits related to root surface area (SA), root volume (RV), and roots with small diameter class (RL005, RL1015) had direct genetic effects, and were connected among each other. As expected, traits related to root volume and area influenced root weight and total root weight density (RW, TRWD). In the reconstruction with , cumulative water transpired (CWT) was affected by stem and leaf weight (SW, LW), and by RL0510, in agreement with the physiological knowledge that water transpiration is influenced by water demand (related to the above-ground biomass) and water supply (related to the roots’ water uptake capacity). The corresponding edges were also present in the reconstruction with , where, however, they could not be oriented because of the denser network (in particular, the presence of ). Root anatomical traits (LMXD, SD, CD, and RD) appeared as a separate module, not related to the plant water dynamics, suggesting that root anatomy had a smaller impact on water uptake compared with root biomass.
Statistical properties of PCgen
We now investigate a number of statistical issues: the assumptions required for asymptotic consistency of PCgen, the assumptions required for faithfulness, and properties of the conditional independence tests. Readers primarily interested in the application of PCgen could skip this section and continue with the Discussion. Proofs of Theorems 1–6 are given in Appendix A.
Consistency:
Asymptotic consistency holds if, for increasing sample size, the probability of finding the correct network converges to 1. Correct in this context means that we recover the CPDAG that contains the underlying DAG. Consistency of the PC-algorithm was shown by Spirtes et al. (2001) (for low dimensions) and Kalisch and Bühlmann (2007) (for high dimensions). These authors distinguished between consistency of the oracle version of PC, where conditional independence information is available without error, and the sample version, where conditional independence is obtained from statistical tests. For PCgen we will focus on the oracle version and consistency of the skeleton, leaving the sample version and the correctness of the orientations for future work.
As for the standard PC-algorithm, consistency of PCgen requires equivalence between conditional independence and d-separation in the graph. Part of this is the Markov property, which states that d-separation of two nodes in the graph, given a set of other nodes, implies conditional independence of the corresponding random variables. The converse (conditional independence implying d-separation) is known as faithfulness. The following result provides the Markov property for SEM with genetic effects. The proof (Appendix A.9) is a straightforward adaptation of Pearl’s proof for general SEMs (Pearl 2009).
Theorem 1 Suppose we have a GSEM as defined by Equation (4), with a graph as defined in the Materials and Methods, and satisfying Assumptions 1–4. Then, the global Markov condition holds for and the joint distribution of In particular, d-separation of and G given implies and d-separation of and given implies for all traits and and subsets
If we now assume faithfulness, the preceding result directly gives the equivalence between conditional independence and d-separation. This, in turn, implies that PCgen will recover the correct skeleton:
Theorem 2 Let denote d-separation in the graph . Suppose we have a GSEM as in Theorem 1, and we make the additional assumptions of faithfulness:
| (9) |
| (10) |
for all traits and and subsets The oracle version of PCgen then gives the correct skeleton.
Faithfulness:
For our first faithfulness condition (expression 9) to hold, it suffices to have faithfulness for the graph without genetic effects:
Theorem 3 Let denote the joint distribution of conditional on the matrix of total genetic effects. Then is equivalent with and is equivalent with Therefore, (9) holds if
| (11) |
Consequently, we can rephrase (9) in terms of a faithfulness assumption for the analogous SEM without genetic effects. A necessary (but not sufficient) condition for this is that contributions from different paths do not cancel out (Appendix A.5).
The second faithfulness statement (10) involves d-separation of and G, and requires that the genetic effects are not collinear. If, for example, we have with and it follows that Consequently, because we find that and are marginally independent, but, in the graph the nodes and G are not d-separated by the empty set, as there are directed paths and Conversely, if and are not perfectly correlated, this violation of faithfulness cannot occur. The following theorem shows that marginal independence always implies d-separation. We conjecture (but could not prove) that (10) also holds for nonempty conditioning sets.
Theorem 4 Suppose we have a GSEM satisfying Assumptions 1–5, and faithfulness for the graph without genetic effects, given by (11). Then implication (10) holds for i.e., marginal independence implies d-separation of and G.
Hence, faithfulness involving and requires (at least) absence of collinearities between genetic effects, as well as faithfulness for the corresponding SEM without genetic effects.
Properties of the tests:
Theorem 2 provides consistency of the oracle version of PCgen, where conditional independence information is available without error. Proving consistency of the sample version is challenging for two reasons. First, the assumptions made for our conditional independence tests may not always hold, introducing approximation errors. Second, even without these errors, the probabilities of type-I and type-II errors still need to converge to zero with increasing sample size. This is well known for the PC-algorithm with independent Gaussian data (Kalisch and Bühlmann 2007), but more difficult to establish in the presence of genetic effects. Here, we address the first issue, leaving the second for future work.
Our tests for conditional independence statements (A) and (B) (i.e., and ) rely on the conditional distributions of, respectively, and given the observed
The normality of these distributions directly follows from the assumed normality of the genetic and residual effects. We made the following assumptions about the form of their covariance and mean:
- The covariance matrix is that of a single-trait mixed model with the same relatedness matrix K assumed in the GSEM, i.e.,
for some variance components and .(12) - The covariance matrix is that of a bivariate MTM, again with the same K assumed in the GSEM:
for some matrices and .(13) - The conditional means and are linear regressions over the conditioning traits:
where is the marginal mean of (see Equation 8), and and are vectors of regression coefficients.(14)
In the following theorems we show that when , the assumptions in Equations 12 and 13 always hold, i.e., they directly follow from our GSEM model.
Theorem 5 When the distribution of has covariance of the form given by Equation 13, i.e., that of a bivariate MTM. Moreover, under faithfulness condition (9), the residual covariance in the MTM is zero if and only if
Theorem 6 Suppose we have a GSEM as described in Theorem 1, with Then, the covariance of is of the form , for any conditioning set S. Moreover, assuming the faithfulness condition (10) and of full rank (Assumption 5), is zero if and only if
Apart from the covariance structure, these theorems address the correctness of our tests. In particular, Theorem 5 shows that the residual covariance in the distribution of is indeed the right quantity to test statement (B). Similarly, the genetic variance in the conditional distribution of is the relevant thing for testing (A). This appears to be true for any conditioning set S, although (in Theorem 6) we could prove it only for the empty conditioning set, because faithfulness is required (which we also established only for ; see Theorem 4).
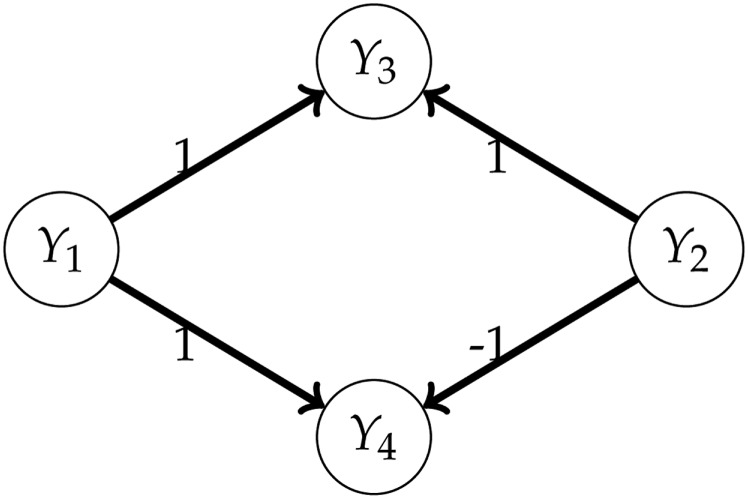
The situation is different for the assumption in Equation 14, regarding the conditional means: even when it holds for certain conditioning sets and not for others. We illustrate this with the following example. Suppose that and with independent vectors and Then, the graph is given by . There is no edge although this is not essential for the example. The distributions are given by
The conditional mean of given is As expected given the graph, the conditional mean is a simple linear regression on However, the conditional mean of given equals
which is a linear transformation, but not a multiple of In summary, our models for and are sometimes misspecified in terms of the mean, although still correct in terms of covariance, provided (Theorems 5 and 6). Despite the approximation error occurring sometimes for the conditional means, our tests still seem to perform reasonably, as shown in the simulations above. The assumption in (14) is more problematic if relations between traits are nonlinear. Suppose, for example, that, for each individual i, and where, for the sake of argument, we assume absence of residual errors. Then, the factor genotype will generally be significant in the ANCOVA with as covariate. For example, there could be two replicates of three genotypes, with genetic effects . Then, clearly, there is some unexplained genetic variance when regressing on .
Finally, we briefly discuss how the approximation could be improved. In general, the conditional mean is a function of the genetic and residual covariances between and . In Appendix A.7 (Equation 23) we derive that has mean Defining for we can write Consequently, our approximation of the conditional mean models as a linear regression on This approximation could probably be improved if we have good estimates of and and set Such estimates, however, require fitting an MTM for traits, which for large S is statistically and computationally challenging, unless pairwise or other approximations are applied (Furlotte and Eskin 2015; Joo et al. 2016). Moreover, it seems unclear how should be incorporated in the tests.
Discussion
Causal inference for data with random genetic effects is challenging because of the covariance between these effects, and because the usual assumption of independent observations is violated. To address these problems we have proposed a model where random genetic effects are part of the causal graph, rather than a nuisance factor that first needs to be eliminated. The resulting distributions and graphs were shown to satisfy the Markov property. This led us to develop the PCgen algorithm, which tests conditional independence between traits in the presence of genetic effects, and also conditional independence between traits and genetic effects. We showed that the presence of a direct genetic effect can be tested, just like the direct (fixed) effect of a QTL can be tested. This is, of course, relative to the observed traits, i.e., for any effect there may always be an unmeasured trait Z, such that
In the linear simulations as well as in the rice data, our tests could indeed identify the absence of many direct genetic effects. By contrast, in the APSIM simulations and maize data all traits had such effects. In the latter case, this could be for biological reasons, i.e., the genetic variance of each trait might really be “unique” to some degree. However, the APSIM results showed that nonlinearities could increase the false positive rate in the edges which may be avoided in future versions with better conditional independence tests. Such tests might also allow for non-Gaussian data.
In our simulations, PCgen also improved the reconstruction of between-trait relations. Part of this improvement is due to phenotypic information on replicates, reducing the number of errors in the tests. Another part is due to the improved orientation, which is a consequence of the additional edges Compared to previous algorithms, PCgen also appeared to be computationally more efficient: depending on the sparseness of the network, it can analyze ∼10–50 traits on a single core, and many more if we limit the maximum size of the conditioning sets, or would parallelize the conditional independence tests.
As for the original PC-algorithm, PCgen is most efficient for sparse graphs, i.e., when each trait is connected to only a few other traits, and when there are few direct genetic effects. But, even when this is not the case, PCgen still has an advantage over existing approaches: by incorporating the genetic effects in the PC-algorithm, we do not need to fit an MTM for all traits simultaneously, but only for bivariate models. Our approach also makes genetic network reconstruction feasible with just two traits, and in the absence of QTL or even no genotypic data at all.
As any causal inference method, PCgen only suggests causal models that are, in some sense, compatible with the data, and cannot validate the existence of a functional relationship, which is possible only through additional experiments. Because of the required assumptions, the identifiability issues and the uncertainty in the estimated networks, it may be better to speak of an algorithm for causal “exploration” than for causal “discovery”. At the same time, analysis of one trait conditional on other traits (e.g., yield given plant height) is a common and natural thing to do (Stephens 2013). From that perspective, PCgen could be seen simply as a tool that performs such analyses systematically, combines them, and visualizes the results. PCgen results for different significance levels could then be reported alongside other “descriptive” statistics like heritability estimates and genetic correlations, suggesting functional hypotheses that are of interest in future research.
Dealing with derived traits
We analyzed the maize and rice datasets using the traits as they were measured, without adding “derived” traits defined by ratios, sums, or differences of the original traits. Because such derived traits are not measured themselves, there is no error associated with them, apart from “copies” of errors in the original trait. For example, if there is much variation in leaf weight but almost none in the total weight of a plant, the derived trait “leaf weight ratio” will be essentially a copy of the original leaf weight trait. This can violate our assumption of faithfulness, and lead to errors in the reconstruction; see Figure 7 in Appendix A for an example. Sometimes, derived traits are biologically highly relevant. It may then be desirable to include them in the analysis, and omit some of the original traits. Alternatively, derived traits may be added after running PCgen. For example, we may extend the reconstructed graph with the node and edges and provided that this makes sense biologically.
Data from different experiments
We assumed traits to be measured on the same individuals in the same experiment, with residuals errors arising from biological variation (Assumption 1). In certain applications, this assumption can indeed be restrictive, but seems to be inevitable. Suppose traits were measured in different experiments, or residual errors would only come from measurement errors. Then there would be no propagation of residual errors, and the reconstruction would rely completely on how the genetic effects are propagated through the network. The GSEM model (Equation 4) would need to be replaced by and where in a sense models direct genetic effects, reminiscent of the genomic networks of Töpner et al. (2017). However, if data are actually generated by Equation (4), these networks provide only partial information about the direct genetic effects, even without any type-I or type-II error in the tests (see File S7.2). Moreover, the use of the PC-algorithm would require the columns of to be independent, which appears to be a rather unrealistic assumption.
Biologically, the genomic networks have a different interpretation: for example, we would assume that the genetic component in high blood pressure causes some cardiovascular disease, rather than high blood pressure itself. The alternative model implies that the observed traits have diagonal residual covariance, instead of the matrix obtained under Assumption 1 (see Equation 5). However, the latter matrix turned out to be essential for network reconstruction (see e.g., Theorems 5–6 above). This is why, without Assumption 1, we would need to rely completely on the genetic effects.
A relevant alternative approach here is that of invariance causal prediction (Peters et al. 2016), which infers causal effects that are consistent across several experiments, but still requires all traits to be measured in each experiment (as well as low genotype-by-environment interaction).
Replicates vs. means
In principle, PCgen allows for any type of genetic relatedness. We have however focused on the case of independent genetic effects, for the following reasons:
Performance under model misspecification: different types of genetic effects could, in theory, be represented by introducing multiple genetic nodes, with conditional independence tests that can distinguish between these effects. But this seems difficult in practice due to the computational requirements and lack of statistical power (Blair et al. 2012; Uhler et al. 2013; Kruijer 2016). For this reason, it seems, previous work on network reconstruction used genotypic means and an additive GRM. For the analysis of a single trait, however, Kruijer (2016) showed that broad-sense heritability estimates (obtained with ) capture any type of genetic effect, while a model assuming only additive effects can produce strongly biased heritability estimates, if the actual genetic effects are, for example, partly epistatic. It seems plausible that this robustness extends to the multivariate models considered here, for example, when direct genetic effects are driven by different sets of QTL, leading to trait-specific relatedness matrices.
Higher power: estimates of (total) genetic variance based on replicates are typically more accurate than marker-based estimates based on genotypic means (Kruijer et al. 2015; Visscher and Goddard 2015), and the use of replicates is therefore also likely to improve hypothesis testing. For the reconstruction of trait-to-trait relations with PCres, our simulations indeed suggest that replicates give more power. Mixed models with both replicates and a GRM might further increase power if the true architecture is really additive (Kruijer et al. 2015), but also these models lead to biased inference if the actual architecture is different (Kruijer 2016).
When the conditional independence statement considered in the RC-test is completely equivalent with (Theorem 5), while for other K it is not, and an alternative test might be required.
Apart from these statistical issues, there is also a computational advantage: the test for can be based on standard ANCOVA, which is many times faster than the LRT for a mixed model. Also the tests for are faster when
Finally, we have not investigated the performance of PCgen for unbalanced designs, but it seems likely that small unbalancedness has only a minor effect. A more fundamental challenge seems to be the presence of incomplete blocks or spatial trends (Flaxman et al. 2015; Rodríguez-Álvarez et al. 2018).
Assessing uncertainty
If one mistakenly rejects the null-hypothesis of conditional independence (type-I error), PCgen leaves the corresponding edge, although it may still be removed at a later stage, with a different conditioning set. If the null-hypothesis is mistakenly not rejected (type-II error), a true edge is removed, and will not be recovered. Moreover, it may affect the remaining tests, since d-separation of and is only tested given conditioning sets contained in or , where the adjacency sets are defined relative to the current skeleton. This is correct in the oracle version, but in the sample version of PC(gen), or may become smaller than the corresponding adjacency sets in the true graph, and the algorithm may therefore not perform an essential independence test. See Colombo and Maathuis (2014) for examples.
Consequently, assessing uncertainty for constraint-based algorithms is difficult, and cannot be achieved by just applying some multiple testing correction to the P-values. To obtain bounds on the expected number of false edges in the skeleton, several authors have used stability selection (Meinshausen and Bühlmann 2010; Stekhoven et al. 2012) or other sample-splitting techniques (Töpner et al. 2017), but these are often too stringent and require an additional exchangeability assumption (Bühlmann et al. 2013; Meinshausen et al. 2016). Moreover, these approaches do not provide a level of confidence for specific edges. For example, an edge present in of all subsamples may appear to be present in the true graph with a probability of 0.6, but there is no justification for such a statement.
Alternatively, uncertainty may be assessed using Bayesian methods, which are, however, computationally very demanding and outside the scope of this work. Moreover, despite the recent progress in Bayesian asymptotics (Ghosal and van der Vaart 2017), there seem to be no results regarding the correct coverage of posteriors in these models.
Genomic prediction
PCgen can select traits with direct genetic effects, which are the most relevant in genomic selection. More generally, the usefulness of structural models for genomic selection depends on whether there is an interest in some kind of intervention (Valente et al. 2013, 2015). Informally speaking, an intervention is an external manipulation that forces some of the traits to have a particular distribution. For example, with a so-called hard intervention on the jth trait, is forced to a constant level c, e.g., when is the expression of a gene that is knocked out. The manipulation or truncated factorization theorem (Spirtes et al. 2001; Pearl 2009) can then predict the joint distribution of the system after the intervention:
| (15) |
where is the set of parents of This is generally different from the distribution
| (16) |
obtained from conditioning on , prior to the intervention [see e.g., Peters et al. (2017)]. In other words, conditioning is not the same as doing (intervening). In a simulated example (File S7), we show that the use of Equation 15 can indeed greatly improve accuracy after an intervention, compared to standard methods ignoring the underlying structure. When, however, the intervention is on a root node, Equations 15 and 16 are the same (see again Peters et al. 2017, p. 110).
The example in File S7.4 is special in the sense that PCgen could correctly infer the complete graph, for most of the simulated datasets. A technical obstacle for a more general use of our networks in genomic prediction is the identifiability issue mentioned in the introduction. PCgen (and the PC-algorithm in general) typically outputs a partially directed graph, several DAGs being compatible with this graph. This is particularly problematic for edges between the traits in D (traits with direct genetic effects). For traits with only indirect genetic effects, it is possible to estimate how much of the genetic variance originates from a particular trait in D, the result being independent of the chosen DAG. This would first require estimates of the (total) genetic covariance among traits in D, obtained either by fitting an MTM, or by an approximation (as in Furlotte and Eskin 2015).
In absence of interventions on the traits, we can think of genomic prediction in terms of an intervention on the node G. Because the latter is a root node by definition, standard genomic prediction methods can, in principle, have optimal performance (Valente et al. 2013). More specifically, genomic prediction usually involves a regression of a target trait on a number of markers, having either fixed or random effects. In either case, it is only the total effect of each underlying locus on the target trait that matters, not through which other traits this effect passes.
Optimal prediction accuracy, however, requires that the regression model contains the true distribution (or a good approximation), and a sufficiently accurate estimate of this distribution. We therefore believe that structural models may sometimes be an appealing alternative, especially if the underlying model is highly nonlinear, or when prior physiological knowledge can be incorporated. The extent to which this can really improve accuracy remains to be investigated.
Open questions and extensions
Although we have shown the Markov property for our model, and studied consistency of PCgen, there are a number of open questions left for future work. First, it may be possible to construct better tests, especially for nonlinear structural models and non-Gaussian error distributions. The recent work of Pfister et al. (2018) seems particularly relevant here. A second issue is the consistency of the orientations: while we have shown PCgen’s consistency in reconstructing the skeleton, we did not address this for the final CPDAG. This is well known for the PC-algorithm without genetic effects (Spirtes et al. 2001; Kalisch and Bühlmann 2007), but more difficult to establish here, as the class of CPDAGs needs to be restricted to those without errors pointing to G. More generally, orientation constraints seem to be of interest for trait-to-trait relationships as well, e.g., one may require that, if there is an edge, the expression of a gene can only affect a metabolite and not the other way round. To the best of our knowledge, current methodology and theory has considered only the forced absence/presence of an edge, leaving the orientation to the algorithm [The pcalg-package (Kalisch et al. 2012) has the addBgKnowledge option to add orientations (‘background knowledge’), in the estimated CPDAG. This is however only done after running PC or a related algorithm, and is only allowed if compatible with the CPDAG]. A final question for future work is whether Theorems 4 and 6 hold for general conditioning sets.
Apart from these open questions, we believe that the idea of explicitly modeling direct genetic effects can be applied more generally. In particular, we hope that the ideas developed here provide a first step toward the more ambitious goal of modeling multiple traits through time, simultaneously for many environments. A first generalization would be to replace the PC-algorithm with other constraint-based algorithms, in particular FCI and RFCI (Spirtes et al. 2001; Colombo et al. 2012). These have the advantage that the causal sufficiency assumption (no latent variables) can be dropped or considerably weakened. The presence or absence of direct genetic effects could also be incorporated in (empirical) Bayesian approaches for genetic network reconstruction, or in invariant causal prediction (Peters et al. 2016). It might also be possible to extend the approach of Stephens (2013), and focus only on the detection of traits with direct genetic effects. Another application of GSEM might be as covariance models in multi-trait GWAS, as an alternative to unstructured (Zhou and Stephens 2014) or low-rank (Millet et al. 2016) models. Finally, the concept of direct and indirect genetic effects may be useful in deep-learning models for high-dimensional phenotypes, observed on genetically diverse individuals.
Acknowledgments
We thank Niteen Kadam for providing the rice data, and Xinyou Yin for useful discussions on the interpretation of the resulting networks. Emilie Millet and François Tardieu are acknowledged for providing and interpreting the maize data. We thank Sach Mukherjee for suggesting the graphical overview shown in Figure 2. Guido Schmeits is acknowledged for Latex support for constructing this figure. W.K. was funded by the Learning from Nature project of the Dutch Technology Foundation (STW), which is part of the Netherlands Organisation for Scientific Research (NWO). M.X.R. was funded by project MTM2017-82379-R (AEI/FEDER, UE), by the Basque Government through the BERC 2018–2021 program and by the Spanish Ministry of Science, Innovation and Universities (BCAM Severo Ochoa accreditation SEV-2017-0718). E.W. acknowledges support from the EU COST Action CA15109.
Author contributions: W.K. developed the PCgen algorithm. P.B. developed the package pcgen, based on code written by W.K. and the E.M.-algorithm contributed by M.X.R. W.K. wrote the paper, with input from F.A.v.E., D.B.-K., E.W., B.Y., P.B., and M.X.R. D.B.-K. simulated data with APSIM, and analyzed the rice data. P.B. visualized the estimated networks for the rice, maize, and APSIM data. WK proved Theorem 1–2 and W.K., E.W., and S.M.M. proved Theorems 3–6.
APPENDIX A. Faithfulness, conditional distributions, and proofs of Theorems 1–6
A.1. Overview of graph theoretic definitions
Definitions of, for example, d-separation and CPDAGs can be found in many books and articles on graphical models and causal inference (see for example, Lauritzen 1996; Spirtes et al. 2001; Kalisch and Bühlmann 2007; Pearl 2009). The following summary was inspired by Shipley (2016) and Maathuis (2014).
Given different nodes and a path from to is a sequence of edges connecting and When all edges are directed and pointing toward the same node, we have a directed path. A path that is not directed is an undirected or nondirected path.
A cycle is a path from to with an additional edge between and A directed cycle is a directed path from to together with a directed edge
A directed acyclic graph (DAG) is a directed graph without any directed cycle. When a graph underlying a SEM is a DAG, the SEM is said to be recursive.
is the set of nodes for which there is a directed edge in this case, is a child of and is a parent of The nodes and are adjacent if there is an edge or
If in a DAG there is a directed path from to then is a descendant of and is an ancestor of
In a DAG with nodes it is always possible to relabel the nodes, such that for each node for all parents in the set Such a relabeling is known as a topological ordering of the DAG. Using this ordering, the root nodes (without any parents) have the lowest labels, the sink nodes (without any children) have the highest labels, and the matrix of path coefficients is upper triangular. Formally, a topological ordering is a function such that the preceding property holds. A topological ordering always exists, and does not need to be unique (Peters et al. 2017).
If, for a given path, two directed edges point into the same node, the latter is a collider. For example, given the DAG C is a collider on the (only) path between A and B. In all other cases (, and ), C is a noncollider. Several different paths can pass through a node, and being a (non)collider is always relative to the path.
In a DAG, a v-structure or immorality is a collection of three nodes (say A, B, and C), such that there are directed edges and but no edge between A and C. In this case B is an unshielded collider. If there is an edge between A and C, it is a shielded collider. Similarly, in an undirected graph, A, B, and C form an unshielded triple if there are edges and but no edge .
The skeleton of a (partially) directed graph is the undirected graph obtained after removing all arrowheads.
Given a directed graph , two nodes A and B, and a (possibly empty) subset of nodes S not containing A and B, a path between A and B is blocked by S if at least one of the following two conditions holds: (i) there exists a collider on the path which is not in S, and also none of its descendants are in S; (ii) there exists a noncollider on the path that is in S.
Nodes A and B are d-separated by a set S if S blocks all paths from A to B.
Given disjoint sets U, V, and S (U and V should be nonempty), U and V are d-separated by S if S blocks all paths from to for all nodes and
Two DAGs are equivalent if they have the same skeleton and the same v-structures.
An equivalence class of DAGs is a set containing all DAGs that are equivalent to one another. For example, given a skeleton there is one equivalence class containing the three DAGs and and one equivalence class with only one DAG Any DAG in the class can be used to represent the class. But instead of picking an arbitrary DAG, an equivalence class can also be represented by a completed partially directed acyclic graph (CPDAG). A partially directed acyclic graph (PDAG) is “a graph where some edges are directed and some are undirected and one cannot trace a cycle by following the direction of directed edges and any direction for undirected edges” (Kalisch and Bühlmann 2007). A PDAG is a CPDAG if (a) every directed edge in the PDAG exists in all DAGs in the equivalence class it represents (b) for every undirected edge in the PDAG, the equivalence class contains at least one DAG with and at least one with . Chickering (2002) showed that CPDAGs represent equivalence classes uniquely.
A.2. Identifiability
In a general GSEM, the parameters in , and are not identifiable, which was pointed out by Gianola and Sorensen (2004). However, when we know a topological ordering of the graph (defined above in Appendix A.1), and Assumption 3 (diagonal ) and faithfulness assumptions (9) and (10) hold, it appears that , and are identifiable from the joint distribution of (see Equation 7). Our approach relies on the decomposition of and is probably known, although we could not find it in the literature. Neither could we find a proof, but the decomposition seemed valid in all examples we considered. It works as follows:
Relabel the nodes (traits) according to a topological ordering. Then, both and are upper triangular. Recall that has zeros on the diagonal. always has ones on the diagonal, i.e., it is unit upper-triangular.
- Now, we use the fact that every positive definite matrix A can be decomposed as with a diagonal matrix D and unit upper-triangular L [see e.g., Petersen et al. (2008), section 5.7]. We apply this result to
(17)
and set equal to D, and equal to L. Let
- 3. Finally, using and L we obtain
(18)
For example, consider the graph with path coefficients equal to one and unit error variances (for simplicity, we ignore the genetic effects in this example). We need to relabel the graph such that After relabeling, we have
as is the identity matrix. In R, the decomposition can be computed as follows:
b <- matrix(c(1,1,1,1,2,2,1,2,3), ncol = 3)
#b <- b[c(3,2,1), c(3,2,1)]
v <- Cholesky(Matrix(b, sparse = T), LDL = T, perm = F)@x
Gamma <- matrix(0,3,3)
Gamma[lower.tri(Gamma, diag = T)] <- v
D <- diag(diag(Gamma))
diag(Gamma) <- 1
Gamma <- t(Gamma)
Lambda <- diag(3) - (solve(Gamma))
We emphasize that the topological ordering is crucial. Without the relabeling (e.g., uncomment the second line in the above R-code), a different is obtained (also after interchanging the first and third row). Although in this example the topological ordering is unique, there may in general be multiple valid orderings; e.g., and in case the graph is Based on all investigated examples, we conjecture (but could not prove) that these orderings lead to the same parameter estimates.
A.3. The matrix Γ expressed as a function of path coefficients
Let denote the DAG over the nodes with edges defined by . For each let denote the union of the set and the set of root traits (i.e., those without parents in ) for which there is a directed path toward For all let denote the set of all directed paths from to For contains only the empty path from to itself. For any directed path π from to let denote the product of the corresponding path coefficients as given by for the empty path we define
Using these definitions, we can decompose the variance of a trait into contributions from different ancestors, as well as its own error variance. To this end, we follow Spirtes et al. (2001) and define the column vector with elements
| (19) |
A.4. The covariance between Yj and Yk as function of path coefficients
Since (Equation 8 in the main text), the covariance between the vectors and can be written in terms of and
| (20) |
for all Consequently, we can express the genetic and residual covariance between traits in terms of quadratic forms, involving and the path coefficients. As a special case of Equation 20, it follows that, without random genetic effects,
is the covariance between the jth and kth trait, for each individual i. See also Spirtes et al. (2001) (Lemma 3.1.6), or Lynch and Walsh (1998) (Appendix 2). Using standard expressions for multivariate Gaussian distributions, this implies that when
| (21) |
A.5. The path coefficients condition
It is well known that faithfulness is violated when contributions from different paths cancel out. For example, in the SEM defined by and with respective path coefficients 1, 1, and , and are marginally independent but not d-separated. Conversely, when faithfulness holds, we know that such cancellations cannot occur, and that the sum in Equation 19 is never zero, i.e., only for We will refer to this as the path coefficients condition.
A.6. The path coefficients condition and faithfulness
The path coefficients condition is a necessary but not a sufficient condition for faithfulness. First, faithfulness can also be violated when contributions from different paths cancel out when summing over a subset of (rather than all) the directed paths; see Example 2.10 in Peters (2012). Second, it is not only the contributions of directed paths that should not cancel out, but also those of treks. A trek between and is any path between these nodes without a collider (Spirtes et al. 2001). Every trek consists of two directed paths, starting at the source of the trek, and going toward and One of these can be the empty path; hence, each directed path is also a trek. Figure 6 provides an example where contributions from different treks cancel out, leading to nonfaithfulness.
Figure 6.
An example of a SEM (without genetic effects) where faithfulness does not hold, because the contributions to the covariance from the treks and cancel out. If and are Gaussian with equal error variances, it follows that for every individual i, Consequently, and are marginally independent, but not d-separated by the empty set.
Another necessary condition for faithfulness is that all error variances are strictly positive. Figure 7 provides an example of nonfaithfulness due to a zero error variance. An extended version of the path coefficients condition (involving sums over subset of treks) together with strictly positive error variances may be sufficient for faithfulness, but we could not find such a result in the literature. However, from Equation 21 it follows that, for a Gaussian linear SEM (again without genetic effects), faithfulness is equivalent with
Figure 7.
An example of a SEM where faithfulness does not hold, because the variance of the error variables is zero. The random vectors and are conditionally independent given but, in the graph, the nodes and are not d-separated by
| (22) |
A.7. Conditional means and covariances
Using the notation to select the columns corresponding to S, and to select both rows and columns, it follows from Equation 7 that is multivariate normal with mean and covariance
| (23) |
| (24) |
where
| (25) |
| (26) |
| (27) |
The matrices , and are the variance-covariance matrix of and respectively, and the covariance between and From Equation 7 in the main text we also obtain the conditional distribution
| (28) |
where and are as in Equation 23, and is the block matrix with diagonal blocks and (defined as in Equation 27), and off-diagonal blocks Similarly, given the matrix with columns and it follows that
is the covariance between and
A.8. Covariance structure of the conditional distributions
When is block-diagonal, with m blocks of ones of dimension on the diagonal, then for any positive constants c and d,
Hence, the inverse of is again a linear combination of and K. This follows from the Woodbury identity (Petersen et al. 2008; Golub and Van Loan 2012)
| (29) |
with and In addition we have and therefore Consequently, any product of matrices of the form or their inverse is a linear combination of and K. Combining Equations 25, 26 and 27, and using that for any matrices of appropriate dimensions, it follows that when in Equation 24 is of the form for some numbers and Similarly, it follows that (Equation 28) is of the form
for some matrices and
A.9. Proofs of Theorems 1 and 2
Pearl (2009) (p. 51) showed that, under quite general assumptions, structural equation models satisfy the global Markov property, which means that d-separation in the graph implies conditional independence. It turns out that, in our case, the required assumption of independent errors applies to the error variables and not to G. The intuition behind this is that G is not just an additional error node, but part of the causal graph, and we can always distinguish between residual (co)variance and genetic (co)variance. We now give the proof of Theorem 1, which only requires minor modifications of the proof given by Pearl for the case without the genetic effects.
Let denote the graph, obtained by extending with the error variables, i.e., for traits we add the node and an edge We first show that the local Markov property holds for , i.e., for any variable Z is conditionally independent of its nondescendants given its parents. This is obvious for we now consider In the set of parents of is where contains G if By construction, is entirely determined by and constant conditional on these variables. Consequently, given it is independent of any and of any that it a nondescendant of [note that if is indeed conditionally independent of any nondescendant; if G cannot be the nondescendant because it is already in the conditioning set]. Therefore, the local Markov property holds for By the lemma below, we find that also the Markov factorization property holds for since for any distribution having a density it is equivalent with the local and global Markov properties. Given the Markov factorization property for and the fact that we can just integrate out the and obtain the Markov factorization property for This concludes the proof of Theorem 1.
Given the result of Theorem 1 and the assumed faithfulness, Theorem 2 now follows directly from the consistency for the general PC-algorithm (Spirtes et al. 2001); see the first part of the proof of their Theorem 5.1 (p. 407).
Markov properties:
The following lemma is taken from Lauritzen (1996) (p. 51), and reformulated with somewhat less general conditions, which, however, suffice for our purpose.
Lemma. Let P be the joint distribution of random variables having a density f, and let be a DAG on these variables. The following properties are equivalent:
- The Markov factorization property: given the parents of each the joint density (f) can be decomposed as
where the are the conditional densities. The local Markov property: any variable is conditionally independent of its nondescendants, given its parents.
The global Markov property: for all disjoint sets d-separation of U and V by S in the graph implies conditional independence of U and V given S. In contrast to U and V, the conditioning set S may be empty here. A definition of d-separation is given in Appendix A.1.
A.10. Proof of Theorems 3 and 5:
We first prove Theorem 3, by showing the equivalence of the left- and right-hand sides of (9) and (11) in the main text. The d-separation statements on the right-hand sides are equivalent, as G can never be a (or descendant of a) collider. Also the left-hand sides ( and ) are equivalent, since
For Theorem 5 we make the additional assumption that being the design matrix for r replicates of m genotypes in a balanced design (with ). The first part of Theorem 5 then follows from the results in Appendix A.8. For the second part, we first recall the equivalence of and Because of the Gaussianity and the assumed faithfulness, the latter conditional independence is equivalent with
| (30) |
where we used Equation 21.
Next, we consider the conditional distribution of given in Equation 28, whose covariance is the block matrix All its four blocks are a linear combinations of K and and it suffices to show that the coefficient of in the off-diagonal blocks is zero if and only if Equation 30 holds. We recall from Equation 26 that
Using the Woodbury identity (Equation 29) with and it follows that, for any positive-definite matrices and we have
| (31) |
Setting and it follows that
| (32) |
Combining this with the expressions for and given in Appendix A.7, we find that has off-diagonal blocks
for and Finally, working out the products in the last display [using that ], we find that all terms involving a Kronecker product with correspond exactly to the left-hand side of Equation 30. Consequently, the residual covariance in the distribution is zero if and only if .
A.11. Proof of Theorem 4
To obtain faithfulness for we need to prove that implies d-separation of and G in the graph Because the conditioning set is empty, it suffices to show that there are no directed paths from G to where we can assume that (otherwise would be nonzero, and because of the noncollinearity, and would not be independent). Because of the assumed Gaussianity, the independence of and implies that
| (33) |
where we used that and, therefore, for all and Since is strictly positive and the submatrix has full rank, Equation 33 implies that for all Finally, we use that the assumed faithfulness implies the path coefficients condition (see Appendices A.3–A.6). Consequently, it follows from that there is no directed path from to Since this is the case for all there can neither be a directed path from G to
A.12. Proof of Theorem 6
Assuming the first part of the theorem follows from the results in Appendix A.8. For the second part, we use that has genetic variance (see Equation 20). Because for traits without a direct genetic effect, rows and columns in are zero, we can rewrite this as Hence, is equivalent with for all , where we used that is of full rank (which is a consequence of Assumption 5). Using the arguments from the proof of Theorem 4 and the assumed faithfulness, it follows that this is equivalent with independence of and
Footnotes
Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.11635392.
Communicating editor: J. Wolf
Literature Cited
- Araus J. L., Serret M. D., and Edmeades G., 2012. Phenotyping maize for adaptation to drought. Front. Physiol. 3: 305 10.3389/fphys.2012.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma P., 2014. The quantitative genetics of indirect genetic effects: a selective review of modelling issues. Heredity 112: 61–69. 10.1038/hdy.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair R. H., Kliebenstein D. J., and Churchill G. A., 2012. What can causal networks tell us about metabolic pathways? PLOS Comput. Biol. 8: e1002458 10.1371/journal.pcbi.1002458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrás L., Westgate M. E., Astini J. P., and Echarte L., 2007. Coupling time to silking with plant growth rate in maize. Field Crops Res. 102: 73–85. 10.1016/j.fcr.2007.02.003 [DOI] [Google Scholar]
- Bühlmann P., Rütimann P., and Kalisch M., 2013. Controlling false positive selections in high-dimensional regression and causal inference. Stat. Methods Med. Res. 22: 466–492. 10.1177/0962280211428371 [DOI] [PubMed] [Google Scholar]
- Calus M. P., and Veerkamp R. F., 2011. Accuracy of multi-trait genomic selection using different methods. Genet. Sel. Evol. 43: 26 10.1186/1297-9686-43-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibub Neto E., Ferrara C. T., Attie A. D., and Yandell B. S., 2008. Inferring causal phenotype networks from segregating populations. Genetics 179: 1089–1100. 10.1534/genetics.107.085167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibub Neto E. C., Broman A. T., Keller M. P., Attie A. D., Zhang B. et al. , 2013. Modeling causality for pairs of phenotypes in system genetics. Genetics 193: 1003–1013. 10.1534/genetics.112.147124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickering D. M., 2002. Learning equivalence classes of bayesian-network structures. J. Mach. Learn. Res. 2: 445–498. [Google Scholar]
- Colombo D., and Maathuis M. H., 2014. Order-independent constraint-based causal structure learning. J. Mach. Learn. Res. 15: 3741–3782. [Google Scholar]
- Colombo D., Maathuis M. H., Kalisch M., and Richardson T. S., 2012. Learning high-dimensional directed acyclic graphs with latent and selection variables. Ann. Stat. 40: 294–321. 10.1214/11-AOS940 [DOI] [Google Scholar]
- Cooper M., Messina C. D., Podlich D., Totir L. R., Baumgarten A. et al. , 2014. Predicting the future of plant breeding: complementing empirical evaluation with genetic prediction. Crop Pasture Sci. 65: 311–336. 10.1071/CP14007 [DOI] [Google Scholar]
- Flaxman S. R., Neill D. B., and Smola A. J., 2015. Gaussian processes for independence tests with non-iid data in causal inference. ACM Trans. Intell. Syst. Technol. 7: 1–23. 10.1145/2806892 [DOI] [Google Scholar]
- Furlotte N. A., and Eskin E., 2015. Efficient multiple-trait association and estimation of genetic correlation using the matrix-variate linear mixed model. Genetics 200: 59–68. 10.1534/genetics.114.171447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., and Cui Y., 2015. Learning directed acyclic graphical structures with genetical genomics data. Bioinformatics 31: 3953–3960. 10.1093/bioinformatics/btv513 [DOI] [PubMed] [Google Scholar]
- Ghosal S., and van der Vaart A., 2017. Fundamentals of Nonparametric Bayesian Inference. (Cambridge Series in Statistical and Probabilistic Mathematics), Cambridge University Press, Cambridge, UK: 10.1017/9781139029834 [DOI] [Google Scholar]
- Gianola D., and Sorensen D., 2004. Quantitative genetic models for describing simultaneous and recursive relationships between phenotypes. Genetics 167: 1407–1424. 10.1534/genetics.103.025734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub G. H., and Van Loan C. F., 2012. Matrix computations, Vol. 3. JHU Press, Baltimore, MA. [Google Scholar]
- Hauser A., and Bühlmann P., 2012. Characterization and greedy learning of interventional Markov equivalence classes of directed acyclic graphs. J. Mach. Learn. Res. 13: 2409–2464. [Google Scholar]
- Hemani G., Tilling K., and Davey Smith G., 2017. Orienting the causal relationship between imprecisely measured traits using gwas summary data. PLoS Genet. 13: e1007081 (erratum: PLoS Genet. 13: e1007149). 10.1371/journal.pgen.1007081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzworth D. P., Huth N. I., Zurcher E. J., Herrmann N. I., McLean G. et al. , 2014. Apsim–evolution towards a new generation of agricultural systems simulation. Environ. Model. Softw. 62: 327–350. 10.1016/j.envsoft.2014.07.009 [DOI] [Google Scholar]
- Joo J. W. J., Kang E. Y., Org E., Furlotte N., Parks B. et al. , 2016. Efficient and accurate multiple-phenotype regression method for high dimensional data considering population structure. Genetics 204: 1379–1390. 10.1534/genetics.116.189712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam N., Tamilselvan A., Lawas L. M. F., Quinones C., Bahuguna R. et al. , 2017. Genetic control of plasticity in root morphology and anatomy of rice in response to water-deficit. Plant Physiol. 174: 2302–2315. 10.1104/pp.17.00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisch M., and Bühlmann P., 2007. Estimating high-dimensional directed acyclic graphs with the PC-algorithm. J. Mach. Learn. Res. 8: 613–636. [Google Scholar]
- Kalisch M., Mächler M., Colombo D., Maathuis M. H., and Bühlmann P., 2012. Causal inference using graphical models with the R package pcalg. J. Stat. Softw. 47: 1–26. 10.18637/jss.v047.i11 [DOI] [Google Scholar]
- Keating B. A., Carberry P. S., Hammer G. L., Probert M. E., Robertson M. J. et al. , 2003. An overview of APSIM, a model designed for farming systems simulation. Eur. J. Agron. 18: 267–288. 10.1016/S1161-0301(02)00108-9 [DOI] [Google Scholar]
- Korte A., Vilhjalmsson B. J., Segura V., Platt A., Long Q. et al. , 2012. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 44: 1066–1071. 10.1038/ng.2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., 2016. Misspecification in mixed-model based association analysis. Genetics 202: 363–366. 10.1534/genetics.115.177212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., Boer M. P., Malosetti M., Flood P. J., Engel B. et al. , 2015. Marker-based estimation of heritability in immortal populations. Genetics 199: 379–398. 10.1534/genetics.114.167916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen S. L., 1996. Graphical Models, (Oxford Statistical Science Series). Oxford University Press, New York, USA. [Google Scholar]
- Lynch M., and Walsh B., 1998. Genetics and Analysis of Quantitative Traits, Ed. 1st Sinauer Associates, Sunderland, MA. [Google Scholar]
- Maathuis M. H., 2014. Graphical models and causality: Directed acyclic graphs (dags) and conditional (in)dependence. Available at: https://stat.ethz.ch/ mmarloes/meetings/slides2.pdf.
- Maathuis, M. H., and P. Nandy, 2016 A Review of Some Recent Advances in Causal Inference, pp. 387–408 in Handbook of Big Data, edited by P. Bühlmann, P. Drineas, M. Kane, and M. van der Laan. CRC Press, Boca Raton, FL. [Google Scholar]
- Maathuis M. H., Colombo D., Kalisch M., and Bühlmann P., 2010. Predicting causal effects in large-scale systems from observational data. Nat. Methods 7: 247–248. 10.1038/nmeth0410-247 [DOI] [PubMed] [Google Scholar]
- McMaster G. S., Wilhelm W., and Frank A., 2005. Developmental sequences for simulating crop phenology for water-limiting conditions. Aust. J. Agric. Res. 56: 1277–1288. 10.1071/AR05068 [DOI] [Google Scholar]
- Meinshausen N., and Bühlmann P., 2010. Stability selection. J. R. Stat. Soc. Series B Stat. Methodol. 72: 417–473. 10.1111/j.1467-9868.2010.00740.x [DOI] [Google Scholar]
- Meinshausen N., Hauser A., Mooij J. M., Peters J., Versteeg P. et al. , 2016. Methods for causal inference from gene perturbation experiments and validation. Proc. Natl. Acad. Sci. USA 113: 7361–7368. 10.1073/pnas.1510493113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet E. J., Welcker C., Kruijer W., Negro S., Coupel-Ledru A. et al. , 2016. Genome-wide analysis of yield in europe: allelic effects vary with drought and heat scenarios. Plant Physiol. 172: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet E. J., Kruijer W., Coupel-Ledru A., Prado S. A., Cabrera-Bosquet L. et al. , 2019. Genomic prediction of maize yield across european environmental conditions. Nat. Genet. 51: 952–956. 10.1038/s41588-019-0414-y [DOI] [PubMed] [Google Scholar]
- Moore A. J., Brodie E. D. III, and Wolf J. B., 1997. Interacting phenotypes and the evolutionary process: I. direct and indirect genetic effects of social interactions. Evolution 51: 1352–1362. 10.1111/j.1558-5646.1997.tb01458.x [DOI] [PubMed] [Google Scholar]
- Pearl J., 2009. Causality, Cambridge university press, Cambridge, UK: 10.1017/CBO9780511803161 [DOI] [Google Scholar]
- Pearl J., and Verma T. et al. , 1991. A theory of inferred causation. 91: 441–452. [Google Scholar]
- Peters, J., 2012 Restricted Structural Equation Models for Causal Inference Ph.D. thesis, ETH Zurich and MPI for Intelligent Systems, 10.3929/ethz-a-007597940. [DOI] [Google Scholar]
- Peters J., Bühlmann P., and Meinshausen N., 2016. Causal inference by using invariant prediction: identification and confidence intervals. J. R. Stat. Soc. Series B Stat. Methodol. 78: 947–1012. 10.1111/rssb.12167 [DOI] [Google Scholar]
- Peters J., Janzing D., and Schölkopf B., 2017. Elements of Causal Inference: Foundations and Learning Algorithms, MIT press, Cambridge, MA. [Google Scholar]
- Petersen K. B., and Pedersen M. S. et al. , 2008. The matrix cookbook. Technical University of Denmark 7: 510. [Google Scholar]
- Pfister N., Bühlmann P., Schölkopf B., and Peters J., 2018. Kernel-based tests for joint independence. J. R. Stat. Soc. Series B Stat. Methodol. 80: 5–31. 10.1111/rssb.12235 [DOI] [Google Scholar]
- Poorter H., Bühler J., van Dusschoten D., Climent J., and Postma J. A., 2012. Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct. Plant Biol. 39: 839–850. 10.1071/FP12049 [DOI] [PubMed] [Google Scholar]
- Reynolds M., and Langridge P., 2016. Physiological breeding. Curr. Opin. Plant Biol. 31: 162–171. 10.1016/j.pbi.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Richardson T., and Spirtes P. et al. , 2002. Ancestral graph markov models. Ann. Stat. 30: 962–1030. 10.1214/aos/1031689015 [DOI] [Google Scholar]
- Rodríguez-Álvarez M. X., Boer M. P., van Eeuwijk F. A., and Eilers P. H., 2018. Correcting for spatial heterogeneity in plant breeding experiments with p-splines. Spat. Stat. 23: 52–71. 10.1016/j.spasta.2017.10.003 [DOI] [Google Scholar]
- Rosa G. J., Valente B. D., de los Campos G., Wu X.-L., Gianola D. et al. , 2011. Inferring causal phenotype networks using structural equation models. Genet. Sel. Evol. 43: 6 10.1186/1297-9686-43-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutari M., Howell P., Balding D. J., and Mackay I., 2014. Multiple quantitative trait analysis using bayesian networks. Genetics 198: 129–137. 10.1534/genetics.114.165704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley B., 2016. Cause and Correlation in Biology: A User’s Guide to Path Analysis, Structural Equations and Causal Inference, Ed. 2nd Cambridge University Press, Cambridge: 10.1017/CBO9781139979573 [DOI] [Google Scholar]
- Spirtes, P., C. Glymour, and R. Scheines, 2001 Causation, Prediction, and Search, Second Edition (Adaptive Computation and Machine Learning). MIT Press, Boston, MA. [Google Scholar]
- Stekhoven D. J., Moraes I., Sveinbjörnsson G., Hennig L., Maathuis M. H. et al. , 2012. Causal stability ranking. Bioinformatics 28: 2819–2823. 10.1093/bioinformatics/bts523 [DOI] [PubMed] [Google Scholar]
- Stephens M., 2013. A unified framework for association analysis with multiple related phenotypes. PLoS One 8: e65245 [corrigenda: PLoS One 14: e0213951 (2019)]. 10.1371/journal.pone.0065245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R., and Meyer K., 1986. A review of theoretical aspects in the estimation of breeding values for multi-trait selection. Livest. Prod. Sci. 15: 299–313. 10.1016/0301-6226(86)90071-0 [DOI] [Google Scholar]
- Töpner K., Rosa G. J. M., Gianola D., and Schön C.-C., 2017. Bayesian networks illustrate genomic and residual trait connections in maize (zea mays l.). G3 (Bethesda) 7: 2779–2789. 10.1534/g3.117.044263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y., Sugimoto K., Ogawa S., Rane J., Ishitani M. et al. , 2013. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Publ. Gr. 45: 1097–1102. [DOI] [PubMed] [Google Scholar]
- Uhler C., Raskutti G., Bühlmann P., Yu B. et al. , 2013. Geometry of the faithfulness assumption in causal inference. Ann. Stat. 41: 436–463. 10.1214/12-AOS1080 [DOI] [Google Scholar]
- Valente B. D., Rosa G. J. M., de los Campos G., Gianola D., and Silva M. A., 2010. Searching for recursive causal structures in multivariate quantitative genetics mixed models. Genetics 185: 633–644. 10.1534/genetics.109.112979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente B. D., Rosa G. J. M., Gianola D., Wu X.-L., and Weigel K., 2013. Is structural equation modeling advantageous for the genetic improvement of multiple traits? Genetics 194: 561–572. 10.1534/genetics.113.151209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente B. D., Morota G., Peñagaricano F., Gianola D., Weigel K. et al. , 2015. The causal meaning of genomic predictors and how it affects construction and comparison of genome-enabled selection models. Genetics 200: 483–494. 10.1534/genetics.114.169490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeuwijk F. A., Bustos-Korts D., Millet E. J., Boer M. P., Kruijer W. et al. , 2019. Modelling strategies for assessing and increasing the effectiveness of new phenotyping techniques in plant breeding. Plant Sci. 282: 23–39. 10.1016/j.plantsci.2018.06.018 [DOI] [PubMed] [Google Scholar]
- Visscher P. M., and Goddard M. E., 2015. A general unified framework to assess the sampling variance of heritability estimates using pedigree or marker-based relationships. Genetics 199: 223–232. 10.1534/genetics.114.171017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1921. Correlation and causation. J. Agric. Res. 20: 557–585. [Google Scholar]
- Zhou X., and Stephens M., 2014. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 11: 407–409. 10.1038/nmeth.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiernik P., Uhler C., and Richards D., 2017. Maximum likelihood estimation for linear gaussian covariance models. J. R. Stat. Soc. Series B Stat. Methodol. 79: 1269–1292. 10.1111/rssb.12217 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The maize and rice data used below can be accessed at https://doi.org/10.15454/IASSTN and https://doi.org/10.6084/m9.figshare.7964357.v1, respectively. Supplemental materials are available at figshare: https://doi.org/10.6084/m9.figshare.11635392.