In this Perspectives article, Ogbunugafor revisits a famous and influential analogy introduced by renowned evolutionary biologist John Maynard Smith in a 1970 manuscript entitled “Natural selection and the concept of protein space (Smith 1970)...
Keywords: evolutionary genetics, history of science, protein evolution
Abstract
In 1970, John Maynard Smith published a letter, entitled “Natural Selection and the Concept of a Protein Space,” that proposed a simple analogy for the incremental process of adaptive evolution. His “Protein Space” analogy contains the substrate for many central ideas in evolutionary genetics, and has motivated important discoveries within several subdisciplines of evolutionary science. In this Perspectives article, I commemorate the 50th anniversary of this seminal work by discussing its unique legacy and by describing its intriguing historical context. I propose that the Protein Space analogy is not only important because of its scientific richness, but also because of what it can teach us about the art of constructing useful and subversive analogies.
Metaphors and analogies have long served as central actors in scientific communication. When effective, they capture the essence of complicated or counterintuitive ideas, can transform or reframe debates, and generate hypotheses.
February 7, 2020 marked the 50th anniversary* of one of the most influential analogies ever proposed in evolutionary genetics, appearing in the 1970 letter to Nature entitled “Natural Selection and the Concept of a Protein Space,” written by Maynard Smith (1970). In this letter, Maynard Smith—by then a well-known theoretical biologist at the University of Sussex—compared natural selection in the context of proteins to a word game where the goal is to convert one word into another by changing one letter at a time:
The model of protein evolution I want to discuss is best understood by analogy with a popular word game (Maynard Smith 1970).
The example he used was in the transformation from “WORD” into “GENE,” using the rules from the word game. He proposed that this could be achieved with the following four-step move:
WORD → WORE → GORE → GONE → GENE
Maynard Smith suggested the path above, as opposed to other four-step moves, for example:
WORD → GORD → GOND → GEND → GENE
WORD → WOND → WEND → WENE → GENE
WORD → WERD → GERD → GERE → GENE
In the context of the word game, the path containing WORD, WORE, GORE, GONE, and GENE would be preferred because it contains viable, sensical English words at every step, unlike the other example four-step moves. Maynard Smith elaborates:
It follows that if evolution by natural selection is to occur, functional proteins must form a continuous network which can be traversed by unit mutational steps without passing through nonfunctional intermediates. In this respect, functional proteins resemble four-letter words in the English language, rather than eight-letter words, for the latter form a series of small isolated islands in a sea of nonsense sequences (Maynard Smith 1970).
His main point: in order for natural selection to “locate” solutions in the vast space of possible protein sequences, incremental solution steps only need to be to other meaningful words, that is, protein forms that are functional. This is possible because functional molecules are not dispersed randomly through spaces of possible sequences, but rather, can be located in a manner analogous to the words in his example: clustered in networks, such that natural selection can serve as an effective search algorithm for locating biophysically viable protein sequences (and by extension, adaptation writ large).
The Many Faces of Maynard Smith
John Maynard Smith has been described as one of the most creative biologists of the post neo-Darwinian synthesis era, with a career defined by boundless courage and curiosity (Charlesworth 2004; Michod 2005). Identifying a specific area or subfield that Maynard Smith is most associated with is the stuff of debate, as he authored seminal texts and foundational treatises on a range of topics, including evolutionary genetics (Maynard Smith 1989), the evolution of sex (Maynard Smith 1978), and game theory (Maynard Smith 1982). The breadth of Maynard Smith’s inquiry is more impressive when we consider that most of his work was characterized by a signature style: simple mathematical formalism applied consistently from problem to problem, and an unpretentious, almost folksy manner of describing complicated ideas.
Given what we know about Maynard Smith, his invention of the Protein Space should not surprise us, even if very little of Maynard Smith’s work—before or after 1970—was about proteins. We can explore the ingenuity of the analogy by investigating the underexplored context in which it was born. His Protein Space features several faces of John Maynard Smith: distinguished theoretician, creationism antagonist, and preternatural science communicator.
Maynard Smith’s Protein Space was not presented as an isolated intellectual exercise written in a vacuum, but as part of a dialectic. In October 1969 (several months before Maynard Smith’s letter was published), Nature published a letter entitled “Natural Selection and the Complexity of the Gene” written by Frank Salisbury, a respected plant physiologist at Utah State University (Salisbury 1969). In it, Salisbury posits:
If life really depends on each gene being as unique as it appears to be, then it is too unique to come into being by chance mutations (Salisbury 1969).
Much of the content of Salisbury’s letter resembles creationist arguments, with the characteristic ethos of intelligent design: biological life is too complex to have been engineered by an undirected process like natural selection. Alternatively, Salisbury’s letter might be charitably interpreted as an honest cross-examination of natural selection’s eminence as an effective algorithm for solving complex adaptive problems. Time would soon reveal that cynical takes on Salisbury’s intentions were well founded: not long after the 1969 letter, Salisbury embraced openly creationist stances, starting with his 1971 article “Doubts about the Modern Synthetic Theory of Evolution,” which criticized the modern evolutionary synthesis and proposed intelligent design as an explanation for complex life (Salisbury 1971). This would begin a long career as an ardent creationist, best captured by two books that he wrote on the topic: The Creation (Salisbury 1976) and The Case for Divine Design (Salisbury 2006).
Maynard Smith’s letter appeared in print shortly after Salisbury’s and makes reference to it in the opening sentence (“Salisbury has argued that there is an apparent contradiction between two fundamental concepts of biology…”). This suggests that work on “Natural Selection and the Concept of a Protein Space” began almost immediately after Maynard Smith read Salisbury’s work. However, this is mere speculation, as it is also possible that Maynard Smith had conceived and developed Protein Space well before Salisbury’s letter. Maybe Maynard Smith already had the structure of Protein Space outlined, had filed it among his (surely voluminous) stack of ideas, and unleashed it only after he saw the need to: at the emergency signal of a creationist idea appearing in a high-profile scientific journal.
The contents of “Natural Selection and the Concept of a Protein Space” don’t provide an answer to the question of why (or when) Maynard Smith developed the abstraction: while it is an unambiguous rebuttal to the arguments of Salisbury (and sympathizers), its tone resembles a standard Maynard Smith musing, striking a balance between scientific precision and casual conversation, rather than an aggressive creationist takedown. This quality may reflect Maynard Smith’s intent: to author a harsh rebuke of creationist ideas that didn’t bring unnecessary attention to those arguments, but rather focused on offering an alternative that readers would appreciate. In that way, the casualness of Maynard Smith’s Protein Space is what made it so subversive.
The Scientific Relevance of Protein Space
The scientific importance of Protein Space resides in its versatility, in that it contains the substrate for many cutting-edge ideas in evolutionary and population genetics. For example, Protein Space shares features with the “fitness landscape” analogy, an abstraction that is connected to prominent figures and ideas from the modern evolutionary synthesis (Provine 1989; Gavrilets 2004; Svensson and Calsbeek 2012). It has roots in Ronald Fisher’s geometrical model, which proposed biological information as existing along a continuum from genotype to phenotype to fitness (Fisher 1930). The fitness landscape as we commonly discuss it (also known as the “adaptive landscape”) was introduced by Sewell Wright, who conceptualized populations existing as points on a multidimensional space corresponding to genotype, with evolution equating to movement across this space (Wright 1932). It has since emerged as one of the most popular concepts in all of evolutionary biology, the subject of thousands of manuscripts and treatises, several of which examine its rich history and modern significance [see Gavrilets (2004), Svensson and Calsbeek (2012), de Visser and Krug (2014), and Yi and Dean (2019)].
The fitness landscape has formed the basis of an entire subfield full of studies that have actualized the concept in empirical, biological systems. Since the mid-2000s, antimicrobial resistance has been instrumental in its widespread adoption, as it linked evolutionary processes to a practical medical problem and was amenable to experimental manipulation. In a seminal study, population geneticists used molecular techniques to engineer a bacterial enzyme with combinations of a small subset of mutations conferring resistance to an antibiotic. From this, they were able to explain that Darwinian evolution can only traverse certain pathways to higher fitness (Weinreich et al. 2006).
Related approaches have allowed evolutionary geneticists to examine empirical fitness landscapes toward the study of a number of important evolutionary phenomena, including transfer RNA evolution (Domingo et al. 2018), transcription factor binding (Aguilar-Rodríguez et al. 2017), and the evolution of toxins in butterflies (Karageorgi et al. 2019). And with modern molecular methods like deep mutational scanning—whereby proteins are engineered such that all possible single-amino acid substitution variants are generated in high throughput (Fowler and Fields 2014)—biologists are better equipped to build larger fitness landscapes than ever before, permitting the exploration of wider segments of Protein Space.
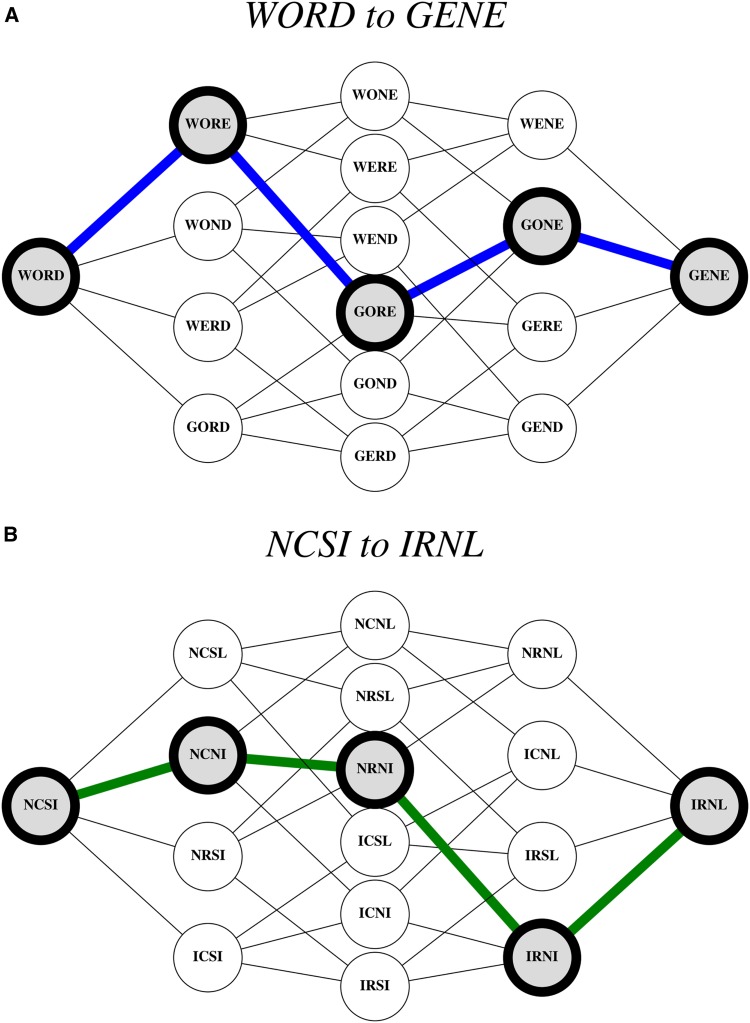
While the fitness landscape and Protein Space analogies have distinct ontogenies, they can each be used to describe certain phenomena. For example, one can depict the Maynard Smith analogy in the form of a hypercube that exhibits all of the intermediate genotypes between WORD and GENE (Figure 1A). For comparison, an identical structure can be applied in the form of a graph depiction of a fitness landscape corresponding to mutations in an enzyme that confer resistance to an antimicrobial drug (Figure 1B). In both cases, there exist “paths” from one word to another on the opposite side of the landscape. In the biological case (Figure 1B), the path corresponds to the most likely evolutionary trajectory from a version of an enzyme that is susceptible to an antimicrobial drug (analogous to WORD) to one that is highly resistant (analogous to GENE). All intermediates in this path correspond to viable, functional forms of the enzyme, equivalent to the sensical words in Maynard Smith’s Protein Space analogy.
Figure 1.
Representations and applications of John Maynard Smith’s Protein Space. (A) A hypercube “fitness graph” representation, where WORD and GENE are the terminal nodes, and there exists a pathway between them (dense blue line, left to right). Importantly, all words along the pathway are sensical English words. (B) A hypercube representation of an empirical fitness landscape corresponding to mutations in dihydrofolate reductase, an enzyme target of drugs in many microbial diseases. Specifically, the mutations are associated with resistance to pyrimethamine, an antimicrobial drug. Letters correspond to single-letter amino acid abbreviations and their substitutions: asparagine (N) to isoleucine (I), cysteine (C) to arginine (R), serine (S) to asparagine (N), and isoleucine to leucine (L). The NCSI node represents the wild type and the IRNL represents the quadruple mutant with much higher resistance to the drug. The dense green line corresponds to the most likely path of enzyme evolution under selection at high concentrations of pyrimethamine: NCSI → NCNI → NRNI → IRNI → IRNL [for more details, see Ogbunugafor and Eppstein (2016)].
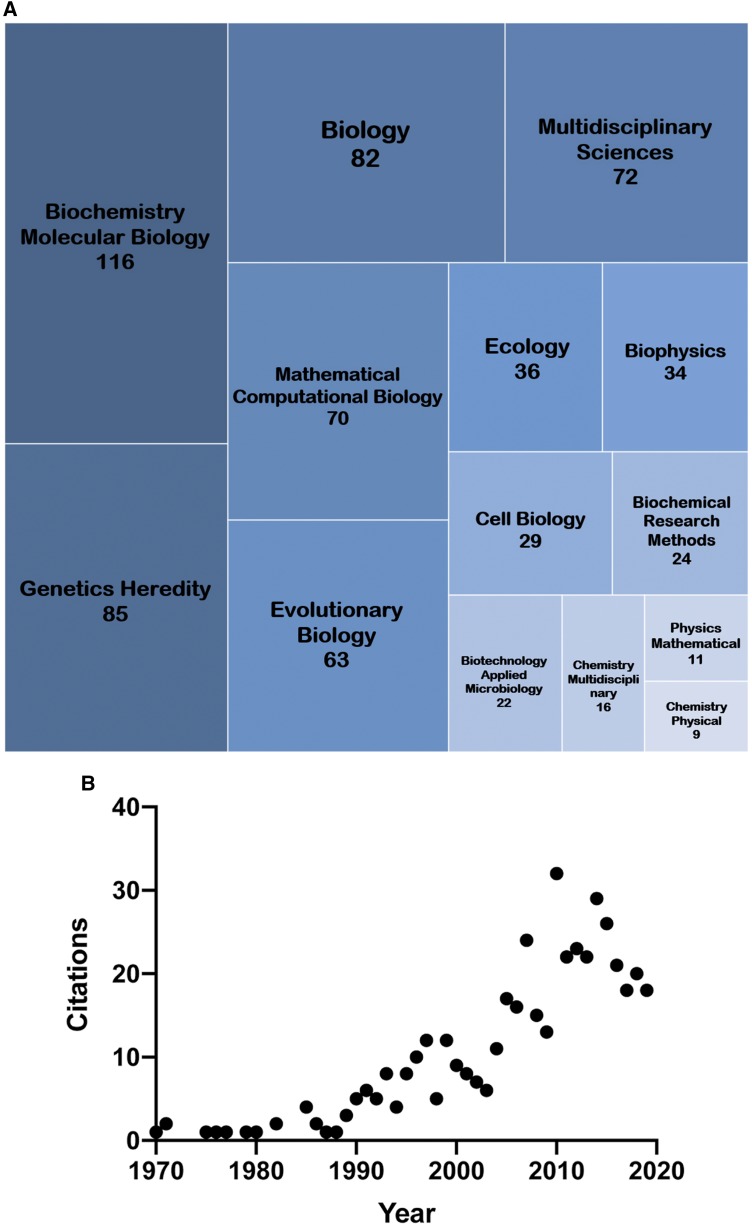
In addition to highlighting conceptual overlap between Protein Space and certain ideas in evolutionary genetics, the analogy’s richness can also be demonstrated through an examination of its citation patterns between 1970 and the last quarter of 2019 (Figure 2). Using data from the Web of Science (https://webofknowledge.com), we observe that papers using “biochemistry and molecular biology” as keywords are the ones that most frequently cite Protein Space (Figure 2A). Also notable is the fact that disparate fields like biophysics and ecology have cited it nearly an equal number of times (34 and 36 times, respectively). In addition, the timescale of the citations tells another important story: Protein Space has become more popular since the year 2000, with year 2010 the one where it was cited the most (Figure 2B). Some very recent examples from the literature highlight its modern reach: it has been cited in papers that focus on cutting-edge topics such as epistasis (Starr and Thornton 2016) and evolvability (Payne and Wagner 2019). Moving forward, there is every reason to believe that the Protein Space analogy will remain relevant. It is compatible with both classical and modern ideas in evolutionary genetics, and is called upon in studies across a breadth of scientific areas.
Figure 2.
The varied influence of Protein Space. Citations of John Maynard Smith’s Protein Space letter as a function of (A) subject area as classified in the Web of Science and (B) year. (A) Image is a TreeMap representation of the number of citations, where the size of the quadrilateral corresponds to the number of citations. Data come from Web of Science (https://webofknowledge.com) as of October 2019. Raw data can found in the Supplemental Material (DOI: https://doi.org/10.25386/genetics.11760213). Note that other data sources (e.g., Google Scholar) may have different values.
Especially noteworthy are the technology-focused papers that cite Protein Space, which indicate that the analogy is even utilized in engineering spheres (Currin et al. 2015; Yang et al. 2019). One high-profile example involves its citation in Frances H. Arnold’s 2018 Nobel Lecture, entitled “Innovation by Evolution: Bringing New Chemistry to Life” (Arnold 2019). The 2018 Nobel Prize in Chemistry—awarded to Frances H. Arnold, George P. Smith, and Gregory P. Winter—stands out because it recognized the role of evolutionary reasoning in the progression of the chemical sciences. In her lecture, Arnold invokes Maynard Smith and Protein Space when describing how she conceptualizes evolutionary innovation:
Consider an ordered space in which any protein sequence is surrounded by neighbors that have a single mutation. For evolution to work, he [Maynard Smith] reasoned, there must exist functional proteins adjacent to one another in this space. Although most sequences do not encode functional proteins, evolution will work even if just a few meaningful proteins lie nearby. Given low levels of random mutation, the filter of natural selection can find those sequences that retain function. In fact, many of today’s proteins are the products of a few billion years of mostly such gradual change (Arnold 2019).
Arnold also connects Protein Space to the fitness landscape analogy, and the directed evolution technology for which she was awarded the Nobel Prize:
Evolution on a rugged [fitness] landscape is difficult, as mutation propels sequences into crevasses of non-function. However, latching onto Maynard Smith’s argument that proteins evolve on a landscape smooth in at least some of its many dimensions, I reasoned that directed evolution could find and follow continuous paths leading to higher fitness (Arnold 2019).
Examples like Arnold’s are a triumph of the Protein Space analogy, and demonstrate its influence on scientists working across disciplines, to sometimes glorious ends.
The Art of the Analogy
Having demonstrated the importance of the analogy for basic science, we can now speculate as to why it has been so successful. Further, we can ask whether there are general lessons to be learned that might be germane to contemporary conversations.
One important feature of Protein Space is its simplicity: it was written by one of the great mathematical biologists in the world, yet only contained a single mathematical relation: fN > 1 (where N is the total number of protein molecules that can be accessed via a mutational step and ƒ the fraction of these that are meaningful). Maynard Smith used this relation to propose the conditions in which Protein Space may exist in networks such that algorithms like natural selection would be able to locate solutions many steps away.
Even though Maynard Smith’s usual choice of reasoning weapon was mathematics, his Protein Space did not require knowledge of higher mathematics to be comprehended and contained no calculations. Alternatively, the article Maynard Smith was responding to (“Natural Selection and the Complexity of the Gene”) did contain calculations, and leaned heavily on the counter intuitiveness of large exponents to drive its Darwinian skepticism: Salisbury guesstimated that the hypothetical primordial soup contained 1085 replicating DNA molecules, and that evolution by natural selection would have a 10−415 chance of producing a hypothetical DNA molecule encoding an essential enzyme in a metabolic pathway. While Salisbury confessed that these were guesses, he hinged much of his argument on the absurdity of these low odds. Maynard Smith’s response didn’t address these guesstimations. Instead, he targeted the flawed logic underlying Salisbury’s claims, which failed to consider how successive adaptive changes undermine the presumptive implausibility of adaptation.
The simplicity of Maynard Smith’s analogy highlights an underappreciated component of the art of modeling complex systems, one that modern scientists often struggle with: all models are analogies of some kind, and not all models require mathematics to be useful. As one of the great scientific pursuits of our time involves ways to communicate complicated ideas to broader audiences, Maynard Smith’s Protein Space offers a canonical example of how to engineer an infectious model or analogy: they should be only as intricate as is necessary to capture the essence of a biological system or phenomenon.
While the impact of Protein Space on various subdisciplines of biological science is easy to understand, part of its original intent—a device used in a rebuttal against creationist-adjacent arguments—remains a salient, if underappreciated, feature. This aspect of Protein Space is consistent with John Maynard Smith’s persona, as he willingly participated in debates against creationists, and was generally consumed by social and scientific questions about the validity of Darwinian evolution (Piel 2019).
Decades after the introduction of Protein Space, creationist ideas persist in large segments of society. If he were alive today, John Maynard Smith would be disappointed by this reality, but also inspired to challenge it. As scientists play an increasing role in engaging the public on scientific matters, communication devices—like Protein Space—are crucial for the continued effort to confront pseudoscientific stances. They offer a way to translate the peculiarities of scientific theories, the ones that relegate comprehension to handfuls of experts and alienate most of the world.
50 years ago, John Maynard Smith offered a blueprint, a powerful way to communicate a key aspect of evolutionary genetics, greatly increasing the depths of our understanding. Through revisiting it, we learn that analogies can help scientists better perform their craft, and navigate the increasingly intertwined expanse between science and society.
Data availability
The authors affirm that all data appearing in Figure 2, originally from the Web of Science (www.webofknowledge.com), can be found at figshare: https://doi.org/10.25386/genetics.11760213.
Acknowledgments
The author thanks R. Rohlfs and three anonymous reviewers for feedback on drafts of the manuscript; Z. Isola, D. Green, L. Zaman, J. Garcia, C. Restrepo, S. Knutie, S. Perkins, R. Olzer, and E. Strombom for seminar and guest lecture invitations, where various iterations of the ideas in the manuscript were discussed and developed; V. Meszaros and M. Miller-Dickson for technical support; and D. Hartl for general mentorship and for conversations on topics related to the ideas in this manuscript.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11760213.
Communicating editor: A. S. Wilkins
*2020 also marks the centennial of John Maynard Smith’s birth (January 6, 1920).
Literature Cited
- Aguilar-Rodríguez J., Payne J. L., and Wagner A., 2017. A thousand empirical adaptive landscapes and their navigability. Nat. Ecol. Evol. 1: 45 10.1038/s41559-016-0045 [DOI] [PubMed] [Google Scholar]
- Arnold F. H., 2019. Innovation by evolution: bringing new chemistry to life (Nobel lecture). Angew. Chem. Int. Ed. Engl. 58: 14420–14426. 10.1002/anie.201907729 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2004. John Maynard Smith: January 6, 1920–April 19, 2004. Genetics 168: 1105–1109. [Google Scholar]
- Currin A., Swainston N., Day P. J., and Kell D. B., 2015. Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem. Soc. Rev. 44: 1172–1239. 10.1039/C4CS00351A [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A. G. M., and Krug J., 2014. Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15: 480–490. 10.1038/nrg3744 [DOI] [PubMed] [Google Scholar]
- Domingo J., Diss G., and Lehner B., 2018. Pairwise and higher-order genetic interactions during the evolution of a tRNA. Nature 558: 117–121. 10.1038/s41586-018-0170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection: A Complete Variorum Edition. Oxford University Press, Oxford: 10.5962/bhl.title.27468 [DOI] [Google Scholar]
- Fowler D. M., and Fields S., 2014. Deep mutational scanning: a new style of protein science. Nat. Methods 11: 801–807. 10.1038/nmeth.3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S., 2004. Fitness Landscapes and The Origin of Species (MPB-41). Princeton University Press, Princeton, NJ. [Google Scholar]
- Karageorgi M., Groen S. C., Sumbul F., Pelaez J. N., Verster K. I. et al. , 2019. Genome editing retraces the evolution of toxin resistance in the monarch butterfly. Nature 574: 409–412. 10.1038/s41586-019-1610-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., 1970. Natural selection and the concept of a protein space. Nature 225: 563–564. 10.1038/225563a0 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., 1978. The Evolution of Sex. Cambridge University Press, Cambridge. [Google Scholar]
- Maynard Smith J., 1982. Evolution and the Theory of Games. Cambridge University Press, Cambridge: 10.1017/CBO9780511806292 [DOI] [Google Scholar]
- Maynard Smith J., 1989. Evolutionary Genetics. Oxford University Press, Oxford. [Google Scholar]
- Michod R. E., 2005. John Maynard Smith. Annu. Rev. Genet. 39: 1–8. 10.1146/annurev.genet.39.040505.114723 [DOI] [PubMed] [Google Scholar]
- Ogbunugafor C. B., and Eppstein M. J., 2016. Competition along trajectories governs adaptation rates towards antimicrobial resistance. Nat. Ecol. Evol. 1: 7 10.1038/s41559-016-0007 [DOI] [PubMed] [Google Scholar]
- Payne J. L., and Wagner A., 2019. The causes of evolvability and their evolution. Nat. Rev. Genet. 20: 24–38. 10.1038/s41576-018-0069-z [DOI] [PubMed] [Google Scholar]
- Piel H., 2019. ‘The most bogus ideas’: science, religion and creationism in the John Maynard Smith archive. Electronic British Library Journal. Accessed September 10, 2019.
- Provine W. B., 1989. Sewall Wright and Evolutionary Biology. University of Chicago Press, Chicago, IL. [Google Scholar]
- Salisbury F. B., 1969. Natural selection and the complexity of the gene. Nature 224: 342–343. 10.1038/224342a0 [DOI] [PubMed] [Google Scholar]
- Salisbury F. B., 1971. Doubts about the modern synthetic theory of evolution. Am. Biol. Teach. 33: 335–354. 10.2307/4443526 [DOI] [Google Scholar]
- Salisbury F. B., 1976. The Creation. Deseret Book Company, Salt Lake City, UT. [Google Scholar]
- Salisbury F. B., 2006. The Case for Divine Design. Cedar Fort, Springville, UT. [Google Scholar]
- Starr T. N., and Thornton J. W., 2016. Epistasis in protein evolution. Protein Sci. 25: 1204–1218. 10.1002/pro.2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson E., and Calsbeek R., 2012. The Adaptive Landscape in Evolutionary Biology. Oxford University Press, Oxford. [Google Scholar]
- Weinreich D. M., Delaney N. F., Depristo M. A., and Hartl D. L., 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312: 111–114. 10.1126/science.1123539 [DOI] [PubMed] [Google Scholar]
- Wright S., 1932. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. Proceedings of the Sixth International Congress of Genetics, Ithaca, New York, Vol. 1, pp. 356–366. [Google Scholar]
- Yang K. K., Wu Z., and Arnold F. H., 2019. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 16: 687–694. 10.1038/s41592-019-0496-6 [DOI] [PubMed] [Google Scholar]
- Yi X., and Dean A. M., 2019. Adaptive landscapes in the age of synthetic biology. Mol. Biol. Evol. 36: 890–907. 10.1093/molbev/msz004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data appearing in Figure 2, originally from the Web of Science (www.webofknowledge.com), can be found at figshare: https://doi.org/10.25386/genetics.11760213.