Abstract
The Transgenic RNAi Project (TRiP), a Drosophila melanogaster functional genomics platform at Harvard Medical School, was initiated in 2008 to generate and distribute a genome-scale collection of RNA interference (RNAi) fly stocks. To date, it has generated >15,000 RNAi fly stocks. As this covers most Drosophila genes, we have largely transitioned to development of new resources based on CRISPR technology. Here, we present an update on our libraries of publicly available RNAi and CRISPR fly stocks, and focus on the TRiP-CRISPR overexpression (TRiP-OE) and TRiP-CRISPR knockout (TRiP-KO) collections. TRiP-OE stocks express single guide RNAs targeting upstream of a gene transcription start site. Gene activation is triggered by coexpression of catalytically dead Cas9 fused to an activator domain, either VP64-p65-Rta or Synergistic Activation Mediator. TRiP-KO stocks express one or two single guide RNAs targeting the coding sequence of a gene or genes. Cutting is triggered by coexpression of Cas9, allowing for generation of indels in both germline and somatic tissue. To date, we have generated >5000 TRiP-OE or TRiP-KO stocks for the community. These resources provide versatile, transformative tools for gene activation, gene repression, and genome engineering.
Keywords: Drosophila, RNAi, CRISPR, Cas9, knockout, overexpression, phenotypes, screens
DROSOPHILA is an exemplary model for genetic studies and has been used for decades to identify genes involved in developmental and cellular processes (Nagy et al. 2003; Venken and Bellen 2005; McGurk et al. 2015; Ugur et al. 2016; Sonoshita and Cagan 2017). Strengths of the Drosophila model include (1) the extraordinary evolutionary conservation between insects and vertebrates of many of the basic processes of development and nearly all of the basic signal transduction mechanisms and transcriptional regulators, and (2) the suite of sophisticated technologies available for manipulating the Drosophila genome and for selection and analysis of mutant phenotypes. The relevance of Drosophila to humans is best illustrated by the observations that >60% of human genes associated with diseases have counterparts in Drosophila (Rubin et al. 2000; Hu et al. 2011), and at least 135 human genes have been shown experimentally to rescue loss-of-function (LOF) mutation of their counterparts in Drosophila (Fernández-Hernández et al. 2016). Moreover, Drosophila is increasingly being used to model a wide range of human diseases, from developmental to metabolic to age-related diseases and more. Notable areas in which Drosophila models are having an impact include a wide variety of neurological diseases (Okray and Hassan 2013; Coll-Tané et al. 2019), cancer (Sonoshita and Cagan 2017), metabolic disease and diabetes (Park et al. 2014), responses to infection by human pathogens (Apidianakis and Rahme 2011), immune disorders (Bergman et al. 2017), heart disease (Viswanathan et al. 2014), and inherited disorders (Wangler et al. 2017; Mishra-Gorur et al. 2019). Drosophila is also increasingly being used to help identify causative variants associated with previously undiagnosed genetic disorders (Wangler et al. 2017; Splinter et al. 2018). Despite the impressive set of tools currently available for Drosophila genetic studies, locating and/or generating disease models in Drosophila can be time consuming, and at the start of our project, well-characterized LOF alleles or validated RNA interference (RNAi) and/or single guide RNA (sgRNA) strains were not available. This is a particularly limiting step for scientists who are not expert at working with Drosophila but have an interest in using this powerful system to gain insight into the biological functions of their genes of interest.
Genetic analysis of mutants relies on two complementary approaches, LOF and gain-of-function (GOF) studies. In LOF studies, the role of a gene is inferred from the phenotype that results from partial or complete absence of the gene product. LOF was classically achieved in vivo in Drosophila using random mutagenesis (St Johnston 2002) and in more recent years, is achieved by knockdown using RNAi (Dietzl et al. 2007), or knockout using CRISPR-Cas9 (Port et al. 2014). Transgenic RNAi is a powerful and straightforward method for analysis of gene function (Perrimon et al. 2010; Heigwer et al. 2018). In Drosophila, RNAi is cell-autonomous, and is used in conjunction with the GAL4/UAS system (Brand and Perrimon 1993) for both spatial and temporal knockdown, providing a powerful approach to the study of genes with pleiotropic functions. CRIPSR/Cas9 can also be used to generate tissue-specific mutations in vivo (Port et al. 2014; Port and Bullock 2016), as CRISPR/Cas9 efficiently generates double-strand breaks (DSBs) that can be used to generate mutations (Bassett et al. 2013; Gratz et al. 2013; Kondo and Ueda 2013; Ren et al. 2013; Yu et al. 2013; Sebo et al. 2014; Lee et al. 2018).
Although powerful, LOF analysis alone is not sufficient to fully annotate gene functions, for example due to the lack of detectable phenotypes for many genes (Rorth et al. 1998). Approximately 48% of protein-coding genes in the genome have no phenotype annotation on FlyBase or are annotated solely as “viable” or “fertile” (Ewen-Campen et al. 2017a). Of these, ∼35% have paralogs, suggesting that the lack of detectable LOF phenotypes for a portion of these genes is due to redundancy (Ewen-Campen et al. 2017a).
To complement LOF studies, GOF studies that use controlled mis- or overexpression of a gene may help elucidate function. In addition, GOF approaches have the potential to provide insights into gene function even in cases of redundancy, and there are many examples of Drosophila genes that, when overexpressed, are associated with phenotypes such as patterning defects, aberrant cell proliferation, neurodegeneration, metabolism abnormalities, etc. Importantly, GOF analyses can also provide insight into diseases associated with mis- or overexpression. For example, genes identified as upregulated in cancer can be tested for oncogenic activity using overexpression studies (Croce 2008). In addition, Drosophila has been used to help uncover specific genes associated with chromosomal duplication disorders (Grossman et al. 2011).
The available reagents for overexpression experiments using GAL4/UAS belong to two categories: (1) random insertions of an element that brings UAS upstream of a specific gene, and (2) transgenic insertions of a UAS sequence fused to a specific open reading frame (ORF). Regarding the insertion lines, many stock collections are available from the Bloomington Drosophila Stock Center (BDSC); namely, the EP (Rorth 1996), EPg (Staudt et al. 2005), EPgy2 (Bellen et al. 2004), and Exelixis (Thibault et al. 2004) collections. These have the potential to misexpress genes that flank the insertion sites in the presence of GAL4, depending on the location and orientation of the insertion. However, only a small number have proven useful to identify overexpression phenotypes as these collections have not been validated systematically to identify which insertions result in overexpression of the nearby gene(s) or to assess other effects of the insertions. To overcome this shortcoming, the Zurich ORFeome project generated fly stocks (whereby the ORF is cloned into a UAS vector, and integrated into the genome using ΦC31 integrase) that cover ∼3100 genes (Bischof et al. 2013; Schertel et al. 2015). However, these resources are limited in scope.
Since its inception in 2008, the Transgenic RNAi Project (TRiP) has generated reagents for in vivo studies in Drosophila. The vectors used for RNAi reagents, approach, and production platform of the lines were described extensively previously (Perkins et al. 2015). Here, we describe an update of the project and focus on novel resources based on CRISPR-Cas9 produced using the same efficient platform. Specifically, we describe (1) currently available reagents for in vivo LOF and GOF studies based on RNAi and CRISPR technologies; (2) large-scale collections of transgenic RNAi fly stocks and more recently, transgenic sgRNA fly stocks for either overexpression or knockout, in particular for studies of orthologs of human disease-associated genes; (3) “toolkit” fly stocks and reagents that facilitate optimal use of RNAi and CRISPR sgRNA fly stock resources; and (4) a database of information on the quality of individual RNAi and sgRNA lines, RSVP Plus (http://fgr.hms.harvard.edu/rsvp).
Materials and Methods
Plasmid cloning
Short hairpin RNAs (shRNAs) for the transgenic RNAi stocks were cloned into the VALIUM series and pNP vectors (see Supplemental Materials). sgRNAs for the TRiP-CRISPR stocks were cloned into pCFD3 (Port et al. 2014), pCFD4 (Port et al. 2014), pCFD6 (Port and Bullock. 2016), U6B-sgRNA2.0 (Jia et al. 2018), and flySAM2.0 (Jia et al. 2018).
Fly genetics and embryo injections
Flies were maintained on standard fly food at 25°, unless otherwise noted. Fly stocks were obtained from the Perrimon laboratory collection or BDSC (indicated with BL#). Stocks used in this study are as follows: y v; attP40 (BL36304); y v; attP2 (BL36303); y v nos-phiC31-int; attP40 (BL25709); y v nos-phiC31-int; attP2 (BL25710); y sc v; Dr e/TM3, Sb (BL32261); y sc v; Gla/CyO (35781); y w; Dmef2-GAL4 (Perrimon); y w; elav-GAL4 (Perrimon); w; ey-GAL4 (BL5534); y w; nub-GAL4 (Perrimon); y w; act5C-GAL4 (Perrimon); w; myo1A-GAL4 (Perrimon); w; Lpp-GAL4 (Perrimon); w; Cg-GAL4 (Perrimon); y w; tub-GAL4 (Perrimon); w; da-GAL4 (Perrimon); PG142-GAL4 (Perrimon); hml-GAL4 (Perrimon); w; UAS-dCas9.VPR; tub-GAL4/T(2;3)TSTL14, SM5: TM6B, Tb (BL67048); w; nub-GAL4; UAS-dCas9.VPR (BL67055); y w; CG6767[MI09551-GFSTF.2] (BL59305); y sc v; TOE.GS01642 = TRiP-OE CG6767 (BL81664); y sc v; TOE.GS00152 = TRiP-OE spz (BL67551); y sc v; TOE.GS00045 = TRiP-OE grk (BL67525); TOE.GS00016 = TRiP-OE spz4 (BL67519); y sc v; TOE.GS00088 = TRiP-OE spz6 (BL67538); y sc v; TOE.GS00062 = TRiP-OE trk (BL67532); y sc v; TOE.GS01583 = TRiP-OE su(r) (BL79460); y sc v; SAM.dCas9.TH12432.S = control guide (Perrimon); and y sc v; SAM.dCas9.TH12427.S = control guide (Perrimon).
For embryo injections, each plasmid was column purified (QIAGEN, Valencia, CA), eluted in injection buffer (100 μM NaPO4, 5 mM KCl), and adjusted to 50 ng/μl. Plasmids were injected individually or as pools into y v nos-phiC31-int; attP40 (for chromosome 2 insertions) or y v nos-phiC31-int; attP2 (for chromosome 3 insertions). Injected G0 flies were crossed with y sc v; Gla/Cyo or y sc v; Dr e/TM3, Sb to identify transformants and remove the integrase from the X chromosome.
flySAM lethality
Virgin females of each GAL4 line were crossed to balanced male control stocks or male flySAM2.0 sgRNA stocks. Vials were stored at 25° and flipped every 3 days. The proportion of GAL4 + sgRNA to GAL4 + Bal were recorded in the resulting progeny (1 = complete viability, 0 = complete lethality). These scores were normalized to the results of the y v; attP2 cross.
Human Disease library screen
We screened ∼1200 RNAi fly stocks corresponding to 670 high-confidence Drosophila orthologs of human disease genes, using ubiquitous and tissue-specific drivers. Female GAL4 lines were crossed to male RNAi lines at 25°. Progeny were scored for lethality and visible phenotypes. Stocks were subcategorized as lethal or viable with ubiquitous drivers. Lines for which ubiquitous expression resulted in lethality were then crossed to a panel of tissue-specific GAL4 drivers and the resulting viability and/or gross morphological phenotypes were recorded. Triglyceride and glucose levels were measured as described below for stocks that were viable with ubiquitous drivers.
Triglyceride, glucose, and glycogen measurement
Six adult male flies (three biological replicates) were homogenized in 200 μl of PBS containing 0.1% Triton-X, heated at 70° for 10 min, and centrifuged at 13,000 × g for 10 min. We used 10 μl of supernatant to measure triglyceride with Serum TG determination kits (TR0100-1KT; Sigma, St. Louis, MO). We treated 10 μl of supernatant with 0.2 μl water or trehalase (E-TREH; Megazyme) to digest trehalose into glucose or 1 μl amyloglucosidase (for glycogen measurement; Sigma) at 37° for 20 min, and glucose was measured by incubation with 150 μl D-Glucose Assay reagent (K-GLUC; Megazyme) at 37° for 5 min. Protein amounts were measured using Bradford Reagent (Sigma). Triglyceride, glucose, and glycogen levels were normalized to protein amount (three technical replicates).
Quantitative PCR
Flies of the genotype w; UAS:dCas9-VPR; tub-Gal4/T(2;3)TSTL14, SM5: TM6B, Tb were crossed to homozygous sgRNA lines, and the F1 generation were maintained for 5 days at 18° to reduce Gal4 activity and thereby lethality. Larvae were then transferred to 27° for 48 hr, and non-Tb larvae were used for quantitative PCR (qPCR). Three to six L3 larvae were flash-frozen on dry ice and homogenized in TRIzol (Thermo Fisher); then, RNA was extracted following the manufacturer’s protocol. RNA was purified using either RNeasy (QIAGEN) or Direct-zol (Zymo) kits, including a 15–20 min DNase treatment, and was then used as template for first-strand synthesis and qPCR. Target genes were compared with the geometric mean of two reference genes (Rp49 and GAPDH), and an annealing temperature of 57° was used. Primers were designed using a precomputed database of Drosophila qPCR primers (Hu et al. 2013). All experiments were conducted in biological duplicates, with technical triplicate qPCR reactions.
Data availability
TRiP stocks are available at the BDSC, National Institute of Genetics Japan, and the TsingHua Fly Center. All vectors are available from the Drosophila Genomics Resource Center. Maps with attributes, complete sequences, and detailed cloning protocols can be found on the TRiP website (https://fgr.hms.harvard.edu/) under Reagents and Protocols. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11857434.
Results and Discussion
The TRiP RNAi collection
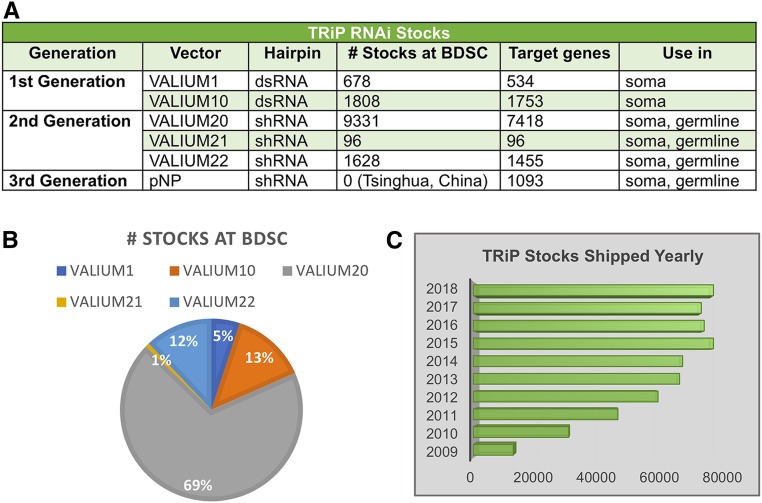
Since the description in 2015 of TRiP RNAi fly stock production and resources (Perkins et al. 2015) (Figure 1 and Supplemental Materials), we have continued to optimize the approach and produce additional new shRNA stocks. To date, TRiP has generated ∼15,800 RNAi stocks covering ∼9800 unique FBgns, or 70% of the protein-coding genes in the fly genome (FlyBase Release 6.30) with 85% coverage of highly conserved genes. Statistics on TRiP reagent gene coverage, vectors, and target regions are shown in Table S1. TRiP lines are regularly used by Drosophila researchers worldwide. Currently, BDSC distributes 13,541 RNAi stocks, and to date have shipped >700,000 TRiP stocks (summarized in Figure 1). In 2018 alone, BDSC sent 78,370 subcultures of TRiP stocks to 1406 different user groups in 45 US states and 47 countries. Additional TRiP shRNA stocks are also available from the National Institute of Genetics, Japan (https://shigen.nig.ac.jp/fly/nigfly/) and the Tsinghua Fly Center, China (http://fly.redbux.cn/). Notably, the latter collection includes 1093 shRNA lines made with the new pNP vector, which functions in both in soma and germline with significantly reduced basal hairpin expression (Qiao et al. 2018).
Figure 1.
TRiP RNAi stocks and distribution. (A) The first-generation VALIUM1 and VALIUM10 TRiP stocks contain long double-strand RNA hairpins. For VALIUM1, higher temperature fly culturing and UAS-Dicer-2 are best to achieve maximum knockdown. VALIUM10 was the best-performing vector among our first series of related vectors, which were generated in an effort to optimize the various features of VALIUM1. The second-generation TRiP stocks contain short hairpins (shRNAs). VALIUM20, VALIUM21 and VALIUM22 have a modified scaffold derived from the microRNA miR-1 flanking the hairpin itself. VALIUM20 works well in the germline and is stronger than VALIUM10 in the soma. VALIUM22 (and VALIUM21) has features optimized for RNAi knockdown in the germline. Specifically, it has a P-transposase promoter instead of the hsp70 basal promoter, and a K10 3′UTR instead of the SV40 3′UTR. (B) Currently, BDSC is distributing ∼13,400 RNAi stocks, and (C) to date have shipped >700,000 TRiP stocks.
TRiP-CRISPR collections
TRiP-CRISPR knockout:
CRISPR/Cas9 efficiently generates DSBs in Drosophila that can be used effectively to generate mutations or for genome engineering (Bassett et al. 2013; Gratz et al. 2013; Kondo and Ueda 2013; Ren et al. 2013; Yu et al. 2013; Sebo et al. 2014). Mutant animals or tissue-specific mosaics can be produced by simply crossing sgRNA-expressing flies to germline-specific Cas9 or somatic tissue-specific GAL4 > Cas9 flies, respectively (Kondo and Ueda 2013; Port et al. 2014; Port and Bullock 2016). A number of recent studies in Drosophila have used transgenic CRISPR for LOF, to identify novel modifiers of tauopathies (Butzlaff et al. 2015; Dourlen et al. 2017) or Parkinson’s disease (Chen et al. 2017; Rousseaux et al. 2018); to model congenital disorders of glycosylation (Parkinson et al. 2016); to identify genes that regulate tumorigenesis (Das et al. 2013; Mishra-Gorur et al. 2019); and to study inflammation and immunity (Lee et al. 2018; Liu et al. 2018).
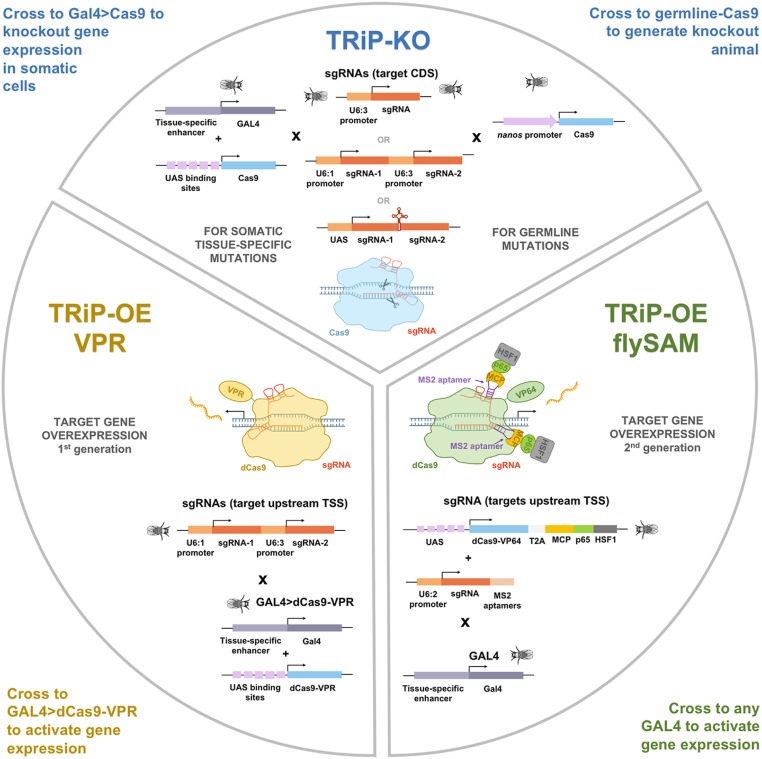
Recently, we and other groups (Meltzer et al. 2019; Port et al. 2020) started to generate libraries of transgenic sgRNA lines to complement existing RNAi collections. Unlike RNAi, which is often limited by incomplete penetrance due to residual gene expression (Echeverri and Perrimon 2006; Ma et al. 2006; Perkins et al. 2015), CRISPR/Cas9 targeted to the beginning of the ORF of a protein-coding gene can generate frameshift mutations that strongly reduce or eliminate gene function. As with TRiP RNAi lines, all TRiP-CRISPR stocks undergo rigorous quality control at the TRiP facility before being shipped to the BDSC for distribution. Available stocks are annotated on the DRSC/TRiP sgRNA Fly Stock Database (https://www.flyrnai.org/tools/grna_tracker/web/) and on FlyBase (Thurmond et al. 2019). As we build this new CRISPR collection, we encourage and receive gene target nominations from the community. As diagrammed in Figure 2, TRiP-CRISPR knockout (TRiP-KO) can be used for:
Germline knockout by Cas9. We produce fly stocks that constitutively express either one or two sgRNAs that target the coding sequence of a single gene. One or two sgRNAs per target are cloned into the pCFD3 and pCFD4 vectors, respectively (Port et al. 2014). To make a mutant fly strain, the sgRNA fly stock is crossed to flies with a germline-specific source of Cas9 (nanos-Cas9). CRISPR/Cas9-induced nonhomologous end joining occurs in the germ cells of the resulting progeny, which are then crossed to a balancer strain. The balanced flies are used to established stocks, which are screened for mutations by PCR.
Somatic tissue mosaic knockout by Cas9. To generate mosaic tissues, an appropriate GAL4 line is used to drive expression of UAS-Cas9 in a specific cell population. When crossed to flies constitutively expressing one or more sgRNA, this results in mutations in the target gene only in the GAL4-expressing cells.
For all TRiP-CRISPR stocks, we used the DRSC Find CRISPR Tool (Ren et al. 2013) to pick the optimal sgRNA designs. With the knockout collection, we selected the sgRNAs targeting the first or second coding exons common to all isoforms with high efficiency scores and specificity scores. The efficiency scores we considered include the Housden efficiency score, which is based on a nucleotide position matrix (Housden et al. 2015), and the frameshift score, which predicts the likelihood of a frameshift happening based on microhomology sequences near the cutting site (Bae et al. 2014). At the same time, we also prioritize the designs with high specificity scores, including the seed score (the length of unique seed region) and off-target score.
Figure 2.
TRiP-CRISPR systems for in vivo gene knockout and overexpression. In the TRiP-CRISPR knockout collection (TRiP-KO), stocks express one or two sgRNAs targeting the coding sequence of a gene or genes. Crossing TRiP-KO stocks to a Gal4 line expressing Cas9 induces cleavage of the target site, nonhomologous end joining (NHEJ) repair and insertion/deletion mutations (indels) in both germline and somatic tissue. Stocks in the TRiP-CRISPR overexpression (TRiP-OE) library express small guide RNAs (sgRNAs) targeting upstream of a gene transcription start site. Gene activation is triggered by co-expression of catalytically dead Cas9 (dCas9) fused to an activator domain, either VP64-p65-Rta (VPR) or Synergistic Activation Mediator (SAM).
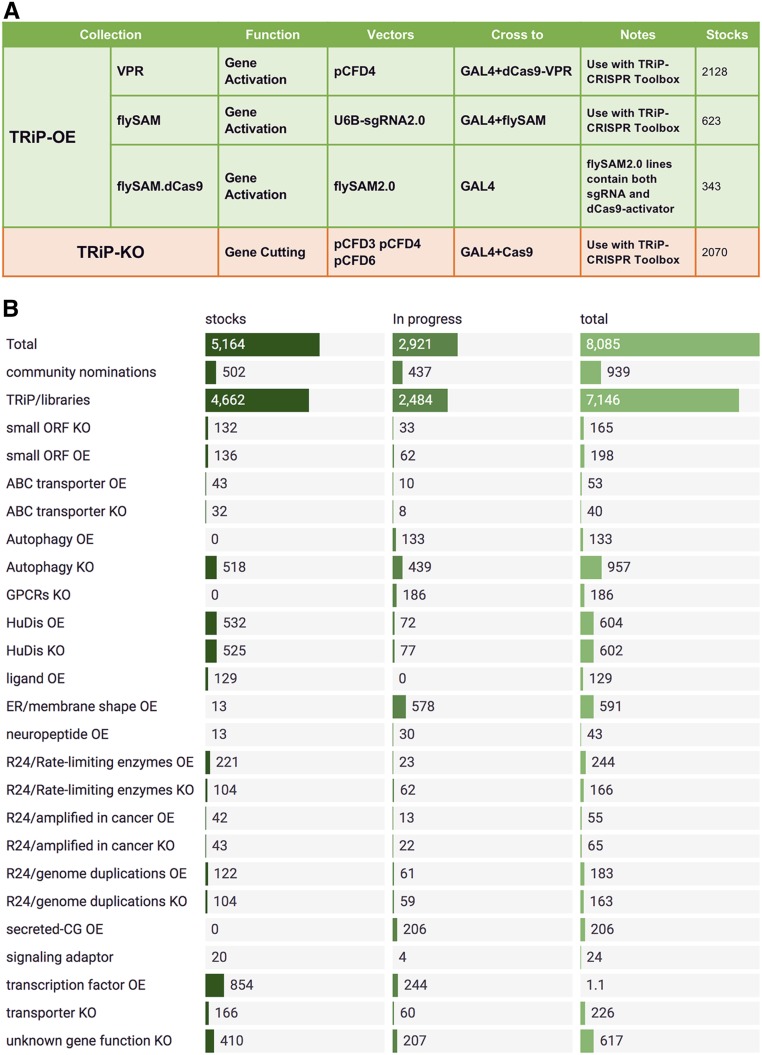
To date, we have produced 2070 TRiP-KO lines targeting community nominations, orthologs of human diseases, genes of unknown function, autophagy-related genes, and other focused gene sets (Figure 3). The first generation of TRiP-KO stocks have been made using the pCFD3 or pCFD4 vectors, in which the sgRNA is driven by the U6 promoter (Port et al. 2014). It was recently shown that in mosaic knockout experiments combining tissue-specific GAL4, UAS-Cas9 with U6 driven guides can result in some DSBs outside the GAL4 expression domain (Port and Bullock 2016). For this reason, TRiP-KO stock production has now shifted to using the pCFD6 vector (Port and Bullock 2016), in which sgRNA expression is under UAS control. In addition, “leaky” cutting outside the GAL4 domain can be mitigated by using the recently described UAS-uORF-Cas9 transgenes to reduce levels of conditional Cas9 expression (Port et al. 2020).
Figure 3.
TRiP-CRISPR libraries. (A) There are three types of TRiP-OE sgRNA stocks: VPR, flySAM guide only, and flySAM.dCas9 with both guide and UAS-dCas9. TRiP-KO stocks are made in the pCFD series of vectors for either constitutive or UAS driven expression of single or double guides. (B) TRiP-CRISPR libraries in progress.
TRiP-CRISPR overexpression:
In contrast to the wealth of reagents available for LOF studies in Drosophila, the development of large-scale resources for GOF studies has lagged behind. This is a major gap in functional genomics, as mis- and overexpression screens provide an important complementary resource for elucidating gene function. Cas9 activators, in which catalytically dead Cas9 (dCas9) recruits transcriptional activation machinery to a DNA sequence upstream of the transcriptional start site (TSS) of a target gene, can potentially overcome these obstacles. Target specificity is conferred by 20-bp protospacer sequences in the sgRNA, such that production of reagents for CRISPR activators (CRISPRa) at genome-wide scale is feasible. We reported the first demonstration of effective CRISPR/Cas9-based transcriptional activation in flies (Lin et al. 2015), and have since developed and optimized two systems for CRISPRa in vivo (Chavez et al. 2016; Jia et al. 2018). Based on these methods, we have now generated a collection of transgenic TRiP-CRISPR overexpression (TRiP-OE) lines for gene activation (Figure 2). TRiP-OE stocks express sgRNAs targeting genes upstream of the TSS. Gene activation is triggered by coexpression of dCas9 fused to an activator domain, either VP64-p65-Rta (VPR) or Synergistic Activation Mediator (SAM). sgRNAs for both systems are designed using the DRSC Find CRISPR Tool (Ren et al. 2013). We select sgRNAs targeting the region of 50–500 bp upstream the TSS of the targeted gene with high specificity scores.
With VPR, the TRiP-OE sgRNA stocks are crossed to a stock in which GAL4 directs expression of dCas9-VPR (Lin et al. 2015; Chavez et al. 2016). In the resulting progeny (GAL4 > dCas9-VPR; sgRNA-gene), the gene of interest is overexpressed in the GAL4 domain. The flySAM method is based on the mammalian engineered protein complex (Konermann et al. 2015). In our version (Jia et al. 2018), a VP64 domain is fused to dCas9 and two additional activator domains, p65 and HSF1, are recruited to the complex via MS2 stem loops in the sgRNA tail. With flySAM, the first set of TRiP-OE sgRNA stocks were made in the U6B-sgRNA2.0 vector (Jia et al. 2018). To activate gene expression, these are crossed to a stock in which GAL4 directs expression of UAS-flySAM. In the more recent flySAM.dCas9 collection, the stocks are made with the flySAM2.0 vector, which contains both the flySAM protein complex and the sgRNAs, thus gene activation is achieved by simply crossing to the GAL4 line of interest. The flySAM method gives considerably greater levels of activation compared to VPR and is comparable to the UAS overexpression system.
Importantly, the CRISPRa system has several advantages over UAS-complementary DNA overexpression as it allows: (1) overexpression of untagged wild-type gene products; (2) overexpression of all of the relevant splice isoforms for a given cell type; (3) preservation of the 3′UTR, which is important as it contains regulatory information such as microRNA binding sites; and (4) overexpression of ORFs that are long, contain repeat sequences, or are otherwise intractable using a complementary DNA approach due to cloning difficulties. Finally, generating a useful genome-wide collection requires that the large majority of the lines generated are functional. To estimate the proportion of TRiP-OE sgRNA lines that function as predicted, we previously created a panel of transgenic fly stocks expressing sgRNAs and analyzed activation using qPCR (Ewen-Campen et al. 2017b). A total of 75% of these sgRNA transgenes led to a greater than threefold increase in transcript levels when combined with dCas9-VPR, and 58% caused a greater than eightfold increase, a success rate comparable to the proportion of transgenes in current RNAi stock collections that confer effective knockdown. In addition, several of the lines that do not appear upregulated when tested via qPCR do in fact produce specific phenotypes, indicating that these success rates are in fact likely to be underestimated.
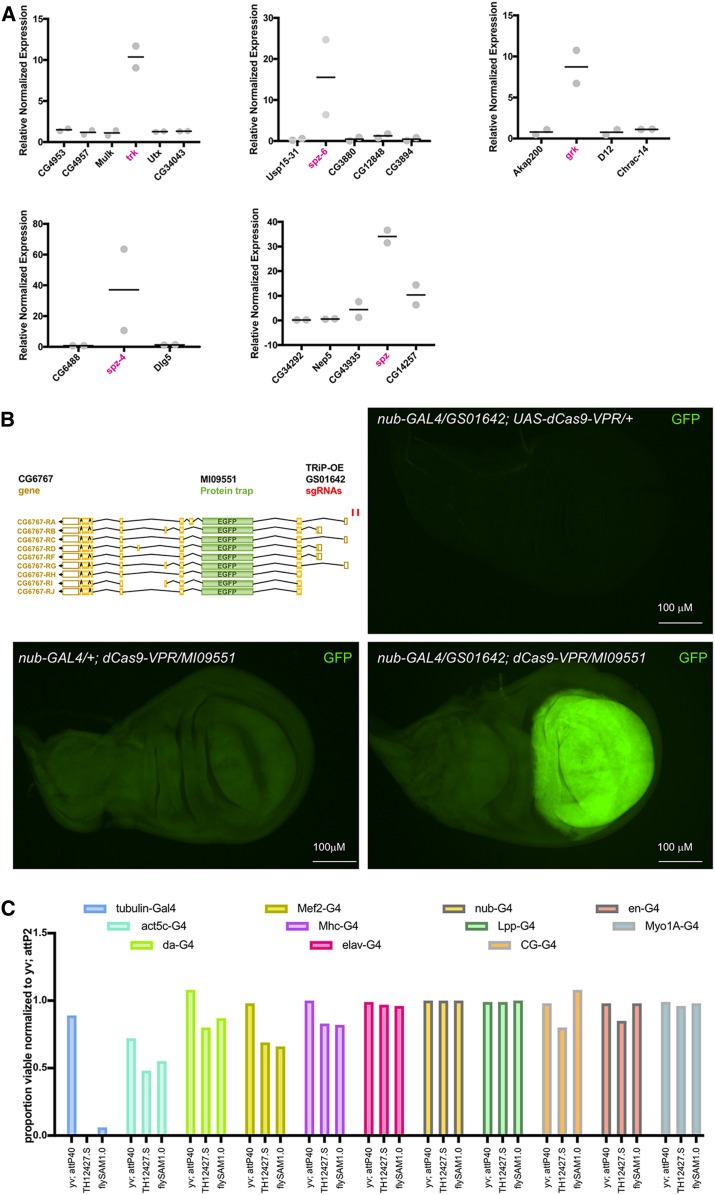
We also tested the stringency of the system with respect to off-target gene activation. This is a concern, since due to the compactness of the Drosophila genome, multiple genes could be within range of activation from a single sgRNA. Using qPCR, sgRNAs were analyzed in vivo for transcriptional activation of the target gene as well as neighboring genes within a ∼20 kb window. For four out of five target genes, there was no off-target activation (Figure 4A). We have previously shown for VPR that sgRNAs are effective between −500 and 0 bp upstream of the target gene in S2R-positive cells, with no bias toward targeting the sense strand (bearing the same sequence as the messenger RNA) or the antisense strand (used as the template for transcription) (Lin et al. 2015). In contrast, for flySAM, the greatest activation efficiency is observed with sgRNAs targeting the antisense strand <450 bp from the TSS of the target gene (J. Sun and J. Ni, personal communication). Thus, when two genes are located “head-to-head” and are transcribed in opposite directions, flySAM mediated activation will be greater for the gene on the opposite strand from the sgRNA target strand. The TRiP follows these guidelines when designing sgRNAs to minimize off-target activation.
Figure 4.
TRiP-OE systems provide robust on-target gene activation. (A) Larvae of the genotype tubulin-GAL4 > UAS-dCas9-VPR, sgRNA were analyzed for transcriptional activation of target gene as well as neighboring genes within a window of ∼20 kb. With one exception (spz), there was no off-target activation. Target genes are indicated in magenta. (B) TRiP-OE activation of tagged endogenous protein. Flies of the genotype nub-GAL4 > UAS-dCas9-VPR were crossed to TRiP-OE sgRNA GS01642 (targeting CG6767), a GFP protein trap in CG6767 (MI09551-GFSTF.2), or both. Strong TRiP-OE induced expression of the GFP-tagged CG6767 protein was observed in the nub-GAL4 domain. (C) A panel of GAL4 drivers were crossed to controls (yv; attp40 and yv; attP2), UAS-flySAM2.0 with nontargeting sgRNA (TH12427.S), or UAS-flySAM1.0 (no guide). The proportion of viable offspring were recorded and normalized to the yv; attP2 control. Normal survival rates and no visible phenotypes for most tissue-specific GAL4 lines. Complete lethality was observed with one ubiquitous driver (tubulin-GAL4) and moderate lethality with some strong tissue-specific drivers (e.g., Mef2-GAL4).
Next, we tested TRiP-OE induced protein expression of a gene containing a GFP protein trap (Figure 4B). We combined the wing-pouch-specific nub-GAL4 driver with UAS-dCas9-VPR, then crossed to the TRiP-OE guide stock targeting CG6767 (GS01642), the GFP MiMIC protein trap in CG6767 (MI09551-GFSTF.2), or both together. When all components were expressed, we observed strong TRiP-OE induced expression of the GFP-tagged CG6767 protein. Importantly, GFP upregulation was strictly confined to the nub-GAL4 domain in the wing pouch, indicating that the CRISPRa system allows tight spatial control of gene expression. Finally, we tested the flySAM system for toxicity. Previously, it had not been possible to apply SAM in vivo in flies because ubiquitous expression of UAS:MCP-p65-HSF1 is lethal in the absence of any sgRNA (Ewen-Campen et al. 2017b). We therefore tested whether flySAM can be expressed in vivo without toxicity by crossing to a panel of GAL4 driver lines with or without control sgRNA (Figure 4C). We observed normal survival rates and no visible phenotypes for most tissue-specific GAL4 lines. However, we did observe complete lethality with one ubiquitous driver (tubulin-GAL4), and we observed moderate lethality and phenotypes with some strong tissue-specific drivers (e.g., Mef2-GAL4 and ey-GAL4). These results suggest that very strong GAL4 drivers should be avoided for flySAM overexpression experiments.
To date, we have produced 3094 TRiP-OE lines targeting transcription factors, small ORFs, orthologs of human diseases, kinases, autophagy-related genes, and other focused libraries (Figure 3 and Figure S2). The majority of TRiP-OE flies were made using the VPR system. However, due to the greater levels of gene activation as well as the ease of use, TRiP-OE stock production has shifted to using the flySAM system exclusively.
Toolbox
In addition to the RNAi stocks, the TRiP, via the BDSC, also provides a “TRiP Toolbox” stock collection (Table S2) that includes injection stocks for laboratories wishing to generate their own RNAi lines and commonly used GAL4 lines with UAS-Dcr2, which is useful to enhance message knockdown when combined with long double-strand RNAs but not shRNAs. Because TRiP-KO and TRiP-OE VPR lines contain only the sgRNA, it is necessary to express Cas9 separately in the tissue of interest. For this reason, we have also produced a TRiP CRISPR/Cas9 toolbox set of GAL4/GAL80ts/UAS stocks that allow spatial and temporal expression of Cas9 or dCas9-VPR (Table S2). A total of 55 TRiP CRISPR/Cas9 toolbox lines are complete and have been shipped to BDSC for distribution.
Human disease-related ortholog RNAi reagent library
To facilitate the use of Drosophila to study human diseases, we assembled and characterized a resource, which we refer to as the Human Disease-TRiP (HuDis-TRiP) resource, consisting of RNAi fly stocks targeting Drosophila orthologs of genes implicated in human disease. This resource will enable the biomedical research community to quickly determine if a Drosophila model is relevant to their studies, and if so, allow them to embark on detailed follow-up analyses.
To identify the set of fly genes orthologous to genes known or suspected to be associated with human diseases, we selected 4850 human disease genes described in the Online Mendelian Inheritance in Man (OMIM) database (Amberger et al. 2015, 2019) as “with known sequence and phenotype” (omim.org/statistics/geneMap). We then mapped the human genes to fly genes using version 3 of our DIOPT tool (Hu et al. 2011) and selected those with DIOPT scores ≥2 (best score, forward or reverse search) or DIOPT score ≥4, identifying 3017 genes (10 was the max score for DIOPT v.3). In an analysis of these genes in the literature, we found that as a group, they are associated with a large number of top-level medical subject heading (MeSH) terms relevant to diseases of interest, e.g., nervous system diseases, musculoskeletal diseases, eye diseases, skin diseases, digestive system diseases, cardiovascular diseases, neoplasms, metabolic diseases, etc. (Figure S1). From this set of ∼3000 genes, we also extracted a set of 670 high-confidence Drosophila orthologs of high-confidence disease-associated human genes using DIOPT v.3 to map OMIM human disease genes to fly with very strong DIOPT score filter of eight or higher. To date we have produced a total of 2487 shRNA stocks targeting the full list of 3017 genes (Figure S1) with >95% coverage of the 670-gene high-confidence set. A sortable list of Human Disease library (HuDis) stocks is available at https://fgr.hms.harvard.edu/hudis-trip-fly-stocks. The HuDis website contains links to the human genes at NCBI, the fly genes at FlyBase, DIOPT results (linked to from orthology scores), BDSC stock IDs, and RNAi screen data in RSVP Plus. The information on the website is meant as a high-confidence starting off point; it is not a comprehensive list of fly gene-human disease gene relationships.
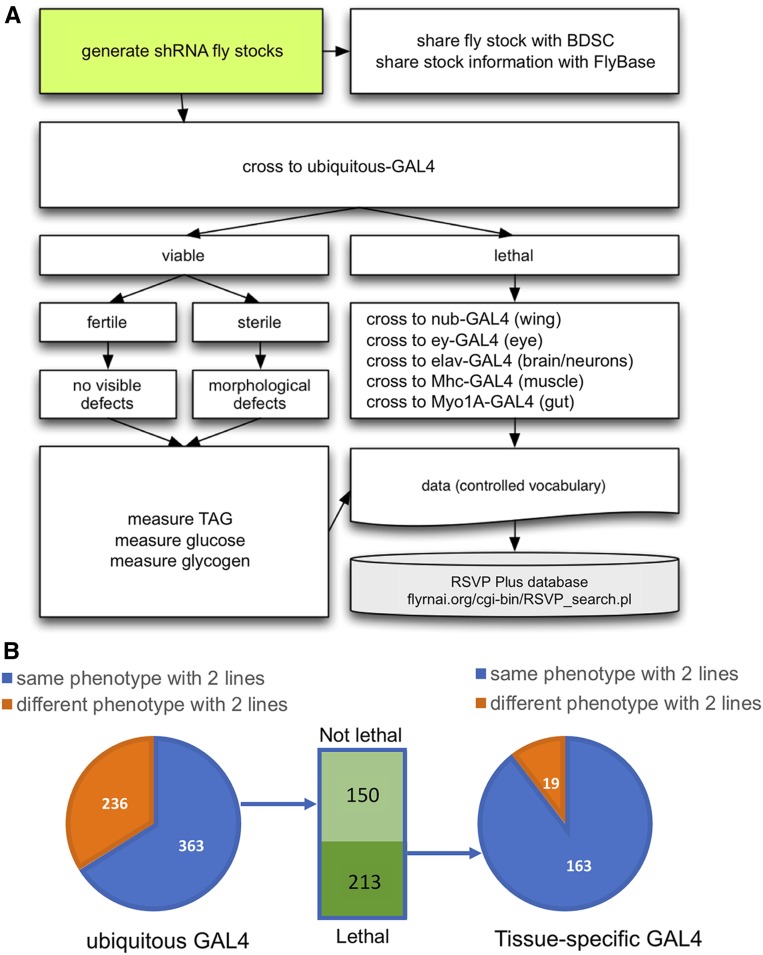
To validate the HuDis RNAi stocks, we first performed an initial characterization of ∼1200 RNAi lines targeting the high-confidence set of 670 genes using two GAL4 driver lines, actin5C-GAL4 and daughterless-GAL4, which are expressed in most or all cells and stages, identifying RNAi knockdown that results in lethality. As shown in our workflow diagram (Figure 5A), we then crossed the RNAi lines to a panel of selected GAL4 drivers to generate phenotypes in specific body parts (muscle and nervous systems, developing adult eye, wing, thorax, etc.) and at specific developmental stages. Flies were examined for visible morphological adult phenotypes, male and female sterility, and locomotor defects. We also measured sugar and triglyceride levels to determine whether the overexpressed gene affects overall metabolism. The full list of morphological and metabolic phenotypes is listed in Table S3. Based on these experiments, we identified 213 genes that were lethal when knocked down by ubiquitous GAL4 with at least two RNAi lines (Figure 5B). Of these, 163 gave the same phenotype with multiple RNAi lines in the follow-up studies with tissue specific GAL4s (Table S3).
Figure 5.
Characterization of HuDis RNAi lines. (A) Workflow diagram. We defined a set of high-confidence orthologs of high-confidence human disease-associated genes and generated a corresponding set of shRNA fly stocks. All available RNAi lines targeting the gene set (∼1200 fly stocks) were screened with ubiquitous GAL4 drivers (act5c and daughterless). Fly stocks were subcategorized as lethal or viable. The former were tested for sterility, gross morphological phenotypes, and metabolic defects. The latter were tested with a panel of tissue-specific GAL4 drivers. All phenotypic data were uploaded to RSVP Plus. (B) For 363 of ∼600 genes, results with two RNAi reagents were concordant, either both lethal or both viable. Knockdown of 213 of the genes resulted in lethality and 163 of these gave concordant phenotypes with multiple RNAi lines using a panel of tissue-specific GAL4 drivers.
HuDis-TRiP stocks were made in the VALIUM20 vector. Typically, ∼15% of shRNA stocks made in VALIUM20 do not homozygose due to leaky expression of the transgene (Qiao et al. 2018). Therefore, we also generated a resource of 1000 lines targeting human disease orthologs using the pNP vector (Qiao et al. 2018). The pNP stocks have reduced basal shRNA expression and RNAi specificity compared to VALIUM series stocks and augment the gene coverage of the collection.
Altogether, the HuDis-TRiP fly stock collection provides a resource for understanding the role of the human disease gene orthologs and for modeling human diseases in flies.
Human disease-related ortholog reagent CRISPR library
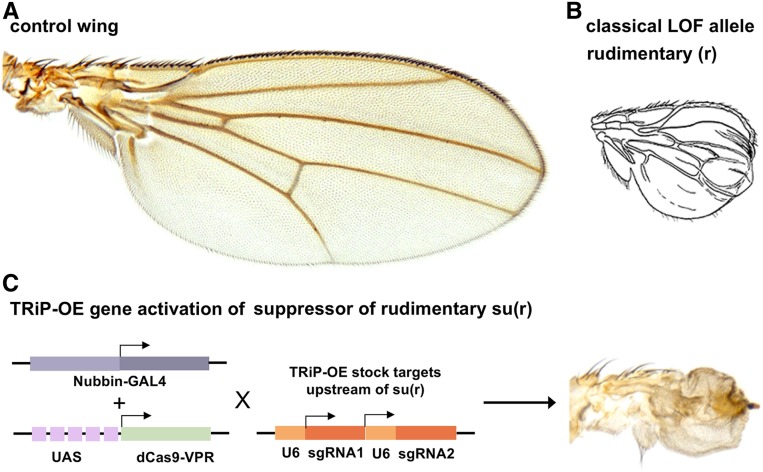
In addition to the HuDis-TRiP RNAi library, we have also assembled TRiP-KO and TRiP-OE libraries targeting the high-confidence HuDis orthologs, as well as putative rate-limiting enzymes, genes that are overexpressed in cancer, and genes mapped to microduplication syndromes (Table S4). To date, we have generated TRiP-OE lines covering 74 and 44%, respectively, of the high-confidence and complete sets of HuDis orthologs for which we were able to design sgRNAs (Figure S2). We have likewise generated TRiP-KO lines covering 76 and 45%, respectively, of the high-confidence and complete sets of HuDis orthologs for which we were able to design sgRNAs (Figure S2). The TRiP-KO stocks serve as a complement to RNAi lines for LOF studies and the TRiP-OE stocks serve as a means to test overexpression models, as might be particularly relevant for human diseases associated with dominant mutations, polynucleotide expansions, translocations, amplification, or duplication syndromes. As noted previously, GOF analysis can also help elucidate gene function when LOF produces no phenotype. As an example, the suppressor of rudimentary gene [su(r)], encodes dihydropyrimidine dehydrogenase, which catalyzes the rate-limiting step of pyrimidine degradation. Human dihydropyrimidine dehydrogenase expression is positively associated with aggressive tumor characteristics in several cancers and is predictive for poor patient prognosis (Mizutani et al. 2001; Horiguchi et al. 2002; Terashima et al. 2003; Zhu et al. 2018). In Drosophila, classical LOF su(r) alleles or RNAi targeting this gene produce no visible phenotypes. Rather, the function of su(r) in pyrimidine metabolism was only identified by suppression of the truncated wing phenotype exhibited by rudimentary (r) mutants in which de novo pyrimidine biosynthesis is blocked (Norby 1970; Rawls and Porter 1979). Using the wing-specific nubbin-GAL4 driver combined with UAS-dCas9-VPR and a TRiP-OE HuDis stock targeting su(r), we were able to generate a strong “rudimentary-like” small wing phenotype (Figure 6).
Figure 6.
TRiP-OE model for dihydropyrimidine dehydrogenase overexpression. (A) Control fly wing. (B) The rudimentary (r) gene encodes the Drosophila carbamoyl-phosphate synthetase 2 (CAD), which catalyzes the rate-limiting step of pyrimidine synthesis. Classical r alleles cause dramatic wing tissue undergrowth, as shown in a line drawing from the “Red Book” (Lindsley et al. 1992). (C) nubbin-GAL4 driving UAS-dCas9-VPR + TRiP-OE sgRNA targeting suppressor of rudimentary gene [su(r)], which encodes dihydropyrimidine dehydrogenase, an enzyme that catalyzes the rate-limiting step of pyrimidine degradation. As expected, overexpression of su(r) phenocopies the LOF of r phenotype.
TRiP data access and curation
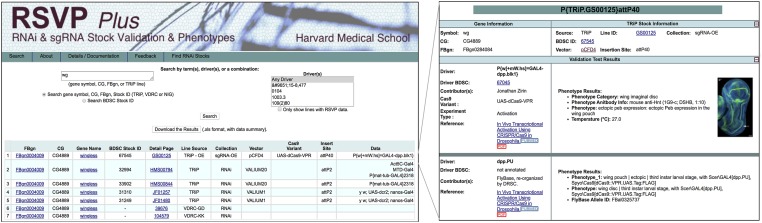
Information about TRiP plasmid vectors, stocks, and/or protocols are available (1) in published papers (Ni et al. 2008, 2009, 2011; Perkins et al. 2015); (2) at our website (https://fgr.hms.harvard.edu/trip-in-vivo-fly-rnai); (3) for RNAi reagents, at our website, as a downloadable Excel file of TRiP fly stocks (see https://fgr.hms.harvard.edu/trip-in-vivo-fly-rnai); (4) at our website, searchable using Gene Lookup (https://www.flyrnai.org/cgi-bin/DRSC_gene_lookup.pl), Updated Targets of RNAi Reagents (https://www.flyrnai.org/up-torr/), or RSVP (see below); (5) for CRISPR sgRNA fly stocks, using or our sgRNA nomination and production tracking database (https://www.flyrnai.org/tools/grna_tracker/web/); (6) at FlyBase; and (7) at the BDSC. In addition, validation and phenotype data for RNAi and CRISPR can be accessed through the RSVP Plus online database (https://www.flyrnai.org/cgi-bin/RSVP_search.pl) (Figure 7). RSVP Plus is an expanded version of our RSVP database (Perkins et al. 2015), modified to accommodate CRISPR data as well as RNAi data. We populate RSVP Plus in the following ways: (1) with unpublished data collected in-house or other research groups, (2) with published data shared in bulk file formats (e.g., Excel spreadsheets) by other research groups, (3) through community input online, and (4) through capture of RNAi or CRISPR fly stock phenotype data curated by FlyBase. Thus, RSVP Plus, which we maintain and regularly update, allows users to search and view information about knockdown, knockout, or overexpression phenotypes, including qPCR data and phenotypes (text or images) for specific reagent-GAL4 combinations. RSVP Plus also includes a modified user interface that lets external users upload data. At RSVP Plus there are currently >11,000 data entries for ∼5500 TRiP lines, representing ∼3900 fly genes. In addition, RSVP Plus contains 23,451 data entries from FlyBase for 17,782 RNAi lines representing 11,346 genes, including lines from the Vienna Drosophila Resource Center and the National Institute of Genetics, Japan. Altogether, researchers will be able to rapidly identify and make use of this data resource to identify effective reagents and design efficient experiments based on published and new data.
Figure 7.
RSVP Plus database of RNAi and CRISPR phenotypes. Shown here are the Search and Details pages for a search with the wingless (wg) gene. RSVP Plus allows users to view information about knockdown, knockout, and overexpression efficiency (qPCR data) and phenotypes (text and, when available, images) for specific combinations of RNAi or sgRNA fly stocks with Gal4 drivers and, when relevant, Cas9 versions (e.g., Cas9 or dCas9-VPR or flySAM). RSVP Plus includes results curated by FlyBase for TRiP reagents or other major stock collections (e.g., Vienna Drosophila Resource Center and Fly Stocks of National Institute of Genetics, Japan).
Concluding remarks
The TRiP platform has been used to generate >20,000 RNAi and CRISPR fly stocks for the research community. These resources, distributed to the community by the BDSC, provide powerful, versatile and transformative tools for gene knockdown, knockout, activation, and genome engineering. Researchers can now easily access fly stocks useful to “dial down” or “dial up” genes covered by the collection, in a given stage or tissue (made possible with the large collection of available Gal4 drivers), and as a result, facilitate a near limitless array of single-gene and multiplexed genetic experiments, as well as facilitate further analysis and integration of the resulting phenotypic data.
Acknowledgments
We thank Filip Port for transgenic Cas9 stocks and cloning advice. We thank the Bloomington Drosophila Stock Center and FlyBase for their support on TRiP fly stock data and resource deposition, as well as for their long-term and continued support of fly stock and data distribution to the Drosophila community. We thank the Harvard Medical School (HMS) Research Computing group for support relevant to our bioinformatics workflows and online resources. We thank the HMS Image Data Management Core and HMS Biopolymers Facility for relevant services and support. We also thank the Dana Farber/Harvard Cancer Center (DF/HCC) in Boston, Massachusetts, for use of the DNA Resource Core Facility. The DF/HCC is supported in part by the National Cancer Institute cancer center support grant NIH 5 P30 CA06516. The TRiP at HMS is supported by NIGMS grant R01-GM084947, with additional support from NCRR/ORIP grants R24-RR032668 and R24-OD021997, and NIGMS grant P41-GM132087. S.E.M. is also supported in part by the DF/HCC and N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11857434.
Communicating editor: K. O’Connor-Giles
Literature Cited
- Amberger J. S., Bocchini C. A., Schiettecatte F., Scott A. F., and Hamosh A., 2015. OMIM.org: online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43: D789–D798. 10.1093/nar/gku1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger J. S., Bocchini C. A., Scott A. F., and Hamosh A., 2019. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 47: D1038–D1043. 10.1093/nar/gky1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apidianakis Y., and Rahme L. G., 2011. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis. Model. Mech. 4: 21–30. 10.1242/dmm.003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S., Kweon J., Kim H. S., and Kim J. S., 2014. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11: 705–706. 10.1038/nmeth.3015 [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., and Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4: 220–228. 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W. et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. 10.1534/genetics.104.026427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman P., Seyedoleslami Esfahani S., and Engstrom Y., 2017. Drosophila as a model for human diseases-focus on innate immunity in barrier epithelia. Curr. Top. Dev. Biol. 121: 29–81. 10.1016/bs.ctdb.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Bischof J., Bjorklund M., Furger E., Schertel C., Taipale J. et al. , 2013. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140: 2434–2442. 10.1242/dev.088757 [DOI] [PubMed] [Google Scholar]
- Brand A. H., and Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Butzlaff M., Hannan S. B., Karsten P., Lenz S., Ng J. et al. , 2015. Impaired retrograde transport by the Dynein/Dynactin complex contributes to Tau-induced toxicity. Hum. Mol. Genet. 24: 3623–3637. 10.1093/hmg/ddv107 [DOI] [PubMed] [Google Scholar]
- Chavez A., Tuttle M., Pruitt B. W., Ewen-Campen B., Chari R. et al. , 2016. Comparison of Cas9 activators in multiple species. Nat. Methods 13: 563–567. 10.1038/nmeth.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xue J., Ruan J., Zhao J., Tang B. et al. , 2017. Drosophila CHIP protects against mitochondrial dysfunction by acting downstream of Pink1 in parallel with Parkin. FASEB J. 31: 5234–5245. 10.1096/fj.201700011R [DOI] [PubMed] [Google Scholar]
- Coll-Tané M., Krebbers A., Castells-Nobau A., Zweier C., and Schenck A., 2019. Intellectual disability and autism spectrum disorders ‘on the fly’: insights from Drosophila. Dis. Model. Mech. 12: dmm039180. 10.1242/dmm.039180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C. M., 2008. Oncogenes and cancer. N. Engl. J. Med. 358: 502–511. 10.1056/NEJMra072367 [DOI] [PubMed] [Google Scholar]
- Das T. K., Sangodkar J., Negre N., Narla G., and Cagan R. L., 2013. Sin3a acts through a multi-gene module to regulate invasion in Drosophila and human tumors. Oncogene 32: 3184–3197. 10.1038/onc.2012.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y. et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Dourlen P., Fernandez-Gomez F. J., Dupont C., Grenier-Boley B., Bellenguez C. et al. , 2017. Functional screening of Alzheimer risk loci identifies PTK2B as an in vivo modulator and early marker of Tau pathology. Mol. Psychiatry 22: 874–883. 10.1038/mp.2016.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C. J., and Perrimon N., 2006. High-throughput RNAi screening in cultured cells: a user’s guide. Nat. Rev. Genet. 7: 373–384. 10.1038/nrg1836 [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B., Mohr S. E., Hu Y., and Perrimon N., 2017a Accessing the phenotype gap: enabling systematic investigation of paralog functional complexity with CRISPR. Dev. Cell 43: 6–9. 10.1016/j.devcel.2017.09.020 [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B., Yang-Zhou D., Fernandes V. R., Gonzalez D. P., Liu L. P. et al. , 2017b Optimized strategy for in vivo Cas9-activation in Drosophila. Proc. Natl. Acad. Sci. USA 114: 9409–9414. 10.1073/pnas.1707635114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Hernández I., Scheenaard E., Pollarolo G., and Gonzalez C., 2016. The translational relevance of Drosophila in drug discovery. EMBO Rep. 17: 471–472. 10.15252/embr.201642080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K. et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman T. R., Gamliel A., Wessells R. J., Taghli-Lamallem O., Jepsen K. et al. , 2011. Over-expression of DSCAM and COL6A2 cooperatively generates congenital heart defects. PLoS Genet. 7: e1002344 10.1371/journal.pgen.1002344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F., Port F., and Boutros M., 2018. RNA interference (RNAi) screening in Drosophila. Genetics 208: 853–874. 10.1534/genetics.117.300077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi J., Takei H., Koibuchi Y., Iijima K., Ninomiya J. et al. , 2002. Prognostic significance of dihydropyrimidine dehydrogenase expression in breast cancer. Br. J. Cancer 86: 222–225. 10.1038/sj.bjc.6600040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden B. E., Valvezan A. J., Kelley C., Sopko R., Hu Y. et al. , 2015. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci. Signal. 8: rs9 10.1126/scisignal.aab3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B. et al. , 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Sopko R., Foos M., Kelley C., Flockhart I. et al. , 2013. FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3 (Bethesda) 3: 1607–1616. 10.1534/g3.113.007021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Xu R. G., Ren X., Ewen-Campen B., Rajakumar R. et al. , 2018. Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc. Natl. Acad. Sci. USA 115: 4719–4724. 10.1073/pnas.1800677115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., and Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M. D., Trevino A. E., Joung J., Abudayyeh O. O. et al. , 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. A., Cho K. C., Kim B., Jang I. H., Nam K. et al. , 2018. Inflammation-modulated metabolic reprogramming is required for DUOX-dependent gut immunity in Drosophila. Cell Host Microbe 23: 338–352.e5. 10.1016/j.chom.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Lin S., Ewen-Campen B., Ni X., Housden B. E., and Perrimon N., 2015. In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics 201: 433–442. 10.1534/genetics.115.181065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Zimm G. G., and Lindsley D. L., 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego. [Google Scholar]
- Liu Y., Gordesky-Gold B., Leney-Greene M., Weinbren N. L., Tudor M. et al. , 2018. Inflammation-induced, STING-dependent autophagy restricts zika virus infection in the Drosophila brain. Cell Host Microbe 24: 57–68.e3. 10.1016/j.chom.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Creanga A., Lum L., and Beachy P. A., 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363. 10.1038/nature05179 [DOI] [PubMed] [Google Scholar]
- McGurk L., Berson A., and Bonini N. M., 2015. Drosophila as an in vivo model for human neurodegenerative disease. Genetics 201: 377–402. 10.1534/genetics.115.179457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer H., Marom E., Alyagor I., Mayseless O., Berkun V. et al. , 2019. Tissue-specific (ts)CRISPR as an efficient strategy for in vivo screening in Drosophila. Nat. Commun. 10: 2113 10.1038/s41467-019-10140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra-Gorur K., Li D., Ma X., Yarman Y., Xue L. et al. , 2019. Spz/Toll-6 signal guides organotropic metastasis in Drosophila. Dis. Model. Mech. 12: dmm039727. 10.1242/dmm.039727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani Y., Wada H., Fukushima M., Yoshida O., Ukimura O. et al. , 2001. The significance of dihydropyrimidine dehydrogenase (DPD) activity in bladder cancer. Eur. J. Cancer 37: 569–575. 10.1016/S0959-8049(00)00440-8 [DOI] [PubMed] [Google Scholar]
- Nagy A., Perrimon N., Sandmeyer S., and Plasterk R., 2003. Tailoring the genome: the power of genetic approaches. Nat. Genet. 33: 276–284. 10.1038/ng1115 [DOI] [PubMed] [Google Scholar]
- Ni J.-Q., Liu L.-P., Binari R., Hardy R., Shim H.-S., Cavallaro A., Booker M., Pfeiffer B. D., Markstein M., Wang H. et al. , 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182: 1089–1100. 10.1534/genetics.109.103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Markstein M., Binari R., Pfeiffer B., Liu L.-P., Villalta C., Booker M., Perkins L., Perrimon N., 2008. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods 5: 49–51. 10.1038/nmeth1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H.-S., Tao R., Handler D., Karpowicz P. et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8: 405–407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norby S., 1970. A specific nutritional requirement for pyrimidines in rudimentary mutants of Drosophila melanogaster. Hereditas 66: 205–214. 10.1111/j.1601-5223.1970.tb02346.x [DOI] [PubMed] [Google Scholar]
- Okray Z., and Hassan B. A., 2013. Genetic approaches in Drosophila for the study neurodevelopmental disorders. Neuropharmacology 68: 150–156. 10.1016/j.neuropharm.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Ludwig M. Z., Tamarina N. A., He B. Z., Carl S. H. et al. , 2014. Genetic complexity in a Drosophila model of diabetes-associated misfolded human proinsulin. Genetics 196: 539–555. 10.1534/genetics.113.157602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson W. M., Dookwah M., Dear M. L., Gatto C. L., Aoki K. et al. , 2016. Synaptic roles for phosphomannomutase type 2 in a new Drosophila congenital disorder of glycosylation disease model. Dis. Model. Mech. 9: 513–527. 10.1242/dmm.022939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R. et al. , 2015. The transgenic RNAi project at harvard medical School: resources and validation. Genetics 201: 843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N., Ni J. Q., and Perkins L., 2010. In vivo RNAi: today and tomorrow. Cold Spring Harb. Perspect. Biol. 2: a003640 10.1101/cshperspect.a003640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., and Bullock S. L., 2016. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13: 852–854. 10.1038/nmeth.3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H. M., Lee T., and Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Strein C., Stricker M., Rauscher B., Heigwer F. et al. , 2020. A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. Elife 9: e53865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H. H., Wang F., Xu R. G., Sun J., Zhu R. et al. , 2018. An efficient and multiple target transgenic RNAi technique with low toxicity in Drosophila. Nat. Commun. 9: 4160 10.1038/s41467-018-06537-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J. M., and Porter L. A., 1979. Organization of the rudimentary wing locus in DROSOPHILA MELANOGASTER. I. The isolation and partial characterization of mutants induced with ethyl methanesulfonate, icr-170 and X rays. Genetics 93: 143–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Sun J., Housden B. E., Hu Y., Roesel C. et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110: 19012–19017. 10.1073/pnas.1318481110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. 10.1073/pnas.93.22.12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P., Szabo K., Bailey A., Laverty T., Rehm J. et al. , 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Rousseaux M. W. C., Vazquez-Velez G. E., Al-Ramahi I., Jeong H. H., Bajic A. et al. , 2018. A druggable genome screen identifies modifiers of alpha-synuclein levels via a tiered cross-species validation approach. J. Neurosci. 38: 9286–9301. 10.1523/JNEUROSCI.0254-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R. et al. , 2000. Comparative genomics of the eukaryotes. Science 287: 2204–2215. 10.1126/science.287.5461.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertel C., Albarca M., Rockel-Bauer C., Kelley N. W., Bischof J. et al. , 2015. A large-scale, in vivo transcription factor screen defines bivalent chromatin as a key property of regulatory factors mediating Drosophila wing development. Genome Res. 25: 514–523. 10.1101/gr.181305.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebo Z. L., Lee H. B., Peng Y., and Guo Y., 2014. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly (Austin) 8: 52–57. 10.4161/fly.26828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita M., and Cagan R. L., 2017. Modeling human cancers in Drosophila. Curr. Top. Dev. Biol. 121: 287–309. 10.1016/bs.ctdb.2016.07.008 [DOI] [PubMed] [Google Scholar]
- Splinter K., Adams D. R., Bacino C. A., Bellen H. J., Bernstein J. A. et al. , 2018. Effect of genetic diagnosis on patients with previously undiagnosed disease. N. Engl. J. Med. 379: 2131–2139. 10.1056/NEJMoa1714458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt N., Molitor A., Somogyi K., Mata J., Curado S. et al. , 2005. Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS Genet. 1: e55 10.1371/journal.pgen.0010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3: 176–188. 10.1038/nrg751 [DOI] [PubMed] [Google Scholar]
- Terashima M., Fujiwara H., Takagane A., Abe K., Irinoda T. et al. , 2003. Prediction of sensitivity to fluoropyrimidines by metabolic and target enzyme activities in gastric cancer. Gastric Cancer 6: 71–81. 10.1007/s10120-003-0221-z [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A. et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S. et al. , 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47: D759–D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur B., Chen K., and Bellen H. J., 2016. Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 9: 235–244. 10.1242/dmm.023762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., and Bellen H. J., 2005. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6: 167–178 (erratum: Nat. Rev. Genet 6: 340). 10.1038/nrg1553 [DOI] [PubMed] [Google Scholar]
- Viswanathan M. C., Kaushik G., Engler A. J., Lehman W., and Cammarato A., 2014. A Drosophila melanogaster model of diastolic dysfunction and cardiomyopathy based on impaired troponin-T function. Circ. Res. 114: e6–e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler M. F., Yamamoto S., Chao H. T., Posey J. E., Westerfield M. et al. , 2017. Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207: 9–27. 10.1534/genetics.117.203067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S. et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. 10.1534/genetics.113.153825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W. P., Liu Z. Y., Zhao Y. M., He X. G., Pan Q. et al. , 2018. Dihydropyrimidine dehydrogenase predicts survival and response to interferon-alpha in hepatocellular carcinoma. Cell Death Dis. 9: 69 10.1038/s41419-017-0098-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
TRiP stocks are available at the BDSC, National Institute of Genetics Japan, and the TsingHua Fly Center. All vectors are available from the Drosophila Genomics Resource Center. Maps with attributes, complete sequences, and detailed cloning protocols can be found on the TRiP website (https://fgr.hms.harvard.edu/) under Reagents and Protocols. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11857434.