Obesity results from a complex interplay of diet, behavior, and genetic background. Our genes are out of our control, but it may be possible to customize our diet to match changes in metabolism resulting from...
Keywords: Spen, Drosophila melanogaster, fat body, metabolism, gene–diet interaction
Abstract
Obesity and its comorbidities are a growing health epidemic. Interactions between genetic background, the environment, and behavior (i.e., diet) greatly influence organismal energy balance. Previously, we described obesogenic mutations in the gene Split ends (Spen) in Drosophila melanogaster, and roles for Spen in fat storage and metabolic state. Lipid catabolism is impaired in Spen-deficient fat storage cells, accompanied by a compensatory increase in glycolytic flux and protein catabolism. Here, we investigate gene–diet interactions to determine if diets supplemented with specific macronutrients can rescue metabolic dysfunction in Spen-depleted animals. We show that a high-yeast diet partially rescues adiposity and developmental defects. High sugar partially improves developmental timing as well as longevity of mated females. Gene–diet interactions were heavily influenced by developmental-stage-specific organismal needs: extra yeast provides benefits early in development (larval stages) but becomes detrimental in adulthood. High sugar confers benefits to Spen-depleted animals at both larval and adult stages, with the caveat of increased adiposity. A high-fat diet is detrimental according to all tested criteria, regardless of genotype. Whereas Spen depletion influenced phenotypic responses to supplemented diets, diet was the dominant factor in directing the whole-organism steady-state metabolome. Obesity is a complex disease of genetic, environmental, and behavioral inputs. Our results show that diet customization can ameliorate metabolic dysfunction underpinned by a genetic factor.
Obesity is a complex disease arising from interactions between environment, behavior, and genetics (Silventoinen et al. 2010; Fryar et al. 2016; Qasim et al. 2018). Metabolism of dietary nutrients into energy and biomass for survival and reproduction is a highly conserved process involving three major classes of macronutrients: proteins, carbohydrates, and lipids. One major contributor to the growing obesity epidemic is Western diet, characterized by overconsumption of calories from processed carbohydrates, fatty animal meats and byproducts, and salt, while lacking in fiber and important minerals and nutrients (Grotto and Zied 2010). Prolonged intake of excess nutrients disrupts organismal metabolic homeostasis and drives a shift from the utilization of nutrients (catabolism) to nutrient storage (anabolism).
Although diet can be a major driver of fat storage, gene–environment interactions play a critical yet poorly understood role in the development of obesity and metabolic syndrome, defined as “clustering of abdominal obesity, dyslipidemia, hyperglycemia, and hypertension” (Giovannucci 2007). Indeed, previous work showed that gene–diet interactions account for a much larger degree of metabolic trait variance than diet alone (Reed et al. 2010). Subsequent work suggested that genetic risk factors alter the effect of lifestyle on obesity predisposition (Nakamura et al. 2016). Decades ago, the increased incidence of diabetes mellitus was attributed to an evolutionarily beneficial “thrifty” genotype that is incompatible with the modern Western diet and lifestyle (Neel 1962). Evidence of genetic adaptation to a diet high in polyunsaturated fatty acids can be found in the genomes of Greenlandic Inuit, in the form of genetic variants in fatty acid desaturases (Fumagalli et al. 2015; Chen et al. 2019). Accordingly, developing a comprehensive understanding of obesity and metabolic syndrome demands examination of the interplay between genetics and diet.
Drosophila melanogaster is an excellent model in which to study gene function in metabolism (Grönke et al. 2003, 2005; Baker and Thummel 2007; Leopold and Perrimon 2007; Schlegel and Stainier 2007; Guo et al. 2008; Beller et al. 2010; Reis et al. 2010; Hazegh et al. 2017; Musselman and Kuhnlein 2018). Metabolic pathways are highly conserved in D. melanogaster (Bernards and Hariharan 2001) and many fat regulatory genes have functional orthologs in mammals (Pospisilik et al. 2010; Reis et al. 2010). D. melanogaster also recapitulate the sensitivity of metabolism to genetic variation within a population while retaining the physiological plasticity to compensate, or buffer, against defects in key processes (Li et al. 2019; Matoo et al. 2019).
We have previously shown that the RNA-binding protein Split ends (Spen), a pleiotropic transcriptional regulator in D. melanogaster (Rebay et al. 2000; Lin et al. 2003; Hazegh et al. 2017), potentiates fat catabolism (Hazegh et al. 2017). When Spen is mutated or knocked down via RNA interference (RNAi) in the fat body (FB; main metabolic organ of the fly) of D. melanogaster larvae, fat catabolism is strongly inhibited, reflected by higher levels of stored triglycerides (Hazegh et al. 2017). FB-specific depletion of Spen (hereafter Spen-KD) also delays larval wandering, consistent with a failure to mobilize energy to fuel development. Impairment of fatty acid catabolism in Spen-KD larvae is accompanied by increased adiposity, decreased lipase expression, depletion of L-carnitine, and evidence of compensatory upregulation of alternative metabolic pathways, such as protein catabolism (Hazegh et al. 2017). Spen depletion also decreased glycogen, pointing to the use of stored carbohydrates as an auxiliary energy source in lieu of fat (Hazegh et al. 2017). Finally, Spen-KD larvae are short-lived under conditions of amino acid deprivation (Hazegh et al. 2017).
Considering the failure to utilize dietary fats and potential upregulation of protein and carbohydrate catabolism, we hypothesized that custom diets designed to suit the unique metabolic demands of Spen-KD larvae may be capable of rescuing their defects in adiposity, development, and longevity. Here we report the effects of such supplemented diets. Our results illustrate gene–diet interactions in the Spen-KD model of genetic obesity in D. melanogaster and support the use of dietary interventions to mitigate the influence of genetic mutations on metabolic functions and physiological outcomes. Since Spen homologs may also regulate energy balance in mammals (Hazegh et al. 2017), our findings provide a foundation for possible dietary interventions in humans struggling with defects in metabolic pathways involving Spen.
Materials and Methods
Fly strains and husbandry
w1118 [Bloomington Drosophila Stock Center stock number (BL) 3605], w1118; dcg > GAL4 (BL 7011), y1sc v1; +; UAS-spen RNAi (BL 33398), and y1 v1; +; UAS-w RNAi (BL 28980) were obtained from the Bloomington Drosophila Stock Center. Animals were reared at 25° and 60% humidity unless otherwise specified and fed a modified Bloomington Drosophila Stock Center media (1L: yeast 15.9 g, soy flour 9.2 g, yellow cornmeal 67.1 g, light malt extract 42.4 g, agar 5.3 g, light corn syrup 90 g, propionic acid 4.4 ml, tegosept in ethanol 8.4 ml). Medium-yeast diet (MYD) contained 35 g yeast per liter. High-yeast diet (HYD) contained 70 g yeast per liter. High-fat diet (HFD) is MYD with an additional 150 g coconut oil per liter (15%). High-sugar diet (HSD) is MYD with an additional 180 g corn syrup per liter (1 M). Eggs were collected on grape plates at 25° and 50 first instar larvae were transferred 22–24 hr later into a vial of the specified diet.
Density assay
Density assays were performed as described previously with 50 larvae per sample (Reis et al. 2010; Hazegh and Reis 2016; Hazegh et al. 2017). Five independent samples were analyzed for each experimental condition and genotype. We used the Kolmogorov–Smirnov test (P > 0.1) to confirm normality and two-way ANOVA to calculate statistical significance between genotypes on the same diet (i.e., Spen-KD|HYD vs. CTRL-KD|HYD) and different diets within genotype (i.e., Spen-KD|HYD vs. Spen-KD|MYD) (*P < 0.05). Prism 8 software (GraphPad Software, La Jolla, CA) was used for this analysis and to generate plots.
Developmental timing
In each trial, 50 first instar larvae were transferred per genotype per diet. Upon hatching larvae were transferred to experimental diets (EXPs), allowed to develop, and then counted three times daily (8:00 am, 12:00 pm, and 4:00 pm ) for developmental progress after reaching the late third instar wandering stage. Three replicates were analyzed per genotype per diet for differences in developmental timing. The sampling intervals were unequal (16 hr overnight, followed by two 4-hr intervals during the day). See below for diet effect data analysis methods.
Longevity
Larvae were reared in standard diet. Upon eclosion, flies were allowed 48 hr to mate, whereupon flies were separated by sex and 20 of same sex were placed into new vials of standard diet or EXPs. Flies were transferred every 2 days into vials of fresh food and scored for deaths once every day. Log-rank test was used to calculate statistical significance with Prism 8 software. This process was repeated three times independently.
Diet effect data analysis
To examine developmental timing differences of a genotype between different diets, a chi-square test was performed using the Mantel–Cox log-rank test in GraphPad Prism 8 software. Next, we determined the effect size of a diet on a given genotype using Cliff’s delta (d). We compared the ratios of distributions between diets in one genetic background (for example, Spen-KD|MYD vs. Spen-KD|EXP) to the ratios of distributions between matched diets in another genetic background (for example, CTRL-KD|MYD vs. CTRL-KD|EXP). These two sets of ratios were then compared to each other using a Wilcoxon rank sum test. Values for Cliff’s delta range between −1 and 1, and larger absolute values indicate stronger effects.
Metabolomics
Metabolomics was performed in triplicate, with 10 larvae per genotype per diet for a cumulative 30 individual larvae per genotype, per diet condition. Briefly, individual larvae, were suspended in 1 ml of methanol/acetonitrile/water (5:3:2, v/v) prechilled to −20° and vortexed continuously for 30 min at 4°. Insoluble material was removed by centrifugation at 10,000 × g for 10 min at 4° and supernatants were isolated for metabolomics analysis by ultra high-performance liquid chromatography and mass spectrometry. Analyses were performed as previously described (Nemkov et al. 2017, 2019) using a Vanquish UHPLC system coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Metabolite assignments, isotopic patterns, and correction for expected natural abundances of deuterium, 13C, and 15N isotopes were performed using MAVEN (Princeton, NJ; Clasquin et al. 2012). To compare differences within both the genotype and between genotypes on the same diet, multivariate analyses including orthogonal partial least squares discriminant analysis (OPLS-DA) were prepared using MetaboAnalyst 4.0 (Chong et al. 2018). Before analysis, data were normalized by metabolite median value and ranged scaled in the MetaboAnalyst workflow. Graphs, heat maps, and statistical analyses (t-test) were performed with GraphPad Prism 8.0.
Data availability
Flies and analysis scripts are available upon request. The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11446206.
Results
HSDs and HYDs dampen the effect of FB-specific spen depletion on fat storage
Spen-KD causes an increased adiposity phenotype on the control diet (MYD) compared to the White-RNAi control (CTRL-KD; Figure 1A, Hazegh et al. 2017). To test if macronutrient supplementation can rescue the increased fat storage in Spen-KD larvae, the standard MYD was supplemented with excess yeast to make an HYD as used in previous studies on diet composition and life span (Lee et al. 2008; Skorupa et al. 2008). Yeast is the main source of protein in standard fly diets, although it also provides other nutrients, such as vitamins and carbohydrates. Supplementation with 1 M sugar produced an HSD as used in previous studies examining D. melanogaster obesity and insulin resistance (Musselman et al. 2011). We used an HFD supplemented with coconut oil, as used in previous obesity pathophysiology research in D. melanogaster (Birse et al. 2010). Larvae were transferred from grape plates into these diets as first instar larvae, at 22–24 hr after egg deposition (AED), and collected at the third instar wandering stage to measure buoyancy (Figure 1). There are sex-specific differences in adiposity; however, the Spen-KD affected both sexes similarly (Hazegh et al. 2017), therefore we analyzed mixed male/female larvae in the buoyancy assays. For all buoyancy assays, progeny from crosses of UAS-spen RNAi, UAS-w RNAi, and dcg > GAL4 to w1118 strains were used as genetic background controls (Supplemental Material, Figure S1; Hazegh et al. 2017).
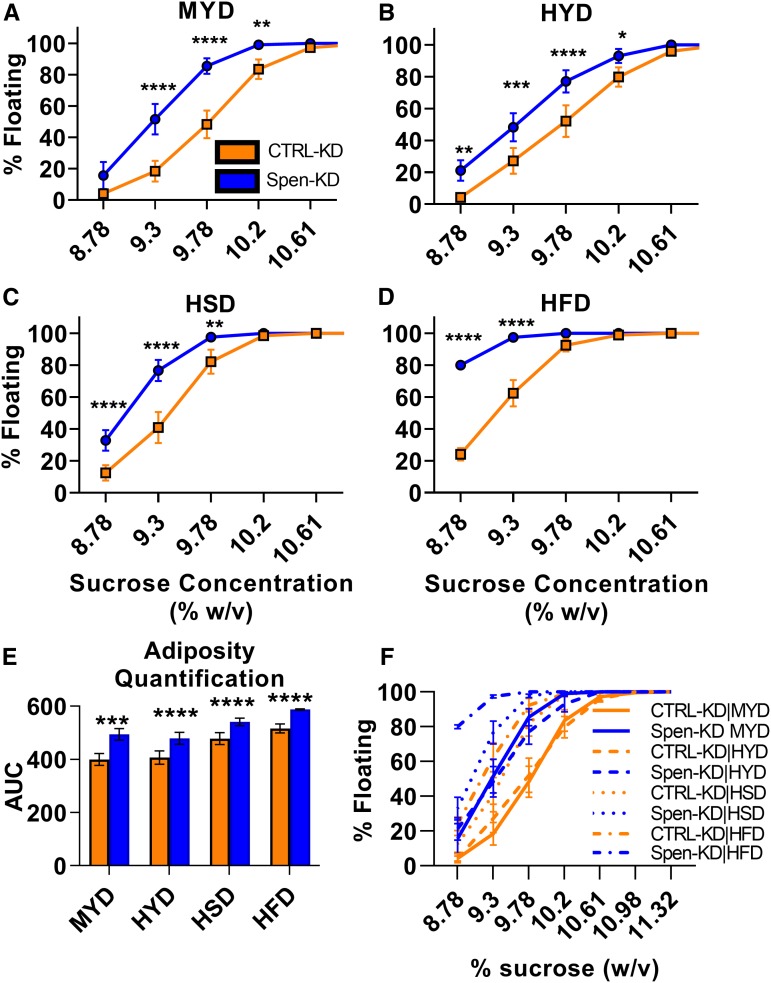
Figure 1.
Dietary intervention affects the adiposity phenotype of Spen-KD. Buoyancy assays were performed on third instar wandering larvae. (A) The percent of floating larvae at different sucrose densities (percent weight/volume) of FB-specific Spen KD (Spen-KD, blue) compared to control KD (CTRL-KD, orange) reared on the control MYD. (B) As in Figure 1A, Spen-KD buoyancy compared to CTRL-KD across a sucrose gradient with larvae reared on HYD, (C) reared on HSD, and (D) reared on HFD. Two-way ANOVA compared CTRL-KD to Spen-KD buoyancy at increasing sucrose concentration; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (E) Quantification of the buoyancy curves by integrating the area under the curve (AUC) represented as arbitrary units (A.U.). (F) The data from A–D combined in one plot. Comparison by two-way ANOVA (***P < 0.001, ****P < 0.0001). Error bars represent SEM. n = 5 replicates, 50 larvae per replicate.
We compared the buoyancy of the larvae on different diets by integrating the area under the curves generated by plots of percentage of floating larvae vs. sucrose concentration and comparing the differences between areas (Figure 1E and Table 1). Spen-KD was significantly fatter than CTRL-KD on all diets (two-way ANOVA, P < 0.05; Figure 1, A–D). MYD drove the largest buoyancy difference between Spen-KD and CTRL-KD [94.7 arbitrary units (A.U.); Figure 1A and Table 1]. Both Spen-KD and CTRL-KD had the largest area under the curve on HFD, indicating the HFD drives the largest increase in adiposity regardless of genotype (Figure 1D and Table 1). The smallest difference between Spen-KD and CTRL-KD was observed on the HSD (Figure 1C and Table 1). While HSD increased the adiposity of Spen-KD and CTRL-KD when compared to MYD matched controls, this diet decreased genotypical adiposity differences (Table 1; see File S1 for all two-way ANOVA comparisons between diet and genotype at each sucrose concentration).
Table 1. Quantification of the area under the curve (AUC) of Spen-KD and CTRL-KD on diet buoyancy assays.
| Diet | CTRL-KD | Spen-KD | Area difference | P-value |
|---|---|---|---|---|
| AUC (A.U.) | AUC (A.U.) | Spen-KD–CTRL-KD | ||
| MYD | 399.3 | 494 | 94.70 ± 11.75 | <0.0001 |
| HYD | 407 | 479 | 72.00 ± 12.71 | <0.001 |
| HSD | 478 | 540.9 | 62.90 ± 10.01 | <0.0001 |
| HFD | 515.9 | 587.4 | 71.50 ± 7.558 | <0.0001 |
Area difference is reported with confidence interval. Two-way ANOVA analysis compared Spen-KD and CTRL-KD in matched diets (see File S1). n = 5, A.U., arbitrary units.
HSDs and HYDs partially rescue previously characterized developmental delays in Spen-KD
Larvae in which Spen is depleted in the FB are delayed in development (Hazegh et al. 2017). To test how diet supplementation affects development, Spen-KD and CTRL-KD larvae were reared in HYD, HFD, or HSD from first instar larvae to eclosion. Vials were assessed three times daily for numbers of wandering larvae, pupal cases, and eclosed flies and compared by two-way ANOVA, Cliff’s delta, chi-square log-rank (Mantel–Cox) to examine gene–diet interaction.
We first assessed the wandering stage, when larvae reach “critical mass” and leave the food substrate to find a suitable environment for pupariation. The timing of third instar wandering stage was more affected by the genotype than by diet composition (Figure 2, A–C and Table 2). The mean wandering time of Spen-KD|MYD larvae was 159.5 hr AED, a significant 24-hr delay from the CTRL-KD|MYD (wandering mean of 135.5 hr AED; two-way ANOVA, P < 0.0001; Figure 2A and Table 2). The delay observed between the Spen-KD|experimental (EXP) and CTRL-KD|EXP diet-matched controls was strongly conserved in the different diets: Spen-KD|HYD was delayed 25.9 hr (two-way ANOVA P < 0.0001; Figure 2A and Table 2), Spen-KD|HSD was delayed 22.6 hr (two-way ANOVA, P < 0.0001; Figure 2B and Table 2), and Spen-KD|HFD was delayed 27.8 hr (two-way ANOVA, P < 0.0001; Figure 2C and Table 2).
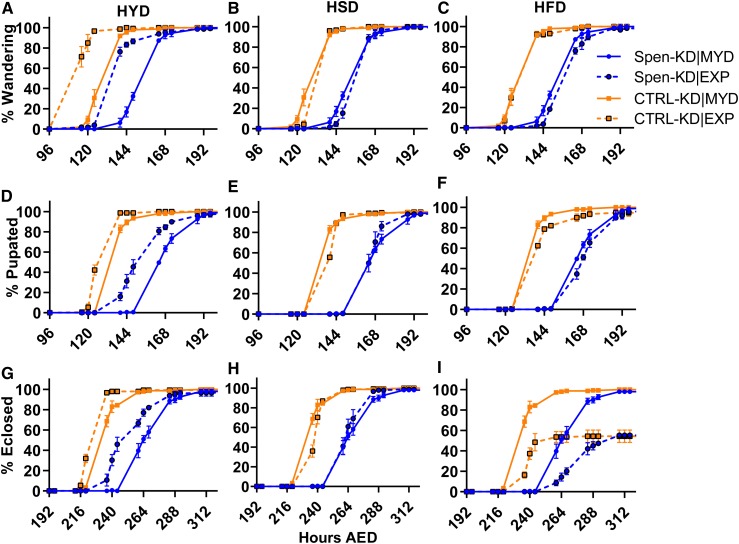
Figure 2.
HYD improves developmental delays in Spen-KD larvae. Eggs were collected using grape plates and at 22–24 hr after egg deposition (AED), 50 larvae per genotype were transferred into either the control MYD, or an EXP: HYD, HSD, or HFD. Vials were monitored at 8:00 am, 12:00 pm, and 4:00 pm for developmental progress. (A–C) Spen-KD|MYD (solid blue) and CTRL-KD|MYD (solid orange) wandering percentage at each time point in hours AED compared to Spen-KD and CTRL-KD larvae reared on the experimental (A) HYD, (B) HSD, and (C) HFD (dashed lines). (D–F) Spen-KD|MYD and CTRL-KD|MYD pupariation percentage at each time point in hours AED compared to Spen-KD and CTRL-KD larvae reared on the EXP: (D) HYD, (E) HSD, and (F) HFD. (G–I) Spen-KD|MYD and CTRL-KD|MYD eclosion percentage at each time point in hours AED compared to Spen-KD and CTRL-KD larvae reared on the experimental, (G) HYD, (H) HSD, and (I) HFD. n = 3 replicates, 50 larvae per replicate.
Table 2. Developmental timing of Spen-KD and CTRL-KD genotypes on diets.
| Diet | CTRL-KD mean (hours AED) | Spen-KD mean (hours AED) | Spen-KD/ CTRL-KD delay (hours) | Two-way ANOVA (P-value) | CTRL-KD (MYD|EXP) chi-square | P-value | Spen-KD (MYD|EXP) chi-square | P-value | Cliff’s delta (d) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Wandering | MYD | 135.5 | 159.5 | 24.0 | <0.0001 | NA | NA | NA | NA | NA |
| HYD | 118.2 | 144.1 | 25.9 | <0.0001 | 208.3 | <0.0001 | 83.47 | <0.0001 | −0.26 | |
| HSD | 139.9 | 162.5 | 22.6 | <0.0001 | 14.95 | 0.0001 | 2.910 | NS | 0.07 | |
| HFD | 136.7 | 164.5 | 27.8 | <0.0001 | 0.7895 | NS | 8.459 | 0.0036 | −0.14 | |
| Pupariation | MYD | 142.6 | 171.5 | 28.9 | <0.0001 | NA | NA | NA | NA | NA |
| HYD | 132.7 | 158.6 | 25.9 | <0.0001 | 77.50 | <0.0001 | 35.55 | <0.0001 | 0.09 | |
| HSD | 142.9 | 168.3 | 25.4 | <0.0001 | 5.153 | 0.0232 | 6.387 | 0.0115 | 0.2 | |
| HFD | 143.6 | 173.2 | 29.6 | <0.0001 | 2.080 | NS | 0.633 | NS | −0.03 | |
| Eclosion | MYD | 239.9 | 272.9 | 33.0 | <0.0001 | NA | NA | NA | NA | NA |
| HYD | 231.1 | 256.2 | 25.1 | <0.0001 | 61.08 | <0.0001 | 52.44 | <0.0001 | 0.2 | |
| HSD | 242.6 | 268.7 | 26.1 | <0.0001 | 10.07 | 0.0015 | 7.247 | 0.0071 | 0.28 | |
| HFD | 243.1 | 280.4 | 37.3 | <0.0001 | 8.648 | 0.0033 | 10.70 | 0.0011 | −0.08 |
Developmental timing is reported in mean hours AED. Cliff’s delta measures the effect size of a diet between two genotypes. Two-way ANOVA compares Spen-KD and CTRL-KD in matched diet conditions. Mean hour after egg deposition (AED). n = 3 replicates, 50 larvae per replicate. Chi-square from log-rank (Mantel–Cox) test.
While genotype has a clear and significant effect on wandering stage, we wanted to determine if Spen-KD is more responsive to dietary changes than CTRL-KD. To do so, we calculated effect size, a quantitative measure of the magnitude of a developmental time difference. A larger absolute value of an effect size indicates a stronger effect between the two variables. We used Cliff’s delta (d) to measure how often the values in one distribution (ratio of Spen-KD|EXP:Spen-KD|MYD) were larger than the values in a second distribution (ratio of CTRL-KD|EXP:CTRL-KD|MYD). Importantly, this does not require assumptions about how the data are distributed. Therefore, the fact that our data collection was not evenly distributed through the 24 hr of each day does not more heavily weight one data point vs. another. d values of 0.11, 0.28, and 0.43 denote small, medium, and large effect size, respectively (Vargha and Delaney 2000).
Both genotypes wandered earlier when reared on the HYD compared to MYD; however, CTRL-KD|HYD (17.3 hr earlier than control diet, chi-square = 208.3, P < 0.0001; Table 2) was more responsive to increased protein than the Spen-KD|HYD (15.4 hr earlier than control diet, chi-square = 83.47, P < 0.0001; Table 2). This conclusion is supported by the effect ratio (d = −0.26), which means that both genotypes were positively affected by extra dietary protein; however, CTRL-KD was more strongly affected than Spen-KD (Table 2). Spen-KD|HSD wandering time was not statistically different from Spen-KD|MYD (3 hr delayed, chi-square = 2.91, P = NS; Table 2); however, CTRL-KD|HSD was significantly delayed 4.4 hr from control (chi-square = 14.95, P = 0.0001; Table 2). HSD had a small effect size (d = 0.07) between Spen-KD and CTRL-KD, indicating that a carbohydrate-enriched diet has only a small effect on wandering time between genotypes (Table 2). Extra fat delayed development of Spen-KD larvae to a greater extent than controls. Spen-KD|HFD was delayed in wandering compared to Spen-KD|MYD (5 hr, chi-square = 8.459, P = 0.0036), whereas CTRL-KD wandering was not significantly different (1.2 hr delayed, chi-square = 0.7895, P = NS; Table 2). The effect size (d = −0.14) confirms that Spen-KD was more negatively affected than CTRL-KD by HFD at the wandering stage (Table 2).
As observed in wandering, pupariation was slower in Spen-KD animals compared to CTRL-KD on all the EXPs (Figure 2, D–F and Table 2). Spen-KD|MYD pupariation occurred at a mean 171.5 hr AED, a 28.9-hr delay from CTRL-KD|MYD pupation (two-way ANOVA, P < 0.0001; Figure 2D and Table 2). Compared to matched-diet CTRL-KD Spen-KD|HYD was delayed 25.9 hr (two-way ANOVA, P < 0.0001; Figure 2D and Table 2), Spen-KD|HSD was delayed 25.4 hr (two-way ANOVA, P < 0.0001; Figure 2E and Table 2), and Spen-KD|HFD was delayed 29.6 hr (two-way ANOVA, P < 0.0001; Figure 2F and Table 2). We next examined the sensitivity of the Spen-KD genotype to macronutrient content and asked if a partial phenotypic rescue is possible through dietary supplementation. Spen-KD|HSD pupariated earlier than Spen-KD|MYD (3.2 hr earlier than control, chi-square = 6.387 P = 0.0115; Table 2). Conversely, CTRL-KD|HSD was delayed compared to CTRL-KD|MYD (0.3 hr from control, chi-square = 5.153, P = 0.0232; Table 2). When comparing the effect of diet between genotypes, we saw that HSD had a larger effect on Spen-KD than CTRL-KD (d = 0.20; Table 2). There was no significant difference in pupariation timing between Spen-KD|HFD and Spen-KD|MYD (1.7 hr, chi-square = 0.633, P = NS; Table 2), or between CTRL-KD|HFD and CTRL-KD|MYD (1 hr, chi-square = 2.080, P = NS; Table 2). The HFD effect size showed almost no difference in the effect of HFD between genotypes (d = −0.03). Spen-KD|HYD pupariated earlier than Spen-KD|MYD (12.9 hr earlier than control, chi-square = 35.55, P < 0.0001). The CTRL-KD|HYD also pupariated earlier than CTRL-KD|MYD (9.9 hr earlier than control, chi-square = 77.50, P < 0.0001; Table 2). While both genotypes responded positively to HYD, HYD had a stronger effect on CTRL-KD (d = 0.09).
Eclosion appeared to be more sensitive to diet than wandering or pupariation (Figure 2, G–I and Table 2). Spen-KD|MYD eclosed a mean 272.9 hr AED, a 33-hr delay relative to CTRL-KD|MYD (two-way ANOVA, P < 0.0001; Figure 2G and Table 2), which could be a cumulative effect from delays in earlier development. Extra sugar accelerated eclosion in Spen-KD animals, with Spen-KD|HSD eclosing an average of 4.2 hr earlier than Spen-KD|MYD (chi-square = 7.247, P = 0.0071; Figure 2H and Table 2). By contrast, extra sugar delayed eclosion in control animals (CTRL-KD|HSD delayed 2.7 hr compared to CTRL-KD|MYD, chi-square = 10.07, P = 0.0015; Figure 2H and Table 2). HSD affected Spen-KD more than CTRL-KD (d = 0.28). The HFD delayed eclosion in both genotypes. In fact, not all pupae eclosed, which is a caveat of our analysis. Compared to MYD-matched controls, on the HFD Spen-KD animals delayed pupariation by 7.5 hr (chi-square = 10.70, P = 0.0011; Figure 2I and Table 2) and CTRL-KD animals were 3.2 hr delayed (chi-square = 8.648, P = 0.0033; Figure 2I and Table 2). The Spen-KD genotype was more adversely affected by HFD than CTRL-KD (d = −0.08). Both Spen-KD|HYD (16.7 hr earlier, chi-square = 52.44, P < 0.0001; Figure 2I and Table 2) and CTRL-KD|HYD (8.8 hr earlier, chi-square = 61.08, P < 0.0001) eclosed earlier than their MYD controls (Figure 2G and Table 2). The effect of the HYD was larger on the Spen-KD genotype compared to the CTRL-KD (d = 0.20), suggesting Spen-KD is more responsive to HYD than CTRL-KD. Despite this, Spen-KD genotype was significantly delayed in all diets. These findings point to a defect in Spen-KD animals in maintaining normal developmental timing but demonstrate that certain enriched diets can partially ameliorate the effect of Spen depletion on a standard diet (i.e., MYD).
HSD partially rescues the early death of Spen-KD flies
Just as defects in metabolism can lead to delays in development, so can they lead to longevity defects (Lushchak et al. 2012; Kitada et al. 2019). Since Spen-KD flies displayed defects in development during the larval stage in conjunction with the larval fat phenotype on the control MYD, we asked whether there was a long-term effect on the life span of adult flies. Dietary effects on life span could be tested in various ways. While it would address an interesting question, measuring the longevity of adults reared on EXPs would be confounded by the developmental delay and/or reduction in viability observed for some diets. Flight allows adults to leave the environment where they developed and explore a broader range of dietary choices than are available to larvae. We thus chose to test the life span of animals reared on a standard diet and switched to an EXP as adults.
CTRL-KD|MYD mated females lived 84% longer than Spen-KD diet- and sex-matched flies (median 68 days, compared to 37 days; Figure 3A and Table 3). However, Spen KD did not curtail the life span of males fed a standard diet. Spen-KD|MYD males lived 3% longer than CTRL-KD diet- and sex-matched flies (median 50 days, compared to 48.5 days; Figure 3D and Table 3). Life span varied significantly between sexes, but the effect depended on genotype. On MYD, CTRL-KD mated females lived longer than CTRL-KD males (median 68 days, compared to 48.5 days; Table 3), whereas Spen-KD mated females had shorter life spans than Spen-KD males (median 37 days, compared to 50 days; Table 3). These data establish the effects of sex and Spen on life span in the context of a standard diet.
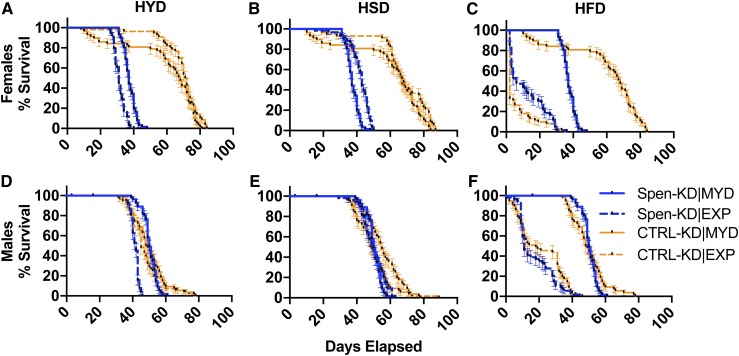
Figure 3.
Macronutrient composition differentially affects life span. Adults were allowed to eclose and mate for 48 hr before being separated by sex and transferred to either MYD (solid lines) or an EXP (dashed lines): HYD, HSD, and HFD. Vials were assessed daily for death events to generate survival curves. (A–C) Mated female percent survival (days) of Spen-KD|MYD (yellow lines) and CTRL-KD|MYD (blue lines) compared to (A) HYD, (B) HSD, and (C) HFD. (D–F) Male percent survival (days) of Spen-KD|MYD and CTRL-KD|MYD compared to (D) HYD, (E) HSD, and (F) HFD. Error bars represent SEM. n = 3 replicates, 20 adults per replicate.
Table 3. Life span, in median days elapsed to death, of male and mated female Spen-KD and CTRL-KD genotypes in control and EXPs.
| Sex | Diet | CTRL-KD survival (median days) | CTRL-KD (MYD|EXP) chi-square | P-value | Spen-KD survival (median days) | Spen-KD (MYD|EXP) chi-square | P-value | Cliff’s delta (d) |
|---|---|---|---|---|---|---|---|---|
| Female | MYD | 68 | NA | NA | 37 | NA | NA | NA |
| HYD | 71.5 | 0.02097 | ns | 31 | 56.81 | <0.0001 | 0.62 | |
| HSD | 67 | 4.349 | 0.037 | 43 | 38.35 | <0.0001 | −0.12 | |
| HFD | 2 | 120.5 | <0.0001 | 6 | 136.7 | <0.0001 | −0.54 | |
| Male | MYD | 48.5 | NA | NA | 50 | NA | NA | NA |
| HYD | 45 | 0.9183 | ns | 42 | 90.88 | <0.0001 | 0.37 | |
| HSD | 55 | 6.227 | 0.013 | 50 | 1.188 | ns | 0.27 | |
| HFD | 18 | 105.0 | <0.0001 | 11 | 129.0 | <0.0001 | 0.12 |
Cliff’s delta (d) shows effect size of diet on genotype. n = 3 replicates, 20 flies per replicate. Chi-square from log-rank (Mantel–Cox) test.
We wished to know if dietary intervention in early adulthood could partially rescue the Spen-KD longevity defects we observed. Because HYD rescued the fat storage phenotype in the larval stage, and HYD and HSD were able to partially rescue various developmental delays, we predicted that altering the diet of adult Spen-KD flies might similarly improve life span. To test this prediction, Spen-KD and CTRL-KD larvae were reared in MYD, allowed to eclose and mate for 48 hr, separated by sex, transferred onto MYD, HYD, HFD, or HSD, and analyzed daily for survival. Chi-square values for EXPs were calculated using the MYD matched control as the predicted value. As detailed below, we observed significant life span differences between Spen-KD and CTRL-KD flies between diets, as well as sex- and genotype-specific responses to diets.
CTRL-KD mated females lived longer than CTRL-KD males on MYD, HYD, and HSD, but not HFD (Figure 3, A–C and Table 3). This trend was the opposite for Spen-KD; Spen-KD mated females died earlier than Spen-KD males on all diets tested (Figure 3 and Table 3). CTRL-KD males significantly outlived Spen-KD males on all diets except for MYD (Figure 3, D–F and Table 3).
High protein (HYD) shortened life span by 16% in Spen-KD mated females compared to MYD (chi-square = 56.8, P < 0.0001; Figure 3A and Table 3), whereas there was little effect on CTRL-KD (chi-square = 0.021, P = NS; Figure 3A and Table 3). Comparing diet-matched conditions, the CTRL-KD|HYD mated females lived 131% longer than Spen-KD|HYD mated females (d = 0.62; Figure 3A and Table 3). Hence, a diet enriched in protein did not promote longevity in Spen-KD mated females.
Longevity in males also did not benefit from extra dietary protein, and it was detrimental to Spen-KD males. Spen-KD|HYD males died 7% earlier than CTRL-KD|HYD males (Figure 3D). Spen-KD|HYD male longevity was 16% shorter than Spen-KD|MYD controls (chi-square = 90.88, P < 0.0001; Figure 3D and Table 3). CTRL-KD|HYD male longevity was not significantly different from CTRL-KD|MYD (chi-square = 0.918, P = NS; Figure 3D and Table 3). Comparing diet-matched conditions, HYD was more deleterious to Spen-KD males than to CTRL-KD (d = 0.37).
Extra sugar extended life span in Spen-KD mated females but not CTRL-KD mated females. CTRL-KD|HSD mated female longevity decreased 1.5% compared to CTRL-KD|MYD (chi-square = 4.349, P = 0.037; Figure 3B and Table 3), and Spen-KD|HSD mated female longevity increased by 16% compared to Spen-KD|MYD controls (chi-square = 38.35, P < 0.0001; Figure 3B and Table 3). CTRL-KD|HSD mated females lived 56% longer than Spen-KD|HSD mated females (d = −0.12) (Figure 3B and Table 3). Thus Spen-KD mated females were more positively affected by HSD than CTRL-KD mated females, even if the extra sugar did not eliminate the effect of Spen depletion.
HSD had no effect on the longevity of Spen-KD males but was beneficial to CTRL-KD males, which lived 13% longer than CTRL-KD|MYD males (chi-square = 6.227, P = 0.013; Figure 3E and Table 3). CTRL-KD|HSD males lived 10% longer than Spen-KD|HSD males (d = 0.27; Figure 3E and Table 3). Thus, in males, depletion of Spen blocked an otherwise life-span-extending effect of extra dietary sugar.
Excess dietary fat was deleterious to longevity regardless of sex or genotype. Spen-KD|HFD mated female life span decreased 84% compared to Spen-KD|MYD controls (chi-square = 136.7, P < 0.0001; Figure 3C and Table 3), and CTRL-KD|HFD mated female life span decreased 97% compared to CTRL-KD|HFD (chi-square = 120.5, P < 0.0001; Figure 3C and Table 3). Interestingly, HFD was more deleterious to CTRL-KD mated females than to Spen-KD mated females: Spen-KD|HFD mated females lived 67% longer than CTRL-KD|HFD mated females (d = −0.54; Figure 3C and Table 3).
By contrast, HFD was more deleterious to Spen-KD males than to CTRL-KD males. Spen-KD|HFD male life span decreased 78% compared to Spen-KD|MYD controls (chi-square = 129, P < 0.0001; Figure 3F and Table 3), and CTRL-KD|HFD male life span decreased 63% compared to CTRL-KD|MYD (chi-square = 105, P < 0.0001; Figure 3C and Table 3). Comparing diet-matched conditions, the CTRL-KD|HFD males lived 64% longer than Spen-KD|HFD males (d = 0.12; Figure 3F and Table 3). These findings illustrate that the effects of diet on longevity vary greatly by diet, genotype, and sex. Among these effects, the apparent benefits of extra dietary sugar to Spen-KD females are consistent with our hypothesis that specific diets may ameliorate the adverse consequences of Spen depletion.
Dietary effects on metabolism measured by steady-state metabolomic profiling
We previously showed major alterations in the metabolomic profiles of Spen-KD larvae compared to CTRL-KD on the standard MYD (Hazegh et al. 2017). These results suggested an increase in protein catabolism and glycogen utilization and led us to investigate dietary intervention as a treatment for the Spen-KD obesity phenotype. A major finding was a decrease in steady-state levels of L-carnitine, which, in conjunction with other published data, supported our hypothesis that Spen-KD causes defects in β-oxidation and upregulation of compensatory pathways.
Impaired ability to mobilize and oxidize free fatty acids through β-oxidation is a substantial contributor to obesity [reviewed in Serra et al. (2013)]. Considering that L-carnitine is necessary for the β-oxidation of fatty acids within the mitochondrial matrix to feed the tricarboxylic acid cycle (TCA), and its production can be rate-limiting for fatty acid metabolism (Fritz and McEwen 1959; Bremer 1983), we measured its levels in response to diet through metabolomics (Figure 4A, Figure S2A; see File S2 for the metabolomics data set). In accordance with our previous results, we observed a decrease in steady-state levels of L-carnitine in Spen-KD|MYD compared to CTRL-KD|MYD (Figure 4A, Figure S2; Hazegh et al. 2017). Similarly, Spen-KD|HSD and Spen-KD|HFD had decreased levels of steady-state L-carnitine compared to matched-diet CTRL-KDs (Figure 4A, Figure S2A). In contrast, HYD restored Spen-KD L-carnitine levels to those of diet-matched CTRL-KD (Figure 4A, Figure S2A).
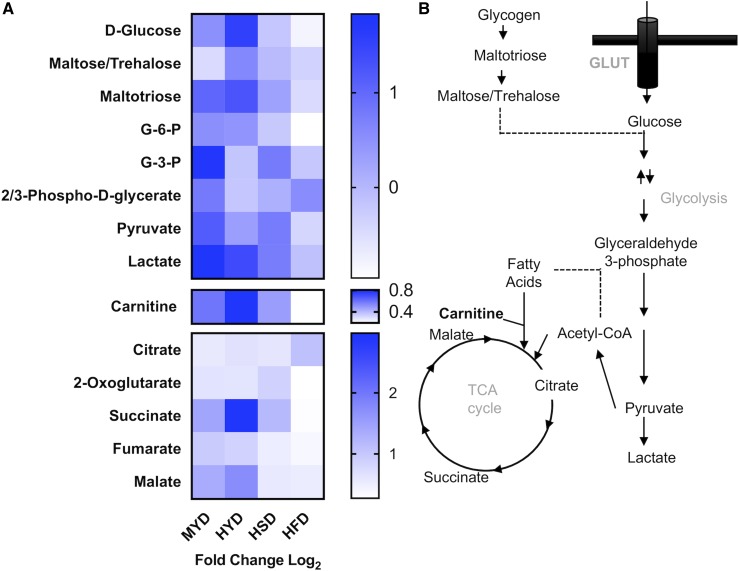
Figure 4.
HYD rescues L-carnitine levels in Spen-KD. (A) Heat map of glycolytic metabolites, L-carnitine, and the citric acid cycle (TCA cycle) metabolites, showing differences between Spen-KD and CTRL-KD on matched diet. MYD, HYD, HSD, and HFD. Heat map shows log2 fold change in metabolite abundance. Carnitine is an essential carrier for fatty acids into the mitochondrial matrix to facilitate β-oxidation of lipids as TCA cycle inputs. (B) Schematic summary of metabolite flux through the metabolic pathways analyzed in the heat map. n = 3 replicates, 10 individual larvae per replicate per genotype per diet (30 measurements per genotype|diet). G-3-P, glyceraldehyde 3-phosphate; G-6-P, glucose 6-phosphate.
Diet influenced the abundance of metabolites involved in glycolysis and the citric acid (TCA) cycle (Figure 4B). Spen-KD|MYD and HYD larvae accumulated more succinate than their CTRL-KD diet-matched counterparts (Figure 4A, Figure S2D). The HFD diminished or abolished metabolite differences between Spen-KD and CTRL-KD TCA cycle intermediates (Figure 4A, Figure S2, B–F). In glycolysis Spen-KD|MYD flies accumulated glyceraldehyde 3-phosphate (G-3-P), pyruvate, and lactate compared to CTRL-KD diet-matched controls (Figure 4A, Figure S3, E, G, and H). Spen-KD|HYD also had more pyruvate and lactate, suggesting alterations in glycolytic activity in these larvae (Figure 4A, Figure S3). In glycolysis, as with the TCA cycle, HFD abolished metabolite differences between Spen-KD and CTRL-KD (Figure 4A, Figures S2 and S3).
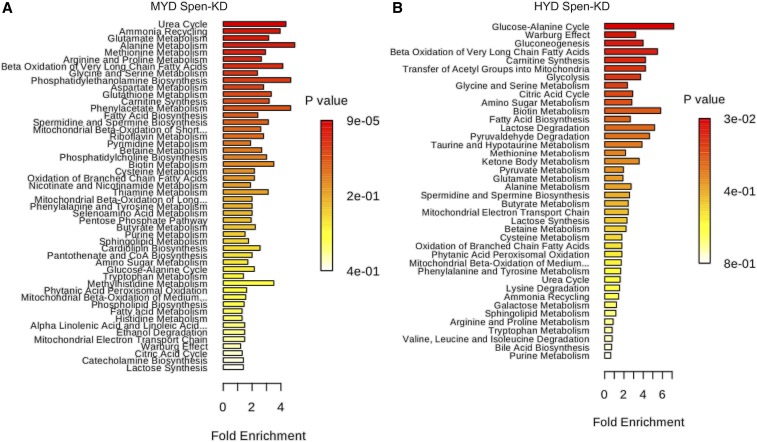
Additionally, metabolite set enrichment analysis (MetaboAnalyst 4.0), using the entire data set of 142 profiled metabolites, revealed changes of various magnitudes in specific metabolic pathways (Figure 5). We considered those with the largest magnitude changes and lowest one-tailed P value (hypergeometric test, adjusted for multiple testing) to be most biologically significant. We observed in Spen-KD|MYD larvae an enrichment compared to CTRL-KD|MYD for pathways in amino acid metabolism and additional atypical metabolic processes (Figure 5A and File S3). Comparing the steady-state metabolome of Spen-KD|HYD to CTRL-KD|HYD, the metabolite set enrichment observed in the MYD was abolished and amino acid metabolism was no longer statistically significantly enriched in the Spen-KD|HYD (Figure 5B). Thus, amelioration of these metabolic steady-state defects could be the basis of the partial rescues we observed for development of Spen-KD raised on HYD. We also observed a large reduction in the metabolite steady-state variation between Spen-KD and CTRL-KD on the HSD and HFD (data not shown), suggesting that the supplemented quantity of sugar and fat within these diets is epistatic to the genetic influence of Spen depletion on metabolism.
Figure 5.
Gene–diet interactions dictate metabolic steady state in third instar larvae. Metabolite set enrichment over representation analysis (ORA) generated by MetaboAnalyst 4.0. (A) Spen-KD|MYD metabolite ORA compared to CTRL-KD|MYD and (B) Spen-KD|HYD metabolite ORA compared to CTRL-KD|HYD. Data were collected from three biological replicates with 10 individual larvae per biological replicate per genotype per diet (30 measurements per genotype|diet).
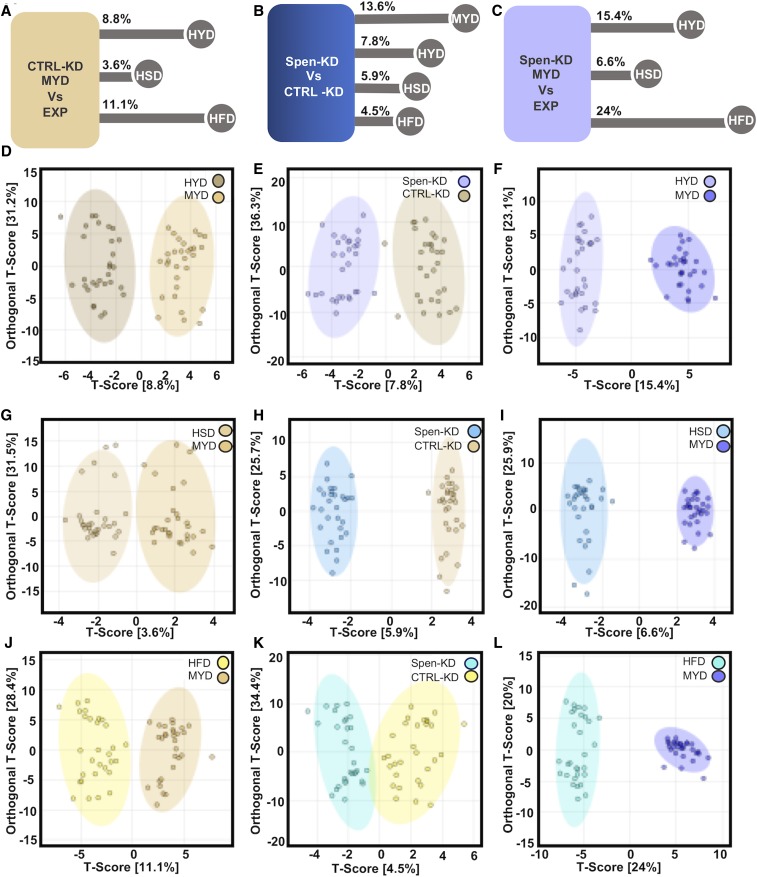
To examine the global metabolomic differences within the same genotype between different diets we applied OPLS-DA to our entire data set (Figure 6 and File S4). The metabolomic difference between Spen-KD on HYD and MYD was 15.4% (Figure 6, C and F). Spen-KD on HSD was the least different from MYD with only 6.6% difference (Figure 6, C and I). HFD drove the largest difference from MYD in Spen-KD at 24% difference (Figure 6, C and L). CTRL-KD followed the same trend (Figure 6A), with HSD being the least different (3.6%; Figure 6, A and D), followed by HYD (8.8%; Figure 6, A and G), and HFD was the most different (11.1%; Figure 6, A and J). While CTRL-KD follows the same trend, the differences induced by diet challenge are much smaller than those observed in Spen-KD (e.g., CTRL-KD|HFD vs. Spen-KD|HFD, 11.1% vs. 24%). Therefore, the different diets induced similar patterns of metabolite changes regardless of genotype, but CTRL-KD was more resistant to change.
Figure 6.
OLPS-DA comparisons of genotypes on diets and between genotypes. (A) The metabolite differences (as percentage of all metabolites assayed) between CTRL-KD|MYD and CTRL-KD|EXP. (D, G, and J) OLPS-DA of CTRL-KD|MYD compared to (D) HYD, (G) HSD, and (J) HFD. (B) Metabolite profile differences between Spen-KD and CTRL-KD on control MYD diet. (E, H, and K) OLPS-DA of CTRL-KD (yellow) and Spen-KD (blue) on (E) HYD, (H) HSD, and (K) HFD. (C) Metabolite differences between Spen-KD|MYD and Spen-KD|EXP. (F, I, and L) OLPS-DA of Spen-KD|MYD compared to Spen-KD| (F) HYD, (I) HSD, and (L) HFD. T-score: variation between groups. Orthogonal T-score: variation within the group. n = 3 replicates, 10 individual larvae per replicate per genotype per diet (30 measurements per genotype|diet).
We hypothesized that the developmental changes observed reflect a rescue of the steady-state metabolism of Spen-KD. To this end, we compared diet-matched Spen-KD and CTRL-KD by OPLS-DA (Figure 6B). The largest difference between genotypes was on MYD (13.6%). HYD decreased the variability between Spen-KD and CTRL-KD to 7.8% (Figure 6, B and F), while HSD shrank it to 5.9% (Figure 6, B and H). The least different was HFD at 4.5% (Figure 6, B and K).
Altogether, our data suggest that diet challenge is epistatic to genotype influences on whole organism metabolism. CTRL-KD had more metabolic plasticity and was better able to resist metabolite change than Spen-KD. HFD, which drove severe developmental delays, provided the largest metabolic challenge to both genotypes and pushed them toward the most similar metabolic profiles (Figure 6, B and K).
Discussion
Here we show that targeted gene–environment interactions can partially mitigate whole organismal pathophysiology. We show that supplementing the diet of Spen-KD individuals with macronutrients that fall within their metabolic capacities partially ameliorates the adiposity, development, and life span defects. We show that in Spen-KD animals a protein-supplemented diet (HYD) partially restores diet-appropriate fat storage, and partially restores the timing of larval development at the wandering, pupariation, and eclosion time points. While a sugar-supplemented diet (HSD) exacerbates the increased fat storage phenotype, it partially restores Spen-KD development during pupariation and eclosion and improves Spen-KD mated female life span. Macronutrient supplementation led to a decrease in metabolic steady-state differences between Spen-KD and CTRL-KD. The most pronounced differences were on MYD control diet, which is optimized for larval development and adult fecundity in wild-type strains. HYD abolishes many of the metabolic steady-state differences between Spen-KD and CTRL-KD and brings the metabolic profile back toward more canonical larval pathways. Larvae upregulate metabolic pathways, using the Warburg effect, to support their rapid development and accumulation of biomass (Tennessen et al. 2011). Spen-KD|HYD are enriched for metabolites involved in the Warburg effect, demonstrating a shift back to “canonical” metabolic programming (Figure 5B).
We previously showed that Spen depletion in the FB causes an obesity phenotype with defects in β-oxidation, increased protein catabolism, and increased glycolytic flux (Hazegh et al. 2017). These metabolic changes, most markedly the inability to properly liberate stored lipids, are accompanied by several developmental and life span defects. Here we demonstrate the successful application of targeted gene–environment interactions by using custom diets to alleviate Spen-derived developmental and life span defects.
Diet-induced obesity has been modeled in mammals by supplementing diet with refined carbohydrates and fats (Levin et al. 1989; Buettner et al. 2007; Bortolin et al. 2018). These studies have shown that diets high in fat and carbohydrates are capable of producing obesity in rat and mouse models with similar pathology to human obesity and produce associated comorbidities and metabolic syndromes. In flies it has also been observed that carbohydrate- and fat-enriched diets induce obesity despite attenuated feeding behaviors, and drive metabolic syndromes and comorbidities (Skorupa et al. 2008; Matzkin et al. 2011; Musselman et al. 2011; Birse et al. 2010). Our findings that the Spen-KD|HSD and HFD groups were fatter than Spen-KD|MYD (Figure 1, C and D and Table 1) demonstrate that Spen-KD larvae follow these same expectations. Extra fat drove the largest increase in fat storage in both Spen-KD and control animals (Figure 1D and Table 1), consistent with the earlier mammalian and D. melanogaster studies (Levin et al. 1989; Buettner et al. 2007; Padmanabha and Baker 2014).
Since Spen-KD larvae cannot properly regulate lipid metabolism, it is not surprising that feeding larvae excess dietary fat had the strongest adverse effect on development (Figure 2C and Table 2; Hazegh et al. 2017). Control animals were also delayed, but the effect size (d = −0.14) shows that excess fat more strongly delayed wandering in Spen-KD. It is likely that the HFD exacerbates the impaired lipid metabolic pathways within Spen-KD larvae, further exaggerating their developmental delays. Earlier studies established that an HFD is extremely deleterious to the life span of both sexes and all genotypes of flies assayed (Driver and Cosopodiotis 1979; Woodcock et al. 2015). Our data agree, although males survived better on HFD than did genotype-matched mated females. Previous studies showed major transcriptome differences between males and mated females reared on HFD: females upregulated fatty acid metabolism and their stress response while males downregulated stress response genes (Heinrichsen et al. 2014; Stobdan et al. 2019). Our data suggests that Spen-KD flies maintain this sex-specific sensitivity to dietary fat intake.
Extra dietary yeast, which represents an increase primarily in protein but also in other macronutrients, induced obesity in control animals (Figure 1B and Table 1). Conversely, on the HYD Spen-KD larvae were less fat than on the standard MYD, suggesting the HYD is protective against obesity in Spen-KD despite the nutrient enrichment (Figure 1B and Table 1). The difference between Spen-KD and CTRL-KD was smaller on the HYD compared to MYD (Table 1), which suggests that the HYD decreases the difference in fat storage between Spen-KD and CTRL-KD. Therefore, HYD partially ameliorates the increased fat storage phenotype observed in Spen-KD larvae and narrows the adiposity differences between Spen-KD and CTRL-KD larvae. HYD also partially rescued delayed pupariation and eclosion in Spen-KD animals (Figure 2, D and G and Table 2), but it became detrimental to the longevity of both male and mated female Spen-KD adults (Figure 3, A and D and Table 3). This is in accordance with previous data suggesting a high-protein diet has negative longitudinal effects on life span (Kitada et al. 2019). Conversely, CTRL-KD mated female and male life spans were not significantly affected by HYD (Figure 3, A and D and Table 3). This result suggests a sex-specific sex–diet–genotype interaction. We hypothesize that this sex difference in CTRL-KD life span is due to the unique metabolic needs of egg production and egg laying sustained by mated females as previous studies have shown that dietary amino acids are crucial to these processes (Lee et al. 2008; Haussmann et al. 2013). We hypothesize that genetic background, food media preparation, and assay conditions contribute to the heterogeneity observed in diet life span studies, further emphasizing the importance of controlling gene–diet–environment interactions in experimental designs.
HYD improved early larval developmental timing in Spen-KD animals to nearly that of controls. Essential amino acids derived from the diet (histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, valine, and arginine) must reach a threshold amount for larvae to commit to pupariation (Sang and King 1961). We hypothesize that the wandering development improvement measured in Spen-KD|HYD is a consequence of enriched essential amino acids. Supplemented protein is broadly beneficial to developing larvae, regardless of genotype. HYD had a larger effect on CTRL-KD than Spen-KD, as denoted by the Cliff’s delta (d = −0.26). This result was surprising, as we initially hypothesized Spen-KD would experience greater benefits from a high-protein diet than would CTRL-KD, owing to the upregulation of protein metabolism we previously observed (Hazegh et al. 2017). While protein catabolism can compensate in part for the decrease in available energy, it cannot fully rescue the metabolic defects observed in Spen-KD. Therefore, the beneficial effect of HYD on Spen-KD was smaller than CTRL-KD because it only addresses one part of organism-wide metabolic dysregulation.
The only diets to significantly improve the pupariation and eclosion rates of Spen-KD larvae were HYD and HSD. While macronutrients are used as fuel for development, they also play an equally vital role as building blocks for biomass to support the 400-fold increase in body size the larvae undergo during their rapid development. We postulate that the increase in protein is insufficient to counteract the “starvation” effect caused by immobilized lipid stores, which are critical mediators of cell growth and division by providing phospholipid, glycolipid, and glycosphingolipid precursors. Glycosphingolipids are necessary for proper cell cytokinesis in mammals, and studies in D. melanogaster have shown proper neuronal expansion and remodeling requires the lipid metabolism pathway SREBP (Ziegler et al. 2017; Huang et al. 2018). We postulate that the persisting developmental delays are consequences of dysregulated lipid metabolism, and while HYD and HSD can improve development, it cannot compensate for this critical failure.
Spen-KD|HSD were still significantly fatter than CTRL-KD|HSD; however, sugar enrichment also narrowed the fat storage differences between Spen-KD and CTRL-KD (Figure 1C and Table 1). This was the smallest difference between diet-matched Spen-KD and CTRL-KD, suggesting that HSD-induced obesity originates from a different metabolic pathway. That is to say, an HSD was epistatic to genotype in this investigation of gene–environment interaction.
HSD negatively affected the developmental timing of both genotypes, but it affected CTRL-KD more than Spen-KD. HSD drove the largest wandering delay in CTRL-KD (4.4 hr; Table 2), suggesting that excessive carbohydrate is the most detrimental macronutrient in early larval development in normal circumstances. Excess sugar is stored as a limited amount of glycogen and then shunted into lipogenesis to be stored as lipids (Acheson et al. 1982; Flatt 1995). We propose that the HSD exacerbates the obesity phenotype and lipid dysregulation through its contribution to increased de novo lipogenesis, but to a lesser degree than HFD because sugar can be shunted to fates other than triglycerides (i.e., glycogen).
Spen-KD displayed increased fat storage compared to CTRL-KD in all diet-matched assays (Figure 1). This result is consistent with our previously purposed model that Spen-KD generates a genetically underpinned obesity phenotype through dysregulated metabolism in FB cells (Hazegh et al. 2017). Most importantly, HYD partially rescued the increased fat storage phenotype in Spen-KD larvae. While Spen-KD|HYD still stored more fat than CTRL-MYD, HYD attenuated the development of genetic obesity and decreased the pathophysiological burden of excessive fat storage in these animals. The specificity of these gene–diet interactions provides support for the application of custom dietary intervention to treat obesity.
Diet composition is a primary variable in the metabolome at steady state
By challenging Spen-KD and CTRL-KD larvae with HSD, HFD, and HYD we were largely able to abolish the influence of genetics on the metabolic steady state of the animals and achieved strikingly similar metabolomic profiles between the two genotypes. The most different metabolite profiles occurred on MYD, with 13.6% of metabolites being expressed differently between Spen-KD and CTRL-KD (Figure 6B).
Dietary extremes appear epistatic to more nuanced genetic regulators of metabolism, such as Spen-KD. Larvae reared on the HFD were least different between Spen-KD and CTRL-KD genotypes, differing in only two of 140 metabolites assessed (acetylcholine and L-Carnitine) with a global 4.5% difference between genotypes. This is an interesting observation when paired with the observation that Spen-KD|HFD were significantly fatter than CTRL-KD|HFD controls. We postulate that Spen-KD is less resilient to diet challenge and develops more severe pathophysiological obesity phenotypes due to their defective lipid mobilization.
The role of Spen in metabolism changes as metabolic needs change through maturation. HYD partially rescued pupation and eclosion timing of Spen-KD larvae but had a significant negative effect on female longevity, which is in agreement with existing literature on the influence of protein in the D. melanogaster life cycle (Lee et al. 2008; Kitada et al. 2019). Spen-KD was more sensitive to the beneficial developmental effects of HYD than CTRL-KD in whole organismal energy homeostasis (Figure 1F and Table 1), pupariation, and eclosion (Table 2).
HSD accelerated pupariation and eclosion during development, as well as partially rescued Spen-KD female longevity. Unlike the high-protein diet, excess carbohydrate is beneficial and confers starvation resistance in mated females and, to a lesser extent, males (Chandegra et al. 2017). As described in our previous study, Spen-KD alters carbohydrate metabolism and changes glycolytic flux (Hazegh et al. 2017). We see in the adult stage that HSD was a more favorable diet composition for longevity of Spen-KD mated females than for CTRL-KD. This is not conserved in males, which is not entirely surprising given the generally lesser benefit of increased sugar conferred to adult males in general (Chandegra et al. 2017). HSD altered the metabolite profiles of Spen-KD and CTRL-KD the least; however, Spen-KD|HSD changed twice as much CTRL-KD|HSD compared to MYD matched controls (Figure 6, A and C).
While we are interested in genetically driven forms of obesity, we seek to understand the functions of these genes in a whole-organism metabolic context to leverage this molecular understanding toward designing diet interventions that ameliorate pathologies. This study demonstrates the feasibility of gene–macronutrient interventions as a treatment for obesity and related comorbidities resulting from a specific genetic perturbation.
Acknowledgments
We would like to thank the Bloomington Drosophila Stock Center for providing fly stocks, and Michael McMurray for his helpful comments on the manuscript. This work was supported by National Institutes of Health (NIH) grant T32-GM008730 and the Victor W. and Earleen D. Bolie Graduate Scholarship to K.E.H.; and NIH grant T32-GM008730 and RNA Scholar of the RNA Bioscience Initiative, University of Colorado School of Medicine to C.M.G. A.D.’A. was supported by funds from the Boettcher Webb-Waring Investigator Award, NIH/NIGMS grant RM1-GM131968 and NIH/NHLBI grant R01-HL146442. J.M.T. is supported by the RNA Bioscience Initiative at the University of Colorado Anschutz Medical Campus and a Webb-Waring Early Career Investigator Award from the Boettcher Foundation (AWD-182937). T.R. is supported by NIH/NIDDK grant R01-DK106177 and a pilot grant from the RNA Bioscience Initiative, University of Colorado School of Medicine.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11446206.
Communicating editor: K. O’Connor-Giles
Literature Cited
- Acheson K. J., Flatt J. P., and Jéquier E., 1982. Glycogen synthesis versus lipogenesis after a 500 gram carbohydrate meal in man. Metabolism 31: 1234–1240. 10.1016/0026-0495(82)90010-5 [DOI] [PubMed] [Google Scholar]
- Baker K. D., and Thummel C. S., 2007. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 6: 257–266. 10.1016/j.cmet.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M., Bulankina A. V., Hsiao H.-H., Urlaub H., Jäckle H. et al. , 2010. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12: 521–532. 10.1016/j.cmet.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Bernards A., and Hariharan I. K., 2001. Of flies and men–studying human disease in Drosophila. Curr. Opin. Genet. Dev. 11: 274–278. 10.1016/S0959-437X(00)00190-8 [DOI] [PubMed] [Google Scholar]
- Birse R. T., Choi J., Reardon K., Rodriguez J., Graham S. et al. , 2010. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 12: 533–544. 10.1016/j.cmet.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin R. C., Vargas A. R., Gasparotto J., Chaves P. R., Schnorr C. E. et al. , 2018. A new animal diet based on human Western diet is a robust diet-induced obesity model: comparison to high-fat and cafeteria diets in term of metabolic and gut microbiota disruption. Int. J. Obes. 42: 525–534. 10.1038/ijo.2017.225 [DOI] [PubMed] [Google Scholar]
- Bremer J., 1983. Carnitine–metabolism and functions. Physiol. Rev. 63: 1420–1480. 10.1152/physrev.1983.63.4.1420 [DOI] [PubMed] [Google Scholar]
- Buettner R., Schölmerich J., and Bollheimer L. C., 2007. High-fat diets: modeling the metabolic disorders of human obesity in rodents. Obesity (Silver Spring) 15: 798–808. 10.1038/oby.2007.608 [DOI] [PubMed] [Google Scholar]
- Chandegra B., Tang J. L. Y., Chi H., and Alic N., 2017. Sexually dimorphic effects of dietary sugar on lifespan, feeding and starvation resistance in Drosophila. Aging (Albany NY) 9: 2521–2528. 10.18632/aging.101335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Estampador A. C., Keller M., Poveda A., Dalla-Riva J. et al. , 2019. The combined effects of FADS gene variation and dietary fats in obesity-related traits in a population from the far north of Sweden: the GLACIER Study. Int. J. Obes. 43: 808–820. 10.1038/s41366-018-0112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Soufan O., Li C., Caraus I., Li S. et al. , 2018. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46: W486–W494. 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasquin M. F., Melamud E., and Rabinowitz J. D., 2012. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics Chapter 14: Unit14.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver C. J. I., and Cosopodiotis G., 1979. The effect of dietary fat on longevity of Drosophila melanogaster. Exp. Gerontol. 14: 95–100. 10.1016/0531-5565(79)90023-8 [DOI] [PubMed] [Google Scholar]
- Flatt J. P., 1995. Use and storage of carbohydrate and fat. Am. J. Clin. Nutr. 61: 952S–959S. 10.1093/ajcn/61.4.952S [DOI] [PubMed] [Google Scholar]
- Fritz I., and McEwen B., 1959. Effects of carnitine on fatty-acid oxidation by muscle. Science 129: 334–335. 10.1126/science.129.3345.334 [DOI] [PubMed] [Google Scholar]
- Fryar C. D., Carroll M. D., and Ogden C. L., 2016. Prevalence of Overweight, Obesity, and Extreme Obesity Among Adults Aged 20 and Over: United States, 1960–1962 Through 2013–2014. U.S. Center for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P. et al. , 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349: 1343–1347. 10.1126/science.aab2319 [DOI] [PubMed] [Google Scholar]
- Giovannucci E., 2007. Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am. J. Clin. Nutr. 86: s836–s842. 10.1093/ajcn/86.3.836S [DOI] [PubMed] [Google Scholar]
- Grönke S., Beller M., Fellert S., Ramakrishnan H., Jäckle H. et al. , 2003. Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 13: 603–606. 10.1016/S0960-9822(03)00175-1 [DOI] [PubMed] [Google Scholar]
- Grönke S., Mildner A., Fellert S., Tennagels N., Petry S. et al. , 2005. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1: 323–330. 10.1016/j.cmet.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Grotto D., and Zied E., 2010. The standard American diet and its relationship to the health status of Americans. Nutr. Clin. Pract. 25: 603–612. 10.1177/0884533610386234 [DOI] [PubMed] [Google Scholar]
- Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G. et al. , 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453: 657–661. 10.1038/nature06928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann I. U., Hemani Y., Wijesekera T., Dauwalder B., and Soller M., 2013. Multiple pathways mediate the sex-peptide-regulated switch in female Drosophila reproductive behaviours. Proc. Biol. Sci. 280: 20131938 10.1098/rspb.2013.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazegh K. E., and Reis T., 2016. A buoyancy-based method of determining fat levels in Drosophila. J. Vis. Exp. (117): 54744. 10.3791/54744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazegh K. E., Nemkov T., D’Alessandro A., Diller J. D., Monks J. et al. , 2017. An autonomous metabolic role for Spen. PLoS Genet. 13: e1006859 10.1371/journal.pgen.1006859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichsen E. T., Zhang H., Robinson J. E., Ngo J., Diop S. et al. , 2014. Metabolic and transcriptional response to a high-fat diet in Drosophila melanogaster. Mol. Metab. 3: 42–54. 10.1016/j.molmet.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Huang S., Di Scala C., Wang Q., Wandall H. H. et al. , 2018. The glycosphingolipid MacCer promotes synaptic bouton formation in Drosophila by interacting with Wnt. eLife 7: e38183. 10.7554/eLife.38183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada M., Ogura Y., Monno I., and Koya D., 2019. The impact of dietary protein intake on longevity and metabolic health. EBioMedicine 43: 632–640. 10.1016/j.ebiom.2019.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., Simpson S. J., Clissold F. J., Brooks R., Ballard J. W. O. et al. , 2008. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc. Natl. Acad. Sci. USA 105: 2498–2503. 10.1073/pnas.0710787105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold P., and Perrimon N., 2007. Drosophila and the genetics of the internal milieu. Nature 450: 186–188. 10.1038/nature06286 [DOI] [PubMed] [Google Scholar]
- Levin B. E., Hogan S., and Sullivan A. C., 1989. Initiation and perpetuation of obesity and obesity resistance in rats. Am. J. Physiol. 256: R766–R771. 10.1152/ajpregu.1989.256.3.R766 [DOI] [PubMed] [Google Scholar]
- Li H., Rai M., Buddika K., Sterrett M. C., Luhur A. et al. , 2019. Lactate dehydrogenase and glycerol-3-phosphate dehydrogenase cooperatively regulate growth and carbohydrate metabolism during Drosophila melanogaster larval development. Development 175315 10.1242/dev.175315 [DOI]
- Lin H. V., Doroquez D. B., Cho S., Chen F., Rebay I. et al. , 2003. Splits ends is a tissue/promoter specific regulator of Wingless signaling. Development 130:3125–31335. 10.1242/dev.00527 [DOI] [PubMed] [Google Scholar]
- Lushchak O. V., Gospodaryov D. V., Rovenko B. M., Glovyak A. D., Yurkevych I. S. et al. , 2012. Balance between macronutrients affects life span and functional senescence in fruit fly Drosophila melanogaster. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 67: 118–125. 10.1093/gerona/glr184 [DOI] [PubMed] [Google Scholar]
- Matoo O. B., Julick C. R., and Montooth K. L., 2019. Genetic variation for ontogenetic shifts in metabolism underlies physiological homeostasis in Drosophila. Genetics 212: 537–552. 10.1534/genetics.119.302052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin L. M., Johnson S., Paight C., Bozinovic G., and Markow T. A., 2011. Dietary protein and sugar differentially affect development and metabolic pools in ecologically diverse Drosophila. J. Nutr. 141: 1127–1133. 10.3945/jn.111.138438 [DOI] [PubMed] [Google Scholar]
- Musselman L. P., and Kuhnlein R. P., 2018. Drosophila as a model to study obesity and metabolic disease. J. Exp. Biol. 221: jeb163881. 10.1242/jeb.163881 [DOI] [PubMed] [Google Scholar]
- Musselman L. P., Fink J. L., Narzinski K., Ramachandran P. V., Hathiramani S. S. et al. , 2011. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis. Model. Mech. 4: 842–849. 10.1242/dmm.007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Narimatsu H., Sato H., Sho R., Otani K. et al. , 2016. Gene-environment interactions in obesity: implication for future applications in preventive medicine. J. Hum. Genet. 61: 317–322. 10.1038/jhg.2015.148 [DOI] [PubMed] [Google Scholar]
- Neel J. V., 1962. Diabetes mellitus: a “Thrifty” genotype rendered detrimental by “Progress”? Am. J. Hum. Genet. 14: 353–362. [PMC free article] [PubMed] [Google Scholar]
- Nemkov T., Hansen K. C., and D’Alessandro A., 2017. A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun. Mass Spectrom. 31: 663–673. 10.1002/rcm.7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemkov T., Reisz J. A., Gehrke S., Hansen K. C., and D’Alessandro A., 2019. High-throughput metabolomics: isocratic and gradient mass spectrometry-based methods. Methods Mol. biol. 1978: 13–26. 10.1007/978-1-4939-9236-2_2 [DOI] [PubMed] [Google Scholar]
- Padmanabha D., and Baker K. D., 2014. Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol. Metab. 25: 518–527. 10.1016/j.tem.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Pospisilik J. A., Schramek D., Schnidar H., Cronin S. J. F., Nehme N. T. et al. , 2010. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of Brown reveals White adipose cell fate. Cell 140: 148–160. 10.1016/j.cell.2009.12.027 [DOI] [PubMed] [Google Scholar]
- Qasim A., Turcotte M., de Souza R. J., Samaan M. C., Champredon D. et al. , 2018. On the origin of obesity: identifying the biological, environmental and cultural drivers of genetic risk among human populations. Obes. Rev. 19: 121–149. 10.1111/obr.12625 [DOI] [PubMed] [Google Scholar]
- Rebay I., Chen F., Hsiao F., Kolodziej P. A., Kuang B. H. et al. , 2000. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154: 695–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. K., Williams S., Springston M., Brown J., Freeman K. et al. , 2010. Genotype-by-diet interactions drive metabolic phenotype variation in Drosophila melanogaster. Genetics 185: 1009–1019. 10.1534/genetics.109.113571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis T., Van Gilst M. R., and Hariharan I. K., 2010. A Buoyancy-Based Screen of Drosophila Larvae for Fat-Storage Mutants Reveals a Role for Sir2 in Coupling Fat Storage to Nutrient Availability. PLoS Genet. 6: e1001206 10.1371/journal.pgen.1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang J. H., and King R. C., 1961. Nutritional requirements of axenically cultured Drosophila melanogaster adults. J. Exp. Biol. 38: 793–809. [Google Scholar]
- Schlegel A., and Stainier D. Y. R., 2007. Lessons from “‘Lower’” organisms: what worms, flies, and zebrafish can teach us about human energy metabolism. PLoS Genet. 3: e199 10.1371/journal.pgen.0030199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra D., Mera P., Malandrino M. I., Mir J. F., and Herrero L., 2013. Mitochondrial fatty acid oxidation in obesity. Antioxid. Redox Signal. 19: 269–284. 10.1089/ars.2012.4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silventoinen K., Rokholm B., Kaprio J., and Sørensen T. I. A., 2010. The genetic and environmental influences on childhood obesity: a systematic review of twin and adoption studies. Int. J. Obes. 34: 29–40. 10.1038/ijo.2009.177 [DOI] [PubMed] [Google Scholar]
- Skorupa D. A., Dervisefendic A., Zwiener J., and Pletcher S. D., 2008. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7: 478–490. 10.1111/j.1474-9726.2008.00400.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobdan T., Sahoo D., Azad P., Hartley I., Heinrichsen E. et al. , 2019. High fat diet induces sex-specific differential gene expression in Drosophila melanogaster. PLoS One 14: e0213474 10.1371/journal.pone.0213474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Baker K. D., Lam G., Evans J., and Thummel C. S., 2011. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 13: 139–148. 10.1016/j.cmet.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargha A., and Delaney H. D., 2000. A critique and improvement of the CL common language effect size statistics of McGraw and Wong. J. Educ. Behav. Stat. 25: 101–132 [Google Scholar]
- Woodcock K. J., Kierdorf K., Pouchelon C. A., Vivancos V., Dionne M. S. et al. , 2015. Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity 42: 133–144. 10.1016/j.immuni.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A. B., Thiele C., Tenedini F., Richard M., Leyendecker P. et al. , 2017. Cell-autonomous control of neuronal dendrite expansion via the fatty acid synthesis regulator SREBP. Cell Rep. 21: 3346–3353. 10.1016/j.celrep.2017.11.069 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Flies and analysis scripts are available upon request. The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11446206.