Abstract
Mapping the chromosomal rearrangements between species can inform our understanding of genome evolution, reproductive isolation, and speciation. Here, we present a novel algorithm for identifying regions of synteny in pairs of genetic maps, which is implemented in the accompanying R package syntR. The syntR algorithm performs as well as previous ad hoc methods while being systematic, repeatable, and applicable to mapping chromosomal rearrangements in any group of species. In addition, we present a systematic survey of chromosomal rearrangements in the annual sunflowers, which is a group known for extreme karyotypic diversity. We build high-density genetic maps for two subspecies of the prairie sunflower, Helianthus petiolaris ssp. petiolaris and H. petiolaris ssp. fallax. Using syntR, we identify blocks of synteny between these two subspecies and previously published high-density genetic maps. We reconstruct ancestral karyotypes for annual sunflowers using those synteny blocks and conservatively estimate that there have been 7.9 chromosomal rearrangements per million years, a high rate of chromosomal evolution. Although the rate of inversion is even higher than the rate of translocation in this group, we further find that every extant karyotype is distinguished by between one and three translocations involving only 8 of the 17 chromosomes. This nonrandom exchange suggests that specific chromosomes are prone to translocation and may thus contribute disproportionately to widespread hybrid sterility in sunflowers. These data deepen our understanding of chromosome evolution and confirm that Helianthus has an exceptional rate of chromosomal rearrangement that may facilitate similarly rapid diversification.
Keywords: chromosomal rearrangement, synteny block, Helianthus, syntR, dot plot
ORGANISMS vary widely in the number and arrangement of their chromosomes, i.e., their karyotype. Interestingly, karyotypic differences are often associated with species boundaries and, therefore, suggest a link between chromosomal evolution and speciation (White 1978; King 1993). Indeed, it is well established that chromosomal rearrangements can contribute to reproductive isolation. Individuals heterozygous for divergent karyotypes are often sterile or inviable (King 1987; Lai et al. 2005; Stathos and Fishman 2014). Apart from directly causing hybrid sterility and inviability, chromosomal rearrangements can also facilitate the evolution of other reproductive barriers by extending genomic regions that are protected from introgression (Noor et al. 2001; Rieseberg 2001), accumulating genetic incompatibilities (Navarro and Barton 2003), and simplifying reinforcement (Trickett and Butlin 1994). Despite its prevalence and potentially important role in speciation, the general patterns of karyotypic divergence are still not well understood. Mapping and characterizing chromosomal rearrangements in many taxa is a critical step toward understanding their evolutionary dynamics.
The genus Helianthus (sunflowers) is well known to have particularly labile genome structure, and is thus a viable system in which to map and characterize a variety of rearrangements. These sunflowers have several paleopolyploidy events in their evolutionary history (Barker et al. 2008, 2016; Badouin et al. 2017), have given rise to three homoploid hybrid species (Rieseberg 1991), and are prone to transposable element activity (Kawakami et al. 2011; Staton et al. 2012). Evidence in the form of hybrid pollen inviability, abnormal chromosome pairings during meiosis, and genetic map comparisons suggests that Helianthus karyotypes are unusually diverse (Heiser 1947, 1951, 1961; Whelan 1979; Chandler et al. 1986; Quillet et al. 1995; Rieseberg et al. 1995; Burke et al. 2004; Heesacker et al. 2009; Barb et al. 2014). In fact, annual sunflowers have one of the highest described rates of chromosomal evolution across all plants and animals (Burke et al. 2004).
Studying chromosomal evolution within any group requires high-density genetic maps. Recently, Barb et al. (2014) built high-density genetic maps for the sunflower species Helianthus niveus ssp. tephrodes and H. argophyllus, and compared them to H. annuus. This analysis precisely mapped previously inferred karyotypes (Heiser 1951; Chandler et al. 1986; Quillet et al. 1995), but only captured a small amount of the chromosomal variation in the annual sunflowers. For example, comparisons of genetic maps with limited marker density suggest that several chromosomal rearrangements differentiate H. petiolaris from H. annuus (Rieseberg et al. 1995; Burke et al. 2004), and evidence from cytological surveys suggests that subspecies within H. petiolaris subspecies carry divergent karyotypes (Heiser 1961). Adding high-density genetic maps of H. petiolaris subspecies to the Barb et al. (2014) analysis will allow us to: (1) precisely track additional rearrangements, (2) reconstruct ancestral karyotypes for the group, and (3) untangle overlapping rearrangements that can be obscured by directly comparing present-day karyotypes.
Another critical part of a multispecies comparative study of chromosome evolution using genetic map data is a systematic and repeatable method for identifying syntenic chromosomal regions (sensu Pevzner and Tesler 2003). These methods are especially important for cases with high marker density because breakpoints between synteny blocks can be blurred by mapping errors, microrearrangements, and paralogy (Hackett and Broadfoot 2003; Choi et al. 2007; Barb et al. 2014; Bilton et al. 2018). In previous studies, synteny blocks have been found by a variety of ad hoc methods, including counting all differences in marker order (Wu and Tanksley 2010), by visual inspection (Burke et al. 2004; Marone et al. 2012; Latta et al. 2019), or by manually applying simple rules like size thresholds (Heesacker et al. 2009; Barb et al. 2014; Rueppell et al. 2016) and Spearman’s rank comparisons (Berdan et al. 2014; Schlautman et al. 2017). However, these methods become intractable and prone to error when applied to very-dense genetic maps. Furthermore, to our knowledge, there is no software available that identifies synteny blocks based on relative marker positions alone (i.e., without requiring reference genomes, sequence data, or markers with known orientations).
Here, with the goal of understanding chromosome evolution in Helianthus and more generally, we aimed to: (1) build high-density genetic maps for two subspecies of H. petiolaris, (2) develop a method and software to systematically and repeatably identify synteny blocks from any number of paired genetic map positions, (3) reconstruct ancestral karyotypes for a subsection of annual sunflowers, and (4) detect general patterns of chromosomal rearrangement in Helianthus.
Materials and Methods
Study system
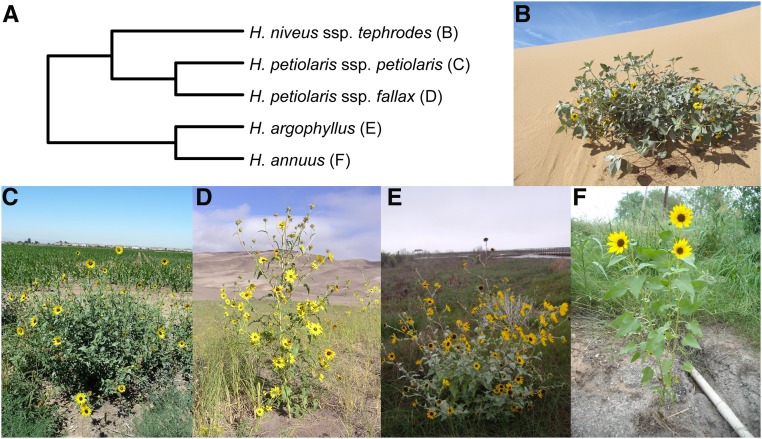
We focused on five closely related diploid (2n = 34) taxa from the annual clade of the genus Helianthus (Figure 1). These sunflowers are native to North America (Supplemental Material, Figure S1; Rogers et al. 1982) and are naturally self-incompatible (domesticated lineages of H. annuus are self-compatible). H. annuus occurs throughout much of the central US states, often in somewhat heavy soils and along roadsides (Heiser 1947). H. petiolaris occurs in sandier soils and is made up of two subspecies: H. petiolaris ssp. petiolaris, which is commonly found in the southern Great Plains, and H. petiolaris ssp. fallax, which is limited to more arid regions in Colorado, Utah, New Mexico, and Arizona (Heiser 1961). Where H. petiolaris and H. annuus are sympatric, gene flow occurs between the species (Strasburg and Rieseberg 2008). H. argophyllus is primarily found along the east coast of Texas, where it also overlaps and hybridizes with H. annuus (Baute et al. 2016). Finally, H. niveus ssp. tephrodes is a facultative perennial that grows in dunes from the southwestern US states into Mexico.
Figure 1.
The sunflower taxa used in this study. (A) Phylogenetic relationships based on Stephens et al. (2015) and Baute et al. (2016). (B) H. niveus ssp. tephrodes. (C) H. petiolaris ssp. petiolaris. (D) H. petiolaris ssp. fallax. (E) H. argophyllus. (F) H. annuus. Photo credits: Brook Moyers (B, C, E, and F) and Rose Andrew (D).
Controlled crosses
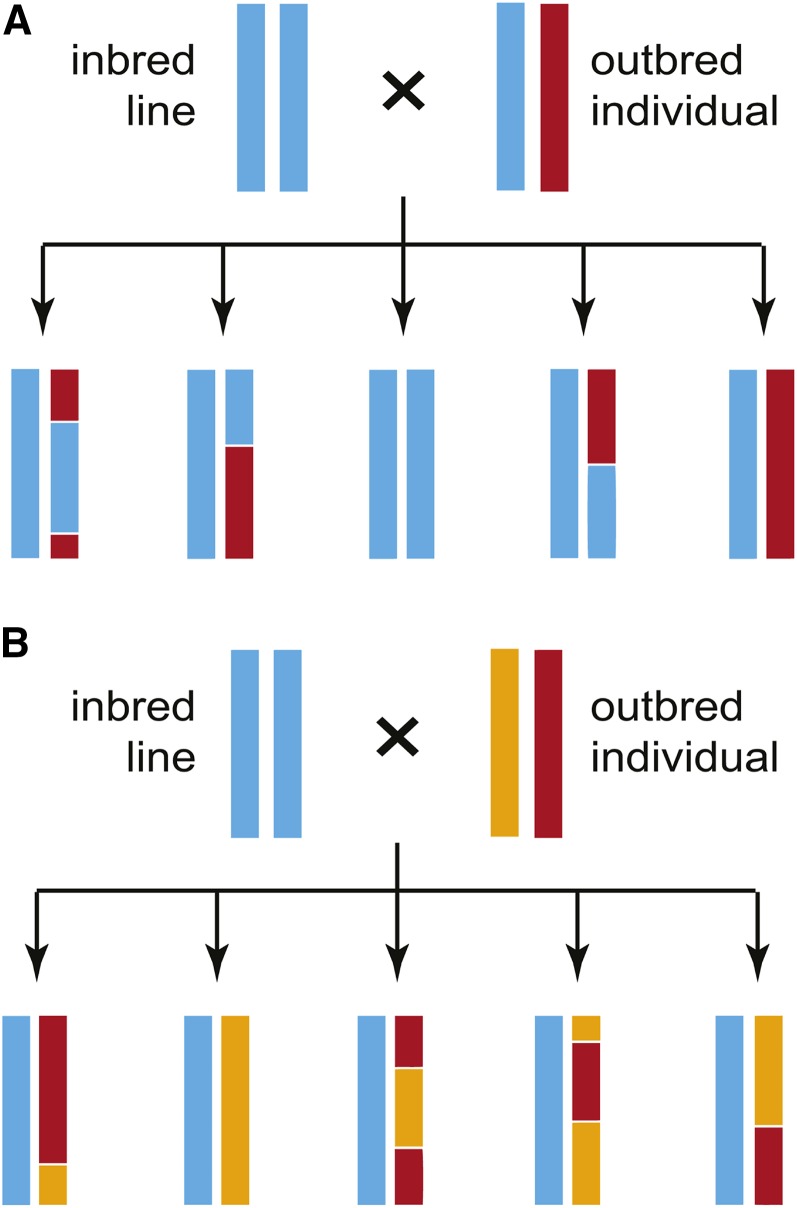
To make genetic maps, we crossed an outbred individual with presumably high heterozygosity from each H. petiolaris subspecies to a homozygous inbred line of domesticated sunflower and genotyped the resulting F1 offspring. This test-cross design allows us to infer where recombination occurred in the heterozygous parents because we can reliably track the segregation of those parents’ alleles against a predictable background (Figure 2).
Figure 2.
Diagram showing how a test cross can be used to map the recombination events in an outbred individual that may (A) or may not (B) share alleles with the inbred line. Each line represents a chromosome, and the colors represent ancestry.
Specifically, we used pollen from a single H. petiolaris ssp. petiolaris plant (PI435836) and a single H. petiolaris ssp. fallax plant (PI435768) to fertilize individuals of a highly inbred and male sterile line of H. annuus (HA89cms). The self-incompatible H. petiolaris accessions were collected in central Colorado (PI435836, 39.741°, −105.342°, Boulder County) and the southeast corner of New Mexico (PI435768, 32.3°, −104.0°, Eddy County; Figure S1), and were maintained at large population sizes by the US Department of Agriculture. When it was originally collected, accession PI435768 was classified as H. neglectus. However, based on the location of the collection (Heiser 1961) and a more recent genetic analysis of the scale of differences between H. petiolaris ssp. fallax and H. neglectus (Raduski et al. 2010), we believe that this accession should be classified H. petiolaris ssp. fallax.
Genotyping
We collected leaf tissue from 116 H. annuus × H. petiolaris ssp. petiolaris F1 seedlings and 132 H. annuus × H. petiolaris ssp. fallax F1 seedlings. We extracted DNA using a modified CTAB protocol (Doyle and Doyle 1987) and prepared individually barcoded genotyping-by-sequencing (GBS) libraries using a version of the Poland et al. (2012) protocol. Our modified protocol includes steps to reduce the frequency of high-copy fragments (e.g., chloroplast and repetitive sequences) based on Shagina et al. (2010) and Matvienko et al. (2013), and steps to select specific fragment sizes for sequencing [see appendix B in Ostevik (2016) for the full protocol].
Briefly, we digested 100 ng of DNA from each individual with restriction enzymes (either PstI-HF or PstI-HF, and MspI), and ligated individual barcodes and common adapters to the digested DNA. We pooled barcoded fragments from up to 192 individuals, cleaned and concentrated the libraries using SeraMag Speed Beads made in-house (Rohland and Reich 2012), and amplified fragments using 12 cycles of PCR. We depleted high-copy fragments based on Todesco et al. (2019) using the following steps: (1) denature the libraries using high temperatures, (2) allow the fragments to rehybridize, (3) digest the double-stranded fragments with duplex-specific nuclease (Zhulidov et al. 2004), and (4) amplify the undigested fragments using another 12 cycles of PCR. We ran the libraries out on a 1.5% agarose gel and extracted 300–800-bp fragments using a Zymoclean Gel DNA Recovery kit (Zymo Research, Irvine, CA). Then, following additional library cleanup and quality assessment, we sequenced paired ends of our libraries on an Illumina HiSeq 2000 (Illumina Inc., San Diego, CA).
To call variants, we used a pipeline that combines the Burrows–Wheeler Aligner version 0.7.15 (BWA) (Li and Durbin 2010) and the Genome Analysis Toolkit version 3.7 (GATK) (McKenna et al. 2010). First, we demultiplexed the data using sabre (Joshi 2011). Next, we aligned reads to the H. annuus reference (HanXRQr1.0-20151230; Badouin et al. 2017) with BWA-mem (Li 2013), called variants with GATK “HaplotypeCaller,” and jointly genotyped all samples within a cross type with GATK “GentypeGVCFs.” We split variants into SNPs and insertions/deletions (indels), and filtered each marker type using hard-filtration criteria suggested in the GATK best practices (DePristo et al. 2011; Van der Auwera et al. 2013). Specifically, we removed SNPs that had quality by depth scores (QD) < 2, strand bias scores (FS) > 60, mean mapping quality (MQ) < 40 or allele mapping bias scores (MQRankSum) < −12.5, and indels that had QD < 2 or FS > 200. After further filtering variants for biallelic and triallelic markers with genotype calls in at least 50% of individuals, we used GATK “VariantsToTable” to merge SNPs and indels into a single variant table for each cross type.
Finally, we converted our variant tables into AB format, such that the heterozygous parents contribute “A” and “B” alleles to offspring, while the H. annuus parent contributes exclusively A alleles. At biallelic markers (Figure 2A), sites with two reference alleles became “AA,” and sites with the reference allele and the alternate allele became “AB.” At triallelic markers (Figure 2B), sites with the reference allele and one alternate allele became AA, and sites with the reference allele and the other alternate allele became “AB.” This method randomly assigns A and B alleles to the homologous chromosomes in each heterozygous parent, so our genetic maps initially consisted of pairs of mirror-imaged linkage groups that we later merged.
Genetic mapping
We used R/qtl (Broman et al. 2003) in conjunction with R/ASMap (Taylor and Butler 2017) to build genetic maps. After excluding markers with < 20% or > 80% heterozygosity, and individuals with < 50% of markers scored, we used the function “mstmap.cross” with a stringent significance threshold (P-value = 1 × 10−16) to form conservative linkage groups. We used the function “plotRF” to identify pairs of linkage groups with unusually high recombination fractions and the function “switchAlleles” to reverse the genotype scores of one linkage group in each mirrored pair. We did this until reversing genotype scores no longer reduced the number of linkage groups.
Using the corrected genotypes, we made new linkage groups with only the most reliable markers. Namely, we used the function “mstmap.cross” (with the parameter values: dist. fun = “kosambi”, P-value = 1 × 10−6, noMap.size = 2, and noMap.dist = 5) on markers with < 10% missing data and without significant segregation distortion. We refined the resulting linkage groups by removing: (1) markers with more than three double crossovers, (2) markers with aberrant segregation patterns (segregation distortion > 2 SD above or below the mean segregation distortion of the nearest 20 markers), and (3) linkage groups made up of less than four markers.
We progressively pushed markers with increasing amounts of segregation distortion and missing data into the maps using the function “pushCross.” After adding each batch of markers, we reordered the linkage groups, and dropped markers and linkage groups as described above. Once all the markers had been pushed back, we used the function “calc.errorlod” to identify possible genotyping errors (error scores > 2) and replaced those genotypes with missing data. We continued to drop linkage groups, markers, and genotypes that did not meet our criteria until none remained.
Finally, we dropped five excess linkage groups, each made up of < 30 markers, from each map. The markers in these linkage groups mapped to regions of the H. annuus genome that were otherwise represented in the final genetic maps but could not be explained by reversed genotypes. Instead, these markers were likely polymorphic in the HA89cms individual used for crosses because of the 2–4% residual heterozygosity in sunflower inbred lines (Mandel et al. 2013).
Development of syntR
To aid in the identification of chromosomal rearrangements, we developed the R package syntR (code and documentation available at http://ksamuk.github.io/syntR). This package implements a heuristic algorithm for systematically detecting synteny blocks from marker positions in two genetic maps. The key innovation of the syntR algorithm is coupling a biologically informed noise-reduction method with a cluster-identification method better suited for detecting linear (as opposed to circular) clusters of data points.
We based the syntR algorithm on the following statistical and biological properties of genetic maps and chromosomal rearrangements:
Synteny blocks appear as contiguous sets of orthologous markers in the same or reversed order in pairs of genetic maps (Pevzner and Tesler 2003; Choi et al. 2007).
The inferred order of markers in individual genetic maps is subject to error due to genotyping errors and missing data (Hackett and Broadfoot 2003). This error manifests as slight differences in the order of nearby markers within a linkage group between maps. This mapping error (which we denote “error rate one”) results in uncertainty in the sequence of markers in synteny blocks.
In genomes with a history of duplication, seemingly orthologous markers can truly represent paralogs. These errors (“error rate two”) look like tiny translocations and also disrupt marker orders within synteny blocks.
When comparing genetic maps derived from genomes without duplications or deletions, every region of each genome will be uniquely represented in the other. Because syntR is made for comparing homoploid genomes with this property, we expect each point in each genetic map to be contained within a single unique synteny block. Therefore, overlaps between synteny blocks are likely errors. Note that this assumption precludes the identification of duplications.
Chromosomal rearrangements can be of any size, but smaller rearrangements are difficult to distinguish from error (Pevzner and Tesler 2003). A key decision in synteny block detection is thus the choice of a detection threshold for small rearrangements, which results in a trade-off between error reduction and the minimum size of detectable synteny blocks.
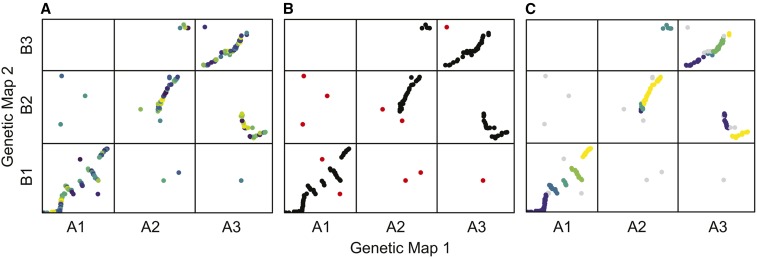
The first step of the syntR algorithm is to smooth over mapping error (error rate one) by identifying highly localized clusters of markers based on a genetic distance threshold (cM) in both maps using hierarchical clustering (Figure 3A). The number of clusters formed is determined by the parameter maximum cluster range (CRmax) that defines the maximum genetic distance (cM) that any cluster can span in either genetic map. After determining these initial clusters, we smooth the maps by collapsing each multimarker cluster down into a single representative point (the centroid of the cluster) for processing in subsequent steps. Next, we address errors introduced by poorly mapped or paralogous markers (error rate two) by flagging and removing outlier clusters that do not have a neighboring cluster within a specified maximum genetic distance (cM), a parameter we denote nearest neighbor distance (NNdist, Figure 3B).
Figure 3.
The stages of the syntR algorithm. Each plot shows the relationship between markers or clusters of markers from three chromosomes in two genetic maps. (A) Highly localized markers are clustered. Each shade represents an individual cluster of markers that will be collapsed into a single representative point. (B) Clusters without another cluster nearby are dropped. Red points represent clusters without a neighbor within 10 cM. (C) Clusters are grouped into synteny blocks based on their rank positions. Gray points represent markers that were dropped in previous steps, and each other color represents a different synteny block.
After the noise-reduction steps, we define preliminary synteny blocks using a method similar to the “friends-of-friends” clustering algorithm (Huchra and Geller 1982). First, we transform the genetic position of each cluster into rank order to minimize the impact of gaps between markers. We then group clusters that are: (1) adjacent in rank position in one of the maps and (2) within two rank positions in the other map (Figure S2). This grouping method further reduces the effect of mapping error by aggregating over pairs (but not triplets) of clusters that have reversed orientations. If a minimum number of clusters per synteny block has been (optionally) defined, we sequentially eliminate blocks that fall below the minimum number of clusters, starting with blocks made up of one cluster and ending with blocks made up of clusters equal to one less than the minimum. After each elimination, we regroup the clusters into new synteny blocks. Finally, we adjust the extents of each synteny block by removing overlapping sections from both synteny blocks so that every position in each genetic map is uniquely represented (Figure 3C).
Assessing the performance of the syntR algorithm
To evaluate the performance of this method and explore the effect of parameter choice on outcomes, we simulated genetic map comparisons with known inversion breakpoints and error rates in R. The genetic map comparisons were made by randomly placing 200 markers at 100 positions along a 100-cM chromosome in two maps, reversing marker positions within a defined inversion region in one map, and then repositioning markers based on simulated mapping noise using the following two error parameters: (1) ER1 is the SD of a normal distribution used to pick the distances markers are pushed out of their correct positions (e.g., when ER1 is 1 cM, 95% of markers will be within 2 cM of their true position) and (2) ER2 is the proportion of markers that are repositioned according to a uniform distribution (i.e., these markers can be moved to any position on the simulated chromosome).
We initially ran syntR using fixed syntR parameters (CRmax = 2 and NNdist = 10) on multiple simulated maps, which were made using variable parameters (inversion size: 2.5–50 cM, ER1: 0–2.0 cM, and ER2: 0–20%), and counted the number of times the known breakpoints were identified within 1 cM (Figure S3). As expected, we find that rearrangement size affects the false-negative rate (i.e., failing to detect known breakpoints), such that smaller inversions are more likely to be missed (Figure S3C), but does not affect the false-positive rate (i.e., detecting breakpoints where there are none). We also find that increasing both types of error in the genetic maps tends to increase both the false-positive and false-negative rates, although ER1 has a much stronger effect on the false-positive rate than any other combination (Figure S3, A and B).
Using the same simulation methods as above but now varying the syntR parameter CRmax, we find that small values of CRmax yield high false-positive rates while large values yield high false-negative rates (Figure S4A). In addition, the ER1 parameter has a strong effect on the relationship between CRmax and the false-positive rate. Higher values of CRmax are needed to reduce the false-positive rate when ER1 is also high (Figure S4B). This means that picking an appropriate CRmax value is key to the accuracy of this method. Although NNdist has a much weaker effect on outcomes than CRmax, it is useful to consider both parameter values carefully.
When the syntR heuristic algorithm is performing well, the final synteny blocks should represent all positions in the two genetic maps being compared (Chen et al. 2009). Based on this characteristic, we developed a method to choose optimal syntR tuning parameters (CRmax and NNdist) that maximize the representation of the genetic maps and markers in synteny blocks. In this method a user: (1) runs syntR with a range of parameter combinations, (2) saves summary statistics about the genetic distance of each map represented in the synteny blocks and the number of markers retained for each run, and (3) finds the parameter combination that maximizes a composite statistic that equally weights these three measures. In cases where there are multiple local maxima, we suggest choosing the local maximum with the smallest value of CRmax to reduce the number of potential false positives.
The “maximize representation” method for choosing syntR parameters has several benefits. First, it does not rely on any additional information (e.g., error rate estimates from the genetic maps compared). Second, when we use this method to choose the best parameters for simulated genetic maps, we find that these parameter values also minimize false-positive and false-negative rates (Figure S5). Third, when we simulate biologically realistic genetic map comparisons, the absolute values of false positives and false negatives are small. For example, when comparing two genetic maps in which ∼95% of markers are within 1 cM of their true position (ER1 = 0.5) and 5% of markers are randomly permuted (ER2 = 0.05), nonexistent breakpoints will be identified 0.1 times and a breakpoint of a 20 cM inversion will be missed 0.04 times. These low error rates also highlight the overall robustness and accuracy of the syntR algorithm.
In addition to performing simulations, we compared the synteny blocks identified by syntR to those identified by other means in a previously published comparison of H. niveus ssp. tephrodes and H. argophyllus maps to H. annuus (Barb et al. 2014). To do this, we formatted the original data sets for input into syntR and used the “maximize representation” method to determine the optimal parameter values for the two comparisons (H. niveus vs. H. annuus: CRmax = 1.5, NNdist = 30; H. argophyllus vs. H. annuus: CRmax = 2, NNdist = 20). We found that syntR was in strong agreement with previous work (Figure S6), recovering all the same translocations and most of the same inversions as the Barb et al. (2014) maps. Most of the cases of mismatches were very small or weakly supported inversions in the Barb et al. (2014) maps that syntR did not identify.
Finding synteny blocks
We used syntR to identify synteny blocks between our newly generated genetic maps and an ultrahigh-density map of H. annuus that was used to build the sunflower genome that we used as a reference (Badouin et al. 2017). This allowed us to easily convert between physical position in the H. annuus reference and position in the H. annuus genetic map. Using this property, we further compared two previously published genetic maps for the closely related sunflower species, H. niveus ssp. tephrodes and H. argophyllus (Barb et al. 2014), to the same H. annuus map. We aligned marker sequences from the published maps to the H. annuus reference using BWA and converted well-aligned markers (MQ > 40) to their positions in the H. annuus genetic map.
Initially, we ran syntR using parameters identified through the “maximize representation” method for each map comparison separately (Table S1). However, varying CRmax revealed rearrangements that were shared between the maps (Figure S7). Therefore, we ran syntR again using a range of CRmax values that included the best fit for each comparison (1.0–3.5 in 0.5 increments) and extracted a curated set of synteny blocks from the output. A synteny block was retained if it fulfilled any of the following criteria (in decreasing order of importance): (1) it was found in another species, (2) it was identified in the majority of syntR runs for a single species, and (3) it maximized the genetic distance represented by synteny blocks. We present this curated set of synteny blocks below, but our results are unchanged if we use the individually fitted synteny blocks.
We named the chromosomes in our genetic maps based on their synteny with the standard order and orientation of H. annuus chromosomes (Tang et al. 2002; Bowers et al. 2012) following Barb et al. (2014) but with shortened prefixes (A = H. annuus, R = H. argophyllus, N = H. niveus ssp. tephrodes, P = H. petiolaris ssp. petiolaris, and F = H. petiolaris ssp. fallax). For example, an H. petiolaris ssp. fallax chromosome made up of regions that are syntenic with H. annuus chromosomes 4 and 7 is called F4–7.
Karyotype reconstruction and analysis
We used our inferred synteny blocks and the software MGR v 2.01 (Bourque and Pevzner 2002) to infer ancestral karyotypes for our five Helianthus taxa and to determine the number of chromosomal rearrangements that occurred along each branch of the species tree. To run the MGR analysis, we needed the order and orientations of synteny blocks in all five maps. However, individual synteny blocks were often missing from one or more of our final maps. We approached this problem in two ways. First, we inferred the likely position of missing synteny blocks based on the location of markers that were too sparse to be grouped by syntR and matched the location of synteny blocks in other maps. In the second case, we dropped any synteny blocks that were not universally represented. Because we already had two sets of synteny blocks for each map (curated and individually optimized), we ran the MGR analyses using three different sets of synteny blocks: set 1 (curated and inferred), set 2 (curated and present in all five maps), and set 3 (individually optimized and present in all five maps).
Data availability
The R program, syntR, is available on GitHub: https://github.com/ksamuk/syntR. The sequences used to generate genetic maps are available at the Sequence Read Archive: http://www.ncbi.nlm.nih.gov/bioproject/598366. All other data and scripts are available on dryad: https://doi.org/10.5061/dryad.7sqv9s4pc. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11819625.
Results
Genetic maps
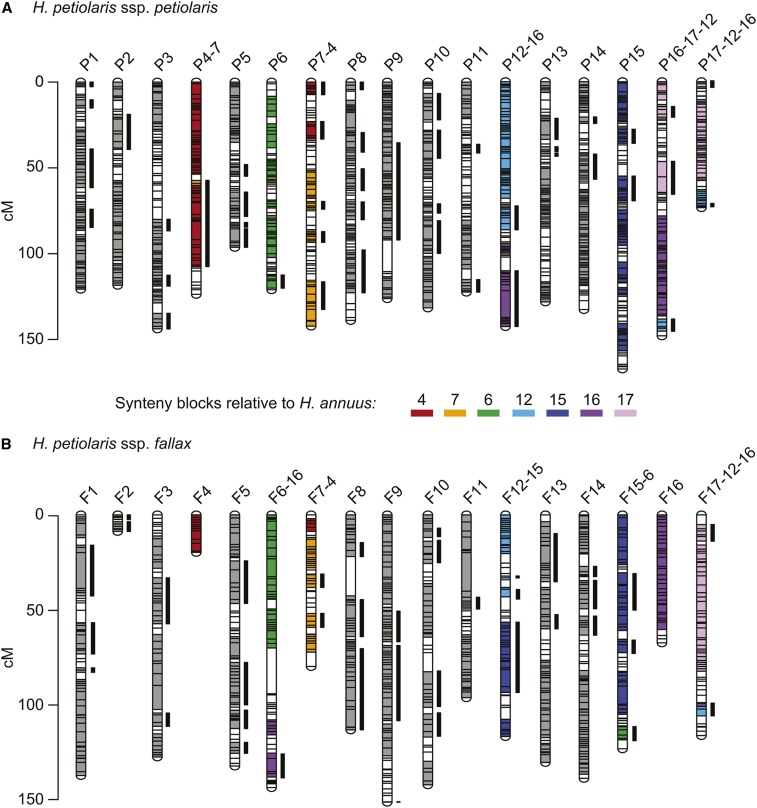
Both H. petiolaris genetic maps are made up of the expected 17 chromosomes and have very high marker densities (Figure 4 and Figure S8). Only 6% of the H. petiolaris ssp. petiolaris map and 10% of the H. petiolaris ssp. fallax map fails to have a marker within 2 cM (Figure S9). Overall, both maps are somewhat longer than the H. petiolaris map reported by Burke et al. (2004). Although this could represent real variation between genotypes, it could also be the result of spurious crossovers that are inferred based on genotyping errors. Because genotyping errors are proportional to the number of markers, maps with high marker densities are more likely to be inflated. Indeed, building maps with variants that are thinned to 1 per 150 bp using vcftools version 0.1.13 (Danecek et al. 2011) yields collinear maps that are closer to the expected lengths (Figure S10 and Table S2,). We present subsequent results based on the full maps to improve our resolution for detecting small rearrangements.
Figure 4.
H. petiolaris genetic maps showing blocks of synteny with H. annuus. Each horizontal bar represents a genetic marker. The thick vertical bars next to chromosomes represent synteny blocks that are inverted relative to the H. annuus genetic map. Where there are no translocations between H. petiolaris and H. annuus chromosomes (e.g., all synteny blocks in P1 and F1 are syntenic with A1), the synteny blocks are shown in gray. Where there are translocations, the synteny blocks are color-coded based on their synteny with H. annuus chromosomes. Regions that are not assigned to a synteny block remain white. The synteny blocks plotted are those curated based on multiple runs of syntR using different parameters. Please see Figure S12 for a labeled version. This figure was made with LinkageMapView (Ouellette et al. 2017).
Despite the general expansion of our maps, we find that chromosomes 2 and 4 in the H. petiolaris ssp. fallax map (F2 and F4) are unexpectedly short (Figure 4). When we look at the distribution of markers for this map relative to the H. annuus reference, we find very few variable sites in the distal half of these chromosomes (Figure S11). That is, this individual was homozygous along vast stretches of F2 and F4. These runs of homozygosity could be explained by recent common ancestry (i.e., inbreeding) or a lack of variation in the population (e.g., because of background selection or a recent selective sweep). Regardless, the lack of variable sites within the H. petiolaris ssp. fallax individual used for crosses explains the shortness of F2 and F4. Notably, we find the same pattern on the distal half of H. annuus chromosome 7 and find that this region is also not represented in the H. petiolaris spp. fallax map.
Synteny blocks
Using syntR, we recovered 97 genetic regions that are syntenic between H. petiolaris ssp. petiolaris and H. annuus, and 79 genetic regions that are syntenic between H. petiolaris ssp. fallax and H. annuus (Figure 4). We also recovered synteny blocks for the H. niveus ssp. tephrodes and H. argophyllus comparisons that are similar to those found previously (Figure S13). In all four comparisons, syntR successfully identified synteny blocks that cover large proportions (63–90%) of each genetic map, even in the face of a very high proportion of markers that map to a different chromosome than their neighbors (Table 1). These “rogue markers” could be the result of very small translocations, poorly mapped markers, or extensive paralogy. Over and above the prevalence of rogue markers, the karyotypes we recovered are substantially rearranged. Only between 32% and 45% of synteny blocks for each map are collinear with the H. annuus genetic map in direct comparisons (Table 1).
Table 1. Properties of the synteny blocks found using a syntR analysis between genetic maps of H. annuus and four other Helianthus taxa.
| Genetic map | Number of synteny blocks | Rogue markers (%) | Map coverage | H. annuus coverage | Collinear | Inverted | Translocated |
|---|---|---|---|---|---|---|---|
| H. petiolaris ssp. petiolaris | 97 | 19 | 80% | 74% | 39% | 36% | 26% |
| H. petiolaris spp. fallax | 79 | 17 | 63% | 65% | 32% | 34% | 34% |
| H. niveus ssp. tephrodes | 43 | 26 | 78% | 75% | 40% | 21% | 39% |
| H. argophyllus | 31 | 20 | 90% | 82% | 45% | 16% | 39% |
The proportion of rogue markers is based only on the chromosomes without translocations in any map (i.e., chromosomes 1–3, 5, 8–10, 11, and 14). For those chromosomes, the majority of markers mapped to a single H. annuus chromosome. The other markers are considered rogue.
Karyotype reconstruction and chromosomal rearrangement
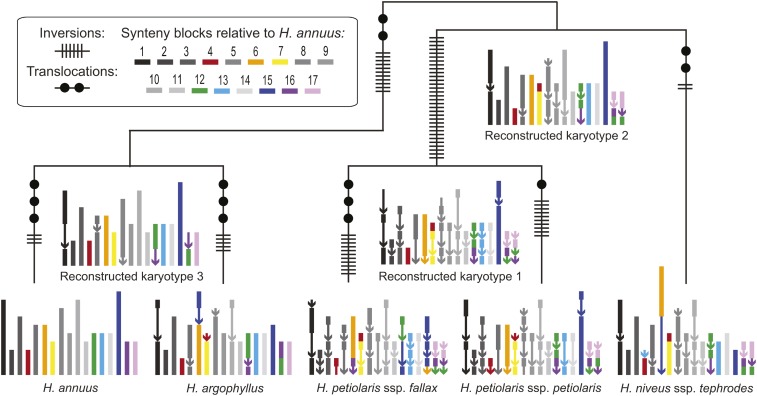
Because nested and shared rearrangements can obscure patterns of chromosome evolution, we use the MGR analyses to predict the most likely sequence of rearrangements in a phylogenetic context before quantifying the rearrangement rate. These MGR analyses identified similar patterns of chromosome evolution regardless of the exact set of synteny blocks that we used (Table S5). Multiple taxa share many rearrangements, and the similarity of karyotypes matches known phylogenetic relationships. Moreover, MGR analyses run without a guide tree inferred the known species tree, and MGR analyses run with all other topologies identified an inflated number of chromosomal rearrangements.
Using the most complete set of synteny blocks (set 1), we find that 88 chromosomal rearrangements occurred across the phylogeny (Figure 5). Then, using the most current divergence time estimates for this group (Todesco et al. 2019) and conservatively assuming that H. niveus ssp. tephrodes diverged at the earliest possible point, we estimate that 7.9 (7.8–8) rearrangements occurred per million years in this clade (Tables S3–S5). To further explore the potential range of rearrangement rates, we considered other estimates of divergence times in sunflower (Sambatti et al. 2012; Mason 2018) and the other sets of synteny blocks. Overall, the lowest rate we identified was 2.6 rearrangements per million years, while the highest rate was indeterminable because some minimum divergence time estimates for the group include 0 (Tables S3–S5).
Figure 5.
Diagram showing the karyotypes of five Helianthus taxa as well as reconstructed ancestral karyotypes and the locations of chromosomal rearrangements. The karyotypes were built using synteny block set 1, and were curated based on multiple syntR runs and inferred when missing. Each synteny block is represented using a line segment that is color-coded based on its position in the H. annuus genome (see Figure S14 for a labeled version). Chromosomes without translocations in any map are plotted in gray, and synteny blocks that are inverted relative to H. annuus are plotted using arrows. Also, note that along some branches the same pair of chromosomes is involved in multiple translocations.
The 88 rearrangements include 74 inversions and 14 translocations that are quite evenly distributed across the phylogeny. However, the excess inversions indicate that it is unlikely that the rate of inversions is equal to the rate of translocation (binomial test, 5.1 × 10−11). Furthermore, we find that only 8 of the 17 chromosomes are involved in the 14 translocations we identified. If translocations were equally likely for all chromosomes, this asymmetry would be very unlikely to have happened by chance (the probability of sampling at most eight chromosomes in 14 translocations is 8.0 × 10−8, Figure S15), suggesting that some chromosomes are more likely to be involved in translocations than other. In line with this observation, we see that some chromosome segments are repeatedly translocated. For example, A4 and A7 are involved in several exchanges, and part of A6 has a different position in almost every map (Figure 5).
Discussion
Large-scale chromosomal changes may be key contributors to the process of adaptation and speciation, yet we still have a poor understanding of rates of chromosomal rearrangement and the evolutionary forces underlying those rates. Here, we devised a novel, systematic method for comparing any pair of genetic maps, and performed a comprehensive analysis of the evolution of chromosomal rearrangements in a clade of sunflowers. We created two new genetic maps for Helianthus species and used our new method to identify a wide range of karyotypic variation in our new maps, as well as previously published maps. Consistent with previous studies, we discovered a high rate of chromosomal evolution in the annual sunflowers. Further, we found that inversions are more common than translocations and that certain chromosomes are more likely to be translocated. Below, we discuss the evolutionary and methodological implications of this work and suggest some next steps in understanding the dynamic process of chromosomal rearrangement.
Identifying rearrangements
Studying the evolution of chromosomal rearrangements requires dense genetic maps and systematic methods to analyze and compare these maps between species. Our new software, syntR, provides an end-to-end solution for systematic and repeatable identification of synteny blocks in pairs of genetic maps with any marker density. Our tests on real and simulated data find that syntR recovers chromosomal rearrangements identified previously by both manual comparisons and cytological studies, suggesting that syntR is providing an accurate view of karyotypic differences between species.
Overall, we believe that syntR will be a valuable tool for the systematic study of chromosomal rearrangements in any species. The only data syntR needs to identify synteny blocks is relative marker positions in two genetic maps. This fact is significant because, although the number of species with whole-genome sequences and methods to detect synteny blocks from those sequences are rapidly accumulating, such as Mauve (Darling et al. 2004), Cinteny (Sinha and Meller 2007), syMAP (Soderlund et al. 2011), SynChro (Drillon et al. 2014), and SyRI (Goel et al. 2019), it is still uncommon to have multiple closely related whole-genome sequences that are of sufficient quality to compare for karyotype differences. At the same time, the proliferation of reduced-representation genome sequencing methods means that it is easy to generate many genetic markers for nonmodel species and produce very dense genetic maps. Furthermore, syntR allows comparisons to include older genetic map data that would otherwise go unused. The simplicity of the syntR algorithm will facilitate rapid karyotype mapping in a wide range of taxa.
We also believe that syntR provides a baseline for the development of further computational and statistical methods for the study of chromosomal rearrangements. One fruitful direction would be to integrate the syntR algorithm for synteny block detection directly into the genetic map-building process (much like GOOGA; Flagel et al. 2019). Another key extension would be to allow syntR to compare multiple genetic maps simultaneously to detect synteny blocks in a group of species (e.g., by leveraging information across species). Finally, formal statistical methods for evaluating the model fit and the uncertainty involved with any set of synteny blocks would be a major (albeit challenging) improvement to all existing methods, including syntR.
The similarity of H. petiolaris maps to previous studies
Compared with previous work, we found more inversions and fewer translocations between H. petiolaris subspecies and H. annuus (Rieseberg et al. 1995; Burke et al. 2004). This is probably due to a combination of factors. First, there appears to be karyotypic variation within some Helianthus species (Heiser 1948, 1961; Chandler et al. 1986). Second, the maps presented here are made up of more markers and individuals, which allowed us to identify small inversions that were previously undetected as well as to eliminate false linkages that can be problematic in small mapping populations. Lastly, we required more evidence to call rearrangements. Although we recovered some of the translocations supported by multiple markers in Rieseberg et al. (1995) and Burke et al. (2004), we did not recover any of the translocations supported by only a single sequence-based marker. Given the high proportion of rogue markers in our maps, it is likely that some of the putative translocations recovered in those earlier comparisons are the result of the same phenomenon.
On the other hand, we found that rearrangements between our H. petiolaris maps match the translocations predicted from cytological studies quite well. Heiser (1961) predicted that H. petiolaris ssp. petiolaris and H. petiolaris ssp. fallax karyotypes would have three chromosomes involved in two translocations that form a ring during pairing at meiosis, as well as the possibility of a second independent rearrangement. This exact configuration is likely to occur at meiosis in hybrids between the H. petiolaris subspecies maps we present here (Figure S16). Also, the most noteworthy chromosome configuration in cytological studies of H. annuus–H. petiolaris hybrids (Heiser 1947; Whelan 1979; Ferriera 1980; Chandler et al. 1986) was a hexavalent (a six-chromosome structure) plus a quadrivalent (a four-chromosome structure). Again, this is the configuration that we would expect in a hybrid between H. annuus and the H. petiolaris ssp. petiolaris individual mapped here. Furthermore, the complicated arrangement and relatively small sizes of A12, A16, and A17 synteny blocks in H. petiolaris might explain why cytological configurations in H. annuus–H. petiolaris hybrids are so variable. Interestingly, the rearrangements identified between H. argophyllus and H. annuus karyotypes here and in Barb et al. (2014) also match the cytological studies better than an earlier comparison of sparse genetic maps (Heesacker et al. 2009). It seems that, in systems with the potential for high proportions of rogue markers, many markers are needed to identify chromosomal rearrangements reliably.
Total rearrangement rates
Our data suggest that annual sunflowers experience ∼7.9 chromosomal rearrangements per million years. This rate overlaps with recent estimates for this group (7.4–10.3, Barb et al. 2014) and is even higher than the estimate that highlighted sunflower as a group with exceptionally fast chromosomal evolution (5.5–7.3, Burke et al. 2004). However, since Burke et al. (2004), chromosomal rearrangements have been tracked in many additional groups, including mammals (Ferguson-Smith and Trifonov 2007; Martinez et al. 2016; Oliveira da Silva et al. 2019), fish (Molina et al. 2014; Ayres-Alves et al. 2017), insects (Rueppell et al. 2016; Corbett-Detig et al. 2019), fungi (Sun et al. 2017), and plants (Yogeeswaran et al. 2005; Schranz et al. 2006; Huang et al. 2009; International Brachypodium Initiative 2010; Latta et al. 2019). Of these analyses, relatively few have systematically studied karyotype evolution across multiple species and estimated total rearrangement rates. Of those that do, most studies report <7.9 chromosomal rearrangements per million years, for example, in Solanum (0.36–1.44; Wu and Tanksley 2010), Drosophila (0.44–2.74; Bhutkar et al. 2008), and mammals (0.05–2.76; Murphy et al. 2005). However, there are exceptions, such as a comparison of genome sequences that revealed up to 35.7 rearrangements per million years in some grass lineages (Dvorak et al. 2018).
At the same time, we are likely underestimating rearrangement rates here for two reasons. First, we used conservative thresholds for calling rearrangements. For example, some proportion of the rogue markers that we identified could be the result of very small but real chromosomal rearrangements. Second, our ability to resolve very small synteny blocks and breakpoints between synteny blocks depends on marker density. Until we have full genome sequences to compare (like for the grass lineages), we could be failing to detect very small rearrangements and falsely inferring that independent rearrangements are shared. However, regardless of just how much we are underestimating the rate, sunflower chromosomes are evolving quickly. This high rate of chromosomal evolution could be a consequence of a higher rate of chromosomal mutation, a decreased chance that chromosomal polymorphisms are lost, or both processes.
Type of rearrangements
We found that inversions and interchromosomal translocations dominate chromosomal evolution in Helianthus. This pattern is common in angiosperm lineages (Weiss-Schneeweiss and Schneeweiss 2012) and fits with the consistent chromosome counts across annual sunflowers (2n = 34; Chandler et al. 1986). In addition, we found more inversions than translocations, which has previously been seen in both plant (Wu and Tanksley 2010; Amores et al. 2014) and animal systems (Rueppell et al. 2016), and echoes general reports that intrachromosomal rearrangements are more common than interchromosomal rearrangements (Pevzner and Tesler 2003). These consistent rate differences are notable because, although both rearrangement types depend on double-strand breaks, two of the major consequences of chromosomal rearrangements, underdominance (i.e., rearrangement heterozygotes are less fit than either homozygote) and recombination modification, might be more common for some types of rearrangements.
Translocations have a more predictable effect on hybrid fertility, while inversions consistently reduce recombination. Reciprocal translocation heterozygotes can affect fertility because missegregation during meiosis can cause one-half of the gametes to be unbalanced and thus inviable (White 1973; King 1993). Although inversion heterozygotes can also produce unbalanced gametes, whether that happens is dependent on the size of the inversion and whether disrupted pairing during meiosis inhibits crossovers (Searle 1993). When inversions are small or have suppressed crossing over, they will not be strongly underdominant. On the other hand, inversions often exhibit reduced recombination, either because recombination is suppressed through disrupted pairing (Searle 1993) or ineffective through the production of inviable gametes (Rieseberg 2001). While interactions between reduced recombination and adaptation with gene flow have been extensively examined in the case of inversions (Kirkpatrick and Barton 2006; Hoffmann and Rieseberg 2008; Yeaman and Whitlock 2011; Yeaman 2013), it is not clear whether the same pattern will be common for translocations [but see Fishman et al. (2013) and Stathos and Fishman (2014) for one example]. Translocations bring together previously unlinked alleles and mispairing at translocation breakpoints could suppress crossing over, but recombination inside reciprocal translocations will not necessarily produce inviable gametes and thus reduce effective recombination.
Although any selective force could be responsible for the evolution of any chromosomal rearrangement, potential differences in the relative magnitude of underdominance vs. recombination suppression may contribute to the evolution of sunflower chromosomes. While many chromosomal rearrangements in sunflowers appear to be strongly underdominant (Chandler et al. 1986, Lai et al. 2005), inversions typically are not (L. Rieseberg, unpublished data). If translocations tend to be more underdominant than inversions, they would be less likely to evolve through drift and more likely to cause reproductive isolation directly. This could explain why translocations are less common than inversions and why pollen viability is accurately predicted by the number of translocations inferred from cytological studies (Chandler et al. 1986). At the same time, recent genomic analyses have identified several extensive regions of very low recombination caused by large inversions segregating in natural sunflower populations (Todesco et al. 2019; Huang et al. 2019). Mutations that segregate for extended periods are unlikely to be strongly underdominant and these inversions are associated with multiple adaptive alleles (Todesco et al. 2019), which is consistent with a role for selection in their origin or maintenance.
Nonrandom chromosomal rearrangement
We also found that some sunflower chromosomes are involved in more translocations than others. This pattern has been observed in wheat (Badaeva et al. 2007) and breakpoint reuse is a common phenomenon in comparative studies of karyotypes (Pevzner and Tesler 2003; Bailey et al. 2004; Murphy et al. 2005; Larkin et al. 2009). Many studies support the idea that chromosomal regions with greater sequence similarity are more likely to recombine and thus potentially generate novel chromosomal arrangements. Some of the clearest examples of this come from the polyploidy literature, where chromosomes with ancestral homology are more likely to recombine (Nicolas et al. 2007; Marone et al. 2012; Mason et al. 2014; Tennessen et al. 2014; Nguepjop et al. 2016). However, centromeres and other repetitive regions can also affect the rate of mutations that cause chromosomal rearrangements (Hardison et al. 2003; Murphy et al. 2005; Raskina et al. 2008; Molnár et al. 2010; Vitte et al. 2014; Ayers-Alves et al. 2017; Li et al. 2017; Corbett-Detig et al. 2019). Given that sunflowers have several genome duplications and a burst of transposable element activity in their evolutionary history (Barker et al. 2008, 2016; Kawakami et al. 2011; Staton et al. 2012; Badouin et al. 2017), it is plausible that ancestral homology or repeat content could be associated with translocation propensity.
Of the above possibilities, an association between repeated translocations and centromeres would be particularly compelling. Beyond the repeat content of centromeres explaining nonrandom mutation [Kawabe et al. (2006) and Sun et al. (2017), but see Lin et al. (2018) and Okita et al. (2019)], the positions and sizes of centromeres on chromosomes are known to affect meiotic drive, and thus the repositioning of centromeres through rearrangement could cause nonrandom fixation of translocations (Kaszás and Birchler 1998, Chmátal et al. 2014; Zanders et al. 2014). The relative placement of centromeres has been associated with chromosome evolution in Brassica (Schranz et al. 2006) and wheat (Badaeva et al. 2007), and associations between meiotic drive and chromosome evolution have been found in several animal taxa (Bidau and Martí 2004; Palestis et al. 2004; Molina et al. 2014; Blackmon et al. 2019). In sunflower, we see some hints that centromeric repeats might be associated with repeated translocation. Using the locations of the centromere-specific retrotransposon sequence HaCEN-LINE (Nagaki et al. 2015) to roughly identify the locations of centromeres in our reference, we find that some rearrangement breakpoints, e.g., the section of A16 with a different position in each map, are close to putative centromeres (Figure S17–S20). Although a more thorough analysis of centromeric repeat locations and their association with rearrangement breakpoints is required to draw firm conclusions about the importance of centromeres to chromosomal evolution in sunflower, the development of reference sequences for wild sunflower species is underway, which will allow those and other associations to be confirmed. Further, it is time to directly test for meiotic drive in this system by examining the transmission of rearrangements that affect centromeres in gametes produced by plants that have heterozygous karyotypes.
Conclusion
Understanding the evolution of chromosomal rearrangements remains a key challenge in evolutionary genetics. By developing new software to systematically detect synteny blocks and building new genetic maps, we show that sunflowers exhibit rapid and nonrandom patterns of chromosomal evolution. These data generate specific and testable hypotheses about chromosomal evolution in sunflower. We believe that our work will spur additional studies of karyotypic evolution and diversity, and ultimately lead to a more comprehensive understanding of the interplay between chromosomal evolution and speciation.
Acknowledgments
We thank Jessica Barb for providing marker sequence data; Marcy Uyenoyama for help with our random walk analysis; Greg Baute for sharing hybrid seed; Chris Grassa for growing seedlings and sharing scripts; Marco Todesco and Nadia Chaidir for help in the laboratory; and Jenn Coughlan, Andrew MacDonald, Brook Moyers, Mariano Alvarez, Dolph Schluter, Darren Irwin, Sally Otto, and three anonymous reviewers for thoughtful discussions and help with earlier drafts of this manuscript. This work was supported by an Natural Sciences and Engineering Research Council (NSERC) of Canada Postgraduate Scholarship awarded to K.L.O. and an NSERC Discovery grant awarded to L.H.R. (327475).
Author contributions: K.L.O. and L.H.R. planned the study. K.L.O. and K.S. designed and built the R package syntR. K.L.O. made genetic maps, carried out data analysis, and drafted the manuscript. All authors read, edited, and approved the final manuscript.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11819625.
Communicating editor: L. Moyle
Literature Cited
- Amores A., Catchen J., Nanda I., Warren W., Walter R. et al. , 2014. A RAD-tag genetic map for the platyfish (Xiphophorus maculatus) reveals mechanisms of karyotype evolution among teleost fish. Genetics 197: 625–641. 10.1534/genetics.114.164293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres-Alves T., Cardoso A. L., Nagamachi C. Y., de Sousa L. M., Pieczarka J. C. et al. , 2017. Karyotypic evolution and chromosomal organization of repetitive DNA sequences in species of Panaque, Panaqolus, and Scobinancistrus (Siluriformes and Loricariidae) from the Amazon Basin. Zebrafish 14: 251–260. 10.1089/zeb.2016.1373 [DOI] [PubMed] [Google Scholar]
- Badaeva E. D., Dedkova O. S., Gay G., Pukhalskyi V. A., Zelenin A. V. et al. , 2007. Chromosomal rearrangements in wheat: their types and distribution. Genome 50: 907–926. 10.1139/G07-072 [DOI] [PubMed] [Google Scholar]
- Badouin H., Gouzy J., Grassa C. J., Murat F., Staton S. E. et al. , 2017. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546: 148–152. 10.1038/nature22380 [DOI] [PubMed] [Google Scholar]
- Bailey J. A., Baertsch R., Kent W. J., Haussler D., and Eichler E. E., 2004. Hotspots of mammalian chromosomal evolution. Genome Biol. 5: R23 10.1186/gb-2004-5-4-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb J. G., Bowers J. E., Renaut S., Rey J. I., Knapp S. J. et al. , 2014. Chromosomal evolution and patterns of introgression in Helianthus. Genetics 197: 969–979. 10.1534/genetics.114.165548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M. S., Kane N. C., Matvienko M., Kozik A., Michelmore R. W. et al. , 2008. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol. 25: 2445–2455. 10.1093/molbev/msn187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M. S., Li Z., Kidder T. I., Reardon C. R., Lai Z. et al. , 2016. Most Compositae (Asteraceae) are descendants of a paleohexaploid and all share a paleotetraploid ancestor with the Calyceraceae. Am. J. Bot. 103: 1203–1211. 10.3732/ajb.1600113 [DOI] [PubMed] [Google Scholar]
- Baute G. J., Owens G. L., Bock D. G., and Rieseberg L. H., 2016. Genome-wide genotyping-by-sequencing data provide a high-resolution view of wild Helianthus diversity, genetic structure, and interspecies gene flow. Am. J. Bot. 103: 2170–2177. 10.3732/ajb.1600295 [DOI] [PubMed] [Google Scholar]
- Berdan E. L., Kozak G. M., Ming R., Rayburn A. L., Kiehart R. et al. , 2014. Insight into genomic changes accompanying divergence: genetic linkage maps and synteny of Lucania goodei and L. parva reveal a Robertsonian fusion. G3 (Bethesda) 4: 1363–1372. 10.1534/g3.114.012096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A., Schaeffer S. W., Russo S. M., Xu M., Smith T. F. et al. , 2008. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179: 1657–1680. 10.1534/genetics.107.086108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidau C. J., and Martí D. A., 2004. B chromosomes and Robertsonian fusions of Dichroplus pratensis (Acrididae): intraspecific support for the centromeric drive theory. Cytogenet. Genome Res. 106: 347–350. 10.1159/000079311 [DOI] [PubMed] [Google Scholar]
- Bilton T. P., Schofield M. R., Black M. A., Chagné D., Wilcox P. L. et al. , 2018. Accounting for errors in low coverage high-throughput sequencing data when constructing genetic maps using biparental outcrossed populations. Genetics 209: 65–76. 10.1534/genetics.117.300627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmon H., Justison J., Mayrose I., and Goldberg E. E., 2019. Meiotic drive shapes rates of karyotype evolution in mammals. Evolution 73: 511–523. 10.1111/evo.13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G., and Pevzner P. A., 2002. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 12: 26–36. [PMC free article] [PubMed] [Google Scholar]
- Bowers J. E., Bachlava E., Brunick R. L., Rieseberg L. H., Knapp S. J. et al. , 2012. Development of a 10,000 locus genetic map of the sunflower genome based on multiple crosses. G3 (Bethesda) 2: 721–729. 10.1534/g3.112.002659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., and Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Burke J. M., Lai Z., Salmaso M., Nakazato T., Tang S. et al. , 2004. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics 167: 449–457. 10.1534/genetics.167.1.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. M., Jan C. C., and Beard B. H., 1986. Chromosomal differentiation among the annual Helianthus species. Syst. Bot. 11: 354–371. 10.2307/2419126 [DOI] [Google Scholar]
- Chen Z., Fu B., Jiang M., and Zhu B., 2009. On recovering syntenic blocks from comparative maps. J. Comb. Optim. 18: 307–318. 10.1007/s10878-009-9233-x [DOI] [Google Scholar]
- Chmátal L., Gabriel S. I., Mitsainas G. P., Martínez-Vargas J., Ventura J. et al. , 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 24: 2295–2300. 10.1016/j.cub.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi V., Zheng C., Zhu Q., and Sankoff D., 2007. Algorithms for the extraction of synteny blocks from comparative maps, pp. 277–288 in International Workshop on Algorithms in Bioinformatics Springer, Berlin, Heidelberg: 10.1007/978-3-540-74126-8_26 [DOI] [Google Scholar]
- Corbett-Detig R. B., Said I., Calzetta M., Genetti M., McBroome J. et al. , 2019. Fine-mapping complex inversion breakpoints and investigating somatic pairing in the Anopheles gambiae species complex using proximity-ligation sequencing. Genetics 213: 1495–1511. 10.1534/genetics.119.302385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C. E., Mau B., Blattner F. R., and Perna N. T., 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14: 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R. et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J., and Doyle J., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Drillon G., Carbone A., and Fischer G., 2014. SynChro: a fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One 9: e92621 10.1371/journal.pone.0092621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J., Wang L., Zhu T., Jorgensen C. M., Deal K. R. et al. , 2018. Structural variation and rates of genome evolution in the grass family seen through comparison of sequences of genomes greatly differing in size. Plant J. 95: 487–503. 10.1111/tpj.13964 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith M. A., and Trifonov V., 2007. Mammalian karyotype evolution. Nat. Rev. Genet. 8: 950–962. 10.1038/nrg2199 [DOI] [PubMed] [Google Scholar]
- Ferriera J. V., 1980. Introgressive hybridization between Helanthus annuus L. and Helianthus petiolaris Nutt. Mendeliana 4: 81–93. [Google Scholar]
- Fishman L., Stathos A., Beardsley P. M., Williams C. F., and Hill J. P., 2013. Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monkey flowers). Evolution 67: 2547–2560. 10.1111/evo.12154 [DOI] [PubMed] [Google Scholar]
- Flagel L. E., Blackman B. K., Fishman L., Monnahan P. J., Sweigart A. et al. , 2019. GOOGA: a platform to synthesize mapping experiments and identify genomic structural diversity. PLoS Comput. Biol. 15: e1006949 10.1371/journal.pcbi.1006949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel M., Sun H., Jiao W. B., and Schneeberger K., 2019. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20: 277 10.1186/s13059-019-1911-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett C. A., and Broadfoot L. B., 2003. Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity 90: 33–38. 10.1038/sj.hdy.6800173 [DOI] [PubMed] [Google Scholar]
- Hardison R. C., Roskin K. M., Yang S., Diekhans M., Kent W. J. et al. , 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13: 13–26. 10.1101/gr.844103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesacker A. F., Bachlava E., Brunick R. L., Burke J. M., Rieseberg L. H. et al. , 2009. Karyotypic evolution of the common and silverleaf sunflower genomes. Plant Genome 2: 233–246. 10.3835/plantgenome2009.05.0015 [DOI] [Google Scholar]
- Heiser C. B., Jr, 1947. Hybridization between the sunflower species Helianthus annuus and H. petiolaris. Evolution 1: 249–262. 10.1111/j.1558-5646.1947.tb02722.x [DOI] [Google Scholar]
- Heiser C. B., Jr, 1948. Taxonomic and cytological notes on the annual species of Helianthus. Bull. Torrey Bot. Club 75: 512–515. 10.2307/2481785 [DOI] [Google Scholar]
- Heiser C. B., Jr, 1951. Hybridization in the annual sunflowers: Helianthus annuus x H. argophyllus. Am. Nat. 85: 65–72. 10.1086/281651 [DOI] [Google Scholar]
- Heiser C. B., Jr, 1961. Morphological and cytological variation in Helianthus petiolaris with notes on related species. Evolution 15: 247–258. 10.1111/j.1558-5646.1961.tb03147.x [DOI] [Google Scholar]
- Hoffmann A. A., and Rieseberg L. H., 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39: 21–42. 10.1146/annurev.ecolsys.39.110707.173532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Li R., Zhang Z., Li L., Gu X. et al. , 2009. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41: 1275–1281. 10.1038/ng.475 [DOI] [PubMed] [Google Scholar]
- Huang K., R. L. Andrew, G. L. Owens, K. L. Ostevik, and L. H. Rieseberg, 2019 Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype. bioRxiv. Available at: https://www.biorxiv.org/content/10.1101/829622v1.full. Accessed: December 30, 2019. doi: 10.1101/829622 10.1101/829622 [DOI] [PubMed]
- Huchra J. P., and Geller M. J., 1982. Groups of galaxies. I. Nearby groups. Astrophys. J. 257: 423–437. 10.1086/160000 [DOI] [Google Scholar]
- International Brachypodium Initiative, et al. , 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- Joshi N. A., 2011. Sabre: A barcode demultiplexing and trimming tool for FastQ files. https://github.com/najoshi/sabre. Accessed: January 27, 2017.
- Kaszás E., and Birchler J. A., 1998. Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150: 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A., Hansson B., Hagenblad J., Forrest A., and Charlesworth D., 2006. Centromere locations and associated chromosome rearrangements in Arabidopsis lyrata and A. thaliana. Genetics 173: 1613–1619. 10.1534/genetics.106.057182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Dhakal P., Katterhenry A. N., Heatherington C. A., and Ungerer M. C., 2011. Transposable element proliferation and genome expansion are rare in contemporary sunflower hybrid populations despite widespread transcriptional activity of LTR retrotransposons. Genome Biol. Evol. 3: 156–167. 10.1093/gbe/evr005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M., 1987. Chromosomal rearrangements, speciation and the theoretical approach. Heredity 59: 1–6. 10.1038/hdy.1987.90 [DOI] [PubMed] [Google Scholar]
- King M., 1993. Species Evolution, Cambridge University Press, Cambridge. [Google Scholar]
- Kirkpatrick M., and Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434 [corrigenda: Genetics 208: 433 (2018)]. 10.1534/genetics.105.047985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Nakazato T., Salmaso M., Burke J. M., Tang S. et al. , 2005. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171: 291–303. 10.1534/genetics.105.042242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin D. M., Pape G., Donthu R., Auvil L., Welge M. et al. , 2009. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 19: 770–777. 10.1101/gr.086546.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta R. G., Bekele W. A., Wight C. P., and Tinker N. A., 2019. Comparative linkage mapping of diploid, tetraploid, and hexaploid Avena species suggests extensive chromosome rearrangement in ancestral diploids. Sci. Rep. 9: 12298 10.1038/s41598-019-48639-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. Available: https://arxiv.org/abs/1303.3997. (accessed January 27, 2017) doi: 1303.3997v2
- Li H., and Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-F., Su T., Cheng G.-Q., Wang B.-X., Li X. et al. , 2017. Chromosome evolution in connection with repetitive sequences and epigenetics in plants. Genes (Basel) 8: 290. 10.3390/genes8100290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-Y., Shukla A., Grady J. P., Fink J. L., Dray E. et al. , 2018. Translocation breakpoints preferentially occur in euchromatin and acrocentric chromosomes. Cancers (Basel) 10: 13. 10.3390/cancers10010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. R., Nambeesan S., Bowers J. E., Marek L. F., Ebert D. et al. , 2013. Association mapping and the genomic consequences of selection in sunflower. PLoS Genet. 9: e1003378 10.1371/journal.pgen.1003378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone D., Laidò G., Gadaleta A., Colasuonno P., Ficco D. B. M. et al. , 2012. A high-density consensus map of A and B wheat genomes. Theor. Appl. Genet. 125: 1619–1638. 10.1007/s00122-012-1939-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P. A., Jacobina U. P., Fernandes R. V., Brito C., Penone C. et al. , 2016. A comparative study on karyotypic diversification rate in mammals. Heredity 118: 366–373. 10.1038/hdy.2016.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason C. M., 2018. How old are sunflowers? A molecular clock analysis of key divergences in the origin and diversification of Helianthus (Asteraceae). Int. J. Plant Sci. 179: 182–191. 10.1086/696366 [DOI] [Google Scholar]
- Mason A. S., Nelson M. N., Takahira J., Cowling W. A., Alves G. M. et al. , 2014. The fate of chromosomes and alleles in an allohexaploid Brassica population. Genetics 197: 273–283. 10.1534/genetics.113.159574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M., Kozik A., Froenicke L., Lavelle D., Martineau B. et al. , 2013. Consequences of normalizing transcriptomic and genomic libraries of plant genomes using a duplex-specific nuclease and tetramethylammonium chloride. PLoS One 8: e55913 10.1371/journal.pone.0055913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K. et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina W. F., Martinez P. A., Bertollo L. A. C., and Bidau C. J., 2014. Evidence for meiotic drive as an explanation for karyotype changes in fishes. Mar. Genomics 15: 29–34. 10.1016/j.margen.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Molnár I., Cifuentes M., Schneider A., Benavente E., and Molnár-Láng M., 2010. Association between simple sequence repeat-rich chromosome regions and intergenomic translocation breakpoints in natural populations of allopolyploid wild wheats. Ann. Bot. (Lond.) 107: 65–76. 10.1093/aob/mcq215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W. J., Larkin D. M., Everts-van der Wind A., Bourque G., Tesler G. et al. , 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617. 10.1126/science.1111387 [DOI] [PubMed] [Google Scholar]
- Nagaki K., Tanaka K., Yamaji N., Kobayashi H., and Murata M., 2015. Sunflower centromeres consist of a centromere-specific LINE and a chromosome-specific tandem repeat. Front. Plant Sci. 6: 912 10.3389/fpls.2015.00912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., and Barton N. H., 2003. Chromosomal speciation and molecular divergence–accelerated evolution in rearranged chromosomes. Science 300: 321–324. 10.1126/science.1080600 [DOI] [PubMed] [Google Scholar]
- Nguepjop J. R., Tossim H.-A., Bell J. M., Rami J.-F., Sharma S. et al. , 2016. Evidence of genomic exchanges between homeologous chromosomes in a cross of peanut with newly synthetized allotetraploid hybrids. Front. Plant Sci. 7: 1635 10.3389/fpls.2016.01635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas S. D., Mignon G. L., Eber F., Coriton O., Monod H. et al. , 2007. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175: 487–503. 10.1534/genetics.106.062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. A., Grams K. L., Bertucci L. A., and Reiland J., 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. 10.1073/pnas.221274498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita A. K., Zafar F., Su J., Weerasekara D., Kajitani T. et al. , 2019. Heterochromatin suppresses gross chromosomal rearrangements at centromeres by repressing Tfs1/TFIIS-dependent transcription. Commun. Biol. 2: 17 10.1038/s42003-018-0251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira da Silva W., Pieczarka J. C., Rodrigues da Costa M. J., Ferguson-Smith M. A., O’Brien P. C. M. et al. , 2019. Chromosomal phylogeny and comparative chromosome painting among Neacomys species (Rodentia, Sigmodontinae) from eastern Amazonia. BMC Evol. Biol. 19: 184 10.1186/s12862-019-1515-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostevik K. L., 2016. The ecology and genetics of adaptation and speciation in dune sunflowers. Ph.D. Thesis, University of Toronto, Toronto.
- Ouellette L. A., Reid R. W., Blanchard S. G., and Brouwer C. R., 2017. LinkageMapView - rendering high resolution linkage and QTL maps. Bioinformatics 34: 306–307. 10.1093/bioinformatics/btx576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palestis B. G., Burt A., Jones R. N., and Trivers R., 2004. B chromosomes are more frequent in mammals with acrocentric karyotypes: support for the theory of centromeric drive. Proc. Biol. Sci. 271: S22–S24. 10.1098/rsbl.2003.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P., and Tesler G., 2003. Genome rearrangements in mammalian evolution: lessons from human and mouse genomes. Genome Res. 13: 37–45. 10.1101/gr.757503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J. A., Brown P. J., Sorrells M. E., and Jannink J. L., 2012. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillet M. C., Madjidian N., Griveau Y., Serieys H., Tersac M. et al. , 1995. Mapping genetic factors controlling pollen viability in an interspecific cross in Helianthus sect. Helianthus. Theor. Appl. Genet. 91: 1195–1202. 10.1007/BF00220929 [DOI] [PubMed] [Google Scholar]
- Raduski A. R., Rieseberg L., and Strasburg J., 2010. Effective population size, gene flow, and species status in a narrow endemic sunflower, Helianthus neglectus, compared to its widespread sister species, H. petiolaris. IJMS 11: 492–506. 10.3390/ijms11020492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskina O., Barber J. C., Nevo E., and Belyayev A., 2008. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet. Genome Res. 120: 351–357. 10.1159/000121084 [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., 1991. Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. Am. J. Bot. 78: 1218–1237. 10.1002/j.1537-2197.1991.tb11415.x [DOI] [Google Scholar]
- Rieseberg L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358. 10.1016/S0169-5347(01)02187-5 [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Linder C. R., and Seiler G. J., 1995. Chromosomal and genic barriers to introgression in Helianthus. Genetics 141: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, C. E., T. E. Thompson, and G. J. Seiler, 1982 Sunflowers Species of the United States. National Sunflower Association, Mandan, ND. [Google Scholar]
- Rohland N., and Reich D., 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22: 939–946. 10.1101/gr.128124.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O., Kuster R., Miller K., Fouks B., Rubio Correa S. et al. , 2016. A new metazoan recombination rate record and consistently high recombination rates in the honey bee genus Apis accompanied by frequent inversions but not translocations. Genome Biol. Evol. 8: 3653–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambatti J. B. M., Strasburg J. L., Ortiz-Barrientos D., Baack E. J., and Rieseberg L. H., 2012. Reconciling extremely strong barriers with high levels of gene exchange in annual sunflowers. Evolution 66: 1459–1473. 10.1111/j.1558-5646.2011.01537.x [DOI] [PubMed] [Google Scholar]
- Schlautman B., Diaz-Garcia L., Covarrubias-Pazaran G., Schlautman N., Vorsa N. et al. , 2017. Comparative genetic mapping reveals synteny and collinearity between the American cranberry and diploid blueberry genomes. Mol. Breed. 38: 9. [Google Scholar]
- Schranz M. E., Mitchell-Olds T., and Lysak M. A., 2006. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542. 10.1016/j.tplants.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Searle J. B., 1993. Chromosomal hybrid zones in eutherian mammals, pp. 309–353 in Hybrid Zones and the Evolutionary Process, edited by Harrison R. G., Oxford University Press on Demand, Oxford. [Google Scholar]
- Shagina I., Bogdanova E., Mamedov I., Lebedev Y., Lukyanov S. et al. , 2010. Normalization of genomic DNA using duplex-specific nuclease. Biotechniques 48: 455–459. 10.2144/000113422 [DOI] [PubMed] [Google Scholar]
- Sinha A. U., and Meller J., 2007. Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinformatics 8: 82 10.1186/1471-2105-8-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C., Bomhoff M., and Nelson W. M., 2011. SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res. 39: e68 10.1093/nar/gkr123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathos A., and Fishman L., 2014. Chromosomal rearrangements directly cause underdominant F1 pollen sterility in Mimulus lewisii-Mimulus cardinalis hybrids. Evolution 68: 3109–3119. 10.1111/evo.12503 [DOI] [PubMed] [Google Scholar]
- Staton S. E., Bakken B. H., Blackman B. K., Chapman M. A., Kane N. C. et al. , 2012. The sunflower (Helianthus annuus L.) genome reflects a recent history of biased accumulation of transposable elements. Plant J. 72: 142–153. 10.1111/j.1365-313X.2012.05072.x [DOI] [PubMed] [Google Scholar]
- Stephens J. D., Rogers W. L., Mason C. M., Donovan L. A., and Malmberg R. L., 2015. Species tree estimation of diploid Helianthus (Asteraceae) using target enrichment. Am. J. Bot. 102: 910–920. 10.3732/ajb.1500031 [DOI] [PubMed] [Google Scholar]
- Strasburg J., and Rieseberg L., 2008. Molecular demographic history of the annual sunflowers Helianthus annuus and H. petiolaris—large effective population sizes and rates of long‐term gene flow. Evolution 62: 1936–1950. 10.1111/j.1558-5646.2008.00415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Yadav V., Billmyre R. B., Cuomo C. A., Nowrousian M. et al. , 2017. Fungal genome and mating system transitions facilitated by chromosomal translocations involving intercentromeric recombination. PLoS Biol. 15: e2002527 10.1371/journal.pbio.2002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Yu J. K., Slabaugh M. B., Shintani D. K., and Knapp S. J., 2002. Simple sequence repeat map of the sunflower genome. TAG Theoretical and Applied Genetics 105: 1124–1136. 10.1007/s00122-002-0989-y [DOI] [PubMed] [Google Scholar]
- Taylor J., and Butler D., 2017. R Package ASMap: efficient genetic linkage map construction and diagnosis. J. Stat. Softw. 79: 1–29. 10.18637/jss.v079.i0630220889 [DOI] [Google Scholar]
- Tennessen J. A., Govindarajulu R., Ashman T. L., and Liston A., 2014. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 6: 3295–3313. 10.1093/gbe/evu261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., G. L. Owens, N. Bercovich, J. S. Légaré, S. Soudi et al. 2019 Massive haplotypes underlie ecotypic differentiation in sunflowers. bioRxiv. Available at: https://www.biorxiv.org/content/10.1101/790279v1. Accessed December 30, 2019. doi: 10.1101/79027 10.1101/79027 [DOI] [PubMed]
- Trickett A. J., and Butlin R. K., 1994. Recombination suppressors and the evolution of new species. Heredity 73: 339–345. 10.1038/hdy.1994.180 [DOI] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G. et al. , 2013. From FastQ data to high-confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C., Fustier M. A., Alix K., and Tenaillon M. I., 2014. The bright side of transposons in crop evolution. Brief. Funct. Genomics 13: 276–295. 10.1093/bfgp/elu002 [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss, H., and G. M. Schneeweiss, 2012 Karyotype diversity and evolutionary trends in angiosperms, pp. 209–230 in Plant Genome Diversity Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes. Edited by I. J. Leitch, J. Greilhuber, J. Doležel, and J. F. Wendel. Springer-Verlag, Heidelberg. [Google Scholar]
- Whelan E. D., 1979. Interspecific hybrids between Helianthus petiolaris Nutt. and H. annuus L.: effect of backcrossing on meiosis. Euphytica 28: 297–308. 10.1007/BF00056586 [DOI] [Google Scholar]
- White M. J. D., 1973. Animal Cytology and Evolution. Cambridge University Press, Cambridge. [Google Scholar]
- White M. J. D., 1978. Modes of Speciation. W. H. Freeman & Co., San Francisco. [Google Scholar]
- Wu F., and Tanksley S. D., 2010. Chromosomal evolution in the plant family Solanaceae. BMC Genomics 11: 182 10.1186/1471-2164-11-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S., 2013. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc. Natl. Acad. Sci. USA 110: E1743–E1751. 10.1073/pnas.1219381110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S., and Whitlock M., 2011. The genetic architecture of adaptation under migration-selection balance. Evolution 65: 1897–1911. 10.1111/j.1558-5646.2011.01269.x [DOI] [PubMed] [Google Scholar]
- Yogeeswaran K., Frary A., York T. L., Amenta A., Lesser A. H. et al. , 2005. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15: 505–515. 10.1101/gr.3436305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S. E., Eickbush M. T., Yu J. S., Kang J. W., Fowler K. R. et al. , 2014. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. Elife 3: e02630 10.7554/eLife.02630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulidov P. A., Bogdanova E. A., Shcheglov A. S., Vagner L. L., Khaspekov G. L. et al. , 2004. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 32: e37 10.1093/nar/gnh031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The R program, syntR, is available on GitHub: https://github.com/ksamuk/syntR. The sequences used to generate genetic maps are available at the Sequence Read Archive: http://www.ncbi.nlm.nih.gov/bioproject/598366. All other data and scripts are available on dryad: https://doi.org/10.5061/dryad.7sqv9s4pc. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11819625.