Abstract
Implanted bioengineered livers have not exceeded three days of continuous perfusion. Here, we show that decellularized whole porcine livers revascularized with human umbilical endothelial cells and implanted heterotopically into immunosuppressed pigs whose spleen has been removed can sustain perfusion for up to 15 days. We identified peak glucose consumption rate as a main predictor of the patency of the revascularized bioengineered livers (rBELs). On heterotopic implantation of the rBELs into pigs in the absence of anticoagulation therapy led to sustained perfusion for 3 days, followed by significant immune responses directed against the human endothelial cells. A 10-day steroid-based immunosuppression protocol and a splenectomy at time of rBEL implantation reduced the immune responses and resulted in continuous perfusion of the rBELs for over two weeks. We also show that the human endothelial cells in the perfused rBELs colonize the liver sinusoids and express sinusoidal endothelial markers similar to those in normal liver tissue. Revascularized liver scaffolds that can maintain blood perfusion at physiological pressures might eventually help overcome the chronic shortage of transplantable human livers.
Liver transplantation currently represents the only treatment for end stage liver disease (ESLD), though a chronic shortage of viable donor organ material continues to limit the number of lifesaving liver transplants that can be performed. Recent advances in tissue engineering methods have accelerated the development of bioengineered organs as an alternative source of donor material, though the challenge of developing functional vasculature to support cellular metabolism, gas-exchange and sustained perfusion in vivo has remained a major barrier to the clinical translation of these technologies1. Bioengineered liver (BEL) constructs have been created through the recellularization of acellular liver scaffolds and have demonstrated in vitro functionality2–8. Short-term in vivo function has also been reported following transplantation in small animal models9, though rapid vascular thrombosis has limited the duration of in vivo graft patency. Passivation methods and high doses of anticoagulants have been employed to extend the in vivo patency of BELs, though these approaches have only modestly extended thromboresistance6,10,11. To date, no reported methods have demonstrated a capacity to exceed three days of continuous BEL perfusion in-vivo.
Decellularized whole liver matrix holds great potential as a technology for developing a theoretically limitless supply of functional BELs12,13. While several groups have demonstrated an ability to seed a variety of liver-specific cell types into decellularized liver constructs12,14–16, reconstituting the endothelial cell lining within the vascular networks in these scaffolds has remained a significant challenge to the development of a therapeutic BEL17.
Here, we report an optimized method for revascularizing a clinically-scaled porcine liver scaffold. Using human umbilical endothelial cells (HUVECs) as a primary cell source, we characterized the kinetics of endothelial cell proliferation within decellularized liver scaffolds during perfusion bioreactor culture and identified glucose consumption rate (GCR) as a reliable metric for non-destructively monitoring cell proliferation, and ultimately predicting graft patency upon acute blood perfusion. Histological characterization of revascularized bioengineered livers (rBELs) revealed that HUVECs localized within sinusoidal regions upregulated expression of LYVE1 and downregulated CD31 expression over time, suggesting a transition toward a liver sinusoidal endothelial cell (LSEC)-like phenotype. Transcript analysis of rBEL samples revealed global changes in gene expression in grafts with high GCRs, including upregulation of additional LSEC markers, and transmission electron microscopy (TEM) revealed the existence of fenestrae-like structures in endothelial cells localized within sinusoidal regions. Finally, up to 15 days of continuous in vivo rBEL perfusion was achieved under a steroid-based immunosuppression protocol following heterotopic implantation in a large animal liver transplant model. Characterization of the recipient animal immune response following graft implantation revealed the presence of HUVEC-reactive antibodies and identified complement-activation as a likely driver of graft rejection and eventual thrombosis. Taken together, this work demonstrates to our knowledge the longest-to-date example of in vivo perfusion of a BEL, and establishes a method for future recellularization studies aimed at bioengineering a functional liver construct for eventual translation into the clinic.
Results
Liver decellularization and perfusion bioreactor culture
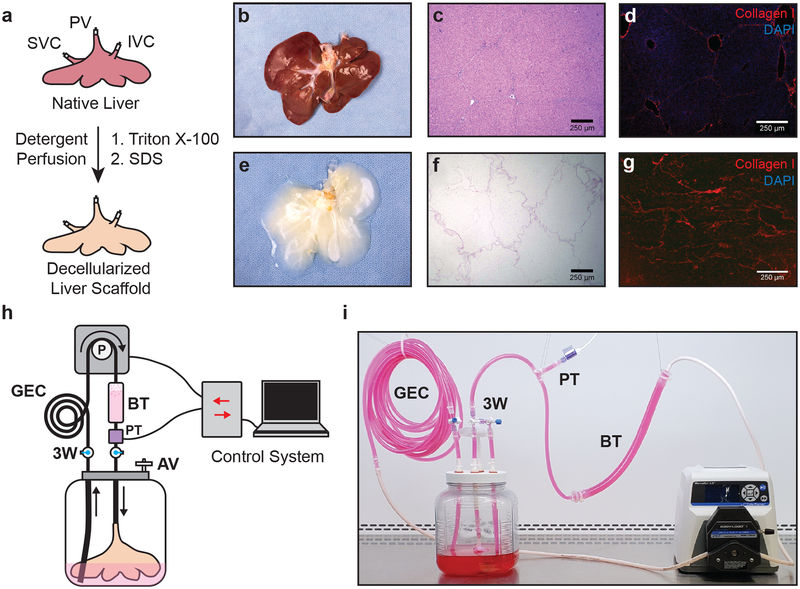
Porcine livers utilized in this study were cannulated on the portal vein (PV), infrahepatic inferior vena cava (IVC), and suprahepatic vena cava (SVC), and decellularized by sequential perfusion with Triton X-100 solutions and sodium dodecyl sulfate (SDS) solutions to remove cellular material while preserving the overall architecture of the scaffold (Fig. 1a, b, e). Histological sectioning from representative decellularized scaffolds confirmed the maintenance of parenchymal liver lobule structures when compared to that of native porcine liver tissue (Fig. 1c, f), as well as retention of Collagen I (Fig. 1d, g).
Figure 1. Porcine liver decellularization and perfusion bioreactor system.
(a) Native porcine livers are cannulated on the PV, IVC and SVC, and decellularized by sequential perfusion with Triton X-100 and SDS solutions. (b-g) Representative photographs, H&E staining and Collagen I immunofluorescence of native (b-d) and decellularized (e-g) porcine livers. (h, i) Schematic (h) and photo (i) of perfusion bioreactor system comprised of a custom bioreactor and a pressure-dependent perfusion control system. The bioreactor includes a pressure transducer (PT) to monitor perfusion pressure, a gas exchange coil (GEC) to allow efficient gas exchange during media perfusion, a bubble trap (BT) to prevent the introduction of bubbles into the rBEL, a 0.22 μm filter air vent (AV), and three-way stopcocks (3W) to enable media exchange and sampling.
Decellularized liver scaffolds were mounted in custom bioreactors (Fig. 1h, i) and perfused with culture media through the SVC at a constant inflow pressure of 12 mmHg at 37°C and 5% CO2. Following 72 h of continuous media perfusion to precondition the scaffold and confirm the absence of viable bioburden, 1.5×108 HUVECs were infused through the perfusion circuit into the SVC vasculature (Fig. 2a). Following 24 h of continuous media perfusion, liver scaffolds were aseptically manipulated and infused with an additional 1.5×108 HUVECs through the PV (Fig. 2a). Prior to seeding, purity of the HUVEC cultures were confirmed by CD31+ flow cytometry (Fig. 2b). Culture media was continuously perfused through the PV at 12 mmHg for the remaining period of bioreactor culture.
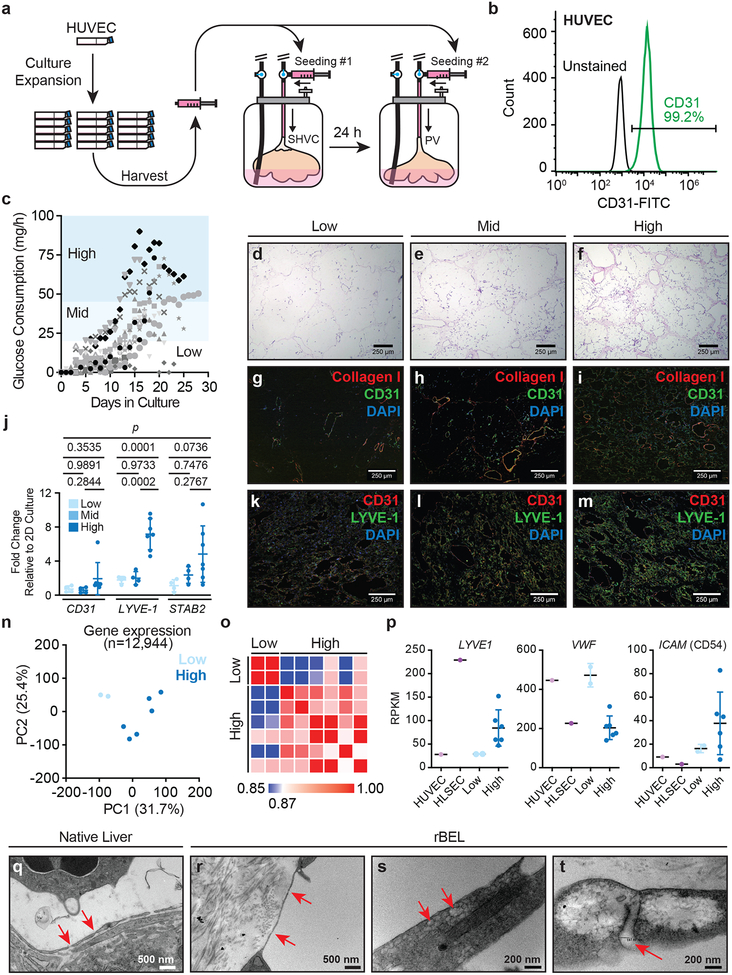
Figure 2. Analysis of rBEL culture kinetics and HUVEC phenotypic plasticity in decellularized liver matrix.
(a) HUVECs are expanded in 2D tissue culture flasks, harvested and seeded through the graft SVC, followed by the PV 24 hours later. (b) Representative CD31+ flow cytometry demonstrating a phenotypically pure HUVEC population immediately prior to graft seeding. (c) Plots of glucose consumption rates over time from independently seeded and cultured rBEL constructs (n=14). Peak glucose consumption rates correlated with total endothelial cell coverage as characterized by H&E staining (d-f) and anti-CD31 immunostaining (g-i). (j) Quantitative RT-PCR analysis of CD31, LYVE1 and STAB2 mRNA levels in rBELs harvested at low (n=4), mid (n=4) and high (n=7) glucose consumption phases Data are plotted as fold change relative to HUVECs in 2D culture. Individual values represent biological replicates. Mean values ± standard deviation are shown. Statistical significance was determined using a one-way ANOVA test from dCT values prior to fold-change normalization. (k-m) CD31 and LYVE-1 immunostaining from rBELs harvested at low, mid and high glucose consumption phases.(n) Principal component analysis of RNA-seq gene expression profiles from rBELs harvested at low (n=2 biological replicates) and high (n=6 biological replicates) glucose consumption phases. (o) Similarity matrix of BEL samples from (n) comparing low and high glucose consumption phase rBEL samples with respect to panel of known liver endothelial cell markers (input genes: F8, CD31, STAB2, LYVE1, CD14, VWF, ENG, ICAM1).(p) RNA-seq expression profiles for liver sinusoidal endothelial markers LYVE1, VWF, and ICAM1 in low (n=2) and high (n=6) glucose consumption phase rBEL samples. HUVECs (n=1) and primary human LSECs (n=1) cultured in 2D are included for comparison. Biological replicates are plotted along with the mean ± standard deviation. (q-t) TEM images from native liver (q) and rBEL (r-t) samples. Red arrows indicate fenestrae-like structures within endothelial cells.
Characterization of HUVEC proliferation
To define quantitative markers for non-invasively monitoring endothelial cell proliferation in the liver scaffold, a panel of metabolites (glucose, lactate, glutamate, and ammonia) were measured daily from a sample of rBEL culture media (data not shown). GCR measured throughout the period of bioreactor culture exhibited sigmoidal kinetics in most rBELs and could be generally characterized by low (<20 mg/h), mid (20–45 mg/h), and high (>45 mg/h) GCR phases (Fig. 2c). Histological examination of representative rBELs with low, mid, and high GCRs correlated with increasing endothelial cell densities as inferred by H&E staining, with evidence of primary engraftment in larger vessels and subsequent expansion and migration into the parenchymal or sinusoidal niche at mid and high GCRs (Fig. 2d–f). As a result, rBEL GCRs were utilized as a metric for estimating the extent of graft re-endothelialization in later parts of this study. During bioreactor culture, media volumes were adjusted and replaced daily to maintain steady state glucose levels above 500 mg/L (>50% of baseline media concentrations) to ensure consistent proliferation kinetics and discourage premature cell senescence due to glucose starvation18. Two rBELs failed to exhibit sigmoidal kinetics and achieve a GCR rate >20 mg/h (Fig. 2c) and demonstrated similar proliferation kinetics to significantly under seeded rBELs (data not shown), likely a result of an undetected hole in the portal or hepatic vein during seeding.
Phenotypic plasticity of HUVECs in rBELs
Immunostaining of rBELs during low, mid and high GCR phases with anti-CD31 and anti-Collagen I antibodies (Fig. 2g–i, Supplementary Fig. S1a) revealed HUVEC localization within vascular structures and overall uniform cell distribution within the decellularized liver matrix. Endothelial cell engraftment was primarily localized within larger vessels during the low GCR phase following seeding, followed by an increase in cell proliferation within sinusoidal regions at mid and high GCR phases. Immunostaining for LYVE1, a marker expressed by LSECs, demonstrated highest expression in the parenchymal sinusoids with little expression in larger vessels (Fig. 2k–m, Supplementary Fig. S1b) consistent with native liver sinusoid staining19. The expression and localization of LYVE1 was weakly detected during the low GCR phase and became progressively stronger in the mid and high GCR phases. Transcript levels of LSEC-associated markers measured by qRT-PCR at high GCR phase demonstrated an upregulation of LYVE1 (7.2-fold +/− 1.8, n=7, mean ± std) and STAB2 (4.8-fold +/− 3.31, n=7) compared to HUVEC cells in 2D culture, as well as a significant increase from low to high phases (p<0.05) for LYVE1, while CD31 was not significantly changed (1.9-fold +/− 1.8, n=7) (Fig. 1j). Global characterization of rBEL samples from low and high GCR phases through RNA-seq analysis revealed significant changes in gene expression profiles over time as demonstrated by a global principle component analysis (Fig. 2n) and targeted similarity analysis using known liver endothelial cell markers20 (Fig. 2o, Supplementary Dataset 3). Further analysis of the RNA-seq datasets confirmed upregulation of LYVE1, and additionally showed downregulation of VWF and upregulation of ICAM1 in high GCR samples (Fig. 2p), revealing additional expression trends that resemble recently reported primary human LSEC transcript profiles20.
A hallmark feature of LSECs in normal liver tissue is the presence of plasma membrane fenestrations which enable diffusion of nutrients and waste products between the capillary vessels and the adjacent parenchymal space. To determine whether endothelial cells localized within sinusoids of rBEL constructs exhibited such features, TEM was performed on samples from native porcine liver tissue (Fig. 2q) and high GCR phase rBELs (Fig. 2r–t). Micrographs from rBELs exhibited fenestrae-like structures similar to those observed in native porcine liver sections. The quantified dimensions of these features were consistent with those of LSEC fenestrations (100‐150 nm)21. Collectively, these results reveal a new dimension of HUVEC phenotypic plasticity22,23 and suggest that distinct microenvironments in decellularized liver matrix may have the capacity to direct phenotypic differentiation of endothelial cells.
Peak GCR in rBELs correlates with patency
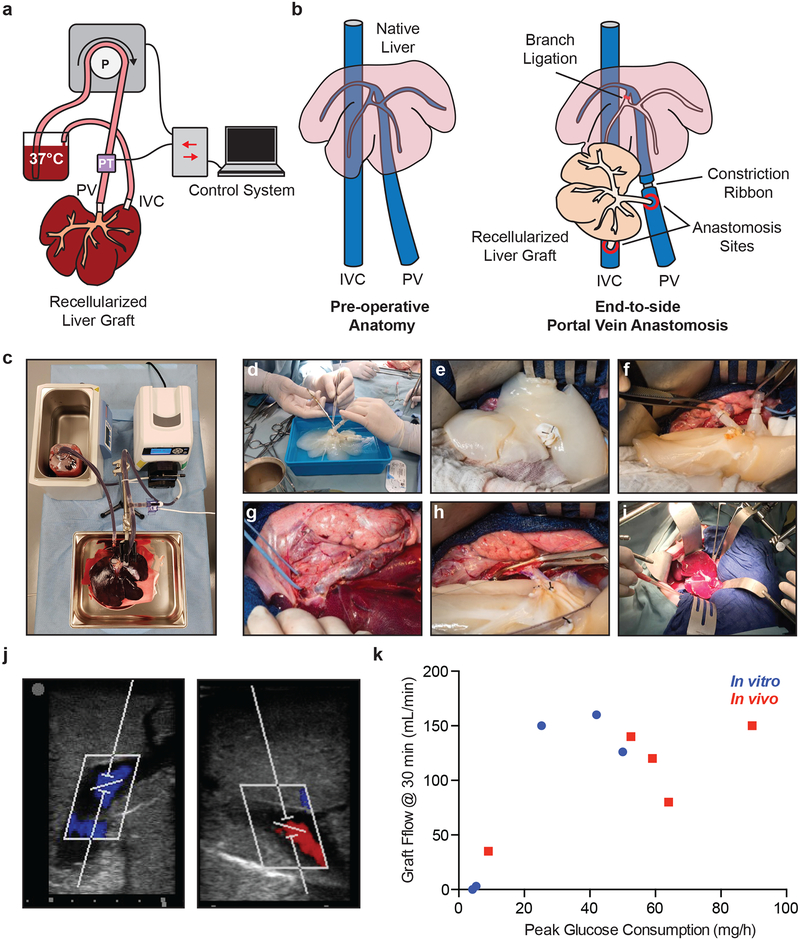
To assess the patency of rBELs, an ex vivo blood loop circuit utilizing fresh heparinized porcine blood was employed (Fig. 3a, c). Using a peristaltic pump, pre-warmed blood (37°C) was perfused through the PV of the rBEL and returned to the blood reservoir following outflow from the IVC. Perfusion was maintained at a constant pressure of 12 mmHg and flow rates were monitored over time. Flow rates <50ml/min after 30 minutes were deemed inadequate for in vivo perfusion. Evaluation of non-seeded decellularized liver scaffolds consistently resulted in flow rates <10 ml/min after 5 minutes and had zero flow after 15 minutes (data not shown). Peak glucose consumption rate (PGCR) is the maximum GCR measured during BEL culture with rates >30 mg/h in rBELs correlated with sustained flow rates >50 ml/min, thereby validating the use of the PGCR as a marker for functional re-endothelialization of rBELs (n=5) (Fig. 3k). Histological evaluation of low glucose consuming grafts following blood perfusion and saline flushing showed blood pooling and compaction within the graft, while grafts with PGCRs >30 mg/h were efficiently cleared with saline (Supplementary Fig. S2).
Figure 3. In vitro and in vivo patency correlates with peak glucose consumption rate.
(a, c) Diagram and setup of the in vitro blood circuit used to evaluate rBEL patency. The circuit perfuses a rBEL with warm, heparinized whole porcine blood and is driven by a peristaltic pump controlled by a pressure-based custom control system. (b) Illustration of in vivo heterotopic liver implant model where the rBEL is anastomosed via the PV and IVC to the native PV and IVC. Partial flow was given to both the rBEL and the native liver by restricting flow to the native liver. (d-i) Representative images of the heterotopic liver implant including graft preparation, anastomosis and reperfusion. (j) Representative ultrasound images of an implanted rBEL demonstrating portal and hepatic veinous flow after 30 min. (k) Flow rates from in vitro (n=5) and in vivo (n=5) perfusion studies. Values from independent rBELs are plotted. Peak glucose consumption of >30 mg/h correlates with >100 mL/min of blood flow in vitro and in vivo.
To determine the value of PGCR in predicting rBEL patency in vivo, a large animal porcine model for auxiliary liver transplantation was established to enable the implantation and patency assessment of rBELs (n=5). To this end, rBELs were implanted with end-to-side anastomoses between the graft’s and recipient pig’s portal veins (Fig. 3b, d–i). Prior to perfusing the rBELs, Portal branches supplying the pig’s native liver were tied off preserving the first portal branch supplying the caudate lobe and the right lateral lobe. A constriction ribbon was also applied to the pig’s portal vein distal to the anastomosis to partially bias flow through the rBEL. These measures were taken to raise the portal pressure and facilitate preferential blood flow to the implanted rBELs. The portal vein surgical model provided mean venous pressure of 8.4 ± 2.2 mmHg (n=5) (mean +/− s.d.) and mean blood flow rates of 414 ± 16.7 ml/min (n=5) (mean +/− s.d.). rBELs were implanted and monitored for 30 minutes with inflow and outflow confirmation via Doppler ultrasound (Fig. 3j). Vascular perfusion was assessed through direct measurement 30 minutes after anastomosis through direct volumetric measurement of outflow blood for 60 seconds, which was subsequently returned to the circuit. PGCR >30 mg/h demonstrated >100 ml/min of perfusion after 30 minutes in 3 of 4 rBELs (Fig. 3k) further confirming the correlation between PGCR and in vivo graft patency.
Long-term in vivo perfusion in an immunosuppressed porcine liver transplant model
To assess long-term patency in vivo, rBELs were implanted utilizing the previously described auxiliary liver transplantation model (Fig. 3b, Supplementary Movie S1) and recipient animals were recovered without the addition of post-operative anti-platelet or anti-coagulation therapies. To determine the impact of a host immune response directed toward the HUVEC-component of the rBELs on eventual graft failure, recipient animals were divided into two cohorts (n=4 per condition) One group underwent an immunosuppressive therapy regimen, and the other received no additional treatment (Excluded were 3 additional non-immunosuppressed implants and one additional immunosuppressed implants that experienced immediate – within 24 hours–graft loss attributable to surgical complications). In the immunosuppressed group prior to rBEL implantation, surgical splenectomy was performed, and intravenous methylprednisolone was administrated at 500 mg with subsequent daily doses of 500 mg, 250 mg, 250 mg, 125 mg, 125 mg, 80 mg, 60 mg, 40 mg, 30 mg and 20 mg (Fig. 4a). CT imaging with intravenous contrast was performed post-operatively on days 0, 1, 3, 7, 10, 15, and 20 to assess the extent of perfusion through the rBELs (Fig. 4a–c). Perfusion following each CT imaging time point was quantified through computed tomography volumetric measurements using SIEMENS MultiModality Workstation Software. Graft volume was manually marked and the perfused area was auto detected through Hounsfield threshold cutoff. 3D reconstruction was performed using TeraRecon medical imaging software as well as the assistance of 3D visualizations created with Analyze24 (Fig. 4b, Supplementary Movie S2). The percentage of the reduction in perfusion of the rBELs from baseline postoperative CT scan was calculated and plotted (Fig. 4d, Supplementary Fig. S4). In the absence of immunosuppression, all four implanted rBELs lost >85% of their initial perfusion by day 7 post-transplant. In contrast, the immunosuppressed group had significantly longer graft perfusion and vascular patency when compared to the group without immunosuppression, 8.5 [7–15] (Median [Range]) versus 3 [1–7] days; p=0.037, and 11 [7–20] versus 3 [1–7] days; p=0.037, respectively (Supplementary Table S1). Complete blood count (CBC), liver function, renal function and coagulation factors were followed pre-operatively as well as at post-operative days 1, 3, 7, 10, 15, and 20 (Supplementary Table S2). One rBEL was harvested from an immunosuppressed recipient animal at day 7 for histological analysis which demonstrated persistence of the HUVEC populations in the graft (Supplementary Fig. S3a, b). The other 3 grafts were monitored by CT imaging until total loss of graft perfusion, which was observed on days 10, 15, and 20, respectively. Total loss of porto-venous flow was seen on days 10, 20 while one graft continued to have some portal-venous flow through the graft despite total loss of parenchymal perfusion (Fig. 4d). Evidence of rBEL perfusion was present in all of the grafts in the immunosuppressed group at day 7, demonstrating a significant increase in sustained perfusion over the non-immune suppressed group (p=0.01).
Figure 4. Long term in vivo perfusion studies in the presence and absence of immunosuppression.
(a) In vivo implants were separated into two groups: no treatment and immunosuppression. The immunosuppression group received a methylprednisolone immunosuppression dose (I.D.) starting at 500 mg on Day 0 and was tapered over ten days. (b) 3D CT reconstruction after graft implantation showing the heterotopic position of the implanted graft below the native liver while demonstrating good vascular perfusion of the implanted graft. (c) Serial contrast enhanced CT images of the implanted bioengineered liver grafts over time. Grafts are highlighted with dotted lines. Yellow dotted lines delineate perfused graft with contrast in white. Red dotted lines reflect no parenchymal perfusion. (d) Quantification of graft perfusion reduction over time in untreated (n=4) and immunosuppressed (n=4) animals. (e) Cytotoxicity of pig sera from untreated (n=4) and immunosuppressed (n=4) animals incubated on in vitro HUVEC cultures following addition of unabsorbed rabbit complement.
Early immune response to rBEL xenotransplantation
The native pig immune response to HUVEC cells was characterized to confirm if the high rate of graft failure between Days 3 and 7 was associated with an immune response to the HUVECs used to revascularize the rBELs. Pig serum was collected at each CT scan and incubated with HUVEC cultures to perform a complement mediated cytotoxicity assay25. Complement mediated cytotoxicity reaction was observed between naïve pig sera and HUVECs at baseline (range 30–85% cell death) demonstrating an inherent immune response to the human-derived cells without graft exposure in both no treatment and immunosuppressed groups (Fig. 4e). Evidence of an in vivo complement activation was observed by C4D deposition on endothelial cells in explanted rBEL samples (Supplementary Fig. S3c).
Cytotoxicity significantly increased in the no treatment by Day 3 (81.7 ± 21.0) (mean +/− s.d.) and remained at >98% following Day 3. In contrast, immunosuppression significantly reduced cytotoxicity at Day 1 and Day 3 25.2 (±29.4), and 8.68 (±8.49) respectively, followed by a notable increase in cytotoxicity at day 7 84.7 (±13.0) and >98% cytotoxicity at Day 10 and Day 15 (Fig 4e). rBEL perfusion in the no treatment and immunosuppression groups correlated to cytotoxicity responses. Decreased rBEL perfusion was preceded by a significant increase in an immune response as seen on Day 3 for no treatment and Day 7 for the immunosuppressed group (Fig 4d, e).
Discussion
The only cure for ESLD, the 8th most frequent cause of death in the United States, is liver transplantation. Unfortunately, the demand for transplantable donor livers continues to exceed the available supply, and as a result, only half of the 11,000 wait-listed patients will receive transplants26. Globally, it is estimated that over 1 million patients could benefit from a liver transplant27. The insufficient supply in transplantable donor livers underscores the need for alternative strategies to generate functional liver tissue for patients suffering from liver failure.
Efforts to overcome the chronic shortage of transplantable human livers include attempts at alternative organ procurement and replacement strategies28. Xenogeneic whole organ transplantation promises a theoretically limitless supply of donor organ material, but immunological incompatibility, fundamental differences in basal metabolism between humans and other species, and coagulopathy remain significant translational barriers6,17,29. Hepatocyte transplantation represents a currently approved alternative to whole organ replacement; however, poor cell engraftment efficiency and deterioration of cell function over time has largely relegated this to a bridge therapy and temporary metabolic support ahead of whole liver transplantation30.
Tissue engineering remains a promising solution to the chronic organ shortage but has been significantly inhibited by lack of functional vasculature required to support implantation of engineered tissue similar to donor grafts. Whole organ perfusion decellularization and recellularization technologies have demonstrated the ability to maintain the native architecture and have demonstrated organ function from the recellularization of liver6,31,32, kidney33,34 and lung35, However, none have demonstrated the ability to sustain long-term in vivo perfusion because of incomplete vasculature, a critical component to move the field forward.
The daily measurement and analysis of metabolic markers, specifically daily GCRs, provided the opportunity for critical adjustments for bioreactor media volume and provided growth parameters that were evaluated as surrogates for re-endothelialization. PGCR not only indicates the level of rBEL endothelialization in culture.36, but is also predictive of in vivo performance. The resulting function of the rBEL was independent of days in culture, but instead dependent upon metabolic activity. It appears that these phases correlated with increased endothelial coverage starting in the larger vessels followed by proliferation and migration into the parenchymal space during mid and high GCR phases. The defined PGCR levels were characterized for 250 to 350g livers and it is expected that moving to larger livers will require a higher minimum PGCR threshold.
During this time, LYVE1 expression and localization was increased in the parenchymal space, with little expression in larger vessels, the inverse was observed with the localization of CD31, suggesting a degree of phenotypic plasticity of HUVECs within the decellularized liver scaffold. Global gene expression via RNA-seq analysis on isolated rBEL sections characterized a shift in gene expression between low and high glucose consuming rBELs and revealed additional gene expression trends consistent with an LSEC-like phenotype. To provide physical evidence of fenestrations, a key hallmark of LSECs, TEM evaluation of rBELs exhibited fenestrae-like features similar in size to those observed in native liver sections. The combined molecular analysis and microscopic examination support the shift towards an LSEC-like phenotype and will need to be further characterized following the addition of hepatocytes and cholangiocytes to the rBELs.
To assess the deleterious effects of a host immune response to human cells, a detection of the xeno-compatibility phenomenon between pig serum and human cells was employed. A non-sophisticated 10-day immunosuppression protocol was used to demonstrate the ability to increase the vascular perfusion of the rBELs from, on average, 3.5 days up to 15 days and extended the patency of the vascular tree beyond 15 days. The immunosuppression protocol used in this study was not intended to provide long-lasting immunosuppression, but rather to determine whether a species-dependent immunological response was contributing to the loss of graft perfusion. Vascular patency was prolonged using the immunosuppression regimen indicating that xeno-incompatibility played a significant role in eventual rBEL thrombosis in vivo. Following immunosuppression withdrawal, similar kinetics for graft thrombosis to the non-immunosuppressed group were observed. Vascular patency in the humanized bioengineered liver grafts would have likely been sustained longer with continued immunosuppressive therapy.
In this study, we demonstrate an ability to produce rBEL constructs with functional vasculature that can retain long-term in vivo vascular patency leading to definitive functional testing of co-seeded whole liver grafts with parenchymal hepatocytes and cholangiocytes. Adequate endothelization is sufficient to prevent vascular thrombosis thus providing a path forward to engineering a fully functional transplantable liver.
Methods
Decellularization of porcine livers.
Whole livers (250 to 350 grams) were excised from cadaveric pigs. The Suprahepatic Vena Cava (SVC), Inferior Vena Cava (IVC), Portal Vein (PV), and Bile Duct were cannulated and flushed with 150 ml of sterile saline. The cannulated livers were perfusion decellularized with 1× Triton X-100 (Amresco, M143) for 2–5 hours followed by 0.6% sodium dodecyl sulfate (Amresco, 0227) for 4–8 hours at a perfusion pressure maintained between 8–12 mmHg. The decellularized livers were disinfected with 1000 ppm peracetic acid (PAA; U.S. Water, BI0032–6). The decellularized grafts were washed with phosphate buffered saline (PBS; Corning 21–040-CMX12) and stored. All decellularization was completed in an ISO 7 cleanroom. Decellularization and recellularization utilized a custom-built perfusion system to automatically adjust flow to maintain a defined pressure utilizing Cole-Palmer peristaltic pumps (7575–30, 77200–60).
Decellularization and recellularization utilized a custom-built perfusion system to automatically adjust flow to maintain a defined pressure utilizing Cole-Palmer peristaltic pumps (7575–30, 77200–60).
HUVEC cell culture and seeding of decellularized liver constructs.
Human umbilical vein endothelial cells (Lonza, C2517A) were cultured in antibiotic-free EGM-2 (Lonza, CC-3162) medium in tissue culture flasks (Falcon) at 37°C and 5% CO2 and passaged with 0.25% trypsin (Thermo, 25200056) at 90–100% confluency according to manufacturer’s protocol. The highest passage used for seeding liver grafts was passage 11.
The medium used for HUVEC culture was also used for seeding and maintaining re-vascularized liver constructs in this study. Decellularized porcine livers were placed in a custom bioreactor containing 800ml of media, connected to the perfusion inlet via the SVC, and perfused at 12 mmHg with culture media prior to seeding. 1.5×108 HUVECs were re-suspended in 100 ml of media and seeded through the SVC followed by 50 ml of fresh media to clear the measured tubing void volume. The infused cell suspension was left under static conditions for one hour and then continuous perfusion was restarted. After 24 hours, perfusion was changed from the SVC to the PV and the seeding protocol was repeated with an additional1.5×108 HUVECs. Re-endothelialized grafts were maintained in a continuous perfusion loop with metabolites (glucose, lactate, glutamine, glutamate and ammonia) monitored daily in collected media samples using a BioProfile FLEX analyzer (Nova Biomedical). Culture media was exchanged and the volume increased depending on the rate of glucose depletion in the circulating medium to ensure 24 hour glucose levels above 500 mg/L. All liver bioreactors were cultured in a humidified 37⁰C incubator with 5% CO2.
Histological Analysis.
Tissue samples analyzed in this study were perfused with PBS and fixed with 10% Neutral Buffered Formalin (VWR 16004–128). Fixed tissues were paraffin embedded, sectioned and stained using standard histologic techniques. Immunofluorescence slides were deparaffinized, rehydrated and retrieval was performed in citrate buffer, pH 6.0 (Abcam AB93678) in a programmable decloaker (Biocare DC2012). Slides were permeabilized with PBS + 0.05% Tween-20 (Sigma P9416) and blocked with Sea Block (Thermo 37527). Primary antibodies used included mouse anti-CD31 (Abcam AB187377), rabbit anti-Collagen I (Abcam AB34710), rabbit anti-LYVE1 (Abcam AB33682), and mouse anti-C4D (Abcam AB90804). Secondary antibodies used were goat anti-rabbit Alexa Fluor 488 (Thermo A11078), goat anti-mouse Alexa Fluor 488 (Thermo A11029), and goat anti-rabbit Alexa Fluor 555 (Thermo A21429). Slides were stained with 4’,6-diamidino-2-phenylindole (Thermo D1306) diluted 1:200 in PBS and mounted using ProLong Antifade Mountant (Thermo P36961). Fluorescence slides were imaged on a Zeiss Axioskop 40 and H&E slides were imaged on an Accuscope 3012.
Transmission Electron Microscopy.
Tissue was fixed with 4% paraformaldehyde + 1% glutaraldehyde fix in phosphate buffered saline, pH 7.2. Following fixation, cells were stained with 1% osmium tetroxide and 2% uranyl acetate, dehydrated through an ethanol series and embedded into Embed 812 resin. After a 24 h polymerization at 60°C, 0.1 μm ultrathin sections were post-stained with lead citrate. Micrographs were acquired using a JEOL1400+ transmission electron microscope (Peabody, MA) operating at 80kV with a Gatan Orius camera and Digital Micrograph software (Pleasanton, CA).
RNA extraction and quantitative reverse-transcription PCR (qRT-PCR).
RNA isolation was performed using TRIzol Reagent (Thermo Fisher) and transcribed to cDNA using the Superscript III First-Strand Synthesis System (Invitrogen). Gene expression analysis was performed using the Platinum SYBR green qRT-PCR supermix-UDG kit (Invitrogen) in a ViiA 7 Real-Time PCR instrument (Thermo Fisher Scientific). Ribosomal protein L19 (RPL19) was used as a housekeeping gene for normalization. The following primer sets were used in this study: RPL19 5’ATTGGTCTCATTGGGGTCTAAC3’, 5’AGTATGCTCAGGCTTCAGAAGA3’; STAB2 5’GCAAGAAGATGTGATAGGAAGTCTC3’, 5’ACAACACCGAGGTTGGAGAT3’, LYVE1 5’TTTGCAGCCTATTGTTACAACTCAT3’, 5’GGGATGCCACCCAGTAGGTA3’ and CD31 5’TCTGCACTG CAGGTATTGACAA, 5’CTGATCGATTCGCAACGGA3’.
RNA-seq analysis.
Tissue samples from low GCR (n=2) and high GCR (n=6), along with HUVECs (n=1) and human LSECs (Cell Systems, ACBRI 566) (n=1) were processed for RNA-seq analysis. RNA isolation was performed using TRIzol Reagent (Thermo Fisher). mRNA isolation for all samples was performed using the Direct-zol RNA Miniprep Kit (Zymo Research) and quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher).
Samples were assessed for RNA integrity (RIN) using the Agilent Bioanalyzer DNA 1000 chip (Invitrogen). Only samples with RIN scores > 6 and DV200 > 50% were selected for sequencing. RNA-sequencing and subsequent primary and secondary data analysis was performed as previously described37,38. In brief, library preparation was performed using the TruSeq RNA library preparation kit (Illumina). Polyadenylated mRNAs were selected using oligo dT magnetic beads. TruSeq Kits were used for indexing to permit multiplex sample loading on the flow cells and paired-end sequencing reads were generated on the Illumina HiSeq 2000 sequencer. Quality control for concentration and library size distribution was performed using an Agilent Bioanalyzer DNA 1000 chip and Qubit fluorometry (Invitrogen). Sequence alignment of reads and determination of normalized gene counts were performed using the MAP-RSeq (v.1.2.1) workflow, utilizing TopHat 2.0.639, and HTSeq40. Normalized read counts were expressed as reads per kilobasepair per million mapped reads (RPKM).
All genes with an average expression > 0.3 RPKM in at least one group (n=12,944) were utilized for subsequent analyses. Principal Component Analysis (PCA) was performed using ClustVis online tool41. Similarity matrix and hierarchical clustering analysis was performed using Morpheus matrix visualization and analysis (Morpheus, https://software.broadinstitute.org/morpheus, Broad Institute). Functional annotation and Gene Ontology (GO) term enrichment scores were calculated using DAVID Bioinformatics Resources 6.8 database (DAVID 6.8)42. Results from the DAVID analysis (Supplementary Dataset 1), a list of upregulated genes (Supplementary Dataset 2), and a list of input genes used for the similarity matrix analysis (Supplementary Dataset 3) are included in the Supplementary Materials.
Acute blood flow studies.
For in vitro blood perfusion studies, each rBEL was connected to a circuit composed of silicone tubing, a pressure transducer, and a peristaltic pump. Recirculation of freshly harvested, 37°C heparinized porcine blood was targeted at 9–12 mmHg to mimic maximum physiologically achievable venous pressure and resulting flow rates were monitored over time.
In vivo acute blood studies were performed using domestic pigs weighing 30–35 kg. rBEL construct were connected to portal venous blood flow using silicone tubing and luer-lock connectors to achieve functional end-to-side anastomoses between the graft’s and recipient animal’s portal veins and IVCs. Luer lock connectors allowed for direct measurement of blood flow with and without the rBEL in the circuit. Flow was assessed through direct measurement of collected outflow blood for 60 seconds, which was subsequently returned to the circuit. Post-test graft venogram through the PV was performed to ensure the patency of the portal vascular tree using 15–20 ml of Omnipaque 3000. Intravenous (IV) heparin was used to maintain an activated clotting time (ACT) > 600 seconds throughout the procedure.
Long-term studies to assess graft perfusion and vascular patency.
Grafts with >30 mg/h GCR were selected for transplantation in all long term in-vivo perfusion studies. All animal experiments were performed in accordance with the Animal Welfare Act and approved by the institutional animal care and use committee at Mayo Clinic. Heterotopic implantation of rBELs in this study relied on end-to-side anastomoses between the graft’s and recipient animal’s PV, and the graft’s and recipient animal’s IVC. All portal branches supplying the animal’s native liver with the exception of the first branch were tied off, thereby preserving blood flow to the caudate lobe and the right lateral lobe. Additionally, a constricting ribbon was applied surgically around the native portal vein distal to the anastomosis to enhance blood flow to the implanted rBEL. This sequence was followed to elevate the portal vein pressure while avoiding hemodynamic instability resulting from host portal vein clamping and abrupt cessation of portal flow.
The surgical procedure was performed under normal hemostasis without the use of systemic heparinization. Side clamping of the pigs’ portal vein and vena cava were performed to allow the anastomoses to be fashioned while minimizing the risk of thrombosis. After the procedure, the pigs were monitored in a recovery cage for the first 24 hours and assessed every 4–6 hours for any signs of bleeding or immediate surgical complications. Pigs were allowed to drink during this period as tolerated. Subsequently they were allowed to return to the regular housing and allowed regular diet as tolerated.
Operative data included OR time, cold ischemia time, ACT, liver function test (LFT), complete blood count (CBC), and coagulation factors. Cytotoxicity profile were followed pre-operatively as well as at post-operative days 1, 3, 7, 10, 15, and 20. To evaluate vascular patency and graft perfusion, contrast enhanced computed tomography (CT) scans were performed serially postoperatively following the same time points listed above. All scans included a dedicated porto-venous phase taken 50–60 seconds after contrast infusion.
All pigs were followed until graft thrombosis was ascertained via contrast enhanced CT scans, except for one pig in the immunosuppressed group which was intentionally euthanized at Post-Operative day 7 for the purpose of obtaining histopathological data of the graft at that time.
Volumetric analysis of the graft perfusion and vascular patency of the large vessels was calculated using Siemens MultiModality Workstation software. Perfused areas were automatically detected through Hounsfield density cutoff threshold. Subsequent loss of perfusion was calculated over time, at day 1, 3, 7, 15 and 20. Loss of graft perfusion was defined by absence of notable perfusion outside the major portal branches and hepatic veins. Complete loss of perfusion and vascular patency was defined by a clear loss of flow in the graft’s portal vein branches. Given the plasticity of the rBEL, the volume was affected in cases of ileus or gastric distension which sometimes occurred in the first 3–5 days after surgery.
Serum cytotoxicity assay.
The complement-based cytotoxicity assay was adapted from a previously described protocol25. Briefly, 48-well tissue culture plates were seeded with 1×105 HUVECs/well and incubated at 37°C until 80–100% confluence was reached (24–48 hours). Following an initial PBS wash, pig sera were diluted 1:32 with EGM-2 media and 200 μl of diluted pig sera were added to each well. Following 30 minutes of incubation at room temperature, wells were washed with PBS, and 200 μl of unabsorbed rabbit complement (Pel-Freez) diluted 1:16 with EGM-2 media was added to each well and incubated at room temperature for 1 hour. 2 μl of 1% Fluoroquench (Thermo Fisher) was added for fluoroscopic assessment of viable and nonviable cells. Cytotoxicity was characterized by the resulting percentage of nonviable cells.
Statistical Analysis.
IBM SPSS Software version 25 was used to conduct the statistical analysis. Descriptive data are presented as mean +/− Standard deviation. For subsets of data that did not meet normality tests, the median and [range] were used. Correlations between graft flow and metabolic parameters were statistically compared using binary correlation and linear logistic regression. Surgical model parameters, duration of graft perfusion, and vascular patency between immunosuppressed and non-immunosuppressed animals were compared using a Student’s t-test. Mann-Whitney test was alternatively used for subsets of data that are not normally distributed with a Shaprio-Wilk’s tests of (P<0.05).
Supplementary Material
Acknowledgments
We would like to thank Catherine Verfaillie and Manoj Kumar from the Stem Cell Institute Leuven, KU Leuven, Leuven, Belgium for the qPCR analysis performed in the manuscript, the Mayo Clinic Microscopy and Cell Analysis Core for experimental and technical support, and the Mayo clinic Biomedical Imaging Resource for creating the 3D visualizations from CT images. We would also like to thank Lindsay Wentz for assistance with cell culture, and Joseph Uzarski and M. Mason Macenski for review and comments on the final manuscript. This work was made possible by the financial support from Miromatrix, Mayo Clinic ILP grants, and the National Institutes of Health (R01DK106667, SLN; R01DK117861 and R03DK113339, RCH)
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are available for research purposes from the corresponding authors on reasonable request.
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-01X-XXXX-X.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Competing interests
Miromatrix Medical Inc. is a privately funded company and owns the patent rights for the perfusion decellularization and recellularization technologies employed in this study. The research was funded by Miromatrix and a Mayo Clinic Innovation grant. JJR, BDA, AY, BGS, and DSD are employees of Miromatrix.
References
- 1.Shirakigawa N, Takei T & Ijima H Base structure consisting of an endothelialized vascular-tree network and hepatocytes for whole liver engineering. J Biosci Bioeng 116, 740–745, doi: 10.1016/j.jbiosc.2013.05.020 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Badylak SF, Taylor D & Uygun K Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng 13, 27–53, doi: 10.1146/annurev-bioeng-071910-124743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soto-Gutierrez A et al. A whole-organ regenerative medicine approach for liver replacement. Tissue Eng Part C Methods 17, 677–686, doi: 10.1089/ten.tec.2010.0698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baptista PM et al. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 53, 604–617, doi: 10.1002/hep.24067 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Barakat O et al. Use of decellularized porcine liver for engineering humanized liver organ. J Surg Res 173, e11–25, doi: 10.1016/j.jss.2011.09.033 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Bao J et al. Construction of a portal implantable functional tissue-engineered liver using perfusion-decellularized matrix and hepatocytes in rats. Cell Transplant 20, 753–766, doi: (2011). [DOI] [PubMed] [Google Scholar]
- 7.Zhou P et al. Decellularized liver matrix as a carrier for the transplantation of human fetal and primary hepatocytes in mice. Liver Transpl 17, 418–427, doi: 10.1002/lt.22270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yagi H et al. Human-scale whole-organ bioengineering for liver transplantation: a regenerative medicine approach. Cell Transplant 22, 231–242, doi: (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uygun BE et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med 16, 814–820, doi: 10.1038/nm.2170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao J et al. Hemocompatibility improvement of perfusion-decellularized clinical-scale liver scaffold through heparin immobilization. Sci Rep 5, 10756, doi: 10.1038/srep10756 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park KM et al. Decellularized Liver Extracellular Matrix as Promising Tools for Transplantable Bioengineered Liver Promotes Hepatic Lineage Commitments of Induced Pluripotent Stem Cells. Tissue Eng Part A 22, 449–460, doi: 10.1089/ten.TEA.2015.0313 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Wang Y et al. Recent Advances in Decellularization and Recellularization for Tissue-Engineered Liver Grafts. Cells Tissues Organs 204, 125–136, doi: 10.1159/000479597 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Uygun BE & Yarmush ML Engineered liver for transplantation. Curr Opin Biotechnol 24, 893–899, doi: 10.1016/j.copbio.2013.05.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirmalek-Sani S-H, Sullivan DC, Zimmerman C, Shupe TD & Petersen BE Immunogenicity of Decellularized Porcine Liver for Bioengineered Hepatic Tissue. The American Journal of Pathology 183, 558–565, doi: 10.1016/j.ajpath.2013.05.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uzarski JS et al. Dual-Purpose Bioreactors to Monitor Noninvasive Physical and Biochemical Markers of Kidney and Liver Scaffold Recellularization. Tissue Engineering. Part C, Methods 21, 1032–1043, doi: 10.1089/ten.tec.2014.0665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson MJ, Soibam B, O’Leary JG, Sampaio LC & Taylor DA Recellularization of rat liver: An in vitro model for assessing human drug metabolism and liver biology. PLoS One 13, e0191892, doi: 10.1371/journal.pone.0191892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazza G, Al-Akkad W, Rombouts K & Pinzani M Liver tissue engineering: From implantable tissue to whole organ engineering. Hepatol Commun 2, 131–141, doi: 10.1002/hep4.1136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers SC, Zhang X, Azhar G, Luo S & Wei JY Exposure to high or low glucose levels accelerates the appearance of markers of endothelial cell senescence and induces dysregulation of nitric oxide synthase. J Gerontol A Biol Sci Med Sci 68, 1469–1481, doi: 10.1093/gerona/glt033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalor PF, Lai WK, Curbishley SM, Shetty S & Adams DH Human hepatic sinusoidal endothelial cells can be distinguished by expression of phenotypic markers related to their specialised functions in vivo. World Journal of Gastroenterology : WJG 12, 5429–5439, doi: 10.3748/wjg.v12.i34.5429 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacParland SA et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 9, 4383, doi: 10.1038/s41467-018-06318-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLeve LD Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology 61, 1740–1746, doi: 10.1002/hep.27376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skovseth DK, Yamanaka T, Brandtzaeg P, Butcher EC & Haraldsen G Vascular morphogenesis and differentiation after adoptive transfer of human endothelial cells to immunodeficient mice. The American journal of pathology 160, 1629–1637, doi: 10.1016/S0002-9440(10)61110-8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach L, Hamilton RD & Foss AJE Phenotypic plasticity of human umbilical vein endothelial cells. British Journal of Ophthalmology 96, 1152 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Robb RA The biomedical imaging resource at Mayo Clinic. IEEE Transactions on Medical Imaging 20, 854–867, doi: 10.1109/42.952724 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Nyberg SL, Amiot B, Hardin J, Baskin-Bey E & Platt JL Cytotoxic immune response to a xenogeneic bioartificial liver. Cell Transplant 13, 783–791 (2004). [DOI] [PubMed] [Google Scholar]
- 26.R. KW et al. OPTN/SRTR 2016 Annual Data Report: Liver. American Journal of Transplantation 18, 172–253, doi:doi: 10.1111/ajt.14559 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Lopez PM & Martin P Update on liver transplantation: indications, organ allocation, and long-term care. Mt Sinai J Med 73, 1056–1066 (2006). [PubMed] [Google Scholar]
- 28.Dhawan A, Puppi J, Hughes RD & Mitry RR Human hepatocyte transplantation: current experience and future challenges. Nat Rev Gastroenterol Hepatol 7, 288–298, doi: 10.1038/nrgastro.2010.44 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Suchy F, Yamaguchi T & Nakauchi H iPSC-Derived Organs In Vivo: Challenges and Promise. Cell Stem Cell 22, 21–24, doi: 10.1016/j.stem.2017.12.003 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Nicolas C, Wang Y & Nyberg SL Cell Therapy in Chronic Liver Disease. Current opinion in gastroenterology 32, 189–194, doi: 10.1097/MOG.0000000000000262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussein KH, Park KM, Kang KS & Woo HM Heparin-gelatin mixture improves vascular reconstruction efficiency and hepatic function in bioengineered livers. Acta Biomater 38, 82–93, doi: 10.1016/j.actbio.2016.04.042 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Ko IK et al. Bioengineered transplantable porcine livers with re-endothelialized vasculature. Biomaterials 40, 72–79, doi: 10.1016/j.biomaterials.2014.11.027 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Mei J et al. The angiogenesis in decellularized scaffold-mediated the renal regeneration. Oncotarget 7, 27085–27093, doi: 10.18632/oncotarget.7785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan Y et al. The effective bioengineering method of implantation decellularized renal extracellular matrix scaffolds. Oncotarget 6, 36126–36138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doi R et al. Transplantation of bioengineered rat lungs recellularized with endothelial and adipose-derived stromal cells. Scientific Reports 7, 8447, doi: 10.1038/s41598-017-09115-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao S, Glorioso J, Elgilani F, De Lorenzo S & Deeds M Sustained In Vivo Perfusion of a Re-Endothelialized Tissue Engineered Porcine Liver. Int J Transplant Res Med 3, 031 (2017). [Google Scholar]
- 37.Dudakovic A et al. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem 115, 1816–1828, doi: 10.1002/jcb.24852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalari KR et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics 15, 224, doi: 10.1186/1471-2105-15-224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36, doi: 10.1186/gb-2013-14-4-r36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metsalu T & Vilo J ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 43, W566–570, doi: 10.1093/nar/gkv468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.