Abstract
Background
Remodeling and spacing factor-1 (RSF-1) is an identified tumor biomarker that is overexpressed in a variety of human cancers, but its effect on radiotherapy remains unclear. In this study, we aimed to explore the effect of RSF-1 siRNA on sensitizing cervical cancer cells to radiation and its underlying mechanism.
Methods
The mRNA and protein expression of RSF-1 in tissue and cells were measured by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting. Cell counting kit-8 (CCK-8) and colony formation assay were used to examine cell proliferation. Flow cytometry was used to analyzed the cell cycle and cell apoptosis. DNA damage was examined by the comet assay. ATM, ATR, CHK1, CHK2, H2AX, γH2AX and phosphorylated ATM, ATR, CHK1 and CHK2 were detected by Western blotting. γH2AX foci were demonstrated by immunofluorescence staining.
Results
RSF-1 was upregulated in cervical cancer tissue and decreased after effective treatment. RSF-1 siRNA in combination with radiation suppressed cell viability, redistributed cell cycles and also induced cell apoptosis in HeLa and SiHa cell lines. Further, knockdown of RSF-1 induced DNA damage by attenuating DNA repair capability, thereby sensitizing cervical cancer cells to radiation.
Conclusions
These data demonstrate that RSF-1 siRNA enhanced the sensitivity of radiotherapy, and targeting RSF-1 may be a promising approach for the development of novel radiosensitizing agents for the treatment of cervical cancer.
Keywords: RSF-1, cervical cancer, radiotherapy, DNA damage
Background
Cervical cancer is the second most common female malignancy around the world. It presents a major health problem for women, with its morbidity and mortality increasing each year. Despite substantial improvements in treatment in the last decades, the prognosis for advanced cervical cancer remains quite poor with a 5-year survival rate ranging from 40% to 50%.1 Radiotherapy and chemotherapy remain the major treatments of cervical cancer. Radiotherapy is the major treatment for patients with advanced cervical cancer, whose efficacy is limited by the intrinsic characteristics and acquired radioresistance. Therefore, exploring new molecular targets and pathways is essential for enhancing the radiosensitivity and improving the prognosis of cervical cancer.
RSF-1, known as HBXAP, is located in the nucleus and interacts with human sucrose nonfermenting protein 2 homologue (hSNF2H) protein to form the RSF chromatin-remodeling complex, in which the SNF2H subunit functions as the nucleosome-dependent ATPase, and RSF-1 protein functions as the histone chaperone. The complex regulates chromatin remodeling and alters the chromatin structure of nucleosomes, which are required for biological processes, including activation or repression of transcription, DNA replication, cell cycle progression and other cellular processes.2,3 RSF-1 is an identified tumor biomarker that is highly expressed in numerous human cancers. RSF-1 overexpression correlates with disease aggressiveness and poor prognosis in various cancers, such as ovarian carcinoma,4 prostate cancer,5 gastric adenocarcinoma,6 hepatocellular carcinoma7 and colon cancer.8 Moreover, RSF1 promotes the malignant behavior of cancer cells, whereas RSF-1 knockdown can effectively inhibit cell proliferation and promote cell apoptosis in several cancer cells. For example, overexpression of RSF-1 can promote malignant cell growth of non-small cell lung cancer (NSCLC) cells via the modulation of cyclinD1 expression and ERK activity.9 RSF-1 knockdown induces growth arrest and subsequent cell apoptosis in breast cancer10 and oral squamous cell carcinoma cells.11 In addition, RSF-1 has been shown to influence the sensitivity of cancer to paclitaxel12 and cisplatin.13 In our previous study, we showed that RSF1 overexpression correlated with increased tumor size, poor differentiation, higher nodal metastasis and advanced clinical stages in cervical cancer, and RSF-1 silencing increased drug sensitivity toward paclitaxel treatment.14 However, the impact of RSF-1 on radiotherapy in cervical cancer remains elusive. The aim of this study was to explore the effect of RSF-1 on radiotherapy and its underlying mechanism in cervical cancer.
Materials and Methods
Patients and Tissue Collection
A total of 16 patients with cervical cancer who underwent adjuvant platinum-based radiochemotherapy between 2016 and 2018 at Shandong Cancer Hospital Affiliated Shandong University (Shandong, China) were enrolled. Cervical biopsy tissues before radiochemotherapy and corresponding cervical cancer tissues by surgery after radiochemotherapy were collected, and all samples were formalin-fixed, paraffin-embedded, and pathologically diagnosed. The clinical stage and histological diagnosis were identified according to the International Federation of Gynecology and Obstetrics (FIGO) classification system. Tumor pathological type and differentiation grade were defined according to World Health Organization criteria. All the patients signed written informed consent for the study, and the study was approved by the Institute Research Ethics Committee of Shandong Cancer Hospital Affiliated to Shandong University.
Cell Culture
The human cervical carcinoma HeLa and SiHa cell lines were obtained from the Chinese Academy of Typical Culture Collection Cell Bank (Shanghai, China). Cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum, 100 IU/mL of penicillin, and 100 μg/mL of streptomycin in a humidified 37°C atmosphere of 5% CO2.
Transfection
siRNA targeting RSF-1 (siRNA-RSF-1) and siRNA negative control (siRNA-NC) were obtained from GenePharma Co. Ltd. (Shanghai, China). The sequences of siRNA-RSF-1 were: sense, 5ʹ-GGAGAAGAAGGGCUAUCUUTT-3ʹ; antisense, 5ʹ-AAGAUAGCCCUUCUUCUCCTT-3ʹ. The sequences of siRNA-NC were: sense, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ; antisense, 5ʹ- ACGUGACACGUUCGGAGAATT-3ʹ. pcDNA3.1 containing RSF-1 plasmid (pcDNA3.1-RSF-1) and the empty vector were purchased from Shanghai Sangon Biotech Co., Ltd (Shanghai, China). siRNA or plasmids were transfected into HeLa and SiHa cells using a Lipofectamine 3000 transfection reagent kit (Invitrogen) according to the manufacturer’s protocol.
Radiation Treatment
Cells were irradiated with 0, 4 and 10Gy of X-rays using a 6-MeV X-ray linear accelerator (Siemens, Munich, Bavaria, Germany) 48h post-transfection. Radiation treatment was performed in triplicate for each condition.
Cell Proliferation Assay
For the CCK-8 assay, cells were seeded into 96-well culture plates with a density of 4000 cells/well. The next day, cells were treated with radiation, and then incubation was continued for 24, 48 or 72h. Subsequently, cells were incubated in the dark for 1 h after adding cell counting solution according to the manufacturer’s protocol (Dojindo Laboratories, Japan). The optical density (OD) of each well was measured at 450 nm using BIOBASE-EL10A.
For colony formation assay, cells were seeded into 12-well culture plates at a density of 1000 cells/well. The next day, cells were treated with radiation and then incubation was continued for up to 2 weeks. Then the colonies were stained with crystal violet (Solarbio, China) for 20 min and the colony count was evaluated with the Image J software.
Apoptosis Analysis
Cells were seeded into 12-well culture plates at a density of 5×104 cells/well. The next day, cells were treated with radiation and then incubation was continued for 24h. Apoptosis was assessed by flow cytometry. Populations of apoptotic cells were measured by staining cells with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide according to the manufacturer’s protocol (annexin V apoptosis kit; Southern Biotech, China). Briefly, the cells and the supernatant were harvested by trypsinization and centrifuged at 1500 r/min for 8 min before being washed twice with cold phosphate-buffered saline (PBS) and resuspended in binding buffer. Then, the cell suspension was stained with annexin V/FITC and PI for 5 min in a dark at room temperature and immediately analyzed by flow cytometry (NovoCyte, ACEABIO, China). Each experiment was performed in triplicate.
Cell Cycle Analysis
Cells were seeded into 12-well culture plates at a density of 5×104 cells/well. The next day, cells were treated with radiation or drugs and then incubation was continued for 24h. Cells were then collected using trypsin and washed twice with pre-chilled PBS. After ethanol precipitation at −4 °C for 12h, the cells were resuspended and stained with propidium iodide (PI) solution for 30 min in the dark at room temperature, and analyzed using flow cytometry (NovoCyte, ACEABIO, China), and cell cycle distribution was analyzed using Modfit software. Each experiment was performed in triplicate.
Comet Assay
Comet assay was performed to evaluate the DNA damage induced by radiation according to the manufacturer’s instructions (Cell Biolabs, USA). Cells were digested by trypsin and washed twice with cold PBS (1x106/mL). Then OxiSelect Comet Slides were covered by the mixture of comet agarose and cells. After lysis, the slides underwent electrophoresis for 30 mins under 1 volt/cm. Cells were stained with Vista Green DNA dye and visualized by the epifluorescence microscopy. Tail length, head length, and tail moment were analyzed by Comet-Assay IV software V4.3 (Perceptive Instruments, UK).
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Aliquots of 1 μg of total RNA extracted from cells were used to synthesize first-strand cDNA using PrimeScriptTM RT Reagent Kit (TaKaRa) according to the manufacturer’s protocol. PCR amplification was carried out using 2 µL of cDNA as a template in a total reaction volume of 20 μL containing 6 µL of deionized water, 10 µL of SYBR® Premix Ex Taq II (Tli RNaseH Plus) (2×), 0.8µL of each primer and 0.4 µL of ROX Reference Dye or Dye II (50×). The primers used were as listed for RSF-1 (forward, 5′-GGCTACACCGGATTGAGACGGATGA-3′; reverse, 5′-AGGGCTCTGTCCATTGGTTGAAGGTAAGT-3′) and GAPDH (forward, 5ʹ-ATGACATCAAGAAGGTGGTGAAGCAGG-3ʹ; reverse, 5ʹ-GCGTCAAAGGTGGAGGAGTGGGT-3ʹ). Following a 30 s denaturation step at 95°C, the reactions were performed for 40 cycles of denaturation for 5 s at 95°C, and annealing at 60°C for 30 s, followed by a final extension at 72°C for 10 min.
Western Blotting
The proteins were extracted with radioimmunoprecipitation assay (RIPA) lysis buffer containing several protease and phosphatase inhibitors. The proteins were separated using SDSsodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore Co., USA). Immunoblotting was carried out using monoclonal antibodies. Anti-ATR, -ATM, -CHK1, -CHK2, and –H2AX and anti-phosphorylated ATM (Ser1981), CHK2 (Thr68), and γH2AX (Ser139) antibodies were all purchased from ImmunoWay (Suzhou, China). Anti-RSF-1, anti-phosphorylated ATR (Ser428) and CHK1 (Ser345) antibodies were purchased from Abcam (Shanghai, China). GAPDH was purchased from Proteintech Group (Wuhan, China). The membranes were then incubated with secondary antibody, detected using an ECL chemiluminescence assay kit (Proteintech, Wuhan, China) and visualized by X-ray film exposure. Protein expression was analyzed semiquantitatively using the ChemiDoc XRS+ System (Bio-Rad, Hercules, CA, USA).
Statistical Analysis
Results shown were representative of at least three separate experiments. Comparisons between groups were performed using Student’s t-test and one-way or two-way analysis of variance (ANOVA) using SPSS 21.0 software. All statistical data are presented as the mean ± SD. In all analyses, P < 0.05 was taken to indicate statistical significance.
Results
RSF-1 Was Upregulated in Cervical Cancer Tissues and Decreased After Effective Treatment
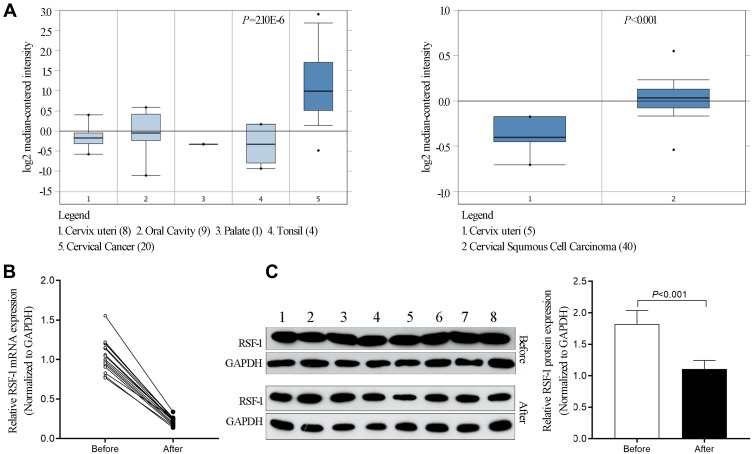
Based on the data obtained from the ONCOMINE database, the expression level of RSF-1 was measured. As Figure 1A showed, the expression level of RSF-1 in cervical cancer tissues was significantly higher than the control group (P<0.001), which is consistent with our previous findings.14 To explore the potential relationship between RSF-1 and radiochemotherapy, we used qRT-PCR and Western blotting to investigate the mRNA and protein level of RSF-1 in 16 pairs of cervical cancer tissues before and after radiochemotherapy. The results showed that RSF-1 mRNA and protein in cervical cancer tissues after radiochemotherapy were significantly decreased in comparison with the level in the matched tissues before radiochemotherapy (Figure 1B and C), indicating that RSF-1 may be a potential biomarker for prognosis of patients.
Figure 1.
RSF-1 was upregulated in cervical cancer tissues and decreased after effective treatment. (A) The expression of RSF-1 mRNA in cervical cancer tissue and normal control from the ONCOMINE database. (B) The expression of RSF-1 mRNA and (C) RSF-1 protein in 16 pairs of tumor samples from cervical cancer patients before and after radiochemotherapy were examined by qRT-PCR and Western blot.
RSF-1 siRNA Enhances the Radiosensitivity of HeLa and SiHa Cells
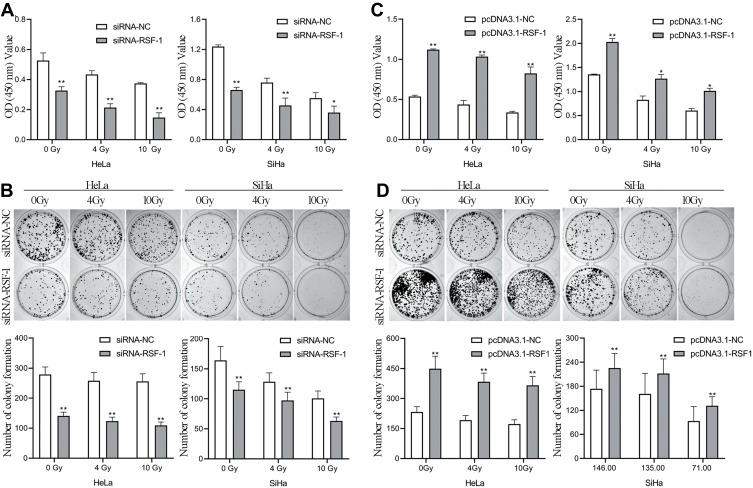
To evaluate the effects of RSF-1 siRNA on radiation efficacy, HeLa and SiHa cells were transfected with RSF-1 siRNA and exposed to different doses of radiation. The CCK-8 and colony formation assays were performed to assess the viability and proliferative capacity of HeLa and SiHa cells. As shown in Figure 2A and B, both RSF-1 siRNA and radiation could attenuate the cancer cell viability and proliferation in both HeLa and SiHa cell lines. Furthermore, RSF-1 siRNA combined with radiation markedly decreased cell viability and proliferation compared with RSF-1 siRNA or radiation alone (P < 0.05). To confirm this result, RSF-1 overexpression was performed. RSF-1 overexpressing cells were more resistant to radiotherapy than control cells (Figure 2C and D). These results suggest that RSF-1 siRNA enhances the radiosensitivity of cervical cancer cells via inhibiting cell viability and proliferation.
Figure 2.
RSF-1 siRNA confers sensitivity to radiation in HeLa and SiHa cells. HeLa and SiHa cells transfected with siRNA-NC or siRNA-RSF-1 were irradiated with 0, 4 and 10Gy and then subjected to (A) CCK-8 assays and (B) colony formation assays. HeLa and SiHa cells transfected with pcDNA3.1-NC or pcDNA3.1-RSF-1, were irradiated with 0, 4 and 10Gy and then subjected to (C) CCK-8 assays and (D) colony formation assays. Data were presented as mean ± SD from three independent experiments. **P<0.01, *P <0.05 as compared with the siRNA-NC or pcDNA3.1-NC groups.
RSF-1 siRNA in Synergy with Radiation Redistributes Cell Cycle and Promotes Apoptosis
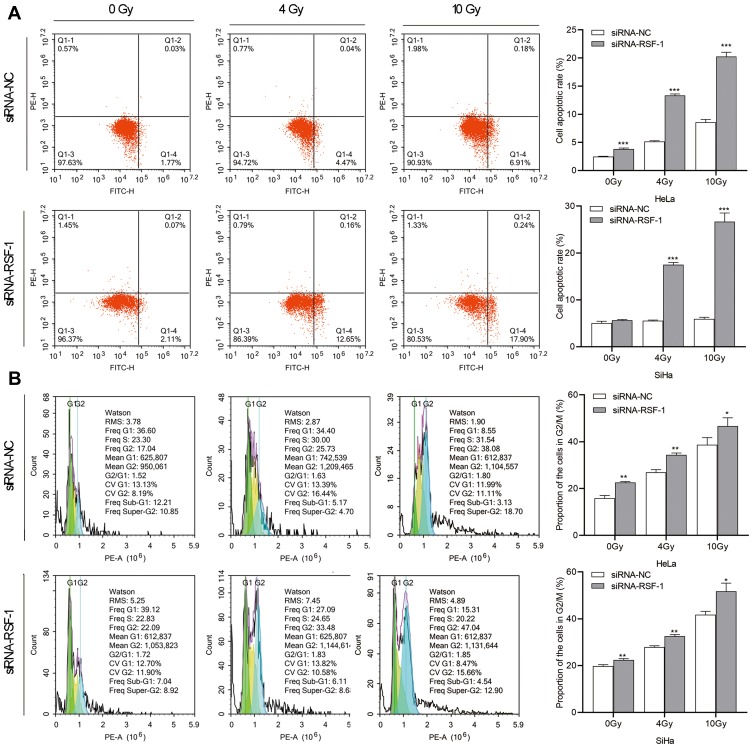
To further investigate whether RSF-1 siRNA enhances the radiosensitivity of HeLa and SiHa cells by changing on radiation-induced cell cycle and apoptosis, the apoptosis rate and cell cycle were analyzed at 24h after exposure to different doses of radiation. As shown in Figure 3A, apoptosis analysis demonstrated that RSF-1 siRNA combined with radiation dramatically increased the apoptotic rate of cells compared with RSF-1 siRNA, radiation and control groups (P < 0.01). In addition, cell cycle analysis showed that the proportion of cells in the G2/M phase was higher in the combination treatment group than those in the RSF-1 siRNA, radiation and control groups (P<0.01, Figure 3B). Whereas RSF-1 overexpression exerted an opposite effect. In conclusion, these findings suggest that RSF-1 siRNA enhances the radiosensitivity of cervical cancer cells by promoting radiation-induced apoptosis and G2/M phase cell cycle arrest.
Figure 3.
RSF-1 siRNA promotes radiation-induced cell cycle redistribution and apoptosis. HeLa and SiHa cells transfected with siRNA-NC or siRNA-RSF-1 were irradiated with 0, 4 and 10Gy and then subjected to (A) cell apoptosis and (B) cell cycle analysis. Data were presented as mean ± SD from three independent experiments. ***P<0.001, **P<0.01, *P <0.05 as compared with the siRNA-NC group.
RSF-1 siRNA Promotes Radiation-Induced DNA Strand Breaks
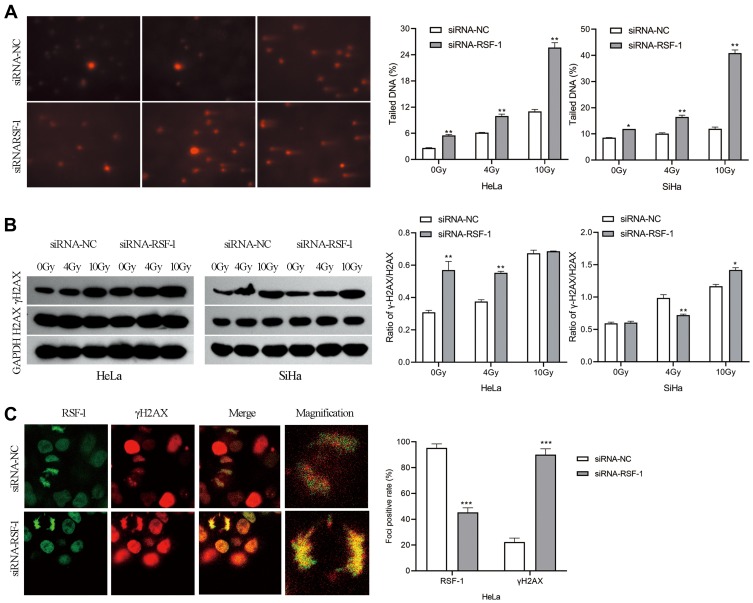
DNA damage, especially DNA strand breaks, are the major cause of radiation-induced cell death. Phosphorylation of H2AX at Ser 139 (γH2AX) was used as a marker of radiation-induced DNA double-strand breaks (DSBs). To evaluate whether RSF-1 siRNA could aggregate radiation-induced DNA strand breaks, the comet assay and γH2AX were analyzed in HeLa and SiHa cells exposed to different doses of radiation. The comet assay showed that HeLa and SiHa cells with RSF-1 siRNA had significantly higher residual DNA damage relative to control cells (Figure 4A). Meanwhile, Western blot analysis showed that γH2AX expression was significantly elevated in HeLa cells transfected with RSF-1 siRNA treated with 4Gy of radiation, compared with the siRNA-NC group (P<0.05, Figure 4B), indicating that RSF-1 siRNA caused much more radiation-induced DSBs in HeLa cells. Visualized immunofluorescence staining also demonstrated that γH2AX foci were significantly increased in the siRNA-RSF-1 group compared to the siRNA-NC group in HeLa cells following 4 Gy of radiation (Figure 4C). These results indicate that RSF-1 siRNA caused greater DNA damage, thereby increasing the sensitivity to radiation.
Figure 4.
RSF-1 siRNA promotes radiation-induced DNA damage. (A) The extent of DNA damage was assessed by Comet assay. The percentage of DNA in comet tail (Tail DNA %) was quantified and graphed for each group. (B) The expression of H2AX and γH2AX proteins were examined by Western blot. The ratio of γH2AX level relative to H2AX after normalization to GAPDH was quantified and graphed for each group. (C) HeLa cells transfected with siRNA-NC or siRNA-RSF-1 were irradiated with 4Gy and then stained with an anti-RSF-1 (green) or an anti-γH2AX (red) antibody. RSF-1- and γH2AX-positively stained cells were scored and showed in right panel. The experiments were repeated three times independently. ***P<0.001, **P<0.01, *P <0.05 as compared with the siRNA-NC group.
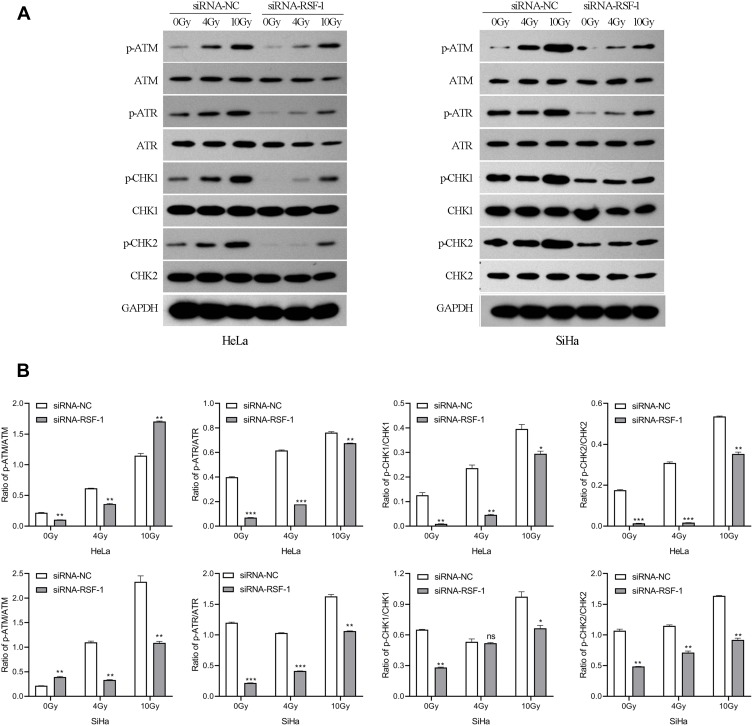
RSF-1 siRNA Impairs the ATM/ATR Signaling Pathway Activated by Radiation
Ataxia telangiectasia mutated (ATM), and Rad3-related (ATR) kinase play important roles in the cellular response to DNA damage, which can phosphorylate numerous downstream targets that are involved in DNA repair and cell cycle arrest, including CHK1 and CHK2. To investigate the effect of RSF-1 siRNA on ATM/ATR signaling pathway activated by radiation, Western blotting was used to measure the protein levels of key pathway components. Protein such as p-ATM, p-ATR, p-CHK1 and p-CHK2 were detectable and variable in both HeLa and SiHa cell lines (Figure 5A). As shown in Figure 5B, phosphorylated ATM and ATR were markedly decreased in both HeLa and SiHa cells transfected with RSF-1 siRNA following 0 or 4Gy doses of radiation compared to those in the siRNA-NC group (P < 0.01). In addition, phosphorylated CHK1 and CHK2 expression were decreased in accordance with the inactivation of ATM/ATR. However, RSF-1 siRNA did not significantly change the protein expression of ATM, ATR, CHK1 and CHK2 in both HeLa and SiHa cells. Taken together, the above findings indicate that RSF-1 siRNA inhibits ATM/ATR signaling pathway activated by radiation.
Figure 5.
RSF-1 siRNA impairs ATM/ATR signaling pathway activated by radiation. (A) HeLa and SiHa cells transfected with siRNA-NC or siRNA-RSF-1 were irradiated with 0, 4 and 10Gy, and the ATM/ATR pathway proteins were examined by Western blot. (B) The relative gray values of p-ATM, p-ATR, p-CHK1 and p-CHK2 were calculated and graphed for each group. The experiments were repeated three times independently. ***P<0.01, **P<0.01, *P <0.05 as compared with the siRNA-NC group.
Discussion
RSF-1 plays an important role in tumor progression. Previous reports show that RSF-1 overexpression contributes to cell survival, growth and metastasis in breast and ovarian cancers,15,16 whereas RSF-1 knockdown in tumor cells with high endogenous RSF-1 expression inhibits cell proliferation and promotes apoptosis in breast and NSCLC.10,17 Moreover, RSF-1 has been shown to influence the sensitivity of cancer cells to chemotherapy. However, the effect of RSF-1 on radiotherapy and its underlying mechanism in cervical cancer remain unclear. In the present study, we confirmed that the expression of RSF-1 was upregulated in cervical cancer tissues and significantly decreased after effective treatment. As expected, cell experiments showed that RSF1 siRNA enhanced the sensitivity of cervical cancer cells to radiation, via inhibiting cell growth, redistributing cell cycle and inducing apoptosis. Further mechanism exploration revealed that RSF-1 may be involved in DNA damage and ATM/ATR signaling pathways.
Overexpression of RSF-1 has been reported in numerous human solid cancers and correlates with advanced stage, poor prognosis and poor therapeutic response in human cancers.11,17-21 Similar to previous study, our data showed that RSF-1 was upregulated in cervical cancer tissues compared to control. In addition, RSF-1 expression levels were decreased significantly after effective treatment, suggesting RSF-1 may be a potential biomarker for monitoring the prognosis of patients.
Radiotherapy is an important clinical treatment for cancers, but it often shows only a modest effect on cell proliferation and apoptosis in most solid tumors.22 A deeper understanding of tumors and their underlying mechanisms is needed to sensitize tumor cells to radiation. Recently, several studies have shown that RSF-1 plays a critical role in regulating drug resistance in cancers.23 For example, overexpression of RSF-1 remarkably promotes paclitaxel resistance, while inhibition of RSF-1 enhances sensitivity to paclitaxel through the formation of remodeling and spacing factor complex in ovarian cancer cells.24 Besides, RSF-1 influences the sensitivity to paclitaxel by regulating NF-κB pathway in non-small cell lung cancer and nasopharyngeal carcinoma.12,25 Additionally, RSF-1 may be involved in the sensitive to cisplatin resistance in nasopharyngeal carcinoma.13 To determine the effect of RSF-1 on radiotherapy in cervical cancer, the cells with or without RSF-1 siRNA were exposed to different doses of radiation, and their proliferative capacity, cell cycle progression and cell apoptosis were assessed by a series of experiments. The results showed that both radiation and RSF-1 siRNA significantly affected the cell proliferation, cell cycle and apoptosis. Compared with radiation alone, combined treatment with RSF-1 siRNA and radiation showed more significant effects on the cell proliferation, cell cycle and cell apoptosis. Therefore, we speculated that RSF-1 siRNA enhanced the sensitivity of cancer cells to radiation via redistributing cell cycle and inducing cell apoptosis and eventually inhibiting cell proliferation.
DNA damage, especially DSB, is one of the main mechanisms by which radiotherapy kills tumor cells. However, radiation not only causes DNA damage, but also activates DNA repair pathway of tumor cells which results in tumor radioresistance.26 Thus, the balance between DNA damage and repair capacity determines the efficacy of radiotherapy. An elevated DNA repair capacity in tumor cells contributes to radioresistance and severely limits the efficacy of radiotherapy. RSF-1 is associated with DNA damage and related DNA damage response (DDR).27 In nontransformed cells with wild-type p53, RSF-1 overexpression can induce DNA damage, activate the ATM-pCHK2-p53-p21 pathway and lead to growth arrest and apoptosis, whereas in cancer cells with TP53 mutation, RSF-1-induced DDR allows tumor cells to proliferate in the presence of DNA strand breaks, resulting in chromosomal aberrations.28,29 Furthermore, RSF-1 can be recruited to DNA damage sites in ATM-dependent manner and promotes homologous recombination repair, while knockdown of RSF-1 attenuates the activation of DNA damage checkpoint signals and cell survival upon DNA damage.30 To further explore the mechanism of RSF-1 on radiotherapy in cervical cancer, DNA damage and DNA damage repair-related markers were analyzed in HeLa and SiHa cells in different circumstances. In the present study, the results revealed that both radiation and RSF-1 siRNA promoted DNA damage, evidenced by comet assay and γH2AX analysis. Radiation activated ATM/ATR signaling pathway in dose-dependent manner. Surprisingly, RSF-1 siRNA could attenuate radiation-induced the activation of ATM/ATR signals. Taken together, these results demonstrate RSF-1 siRNA enhanced the sensitivity of cancer cells to radiation through blocking ATM/ATR signaling pathway and promoting DNA damage and ultimately leading to cell arrest and apoptosis.
Conclusions
In conclusion, RSF-1 siRNA enhances tumor radiosensitivity in cervical cancer via enhanced DNA damage, cell cycle redistribution, and promotion of apoptosis through inactivation of ATM/ATR signaling pathway. Therefore, RSF-1 may be a potential therapeutic target for increasing radiosensitivity in cervical cancer.
Funding Statement
The present study was supported by grants from The National Natural Science Foundation of China (grant no. 81671433), the Province Key Research and Development Project of Shandong (grant no. 2018GSF118238), the CSCO-HengRui Tumor Research Fund (grant no. Y-HR2018-200), the Medical Science and Technology Project of Shandong (grants nos. 2016WS0562 and 2011HZ097) and the Natural Science Foundation of Shandong (grant no. ZR2012HM010), China Medical University Hospital, Taiwan (DMR109-088).
Abbreviations
RSF-1, Remodeling and spacing factor-1; qRT-PCR, Quantitative real-time polymerase chain reaction; hSNF2H, Human sucrose nonfermenting protein 2 homologue; CCK-8, Cell counting kit-8 assay; PBS, Phosphate-buffered saline; PI, propidium iodide; SDS-PAGE, SDSsodium dodecyl sulfate-polyacrylamide gel electrophoresis; RIPA, Radioimmunoprecipitation assay; FITC, Fluorescein isothiocyanate; PVDF, Polyvinylidene difluoride; ANOVA, Analysis of variance; ATM, Ataxia telangiectasia mutated; ATR, Ataxia telangiectasia and Rad3 related; DSB, DNA double strand breaks; DDR, DNA damage response.
Data Sharing Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Ethics and Consent Statement
The study was approved by the Institute Research Ethics Committee of Shandong Cancer Hospital Affiliated to Shandong University (No. 2016-24, date: 20160206). All protocols and methods were in accordance with the guidelines and regulations. Written informed consent was obtained from all patients.
Consent for Publication
All authors approved publication of the manuscript.
Author Contributions
JT, EK, XW, ZX, CYYC, JJCS, QH, LS contributed to experimental design, procedure and data acquisition. JT, EK, XW, ZX and LS contributed to data interpretation as well as manuscript drafting or revision. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Disclosure
All authors declare that there is no conflict of interest in this work.
References
- 1.Wright JD, Chen L, Tergas AI, et al. Population-level trends in relative survival for cervical cancer. Am J Obstet Gynecol. 2015;213(5):670 e671–677. doi: 10.1016/j.ajog.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loyola A, Huang JY, LeRoy G, et al. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23(19):6759–6768. doi: 10.1128/MCB.23.19.6759-6768.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185(3):397–407. doi: 10.1083/jcb.200903088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shih IM, Sheu JJ, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci U S A. 2005;102(39):14004–14009. doi: 10.1073/pnas.0504195102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Zhang Y, Zhang Y, Bai X, Peng Y, He P. Rsf-1 overexpression in human prostate cancer, implication as a prognostic marker. Tumour Biol. 2014;35(6):5771–5776. doi: 10.1007/s13277-014-1766-7 [DOI] [PubMed] [Google Scholar]
- 6.Hu B-S, Yu H-F, Zhao G, Zha T-Z. High RSF-1 expression correlates with poor prognosis in patients with gastric adenocarcinoma. Int J Clin Exp Pathol. 2012;5(7):668–673. [PMC free article] [PubMed] [Google Scholar]
- 7.Xie C, Fu L, Xie L, Liu N, Li Q. Rsf-1 overexpression serves as a prognostic marker in human hepatocellular carcinoma. Tumour Biol. 2014;35(8):7595–7601. doi: 10.1007/s13277-014-2008-8 [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Dong Q, Wang E. Rsf-1 overexpression correlates with poor prognosis and cell proliferation in colon cancer. Tumour Biol. 2012;33(5):1485–1491. doi: 10.1007/s13277-012-0399-y [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Dong Q, Wang E. Rsf-1 is overexpressed in non-small cell lung cancers and regulates cyclinD1 expression and ERK activity. Biochem Biophys Res Commun. 2012;420(1):6–10. doi: 10.1016/j.bbrc.2012.02.095 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Gai J, Fu L, Zhang X, Wang E, Li Q. Effects of RSF-1 on proliferation and apoptosis of breast cancer cells. Oncol Lett. 2018;16(4):4279–4284. doi: 10.3892/ol.2018.9172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang FM, Li CF, Huang HY, et al. Overexpression of a chromatin remodeling factor, RSF-1/HBXAP, correlates with aggressive oral squamous cell carcinoma. Am J Pathol. 2011;178(5):2407–2415. doi: 10.1016/j.ajpath.2011.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Sun X, Guan J, et al. Rsf-1 influences the sensitivity of non-small cell lung cancer to paclitaxel by regulating NF-kappaB pathway and its downstream proteins. Cell Physiol Biochem. 2017;44(6):2322–2336. doi: 10.1159/000486116 [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Tai Y, Ma J. LncRNA NEAT1/let-7a-5p axis regulates the cisplatin resistance in nasopharyngeal carcinoma by targeting Rsf-1 and modulating the Ras-MAPK pathway. Cancer Biol Ther. 2018;19(6):534–542. doi: 10.1080/15384047.2018.1450119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Sheu JJ, Lai MT, et al. RSF-1 overexpression determines cancer progression and drug resistance in cervical cancer. Biomedicine (Taipei). 2018;8(1):4. doi: 10.1051/bmdcn/2018080104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheu JJ, Choi JH, Yildiz I, et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68(11):4050–4057. doi: 10.1158/0008-5472.CAN-07-3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Garcia F, Villagrasa P, Matsui J, et al. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014;159(6):1461–1475. doi: 10.1016/j.cell.2014.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Fu L, Xue D, et al. Overexpression of Rsf-1 correlates with poor survival and promotes invasion in non-small cell lung cancer. Virchows Arch. 2017;470(5):553–560. doi: 10.1007/s00428-017-2102-6 [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Hu L, Wu F, He T. Prognostic value of rsf-1/hbxap in human solid tumors: a meta-analysis of cohort studies. Int J Clin Exp Med. 2015;8(2):1944–1955. [PMC free article] [PubMed] [Google Scholar]
- 19.Tai HC, Huang HY, Lee SW, et al. Associations of Rsf-1 overexpression with poor therapeutic response and worse survival in patients with nasopharyngeal carcinoma. J Clin Pathol. 2012;65(3):248–253. doi: 10.1136/jclinpath-2011-200413 [DOI] [PubMed] [Google Scholar]
- 20.Lin CY, Tian YF, Wu LC, et al. Rsf-1 expression in rectal cancer: with special emphasis on the independent prognostic value after neoadjuvant chemoradiation. J Clin Pathol. 2012;65(8):687–692. doi: 10.1136/jclinpath-2012-200786 [DOI] [PubMed] [Google Scholar]
- 21.Liang PI, Wu LC, Sheu JJ, et al. Rsf-1/HBXAP overexpression is independent of gene amplification and is associated with poor outcome in patients with urinary bladder urothelial carcinoma. J Clin Pathol. 2012;65(9):802–807. doi: 10.1136/jclinpath-2012-200897 [DOI] [PubMed] [Google Scholar]
- 22.Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31(4):363–372. doi: 10.1007/s13277-010-0042-8 [DOI] [PubMed] [Google Scholar]
- 23.Zhao XC, An P, Wu XY, et al. Overexpression of hSNF2H in glioma promotes cell proliferation, invasion, and chemoresistance through its interaction with Rsf-1. Tumour Biol. 2016;37(6):7203–7212. doi: 10.1007/s13277-015-4579-4 [DOI] [PubMed] [Google Scholar]
- 24.Choi JH, Sheu JJ, Guan B, et al. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69(4):1407–1415. doi: 10.1158/0008-5472.CAN-08-3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Li G, Liu C, Tang Y, Zhang S. RSF1 regulates the proliferation and paclitaxel resistance via modulating NF-kappaB signaling pathway in nasopharyngeal carcinoma. J Cancer. 2017;8(3):354–362. doi: 10.7150/jca.16720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Ejeh F, Kumar R, Wiegmans A, Lakhani SR, Brown MP, Khanna KK. Harnessing the complexity of DNA-damage response pathways to improve cancer treatment outcomes. Oncogene. 2010;29(46):6085–6098. doi: 10.1038/onc.2010.407 [DOI] [PubMed] [Google Scholar]
- 27.Kshirsagar M, Jiang W, Shih IM. DNA damage response is prominent in ovarian high-grade serous carcinomas, especially those with Rsf-1 (HBXAP) overexpression. J Oncol. 2012;2012:621685. doi: 10.1155/2012/621685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheu JJ, Guan B, Choi JH, et al. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285(49):38260–38269. doi: 10.1074/jbc.M110.138735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheu JJ, Choi JH, Guan B, et al. Rsf-1, a chromatin remodelling protein, interacts with cyclin E1 and promotes tumour development. J Pathol. 2013;229(4):559–568. doi: 10.1002/path.4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Min S, Jo S, Lee HS, et al. ATM-dependent chromatin remodeler Rsf-1 facilitates DNA damage checkpoints and homologous recombination repair. Cell Cycle. 2014;13(4):666–677. doi: 10.4161/cc.27548 [DOI] [PubMed] [Google Scholar]