Abstract
Purpose
Circular RNAs (circRNAs) are novel type of noncoding RNAs that play important roles and serve as noninvasive biomarkers in various cancers. In the present study, we focused on circFoxO3a and aimed to investigate its prognostic value as a novel serum biomarker for squamous cervical cancer (SCC).
Patients and Methods
Our study included 103 SCC patients from Obstetrics and Gynecology Hospital of Fudan University. Expression levels of circFoxO3a in the serum of patients with SCC were examined by reverse transcription‑quantitative PCR (RT‑qPCR). The correlation between serum circFoxO3a expression and clinicopathologic factors was analyzed. The Kaplan–Meier method and multivariate Cox regression analysis were applied to evaluate the independent prognostic factors for SCC. A prognostic predictive nomogram was constructed using R software.
Results
Levels of serum circFoxO3a were decreased in SCC patients compared with controls. Low expression of circFoxO3a was correlated with deeper stromal invasion and positive lymph node metastasis. Moreover, SCC patients with lower expression of serum circFoxO3a showed poorer prognosis, including both overall survival (OS) and recurrence-free survival (RFS). Multivariate Cox analysis indicated low serum circFoxO3a levels to be an unfavorable prognostic factor for both OS and RFS, independent of positive lymph node metastasis. Notably, the predictive nomogram we established further confirmed that serum circFoxO3a is a useful tool for predicting survival in SCC.
Conclusion
Altogether, our findings demonstrated that serum circFoxO3a could serve as a potential novel noninvasive predictive prognostic biomarker and therapeutic target for SCC.
Keywords: circular RNA, FoxO3a, squamous cervical cancer, biomarker
Introduction
Cervical cancer is the fourth most common and leading cause of cancer-related death in women, with an estimated 570,000 cases and 311,000 deaths worldwide in 2018.1 Squamous cervical cancer (SCC), which is the predominant histological type, accounts for approximately 80% of all cervical cancer cases.2 Despite the widespread implementation of screening tests and advancements in treatment for SCC, the prognosis of SCC patients remains poor.3,4 Regarding prognostic factors, certain traditional factors such as International Federation of Gynecology and Obstetrics (FIGO) stage, lymph node metastasis, lymph-vascular space invasion (LVSI) and deep stromal infiltration are usually considered.5 However, assessments of these clinicopathological factors are invasive because they are obtained from postoperative tissues; moreover, they cannot precisely predict prognosis preoperatively.6 Therefore, noninvasive prognosis prediction is crucial, which can help decision-making and provide therapeutic guidance for SCC to a certain degree.
Currently, different types of serum biomarkers are used for the diagnosis and prognosis of various cancers because of their noninvasiveness and reliability, such as prostate-specific antigen (PSA) for prostate cancer and carcinoembryonic antigen (CEA) for colorectal cancer.7,8 Although certain serum biomarkers are also used for CC, their sensitivity and specificity are not satisfactory.9 For example, SCC antigen (SCC-Ag), which is currently the most commonly used biomarker for SCC, can also be elevated in squamous cell carcinoma of the lungs,10 esophagus,11 head and neck.12 Moreover, SCC-Ag levels are also increased in nontumorous diseases such as psoriasis,13 nephritic syndrome14 and allergic asthma.15 Hence, there is an urgent need to explore novel noninvasive serum biomarkers for a more accurate prognosis prediction of SCC.
Circular RNAs (circRNAs) are a new class of noncoding RNAs that are characterized by covalently closed loops without 3ʹ- and 5ʹ- ends.16 In recent years, numerous studies have revealed that circRNAs are widely expressed in various tissues and play important roles in tumor development and progression.17 Expression of circRNAs is tissue- and developmental stage-specific.18 Furthermore, their closed circular structure renders circRNAs relatively resistant to RNA enzymes, and they are stable and enriched in peripheral blood and/or body fluids.16 Therefore, circular RNAs have a distinct advantage in the development and application as novel clinical biomarkers. For example, Fan et al found that circMAN1A2 may serve as a serum biomarker for malignant tumors, providing important insights into diagnostic approaches for malignant tumors such as gastrointestinal cancer, thyroid cancer, ovarian cancer, and lung cancer.19 In plasma and tissues obtained from patients with gastric cancer, has_circ_0001017 and has_circ_0061276 were evidently decreased and this reduction was significantly associated with distal metastasis. The combination of has_circ_0001017 and has_circ_0061276 showed a sensitivity and specificity of 95.5% and 95.7% respectively, for the diagnosis of gastric cancer.20 Despite these findings, however, research related to circRNAs as noninvasive biomarkers in SCC is limited.
Circular Forkhead box O3a (circFoxO3a), encoded by the FoxO3 gene which is a well-known tumor suppressor, is receiving increasing attention.21–23 For example, circFoxO3a was reported to be downregulated in various cancer cells, and is involved in cell proliferation and cell cycle progression.24 A recent study also found that circFoxO3a has a tumor suppressive activity and may serve as a novel biomarker for the early diagnosis, treatment monitoring and prognosis of patients with non-small cell lung cancer (NSCLC).25 However, to the best of our knowledge, the specific clinical significance of circFoxO3a and whether it can serve as a novel biomarker in SCC have not been investigated.
In this study, we first detected circFoxO3a in sera from SCC patients and then investigated the correlation between the expression of serum circFoxO3a and clinicopathological factors. Furthermore, we analyzed the prognostic value of circFoxO3a and attempted to establish a predictive model for SCC.
Materials and Methods
Patients
One hundred and three SCC patients who underwent surgery at the Obstetrics and Gynecology Hospital of Fudan University between December 2013 and October 2014 were enrolled in this study. The inclusion criteria were as follows: 1) the diagnosis of SCC was confirmed by the pathological report; 2) patients without any preoperative radiotherapy or chemotherapy; and 3) patients without any other malignancies or severe chronic disease. Thirty healthy women, as confirmed by their routine physical examination reports were enrolled as a control group. All participants were well informed and signed the written informed consent form. The study was approved by the Medical Ethics Committee of Obstetrics and Gynecology Hospital of Fudan University. Regular follow-up was conducted with a median period of 49 months (range: 8–60 months). Preoperative blood samples were stored at −80°C in the tissue bank of our hospital. Clinical data were extracted from medical records, including FIGO stage, tumor size, depth of invasion, lymph node metastasis and LVSI status. The depth of stromal invasion was defined as the percentage of the involved cervical stroma, and the tumor size was a direct measurement of the greatest dimension. The clinicopathological characteristics of the patients are listed in Table 1.
Table 1.
Baseline Characteristics of 103 Patients with Squamous Cervical Cancer
| Variable | Number (%) |
|---|---|
| Age, y (mean ± SD) | 51.1±9.41 |
| ≤45 | 29 (28.2%) |
| >45 | 74 (71.8%) |
| Stage | |
| IA2 | 2 (1.9%) |
| IB1-IB2 | 70 (68.0%) |
| IIA1-IIA2 | 31 (30.1%) |
| Surgery Type | |
| Laparoscopy | 96(93.2%) |
| Laparotomy | 7 (6.8%) |
| Tumor size | |
| ≤4cm | 71(68.9%) |
| >4cm | 32(31.3%) |
| Lymphovascular invasion | |
| Negative | 44(42.7%) |
| Positive | 59(57.3%) |
| Stromal invasion depth | |
| <1/2 | 38(36.9%) |
| ≥1/2 | 65(63.1%) |
| Lymph node metastasis | |
| Negative | 71(68.9%) |
| Positive | 32(31.1%) |
| Parametrial invasion | |
| Negative | 86(83.5%) |
| Positive | 17(16.5%) |
| Margin | |
| Negative | 98(95.1%) |
| Positive | 5(4.9%) |
| HPV infection | |
| Negative | 12(11.7%) |
| Positive | 91(88.3%) |
| Ki-67 | |
| Low (≤35%) | 27(26.2%) |
| Median (≤70%) | 64(62.1%) |
| High (>70%) | 12(11.7%) |
RNA Preparation and RNase R Digestion
Total serum RNA was extracted and purified using an RNA Isolation Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The RNA concentration was assessed with NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Total RNA (2 µg) was incubated at 37°C for 15 min with 3 U/µg RNase R (Epicenter Biotechnologies, Shanghai, China) to remove linear RNA and purify the circRNAs.
Reverse Transcription‑Quantitative Polymerase Chain Reaction (RT‑qPCR)
After digestion with RNase R, expression of circFoxO3a was detected by RT-qPCR. Quantitative RT-qPCR was performed using an ABI 7500 Fast Real‐time PCR system (Applied Biosystems, Germany) with the SYBR-Green PCR Master Mix kit (Takara, Dalian, China) according to the manufacturer’s instructions. The primers were purchased from Sangon Biotech (Shanghai, China) and the sequences were as follows: circFoxO3a forward, 5ʹ-ATTGTCCATGGAGACAGGCCCGCCG-3ʹ and reverse, 5ʹ-GTGGGGAACTTCACTGGTGCTAAG-3ʹ; and GAPDH forward, 5ʹ-GTCTCCTCTGACTTCAACAGCG-3ʹ and reverse, 5ʹ- ACCACCCTGTTGCTGTAGCCAA-3ʹ. GAPDH was used as an internal reference, and relative expression of circFoxO3a was calculated using the 2−ΔΔCt method.
Statistical Analysis
Statistical analysis was performed with SPSS 22.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Student’s t-test was used to compare the expression of circFoxO3a between SCC patients and healthy women, and the Chi square test and Fisher’s exact test were employed to analyze associations between expression of serum circFoxO3a and clinicopathological factors. Overall survival (OS) and recurrence-free survival (RFS) were defined as the time interval from the date of surgery to the date of death or recurrence, respectively, or final contact. The Kaplan–Meier method along with the Log rank test were applied for survival analysis. Multivariate Cox regression analysis was then carried out to investigate the prognostic value of the clinicopathological factors and circFoxO3a expression. Based on the results of multivariate Cox analysis, nomograms for OS and RFS were conducted in R version 3.5.3 (https://www.r-project.org/) as described in previous studies.26 P<0.05 was considered statistically significant. The related computerized programs for the nomograms are shown in Supplementary Materials.
Results
Expression of Serum circFoxO3a Was Decreased in SCC Patients
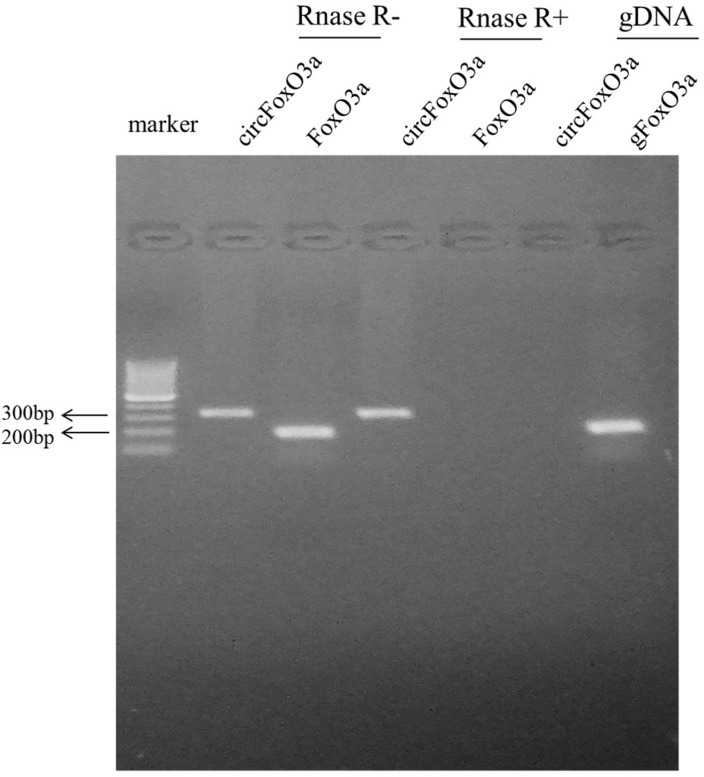
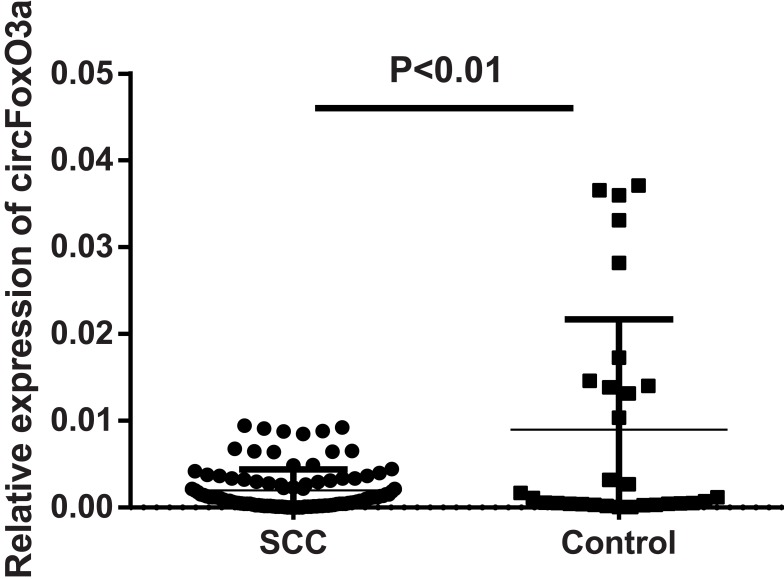
The presence of serum circFoxO3a was detected using convergent or divergent primers by qRT-PCR and validated by gel electrophoresis (Figure 1). As shown in Figure 2, the expression of serum circFoxO3a in SCC patients was significantly lower than that of normal healthy women (P<0.01), indicating that decreased serum circFoxO3a levels might be correlated with the malignant behavior of SCC.
Figure 1.
The existence of circFoxO3a was validated by Gel electrophoresis. The divergent primers could detect circular RNAs in cDNA with or without RNase R, which demonstrate that they are truly circular not linear, but these divergent primers could not amplify any product in genomic DNA. The convergent primers amplified PCR production in linear FoxO3a mRNA, which disappeared after RNase R treatment.
Figure 2.
Relative expression of serum circFoxO3a in SCC patients (n=103) and normal controls (n=30).
Low Expression of Serum circFoxO3a Was Correlated with Deeper Stromal Invasion, and Positive Lymph Node Metastasis
Next, all 103 SCC patients were divided into two groups based on the median expression levels of circFoxO3a: high (n=52) and low (n=51) groups. We further investigated whether the expression of circFoxO3a was correlated with several clinicopathological factors of SCC (Table 2). The results showed that a low expression of serum circFoxO3a was significantly associated with deeper stromal invasion depth (P=0.005) and positive lymph node metastasis (P=0.008). Therefore, these findings suggest that decreased circFoxO3a levels are associated with the aggressive behaviors of SCC.
Table 2.
The Correlation Between the Expression of Serum circFoxO3a and Clinicopathological Factors
| Variable | Low Expression of circFoxO3a (n=51) |
High Expression of circFoxO3a (n=52) |
P |
|---|---|---|---|
| Age, y (mean ± SD) | 0.105 | ||
| ≤45 | 11 | 18 | |
| >45 | 40 | 34 | |
| Stage | 0.881 | ||
| I | 36 | 36 | |
| II | 15 | 16 | |
| Surgery Type | 0.652 | ||
| Laparoscopy | 48 | 49 | |
| Laparotomy | 3 | 3 | |
| Tumor size | 0.059 | ||
| ≤4cm | 31 | 40 | |
| >4cm | 20 | 12 | |
| Lymphovascular invasion | 0.181 | ||
| Negative | 19 | 25 | |
| Positive | 32 | 27 | |
| Stromal invasion depth | 0.005 | ||
| <1/2 | 12 | 26 | |
| ≥1/2 | 39 | 26 | |
| Lymph node metastasis | 0.008 | ||
| Negative | 29 | 42 | |
| Positive | 22 | 10 | |
| Parametrial invasion | 0.347 | ||
| Negative | 46 | 49 | |
| Positive | 5 | 3 | |
| Margin | 0.491 | ||
| Negative | 48 | 50 | |
| Positive | 3 | 2 | |
| HPV infection | 0.188 | ||
| Negative | 4 | 8 | |
| Positive | 47 | 44 | |
| Ki-67 | 0.166 | ||
| Low (≤35%) | 13 | 14 | |
| Median (≤70%) | 29 | 35 | |
| High (>70%) | 9 | 3 |
Serum circFoxO3a Was an Independent Prognostic Factor in SCC
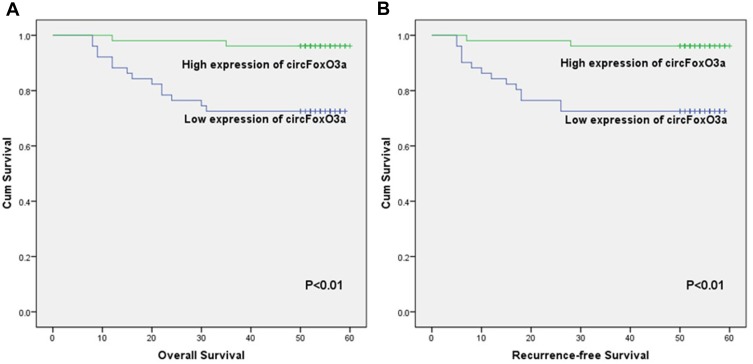
We conducted the Kaplan-Meier method and Log rank test to evaluate the prognostic value of serum circFoxO3a in SCC. The median OS time in SCC patients with a low expression of serum cicrFoxO3a and positive lymph node metastasis was significantly shorter than that in patients with a high expression (47.47±2.68 vs 58.60±1.02, 43.44±3.85 vs 57.01±1.01, respectively, all P<0.05, Table 3); similar findings were obtained in the analysis of RFS. Additionally, as shown in Figure 3A and B, a low expression of serum circFoxO3a was associated with poor survival outcomes including both OS and RFS. Moreover, univariate and multivariate analyses (Table 4) identified low serum circFoxO3a, independent of positive lymph node metastasis, to be an unfavorable prognostic factor for both OS and RFS (P<0.05). Altogether, these findings suggest that serum circFoxO3a can serve as a novel prognostic biomarker for SCC.
Table 3.
The Correlation Between Clinicopathological Characteristics and Overall Survival, Recurrence-Free Survival Using Kaplan–Meier Method
| Variable | Overall Survival (Months) Mean ± SE |
P | Recurrence-Free Survival (Months) Mean ± SE |
P |
|---|---|---|---|---|
| Age, y (mean ± SD) | 0.146 | 0.150 | ||
| ≤45 | 55.66±2.28 | 55.35±2.50 | ||
| >45 | 52.22±1.92 | 51.37±2.12 | ||
| Stage | 0.124 | 0.134 | ||
| I | 56.18±1.41 | 55.66±1.59 | ||
| II | 49.31±3.23 | 48.55±3.50 | ||
| Surgery Type | 0.229 | 0.299 | ||
| Laparoscopy | 48.43±14.42 | 47.68±15.90 | ||
| Laparotomy | 55.14±3.02 | 55.14±3.02 | ||
| Tumor size | 0.086 | 0.094 | ||
| ≤4cm | 54.85±1.73 | 54.31±1.91 | ||
| >4cm | 49.59±1.54 | 48.53±3.29 | ||
| Lymphovascular invasion | 0.342 | 0.352 | ||
| Negative | 53.80±2.20 | 53.27±2.42 | ||
| Positive | 52.53±2.10 | 51.70±2.30 | ||
| Stromal invasion depth | 0.314 | 0.323 | ||
| <1/2 | 55.03±2.37 | 54.58±2.58 | ||
| ≥1/2 | 51.71±1.96 | 50.86±2.16 | ||
| Lymph node metastasis | <0.001 | <0.001 | ||
| Negative | 57.01±1.01 | 56.75±1,12 | ||
| Positive | 43.44±3.85 | 41.78±4.21 | ||
| Parametrial invasion | 0.066 | 0.073 | ||
| Negative | 54.13±1.54 | 53.52±1.69 | ||
| Positive | 43.50±6.61 | 41.75±7.23 | ||
| Margin | 0.083 | 0.078 | ||
| Negative | 54.03±1.51 | 53.38±1.67 | ||
| Positive | 40.20±9.41 | 38.60±10.22 | ||
| HPV infection | 0.943 | 0.950 | ||
| Negative | 52.17±3.86 | 51.50±4.28 | ||
| Positive | 53.40±1.66 | 52.69±1.82 | ||
| Ki-67 | 0.732 | 0.736 | ||
| Low | 51.70±3.42 | 50.96±3.70 | ||
| Median | 52.92±1.81 | 52.16±2.02 | ||
| High | 53.45±1.54 | 54.50±4.31 | ||
| CircFoxO3a | 0.001 | 0.001 | ||
| Low | 47.47±2.68 | 46.29±2.94 | ||
| High | 58.60±1.02 | 58.37±1.17 |
Figure 3.
Kaplan–Meier survival curves for overall survival (A) and recurrence-free survival (B) of SCC patients according to expression levels of serum circFoxO3a.
Table 4.
Univariate and Multivariate Cox Regression Analysis of Overall and Recurrence-Free Survival for Patients with Squamous Cervical Cancer
| Variable | Recurrence-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |||||
| HR (95% CI) | P | HR (95% CI) | HR (95% CI) | P | HR (95% CI) | P | ||
| Age | 2.822(0.641±12.417) | 0.170 | 2.848(0.647±12.532) | 0.166 | ||||
| Stage | 2.242(0.753±6.675) | 0.147 | 2.292(0.770±6.821) | 0.136 | ||||
| Surgery Type | 0.044(0.000±179.912) | 0.462 | 0.044(0.000±179.843) | 0.462 | ||||
| Tumor size | 2.254(0.846±6.007) | 0.104 | 2.295(0.861±6.117) | 0.097 | ||||
| Lymphovascular invasion | 1.640(0.570±4.721) | 0.359 | 1.658(0.576±4.772) | 0.349 | ||||
| Stromal invasion depth | 1.752(0.565±5.432) | 0.332 | 1.771(0.571±5.492) | 0.322 | ||||
| Lymph node metastasis | 8.154(2.624±25.342) | 0.000 | 5.940(1.883±18.744) | 0.002 | 8.157(2.625±25.353) | 0.000 | 5.926 (1.878±18.705) | 0.002 |
| Parametrial invasion | 2.971(0.846±10.431) | 0.089 | 3.054(0.870±10.723) | 0.081 | ||||
| Margin | 3.472(0.788±15.297) | 0.100 | 3.411(0.774±15.025) | 0.105 | ||||
| HPV infection | 0.954(0.217±4.199) | 0.951 | 0.948(0.215±4.171) | 0.944 | ||||
| Ki-67 | 0.727(0.316±1.671) | 0.452 | 0.724(0.314±1.665) | 0.447 | ||||
| CircFoxO3a | 0.123(0.028±0.541) | 0.006 | 0.180(0.040±0.809) | 0.025 | 0.122(0.028±0.539) | 0.005 | 0.181(0.040±0.815) | 0.026 |
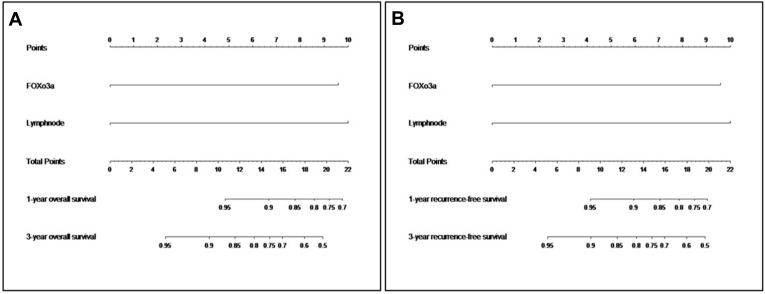
A Nomogram for Prognosis Prediction of SCC Was Established
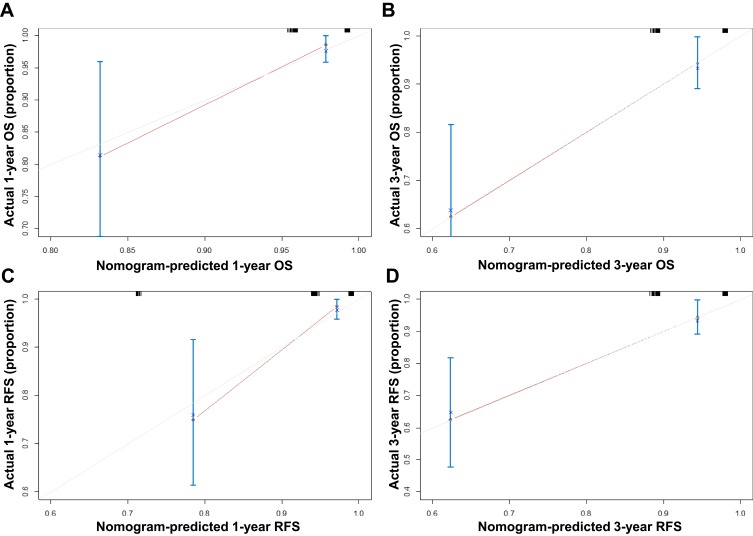
Based on the above findings, we established a nomogram using R version 3.5.3 (Figure 4A and B). Different scores were assigned to different statuses of circFoxO3a expression and lymph node metastasis. By summing the corresponding score, we were able to directly and easily predict the probability of survival at different time points. The c-index for the established nomogram to predict OS and RFS were both 0.808 (95% CI, 0.616 to 0.990). The calibration curves for the nomogram-predicted probability of 1- and 3-year OS/RFS showed good agreement between the actual observation and nomogram prediction (Figure 5). These results confirm that the nomogram combining expression of serum circFoxO3a and status of lymph node metastasis is a useful tool for predicting the survival of SCC patients.
Figure 4.
Nomogram for predicting overall survival (A) and recurrence-free survival (B) in SCC patients.
Figure 5.
The calibration curves of nomogram for predicting 1-year (A), 3-year (B) overall survival and 1-year (C), 3-year (D) recurrence-free survival.
Discussion
Circular RNAs, as a novel type of noncoding RNA, were originally considered nonfunctional accidental byproducts of aberrant splicing.17 However, emerging evidence indicates that circRNAs can play vital roles via miRNA sponge-like effects or circRNA-protein interactions in tumorigenesis and metastasis, such as in breast cancer,27 hepatocellular carcinoma,28 colorectal cancer,29 and lung cancer.30 Furthermore, an increasing number of studies have shown that circRNAs can serve as effective diagnostic and prognostic biomarkers for cancer.31 Nonetheless, studies focusing on circular RNAs and SCC are limited.
Forkhead box (Fox) proteins are a well-known family of transcription factors. Among them, FoxO3a, also known as FoxO3, has been extensively investigated in various types of cancers. Most of these studies reported a tumor-suppressive function of FoxO3a. In addition, tumors with high FoxO3a expression usually indicates better patient survival.32 With the increasing focus on circRNAs, circFoxO3a has also attracted researchers’ attention. Du et al demonstrated that circFoxO3a is highly expressed in noncancer cells and associated with cell cycle progression.33 Zhang et al reported that circFoxO3a appeared to have tumor-suppressive activity by sponging miR‑155 in NSCLC and that it may serve as a novel biomarker for the early diagnosis and prognosis prediction of NSCLC.25 However, the involvement of circFoxO3a in SCC remains unclear.
In this study, we first noted that serum circFoxO3a was decreased in SCC patients compared with in normal healthy women. We next found that a low expression of serum circFoxO3a was correlated with deeper stromal invasion and positive lymph node metastasis, suggesting an association between a low expression of serum circFoxO3a and aggressive tumor behavior. Further survival analysis revealed that SCC patients with a low expression of serum circFoxO3a and positive lymph node metastasis displayed poorer outcomes, including both OS and RFS. In multivariate Cox analysis, circFoxO3a expression in addition to lymph node metastasis was an independent factor of survival in SCC patients. As all SCC patients with recurrence died due to recurrence, multivariate analysis for OS and RFS yielded similar results. Taken together, these results indicated that low expression of serum circFoxO3a is associated with the aggressive behavior of SCC and that serum circFoxO3a may serve as an independent prognostic factor for SCC.
In recent years, nomograms, as a tool for prognosis prediction, have been well developed and proven to be more accurate than conventional staging systems in some cancers.34,35 Based on the results of multivariate Cox analysis, we successfully established a nomogram for the prognosis prediction of SCC. The nomogram performed well in predicting both OS and RFS with a c-index of 0.808. The c-index is the most widely used index to assess the predictive efficiency of nomograms. Theoretically, prognostic prediction of the nomogram is more accurate with a larger c-index. Additionally, calibration curves confirmed the optimal predictive efficiency of the nomograms. Based on this nomogram model, we conclude that serum circFoxO3a can be utilized as a novel biomarker for prognosis prediction of SCC in clinical practice.
There are several limitations of this study. First, the sample size was relatively small and all patients were from a single medical center in China, which may have led to selection bias. Second, the detailed molecular mechanism of circFoxO3a in SCC has not been explored and needs further investigation. Last, this study focused on the expression and prognostic value of circFoxO3a in SCC. In the future, we will enroll cervical cancer patients with other histology such as adenocarcinoma and adenosquamous carcinoma, and enlarge the sample size to further explore the role of the circFoxO3a in cervical cancer and enhance its clinical application.
Conclusion
In summary, we reported decreased expression of serum circFoxO3a in SCC patients for the first time. Furthermore, our study demonstrated that lower expression of circFoxO3a was associated with deeper stromal invasion and positive lymph node metastasis in SCC patients. Additionally, there was a strong correlation between expression level of serum circFoxO3a and prognosis in SCC patients. SCC patients with lower expression of serum circFoxO3a showed shorter OS and RFS. A predictive nomogram model utilizing serum circFoxO3a was successfully established and showed good predictive ability for the OS and RFS of SCC patients. Altogether, our findings indicate that serum circFoxO3a can potentially serve as a novel predictive prognostic biomarker and therapeutic target for SCC.
Funding Statement
This project was supported by funding from the National Natural Science Foundation of China (No. 81971361; to Jun-jun Qiu), the Natural Science Foundation of Shanghai Science and Technology (No. 19ZR1406900; to Jun-jun Qiu), and shanghai “Rising Stars of Medical Talent” Youth Development Program to Jun-jun Qiu, the Research and Innovation Project of Shanghai Municipal Education Commission (No. 2019-01-07-00-07-E00050; to Ke-qin Hua), and the artificial Intelligence Innovation Project of Shanghai Municipal Commission of Economy and Informatization (No. 2018-RGZN-02041; to Ke-qin Hua).
Abbreviations
SCC, squamous cervical cancer; FIGO, International Federation of Gynecology and Obstetrics; LVSI, lymph-vascular space invasion; CircRNAs, circular RNAs; CircFoxO3a, circular Forkhead box O3a; OS, overall survival; RFS, recurrence-free survival; RT‑qPCR, reverse transcription‑quantitative polymerase chain reaction; NSCLC, non-small cell lung cancer; PSA, prostate-specific antigen; CEA, carcinoembryonic antigen; SCC-Ag, squamous cell carcinoma antigen.
Consent for Publication
This manuscript has been approved by all authors for publication. I declare on behalf of my coauthors that the work described is original research that has not been published previously and is not under consideration for publication elsewhere, either in whole or in part. All of the authors have approved the enclosed manuscript.
Data Sharing Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Ethics and Consent Statement
Written consent was obtained from each participant. The study protocol was approved by the Medical Ethics Committee of Obstetrics and Gynecology Hospital of Fudan University and conducted in accordance with the Declaration of Helsinki.
Author Contributions
Xiaoyan Tang, Songping Liu, Yan Ding are the co-first authors to this article and contributed equally to this research project. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Ghasemi F, Shafiee M, Banikazemi Z, et al. Curcumin inhibits NF-kB and Wnt/beta-catenin pathways in cervical cancer cells. Pathol Res Pract. 2019;215(10):152556. doi: 10.1016/j.prp.2019.152556 [DOI] [PubMed] [Google Scholar]
- 3.Shafabakhsh R, Reiter RJ, Mirzaei H, Teymoordash SN, Asemi Z. Melatonin: a new inhibitor agent for cervical cancer treatment. J Cell Physiol. 2019;234(12):21670–21682. doi: 10.1002/jcp.v234.12 [DOI] [PubMed] [Google Scholar]
- 4.Sadri Nahand J, Moghoofei M, Salmaninejad A, et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: a review. Int J Cancer. 2020;146(2):305–320. doi: 10.1002/ijc.v146.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonardi S, Buttarelli M, De Stefano I, et al. The relevance of prelamin A and RAD51 as molecular biomarkers in cervical cancer. Oncotarget. 2017;8(55):94247–94258. doi: 10.18632/oncotarget.v8i55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasari S, Wudayagiri R, Valluru L. Cervical cancer: biomarkers for diagnosis and treatment. Clin Chim Acta. 2015;445:7–11. doi: 10.1016/j.cca.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 7.Troyer DA, Mubiru J, Leach RJ, Naylor SL. Promise and challenge: markers of prostate cancer detection, diagnosis and prognosis. Dis Markers. 2004;20(2):117–128. doi: 10.1155/2004/509276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baqar AR, Wilkins S, Staples M, Angus Lee CH, Oliva K, McMurrick P. The role of preoperative CEA in the management of colorectal cancer: a cohort study from two cancer centres. Int J Surg. 2019;64:10–15. doi: 10.1016/j.ijsu.2019.02.014 [DOI] [PubMed] [Google Scholar]
- 9.Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234(10):17064–17099. doi: 10.1002/jcp.28457 [DOI] [PubMed] [Google Scholar]
- 10.Tao H, Soh J, Yamamoto H, et al. Restrictive ventilatory impairment is associated with poor outcome in patients with cT1aN0M0 peripheral squamous cell carcinoma of the lung. J Thorac Dis. 2017;9(11):4325–4335. doi: 10.21037/jtd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng JF, Chen S, Yang X. Combination of c-reactive protein and squamous cell carcinoma antigen in predicting postoperative prognosis for patients with squamous cell carcinoma of the esophagus. Oncotarget. 2017;8(38):63132–63139. doi: 10.18632/oncotarget.18667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Travassos DC, Fernandes D, Massucato EMS, Navarro CM, Bufalino A. Squamous cell carcinoma antigen as a prognostic marker and its correlation with clinicopathological features in head and neck squamous cell carcinoma: systematic review and meta-analysis. J Oral Pathol Med. 2018;47(1):3–10. doi: 10.1111/jop.12600 [DOI] [PubMed] [Google Scholar]
- 13.El-Rachkidy RG, Young HS, Griffiths CE, Camp RD. Humoral autoimmune responses to the squamous cell carcinoma antigen protein family in psoriasis. J Invest Dermatol. 2008;128(9):2219–2224. doi: 10.1038/jid.2008.71 [DOI] [PubMed] [Google Scholar]
- 14.Yang D, Wang J, Zhang L. Serum SCCA levels in patients suffering cancers or other diseases. Prog Mol Biol Transl Sci. 2019;162:165–175. [DOI] [PubMed] [Google Scholar]
- 15.Ray R, Choi M, Zhang Z, Silverman GA, Askew D, Mukherjee AB. Uteroglobin suppresses SCCA gene expression associated with allergic asthma. J Biol Chem. 2005;280(11):9761–9764. doi: 10.1074/jbc.C400581200 [DOI] [PubMed] [Google Scholar]
- 16.Gao YL, Zhang MY, Xu B, et al. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8(49):86625–86633. doi: 10.18632/oncotarget.21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shabaninejad Z, Vafadar A, Movahedpour A, et al. Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res. 2019;12(1):84. doi: 10.1186/s13048-019-0558-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Teng S, Xu J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2018;20(4):1420–1433. [DOI] [PubMed] [Google Scholar]
- 19.Fan CM, Wang JP, Tang YY, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019;110(7):2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96(1):85–96. doi: 10.1007/s00109-017-1600-y [DOI] [PubMed] [Google Scholar]
- 21.Gao Z, Li Z, Liu Y, Liu Z. Forkhead box O3 promotes colon cancer proliferation and drug resistance by activating MDR1 expression. Mol Genet Genomic Med. 2019;7(3):e554. doi: 10.1002/mgg3.2019.7.issue-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120(Pt 15):2479–2487. doi: 10.1242/jcs.001222 [DOI] [PubMed] [Google Scholar]
- 23.Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O’Brien R, Granner DK. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J Biol Chem. 2000;275(39):30169–30175. doi: 10.1074/jbc.M004898200 [DOI] [PubMed] [Google Scholar]
- 24.Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Zhao H, Zhang L. Identification of the tumor-suppressive function of circular RNA FOXO3 in nonsmall cell lung cancer through sponging miR155. Mol Med Rep. 2018;17(6):7692–7700. doi: 10.3892/mmr.2018.8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol. 2015;33(8):861–869. doi: 10.1200/JCO.2014.56.6661 [DOI] [PubMed] [Google Scholar]
- 27.Chen N, Zhao G, Yan X, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218. doi: 10.1186/s13059-018-1594-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–316. [DOI] [PubMed] [Google Scholar]
- 29.Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi: 10.1158/0008-5472.CAN-16-1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan S, Gou Q, Pu W, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018;28(6):693–695. doi: 10.1038/s41422-018-0033-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Zeng X, He J, et al. Circular RNA as a biomarker for cancer: a systematic meta-analysis. Oncol Lett. 2018;16(3):4078–4084. doi: 10.3892/ol.2018.9125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullock MD, Bruce A, Sreekumar R, et al. FOXO3 expression during colorectal cancer progression: biomarker potential reflects a tumour suppressor role. Br J Cancer. 2013;109(2):387–394. doi: 10.1038/bjc.2013.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sternberg CN. Are nomograms better than currently available stage groupings for bladder cancer? J Clin Oncol. 2006;24(24):3819–3820. doi: 10.1200/JCO.2006.07.1290 [DOI] [PubMed] [Google Scholar]
- 35.Fornaro L, Vivaldi C, Parnofiello A, et al. Validated clinico-pathologic nomogram in the prediction of HER2 status in gastro-oesophageal cancer. Br J Cancer. 2019;120(5):522–526. doi: 10.1038/s41416-019-0399-4 [DOI] [PMC free article] [PubMed] [Google Scholar]