Abstract
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most pathogens associated with health care. Molecular typing methods are vital for outbreak investigations of MRSA. The aim of this study was characterization of SCCmec, spa types and multidrug resistant of methicillin-resistant Staphylococcus aureus isolates in Ahvaz, Iran.
Methods
A total of 50 MRSA isolates were determined by using the phenotypic method and mecA gene. Antibiotic resistance profile and SCCmec types were screened using disc diffusion method and PCR, respectively. For spa typing of MRSA isolates, two molecular typing methods including the PCR-sequencing and high-resolution melting (HRM) analysis were used.
Results
In the present study, the highest sensitivity of MRSA was to vancomycin and linezolid and the lowest to clindamycin. In the MRSA isolates, 22% were XDR and 78% were MDR. SCCmec type III was found commonly among MRSA. Based on PCR-sequencing and HRM results, 10 different spa types were identified. The spa types t037 and t030 were the most common in this study.
Conclusion
This study emphasizes the spa variation among MRSA isolates, which may be considered as an important criterion when treating staphylococcal infections. Accurate and early detection of MDR, XDR, or even PDR MRSA isolates strains must be commenced by all clinical microbiology laboratories to reduce the menace of antimicrobial resistance.
Keywords: high-resolution melting, HRM, spa-typing, multidrug resistant Staphylococcus aureus, Antibiotic resistance
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is still a major cause of community and healthcare-associated infections worldwide.1,2 In Iran, the frequency of MRSA infection has increased over the past decade. In a recently published meta-analysis, MRSA infections were reported to be highly prevalent (43.0%) among confirmed S. aureus isolates, underscoring the increasing prevalence of MRSA isolates over the recent years.3 The emergence of MRSA strains is associated with the presence of the mecA gene, which encodes an alternative 78 kDa penicillin-binding protein (PBP2ʹ) that has decreased affinity for β-lactam antibiotics.
MRSA has been detected by disk diffusion method, agar dilution method and oxacillin screen agar test as recommended by Clinical Laboratory Standard Institution. In a number of clinical microbiology laboratories, the routine procedure for MRSA detection was based on phenotypic assays such as disk diffusion and broth microdilution. Culture requires a long turnaround time of approximately 18–48 hour with low sensitivity and 100% specificity. Whereas detection of mecA gene is the “Gold Standard” offering high sensitivity and rapid results but these methods are not always been possible in many facilities constrains laboratories.4
The mecA gene is located on a mobile genetic element called the staphylococcal cassette chromosome mec (SCCmec). Moreover, SCCmec is comprised of two main components, namely, ccr gene complex ccr and mec gene complex. The combination of ccr allotypes with the mec gene complex, 11 types (I–XI) SCCmec has already been reported. According to previous studies, SCCmec types I, II, and III are related to hospital-acquired MRSA.5
The molecular typing of MRSA is crucial for outbreak investigation and epidemiological studies.6 The techniques most widely employed for MRSA typing are pulsed-field gel electrophoresis (PFGE), staphylococcal cassette chromosome mec (SCC mec) typing, sequence-based typing methods such as S. aureus protein A (spa) typing and multi-locus sequence typing (MLST).7 In the last decade, the spa gene is the most widely used for S. aureus typing, which is based on repeats of the hypervariable X region in the spa gene.8 The Ridom spa server database currently contains 18857 spa types with data from 141 countries worldwide (http://www.spaserver.ridom.de, 2019). Although sequencing technology is currently very effective, non-sequence-based genotyping methods can also offer advantages. In particular, the real-time PCR platform supports single step and closed tube genotyping methods and can potentially be carried out simultaneously with diagnosis and interrogate different classes of genetic polymorphisms.
These features provide real advantages for the clinical microbiology laboratory. The high-resolution DNA melting analysis (HRMA) has been recently assessed for spa typing in comparison with DNA sequencing of the spa gene.9 This rapid, simple, and cost-effective assay is able to differentiate the PCR products with minimum differences in GC content, length, and compositions, resulting in melting temperature (TM) variation.10
In the previous studies performing HRM and spa typing, certain spa types could not be separated from each other based on their Tm.9,11
The present study had three objectives: i) characterization of SCCmec, spa types, and multidrug resistance of methicillin-resistant Staphylococcus aureus isolates in Ahvaz, Iran; ii) application of high-resolution melting curve analysis for a rapid identification of endemic spa types in multidrug resistant staphylococcus aureus.
Materials and Methods
Ethics Statement
The study was approved by the Research Ethics Committee (REC) of the Ahvaz Jundishapur University of Medical Sciences (No: IR.AJUMS.REC.1395.187), Ahvaz, Iran, and all patients provided written informed consent. The REC has a mission to protect the dignity, rights, safety, and well-being of subjects who participate in biomedical research and to offer public accountability through the publication of their decisions.
Sample Collection and Bacterial Isolates
A total of 146 non-duplicated S. aureus isolates collected from patients admitted to Golestan, Taleghani and Razi teaching hospitals, were recorded from January to December 2018. We have described the aim of the study and its methods to the individuals, and if they consented to participate in the study, the data information of each person was entered into a previously prepared form. The participants have included the personnel of Surgery, Emergency, ICU, Women, pediatrics, pediatrics ICU, nephrology and orthopedic wards of the hospital due to the higher prevalence of infection in these wards. The S. aureus was isolated from various clinical samples including blood, urine, wound, synovial fluid, and abscesses. The collected samples were immediately transferred to the Department of Microbiology of Ahvaz Jundishapur University of Medical Sciences. First, the isolates were subcultured on Blood Agar (Merck, Germany), and the single colony was inoculated on Mannitol Salt Agar (Merck, Germany) at 37°C for 24 h. The suspicious colonies were subjected to biochemical tests including Gram staining, catalase, tube-Coagulase and Dnase.12 The S. aureus ATCC 29213 strain was used as the reference strain. Finally, S. aureus isolates were inoculated in Trypticase soy broth (TSB) (Merck, Germany) with 20% glycerol and were kept at −80°C until use.
Investigation of Susceptibility to Antimicrobial Agents
Antibiotic susceptibility testing was performed for 11 drugs covering all the 11 antimicrobial categories comprising aminoglycosides, ansamycins, fluoroquinolones, folate pathway inhibitors, tetracyclines, glycopeptides, oxazolidinones, phenicols, macrolides, lincosamides and penicillinase-stable penicillins were determined using the disc diffusion susceptibility test according to clinical and laboratory standards institute (CLSI) guidelines. Commercial antibiotic discs of rifampin (5 μg), linezolid (30 μg), ciprofloxacin (5 μg), tetracycline (10 μg), gentamycin (10 μg), erythromycin (15 μg), clindamycin (2 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), cefoxitin (30 μg) and chloramphenicol (30 μg) (MAST Diagnostics, Merseyside UK) were used in disc diffusion test. Then, MDR/XDR/PDR phenotype of these isolates was established according to the results obtained from the disc diffusion test.13
Criteria for Defining MDR, XDR and PDR in S. aureus
As per standardized international terminology created by European Centre for Disease Control (ECDC) and Centre for Disease Control & Prevention (CDC), the multidrug-resistant (MDR), extensively drug-resistant (XDR), and pandrug-resistant (PDR) bacteria have been well defined. Multidrug-resistant (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories. Extensively drug-resistant (XDR) was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e., bacterial isolates remain susceptible to only one or two antimicrobial categories). Pandrug resistance (PDR) was defined as non-susceptibility to all agents in all antimicrobial categories. According to reference, MRSA isolates should be considered MDR.14
Screening for Methicillin Resistance by Cefoxitin Disc Diffusion Test
All of S. aureus isolates were tested for susceptibility to methicillin using a cefoxitin (30 µg) disc. Results were interpreted according to the criteria established by the CLSI. S. aureus ATCC 29213 strain were used as a positive control and distilled water as a negative control.13
Screening for Vancomycin Resistance
Resistance to vancomycin (MAST Diagnostics) was prepared by Mueller–Hinton agar (Merck, Germany) containing vancomycin 6 μg/mL and 4% NaCl. The plates were incubated at 37°C for 24 hrs according to CLSI guidelines. Any visible growth after 24 h was considered vancomycin-resistant.13
Molecular Identification
DNA Extraction
Genomic DNA was extracted from pure colonies of MRSA strains using the high pure PCR template preparation kit (QIAGEN, Germany) according to the manufacturer’s procedure. The concentration of extracted DNA was measured at 260 nm, using a Nanodrop instrument (Thermo Scientific, USA) and gel electrophoresis. The samples were stored at −20°C until PCR amplification.
Detection of mecA Gene
The presence of the mecA gene was evaluated using the PCR amplification. An approximately 359 bp fragment of mecA gene was amplified by two specific primers mecA F (5′- AGA AGA TGG TAT GTG GAA GTT AG-3′) and mecA R (5′- ATG TAT GTG CGA TTG TAT TGC -3′) as previously described.15 S. aureus ATCC 33591 strain was used as a positive control and S. aureus ATCC 25923 strain as the negative control.
Screening for of SCCmec Elements
The presence of SCCmec genes in S. aureus isolates was checked using PCR amplification, as previously explained by Moosavian et al.16 Five MRSA strains, NCTC10442 (SCCmec I), NCTC N315 (SCCmec II), NCTC 85/2082 (SCCmec III), NCTC CA05 (SCCmec IVa), and JCSC3624 (SCCmec V) were used as a positive control and uninoculated medium as negative control.
Conventional Spa Typing (Sequence-Based Method)
All MRSA isolates were subjected to spa typing according to the method of Oliveira et al17 The polymorphic X regions of the spa gene were amplified and sequenced using the primers spa 1113F (5ʹ- TAA AGA CGA TCC TTC GGT GAG C -3ʹ) and spa-1514R (5ʹCAG CAG TAG TGC CGT TTG CTT -3ʹ).The sequences of the products were determined using the ABI PRISM} 7700 Sequence Detection System (Applied Biosystems, Foster City, Calif., USA) following the standard protocol of the supplier. The Ridom Spa Server database (http://www.spaserver.ridom.de) (Ridom, Wurzburg, Germany) was used to assign the edited sequences to particular spa types.
High-Resolution Melting Curve Analysis (HRMA)
The polymorphic X region of the spa gene was amplified on the Rotor-gene Q instrument (Qiagen, Hilden, Germany) using the same DNA extract and primer pair (Spa1113F and Spa1514R). In a final volume of 10 μL, the HRM PCR reaction mix contained 5 ng of sample gDNA, 0.3 pmol of each primer, and the Type-it HRM PCR Kit (QIAGEN, Germany) using EvaGreen as the fluorescent dye. The settings for HRM were used as recommended by the manufacturer. Briefly, on the Rotor-Gene Q, PCR conditions were 10 min activation at 94°C, 42 cycles 94°C for 7 s, 60°C for 30s and a final step at 95°C for 1 min and cooling to 40°C for 1min. HRM was performed from 61°C to 87°C with a ramp rate of 0.01°C/s and 100 reads/s. Reactions were performed in duplicate. The reproducibility of the assay was tested by three different persons and all operators gained similar results. For each separate run, 4 control samples with known spa types (t037, t030, t034 and t304) were included as melting curve standards. The curves obtained from each type were considered to determine a standard curve for the rest of the procedure. The melting curves of all samples were generated automatically by the Rotor Gene Q Series Software 2.1.0 and were normalized by using the default setting. HRM difference plot was also generated by the software, which could give a better comparison of melting temperatures. In the experiment, HRM analysis was performed 3 times on the 50 isolates, and the results proved to be completely reproducible concerning calling the spa types.
Statistical Analysis
Differences of MRSA isolates frequencies among hospitals and specimens and relationship between antibiotic resistance and spa typing, we performed the chi-square test and Fisher’s exact test using the SPSS version 22 was used to analyze data (SPSS, Chicago, IL, USA). A P ˂ 0.05 was considered statistically significant.
Results
We confirmed a total of 146 clinical S. aureus isolates from various clinical specimens based on culture and standard biochemical criteria. Out of the 146 clinical S. aureus isolates (n= 50, 34%) were resistant to methicillin by cefoxitin disc diffusion test. As indicated by PCR, all 50 isolates were positive for mecA, leading to MRSA and MSSA (methicillin-susceptible S. aureus) prevalence rates of (n = 50, 34%) and (n= 96, 66%), respectively. In the study group, (n= 29/50, 58%) of the patients were women and (n= 21/50, 42%) were men. The mean age of the patients included in the study was 45.06 years and mean ± standard deviation (SD) was 11.59 (Table 1). Our results revealed that Razi Hospital had a higher prevalence of MRSA (n=29,58%) while Taleghani had the lowest prevalence (n=3,6%). Table 2 shows the total distribution of MRSA in various types of clinical specimens in three major hospitals. The prevalence rate of MRSA in different specimens was (n=13/50, 26%) blood, (n=14/50, 28%) wound, (n=15/50, 30%) urine, (n =4/50, 8%) synovial fluid and (n= 4/50, 6%) abscesses in clinical samples. MRSA isolates were subjected to disk diffusion to assess the antibiotic resistance pattern.
Table 1.
Characteristics of MRSA isolates from Patients Seen at the University Hospitals of Ahvaz, Iran
| MSRA | Age | Gender | Hospital | Source | Ward | HRM-Based spa-Typing | PCR-Based spa-Typing | ARPs | MDR | XDR | SCCmec Typing |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 29 | F | Razi | Blood | ICU | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
| 2 | 33 | M | Golestan | Urine | Nephrology | t037 | t037 | CD,E,T,FOX | + | − | II |
| 3 | 39 | F | Golestan | Blood | Surgery | t030 | t030 | CIP,CD,E,GN,RP,T,FOX | + | − | II |
| 4 | 54 | F | Razi | Urine | ICU | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | IV |
| 5 | 63 | F | Golestan | Urine | Pediatric | t030 | t030 | CIP,CD,RP,T,FOX | + | − | III |
| 6 | 51 | F | Golestan | Blood | Pediatrics ICU | t030 | t030 | CIP,CD,E,GN,RP,T,FOX | + | − | II |
| 7 | 45 | M | Razi | Blood | Women | t037 | t037 | CD,E,RP,T,FOX | + | − | NT |
| 8 | 64 | F | Razi | Abscesses | Surgery | t037 | t037 | SXT,CD,RP,T,FOX | + | − | II |
| 9 | 54 | F | Razi | Urine | Emergency | t363 | t363 | CIP,CD,T,FOX | + | − | II |
| 10 | 42 | M | Taleghani | Urine | Emergency | t037 | t037 | CIP,CD,E,RP,T,FOX | + | − | I |
| 11 | 47 | F | Golestan | Abscesses | Pediatrics ICU | t030 | t030 | CIP,CD,E,RP,T,FOX | + | − | NT |
| 12 | 43 | F | Razi | Urine | Women | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
| 13 | 52 | F | Razi | Urine | Emergency | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
| 14 | 28 | M | Razi | Blood | ICU | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
| 15 | 32 | F | Golestan | Abscesses | ICU | t030 | t030 | CIP,CD,E,T,FOX | + | − | I |
| 16 | 36 | F | Taleghani | Wound | ICU | t030 | t030 | CIP,CD,T,FOX | + | − | II |
| 17 | 21 | F | Razi | Blood | ICU | t037 | t037 | CIP,CD,GN,RP,T,FOX | + | − | II |
| 18 | 51 | F | Golestan | Synovial fluid | Surgery | t030 | t030 | CIP,CD,GN,RP,FOX | + | − | II |
| 19 | 53 | M | Taleghani | Blood | ICU | t037 | t037 | SXT,C,CD,GN,RP,T,FOX | + | − | IV |
| 20 | 36 | F | Razi | Wound | ICU | t037 | t037 | CIP,CD,GN,RP,T,FOX | + | − | IV |
| 21 | 37 | F | Golestan | Blood | ICU | t030 | t030 | SXT,C,RP,FOX | + | − | II |
| 22 | 51 | F | Razi | Wound | Women | t030 | t030 | CD,E,GN,RP,FOX | + | − | I |
| 23 | 62 | M | Razi | Blood | Surgery | t275 | t275 | CD,E,FOX | + | − | II |
| 24 | 41 | M | Golestan | Wound | Pediatrics ICU | t037 | t037 | C,CD,FOX | + | − | III |
| 25 | 48 | M | Razi | Blood | ICU | t030 | t030 | SXT,C,CD,E,GN,RP,T,FOX | + | − | III |
| 26 | 58 | M | Razi | Wound | Surgery | t034 | t034 | CD,E,FOX | + | − | III |
| 27 | 54 | F | Razi | Wound | ICU | t037 | t037 | CD,E,GN,FOX | + | − | III |
| 28 | 27 | M | Golestan | Blood | Pediatric | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
| 29 | 41 | M | Razi | Blood | ICU | t030 | t030 | CIP,CD,E,GN,FOX | + | − | III |
| 30 | 52 | F | Golestan | Blood | Surgery | t037 | t037 | C,E,GN,FOX | + | − | II |
| 31 | 36 | M | Razi | Synovial fluid | Orthopedic | t030 | t030 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
| 32 | 63 | F | Razi | Wound | ICU | t459 | t459 | CIP,GN,FOX | + | − | III |
| 33 | 30 | M | Golestan | Wound | Pediatric | t037 | t037 | SXT,C,CD,E,GN,RP,T,FOX | + | − | III |
| 34 | 63 | M | Razi | Urine | Surgery | t030 | t030 | CIP,E,GN,FOX | + | − | III |
| 35 | 61 | F | Golestan | Wound | ICU | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | II |
| 36 | 39 | M | Razi | Wound | Surgery | t363 | t363 | CIP,CD,E,GN,FOX | + | − | IV |
| 37 | 47 | F | Golestan | Wound | ICU | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | II |
| 38 | 46 | M | Razi | Wound | Surgery | t304 | t304 | CIP,FOX | + | − | II |
| 39 | 52 | F | Razi | Wound | Women | t030 | t030 | CIP,E,GN,T,FOX | + | − | III |
| 40 | 35 | M | Razi | Wound | ICU | t030 | t030 | E,GN,T,CIP,FOX | + | − | III |
| 41 | 42 | M | Razi | Synovial fluid | Orthopedic | t030 | t030 | SXT,C,CD,E,GN,T,FOX | + | − | III |
| 42 | 61 | F | Golestan | Urine | Nephrology | t037 | t037 | CIP,CD,E,GN,RP,T,FOX | + | − | III |
| 43 | 37 | M | Razi | Synovial fluid | ICU | t459 | t459 | CD,E,FOX | + | − | II |
| 44 | 33 | M | Razi | Urine | Emergency | t189 | t189 | CD,E,GN,RP,T,FOX | + | − | IV |
| 45 | 46 | F | Razi | Urine | Emergency | t030 | t030 | SXT,CIP,CD,E,GN,FOX | + | − | III |
| 46 | 28 | M | Razi | Abscesses | Surgery | t044 | t044 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
| 47 | 53 | F | Golestan | Urine | ICU | t037 | t037 | CIP,CD,E,GN,FOX | + | − | II |
| 48 | 27 | F | Golestan | Urine | ICU | t037 | t037 | SXT,CIP,CD,E,GN,RP,FOX | + | − | II |
| 49 | 48 | F | Razi | Urine | Emergency | t030 | t030 | CIP,CD,E,GN,FOX | + | − | IV |
| 50 | 62 | F | Golestan | Urine | Emergency | t037 | t037 | SXT,C,CIP,CD,E,GN,RP,T,FOX | − | + | III |
Table 2.
Total Distribution of MRSA Among Different Clinical Specimens of Three Major Hospitals
| Hospital | ||||||
|---|---|---|---|---|---|---|
| Clinical Specimens | Razi Hospital | Golestan Hospital | Taleghani Hospital | |||
| MRSA (29) | MSSA(49) | MRSA (18) | MSSA (20) | MRSA (3) | MSSA (27) | |
| Blood | 7 | 18 | 5 | 10 | 1 | 14 |
| Wound | 9 | 12 | 4 | 5 | 1 | 9 |
| Synovial fluid | 3 | 3 | 1 | 1 | – | 1 |
| Urine | 8 | 11 | 6 | 4 | 1 | 3 |
| Abscesses | 2 | 5 | 2 | – | – | – |
Tables 1 and 3 present the phenotypic pattern of antibiotic resistance in MRSA isolates. A high rate of resistance was detected against erythromycin (n=38/50,76%), chloramphenicol (n=18/50,36%), gentamicin (n=35/69%), clindamycin (n=43/50,86%), ciprofloxacin (n=34/50,68%), and tetracycline (n=32/50,64%) antibiotic agents. MRSA isolates had the lowest prevalence of resistance against rifampin (n=29/50,58%), trimethoprim-sulfamethoxazole (n= 19/50,38%), and antibiotic agents.
Table 3.
Results of Antimicrobial Resistance Tests by Disk Diffusion Method for MRSA
| Antimicrobial Category | Antimicrobial Agent | Negative | Positive | Pvalue for χ2 |
|---|---|---|---|---|
| Oxazolidinones | Linezolid | 50(100%) | – | |
| Folat pathway inhibitors | Trimethoprim-sulphamethoxazole | 31(62%) | 19(38%) | 0.090 |
| Phenicols | Chloramphenicol | 32(64%) | 18(36%) | 0.048* |
| Fluoroquinolones | Ciprofloxacin | 16(32%) | 34(68%) | 0.011* |
| Lincosamides | Clindamycin | 7(14%) | 43(86%) | <0.0001* |
| Macrolides | Erythromycin | 12(24%) | 38(76%) | <0.0001* |
| Aminoglycosides | Gentamycin | 15(31%) | 35(69%) | 0.005* |
| Ansamycins | Rifampin | 21(42%) | 29(58%) | 0.258 |
| Tetracyclines | Tetracycline | 18(36%) | 32(64%) | 0.048* |
| Glycopeptides | Vancomycin | 50(100%) | – | |
| Penicillinase-stable penicillins | Cefoxitin | – | 50(100%) |
Note: *Denotes statistically significant difference between resistance and susceptible MRSA isolated.
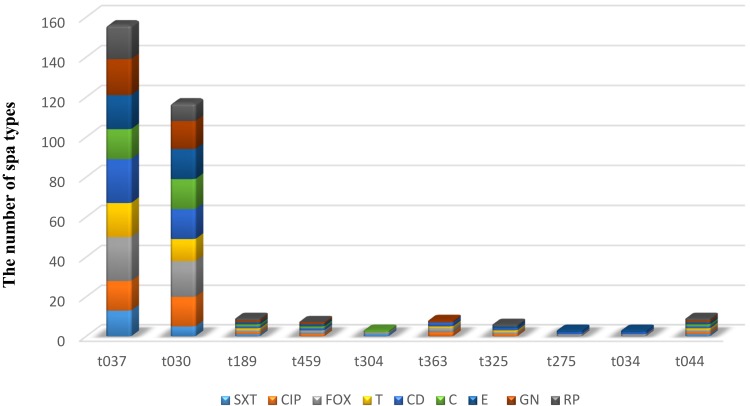
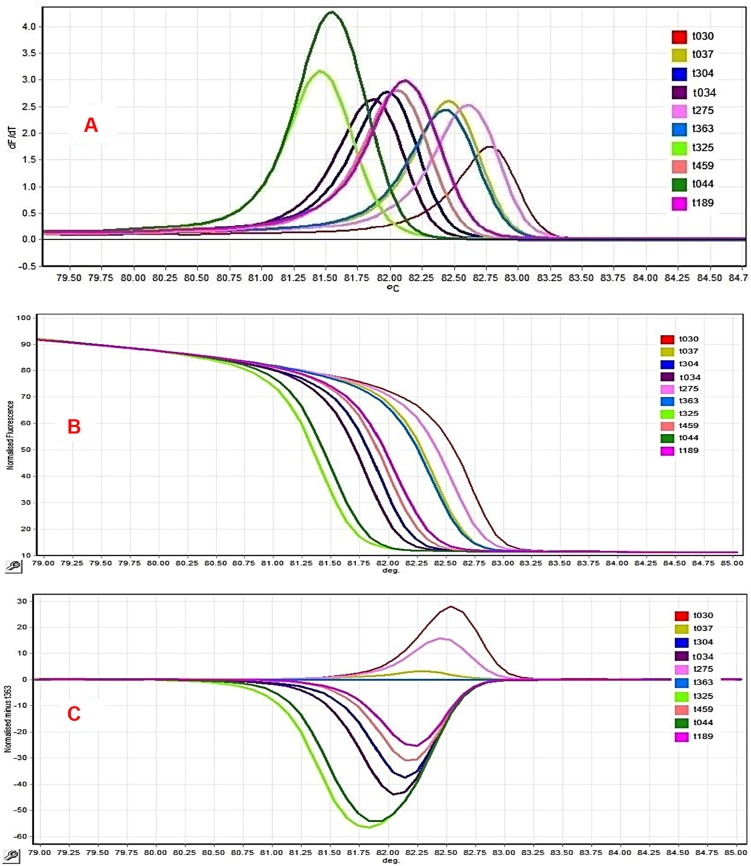
All MRSA isolates were susceptible to vancomycin and linezolid. In the MRSA isolates, (n=11/50,22%) were XDR and (n=39/50,78%) were MDR. PDR strain was not observed. Based on our results, Razi Hospital had a higher prevalence of MDR (n=22/50,44%) while Taleghani had the lowest prevalence (n =3/50,6%). Statistically significant differences were found among different hospitals regarding the prevalence of MDR (χ2=14.1, P=0.001). Our findings showed that Razi Hospital had a higher prevalence of XDR (n=7/50,14%) while Golestan had the lowest prevalence (n =4/50,8%). Statistically non-significant differences were detected among different hospitals regarding the prevalence of XDR (χ2=0.82, P=0.366). The prevalence rate of MDR in different specimens was (n =10/50, 26%) in blood, (n =12/50, 28%) in wound, (n=11/50, 30%) in urine, (n =3/50, 8%) in synovial fluid, and (n= 3/50, 6%) in the abscesses of clinical samples. Statistically significant differences were seen as far as the prevalence of MDR and type of specimens (χ2=10.1, P=0.039) are concerned. The prevalence rate of XDR in different specimens was (n =3/50, 26%) in blood, (n =2/50, 28%) in wound, (n=4/50, 30%) in urine, (n =1/50, 8%) in synovial fluid, and (n=1/50, 6%) in the abscesses of the clinical samples. Statistically non-significant differences existed regarding the prevalence of XDR and type of specimens (P=0.664). The predominant simultaneous resistance patterns were (n=11/50, 22%) for nine antibiotics, (n=2/50, 4%) for eight antibiotics, (n=6/50,12%) for seven antibiotics, (n=7/50,14%) for six antibiotics, (n=11/50,22) for five antibiotics, (n= 7/50,14%) for four antibiotics, and (n=5/50,10%) for three antibiotics (Table 4). Of the 50 MRSA isolates, SCCmec type I (n=3/50,6%), type II (n=17/50, 34%), type III (n=22/50,44%), and type IV (n=6/50,12%) were the most prevalent. None of the tested isolates were type V and two isolates were untypable by the routine PCR assays utilized (Table 1). The frequency of MDR and XDR among MRSA isolates carrying the SCCmec types II and III was significantly higher than other types, although the differences were statistically significant, respectively (X2=19.84, P=0.001). Using PCR-sequencing as a reference method, a total of 10 different spa types were detected among the 50 MRSA isolates. The 10 spa types were: t037 (n=22,46%), t030 (n=18,36%), t363 (n=2,0.04%), t459 (n=2,0.04%), t304 (n=1, 0.02%), t034 (n=1,0.02%), t275 (n=1,0.02%), t325 (n=1,0.02%), t044 (n=1, 0.02%), and t189 (n=1, 0.02%). Spa type t037 was the major spa type circulating in the study region. Razi Hospital isolates were fully diverse, with six spa-types detected among the 29 MRSA isolates. Statistically non-significant differences were detected regarding the type of hospital and spa types (P=0.902). Table 1 shows the distribution of the spa types isolatesfrom the clinical sources. Figure1 compares the occurrence of antimicrobial resistance among the investigated MRSA isolates concerning different spa types. There were statistically significant differences regarding the prevalence of MDR and spa types (P<0.0001). Statistically significant differences also existed in the prevalence of XDR and spa types (P=0.004). Table 5 presents the distribution of SCCmec types in spa types. Statistically non-significant differences were further detected as regards the distribution of SCCmec types in spa types (P=0.841). Following sequencing comparison, the 50 clinical MRSA isolates were subjected to HRM analysis. HRM analysis revealed 10 unique spa types out of 10 with melting temperatures ranging between 81.38°C and 82.70°C (Table 5 and Figure 2). Four distinct melting curves and HRM difference plots were obtained by the Rotor-gene Q real-time instrument. All the four control spa types showed a minimum of 0.2°C temperature difference (t037: 82:50°C, t030: 82:30°C, t0190: 81:80 and t304:82:00 °C). These empirically obtained criteria were employed to name the melting curves “the same” or “different” using difference graphs. The melting curves were called “the same” as the defined control which the difference graph within ±0.2 relative to the x-axis and did not display reproducible differences such as double peaks or crossing the x-axis more than twice in both replicates.
Table 4.
Multidrug Resistance Patterns Among MRSA Isolates
| Number of Resistant Antibiotics | Antibiotics | Number of Species |
|---|---|---|
| 9 | SXT,C,CIP,CD,E,GN,RP,T,FOX | 11 |
| 8 | SXT,C,CD,E,GN,RP,T,FOX | 2 |
| 7 | CIP,CD,E,GN,RP,T,FOX | 3 |
| SXT,C,CD,GN,RP,T,FOX | 1 | |
| SXT,C,CD,E,GN,T,FOX | 1 | |
| SXT,CIP,CD,E,GN,RP,FOX | 1 | |
| 6 | CIP,CD,RP,T,CIP,FOX | 1 |
| CIP,CD,E,RP,T,FOX | 2 | |
| CIP,CD,GN,RP,T,FOX | 2 | |
| CD,E,GN,RP,T,FOX | 1 | |
| SXT,CIP,CD,E,GN,FOX | 1 | |
| 5 | CD,E,RP,T,FOX | 1 |
| SXT,CD,RP,T,FOX | 1 | |
| CIP,CD,E,T,FOX | 1 | |
| CIP,CD,GN,RP,FOX | 1 | |
| CD,E,GN,RP,FOX | 1 | |
| CIP,CD,E,GN,FOX | 4 | |
| CIP,E,GN,T,FOX | 2 | |
| 4 | CD,E,T,FOX | 2 |
| CIP,CD,T,FOX | 2 | |
| SXT,C,RP,FOX | 1 | |
| C,E,GN,FOX | 1 | |
| CIP,E,GN,FOX | 1 | |
| 3 | CD,E,FOX | 3 |
| C,CD,FOX | 1 | |
| CIP,GN,FOX | 1 | |
| 2 | CIP,FOX | 1 |
Figure 1.
Distribution of antibiotic resistance in MRSA with spa types.
Table 5.
HRM and Spa Sequence Types of the 50 MRSA Isolates, and the Frequencies in the Hospital of Origin
| HRM | Tm | Spa-Type | Repeat of Spa Type | Size, bp | CG% | Number of Isolates |
|---|---|---|---|---|---|---|
| 1 | 82.30 | T030 | 15-12-16-02-24-24 | 144 | 45 | 18 |
| 2 | 82.50 | T037 | 15-12-16-02-25-17-24 | 168 | 45.2 | 22 |
| 3 | 82.55 | T363 | 15-16-02-25-17-24 | 144 | 45.8 | 2 |
| 4 | 82.15 | T459 | 15-12-16-02-24 | 120 | 44 | 2 |
| 5 | 82.23 | T189 | 07-23-12-12-21-17-34 | 168 | 43 | 1 |
| 6 | 82.00 | T304 | 11-10-21-17-34-24-34-22-25 | 216 | 43.5 | 1 |
| 7 | 81.80 | T034 | 11-17-34-24-34-22-25 | 168 | 44 | 1 |
| 8 | 82.75 | T275 | 15-12-16-02-25-17-24-24 | 192 | 44 | 1 |
| 9 | 81.38 | T325 | 7-12-21-17-34-13-34-34-33-34 | 240 | 40.4 | 1 |
| 10 | 81.60 | T044 | 7-23-12-34-34-33-34 | 168 | 41.7 | 1 |
Abbreviations: HRM, high-resolution melting; Tm, melting temperature.
Figure 2.
Comparison of different spa polymorphic region X HRM curves obtained from MRSA isolates. (A) Negative derivative of fluorescence over temperature (df/dt) plots displaying 10 HRM profiles. (B) Normalization data curve shows the decreasing fluorescence vs increasing temperature. (C) Difference graph demonstrating the accurate reproduction of eight spa HRM profiles in a run experiment. Isolates with difference plots that fall within the ±0.2 relative fluorescence unit (RFU) cutoffs were considered as the “same” type, while the isolates that lie outside of the ±0.2 RFU cutoffs were denoted as “different”.
Discussion
In 2018, the Center for Disease Control and Prevention (CDC) estimated that more than 70,000 invasive infections and more than 9000 deaths were caused by MRSA.18 In Iran, the total number of MRSA infection cases has increased, thereby introducing a serious problem in the form of nosocomial infections.16
A previous study on MRSA in Khuzestan province revealed a dramatic annual increase in the proportion of isolates resistant to methicillin during the recent years, rising from 60% in 2017 and 2018 to a peak of 77% in 2019.19–22 Such differences in the prevalence of MRSA infections in our region might reflect the fact that in these areas, different policies are considered for infection control and other factors.23
There is a continuous increase in the global prevalence of MRSA in European countries. In Belgium and Spain, 19% and 29% of the isolates S. aureus strains are methicillin-resistant, respectively.24,25 Similar antibiotic resistance patterns of MRSA strains have also been reported against aminoglycosides, glycopeptides, penicillins, macrolides, tetracyclines, fluoroquinolones, lincosamides, folate inhibitors, and ansamycins groups of antibiotics.26–28 In the present study, MRSA had the highest sensitivity to vancomycin and linezolid (100%) and the lowest to clindamycin (86%). This limits the use of this drug and often warrants prolonged treatment. Results obtained by Huang et al showed that MRSA had no resistance to vancomycin and 91% resistance to clindamycin.29
The present study highlighted that a large number of MRSA were MDR, being in line with the previous findings reporting a high rate of multidrug resistance in MRSA.30 The observed MDR and XDR rates in hospitals indicate that antibiotics resistance is increasing at an alarming rate and pathogenic bacteria circulating in hospitals are becoming more resistant to all available antibiotics.
The first reason might be associated with the lack of an antibiotics resistance surveillance and stewardship program in Iran. There is sufficient evidence indicating that an antibiotics resistance surveillance and stewardship program contributes to understanding the pattern of resistance and improving the utilization of antibiotics to prevent antibiotic resistance. The second reason might be the lack of comprehensive national antibiotic policies and problems associated with their implementation. In Iran, it is a common practice to buy any antibiotic from private drug vendors and pharmacies without any prescription.31 Studies in other countries have reported a lower prevalence. For instance, Ullah et al found that the prevalence of MDR and XDR was 15.84% and 6.93%, respectively.32 A major concern for establishing a reliable infection control system in hospitals is the time to perform molecular typing tests. Monitoring the spread of MRSA strains in a hospital setting requires the use of accurate and rapid typing methods. The region X of the spa gene as a genetic marker allows for the global investigation outbreak by analyzing a single locus.33 The conventional PCR-sequencing typing method has been extensively used worldwide. Based on the results of PCR-sequencing method, the t037spa type was the most prevalent among Iranian patients, accounting for 44% of the isolates and their transfer from the community to hospitals.
Our results are similar to those of the previous studies conducted in Iran and other parts of the world.9,34 All the 50 MRSA isolates were then subjected to HRMA. Among the 10 different spa types belonging to the 50 clinical MRSA isolates, 10 specific spa types could be assigned by being matched with the control curves. Contrary to our study, Mayerhofer et al found that certain spa types could not be separated from each other based on their Tm.8,9,11 However, the major advantage of this method is that it is single-step and closed-tube, performed on a moderately priced and generic piece of laboratory equipment. This technique offsets the occasional inability to differentiate between spa alleles because spa interrogation could be simply combined with the real-time PCR interrogation of clonal-complex specific single-nucleotide polymorphisms and toxin encoding genes for instance. This approach is much less expensive than full spa sequencing; each sequencing reaction in our institution costs $20 while each HRM runs costs$0.50 for reagents.35 In our study, initial HRM results showed some similar melting temperatures and curve shapes; however, optimizing several methods and materials such as DNA extraction method and HRM Master mix, a real-time PCR-based HRM assay was developed to differentiate between S. aureus isolates, particularly major MRSA spa types (t037 and t030), present in Iran. With the standardization and improvements in the HRM method in the present study, predominant spa types in Iran such as t030 and t037 could be differentiated from each other.
Myriad factors (band size, G + C content, real-time thermal cycler, HRM software, DNA and primer concentrations, and DNA quality) can affect HRM results; therefore, this technique can be used as a screening approach, along with other genotyping methods, to increase the discrimination power in molecular typing, reduce the expense, and minimize the risk of sample contamination. This study showed that the presence of (SCCmecIII-t030) and (SCCmecIII-t037) was largely disseminated in our region. Interestingly, spa types t037and t030, identified in 41 MRSA isolates, were MDR and XDR, including isolates causing severe human infections. Similar to our results, the most common MRSA in Turkey was found to be (91.4%), followed by SCCmec type III (91.4%), and t030 (85.1%).36 Use of genomic methods such as MLST analysis was a limitation of our study; in this regard, clonal complex for correlation with spa typing should be designed. In regard to our small sample size and the large number of spa types registered in the SpaServer database, the use of HRM curve profiling replacing sequence-based spa typing is not promising, hence the necessity of future studies.
Conclusion
We found a regionally divergent distribution of MRSA in the largest population of Iranian patients with distinct occurrences of single spa types. We hereby conclude that early detection and close monitoring of MDR, XDR, or even PDR MRSA isolates strains must be commenced by all clinical microbiology laboratories to reduce the menace of antimicrobial resistance as a global issue. This study emphasizes the spa variation among different S. aureus isolates, which might be considered as an important criterion when treating staphylococcal infections. Additionally, two common types of spa had an alarming rate of antibiotic resistance, which should be taken into consideration by hospital settings. Furthermore, the HRM-based spa typing is extremely rapid, easy to perform, and cost-effective but has to be standardized for different regions, spa types, and real-time machinery. In contrast to more expensive and labor-intensive techniques, this method could be used for active screening and rapid detection of common endemic spa types. Moreover, the large number of spa types registered in the SpaServer database indicates that HRM curve profiling cannot replace sequence-based spa typing. However, HRM can be used as a superior screening method for the detection of endemic strains and surveillance of non-endemic strains through detecting new curve profiles. Accordingly, to definitively specify rare types or non-endemic spa types, it is inevitable to use a reference method such as DNA sequencing.
Funding Statement
This work is a part of the MSc. thesis of Paria Baratian Dehkordi, which has been approved in the Department of Microbiology of Ahvaz Jundishapur University of Medical Sciences. The authors thank the Department of Microbiology, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran and Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, for financial support of this project (Grant No. 1395.187). The study was sponsored by the authors. The authors received no funding from any other individual or institution.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Struelens MJ, Hawkey PM, French GL, Witte W, Tacconelli E. Laboratory tools and strategies for methicillin-resistant Staphylococcus aureus screening, surveillance and typing: state of the art and unmet needs. Clin Microbiol Infect. 2009;15(2):112–119. doi: 10.1111/j.1469-0691.2009.02698.x [DOI] [PubMed] [Google Scholar]
- 2.Goetghebeur M, Landry PA, Han D, Vicente C. Methicillin-resistant Staphylococcus aureus: a public health issue with economic consequences. Can J Infect Dis Med Microbiol. 2007;18(1):27–34. doi: 10.1155/2007/253947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dadashi M, Nasiri MJ, Fallah F, et al. Methicillin-resistant Staphylococcus aureus (MRSA) in Iran: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2018;12:96–103. doi: 10.1016/j.jgar.2017.09.006 [DOI] [PubMed] [Google Scholar]
- 4.Ghanwate N, Thakare P, Bhise PR, Gawande S. Colorimetric method for rapid detection of Oxacillin resistance in Staphylococcus aureus and its comparison with PCR for mec A gene. Sci Rep. 2016;6(1):1–5. doi: 10.1038/srep23013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boye K, Bartels MD, Andersen IS, Moeller JA, Westh H. A new multiplex PCR for easy screening of methicillin‐resistant Staphylococcus aureus SCCmec types I–V. Clin Microbiol Infect. 2007;13(7):725–727. doi: 10.1111/j.1469-0691.2007.01720.x [DOI] [PubMed] [Google Scholar]
- 6.Chung M, De Lencastre H, Matthews P, et al; Multilaboratory Project Collaborators. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6(3):189–198. DOI: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 7.David MZ, Taylor A, Lynfield R, et al. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a US medical center. J Clin Microbiol. 2013;51(3):814–819. doi: 10.1128/JCM.02429-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayerhofer B, Stöger A, Pietzka AT, et al. Improved protocol for rapid identification of certain spa types using high resolution melting curve analysis. PLoS One. 2015;10(3):e0116713. doi: 10.1371/journal.pone.0116713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fasihi Y, Fooladi S, Mohammadi MA, Emaneini M, Kalantar-Neyestanaki D. The spa typing of methicillin-resistant Staphylococcus aureus isolates by high resolution melting (HRM) analysis. J Med Microbiol. 2017;66(9):1335–1337. doi: 10.1099/jmm.0.000574 [DOI] [PubMed] [Google Scholar]
- 10.Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245(2):154–160. doi: 10.1006/abio.1996.9916 [DOI] [PubMed] [Google Scholar]
- 11.Mazi W, Sangal V, Sandstrom G, Saeed A, Yu J. Evaluation of spa-typing of methicillin-resistant Staphylococcus aureus using high-resolution melting analysis. Int J Infect Dis. 2015;38:125–128. doi: 10.1016/j.ijid.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 12.Tille P. Bailey & Scott’s Diagnostic Microbiology-E-Book. St. Louis, MO: Elsevier Health Sciences; 2015:307–327. [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Sixteenth Informational Supplement; 2019 [Google Scholar]
- 14.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 15.Havaei SA, Azimian A, Fazeli H, et al. Genetic characterization of methicillin resistant and sensitive, vancomycin intermediate Staphylococcus aureus strains isolated from different Iranian Hospitals. ISRN Microbiol. 2012;20:2012. doi: 10.5402/2012/215275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moosavian M, Shahin M, Navidifar T, Torabipour M. Typing of staphylococcal cassette chromosome mec encoding methicillin resistance in Staphylococcus aureus isolates in. New Microbes New Infect. 2018;21:90–94. DOI: 10.1016/j.nmni.2017.11.006 Ahvaz, Iran. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira DC, Crisóstomo I, Santos-Sanches I, et al. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J of clin microbiolo. 2001;39(2):574–580. doi: 10.1128/JCM.39.2.574-580.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MRSA, CDC. Available from: https://www.cdc.gov/mrsa/index.html. Accessed August4, 2019.
- 19.Abbasi-Montazeri E, Khosravi AD, Feizabadi MM, et al. The prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolates with high-level mupirocin resistance from patients and personnel in a burn center. Burns. 2013;39(4):650–654. doi: 10.1016/j.burns.2013.02.005 [DOI] [PubMed] [Google Scholar]
- 20.Khosravi AD, Jenabi A, Montazeri EA. Distribution of genes encoding resistance to aminoglycoside modifying enzymes in methicillin-resistant Staphylococcus aureus (MRSA) strains. Kaohsiung J Med Sci. 2017;33(12):587–593. doi: 10.1016/j.kjms.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoshnood S, Shahi F, Jomehzadeh N, et al. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among methicillin-resistant Staphylococcus aureus strains isolated from burn patients. Acta Microbiol Immunol Hung. 2019;17:1–2. doi: 10.1556/030.66.2019.015 [DOI] [PubMed] [Google Scholar]
- 22.Nikfar R, Shamsizadeh A, Kajbaf TZ, Panah MK, Khaghani S, Moghddam M. Frequency of methicillin-resistant Staphylococcus aureus nasal carriage in healthy children. Iran J Microbiol. 2015;7:67. [PMC free article] [PubMed] [Google Scholar]
- 23.Turlej AG, Hryniewicz WA, Empel J. Staphylococcal cassette chromosome mec (Sccmec) classification and typing methods: an overview. Pol J Microbiol. 2011;60(2):95–103. doi: 10.33073/pjm- [DOI] [PubMed] [Google Scholar]
- 24.Denis O, Jans B, Deplano A, et al. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) among residents of nursing homes in Belgium. J Antimicrob Agents. 2009;64(6):1299–1306. doi: 10.1093/jac/dkp345 [DOI] [PubMed] [Google Scholar]
- 25.Vindel A, Cuevas O, Cercenado E, et al. Methicillin-resistant Staphylococcus aureus in spain: molecular epidemiology and utility of different typing methods. J Clin Microbiol. 2009;47(6):1620–1627. doi: 10.1128/JCM.01579-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saba CK, Amenyona JK, Kpordze SW. Prevalence and pattern of antibiotic resistance of Staphylococcus aureus isolated from door handles and other points of contact in public hospitals in Ghana. Antimicrob Resist Infect Control. 2017;6(1):44. doi: 10.1186/s13756-017-0203-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husain A, Rawat V, Umesh MK, Verma PK. Vancomycin, linezolid and daptomycin susceptibility pattern among clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA) from Sub- Himalayan Center. J Lab Physicians. 2018;10(02):145. doi: 10.4103/JLP.JLP_92_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdolmaleki Z, Mashak Z, Dehkordi FS. Phenotypic and genotypic characterization of antibiotic resistance in the methicillin-resistant Staphylococcus aureus strains isolated from hospital cockroaches. Antimicrob Resist Infect Control. 2019;8(1):54. doi: 10.1186/s13756-019-0505-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YC, Hwang KP, Chen PY, Chen CJ, Lin TY. Prevalence of methicillin-resistant Staphylococcus aureus nasal colonization among Taiwanese children in 2005 and 2006. J Clin Microbiol. 2007;45(12):3992–3995. doi: 10.1128/JCM.01202-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan SU, Mahmud MN, Chowdhury MA, Hakim MA. Prevalence of multidrug resistant Staphylococcus aureus isolates in clinical specimens collected from local patients of Chittagong, Bangladesh. Chittagong Univ J Biol Sci. 2011;61:75–85. [Google Scholar]
- 31.Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ullah A, Qasim M, Rahman H, et al. High frequency of methicillin-resistant Staphylococcus aureus in Peshawar Region of Pakistan. Springerplus. 2016;5(1):600. doi: 10.1186/s40064-016-2277-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn G, Francioli P, Blanc DS. Double-locus sequence typing using clfB and spa, a fast and simple method for epidemiological typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45(1):54–62. doi: 10.1128/JCM.01457-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang FD, Wu PF, Chen SJ. Distribution of virulence genes in bacteremic methicillin-resistant Staphylococcus aureus isolates from various sources. J Microbiol Immunol Infect. 2019;52(3):426–432. doi: 10.1016/j.jmii.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 35.Tong SY, Lilliebridge RA, Holt DC, McDonald MI, Currie BJ, Giffard PM. High-resolution melting analysis of the spa locus reveals significant diversity within sequence type 93 methicillin-resistant Staphylococcus aureus from northern Australia. Clin Microbiol Infect. 2009;15(12):1126–1131. doi: 10.1111/j.1469-0691.2009.02732.x [DOI] [PubMed] [Google Scholar]
- 36.Bozdoğan B, Yıldız O, Oryaşın E, et al. t030 is the most common spa type among methicillin-resistant Staphylococcus aureus strains isolated from Turkish hospitals. Mikrobiyol Bul. 2013;47(4):571–581. doi: 10.5578/mb.5770 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- MRSA, CDC. Available from: https://www.cdc.gov/mrsa/index.html. Accessed August4, 2019.