Abstract
The aim of this study was to explore the underlying mechanism and function of dexmedetomidine (Dex)-regulated long non-coding RNAs (lncRNAs) in improving postoperative cognitive dysfunction (POCD) in rats. The established POCD model, Dex treatment model in rats, Morris water maze testing, and HE staining assays were used to evaluate the efficacy of Dex in POCD treatment in rats. Hippocampus samples of rats from the POCD group and the Dex group were used for lncRNA sequencing. The expression of five differentially expressed lncRNAs (DElncRNAs) was verified by quantitative reverse transcription PCR (qRT-PCR). Competing endogenous RNAs (ceRNA) network was constructed using Cytoscape. The concentration of inflammatory cytokines were measured by ELISA. Microglia proliferation and apoptosis were assessed using CCK-8 assay and flow cytometry, respectively. In the Dex group, the escape latency was shorter, neuron cell injury levels were alleviated, and the expression levels of TNF-α and IL-1β were significantly down-regulated compared with the POCD group. A total of 60 DE lncRNAs were identified, including 16 up- and 44 down-regulated lncRNAs in the Dex group. KEGG pathway analysis revealed that DElncRNAs were significantly enriched in cytokine–cytokine receptor interactions, the p53 signaling pathway, and the NF-kappa B signaling pathway. The qRT-PCR results and ceRNA network suggested that the lncRNA LOC102546895 may play a key role in POCD. LOC102546895 inhibited proliferation while promoting apoptosis in microglial cells and promoted the mRNA and protein expression of the target gene Npas4. Our findings showed that Dex alleviated POCD in rats and regulated lncRNAs expression profile in the hippocampus tissues of rats with POCD. In conclusion, our study outcome proposes that Dex-regulated lncRNA LOC102546895 may play a role in rats with POCD through targeting Npas4.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02190-9) contains supplementary material, which is available to authorized users.
Keywords: Postoperative cognitive dysfunction, Dexmedetomidine, lncRNA LOC102546895, RNA sequencing
Introduction
Postoperative cognitive dysfunction (POCD) loosely refers to the phenomenon where individuals with no mental illness experience a decline in fundamental mental functions such as memory, thought, attention, information processing, perception and the sleep–wake cycle after surgery(Bharadwaj and Kamath 2019). POCD negatively impacts patients by causing mental, physical and economic secondary damage. The primary impact on patients is increased cost and reduced quality of recovery and this occurs primarily among the elderly. Some people diagnosed with POCD are forced into early retirement and become dependent upon the social security system (Jacob et al. 2009). The mechanisms underlying POCD are associated with neurogenesis aberrations (Hem et al. 2016), excessive nerve cell apoptosis (Jevtovic-Todorovic et al. 2003), and systemic inflammation (Mario et al. 2010). POCD is a type of clinically complicated syndrome that can be affected by a number of factors that commonly include advanced age, cognitive impairment at baseline, DM/metabolic syndrome, renal insufficiency, and cardiovascular disease (Green and Schaffer 2019). Preventive measures can be taken to protect against POCD, and these include physical prevention methods such as cerebral monitoring (Deiner et al. 2015) and avoidance of general anesthetics (Zywiel et al. 2014) and pharmacologic strategies that incorporate the use of steroids (Fang et al. 2014), ketamine (Hudetz et al. 2009), and dexmedetomidine (Dex) (Lu et al. 2017). Unfortunately, the understanding of the treatment and pathogenesis of POCD is far less advanced than the understanding of the symptoms and preventative measures. Currently, the treatment of POCD primarily aims to inhibit brain excitation by reducing the secretion levels of dopamine and acetylcholine (Dawson et al. 2011).
Dexmedetomidine (Dex) is a selective α2 adrenal receptor agonist that can influence the effects of anesthesia and analgesia, inhibit sympathetic activity, and can provide cerebral protection (Qian et al. 2015). Studies have found that dexmedetomidine converts disordered electroencephalogram waves in patients with sleep disorder into normal non-rapid eye waves (Huupponen et al. 2010). Post-operative inflammatory cytokine release and BDNF (brain derived growth factor) expression can be modulated by Dex and this leads to improved long-term spatial memory after surgery and promotes neurogenesis involving restored p-CREB/CREB (cAMP response element binding protein) and PKA (proteins of kinase A) expression one week postoperatively. These events were regulated by intraneuronal p-P38-MAPK pathways (Wang et al. 2018). Preconditioning with parecoxib sodium in combination with Dex reduced the incidence of early postoperative cognitive dysfunction in elderly patients and may be related to improved postoperative analgesia and brain oxygen metabolism (Lu et al. 2017). Splenectomy can significantly increase the expression of TNF-α, Bax, IL-1β, and caspase-3 in hippocampal tissues to exacerbate POCD, and Dex administration significantly decreases the expression levels of inflammatory factors (Qian et al. 2015). However, the mechanisms underlying the function of Dex in the treatment of POCD remain unclear.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules that do not encode proteins and these molecules possess a length ranging from 200 nucleotides to 100 kb. LncRNAs are involved in the regulation of a number of diseases such as prostate cancer (Ren et al. 2012), leukemia (Huang et al. 2013), and lung cancer (Sui et al. 2016). Li et al. (2019) identified and analyzed the expression profiles of mRNAs and lncRNAs in the context of the macrophage inflammatory response, and they found that the abnormally expressed mRNAs and lncRNAs were primarily involved in the inflammatory and immune response. Another example involves the progressive neurodegenerative disease Alzheimer’s, in which silencing of the lncRNA SOX21-AS1 can attenuate neuronal oxidative stress and inhibit neuronal apoptosis by up-regulating FZD3/5 and activating the Wnt signaling pathway to ultimately improve disease symptoms (Zhang et al. 2019). Currently, basic and clinical studies indicate that POCD in the early postoperative period is associated with long-term Alzheimer’s disease (AD). Pathophysiological changes associated with POCD can lead to AD, and the clinical manifestations of moderate and severe POCD are considered to be indicative of the early stage of AD. Therefore, it has been speculated that a class of lncRNAs may be involved in the regulation of POCD.
In this study, we performed behavioral tests on rats, and we also utilized Hematoxylin–Eosin (HE) staining on hippocampus tissues isolated from these rats and analyzed inflammatory factors within the peripheral blood of rats to assess the treatment effects of Dex in regard to POCD in elderly rats. The hippocampus tissues from the POCD group and Dex group were also collected and subjected to lncRNA sequencing, and differentially expressed genes were selected for PCR verification. Finally, the functional influence of key lncRNAs on microglial cells was detected by CCK-8 analysis and flow cytometry, and the expression of downstream target genes was assessed. Our study provides insights into the mechanisms underlying Dex-mediated improvement of POCD by identifying the functions of various lncRNAs in this process, and our results lay a foundation for future research examining POCD.
Materials and methods
Rats and samples
All rat experiments completely adhered to the Guide for the Care and Use of Laboratory Animals (National Institutes of Health). All the experiments were approved by the Institutional Animal Care and Use Committee of the second affiliated hospital of Nanchang university. For all studies, 18-month-old Sprague–Dawley (SD) male rats weighing 220–280 g were purchased from JRDUN Biotechnology (Shanghai) Co., Ltd. All rats were provided with free access to food and water and were housed under controlled environmental conditions at 25 °C temperature. All efforts were made to minimize the suffering of the rats.
Establishment of the rat model and the treatment model
SD rats were randomly divided into four equal groups comprised of four rats each. These groups included the control group, sham operation group, POCD group, and Dex group. In the control group, all SD rats were intraperitoneally injected (ip) with 2 mL of normal saline (NS) at 30 min prior to surgery and during surgery. In the sham operation group, rats were not subjected to spleen removal, and they received 2 mL NS ip at 30 min prior to surgery and 2% pentobarbital sodium (50 mg/kg) during surgery. In the POCD group, rats were given 2 mL NS ip at 30 min prior to surgery and 2% pentobarbital sodium (50 mg/kg) during surgery, and the splenectomy was then performed under anesthesia. The skin of rats was prepared at the surgical incision and disinfected using iodophor. A small transverse incision was made along below the left rib margin at approximately 1 cm and the incision length was approximately 2 cm. The subcutaneous tissue was then bluntly separated layer by layer, and the abdominal cavity was finally exposed. Next, the spleen-related blood vessels were freely ligated and separated, and the spleen was removed. After confirming the absence of bleeding, the abdomen was closed by suture and disinfected using iodophor at the wound site. The entire surgical procedure was performed under sterile conditions. In the Dex group, all older SD rats received ip Dex (20 μg/kg) at 30 min prior to surgery, and the operating procedure was the same as that used for the POCD group.
Morris water maze testing
Behavioral tests were performed on the four rat groups (Each group possessed N = 3) to evaluate learning and memory ability. Behavioral tests including the Morris water maze test and the spatial acquisition experiment were performed on the 1st, 3rd, 7th, and 14th day after injection. The device was filled with water (25 ± 1 °C), and the animal was familiarized with the environment prior to the start of the experiment. The software used for this experiment was SuperMaze Version 3.3.0.0. The first experimental stage involved positioning navigation. The rats were successively placed in the four quadrant water maze at an orientation where their heads faced the wall, and when the rats finished their original quadrant they were moved to the next quadrant. If the animal boarded the platform area (located in the middle of the 4th quadrant) within 90 s, the recording was stopped. If the animal did not board the platform area within 90 s, the rats were guided to the platform using a stick and held there for 30 s. Each rat was trained four times per day for 5 days. The second stage involved space exploration. After all the first through fourth quadrants were completed, the platform in the water maze was removed, and the rats were placed in the same quadrant with their heads facing the wall. The action trajectory that occurred within 90 s of rats within quadrants was observed.
HE staining
Hippocampus tissues from each group of rats (each group possessed N = 3) were selected for HE staining to detect the loss of neuronal cells. Brain tissues were fixed in 10% formalin and embedded in paraffin, cut into pieces (1.5 cm × 1.5 cm × 0.3 cm), mounted onto sides, hematoxylin and eosin stained, and finally, imaged for analysis. All images are original magnification × 200.
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of tumor necrosis factor (TNF)-α and interleukin (IL)-1β in hippocampus tissue per rat among the four group were measured using a commercial ELISA Kit (Boster Biological Technology, China) according to the manufacturer’s instructions. The absorbance at 450 nm was determined using a microplate reader. The TNF-α and IL-1β concentrations were calculated using TNF-α and IL-1β standards.
RNA isolation, library construction, and lncRNA sequencing
Hippocampal tissues from the POCD group and the Dex group after successful modeling were used as materials for total RNA extraction using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and all extractions were performed in triplicate. The quality and concentration of total RNA was detected using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc.).
All sample RNA was separated from ribosomal RNA through the use of a Ribo-zero Gold rRNA Removal kit (Illumina, USA), and the subsequent library was constructed using a TruSeq Stranded RNA sample preparation kit (Illumina, USA). Ribosomal-depleted RNA was randomly fragmented using divalent cations and then reverse transcribed into first-strand cDNA. Then, adaptors were ligated onto both ends of the DNA fragments the 300-bp fragments were sorted and preserved, and then, amplification was performed to obtain the library. Library quality was assessed using an Agilent Bioanalyzer 2100 system according to the manufacturer’s instructions. The DNA fragment libraries were used for lncRNA sequencing on a Hiseq™ 2500 platform (Illumina, USA) that incorporated a paired-end 150 bp read run.
Functional enrichment analysis
The raw reads from each sample were qualified by FastQC. Fragments per kilobase of exon per million fragments mapped (FPKM) was used to calculate and normalize the read abundance. The basic screening and test criteria for the identification of significant differentially expressed genes were |log2FC| > 1 and FDR < 0.05, and the higher values of log2FC were selected for false-positive findings based on low number of samples. Predicted target genes were identified based on the above results. To understand the function and pathway influences of differentially expressed lncRNA, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) and Gene Ontology (GO) Consortium platforms at a threshold of FDR < 0.05 to conduct functional enrichment analysis. Our analysis incorporated hypergeometric distribution based on the Kyoto encyclopedia of genes and genomes (KEGG) with a p < 0.05 and an enrichment score of > 1.5 to determine the significant pathways.
Quantitative reverse transcription PCR (qRT-PCR) verification
We selected five differentially expressed lncRNAs that exhibited large differences and high expression levels to verify the reliability of our sequencing data. Total RNA was isolated using TRIzol, and the concentration and purity were assessed using a microspectrophotometer (Tiangen Biotech Co., Ltd.). All primers were purchased from Sangon Biotech (Shanghai, China), and they are listed in Supplement Table 1. qRT-PCR was conducted using a two-step process, where first-strand cDNA was created using the RevertAid First-Strand cDNA synthesis kit (Thermo Fisher Scientific, Inc.) followed by FastStart Universal SYBR Green Master mix to amplify cDNA using a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. GAPDH was used as an internal reference gene, and the 2−ΔΔCq method was used to determine the relative gene expression (Livak and Schmittgen 2001).
The regulatory network of lncRNA-miRNA-mRNA
Based on KEGG pathway analysis and qRT-PCR validation, we selected two lncRNAs for use in constructing regulatory networks. This network was used to explore the relationship between RNA expression and the co-expression regulation of target genes. The network was visualized using version 3.6.1 of Cytoscape.
Cell culture and transfection
BV-2 cells (purchased from ProcellLifeScience, Wuhan) are a murine immortalized microglia cell line (Blasi et al. 1990). The BV-2 cells were cultured in Dulbecco Modified Eagle Medium (DMEM, Corning 10-013-CVR) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin–streptomycin. The BV-2 cells were grown in a humidified atmosphere under 5% CO2 at 37 °C. The siRNA sequences of LOC102546895 were synthesized by GenePharma (Shanghai, China) and are provided in Supplement Table 1. The BV-2 cells were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
CCK-8 assay
For the cell proliferation assay, 1 × 104 cells were seeded into 96-well plates with 100 μL of complete culture media for five different time periods. These cells were then incubated with 10 μL of CCK-8 (Beyotime Biotechnology) assay solution for 1 h. An enzyme immunoassay analyzer (Infinite M1000, TECAN) was used to detect absorbance values at 450 nm.
Western blotting
The cells were lysed using RIPA (Thermo Scientific) buffer to release the total protein according to the manufacturer’s instructions. Equal amounts of protein (20 μg) were separated using 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) gels and then transferred onto PVDF membranes. After blocking with 5% nonfat milk in TBST to reduce nonspecific binding, the membranes were immunoblotted with primary antibodies specific for Npas4 (1:1000 dilution, SMC-495, SMC) and GAPDH (1:1000 dilution, 60004-1-Ig, Proteintech) at 4 °C overnight. After washing, the membrane was exposed to secondary antibody specific for HRP-conjugated goat anti-mouse (1:1000 dilution, A0216, Beyotime). Bands were detected using an ECL detection system (Thermo Scientific).
Flow cytometry
To test microglial cell apoptosis by the flow cytometry, the cell culture medium was collected, and the supernatant was removed by high-speed centrifugation. The cells were resuspended in 1X Binding Buffer to a cell density of 2 ~ 5 × 105 cells/mL. Annexin V-FITC solution was then added and the reactions were incubated for 15 min at room temperature in the dark. The cells were again resuspended, and propidium iodide was added to allow for flow cytometry detection. To prevent fluorescence decay, the experiment was performed within 4 h after staining, and the experiment was repeated three times.
Statistical analysis
All data are expressed as mean ± (SD) using at least three independent experiments, and statistical analyses were performed using SPSS 16 (Chicago, IL, USA). The escape latency time of rats were analyzed by Post Pairwise Comparisons using Bonferroni method. One-way ANOVA followed by Tukey’s post hoc test was used to analyze the significance between three or four groups, and a t test was used to analyze the significance between two groups. A value of p < 0.05 was defined as statistically significant.
Results
Dex attenuate cognitive deficits
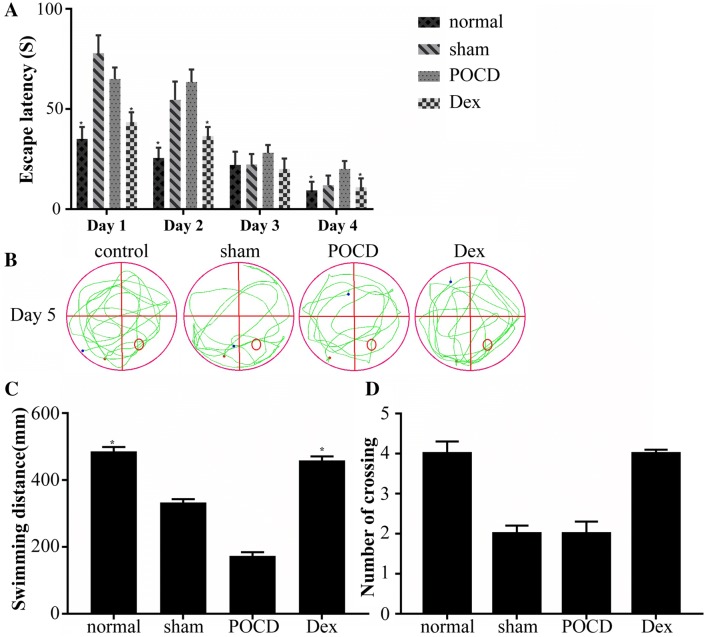
In this study, the Morris water maze test was applied to assess spatial learning and memory (Walsh et al. 2011). In all rats, the escape latency was shortened after they were trained for 4 days (Fig. 1a). The escape latencies of the sham operation group and the POCD group rats were significantly higher than those of the Dex group rats at day 1, and the escape latencies of rats in the Dex group were significantly lower than those of the POCD group on day 2 and day 4. Additionally, the escape latency changes in the Dex group were not significant compared to those in the POCD group, where a dramatic decrease was observed. On the last day of the experiment, the platform was removed, the traveling trajectories were mapped, and the total distance and the number of times the original platform position was crossed were recorded for all rats in the Morris water maze (Fig. 1b–d). We found that Dex treatment significantly increased the crossing number and the swimming distance of rats with POCD. Based on these observations, Dex treatment can improve POCD in rats.
Fig. 1.
Results of Morris Water Maze test among rats from control group (N = 3), sham group (N = 3), postoperative cognitive dysfunction (POCD) group (N = 3) and dexmedetomidine (Dex) group (N = 3). a Escape latency time (the time from launch to the first landing on the platform) of rats in each group. The escape latencies of rats in the Dex group were significantly lower than those of the POCD group on day 2 and day 4. b The maps of computer printouts of the swimming trajectories in the probe test day of each group. c The total swimming distance by rats in the water for 90 s. The swimming distance of the rats treated with Dex was increased compared with those of POCD group. d Number of crossings over the original platform position by rats. Dex treatment significantly increased the crossing number compared with POCD group. One-way ANOVA was used to analyze the data and this analysis was followed by Tukey’s analysis. *Indicates a significant difference of p < 0.05 relative to the sham group
Dex protects against neuron injury from POCD in the hippocampus of rats
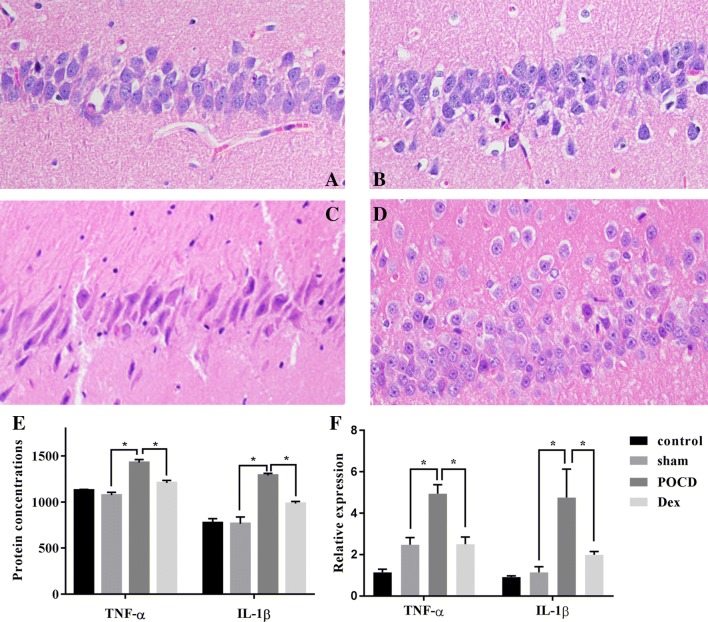
To explore the effect of Dex on neurons, the density of the neurons within the hippocampus of rats was observed using HE staining. As shown in Fig. 2a–d, in the control group and sham operation group, the cells were arranged in a compact, continuous, and uniform shape. The nucleoli of all cells were clearly visible and primarily located in the nuclear center. In the POCD group, however, neuron cells were severely damaged and exhibited a disordered arrangement and unclear cell structure. Nerve cells were observed to be decreased in density, deeply stained, and to exhibit karyopyknosis. In the Dex group, there were a greater number of neurons in the hippocampus that exhibited clear nucleoli structure and slightly sparse arrangement. Additionally, the nucleolus staining was shallow and no obvious karyopyknosis was present.
Fig. 2.
The survival of hippocampal neuron in rats was detected by HE staining. Dex alleviated hippocampal neuron injury in rats with POCD. a Normal group. b Sham group. c POCD group. d Dex group. e, f Using ELISA and qRT-PCR, the protein concentration and mRNA expression levels were detected, respectively, for TNF-α and IL-1β, and were found to be significantly down-regulated in the Dex group compared with the POCD group. Gene expression was normalized to actin transcript levels. Significant comparisons were assessed by t test. *Indicates significant differences where p < 0.05. All images are original magnification × 200
We also examined if changes in cognitive function could be associated with the inflammatory response in the hippocampus, and we selected two pro-inflammatory factors (TNF-α and IL-1β) as indicators for ELISA and qRT-PCR verification. As shown in Fig. 2e and f, the mRNA and protein levels of TNF-α were significantly up-regulated in the POCD group compared to those of the sham operation group. As expected, the expression level of TNF-α was significantly down-regulated in the Dex group compared to that of the POCD group. Analysis of another important pro-inflammatory factor (IL-1β) yielded similar results. The expression of IL-1β was significantly up-regulated in the POCD group compared to that of the sham operation group and the Dex group. In conclusion, these findings indicate that Dex regulates the inflammatory response associated with POCD and protects against neuron injury caused by POCD in the hippocampus of rats.
Analysis of differently expressed mRNAs and lncRNAs
A total of 81,534,914 ~ 94,301,368 clean reads were obtained from raw reads, and the average filter rate was above 96.3% (Supplement Table 2). Additionally, the GC and mapped rates were greater than 43% and 92.7%, respectively. These results indicate that the quality of sequencing is sufficient for subsequent analysis.
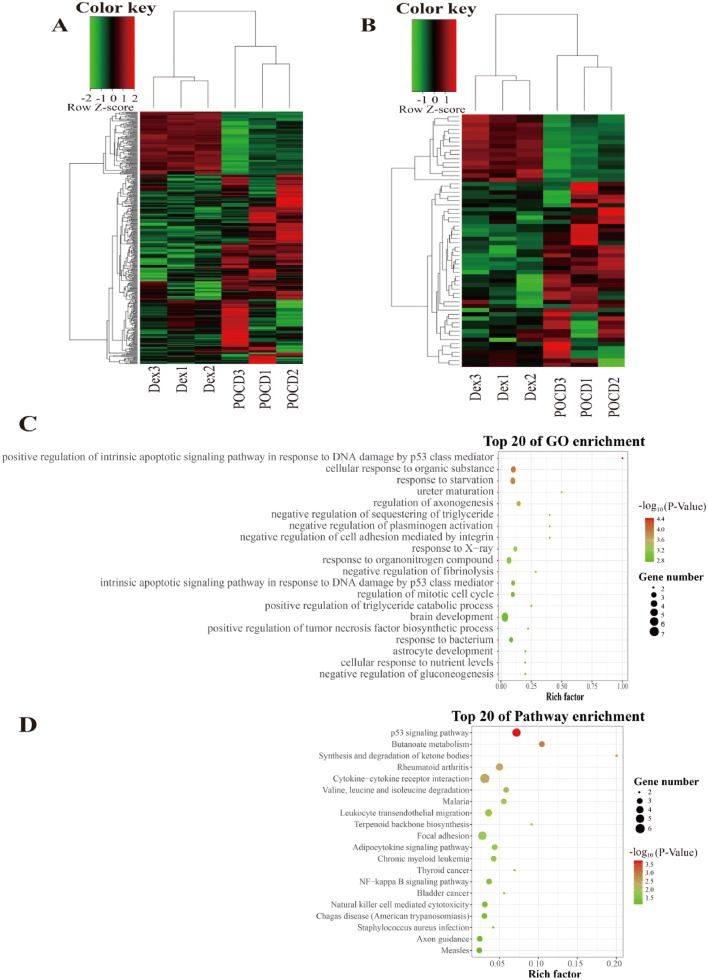
To investigate the mechanisms underlying the Dex-mediated regulation of inflammation in the context of POCD, we performed lncRNA sequencing and screened differently expressed genes (DEGs) between the Dex group and the POCD group. A total of 23,422 mRNAs and 5897 lncRNAs were obtained (Supplement Table 3). Notably, we identified 60 significantly differentially expressed lncRNAs (DE lncRNAs) that included 44 up-regulated and 16 down-regulated lncRNAs in the Dex group compared to the POCD group (Supplement Table 3). Additionally, 397 genes were significantly differentially expressed, and these mRNAs were collected for 99 genes that were up-regulated and 298 genes that were down-regulated (Supplement Table 3). A subsequent heat map analysis was performed on these DEGs (Fig. 3a, b). For both mRNA and lncRNA, it was clear that the number of down-regulated DEGs was more than twofold higher than the number of up-regulated DEGs. The specific information detailing DE lncRNAs and DE mRNAs are provided in Supplement Table 4 and Supplement Table 5, respectively.
Fig. 3.
Cluster heatmaps of significantly differentially expressed (DE) mRNAs and lncRNAs in POCD rats treated with or without Dex. a Heatmaps of DE mRNAs, b Heat maps of DE lncRNAs. Each column represents a sample. The row shaded in red represents up-regulated genes, and the row shaded in green represents down-regulated genes. c The top 20 GO enrichments based on differentially expressed lncRNA-target genes. d The top 20 KEGG enrichments based on differentially expressed lncRNA-target genes. The left represents the GO term or KEGG pathway, the right represents enrichment, and the size of solid circle indicates the number of genes
To investigate the role of lncRNA in regulating inflammation, we attempted to identify potential target genes associated with inflammation that were directly or indirectly regulated by lncRNAs. GO and KEGG pathway analyses were employed to analyze lncRNA-target genes (Fig. 3c, d). The results revealed that DElncRNAs were enriched during brain development, and the positive regulation of the intrinsic apoptotic signaling pathway mediated by p53 in response to DNA damage was the most significant. The GO terms for cellular response to organic substance and for response to starvation indicated higher gene enrichment and a higher significance. Moreover, KEGG pathway analysis revealed that cytokine–cytokine receptor interaction was the most highly enriched while the p53 signaling pathway was the most significant. The second most significant pathway was butanoate metabolism, and this was followed by synthesis and degradation of ketones and then rheumatoid arthritis pathways. We also found that the NF-kappa B signaling pathway is one of the top 20 enrichment pathways.
Validation of the sequencing data by qRT-PCR
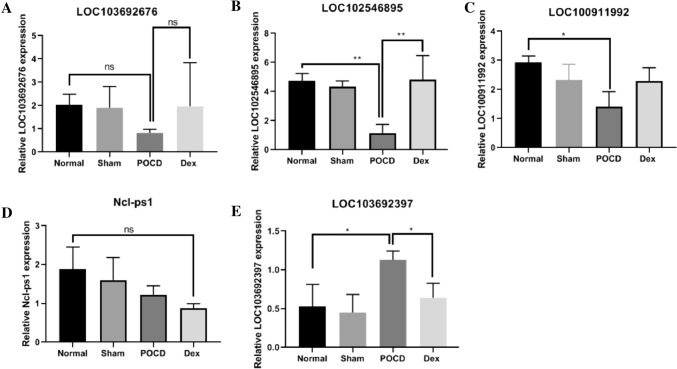
A total of 5 DE lncRNAs that exhibited large differences and high expression levels were used to verify the reliability of sequencing data. The expression patterns of the five lncRNAs indicated that the sequencing data were reliable (Fig. 4). LOC103692676, LOC102546895, and LOC100911992 were up-regulated in the Dex group, and LOC103692397 and Ncl-ps1 were down-regulated compared to levels in the POCD group. The LOC102546895 and LOC103692397 expression according to qRT-PCR data, however, exhibited significant differences between the POCD and Dex groups.
Fig. 4.
Validation of five selected DE lncRNAs performed in triplicate using qRT-PCR. a–e Expression of LOC103692676, LOC102546895, LOC100911992, Ncl-ps1, and LOC103692397 in hippocampus tissues of normal group, sham group, POCD group, and Dex treatment group. Gene expression was normalized to GAPDH transcript levels. Significance was determined using one-way ANOVA. ns no significant, *p < 0.05, **p < 0.01
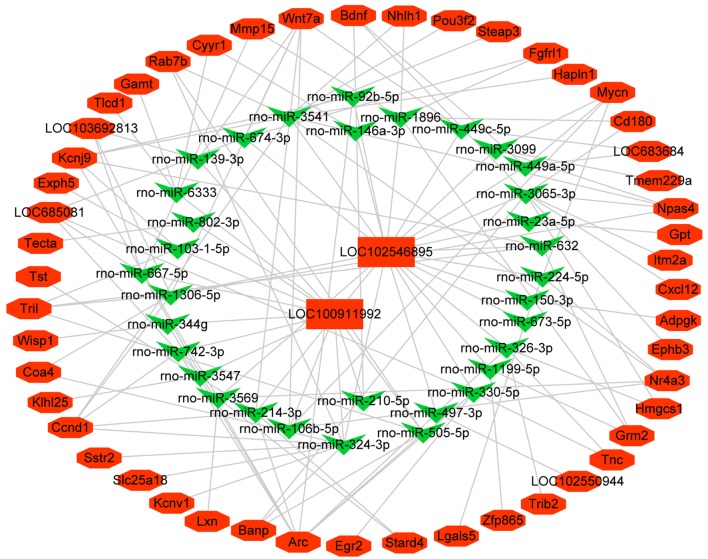
The ceRNA network of lncRNA–miRNA–mRNA
The ceRNA network is presented in Fig. 5, and this network included two lncRNAs (LOC102546895 and LOC100911992), 33 miRNAs, and 49 mRNAs. The co-expression relationship between the expression of lncRNAs and the regulation of mRNA levels was observed within the ceRNA network (Fig. 5). The ceRNA network revealed that the specific lncRNAs LOC102546895 and LOC100911992 interacted with 33 miRNAs to regulate 49 mRNAs. LOC102546895 and LOC100911992 exhibited a high correlation with the co-expressed mRNAs, suggesting that LOC102546895 and LOC100911992 may play an important role in response to Dex treatment.
Fig. 5.
The ceRNA network of lncRNA-miRNA-mRNA. Red indicates up-regulated genes and green indicates down-regulated genes. Rectangles indicate lncRNAs, V indicate miRNA, and hexagons indicate mRNAs
LOC102546895 inhibits proliferation and promotes apoptosis of microglial cells
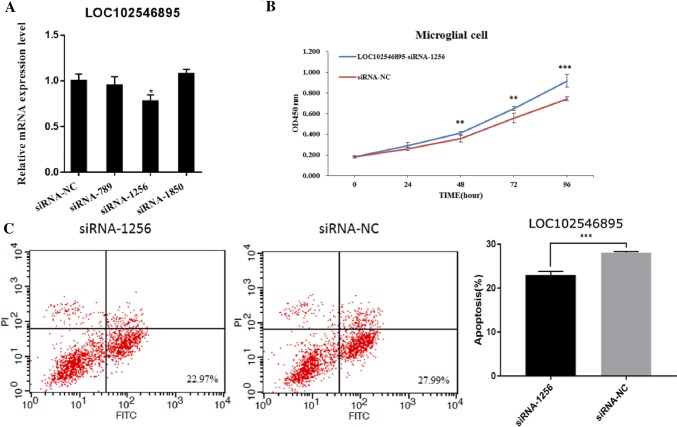
To investigate the function of LOC102546895, we constructed three short interfering RNAs (LOC102546895-siRNA-789, LOC102546895-siRNA-1256, and LOC102546895-siRNA-1850) that all specifically targeted LOC102546895. The LOC102546895-siRNA transfection efficiency was analyzed by qRT-PCR (Fig. 6a). We observed that compared to the control, the mRNA expression level of LOC102546895 exhibited a significant down-regulation only after transfection with LOC102546895-siRNA-1256, indicating that LOC102546895-siRNA-1256 possessed the best transfection efficiency and was suitable for use in subsequent experiments.
Fig. 6.
Effect of LOC102546895 on proliferation and apoptosis in microglial cells. a Interference efficiency of three siRNAs specific to LOC102546895. b LOC102546895-siRNA-1256 promoted the proliferation of microglial cells as assessed by CCK-8 assay. c LOC102546895-siRNA-1256 inhibited apoptosis in microglial cell as assessed by flow cytometry. Significance was determined using t tests. *Indicates a significant difference where p < 0.05, **indicates a significant difference where p < 0.01, ***indicates a significant differences where p < 0.001
To study the role of LOC102546895 in microglial cell proliferation and apoptosis, cell proliferation and apoptosis in microglial cells were detected using CCK-8 analysis and flow cytometry, respectively, after knock-down using LOC102546895-siRNA-1256. Our results indicated that the cell proliferation and viability in the LOC102546895-siRNA-1256 group was significantly increased compared to that of the siRNA-NC group (Fig. 6b), suggesting that LOC102546895 inhibits microglial cell proliferation. Conversely, apoptosis in microglia cells decreased after transfection with LOC102546895-siRNA-1256 (Fig. 6c), suggesting that LOC102546895 promotes microglial cell apoptosis. Based on these results, we conclude that LOC102546895 acts as a negative modulator of survival in microglial cells.
LOC102546895 promotes the expression of Npas4 in microglial cells
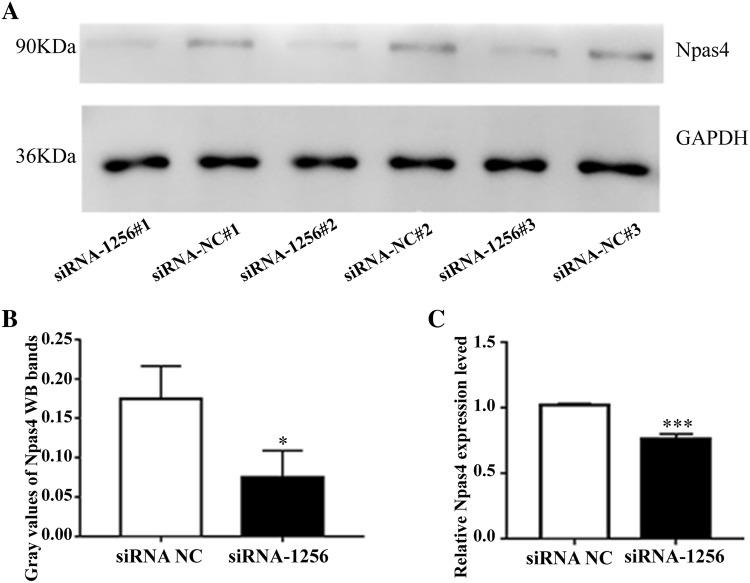
Neuronal Per-Arnt-Sim domain protein 4 (Npas4) is a highly protective neuron transcription factor that is involved not only in the transcriptional regulation of neurons and synaptic development genes, but also in the inhibition of neuroinflammatory responses (Choy et al. 2015). Interestingly, the target gene Npas4 was demonstrated to be regulated by LOC102546895 according to the ceRNA network. Therefore, we selected Npas4 for further studies where we explored the effect of LOC102546895 on Npas4. Specifically, we examined the mRNA and protein expression levels of Npas4 after the microglial cells were transfected with LOC102546895-siRNA-1256. The results of qRT-PCR and western blotting (WB) analyses revealed that LOC102546895 knockdown using LOC102546895-siRNA-1256 significantly reduced the level of Npas4 mRNA and protein in the microglial cells compared to those in cells transfected with siRNA-NC (Fig. 7), suggesting that LOC102546895 may promote the expression of Npas4.
Fig. 7.
Effect of LOC102546895 on the mRNA expression and protein levels of target gene Npas4 in microglia cells. a, b WB assays and gray values analysis showed that LOC102546895-siRNA-1256 inhibited the protein levels of Npas4. c LOgC102546895-siRNA-1256 inhibited the mRNA expression levels of Npas4 via qRT-PCR result analysis. Statistical significance was determined using t test. *Indicates a significant difference where p < 0.05, ***indicates a significant difference where p < 0.001
Discussion
POCD negatively impacts life expectancy and quality of life in patients, particularly in elderly individuals that exhibit low tolerance to surgical stress (Bharadwaj and Kamath 2019). Studies have shown that Dex treatment improves POCD in patients; however, the exact mechanisms underlying Dex function remain undefined (Carr et al. 2018; Liu et al. 2016; Lu et al. 2017). Although Dex plays a function role in repairing nerves, the drug has side effects in clinical application. Also, a massive body of clinical drugs have the same problem. However, the primary aim of this study was to explore the mechanisms regulating lncRNA in the process of Dex-mediated POCD improvement and to find a better therapeutic target. These results would further provide a theoretical basis for the subsequent clinical application of Dex in the treatment of POCD.
Alpha-2 adrenoceptors are widely found throughout the central and peripheral nervous systems of mammals, and these receptors are involved in a number of complicated regulatory functions of the norepinephrine system such as nociception, attention, and emotion (Dawson et al. 2011). Dex is a strongly selective agonist of the alpha-2 adrenoceptor, and thus, Dex can be used for the treatment of POCD (Carr et al. 2018). In this study, we found that Dex could improve spatial memory ability and neuronal damage in hippocampal tissue as assessed by Morris water maze testing and HE staining. Pro-inflammatory cytokines such as TNF-α and IL-1β disrupt blood–brain barrier (BBB) integrity, and therefore, TNF-α and IL-1β may act as important risk factors for POCD after surgery (Terrando et al. 2010). In the present study, according to the expression levels of TNF-α and IL-1β, we found that Dex treatment could inhibit the expression of pro-inflammatory factors after splenectomy in rats and that sham operation alone was not enough to trigger cognitive dysfunction. Our conclusions were consistent with those of previous research that found Dex significantly decreased the expression of TNF-α and IL-1β (Qian et al. 2015) by reducing the LPS-induced systemic release of TNF-α and IL-1β (Xiang et al. 2014).
Currently, research is lacking regarding Dex treatment of POCD at the lncRNA sequencing level. In the present study, we identified 5897 lncRNAs and 23,422 mRNAs using high-throughput sequencing techniques, and of these, only 0.8% (60) and 1.67% (397) have been identified as DElncRNAs and DEGs, respectively. This is a common phenomenon in transcriptome sequencing and suggests that only partial genes are specifically involved in different stages of life (Yan et al. 2018). These DElncRNAs were preferentially clustered within the p53 signaling pathway and the NF-kappa-B signaling pathway. It is currently understood that the p53 signaling pathway is activated by multiple signals when cells are exposed to the various stresses such as genotoxic damage, hypoxia, heat shock, and oncogenic assault (Vousden and Lane 2007). Once activated, the p53 signaling pathway will enable DNA repair or advance cellular death programs to provoke transient or permanent growth arrest to prevent cancer development (Stegh 2012). Activation of the NF-kappa-B signaling pathway stimulates the excessive production of cytotoxic factors such as the inflammatory mediators TNF-α and IL-1β in microglial cells to promote cell death (Dresselhaus and Meffert 2019). In this study, knock-down of DElncRNA LOC102546895 expression inhibited the proliferative capacity of microglial cells while increasing their apoptotic potential, which is consistent with our study prediction. It is possible that splenectomy caused brain stress in rats and this was accompanied by neuron damage that triggered POCD and activated the p53 pathway. In the process of Dex-mediated POCD treatment, the NF-kappa-B signaling pathway responds to the regulation of p53 and inhibits microglial activation and neuroinflammation, ultimately hindering the aggravation of cognitive impairment.
Npas4 is a highly protective neuronal transcription factor that has been demonstrated to play a role in synapse formation and neuronal survival (Choy et al. 2015). Studies have shown that Npas4 is directly related to learning and memory formation in the hippocampus and it is also related to cognitive and social neural behaviors (Ramamoorthi et al. 2011; Coutellier et al. 2012). Knock-down of Npas4 resulted in the activation of astrocytes and microglia and accelerated the expression of the pro-inflammatory cytokines IL-6 and TNF-α, ultimately leading to a significant deterioration in stroke lesion size, neuronal necrosis, and neurodegeneration in mice (Choy et al. 2016). Consistent with previous studies, our results show that the down-regulation of the expression of LOC102546895 resulted in a decrease in the transcription level and protein level of Npas4, an increase in the proliferation of microglial cells, and a reduction in apoptosis. These results suggest that during the Dex-mediated treatment of POCD, LOC102546895 may inhibit neuroglial activation and inflammation by targeting Npas4 to improve cognitive function in rats with POCD.
Conclusions
In conclusion, we demonstrated that Dex improved POCD in rats by using Morris water maze and HE staining experiments. ELISA and qRT-PCR analyses revealed that Dex-mediated treatment of POCD inhibited the expression levels of the pro-inflammatory factors TNF-α and IL-1β. Additionally, lncRNA sequencing revealed that DElncRNAs were primarily involved in the p53 signaling pathway and the NF-kappa-B signaling pathway. Furthermore, DElncRNA LOC102546895 may target Npas4 to inhibit proliferation of microglial cells while promoting apoptosis. Our study showed that Dex regulated lncRNAs expression profile of hippocampus tissues from rats, and the LOC102546895 regulated by Dex may play a role in in rats with POCD through targeting Npas4.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Table 4. The specific information detailing DE lncRNAs (XLSX 15 kb)
Supplement Table 5. The specific information detailing DE mRNAs (XLSX 57 kb)
Author contributions
Conceptualization: ZZ and GX. Data curation: ZZ and GX. Formal analysis: FD and LC. Funding acquisition: ZZ and GX. Investigation: FD, LC and BZ. Methodology: FD, LC and BZ. Software: FD. Writing—original draft: FD and LC. Writing—review and editing: FD, LC and BZ. All authors read and approved the final manuscript.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics approval and consent to participate
All rat experiments completely adhered to the Guide for the Care and Use of Laboratory Animals (National Institutes of Health). All the experiments were approved by the Institutional Animal Care and Use Committee of the second affiliated hospital of Nanchang university.
Contributor Information
Zhidong Zhou, Email: 45285184@qq.com.
GuoHai Xu, Email: xuguohai@sina.com.
References
- Bharadwaj S, Kamath S. Postoperative cognitive dysfunction. Textb Neuroanesthesia Neurocrit Care. 2019 doi: 10.1007/978-981-13-3387-3_34. [DOI] [Google Scholar]
- Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol. 1990;27(2):229–237. doi: 10.1016/0165-5728(90)90073-v. [DOI] [PubMed] [Google Scholar]
- Carr ZJ, Cios TJ, Potter KF, Swick JT. Does dexmedetomidine ameliorate postoperative cognitive dysfunction? A brief review of the recent literature. Curr Neurol Neurosci Rep. 2018;18(10):64. doi: 10.1007/s11910-018-0873-z. [DOI] [PubMed] [Google Scholar]
- Choy FC, Klaric TS, Koblar SA, Lewis MD. The role of the neuroprotective factor Npas4 in cerebral ischemia. Int J Mol Sci. 2015;16(12):29011–29028. doi: 10.3390/ijms161226144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy FC, Klaric TS, Leong WK, Koblar SA, Lewis MD. Reduction of the neuroprotective transcription factor Npas4 results in increased neuronal necrosis, inflammation and brain lesion size following ischaemia. J Cereb Blood Flow Metab. 2016;36(8):1449–1463. doi: 10.1177/0271678X15606146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutellier L, Beraki S, Ardestani PM, Saw NL, Shamloo M. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS ONE. 2012;7(9):e46604. doi: 10.1371/journal.pone.0046604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson LF, Phillips JK, Finch PM, Inglis JJ, Drummond PD. Expression of α 1-adrenoceptors on peripheral nociceptive neurons. Neuroscience. 2011;175:300–314. doi: 10.1016/j.neuroscience.2010.11.064. [DOI] [PubMed] [Google Scholar]
- Deiner S, Luo X, Silverstein JH, Sano M. Can intraoperative processed EEG predict postoperative cognitive dysfunction in the elderly? Clin Ther. 2015;37(12):2700–2705. doi: 10.1016/j.clinthera.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus EC, Meffert MK. Cellular specificity of NF-kappaB function in the nervous system. Front Immunol. 2019;10:1043. doi: 10.3389/fimmu.2019.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q, Qian X, An J, Wen H, Cope DK, Williams JP. Higher dose dexamethasone increases early postoperative cognitive dysfunction. J Neurosurg Anesthesiol. 2014;26(3):220–225. doi: 10.1097/ANA.0000000000000024. [DOI] [PubMed] [Google Scholar]
- Green CM, Schaffer SD. Postoperative cognitive dysfunction in noncardiac surgery: a review. Trends Anaesth Crit Care. 2019;24:40–48. doi: 10.1016/j.tacc.2018.08.003. [DOI] [Google Scholar]
- Hem S, Albite R, Loresi M, Rasmussen J, Ajler P, Yampolsky C, Chabot JD, Gerszten PC, Goldschmidt E. Pathological changes of the hippocampus and cognitive dysfunction following frontal lobe surgery in a rat model. Acta Neurochir. 2016;158(11):1–9. doi: 10.1007/s00701-016-2938-6. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wang JP, Yu XL, Wang ZB, Xu TS, Cheng XC. Non-coding RNAs and diseases. Mol Biol+ 2013;47(4):465–475. doi: 10.1134/s0026893313040171. [DOI] [PubMed] [Google Scholar]
- Hudetz JA, Iqbal Z, Gandhi SD, Patterson KM, Byrne AJ, Hudetz AG, Pagel PS, Warltier DC. Ketamine attenuates post-operative cognitive dysfunction after cardiac surgery. Acta Anaesthesiol Scand. 2009;53(7):864–872. doi: 10.1111/j.1399-6576.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- Huupponen E, Maksimow A, Lapinlampi P, Särkelä M, Saastamoinen A, Snapir A, Scheinin H, Scheinin M, Meriläinen P, Himanen SL, Jääskeläinen S. Electroencephalogram spindle activity during dexmedetomidine sedation and physiological sleep. Acta Anaesthesiol Scand. 2010;52(2):289–294. doi: 10.1111/j.1399-6576.2007.01537.x. [DOI] [PubMed] [Google Scholar]
- Jacob S, Karl Bang C, Thomas L, Nicolai L, Rasmussen LS. Long-term consequences of postoperative cognitive dysfunction. Anesthesiology. 2009;110(3):548–555. doi: 10.1097/ALN.0b013e318195b569. [DOI] [PubMed] [Google Scholar]
- Jevtovic-Todorovic V, Beals J, Benshoff N, Olney JW. Prolonged exposure to inhalational anesthetic nitrous oxide kills neurons in adult rat brain. Neuroscience. 2003;122(3):609–616. doi: 10.1016/j.neuroscience.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Li L, Zhang Y, Luo H, Huang C, Li S, Liu A, Jiang Y. Systematic identification and analysis of expression profiles of mRNAs and incrnas in macrophage inflammatory response. Shock. 2019;51(6):770–779. doi: 10.1097/SHK.0000000000001181. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ma L, Gao M, Guo W, Ma Y. Dexmedetomidine reduces postoperative delirium after joint replacement in elderly patients with mild cognitive impairment. Aging Clin Exp Res. 2016;28(4):729–736. doi: 10.1007/s40520-015-0492-3. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu J, Chen G, Zhou H, Zhou Q, Zhu Z, Wu C. Effect of parecoxib sodium pretreatment combined with dexmedetomidine on early postoperative cognitive dysfunction in elderly patients after shoulder arthroscopy: a randomized double blinded controlled trial. J Clin Anesth. 2017;41:30–34. doi: 10.1016/j.jclinane.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mario C, Antonio Rei F, Niccolò T, Daqing M, Claudia M, Marc F, Masao T, Lever IJ, Jagdeep N, Fanselow MS. Role of interleukin-1β in postoperative cognitive dysfunction. Ann Neurol. 2010;68(3):360–368. doi: 10.1002/ana.22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian XL, Zhang W, Liu MZ, Zhou YB, Zhang JM, Han L, Peng YM, Jiang JH, Wang QD. Dexmedetomidine improves early postoperative cognitive dysfunction in aged mice. Eur J Pharmacol. 2015;746:206–212. doi: 10.1016/j.ejphar.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, Lin Y. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334(6063):1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, Tian Z, Guan Y, Tang L, Xu C, Wang L, Gao X, Tian W, Wang J, Yang H, Wang J, Sun Y. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22(5):806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegh AH. Targeting the p53 signaling pathway in cancer therapy - the promises, challenges and perils. Expert Opin Ther Targets. 2012;16(1):67–83. doi: 10.1517/14728222.2011.643299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao WZ, Peng H, Hong WW, Yin LH, Pu YP, Liang GY. Integrated analysis of long non-coding RNAassociated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49(5):2023–2036. doi: 10.3892/ijo.2016.3716. [DOI] [PubMed] [Google Scholar]
- Terrando N, Monaco C, Ma D, Foxwell BM, Feldmann M, Maze M. Tumor necrosis factor-alpha triggers a cytokine cascade yielding postoperative cognitive decline. Proc Natl Acad Sci USA. 2010;107(47):20518–20522. doi: 10.1073/pnas.1014557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Walsh CM, Booth V, Poe GR. Spatial and reversal learning in the Morris water maze are largely resistant to six hours of REM sleep deprivation following training. Learn Mem. 2011;18(7):422–434. doi: 10.1101/lm.2099011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wu Q, Liang SS, Zhang XK, Hu Q, Chen QH, Huang HJ, Xu L, Lou FQ. Dexmedetomidine promotes the recovery of neurogenesis in aged mouse with postoperative cognitive dysfunction. Neurosci Lett. 2018;677:110–116. doi: 10.1016/j.neulet.2018.03.043. [DOI] [PubMed] [Google Scholar]
- Xiang H, Hu B, Li Z, Li J. Dexmedetomidine controls systemic cytokine levels through the cholinergic anti-inflammatory pathway. Inflammation. 2014;37(5):1763–1770. doi: 10.1007/s10753-014-9906-1. [DOI] [PubMed] [Google Scholar]
- Yan Y, Ye W, Chen Q, Yang L, Zhang L, Liu Y, Zhou X, Wang G. Differential expression profile of long non-coding RNA in the stenosis tissue of arteriovenous fistula. Gene. 2018;664:127–138. doi: 10.1016/j.gene.2018.04.028. [DOI] [PubMed] [Google Scholar]
- Zhang L, Fang Y, Cheng X, Lian YJ, Xu HL. Silencing of long noncoding RNA SOX21-AS1 relieves neuronal oxidative stress injury in mice with alzheimer’s disease by upregulating FZD3/5 via the Wnt signaling pathway. Mol Neurobiol. 2019;56(5):3522–3537. doi: 10.1007/s12035-018-1299-y. [DOI] [PubMed] [Google Scholar]
- Zywiel MG, Prabhu A, Perruccio AV, Gandhi R. The influence of anesthesia and pain management on cognitive dysfunction after joint arthroplasty: a systematic review. Clin Orthop Related Res. 2014;472(5):1453–1466. doi: 10.1007/s11999-013-3363-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Table 4. The specific information detailing DE lncRNAs (XLSX 15 kb)
Supplement Table 5. The specific information detailing DE mRNAs (XLSX 57 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.