Abstract
The screening and testing of extracts against a variety of pharmacological targets in order to benefit from the immense natural chemical diversity is a concern in many laboratories worldwide. And several successes have been recorded in finding new actives in natural products, some of which have become new drugs or new sources of inspiration for drugs. But in view of the vast amount of research on the subject, it is surprising that not more drug candidates were found. In our view, it is fundamental to reflect upon the approaches of such drug discovery programs and the technical processes that are used, along with their inherent difficulties and biases. Based on an extensive survey of recent publications, we discuss the origin and the variety of natural chemical diversity as well as the strategies to having the potential to embrace this diversity. It seemed to us that some of the difficulties of the area could be related with the technical approaches that are used, so the present review begins with synthetizing some of the more used discovery strategies, exemplifying some key points, in order to address some of their limitations. It appears that one of the challenges of natural product-based drug discovery programs should be an easier access to renewable sources of plant-derived products. Maximizing the use of the data together with the exploration of chemical diversity while working on reasonable supply of natural product-based entities could be a way to answer this challenge. We suggested alternative ways to access and explore part of this chemical diversity with in vitro cultures. We also reinforced how important it was organizing and making available this worldwide knowledge in an “inventory” of natural products and their sources. And finally, we focused on strategies based on synthetic biology and syntheses that allow reaching industrial scale supply. Approaches based on the opportunities lying in untapped natural plant chemical diversity are also considered.
Keywords: secondary metabolites, drug production, natural products, renewal sources, plant chemodiversity, synthetic biology, in vitro cultures, medicinal chemistry
Background on Natural Compounds in Drug Discovery
Drugs and Natural Products
Several reviews, like the updated survey from Newman and Cragg (2016), pointed to the fact that many drugs on the market are from natural origin; these authors stated that, out of the 1,328 new chemical entities approved as drugs between 1981 and 2016, only 359 were purely of synthetic origin. From the remaining ones, 326 were “biological” entities (peptides of more than 50 residues, including therapeutic antibodies), and 94 were vaccines. A little less than half of those new drugs (549, exactly) were from natural origin or derived inspired from natural compounds. Furthermore, in the anticancer area, out of the 136 approved nonbiological compounds from the same period (1981–2014), only 23 were purely synthetic (i.e. not derived from natural compounds nor natural compounds themselves) (Newman and Cragg, 2016). Natural origin can have different definitions, and these authors accounted for three categories: unaltered natural (pure) products; defined mixture of natural products (NP) and natural product derivatives isolated from plants or other living organisms as fungi, sponges, lichens, or microorganisms; and products modified by medicinal chemistry. There are many examples: anticancer drugs such as docetaxel (Taxotere™), paclitaxel (Taxol™), vinblastine, podophyllotoxin (Condylin™), or etoposide; steroidal hormones such as progesterone, norgestrel, or cortisone; cardiac glycosides such as digitoxigenin; antibiotics like penicillin, streptomycin, and cephalosporins [see IA Ross for more examples (1999)]. Furthermore, Rodrigues et al. (2016) pointed to the fact that fragments derived from natural structures are a source of diverse molecules from which new drugs can be designed, thanks to the fragment-based drug discovery approach (Erlanson et al., 2016; Mortenson et al., 2018; Yñigez-Gutierrez and Bachmann, 2019).
Screening for New Drugs and Discovery Approaches
Besides the understanding of pathological processes, the source of molecules has been a main concern for the pharmaceutical industry. Vast libraries of compounds have been established in order to feed the research. For example, in midsize pharmaceutical companies, it is common to find libraries from 30,000 up to 500,000 compounds, while for big pharmas, the numbers are more in the 500,000 to several million ranges (Macarron et al., 2011). To our knowledge this is also the case for the National Chinese Compound Library in Shanghai, China (http://en.cncl.org.cn/). Finally, national or transnational efforts have been reported to create such depositories of compounds for the use of screening programs from the Academy: see Horvath et al. (2014) in Europe and Thornburg et al. (2018) for the NIH/NCI effort. In addition, vendors are also selling libraries of compounds composed of a “large” diversity that they build according to different principles (Boss et al., 2017). Several publications deal with how the compounds are chosen (Langer et al., 2009), if they are following Lipinski rules (Lipinski et al., 2001; Lipinski, 2003) or not, if they are virtual (Glaab, 2016) or real, if they are systematically tested on all the targets, how they can be organized in subclasses of compounds designed to potentially interact with channels, receptors, or enzymes, etc. Furthermore, the composition of the library in relation with the main categories of molecules—small synthetic compounds, drug-like organic compounds, peptides, proteins, sugars, nucleosides, or natural compounds—can greatly vary in function of the “Pharma company culture”, that is to say, the compounds that have already been synthesized in a given company as well as the “sensitivity” of the medicinal chemists and screening people.
When the decision to incorporate natural products is made, pure well-defined compounds or extracts are selected according to different criteria: pharmacognosy, ethnopharmacology, or even traditional knowledge. Because of the traditions existing in the uses of botanicals and medicinal plants, this empirical knowledge has accumulated for ages and passed through generations. Modern pharmacology has explored and validated probably only a minor part of this knowledge through attempts of rationalizing the use of plants as sources of drugs. This first possible approach would be a way to guide some drug discovery projects. Another totally different approach based on the use of high-throughput screening (HTS) emerged one or two decades ago. It aimed at exploring systematically the immense chemical diversity in secondary metabolites and was based on the technological developments of discovery tools such as miniaturization and automation (Atanasov et al., 2015). In this sense, the increase of the amount of compounds was tested simultaneously, and the scientific rationalization of the selection of those compounds, thanks to the growing capacities of chemo-informatic approaches (Larsson et al., 2005; Larsson et al., 2007), emerged as trends in HTS.
Strategies for Identification of Bioactive Compounds From Crude Extracts
When reviewing the literature on how discovery of plant-derived actives is performed, the following pattern emerges (Weller, 2012; Sharma and Gupta, 2015, Heinrich et al., 2018). Plants are collected in precise geo-localized sites. The collection may include the whole plant or any part such as leaves, stems, bark, seeds, or roots. Then, the botanical material is dried and powdered using mechanical means, such as grounders. Those powders are then extracted with solvents at different temperatures or of increasing polarities with sequential extraction procedures in the cases when, for example, the chemistry of the active compounds is unknown. These first steps are important to consider as the extraction method might influence the chemical composition of the extract and consequently, its biological activity.
Then, the extracts are dried under low pressure, and the final solid residue is suspended in a solution comprising the minimal amount of a biological-compatible solvent, often DMSO. The next steps would include the testing of the extracts in 96 (or 384) well plates against the biological targets of the program. These biological tests are often a cloned enzyme catalytic assay, a receptor binding assay, a protein–protein interaction assay or even a whole pathway (Ishibashi and Ohtsuki, 2008; Xie et al., 2018), to name but a few. Biological testing targeting whole-cells was also reported against cancer cells (Mazzio et al., 2014; Kant et al., 2016), virus-infected cells (Yi et al., 2004; Dai et al., 2012), or microorganisms (Correia et al., 2008; Figueroa-López et al., 2014; St-Pierre et al., 2018). Extracts showing activity on those tests are selected and submitted to fractionation by chromatography. Each fraction (typically around ten) will be tested in turn in the same assay, and the active fractions are subjected to one or several extra fractionations, often using alternative chromatographic conditions.
Some of these experiments are based on an HTS environment. HTS methodologies provided us with impressive progresses in terms of increased speed of assay and lowering of price. Indeed, robots can handle several thousands of tests during a workday. However, the initial enthusiasm for HTS (Harvey and Cree, 2010; Sarker and Nahar, 2012) when applied to crude extract libraries in targeted assays systems has been facing several issues as stated recently by Thornburg et al. (2018). In fact, HTS techniques do not really modify the discovery process itself. For example, starting with 2,000 extracts—which is a modest number considering (a) the number of plants species available and (b) the number of compounds in an HTS campaign—might result into 10% actives, in the best case. Thus, the next step would be 200 hydrophobic columns, with the collection of about 10 fractions per chromatography, and then 2,000 tests. As exemplified, the real bottleneck in this whole process is the parallel fractionations of the actives; as of today, only partial progresses in their automatization have been reported over the last years (Sharma and Gupta, 2015).
Other strategies have been developed that delivered results. In their detailed review, Atanasov et al. (2015) classify into five groups the strategies to identify bioactive compounds from plant extracts: the bioactivity-guided fractionation strategy previously mentioned, the similar synergy-directed fractionation strategy, the metabolic profiling strategy, the metabolism-directed (biotransformation focused) strategy, and finally the direct phytochemical isolation strategy. The group of metabolic profiling approaches has a more detached relationship with bioactivity as they are not focused on compounds. They were developed during the last 10–15 years in plant natural products. Using highly sensitive and reproducible analytical methods, they allow the correlation between chemical profile (qualitative or quantitative) and bioactivity data (Zampieri et al., 2017; Parrot et al., 2018; Wolfender et al., 2019) with recent progress in the field of data analysis and integration at the extract level (Wolfender et al., 2019). Finally, the group of direct phytochemical isolation approaches focuses on the comprehensive chemical characterization of the plant extract and the isolation of novel scaffolds without immediately evaluating their bioactivity.
Successes and Limitations
Some of the successes in terms of new drug development have already been mentioned in Drugs and Natural Products. But in terms of screening and strategy for finding an active compound in an extract as an enzyme inhibitor or a protein/protein interaction inhibitor, many successes have also been reported. Some examples of screening results of extracts with those approaches (Atanasov et al., 2015) can be mentioned here, but being exhaustive is impossible, as literally hundreds of such tests were performed [e.g. from our laboratories (Bousserouel et al., 2005; Lautié et al., 2008; Litaudon et al., 2009a; Litaudon et al., 2009b; Columba-Palomares et al., 2015; Catteau et al., 2017; Olivon et al., 2018) and from hundreds of others]. Some examples reported recently are given in Table 1 of the inhibitory activities of plant extracts on specific enzymes. Of note, the panel of enzyme activities is large and diverse; the examples in Table 1 reflect the current trend in terms of enzyme of origin for this type of work: they are mainly coming from cancer and diabetes/obesity fields. It is important to say that an unknown number of failing attempts remain unpublished.
Table 1.
Some examples of enzyme inhibiting extracts from natural origin.
| Enzyme | Plant extract | Reference |
|---|---|---|
| Carbohydrase | Hibiscus sabdariffa | (Ifie et al., 2016) |
| Cholinesterases | Jatropha gossypifolia | (Saleem et al., 2016) |
| COX-1, COX-2 | Onopordum acanthium | (Lajter et al., 2015) |
| Cytochrome P450 3A | Aframomum melegueta, Dennettia tripelata | (Nduka et al., 2017) |
| Cytochrome P450 1B1 | Glycyrrhiza glabra | (Sharma et al., 2017) |
| DNA polymerase | Sorbus acuparia | (Turumtay et al., 2017) |
| Elastase | Tagetes erecta | (Vallisuta et al., 2014) |
| Lipoxygenase, urease | Polygonatum verticillatum | (Khan et al., 2015) |
| Metalloproteinase | Citrus paradisii, Citrus sinensis, Citrus maxima | (Ademosun et al., 2015) |
| Monoamine oxidase A | Hypericum perforatum, Paganum harmal | (Herraiz and Guillen, 2018) |
| Angiotensin converting enzyme | Vernonia amygdalina, Gongronema latifolium | (Ajibola et al., 2011) |
| N-myriostoyltransferase | Terminalia bentzoë | (Apel et al., 2018) |
| Protein tyrosine phosphatase | Eugenia jambolana | (Liu et al., 2017) |
| Tyrosinase | Gynotroches axillaris | (Abed et al., 2016) |
| Tyrosinase | Phaleria macrocarpa | (Nadri et al., 2014) |
| Urease | Rumex nervosus | (Khan et al., 2014) |
| Xanthine oxidase | Fraxinus angustifolia, Pistacia lentiscus | (Berboucha et al., 2010) |
| α-Amylase | Urtica dioica, Juglans regia | (Rahimzadeh et al., 2014) |
| α-Glucosidase | Terminalia macroptera | (Pham et al., 2014) |
| β-ketoacyl-ACP reductase | Alpinia officinarum | (Huang et al., 2008) |
| Xanthine oxidase | Puertorican plants* | (Guerrero and Guzman, 1998) |
| Epoxide hydrolase | Chinese herbs* | (Shi et al., 2008) |
| HIV-1 integrase | Chinese herbs* | (Au et al., 2001) |
| Acetylcholinesterase | Marine fungi**,* | (Liu et al., 2014) |
| Lipase | Chlorella sp.*** | (Sibi, 2015) |
| Lipase | Ascophyllum nodosum*** | (Austin et al., 2018) |
*: unspecified genuses; **: fungi; ***: algae
For the last thirty years screening for new molecules—whether candidate drugs per se or hit compound that should be modified—is one of the two main sources of drugs for the pharmaceutical industry. These screening strategies delivered mixed results, and several factors complicated dramatically the analyses of these results. The main ones are in our opinion: the way diseases can be simplified—or not—to a molecular target that is amenable to HTS; the choices of the molecules which can be screened into these assays; the quality of the assays and/or their sensitivity; the statistical considerations to determine the threshold at which a compound is recognized as an active; and finally, the source of the screened molecules. We (Boutin et al., 2018) and others (Schmid et al., 1999; Dufresne, 2000; Thiericke, 2000; Mayr and Bojanic, 2009; Bodle et al., 2017) exposed our screening strategies at several occasions, and considering the diversity of the potential problems, it can be hard to identify a universal approach. This is a fact that HTS promising techniques have not delivered as many drugs as expected with regard to the price and the ambition of the solutions/organizations involved (Newman and Cragg, 2016).
Several Limitations in the “Classical” Strategies to Identify Bioactive Natural Products
The first type of limitation would be related to the collections of living organisms and access to plant species, for example. The probability still exists that a plant collected on a given location would not be there the next time around, especially the whole plant. If bioactivities associated with this species are discovered during the process, it could be hard to find again the same plant population. In those cases, two choices are possible: to look for this species in another location or to look in the same location for a closely related species with the help of botanists. Neither solution is entirely satisfactory: the change of harvest location can imply change(s) in secondary metabolite production as their biosynthesis generally depends on the control by both biotic and abiotic factors. Furthermore, the other species of the same genus might very well express different genes coding for proteins acting slightly differently in biosynthetical pathways leading to different compounds or to the same compounds synthetized in different proportions (see below for discussion on diversity). Moreover, if the collected species are not properly reported following the correct taxonomy, the harvest might be even more difficult to reproduce: thereby, a table of correspondence between the names mentioned in this review (and used in the articles of origin) and the accepted plant species names is provided in the supplementary data ( Table S1 ).
Another type of limitation is related with the fact that groups performing bioguided discovery of new actives are often confronted with similar problems: (a) the active extracts, once fractionated, do not maintain the level of activity; (b) the whole long and difficult isolation process leads to a molecule that is mundane or known for decades; (c) the final product quantity in the remaining active fraction is too small to hope for a structure elucidation; (d) going back to the location of origin, the same plant population is not found anymore or if the plant is found, the same extraction treatment led to an apparent lack of active fraction(s) on the same target. Most of those events are not discussed in publications, obviously. Therefore, this bioguided approach, existing with small variations––mostly technical––is paved with difficulties that are more than often discouraging at least for an industry or for big programs (Nothias et al., 2018).
As discussed in many instances and particularly well summarized by Atanasov et al. (2015), one of the key starting points is the choice of the pharmacological assay used for the screening. The fact that the targeted disease is obviously more complex than the molecular target it has been simplified to, is universally recognized, but alternative approaches remain elusive or still difficult to set up. For example, the disease could also be simplified by a phenotypic change of a cell originating from a diseased organ using the stem cell differentiation approaches (Bedut et al., 2016). After treating such a model with NP extracts—having potentially hundreds of compounds—reverse pharmacology should allow recognizing the actual target of the NP compound as the lysed cell extract is chromatographed to retain the target proteins (Raut et al., 2017). Nevertheless, cellular phenotypic changes are extremely complex to understand, and this approach has been more than once extremely frustrating despite some successes, outside of the NP recognition area (Nosjean et al., 2000; Graves et al., 2002). Actually, another limitation of this approach is linked with working with complex mixtures in NP discovery as sometimes several compounds could contribute synergistically to the global action on a disease: like for example the compounds (at least three of them) derived from leaves of Salvia miltiorrhiza (red sage) that are reported to act at many different levels of liver fibrosis (Ma et al., 2019). This kind of observation would strongly argue against the strategy described in here aiming at the discovery of the main pure drug-like compound in a plant extract. In summary, such approaches (phenotype screening and reverse pharmacology) would require disease models that are far from being the current available systems.
Limitations Related to the Need of Market-Compatible Amounts
In the domain of drug production, a fact is that around 60,000 tons per year of salicylic acid are produced synthetically worldwide (https://ihsmarkit.com/products/chemical-technology-pep-reviews-salicylic-acid-2003.html), and this figure translates to 80 billions of pills. That is to say, that in order to benefit patients, critical amounts of a potential drug need to be considered, and the access to the identified NP as well as its availability are critical. For example, at the start of the Taxol™ story, the compound was isolated from Taxus brevifolia bark. Ten kilograms of bark was necessary to obtain 2 g of the pure compound needed for the treatment of one patient. For the clinical study, 12,000 trees were cut down to obtain the 2 kg necessary for the studies leading to the approval of the compound. In fact, the antineoplastic Taxotere™ is now produced by hemi-synthesis from the precursor 10-deacetyl-baccatine III, which is isolated from leaves of Taxus baccata. In other words, an alternative had to be found to the initial procedure taking a too heavy toll on trees of the initial species. Another interesting figure dealing this time with herbals is the annual demand of ginseng roots around 50,000 tons (Mathur et al., 1999). This kind of figures embodies a major limitation to new drugs originating from NP that is sometimes disregarded by research laboratories: there is a real challenge for the use of NP in reaching a tonnage compatible with the drug market.
Another reason for which NP based projects might be challenged for consideration by the industry is the small amount in which natural compounds generally accumulate in plant tissues. Indeed, structural characterization as well as biological testing can be made more difficult. Nevertheless, many progresses have been done in this area: in relation with structural characterization, for example, considering the growing sensitivity of the analytical methods and instruments, it will not be long until the barrier of the microgram for the determination of the structure of a compound would be reached. The analytical techniques for structural elucidation of an unknown compound are based on a mixture of spectroscopy (infrared, UV, Raman), mass spectrometry, and nuclear magnetic resonance (proton and C13 NMR). Nice examples of such task are described in various publications (Baker et al., 1990; El-Elimat et al., 2013) while difficulties are also discussed by Amagata (2010). Advances in NMR techniques were reviewed (Breton and Reynolds, 2013; Harvey et al., 2015; Gomes et al., 2018), and several examples of compounds for which structures were solved with mass of pure compounds lower than 100 µg were reported. In fact, the situation could be different from one plant to another or from one organ/tissue to another, even in a single plant species. Therefore, even if we can identify the many compounds in each sample while being able to establish their structure, it would be more straightforward to consider the feasibility of the systematic compendium of all the plant chemical components.
A careful understanding of the metabolic pathways of the tissues of the plant species from which the product has been isolated is also necessary. It is especially the case when hemi-synthesis is to be used to perform the NP’s industrial production. Instead of using a vital organ of the plant in which the compound is in fair concentration, one tries to find a precursor of the product in a renewable part of the plant, like the leaves. For the hemi-synthesis of Taxol™, 30 km2 of T. brevifolia fields in the Yunan province (China) consists in the main source of precursor for (http://www.yewcare.com/index.php cited in Malik et al., 2011). Certainly, the use of a renewable source of the natural product is desirable, and often enough that such sustainable and profitable solutions are found, as proven by the 40% of the available drugs in the Pharmacopeia that are from natural origin (Cragg et al., 1997; Newman et al., 2003; Newman and Cragg, 2007; Da Silva and Meijer, 2012; Newman and Cragg, 2012; Newman and Cragg, 2016). Many examples of drugs from natural origin exist, one being found in the still common use of plant sapogenins, alkaloids, or sterols for the production of steroids for human drugs (sex hormones, corticosteroids, contraceptive drugs, etc.). Other examples involving hemi-syntheses modifying the NP can be given: quinine (at least 100 tons per year), camptothecin (Asano et al., 2009), cocaine, camphor, vitamin B12, etc. Interestingly, at the other end of the spectrum, extremely simple molecules also of NP origin like salicylic acid for pain (J. B. Jin et al., 2017) or metformin for diabetes (Bailey, 2017) see their production reach industrial scales by total syntheses using standard organic chemistry.
The Need for Easy Access to Chemically Diverse Compounds
Testing as many compounds as possible with chemically diverse structures is important in order to have better chances to discover new drugs (Firn and Jones, 2003). Indeed, the affinity of a drug to a target is the result of shape and electrostatic potential complementarity between the drug and the binding site (Bauer and Mackey, 2019) as well as binding kinetic related properties such as desolvation and conformation changes upon binding. Ligand flexibility therefore plays an important role in identifying partially fitting chemicals as starting points further optimized by medicinal chemistry.
Screened compounds need to be diverse in shape and electrostatics to match any of the 3,300+ binding site, as listed in the pocketome (Kufareva et al., 2012), as well as diverse in structure to allow thorough optimization. It is noteworthy that the probability of identifying a hit considerably decreases with the increasing complexity of the ligand (Hann et al., 2001). The complexity of a molecule increases with the size and the atom connectivity. As a result, the more complex they are, the greater the number of molecules one should screen. Statistical analyses have been performed on natural products to study their chemical diversity (Firn and Jones, 2003; Hong, 2011), molecular properties (Feher and Schmidt, 2003; Quinn et al., 2008), scaffold diversity (Lee and Schneider, 2001; Grabowski et al., 2008; Yongye et al., 2012), and coverage of the chemical space (Rosén et al., 2009a). Comparisons between NP and other compound collections (Henkel et al., 1999; Lee and Schneider, 2001; Grabowski and Schneider, 2007; Rosén et al., 2009a) have also been performed, showing that NP differ from drugs and synthetic compounds in several aspects. NP are considered complex due to their number of asymmetric centers, their number of Sp3 carbon ratio in rings, and their number of ring junctions. Indeed, NP display in general more chiral centers than drugs, although their number of chiral centers to number to carbon atoms ratio may be lower (Gu et al., 2013; Skinnider et al., 2017). Although there is no clear evidence that this complexity is necessary for their biological activity (Firn and Jones, 2003), it greatly impacts their specificity (Clemons et al., 2011). NP also display a great diversity in their scaffolds (Lee and Schneider, 2001; Grabowski et al., 2008; Yongye et al., 2012). For instance, the GreenPharma database (Do et al., 2015; Gally et al., 2017) contains 55,185 Murcko frameworks (Bemis and Murcko, 1996) for 302,000 natural products (18.3%), whereas NCI and ChEMBL databases contain a little of less NP-derived scaffolds: 13.1 and 13.6% respectively.
An interesting representation of the divergent diversity between bioactive NP and synthetic compounds can be seen in Figure 1 . This graph reported by Rosén et al. (2009a) shows, on a small sample of compounds (126,140 natural compounds versus 178,210 medicinal chemistry-issued compounds), the difference in repartition between these two populations after a principal component analysis. NP are also considered biologically diverse compounds that can hit a vast diversity of biological targets (Hong, 2011). Most of them have a single known target, with a mean of 2.66 targets and only few highly promiscuous compounds. However, biological activity is reported for only 2% of the NP (Gu et al., 2013). An extended study using docking experiments of NP in 332 targets showed a mean of 2.14 targets per natural products, while in comparison, drugs interact in average with 3–6 targets, and 50% of all drugs might exhibit activity against more than five targets (Mestres et al., 2008; Jalencas and Mestres, 2013; Hu et al., 2014).
Figure 1.
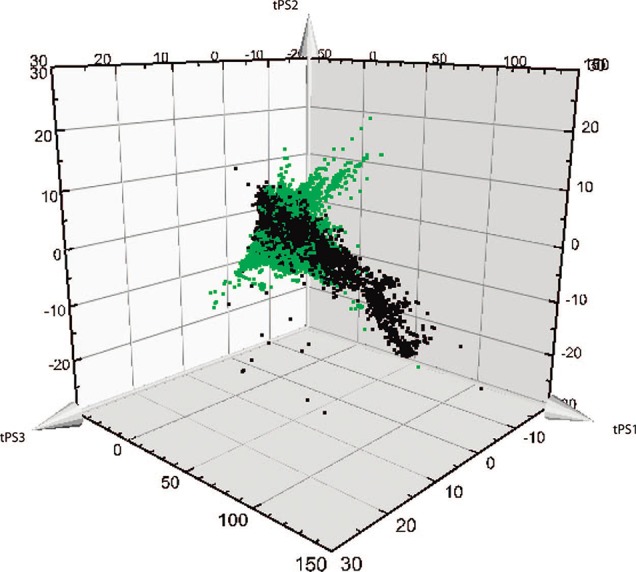
Representation of the divergent diversity between natural and synthetic compounds. (adapted with permission from Rosén et al. (2009a). Copyright 2009 American Chemical Society). Biologically relevant chemical space covered by natural products (in green) or by bioactive medicinal chemistry compounds from the database WOMBAT (in black) in the first three principal components.
For all these reasons, there is a strong need for an easy and organized access to chemically diverse structures in the form of libraries of compounds. It is necessary to keep a good balance between diversity, defined as the mean of compound’s pairwise dissimilarity and structural redundancy to ensure greater diversity of hits at an overall hit rate in the range of 0.5 to 2% and 20–40% within an active chemical series. In addition, these compound libraries are required to be constantly enriched with compounds filling the gaps in the molecular space. Interestingly, Harvey et al. (2015) stated: “diversity within biologically relevant chemical space is more important than library size”, this biologically relevant chemical space being defined by protein-binding sites for potential ligands. Another point emphasized by these authors is that while the commercially available chemical space is wider than the explored natural product universe—the first one is evaluated to 5.109 compounds, while the second only represents 3.105 compounds (Banerjee et al., 2015)—NP are intrinsically more diverse. A major component of this comparison is that NP are more complex, from a chemical point of view, leading to greater shape diversity. However, NP might not be the best compounds to screen in HTS campaigns due to their complexity (“best” in a drug-discovery perspective). Indeed, their probability of being active on a random target is lower, and their chemical tractability is far from optimal. Although NP decently occupy the biologically relevant chemical space, and 80% meet the criteria to be considered as drug like compounds (Harvey et al., 2015), their complexity might be the bottleneck during the optimization phase of the lead compounds.
Thus, acquiring as many diverse compounds as possible is necessary in drug discovery programs, and the exploration of natural existing chemical diversity would be a must. Nevertheless, not only the design of those libraries, but also the design of the assays and the understanding of the pathways that are responsible for the targeted pathologies need to be relevant and well understood. In summary, the main idea is that if compounds as chemically diverse as possible are necessary to feed drug discovery programs, then a great deal of efforts should still be invested in the exploration of natural chemical diversity because, in this particular context, natural chemistry is more advanced than organic chemistry. After this introduction on the background of NP in the area of drug discovery, we aim at discussing and showing the extent of natural chemical diversity (Natural Chemical Diversity) as well as the strategies of having the potential to embrace this diversity (Different Strategies to Benefit From This Diversity) based on an extensive survey of recent publications. A preliminary survey on the main origins of recently reported natural compound structures’ literature was performed by taking a complete volume from a journal that is considered as a gold standard in NP (J. Natural Products, 2018, vol 81). Then three different tables mentioning the species of origin and the type of compounds newly reported in 2017 and 2018 in several journals selected for their specialization in the area (like for example Journal of natural products, Planta medica, Tetrahedron Letters or Natural products communications) were built upon this sampling. Finally, Different Strategies to Benefit From This Diversity offers some strategic and technical proposals based on our interpretations of the trends of the field supported by recent ad hoc literature.
Natural Chemical Diversity
The chemical diversity of NP is known to be extremely wide, and it can be divided in several types representing a challenge for the chemist and the biologist.
Diversity of Origins in Natural Compounds
Diversity can be measured and quantified by many methods some of which have been used for decades to qualify the product library diversity (Langer et al., 2009). Our first survey showed the main origins of the molecules reported as natural compounds in 2018: they were classified between those coming from animals (2.6%), from fungi and lichens (9.3%), from microorganisms (12.9%), from marine organisms in the broadest sense of the term (13.2%), and from plants (44.1%), while the papers dealing with partial or total syntheses of natural compounds (19) and about 20 others describing various methodologies of analyses were put aside. As shown, in 2018 plants were still the main source of “new” natural compounds.
Then, by sampling the recent ad hoc literature, three different tables were constructed gathering the origin of new compounds reported in 2017 and 2018 in several journals selected for their specialization in the area. Although plants were our main focus, we thought it might be useful to also consider other sources of diversity from other living kingdoms as they also consist of very interesting sources to explore and because some of them are somehow related to plants: it is especially the case for compounds which functions in plants are related with defense (Bednarek and Osbourn, 2009) and other ecological interactions as compounds “optimized by evolution” according to Gunatilaka in its review on natural products from plant-associated microorganisms (Gunatilaka, 2006). Among the 240 papers that were reviewed, a handful described compounds isolated from venoms and toxins originating from marine organisms and animals (insects, snakes, and alike). Then, a few other papers described compounds coming from microorganisms as can be seen in Table 2 . Microorganisms, such as bacteria and yeasts, were traditionally important sources of antibiotics and are still a source of new peptides (Xue et al., 2018), but here compounds other than peptides can also be found that are close to secondary metabolites naturally synthesized in those cells. New peptides are more often described in journals specialized in peptide chemistry and pharmacology, even if their source is a living organism. Table 3 gives examples of compounds isolated from lichen, fungi, and sponges. The chemical diversity of these compounds is interesting but, as of today, growing lichen or sponges is not so much reported and might turn out to be difficult on a larger scale. On the other hand, secondary metabolites synthetized by microorganisms growing in extreme conditions might also be difficult to obtain in large quantities. It would be therefore interesting to dig deeper in this area, as it has been done elsewhere for other purposes: biotechnological (Kruger et al., 2018; Mokashe et al., 2018), industrial (Sarmiento et al., 2015), pharmaceutical (Irwin, 2010; Karker et al., 2016; Patel, 2016; Oliveira et al., 2018), or ecological (Casillo et al., 2018; Orellana et al., 2018). Finally, Table 4 gives many examples of compounds isolated from different plant species and different tissues from these species.
Table 2.
Some examples of compounds isolated from microorganisms.
| Species | Compound | Reference |
|---|---|---|
| Hypoxylon. Monticulosum | Sporothriolide derivatives | (Leman-Loubiere et al., 2017) |
| Malbranchea flavorosea | X (*) | (Verastegui-Omana et al., 2017) |
| Micromonospora. Harpali | Spirotetronate | (Gui et al., 2017) |
| Moorea. Producens | Cryptomaldamide | (Kinnel et al., 2017) |
| Okeania sp. | Kohamamides | (Iwasaki et al., 2017) |
| Paecilomyces sp. | β-Resorcylic Acid Lactones | (Xu L. et al., 2017) |
| Palicourea sessilis | Monoterpene indole alkaloids | (Klein-Junior et al., 2017) |
| Penicillium commune | Cyclopiane diterpenes | (Niu et al., 2018) |
| Plasmodium falciparum | Octaminomycins | (Jang et al., 2017) |
| Pseudonocardia carboxydivorans | Branimycins | (Brana et al., 2017) |
| Pseudovibrio denitrificans | Alkaloids | (Nicacio et al., 2017) |
| Rhytismataceae sp. | X (*) | (McMullin et al., 2017) |
| Serratia plymuthica | Plymuthipyranones | (Bjerketorp et al., 2017) |
| Streptomyces misionensis | Streptenols | (Tarazona et al., 2017) |
| Streptomyces sp. | Spoxazomicin D | (Shaaban et al., 2017) |
| Streptomyces sp. | Dibohemamines | (Jiang et al., 2017) |
| Symploca sp. | Caracolamide | (Naman et al., 2017) |
| Thalassospira profundimaris | Thalassosamide | (Zhang et al., 2017) |
| Thermoactinomyces. Vulgaris | Thermoactinoamide A | (Teta et al., 2017) |
| Trichoderma sp. | Neomacrophorin X | (Kusakabe et al., 2017) |
(*) those papers described many different compounds in those microorganisms.
Table 3.
Some examples of compounds isolated from fungi, sponges and lichens.
| Species | Compound(s) found in Fungus, Sponge, Lichen | Reference |
|---|---|---|
| Acanthostrongylophora sp. | Manzamine Alkaloids | (Kim et al., 2017) |
| Amphimedon sp. | Zamamidine D | (Kubota et al., 2017) |
| Annulohypoxylon truncatum | Isochromans | (Li et al., 2017f) |
| Antrodia cinnamomea | Antroquinonol derivatives | (Chen et al., 2017) |
| Aspergillus ochraceopetaliformis | Ochracenes | (Wang et al., 2017b) |
| Aspergillus sp. | Protulactone A | (Markovic et al., 2017) |
| Auxarthron pseudauxarthron | Phenalenones | (Li et al., 2017) |
| Botrysphaeria laricina | Botrysphones | (Zhang et al., 2017) |
| Chondrostereum sp. | Hirsutane Sesquiterpenes | (Hsiao et al., 2017) |
| Clathria gombawuiensis | Gombasterols | (Woo et al., 2017) |
| Cochliobolus australiensis | Chloromonilinic Acids | (Masi et al., 2017) |
| Delitschia. Confertaspora | Fimetarone | (Jayanetti et al., 2017) |
| Diaporthe toxica | Phomopsin A | (Schloss et al., 2017) |
| Emericella sp. | Emervaridones | (He et al., 2017) |
| Ganoderma applanatum | Spiroapplanatumines | (Luo et al., 2017) |
| Glomerella cingulata | Sesquiterpenes | (Liu et al., 2017a) |
| Hyrtios sp. | Meroterpenoids | (Wang et al., 2017c) |
| Ircinia oros | Furanosesterterpenoids | (Chianese et al., 2017) |
| Lamellodysidea herbácea | Lamellodysidines | (Torii et al., 2017) |
| Lobaria orientalis | Lobarientalones | (Nguyen et al., 2017) |
| Monanchora unguiculata | Unguiculin A | (Campos et al., 2017) |
| Montagnulaceae sp. | Montagnuphilones | (Luo et al., 2017) |
| Oscarella stillans | Oscarellin | (Kwon et al., 2017) |
| Paecilomyces gunnii | Gunnilactams | (Zheng et al., 2017) |
| Paraconiothyrium sp. | Versiol derivatives | (Cho et al., 2017) |
| Penicillium concentricum | Halogenated compounds | (Ali et al., 2017) |
| Penicillium janthinellum | Penicilones | (Chen et al., 2017) |
| Penicillium janthinellum | Penisulfuranols | (Zhu et al., 2017) |
| Phaeoacremonium sp. | Isoaigialones | (Silva et al., 2017) |
| Preussia similis | Preussilides | (Noumeur et al., 2017) |
| Preussia. Minimoides | Minimoidiones | (Rangel-Grimaldo et al., 2017) |
| Psammocinia sp. | Sulawesins | (Afifi et al., 2017) |
| Spongia ceylonensis | Ceylonins | (El-Desoky et al., 2017) |
| Spongia pertusa | Sesquiterpene quinones | (Li et al., 2017a) |
| Stachybiotrys chartarum | Stachybotrysins | (Zhao et al., 2017) |
| Stereocaulon paschale | Dibenzofuranes | (Carpentier et al., 2017) |
| Talaromyces aculeatus | Azaphilone derivatives | (Ren et al., 2017) |
| Talaromyces islandicus | Hydroanthraquinones | (Li et al., 2017) |
| Talaromyces sp. | Talarazines | (Kalansuriya et al., 2017) |
| Theonella swinhoei | Cyclotheonellazoles | (Issac et al., 2017) |
| Topsentia sp. | Tulongicin | (Liu et al., 2017) |
| Trichoderma gamsii | Trichoderpyorone | (Chen et al., 2017) |
| Xylaria nigripes | X (*) | (Chang et al., 2017) |
Compounds originating from fungus in black, from sponge in red, and from lichen in green; (*) this paper described many different compounds in this fungus.
Table 4.
Compounds from plant parts.
| Species | Plant leaf | Whole aerial plant and stems | Plant rhizomes and roots | Plant fruit and flowers | Plant bark, wood and seeds | Ref |
|---|---|---|---|---|---|---|
| Acacia ligulata | Ligulatasides | (Jaeger et al., 2017) | ||||
| Acorus tatarinowii | Asarone derivatives | (Gao et al., 2017) | ||||
| Aconitum apetalum | Apetaldines | (Zhang et al., 2017) | ||||
| Allanblackia floribunda | Xanthones | (Mountessou et al., 2018) | ||||
| Alnus viridis | Hydroxyalphitolic acid derivatives | (Novakovic et al., 2017) | ||||
| Althaea officinalis | X (*) | (Sendker et al., 2017) | ||||
| Amorpha fruticosa | Amorphispironones | (Muharini et al., 2017) | ||||
| Anabasis articulata | nor-Triterpenoidal Saponins | (Salah El Dine et al., 2019) | ||||
| Ancistrocladus ileboensis | Jozilebomines | (Li et al., 2017c) | ||||
| Ancistrocladus ileboensis | Dioncophyllines | (Li et al., 2017d) | ||||
| Anigozanthos sp. | X (*) | (Hendra and Keller, 2017) | ||||
| Anthemis nobilis | Sesquiterpene lactones | (Mieri et al., 2017) | ||||
| Aphanamixis polystachya | Limonoids | (Camero et al., 2018) | ||||
| Aquilaria malaccensis | Aquilanols | (Ma et al., 2017) | ||||
| Aquilaria malaccensis | Phorbol esters | (Wagh et al., 2017) | ||||
| Aquilaria sinensis | chromen-4-one | (Wang S.-L. et al., 2018) | ||||
| Aristolochia orbicularis | Aristoloxazines | (Rios et al., 2017) | ||||
| Artocarpus rigida | Artocarmins | (Nguyen et al., 2017) | ||||
| Atalantia monophylla | Atalantums | (Sribuhom et al., 2017) | ||||
| Azadirachta indica | Tamarixetin glucopyranoside | (Yadav et al., 2017) | ||||
| Baeckea frutescens | Phloroglucinol meroterpenoids | Phloroglucinol meroterpenoids | (Qin et al., 2018) | |||
| Baeckea frutescens | Frutescones | (Hou et al., 2017) | ||||
| Balthasaria mannii | Pyran-2-ones derivatives | (Ahoua et al., 2018) | ||||
| Belamcanda chinensis | Belamcandanes | (Ahoua et al., 2018; Ni et al., 2018) | ||||
| Berchemia berchemiifolia | Berchemiosides | (Kang et al., 2017b) | ||||
| Betula pendula | Betulin derivatives | (Tsepaeva et al., 2017) | ||||
| Betula pubescens | Suberin fatty acids | (Heinamaki et al., 2017) | ||||
| Boesenbergia pandurata | Cyclohexene chalcones | (Nguyen et al., 2017b) | ||||
| Bougainvillea spectabilis | Bougainvinones | (Do et al., 2018) | ||||
| Bowdichia virgilioides | Sucupiranines | (Endo et al., 2017) | ||||
| Buddleja asiatica | Iridoid glycosides | (Hien et al., 2018) | ||||
| Bupleurum fruticosum | Phenylpropenoids | (Fois et al., 2017) | ||||
| Calotropis gigantea | Uscharin | (Yoneyama et al., 2017) | ||||
| Camellia crapnelliana | Camellianols | Camellianols | (Xiong et al., 2017) | |||
| Carpesium cernuum | Carpescernolides | (Yan et al., 2018)) | ||||
| Caryopteris Nepetaefolia | Nepetaefolins | (Zhang et al., 2017) | ||||
| Catalpa ovata | Catalpol derivatives | (Kil et al., 2017) | ||||
| Celastrus subspicata | Celastrofurans | (Wibowo et al., 2017) | ||||
| Cephalotaxus fortunei | Cephalotaxus troponoids | (Zhao et al., 2017) | ||||
| Cephalotaxus sinensis | Cephanolides | Cephanolides | (Fan et al., 2017) | |||
| Ceratodon purpureus | Biflavonoids | (Waterman et al., 2017) | ||||
| Chaenomeles sinensis | Sinenic acid A analogues | (Kim et al., 2017b) | ||||
| Chrysanthemum morifolium | Caffeoylquinic acid | (Yang et al., 2017) | ||||
| Chrysanthemum indicum | Chrysanthemumins | (Liu et al., 2017) | ||||
| Cimicifuga dahurica | Cimiricaside B | (Thao et al., 2017) | ||||
| Cinnamomum cassia | Caffeic acid phenethyl ester | (Shin et al., 2017) | ||||
| Clausena anisumolens | Anisucoumaramide | (Wang et al., 2017) | ||||
| Cleistochlamys kirkii | Cleistodienol derivatives | (Nyandoro et al., 2017a) | ||||
| Codonopsis pilosula | Piluloside | (Zheng et al., 2018) | ||||
| Coleonema album | Coumarins | (Lima et al., 2017) | ||||
| Cornus officinalis | Cornusides | (Ye et al., 2017) | ||||
| Curcuma aromatica | Sesquiterpenoids | (Dong et al., 2017) | ||||
| Curcuma longa | Cyclocurcumin | (Kim et al., 2017) | ||||
| Cyclopia intermedia | Polyphenols | (Jack et al., 2018) | ||||
| Cynomorium songaricum | X (*) | (Ma et al., 2018) | ||||
| Dasymaschalon echinatum | Aristolactams | (Bunteang et al., 2018) | ||||
| Eremophila longifolia | Nerol cinnamates | (Galappathie et al., 2017; Nyandoro et al., 2017b)) | ||||
| Erythrina schliebenii | Isoflavones | (Nyandoro et al., 2017b) | ||||
| Euphorbia fischeriana | ent-Abietane derivatives | (Wang et al., 2017) | ||||
| Euphorbia gaditana | Gaditanone | (Flores-Giubi et al., 2017) | ||||
| Euphorbia kansui | Ingenane Diterpenoids | (Zhang et al., 2018) | ||||
| Euphorbia pithyusa | Premyrsinane | (Esposito et al., 2017) | ||||
| Euphorbia semiperfoliata | Jatrophane esters | (Nothias et al., 2017) | ||||
| Euphorbia soongarica | Sooneuphoramine | (Gao and Aisa, 2017) | ||||
| Euphorbia taurinensis | Diterpenes | (Rédei et al., 2018) | ||||
| Euphorbia ebracteolata | Abietane derivatives | (Wei et al., 2017) | ||||
| Excoecaria agallocha | nor-Oleanane triterpenes | (Mo et al., 2018) | ||||
| Ficus fistulosa | Phenanthroindolizidine Alkaloid | (Al-Khdhairawi et al., 2017) | ||||
| Fissistigma latifolium | Ecarlottones | (Geny et al., 2017) | ||||
| Flindersia pimenteliana | Pimentelamines | (Robertson et al., 2017) | ||||
| Forsythia suspensa | X (*) | (Kuo et al., 2017) | ||||
| Garcinia propinqua | Xanthones | (Sriyatep et al., 2017) | ||||
| Gardenia ternifolia | Gardenifolins | (Tshitenge et al., 2017) | ||||
| Gloriosa superba | Gloriodiside | (Zarev et al., 2017) | ||||
| Glycyrrhiza glabra | Glycybridins | (Li et al., 2017) | ||||
| Hoya kerrii | Epoxypregnane | (Seeka et al., 2017) | ||||
| Humulus japonicus | Humulusides | (Yang et al., 2018) | ||||
| Humulus lupulus | Xanthohumol | (Liu et al., 2017) | ||||
| Humulus lupulus | α-acid derivatives | (Li et al., 2017b) | ||||
| Hunteria zeylanica | Hunterizeylines | (Bao et al., 2017) | ||||
| Hypericum henryi | Hyperhenones | (Duan et al., 2018) | ||||
| Hypericum perforatum | Acylphloglucinols | (Guo et al., 2017) | ||||
| Hyptis brevipes | Brevipolides | (Suarez-Ortiz et al., 2017) | ||||
| Impatiens balsamina | Balsamisides | (Kim et al., 2017a) | ||||
| Indigofera stachyodes | Stachyodin A | (Zhang et al., 2018) | ||||
| Iris tectorum | Polycycloiridals | (Zhang et al., 2017) | ||||
| Isodon pharicus | Pharicins | (Hu et al., 2018) | ||||
| Isodon scoparius | ent-kaurane derivatives | (Jiang et al., 2017) | ||||
| Jatropha dioica | Riolozane derivatives | (Melchor-Martinez et al., 2017) | ||||
| Juglans regia | Azacyclo-indoles | (Li et al., 2017) | ||||
| Justicia gendarussa | Patentiflorin A | Patentiflorin A | (Zhang et al., 2017a) | |||
| Kopsia officinalis | Rhazinilam | (Zeng et al., 2017) | ||||
| Laetia corymbulosa | Corymbulosins | (Suzuki et al., 2017) | ||||
| Lepidozia reptans | Terpenoids | (Li S. et al., 2018) | ||||
| Leplaea mayombensis | Leplaeric acid derivatives | (Sidjui et al., 2017) | ||||
| Ligularia fischeri | Diterpenoids | (Gobu et al., 2017) | ||||
| Liquidambar formosana | Isorugosins | (Shimozu et al., 2017) | ||||
| Lithocarpus litseifolius | Triterpenoids | Triterpenoids | (Cheng et al., 2018) | |||
| Litsea cubeba | Glycosides | (Wang et al., 2017) | ||||
| Litsea cubeba | Isoquinoline alkaloids | Isoquinoline alkaloids | (Yang et al., 2018) | |||
| Macaranga tanarius | Schweinfurthins | (Peresse et al., 2017) | ||||
| Mammea harmandii | Xanthones | (Liangsakul et al., 2018) | ||||
| Millettia oblata | Rotenoids | (Deyou et al., 2017) | ||||
| Momordica balsamina | Triterpenoids | (Ramalhete et al., 2018) | ||||
| Momordica charantia | Cucurbitanes | (Tuan et al., 2017) | ||||
| Nauclea orientalis | Indole alkaloids | (Surapinit et al., 2018) | ||||
| Ongokea gore | Hydroxy diynes | (Ntumba et al., 2018) | ||||
| Panax ginseng | Malonylginsenosides | (Qiu et al., 2017) | ||||
| Paramignya trimera | Quinoliniumolate | (Nguyen et al., 2017a) | ||||
| Paulownia tomentosa | Tomentins | (Ryu et al., 2017) | ||||
| Peganum harmala | Peganine derivatives | (Wang et al., 2017) | ||||
| Pentalinon andrieuxii | Pentalinonsterol | (Oghumu et al., 2017) | ||||
| Pentarhizidium orientale | Matteuorienates | (Huh et al., 2017) | ||||
| Peperomia obtusifolia | Orsellinic acid derivatives | Orsellinic acid derivatives | Orsellinic acid derivatives | (Batista et al., 2017) | ||
| Perovskia abrotanoides | Diterpenes | (Tabefam et al., 2018) | ||||
| Peucedanum japonicum | Khellactone Esters | ((Hong and Kim, 2017) | ||||
| Phoradendron vernicosum | Lupane derivatives | (Valencia-Chan et al., 2017) | ||||
| Phyllanthus acidus | t-Muurolol | (Pérez-Colmenares et al., 2018) | ||||
| Phyllanthus flexuosus | Purine derivative | (Zhu et al., 2018) | ||||
| Physalis peruviana | Whithanolides | (Xu Y.-M et al., 2017) | ||||
| Plectranthus africanus | Abietane-derivatives | (Nzogong et al., 2018) | ||||
| Podocarpus nagi | Nagilactone derivatives | (Feng et al., 2017) | ||||
| Polygala flavescens | X (*) | (Leo et al., 2017) | ||||
| Pongamia pinnata | Furanoflavones | (Sharma et al., 2018) | ||||
| Poupartia borbonica | Poupartones | (Ledoux et al., 2017)) | ||||
| Prangos haussknechtii | Prenylated coumarins | (Dissanayake et al., 2017) | ||||
| Pulicaria undulata | Asteriscunolides | (Boumaraf et al., 2017) | ||||
| Radula sumatrana | Rasumatranins | (Wang et al., 2017) | ||||
| Raphanus sativus | Monoglyceride | (Ogihara et al., 2017) | ||||
| Rhodocodon campanulatus | Bufadienolides | (Schwikkard et al., 2017) | ||||
| Rhodomyrtus tomentosa | Tomentodiones | (Zhang et al., 2017) | ||||
| Salvia chamaedryoides | Diterpenoids | (Bisio et al., 2017) | ||||
| Salvia circinata | Neoclerodane glucosides | (Flores-Bocanegra et al., 2017) | ||||
| Salvia miltiorrhiza | Salvianans | Salvianans | (Zhang et al., 2017) | |||
| Salvia plebeia | epi-Eudebeiolides | (Jang et al., 2017) | ||||
| Salvia polystachya | Polystachynes | (Bautista et al., 2017) | ||||
| Sambucus williamsii | Iridoid glycosides | (Suh et al., 2017) | ||||
| Saxifraga spinulosa | Galloyl glycosides | (Badral et al., 2017) | ||||
| Schisandra bicolor | Shibitubins | (Liu et al., 2017b) | ||||
| Scutellaria barbata | neo-Clerosane diterpenoids | (Yang et al., 2018) | ||||
| Selaginella pulvinata | Diselaginellin | (Cao et al., 2017) | ||||
| Siegesbeckia pubescens | ent-Strobane | (Wang et al., 2017a) | ||||
| Sinningia reitzii | Dunnione derivatives | (Soares et al., 2017) | ||||
| Siraitia grosvenorii | Cucurbitane | (Niu et al., 2017) | ||||
| Streblus asper | Kamaloside derivatives | (Ren et al., 2017) | ||||
| Strychnos icaja | Strychnogucine B | (Beaufay et al., 2018) | ||||
| Tabernaemontana bufalina | Bisendol alkaloids | (Zhou et al., 2018) | ||||
| Taxus wallichiana | Wallitaxanes | (Dang et al., 2017a) | ||||
| Taxus wallichiana | Lignans | (Dang et al., 2017b) | ||||
| Teucrium yemense | Neoclerodane Diterpenoids | (Nur-E-Alam et al., 2017) | ||||
| Thalictrum cultratum | Aporphinoid alkaloids | (Li et al., 2017) | ||||
| Tinospora sagittata | Bistinospinosides | (Li et al., 2017e) | ||||
| Tinospora sinensis | Tinosinenosides | (Jiang et al., 2017) | ||||
| Trichospira verticillata | Sesquiterpenoid Lactones | (Du et al., 2017) | ||||
| Typhonium giganteum | Cerebrosides | (Jin et al., 2017) | ||||
| Uncaria rhynchophylla | Dimeric isoechinulin | (Geng et al., 2017) | ||||
| Uvaria alba | Albanols | (Macabeo et al., 2017) | ||||
| Vaccinium ashei | Anthocyanins | (Huang et al., 2018)) | ||||
| Vallaris glabra | Cardenolide glycosides | (Kruakaew et al., 2017) | ||||
| Verbesina lanata | Eudesmane sesquiterpenes | (Ramseyer et al., 2017) | ||||
| Vernonia cinerea | Hirsutinolide analogues | (Kuo et al., 2018) | ||||
| Vitex trifolia | Vitepyrroloids | (Luo et al., 2017) | ||||
| Xylocarpus granatum | Xylomexicanins | (Wu et al., 2017) | ||||
| Xylocarpus rumphii | Xylorumphiins | (Waratchareeyakul et al., 2017) | ||||
| Zephyranthes carinata | Plicamine derivatives | (Zhan et al., 2017) | ||||
| Zizyphus jujuba | Epicatechinoceanothic acids | (Kang et al., 2017a) |
Compound names in bold characters are those that are exemplified in Figure 1 . Compounds in green were extracted from stems or seeds; in red from roots; in blue from flowers or wood; and in black from leaves, hole aerial part, rhizomes, fruit or bark; (*) those papers described many different compounds in those plant parts. Lines overlaid in gray exemplify similar genuses expressing different compounds.
As plants are one of the major sources of “new” compounds, the following sections will deal with their diversity.
Diversity of Plant Species
As mentioned, the compounds reported from plants ( Table 4 of our survey) originated from 165 different plant species, from trees to ornamental plants and harvested in very diverse areas from Australia to Antarctica.
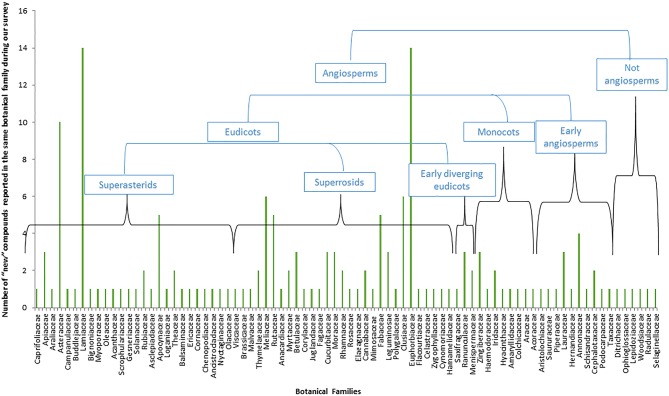
In fact, a total of 146 different genera were represented from 82 different botanical families, only six of which are not Angiospems (two ferns, two liverworts, a moss, and a spikemoss). Most of them, around 93%, are thus from the flowering plant group ( Figure 2 ). Furthermore, it is interesting to observe that the remaining 76 families from which these new compounds were reported represent a small part of the total 416 botanical families defined in the last 2016 update of the Angiosperms Phylogeny Group (The Angiosperm Phylogeny Group et al., 2016). As illustrated, most of the new compounds of our survey were isolated from the superasterids and superrosids, major clades from the eudicots. Indeed, this small survey illustrates how a large proportion of plant species still remains underexplored, not only in the Angiosperms but between all taxa of Plant kingdom.
Figure 2.
Representation of the relative weight of the different phylogenetic groups in the number of “new” compounds reported based on our survey ( Table 4 ).
Furthermore, it is important to consider that new species of plant are discovered quite often: for example, 2,034 new plant species were recorded in 2014, including at least one tree species (Mancuso, 2018). So far, 310,000 plant species have already been described, among which authors estimate that only 6% have been investigated pharmacologically and 15% phytochemically (Atanasov et al., 2015). It clearly indicates that even more compounds remain to be found in plants.
It is also interesting to observe that reviews such as Solyomváry et al. (2017) teach us that plants from different taxa can also synthesize identical although rather complex compounds.
Gene duplication and neo-functionalization leading to the extension of the existing metabolic pathways are both part of the mechanisms that have been identified in plants as responsible for diversification of secondary metabolites together with the influence of ecological factors: for example, it has been suggested that, from a small group of precursors, plants would synthetize a full range of highly diverse compounds rapidly changing that are “screened” for their biological activity afterwards as mechanisms of adaptation that would help plants cope better with biotic and abiotic pressures (Moore et al., 2014). This ecological understanding of plant secondary metabolite diversification also contributes to the anticipation that the more plants species are described, the more diversity is to be found in the end-products of these pathways along with possibly valuable “new” compounds. A recent interesting study (Henz Ryen and Backlund, 2019) discussed the chemical Angiosperms’ diversity among natural products and some of these evolutionary mechanisms leading to diversification of chemical structures in function of the group of secondary metabolites (flavonoids, tropane alkaloids, sesquiterpene lactones, and betalains). They use the ChemGPS-NP developed previously by their group (Rosén et al., 2009b) as tool to localize compounds in the chemical property space, “measuring” this way the chemical diversity.
In other words, the preservation of ecological niches and plant biodiversity will also serve our interest in terms of chemical diversity for possible applications. And, interestingly, at higher trophic levels, it will also contribute to biodiversity (Schuman et al., 2016), creating in turn other biotic pressures on plants that may adapt by synthetizing other compounds!
Chemical Diversity in Plants Related to Space and Time
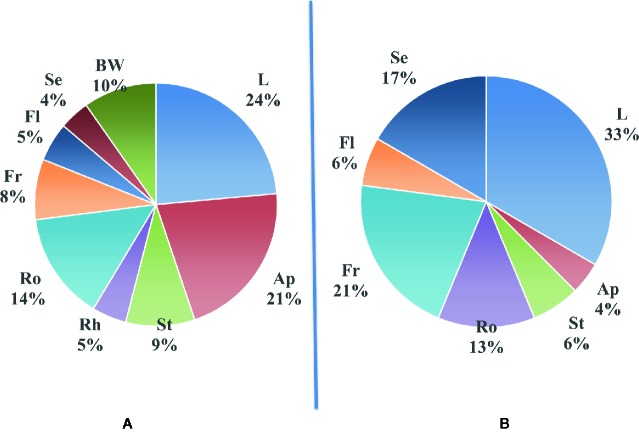
As plants are multicellular organisms with organs specialized for different functions, it seems logical to think that some biosynthetic pathways could be turned on or off depending on the part of the plant studied and that a certain level of diversity can exist within the plant tissues. The main parts of the plants that are classically separated are roots, twigs (stems), leaves, flowers, fruits, and seeds. Of course, constraints of collection make some of these parts more suitable than others. Our survey in Table 4 details for each one of the 165 reported plant species the plant part from which it was isolated. Figure 3A summarizes the proportion of new compounds reported in different plant parts in our survey. For comparison, Figure 3B shows similarly the organs/parts of origin of the active compounds reported from the 49 plants described in Ross work (Ross, 1999; Ross, 2001). It seems that in both cases, most of the studies are done on leaves. On the other hand, it is not clear, on a systematic basis, if what was found in the other organs can be found as well—even in small quantities—in the leaves. Nevertheless, leaves are the most accessible part of any plant, and they are renewable which allows preserving the whole plant. Obviously, it is also valid for fruits or flowers, but their availability may be subjected to seasonality.
Figure 3.
Distribution of newly reported compounds according to the part of plants from which they were extracted. (A): Proportion of new compounds reported in different parts from the plant (from our survey); (B): Proportion of active compounds in different plant parts reported in Ross work (Ross, 1999; Ross, 2001). Ap, aerial parts; BW, bark and wood; Fl, flowers; L, leaves; Fr, fruit; Rh, rhizome; Ro, roots; Se, seeds; St, stems.
Indeed, often enough, a compound is described from a part of a given plant. It seems interesting to emphasize that it does not necessarily mean that it is the main component or a major component of this plant part. It can be accumulated in superior amounts in other tissues of the same plant. Indeed, in plant descriptions such as those in Ross work (Ross, 1999; Ross, 2001), the author described the way plants are used in traditional medicine. The different parts of a given genus can be used alone or in mixtures in preparations ranging from infusion, maceration, decoction, juice, dried powder, or even fresh organs (fruits, leaves) ingested as such. Astonishingly, those recipes were collected from scattered geographic places. This last observation suggests that traditional medicines around the world independently found similar remedies to similar diseases and sometimes with the use of related plant species.
Another interesting source of intraspecific chemical diversity is the environment of the plant cells biosynthesizing the chemical compounds of interest: it is well known that compounds produced by the same plant species can vary in nature or quantity depending on the environment (localization or time of the year) or the part of the plant where it has been extracted from. Moore et al. (2014) pointed out in particular the plant ontogeny, but also genetic and environmental variations as major sources of diversity for plant secondary metabolites. The existence of chemotypes is another example of this intraspecific chemo-diversity well described for example in plants producing essential oils. Factors like moisture, salinity, temperature, or nutrition levels are known to influence the essential oil production (Sangwan et al., 2001), and the genotype could also significantly influence the chemotype as it was shown recently for Valeriana jatamansi Jones (He et al., 2018). The biosynthesis of natural products can also differ in function of the different individuals from the same population. It is the case, for example, when these compounds are related to antimicrobial activity as summarized by Bednarek and Osbourn (2009) in their perspective article on chemical diversity linked with plant defense: these compounds can be synthetized constitutively as part of normal plant development—and stored in specialized tissues—or synthetized in response to pathogenic challenges through the activation of the transcription of specific genes of the corresponding biosynthetic pathways.
Diversity of Chemical Skeletons and Structures
Based on the number of genes, it has been estimated that the plant kingdom contains more than 200,000 different metabolites with values for single species ranging between 5,000 and 15,000 (Trethewey, 2004; Fernie, 2007), values that are significantly greater than those of microorganisms (∼1,500) and animals (∼2,500) (Oksman-Caldentey and Saito, 2005). But it is not only the global absolute value of chemically diverse compounds that is interesting. Such diversity and such dynamicity are indeed a wonderful wealth of chemical structures, source of inspiration for medicinal chemist, once the structure is carefully identified, and the related pharmacological activity is screened. But it can also become a source of complexity for the phytochemist working on the structure or on the structure/activity relation.
Classically, within this chemical diversity in plant secondary metabolites, the nomenclature used by pharmacologists to attempt classifying several families of natural compounds such as polyketides, phenylpropanoids, terpenoids, steroids, or alkaloids is based more on their biogenesis and the pathway they originate from (acetate, shikimate, mevalonate or methylerythritol phosphate pathways) or their combination, than their structure itself. And even within a defined group, the diversity can be impressive: for example, the terpenoid family is suspected to contain at least 50,000 different molecules (Kirby and Keasling, 2009) while at least 12,000 flavonoids have been described (Henz Ryen and Backlund, 2019). Certainly, how “different” these molecules are could be further commented, but as discussed below, apparently minor differences (for example a methyl or a hydroxyl moiety) might dramatically change the molecule’s pharmacological properties.
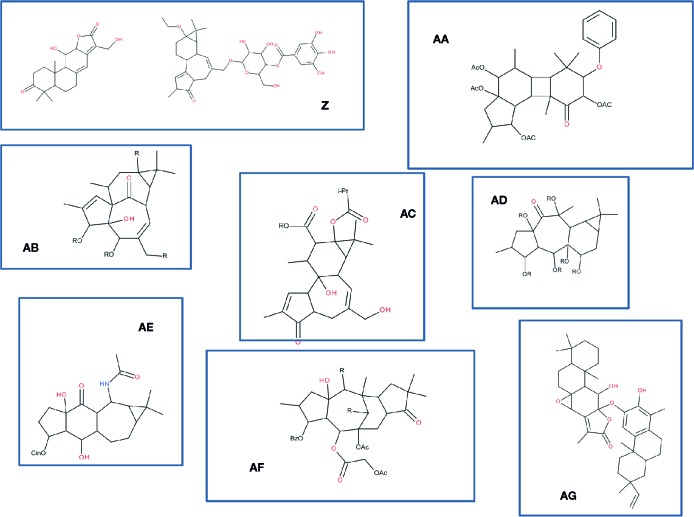
In the data gathered in Table 4 , as previously commented, it can be observed that even in the same plant species and same organ, different compounds have been characterized like jozilebomines and dioncophyllines in Ancistrocladus ileboensis leaves (J. Li et al., 2017d; J. Li et al., 2017c) or xanthohumol and α-acid derivatives such as humulones in Humulus lupulus flowers (J. Li et al., 2017b; R. Liu et al., 2017). Nevertheless, these compounds might only be slightly different from each other in terms of skeletons as illustrated in the following example: the compounds which structures are shown in Figure 4 were recently isolated from six different species of Euphorbia. They are all diterpenoids that slightly differ in their structures: ent-abietane derivatives (structures Z1 and Z2) (C.-J. Wang et al., 2017), gaditanone (structure AA) (Flores-Giubi et al., 2017), ingenane derivatives (structure AB) (Zhang et al., 2018), premyrsinane and tigliane derivatives (structure AC) (Nothias et al., 2017), dideoxyphorbol ester (structure AD) (Esposito et al., 2017), sooneuphoramine (structure AE) (Gao and Aisa, 2017), jatrophane analog (structure AF) (Rédei et al., 2018), and other abietane derivative (structure AG) (Wei et al., 2017). These compounds all originated from the same biosynthetic pathways where the cyclization reactions of the precursor geranylgeranyl diphosphate and several rearrangements allow many structural variants of diterpenoids to be produced. It is interesting to notice that despite a homology in term of basic scaffold (a phorbol ring system with some rearrangements), all the compounds are extremely different from each other from a chemical point of view.
Figure 4.
Variations around diterpenoids found in six species of Euphorbia. Compounds were drawn from the works on Euphorbia. Z : ent-abietane-type diterpenoid (Z1) and a tiglane (Z2) (Wang et al., 2017); AA: gaditanone (Flores-Giubi et al., 2017); AB: ingenane-type diterpenoid, euphorkan A (Zhang et al., 2018); AC: jatrophane-type ester (Nothias et al., 2017); AD: dideoxyphorbol ester (Esposito et al., 2017); AE: diterpenoid alkaloid sooneuphoramine (Gao and Aisa, 2017); AF: jatrophane-type diterpenoid (Rédei et al., 2018); AG: abietane-lactone- and nor-rosane-based heterodimeric terpenoids (Wei et al., 2017). Note that the stereospecificity of the compounds was not given, as many different compounds were described in each of those references.
This last statement requires underlining that the difference between two chemicals can be dramatic concerning their biological activities while minimal concerning their chemistry. For example, the chemical difference between testosterone and estrone is a saturation of the A cycle of the cholesterol backbone. These minor chemical differences leading to massive differences in pharmacological potential have been the source of a never ending debate among screeners in the pharmaceutical industry on what should populate chemical libraries: should minor variations of basic skeletons be included or not (in the primary screening) knowing that a missing methyl could lead to a nonactivity and vice-versa? In our view, it seems important to gather as many compounds as possible inside a chemical series, even if the diversity seems to be futile, because, by setting the minimum results at a poor but significant level, such as 1 to 10 µM (depending on the molecular target) hits could be found, and even minor differences in structures can lead to new leads. In this sense, working with phytochemical diversity as shown in Figure 5 becomes meaningful.
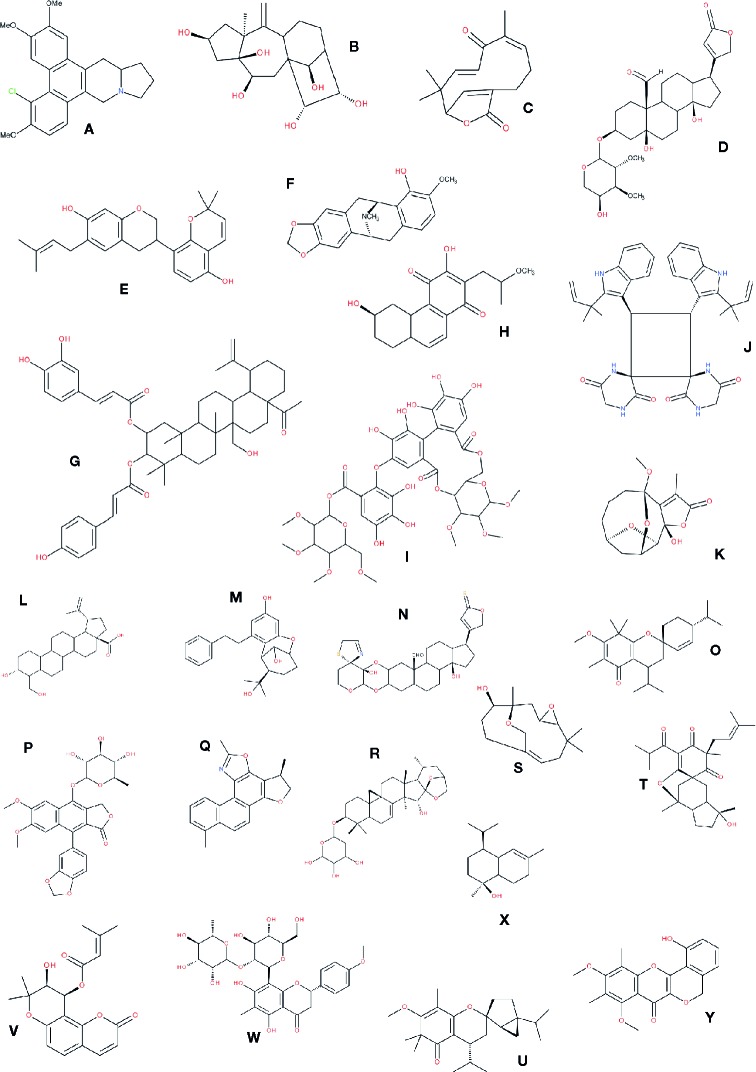
Figure 5.
Different types of molecules isolated from randomly chosen plants. Those molecules correspond to examples in Table 3 where the entries are in bold characters. (A) chlorinated phenanthroindolizidine (Al-Khdhairawi et al., 2017); (B) 3,4-seco-lupane-type triterpenoid (Cheng et al., 2018); (C) asteriscunolide C (Boumaraf et al., 2017); (D) (+)-strebloside (Ren et al., 2017); (E) Glycybridin D (Li et al., 2017); (F) (-)-neocaryachine (Suzuki et al., 2017); (G) plectranthroyleanone A (Nzogong et al., 2018); (H) uncarilin A (Geng et al., 2017); (I) 27-hydroxyalphitolic acid derivative (Novakovic et al., 2017); (J) Isorugosin (Shimozu et al., 2017); (K) carpescernolide A (Yan et al., 2018); (L) 3α,24-dihydroxylup-20(29)-en-28-oic acid (Valencia-Chan et al., 2017); (M) rasumatranin A (Wang et al., 2017); (N) uscharin (Yoneyama et al., 2017); (O) baeckfrutone A (Qin et al., 2018); (P) patentiflorin A (Zhang et al., 2017b); (Q) salvianan A (Zhang et al., 2017); (R) cimiricaside B (Thao et al., 2017); (S) aquilanol A (Ma et al., 2017); (T) hyperhenone G (Duan et al., 2018); (U) frustescone O(Hou et al., 2017); (V) dihydropyrano-coumarin derivative (Hong and Kim, 2017); (W) matteuorienate A (Huh et al., 2017); (X) t-muurolol (Pérez-Colmenares et al., 2018); (Y) bougainvinones I (Do et al., 2018). Note: for most of those compounds, the stereospecificity was represented.
Furthermore, most of these compounds would be difficult to obtain by conventional organic chemistry methods, not only because of the presence of several intramolecular bridges, but also because of the stereochemistry of the final product [see for example discussion on isoprenoids (Bouvier et al., 2005)]. Secondary metabolites are the results of multienzymatic pathways, and all these enzymes have a strict stereo-specificity. These multiple possibilities in terms of spatial arrangement in compounds contribute to the wide range of pharmacophores lying in natural products.
It seems interesting not only to mention the plant chemical diversity but also to show it with some interesting and diverse structures: Figure 5 already presented about 20 different chemical skeletons of NP that are reported in Table 4 (compound appearing in bold cases). More diversity is presented in Figure 6 . To be noticed that compounds issued from microorganisms are often—but not always—peptide-derived structures, often macrocyclic compounds (Newman and Cragg, 2015), such as the families of antibiotics found in Penicillium and the like, and thus are not the main purpose here.
Figure 6.
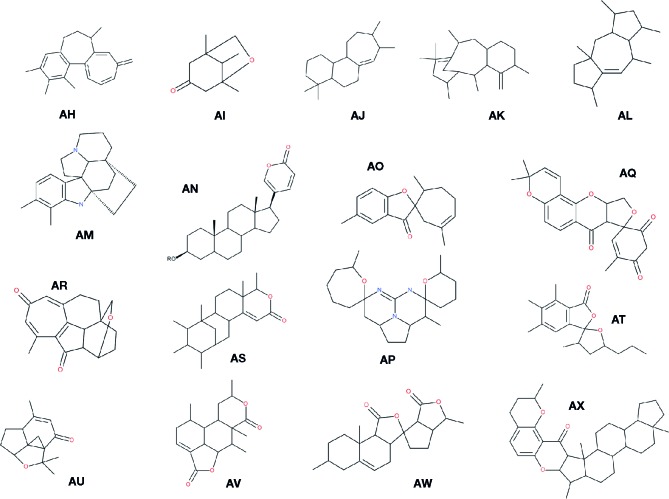
Examples of skeletons of natural compounds described in the present survey. The following chemical names were given when the structures were fairly simple. For other more complex skelettons (AM, AP, AQ, AR, AU and AV), common names were given that correspond to the global structure of the core molecule. We simplified the structures in order to give a general idea of the various natural structures met in natural compounds. Many variations exist of those families of compounds. AH, 1,2,3,7-tetramethyl-9-methylene-6,7-dihydro-5H-benzo[a]heptalene (Zarev et al., 2017); AI, 1,5,8-trimethyl-6-oxabicyclo[3.2.1]octan-3-one (Jiang et al., 2017); AJ, 4,4,8,9-tetramethyl-1,2,3,4a,5,6,8,9,10,11,11a,11b-dodecahydrocyclohepta[a]naphthalene (Wang et al., 2017a); AK, taxadiene (Dang et al., 2017a); AL, 1,3,4,6,8a-pentamethyl-1,2,3,3a,4,6,7,8,9,9a-decahydrocyclopenta [f]azulene (Zheng et al., 2017); AM, kopsine (Zeng et al., 2017); AN, 5-[(3S,10S,13S,17S)-3-hydroxy-10,13-dimethyl-2,3,4,5,6, 7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pyran-2-one (Schwikkard et al., 2017); AO, 1′,5,5′-trimethylspiro[benzofuran-2,6′-cycloheptene]-3-one (Luo et al., 2017); AP: amorphispirone (Muharini et al., 2017); AQ, cephalotane (Zhao et al., 2017); AR, xylorumphiin (Waratchareeyakul et al., 2017); AS, 5-ethyl-1,8a-dimethyl-6-(1,2,3,4-tetramethylcyclohexyl)-5,6,7,8-tetrahydro-1H-isochromen-3-one (Campos et al., 2017); AT, 3′,5,6,7-tetramethyl-5′-propyl-spiro[isobenzo furan-3,2′-tetrahydrofuran]-1-one (Xiao et al., 2018); AU, aromaticane (Dong et al., 2017); AV, clerodane (Bisio et al., 2017); AW, 1,7′,9′a-trimethyl spiro[3a,5,6,6a-tetrahydro-1H-cyclopenta[c]furan-4,3′-4,6,7,8,9,9b-hexahydro-3aH-benzo[g] isobenzofuran]-1′,3-dione (Seeka et al., 2017); AX, catechin-bound ceanothane-type triterpenoid (Kang et al., 2017a).
However, it is a fact that the structures presented in Figure 6 , even if they are almost randomly chosen, are different from what provides the current state of the art in medicinal chemistry. This is not really surprising when considering that 83% of core ring scaffolds shown in natural products cannot be found in libraries of synthetic compounds (Harvey et al., 2015). It should be reminded that part of the interest in “discovering” such structures, if they have any biological activities, resides in our capacity to use medicinal chemistry to translate such complex structures in simpler molecules amenable to the industrial production. At the same time, those structures might bear activity towards proteins, the inhibition of which has not been reached yet, with our current access to chemical synthons. This has been reported and discussed according to two points of view: (a) the natural compound-derived fragments (Rodrigues et al., 2016) and (b) the list of compounds issued from natural skeletons: what Newman and Cragg (2016) called NP derivatives in which 268 out of 1,328 new drugs can be found. Among the classical examples, there are vincristine and vinblastine as part of vinca-alkaloids (Zhou and Rahmani, 1992), statins (Sirtori, 2014), glifozins (Burson and Moran, 2015), and ingenol mebutate (Alchin, 2014). It should be reminded here that the current process is moving more from “simple” hits—from whatever origin—to more complex molecules by means of medicinal chemistry decoration of these synthons. The process of lead hopping, as defined by Krueger et al. (2009) and Chakka et al. (2017), should theoretically permit to mimic one complex structure by another, simpler one. All this body of techniques and theories should be put together at work in order to gain new powerful compounds from new, NP-based approaches (Yñigez-Gutierrez and Bachmann, 2019).
In fact, a survey of the data indicates two important features of NP, on the basis of this selection: 1/compounds are very diverse even if they include some common features (like for instance the particular cycloheptanic structure found in some of the main compounds from Euphorbia—see Figure 4 ) and 2/the high number of asymmetric carbons render their synthesis by standard organic chemistry difficult if not impossible. For instance, some examples of those numbers are given in Table 5 . Considering that the number of theoretically possible isomers is 2n, n being the number of asymmetric carbons, in some cases, the total number of possible isomers is in the several thousand ranges (see numbers in Table 5 ). Nevertheless, a series of impressive chemical papers reported complete syntheses of such compounds by “standard” synthetic organic chemistry, even if the up-scaling of such tour de forces remains to be addressed (Kuttruff et al., 2014).
Table 5.
Examples of asymmetric carbons in natural products.
| Molecules | Number of asymmetric carbons | (Number of possible isomers) | Reference | |
|---|---|---|---|---|
| Bufadienolide | 6 | (64) | (Schwikkard et al., 2017) | |
| 3α,24-dihydroxylup-20(29)-en-28-oic acid | 7 | (128) | (Valencia-Chan et al., 2017) | |
| Xylomexicanin I | 7 | (Wu et al., 2017) | ||
| Salvinorin A | 7 | (Yilmaz et al., 2017) | ||
| Sucupiranin A | 8 | (256) | (Endo et al., 2017) | |
| 17-nor-cephalotane-type diterpenoids | 9 | (512) | (Zhao et al., 2017) | |
| Pharicin C | 9 | (Hu et al., 2018) | ||
| Scalaradial | 9 | (Starkus et al., 2017) | ||
| Oridonin | 9 | (Xu S. et al., 2017) | ||
| Xylorumphiin E | 11 | (1024) | (Waratchareeyakul et al., 2017) | |
| Cimiricaside B | 12 | (2048) | (Thao et al., 2017) | |
| Bistinospinoside A | 15 | (16384) | (Li et al., 2017e) |
Different Strategies to Benefit From This Diversity
As previously mentioned, a deep interest lies in searching and exploring the immense plant chemical diversity for drug discovery purposes, but the strategies to do so need to be reevaluated. Indeed, most of the natural secondary metabolites mentioned herein are not—so far and by far—easily synthesized. It is still through harvesting that we can use plants for discovery and development purposes or for industrial scale production. Compounds with a superior pharmacological activity isolated from a plant part must be isolated from plant extracts where they lay in minor amounts. Certainly, as previously discussed, this is a main bottleneck for many applications, as it is time-consuming, the “superior activity” can vanish in the process for several reasons, or a compound of already known activity can be rediscovered at the end of the process.
In our view these limitations can be overcome by developing alternative strategies. Among these strategies, it seems that the following ones could help solve some of the difficulties linked to NP discovery programs. The first one focuses on gaining systematic access to compounds from plant parts: it deals with the use of scalable cultures of plant cells remaining capable of producing these specific chemicals. It would allow to bypass repetitive massive in natura harvests and to have the opportunity to go back to the biological material for further chemical explorations of the remaining compounds, if some of its own limitations would be overcome. The second strategy aims at the appreciation of the known plant chemical diversity, through an “inventory” of NP sources. The third strategy focuses on the industrial scale production through synthetic biology, a branch of biotechnology by which one can construct inside a microorganism the enzymatic pathways leading to a given compound (Zhao et al., 2019). This last body of techniques leads to the possibility to grow thousands of liters of such microorganisms and thus, to obtain large amounts of the desired product(s).
“Research and Discovery”: In Vitro Culture
Regarding plant cell culture, several recent reviews bring a new light to in vitro culture, for investigation purposes as well as for its uses as a valuable platform for high-value metabolite production (Wilson and Roberts, 2012; Moscatiello et al., 2013; Ochoa-Villarreal et al., 2015; Eibl et al., 2018). Examples are scattered in the literature with in vitro cultures producing compounds of interest either using dedifferentiated cells from callus or undifferentiated cells from meristematic cambial cells. From the calli, systems of plant suspension cell cultures can be generated like for example for acteoside production from Scrophularia stiata (Khanpour-Ardestani et al., 2015), rosmarinic acid from Satureja khuzistanica (Sahraroo et al., 2014), or carotenoid from Tagetes erecta (Benítez-García et al., 2014).
On the other hand, tissue cultures (i.e. hairy roots) can also be developed from already differentiated cells. All these types of culture are generally developed for a particular purpose, often very restricted to a given compound in a given plant, such as for camptothecin production by Ophiorrhiza species (Asano et al., 2009), Schisandra chinensis lignans production (Szopa et al., 2017; Szopa et al., 2018) or boeravinone Y by Abronia nana (Lee et al., 2018). Several examples of cultures at a commercial scale have also been described validating its feasibility and scalability from lab-scale to large-scale (paclitaxel form Taxus spp. cultures, rosmarinic acid from Coleus blumei cultures, scopolamine from Duboisia spp. cultures (Wilson and Roberts, 2012) to name only but a few).
Nevertheless, if the literature provides us with some very well-described examples of such tasks, it remains to be seen if these techniques are universal. In other words, what has been described to be possible to obtain a “large scale” plant cell culture is not necessarily applicable to the next cell culture of a different plant, and a fortiori, of a different organ of the same plant species. What has been considered as a promising approach remains in some cases a challenge, as no experimental process has been developed—or published—with a general usage purpose. Thus, the perfectly described process to obtain stem-derived callus or leaf-derived callus producing anticancer phenolic compounds from Fagonia indica (Khan et al., 2016) is only partially similar to the process to obtain callus from fruit pulp of varieties of apple producing high triterpenic acids (Verardo et al., 2017) or the process to obtain callus originating from seeds of Abronia nana (Kim et al., 2014), a desert plant found in North America to produce massive amount of boeravinone Y (Lee et al., 2018) or even callus from Scrophularia striata for the production of acteoside (Khanpour-Ardestani et al., 2015). As seen above, many examples can be found in the literature, but the remaining question is how much the methodology varied from one example to the next in order to obtain such callus and then to obtain such cell suspensions producing the desired compound. Some general procedures for the establishment of dedifferentiated plant cell suspension cultures exist (Mustafa et al., 2011; Eibl et al., 2018) but with many specific adaptations in the function of species, organs of origins, and culture conditions. Probably because the authors concentrated mainly on the productivity and yield of the targeted compound and not on a larger picture applicable to a more general view of accessing and testing the chemical diversity of plants. If one considers the natural diversity lying in the plant species from our environment as a source of “new” chemicals, it would be wonderful to rely on a methodology universal enough to collect the biological material just once (or a very few times) and then, to rationalize the culture of the cells originating from the specific plant organ (leaves, stems, roots, etc.). The interest on these cell cultures is that the cells would be able to biosynthesize a large variety of compounds in quantities large enough to isolate and identify compounds with pharmacological activity and completely characterize them. At that stage, the culture size can be customized by expanding from several liters to several tens of liters of cell culture (hundreds if the initial results are promising and more biomass is needed to go on with testing). Finally, even modifications of culture conditions could work at enhancing chemical diversity (Jozwiak et al., 2013) or at least variations within the proportions of different secondary metabolites (Jozwiak et al., 2013; Akbari et al., 2018; Saad et al., 2018).
The originality of our approach thus resides in using plant tissue and cell culture not for the production scale for which some limitations exist but to attempt facilitating the access to plant chemical diversity. With these perspectives, a repository associated with plant cell culture would be a valuable tool. And the versatility in terms of scaling the in vitro cultures would also allow bridging the gap between drug discovery and the first stages of development.
At the Development Stage: Systematic Inventories of Natural Products and Their Sources
Classically, the approaches used for drug discovery—detailed above for some of them—are quite specific: trying to find a compound in a plant organ that has some specific activity against a particular enzyme, receptor, or pathway. On the contrary, a systematic “inventory” of the NP existing in living organisms, in plant parts for example, would be of great interest both for the drug discovery aspects as well as the development aspects, for the discovery aspects because it would allow a better use of the known natural chemical diversity and for the drug development aspects, because it would allow to change the sourcing of the NP keeping in mind how important the supply of the compound of natural origin is for a company.
As shown for a few examples in Tables 2 – 4 which consist in a sampling of recently reported works on natural compounds, this “Systematism” is already used by a few groups who catalogued the chemical compounds in a given organ of a given plant (Batista et al., 2017; Sendker et al., 2017; Ma et al., 2018; Sharma et al., 2018), in a fungus (Chang et al., 2017), or in a microorganism (McMullin et al., 2017; Verastegui-Omana et al., 2017). In such cases, an idea of the possible diversity of those sources is given. A compendium of some plant compositions had been done by I.A. Ross (Ross, 1999; Ross, 2001). Such inventory organized in a global database would greatly facilitate the access to the diversity of secondary metabolites of plants, for example. It would somewhat ease any strategy based on chemotaxonomy by describing better the filiations/relationships between the biosynthetical pathways in a different genus and by facilitating the access to some types of chemical skeletons in renewable naturally producing sources such as leaves or fruits. Furthermore, Table 4 indicates, in our view, the way the literature can be compiled from all the available sources to build a database based only on published articles describing one or several compounds from plant parts. In this line, the remarkable paper of Solyomváry et al. (2017) reviews the available literature on the compounds from the dibenzylbutyrolactone lignan family and describes some 91 compounds of this chemical family from their origin in terms of plant species and plant parts.
For many reasons, such a systematic inventory of plants chemical components related to the tissues and the species from which they have been extracted, would be of great use but could be difficult to complete, even with modern and fast analytical tools. Indeed, it is the completion of such a compendium that is the real challenge, and the experience of the few already existing NP databases exemplifies that challenge. For example the NAPRALERT experience (https://napralert.org) gathering data from more than 200,000 scientific papers is very informative: comprehensive coverage is claimed from 1975 to 2004 while only 20% of the global data is covered from 2005 due to budgetary constraints. Organisms, compounds, activities, or authors can be searched. In fact, the last decade has seen the development of several databases providing systematic collection of information that focuses on natural compounds themselves, offering the possibility of searching structure, source, and mechanisms of action of the searched compounds. For example, DEREP-NP is a database that compiles structural data (Zani and Carroll, 2017). An interesting review from Xie et al. (2015) allows the comparison of fourteen of these databases focusing on NP, balancing their advantages and disadvantages. Among them, the updated version of a 2006 database SuperNatural II is a public resource (http://bioinformatics.charite.de/supernatural) with more than 325,500 natural compounds, offering 3D structure and conformers (Banerjee et al., 2015) which seems to outperform many others (Harvey et al., 2015; Xie et al., 2015). Another source of natural compounds is also the Greenpharma collection (www.greenpharma.com/products/compound-librairies/#GPNDB) (Do et al., 2015; Gally et al., 2017).
Industrial Scale Production: Synthetic Biology and Organic Syntheses
The use of plant cell and organ culture for the production at the industrial scale of compounds with superior added value has been reported in various reviews (Wilson and Roberts, 2012; Imseng et al., 2014; Eibl et al., 2018). These reviews cited a large range of applications from the pharmaceutical area (suspension cells of Pacific yew in 75m3 stirred bioreactors delivering 500kg/year of paclitaxel) to the cosmetic and food industries like cell cultures of Malus domestica grown in 50 to 100 L production bioreactors. But still, it is acknowledged that some limitations exist: mainly the fact that time-consuming processes are involved, with possibly low titers, and the possibility of somaclonal variations appearing in the selected top producing cell lines. Several solutions to try to avoid these kinds of limitation can be assessed (Trosset and Carbonell, 2015), but in our opinion, general strategies should consider other alternatives for the large industrial production scale depending on the kind of applications. For some specific NP of pharmacological interest like podophyllotoxin, artemisinin or plumbagin for example, a whole set of biotechnological approaches has been developed and described for the production at larger scale of these valuable compounds (Lautié et al., 2010; Kayani et al., 2018; Roy and Bharadvaja, 2018). But once more, these strategies are quite specific, driven only in one identified compound and its specific biosynthetical pathway.
Synthetic Biology
As previously mentioned, one of the major requirements to this approach is to understand the pathways through which a particular compound is biosynthesized, thanks to the activity of a series of enzymes involved in a particular plant part. Within this last approach, the focus in plants is more on their capacity to synthetize unique scaffolds than on the end-products themselves. Indeed, the use of the recent integrative approaches based on “Omic” analyses (metabolomics, proteomics, transcriptomic, and genomics) can be of great value. Indeed, knowing the precursors and intermediates through the biochemical status of a tissue, identifying the key enzymes and the limiting steps of the pathways, monitoring indirectly the function of the genes involved in these pathways and their regulation will contribute to decipher the biosynthetic routes in planta (Cheallaigh et al., 2018; Scossa et al., 2018). The relative ease at which one can now obtain large-scale data has facilitated the analyses at the level of the whole metabolic network (Paddon and Keasling, 2014; Ikram and Simonsen, 2017). For example, large amounts of transcriptomic data are now easier to access as stated by Owen et al. (2017), making possible the identification of multistep pathways by coexpression analyses or untargeted metabolomics. Furthermore, the discovery that genes linked to biosynthetic pathways are organized in clusters has opened new opportunities by adapting methodologies developed initially for microorganisms to plants like systematic cluster mining algorithms (Owen et al., 2017). Scossa et al. (2018) reviewed recently the progresses made in the understanding of plant biosynthetic pathways with the integration of metabolomics and next-generation sequencing based on various families of compounds: for example, benzoisoquinoline and monoterpenoid indole alkaloids, cannabinoids, ginsenosides, or withanolides. They also emphasized the new insight that this area can bring in the field of synthesis of NP. They mention for example, the intriguing case of caffeine biosynthesis that evolved independently in several orders of eudicots: at least three metabolic pathways evolved separately coopting genes from different gene families illustrating how biosynthetic pathways can evolve with land plant diversification (Scossa et al., 2018).
After having identified the genes involved, the reconstitution of the biosynthetic pathways of interest can be realized thanks to novel DNA construction technologies. It can be realized in a foreign host which enables the increase of product yields. The choice of this host organism is key as the goal is to develop an efficient platform for heterologous gene expression. Microbial hosts are generally considered more amenable than plants to fermentation process (Atanasov et al., 2015). Among them, classical work horses like E. coli or S. cerevisiae, or newcomers like Bacillus subtilis (Abdallah et al., 2019) and Pseudomonas putidaare (Nikel et al., 2014; Loeschcke and Thies, 2015; Choi and Lee, 2020) can be cited. Another interesting example is the recent high-cell-density fermentation strategies developed for heterologous production in Pichia pastoris (W.-C. Liu et al., 2019). Then, basically, the strategy will consist in cloning the genes of the enzymes of the pathway that have been identified; constructing large plasmid (or family of plasmids) encoding for those enzymes; transfecting with the plasmid a microorganism that will be grown afterwards; and purifying the product (Xiao et al., 2018). Several recent reviews detail how the technical advances in synthetic biology and multiplexed genome engineering allow for optimizing the design and synthesis of the pathways involved in NP production (Awan et al., 2016; Breitling and Takano, 2016; Carbonell et al., 2016; Smanski et al., 2016; Moses et al., 2017). Many such examples can be found such as for curcumin synthesis reconstitution in E. coli (Kang et al., 2018), polyunsaturated fatty acids production in the fungus Ashbya gossypii (Ledesma-Amaro et al., 2018), α-amyrin, lupulones ou ginsenosides synthesis in S. cerevisiae (Dai et al., 2014; Yu et al., 2018; Guo et al., 2019), or the diversification of the carotenoid biosynthetic pathways (Umeno et al., 2005). But probably the best example of economically feasible process is reported for the production of artemisin at an industrial scale (Paddon and Keasling, 2014; Ikram and Simonsen, 2017).
Alternatively, the developments in plant transformation and transfection technology offering rapid and scalable biosynthesis allow for considering more and more the use of plant-based expression platforms like Nicotiana or Arabidopsis spp.(Fuentes et al., 2016; Lu et al., 2017; Reed et al., 2017; Appelhagen et al., 2018). Indeed they are considered genetically more flexible than the native plant sources and offer in some cases several advantages even over microbial hosts that can lack the endogenous biosynthetic precursors of these NP or intracellular compartments as endoplasmic reticulum related with the implementation of enzymes like cytochrome P450s (Appelhagen et al., 2018).
These advances in plant synthetic biology will increase the access to NP through new synthetic routes (Reed et al., 2017) but will also allow the synthesis of new-to-nature molecules and so, expand the natural plant chemical diversity.
Organic Syntheses
A way to analyze the total syntheses that have been recently produced in the literature is to use simple criteria in order to evaluate the feasibility of such approach in case of similar compounds finding their way to the clinic. We evaluated a set of recent publications dealing with “total synthesis” of NP according to the simple criteria: number of cycles in the compounds, number of carbons and heteroatoms—including sulfur—in those cycles and number of asymmetric carbons in these structures ( Table 6 ). In this nonexhaustive set of publications, it was decided not to consider peptides and peptide-derived macrocycles (about a dozen structures). The next observation was that there were a surprising high number of bacteria-derived compounds, a feature that we did not notice in our previous surveys ( Tables 2 – 4 ). Another parameter that allows for judging the feasibility and scalability of the processes is the yield and the number of steps. In that sense, most of those works are exquisitely delicate enterprises. The success of those publications in terms of tour de force is obvious, but they also allow for emphasizing the necessity to obtain such general synthetic routes, as most of those were used then to provide analogs to the desired NP in each publication.
Table 6.
Some examples of total synthesis of natural products.
| Compound (reference) | Origin* | Number of cycles | Number of carbons ** | Number of heteroatoms** | Number of asymetric centers | number of steps*** | yield (%)*** |
|---|---|---|---|---|---|---|---|
| cerorubenic acid III (Liu et al., 2019) | Ceroplastes rubens (insect) | 4 | 15 | 0 | 7 | ~20 | |
| (−)-ambiguine P (Johnson et al., 2019) | Fischerella ambigua (cyanobacteria) | 5 | 18 | 1 | 4 | 20 | |
| actinorhodin (Ninomiya et al., 2019) | Streptomyces coelicolor (actinobacteria) | 2 × 3 | 2 × 13 | 2 × 1 | 2 X1 | 16 | 1,6 |
| (+)-nivetetracyclate A (Blitz et al., 2019) | Streptomyces niveus (actinobacteria) | 4 | 18 | 0 | 3 | 17 | < 2 |
| nogalamycin (Peng and VanNieuwenhze, 2019) | Streptomyces nogalator (actinobacteria) | 6 | 23 | 2 | 5 | > 20 | |
| albomycins δ1 (Lin et al., 2018) | Streptomyces griseus (actinobacteria) | 2 | 8 | 3 | 10 | 13 | |
| (−)-omuralide (Rulliere et al., 2018) | Streptomyces lactacystenaeus (actinobacteria) | 2 | 5 | 3 | 4 | 19 | 2,6 |
| (+)-guadinomic acid (Reid et al., 2018) | Streptomyces sp. (actinobacteria) | 1 | 3 | 2 | 2 | 7 | 0,6 |
| (−)-(3R)inthomycin C (Balcells et al., 2018) | Streptomyces sp. (actinobacteria) | 1 | 3 | 2 | 2 | 10 | 15 |
| (±)-vibralactone (Nistanaki et al., 2019) | Boreostereum vibrans (fungus) | 2 | 6 | 1 | 2 | 6 | 4,3 |
| epi-trichosetin (Kauhl et al., 2018) | Fusarium oxysporum (fungus) | 3 | 15 | 1 | 6 | 8 | 4,1 |
| tryptoquivaline (Wei and Dixon, 2018) | Aspergillus fumigatus (fungus) | 5 | 19 | 3 | 2 | ~20 | 11 |
| (−)-chaetominine (Geng and Huang, 2019) | Chaetomium sp. (fungus) | 6 | 21 | 4 | 4 | 4 | 33,4 |
| repraesentin F (Ferrer and Echavarren, 2018) | Lactarius repraesentaneus (fungus) | 3 | 10 | 0 | 6 | 16 | 2 |
| epicolactone (Kravina and Carreira, 2018) | Epicoccum nigrum (fungus) | 5 | 15 | 2 | 17 | ||
| (−)-6,7-dideoxysqualestatin H5 (Almohseni et al., 2018) | Lepzodontium elarius (fungus) | 3 | 12 | 2 | 1 | 11 | 5,5 |
| delitschiapyrone A (Kurasawa et al., 2018) | Delitschia sp. (fungus) | 5 | 18 | 2 | 5 | 8 | 33 |
| phyllostictine A (Riemer et al., 2018) | Phyllosticta cirsii (fungus) | 2 | 7 | 2 | 3 | 13 | 4 |
| spiromamakone A (Tsukamoto et al., 2018) | Preussia sp. (fungus) | 4 | 19 | 0 | 1 | 16 | |
| penicophenone A (Pantin et al., 2018) | Penicilium dipodomycola (fungus) | 3 | 13 | 2 | 3 | 14 | 6 |
| aurofusarin (Qi et al., 2018) | Fusarium graminearum (fungus) | 2 x 3 | 26 | 2 x 1 | 0 | 10 | 10 |
| (±)-antroquinonol (Wang et al., 2019) | Antrodia camphorata (mushroom) | 1 | 6 | 0 | 3 | 10 | 29 |
| Suillusin (Zhang et al., 2018) | Suillus granulatus (mushroom) | 4 | 17 | 1 | 2 | 8 | 11 |
| conosilane A (Yuan et al., 2018) | Conocybe siliginea (mushroom) | 4 | 13 | 2 | 4 | 10 | |
| Isopanepoxydone (Man et al., 2018) | Lentinus strigellus (mushroom) | 2 | 6 | 1 | 3 | 9 | 21 |
| cochlearoid B (Zhang et al., 2018) | Ganoderma cochlear (mushroom) | 5 | 18 | 2 | 3 | 7 | 12 |
| boletopsin 11 (Zhang and Barrow, 2018) | Boletopsis sp. (mushroom) | 4 | 18 | 1 | 0 | 9 | 6 |
| (+)-dimericbiscognienyne A (Kim et al., 2018) | Biscogniauxia sp. (mushroom) | 7 | 18 | 3 | 12 | 7 | |
| (−)-mitrephorone A (Richter et al., 2018) | Mitrephora glabra (tree) | 5 | 17 | 1 | 5 | 7 | 26 |
| (+)-chamuvarinin (Samala et al., 2018) | Uvaria chamae (plant) | 3 × 1 | 12 | 3 | 7 | 20 | 3 |
| (±)-aspidofractinine (Saya et al., 2018) | Aspidosperma cylindrocarpon (tree) | 6 | 17 | 2 | 4 | 8 | < 5 |
| (+)-leucomidine A (Yao et al., 2018) | Leuconotis griffithii (plant) | 5 | 17 | 3 | 2 | 8 | 31 |
| larreatricin 1 (Martin et al., 2018) | Larrea tridentata (plant) | 3 | 16 | 1 | 4 | 6 | 40 |
| (±)−exotine B (Cheng et al., 2018) | Murraya exotica (plant) | 5 | 22 | 2 | 2 | 6 | |
| lanceolactone A (Acharyya and Nanda, 2018) | Illicium lanceolatum (plant) | 2 | 7 | 2 | 2 | 4 | 44 |
| bussealin E (Twigg et al., 2018) | Bussea sakalava (plant) | 4 | 15 | 1 | 0 | 11 | 14 |
| polyflavanostilbene B (Wang X. et al., 2018) | Polygonum cuspidatum(plant) | 9 | 40 | 2 | 7 | ||
| “Unnamed alkaloid” (Davison et al., 2018) | Isatis indigotica (plant) | 4 | 14 | 5 | 2 | 6 | |
| 2-epi-narciclasine (Borra et al., 2018) | Narcissus sp. (plant) | 4 | 13 | 3 | 4 | 9 | 4,5 |
| (+)-psiguadial B (Chapman et al., 2018) | Psidium guajava (plant) | 6 | 23 | 1 | 7 | 15 | 1,3 |
| Parvineostemonine (Gerlinger et al., 2018) | Stemona parviflora (plant) | 4 | 14 | 2 | 4 | 5 | 17 |
| Arboridinine (Gan et al., 2018) | Kopsia arborea (plant) | 5 | 16 | 2 | 3 | 16 | |
| adunctin B (Dethe and Dherange, 2018) | Piper adunctum (plant) | 4 | 18 | 1 | 3 | 6 | 23 |
| (±)-deguelin (Xu et al., 2018) | Tephrosia vogelii (plant) | 5 | 19 | 3 | 2 | 4 | 62 |
| englerin A (Hatakeyama, 2018) | Phyllanthus engleri (plant) | 5 | 19 | 2 | 6 | 23 | 13 |
| houttuynoid A (Jian et al., 2018) | Houttuynia cordata (plant) | 5 | 22 | 3 | 4 | 7 | 24 |
| (−)-mucosin (Nolsoe et al., 2018) | Reniera mucosa (sponge) | 2 | 8 | 0 | 2 | 15 | |
| (rac)-renieramycin T (Kimura and Saito, 2018) | Reniera sp. (sponge) | 6 | 20 | 4 | 5 | 19 | 1,8 |
| (+)-frondosin B (Zong et al., 2018) | Dysidea frondosa (sponge) | 4 | 16 | 1 | 1 | 15 | 12 |
| shishijimicin A (Nicolaou et al., 2018b) | Theonella sp. (sponge) | 7 | 36 | 4 | 8 | > 20 | 16 |
| ulbactin F (Shapiro et al., 2018) | Brevibacillus sp. (sponge) | 5 | 16 | 6 | 5 | 7 | 12 |
| spongosoritin A (Alcock et al., 2018) | Spongosorites sp.(sponge) | 1 | 4 | 1 | 2 | 11 | 1,8 |
| Namenamicin (Nicolaou et al., 2018a) | Polysyncraton lithostrotum (ascidian) | 5 | 29 | 3 | 11 | > 20 | 14 |
*The origin of the compounds is color-coded: light red: insect; black: bacteria; light blue: fungus; violet: mushroom; green: plants and trees; dark red, sponge; dark blue: ascidian. ** The numbers of carbon atoms and of heteroatoms were those included in the cycles. ***Yellow cells: approx. calculations by the present authors. Gray cells: This information is too deeply embedded in the paper.
Some of those compounds are devoid of asymmetric carbons, rendering the synthesis ‘easier’, but still a challenge requiring several steps, with an overall poor yield. At the other end of the spectrum are compounds with a considerable number of asymmetric carbons, such as (+)-dimericbiscognienyne A with 12 asymmetric carbons (Kim et al., 2018), or namenamicin with 11 (Nicolaou et al., 2018a) and/or with a high number of cycles, even if they were not always fused with each other. Indeed, a series of three furan-based cycles (Samala et al., 2018) separated by alkyl carbon chains would not be a considerable difficulty to synthesize, depending on the decorations of those cycles that introduce notions of asymmetries and thus difficulties to perform.
Those data, when compared to the ones gathered in Table 5 , show that in these particular cases, the access by chemistry of all the possible optical isomers would be simply impossible. These observations cast some shadows on the possibility of using those synthetic routes at the industrial scale. On the other hand, the mastering of some steps, particularly the stereo-controlled ones, are key in the cases where alternative hemi-syntheses solutions are adopted from a most abundant intermediary (natural) compound. Finally, another point is certainly the growing numbers of synthetic routes that are explored, assessed, and validated to access some “common” features from those natural compounds. A review of this literature can be found in Li L. et al. (2018). Even if partial by essence, it shows the considerable number of routes that has been set up and that permits access to some of the main fused cycles found in compounds coming from different natural sources. Another review summarized the way spiroacetal can be accessed, another common feature of many natural compounds (Zhang et al., 2018). This last point strongly emphasizes the common nature throughout the living world of the basic enzymatic systems aiming at producing secondary metabolites from the same fundamental bricks such as mevalonate or other isoprenoids.
Nevertheless, it is also clear from that survey that chemistry is not, at the present time, the solution to the problem of scalability of NP productions to an industrial level, even if these compounds were extremely active on a given disease and even if a large panel of examples in which complete syntheses of NP are presented (Kuttruff et al., 2014).
As pointed out earlier in the present assay, at the research level and even at the level of exemplifications of chemical analogs of a given active, these approaches are necessary and important. Indeed, deciphering the various routes to some of those compounds might help design and simplify the overall structures, as it is the case in standard, organic chemistry-based, medicinal chemistry.
Conclusions
For decades, the interplay between the search for “new” drugs and NP has been strong, to a point where some fear that the destruction of native forests, leading to a reduction of plant diversity would jeopardize our finding of new cures for old and new diseases. The present essay aimed to offer a global overview of the extent of the known chemical diversity, its access, and its use. Several approaches to chemical diversity were also discussed maximizing, in our view, the possibilities of finding useful compounds for human unmet medical needs.
As illustrated, plant natural chemical diversity is indeed immense. And the knowledge we gathered on plants is only the tip of the iceberg as exemplified in IA Ross’s books (Ross, 1999; Ross, 2001), in which he gathered all the chemical components found in some 40 plants and their many different components. And this knowledge is certainly scattered all over the world. A unifying work should be done, under a simple format, that could be like the one presented in Table 3 and completed by following the example of Solyomváry et al. (2017). This kind of tool would incredibly ease the access to plant natural chemical diversity and should ideally be comprehensive, organized and include data from worldwide plant species, from past to recent studies. Such a globalized database could furthermore be integrated to other ones like genomic, phylogenic, species occurrence, biosynthetic pathway, biological activity, or chemical classification (Allen et al., 2019) allowing researchers to mine the resources and correlate the information, hence empowering all kind of research studies. This trend has been emphasized by several authors in their recent reviews (Atanasov et al., 2015; Harvey et al., 2015) stating that drug discovery from plants requires multidisciplinary approaches. Experiences from the past tell us how important it is, for drug discovery purposes, to access this wide diversity lying in the Plant kingdom, especially because it may be shrinking due to the rapid alterations of the biosphere. In order to fully access the whole chemical diversity without jeopardizing plant biodiversity, alternative ways to collect and store plant tissues can be explored, as for example the use of in vitro culture techniques allowing a renewable and sustainable access to plant chemical diversity. As the final purpose is giving access to workable quantities of therapeutic compound(s), we suggested that the advances in synthetic biology coupled with genomics and bioinformatics can pave the way to possible future strategies of productions of the compounds originating from this diversity. But the chemical diversity in the scaffolds of plant natural compounds is so wide that there is still some space from different strategies for large-scale production: from organic total synthesis for the simpler scaffolds like ephedrine or metformin that are able to be synthetized in few steps, that is to say, at a reasonable cost or, at the other end of the spectrum, heterologous (plant)? production for compounds with more complex scaffolds like taxanes and multistep biosynthesis, and in between even hybrids (multihosts)? semisynthetic strategies can be imagined and developed.
Author Contributions
EL and JB wrote the review with the help of PD (modelization) and OR (chemistry).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Ms. Luana Gessica do Carmo da Silva for her help in preparing Figure 2 and Dr. Natalia Sayuri Muto for her help in the language edition of the manuscript. The Center of Agro-food, Pharmaceutical and Cosmetic Valorization of Amazonian Bioactive Compounds (CVACBA) and Prof. Hervé Rogez are also acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.00397/full#supplementary-material
References
- Abdallah I. I., Pramastya H., van Merkerk R., Sukrasno, Quax W. J. (2019). Metabolic Engineering of Bacillus subtilis Toward Taxadiene Biosynthesis as the First Committed Step for Taxol Production. Front. Microbiol. 10, 218. 10.3389/fmicb.2019.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed S. A., Sirat H. M., Taher M. (2016). Tyrosinase inhibition, anti-acetylcholinesterase, and antimicrobial activities of the phytochemicals from Gynotroches axillaris Blume. Pakistan J. Pharm. Sci. 29, 2071–2078. [PubMed] [Google Scholar]
- Acharyya R. K., Nanda S. (2018). Asymmetric total synthesis of naturally occurring spirocyclic tetranorsesquiterpenoid lanceolactone A. Org. Biomolec. Chem. 16, 5027–5035. 10.1039/c8ob01328d [DOI] [PubMed] [Google Scholar]
- Ademosun A. O., Oboh G., Passamonti S., Tramer F., Ziberna L., Boligon A. A., et al. (2015). Inhibition of metalloproteinase and proteasome activities in colon cancer cells by citrus peel extracts. J. Basic Clin. Physiol. Pharmacol. 26, 471–477. 10.1515/jbcpp-2013-0127 [DOI] [PubMed] [Google Scholar]
- Afifi A. H., Kagiyama I., El-Desoky A. H., Kato H., Mangindaan R. E. P., Voogd N. J., et al. (2017). Sulawesins A-C, Furanosesterterpene Tetronic Acids That Inhibit USP7, from a Psammocinia sp. Marine Sponge. J. Natural Prod. 80, 2045–2050. 10.1021/acs.jnatprod.7b00184 [DOI] [PubMed] [Google Scholar]
- Ahoua A. R. C., Monteillier A., Borlat F., Ciclet O., Marcourt L., Nejad Ebrahimi S., et al. (2018). Anti-inflammatory and Quinone Reductase-Inducing Compounds from Beilschmiedia mannii. Planta Med. 85, 379–384. 10.1055/a-0798-3155 [DOI] [PubMed] [Google Scholar]
- Ajibola C. F., Eleyinmi A. F., Aluko R. E. (2011). Kinetics of the Inhibition of Renin and Angiotensin I Converting Enzyme by Polar and Non-polar Polyphenolic Extracts of Vernonia Amygdalina and Gongronema Latifolium Leaves. Plant Foods Hum. Nutr. 66, 320–327. 10.1007/s11130-011-0257-x [DOI] [PubMed] [Google Scholar]
- Akbari F., Arminian A., Kahrizi D., Fazeli A., Ghaheri M. (2018). Effect of nitrogen sources on gene expression of Stevia rebaudiana (Bertoni) under in vitro conditions. Cell. Mol. Biol. (Noisy-le-grand) 64, 11–16. 10.14715/cmb/2018.64.2.3 [DOI] [PubMed] [Google Scholar]
- Alchin D. R. (2014). Ingenol Mebutate: A Succinct Review of a Succinct Therapy. Dermatol. Ther. (Heidelb) 4, 157–164. 10.1007/s13555-014-0061-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock L. J., Norris M. D., Perkins M. V. (2018). Total synthesis and structural elucidation of spongosoritin A. Org. Biomolec. Chem. 16, 1351–1358. 10.1039/c7ob03150e [DOI] [PubMed] [Google Scholar]
- Ali T., Inagaki M., Chai H.-B., Wieboldt T., Rapplye C., Rakotondraibe L. H. (2017). Halogenated Compounds from Directed Fermentation of Penicillium concentricum, an Endophytic Fungus of the Liverwort Trichocolea tomentella. J. Natural Prod. 80, 1397–1403. 10.1021/acs.jnatprod.6b01069 [DOI] [PubMed] [Google Scholar]
- Al-Khdhairawi A. A. Q., Krishnan P., Mai C.-W., Chung F. F.-L., Leong C.-O., Yong K.-T., et al. (2017). A Bis-benzopyrroloisoquinoline Alkaloid Incorporating a Cyclobutane Core and a Chlorophenanthroindolizidine Alkaloid with Cytotoxic Activity from Ficus fistulosa var. tengerensis. J. Natural Prod. 80, 2734–2740. 10.1021/acs.jnatprod.7b00500 [DOI] [PubMed] [Google Scholar]
- Allen J. M., Folk R. A., Soltis P. S., Soltis D. E., Guralnick R. P. (2019). Biodiversity synthesis across the green branches of the tree of life. Nat. Plants 5, 11–13. 10.1038/s41477-018-0322-7 [DOI] [PubMed] [Google Scholar]
- Almohseni H. A. A., Al Mamari H. H., Valade A., Sintim H. O., Hodgson D. M. (2018). Alkene protection against acid using a bromide substituent: application in a total synthesis of (-)-6,7-dideoxysqualestatin H5. Chem. Commun. (Cambridge England) 54, 5354–5356. 10.1039/c8cc02690d [DOI] [PubMed] [Google Scholar]
- Amagata T. (2010). Missasigned Structures: Case Examples from the Past Decade. Compr. Natural Products II, eds Mander L., Liu H.-W. Oxford: Elsevier,. 581–621. 10.1016/B978-008045382-8.00053-8 [DOI] [Google Scholar]
- Apel C., Bignon J., Garcia-Alvarez M. C., Ciccone S., Clerc P., Grondin I., et al. (2018). N-myristoyltransferases inhibitory activity of ellagitannins from Terminalia bentzoë (L.) L. f. subsp. bentzoë. Fitoterapia 131, 91–95. 10.1016/j.fitote.2018.10.014 [DOI] [PubMed] [Google Scholar]
- Appelhagen I., Wulff-Vester A. K., Wendell M., Hvoslef-Eide A.-K., Russell J., Oertel A., et al. (2018). Colour bio-factories: Towards scale-up production of anthocyanins in plant cell cultures. Metab. Eng. 48, 218–232. 10.1016/j.ymben.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Sudo H., Yamazaki M., Saito K. (2009). Camptothecin production by in vitro cultures and plant regeneration in Ophiorrhiza species. Methods Mol. Biol. (Clifton N.J.) 547, 337–345. 10.1007/978-1-60327-287-2_27 [DOI] [PubMed] [Google Scholar]
- Atanasov A. G., Waltenberger B., Pferschy-Wenzig E.-M., Linder T., Wawrosch C., Uhrin P., et al. (2015). Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 33, 1582–1614. 10.1016/j.biotechadv.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au T. K., Lam T. L., Ng T. B., Fong W. P., Wan D. C. (2001). A comparison of HIV-1 integrase inhibition by aqueous and methanol extracts of Chinese medicinal herbs. Life Sci. 68, 1687–1694. 10.1016/S0024-3205(01)00945-6 [DOI] [PubMed] [Google Scholar]
- Austin C., Stewart D., Allwood J. W., McDougall G. J. (2018). Extracts from the edible seaweed, Ascophyllum nodosum, inhibit lipase activity in vitro: Contributions of phenolic and polysaccharide components. Food Funct. 9, 502–510. 10.1039/c7fo01690e [DOI] [PubMed] [Google Scholar]
- Awan A. R., Shaw W. M., Ellis T. (2016). Biosynthesis of therapeutic natural products using synthetic biology. Adv. Drug Deliv. Rev. 105, 96–106. 10.1016/j.addr.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Badral D., Odonbayar B., Murata T., Munkhjargal T., Tuvshintulga B., Igarashi I., et al. (2017). Flavonoid and Galloyl Glycosides Isolated from Saxifraga spinulosa and Their Antioxidative and Inhibitory Activities against Species That Cause Piroplasmosis. J. Natural Prod. 80, 2416–2423. 10.1021/acs.jnatprod.7b00142 [DOI] [PubMed] [Google Scholar]
- Bailey C. J. (2017). Metformin: historical overview. Diabetologia 60, 1566–1576. 10.1007/s00125-017-4318-z [DOI] [PubMed] [Google Scholar]
- Baker G. H., Dorgan R. J., Everett J. R., Hood J. D., Poulton M. E. (1990). A novel series of milbemycin antibiotics from Streptomyces strain E225. II. Isolation, characterization, structure elucidation and solution conformations. J. Antibiot. 43, 1069–1076. 10.7164/antibiotics.43.1069 [DOI] [PubMed] [Google Scholar]
- Balcells S., Haughey M. B., Walker J. C. L., Josa-Cullere L., Towers C., Donohoe T. J. (2018). Asymmetric Total Synthesis of (-)-(3 R)-Inthomycin C. Org. Lett. 20, 3583–3586. 10.1021/acs.orglett.8b01370 [DOI] [PubMed] [Google Scholar]
- Banerjee P., Erehman J., Gohlke B.-O., Wilhelm T., Preissner R., Dunkel M. (2015). Super Natural II—a database of natural products. Nucleic Acids Res. 43, D935–D939. 10.1093/nar/gku886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M.-F., Zeng C.-X., Liu Y.-P., Zhang B.-J., Ni L., Luo X.-D., et al. (2017). Indole Alkaloids from Hunteria zeylanica. J. Natural Prod. 80, 790–797. 10.1021/acs.jnatprod.5b01035 [DOI] [PubMed] [Google Scholar]
- Batista A. N. L., dos Santos-Pinto J. R., Batista J. M., Souza-Moreira T. M., Santoni M. M., Zanelli C. F., et al. (2017). The Combined Use of Proteomics and Transcriptomics Reveals a Complex Secondary Metabolite Network in Peperomia obtusifolia. J. Natural Prod. 80, 1275–1286. 10.1021/acs.jnatprod.6b00827 [DOI] [PubMed] [Google Scholar]
- Bauer M. R., Mackey M. D. (2019). Electrostatic Complementarity as a Fast and Effective Tool to Optimize Binding and Selectivity of Protein-Ligand Complexes. J. Med. Chem. 62, 3036–3050. 10.1021/acs.jmedchem.8b01925 [DOI] [PubMed] [Google Scholar]
- Bautista E., Ortiz-Pastrana N., Pastor-Palacios G., Montoya-Contreras A., Toscano R. A., Morales-Jimenez J., et al. (2017). neo-Clerodane Diterpenoids from Salvia polystachya Stimulate the Expression of Extracellular Matrix Components in Human Dermal Fibroblasts. J. Natural Prod. 80, 3003–3009. 10.1021/acs.jnatprod.7b00591 [DOI] [PubMed] [Google Scholar]
- Beaufay C., Ledoux A., Jansen O., Bordignon A., Zhao S., Teijaro C. N., et al. (2018). In vivo Antimalarial and Antitrypanosomal Activity of Strychnogucine B, a Bisindole Alkaloid from Strychnos icaja. Planta Med. 84, 881–885. 10.1055/a-0644-2723 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Osbourn A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Sci. (New York N.Y.) 324, 746–748. 10.1126/science.1171661 [DOI] [PubMed] [Google Scholar]
- Bedut S., Seminatore-Nole C., Lamamy V., Caignard S., Boutin J. A., Nosjean O., et al. (2016). High-throughput drug profiling with voltage- and calcium-sensitive fluorescent probes in human iPSC-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 311, H44–H53. 10.1152/ajpheart.00793.2015 [DOI] [PubMed] [Google Scholar]
- Bemis G. W., Murcko M. A. (1996). The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893. 10.1021/jm9602928 [DOI] [PubMed] [Google Scholar]
- Benítez-García I., Vanegas-Espinoza P. E., Meléndez-Martínez A. J., Heredia F. J., Paredes-López O., Del Villar-Martínez A. A. (2014). Callus culture development of two varieties of Tagetes erecta and carotenoid production. Electron. J. Biotechnol. 17, 107–113. 10.1016/j.ejbt.2014.01.004 [DOI] [Google Scholar]
- Berboucha M., Ayouni K., Atmani D., Atmani D., Benboubetra M. (2010). Kinetic study on the inhibition of xanthine oxidase by extracts from two selected Algerian plants traditionally used for the treatment of inflammatory diseases. J. Med. Food 13, 896–904. 10.1089/jmf.2009.0164 [DOI] [PubMed] [Google Scholar]
- Bisio A., Mieri M., Milella L., Schito A. M., Parricchi A., Russo D., et al. (2017). Antibacterial and Hypoglycemic Diterpenoids from Salvia chamaedryoides. J. Natural Prod. 80, 503–514. 10.1021/acs.jnatprod.6b01053 [DOI] [PubMed] [Google Scholar]
- Bjerketorp J., Levenfors J. J., Sahlberg C., Nord C. L., Andersson P. F., Guss B., et al. (2017). Antibacterial 3,6-Disubstituted 4-Hydroxy-5,6-dihydro-2H-pyran-2-ones from Serratia plymuthica MF371-2. J. Natural Prod. 80, 2997–3002. 10.1021/acs.jnatprod.7b00565 [DOI] [PubMed] [Google Scholar]
- Blitz M., Heinze R. C., Harms K., Koert U. (2019). Total Synthesis of (+)-Nivetetracyclate A. Org. Lett. 21, 785–788. 10.1021/acs.orglett.8b04044 [DOI] [PubMed] [Google Scholar]
- Bodle C. R., Mackie D. I., Hayes M. P., Schamp J. H., Miller M. R., Henry M. D., et al. (2017). Natural Products Discovered in a High-Throughput Screen Identified as Inhibitors of RGS17 and as Cytostatic and Cytotoxic Agents for Lung and Prostate Cancer Cell Lines. J. Natural Prod. 80, 1992–2000. 10.1021/acs.jnatprod.7b00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra S., Lapinskaite R., Kempthorne C., Liscombe D., McNulty J., Hudlicky T. (2018). Isolation, Synthesis, and Semisynthesis of Amaryllidaceae Constituents from Narcissus and Galanthus sp. De Novo Total Synthesis of 2- epi-Narciclasine. J. Natural Prod. 81, 1451–1459. 10.1021/acs.jnatprod.8b00218 [DOI] [PubMed] [Google Scholar]
- Boss C., Hazemann J., Kimmerlin T., Korff M., Lüthi U., Peter O., et al. (2017). The Screening Compound Collection: A Key Asset for Drug Discovery. Chimia 71, 667–677. 10.2533/chimia.2017.667 [DOI] [PubMed] [Google Scholar]
- Boumaraf M., Carbone M., Ciavatta M. L., Benyahia S., Ameddah S., Menad A., et al. (2017). Exploring the Bioactive Terpenoid Content of an Algerian Plant of the Genus Pulicaria: The ent-Series of Asteriscunolides. J. Natural Prod. 80, 82–89. 10.1021/acs.jnatprod.6b00717 [DOI] [PubMed] [Google Scholar]
- Bousserouel H., Litaudon M., Morleo B., Martin M.-T., Thoison O., Nosjean O., et al. (2005). New biologically active linear triterpenes from the bark of three new-caledonian Cupaniopsis species. Tetrahedron 61, 845–851. 10.1016/j.tet.2004.11.036 [DOI] [Google Scholar]
- Boutin J. A., Nosjean O., Ferry G. (2018). “On the Organization of a Drug Discovery Platform,” in Drug Discovery - Concepts to Market. Ed. Bobbarala V. London, UK InTechOpen Limited. 10.5772/intechopen.73170 [DOI] [Google Scholar]
- Bouvier F., Rahier A., Camara B. (2005). Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44, 357–429. 10.1016/j.plipres.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Brana A. F., Sarmiento-Vizcaino A., Perez-Victoria I., Otero L., Fernandez J., Palacios J. J., et al. (2017). Branimycins B and C, Antibiotics Produced by the Abyssal Actinobacterium Pseudonocardia carboxydivorans M-227. J. Natural Prod. 80, 569–573. 10.1021/acs.jnatprod.6b01107 [DOI] [PubMed] [Google Scholar]
- Breitling R., Takano E. (2016). Synthetic Biology of Natural Products. Cold Spring Harbor Perspect. Biol. 8 a023994, 10.1101/cshperspect.a023994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton R. C., Reynolds W. F. (2013). Using NMR to identify and characterize natural products. Nat. Prod. Rep. 30, 501–524. 10.1039/c2np20104f [DOI] [PubMed] [Google Scholar]
- Bunteang S., Chanakul W., Hongthong S., Kuhakarn C., Chintakovid W., Sungchawek N., et al. (2018). Anti-HIV Activity of Alkaloids from Dasymaschalon echinatum. Natural Prod. Commun. 13, 29–32. 10.1177/1934578X1801300110 [DOI] [Google Scholar]
- Burson R., Moran K. J. (2015). Gliflozins. Home Healthc. Now 33, 281. 10.1097/NHH.0000000000000238 [DOI] [PubMed] [Google Scholar]
- Camero C. M., Vassallo A., Leo M., Temraz A., Tommasi N., Braca A. (2018). Limonoids from Aphanamixis polystachya Leaves and Their Interaction with Hsp90. Planta Med. 84, 964–970. 10.1055/a-0624-9538 [DOI] [PubMed] [Google Scholar]
- Campos P.-E., Wolfender J.-L., Queiroz E. F., Marcourt L., Al-Mourabit A., Frederich M., et al. (2017). Unguiculin A and Ptilomycalins E-H, Antimalarial Guanidine Alkaloids from the Marine Sponge Monanchora unguiculata. J. Natural Prod. 80, 1404–1410. 10.1021/acs.jnatprod.6b01079 [DOI] [PubMed] [Google Scholar]
- Cao Y., Zhao M., Zhu Y., Zhu Z.-H., Oberer L., Duan J.-A. (2017). Diselaginellin B, an Unusual Dimeric Molecule from Selaginella pulvinata, Inhibited Metastasis and Induced Apoptosis of SMMC-7721 Human Hepatocellular Carcinoma Cells. J. Natural Prod. 80, 3151–3158. 10.1021/acs.jnatprod.7b00404 [DOI] [PubMed] [Google Scholar]
- Carbonell P., Currin A., Jervis A. J., Rattray N. J. W., Swainston N., Yan C., et al. (2016). Bioinformatics for the synthetic biology of natural products: integrating across the Design-Build-Test cycle. Nat. Prod. Rep. 33, 925–932. 10.1039/c6np00018e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier C., Queiroz E. F., Marcourt L., Wolfender J.-L., Azelmat J., Grenier D., et al. (2017). Dibenzofurans and Pseudodepsidones from the Lichen Stereocaulon paschale Collected in Northern Quebec. J. Natural Prod. 80, 210–214. 10.1021/acs.jnatprod.6b00831 [DOI] [PubMed] [Google Scholar]
- Casillo A., Lanzetta R., Parrilli M., Corsaro M. M. (2018). Exopolysaccharides from Marine and Marine Extremophilic Bacteria: Structures, Properties, Ecological Roles and Applications. Mar. Drugs 16, 69. 10.3390/md16020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catteau L., Reichmann N., Olson J., Pinho M., Nizet V., van Bambeke F., et al. (2017). Synergy between Ursolic and Oleanolic Acids from Vitellaria paradoxa Leaf Extract and β-Lactams against Methicillin-Resistant Staphylococcus aureus: In Vitro and In Vivo Activity and Underlying Mechanisms. Molecules 22, 2245. 10.3390/molecules22122245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakka N., Andrews K. L., Berry L. M., Bregman H., Gunaydin H., Huang L., et al. (2017). Applications of parallel synthetic lead hopping and pharmacophore-based virtual screening in the discovery of efficient glycine receptor potentiators. Eur. J. Med. Chem. 137, 63–75. 10.1016/j.ejmech.2017.05.036 [DOI] [PubMed] [Google Scholar]
- Chang J.-C., Hsiao G., Lin R.-K., Kuo Y.-H., Ju Y.-M., Lee T.-H. (2017). Bioactive Constituents from the Termite Nest-Derived Medicinal Fungus Xylaria nigripes. J. Natural Prod. 80, 38–44. 10.1021/acs.jnatprod.6b00249 [DOI] [PubMed] [Google Scholar]
- Chapman L. M., Beck J. C., Lacker C. R., Wu L., Reisman S. E. (2018). Evolution of a Strategy for the Enantioselective Total Synthesis of (+)-Psiguadial B. J. Org. Chem. 83, 6066–6085. 10.1021/acs.joc.8b00728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheallaigh A. N., Mansell D. J., Toogood H. S., Tait S., Lygidakis A., Scrutton N. S., et al. (2018). Chemoenzymatic Synthesis of the Intermediates in the Peppermint Monoterpenoid Biosynthetic Pathway. J. Natural Prod. 81, 1546–1552. 10.1021/acs.jnatprod.7b01026 [DOI] [PubMed] [Google Scholar]
- Chen L., Niu S.-B., Li L., Ding G., Yu M., Zhang G.-S., et al. (2017). Trichoderpyrone, a Unique Polyketide Hybrid with a Cyclopentenone-Pyrone Skeleton from the Plant Endophytic Fungus Trichoderma gamsii. J. Natural Prod. 80, 1944–1947. 10.1021/acs.jnatprod.7b00190 [DOI] [PubMed] [Google Scholar]
- Chen M., Shen N.-X., Chen Z.-Q., Zhang F.-M., Chen Y. (2017). Penicilones A-D, Anti-MRSA Azaphilones from the Marine-Derived Fungus Penicillium janthinellum HK1-6. J. Natural Prod. 80, 1081–1086. 10.1021/acs.jnatprod.6b01179 [DOI] [PubMed] [Google Scholar]
- Chen M.-C., Cho T.-Y., Kuo Y.-H., Lee T.-H. (2017). Meroterpenoids from a Medicinal Fungus Antrodia cinnamomea. J. Natural Prod. 80, 2439–2446. 10.1021/acs.jnatprod.7b00223 [DOI] [PubMed] [Google Scholar]
- Cheng B., Volpin G., Morstein J., Trauner D. (2018). Total Synthesis of (+/-)-Exotine B. Org. Lett. 20, 4358–4361. 10.1021/acs.orglett.8b01817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.-B., Liu F.-J., Wang C.-H., Hwang T.-L., Tsai Y.-F., Yen C.-H., et al. (2018). Bioactive Triterpenoids from the Leaves and Twigs of Lithocarpus litseifolius and L. corneus. Planta Med. 84, 49–58. 10.1055/s-0043-113826 [DOI] [PubMed] [Google Scholar]
- Chianese G., Silber J., Luciano P., Merten C., Erpenbeck D., Topaloglu B., et al. (2017). Antiprotozoal Linear Furanosesterterpenoids from the Marine Sponge Ircinia oros. J. Natural Prod. 80, 2566–2571. 10.1021/acs.jnatprod.7b00543 [DOI] [PubMed] [Google Scholar]
- Cho N., Ransom T. T., Sigmund J., Tran T., Cichewicz R. H., Goetz M., et al. (2017). Growth Inhibition of Colon Cancer and Melanoma Cells by Versiol Derivatives from a Paraconiothyrium Species. J. Natural Prod. 80, 2037–2044. 10.1021/acs.jnatprod.7b00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. R., Lee S. Y. (2020). Protocols for RecET-based markerless gene knockout and integration to express heterologous biosynthetic gene clusters in Pseudomonas putida. Microb. Biotechnol. 13, 199–209. 10.1111/1751-7915.13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons P. A., Wilson J. A., Dančík V., Muller S., Carrinski H. A., Wagner B. K., et al. (2011). Quantifying structure and performance diversity for sets of small molecules comprising small-molecule screening collections. Proc. Natl. Acad. Sci. U.S.A. 108, 6817–6822. 10.1073/pnas.1015024108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columba-Palomares M. F. M. C., Villareal D. M., Acevedo Quiroz M. C. M. E., Marquina Bahena M. C. S., Álvarez Berber D. L., Rodríguez-López D. V. (2015). Anti-inflammatory and cytotoxic activities of Bursera copallifera. Phcog. Mag. 11, 322. 10.4103/0973-1296.166067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia A. F., Segovia J. F. O., Gonçalves M. C. A., Oliveira V. L., Silveira D., Carvalho J. C. T., et al. (2008). Amazonian plant crude extract screening for activity against multidrug-resistant bacteria. Eur. Rev. Med. Pharmacol. Sci. 12, 369–380. [PubMed] [Google Scholar]
- Cragg G. M., Newman D. J., Snader K. M. (1997). Natural products in drug discovery and development. J. Natural Prod. 60, 52–60. 10.1021/np9604893 [DOI] [PubMed] [Google Scholar]
- Da Silva P. P. J., Meijer L. (2012). Recherche de substances naturelles à activité thérapeutique: Pierre Potier, (1934-2006). Med. Sci. : M/S 28, 534–542. 10.1051/medsci/2012285020 [DOI] [PubMed] [Google Scholar]
- Dai J., Wang G., Li W., Zhang L., Yang J., Zhao X., et al. (2012). High-Throughput Screening for Anti–Influenza A Virus Drugs and Study of the Mechanism of Procyanidin on Influenza A Virus–Induced Autophagy. J. Biomol. Screen 17, 605–617. 10.1177/1087057111435236 [DOI] [PubMed] [Google Scholar]
- Dai Z., Wang B., Liu Y., Shi M., Wang D., Zhang X., et al. (2014). Producing aglycons of ginsenosides in bakers’ yeast. Sci. Rep. 4, 3698. 10.1038/srep03698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang P. H., Nguyen H. X., Duong T. T. T., Tran T. K. T., Nguyen P. T., Vu T. K. T., et al. (2017. a). alpha-Glucosidase Inhibitory and Cytotoxic Taxane Diterpenoids from the Stem Bark of Taxus wallichiana. J. Natural Prod. 80, 1087–1095. 10.1021/acs.jnatprod.7b00006 [DOI] [PubMed] [Google Scholar]
- Dang P. H., Nguyen H. X., Nguyen H. H. T., Vo T. D., Le T. H., Phan T. H. N., et al. (2017. b). Lignans from the Roots of Taxus wallichiana and Their alpha-Glucosidase Inhibitory Activities. J. Natural Prod. 80, 1876–1882. 10.1021/acs.jnatprod.7b00171 [DOI] [PubMed] [Google Scholar]
- Davison E. K., Hume P. A., Sperry J. (2018). Total Synthesis of an Isatis indigotica-Derived Alkaloid Using a Biomimetic Thio-Diels-Alder Reaction. Org. Lett. 20, 3545–3548. 10.1021/acs.orglett.8b01321 [DOI] [PubMed] [Google Scholar]
- Dethe D. H., Dherange B. D. (2018). Total Synthesis of Adunctin B. J. Org. Chem. 83, 3392–3396. 10.1021/acs.joc.8b00015 [DOI] [PubMed] [Google Scholar]
- Deyou T., Marco M., Heydenreich M., Pan F., Gruhonjic A., Fitzpatrick P. A., et al. (2017). Isoflavones and Rotenoids from the Leaves of Millettia oblata ssp. teitensis. J. Natural Prod. 80, 2060–2066. 10.1021/acs.jnatprod.7b00255 [DOI] [PubMed] [Google Scholar]
- Dissanayake A. A., Ameen B. A. H., Nair M. G. (2017). Lipid Peroxidation and Cyclooxygenase Enzyme Inhibitory Compounds from Prangos haussknechtii. J. Natural Prod. 80, 2472–2477. 10.1021/acs.jnatprod.7b00322 [DOI] [PubMed] [Google Scholar]
- Do Q. T., Medina-Franco J. L., Scior T., Bernard P. (2015). How to Valorize Biodiversity? Let’s Go Hashing, Extracting, Filtering, Mining, Fishing. Planta Med. 81, 436–449. 10.1055/s-0034-1396314 [DOI] [PubMed] [Google Scholar]
- Do L. T. M., Aree T., Siripong P., Vo N. T., Nguyen T. T. A., Nguyen P. K. P., et al. (2018). Cytotoxic Flavones from the Stem Bark of Bougainvillea spectabilis Willd. Planta Med. 84, 129–134. 10.1055/s-0043-118102 [DOI] [PubMed] [Google Scholar]
- Dong S., Li B., Dai W., Wang D., Qin Y., Zhang M. (2017). Sesqui- and Diterpenoids from the Radix of Curcuma aromatica. J. Natural Prod. 80, 3093–3102. 10.1021/acs.jnatprod.6b01100 [DOI] [PubMed] [Google Scholar]
- Du Y., Pearce K. C., Dai Y., Krai P., Dalal S., Cassera M. B., et al. (2017). Antiplasmodial Sesquiterpenoid Lactones from Trichospira verticillata: Structure Elucidation by Spectroscopic Methods and Comparison of Experimental and Calculated ECD Data. J. Natural Prod. 80, 1639–1647. 10.1021/acs.jnatprod.7b00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y.-T., Zhang J., Lao Y.-Z., Tan H.-S., Ye Y.-S., Yang X.-W., et al. (2018). Spirocyclic polycyclic polyprenylated acylphloroglucinols from the ethyl acetate fraction of Hypericum henryi. Tetrahedron Lett. 59, 4067–4072. 10.1016/j.tetlet.2018.09.071 [DOI] [Google Scholar]
- Dufresne C. (2000). The planning and establishment of a sample preparation laboratory for drug discovery. J. Autom. Methods Manage. Chem. 22, 175–179. 10.1155/S1463924600000316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibl R., Meier P., Stutz I., Schildberger D., Hühn T., Eibl D. (2018). Plant cell culture technology in the cosmetics and food industries: current state and future trends. Appl. Microbiol. Biotechnol. 102, 8661–8675. 10.1007/s00253-018-9279-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Desoky A. H., Kato H., Kagiyama I., Hitora Y., Losung F., Mangindaan R. E. P., et al. (2017). Ceylonins A-F, Spongian Diterpene Derivatives That Inhibit RANKL-Induced Formation of Multinuclear Osteoclasts, from the Marine Sponge Spongia ceylonensis. J. Natural Prod. 80, 90–95. 10.1021/acs.jnatprod.6b00725 [DOI] [PubMed] [Google Scholar]
- El-Elimat T., Figueroa M., Ehrmann B. M., Cech N. B., Pearce C. J., Oberlies N. H. (2013). High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Natural Prod. 76, 1709–1716. 10.1021/np4004307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Kasahara T., Harada K., Kubo M., Etoh T., Ishibashi M., et al. (2017). Sucupiranins A-L, Furanocassane Diterpenoids from the Seeds of Bowdichia virgilioides. J. Natural Prod. 80, 3120–3127. 10.1021/acs.jnatprod.7b00249 [DOI] [PubMed] [Google Scholar]
- Erlanson D. A., Fesik S. W., Hubbard R. E., Jahnke W., Jhoti H. (2016). Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discovery 15, 605–619. 10.1038/nrd.2016.109 [DOI] [PubMed] [Google Scholar]
- Esposito M., Nothias L.-F., Retailleau P., Costa J., Roussi F., Neyts J., et al. (2017). Isolation of Premyrsinane, Myrsinane, and Tigliane Diterpenoids from Euphorbia pithyusa Using a Chikungunya Virus Cell-Based Assay and Analogue Annotation by Molecular Networking. J. Natural Prod. 80, 2051–2059. 10.1021/acs.jnatprod.7b00233 [DOI] [PubMed] [Google Scholar]
- Fan Y.-Y., Xu J.-B., Liu H.-C., Gan L.-S., Ding J., Yue J.-M. (2017). Cephanolides A-J, Cephalotane-Type Diterpenoids from Cephalotaxus sinensis. J. Natural Prod. 80, 3159–3166. 10.1021/acs.jnatprod.7b00412 [DOI] [PubMed] [Google Scholar]
- Feher M., Schmidt J. M. (2003). Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 43, 218–227. 10.1021/ci0200467 [DOI] [PubMed] [Google Scholar]
- Feng Z.-L., Zhang L.-L., Zheng Y.-D., Liu Q.-Y., Liu J.-X., Feng L., et al. (2017). Norditerpenoids and Dinorditerpenoids from the Seeds of Podocarpus nagi as Cytotoxic Agents and Autophagy Inducers. J. Natural Prod. 80, 2110–2117. 10.1021/acs.jnatprod.7b00347 [DOI] [PubMed] [Google Scholar]
- Fernie A. R. (2007). The future of metabolic phytochemistry: larger numbers of metabolites, higher resolution, greater understanding. Phytochemistry 68, 2861–2880. 10.1016/j.phytochem.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Ferrer S., Echavarren A. M. (2018). Total Synthesis of Repraesentin F and Configuration Reassignment by a Gold(I)-Catalyzed Cyclization Cascade. Org. Lett. 20, 5784–5788. 10.1021/acs.orglett.8b02478 [DOI] [PubMed] [Google Scholar]
- Figueroa-López A. M., Cordero-Ramírez J. D., Quiroz-Figueroa F. R., Maldonado-Mendoza I. E. (2014). A high-throughput screening assay to identify bacterial antagonists against Fusarium verticillioides. J. Basic Microbiol. 54, S125–S133. 10.1002/jobm.201200594 [DOI] [PubMed] [Google Scholar]
- Firn R.D., Jones C. G. (2003). Natural products ? a simple model to explain chemical diversity. Nat. Prod. Rep. 20, 382. 10.1039/b208815k [DOI] [PubMed] [Google Scholar]
- Flores-Bocanegra L., Gonzalez-Andrade M., Bye R., Linares E., Mata R. (2017). alpha-Glucosidase Inhibitors from Salvia circinata. J. Natural Prod. 80, 1584–1593. 10.1021/acs.jnatprod.7b00155 [DOI] [PubMed] [Google Scholar]
- Flores-Giubi M. E., Duran-Pena M. J., Botubol-Ares J. M., Escobar-Montano F., Zorrilla D., Macias-Sanchez A. J., et al. (2017). Gaditanone, a Diterpenoid Based on an Unprecedented Carbon Skeleton Isolated from Euphorbia gaditana. J. Natural Prod. 80, 2161–2165. 10.1021/acs.jnatprod.7b00332 [DOI] [PubMed] [Google Scholar]
- Fois B., Bianco G., Sonar V. P., Distinto S., Maccioni E., Meleddu R., et al. (2017). Phenylpropenoids from Bupleurum fruticosum as Anti-Human Rhinovirus Species A Selective Capsid Binders. J. Natural Prod. 80, 2799–2806. 10.1021/acs.jnatprod.7b00648 [DOI] [PubMed] [Google Scholar]
- Fuentes P., Zhou F., Erban A., Karcher D., Kopka J., Bock R. (2016). A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. Elife 5, e13664. 10.7554/eLife.13664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galappathie S., Edwards D. J., Elliott A. G., Cooper M. A., Palombo E. A., Butler M. S., et al. (2017). Antibacterial Nerol Cinnamates from the Australian Plant Eremophila longifolia. J. Natural Prod. 80, 1178–1181. 10.1021/acs.jnatprod.6b00888 [DOI] [PubMed] [Google Scholar]
- Gally J.-M., Bourg S., Do Q.-T., Aci-Sèche S., Bonnet P. (2017). VSPrep: A General KNIME Workflow for the Preparation of Molecules for Virtual Screening. Mol. Inform. 36, 10.1002. 10.1002/minf.201700023 [DOI] [PubMed] [Google Scholar]
- Gan P., Pitzen J., Qu P., Snyder S. A. (2018). Total Synthesis of the Caged Indole Alkaloid Arboridinine Enabled by aza-Prins and Metal-Mediated Cyclizations. J. Am. Chem. Soc. 140, 919–925. 10.1021/jacs.7b07724 [DOI] [PubMed] [Google Scholar]
- Gao J., Aisa H. A. (2017). Terpenoids from Euphorbia soongarica and Their Multidrug Resistance Reversal Activity. J. Natural Prod. 80, 1767–1775. 10.1021/acs.jnatprod.6b01099 [DOI] [PubMed] [Google Scholar]
- Gao E., Zhou Z.-Q., Zou J., Yu Y., Feng X.-L., Chen G.-D., et al. (2017). Bioactive Asarone-Derived Phenylpropanoids from the Rhizome of Acorus tatarinowii Schott. J. Natural Prod. 80, 2923–2929. 10.1021/acs.jnatprod.7b00457 [DOI] [PubMed] [Google Scholar]
- Geng H., Huang P.-Q. (2019). Rapid Generation of Molecular Complexity by Chemical Synthesis: Highly Efficient Total Synthesis of Hexacyclic Alkaloid (-)-Chaetominine and Its Biosynthetic Implications. Chem. Rec. (New York N.Y.) 19, 523–533. 10.1002/tcr.201800079 [DOI] [PubMed] [Google Scholar]
- Geng C.-A., Huang X.-Y., Ma Y.-B., Hou B., Li T.-Z., Zhang X.-M., et al. (2017). (+/-)-Uncarilins A and B, Dimeric Isoechinulin-Type Alkaloids from Uncaria rhynchophylla. J. Natural Prod. 80, 959–964. 10.1021/acs.jnatprod.6b00938 [DOI] [PubMed] [Google Scholar]
- Geny C., Abou Samra A., Retailleau P., Iorga B. I., Nedev H., Awang K., et al. (2017). (+)- and (-)-Ecarlottones, Uncommon Chalconoids from Fissistigma latifolium with Pro-Apoptotic Activity. J. Natural Prod. 80, 3179–3185. 10.1021/acs.jnatprod.7b00494 [DOI] [PubMed] [Google Scholar]
- Gerlinger C. K. G., Kruger S., Gaich T. (2018). Total Synthesis of Parvineostemonine by Structure Pattern Recognition: A Unified Approach to Stemona and Sarpagine Alkaloids. Chem. (Weinheim an der Bergstrasse Germany) 24, 3994–3997. 10.1002/chem.201800365 [DOI] [PubMed] [Google Scholar]
- Glaab E. (2016). Building a virtual ligand screening pipeline using free software: A survey. Briefings Bioinf. 17, 352–366. 10.1093/bib/bbv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobu F.-R., Chen J.-J., Zeng J., Wei W.-J., Wang W.-F., Lin C.-J., et al. (2017). Isolation, Structure Elucidition, and Immunosuppressive Activity of Diterpenoids from Ligularia fischeri. J. Natural Prod. 80, 2263–2268. 10.1021/acs.jnatprod.7b00198 [DOI] [PubMed] [Google Scholar]
- Gomes N. G. M., Pereira D. M., Valentão P., Andrade P. B. (2018). Hybrid MS/NMR methods on the prioritization of natural products: Applications in drug discovery. J. Pharm. Biomed. Anal. 147, 234–249. 10.1016/j.jpba.2017.07.035 [DOI] [PubMed] [Google Scholar]
- Grabowski K., Schneider G. (2007). Properties and Architecture of Drugs and Natural Products Revisited. Curr. Chem. Biol. 1, 115–127. 10.2174/187231307779814066 [DOI] [Google Scholar]
- Grabowski K., Baringhaus K.-H., Schneider G. (2008). Scaffold diversity of natural products: inspiration for combinatorial library design. Nat. Prod. Rep. 25, 892–904. 10.1039/b715668p [DOI] [PubMed] [Google Scholar]
- Graves P. R., Kwiek J. J., Fadden P., Ray R., Hardeman K., Coley A. M., et al. (2002). Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol. Pharmacol. 62, 1364–1372. 10.1124/mol.62.6.1364 [DOI] [PubMed] [Google Scholar]
- Gu J., Gui Y., Chen L., Yuan G., Lu H.-Z., Xu X. (2013). Use of natural products as chemical library for drug discovery and network pharmacology. PloS One 8, e62839. 10.1371/journal.pone.0062839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero R. O., Guzman A. L. (1998). Inhibition of xanthine oxidase by Puerto Rican plant extracts. Puerto Rico Health Sci. J. 17, 359–364. [PubMed] [Google Scholar]
- Gui C., Zhang S., Zhu X., Ding W., Huang H., Gu Y.-C., et al. (2017). Antimicrobial Spirotetronate Metabolites from Marine-Derived Micromonospora harpali SCSIO GJ089. J. Natural Prod. 80, 1594–1603. 10.1021/acs.jnatprod.7b00176 [DOI] [PubMed] [Google Scholar]
- Gunatilaka A. A. L. (2006). Natural products from plant-associated microorganisms: distribution, structural diversity, bioactivity, and implications of their occurrence. J. Natural Prod. 69, 509–526. 10.1021/np058128n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zhang N., Chen C., Huang J., Li X.-N., Liu J., et al. (2017). Tricyclic Polyprenylated Acylphloroglucinols from St John’s Wort, Hypericum perforatum. J. Natural Prod. 80, 1493–1504. 10.1021/acs.jnatprod.6b01178 [DOI] [PubMed] [Google Scholar]
- Guo X., Shen H., Liu Y., Wang Q., Wang X., Peng C., et al. (2019). Enabling Heterologous Synthesis of Lupulones in the Yeast Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 188, 787–797. 10.1007/s12010-019-02957-8 [DOI] [PubMed] [Google Scholar]
- Hann M. M., Leach A. R., Harper G. (2001). Molecular complexity and its impact on the probability of finding leads for drug discovery. J. Chem. Inf. Comput. Sci. 41, 856–864. 10.1021/ci000403i [DOI] [PubMed] [Google Scholar]
- Harvey A. L., Cree I. A. (2010). High-throughput screening of natural products for cancer therapy. Planta Med. 76, 1080–1086. 10.1055/s-0030-1250162 [DOI] [PubMed] [Google Scholar]
- Harvey A. L., Edrada-Ebel R., Quinn R. J. (2015). The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discovery 14, 111–129. 10.1038/nrd4510 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S. (2018). Stereocontrolled Total Synthesis of Biologically Active Natural Products. Yakugaku Zasshi : J. Pharm. Soc. Jpn. 138, 191–209. 10.1248/yakushi.17-00187 [DOI] [PubMed] [Google Scholar]
- He Y., Hu Z., Li Q., Huang J., Li X.-N., Zhu H., et al. (2017). Bioassay-Guided Isolation of Antibacterial Metabolites from Emericella sp. TJ29. J. Natural Prod. 80, 2399–2405. 10.1021/acs.jnatprod.7b00077 [DOI] [PubMed] [Google Scholar]
- He X., Wang S., Shi J., Sun Z., Lei Z., Yin Z., et al. (2018). Genotypic and Environmental Effects on the Volatile Chemotype of Valeriana jatamansi Jones. Front. Plant Sci. 9, 61. 10.3389/fpls.2018.01003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinamaki J., Pirttimaa M. M., Alakurtti S., Pitkanen H. P., Kanerva H., Hulkko J., et al. (2017). Suberin Fatty Acids from Outer Birch Bark: Isolation and Physical Material Characterization. J. Natural Prod. 80, 916–924. 10.1021/acs.jnatprod.6b00771 [DOI] [PubMed] [Google Scholar]
- Heinrich M., Barnes J., Prieto-Garcia J., Gibbons S., Williamson E. (2018). Fundamentals of Pharmacognosy and Phytotherapy. 3rd Edition (London: Elsevier Health Sciences; )9780702070075. [Google Scholar]
- Hendra R., Keller P. A. (2017). Phytochemical Studies on Two Australian Anigozanthos Plant Species. J. Natural Prod. 80, 2141–2145. 10.1021/acs.jnatprod.7b00063 [DOI] [PubMed] [Google Scholar]
- Henkel T., Brunne R. M., Müller H., Reichel F. (1999). Statistical Investigation into the Structural Complementarity of Natural Products and Synthetic Compounds. Angew. Chem. Int. Ed. Engl. 38, 643–647. [DOI] [PubMed] [Google Scholar]
- Henz Ryen A., Backlund A. (2019). Charting Angiosperm Chemistry: Evolutionary Perspective on Specialized Metabolites Reflected in Chemical Property Space. J. Nat. Prod. (Gorakhpur) 82, 798–812. 10.1021/acs.jnatprod.8b00767 [DOI] [PubMed] [Google Scholar]
- Herraiz T., Guillen H. (2018). Monoamine Oxidase-A Inhibition and Associated Antioxidant Activity in Plant Extracts with Potential Antidepressant Actions. BioMed. Res. Int. 2018, 4810394. 10.1155/2018/4810394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien T. T. T., Quang T. H., Tai B. H., Nhiem N. X., Yen P. H., Yen D. T. H., et al. (2018). Iridoid Glycosides and Phenolic Glycosides from Buddleja asiatica with Anti-inflammatory and Cytoprotective Activities. Natural Prod. Commun. 13, 1–4. 10.1177/1934578X1801300102 [DOI] [Google Scholar]
- Hong M. J., Kim J. (2017). Determination of the Absolute Configuration of Khellactone Esters from Peucedanum japonicum Roots. J. Natural Prod. 80, 1354–1360. 10.1021/acs.jnatprod.6b00947 [DOI] [PubMed] [Google Scholar]
- Hong J. (2011). Role of natural product diversity in chemical biology. Curr. Opin. Chem. Biol. 15, 350–354. 10.1016/j.cbpa.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath D., Lisurek M., Rupp B., Kühne R., Specker E., Kries J., et al. (2014). Design of a general-purpose European compound screening library for EU-OPENSCREEN. ChemMedChem 9, 2309–2326. 10.1002/cmdc.201402126 [DOI] [PubMed] [Google Scholar]
- Hou J.-Q., Guo C., Zhao J.-J., Dong Y.-Y., Hu X.-L., He Q.-W., et al. (2017). Anti-inflammatory Meroterpenoids from Baeckea frutescens. J. Natural Prod. 80, 2204–2214. 10.1021/acs.jnatprod.7b00042 [DOI] [PubMed] [Google Scholar]
- Hsiao G., Chi W.-C., Pang K.-L., Chen J.-J., Kuo Y.-H., Wang Y.-K., et al. (2017). Hirsutane-Type Sesquiterpenes with Inhibitory Activity of Microglial Nitric Oxide Production from the Red Alga-Derived Fungus Chondrostereum sp. NTOU4196. J. Natural Prod. 80, 1615–1622. 10.1021/acs.jnatprod.7b00196 [DOI] [PubMed] [Google Scholar]
- Hu Y., Gupta-Ostermann D., Bajorath J. (2014). Exploring Compound Promiscuity Patterns and Multi-Target Activity Spaces. Comput. Struct. Biotechnol. J. 9 ,e201401003. 10.5936/csbj.201401003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.-X., Liu M., Wang W.-G., Li X.-N., Hu K., Li X.-R., et al. (2018). 7alpha,20-Epoxy-ent-kaurane Diterpenoids from the Aerial Parts of Isodon pharicus. J. Natural Prod. 81, 106–116. 10.1021/acs.jnatprod.7b00723 [DOI] [PubMed] [Google Scholar]
- Huang H., Wu D., Tian W.-X., Ma X.-F., Wu X.-D. (2008). Antimicrobial effect by extracts of rhizome of Alpinia officinarum Hance may relate to its inhibition of β-ketoacyl-ACP reductase. J. Enzyme Inhib. Med. Chem. 23, 362–368. 10.1080/14756360701622099 [DOI] [PubMed] [Google Scholar]
- Huang W.-Y., Wang X.-N., Wang J., Sui Z.-Q. (2018). Malvidin and its glycosides from vaccinium ashei improve endothelial function by anti-inflammatory and angiotensin I-converting enzyme inhibitory effects. Natural Prod. Commun. 13, 49–52. 10.1177/1934578X1801300115 [DOI] [Google Scholar]
- Huh J., Ha T. K. Q., Kang K. B., Kim K. H., Oh W. K., Kim J., et al. (2017). C-Methylated Flavonoid Glycosides from Pentarhizidium orientale Rhizomes and Their Inhibitory Effects on the H1N1 Influenza Virus. J. Natural Prod. 80, 2818–2824. 10.1021/acs.jnatprod.7b00677 [DOI] [PubMed] [Google Scholar]
- Ifie I., Marshall L. J., Ho P., Williamson G. (2016). Hibiscus sabdariffa (Roselle) Extracts and Wine: Phytochemical Profile, Physicochemical Properties, and Carbohydrase Inhibition. J. Agric. Food Chem. 64, 4921–4931. 10.1021/acs.jafc.6b01246 [DOI] [PubMed] [Google Scholar]
- Ikram N. K. B. K., Simonsen H. T. (2017). A Review of Biotechnological Artemisinin Production in Plants. Front. Plant Sci. 8, 1966. 10.3389/fpls.2017.01966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imseng N., Schillberg S., Schürch C., Schmid D., Schütte K., Gorr G., et al. (2014). ““Suspension Culture of Plant Cells Under Heterotrophic Conditions,” in Industrial scale suspension culture of living cells. Eds. Meyer H.-P., Schmidhalter D. R. (Weinheim, Germany: Wiley Blackwell; ), 224–258. [Google Scholar]
- Irwin J. A. (2010). Extremophiles and their application to veterinary medicine. Environ. Technol. 31, 857–869. 10.1080/09593330.2010.484073 [DOI] [PubMed] [Google Scholar]
- Ishibashi M., Ohtsuki T. (2008). Studies on search for bioactive natural products targeting TRAIL signaling leading to tumor cell apoptosis. Med. Res. Rev. 28, 688–714. 10.1002/med.20123 [DOI] [PubMed] [Google Scholar]
- Issac M., Aknin M., Gauvin-Bialecki A., Voogd N., Ledoux A., Frederich M., et al. (2017). Cyclotheonellazoles A-C, Potent Protease Inhibitors from the Marine Sponge Theonella aff. swinhoei. J. Natural Prod. 80, 1110–1116. 10.1021/acs.jnatprod.7b00028 [DOI] [PubMed] [Google Scholar]
- Iwasaki A., Shiota I., Sumimoto S., Matsubara T., Sato T., Suenaga K. (2017). Kohamamides A, B, and C, Cyclic Depsipeptides from an Okeania sp. Marine Cyanobacterium. J. Natural Prod. 80, 1948–1952. 10.1021/acs.jnatprod.7b00256 [DOI] [PubMed] [Google Scholar]
- Jack B. U., Malherbe C. J., Willenburg E. L., Beer D., Huisamen B., Joubert E., et al. (2018). Polyphenol-Enriched Fractions of Cyclopia intermedia Selectively Affect Lipogenesis and Lipolysis in 3T3-L1 Adipocytes. Planta Med. 84, 100–110. 10.1055/s-0043-119463 [DOI] [PubMed] [Google Scholar]
- Jaeger D., Ndi C. P., Crocoll C., Simpson B. S., Khakimov B., Guzman-Genuino R. M., et al. (2017). Isolation and Structural Characterization of Echinocystic Acid Triterpenoid Saponins from the Australian Medicinal and Food Plant Acacia ligulata. J. Natural Prod. 80, 2692–2698. 10.1021/acs.jnatprod.7b00437 [DOI] [PubMed] [Google Scholar]
- Jalencas X., Mestres J. (2013). On the origins of drug polypharmacology. Medchemcomm 4, 80–87. 10.1039/C2MD20242E [DOI] [Google Scholar]
- Jang H.-J., Lee S., Lee S.-J., Lim H.-J., Jung K., Kim Y. H., et al. (2017). Anti-inflammatory Activity of Eudesmane-Type Sesquiterpenoids from Salvia plebeia. J. Natural Prod. 80, 2666–2676. 10.1021/acs.jnatprod.7b00326 [DOI] [PubMed] [Google Scholar]
- Jang J.-P., Nogawa T., Futamura Y., Shimizu T., Hashizume D., Takahashi S., et al. (2017). Octaminomycins A and B, Cyclic Octadepsipeptides Active against Plasmodium falciparum. J. Natural Prod. 80, 134–140. 10.1021/acs.jnatprod.6b00758 [DOI] [PubMed] [Google Scholar]
- Jayanetti D. R., Li Y., Bartholomeusz G. A., Bills G. F., Gloer J. B. (2017). Benzophenone and Fimetarone Derivatives from the Coprophilous Fungus Delitschia confertaspora. J. Natural Prod. 80, 707–712. 10.1021/acs.jnatprod.6b01091 [DOI] [PubMed] [Google Scholar]
- Jian J., Fan J., Yang H., Lan P., Li M., Liu P., et al. (2018). Total Synthesis of the Flavonoid Natural Product Houttuynoid A. J. Natural Prod. 81, 371–377. 10.1021/acs.jnatprod.7b00791 [DOI] [PubMed] [Google Scholar]
- Jiang B., Zhao W., Li S., Liu H., Yu L., Zhang Y., et al. (2017). Cytotoxic Dibohemamines D-F from a Streptomyces Species. J. Natural Prod. 80, 2825–2829. 10.1021/acs.jnatprod.7b00136 [DOI] [PubMed] [Google Scholar]
- Jiang H., Zhang G.-J., Liu Y.-F., Wang H.-S., Liang D. (2017). Clerodane Diterpenoid Glucosides from the Stems of Tinospora sinensis. J. Natural Prod. 80, 975–982. 10.1021/acs.jnatprod.6b00976 [DOI] [PubMed] [Google Scholar]
- Jiang H.-Y., Wang W.-G., Tang J.-W., Liu M., Li X.-R., Hu K., et al. (2017). Structurally Diverse Diterpenoids from Isodon scoparius and Their Bioactivity. J. Natural Prod. 80, 2026–2036. 10.1021/acs.jnatprod.7b00163 [DOI] [PubMed] [Google Scholar]
- Jiang L., Liu X., Yuan P., Zhang Y., Chen X. (2017). Stereoselective Synthesis of (+)-Annuionone A and (-)-Annuionone B. J. Natural Prod. 80, 805–812. 10.1021/acs.jnatprod.6b00522 [DOI] [PubMed] [Google Scholar]
- Jin J. B., Cai B., Zhou J.-M. (2017). “Salicylic acid,” in Hormone Metabolism and Signaling in Plants (London: Academic Press; ), 273–289. [Google Scholar]
- Jin Y., Fan J.-T., Gu X.-L., Zhang L.-Y., Han J., Du S.-H., et al. (2017). Neuroprotective Activity of Cerebrosides from Typhonium giganteum by Regulating Caspase-3 and Bax/Bcl-2 Signaling Pathways in PC12 Cells. J. Natural Prod. 80, 1734–1741. 10.1021/acs.jnatprod.6b00954 [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Ree H., Hartmann M., Lang L., Sawano S., Sarpong R. (2019). Total Synthesis of Pentacyclic (-)-Ambiguine P Using Sequential Indole Functionalizations. J. Am. Chem. Soc. 141, 2233–2237. 10.1021/jacs.8b13388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwiak A., Ples M., Skorupinska-Tudek K., Kania M., Dydak M., Danikiewicz W., et al. (2013). Sugar availability modulates polyisoprenoid and phytosterol profiles in Arabidopsis thaliana hairy root culture. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1831, 438–447. 10.1016/j.bbalip.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Kalansuriya P., Quezada M., Esposito B. P., Capon R. J. (2017). Talarazines A-E: Noncytotoxic Iron(III) Chelators from an Australian Mud Dauber Wasp-Associated Fungus, Talaromyces sp. (CMB-W045). J. Natural Prod. 80, 609–615. 10.1021/acs.jnatprod.6b00889 [DOI] [PubMed] [Google Scholar]
- Kang K. B., Kim H. W., Kim J. W., Oh W. K., Kim J., Sung S. H. (2017. a). Catechin-Bound Ceanothane-Type Triterpenoid Derivatives from the Roots of Zizyphus jujuba. J. Natural Prod. 80, 1048–1054. 10.1021/acs.jnatprod.6b01103 [DOI] [PubMed] [Google Scholar]
- Kang K. B., Park E. J., Kim J., Sung S. H. (2017. b). Berchemiosides A-C, 2-Acetoxy-omega-phenylpentaene Fatty Acid Triglycosides from the Unripe Fruits of Berchemia berchemiifolia. J. Natural Prod. 80, 2778–2786. 10.1021/acs.jnatprod.7b00602 [DOI] [PubMed] [Google Scholar]
- Kang S.-Y., Heo K. T., Hong Y.-S. (2018). Optimization of Artificial Curcumin Biosynthesis in E. coli by Randomized 5′-UTR Sequences To Control the Multienzyme Pathway. ACS Synth. Biol. 7, 2054–2062. 10.1021/acssynbio.8b00198 [DOI] [PubMed] [Google Scholar]
- Kant R., Yen C.-H., Lu C.-K., Lin Y.-C., Li J.-H., Chen Y.-M. (2016). Identification of 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside as a Glycine N-Methyltransferase Enhancer by High-Throughput Screening of Natural Products Inhibits Hepatocellular Carcinoma. IJMS 17, 669. 10.3390/ijms17050669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karker M., Tommasi N., Smaoui A., Abdelly C., Ksouri R., Braca A. (2016). New Sulphated Flavonoids from Tamarix africana and Biological Activities of Its Polar Extract. Planta Med. 82, 1374–1380. 10.1055/s-0042-111520 [DOI] [PubMed] [Google Scholar]
- Kauhl U., Andernach L., Opatz T. (2018). Total Synthesis of epi-Trichosetin. J. Org. Chem. 83, 15170–15177. 10.1021/acs.joc.8b02450 [DOI] [PubMed] [Google Scholar]
- Kayani W. K., Kiani B. H., Dilshad E., Mirza B. (2018). Biotechnological approaches for artemisinin production in Artemisia. World J. Microbiol. Biotechnol. 34, 2542. 10.1007/s11274-018-2432-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R., Quradha M. M., Saif A. Q., Ali J., Rauf A., Khan A. (2014). Comparative urease enzyme inhibition profile of leaves and stems of Rumex nervosus vahl. Natural Prod. Res. 28, 2355–2357. 10.1080/14786419.2014.940346 [DOI] [PubMed] [Google Scholar]
- Khan H., Saeed M., Muhammad N., Gaffar R., Gul F., Raziq N. (2015). Lipoxygenase and urease inhibition of the aerial parts of the Polygonatum verticillatum. Toxicol. Ind. Health 31, 758–763. 10.1177/0748233713483197 [DOI] [PubMed] [Google Scholar]
- Khan T., Abbasi B. H., Khan M. A., Shinwari Z. K. (2016). Differential Effects of Thidiazuron on Production of Anticancer Phenolic Compounds in Callus Cultures of Fagonia indica. Appl. Biochem. Biotechnol. 179, 46–58. 10.1007/s12010-016-1978-y [DOI] [PubMed] [Google Scholar]
- Khanpour-Ardestani N., Sharifi M., Behmanesh M. (2015). Establishment of callus and cell suspension culture of Scrophularia striata Boiss. an in vitro approach for acteoside production. Cytotechnology 67, 475–485. 10.1007/s10616-014-9705-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil Y.-S., Kim S. M., Kang U., Chung H. Y., Seo E. K. (2017). Peroxynitrite-Scavenging Glycosides from the Stem Bark of Catalpa ovata. J. Natural Prod. 80, 2240–2251. 10.1021/acs.jnatprod.7b00139 [DOI] [PubMed] [Google Scholar]
- Kim S.-I., Yang E.-J., Park S.-H., Kim C.-K., Song K.-S. (2014). A β-secretase (BACE1)-inhibiting C-methylrotenoid induced by yeast elicitation in Abronia nana suspension cultures. Appl. Biochem. Biotechnol. 172, 3529–3537. 10.1007/s12010-014-0762-0 [DOI] [PubMed] [Google Scholar]
- Kim C.-K., Riswanto R., Won T. H., Kim H., Elya B., Sim C. J., et al. (2017). Manzamine Alkaloids from an Acanthostrongylophora sp. Sponge. J. Natural Prod. 80, 1575–1583. 10.1021/acs.jnatprod.7b00121 [DOI] [PubMed] [Google Scholar]
- Kim K., Kim J.-J., Jung Y., Noh J.-Y., Syed A. S., Kim C. Y., et al. (2017). Cyclocurcumin, an Antivasoconstrictive Constituent of Curcuma longa (Turmeric). J. Natural Prod. 80, 196–200. 10.1021/acs.jnatprod.6b00331 [DOI] [PubMed] [Google Scholar]
- Kim C. S., Bae M., Oh J., Subedi L., Suh W. S., Choi S. Z., et al. (2017. a). Anti-Neurodegenerative Biflavonoid Glycosides from Impatiens balsamina. J. Natural Prod. 80, 471–478. 10.1021/acs.jnatprod.6b00981 [DOI] [PubMed] [Google Scholar]
- Kim C. S., Subedi L., Oh J., Kim S. Y., Choi S. U., Lee K. R. (2017. b). Bioactive Triterpenoids from the Twigs of Chaenomeles sinensis. J. Natural Prod. 80, 1134–1140. 10.1021/acs.jnatprod.7b00111 [DOI] [PubMed] [Google Scholar]
- Kim G., Kim M. J., Chung G., Lee H.-Y., Han S. (2018). (+)-Dimericbiscognienyne A: Total Synthesis and Mechanistic Investigations of the Key Heterodimerization. Org. Lett. 20, 6886–6890. 10.1021/acs.orglett.8b03025 [DOI] [PubMed] [Google Scholar]
- Kimura S., Saito N. (2018). Construction of the Pentacyclic Core and Formal Total Synthesis of (rac)-Renieramycin T. ChemistryOpen 7, 764–771. 10.1002/open.201800112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnel R. B., Esquenazi E., Leao T., Moss N., Mevers E., Pereira A. R., et al. (2017). A Maldiisotopic Approach to Discover Natural Products: Cryptomaldamide, a Hybrid Tripeptide from the Marine Cyanobacterium Moorea producens. J. Natural Prod. 80, 1514–1521. 10.1021/acs.jnatprod.7b00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J., Keasling J. D. (2009). Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu. Rev. Plant Biol. 60, 335–355. 10.1146/annurev.arplant.043008.091955 [DOI] [PubMed] [Google Scholar]
- Klein-Junior L. C., Cretton S., Allard P.-M., Genta-Jouve G., Passos C. S., Salton J., et al. (2017). Targeted Isolation of Monoterpene Indole Alkaloids from Palicourea sessilis. J. Natural Prod. 80, 3032–3037. 10.1021/acs.jnatprod.7b00681 [DOI] [PubMed] [Google Scholar]
- Kravina A. G., Carreira E. M. (2018). Total Synthesis of Epicolactone. Angew. Che. (Int. Ed. English) 57, 13159–13162. 10.1002/anie.201807709 [DOI] [PubMed] [Google Scholar]
- Kruakaew S., Seeka C., Lhinhatrakool T., Thongnest S., Yahuafai J., Piyaviriyakul S., et al. (2017). Cytotoxic Cardiac Glycoside Constituents of Vallaris glabra Leaves. J. Natural Prod. 80, 2987–2996. 10.1021/acs.jnatprod.7b00554 [DOI] [PubMed] [Google Scholar]
- Krueger B. A., Dietrich A., Baringhaus K.-H., Schneider G. (2009). Scaffold-hopping potential of fragment-based de novo design: the chances and limits of variation. Comb. Chem. High Throughput Screen. 12, 383–396. 10.2174/138620709788167971 [DOI] [PubMed] [Google Scholar]
- Kruger A., Schafers C., Schroder C., Antranikian G. (2018). Towards a sustainable biobased industry - Highlighting the impact of extremophiles. New Biotechnol. 40, 144–153. 10.1016/j.nbt.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Kubota T., Nakamura K., Kurimoto S.-I., Sakai K., Fromont J., Gonoi T., et al. (2017). Zamamidine D, a Manzamine Alkaloid from an Okinawan Amphimedon sp. Marine Sponge. J. Natural Prod. 80, 1196–1199. 10.1021/acs.jnatprod.6b01110 [DOI] [PubMed] [Google Scholar]
- Kufareva I., Ilatovskiy A. V., Abagyan R. (2012). Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic Acids Res. 40, D535–D540. 10.1093/nar/gkr825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo P.-C., Hung H.-Y., Nian C.-W., Hwang T.-L., Cheng J.-C., Kuo D.-H., et al. (2017). Chemical Constituents and Anti-inflammatory Principles from the Fruits of Forsythia suspensa. J. Natural Prod. 80, 1055–1064. 10.1021/acs.jnatprod.6b01141 [DOI] [PubMed] [Google Scholar]
- Kuo L.-M. Y., Tseng P.-Y., Lin Y.-C., Liaw C.-C., Zhang L.-J., Tsai K.-C., et al. (2018). New Hirsutinolide-Type Sesquiterpenoids from Vernonia cinerea Inhibit Nitric Oxide Production in LPS-Stimulated RAW264.7 Cells. Planta Med. 84, 1348–1354. 10.1055/a-0647-1901 [DOI] [PubMed] [Google Scholar]
- Kurasawa K., Kwon E., Kuwahara S., Enomoto M. (2018). Bioinspired Total Synthesis of Delitschiapyrone A. Org. Lett. 20, 4645–4648. 10.1021/acs.orglett.8b01932 [DOI] [PubMed] [Google Scholar]
- Kusakabe K., Honmura Y., Uesugi S., Tonouchi A., Maeda H., Kimura K.-I., et al. (2017). Neomacrophorin X, a 4.4.3Propellane-Type Meroterpenoid from Trichoderma sp. 1212-03. J. Natural Prod. 80, 1484–1492. 10.1021/acs.jnatprod.6b01177 [DOI] [PubMed] [Google Scholar]
- Kuttruff C. A., Eastgate M. D., Baran P. S. (2014). Natural product synthesis in the age of scalability. Nat. Prod. Rep. 31, 419–432. 10.1039/c3np70090a [DOI] [PubMed] [Google Scholar]
- Kwon I.-S., Kwak J. H., Pyo S., Lee H.-W., Kim A., Schmitz F. J. (2017). Oscarellin, an Anthranilic Acid Derivative from a Philippine Sponge, Oscarella stillans, as an Inhibitor of Inflammatory Cytokines in Macrophages. J. Natural Prod. 80, 149–155. 10.1021/acs.jnatprod.6b00787 [DOI] [PubMed] [Google Scholar]
- Lajter I., Pan S.-P., Nikles S., Ortmann S., Vasas A., Csupor-Löffler B., et al. (2015). Inhibition of COX-2 and NF-κB1 Gene Expression, NO Production, 5-LOX, and COX-1 and COX-2 Enzymes by Extracts and Constituents of Onopordum acanthium. Planta Med. 81, 1270–1276. 10.1055/s-0035-1546242 [DOI] [PubMed] [Google Scholar]
- Langer T., Hoffmann R., Bryant S., Lesur B. (2009). Hit finding: Towards ‘smarter’ approaches. Curr. Opin. Pharmacol. 9, 589–593. 10.1016/j.coph.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Larsson J., Gottfries J., Bohlin L., Backlund A. (2005). Expanding the ChemGPS chemical space with natural products. J. Natural Prod. 68, 985–991. 10.1021/np049655u [DOI] [PubMed] [Google Scholar]
- Larsson J., Gottfries J., Muresan S., Backlund A. (2007). ChemGPS-NP: Tuned for navigation in biologically relevant chemical space. J. Natural Prod. 70, 789–794. 10.1021/np070002y [DOI] [PubMed] [Google Scholar]
- Lautié E., Quintero R., Fliniaux M.-A., Villarreal M.-L. (2008). Selection methodology with scoring system: Application to Mexican plants producing podophyllotoxin related lignans. J. Ethnopharmacol. 120, 402–412. 10.1016/j.jep.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Lautié E., Fliniaux M.-A., Villarreal M. L. (2010). Updated biotechnological approaches developed for 2,7′-cyclolignan production. Biotechnol. Appl. Biochem. 55, 139–153. 10.1042/BA20090253 [DOI] [PubMed] [Google Scholar]
- Ledesma-Amaro R., Jiménez A., Revuelta J. L. (2018). Pathway Grafting for Polyunsaturated Fatty Acids Production in Ashbya gossypii through Golden Gate Rapid Assembly. ACS Synth. Biol. 7, 2340–2347. 10.1021/acssynbio.8b00287 [DOI] [PubMed] [Google Scholar]
- Ledoux A., St-Gelais A., Cieckiewicz E., Jansen O., Bordignon A., Illien B., et al. (2017). Antimalarial Activities of Alkyl Cyclohexenone Derivatives Isolated from the Leaves of Poupartia borbonica. J. Natural Prod. 80, 1750–1757. 10.1021/acs.jnatprod.6b01019 [DOI] [PubMed] [Google Scholar]
- Lee M. L., Schneider G. (2001). Scaffold architecture and pharmacophoric properties of natural products and trade drugs: application in the design of natural product-based combinatorial libraries. J. Comb. Chem. 3, 284–289. 10.1021/cc000097l [DOI] [PubMed] [Google Scholar]
- Lee W., Lee D., Lee Y., Lee T., Song K.-S., Yang E.-J., et al. (2018). Isolation, Synthesis, and Antisepsis Effects of a C-Methylcoumarinochromone Isolated from Abronia nana Cell Culture. J. Natural Prod. 81, 1173–1182. 10.1021/acs.jnatprod.7b00826 [DOI] [PubMed] [Google Scholar]
- Leman-Loubiere C., Le Goff G., Retailleau P., Debitus C., Ouazzani J. (2017). Sporothriolide-Related Compounds from the Fungus Hypoxylon monticulosum CLL-205 Isolated from a Sphaerocladina Sponge from the Tahiti Coast. J. Natural Prod. 80, 2850–2854. 10.1021/acs.jnatprod.7b00714 [DOI] [PubMed] [Google Scholar]
- Leo M., Peruzzi L., Granchi C., Tuccinardi T., Minutolo F., Tommasi N., et al. (2017). Constituents of Polygala flavescens ssp. flavescens and Their Activity as Inhibitors of Human Lactate Dehydrogenase. J. Natural Prod. 80, 2077–2087. 10.1021/acs.jnatprod.7b00295 [DOI] [PubMed] [Google Scholar]
- Li D.-H., Li J.-Y., Xue C.-M., Han T., Sai C.-M., Wang K.-B., et al. (2017). Antiproliferative Dimeric Aporphinoid Alkaloids from the Roots of Thalictrum cultratum. J. Natural Prod. 80, 2893–2904. 10.1021/acs.jnatprod.7b00387 [DOI] [PubMed] [Google Scholar]
- Li H.-L., Li X.-M., Li X., Wang C.-Y., Liu H., Kassack M. U., et al. (2017). Antioxidant Hydroanthraquinones from the Marine Algal-Derived Endophytic Fungus Talaromyces islandicus EN-501. J. Natural Prod. 80, 162–168. 10.1021/acs.jnatprod.6b00797 [DOI] [PubMed] [Google Scholar]
- Li K., Ji S., Song W., Kuang Y., Lin Y., Tang S., et al. (2017). Glycybridins A-K, Bioactive Phenolic Compounds from Glycyrrhiza glabra. J. Natural Prod. 80, 334–346. 10.1021/acs.jnatprod.6b00783 [DOI] [PubMed] [Google Scholar]
- Li Q., Deng A.-J., Li L., Wu L.-Q., Ji M., Zhang H.-J., et al. (2017). Azacyclo-indoles and Phenolics from the Flowers of Juglans regia. J. Natural Prod. 80, 2189–2198. 10.1021/acs.jnatprod.6b00887 [DOI] [PubMed] [Google Scholar]
- Li Y., Yue Q., Jayanetti D. R., Swenson D. C., Bartholomeusz G. A., An Z., et al. (2017). Anti-Cryptococcus Phenalenones and Cyclic Tetrapeptides from Auxarthron pseudauxarthron. J. Natural Prod. 80, 2101–2109. 10.1021/acs.jnatprod.7b00341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Gu B.-B., Sun F., Xu J.-R., Jiao W.-H., Yu H.-B., et al. (2017. a). Sesquiterpene Quinones/Hydroquinones from the Marine Sponge Spongia pertusa Esper. J. Natural Prod. 80, 1436–1445. 10.1021/acs.jnatprod.6b01105 [DOI] [PubMed] [Google Scholar]
- Li J., Li N., Li X., Chen G., Wang C., Lin B., et al. (2017. b). Characteristic alpha-Acid Derivatives from Humulus lupulus with Antineuroinflammatory Activities. J. Natural Prod. 80, 3081–3092. 10.1021/acs.jnatprod.6b00921 [DOI] [PubMed] [Google Scholar]
- Li J., Seupel R., Bruhn T., Feineis D., Kaiser M., Brun R., et al. (2017. c). Jozilebomines A and B, Naphthylisoquinoline Dimers from the Congolese Liana Ancistrocladus ileboensis, with Antiausterity Activities against the PANC-1 Human Pancreatic Cancer Cell Line. J. Natural Prod. 80, 2807–2817. 10.1021/acs.jnatprod.7b00650 [DOI] [PubMed] [Google Scholar]
- Li J., Seupel R., Feineis D., Mudogo V., Kaiser M., Brun R., et al. (2017. d). Dioncophyllines C2, D2, and F and Related Naphthylisoquinoline Alkaloids from the Congolese Liana Ancistrocladus ileboensis with Potent Activities against Plasmodium falciparum and against Multiple Myeloma and Leukemia Cell Lines. J. Natural Prod. 80, 443–458. 10.1021/acs.jnatprod.6b00967 [DOI] [PubMed] [Google Scholar]
- Li W., Huang C., Liu Q., Koike K. (2017. e). Bistinospinosides A and B, Dimeric Clerodane Diterpene Glycosides from Tinospora sagittata. J. Natural Prod. 80, 2478–2483. 10.1021/acs.jnatprod.7b00324 [DOI] [PubMed] [Google Scholar]
- Li W., Lee C., Bang S. H., Ma J. Y., Kim S., Koh Y.-S., et al. (2017. f). Isochromans and Related Constituents from the Endophytic Fungus Annulohypoxylon truncatum of Zizania caduciflora and Their Anti-Inflammatory Effects. J. Natural Prod. 80, 205–209. 10.1021/acs.jnatprod.6b00698 [DOI] [PubMed] [Google Scholar]
- Li L., Chen Z., Zhang X., Jia Y. (2018). Divergent Strategy in Natural Product Total Synthesis. Chem. Rev. 118, 3752–3832. 10.1021/acs.chemrev.7b00653 [DOI] [PubMed] [Google Scholar]
- Li S., Niu H., Qiao Y., Zhu R., Sun Y., Ren Z., et al. (2018). Terpenoids isolated from Chinese liverworts Lepidozia reptans and their anti-inflammatory activity. Bioorg. Med. Chem. 26, 2392–2400. 10.1016/j.bmc.2018.03.040 [DOI] [PubMed] [Google Scholar]
- Liangsakul P., Kuhakarn C., Hongthong S., Jariyawat S., Suksen K., Akkarawongsapat R., et al. (2018). Anti-HIV 1 Activity of Xanthones from the Bark of Mammea harmandii. Natural Prod. Commun. 13, 53–56. 10.1177/1934578X1801300116 [DOI] [Google Scholar]
- Lima R. D. C. L., Gramsbergen S. M., van Staden J., Jager A. K., Kongstad K. T., Staerk D. (2017). Advancing HPLC-PDA-HRMS-SPE-NMR Analysis of Coumarins in Coleonema album by Use of Orthogonal Reversed-Phase C18 and Pentafluorophenyl Separations. J. Natural Prod. 80, 1020–1027. 10.1021/acs.jnatprod.6b01020 [DOI] [PubMed] [Google Scholar]
- Lin Z., Xu X., Zhao S., Yang X., Guo J., Zhang Q., et al. (2018). Total synthesis and antimicrobial evaluation of natural albomycins against clinical pathogens. Nat. Commun. 9, 3445. 10.1038/s41467-018-05821-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26. 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- Lipinski C. A. (2003). Chris Lipinski discusses life and chemistry after the Rule of Five. Drug Discovery Today 8, 12–16. 10.1016/S1359-6446(02)02556-4 [DOI] [PubMed] [Google Scholar]
- Litaudon M., Bousserouel H., Awang K., Nosjean O., Martin M.-T., Dau M. E. T. H., et al. (2009. a). A Dimeric sesquiterpenoid from a Malaysian Meiogyne as a new inhibitor of Bcl-xL/BakBH3 domain peptide interaction. J. Natural Prod. 72, 480–483. 10.1021/np8006292 [DOI] [PubMed] [Google Scholar]
- Litaudon M., Jolly C., Le Callonec C., Cuong D. D., Retailleau P., Nosjean O., et al. (2009. b). Cytotoxic pentacyclic triterpenoids from Combretum sundaicum and Lantana camara as inhibitors of Bcl-xL/BakBH3 domain peptide interaction. J. Natural Prod. 72, 1314–1320. 10.1021/np900192r [DOI] [PubMed] [Google Scholar]
- Liu Y.-W., Shi D.-H., Chen A.-J., Zhu Q., Xu J.-T., Zhang X.-X. (2014). Acetylcholinesterase inhibition effects of marine fungi. Pharm. Biol. 52, 539–543. 10.3109/13880209.2013.850516 [DOI] [PubMed] [Google Scholar]
- Liu F., Yuan T., Liu W., Ma H., Seeram N. P., Li Y., et al. (2017). Phloroglucinol Derivatives with Protein Tyrosine Phosphatase 1B Inhibitory Activities from Eugenia jambolana Seeds. J. Natural Prod. 80, 544–550. 10.1021/acs.jnatprod.6b01073 [DOI] [PubMed] [Google Scholar]
- Liu H.-B., Lauro G., O’Connor R. D., Lohith K., Kelly M., Colin P., et al. (2017). Tulongicin, an Antibacterial Tri-Indole Alkaloid from a Deep-Water Topsentia sp. Sponge. J. Natural Prod. 80, 2556–2560. 10.1021/acs.jnatprod.7b00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-L., Ha T. K. Q., Ha W., Oh W. K., Yang J.-L., Shi Y.-P. (2017). Sesquiterpenoids with Various Carbocyclic Skeletons from the Flowers of Chrysanthemum indicum. J. Natural Prod. 80, 298–307. 10.1021/acs.jnatprod.6b00694 [DOI] [PubMed] [Google Scholar]
- Liu R., Heiss E. H., Schachner D., Jiang B., Liu W., Breuss J. M., et al. (2017). Xanthohumol Blocks Proliferation and Migration of Vascular Smooth Muscle Cells in Vitro and Reduces Neointima Formation in Vivo. J. Natural Prod. 80, 2146–2150. 10.1021/acs.jnatprod.7b00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li Y., Liu Z., Li L., Qu J., Ma S., et al. (2017. a). Sesquiterpenes from the Endophyte Glomerella cingulata. J. Natural Prod. 80, 2609–2614. 10.1021/acs.jnatprod.7b00054 [DOI] [PubMed] [Google Scholar]
- Liu Y., Yu H.-Y., Wang Y.-M., Tian T., Wu W.-M., Zhou M., et al. (2017. b). Neuroprotective Lignans from the Fruits of Schisandra bicolor var. tuberculata. J. Natural Prod. 80, 1117–1124. 10.1021/acs.jnatprod.7b00035 [DOI] [PubMed] [Google Scholar]
- Liu W.-C., Inwood S., Gong T., Sharma A., Yu L.-Y., Zhu P. (2019). Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit. Rev. Biotechnol. 39, 258–271. 10.1080/07388551.2018.1554620 [DOI] [PubMed] [Google Scholar]
- Liu X., Liu J., Wu J., Huang G., Liang R., Chung L. W., et al. (2019). Asymmetric Total Synthesis of Cerorubenic Acid-III. J. Am. Chem. Soc. 141, 2872–2877. 10.1021/jacs.8b12647 [DOI] [PubMed] [Google Scholar]
- Loeschcke A., Thies S. (2015). Pseudomonas putida-a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 99, 6197–6214. 10.1007/s00253-015-6745-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Stegemann S., Agrawal S., Karcher D., Ruf S., Bock R. (2017). Horizontal Transfer of a Synthetic Metabolic Pathway between Plant Species. Curr. Biol. 27, 3034–3041.e3. 10.1016/j.cub.2017.08.044 [DOI] [PubMed] [Google Scholar]
- Luo J.-G., Xu Y.-M., Sandberg D. C., Arnold A. E., Gunatilaka A. A. L. (2017). Montagnuphilones A-G, Azaphilones from Montagnulaceae sp. DM0194, a Fungal Endophyte of Submerged Roots of Persicaria amphibia. J. Natural Prod. 80, 76–81. 10.1021/acs.jnatprod.6b00714 [DOI] [PubMed] [Google Scholar]
- Luo P., Xia W., Morris-Natschke S. L., Lee K.-H., Zhao Y., Gu Q., et al. (2017). Vitepyrroloids A-D, 2-Cyanopyrrole-Containing Labdane Diterpenoid Alkaloids from the Leaves of Vitex trifolia. J. Natural Prod. 80, 1679–1683. 10.1021/acs.jnatprod.6b01195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Wei X.-Y., Yang J., Luo J.-F., Liang R., Tu Z.-C., et al. (2017). Spiro Meroterpenoids from Ganoderma applanatum. J. Natural Prod. 80, 61–70. 10.1021/acs.jnatprod.6b00431 [DOI] [PubMed] [Google Scholar]
- Ma C. T., Eom T., Cho E., Wu B., Kim T. R., Oh K. B., et al. (2017). Aquilanols A and B, Macrocyclic Humulene-Type Sesquiterpenoids from the Agarwood of Aquilaria malaccensis. J. Natural Prod. 80, 3043–3048. 10.1021/acs.jnatprod.7b00462 [DOI] [PubMed] [Google Scholar]
- Ma X., Liu J., Yang L., Zhang B., Dong Y., Zhao Q. (2018). Cynomorium songaricum prevents bone resorption in ovariectomized rats through RANKL/RANK/TRAF6 mediated suppression of PI3K/AKT and NF-κB pathways. Life Sci. 209, 140–148. 10.1016/j.lfs.2018.08.008 [DOI] [PubMed] [Google Scholar]
- Ma L., Tang L., Yi Q. (2019). Salvianolic Acids: Potential Source of Natural Drugs for the Treatment of Fibrosis Disease and Cancer. Front. Pharmacol. 10, 97. 10.3389/fphar.2019.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macabeo A. P. G., Letada A. G., Budde S., Faderl C., Dahse H.-M., Franzblau S. G., et al. (2017). Antitubercular and Cytotoxic Chlorinated seco-Cyclohexenes from Uvaria alba. J. Natural Prod. 80, 3319–3323. 10.1021/acs.jnatprod.7b00679 [DOI] [PubMed] [Google Scholar]
- Macarron R., Banks M. N., Bojanic D., Burns D. J., Cirovic D. A., Garyantes T., et al. (2011). Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discovery 10, 188–195. 10.1038/nrd3368 [DOI] [PubMed] [Google Scholar]
- Malik S., Cusidó R. M., Mirjalili M. H., Moyano E., Palazón J., Bonfill M. (2011). Production of the anticancer drug taxol in Taxus baccata suspension cultures: A review. Process Biochem. 46, 23–34. 10.1016/j.procbio.2010.09.004 [DOI] [Google Scholar]
- Man Y., Fu S., Chen J., Liu B. (2018). Total synthesis and structural revision of an isopanepoxydone analog isolated from Lentinus strigellus. Org. Biomolec. Chem. 16, 5043–5049. 10.1039/c8ob01168k [DOI] [PubMed] [Google Scholar]
- Mancuso S. (2018). The revolutionary genius of plants: A new understanding of plant intelligence and behavior (New York NY: Atria Books an imprint of Simon & Schuster Inc.). [Google Scholar]
- Markovic M., Koos P., Carny T., Sokoliova S., Bohacikova N., Moncol J., et al. (2017). Total Synthesis, Configuration Assignment, and Cytotoxic Activity Evaluation of Protulactone A. J. Natural Prod. 80, 1631–1638. 10.1021/acs.jnatprod.7b00212 [DOI] [PubMed] [Google Scholar]
- Martin H. J., Kampatsikas I., Oost R., Pretzler M., Al-Sayed E., Roller A., et al. (2018). Total Synthesis, Stereochemical Assignment, and Divergent Enantioselective Enzymatic Recognition of Larreatricin. Chem. (Weinheim an der Bergstrasse Germany) 24, 15756–15760. 10.1002/chem.201803785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi M., Meyer S., Clement S., Pescitelli G., Cimmino A., Cristofaro M., et al. (2017). Chloromonilinic Acids C and D, Phytotoxic Tetrasubstituted 3-Chromanonacrylic Acids Isolated from Cochliobolus australiensis with Potential Herbicidal Activity against Buffelgrass (Cenchrus ciliaris). J. Natural Prod. 80, 2771–2777. 10.1021/acs.jnatprod.7b00583 [DOI] [PubMed] [Google Scholar]
- Mathur A., Mathur A. K., Pal M., Uniyal G. C. (1999). Comparison of qualitative and quantitative in vitro ginsenoside production in callus cultures of three Panax species. Planta Med. 65, 484–486. 10.1055/s-2006-960823 [DOI] [PubMed] [Google Scholar]
- Mayr L. M., Bojanic D. (2009). Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 9, 580–588. 10.1016/j.coph.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Mazzio E., Badisa R., Mack N., Deiab S., Soliman K. F. A. (2014). High Throughput Screening of Natural Products for Anti-mitotic Effects in MDA-MB-231 Human Breast Carcinoma Cells. Phytother. Res. 28, 856–867. 10.1002/ptr.5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin D. R., Green B. D., Prince N. C., Tanney J. B., Miller J. D. (2017). Natural Products of Picea Endophytes from the Acadian Forest. J. Natural Prod. 80, 1475–1483. 10.1021/acs.jnatprod.6b01157 [DOI] [PubMed] [Google Scholar]
- Melchor-Martinez E. M., Silva-Mares D. A., Torres-Lopez E., Waksman-Minsky N., Pauli G. F., Chen S.-N., et al. (2017). Stereochemistry of a Second Riolozane and Other Diterpenoids from Jatropha dioica. J. Natural Prod. 80, 2252–2262. 10.1021/acs.jnatprod.7b00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestres J., Gregori-Puigjané E., Valverde S., Solé R. V. (2008). Data completeness—the Achilles heel of drug-target networks. Nat. Biotechnol. 26, 983–984. 10.1038/nbt0908-983 [DOI] [PubMed] [Google Scholar]
- Mieri M., Monteleone G., Ismajili I., Kaiser M., Hamburger M. (2017). Antiprotozoal Activity-Based Profiling of a Dichloromethane Extract from Anthemis nobilis Flowers. J. Natural Prod. 80, 459–470. 10.1021/acs.jnatprod.6b00980 [DOI] [PubMed] [Google Scholar]
- Mo D.-J., Li J., Li M.-Y. (2018). A New 28-Nor-oleanane Triterpene from Excoecaria agallocha. Natural Prod. Commun. 13, 17–20. 10.1177/1934578X1801300107 [DOI] [Google Scholar]
- Mokashe N., Chaudhari B., Patil U. (2018). Operative utility of salt-stable proteases of halophilic and halotolerant bacteria in the biotechnology sector. Int. J. Biol. Macromol. 117, 493–522. 10.1016/j.ijbiomac.2018.05.217 [DOI] [PubMed] [Google Scholar]
- Moore B. D., Andrew R. L., Külheim C., Foley W. J. (2014). Explaining intraspecific diversity in plant secondary metabolites in an ecological context. New Phytol. 201, 733–750. 10.1111/nph.12526 [DOI] [PubMed] [Google Scholar]
- Mortenson P. N., Erlanson D. A., Esch I. J. P. de, Jahnke W., Johnson C. N. (2018). Fragment-to-Lead Medicinal Chemistry Publications in 2017. J. Med. Chem. 62, 3857–3872. 10.1021/acs.jmedchem.8b01472 [DOI] [PubMed] [Google Scholar]
- Moscatiello R., Baldan B., Navazio L. (2013). Plant cell suspension cultures. Methods Mol. Biol. (Clifton N.J.) 953, 77–93. 10.1007/978-1-62703-152-3_5 [DOI] [PubMed] [Google Scholar]
- Moses T., Mehrshahi P., Smith A. G., Goossens A. (2017). Synthetic biology approaches for the production of plant metabolites in unicellular organisms. J. Exp. Bot. 68, 4057–4074. 10.1093/jxb/erx119 [DOI] [PubMed] [Google Scholar]
- Mountessou B. Y. G., Tchamgoue J., Paul Dzoyem J., Tchuenguem R. T., Surup F., Choudhary M. I., et al. (2018). Two xanthones and two rotameric (3⟶8) biflavonoids from the Cameroonian medicinal plant Allanblackia floribunda Oliv. (Guttiferae). Tetrahedron Lett. 59, 4545–4550. 10.1016/j.tetlet.2018.11.035 [DOI] [Google Scholar]
- Muharini R., Diaz A., Ebrahim W., Mandi A., Kurtan T., Rehberg N., et al. (2017). Antibacterial and Cytotoxic Phenolic Metabolites from the Fruits of Amorpha fruticosa. J. Natural Prod. 80, 169–180. 10.1021/acs.jnatprod.6b00809 [DOI] [PubMed] [Google Scholar]
- Mustafa N. R., Winter W., van Iren F., Verpoorte R. (2011). Initiation, growth and cryopreservation of plant cell suspension cultures. Nat. Protoc. 6, 715–742. 10.1038/nprot.2010.144 [DOI] [PubMed] [Google Scholar]
- Nadri M. H., Salim Y., Basar N., Yahya A., Zulkifli R. M. (2014). Antioxidant activities and tyrosinase inhibition effects of Phaleria macrocarpa extracts. Afr. J. Tradit. Complement. Altern. Medicines : AJTCAM 11, 107–111. 10.4314/ajtcam.v11i3.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naman C. B., Almaliti J., Armstrong L., Caro-Diaz E. J., Pierce M. L., Glukhov E., et al. (2017). Discovery and Synthesis of Caracolamide A, an Ion Channel Modulating Dichlorovinylidene Containing Phenethylamide from a Panamanian Marine Cyanobacterium cf. Symploca Species. J. Natural Prod. 80, 2328–2334. 10.1021/acs.jnatprod.7b00367 [DOI] [PubMed] [Google Scholar]
- Nduka S. O., Okonta M. J., Ajaghaku D. L., Ukwe C. V. (2017). In vitro and in vivo cytochrome P450 3A enzyme inhibition by Aframomum melegueta and Denniettia tripetala extracts. Asian Pacific J. Trop. Med. 10, 576–581. 10.1016/j.apjtm.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2007). Natural products as sources of new drugs over the last 25 years. J. Natural Prod. 70, 461–477. 10.1021/np068054v [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Natural Prod. 75, 311–335. 10.1021/np200906s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2015). Endophytic and epiphytic microbes as “sources” of bioactive agents. Front. Chem. 3, 34. 10.3389/fchem.2015.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2016). Natural Products as Sources of New Drugs from 1981 to 2014. J. Natural Prod. 79, 629–661. 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M., Snader K. M. (2003). Natural products as sources of new drugs over the period 1981-2002. J. Natural Prod. 66, 1022–1037. 10.1021/np030096l [DOI] [PubMed] [Google Scholar]
- Nguyen D. M. T., Do L. M. T., Nguyen V. T., Chavasiri W., Mortier J., Nguyen P. P. K. (2017). Phenolic Compounds from the Lichen Lobaria orientalis. J. Natural Prod. 80, 261–268. 10.1021/acs.jnatprod.6b00465 [DOI] [PubMed] [Google Scholar]
- Nguyen M. T. T., Le T. H., Nguyen H. X., Dang P. H., Do T. N. V., Abe M., et al. (2017). Artocarmins G-M, Prenylated 4-Chromenones from the Stems of Artocarpus rigida and Their Tyrosinase Inhibitory Activities. J. Natural Prod. 80, 3172–3178. 10.1021/acs.jnatprod.7b00453 [DOI] [PubMed] [Google Scholar]
- Nguyen N. T., Dang P. H., Vu N. X. T., Le T. H., Nguyen M. T. T. (2017. a). Quinoliniumolate and 2H-1,2,3-Triazole Derivatives from the Stems of Paramignya trimera and Their alpha-Glucosidase Inhibitory Activities: In Vitro and in Silico Studies. J. Natural Prod. 80, 2151–2155. 10.1021/acs.jnatprod.7b00289 [DOI] [PubMed] [Google Scholar]
- Nguyen N. T., Nguyen M. T. T., Nguyen H. X., Dang P. H., Dibwe D. F., Esumi H., et al. (2017. b). Constituents of the Rhizomes of Boesenbergia pandurata and Their Antiausterity Activities against the PANC-1 Human Pancreatic Cancer Line. J. Natural Prod. 80, 141–148. 10.1021/acs.jnatprod.6b00784 [DOI] [PubMed] [Google Scholar]
- Ni G., Li J.-Y., Mai Z.-P., Yu D.-Q. (2018). Belamcandanes A and B, two unprecedented tricyclic-iridal triterpenoids from Belamcanda chinensis. Tetrahedron Lett. 59, 151–155. 10.1016/j.tetlet.2017.12.013 [DOI] [Google Scholar]
- Nicacio K. J., Ioca L. P., Froes A. M., Leomil L., Appolinario L. R., Thompson C. C., et al. (2017). Cultures of the Marine Bacterium Pseudovibrio denitrificans Ab134 Produce Bromotyrosine-Derived Alkaloids Previously Only Isolated from Marine Sponges. J. Natural Prod. 80, 235–240. 10.1021/acs.jnatprod.6b00838 [DOI] [PubMed] [Google Scholar]
- Nicolaou K. C., Li R., Lu Z., Pitsinos E. N., Alemany L. B. (2018. a). Total Synthesis and Full Structural Assignment of Namenamicin. J. Am. Chem. Soc. 140, 8091–8095. 10.1021/jacs.8b04592 [DOI] [PubMed] [Google Scholar]
- Nicolaou K. C., Li R., Lu Z., Pitsinos E. N., Alemany L. B., Aujay M., et al. (2018. b). Streamlined Total Synthesis of Shishijimicin A and Its Application to the Design, Synthesis, and Biological Evaluation of Analogues thereof and Practical Syntheses of PhthNSSMe and Related Sulfenylating Reagents. J. Am. Chem. Soc. 140, 12120–12136. 10.1021/jacs.8b06955 [DOI] [PubMed] [Google Scholar]
- Nikel P. I., Martínez-García E., Lorenzo V. (2014). Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 12, 368–379. 10.1038/nrmicro3253 [DOI] [PubMed] [Google Scholar]
- Ninomiya M., Ando Y., Kudo F., Ohmori K., Suzuki K. (2019). First Total Synthesis of Actinorhodin. Angew. Che. (Int. Ed. English). 58, 4264–4270. 10.1002/anie.201814172 [DOI] [PubMed] [Google Scholar]
- Nistanaki S. K., Boralsky L. A., Pan R. D., Nelson H. M. (2019). A Concise Total Synthesis of (+/-)-Vibralactone. Angew. Che. (Int. Ed. English) 58, 1724–1726. 10.1002/anie.201812711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B., Ke C.-Q., Li B.-H., Li Y., Yi Y., Luo Y., et al. (2017). Cucurbitane Glucosides from the Crude Extract of Siraitia grosvenorii with Moderate Effects on PGC-1alpha Promoter Activity. J. Natural Prod. 80, 1428–1435. 10.1021/acs.jnatprod.6b01086 [DOI] [PubMed] [Google Scholar]
- Niu S., Fan Z., Tang X., Liu Q., Shao Z., Liu G., et al. (2018). Cyclopiane-type diterpenes from the deep-sea-derived fungus Penicillium commune MCCC 3A00940. Tetrahedron Lett. 59, 375–378. 10.1016/j.tetlet.2017.12.045 [DOI] [Google Scholar]
- Nolsoe J. M. J., Antonsen S., Gorbitz C. H., Hansen T. V., Nesman J. I., Rohr A. K., et al. (2018). Total Synthesis of (-)-Mucosin and Revision of Structure. J. Org. Chem. 83,15066–15076. 10.1021/acs.joc.8b02318 [DOI] [PubMed] [Google Scholar]
- Nosjean O., Ferro M., Coge F., Beauverger P., Henlin J. M., Lefoulon F., et al. (2000). Identification of the melatonin-binding site MT3 as the quinone reductase 2. J. Biol. Chem. 275, 31311–31317. 10.1074/jbc.M005141200 [DOI] [PubMed] [Google Scholar]
- Nothias L.-F., Boutet-Mercey S., Cachet X., La Torre E., Laboureur L., Gallard J.-F., et al. (2017). Environmentally Friendly Procedure Based on Supercritical Fluid Chromatography and Tandem Mass Spectrometry Molecular Networking for the Discovery of Potent Antiviral Compounds from Euphorbia semiperfoliata. J. Natural Prod. 80, 2620–2629. 10.1021/acs.jnatprod.7b00113 [DOI] [PubMed] [Google Scholar]
- Nothias L.-F., Nothias-Esposito M., da Silva R., Wang M., Protsyuk I., Zhang Z., et al. (2018). Bioactivity-Based Molecular Networking for the Discovery of Drug Leads in Natural Product Bioassay-Guided Fractionation. J. Natural Prod. 81, 758–767. 10.1021/acs.jnatprod.7b00737 [DOI] [PubMed] [Google Scholar]
- Noumeur S. R., Helaly S. E., Jansen R., Gereke M., Stradal T. E. B., Harzallah D., et al. (2017). Preussilides A-F, Bicyclic Polyketides from the Endophytic Fungus Preussia similis with Antiproliferative Activity. J. Natural Prod. 80, 1531–1540. 10.1021/acs.jnatprod.7b00064 [DOI] [PubMed] [Google Scholar]
- Novakovic M., Nikodinovic-Runic J., Veselinovic J., Ilic-Tomic T., Vidakovic V., Tesevic V., et al. (2017). Bioactive Pentacyclic Triterpene Ester Derivatives from Alnus viridis ssp. viridis Bark. J. Natural Prod. 80, 1255–1263. 10.1021/acs.jnatprod.6b00805 [DOI] [PubMed] [Google Scholar]
- Ntumba J. K., Tshiongo C. M., Mifundu M. N., Robiette R., Taba K. M. (2018). Effective Antimalarial Activities of α-Hydroxy Diynes Isolated from Ongokea gore. Planta Med. 84, 806–812. 10.1055/s-0043-124974 [DOI] [PubMed] [Google Scholar]
- Nur-E-Alam M., Yousaf M., Ahmed S., Al-Sheddi E. S., Parveen I., Fazakerley D. M., et al. (2017). Neoclerodane Diterpenoids from Reehal Fatima, Teucrium yemense. J. Natural Prod. 80, 1900–1908. 10.1021/acs.jnatprod.7b00188 [DOI] [PubMed] [Google Scholar]
- Nyandoro S. S., Munissi J. J. E., Gruhonjic A., Duffy S., Pan F., Puttreddy R., et al. (2017. a). Polyoxygenated Cyclohexenes and Other Constituents of Cleistochlamys kirkii Leaves. J. Natural Prod. 80, 114–125. 10.1021/acs.jnatprod.6b00759 [DOI] [PubMed] [Google Scholar]
- Nyandoro S. S., Munissi J. J. E., Kombo M., Mgina C. A., Pan F., Gruhonjic A., et al. (2017. b). Flavonoids from Erythrina schliebenii. J. Natural Prod. 80, 377–383. 10.1021/acs.jnatprod.6b00839 [DOI] [PubMed] [Google Scholar]
- Nzogong R. T., Nganou B. K., Tedonkeu A. T., Awouafack M. D., Tene M., Ito T., et al. (2018). Three New Abietane-Type Diterpenoids from Plectranthus africanus and Their Antibacterial Activities. Planta Med. 84, 59–64. 10.1055/s-0043-114426 [DOI] [PubMed] [Google Scholar]
- Ochoa-Villarreal M., Howat S., Jang M. O., Kim I. S., Jin Y.-W., Lee E.-K., et al. (2015). Cambial meristematic cells: a platform for the production of plant natural products. New Biotechnol. 32, 581–587. 10.1016/j.nbt.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Oghumu S., Varikuti S., Saljoughian N., Terrazas C., Huntsman A. C., Parinandi N. L., et al. (2017). Pentalinonsterol, a Constituent of Pentalinon andrieuxii, Possesses Potent Immunomodulatory Activity and Primes T Cell Immune Responses. J. Natural Prod. 80, 2515–2523. 10.1021/acs.jnatprod.7b00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara T., Amano N., Mitsui Y., Fujino K., Ohta H., Takahashi K., et al. (2017). Determination of the Absolute Configuration of a Monoglyceride Antibolting Compound and Isolation of Related Compounds from Radish Leaves (Raphanus sativus). J. Natural Prod. 80, 872–878. 10.1021/acs.jnatprod.6b00746 [DOI] [PubMed] [Google Scholar]
- Oksman-Caldentey K.-M., Saito K. (2005). Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr. Opin. Biotechnol. 16, 174–179. 10.1016/j.copbio.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Oliveira M., Rodrigues M. J., Pereira C., Neto R., Junior P. A. S., Neng N., et al. (2018). First report of the in vitro antileishmanial properties of extremophile plants from the Algarve Coast. Natural Prod. Res. 32, 600–604. 10.1080/14786419.2017.1326489 [DOI] [PubMed] [Google Scholar]
- Olivon F., Nothias L.-F., Dumontet V., Retailleau P., Berger S., Ferry G., et al. (2018). Natural Inhibitors of the RhoA-p115 Complex from the Bark of Meiogyne baillonii. J. Natural Prod. 81, 1610–1618. 10.1021/acs.jnatprod.8b00209 [DOI] [PubMed] [Google Scholar]
- Orellana R., Macaya C., Bravo G., Dorochesi F., Cumsille A., Valencia R., et al. (2018). Living at the Frontiers of Life: Extremophiles in Chile and Their Potential for Bioremediation. Front. Microbiol. 9, 2309. 10.3389/fmicb.2018.02309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C., Patron N. J., Huang A., Osbourn A. (2017). Harnessing plant metabolic diversity. Curr. Opin. Chem. Biol. 40, 24–30. 10.1016/j.cbpa.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J., Keasling J. D. (2014). Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 12, 355–367. 10.1038/nrmicro3240 [DOI] [PubMed] [Google Scholar]
- Pantin M., Brimble M. A., Furkert D. P. (2018). Total Synthesis of (-)-Peniphenone A. J. Org. Chem. 83, 7049–7059. 10.1021/acs.joc.7b03231 [DOI] [PubMed] [Google Scholar]
- Parrot D., Papazian S., Foil D., Tasdemir D. (2018). Imaging the Unimaginable: Desorption Electrospray Ionization - Imaging Mass Spectrometry (DESI-IMS) in Natural Product Research. Planta Med. 84, 584–593. 10.1055/s-0044-100188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. (2016). Salicornia: Evaluating the halophytic extremophile as a food and a pharmaceutical candidate. 3 Biotech. 6, 104. 10.1007/s13205-016-0418-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R., VanNieuwenhze M. S. (2019). Studies towards the Total Synthesis of Nogalamycin: Construction of the Complete ABCDEF-ring System via a Convergent Hauser Annulation. J. Org. Chem. 84, 760–768. 10.1021/acs.joc.8b02602 [DOI] [PubMed] [Google Scholar]
- Peresse T., Jezequel G., Allard P.-M., Pham V.-C., Huong D. T. M., Blanchard F., et al. (2017). Cytotoxic Prenylated Stilbenes Isolated from Macaranga tanarius. J. Natural Prod. 80, 2684–2691. 10.1021/acs.jnatprod.7b00409 [DOI] [PubMed] [Google Scholar]
- Pérez-Colmenares A., Obregón-Díaz Y., Rojas-Fermín L., Aparicio-Zambrano R., Carmona-Arzola J., Usubillaga A. (2018). Chemical Composition of the Essential Oil of Phyllanthus acidus. Natural Prod. Commun. 13, 97–98. 10.1177/1934578X1801300128 [DOI] [Google Scholar]
- Pham A. T., Malterud K. E., Paulsen B. S., Diallo D., Wangensteen H. (2014). alpha-Glucosidase inhibition, 15-lipoxygenase inhibition, and brine shrimp toxicity of extracts and isolated compounds from Terminalia macroptera leaves. Pharm. Biol. 52, 1166–1169. 10.3109/13880209.2014.880486 [DOI] [PubMed] [Google Scholar]
- Qi C., Wang W., Reichl K. D., McNeely J., Porco J. A. J. R. (2018). Total Synthesis of Aurofusarin: Studies on the Atropisomeric Stability of Bis-Naphthoquinones. Angew. Che. (Int. Ed. English) 57, 2101–2104. 10.1002/anie.201711535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.-J., Zhi Y.-E., Yan H., Zhang Y., Liu H., Yu Q., et al. (2018). Baeckfrutones A–L, polymethylated phloroglucinol meroterpenoids from the twigs and leaves of Baeckea frutescens. Tetrahedron 74, 6658–6666. 10.1016/j.tet.2018.09.050 [DOI] [Google Scholar]
- Qiu S., Yang W.-Z., Yao C.-L., Shi X.-J., Li J.-Y., Lou Y., et al. (2017). Malonylginsenosides with Potential Antidiabetic Activities from the Flower Buds of Panax ginseng. J. Natural Prod. 80, 899–908. 10.1021/acs.jnatprod.6b00789 [DOI] [PubMed] [Google Scholar]
- Quinn R. J., Carroll A. R., Pham N. B., Baron P., Palframan M. E., Suraweera L., et al. (2008). Developing a drug-like natural product library. J. Nat. Prod. (Gorakhpur) 71, 464–468. 10.1021/np070526y [DOI] [PubMed] [Google Scholar]
- Rédei D., Kúsz N., Sátori G., Kincses A., Spengler G., Burián K., et al. (2018). Bioactive Segetane, Ingenane, and Jatrophane Diterpenes from Euphorbia taurinensis. Planta Med. 84, 729–735. 10.1055/a-0589-0525 [DOI] [PubMed] [Google Scholar]
- Rahimzadeh M., Jahanshahi S., Moein S., Moein M. R. (2014). Evaluation of alpha- amylase inhibition by Urtica dioica and Juglans regia extracts. Iran. J. Basic Med. Sci. 17, 465–469. [PMC free article] [PubMed] [Google Scholar]
- Ramalhete C., Mulhovo S., Lage H., Ferreira M.-J. U. (2018). Triterpenoids from Momordica balsamina with a Collateral Sensitivity Effect for Tackling Multidrug Resistance in Cancer Cells. Planta Med. 84, 1372–1379. 10.1055/a-0651-8141 [DOI] [PubMed] [Google Scholar]
- Ramseyer J., Thuerig B., Mieri M., Scharer H.-J., Oberhansli T., Gupta M. P., et al. (2017). Eudesmane Sesquiterpenes from Verbesina lanata with Inhibitory Activity against Grapevine Downy Mildew. J. Natural Prod. 80, 3296–3304. 10.1021/acs.jnatprod.7b00868 [DOI] [PubMed] [Google Scholar]
- Rangel-Grimaldo M., Rivero-Cruz I., Madariaga-Mazon A., Figueroa M., Mata R. (2017). alpha-Glucosidase Inhibitors from Preussia minimoides double dagger. J. Natural Prod. 80, 582–587. 10.1021/acs.jnatprod.6b00574 [DOI] [PubMed] [Google Scholar]
- Raut A. A., Chorghade M. S., Vaidya A. D. B. (2017). “Reverse Pharmacology,” in Innovative approaches in drug discovery: Ethnopharmacology, systems biology and holistic targeting. Eds. Patwardhan B., Chaguturu R. (London: Academic Press; ), 89–126. [Google Scholar]
- Reed J., Stephenson M. J., Miettinen K., Brouwer B., Leveau A., Brett P., et al. (2017). A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab. Eng. 42, 185–193. 10.1016/j.ymben.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B. T., Mailyan A. K., Zakarian A. (2018). Total Synthesis of (+)-Guadinomic Acid via Hydroxyl-Directed Guanidylation. J. Org. Chem. 83, 9492–9496. 10.1021/acs.joc.8b01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Ding S.-S., Zhu A., Cao F., Zhu H.-J. (2017). Bioactive Azaphilone Derivatives from the Fungus Talaromyces aculeatus. J. Natural Prod. 80, 2199–2203. 10.1021/acs.jnatprod.7b00032 [DOI] [PubMed] [Google Scholar]
- Ren Y., Chen W.-L., Lantvit D. D., Sass E. J., Shriwas P., Ninh T. N., et al. (2017). Cardiac Glycoside Constituents of Streblus asper with Potential Antineoplastic Activity. J. Natural Prod. 80, 648–658. 10.1021/acs.jnatprod.6b00924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M. J. R., Schneider M., Brandstatter M., Krautwald S., Carreira E. M. (2018). Total Synthesis of (-)-Mitrephorone A. J. Am. Chem. Soc. 140, 16704–16710 10.1021/jacs.8b09685 [DOI] [PubMed] [Google Scholar]
- Riemer M., Uzunova V. V., Riemer N., Clarkson G. J., Pereira N., Napier R., et al. (2018). Phyllostictine A: total synthesis, structural verification and determination of substructure responsible for plant growth inhibition. Chem. Commun. (Cambridge England) 54, 7211–7214. 10.1039/c8cc03349h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios M. Y., Navarro V., Ramirez-Cisneros M. A., Salazar-Rios E. (2017). Sulfur-Containing Aristoloxazines and Other Constituents of the Roots of Aristolochia orbicularis. J. Natural Prod. 80, 3112–3119. 10.1021/acs.jnatprod.7b00226 [DOI] [PubMed] [Google Scholar]
- Robertson L. P., Duffy S., Wang Y., Wang D., Avery V. M., Carroll A. R. (2017). Pimentelamines A-C, Indole Alkaloids Isolated from the Leaves of the Australian Tree Flindersia pimenteliana. J. Natural Prod. 80, 3211–3217. 10.1021/acs.jnatprod.7b00587 [DOI] [PubMed] [Google Scholar]
- Rodrigues T., Reker D., Schneider P., Schneider G. (2016). Counting on natural products for drug design. Nat. Chem. 8, 531–541. 10.1038/nchem.2479 [DOI] [PubMed] [Google Scholar]
- Rosén J., Gottfries J., Muresan S., Backlund A., Oprea T. I. (2009. a). Novel chemical space exploration via natural products. J. Med. Chem. 52, 1953–1962. 10.1021/jm801514w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosén J., Lövgren A., Kogej T., Muresan S., Gottfries J., Backlund A. (2009. b). ChemGPS-NP(Web): chemical space navigation online. J. Comput. Aided Mol. Des. 23, 253–259. 10.1007/s10822-008-9255-y [DOI] [PubMed] [Google Scholar]
- Ross I. A. (1999). Medicinal plants of the world Part 1: Chemical constituents, traditional and modern medicinal uses (Totowa, New Jersey: Humana Press; ). [Google Scholar]
- Ross I. A. (2001). Medicinal plants of the world Part 2: Chemical constituents, traditional and modern medicinal uses (Totowa (N.J.): Humana Press; ). [Google Scholar]
- Roy A., Bharadvaja N. (2018). Biotechnological Approaches for the Production of Pharmaceutically Important Compound: Plumbagin. CPB 19, 372–381. 10.2174/1389201019666180629143842 [DOI] [PubMed] [Google Scholar]
- Rulliere P., Cannillo A., Grisel J., Cividino P., Sebastien C., Poisson J.-F. (2018). Total Synthesis of Proteasome Inhibitor (-)-Omuralide through Asymmetric Ketene 2 + 2-Cycloaddition. Org. Lett. 20, 4558–4561. 10.1021/acs.orglett.8b01851 [DOI] [PubMed] [Google Scholar]
- Ryu H. W., Park Y. J., Lee S. U., Lee S., Yuk H. J., Seo K.-H., et al. (2017). Potential Anti-inflammatory Effects of the Fruits of Paulownia tomentosa. J. Natural Prod. 80, 2659–2665. 10.1021/acs.jnatprod.7b00325 [DOI] [PubMed] [Google Scholar]
- Saad K. R., Parvatam G., Shetty N. P. (2018). Medium composition potentially regulates the anthocyanin production from suspension culture of Daucus carota. 3 Biotech. 8, 134. 10.1007/s13205-018-1146-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahraroo A., Babalar M., Mirjalili M. H., Fattahi Moghaddam M. R., Nejad Ebrahimi S. (2014). In-vitro Callus Induction and Rosmarinic Acid Quantification in Callus Culture of Satureja khuzistanica Jamzad (Lamiaceae). Iran. J. Pharm. Res. : IJPR 13, 1447–1456. [PMC free article] [PubMed] [Google Scholar]
- Salah El Dine R., Abdallah H. M., Kandil Z. A., Zaki A. A., Khan S. I., Khan I. A. (2019). PPARα and γ Activation Effects of New Nor-triterpenoidal Saponins from the Aerial Parts of Anabasis articulata. Planta Med. 85, 274–281. 10.1055/a-0762-0885 [DOI] [PubMed] [Google Scholar]
- Saleem H., Ahmad I., Shahid M. N., Gill M. S. A., Nadeem M. F., Mahmood W., et al. (2016). In Vitro Acetylcholinesterase And Butyrylcholinesterase Inhibitory Potentials Of Jatropha Gossypifolia Plant Extracts. Acta Pol. Pharm. 73, 419–423. [PubMed] [Google Scholar]
- Samala M., Lu T. N., Mandava S., Hwang J., Bogonda G., Kim D., et al. (2018). Stereoselective Protection-Free Asymmetric Total Synthesis of (+)-Chamuvarinin, a Potent Anticancer and Antitrypanosomal Agent: Substrate-Controlled Construction of the Adjacently Linked Oxatricyclic Core by Internal Alkylation. Org. Lett. 20, 6398–6402. 10.1021/acs.orglett.8b02706 [DOI] [PubMed] [Google Scholar]
- Sangwan N. S., Farooqi A. H. A., Shabih F., Sangwan R. S. (2001). Regulation of essential oil production in plants. Plant Growth Regul. 34, 3–21. 10.1023/A:1013386921596 [DOI] [Google Scholar]
- Sarker S. D., Nahar L. (2012). Natural Products Isolation (Totowa, NJ: Humana Press; ). [Google Scholar]
- Sarmiento F., Peralta R., Blamey J. M. (2015). Cold and Hot Extremozymes: Industrial Relevance and Current Trends. Front. Bioeng. Biotechnol. 3, 148. 10.3389/fbioe.2015.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saya J. M., Roose T. R., Peek J. J., Weijers B., Waal D. E, Thomas J. S., et al. (2018). Iodospirocyclization of Tryptamine-Derived Isocyanides: Formal Total Synthesis of Aspidofractinine. Angew. Che. (Int. Ed. English) 57, 15232–15236. 10.1002/anie.201809678 [DOI] [PubMed] [Google Scholar]
- Schloss S., Hackl T., Herz C., Lamy E., Koch M., Rohn S., et al. (2017). Detection of a Toxic Methylated Derivative of Phomopsin A Produced by the Legume-Infesting Fungus Diaporthe toxica. J. Natural Prod. 80, 1930–1934. 10.1021/acs.jnatprod.6b00662 [DOI] [PubMed] [Google Scholar]
- Schmid, Sattler, Grabley, Thiericke (1999). Natural Products in High Throughput Screening: Automated High-Quality Sample Preparation. J. Biomol. Screen 4, 15–25. 10.1177/108705719900400104 [DOI] [PubMed] [Google Scholar]
- Schuman M. C., van Dam N. M., Beran F., Harpole W. S. (2016). How does plant chemical diversity contribute to biodiversity at higher trophic levels? Curr. Opin. Insect Sci. 14, 46–55. 10.1016/j.cois.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Schwikkard S., Alqahtani A., Knirsch W., Wetschnig W., Jaksevicius A., Opara E. I., et al. (2017). Phytochemical Investigations of Three Rhodocodon (Hyacinthaceae Sensu APG II) Species. J. Natural Prod. 80, 30–37. 10.1021/acs.jnatprod.6b00240 [DOI] [PubMed] [Google Scholar]
- Scossa F., Benina M., Alseekh S., Zhang Y., Fernie A. R. (2018). The Integration of Metabolomics and Next-Generation Sequencing Data to Elucidate the Pathways of Natural Product Metabolism in Medicinal Plants. Planta Med. 84, 855–873. 10.1055/a-0630-1899 [DOI] [PubMed] [Google Scholar]
- Seeka C., Prabpai S., Kongsaeree P., Tewtrakul S., Lhinhatrakool T., Sutthivaiyakit S. (2017). Anti-inflammatory 12,20-Epoxypregnane and 11,12-seco-Pregnane Glycosides from the Stems of Hoya kerrii. J. Natural Prod. 80, 1714–1724. 10.1021/acs.jnatprod.6b00730 [DOI] [PubMed] [Google Scholar]
- Sendker J., Boker I., Lengers I., Brandt S., Jose J., Stark T., et al. (2017). Phytochemical Characterization of Low Molecular Weight Constituents from Marshmallow Roots (Althaea officinalis) and Inhibiting Effects of the Aqueous Extract on Human Hyaluronidase-1. J. Natural Prod. 80, 290–297. 10.1021/acs.jnatprod.6b00670 [DOI] [PubMed] [Google Scholar]
- Shaaban K. A., Saunders M. A., Zhang Y., Tran T., Elshahawi S. I., Ponomareva L. V., et al. (2017). Spoxazomicin D and Oxachelin C, Potent Neuroprotective Carboxamides from the Appalachian Coal Fire-Associated Isolate Streptomyces sp. RM-14-6. J. Natural Prod. 80, 2–11. 10.1021/acs.jnatprod.6b00948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Morrison K. R., Chodisetty S. S., Musaev D. G., Wuest W. M. (2018). Biologically Inspired Total Synthesis of Ulbactin F, an Iron-Binding Natural Product. Org. Lett. 20, 5922–5926. 10.1021/acs.orglett.8b02599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. B., Gupta R. (2015). Drug development from natural resource: a systematic approach. Mini Rev. Med. Chem. 15, 52–57. 10.2174/138955751501150224160518 [DOI] [PubMed] [Google Scholar]
- Sharma R., Gatchie L., Williams I. S., Jain S. K., Vishwakarma R. A., Chaudhuri B., et al. (2017). Glycyrrhiza glabra extract and quercetin reverses cisplatin resistance in triple-negative MDA-MB-468 breast cancer cells via inhibition of cytochrome P450 1B1 enzyme. Bioorg. Med. Chem. Lett. 27, 5400–5403. 10.1016/j.bmcl.2017.11.013 [DOI] [PubMed] [Google Scholar]
- Sharma R., Williams I. S., Gatchie L., Sonawane V. R., Chaudhuri B., Bharate S. B. (2018). Furanoflavones pongapin and lanceolatin B blocks the cell cycle and induce senescence in CYP1A1-overexpressing breast cancer cells. Bioorg. Med. Chem. 26, 6076–6086. 10.1016/j.bmc.2018.11.013 [DOI] [PubMed] [Google Scholar]
- Shi D.-H., Xu C., Guo B.-X., Wang X.-T., Chen Y.-X., Tan R.-X. (2008). Inhibition of soluble epoxide hydrolase by extracts derived from inflammation-treating Chinese medicinal herbs. Phytother. Res. : PTR 22, 1264–1268. 10.1002/ptr.2326 [DOI] [PubMed] [Google Scholar]
- Shimozu Y., Kuroda T., Tsuchiya T., Hatano T. (2017). Structures and Antibacterial Properties of Isorugosins H-J, Oligomeric Ellagitannins from Liquidambar formosana with Characteristic Bridging Groups between Sugar Moieties. J. Natural Prod. 80, 2723–2733. 10.1021/acs.jnatprod.7b00496 [DOI] [PubMed] [Google Scholar]
- Shin S. H., Lee S. R., Lee E., Kim K. H., Byun S. (2017). Caffeic Acid Phenethyl Ester from the Twigs of Cinnamomum cassia Inhibits Malignant Cell Transformation by Inducing c-Fos Degradation. J. Natural Prod. 80, 2124–2130. 10.1021/acs.jnatprod.7b00433 [DOI] [PubMed] [Google Scholar]
- Sibi G. (2015). Inhibition of lipase and inflammatory mediators by Chlorella lipid extracts for antiacne treatment. J. Adv. Pharm. Technol. Res. 6, 7–12. 10.4103/2231-4040.150364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidjui L. S., Eyong K. O., Hull K. G., Folefoc G. N., Leddet V. M., Herbette G., et al. (2017). Bioactive Seco-Lanostane-Type Triterpenoids from the Roots of Leplaea mayombensis. J. Natural Prod. 80, 2644–2651. 10.1021/acs.jnatprod.7b00210 [DOI] [PubMed] [Google Scholar]
- Silva G. H., Zeraik M. L., Oliveira C. M., Teles H. L., Trevisan H. C., Pfenning L. H., et al. (2017). Lactone Derivatives Produced by a Phaeoacremonium sp., an Endophytic Fungus from Senna spectabilis. J. Natural Prod. 80, 1674–1678. 10.1021/acs.jnatprod.5b00828 [DOI] [PubMed] [Google Scholar]
- Sirtori C. R. (2014). The pharmacology of statins. Pharmacol. Res. 88, 3–11. 10.1016/j.phrs.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Skinnider M. A., Dejong C. A., Franczak B. C., McNicholas P. D., Magarvey N. A. (2017). Comparative analysis of chemical similarity methods for modular natural products with a hypothetical structure enumeration algorithm. J. Cheminform. 9, 46. 10.1186/s13321-017-0234-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski M. J., Zhou H., Claesen J., Shen B., Fischbach M. A., Voigt C. A. (2016). Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 14, 135–149. 10.1038/nrmicro.2015.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares A. S., Barbosa F. L., Rudiger A. L., Hughes D. L., Salvador M. J., Zampronio A. R., et al. (2017). Naphthoquinones of Sinningia reitzii and Anti-inflammatory/Antinociceptive Activities of 8-Hydroxydehydrodunnione. J. Natural Prod. 80, 1837–1843. 10.1021/acs.jnatprod.6b01186 [DOI] [PubMed] [Google Scholar]
- Solyomváry A., Beni S., Boldizsar I. (2017). Dibenzylbutyrolactone Lignans - A Review of Their Structural Diversity, Biosynthesis, Occurrence, Identification and Importance. Mini Rev. Med. Chem. 17, 1053–1074. 10.2174/1389557516666160614005828 [DOI] [PubMed] [Google Scholar]
- Sribuhom T., Boueroy P., Hahnvajanawong C., Phatchana R., Yenjai C. (2017). Benzoyltyramine Alkaloids Atalantums A-G from the Peels of Atalantia monophylla and Their Cytotoxicity against Cholangiocarcinoma Cell Lines. J. Natural Prod. 80, 403–408. 10.1021/acs.jnatprod.6b00908 [DOI] [PubMed] [Google Scholar]
- Sriyatep T., Andersen R. J., Patrick B. O., Pyne S. G., Muanprasat C., Seemakhan S., et al. (2017). Scalemic Caged Xanthones Isolated from the Stem Bark Extract of Garcinia propinqua. J. Natural Prod. 80, 1658–1667. 10.1021/acs.jnatprod.7b00240 [DOI] [PubMed] [Google Scholar]
- Starkus J. G., Poerzgen P., Layugan K., Kawabata K. G., Goto J.-I., Suzuki S., et al. (2017). Scalaradial Is a Potent Inhibitor of Transient Receptor Potential Melastatin 2 (TRPM2) Ion Channels. J. Natural Prod. 80, 2741–2750. 10.1021/acs.jnatprod.7b00515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre A., Blondeau D., Lajeunesse A., Bley J., Bourdeau N., Desgagné-Penix I. (2018). Phytochemical Screening of Quaking Aspen (Populus tremuloides) Extracts by UPLC-QTOF-MS and Evaluation of their Antimicrobial Activity. Molecules 23, 1739. 10.3390/molecules23071739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Ortiz G. A., Cerda-Garcia-Rojas C. M., Fragoso-Serrano M., Pereda-Miranda R. (2017). Complementarity of DFT Calculations, NMR Anisotropy, and ECD for the Configurational Analysis of Brevipolides K-O from Hyptis brevipes. J. Natural Prod. 80, 181–189. 10.1021/acs.jnatprod.6b00953 [DOI] [PubMed] [Google Scholar]
- Suh W. S., Kim C. S., Subedi L., Kim S. Y., Choi S. U., Lee K. R. (2017). Iridoid Glycosides from the Twigs of Sambucus williamsii var. coreana and Their Biological Activities. J. Natural Prod. 80, 2502–2508. 10.1021/acs.jnatprod.7b00410 [DOI] [PubMed] [Google Scholar]
- Surapinit S., Sichaem J., Tip-Pyang S. (2018). Non-redox Lipoxygenase Inhibitors from Nauclea orientalis. Natural Prod. Commun. 13, 33–36. 10.1177/1934578X1801300111 [DOI] [Google Scholar]
- Suzuki A., Saito Y., Fukuyoshi S., Goto M., Miyake K., Newman D. J., et al. (2017). Corymbulosins D-H, 2-Hydroxy- and 2-Oxo-clerodane Diterpenes from the Bark of Laetia corymbulosa. J. Natural Prod. 80, 1065–1072. 10.1021/acs.jnatprod.6b01151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szopa A., Kokotkiewicz A., Luczkiewicz M., Ekiert H. (2017). Schisandra lignans production regulated by different bioreactor type. J. Biotechnol. 247, 11–17. 10.1016/j.jbiotec.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Szopa A., Klimek-Szczykutowicz M., Kokotkiewicz A., Maślanka A., Król A., Luczkiewicz M., et al. (2018). Phytochemical and biotechnological studies on Schisandra chinensis cultivar Sadova No. 1-a high utility medicinal plant. Appl. Microbiol. Biotechnol. 102, 5105–5120. 10.1007/s00253-018-8981-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabefam M., Farimani M. M., Danton O., Ramseyer J., Kaiser M., Ebrahimi S. N., et al. (2018). Antiprotozoal Diterpenes from Perovskia abrotanoides. Planta Med. 84, 913–919. 10.1055/a-0608-4946 [DOI] [PubMed] [Google Scholar]
- Tarazona G., Schleissner C., Rodriguez P., Perez M., Canedo L. M., Cuevas C. (2017). Streptenols F-I Isolated from the Marine-Derived Streptomyces misionensis BAT-10-03-023. J. Natural Prod. 80, 1034–1038. 10.1021/acs.jnatprod.6b01057 [DOI] [PubMed] [Google Scholar]
- Teta R., Marteinsson V. T., Longeon A., Klonowski A. M., Groben R., Bourguet-Kondracki M.-L., et al. (2017). Thermoactinoamide A, an Antibiotic Lipophilic Cyclopeptide from the Icelandic Thermophilic Bacterium Thermoactinomyces vulgaris. J. Natural Prod. 80, 2530–2535. 10.1021/acs.jnatprod.7b00560 [DOI] [PubMed] [Google Scholar]
- Thao N. P., Luyen B. T. T., Lee J. S., Kim J. H., Dat N. T., Kim Y. H. (2017). Inhibition Potential of Cycloartane-Type Glycosides from the Roots of Cimicifuga dahurica against Soluble Epoxide Hydrolase. J. Natural Prod. 80, 1867–1875. 10.1021/acs.jnatprod.7b00166 [DOI] [PubMed] [Google Scholar]
- The Angiosperm Phylogeny Group. Chase M. W., Christenhusz M. J. M., Fay M. F., Byng J. W., Judd W. S., et al. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc 181, (1), 1–20. 10.1111/boj.12385 [DOI] [Google Scholar]
- Thiericke R. (2000). Drug discovery from nature: Automated high-quality sample preparation. J. Autom. Methods Manage. Chem. 22, 149–157. 10.1155/S1463924600000249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg C. C., Britt J. R., Evans J. R., Akee R. K., Whitt J. A., Trinh S. K., et al. (2018). NCI Program for Natural Product Discovery: A Publicly-Accessible Library of Natural Product Fractions for High-Throughput Screening. ACS Chem. Biol. 13, 2484–2497. 10.1021/acschembio.8b00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii M., Kato H., Hitora Y., Angkouw E. D., Mangindaan R. E. P., Voogd N. J., et al. (2017). Lamellodysidines A and B, Sesquiterpenes Isolated from the Marine Sponge Lamellodysidea herbacea. J. Natural Prod. 80, 2536–2541. 10.1021/acs.jnatprod.7b00610 [DOI] [PubMed] [Google Scholar]
- Trethewey R. N. (2004). Metabolite profiling as an aid to metabolic engineering in plants. Curr. Opin. Plant Biol. 7, 196–201. 10.1016/j.pbi.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Trosset J.-Y., Carbonell P. (2015). Synthetic biology for pharmaceutical drug discovery. Drug Des. Dev. Ther. 9, 6285–6302. 10.2147/DDDT.S58049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsepaeva O. V., Nemtarev A. V., Abdullin T. I., Grigor’eva L. R., Kuznetsova E. V., Akhmadishina R. A., et al. (2017). Design, Synthesis, and Cancer Cell Growth Inhibitory Activity of Triphenylphosphonium Derivatives of the Triterpenoid Betulin. J. Natural Prod. 80, 2232–2239. 10.1021/acs.jnatprod.7b00105 [DOI] [PubMed] [Google Scholar]
- Tshitenge D. T., Feineis D., Awale S., Bringmann G. (2017). Gardenifolins A-H, Scalemic Neolignans from Gardenia ternifolia: Chiral Resolution, Configurational Assignment, and Cytotoxic Activities against the HeLa Cancer Cell Line. J. Natural Prod. 80, 1604–1614. 10.1021/acs.jnatprod.7b00180 [DOI] [PubMed] [Google Scholar]
- Tsukamoto H., Hanada S., Nomura Y., Doi T. (2018). Total Synthesis of Spiromamakone A and Structure Revision of Spiropreussione A. J. Org. Chem. 83, 9430–9441. 10.1021/acs.joc.8b01075 [DOI] [PubMed] [Google Scholar]
- Tuan N. Q., Lee D.-H., Oh J., Kim C. S., Heo K.-S., Myung C.-S., et al. (2017). Inhibition of Proliferation of Vascular Smooth Muscle Cells by Cucurbitanes from Momordica charantia. J. Natural Prod. 80, 2018–2025. 10.1021/acs.jnatprod.7b00151 [DOI] [PubMed] [Google Scholar]
- Turumtay H., Midilli A., Turumtay E. A., Demir A., Selvi E. K., Budak E. E., et al. (2017). Gram (-) microorganisms DNA polymerase inhibition, antibacterial and chemical properties of fruit and leaf extracts of Sorbus acuparia and Sorbus caucasica var. yaltirikii. Biomed. Chromatogr. BMC 31. 10.1002/bmc.3901 [DOI] [PubMed] [Google Scholar]
- Twigg D. G., Baldassarre L., Frye E. C., Galloway, Warren R. J. D., Spring D. R. (2018). Bioinspired Total Synthesis of Bussealin E. Org. Lett. 20, 1597–1599. 10.1021/acs.orglett.8b00340 [DOI] [PubMed] [Google Scholar]
- Umeno D., Tobias A. V., Arnold F. H. (2005). Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. : MMBR 69, 51–78. 10.1128/mmbr.69.1.51-78.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Chan L. S., Garcia-Camara I., Torres-Tapia L. W., Moo-Puc R. E., Peraza-Sanchez S. R. (2017). Lupane-Type Triterpenes of Phoradendron vernicosum. J. Natural Prod. 80, 3038–3042. 10.1021/acs.jnatprod.7b00177 [DOI] [PubMed] [Google Scholar]
- Vallisuta O., Nukoolkarn V., Mitrevej A., Sarisuta N., Leelapornpisid P., Phrutivorapongkul A., et al. (2014). In vitro studies on the cytotoxicity, and elastase and tyrosinase inhibitory activities of marigold (Tagetes erecta L.) flower extracts. Exp. Ther. Med. 7, 246–250. 10.3892/etm.2013.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardo G., Gorassini A., Ricci D., Fraternale D. (2017). High Triterpenic Acids Production in Callus Cultures from Fruit Pulp of Two Apple Varieties. Phytochem. Anal. 28, 5–15. 10.1002/pca.2638 [DOI] [PubMed] [Google Scholar]
- Verastegui-Omana B., Rebollar-Ramos D., Perez-Vasquez A., Martinez A. L., Madariaga-Mazon A., Flores-Bocanegra L., et al. (2017). alpha-Glucosidase Inhibitors from Malbranchea flavorosea. J. Natural Prod. 80, 190–195. 10.1021/acs.jnatprod.6b00977 [DOI] [PubMed] [Google Scholar]
- Wagh V. D., Korinek M., Lo I.-W., Hsu Y.-M., Chen S.-L., Hsu H.-Y., et al. (2017). Inflammation Modulatory Phorbol Esters from the Seeds of Aquilaria malaccensis. J. Natural Prod. 80, 1421–1427. 10.1021/acs.jnatprod.6b01096 [DOI] [PubMed] [Google Scholar]
- Wang C.-J., Yan Q.-L., Ma Y.-F., Sun C.-P., Chen C.-M., Tian X.-G., et al. (2017). ent-Abietane and Tigliane Diterpenoids from the Roots of Euphorbia fischeriana and Their Inhibitory Effects against Mycobacterium smegmatis. J. Natural Prod. 80, 1248–1254. 10.1021/acs.jnatprod.6b00786 [DOI] [PubMed] [Google Scholar]
- Wang K.-B., Li D.-H., Bao Y., Cao F., Wang W.-J., Lin C., et al. (2017). Structurally Diverse Alkaloids from the Seeds of Peganum harmala. J. Natural Prod. 80, 551–559. 10.1021/acs.jnatprod.6b01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-Y., Chen M.-H., Wu J., Sun H., Liu W., Qu Y.-H., et al. (2017). Bioactive Glycosides from the Twigs of Litsea cubeba. J. Natural Prod. 80, 1808–1818. 10.1021/acs.jnatprod.6b01189 [DOI] [PubMed] [Google Scholar]
- Wang X., Li L., Zhu R., Zhang J., Zhou J., Lou H. (2017). Bibenzyl-Based Meroterpenoid Enantiomers from the Chinese Liverwort Radula sumatrana. J. Natural Prod. 80, 3143–3150. 10.1021/acs.jnatprod.7b00394 [DOI] [PubMed] [Google Scholar]
- Wang Y.-S., Li B.-T., Liu S.-X., Wen Z.-Q., Yang J.-H., Zhang H.-B., et al. (2017). Anisucoumaramide, a Bioactive Coumarin from Clausena anisum-olens. J. Natural Prod. 80, 798–804. 10.1021/acs.jnatprod.6b00391 [DOI] [PubMed] [Google Scholar]
- Wang J., Duan H., Wang Y., Pan B., Gao C., Gai C., et al. (2017. a). ent-Strobane and ent-Pimarane Diterpenoids from Siegesbeckia pubescens. J. Natural Prod. 80, 19–29. 10.1021/acs.jnatprod.6b00150 [DOI] [PubMed] [Google Scholar]
- Wang J., He W., Kong F., Tian X., Wang P., Zhou X., et al. (2017. b). Ochracenes A-I, Humulane-Derived Sesquiterpenoids from the Antarctic Fungus Aspergillus ochraceopetaliformis. J. Natural Prod. 80, 1725–1733. 10.1021/acs.jnatprod.6b00810 [DOI] [PubMed] [Google Scholar]
- Wang J., Mu F.-R., Jiao W.-H., Huang J., Hong L.-L., Yang F., et al. (2017. c). Meroterpenoids with Protein Tyrosine Phosphatase 1B Inhibitory Activity from a Hyrtios sp. Marine Sponge. J. Natural Prod. 80, 2509–2514. 10.1021/acs.jnatprod.7b00435 [DOI] [PubMed] [Google Scholar]
- Wang S.-L., Liao H.-R., Cheng M.-J., Shu C.-W., Chen C.-L., Chung M.-I., et al. (2018). Four New 2-(2-Phenylethyl)-4H-chromen-4-one Derivatives from the Resinous Wood of Aquilaria sinensis and Their Inhibitory Activities on Neutrophil Pro-Inflammatory Responses. Planta Med. 84, 1340–1347. 10.1055/a-0645-1437 [DOI] [PubMed] [Google Scholar]
- Wang X., Liu F., Yun J., Feng Z., Jiang J., Yang Y., et al. (2018). Iron-Catalyzed Synthesis of the Hexahydrocyclopentacfuran Core and Concise Total Synthesis of Polyflavanostilbene B. Angew. Che. (Int. Ed. English) 57, 10127–10131. 10.1002/anie.201804329 [DOI] [PubMed] [Google Scholar]
- Wang X., Du C., Hong B., Lei X. (2019). Total synthesis of (+/-)-antroquinonol. Org. Biomolec. Chem. 17, 1754–1757. 10.1039/c8ob02494d [DOI] [PubMed] [Google Scholar]
- Waratchareeyakul W., Hellemann E., Gil R. R., Chantrapromma K., Langat M. K., Mulholland D. A. (2017). Application of Residual Dipolar Couplings and Selective Quantitative NOE to Establish the Structures of Tetranortriterpenoids from Xylocarpus rumphii. J. Natural Prod. 80, 391–402. 10.1021/acs.jnatprod.6b00906 [DOI] [PubMed] [Google Scholar]
- Waterman M. J., Nugraha A. S., Hendra R., Ball G. E., Robinson S. A., Keller P. A. (2017). Antarctic Moss Biflavonoids Show High Antioxidant and Ultraviolet-Screening Activity. J. Natural Prod. 80, 2224–2231. 10.1021/acs.jnatprod.7b00085 [DOI] [PubMed] [Google Scholar]
- Wei T., Dixon D. J. (2018). Catalytic stereoselective total synthesis of a spiro-oxindole alkaloid and the pentacyclic core of tryptoquivalines. Chem. Commun. (Cambridge England) 54, 12860–12862. 10.1039/c8cc07479h [DOI] [PubMed] [Google Scholar]
- Wei Y., Wang C., Cheng Z., Tian X., Jia J., Cui Y., et al. (2017). Heterodimeric Diterpenoids Isolated from Euphorbia ebracteolata Roots and Their Inhibitory Effects on alpha-Glucosidase. J. Natural Prod. 80, 3218–3223. 10.1021/acs.jnatprod.7b00595 [DOI] [PubMed] [Google Scholar]
- Weller M. G. (2012). A unifying review of bioassay-guided fractionation, effect-directed analysis and related techniques. Sensors 12, 9181–9209. 10.3390/s120709181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo M., Wang Q., Holst J., White J. M., Hofmann A., Davis R. A. (2017). Celastrofurans A-G: Dihydro-beta-agarofurans from the Australian Rainforest Vine Celastrus subspicata and Their Inhibitory Effect on Leucine Transport in Prostate Cancer Cells. J. Natural Prod. 80, 1918–1925. 10.1021/acs.jnatprod.7b00220 [DOI] [PubMed] [Google Scholar]
- Wilson S. A., Roberts S. C. (2012). Recent advances towards development and commercialization of plant cell culture processes for the synthesis of biomolecules. Plant Biotechnol. J. 10, 249–268. 10.1111/j.1467-7652.2011.00664.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfender J.-L., Nuzillard J.-M., van der Hooft J. J. J., Renault J.-H., Bertrand S. (2019). Accelerating metabolite identification in natural product research: Toward an ideal combination of LC-HRMS/MS and NMR profiling, in silico databases and chemometrics. Analyt. Chem. 91, 704–742. 10.1021/acs.analchem.8b05112 [DOI] [PubMed] [Google Scholar]
- Woo J.-K., Ha T. K. Q., Oh D.-C., Oh W.-K., Oh K.-B., Shin J. (2017). Polyoxygenated Steroids from the Sponge Clathria gombawuiensis. J. Natural Prod. 80, 3224–3233. 10.1021/acs.jnatprod.7b00651 [DOI] [PubMed] [Google Scholar]
- Wu Y.-B., Wang Y.-Z., Ni Z.-Y., Qing X., Shi Q.-W., Sauriol F., et al. (2017). Xylomexicanins I and J: Limonoids with Unusual B/C Rings from Xylocarpus granatum. J. Natural Prod. 80, 2547–2550. 10.1021/acs.jnatprod.7b00305 [DOI] [PubMed] [Google Scholar]
- Xiao F., Li H., Xu M., Li T., Wang J., Sun C., et al. (2018). Staurosporine Derivatives Generated by Pathway Engineering in a Heterologous Host and Their Cytotoxic Selectivity. J. Natural Prod. 81, 1745–1751. 10.1021/acs.jnatprod.8b00103 [DOI] [PubMed] [Google Scholar]
- Xie T., Song S., Li S., Ouyang L., Xia L., Huang J. (2015). Review of natural product databases. Cell Proliferat. 48, 398–404. 10.1111/cpr.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Wang Y., Jiang W.-W., Luo X.-F., Dai T.-Y., Peng L., et al. (2018). Moringa oleifera Leaf Petroleum Ether Extract Inhibits Lipogenesis by Activating the AMPK Signaling Pathway. Front. Pharmacol. 9, 1447. 10.3389/fphar.2018.01447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Wan J., Ding J., Wang P.-P., Ma G.-L., Li J., et al. (2017). Camellianols A-G, Barrigenol-like Triterpenoids with PTP1B Inhibitory Effects from the Endangered Ornamental Plant Camellia crapnelliana. J. Natural Prod. 80, 2874–2882. 10.1021/acs.jnatprod.7b00241 [DOI] [PubMed] [Google Scholar]
- Xu L., Wu P., Xue J., Molnar I., Wei X. (2017). Antifungal and Cytotoxic beta-Resorcylic Acid Lactones from a Paecilomyces Species. J. Natural Prod. 80, 2215–2223. 10.1021/acs.jnatprod.7b00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Yao H., Hu M., Li D., Zhu Z., Xie W., et al. (2017). 6,7-Seco-ent-Kauranoids Derived from Oridonin as Potential Anticancer Agents. J. Natural Prod. 80, 2391–2398. 10.1021/acs.jnatprod.7b00057 [DOI] [PubMed] [Google Scholar]
- Xu Y.-M., Wijeratne E. M. K., Babyak A. L., Marks H. R., Brooks A. D., Tewary P., et al. (2017). Withanolides from Aeroponically Grown Physalis peruviana and Their Selective Cytotoxicity to Prostate Cancer and Renal Carcinoma Cells. J. Natural Prod. 80, 1981–1991. 10.1021/acs.jnatprod.6b01129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wang G., Xu F., Li W., Lin A., Yao H., et al. (2018). Concise Total Synthesis of (+/-)-Deguelin and (+/-)-Tephrosin Using a Vinyl Iodide as a Key Building Block. J. Natural Prod. 81, 1055–1059. 10.1021/acs.jnatprod.7b00794 [DOI] [PubMed] [Google Scholar]
- Xue Y., Zhao P., Quan C., Zhao Z., Gao W., Li J., et al. (2018). Cyanobacteria-derived peptide antibiotics discovered since 2000. Peptides 107, 17–24. 10.1016/j.peptides.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Yñigez-Gutierrez A. E., Bachmann B. O. (2019). Fixing the Unfixable: The Art of Optimizing Natural Products for Human Medicine. J. Med. Chem. 62, 8412–8428. 10.1021/acs.jmedchem.9b00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav D. K., Bharitkar Y. P., Hazra A., Pal U., Verma S., Jana S., et al. (2017). Tamarixetin 3-O-beta-d-Glucopyranoside from Azadirachta indica Leaves: Gastroprotective Role through Inhibition of Matrix Metalloproteinase-9 Activity in Mice. J. Natural Prod. 80, 1347–1353. 10.1021/acs.jnatprod.6b00957 [DOI] [PubMed] [Google Scholar]
- Yan C., Zhang W.-Q., Sun M., Liang W., Wang T.-Y., Zhang Y.-D., et al. (2018). Carpescernolides A and B, rare oxygen bridge-containing sesquiterpene lactones from Carpesium cernuum. Tetrahedron Lett. 59, 4063–4066. 10.1016/j.tetlet.2018.09.067 [DOI] [Google Scholar]
- Yang P.-F., Feng Z.-M., Yang Y.-N., Jiang J.-S., Zhang P.-C. (2017). Neuroprotective Caffeoylquinic Acid Derivatives from the Flowers of Chrysanthemum morifolium. J. Natural Prod. 80, 1028–1033. 10.1021/acs.jnatprod.6b01026 [DOI] [PubMed] [Google Scholar]
- Yang G.-C., Hu J.-H., Li B.-L., Liu H., Wang J.-Y., Sun L.-X. (2018). Six New neo-Clerodane Diterpenoids from Aerial Parts of Scutellaria barbata and Their Cytotoxic Activities. Planta Med. 84, 1292–1299. 10.1055/a-0638-8255 [DOI] [PubMed] [Google Scholar]
- Yang H. H., Oh K.-E., Jo Y. H., Ahn J. H., Liu Q., Turk A., et al. (2018). Characterization of tyrosinase inhibitory constituents from the aerial parts of Humulus japonicus using LC-MS/MS coupled online assay. Bioorg. Med. Chem. 26, 509–515. 10.1016/j.bmc.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Yang X., Gao X., Cao Y., Guo Q., Li S., Zhu Z., et al. (2018). Anti-Inflammatory Effects of Boldine and Reticuline Isolated from Litsea cubeba through JAK2/STAT3 and NF-κB Signaling Pathways. Planta Med. 84, 20–25. 10.1055/s-0043-113447 [DOI] [PubMed] [Google Scholar]
- Yao X., Shan X., Zu L. (2018). Divergent Coupling of 2-Carbonyl-anilines and Diazo-cyclopentanones: Asymmetric Total Synthesis of (+)-Leucomidine A. Org. Lett. 20, 6498–6501. 10.1021/acs.orglett.8b02823 [DOI] [PubMed] [Google Scholar]
- Ye X.-S., He J., Cheng Y.-C., Zhang L., Qiao H.-Y., Pan X.-G., et al. (2017). Cornusides A-O, Bioactive Iridoid Glucoside Dimers from the Fruit of Cornus officinalis. J. Natural Prod. 80, 3103–3111. 10.1021/acs.jnatprod.6b01127 [DOI] [PubMed] [Google Scholar]
- Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., et al. (2004). Small Molecules Blocking the Entry of Severe Acute Respiratory Syndrome Coronavirus into Host Cells. J. Virol. 78, 11334–11339. 10.1128/JVI.78.20.11334-11339.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A., Crowley R. S., Sherwood A. M., Prisinzano T. E. (2017). Semisynthesis and Kappa-Opioid Receptor Activity of Derivatives of Columbin, a Furanolactone Diterpene. J. Natural Prod. 80, 2094–2100. 10.1021/acs.jnatprod.7b00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama T., Arai M. A., Akamine R., Koryudzu K., Tsuchiya A., Sadhu S. K., et al. (2017). Notch Inhibitors from Calotropis gigantea That Induce Neuronal Differentiation of Neural Stem Cells. J. Natural Prod. 80, 2453–2461. 10.1021/acs.jnatprod.7b00282 [DOI] [PubMed] [Google Scholar]
- Yongye A. B., Waddell J., Medina-Franco J. L. (2012). Molecular scaffold analysis of natural products databases in the public domain. Chem. Biol. Drug Des. 80, 717–724. 10.1111/cbdd.12011 [DOI] [PubMed] [Google Scholar]
- Yu Y., Chang P., Yu H., Ren H., Hong D., Li Z., et al. (2018). Productive Amyrin Synthases for Efficient α-Amyrin Synthesis in Engineered Saccharomyces cerevisiae. ACS Synth. Biol. 7, 2391–2402. 10.1021/acssynbio.8b00176 [DOI] [PubMed] [Google Scholar]
- Yuan Z., Hu X., Zhang H., Liu L., Chen P., He M., et al. (2018). Total synthesis of conosilane A via a site-selective C-H functionalization strategy. Chem. Commun. (Cambridge England) 54, 912–915. 10.1039/c7cc09367e [DOI] [PubMed] [Google Scholar]
- Zampieri M., Sekar K., Zamboni N., Sauer U. (2017). Frontiers of high-throughput metabolomics. Curr. Opin. Chem. Biol. 36, 15–23. 10.1016/j.cbpa.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Zani C. L., Carroll A. R. (2017). Database for Rapid Dereplication of Known Natural Products Using Data from MS and Fast NMR Experiments. J. Natural Prod. 80, 1758–1766. 10.1021/acs.jnatprod.6b01093 [DOI] [PubMed] [Google Scholar]
- Zarev Y., Foubert K., Ionkova I., Apers S., Pieters L. (2017). Isolation and Structure Elucidation of Glucosylated Colchicinoids from the Seeds of Gloriosa superba by LC-DAD-SPE-NMR. J. Natural Prod. 80, 1187–1191. 10.1021/acs.jnatprod.6b01024 [DOI] [PubMed] [Google Scholar]
- Zeng T., Wu X.-Y., Yang S.-X., Lai W.-C., Shi S.-D., Zou Q., et al. (2017). Monoterpenoid Indole Alkaloids from Kopsia officinalis and the Immunosuppressive Activity of Rhazinilam. J. Natural Prod. 80, 864–871. 10.1021/acs.jnatprod.6b00697 [DOI] [PubMed] [Google Scholar]
- Zhan G., Zhou J., Liu J., Huang J., Zhang H., Liu R., et al. (2017). Acetylcholinesterase Inhibitory Alkaloids from the Whole Plants of Zephyranthes carinata. J. Natural Prod. 80, 2462–2471. 10.1021/acs.jnatprod.7b00301 [DOI] [PubMed] [Google Scholar]
- Zhang M. Y., Barrow R. A. (2018). Total Synthesis of Boletopsin 11 Enabled by Directed ortho-C(sp(2))-H Arylation. J. Org. Chem. 83, 6776–6782. 10.1021/acs.joc.8b00792 [DOI] [PubMed] [Google Scholar]
- Zhang C.-G., Chou G.-X., Mao X.-D., Yang Q.-S., Zhou J.-L. (2017). Nepetaefolins A-J, Cytotoxic Chinane and Abietane Diterpenoids from Caryopteris nepetaefolia. J. Natural Prod. 80, 1742–1749. 10.1021/acs.jnatprod.6b00972 [DOI] [PubMed] [Google Scholar]
- Zhang C.-L., Hao Z.-Y., Liu Y.-F., Wang Y., Shi G.-R., Jiang Z.-B., et al. (2017). Polycycloiridals with a Cyclopentane Ring from Iris tectorum. J. Natural Prod. 80, 156–161. 10.1021/acs.jnatprod.6b00796 [DOI] [PubMed] [Google Scholar]
- Zhang D., Guo J., Zhang M., Liu X., Ba M., Tao X., et al. (2017). Oxazole-Containing Diterpenoids from Cell Cultures of Salvia miltiorrhiza and Their Anti-HIV-1 Activities. J. Natural Prod. 80, 3241–3246. 10.1021/acs.jnatprod.7b00659 [DOI] [PubMed] [Google Scholar]
- Zhang F., Barns K., Hoffmann F. M., Braun D. R., Andes D. R., Bugni T. S. (2017). Thalassosamide, a Siderophore Discovered from the Marine-Derived Bacterium Thalassospira profundimaris. J. Natural Prod. 80, 2551–2555. 10.1021/acs.jnatprod.7b00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-F., Chen L., Huang S., Shan L.-H., Gao F., Zhou X.-L. (2017). Diterpenoid Alkaloids from Two Aconitum Species with Antifeedant Activity against Spodoptera exigua. J. Natural Prod. 80, 3136–3142. 10.1021/acs.jnatprod.7b00380 [DOI] [PubMed] [Google Scholar]
- Zhang P.-L., Han Y., Zhang L.-T., Wang X.-L., Shen T., Ren D., et al. (2017). Botrysphones A-C and Botrysphins A-F, Triketides and Diterpenoids from the Fungus Botrysphaeria laricina. J. Natural Prod. 80, 1791–1797. 10.1021/acs.jnatprod.6b01196 [DOI] [PubMed] [Google Scholar]
- Zhang Y.-L., Zhou X.-W., Wu L., Wang X.-B., Yang M.-H., Luo J., et al. (2017). Isolation, Structure Elucidation, and Absolute Configuration of Syncarpic Acid-Conjugated Terpenoids from Rhodomyrtus tomentosa. J. Natural Prod. 80, 989–998. 10.1021/acs.jnatprod.6b01005 [DOI] [PubMed] [Google Scholar]
- Zhang H.-J., Rumschlag-Booms E., Guan Y.-F., Wang D.-Y., Liu K.-L., Li W.-F., et al. (2017. a). Correction to Potent Inhibitor of Drug-Resistant HIV-1 Strains Identified from the Medicinal Plant Justicia gendarussa. J. Natural Prod. 80, 2390. 10.1021/acs.jnatprod.7b00562 [DOI] [PubMed] [Google Scholar]
- Zhang H.-J., Rumschlag-Booms E., Guan Y.-F., Wang D.-Y., Liu K.-L., Li W.-F., et al. (2017. b). Potent Inhibitor of Drug-Resistant HIV-1 Strains Identified from the Medicinal Plant Justicia gendarussa. J. Natural Prod. 80, 1798–1807. 10.1021/acs.jnatprod.7b00004 [DOI] [PubMed] [Google Scholar]
- Zhang F.-M., Zhang S.-Y., Tu Y.-Q. (2018). Recent progress in the isolation, bioactivity, biosynthesis, and total synthesis of natural spiroketals. Nat. Prod. Rep. 35, 75–104. 10.1039/c7np00043j [DOI] [PubMed] [Google Scholar]
- Zhang J.-S., Weng H.-Z., Huang J.-L., Tang G.-H., Yin S. (2018). Anti-inflammatory Ingenane Diterpenoids from the Roots of Euphorbia kansui. Planta Med. 84, 1334–1339. 10.1055/a-0646-4306 [DOI] [PubMed] [Google Scholar]
- Zhang M. Y., Malins L. R., Ward J. S., Barrow R. A. (2018). Total Synthesis of Suillusin. Org. Lett. 20, 7304–7307. 10.1021/acs.orglett.8b03234 [DOI] [PubMed] [Google Scholar]
- Zhang W., Xiao D., Wang B. (2018). A concise total synthesis of cochlearoid B. Org. Biomolec. Chem. 16, 3358–3361. 10.1039/c8ob00615f [DOI] [PubMed] [Google Scholar]
- Zhang Y.-F., Zhu Z.-X., Sun H., Yao H.-N., Chen X.-N., Liu L., et al. (2018). Stachyodin A, a pterocarpan derivative with unusual spirotetrahydrofuran ring from the roots of Indigofera stachyodes. Tetrahedron Lett. 59, 4514–4516. 10.1016/j.tetlet.2018.11.024 [DOI] [Google Scholar]
- Zhao J., Feng J., Tan Z., Liu J., Zhao J., Chen R., et al. (2017). Stachybotrysins A-G, Phenylspirodrimane Derivatives from the Fungus Stachybotrys chartarum. J. Natural Prod. 80, 1819–1826. 10.1021/acs.jnatprod.7b00014 [DOI] [PubMed] [Google Scholar]
- Zhao J.-X., Fan Y.-Y., Xu J.-B., Gan L.-S., Xu C.-H., Ding J., et al. (2017). Diterpenoids and Lignans from Cephalotaxus fortunei. J. Natural Prod. 80, 356–362. 10.1021/acs.jnatprod.6b00802 [DOI] [PubMed] [Google Scholar]
- Zhao X., Park S. Y., Yang D., Lee S. Y. (2019). Synthetic Biology for Natural Compounds. Biochemistry 58, 1454–1456. 10.1021/acs.biochem.8b00569 [DOI] [PubMed] [Google Scholar]
- Zheng D., Han L., Qu X., Chen X., Zhong J., Bi X., et al. (2017). Cytotoxic Fusicoccane-Type Diterpenoids from Streptomyces violascens Isolated from Ailuropoda melanoleuca Feces. J. Natural Prod. 80, 837–844. 10.1021/acs.jnatprod.6b00676 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Zhang J., Wei L., Shi M., Wang J., Huang J. (2017). Gunnilactams A-C, Macrocyclic Tetralactams from the Mycelial Culture of the Entomogenous Fungus Paecilomyces gunnii. J. Natural Prod. 80, 1935–1938. 10.1021/acs.jnatprod.7b00060 [DOI] [PubMed] [Google Scholar]
- Zheng T., Cheng L.-Z., Yan Y.-M., Xu F.-R., Tang X.-L., Li X.-Z., et al. (2018). Compounds from the Roots of Codonopsis pilosula and Their SIRT1 Regulatory Activity. Natural Prod. Commun. 13, 37–40. 10.1177/1934578X1801300112 [DOI] [Google Scholar]
- Zhou X.-J., Rahmani R. (1992). Preclinical and Clinical Pharmacology of Vinca Alkaloids. Drugs 44, 1–16. 10.2165/00003495-199200444-00002 [DOI] [PubMed] [Google Scholar]
- Zhou S.-Y., Zhou T.-L., Qiu G., Huan X., Miao Z.-H., Yang S.-P., et al. (2018). Three New Cytotoxic Monoterpenoid Bisindole Alkaloids from Tabernaemontana bufalina. Planta Med. 84, 1127–1133. 10.1055/a-0608-4988 [DOI] [PubMed] [Google Scholar]
- Zhu M., Zhang X., Feng H., Dai J., Li J., Che Q., et al. (2017). Penicisulfuranols A-F, Alkaloids from the Mangrove Endophytic Fungus Penicillium janthinellum HDN13-309. J. Natural Prod. 80, 71–75. 10.1021/acs.jnatprod.6b00483 [DOI] [PubMed] [Google Scholar]
- Zhu G., Wang H., Gan K., Gao H., Li W. (2018). A New Purine Derivative from the Roots of Phyllanthus flexuosus. Natural Prod. Commun. 13, 41–44. 10.1177/1934578X1801300113 [DOI] [Google Scholar]
- Zong Y., Wang W., Xu T. (2018). Total Synthesis of Bioactive Marine Meroterpenoids: The Cases of Liphagal and Frondosin B. Mar. Drugs 16, 155. 10.3390/md16040115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.