Abstract
AIM
To construct an immune-related prognostic signature (IPS) that can distinguish and predict prognosis in uveal melanoma (UM).
METHODS
The transcriptomic data and clinicopathological information of 80 UM patients were extracted from the TCGA database. These patients were randomly assigned to a training and a testing set.
RESULTS
Lasso Cox analysis was performed for the prognostic immune-related genes to develop an IPS for UM in the training set. The signature was validated in both the testing set and entire cohort. We confirmed the prognostic value of our IPS in distinct subgroups and found its association with the T stage and basal diameter of the tumor. Tumor Immune Estimation Resource database analysis revealed that the IPS was negatively correlated with the infiltration of neutrophils and dendritic cells, but positively correlated with the infiltration level of CD8+ T cells. In addition, we demonstrated that immune checkpoints containing PD-1, CTLA-4, IDO, and TIGIT were moderately associated with the IPS.
CONCLUSION
This is the first study to develop and validate an immune-related signature with the ability of predicting prognosis for UM patients. Further studies are needed to validate its prediction accuracy.
Keywords: uveal melanoma, TCGA, prognosis, immune-related signature
INTRODUCTION
Uveal melanoma (UM) is a rare subset of melanoma, but the most common primary intraocular malignancy in adults, arising from melanocytes of the uveal tract (contains iris, choroid, and stroma of the ciliary body) of the eye[1]–[2]. Approximately 40% of UM patients develop metastatic disease with liver being the most common site, leading to a high mortality rate[3]–[4]. Treatment of the primary disease is surgical removal of the tumor, or a more conservative radiotherapy to preserve the affected eye[5]. Despite advancement made in diagnosis and treatment of primary UM, survival rate has not significantly improved during the past three decades[6].
Although the emerging and promising immunotherapy using checkpoint inhibitors has improved the outcomes of many solid tumors, the eye, as an immune privileged region, is associated with a number of immune-suppressive and immune-evasive mechanisms affecting the efficacy of current immune therapies[7]. Accumulating evidence have shown the high immunogenicity of UM by analyzing tumor mutational load, tumor antigen expression, endogenous anti-tumor immunologic reactivity, and the effect of transferring, tumor-infiltrating lymphocytes[8]–[9], highlighting the importance of tumor immune environment characterization. However, few studies have systematically explored the immune microenvironment of UM, and feasible immune biomarkers to predict survival for UM patients are lacking. Thus, it is necessary to develop a stable immune-related signature that can be used as a prognosis predictor and screening tool to identify patients who are more likely to benefit from immunotherapy.
To that end, we extracted transcriptome data from the Cancer Genome Atlas (TCGA) to establish and validate an immune-related prognostic signature (IPS). Combined with the clinicopathological characteristics of patients, we investigated the potential clinical utility of our IPS indifferent UM subgroups. Furthermore, we analyzed the correlation of the IPS with immune cell infiltration and immune checkpoints to gain insight into the tumor immune microenvironment.
MATERIALS AND METHODS
Ethical Approval
Because the TCGA database contains only anonymous data and is accessed publicly, no additional Institutional Review Board or Ethics Committee was required for this study.
Gene Expression Dataset and Immune-Related Genes
Transcriptomic data and clinicopathological information of 80 UM cases were extracted from TCGA website (https://portal.gdc.cancer.gov/repository). We chose the transcriptome profiling of RNA expression with fragments per kilobase of exon per million fragments mapped (FPKM) values and performed log2-based transformation for normalization. We removed genes with FPKM value of 0 in more than 50% of samples to ensure detection reliability. Using ImmPort database (https://immport.niaid.nih.gov), we developed a prognostic signature by focusing on immune-related genes (IRGs)[10]. The ImmPort database contains a variety of immune categories according to different molecular functions such as antigen processing and presentation, B-cell receptor signaling pathway, chemokine, chemokine receptors, cytokines, cytokines receptors, T-cell receptor signaling pathway, and TNF family receptors.
Immune-Related Prognostic Signature Development and Validation
The patients were randomly assigned (preset ratio of 7:3) into training (n=56) and testing (n=24) groups. The training group was employed to identify IRGs and construct prediction model. The testing group was utilized to validate the final model. Kaplan-Meier analysis and log-rank test were used to identify IRGs with prognostic ability. The Cox regression analysis with lasso penalty was conducted using “glmnet” R package to identify the best model for predicting the overall survival (OS) in UM[11]–[12]. The IPS was constructed based on the Cox regression coefficients. Then, risk score was calculated for each patient and the median risk value was set as the cutoff point to assign patents to high- or low-risk groups. We conducted time-dependent receiver-operating characteristic (ROC) curve analysis using “timeROC” R package to evaluate the specificity and sensitivity of the IPS[13]. Area under the curve (AUC) values were calculated from ROC curves. Univariate and multivariate Cox regression analyses were performed to determine independent prognostic role of the IPS.
Functional Enrichment Analysis
Functional enrichment analysis for the prognostic IRGs was conducted using Metascape (http://metascape.org), a web-based tool, designed to provide a comprehensive gene list annotation and analysis resource[14]. We used Metascape to further explore potential molecular mechanisms of the prognostic IRGs. Three categories of Gene Ontology (GO) terms including biological process, cellular component and molecular function were enriched. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were also enriched.
Tumor Immune Estimation Resource Database Analysis
TIMER is a web server for comprehensive analysis of tumor-infiltrating immune cells in different tumor types (https://cistrome.shinyapps.io/timer/)[15]. The TIMER database contains 10 897 samples across 32 tumor types from TCGA to estimate the abundance of tumor-infiltrating immune cells. We downloaded data of immune infiltrate levels of UM patients and investigated the association of IPS with the abundances of tumor-infiltrating immune cells, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells.
Statistical Analysis
Differences in clinicopathologic features between groups were tested using Student's t-test. SPSS software (version 24.0) and R software (version 3.5.3) were used for all statistical analysis. Statistical significance was indicated by a 2-sided P-value<0.05.
RESULTS
Prognostic Immune-Related Genes Identification
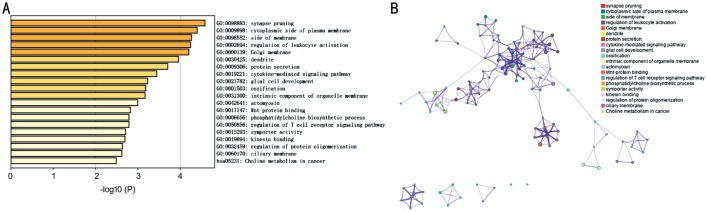
Fifty-sex patients in training set were used for construction of the IPS, and 24 patients in testing group for validation. Using Kaplan-Meier analysis, 149 genes from 1534 immune-related genes, were identified with prognosis predicting ability. The prognostic IRGs mainly enriched in synapse pruning, regulation of leukocyte activation, dendrite, cytokine-mediated signaling pathway, regulation of T cell receptor signaling pathway (GO), and Choline metabolism in cancer (KEGG; Figure 1A and 1B).
Figure 1. Functional analysis of 149 immune-related prognostic genes.
A: Heatmap of enriched terms across input gene lists, colored by P-values; B: Network of enriched terms, colored by cluster ID, where nodes that share the same cluster ID are typically close to each other.
Immune-Related Prognostic Signature Development
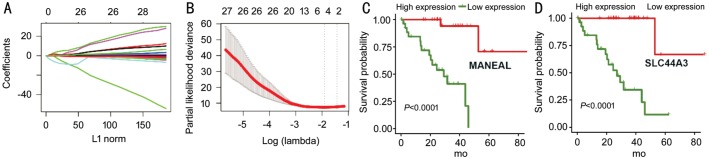
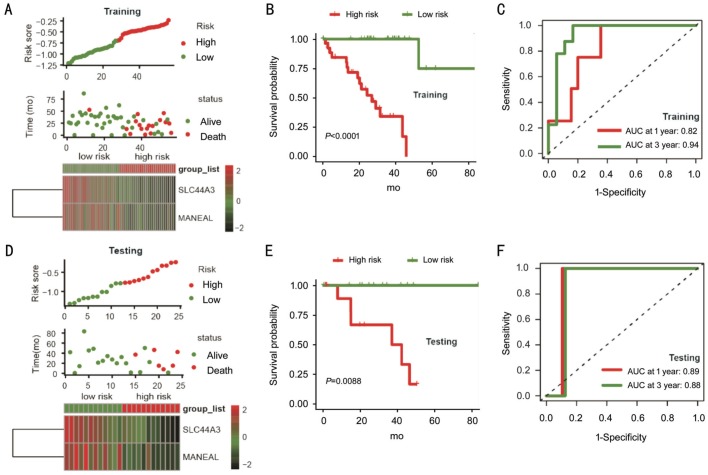
Based on prognostic IRGs, we applied a Cox regression model with lasso penalty for selecting genes with the best prognostic value. This procedure finally built an IPS containing two genes: MANEAL and SLC44A3 (Figure 2A and 2B). The expressions of MANEAL and SLC44A3 were associated with better survival and indicated their protective roles (Figure 2C and 2D). Then, risk score was calculated for each sample based on gene expression level and regression coefficients (risk score =[-0.119*MANEAL] + [-0.176*SLC44A3]). Using the median risk score as cutoff point, UM patients in training group were assigned into high- and low-risk groups. Risk score distribution, patient survival status, and gene expression pattern are demonstrated in Figure 3A. Patients in high-risk group were related to worse outcomes compared with the low-risk group (P<0.001; Figure 3B). Then, we performed time-dependent ROC curve analysis indicating promising prognostic ability of IPS for UM-specific OS (1-year AUC=0.82, 3-year AUC=0.94; Figure 3C).
Figure 2. Development of the IPS.
A, B: LASSO Cox analysis identified 2 immune-related genes most correlated to survival in training set; C, D: Kaplan-Meier survival curves were generated for selected genes by LASSO Cox analysis. IPS: Immune-related prognostic signature.
Figure 3. The IPS-based risk score predicted the OS of UM patients in the training and testing sets.
A, D: Risk score distribution, survival status of each patient, and expression distribution of selected genes in training and testing cohorts. B, E: Kaplan-Meier survival curve of UM patients in the high- and low-risk subgroups of the training and testing sets. C, F: Time-dependent ROC curve analysis of the IPS in training and testing sets.
Immune-Related Prognostic Signature Validation
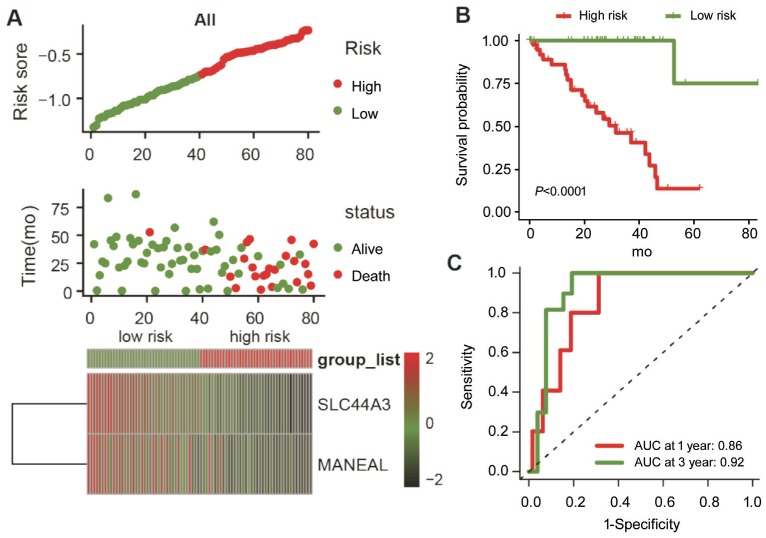
To confirm robust prognostic value of the IPS, the same formula derived from the training set was applied to both testing set and entire cohort. Patients were classified into high- and low-risk groups according to median risk score of the corresponding set. The risk score distribution, patient survival status, and gene expression pattern are plotted in Figure 3D. Patients in high-risk group consistently showed poor survival than patients in low-risk group (P=0.0088; Figure 3E). Analysis of the entire cohort returned similar finding (Figure 4A and 4B). The time-dependent ROC analysis in both testing and entire cohort indicated stable predictive power of the IPS. The 1-year and 3-year AUC values were 0.89 and 0.88 for testing set, and 0.86 and 0.92 for the entire set (Figure 3F and 4C), respectively. These results demonstrated the powerful prognosis predicting ability of the IPS.
Figure 4. The IPS-based risk score predicted the OS of UM patients in entire cohort.
A: Risk score distribution, survival status of each patient, and expression distribution of selected genes; B: Kaplan-Meier survival curve of UM patients in the high- and low-risk subgroups; C: Time-dependent ROC curve analysis of the IPS.
Subgroup Analyses
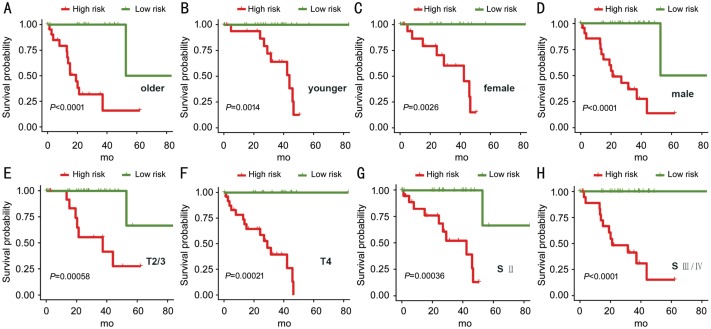
We further investigated the prognostic value of IPS in distinct subgroups. We demonstrated that IPS can predict OS of UM subgroups, including patients with older age (Figure 5A; P<0.0001), younger age (Figure 5B; P=0.0014), females (Figure 5C; P=0.0026), males (Figure 5D; P<0.0001), T2/3 stage (Figure 5E; P=0.00058), T4 stage (Figure 5F; P=0.00021), stage II (Figure 5G; P=0.00036), and stage III/IV (Figure 5H; P<0.0001).
Figure 5. The Kaplan-Meier analysis of the IPS grouping according to patients with older age (A), younger age (B), female (C), male (D), T2/3 stage (E), T4 stage (F), stage II (G), and stage III/IV (H).
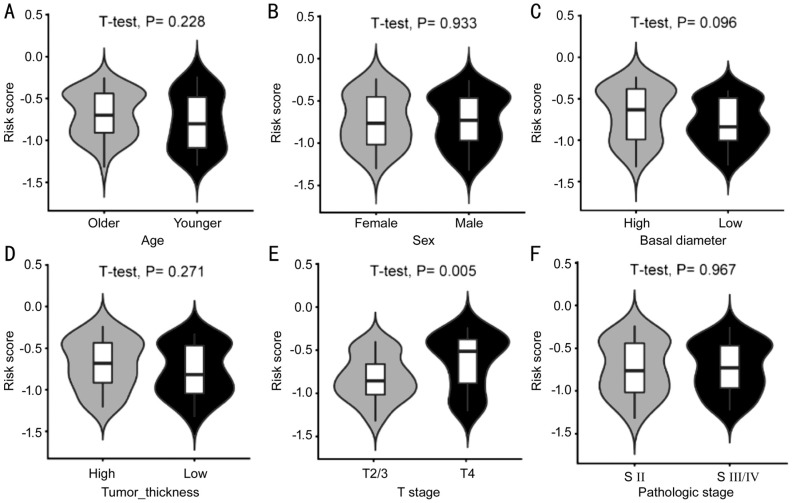
We evaluated the difference in distribution of clinicopathologic features between patents in high- and low-risk groups. The risk score distributed differently in stratified patients indicating their association with the IPS. Patients with higher level of T stage had higher risk score. Basal diameter of UM also seemed to be associated with IPS, with borderline statistical significance (Figure 6C and 6E). However, most features were not found significantly different between the two risk groups, including age at diagnosis, tumor thickness, and pathologic stage (Figure 6A, 6B, 6D, and 6F).
Figure 6. Distribution of the IPS-based risk score in stratified patients by age (A), sex (B), basal diameter (C), tumor thickness (D), T stage (E), and pathologic stage (F).
Immune-Related Prognostic Signature Independent of Clinicopathological Characteristics
To see if the IPS was an independent prognostic factor for UM patients, univariate Cox analysis was conducted and revealed that IPS was significantly related to OS [hazard ratio (HR)=33.437, 95% confidence interval (95%CI)=4.480-249.54, P=0.001; Table 1]. Variables that exhibited P<0.1 in univariate Cox analysis were assembled into a multivariate analysis to determine the independent role of IPS. Consistently, IPS was significantly associated with the OS, confirming its power to independently predict prognosis (HR=5.43, 95%CI=1.820-16.199, P=0.002; Table 1).
Table 1. Univariate and multivariate Cox regression analyses.
| Parameters variable | Univariate analysis |
Multivariate analysis |
||||
| HR | 95%CI | P | HR | 95%CI | P | |
| Age (continued) | 1.046 | 1.008-1.085 | 0.019a | 1.035 | 0.993-1.078 | 0.101 |
| Sex (M/F) | 1.542 | 0.651-3.652 | 0.325 | |||
| Basal diameter (high/low) | 2.484 | 0.999-6.179 | 0.050a | 1.609 | 0.528-4.908 | 0.403 |
| Tumor thickness (high/low) | 1.643 | 0.703-3.838 | 0.251 | |||
| T stage (T4/T2+T3) | 2.252 | 0.945-5.365 | 0.067a | 1.347 | 0.436-4.163 | 0.604 |
| Pathologic stage (III+IV/II) | 1.172 | 0.513-2.677 | 0.706 | |||
| Risk score (high/low) | 33.437 | 4.480-249.54 | 0.001a | 5.430 | 1.820-16.199 | 0.002a |
aStatistical significance.
Correlation of Immune-Related Prognostic Signature with Immune Cell Infiltration and Immune Checkpoints
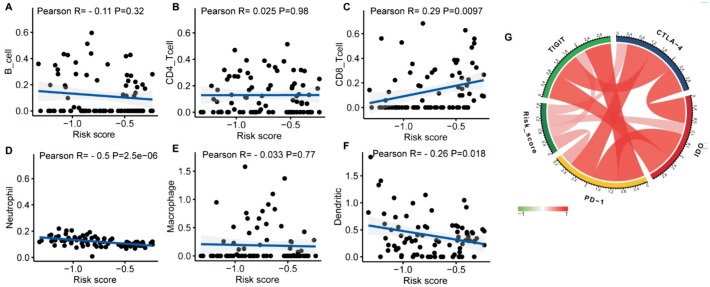
We attempted to explore the association between immune cell infiltration and IPS for a better understanding of tumor-immune interactions. Based on Tumor Immune Estimation Resource (TIMER) database, we found that IPS was significantly negatively correlated to the infiltration of neutrophils and dendritic cells, but positively correlated to the infiltration level of CD8+ T cells (Figure 7C, 7D, and 7F). There was no association between IPS and B cells, CD4+ T cells, and macrophages (Figure 7A, 7B, and 7E). Additionally, we investigated the relationship between IPS and crucial immune checkpoints containing PD-1, CTLA-4, IDO and TIGIT, to further explore immune mechanisms of the IPS. We found that these immune checkpoints were strongly interrelated and moderately associated with IPS (Figure 7G).
Figure 7. Correlations of the risk signature with infiltrating immune cell proportions and immune checkpoints.
A: B cells; B: CD4 T cells; C: CD8 T cell; D: Neutrophils; E: Macrophages; F: Dendritic cells; G: Immune checkpoints.
DISCUSSION
The 5-year survival rate of UM patients is around 60% and the prognosis after metastasis is usually poor[6],[16]–[17]. Despite diagnostic and therapeutic advances, the 5-year mortality rate of UM patients has not significantly reduced since 1973[18]. Therefore, a prognostic signature with a broad scope of application is needed to accurately identify patients with poor prognosis to improve treatment selection. Using TCGA database, we developed an immune-related signature related to OS in UM patients, confirmed its prognosis predicting ability in stratified patients, and demonstrated that IPS independently predicted survival.
Many studies have attempted to identify prognostic signatures for UM patients. Xin et al[19] constructed a 9 miRNA-based model with great predictive ability of OS in UM based on miRNA expression profiling.
Yi and Zou[20] proposed a prognostic signature containing four snoRNAs via Cox proportional regression models. Shi et al[21] also found four hub genes which might play a vital role in progress of UM. These studies allowed delineation of high-risk patients that may benefit from therapy efficacy intensification via molecular diagnostic method. However, there is no prediction model established focusing on IRGs from the perspective of tumor immunology.
Previous studies have indicated that immune system plays a crucial role in cancer initiation and progression[22]. UM has been reported to escape immune attacks employing various mechanisms, including inhibiting the immune-stimulatory function of dendritic cells, impairing the PD-1/PD-L1 axis, and secretion of Fasligand[9],[23]–[25]. Consequently, dramatically improved prognosis through developmentsin immunotherapy, as seen in advanced cutaneous melanoma, is not been observed in UM[16],[26]–[27]. Indeed, research on tumor immune microenvironment is an essential buttress to investigations into immunotherapeutic UM management. We conducted an immune genomic study to improve UM prognosis. This is the first study to propose an IPS for UM patients.
Our IPS, based on two IRGs, revealed favorable predictive ability and clinical utility. Our data demonstrated that IPS was significantly associated to tumor T stage. Basal diameter of UM also seemed to be associated with IPS, although not reaching statistical significance, possibly due to small sample size. These results indicated that the IPS not only predicted prognosis, but also served as an indicator of tumor progression. Moreover, multivariate Cox analysis showed that IPS was independent of these known prognostic clinicopathological variables and further confirmed its prognostic value in UM. Therefore, this signature could serve as a promising tool for predicting prognosis in UM.
We employed TIMER database to uncover relationships between IPS and immune cell infiltration and reflect the status of immune microenvironment in UM. We found that infiltrations of neutrophils and dendritic cells were significantly negatively correlated to the IPS, while the infiltration level of CD8+ T cells was positively correlated. These results revealed that high-risk patients might have lower infiltration levels of neutrophils and dendritic cells, and higher infiltration levels of CD8+ T cells. The IPS can also serve as a predictor for immune cells infiltration. Considering the limitation of immune checkpoint inhibitors in the management of UM, we attempted to explore the correlation of IPS with the expression of critical immune checkpoints, including PD-1, CTLA-4, IDO, and TIGIT. Up regulation of these immunosuppressive factors may partly contribute to the worse outcomes of high-risk patients. Notably, theses immune checkpoints were also strongly positively interrelated suggesting a demand for combination immune therapies.
Our study had limitations. First, the IPS was constructed using retrospective data and prospective studies are needed to confirm the efficacy of the IPS. Limited sample size also made the results potentially inconclusive. Second, we failed to confirm the association between IPS and response to immunotherapy, due to lacking information on patients treated with immune checkpoint inhibitors.
In conclusion, the current study was the first to develop and validate an immune-related gene-based signature for predicting survival in UM. This signature can be clinically used to distinguish and predict prognosis, and improve individualized management for UM patients. Further studies are needed to improve the IPS in large cohorts.
Acknowledgments
The authors would like to thank the ImmPort, TIMER, TCGA databases for the availability of the data.
Conflicts of Interest: Li YZ, None; Huang Y, None; Deng XY, None; Tu CS, None.
REFERENCES
- 1.McLaughlin CC, Wu XC, Jemal A, Martin HJ, Roche LM, Chen VW. Incidence of noncutaneous melanomas in the US. Cancer. 2005;103(5):1000–1007. doi: 10.1002/cncr.20866. [DOI] [PubMed] [Google Scholar]
- 2.Chua V, Aplin AE. Novel therapeutic strategies and targets in advanced uveal melanoma. Curr Opin Oncol. 2018;30(2):134–141. doi: 10.1097/CCO.0000000000000425. [DOI] [PubMed] [Google Scholar]
- 3.Slater K, Hoo PS, Buckley AM, Piulats JM, Villanueva A, Portela A, Kennedy BN. Evaluation of oncogenic cysteinyl leukotriene receptor 2 as a therapeutic target for uveal melanoma. Cancer Metastasis Rev. 2018;37(2-3):335–345. doi: 10.1007/s10555-018-9751-z. [DOI] [PubMed] [Google Scholar]
- 4.Ponti A, Denys A, Digklia A, Schaefer N, Hocquelet A, Knebel JF, Michielin O, Dromain C, Duran R. First-line selective internal radiation therapy in patient with uveal melanoma liver metastases. J Nucl Med. 2019:jnumed.119.230870. doi: 10.2967/jnumed.119.230870. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Manson DK, Marr BP, Carvajal RD. Treatment of uveal melanoma: where are we now? Ther Adv Med Oncol. 2018;10:1758834018757175. doi: 10.1177/1758834018757175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118(9):1881–1885. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Komatsubara KM, Carvajal RD. Immunotherapy for the treatment of uveal melanoma: current status and emerging therapies. Curr Oncol Rep. 2017;19(7):45. doi: 10.1007/s11912-017-0606-5. [DOI] [PubMed] [Google Scholar]
- 8.Chandran SS, Somerville RPT, Yang JC, Sherry RM, Klebanoff CA, Goff SL, Wunderlich JR, Danforth DN, Zlott D, Paria BC, Sabesan AC, Srivastava AK, Xi LQ, Pham TH, Raffeld M, White DE, Toomey MA, Rosenberg SA, Kammula US. Treatment of metastatic uveal melanoma with adoptive transfer of tumour-infiltrating lymphocytes: a single-centre, two-stage, single-arm, phase 2 study. Lancet Oncol. 2017;18(6):792–802. doi: 10.1016/S1470-2045(17)30251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang C, Peng LY, Zeng SX, Zhao Q, Tang LQ, Jiang XS, Zhang JJ, Yan NH, Chen YY. Investigation of indoleamine 2, 3-dioxygenase 1 expression in uveal melanoma. Exp Eye Res. 2019;181:112–119. doi: 10.1016/j.exer.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, Thomson E, Monteiro R, Guimaraes P, Walters B, Wiser J, Butte AJ. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2-3):234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 11.Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52(1):70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- 12.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16(4):385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou YY, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li TW, Fan JY, Wang BB, Traugh N, Chen QM, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jindal V. Role of immune checkpoint inhibitors and novel immunotherapies in uveal melanoma. Chin Clin Oncol. 2018;7(1):8. doi: 10.21037/cco.2018.01.05. [DOI] [PubMed] [Google Scholar]
- 17.Chew AL, Spilsbury K, Isaacs TW. Survival from uveal melanoma in Western Australia 1981-2005. Clin Exp Ophthalmol. 2015;43(5):422–428. doi: 10.1111/ceo.12490. [DOI] [PubMed] [Google Scholar]
- 18.Bakalian S, Marshall JC, Logan P, Faingold D, Maloney S, Di Cesare S, Martins C, Fernandes BF, Burnier MN., Jr Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin Cancer Res. 2008;14(4):951–956. doi: 10.1158/1078-0432.CCR-06-2630. [DOI] [PubMed] [Google Scholar]
- 19.Xin XY, Zhang Y, Ling F, Wang LM, Sheng XH, Qin LR, Zhao X. Identification of a nine-miRNA signature for the prognosis of uveal melanoma. Exp Eye Res. 2019;180:242–249. doi: 10.1016/j.exer.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Yi Q, Zou WJ. A novel four-snoRNA signature for predicting the survival of patients with uveal melanoma. Mol Med Rep. 2019;19(2):1294–1301. doi: 10.3892/mmr.2018.9766. [DOI] [PubMed] [Google Scholar]
- 21.Shi K, Bing ZT, Cao GQ, Guo L, Cao YN, Jiang HO, Zhang MX. Identify the signature genes for diagnose of uveal melanoma by weight gene co-expression network analysis. Int J Ophthalmol. 2015;8(2):269–274. doi: 10.3980/j.issn.2222-3959.2015.02.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology—analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8(12):711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J, Usui Y, Takeuchi M, Okunuki Y, Kezuka T, Zhang LN, Mizota A, Goto H. Human uveal melanoma cells inhibit the immunostimulatory function of dendritic cells. Exp Eye Res. 2010;91(4):491–499. doi: 10.1016/j.exer.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 24.Yang WH, Chen PW, Li HC, Alizadeh H, Niederkorn JY. PD-L1: PD-1 interaction contributes to the functional suppression of T-cell responses to human uveal melanoma cells in vitro. Invest Ophthalmol Vis Sci. 2008;49(6):2518–2525. doi: 10.1167/iovs.07-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallermalm K, De Geer A, Kiessling R, Levitsky V, Levitskaya J. Autocrine secretion of Fas ligand shields tumor cells from Fas-mediated killing by cytotoxic lymphocytes. Cancer Res. 2004;64(18):6775–6782. doi: 10.1158/0008-5472.CAN-04-0508. [DOI] [PubMed] [Google Scholar]
- 26.Heppt MV, Heinzerling L, Kähler KC, et al. Prognostic factors and outcomes in metastatic uveal melanoma treated with programmed cell death-1 or combined PD-1/cytotoxic T-lymphocyte antigen-4 inhibition. Eur J Cancer. 2017;82:56–65. doi: 10.1016/j.ejca.2017.05.038. [DOI] [PubMed] [Google Scholar]
- 27.Rothermel LD, Sabesan AC, Stephens DJ, Chandran SS, Paria BC, Srivastava AK, Somerville R, Wunderlich JR, Lee CC, Xi LQ, Pham TH, Raffeld M, Jailwala P, Kasoji M, Kammula US. Identification of an immunogenic subset of metastatic uveal melanoma. Clin Cancer Res. 2016;22(9):2237–2249. doi: 10.1158/1078-0432.CCR-15-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]