Abstract
In December 2019, pneumonia of unknown cause occurred in Wuhan, Hubei Province, China. On 7 January 2020, a novel coronavirus, named as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), was identified in the throat swab sample of one patient. The World Health Organization (WHO) announced the epidemic disease caused by SARS-CoV-2 as coronavirus disease 2019 (COVID-19). Currently, COVID-19 has spread widely around the world, affecting more than seventy countries. China, with a huge burden of this disease, has taken strong measures to control the spread and improve the curative rate of COVID-19. In this review, we summarized the epidemiological characteristics, clinical features, diagnosis, treatment, and prognosis of COVID-19. A comprehensive understanding will help to control the disease.
Keywords: Severe acute respiratory syndrome coronavirus-2, Coronavirus disease 2019, Epidemiology, Clinical, Pathology, Treatment
Introductions
At the end of 2019, several cases of pneumonia with unknown etiology emerged in Wuhan, Hubei Province, China [1–3]. The pneumonia spread quickly to other provinces of China and overseas. At early stage, it was reported that most patients had the contact history with Huanan seafood market [1–3]. After that, more and more patients had fever and cough symptoms. On 7 January 2020, a novel coronavirus was identified in the throat swab sample of one patient by the Chinese Center for Disease Control and Prevention (CDC), and was subsequently named as 2019nCoV by World Health Organization (WHO) [1, 2]. As the situation got worse, the WHO declared the outbreak as the public health emergency of international concern (PHEIC) [4]. On 11 February 2020, the International Committee on Taxonomy of Viruses renamed the virus as severe acute respiratory syndrome coronaviruse-2(SARS-CoV-2) [5]. And WHO announced the epidemic disease caused by SARS-CoV-2 as coronavirus disease 2019 (COVID-19) [6].
This is the third coronavirus pneumonia in the past 20 years around the world. In November 2002, a novel betacoronavirus called severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in Guangdong, China, and resulted in more than 8000 infections and 774 deaths in 37 countries. In 2012, Middle East respiratory syndrome coronavirus (MERS-CoV), which was first detected in Saudi Arabia, affected 2494 individuals and caused 858 fatalities [7, 8]. After the outbreak of COVID-19, the Chinese government has initiated a level-1 public health response to prevent the spread of the disease on 26 January 2020 [9]. As of 24:00 on 3 March 2020 (Beijing time), the SARS-CoV-2 has resulted in 80,270 laboratory and clinical confirmed cases in the mainland of China, and 2981 patient deaths [10].
Currently, there are many studies of SARS-CoV-2 and COVID-19. This review makes a comprehensive introduction about this disease, including the genome structure and receptor of SARS-CoV-2, epidemiology, clinical features, diagnosis, treatment, and prognosis of COVID-19. We hope our work can provide more information in understanding this disease, and more research findings are needed to help to limit spread of the disease and to invent vaccine and specific drugs.
The genome structure and receptor of human cells of SARS-CoV-2
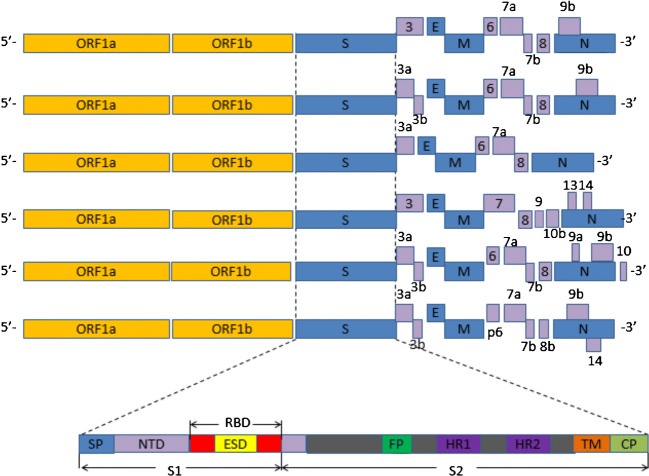
Coronaviruses are the largest, enveloped, single-stranded positive-sense RNA viruses, including 4 genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Alpha- and Betacoronaviruses mainly infect mammals; the rest of two primarily infect birds [11, 12]. Seven coronaviruses that related to human disease had been identified [12]. Four human coronaviruses (HCoV 229E, NL63, OC43, and HKU1) had been endemic globally and just resulted in upper respiratory tract infections in adults. The SARS-CoV, MERS-CoV, and SARS-CoV-2 are the most severe type that can lead to lower respiratory tract infections [11, 13] and acute respiratory distress syndrome (ARDS), which can cause patient deaths. At first, Wei et al. [14] found that the virus appeared to be recombinant by a codon usage analyses. However, this opinion was rejected by Paraskevis’s full-genome evolutionary analysis and Chen’s simplot analysis [14, 15]. Currently, the SARS-CoV-2 was found to be a novel positive-sense RNA virus, which belonged to the Betacoronavirus genus in the Coronaviridae family [7, 15, 16]. Similar to the SARS-CoV and MERS-CoV, the SARS-CoV-2 genome contains two untranslated regions (UTRs): 5′-cap structure and 3′-poly-A tail, and a single open reading frame (ORF) encoding a polyprotein [16, 17]. The SARS-CoV-2 genome is ordered by 5′- the viral replicase (ORF 1a and ORF1b)-structural proteins [Spike(S)-Envelope(E)-Membrane (M)-Nucleocapsid(N)]-3′; some genes of accessory proteins, such as ORF 3a, 7, and 8, are inserted in genes of structural proteins [7, 15–19]. Figure 1 shows the genome structure of SARS-CoV-2 in different studies. In the genome of coronaviruses, the gene of ORF1a and ORF1b occupies about two thirds of the overall genome, encoding 16 non-structural proteins (nsps), while the remaining one-third encodes accessory proteins and structural proteins [12, 16]. As shown in Fig. 1, there are some slight differences in the reported genome structure, mainly in accessory proteins [7, 12, 15, 17–19]. For example, the significant difference of two accessory proteins (ORF3b and ORF8) on the gene sequence between SARS-CoV-2 and SARS-CoV was reported by several studies [12, 16]. Using the different genome sequences as comparison can partly account for the results. As for the novel proteins of SARS-CoV-2, whether involving in pathogenesis of the virus or not is unclear.
Fig. 1.
Genome organization of SARS-CoV-2 in different studies [7, 12, 15, 17–19]. ORF = open reading frame (orange). Structural proteins including S, E, M, N (blue) (S = spike, E = envelope, M = membrane, N = nucleocapsid). Accessory proteins including 3, 3a, 3b, 6, 7, 8, 9a, 9b, 10b, 13, 14 (purple). SP = signal peptide. S1 = subunit 1. S2 = subunit 2. NTD = N-terminal domain. RBD = receptor binding domain. ESD = external subdomain. FP = fusion peptide. HR1 = heptad repeat 1. HR2 = heptad repeat 2. TM = transmembrane domain. CP = cytoplasmic domain. The length of genes is not drawn in scale
Inspired by the pathogenesis of SARS-CoV, the SARS-CoV-2 was presumed to infect the human cells by spike glycoprotein binding to its cellular receptor, angiotensin-converting enzyme 2 (ACE2). In fact, current evidence supports this idea. As for the spike protein of SARS-CoV-2, it contains two regions, S1 subunit and S2 subunit, which consists of 1253 amino acids [20]. The amino acid identity of spike protein between SARS-CoV-2 and SARS-CoV was about 75% [21, 22]. Generally, S1 domain is linked to receptor binding; S2 domain is linked to cell membrane fusion. Similar to SARS-CoV, S1 contains the N-terminal domain (NTD) and a receptor-binding domain (RBD) which contains core domain and external subdomain (ESD). S2 contains three functional domains, fusion peptide (FP), and heptad repeat (HR) 1 and 2 (Fig. 1). Whether SARS-CoV-2 can combine with host cells or not is determined by the affinity between the viral RBD and ACE2 of human cells [22]. Once RBD binds to the receptor, the S2 changes conformation to facilitate the membrane fusion by three functional domains. Although SARS-CoV is not the closest to SARS-CoV-2 at the whole-genome level, the RBD of SARS-CoV-2 is closer to that of SARS-CoV, and 72–74.9% amino acid sequences of RBD in both are identical [7, 19, 23]. Several critical residues in SARS-CoV-2 RBD have good interactions with human ACE2. Most residues of RBD interacting with ACE2 are fully conserved [22, 24]. As for function domains of S2, there is no difference between SARS-CoV-2 and SARS-CoV except some non-critical amino acids in HR1 region [22]. Another strong piece of evidence supporting ACE2 as a receptor of cells is that HR1 and HR2 domain of SARS-CoV-2 can fuse with each other to form 6-HB following SARS-CoV’s fusion mechanism [25]. Despite this, several studies speculated SARS-CoV-2 has less affinity with ACE2 than SARS-CoV [20, 21, 26]. On the contrary, Chen et al. [23] reported SARS-CoV-2 provided a stronger interaction with ACE2 than SARS-CoV by structure analysis of the receptor binding of SARS-CoV-2. By elucidating cryo-EM structure of SARS-CoV-2 spike protein, Wrapp et al. [27] also found that compared with SARS-CoV, SARS-CoV-2 bound to ACE2 with 10- to 20-fold higher affinity. In order to identify the suppose of entering cellular by ACE2, Peng et al. [28] found that SARS-CoV-2 was able to use ACE2 protein of many kinds of cells, including human cells, as an entry receptor in the ACE2-expressing cells, but not cells without ACE2. It also could not use aminopeptidase N and dipeptidyl peptidase which are other coronaviruses’ receptors. In most recent studies, the results provided direct evidence to support ACE2 as the receptor. Yan et al. [29] reported the ACE2-B0AT1 (a neutral amino acid transporter) complex can combine with two spike proteins by structural modeling in a structure analysis of full-length human ACE2, and the extracellular peptidase domain (PD) of ACE2 has the direct interaction with polar residues of RBD [29, 30].
Overall, there is sufficient evidence to support that SARS-CoV-2 infects cells by using the human ACE2. As the cryo-EM structure of spike protein and human ACE2 were revealed successfully, we have more opportunity to clarify the detailed process of entering cells [27, 29, 30].
Epidemiology of COVID-19
In early studies, 49–66% patients had the contact history of Huanan seafood market, where various kinds of living wild animals were on sale, including poultry, bats, and marmots [1, 2, 31]. It is currently speculated that the outbreak of COVID-19 in Wuhan is associated with wild animals. According to WHO, the environmental samples taken from Huanan seafood market were tested positive for SARS-CoV-2 [21], but the specific animals associated with the virus have not been identified. Based on previous evidence, the bats, the host of more than 30 coronaviruses [32], may be the origin of COVID-19. The bats are the natural reservoir of SARS-CoV and MERS-CoV, and spread to human through the palm civets and dromedary camels, respectively [8]. The RaTG13, which is a short RNA-dependent RNA polymerase (RdRp) region from a bat coronavirus, was closest to SARS-CoV-2 with 96.2–98.7% identity in whole-genome sequence [11, 15, 16, 28]. The other two bat-derived SARS-like coronaviruses, bat-SL-CoVZXC21 and bat-SL-CoVZC45, were closer to SARS-CoV-2 than SARS-CoV and MERS-CoV, which have approximately 88% nucleotide identity [7, 15–18]. As for the intermediate hosts of SARS-CoV-2, recent studies suggested pangolins were the most probable animal. Two sub-lineages of SARS-CoV-2 were found in organs of pangolins obtained from anti-smuggling activities in Guangdong and Guangxi Province of China by metagenomic sequence [33]. Xiao et al. [34] reported the SARS-CoV-2 was derived from the reorganization of pangolin-CoV-like virus and a Bat-CoV-RaTG13-like virus. However, the pangolin may not be the only intermediate reservoir, because SARS-CoV-2 was not originated from pangolin-CoV-like virus directly, which was demonstrated by the molecular and phylogenetic analyses in Liu’s study [35].
To sum up, the bats are the most probable original reservoir based on the current evidence. However, it is notable that Wuhan Huanan seafood market may not be the only source of SARS-CoV-2 spreading globally. Cohen pointed out Wuhan Huanan seafood market was not the only origin of SARS-CoV-2 by analyzing the epidemiology of 41 cases in the earliest study [36] . Pangolins may act as one of intermediate hosts. More work is needed to provide more precise information about original reservoir and intermediate hosts of SARS-CoV-2. Table 1 lists some information of studies about the genome sequence identity with SARS-CoV-2.
Table 1.
Genome identity between SARS-CoV-2 and other bat-like coronavirus
| Study | Sample size | Sequence identity of sample (%) | Source of sample | Huanan seafood market | Lung lesion | Approach | Genome length | Type of coronavirus | Gene sequence identity (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RdRp gene* | bat-SL-CoV ZC45 | bat-SL-CoV ZXC21 | SARS-CoV | MERS-CoV | |||||||||
| Lu [7]a | 9 | > 99.98 | BALF or throat swabs | 8/9 | Yes | NGS | - | β-Cov | - | 87.99 | 87.23 | 79.0 | 50.0 |
| Wu [12] | 3 | Almost identity | Obtain from ViPR and NCBI | - | - | In-depth genome annotation | 29.8 kb | β-Cov | - | Closer | Closer | - | Distant |
| D. Paraskevis [15] | 1 | - | Downloaded from NCBI | - | - | NGS | - | β-Cov | 96.3 | - | - | - | - |
| Chen [16] | 2 | - | BALF | 2/2 | Yes | mNGS | 29,881nt | β-Cov | 98.7 | 87.9 | 87.9 | 79.7 | - |
| Chan [17] | 1 | - | Obtain from GenBank | - | - | Phylogenetic analysis | 29,891nt | β-Cov | - | 89.0 | 89.0 | 82.0 | - |
| Chan [18] | 6 | 99.99 | Respiratory samples | 0/5 | Yes | RT-PCR sequence | 29.8 kb | β-Cov | - | 89.0 | - | - | - |
| Wu [19] | 1 | - | BALF | 1/1 | Yes | mNGS | 29,903 nt | β-Cov | - | 89.1 | - | - | - |
| Zhou [28]a | 7 | 99.9 | BALF | 6/7 | Yes | mNGS | 29.9Kb | β-Cov | 96.2 | 88.1 | 88.0 | 79.5 | - |
[7]aEight complete and two partial genome sequences of SARS-CoV-2 were obtained. [28]aFive samples were tested positive for SARS-CoV-2. RdRp gene*, RNA-dependent RNA polymerase gene; CoV, coronavirus; SARS, severe acute respiratory syndrome; MERS, Middle East respiratory syndrome; nt, nucleotide; kb = kilobase; BALF, bronchoalveolar lavage fluid; mNGS, metagenomic next-generation sequencing testing; NCBI, National Center for Biotechnology Information; ViPR, virus pathogen database and analysis resource
Transmissibility is an important factor of an epidemic [37]. Data as reported by 10 AM CET on 3 March 2020, SARS-CoV-2 has been responsible for 90,870 confirmed cases with 3112 (3.4%) deaths around the world [38]. The reported median age of patients was ranged from 41 to 57 years [1, 2, 39]. Male made up the majority of patients with the proportion of 50–75% [1–3, 39]. Due to different data sources, the infection rate of medical staff has a huge difference, with 2.1–29% [3, 40]. Approximately 25.2–50.5% SARS-CoV-2infected patients had one or more underlying diseases (Table 2), including hypertension, diabetes, chronic obstructive pulmonary disease, cardiovascular disease, and malignancy [1–3, 39, 41]. The COVID-19’s median incubation period from exposure to illness onset was 3.0 days in a 1099-case cohort and 4.0 days in a 62-case cohort [39, 41]. It was also reported the longest incubation period was 24 days [41]. The percentage of patients exposed to Huanan seafood market varies between 8.7% and 66% [1–3, 42]. Patients with no contact history of Huanan seafood market and medical staff infection [18, 21, 43] indicated human-to-human transmission mainly via droplets from coughing or sneezing or direct contact [18, 44–47]. In addition, several studies reported fecal-mouth pathway may also be a potential way for the transmission of SARS-CoV-2 [23, 48], and the SARS-CoV-2 was also isolated from urine of a patient in a recent new [49]. But it is unclear whether human-to-human transmission can be implemented by the above routes.
Table 2.
| Common symptoms | Laboratory findings | ||
| Fever | 77.4–98.6% | Lymphopenia | 35.3–82.1% |
| Cough | 59.4–81.8% | Thrombocytopenia | 5.0–36.2% |
| Fatigue | 38.1–69.6% | Leukopenia | 9.1–33.7% |
| Dyspnea | 3.2–55.0% | Increased CRP | 60.7–86.3% |
| Myalgia | 11.1–34.8% | Increased d-dimer | 36.4–46.4% |
| Sputum production | 28.2–56.5% | Increased LDH | 27.4–75.8% |
| Headache | 6.5–33.9% | Increased CK | 8.0–32.5% |
| Underlying diseases | 25.2–50.5% | Prolonged prothrombin time | 58.0% |
| Complication | Increased ALT | 16.1–28.3% | |
| ARDS | 3.4–29.3% | Increased AST | 22.2–36.7% |
| Shock | 1.0–8.7% | Increased interleulin-6 | 51.5% |
| Acute renal injury | 0.5–7.3% | Increased serum fenitin | 62.6% |
| Acute cardiac injury | 7.2–12.2% | Increased ESR | 84.8% |
| Secondary infections | 9.8% | Increased procalcitonin | 5.5–11.3% |
| Increased troponin I | 12.2% | ||
| Increased creatinine | 1.9–9.8% | ||
ARDS, acute respiratory distress syndrome; CRP, C-reactive protein; LDH, lactose dehydrogenase; CK, creatinine kinase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; ESR, erythrocyte sedimentation rate
The basic reproduction number R0, the important property of transmission, is commonly used to estimate the average number of secondary cases generated by an infectious case in a fully susceptible population during the early phase of the outbreak [37, 50]. If the R0 is more than 1, human-to-human transmission may persist. A range from 1.4 to 6.49 of R0 was estimated by various methods [44, 51–53]. The different values of estimated R0 can attribute to different data and different modeling methods [53]. Liu et al. [53] came to conclusion that the average of estimate R0 was 3.28 with a median of 2.79 by an analysis of 12 studies. So, around 2–3 is indeed a reliable range, suggesting the potential of sustained human-to-human transmission [50, 53]. Additionally, so many factors can affect the value of R0, including estimation period, utilized models, and datasets [54]. As various aspects of measures have taken effect, the estimated R0 is mutable. As David said, it is impossible to know what will happen so early in this SARS-CoV-2 epidemic [37]. The initial R0 estimation for SARS-CoV was more than 2.0, but the predicted large outbreak did not occur [8, 37]. It is also notable that asymptomatic patients can also be a source of infection [43, 48]. The super communicator is worthy of notice, who can infect more than 100 individuals [52]. The immediate priority is to clarify all potential routes of transmission. After all, superspreading has a huge impact on the epidemic. Anyway, it is necessary for us to take effective measures and keep alert to control the epidemic. Fortunately, the number of confirmed cases maintains at a lower level in mainland of China with 119 confirmed cases on 3 March 2020 [10]. At this critical period, it is apparently wise to continue taking steps to control the outbreak.
Clinical features and diagnosis
Clinical manifestations
The clinical manifestations of SARS-CoV-2-infected patients ranged from mild non-specific symptoms to severe pneumonia with organ function damage. The common symptoms were fever (77.4–98.6%), cough (59.4–81.8%), fatigue (38.1–69.6%), dyspnea (3.2–55.0%), myalgia (11.1–34.8%), sputum production (28.2–56.5%), and headache (6.5–33.9%) [1–3, 39, 41] (Table 2). Sore throat, rhinorrhea, chest pain, hemoptysis, conjunctival congestion, diarrhea, nausea, and vomiting were less common [1–3, 39, 41]. But one study showed 39.6% of 140 confirmed COVID-19 patients had gastrointestinal symptoms [40], and 10.1% patients presented with gastrointestinal discomfort at onset in Wang’s study [3]. Patients did not necessarily have fever at onset, some patients developed after hospitalization [41], and some severe patients even did not have fever. SARS-CoV-2, SARS-CoV, and MERS-CoV infections share many similar clinical symptoms [8], including fever, cough, myalgia, and dyspnea. However, patients with SARS and MERS have more gastrointestinal involvement (about one-third) than COVID-19 patients [55]. And MERS has a high incidence of renal failure, which is a typical characteristic not often found in other human coronavirus infections [56, 57].
Laboratory and radiologic characteristics
Of confirmed patients, 35.3–82.1%, 5.0–36.2%, and 9.1–33.7% had lymphopenia, thrombocytopenia, and leukopenia, respectively [1–3, 39, 41] (Table 2). C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), serum ferritin, and interleukin-6 (IL6) elevated prominently in Chen’s research [2]. Many patients also had increased levels of d-dimer, lactate dehydrogenase (LDH), creatine kinase (CK), prolonged prothrombin time, alanine aminotransferase (ALT), and aspartate aminotransferase (AST) [1–3, 39, 41, 58]. However, elevated levels of procalcitonin, troponin I, and creatinine were uncommon [1, 39, 41].
The typical imaging features of chest computed tomography (CT) for novel coronavirus pneumonia (NCP) included ground-glass opacity, bilateral patchy shadows, and subsegmental areas of consolidation, sometimes with a rounded morphology and a peripheral lung distribution [1–3, 40, 41, 59]. Changes in the disease were accompanied by changes in CT imaging, reflecting the severity of the disease [58]. Pan et al. [60] analyzed 21 NCP patients from initial diagnosis until recovery (without severe respiratory distress during a hospital stay), and they found chest CT scan showed that the lung abnormalities were the most severe about 10 days after the initial symptoms onset. Overall, the CT manifestations of NCP were diverse and fast-changing [61]. However, a normal chest CT image cannot exclude the diagnosis of SARS-CoV-2 infection [62].
Pathological finding
By obtaining biopsy samples at autopsy of one COVID-19 patient [63], the lung biopsy specimens showed bilateral diffuse alveolar damage with cellular fibromyxoid exudates. Bilateral lung tissue indicated pulmonary edema with hyaline membrane formation, reflecting ARDS. Meanwhile, flow cytometric analysis of peripheral blood suggested the reduced counts of peripheral CD4 and CD8 T cells but a hyperactivated status. A report on systemic anatomy at autopsy from the other patient [64] indicated gray white patchy lesions in lungs, gray white viscous fluid overflow in the lung section, and pulmonary fiber bands, reflecting that COVID-19 caused an inflammatory response characterized by deep airway and alveolar damage. The pathological results of the first one resembled those seen in SARS-CoV and MERS-CoV infection [63]. However, the latter found pulmonary fibrosis and consolidation were less severe than SARS, but exudation was more obvious [64]. These findings, with more pathological research, will make a great importance in understanding the pathogenesis and making therapeutic strategy for COVID-19.
Diagnosis
The COVID-19 patients around the world were diagnosed based on World Health Organization interim guidance [65], and China updated the novel coronavirus pneumonia diagnosis and treatment program (trial version) (in Chinese) according to epidemic situation and improved awareness of disease. A laboratory confirmed case with SARS-CoV-2 infection was defined as a positive result to high-throughput sequencing or real-time reverse-transcriptase polymerase-chain-reaction (RT-PCR) assay for SARS-CoV-2 [65, 66]. Chest CT, as a diagnostic method for COVID-19 with a high sensitivity, is being given more important value for diagnosis [67]. In the fifth trial version of novel coronavirus pneumonia diagnosis and treatment program (in Chinese) [68], clinical diagnosis was proposed for cases in Hubei Province, who had epidemiology history, the above clinical features along with typical chest CT imaging. And more than 10,000 patients for clinical diagnosis got early treatment [69]. The sixth trial version (in Chinese) removed the clinical diagnosis, for reduced suspected cases and improved nucleic acid detection capability [66].
Treatment
In several clinical studies of confirmed cases, strategies for COVID-19 patients included antiviral treatment, empirical antibiotic treatment, corticosteroid, intravenous immunoglobulin therapy, oxygen support (nasal cannula, mask oxygen inhalation, non-invasive ventilation, invasive mechanical ventilation), continuous renal replacement therapy (CRRT), and extracorporeal membrane oxygenation (ECMO) [1–3, 39, 41].
Due to the absence of clinical evidence, there were no approved drugs for antiviral therapy against SARS-CoV-2. Of three clinical cohort studies, oseltamivir was used for antiviral therapy in 35.8% (393/1099) patients, 89.9% (124/138) patients, and 92.7% (38/41) patients, respectively [1, 2, 41]. Another research [2] included 99 COVID-19 patients, in which 76.0% patients received antiviral treatment, including oseltamivir, ganciclovir, and lopinavir and ritonavir tablets, and the duration of antiviral treatment was 3–14 days. Though oseltamivir had high application rate in the early cohort studies, its drug efficacy on COVID-19 was not obvious, and the sixth trial version of novel coronavirus pneumonia diagnosis and treatment program (in Chinese) did not recommend it [66]. Case report implied that lopinavir and ritonavir therapy may be beneficial for COVID-19 cases [70, 71]. Remdesivir, a nucleotide analogue RNA polymerase inhibitor with broad-spectrum antiviral activity, was demonstrated that it could be against Ebola virus in rhesus monkeys [72]. Wang et al. [73] found that remdesivir and chloroquine were highly effective in the control of SARS-CoV-2 infection in vitro. What’s more, the first reported COVID-19 case of the USA was cured by intravenous remdesivir [58] and other supportive care. More researches from cells, animals, and clinical need to explore the effect of remdesivir on SARS-CoV-2. Other methods of antiviral drug administration may also have some certain function. It was reported that different combinations of interferon alpha inhalation, lopinavir/ritonavir, and arbidol may have some effects [39]. Furthermore, combined Chinese and Western medicine treatment, including lopinavir/ritonavir, arbidol, and Shufeng Jiedu Capsule (a traditional Chinese medicine) may also be beneficial to treatment which deserved further study [74].
As for the corticosteroid therapy for SARS-CoV-2, current interim guidance from WHO on clinical management of severe acute respiratory infection, when SARS-CoV-2 infection is suspected (released Jan 28, 2020), suggests not routinely using systemic corticosteroids unless indicated for another reason [65], and there were contradictory opinions from professors [75, 76]. Corticosteroid therapy was used in approximately 20–44.9% COVID-19 patients [1–3, 39, 41]. Compared with non-severe patients, severe patients got more corticosteroid therapy (44.5% vs. 13.7%) with the median of maximal daily dose up to 30.0 (1.0–40.0) (mg/kg) [41]. According to pathological findings of one COVID-19 patient, proper use of corticosteroid together with other support care should be considered for the severe patients to prevent ARDS development [63].
There are more than 80 running or pending clinical trials on potential treatments for COVID-19 in China [77]. Studies of recombinant human angiotensin-converting enzyme 2 (rhACE2), mesenchymal stem cell, PD-1 blocking antibody, bevacizumab injection, and immunoglobulin of cured patients are registered in the website of Clinical Trial [78], and some are recruiting patients. Safe and effective clinical trials will find more therapeutic possibilities for COVID-19 patients.
Prognosis
Many patients infected withSARS-CoV-2, especially for severe patients, had complications (Table 2), including ARDS, shock, acute renal injury, acute cardiac injury, and secondary infection [1–3, 41].
The mortality rate of COVID-19 ranged from 0 to 14.6% [1–3, 39, 41, 79]. However, Yang et al. [42] reported 32 of 52 (61.5%) critically ill adult ICU patients had died at 28 days. It is not hard to know the disease severity is an independent predictor of poor prognosis [41, 42]. Compared with non-ICU patients, the ICU patients were older with a greater number of comorbid conditions and had more common symptoms of dyspnea, abdominal pain, and anorexia [3]. Meanwhile, it was reported that ICU patients had higher plasma cytokine and chemokine levels of IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and TNFα [1]. Non-survivors had more severe lymphopenia and higher blood cell counts, neutrophil counts, d-dimer and fibrin degradation product than survivors [3, 63]. Wang et al. [80] analyzed the first 17 deaths up to 22 January 2020, announced by The China National Health Commission found that the median days from first symptom to death were 14.0 (range 6–41) days, and seemed to be shorter among old people [80]. In Yang’s study [42], the survival time of the non-survivors is likely to be within 1–2 weeks after ICU admission.
In summary, severe patients or ICU patients have a relatively higher mortality [41, 42]. Age, comorbidity, some symptoms (dyspnea, abdominal pain, etc.), and more prominent laboratory abnormalities (lymphopenia, elevated d-dimer, etc.) may be risk factors for poor outcome [3, 40, 42, 80].
According to the latest data released at 24:00 on 3 March 2020 (Beijing time), the fatality is 3.7% and 4.6% for the mainland of China and Wuhan, Hubei Province, respectively [10]. Overall, SARS-CoV-2 has a relatively lower mortality rate than SARS-CoV and MERS-CoV (9.6% and 40.0%, respectively) [55].
Conclusion
COVID-19 has spread to 73 countries, territories, or areas around the world, and is responsible for 90,870 patients as of 10 AM CET on 3 March 2020. The virus may be related to bats, but Wuhan Huanan seafood market may not be its sole origin. Whatever the case, banning of wildlife sales and removing them from wet markets are beneficial to control the epidemic. Except several genes of accessory proteins, SARS-CoV-2 is almost identical to SARS-CoV in genome organization. Hence, we obtain the fact that ACE2 is the receptor of SARS-CoV-2 entering cells. Human to human transmission can be realized mainly by droplets from coughing or sneezing or direct contact. Fever and cough are the main symptoms. Chest CT examination is an important tool for diagnosis, and confirmed cases are diagnosed by detecting SARS-CoV-2 of specimens taken from the upper respiratory tract and lower respiratory tract. As for the treatment, there are no specific drugs for the infection, and many therapies, with preliminary good clinical response, are being tested in clinical trials. We hope that increased awareness of the virus and ongoing clinical trials can help to find effective treatment against SARS-CoV-2. Though the fatality of SARS-CoV-2 is lower than SARS-CoV and MERS-CoV, the overall mortality of SARS-CoV-2 remains to be established in the future, because a large number of confirmed and suspected cases are still in hospital; even worse, the confirmed cases are increasing in other countries, like Korea and Japan. Further study is necessary for us to control this epidemic disease.
Authors’ contribution
Huipeng Ge, Xiufen Wang: manuscript writing, bibliographic retrieval, making tables and figure.
Xiangning Yuan, Gong Xiao, Chengzhi Wang, Tianci Deng: manuscript writing.
Qiongjing Yuan, Xiangcheng Xiao: manuscript editing and critical review.
Funding information
This work was supported by the National Natural Science Foundation of China under Grant no. 81770743, and by the Natural Science Foundation of Hunan province China under Grant no. 2018JJ2654.
Compliance with ethical standards
Conflict of interest
All authors declared that they had no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Huipeng Ge and Xiufen Wang contributed equally to this work.
Contributor Information
Qiongjing Yuan, Email: yuanqiongjing@csu.edu.cn.
Xiangcheng Xiao, Email: xiaoxc@csu.edu.cn.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J et al (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed]
- 4.Zarocostas J. What next for the coronavirus response? Lancet. 2020;395:401. doi: 10.1016/S0140-6736(20)30292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Committee on Taxonomy of Virus. Naming the 2019 Coronavirus (2020) https://talk.ictvonline.org/. Accessed 2 Mar 2020
- 6.Notice of the National Health Commission of the People’s Republic of China on revising the english name of novel coronavirus pneumonia(2020) http://www.nhc.gov.cn/yzygj/s7653p/202002/33393aa53d984ccdb1053a52b6bef810.shtml. Accessed 29 Feb 2020 (in chinese)
- 7.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng SQ, Peng HJ (2020) Characteristics of and public health responses to the coronavirus disease 2019 outbreak in China. J Clin Med 9:575. 10.3390/jcm9020575 [DOI] [PMC free article] [PubMed]
- 10.National Health Commission of the People’s Republic of China. Update on the outbreak of novel coronavirus pneumonia as of 24:00 on 3 March 2020(2020) http://www.nhc.gov.cn/xcs/yqfkdt/202003/7a5f57b3f1b94954b1fc25f81dacc874.shtml. Accessed 4 Mar 2020 (in chinese)
- 11.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P et al (2020) Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed]
- 13.Paules CI, Marston HD, Fauci AS (2020) Coronavirus infections-more than just the common cold. JAMA. 10.1001/jama.2020.0757 [DOI] [PubMed]
- 14.Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microb Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JF, Kok KH, Zhu Z, Chu H, To KK. Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infec. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JF, Yuan S, Kok KH, To KK. Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG et al (2020) A new coronavirus associated with human respiratory disease in China. Nature. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed]
- 20.Morse JS, Lalonde T, Xu S, Liu WR (2020) Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed]
- 21.Gralinski LE, Menachery VD (2020) Return of the coronavirus: 2019-nCoV. Viruses 12:135. 10.3390/v12020135 [DOI] [PMC free article] [PubMed]
- 22.Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 10.1128/jvi.00127-20 [DOI] [PMC free article] [PubMed]
- 23.Chen Y, Guo Y, Pan Y, Zhao ZJ (2020) Structure analysis of the receptor binding of 2019-nCoV. Biochemi Bioph Rese Co. 10.1016/j.bbrc.2020.02.071 [DOI] [PMC free article] [PubMed]
- 24.Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y (2020) Protein structure and sequence re-analysis of 2019-nCoV genome does not indicate snakes as its intermediate host or the unique similarity between its spike protein insertions and HIV-1. bioRxiv 2020 [DOI] [PMC free article] [PubMed]
- 25.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y et al (2020) Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 10.1038/s41423-020-0374-2 [DOI] [PMC free article] [PubMed]
- 26.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X et al (2020) Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed]
- 27.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O et al (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, NY). 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed]
- 28.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed]
- 29.Yan R, Zhang Y, Li Y, Xia L, Zhou Q (2020) Structure of dimeric full-length human ACE2 in complex with B0AT1. bioRxiv
- 30.Yan R, Zhang Y, Guo Y, Xia L, Zhou Q (2020) Structural basis for the recognition of the 2019-nCoV by human ACE2. bioRxiv [DOI] [PMC free article] [PubMed]
- 31.Jiang S, Du L, Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infec. 2020;9:275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan T. Novel coronavirus from discovery to clinical diagnostics. Infect Genet Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam TTY, Shum MHH, Zhu HC, Tong YG, Ni XB, Liao YS et al (2020) Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv
- 34.Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ et al (2020) Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. bioRxiv
- 35.Liu P, Jiang JZ, Wan XF, Hua Y, Wang X, Hou F et al (2020) Are pangolins the intermediate host of the 2019 novel coronavirus (2019-nCoV) ? bioRxiv [DOI] [PMC free article] [PubMed]
- 36.Cohen J (2020) Wuhan seafood market may not be source of novel virus spreading globally. Science. https://www.sciencemag.org/news/2020/01/wuhan-seafood-market-may-not-be-source-novel-virus-spreading-globally. Accessed 2 Mar 2020
- 37.Swerdlow DL, Finelli L (2020) Preparation for possible sustained transmission of 2019 novel coronavirus: lessons from previous epidemics. JAMA. 10.1001/jama.2020.1960 [DOI] [PubMed]
- 38.WHO (2020) Coronavirus disease 2019 (COVID-19) Situation Report – 43. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200303-sitrep-43-covid-19.pdf?sfvrsn=2c21c09c_2. Accessed 4 Mar 2020
- 39.Xu XW, Wu XX, Jiang XG, Xu KJ, Ying LJ, Ma CL, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ (Clin Res Ed) 2020;368:m792. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ et al (2020) Clinical characteristics of 140 patients infected by SARS-CoV-2 in Wuhan, China. Allergy. 10.1111/all.14238 [DOI] [PubMed]
- 41.Guan Wj, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX et al (2020) Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 10.1101/2020.02.06.20020974:2020.02.06.20020974
- 42.Yang X, Yu Y, Xu J,Shu H, Xia J, Liu H et al (2020) Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 10.1016/s2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed]
- 43.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C et al (2020) Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. New Engl J Med. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed]
- 44.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y et al (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. New Engl J Med. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed]
- 45.Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S. Novel Wuhan (2019-nCoV) coronavirus. Am J Resp Crit Care. 2020;201:P7–p8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- 46.Wang FS, Zhang C. What to do next to control the 2019-nCoV epidemic? Lancet. 2020;395:391–393. doi: 10.1016/S0140-6736(20)30300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J (2020) Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 10.1016/j.micinf.2020.01.004 [DOI] [PMC free article] [PubMed]
- 48.Wang G, Jin X (2020) The progress of 2019 novel coronavirus event in China. J Med Virol. 10.1002/jmv.25705 [DOI] [PMC free article] [PubMed]
- 49.Huanqiu.com Zhong Nanshan’s team: Isolating virus from urine of the patient with novel coronavirus pneumonia (2020) https://china.huanqiu.com/article/9CaKrnKpwOu. Accesseed 2 March 2020 (in chinese)
- 50.Riou J, Althaus CL (2020) Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill 25. 10.2807/1560-7917.ES.2020.25.4.2000058 [DOI] [PMC free article] [PubMed]
- 51.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu JT, Leung K, Leung GM (2020) Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 10.1016/s0140-6736(20)30260-9 [DOI] [PMC free article] [PubMed]
- 53.Liu Y, Gayle AA, Wilder-Smith A, Rocklov J (2020) The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 10.1093/jtm/taaa021 [DOI] [PMC free article] [PubMed]
- 54.Tang B, Bragazzi NL, Li Q, Tang S, Xiao Y, Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect Dis Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet (London, England) 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Omari AA, Rabaan AA, Salih S, Al-Tawfiq JA, Memish ZA. MERS coronavirus outbreak: implications for emerging viral infections. Diagn Microbiol Infect Dis. 2019;93:265–285. doi: 10.1016/j.diagmicrobio.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeung ML, Yao Y, Jia L, Chan JF, Chan KH, Cheung KF, et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol. 2016;1:16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H et al (2020) First case of 2019 novel coronavirus in the United States. N Engl J Med. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed]
- 59.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X et al (2020) CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology. 10.1148/radiol.2020200230:200230 [DOI] [PMC free article] [PubMed]
- 60.Pan F, Ye T, Sun P, Gui S, Liang B, Li L et al (2020) Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 10.1148/radiol.2020200370:200370
- 61.Pan Y, Guan H, Zhou S, Wang Y, Li Q, Zhu T et al (2020) Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 10.1007/s00330-020-06731-x [DOI] [PMC free article] [PubMed]
- 62.Kanne JP (2020) Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: key points for the radiologist. Radiology. 10.1148/radiol.2020200241:200241 [DOI] [PMC free article] [PubMed]
- 63.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C et al (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 10.1016/s2213-2600(20)30076-x [DOI] [PMC free article] [PubMed]
- 64.Liu Q, Wang RS, Qu GQ, Wang YY, Liu P, Zhu YZ et al (2020) Xin xing guan zhuang bing du fei yan si wang shi ti xi tong jie pou da ti guan cha bao gao. J Forensic Med. 10.12116/j.issn.1004-5619.2020.01.00 (in chinese)
- 65.WHO (2020) Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: interim guidance. https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected. Accessed 2 Mar 2020
- 66.National Health Commission of the People's Republic of China. novel coronavirus pneumonia diagnosis and treatment program (6th trial version) (in Chinese) (2020) http://www.nhc.gov.cn/xcs/zhengcwj/202002/8334a8326dd94d329df351d7da8aefc2.shtml. Accessed 2 Mar 2020 (in chinese)
- 67.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W et al (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 10.1148/radiol.2020200642:200642 [DOI] [PMC free article] [PubMed]
- 68.National Health Commission of the People's Republic of China. novel coronavirus pneumonia diagnosis and treatment program (5th trial version) (in Chinese) (2020) http://www.nhc.gov.cn/xcs/zhengcwj/202002/d4b895337e19445f8d728fcaf1e3e13a.shtml. Accessed 2 Mar 2020 (in chinese)
- 69.National Health Commission of the People's Republic of China. Update on the outbreak of novel coronavirus pneumonia as of 24:00 on 12 February 2020 (2020) http://www.nhc.gov.cn/xcs/yqtb/202002/26fb16805f024382bff1de80c918368f.shtml. Accessed 2 Mar 2020 (in chinese)
- 70.Han W, Quan B, Guo Y, Zhang J, Lu Y, Feng G et al (2020) The course of clinical diagnosis and treatment of a case infected with coronavirus disease 2019. J Med Virol. 10.1002/jmv.25711 [DOI] [PMC free article] [PubMed]
- 71.Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, Lee WJ, et al. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J Korean Med Sci. 2020;35:e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M et al (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed]
- 74.Wang Z, Chen X, Lu Y, Chen F, Zhang W (2020) Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci Trends. 10.5582/bst.2020.01030 [DOI] [PubMed]
- 75.Shang L, Zhao J, Hu Y, Du R, Cao B. On the use of corticosteroids for 2019-nCoV pneumonia. Lancet. 2020;395:683–684. doi: 10.1016/S0140-6736(20)30361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russell CD, Millar JE, Baillie JK (2020) Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet (London, England). 10.1016/s0140-6736(20)30317-2 [DOI] [PMC free article] [PubMed]
- 77.Maxmen A. More than 80 clinical trials launch to test coronavirus treatments. Nature. 2020;578:347–348. doi: 10.1038/d41586-020-00444-3. [DOI] [PubMed] [Google Scholar]
- 78.ClinicalTrials.gov (2020) https://clinicaltrials.gov/ct2/results?cond=COVID-19&term=&cntry=&state=&city=&dist=. Accessed 2 Mar 2020
- 79.Tang N, Deng L, Wang X, Sun Z (2020) Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 10.1111/jth.14768 [DOI] [PMC free article] [PubMed]
- 80.Wang W, Tang J, Wei F (2020) Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol 92:441–447 [DOI] [PMC free article] [PubMed]