Abstract
Background
Key pathogenetic mechanisms underlying renal disease progression are unaffected by current treatment. Metabolite profiling has significantly contributed to a deeper understanding of the biochemical metabolic networks and pathways in disease, but the biochemical details in maintenance hemodialysis (MHD) patients remain largely undefined.
Methods
The metabolic fingerprinting of plasma samples from 19 MHD patients and 12 healthy controls was characterized using liquid chromatography quadrupole time-of-flight mass spectrometry. Principal component analysis (PCA) and orthogonal partial least squares-discriminant analysis (OPLS-DA) were applied to analyze the metabolic data.
Results
The plasma metabolite profile distinguished the MHD patients from the healthy controls successfully by using both PCA and OPLS-DA models. Sixty-three metabolites were identified as the key metabolites to discriminate the MHD patients from healthy controls, involving several metabolic pathways (all p < 0.05). An increase in plasma levels of D-glucose, hippuric acid, androsterone glucuronide, indolelactic acid, and a reduction in plasma levels of glycerophosphocholine, serotonin, L-lactic acid, phytosphingosine, and several lysophosphatidylcholine were observed in MHD patients compared to healthy subjects. Metabolomics analysis combined with KEGG pathway enrichment analysis revealed that non-alcoholic fatty liver disease, choline metabolism in cancer, the forkhead box O signaling pathway, and the hypoxia-inducible factor-1 signaling pathway in MHD patients were significantly changed (p < 0.05).
Conclusion
The identification of a novel signaling pathway and key metabolite markers in MHD patients provides insights into potential pathogenesis and valuable pharmacological targets for end-stage renal disease.
Keywords: End-stage renal disease, Metabolic profiling, Liquid chromatography quadrupole time-of-flight mass spectrometry
Introduction
End-stage renal disease (ESRD) is a state of disturbed metabolism, although important features such as dyslipidemia [1] and catabolism [2] remain poorly understood. These factors are believed to contribute to mortality in ESRD, which is much higher than in the general population [3, 4]. Furthermore, ESRD or cardiovascular disease (CVD) develop in a substantial proportion of patients with chronic kidney disease (CKD) receiving standard-of-care therapy, and all-cause mortality remains unchanged in the CKD population [5]. These data suggest that key pathogenetic mechanisms underlying renal disease progression are unaffected by current treatment and prompt the search for easily identifiable risk factors and novel pharmacological targets.
Current metabolite profiling (or metabolomics) technologies enable high-throughput, high-resolution phenotyping of human plasma and are able to identify novel disease biomarkers and their underlying metabolic pathways in well-characterized epidemiologic cohorts [6, 7]. Metabolomics studies have demonstrated metabolic alterations in clinical samples, including blood and urine samples of CKD [8, 9] and diabetic kidney disease [10]. Wang et al. [11] applied liquid chromatography-mass spectrometry-based metabolite profiling to plasma obtained from participants in the Framingham Heart Study to identify novel predictors of future type 2 diabetes and CKD. In this pilot study, non-targeted, mass spectrometry-based profiling of plasma from maintenance hemodialysis (MHD) patients and healthy controls demonstrated that a large number of chemical species found in systemic circulation change significantly. The aim of this pilot study was to demonstrate that the systemic metabolic alterations associated with ESRD could be evaluated using a rapid metabolomics screening technique. As such, the identification of potential signaling pathway and key metabolite markers could provide useful information not only about potential new pathogenesis of ESRD, but also about valuable therapeutic targets for ESRD-associated comorbidities.
Materials and Methods
Patients
The study population consisted of 19 patients aged over 30 years with ESRD receiving MHD for at least 3 months from one clinical center, and 12 healthy controls from one health examination center. The blood samples of each patient were collected before hemodialysis, while one blood sample was obtained from each control.
Sample Preparation Procedures
Blood plasma samples were drawn before dialysis into Vacutainer tubes containing ethylenediaminetetraacetic acid for plasma separation. Plasma samples were transported on ice and centrifuged at 4°C at 3,000 rpm for 15 min. Supernatants were stored in aliquots at −80°C for subsequent metabolomics analysis.
The frozen plasma samples were thawed slowly. The correct amount of cold methanol (precooled to −20°C) was added to the samples to make a final 80% (vol./vol.) methanol solution. The samples were gently shaken to mix and incubated for 6–8 h at −20°C, then centrifuged at 12,000 g for 10 min at 4°C. Supernatants were collected, transferred to a new 1.5-mL Eppendorf tube, and evaporated to dryness at room temperature in a SpeedVac concentrator (Thermo Scientific) with no heat. The dried samples were reconstituted in 100 µL of 60% methanol with 0.1% formic acid, and clarified by centrifugation for 5 min at 12,000 g. The clarified samples were transferred to plastic HPLC vials for liquid chromatography quadrupole time-of-flight mass spectrometry (LC-QTOF-MS) measurements.
Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry Measurements
The LC-QTOF-MS measurements were performed using an Agilent 1260 series LC binary pump and well plate autosampler coupled to a 6,520 accurate-mass Q-TOF LC-MS system equipped with a dual electrospray ionization (ESI) source (Agilent Technologies, Santa Clara, CA, USA). A Cogent Diamond HydrideTM (MicroSol, Eatontown, NJ, USA) aqueous normal phase column (4 μm, 2.1 × 100 mm) was used for the separation of metabolites. The LC parameters were as follows: autosampler temperature, 4°C; injection volume, 5 μL; column temperature, 35°C; flow rate, 0.4 mL/min. The solvents and optimized gradient conditions for LC were: solvent A, 50% methanol/50% water/0.05% formic acid; solvent B, 90% acetonitrile with 5 mM ammonium acetate; elution gradient, 0–20 min in 100% B, 20–25 min in 40% B; post-run time for equilibration, 10 min in 100% B. A blank injection was run after every 3 samples. The optimized ESI Q-TOF parameters for MS experiments were: ion polarity, positive and negative; gas temperature, 325°C; drying gas, 10 L/min; nebulizer pressure, 45 psig; capillary voltage, 4,000 V; fragmentor, 140 V; skimmer, 65 V; mass range, 70–1,100 m/z; acquisition rate, 1.5 spectra/s; instrument state, extended dynamic range (1,700 m/z, 2 GHz). Spectra were internally mass calibrated in real time by continuous infusion of a reference mass solution using an isocratic pump connected to a dual sprayer feeding into an ESI source. Data were collected in both the positive and negative ESI mode. Data were acquired with MassHunter Acquisition software (Agilent Technologies).
Data Processing and Metabolomics Data Analysis
Following LC-QTOF-MS data acquisition, the acquired raw data files were processed with Agilent MassHunter Qualitative Analysis software (version 5.0). Reproducibility of chromatograms was first inspected by overlaying the total ion chromatograms of all samples. Data files that showed extraneous peaks were excluded for further processing. The positive and negative data were combined. The normalization procedure was confirmed by a comparison of the total ion intensity of peaks in MS profiles. Initial, putative metabolite identification was achieved by searching the accurate m/z values of the peaks against an in-house database derived from HMDB, METLIN, and other public databases. At the same time, the extracted ion chromatograms for these matched putative metabolites were generated by performing find by formula function integrated into the software. The abundance of the extracted ion chromatograms was calculated by summing the intensities of all compound-related peaks (e.g., isotopic peaks, adduct peaks, etc.). The preprocessed data files were imported into Agilent Mass Profiler Professional software (version 12.1) for further statistical analysis. MS/MS spectra and retention times acquired from reference metabolites were used for confirmation of the identification of statistically significant metabolites. More specifically, the exact m/z values and intensities of fragment ions from the acquired MS/MS spectra of putative metabolites must have a reasonable match with that of reference metabolites or the fragment ions from public databases (e.g., METLIN, MassBank), if available. Unsupervised principal component analysis (PCA) and supervised orthogonal partial least squares-discriminant analysis (OPLS-DA) models were employed to evaluate the difference between 2 tested groups by using SIMCA 14.1 software (Umetrics AB, Umeå, Sweden). Also, the permutation tests (the number of permutations was 200) were used to validate each group in the OPLS-DA models. S-plot and the variable importance for projection (VIP) values were used to figure out the key mediators that contribute significantly to the discrimination of the 2 groups. Those variables with a VIP >1.0 were considered relevant for group discrimination. A heatmap of the different metabolites in this process was constructed using the pheatmap package in R (3.40).
Statistical Data Analysis
The Kolmogorov-Smirnov test was used to check the normality of the data distribution. Numerical variables were expressed as the mean ± SE for normally distributed parameters or median (interquartile range) for skewed distributions, while categorical variables were expressed as counts (%) or as a ratio. To identify statistically significant differences between plasma samples from the healthy controls and MHD patients, the processed data files were imported into Agilent Mass Profiler Professional software version 12.1 (Agilent Technologies) for statistical analysis. The intensities of the identified metabolites in the plasma samples from healthy controls and MHD patients were compared using a t test followed by a Benjamini-Hochberg multiple testing correction.
Results
Patient Characteristics
A total of 31 subjects were enrolled in this study and 19 of them were ESRD patients receiving MHD treatment. Table 1 lists the clinical information for all of the 31 subjects, including clinical chemistry values, as well as their disease history as diagnosed by the attending physician or by previous medical histories. Twelve volunteers without documented kidney disease were used as healthy controls.
Table 1.
Clinical characteristics of the study population and controls
| Controls | MHD patients | |
|---|---|---|
| n | 12 | 19 |
| Male | 6 (50) | 12 (63) |
| Age, years | 42 (39–50) | 56 (45–57)* |
| Disease history | ||
| Myocardial infarction | 0 (0) | 0 (0) |
| Hypertension | 0 (0) | 17 (89)* |
| Diabetes mellitus | 0 (0) | 2 (11) |
| Hypercholesterolemia | 0 (0) | 2 (11) |
| NAFLD | 0 (0) | 1 (5) |
| Systolic blood pressure, mm Hg | 123.1±15.6 | 143.3±28.1* |
| Diastolic blood pressure, mm Hg | 81.0±12.1 | 70.8±22.6* |
| Laboratory values | ||
| Hemoglobin, g/L | − | 100.4±15.5 |
| Parathyroid hormone, pg/mL | − | 222.3±196.1 |
| Calcium, mmol/L | − | 2.3±0.2 |
| Phosphorus, mmol/L | − | 1.6±0.4 |
| Medication | ||
| Use of CCB | − | 11 (58) |
| Use of ARB | − | 2 (11) |
| Use of ACEI | − | 5 (26) |
| Use of β-receptor blocker | − | 10 (53) |
| Use of aspirin | − | 1 (5) |
| Statins | − | 6 (32) |
| Weekly dose of epoetin, IU | − | 3,000 (3,000–6,000) |
Values expressed as n (%), mean ± SE, or median (IQR). * p < 0.05, compared with healthy controls. Comparisons between 2 groups were performed using the Mann-Whiteny U test. MHD, maintenance hemodialysis; NAFLD, non-alcoholic fatty liver disease; CCB, calcium-channel blocker; ACEI, angiotensin-converting enzyme inhibitor; ARB, angiotensin II type 1 receptor blocker.
Metabolic Differences between Healthy Controls and MHD Patients
The plasma samples from both individual MHD patients and healthy controls were subjected to methanol protein precipitation and subsequent centrifugation, because this technique was previously reported to be the most effective, straightforward, and reproducible extraction method for such samples [12].
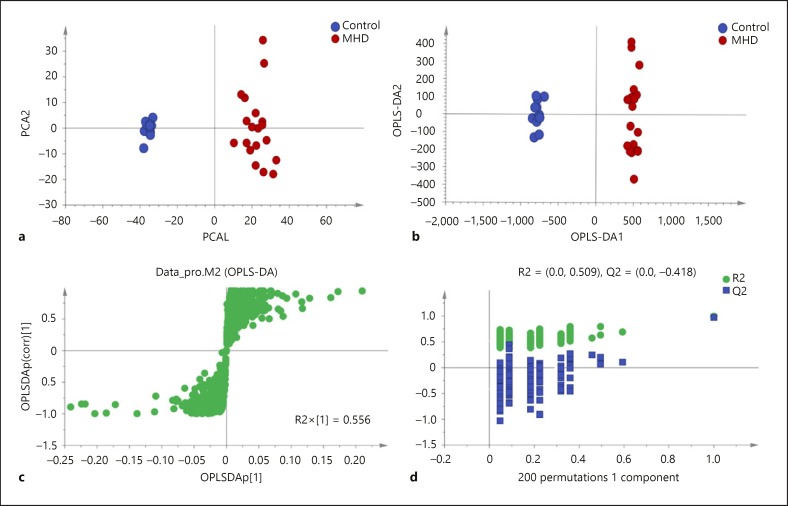
To study the statistically significant metabolic alterations in the plasma between the 2 groups in more depth, PCA, an unsupervised multivariate statistical method, was performed based on the ion intensities of all the identified metabolites in the plasma samples of the healthy controls and MHD patients. As seen in Figure 1a, the results of the PCA (plotting principle component 1 vs. principle component 2) shows an excellent separation of the MHD patients and healthy controls (R2X = 0.459, Q2 = 0.361).
Fig. 1.
Plasma metabolic profiles of LSMs for the MHD patients and healthy controls. a Two-dimensional PCA score plots show visual separation of MHD patients (red dots) from the healthy controls (blue dots). R2X = 0.459, Q2 = 0.361. The percentage variation in the plotted principal components is marked on the axes. Each spot represents one sample, and each group is indicated by a different color. b Two-dimensional OPLS-DA score plots highlight the difference between the patients (red dots) and healthy controls (blue dots). R2X = 0.598, R2Y = 0.995, Q2 = 0.976. c S-plots of the OPLS-DA models for the processed data. d Permutation plots for the MHD patients and controls all show that the OPLS-DA models used above are valid. The green dots represent R2 while blue dots represent Q2. The number of permutations for each plot was 200.
Then, OPLS-DA, a supervised multivariate statistical method, provided valuable insight into group relationships from simple visual inspection of scores-space clustering patterns. One predictive component and one orthogonal components model were constructed with a 2-dimensional score plot illustrated in Figure 1b (R2X = 0.598, R2Y = 0.995, Q2 = 0.976), which shows visual separation of the 2 groups in the t1-axis. S plots of the (O) PLS-DA performed to verify the differentiated metabolites between the 2 groups and supervise the multivariate analysis are shown in Figure 1c.
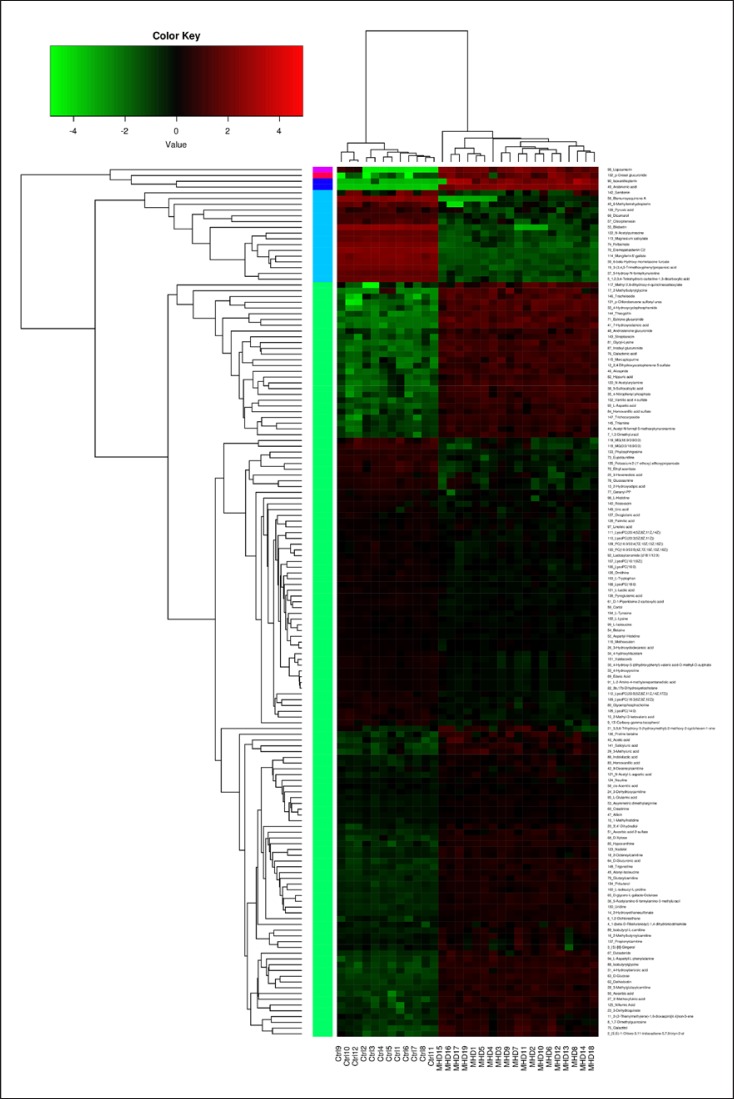
As shown in Figure 2, heatmap analysis revealed distinct patterns of metabolites in the MHD and control groups, with 151 metabolites detected as being significantly changed. Also, the VIP values for each variable were generated and are presented in Table 2, which were all >1.00 (p < 0.05). Of 63 key metabolites, 30 were increased, while 33 were reduced in the plasma of MHD patients compared to those in healthy controls. This showed that the plasma levels of D-glucose, hippuric acid, androsterone glucuronide, and indolelactic acid were increased, and the plasma levels of glycerophosphocholine (GPC), serotonin, L-lactic acid, phytosphingosine, and several lysophosphatidylcholines were decreased in the MHD patients compared to those of controls.
Fig. 2.
Heatmap analysis of the blood metabolite patterns in MHD patients and healthy controls. The heatmap shows the significantly changed metabolites between the 2 groups. Each column in the heatmap represents one sample, and each row represents one metabolite. The color bar showing green to red indicates the relative content of metabolites.
Table 2.
Identification of significant differential metabolites in plasma samples by comparison of the MHD patients and healthy controls using a VIP threshold of 1 (p < 0.05)
| Compound | VIP | FC | Change | Pathway name |
|---|---|---|---|---|
| <smallcaps>l</smallcaps>-Lactic acid | 10.22810 | 0.43789 | Down | HIF-1 signaling pathway |
| LysoPC (16:0) | 9.98057 | 0.48435 | Down | Choline metabolism in cancer |
| Creatinine | 9.58230 | 2.45733 | Up | Arginine and proline metabolism |
| Felbamate | 9.28300 | 0.0003567 | Down | Drug metabolism − cytochrome P450 |
| Hippuric acid | 5.30880 | 58.94476 | Up | Phenylalanine metabolism |
| Acetyl-N-formyl-5-methoxykynurenamine | 3.84561 | 54.30022 | Up | Tryptophan metabolism |
| Geranyl-PP | 3.78926 | 0.33354 | Down | Phosphonate and phosphinate metabolism |
| Pyroglutamic acid | 3.72451 | 0.45121 | Down | <smallcaps>d</smallcaps>-Glutamine and <smallcaps>d</smallcaps>-glutamate metabolism |
| Hypoxanthine | 3.62224 | 13.02824 | Up | Purine metabolism |
| Glycerophosphocholine | 3.54836 | 0.33648 | Down | Choline metabolism in cancer |
| <smallcaps>d</smallcaps>-Glucuronic acid | 3.21844 | 17.55045 | Up | Amino sugar and nucleotide sugar metabolism |
| <smallcaps>d</smallcaps>-Glucose | 3.13755 | 49.69100 | Up | NAFLD |
| 1-Methylhistidine | 3.10201 | 2.65084 | Up | Histidine metabolism |
| LysoPC (18:0) | 2.95710 | 0.66651 | Down | Choline metabolism in cancer |
| Glucosamine | 2.91254 | 0.23987 | Down | Amino sugar and nucleotide sugar metabolism |
| LysoPC (18:3 [6Z, 9Z, 12Z]) | 2.83799 | 0.26197 | Down | Choline metabolism in cancer |
| Thiamine | 2.73887 | 39.48890 | Up | Thiamine metabolism |
| Uridine | 2.62345 | 12.56130 | Up | Chemical carcinogenesis |
| <smallcaps>d</smallcaps>-Xylose | 2.56559 | 12.33261 | Up | ABC transporters |
| Galactonic acid | 2.40013 | 287.15969 | Up | Galactose metabolism |
| Lactosylceramide (d18:1/12:0) | 2.38472 | 0.56823 | Down | Sphingolipid metabolism |
| Trigonelline | 2.34204 | 15.59125 | Up | Nicotinate and nicotinamide metabolism |
| <smallcaps>n</smallcaps>-acetylputrescine | 2.32334 | 0.0014299 | Down | Arginine and proline metabolism |
| Uric acid | 2.23204 | 0.52909 | Down | Bile secretion |
| Mercaptopurine | 2.15339 | 91.55563 | Up | Bile secretion |
| 5-Acetylamino-6-formylamino-3-methyluracil | 2.0563 | 11.81809 | Up | Caffeine metabolism |
| <smallcaps>l</smallcaps>-Isoleucine | 1.97048 | 0.62502 | Down | Central carbon metabolism in cancer |
| <smallcaps>l</smallcaps>-Histidine | 1.88933 | 0.47130 | Down | Central carbon metabolism in cancer |
| LysoPC (20:4 [5Z, 8Z, 11Z, 14Z]) | 1.88369 | 0.59427 | Down | Choline metabolism in cancer |
| Galactitol | 1.87569 | 46.61731 | Up | Galactose metabolism |
| <smallcaps>l</smallcaps>-Tryptophan | 1.81269 | 0.31861 | Down | Central carbon metabolism in cancer |
| 2-Hydroxyethanesulfonate | 1.81057 | 7.84180 | Up | Taurine and hypotaurine metabolism |
| Lysine | 1.78813 | 0.50828 | Down | Protein digestion and absorption |
| 3-Dehydroquinate | 1.77531 | 16.16455 | Up | Biosynthesis of amino acids |
| 5-Hydroxy-N-formylkynurenine | 1.72359 | 0.002373 | Down | Tryptophan metabolism |
| Cortol | 1.70001 | 0.53788 | Down | Steroid hormone biosynthesis |
| Betaine | 1.69081 | 0.66840 | Down | ABC transporters |
| LysoPC (20:3 [5Z, 8Z, 11Z]) | 1.63686 | 0.52152 | Down | Choline metabolism in cancer |
| <smallcaps>l</smallcaps>-Tyrosine | 1.62937 | 0.56811 | Down | Cocaine addiction |
| <smallcaps>l</smallcaps>-Glutamic acid | 1.55062 | 2.70260 | Up | Cocaine addiction |
| Linoleic acid | 1.54579 | 0.63493 | Down | Linoleic acid metabolism |
| <smallcaps>d</smallcaps>-1-Piperideine-2-carboxylic acid | 1.52247 | 0.40154 | Down | Lysine degradation |
| cis-Aconitic acid | 1.52231 | 1.59038 | Up | Citrate cycle |
| Homovanillic acid | 1.49443 | 35.46233 | Up | Tyrosine metabolism |
| LysoPC (20:5 [5Z, 8Z, 11Z, 14Z, 17Z]) | 1.49287 | 0.28354 | Down | Choline metabolism in cancer |
| Pyruvic acid | 1.41018 | 0.08689 | Down | HIF-1 signaling pathway, citrate cycle |
| 4-Hydroxycyclophosphamide | 1.37597 | 773.31431 | Up | Drug metabolism − cytochrome P450 |
| LysoPC (16:1 [9Z]) | 1.35209 | 0.40768 | Down | Choline metabolism in cancer |
| Hydroxybenzoic acid | 1.33573 | 48.97962 | Up | Phenylalanine metabolism |
| Androsterone glucuronide | 1.29119 | 70.87835 | Up | Steroid hormone biosynthesis |
| <smallcaps>l</smallcaps>-Aspartic acid | 1.28381 | 143.33270 | Up | Central carbon metabolism in cancer |
| Ascorbic acid | 1.28257 | 34.16483 | Up | HIF-1 signaling pathway |
| Phytosphingosine | 1.25514 | 0.09685 | Down | Sphingolipid metabolism |
| Oxoglutaric acid | 1.22487 | 0.51611 | Down | HIF-1 signaling pathway |
| Ornithine | 1.22003 | 0.43673 | Down | Arginine biosynthesis |
| Estrone glucuronide | 1.15291 | 3,320.94099 | Up | Steroid hormone biosynthesis |
| N-Acetyl-<smallcaps>l</smallcaps>-aspartic acid | 1.14337 | 5.31335 | Up | Alanine, aspartate and glutamate metabolism |
| Hydroxyproline | 1.12281 | 0.44768 | Down | Arginine and proline metabolism |
| Palmitic acid | 1.11319 | 0.60753 | Down | Biosynthesis of unsaturated fatty acids |
| Dethiobiotin | 1.08593 | 26.11047 | Up | Biotin metabolism |
| Indolelactic acid | 1.06475 | 4.19283 | Up | Tryptophan metabolism |
| Serotonin | 1.04025 | 0.057756 | Down | Bile secretion |
| Acetic acid | 1.00395 | 20.11659 | Up | Glycosaminoglycan biosynthesis − heparan sulfate/ |
| heparin |
MHD, maintenance hemodialysis; VIP, variable importance for projection; FC, fold change (MHD patients vs. healthy controls); NAFLD, non-alcoholic fatty liver disease.
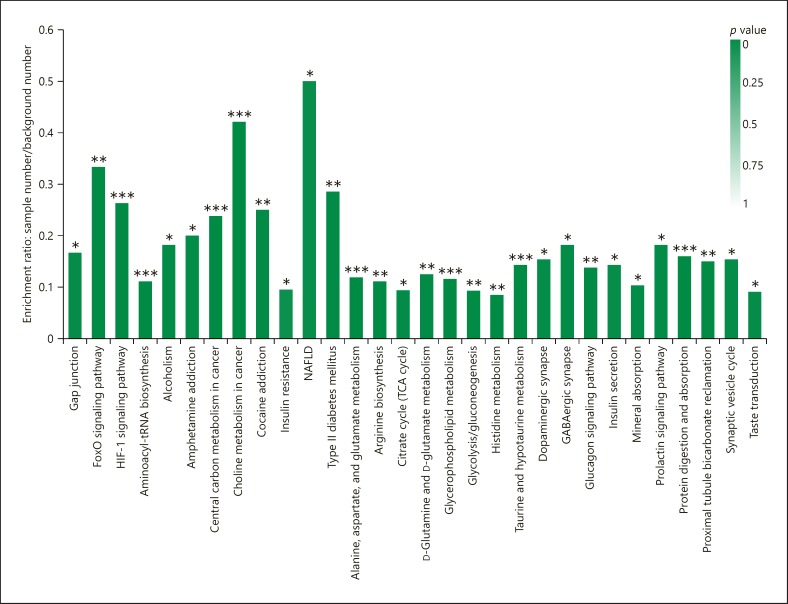
The KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis based on the changed metabolites showed that 47 biological pathways were significantly changed, and that these changes were primarily linked to non-alcoholic fatty liver disease (NAFLD), choline metabolism in cancer, the forkhead box O (FoxO) signaling pathway, and the hypoxia-inducible factor-1 (HIF-1) signaling pathway (Fig. 3).
Fig. 3.
KEGG pathway enrichment analysis based on the changed metabolites. * p < 0.05, ** p < 0.01, *** p < 0.001, Fisher's exact test with BH correction.
Discussion
A broad, untargeted, LC-QTOFMS-based profiling of plasma distinguished the MHD patients from the healthy controls successfully by using both PCA and OPLS-DA models. As seen in Figure 1, PCA shows an excellent separation of the MHD patients and healthy controls. We next turned our focus toward the identification of those features that showed significant differences in levels between the MHD and healthy control sample sets. Sixty-three key metabolites (VIP score >1) were significantly altered in plasma samples of MHD patients, involving several key metabolic pathways (all p < 0.05), including the NAFLD, choline metabolism in cancer, FoxO signaling pathway, and HIF-1 signaling pathway, and so on.
In the kidney, renal fibrosis is a poor prognostic indicator and represents a common final pathway in the development of ESRD irrespective of the underlying cause. Morphologically, tubulointerstitial fibrosis is associated with tubular atrophy, glomerulosclerosis, interstitial inflammation, and loss of peritubular capillaries. Our data demonstrated that the FoxO and HIF-1 signaling pathways were significantly changed in MHD patients, which are all involved in renal fibrosis. HIF-1 and HIF-2 are basic helix-loop-helix transcription factors that allow cells to survive in a low oxygen environment by regulating energy metabolism, vascular remodeling, erythropoiesis, cellular proliferation, and apoptosis. Recent studies suggest that HIF activation promotes epithelial to mesenchymal transition (EMT) and renal fibrogenesis. Hypoxia plays an active role in the progression of fibrotic kidney disease and identifies epithelial HIF-1 as an oxygen-regulated transcription factor that is capable of promoting fibrogenesis through increased expression of extracellular matrix modifying factors, lysyl oxidase genes, and by facilitating EMT [13]. On the other hand, pharmacological targeting of HIF-α degradation by prolyl-hydroxylase inhibition increased serum epoetin levels in humans and improved anemia of chronic disease and inflammation in patients and animal models [14, 15, 16].
Medial vascular calcification is a common complication of CKD or ESRD. Mokas et al. [17] found that HIF-1, the key hypoxic transcription factor, was essential for enhanced vascular smooth muscle cell calcification in CKD-relevant conditions. Elevated inorganic phosphate rapidly activates HIF-1, even in normal oxygenation, and the enhance stimulates vascular smooth muscle cell osteogenic transdifferentiation and calcification.
Recent research showed that FoxOs exerted redundant effects and divergent functions in many diseases, including fibrosis, CVDs, and cancer EMT. FoxO is a potent transcription factor for TGF-β-induced regulatory T cells, which are central to TGF-β anti-inflammatory function [18]. Hypoxia-activated HIF-1α contributed to FoxO3 activation and functioned to protect the kidneys, as tubular deletion of HIF-1α decreased hypoxia-induced FoxO3 activation and resulted in more severe tubular injury and interstitial fibrosis following ischemic injury. Strikingly, tubular deletion of FoxO3 during the AKI to CKD transition aggravated renal structural and functional damage, leading to a more profound CKD phenotype [19]. Additionally, FoxO expression progressively increased in aging human and mouse brains. The nervous system-specific deletion of FoxO transcription factors in mice accelerates aging-related axonal tract degeneration. FoxOs exert both detrimental and protective effects on neurodegenerative diseases [20]. More work is required before FoxO proteins can be considered clinically valuable therapeutic targets for reversing renal fibrosis and ESRD-associated comorbidities.
KEGG pathway enrichment analysis based on the changed metabolites demonstrated that NAFLD pathways were significantly changed in MHD patients. The presence and severity of NAFLD has been related to the incidence and stage of CKD independently of traditional CKD risk factors; conversely, the presence of CKD increases overall mortality in patients with NAFLD compared with the general population [21, 22]. Further supporting a pathogenic link between NAFLD and CKD, non-alcoholic steatohepatitis-related cirrhosis carries a higher risk of renal failure than other etiologies of cirrhosis, is an increasing indication for simultaneous liver-kidney transplantation, and is an independent risk factor for kidney graft loss and CVD [23, 24]. Collectively, these data suggest that common pathogenic mechanisms underlie both liver and kidney injury. Therefore, additional research is needed to explore the possible mechanisms.
Additionally, our KEGG analysis suggested that the choline metabolism in cancer metabolic pathways was altered significantly in MHD patients, involving the key metabolites GPC. Besides, GPC in the plasma of healthy controls was 2-fold higher than that in the plasma of MHD patients, which has been reported to be correlated with neurodegeneration, and could improve metabolic health [25]. GPC is an orthomolecular water-phase, non-membrane phospholipid. It has been clinically validated in 21 clinical trials for attention, immediate recall, and other cognitive functions, as well as for Alzheimer's and vascular dementia, stroke recovery, and recovery from brain trauma. Although not directly incorporated into cell membranes, GPC is readily coupled with the omega-3 fatty acid docosahexaenoic acid (DHA) by enzymes specialized for this task. The resulting phosphatidylcholine molecules containing DHA are highly fluidizing for the new nerve cell membrane. Little energy is required for this conversion, making it highly facile in the brain [26]. Also, GPCs are believed to protect the tissues from urea by stabilizing macromolecules, and thus counteracting the actions of urea [27], while high concentrations of urea are present in the mammalian renal medulla. Urea generally destabilizes biological macromolecules, altering their structure and function. Such effects are expected to be deleterious.
Our study has some limitations that should be considered. Firstly, only a limited number of MHD patients and healthy control plasma samples were examined here. In addition, MHD patients differed from controls in terms of age, cardiovascular history, and blood pressure and treatment. More subjects need to be involved in our future study to discover novel metabolomics biomarkers and signaling pathways of MHD patients.
In conclusion, our study demonstrates striking differences from metabolomics studies between MHD patients and healthy controls, and may provide clues to potential functional pathways and valuable pharmacological targets for reversing renal fibrosis and ESRD-associated comorbidities.
Statement of Ethics
The present study was approval by the responsible Ethics Committee of Nanjing Medical University (permit No. KY2013019). Written informed consent was provided by the patients before the study. Patient consent for publication was not applicable.
Disclosure Statement
The authors declare no competing financial interests.
Funding Sources
This study was supported by the National Natural Science Foundation of China (30801214) and the Natural Science Foundation of Shanghai Municipality (16ZR1426900), with grants from the Scientific and technological Innovation Project of Traditional Chinese Medicine of Shanghai (ZYKC201601030). The funding agencies were not involved in the study design, interpretation of the data, or writing of the paper.
Author Contributions
J.N., J.Y., and Y.C. conceived the current study. Y.C. and P.W. performed the experiments. Y.C. contributed to the statistical analysis. Y.C. and J.N. drafted the manuscript. All authors reviewed and approved the final manuscript.
Availability of Data and Materials
All datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Acknowledgments
We are grateful to the patients who took part. The authors would like to thank Mr. Zhao Liang (Center for Alternatives to Animal Testing, Department of Environmental Health Sciences Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, USA) for his technical support in metabonomic measurements.
References
- 1.Vaziri ND. Dyslipidemia of chronic renal failure: the nature, mechanisms, and potential consequences. Am J Physiol Renal Physiol. 2006 Feb;290((2)):F262–72. doi: 10.1152/ajprenal.00099.2005. [DOI] [PubMed] [Google Scholar]
- 2.Raj DS, Sun Y, Tzamaloukas AH. Hypercatabolism in dialysis patients. Curr Opin Nephrol Hypertens. 2008 Nov;17((6)):589–94. doi: 10.1097/MNH.0b013e32830d5bfa. [DOI] [PubMed] [Google Scholar]
- 3.Shinohara K, Shoji T, Emoto M, Tahara H, Koyama H, Ishimura E, et al. Insulin resistance as an independent predictor of cardiovascular mortality in patients with end-stage renal disease. J Am Soc Nephrol. 2002 Jul;13((7)):1894–900. doi: 10.1097/01.asn.0000019900.87535.43. [DOI] [PubMed] [Google Scholar]
- 4.Kopple JD. Effect of nutrition on morbidity and mortality in maintenance dialysis patients. Am J Kidney Dis. 1994 Dec;24((6)):1002–9. doi: 10.1016/s0272-6386(12)81075-4. [DOI] [PubMed] [Google Scholar]
- 5.Palmer SC, Mavridis D, Navarese E, Craig JC, Tonelli M, Salanti G, et al. Comparative efficacy and safety of blood pressure-lowering agents in adults with diabetes and kidney disease: a network meta-analysis. Lancet. 2015 May;385((9982)):2047–56. doi: 10.1016/S0140-6736(14)62459-4. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999 Nov;29((11)):1181–9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 7.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol. 2002 Jan;48((1-2)):155–71. [PubMed] [Google Scholar]
- 8.Rhee EP, Souza A, Farrell L, Pollak MR, Lewis GD, Steele DJ, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010 Jun;21((6)):1041–51. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyohara T, Akiyama Y, Suzuki T, Takeuchi Y, Mishima E, Tanemoto M, et al. Metabolomic profiling of uremic solutes in CKD patients. Hypertens Res. 2010 Sep;33((9)):944–52. doi: 10.1038/hr.2010.113. [DOI] [PubMed] [Google Scholar]
- 10.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, Saito R, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013 Nov;24((11)):1901–12. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011 Apr;17((4)):448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Want EJ, O'Maille G, Smith CA, Brandon TR, Uritboonthai W, Qin C, et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal Chem. 2006 Feb;78((3)):743–52. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- 13.Higgins DF, Kimura K, Iwano M, Haase VH. Hypoxia-inducible factor signaling in the development of tissue fibrosis. Cell Cycle. 2008 May;7((9)):1128–32. doi: 10.4161/cc.7.9.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haase VH. Hypoxia-inducible factors in the kidney. Am J Physiol Renal Physiol. 2006 Aug;291((2)):F271–81. doi: 10.1152/ajprenal.00071.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen N, Hao C, Liu BC, Lin H, Wang C, Xing C, et al. Roxadustat treatment for anemia in patients undergoing long-term dialysis. N Engl J Med. 2019 Sep;381((11)):1011–22. doi: 10.1056/NEJMoa1901713. [DOI] [PubMed] [Google Scholar]
- 16.Chen N, Hao C, Peng X, Lin H, Yin A, Hao L, et al. Roxadustat for anemia in patients with kidney disease not receiving dialysis. N Engl J Med. 2019 Sep;381((11)):1001–10. doi: 10.1056/NEJMoa1813599. [DOI] [PubMed] [Google Scholar]
- 17.Mokas S, Larivière R, Lamalice L, Gobeil S, Cornfield DN, Agharazii M, et al. Hypoxia-inducible factor-1 plays a role in phosphate-induced vascular smooth muscle cell calcification. Kidney Int. 2016 Sep;90((3)):598–609. doi: 10.1016/j.kint.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 18.Qiao X, Rao P, Zhang Y, Liu L, Pang M, Wang H, et al. Redirecting TGF-β signaling through the b-Catenin/Foxo complex prevents kidney fibrosis. J Am Soc Nephrol. 2018 Feb;29((2)):557–70. doi: 10.1681/ASN.2016121362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Kang H, Zhang Q, D'Agati VD, Al-Awqati Q, Lin F. FoxO3 activation in hypoxic tubules prevents chronic kidney disease. J Clin Invest. 2019 Mar;129((6)):2374–89. doi: 10.1172/JCI122256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang I, Oh H, Santo E, Kim DY, Chen JW, Bronson RT, et al. FOXO protects against age-progressive axonal degeneration. Aging Cell. 2018 Feb;17((1)):e12701. doi: 10.1111/acel.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musso G, Gambino R, Tabibian JH, Ekstedt M, Kechagias S, Hamaguchi M, et al. Association of non-alcoholic fatty liver disease with chronic kidney disease: a systematic review and meta-analysis. PLoS Med. 2014 Jul;11((7)):e1001680. doi: 10.1371/journal.pmed.1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musso G, Cassader M, Cohney S, De Michieli F, Pinach S, Saba F, et al. Fatty liver and chronic kidney disease: novel mechanistic insights and therapeutic opportunities. Diabetes Care. 2016 Oct;39((10)):1830–45. doi: 10.2337/dc15-1182. [DOI] [PubMed] [Google Scholar]
- 23.Singal AK, Salameh H, Kuo YF, Wiesner RH. Evolving frequency and outcomes of simultaneous liver kidney transplants based on liver disease etiology. Transplantation. 2014 Jul;98((2)):216–21. doi: 10.1097/TP.0000000000000048. [DOI] [PubMed] [Google Scholar]
- 24.Mikolasevic I, Racki S, Zaputovic L, Lukenda V, Sladoje-Martinovic B, Orlic L. Nonalcoholic fatty liver disease (NAFLD) and cardiovascular risk in renal transplant recipients. Kidney Blood Press Res. 2014;39((4)):308–14. doi: 10.1159/000355808. [DOI] [PubMed] [Google Scholar]
- 25.Kidd PM. Neurodegeneration from mitochondrial insufficiency: nutrients, stem cells, growth factors, and prospects for brain rebuilding using integrative management. Altern Med Rev. 2005 Dec;10((4)):268–93. [PubMed] [Google Scholar]
- 26.Barbagallo Sangiorgi G, Barbagallo M, Giordano M, Meli M, Panzarasa R. alpha-Glycerophosphocholine in the mental recovery of cerebral ischemic attacks. An Italian multicenter clinical trial. Ann N Y Acad Sci. 1994 Jun;717((1)):253–69. doi: 10.1111/j.1749-6632.1994.tb12095.x. [DOI] [PubMed] [Google Scholar]
- 27.Burg MB, Peters EM. Effects of glycine betaine and glycerophosphocholine on thermal stability of ribonuclease. Am J Physiol. 1998 Apr;274((4)):F762–5. doi: 10.1152/ajprenal.1998.274.4.F762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.