Abstract
Background
Delays in the diagnosis of pediatric osteoarticular infections (OAIs) can cause associated acute complications or long-term morbidity. This study attempts to develop a multiplex PCR-based assay that can rapidly and accurately detect the main pathogens involved in pediatric OAIs, namely, methicillin-sensitive or methicillin-resistant Staphylococcus aureus, Streptococcus pyogenes and Pseudomonas aeruginosa.
Methods
A set of four gene-specific primers suitable for use in a one-tube PCR assay was designed to detect four common pathogens involved in pediatric OAIs, namely, nuc for methicillin-sensitive Staphylococcus aureus, nuc and mecA for methicillin-resistant Staphylococcus aureus, spyM for Streptococcus pyogenes and orpI for Pseudomonas aeruginosa. The multiplex PCR was first evaluated with 39 isolated clinical strains and further with 41 specimens collected from patients suspected of having OAIs.
Results
Specific primer pairs were successfully designed, and the targeted genes were simultaneously amplified. The product sizes in the assay for nuc, mecA, spyM and oprI were 233, 158, 336 and 109 bp, respectively. Evaluation of the multiplex PCR with 39 isolated clinical strains and 41 specimens revealed 100% sensitivity and 100% specificity. The limit of detection of the multiplex PCR assay was approximately 1×103 CFU at the bacterial cell level.
Conclusions
This newly developed multiplex PCR assay, without sequencing, enables a rapid and accurate diagnosis of the major bacterial species in children with OAIs and might serve as an additional diagnostic approach for urgent pathogen determination.
Keywords: Molecular diagnosis, multiplex PCR, pediatric osteoarticular infections (pediatric OAIs), pathogen
Introduction
Osteoarticular infections (OAIs) in children are relatively rare but must be diagnosed and treated urgently due to the risk of prolonged morbidity and long-term functional sequelae. OAIs in children comprise a spectrum of disorders depending on the localization of infection, such as osteomyelitis, arthritis and a combination of both. The reported incidence of osteomyelitis in developed countries varies between 1 and 13 per 100,000 population, with higher values of up to 200 per 100,000 being reported for developing countries (1). Pediatric articular infections are rare, with an incidence of 1 per 100,000 being reported in the developed world over the past few decades (2) and much higher rates in developing regions (3). Due to the risk of severe acute complications or long-term functional sequelae without early appropriate treatment, OAIs could be important diseases to identify.
Rapid identification of the causative pathogens can contribute to the early rational use of antibiotics, shorten healing time, reduce cost, and minimize iatrogenically caused bacterial resistance. The common and important causative pathogens for OAIs are Staphylococcus aureus (S. aureus), Kingella kingae (K. kingae), Streptococcus pyogenes (S. pyogenes) and Pseudomonas aeruginosa (P. aeruginosa) (1,4-7). S. aureus has been reported as the most common pathogen in pediatric OAIs, being detected in 70–90% of the culture-positive cases (1), among which methicillin-resistant S. aureus (MRSA) accounts for approximately 50% of the cases (8). In Western countries, K. kingae is a common cause of OAIs, but it is rarely detected in Chinese patients (7,9-11). S. pyogenes is also the commonly identified causative bacterium in older children and is considered exquisitely susceptive to antibiotics, resulting in a low culture-positive rate in antibiotic-pretreated patients (12). P. aeruginosa is a relatively underreported gram-negative pathogen in pediatric osteomyelitis but is virulent and likely to develop antibiotic resistance (6,13).
Routine culture remains the gold standard for the pathogen diagnosis in OAIs (14). However, it is always time consuming and has low sensitivity, particularly for fastidious, slow-growing or uncultured bacteria or bacteria from patients pretreated with antibiotics. Molecular tests such as conventional 16S rDNA polymerase chain reaction (PCR) combined with DNA sequencing could provide a faster and more accurate result in this scenario and have been widely used in Western countries (14-17). For centers capable of DNA sequencing in house, the turnaround time (TAT) for bacterium-positive samples can be controlled within 24 hr. However, for centers without sequencers, for which the samples need to be sent out for sequencing, TAT may vary from 24 to 72 hr, depending on the location of the center. Therefore, a more rapid, convenient and specific molecular method is urgently needed.
In the present study, we developed a multiplex PCR-based diagnostic assay, which was rapid and gave results within 4 hr, for the major pathogens, namely, MSSA, MRSA, S. pyogenes and P. aeruginosa, in pediatric OAIs according to the literature and evaluated this new assay in a series of isolated clinical strains and specimens collected from children hospitalized for OAIs.
Methods
Bacterial strains and clinical specimens
The isolated bacterial strains used in the present study were obtained from the Microbiology Department at Shanghai Children's Medical Center (SCMC, Shanghai, China). These isolated strains were inoculated onto blood agar or chocolate agar and incubated at 37 °C (with 5% CO2 for S. pyogenes) for 24 hr.
A total of 41 clinical specimens (including bone tissue, pus, synovial fluid and bone marrow) were collected from patients suspected of having OAIs (with fever, swelling, pus, bone pain, and/or limited limb movement, the increase of CRP, ESR and leukocyte, the abscess and effusion of the joint by MRI) and admitted to the SCMC between December 2015 and December 2017. Two samples were generally obtained; one was subjected to the Microbiology Department of SCMC for conventional bacterial tests, including culture, Gram stain, colony morphology, biochemical profile and antimicrobial susceptibility, and the genome of the other was extracted for 16S rDNA PCR assay and stored at –20 °C for subsequent multiplex PCR analysis.
16S rDNA PCR analysis
The sequences 5'-AGAGTTTGATCCTGGCTCAG-3' and 5'-GGTTACCTTGTTACGACTT-3' were used as the forward and reverse primers, respectively, for 16S rDNA PCR amplification. The PCR mixture consisted of 1× Ex Taq buffer (with 20 mM MgCl2), 5 mM each dNTP, 10 pmol of each primer, and 1 U of DNA polymerase (Ex Taq HS DNA polymerase, Takara, Komatsu, Japan). The thermal cycling conditions used were the following: 94 °C for 5 min; 35 cycles of 94 °C for 20 s, 60 °C for 20 s, and 72 °C for 90 s; and 72 °C for 5 min. The resultant amplicon was 1.5 kb. The PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and then sent for sequencing (Beijing Genomics Institute, Beijing, China). The sequences obtained were aligned with sequences in the GenBank database for species assignment.
Primer design for the multiplex PCR assay
In the present study, a new multiplex PCR assay was designed to simultaneously detect the four major pathogens in pediatric OAIs: MSSA, MRSA, S. pyogenes and P. aeruginosa. The nuc, spyM and oprI genes were used to target S. aureus, S. pyogenes and P. aeruginosa, respectively, while the mecA gene was used to differentiate MRSA from MSSA (18-21). Specific primer sets (synthesized by the Beijing Genomics Institute, Beijing, China) targeting the conserved regions of nuc, mecA, spyM or oprI were designed (Primer Premier Ver.5.0, Canada) based on the sequences obtained from the GenBank database, yielding products of 233, 336, 109 and 158 bp, respectively. The specificity of the designed primers was confirmed by BLAST at the GenBank website (http://www.ncbi.nlm.nih.gov/BLAST). The detailed information is listed in Table 1.
Table 1. Sequences of primers used for multiplex PCR assays.
| Bacteria | Gene | Forward primer sequence (5'-3') | Reverse primer sequence (5'-3') | Product size (bp) |
|---|---|---|---|---|
| MSSA | nuc | 5'-AGATAACGGCGTAAATAGAAGTGG-3' | 5'-TAAATGCACTTGCTTCAGGACCATA-3' | 233 |
| MRSA | mecA | 5'-GTAGAAATGACTGAACGTCCGATGA-3' | 5'-ACGTTGCGATCAATGTTACCG-3' | 158 |
| S. pyogenes | spyM | 5'-CCACGACTAATGTCAGCTTGCTCA-3' | 5'-GGGACTTGATGTTGGCTCGAAAACG-3' | 336 |
| P. aeruginosa | oprI | 5'-AGCAGCCACTCCAAAGAAACC-3' | 5'-CCAGAGCTTCGTCAGCCTTG-3' | 109 |
PCR, polymerase chain reaction; MSSA, methicillin sensitive Staphylococcus aureus; MRSA, methicillin resistant Staphylococcus aureus.
Multiplex PCR assay
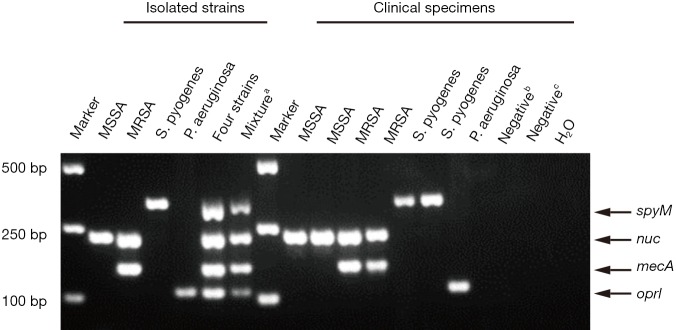
A mixture of DNAs from four clinical strains, namely, MSSA (nuc), MRSA (nuc and mecA), S. pyogenes (spyM) and P. aeruginosa (oprI), that contained the four genes of interest was used as a positive control. DNase-free distilled water was used as a negative control (Figure 1).
Figure 1.
A representative result of the agarose gel electrophoresis showing multiplex PCR amplification products from isolated strains and partial clinical specimens. a, amplicons for nuc, mecA, spyM and oprI were first amplified respectively and then mixed in the same tube; b, specimen detected with other bacteria; c, specimen detected as bacteria negative. PCR, polymerase chain reaction.
Nucleic acid was extracted from each strain or specimen using a QIAamp DNA mini kit (Qiagen, Hilden, Germany). The optimized concentration of primers for each gene (0.3 µM for nuc, 0.4 µM for mecA, 0.15 µM for spyM and 0.3 µM for oprI) was used in the multiplex PCR. The other components used in the PCR were 1× Multiplex PCR Buffer (with Mg2+ and dNTPs) and 1 U of Multiplex PCR Enzyme Mix (Multiplex PCR Assay Kit Ver.2, Takara, Komatsu, Japan) in a 25 µL PCR system. PCR was carried out using a Verity Thermal Cycler (Applied Biosystems, Foster, US), and the conditions used were the following: 94 °C for 5 min; 35 cycles of 94 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s; and then 72 °C for 5 min. The PCR products were analyzed by electrophoresis on 1.5% Tris-acetate-EDTA (TAE) agarose gels. PCR products were visualized under UV illumination and photographed using an image analyzer (Tanon, Shanghai, China).
Evaluation of the multiplex PCR assay
The specificity of multiplex PCR was first analyzed using the genomic DNA extracted from 39 isolated clinical strains and 3 human genome samples. The limit of detection (LoD) was evaluated using various concentrations of a bacterial suspension, ranging from 10 CFU to 105 CFU, obtained from isolated strains. The multiplex PCR was further evaluated with 41 specimens with suspected OAIs and then compared with the results from conventional bacterial culture and 16S rDNA PCR.
Discrepancy analysis
All samples were analyzed using multiplex PCR, culture and 16S rDNA PCR. A multiplex PCR result was considered a true positive (TP) or true negative (TN) only when it agreed with the results from bacterial culture or 16S rDNA PCR. Otherwise, the result was judged to be a false positive (FP) or false negative (FN).
Statistical analysis
Statistical analysis was performed using SAS statistical software (Ver.8.2). The overall percentage of agreement (OPA) was calculated as previously described (22). Briefly, the OPA was calculated as [(TP + TN)/(TP + TN + FP + FN)] ×100%. The sensitivity, calculated as TP/(TP + FN), and specificity, calculated as TN/(TN + FP), were compared for all tests.
Results
Establishment and optimization of a multiplex PCR assay
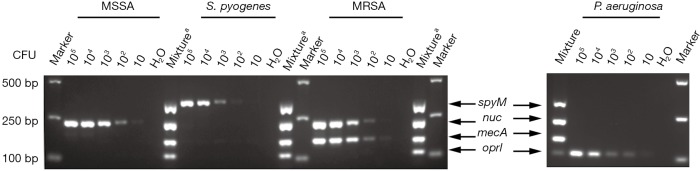
In the present study, stepwise optimization of primer concentration, annealing temperature and different multiplex PCR assay kits was carried out. The optimized concentrations of primers were 0.3, 0.4, 0.15 and 0.3 µM for nuc, mecA, spyM and oprI, respectively. The multiplex PCR assay showed the best results at a 60 °C annealing temperature when the Takara Multiplex PCR Assay Kit Ver.2 was used. No nonspecific products were amplified from the genomic DNA of the four target pathogens (Figure 1). Further investigation showed that the LoD of the assay was approximately 1×103 CFU at the bacterial cell level (Figure 2).
Figure 2.
Detection limit for CFU by multiplex PCR. The limit of detection for MSSA, MRSA, Streptococcus pyogenes and Pseudomonas aeruginosa was evaluated using various concentrations of bacterial suspension starting from 10 CFU to 105 CFU respectively. a, amplicons for nuc, mecA, spyM and oprI were first amplified respectively and then mixed in the same tube. PCR, polymerase chain reaction; MSSA, methicillin sensitive Staphylococcus aureus; MRSA, methicillin resistant Staphylococcus aureus.
Verification of the multiplex PCR assay with isolated clinical strains
A collection of 39 isolated clinical strains identified by the Microbiology Department, namely, 8 MRSA strains, 8 MSSA strains, 2 Staphylococcus epidermidis (S. epidermidis) strains, 2 Staphylococcus haemolyticus (S. haemolyticus) strains, 2 Streptococcus pneumoniae (S. pneumoniae) strains, 7 S. pyogenes strains and 10 P. aeruginosa strains, were used to verify the specificity of the multiplex PCR assay. To test cross-reactivity, negative controls involving 6 other commensal or pathogenic bacteria and 3 human DNA samples were included. All of the isolated clinical MSSA strains were positive for nuc by the multiplex PCR, and all of the MRSA strains were simultaneously positive for nuc and mecA. One S. epidermidis strain and both S. haemolyticus strains showed positive results for the mecA gene, but given the negative results for the nuc gene, these strains could be distinguished from MRSA. All the S. pyogenes and P. aeruginosa strains were positive for oprI and spyM, respectively. The remaining 3 clinical strains and human DNA were negative (Table 2). Overall, the multiplex PCR assay showed 100% analytical specificity in detecting the isolated clinical strains.
Table 2. Bacterial species and strains used in this study and results of the multiplex PCR assay.
| Strain No. | Reference straina | Multiplex PCR result for | |||
|---|---|---|---|---|---|
| nucb | mecAc | spyMd | oprIe | ||
| 1 | MRSAf | + | + | − | − |
| 2 | MRSA | + | + | − | − |
| 3 | MRSA | + | + | − | − |
| 4 | MRSA | + | + | − | − |
| 5 | MRSA | + | + | − | − |
| 6 | MRSA | + | + | − | − |
| 7 | MRSA | + | + | − | − |
| 8 | MRSA | + | + | − | − |
| 9 | MSSA | + | − | − | − |
| 10 | MSSA | + | − | − | − |
| 11 | MSSA | + | − | − | − |
| 12 | MSSA | + | − | − | − |
| 13 | MSSA | + | − | − | − |
| 14 | MSSA | + | − | − | − |
| 15 | MSSA | + | − | − | − |
| 16 | MSSA | + | − | − | − |
| 17 | Staphylococcus epidermidis | − | + | − | − |
| 18 | Staphylococcus epidermidis | − | − | − | − |
| 19 | Staphylococcus haemolyticus | − | + | − | − |
| 20 | Staphylococcus haemolyticus | − | + | − | − |
| 21 | Streptococcus pneumoniae | − | − | − | − |
| 22 | Streptococcus pneumoniae | − | − | − | − |
| 23 | Streptococcus pyogenes | − | − | + | − |
| 24 | Streptococcus pyogenes | − | − | + | − |
| 25 | Streptococcus pyogenes | − | − | + | − |
| 26 | Streptococcus pyogenes | − | − | + | − |
| 27 | Streptococcus pyogenes | − | − | + | − |
| 28 | Streptococcus pyogenes | − | − | + | − |
| 29 | Streptococcus pyogenes | − | − | + | − |
| 30 | Pseudomonas aeruginosa | − | − | − | + |
| 31 | Pseudomonas aeruginosa | − | − | − | + |
| 32 | Pseudomonas aeruginosa | − | − | − | + |
| 33 | Pseudomonas aeruginosa | − | − | − | + |
| 34 | Pseudomonas aeruginosa | − | − | − | + |
| 35 | Pseudomonas aeruginosa | − | − | − | + |
| 36 | Pseudomonas aeruginosa | − | − | − | + |
| 37 | Pseudomonas aeruginosa | − | − | − | + |
| 38 | Pseudomonas aeruginosa | − | − | − | + |
| 39 | Pseudomonas aeruginosa | − | − | − | + |
| 40 | Human genome 1 | − | − | − | − |
| 41 | Human genome 2 | − | − | − | − |
| 42 | Human genome 3 | − | − | − | − |
a, clinical isolates obtained from the Microbiology Department of SCMC; b, to detect S. aureus; c, to identify methicillin-resistance; d, to detect S. pyogenes; e, to detect P. aeruginosa; f, only be confirmed when both nuc and mecA are positive; +: positive result; −: negative result. PCR, polymerase chain reaction; MSSA, methicillin sensitive Staphylococcus aureus; MRSA, methicillin resistant Staphylococcus aureus.
Verification of the multiplex PCR assay with clinical specimens
In addition to testing with the isolated clinical strains, the assay was further retrospectively validated with 41 randomly selected specimens collected from patients suspected of having OAIs, the genomes of which were stored at −20 °C. Among the 41 specimens, the multiplex PCR assay yielded 18 positive results (43.9%), 3 MSSA (7.3%), 9 MRSA (22.0%), 5 S. pyogenes (12. 2%) and 1 P. aeruginosa (2.4%). In comparison, the overall positive rate of standard bacterial culture was 31.7% (13/41), with the same detection rate for MSSA, MRSA and P. aeruginosa, but no detection for S. pyogenes (Table 3).
Table 3. Agreement analysis between multiplex PCR and culture in detecting MSSA, MRSA, S. pyogenes and P. aeruginosa obtained from clinical samples.
| Pathogen identified | No. of resultsa | OPA (%) (95% CI) | |||
|---|---|---|---|---|---|
| C+/M− | C−/M+ | C+/M+ | C−/M− | ||
| MSSA | 0 | 0 | 3 | 38 | 100 (100–100) |
| MRSA | 0 | 0 | 9 | 32 | 100(100–100) |
| S. pyogenes | 0 | 5 | 0 | 36 | 87.8 (77.8–97.8) |
| P. aeruginosa | 0 | 0 | 1 | 40 | 100 (100–100) |
| Total | 0 | 5 | 13 | 23 | 87.8 (77.8–97.8) |
aC, culture; M, multiplex PCR. OPA, overall percentage of agreement; +: positive result; −: negative result. PCR, polymerase chain reaction; CI, confident interval; MSSA, methicillin sensitive Staphylococcus aureus; MRSA, methicillin resistant Staphylococcus aureus.
A total of 164 individual multiplex PCR analyte tests were performed for the 41 samples (for each sample, 4 bacterial tests were included: MSSA, MRSA, S. pyogenes and P. aeruginosa). The OPA between the multiplex PCR and culture was 97% (159/164). For the individual targets, relative to culture, the multiplex PCR had a 100% OPA for MSSA, MRSA and P. aeruginosa and an 87.8% OPA for S. pyogenes (Table 3).
For the targeted bacterial species included in the assay, 5 out of the 41 samples showed discrepant results between the multiplex PCR assay and standard culture. All 5 of these discrepant samples were detected as S. pyogenes positive by multiplex PCR and 16S rDNA PCR but negative by culture. Therefore, these 5 S. pyogenes-positive samples were considered true positive cases (Table 4). The diagnostic accuracy of the multiplex PCR for the four target bacterial species among the 39 isolated clinical strains and 41 clinical specimens was found to have 100% sensitivity and 100% specificity.
Table 4. Discrepant analysis for clinical samples with discordant results.
| No. | Multiplex PCR | Culture | 16S rDNA PCR | Clinical diagnosis from medical records | Final resolution of multiplex PCR |
|---|---|---|---|---|---|
| 1 | S. pyogenes | Negative | S. pyogenes | Arthritis | True positive |
| 2 | S. pyogenes | Negative | S. pyogenes | Arthritis | True positive |
| 3 | S. pyogenes | Negative | S. pyogenes | Arthritis | True positive |
| 4 | S. pyogenes | Negative | S. pyogenes | Arthritis | True positive |
| 5 | S. pyogenes | Negative | S. pyogenes | Arthritis | True positive |
PCR, polymerase chain reaction.
Discussion
To rapidly diagnose the most important causative pathogens in Chinese pediatric OAI cases, we developed a multiplex PCR assay for MSSA, MRSA, S. pyogenes and P. aeruginosa in the present study. The LoD of the assay was 1×103 CFU at the bacterial cell level, illustrating the high analytical sensitivity of the assay (23). In addition, differentiation of MSSA and MRSA by the multiplex PCR assay, which typically takes more than one day for routine culture and cannot be achieved by 16S rDNA PCR, will also contribute to the early rational use of antibiotics and surgical debridement. Therefore, this multiplex PCR-based assay can identify the bacteria causing pediatric OAIs, with the advantages of rapidity and specificity.
In this study, the detection results from the multiplex PCR assay were compared to the results obtained by routine culture, and the discrepant results between the two methods were analyzed. Five S. pyogenes-negative samples according to culture were detected as positive by multiplex PCR assay and 16S rDNA PCR; therefore, these 5 pyogenes-positive samples according to multiplex PCR assay were considered true positive. Overall, our data suggest a higher sensitivity of molecular diagnosis than that of routine culture, which is consistent with previous study (23). In addition to the four target bacterial species, 1 sample was also detected as Enterobacter cloacae positive by culture, which is not analyzed in the multiplex PCR assay. In total, 94.7% (18 of 19) of bacteria could be identified using the multiplex PCR assay. Additionally, the cost of the multiplex PCR assay was approximately USD 10, which is comparable to that of culture.
It is worth noting that the bacterial spectrum involved in OAIs may vary in different regions and time periods. Lemaître et al. reported a decreased incidence of OAIs caused by S. pneumonia in the Paris area over a 10-year period after the introduction of vaccination in France (24). Haemophilus influenza was the second most common cause of acute bacterial arthritis before an effective vaccine, but it is now rarely reported in well-immunized populations (25). Some studies have demonstrated that K. kingae is also a common cause of OAIs in children from Western countries, but it is rarely detected in pediatric OAIs in some countries, such as China and Africa (9-11,26). Consistently, no K. kingae has been detected by either using 16S rDNA PCR or conventional PCR targeting the specific gene cpn60 of K. kingae in our 41 clinical samples in the present study (data not shown). Therefore, the multiplex PCR assay in the present study is designed as a flexible system in which the primer pairs can be optionally added or removed according to the bacterial species and adjusted with regions and times. Additionally, the assay is an auxiliary test for early diagnosis rather than a replacement method for routine culture.
The present study also has some limitations. First, our assay only detects four clinically significant bacteria. Other bacteria that have been detected in the clinic, although not frequently, such as Salmonella enterica and Enterobacter cloacae, are not included in the assay. Second, due to the low incidence of OAIs, the sample size in the present study is relatively small. The clinical application of the assay should be further validated in additional OAI patients.
Conclusions
In summary, our study demonstrates the development of a multiplex PCR assay that can simultaneously and rapidly identify the four most common bacterial species causing pediatric OAIs at the genus level in a single test. The assay was highly sensitive and specific and could provide results within 4 hr, thus allowing clinicians to initiate precise early antibiotic treatment. Therefore, the assay could be used as an effective supplemental diagnostic tool for bacterial detection in OAIs. Further studies are needed to evaluate the cost effectiveness of the present test and, in the long term, the impact of this assay on the clinical outcomes of OAI patients.
Acknowledgments
All authors state that they have read and approved the manuscript. It has not been published elsewhere nor is it under consideration for publication elsewhere. We thank the Microbiology Department for providing the isolated clinical strains.
Funding: This work was financially supported by the Key Developing Disciplines Project from Shanghai Municipal Health Commission (2016ZB0104, 2019SY049), the Collaborative Innovation Center for Translational Medicine at Shanghai Jiao Tong University School of Medicine (TM201616), the Love Charity Foundation Research Project in Shanghai Children’s Medical Center (2017SCMC-AY004), and the Natural Science Foundation of Shanghai (Grand No. 16ZR1421600).
Ethical approval: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Publication of the present manuscript was approved by the Institutional Review Board and the Ethics Committee of Shanghai Children’s Medical Center (SCMCIRB-W2018006), and formal written informed consent is not required for this type of study.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Yeo A, Ramachandran M. Acute haematogenous osteomyelitis in children. BMJ 2014;348:g66. 10.1136/bmj.g66 [DOI] [PubMed] [Google Scholar]
- 2.Gafur OA, Copley LA, Hollmig ST, et al. The impact of the current epidemiology of pediatric musculoskeletal infection on evaluation and treatment guidelines. J Pediatr Orthop 2008;28:777-85. 10.1097/BPO.0b013e318186eb4b [DOI] [PubMed] [Google Scholar]
- 3.Lavy CB, Peek AC, Manjolo G. The incidence of septic arthritis in Malawian children. Int Orthop 2005;29:195-6. 10.1007/s00264-005-0643-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stott NS. Paediatric bone and joint infection. J Orthop Surg (Hong Kong) 2001;9:83-90. 10.1177/230949900100900116 [DOI] [PubMed] [Google Scholar]
- 5.Zhorne D, Bradford KK, Jhaveri R. Review of Pediatric Osteoarticular Infections. Rev Recent Clin Trials 2017;12:260-8. 10.2174/1574887112666170828120040 [DOI] [PubMed] [Google Scholar]
- 6.Saavedra-Lozano J, Mejias A, Ahmad N, et al. Changing trends in acute osteomyelitis in children: impact of methicillin-resistant Staphylococcus aureus infections. J Pediatr Orthop 2008;28:569-75. 10.1097/BPO.0b013e31817bb816 [DOI] [PubMed] [Google Scholar]
- 7.Al-Qwbani M, Jiang N, Yu B. Kingella kingae–Associated Pediatric Osteoarticular Infections. Clinical Pediatrics 2016;55:1328-37. 10.1177/0009922816629620 [DOI] [PubMed] [Google Scholar]
- 8.Arnold JC, Bradley JS. Osteoarticular Infections in Children. Infect Dis Clin North Am 2015;29:557-74. 10.1016/j.idc.2015.05.012 [DOI] [PubMed] [Google Scholar]
- 9.Yagupsky P. Kingella kingae: carriage, transmission, and disease. Clin Microbiol Rev 2015;28:54-79. 10.1128/CMR.00028-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Houmami N, Ceroni D, Codjo Seignon K, et al. Acute Septic Arthritis of the Knee Caused by Kingella kingae in a 5-Year-Old Cameroonian Boy. Front Pediatr 2017;5:230. 10.3389/fped.2017.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzumoto K, Kubota N, Saito Y, et al. A case of osteomyelitis due to Kingella kingae. Kansenshogaku Zasshi 2013;87:207-10. 10.11150/kansenshogakuzasshi.87.207 [DOI] [PubMed] [Google Scholar]
- 12.DeRonde KJ, Girotto JE, Nicolau DP. Management of Pediatric Acute Hematogenous Osteomyelitis, Part I: Antimicrobial Stewardship Approach and Review of Therapies for Methicillin-susceptible Staphylococcus aureus, Streptococcus pyogenes, and Kingella kingae. Pharmacotherapy 2018;38:947-66. 10.1002/phar.2160 [DOI] [PubMed] [Google Scholar]
- 13.Neut D, Hendriks JG, van Horn JR, et al. Pseudomonas aeruginosa biofilm formation and slime excretion on antibiotic-loaded bone cement. Acta Orthop 2005;76:109-14. 10.1080/00016470510030427 [DOI] [PubMed] [Google Scholar]
- 14.Rosey AL, Abachin E, Quesnes G, et al. Development of a broad-range 16S rDNA real-time PCR for the diagnosis of septic arthritis in children. J Microbiol Methods 2007;68:88-93. 10.1016/j.mimet.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 15.Pongsachareonnont P, Honglertnapakul W, Chatsuwan T. Comparison of methods for identifying causative bacterial microorganisms in presumed acute endophthalmitis: conventional culture, blood culture, and PCR. BMC Infect Dis 2017;17:165. 10.1186/s12879-017-2264-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761-4. 10.1128/JCM.01228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marín M, Garcia-Lechuz JM, Alonso P, et al. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J Clin Microbiol 2012;50:583-9. 10.1128/JCM.00170-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol 1992;30:1654-60. 10.1128/JCM.30.7.1654-1660.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Roth E, Claverie-Martin F, Villar J, et al. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol 2001;39:4037-41. 10.1128/JCM.39.11.4037-4041.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Hollingshead S, Swiatlo E, et al. Rapid identification of Streptococcus pyogenes with PCR primers from a putative transcriptional regulator gene. Res Microbiol 2005;156:564-7. 10.1016/j.resmic.2005.01.010 [DOI] [PubMed] [Google Scholar]
- 21.Hancock RE, Siehnel R, Martin N. Outer membrane proteins of Pseudomonas. Mol Microbiol 1990;4:1069-75. 10.1111/j.1365-2958.1990.tb00680.x [DOI] [PubMed] [Google Scholar]
- 22.Du B, Hua C, Xia Y, et al. Evaluation of the BioFire FilmArray meningitis/encephalitis panel for the detection of bacteria and yeast in Chinese children. Ann Transl Med 2019;7:437. 10.21037/atm.2019.08.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Talib H, Latif B, Mohd-Zain Z. Pentaplex PCR assay for detection of hemorrhagic bacteria from stool samples. J Clin Microbiol 2014;52:3244-9. 10.1128/JCM.00891-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaître C, Ferroni A, Doit C, et al. Pediatric osteoarticular infections caused by Streptococcus pneumoniae before and after the introduction of the heptavalent pneumococcal conjugate vaccine. Eur J Clin Microbiol Infect Dis 2012;31:2773-81. 10.1007/s10096-012-1627-5 [DOI] [PubMed] [Google Scholar]
- 25.Dodwell ER. Osteomyelitis and septic arthritis in children: current concepts. Curr Opin Pediatr 2013;25:58-63. 10.1097/MOP.0b013e32835c2b42 [DOI] [PubMed] [Google Scholar]
- 26.Long SS. Recognition of Kingella kingae as a major cause of osteoarticular infections in young children. J Pediatr 2018;194:3. 10.1016/j.jpeds.2018.01.006 [DOI] [PubMed] [Google Scholar]