Summary
-
●
Nitrogen‐fixing nodulation occurs in 10 taxonomic lineages, with either rhizobia or Frankia bacteria. To establish such an endosymbiosis, two processes are essential: nodule organogenesis and intracellular bacterial infection. In the legume–rhizobium endosymbiosis, both processes are guarded by the transcription factor NODULE INCEPTION (NIN) and its downstream target genes of the NUCLEAR FACTOR Y (NF‐Y) complex.
-
●
It is hypothesized that nodulation has a single evolutionary origin c. 110 Ma, followed by many independent losses. Despite a significant body of knowledge of the legume–rhizobium symbiosis, it remains elusive which signalling modules are shared between nodulating species in different taxonomic clades. We used Parasponia andersonii to investigate the role of NIN and NF‐YA genes in rhizobium nodulation in a nonlegume system.
-
●
Consistent with legumes, P. andersonii PanNIN and PanNF‐YA1 are coexpressed in nodules. By analyzing single, double and higher‐order CRISPR‐Cas9 knockout mutants, we show that nodule organogenesis and early symbiotic expression of PanNF‐YA1 are PanNIN‐dependent and that PanNF‐YA1 is specifically required for intracellular rhizobium infection.
-
●
This demonstrates that NIN and NF‐YA1 have conserved symbiotic functions. As Parasponia and legumes diverged soon after the birth of the nodulation trait, we argue that NIN and NF‐YA1 represent core transcriptional regulators in this symbiosis.
Keywords: evolution, intracellular infection, NF‐YA1, NODULE INCEPTION (NIN), nodulation, Parasponia, rhizobium
Introduction
Nitrogen (N) is an essential element for plant growth. To cope with N limitation, some plant species engage with N2‐fixing rhizobium or Frankia bacteria. These bacteria colonize cells of specialized root organs, called nodules. Inside nodule cells, the bacteria convert atmospheric N into ammonium which can be exploited by the plant. Plant species capable of forming N2‐fixing nodules all belong to one of the four orders, Fabales, Fagales, Cucurbitales and Rosales, that together form the so‐called N‐fixing clade (Soltis et al., 1995; Doyle, 2011). Within this clade, nodulation is limited to 10 lineages, of which eight nodulate with Frankia and two with rhizobia (Geurts et al., 2012). The nodulating lineages within the N‐fixing clade are interspersed among tens of nonnodulating lineages. The current hypothesis is that this scattered distribution originates from a single evolutionary gain of nodulation in the ancestor to the N2‐fixing clade, and subsequent loss of this trait in many descending species (Griesmann et al., 2018; van Velzen et al., 2018, 2019). Such a scenario implies that the nodulation trait in all 10 lineages is based on conserved genetic networks.
Rhizobium‐induced nodulation occurs in two lineages; Parasponia (Cannabaceae, Rosales) and legumes (Fabaceae, Fabales). These lineages diverged > 100 Ma and even though the capacity to live in endosymbiosis with diazotrophic bacteria may have been the result of a shared evolutionary event, Parasponia and legumes probably acquired rhizobium as a microsymbiont in parallel (van Velzen et al., 2018, 2019). The molecular and genetic aspects of rhizobium‐induced nodulation have been extensively studied in a number of legume species, for example pea (Pisum sativum), Medicago truncatula and Lotus japonicus, whereas some data are also available for Parasponia. To initiate symbiosis, most rhizobium bacteria excrete lipo‐chitooligosaccharide (LCO) signals that are perceived by plant LysM‐type receptor kinases (Lerouge et al., 1990; Dénarié et al., 1996; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Op den Camp et al., 2011). LCO perception activates the so‐called ‘common symbiosis signalling pathway’, which is coopted from arbuscular mycorrhizal symbiosis (Oldroyd, 2013). Downstream of the common symbiosis signalling pathway, it culminates in the activation of a suite of transcriptional regulators (Soyano & Hayashi, 2014). Among these are NODULE INCEPTION (NIN) and its downstream targets of the NUCLEAR FACTOR Y (NF‐Y) complex that are essential for nodule organogenesis and rhizobium infection and among the first genes transcriptionally induced (Schauser et al., 1999; Combier et al., 2006; Marsh et al., 2007; Soyano et al., 2013; Rípodas et al., 2014; Vernié et al., 2015).
NUCLEAR FACTOR Y complexes are heterotrimeric transcription factors composed of the NF‐YC, NF‐YB and NF‐YA subunits, of which the latter determines the DNA‐binding specificity (Baudin et al., 2015; Myers & Holt, 2018). In plants, each of these subunits is encoded by a small family and in legumes several NF‐Y‐encoding genes display a nodule‐enhanced expression profile (Laloum et al., 2013; Baudin et al., 2015). Mutant analysis in L. japonicus and M. truncatula revealed that NF‐YA1 is required for nodule development (Combier et al., 2006; Soyano et al., 2013; Laloum et al., 2014; Laporte et al., 2014; Hossain et al., 2016). In L. japonicus nf‐ya1 mutants, most nodules do not progress beyond the primordial stage, whereas M. truncatula nf‐ya1 mutants develop nodules of variable size, but all remain substantially smaller than wild‐type nodules (Combier et al., 2006; Hossain et al., 2016). The latter is most probably a result of disturbed formation of the nodule apical meristem (Combier et al., 2006; Laloum et al., 2014; Laporte et al., 2014; Xiao et al., 2014). Besides problems in nodule organogenesis, M. truncatula nf‐ya1 mutants are also affected in the formation of intracellular infection threads (Laporte et al., 2014). These infection threads initiate at the tip of a root hair and function to guide rhizobium bacteria to the underlying nodule primordium, which is formed in the root cortex. In M. truncatula nf‐ya1 mutants, infection thread progression is hampered and infection thread growth is frequently arrested in the epidermal layer (Laporte et al., 2014). In L. japonicus, Ljnf‐ya1 knockdown lines display only a very weak infection phenotype (Soyano et al., 2013; Hossain et al., 2016). Taken together, this shows that in legumes NF‐YA genes function during rhizobia infection and nodule organogenesis.
In legumes, NIN is among the first genes transcriptionally activated upon rhizobium LCO signalling, which is acting downstream of the common symbiosis signalling pathway, and is essential as well as sufficient to initiate nodule organogenesis (Schauser et al., 1999; Borisov et al., 2003; Marsh et al., 2007; Soyano et al., 2013). NIN belongs to a small family of NIN‐Like proteins (NLPs), of which, in Arabidopsis thaliana, several members are involved in nitrate signalling (Schauser et al., 2005; Castaings et al., 2009; Konishi & Yanagisawa, 2013). Orthologues of NIN are found across eudicots, but within the N‐fixation clade functional copies of this gene have been repeatedly lost from the genomes of nonnodulating species (Griesmann et al., 2018; van Velzen et al., 2018). This suggests that within the N2‐fixation clade, NIN predominantly performs a nodulation‐specific function. The first indication that this is indeed the case is obtained from Agrobacterium tumefaciens‐mediated stable transformation knockdown studies in Casuarina glauca, which resulted in a reduced nodulation efficiency when inoculated with Frankia (Clavijo et al., 2015). However, such functional studies to prove that NIN – and its subsequent NF‐YA target genes – has key symbiotic roles in nodulating lineages other than legumes remain scarce.
We aimed to use Parasponia to investigate the extent to which NIN and NF‐YA transcriptional regulators have conserved functions in root nodule formation. Previous studies have shown that NIN and NF‐YA1 are transcriptionally induced in Parasponia andersonii nodules (van Velzen et al., 2018). By creating a series of CRISPR‐Cas9 knockout mutants, we provide evidence that PanNIN is essential for nodule initiation in the nonlegume P. andersonii. Furthermore, we show that PanNF‐YA1 is specifically required for intracellular rhizobium infection, whereas nodule organogenesis is controlled by a genetically redundant network of NF‐YA genes. Taken together, this suggests that NIN and NF‐YA1 are part of a core genetic network essential for rhizobium symbiosis in legumes and nonlegume species.
Materials and Methods
Plant materials and growth conditions
All experiments were done using P. andersonii WU1.14 (van Velzen et al., 2018; Wardhani et al., 2019). Plants were maintained as described previously (van Zeijl et al., 2018; Wardhani et al., 2019). Young plantlets for nodulation assays were vegetatively propagated in vitro, rooted, and inoculated with Mesorhizobium plurifarium BOR2 at an OD600 = 0.03 (van Velzen et al., 2018; van Zeijl et al., 2018; Wardhani et al., 2019). For early induction of symbiotic genes, we made use of Rhizobium tropici CIAT899 transformed with pMP604 (OD600 = 0.03–0.05) (Martínez et al., 1985; Spaink et al., 1989). Nodulation efficiencies were calculated by determining the average nodule number per plant. Nodule size estimates were determined by measuring the two‐dimensional nodule surface area using imagej (Abràmoff et al., 2004). Comparisons were made based on the average nodule size per plant using at least four replicate plants. Acetylene reductase assays (ARAs) were conducted as described previously (van Velzen et al., 2018). Mycorrhization experiments were conducted using 250 spores of Rhizophagus irregularis strain DOAM197198, as described previously (van Velzen et al., 2018; Wardhani et al., 2019).
Lateral root growth assay
Similar‐sized rooted plantlets were grown on EKM‐plates (1% Daishin agar) (Duchefa, Haarlem, the Netherlands) in between two cellophane layers cut to 12 × 8 cm (gel drying frames; Sigma Aldrich) (van Velzen et al., 2018; Wardhani et al., 2019). Plants were grown vertically at a 60º angle for 20 d at 28°C, in a 16 h : 8 h, light : dark regime. The main roots were determined as all roots directly attached to the shoot that were present at the start of the experiment. Per plantlet, root length and lateral root number per root were determined. Total ‘main’ root length per shoot and lateral root density in lateral roots mm–1 root were plotted per plant. Statistical testing was based on a Mann–Whitney U‐test with a significance level of P < 0.05.
Vectors and constructs
Single‐guide RNAs (sgRNAs) were designed using the ‘Find CRISPR Targets’ function implemented in geneious 9.1.5 (Biomatters, Auckland, New Zealand) and subsequently checked against the P. andersonii genome for high‐identity off‐targets. To mutate genes, up to three sgRNAs were used to target either the first or the second coding exon (Supporting Information Table S1). Selected sgRNAs were amplified using sequence‐specific forward primers and a universal reverse primer (Table S2), using Addgene plasmid no. 46966 as template (Nekrasov et al., 2013). Constructs for CRISPR/Cas9‐mediated mutagenesis were assembled as described previously (van Zeijl et al., 2018; Wardhani et al., 2019). To allow golden gate cloning of β‐glucuronidase (GUS) reporter constructs, the BpiI and BsaI restriction sites in putative promoter sequences of PanNF‐YA1, PanNF‐YA3 and PanNF‐YA6 were mutated by introducing single nucleotide substitutions (Engler et al., 2014). The putative promoter sequences are provided in Table S3.
The Gene Identifiers and GenBank accession nos. of the used P. andersonii genes are: PanNIN: PanWU01x14_111140, PON66248.1; PanNF‐YA1: PanWU01x14_284830, PON42093.1; PanNF‐YA3: PanWU01x14_246880, PON47071.1; and PanNF‐YA6: PanWU01x14_192330, PanWU01x14_192330.
Plant transformation
Agrobacterium tumefaciens‐mediated transformation and genotyping were done as previously described (van Zeijl et al., 2018; Wardhani et al., 2019). Primers used for genotyping are listed in Table S2. For promoter‐GUS reporter studies, we investigated five independent lines for each construct.
Histochemical analysis, microtome sectioning and microscopy
Root and nodule samples of the PanNF‐YApro:GUS lines were incubated in GUS buffer (3% (w/v) sucrose, 10 mM EDTA, 2 mM k‐ferrocyanide, 2 mM k‐ferricyanide, and 0.5 mg ml−1 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐glucuronic acid, cyclohexylammonium salt (X‐Gluc) in 0.1 M phosphate buffer (pH 7.2)) at 37°C for 2 and 5 h, respectively. For whole mount sections, GUS‐stained samples were embedded in 6% low‐melting‐point agarose (in PBS). Sections (70 µm thick) were made using a vibratome, and were imaged using Nomarski microscopy. For plastic sections, root segments and nodules were fixed in 4% paraformaldehyde (w/v), 5% glutaraldehyde (v/v) in 50 mM phosphate buffer (pH 7.2) at 4°C for 24 h. Subsequently, the samples were dehydrated using an ethanol series and embedded in Technovit 7100 (Heraeus Kulzer, Hanau, Germany) according to the manufacturer's instructions. Semithin sections were cut using a Leica Ultracut microtome (Leica Microsystems, Wetzlar, Germany) to 4 µm thickness for nodules formed on CRISPR mutant lines and 7 µm thickness for GUS‐stained samples. Sections were stained with 0.05% Toluidine Blue or 0.1% Rethudium Red. Images were photographed using a Leica DM5500B microscope equipped with a DFC425C camera (Leica Microsystems). Samples for electron microscopy were fixed in MTSB buffer (Pasternak et al., 2015) containing 2.5% glutaraldehyde, postfixed in aqueous 1% OsO4 solution, and stained in bloc with 1% uranyl acetate. After dehydration in increasing EtOH concentrations, samples were embedded in epoxy resin. Ultrathin (70 nm) sections were poststained with 2% uranyl acetate and observed in a Philips CM‐10 TEM (Thermo Fisher Scientific, Hillsboro, OR, USA). Images were taken using a Gatan BioScan 792 camera (Gatan, Pleasanton, CA, USA).
In situ hybridization
Parasponia andersonii nodules were fixed with 4% paraformaldehyde, 3% glutaraldehyde in 50 mM phosphate buffer (pH 7.4) and embedded in paraffin (Paraplast X‐tra; Leica Biosystems, Wetzlar, Germany). Root sections of 7 μm were prepared using an RJ2035 microtome (Leica Microsystems). RNA in situ hybridization (ISH) was conducted using Invitrogen ViewRNA™ ISH Tissue 1‐ Plex Assay kits (Thermo Fisher Scientific, Waltham, MA USA) according to a protocol previously developed for M. truncatula (Kulikova et al., 2018). In short, mRNA detection is based on branched (b)DNA signal amplification technology. A mRNA probe set contains c. 20 synthetic adjacent oligonucleotide pairs. Each oligonucleotide is composed of a 20 bp primary sequence to target the sequence of interest and a secondary extended sequence serving as a template for hybridization of a preamplifier oligonucleotide. The preamplifier can hybridize to two adjacent probes. An additional sequence of the preamplifier is designed to hybridize to multiple bDNA amplifier molecules that create a branched structure. Finally, alkaline phosphatase (AP)‐labelled oligonucleotides, which are complementary to bDNA amplifier sequences, bind to the bDNA molecule by hybridization. By adding Fast Red substrate (ThermoFisher Scientific), red punctuated precipitates are formed that can be detected by light microscopy. RNA ISH probe sets were designed and synthesized on request by ThermoFisher Scientific. Catalogue numbers of probes are VF1‐6000380 for PanNIN, VF1‐6000400 for PanNF‐YA1, VF1‐6000767 for PanNF‐YA3, and VF‐6000766 for PanNF‐YA6. Images were taken with an DM5500B microscope equipped with a DFC425C camera (Leica Microsystems).
Phylogenetic reconstruction
Protein sequences of L. japonicus (Lj3.0, Lotus Base (REF; Mun et al., 2016); Sato et al., 2008), Glycine max (Wm82.a2.v1; Sato et al., 2008; Schmutz et al., 2010), Phaseolus vulgaris (Pvulgaris v.2.1; Schmutz et al., 2014), Morus notabilis (Genbank ATGF00000000.1; He et al., 2013), Prunus persica (Ppersica v.2.1; International Peach Genome Initiative et al., 2013) Fragaria vesca (Fvesca v.1.1; Shulaev et al., 2011) were retrieved from Phytozome 12 (http://phytozome.jgi.doe.gov/), unless stated otherwise. Casuarina glauca and Datisca glomerata assemblies were downloaded and set up as custom Blast database in geneious 8.1.9 (Griesmann et al., 2018; van Velzen et al., 2018). Sequences from diploid Peanut Arachis duranensis were retrieved from NCBI (Bertioli et al., 2016). Protein sequences of P. andersonii (PanWU01x14) and Trema orientalis (TorRG33x02) were obtained from http://www.parasponia.org (van Velzen et al., 2018; Holmer et al., 2019). These sequences were mined using sequences from A. thaliana (Tair10; Lamesch et al., 2012) and M. truncatula (Mt4.0v1; Young et al., 2011; Tang et al., 2014). Protein sequences were aligned using mafft v.7.017 (parameter settings: algorithm, auto; scoring matrix, Blosum62; gap open penalty, 1.53; offset value, 0.123; Katoh et al., 2002; Katoh & Standley, 2013; Table S4) implemented in geneious 8.1.9. Bayesian phylogeny was reconstructed using mrbayes 3.2.6. (Ronquist & Huelsenbeck, 2003) implemented in geneious 8.1.9. (parameter settings: rate matrix, poisson; rate variation, gamma; gamma categories, 4; chain length, 5100 000; heated chains, 4; heated chain temp, 0.2; subsampling freq, 1000; burn‐in length, 100 000; random seed, 8681). Midpoint rooting was applied for better tree visualization using figtree v.1.4.2. (http://tree.bio.ed.ac.uk/software/figtree).
RNA isolation and qRT‐PCR analysis
RNA was isolated from snap‐frozen root segments of c. 0.5 cm, which includes the elongation zone and the newly formed differentiation zone. cDNA was prepared from 1 μg of total RNA using the i‐script cDNA synthesis kit (Bio‐Rad), following the manufacturer's instructions. Ten microlitre quantitative reverse transcription polymerase chain reaction (qRT‐PCR) reactions were set up using 2× iQ SYBR Green Supermix (Bio‐Rad) and 5 ng template DNA. Quantification was performed using a CFX Connect optical cycler, according to the manufacturer's protocol (Bio‐Rad). Normalization was performed based on the stably expressed reference gene ELONGATION FACTOR 1α (PanEF1α; van Zeijl et al., 2018). Primers used for qPR‐PCR analysis are listed in Table S2.
Results
P. andersonii NIN and NF‐YA1 are coexpressed during nodule formation
Previously conducted transcriptome studies revealed that PanNIN and PanNF‐YA1 have a nodule‐enhanced expression profile in P. andersonii (van Velzen et al., 2018). To obtain a first insight into the spatiotemporal expression pattern of both genes, we conducted promoter reporter and/or ISH experiments. To this end, a 3.8 kb sequence upstream of the translational start site of PanNF‐YA1, containing the putative promoter sequence and the 5'‐UTR that includes the first intron, was fused to a GUS‐encoding sequence. The resulting construct was introduced into the P. andersonii genome using A. tumefaciens‐mediated stable transformation (van Zeijl et al., 2018). Five lines were selected, for which we compared the GUS reporter activity under symbiotic and nonsymbiotic conditions. Four of these lines yielded comparable results, and therefore one of these lines (line 1E5) was selected for detailed characterization.
Under sterile conditions, activity of the PanNF‐YA1pro:GUS was observed around the vasculature of differentiated root tissue (Fig. S1a,b). Root sections revealed that GUS staining is restricted to the pericycle cells opposite to the protoxylem, but absent from lateral root primordia (Fig. S1b–d). In M. truncatula, similar promoter‐GUS studies using a 2.2 kb upstream region revealed that MtNF‐YA1 is induced in root hairs of the preinfection zone and in the root pericycle upon rhizobium inoculation (Laporte et al., 2014; Liu et al., 2019). We questioned whether this is also the case for P. andersonii. To determine this, transgenic plantlets expressing the PanNF‐YA1pro:GUS reporter were grown in vitro on N‐poor medium (0.375 mM NH4NO3) and inoculated with Mesorhizobium plurifarium BOR2. In contrast to legumes like M. truncatula and L. japonicus, Parasponia species are not infected via curled root hairs. Instead, rhizobia enter apoplastically via cracks that are formed upon cell divisions in the epidermis and outer cortex and only infect intracellularly when a nodule primordium is formed (Lancelle & Torrey, 1984, 1985). At 2 d post‐inoculation (dpi), PanNF‐YA1pro:GUS activity was observed in epidermal and cortical cells located just above the root elongation zone (Fig. S1e). PanNF‐YA1pro:GUS is active in clumps of multicellular root hairs and adjacent cortical cells as well as dividing pericycle‐derived cells (Figs 1a, S1f,g). The formation of multicellular root hairs is one of the earliest responses associated with nodule initiation in Parasponia species and is not observed in noninoculated roots (Lancelle & Torrey, 1984, 1985). In young nodule primordia that are visible as small bumps on the root (5 dpi), the PanNF‐YA1pro:GUS reporter was highly active in clusters of dividing cells (Fig. 1b). Additionally, activity was observed in dividing pericycle cells that flank the developing nodule vascular bundle (Fig. 1b). In young nodules, PanNF‐YA1pro:GUS activity is observed in the central region of the nodule lobes, where intracellular infection by rhizobium will occur (Fig. 1c). In mature nodules, PanNF‐YA1pro:GUS activity was mostly confined to the infection zone (Fig. 1d). Additionally, weaker activity is observed in the cell layers surrounding the nodule vascular bundle (Fig. 1c,d). Taken together, the expression pattern of the PanNF‐YA1pro:GUS reporter suggests a symbiotic role of PanNF‐YA1.
Figure 1.
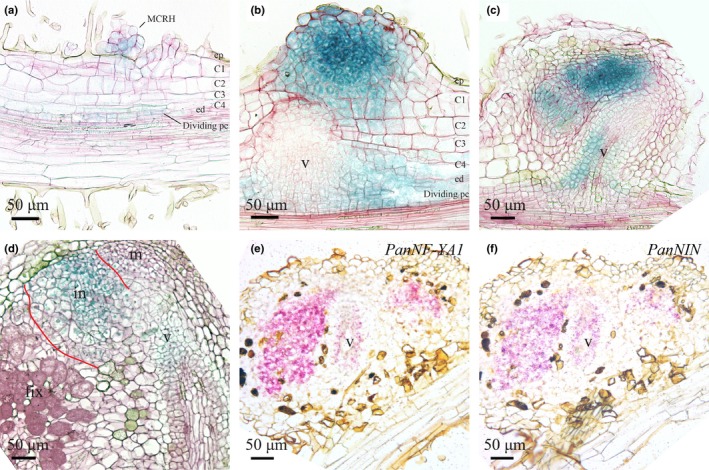
Spatiotemporal expression pattern of PanNF‐YA1 and PanNIN in developing Parasponia andersonii root nodules. (a–d) Spatiotemporal expression pattern of PanNF‐YA1pro:GUS in nodules of different developmental stages. (e, f) Spatiotemporal expression pattern of PanNF‐YA1 and PanNIN visualized by in situ hybridization on consecutive sections of a young P. andersonii nodule primordium. (a) PanNF‐YA1pro:GUS activity in clustered root hairs that are associated with dividing epidermal, outer cortical and pericycle cells. (b) PanNFYA1pro:GUS activity in a young but not yet intracellularly infected nodule and in the pericycle‐derived cells flanking the developing nodule vasculature. (c) PanNF‐YA1pro:GUS activity in the infection zone of young nodules, and in the basal part of the nodule vasculature. (d) PanNF‐YA1pro:GUS activity in a mature nodule is restricted to the infection zone (marked with red lines) and nodule vasculature. (e, f) PanNF‐YA1 (e) and PanNIN (f) transcripts are detected in the infection zone and nodule vasculature by in situ hybridization on consecutive sections. MCRH, multicellular root hairs; ep, epidermis; C1‐C4, first to fourth cortical cell layer; ed, endodermis; pc, pericycle; m, nodule meristem; in, infection zone; fix, fixation zone; v, nodule vasculature. In (a)–(d), sections (7 µm) were counterstained with Ruthenium Red. Nodules were isolated 4 wk post‐inoculation with Mesorhizobium plurifarium BOR2.
Next, we determined whether PanNF‐YA1 is coexpressed with PanNIN in P. andersonii nodules. As regulation of NIN in legumes has been shown to be highly complex and determined by distant cis‐regulatory elements (Heckmann et al., 2011; Kosuta et al., 2011; Popp & Ott, 2011; Soyano et al., 2014; Yoro et al., 2014; Liu et al., 2019), we decided to use RNA ISH. This method showed the accumulation of the PanNIN transcripts in the central region of the lobes where rhizobium infection will take place and in the pericycle/endodermis of the vasculature of young nodules (Fig. 1f). ISH on a consecutive section of the same nodule showed that the PanNF‐YA1 transcripts are present in the same cells as PanNIN (Fig. 1e,f), and that transcript accumulation is consistent with the activity of the PanNF‐YA1pro:GUS reporter in a nodule of a similar developmental stage (Fig. 1c). Therefore, we conclude that PanNIN and PanNF‐YA1 are coexpressed in young nodules.
PanNIN is essential for nodule formation and symbiotic expression of PanNF‐YA1
To determine whether PanNIN is essential for nodule formation in P. andersonii, we created Pannin knockout mutants using CRISPR/Cas9‐mediated mutagenesis. The NIN gene in Parasponia species produces two alternative transcript variants: using a transcriptional initiation site at the 5'‐end of the gene (PanNIN.1); and an alternative transcriptional initiation site for PanNIN.2 located in the second intron of the gene (Fig. S2a; van Velzen et al., 2018). Quantification of RNAseq reads revealed that both transcripts are expressed in roots, whereas only expression of the long transcript (PanNIN.1) encoding a canonical NIN protein is enhanced in nodules (Fig. S2b). Therefore, we decided to create CRISPR‐Cas9 mutants exclusively mutated in the long NIN transcript (PanNIN.1). Two knockout mutant lines (named B1 and B3) were obtained by targeting the first coding‐exon using three sgRNAs (Fig. S2c). These mutants contain premature stop codons at amino acid positions 90 (line B1) and 70 (line B3), respectively (Fig. S2d). Inoculation with M. plurifarium BOR2 showed that both lines are unable to form root nodules or even nodule primordia (Fig. 2c,d), whereas a transgenic control line (CTR44) was well nodulated (Fig. 2a,b). This demonstrates that the PanNIN.1 transcript is essential for nodule organogenesis in P. andersonii.
Figure 2.
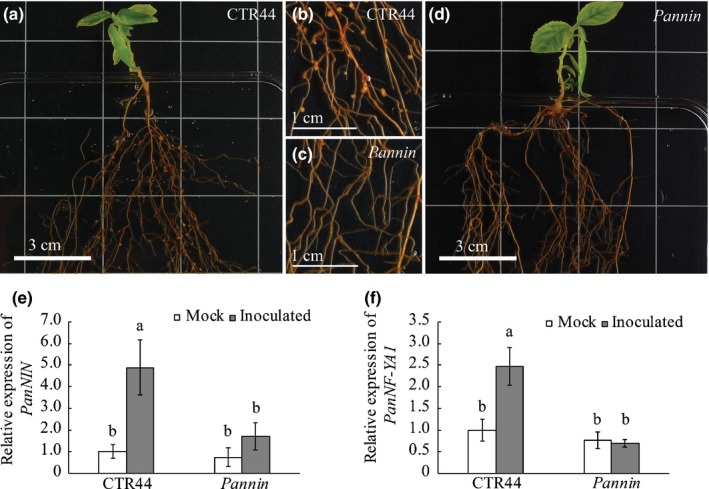
Symbiotic phenotype of the Parasponia andersonii nin mutant. Shown are (a, b) a transgenic control (CTR44) and (c, d) a Pannin knockout mutant (line B3) at 4 wk post‐inoculation with Mesorhizobium plurifarium BOR2. Note that nodules are present on roots of the control (a, b), but not on Pannin mutant roots (n = 50) (c, d). These images are representative results obtained from three independent experiments, with > 20 plants combined for each line. (e) Relative expression of PanNIN in noninoculated and inoculated transgenic control (CTR44) and Pannin mutant (line B3) roots. (f) Relative expression of PanNF‐YA1 in noninoculated and inoculated transgenic control (CTR44) and Pannin mutant (line B3) roots. RNA was isolated from root segments encompassing the elongation and part of the differentiation zone at 1 d post‐inoculation (dpi) with Rhizobium tropici CIAT899 pMP604. Data represent means of two independent experiments with a total of five biological replicates each ± SE. Data were normalized against the mock‐treated CTR44 sample. Different letters indicate statistical significance (Student's t‐test, P < 0.05).
To determine whether rhizobium‐induced PanNF‐YA1 expression is dependent on a functional PanNIN.1 protein, we conducted qRT‐PCR experiments. Root RNA was isolated from a c. 0.5 cm region encompassing part of the root elongation and differentiation zone at 1 dpi with a compatible rhizobium strain that harbours a dominant active NodD protein that transcriptionally activates LCO biosynthesis genes (Rhizobium tropici CIAT899 pMP604; Spaink et al., 1989; Op den Camp et al., 2012; Fig. S3). In roots of transgenic control line CTR44, expression of PanNIN.1 and PanNF‐YA1 was induced five‐ and 2.5‐fold following inoculation, respectively (Fig. 2e,f). By contrast, such induction of PanNF‐YA1 is not observed in Pannin mutant roots (Fig. 2e,f). This indicates that the early symbiotic induction of PanNF‐YA1 is downstream of PanNIN.1.
PanNF‐YA1 is essential for rhizobium intracellular infection
To determine the symbiotic role of PanNF‐YA1, we mutated this gene using CRISPR/Cas9. To this end, sgRNAs were designed that target the first coding‐exon of PanNF‐YA1 (Table S1; Fig. S4a). This allowed the isolation of Pannf‐ya1 knockout mutant line (Fig. S4b).
We noted that Pannf‐ya1 mutant shoots were somewhat more difficult to root (Fig. S5a,b), a phenotype we did not observe with transgenic control or Pannin mutant shoots. As it was reported previously that NF‐YA1 orthologous genes may function in root growth and lateral root formation (Soyano et al., 2013; Sorin et al., 2014), we quantified root development in the Pannf‐ya1‐1 mutant line and transgenic control. This revealed that the Pannf‐ya1‐1 mutant formed less lateral roots when compared with transgenic controls (Fig. S5c–f).
To determine the nodulation phenotype, the Pannf‐ya1‐1 mutant line plants were grown in Perlite and inoculated with M. plurifarium BOR2. This showed that Pannf‐ya1‐1 can be nodulated at least as efficiently as control plants (Fig. S6a). However, quantification of Nitrogenase activity using the ARA indicated that Pannf‐ya1‐1 nodules are unable to fix N2 (Fig. S6b).
Next, we studied the cytoarchitecture of Pannf‐ya1‐1 nodules using light microscopy as well as transmission electron microscopy. In wild‐type Parasponia, rhizobium bacteria first colonize the apoplast of the nodule infection zone, after which they enter nearby cells through infection threads (Lancelle & Torrey, 1984; Fig. 3a,b). Parasponia andersonii nf‐ya1‐1 mutant nodules display a wild‐type cytology, but cells in the infection zone are devoid of intracellular infection threads (Fig. 3d,e). Instead, large apoplastic colonies of rhizobium can be seen that occasionally occupy dead host cells (Fig. 3e). Transmission electron microscopy showed that apoplastic rhizobia in wild‐type nodules are embedded in a thin layer of secreted matrix material from where intracellular infection can occur (Fig. 3c; Trinick, 1979). By contrast, no such intracellular infections were observed in Pannf‐ya1‐1 mutant nodules. Instead, rhizobium formed large apoplastic colonies embedded in a secreted matrix (Fig. 3f). This infection phenotype was confirmed in two additional Pannf‐ya1 mutant lines (Figs S4c,d, S6c,d). Based on these results, we conclude that PanNF‐YA1 has an essential role in intracellular infection thread formation in Parasponia nodules.
Figure 3.
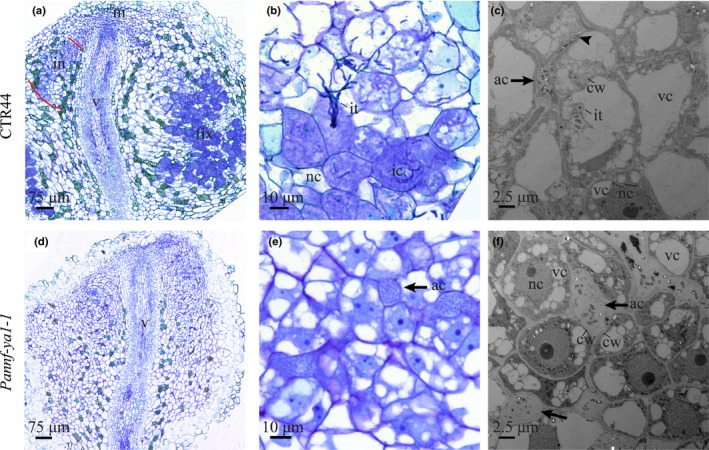
PanNF‐YA1 is essential for intracellular rhizobium infection. (a, b) Nodule cytoarchitecture of Parasponia andersonii transgenic control (CTR44) plants studied by light microscopy. (a) Sections of a mature transgenic control nodule. The infection zone (in) in one lobe is marked with red lines. (b) Formation of intracellular infection threads. Shown is a close‐up of the infection zone of a mature nodule. (c) Transmission electron microscopy image of apoplastic rhizobium infection (arrow) and initiation of intracellular infection (arrowhead) in a transgenic control nodule. (d, e) Cytoarchitecture of a Pannf‐ya1 nodule studied by light microscopy. Pannf‐ya1 mutant nodules lack intracellular infection threads (d). In mature Pannf‐ya1‐1 nodules (e), apoplastic colonies of rhizobium can be detected (arrow). (f) Transmission electron microscopy image of large apoplastic rhizobium colonies (arrows) in a Pannf‐ya1 mutant nodule. Plastic sections (a, b, d, e) were stained using Toluidine Blue. m, nodule meristem; in, infection zone; fix, fixation zone; v, nodule vasculature; it, intracellular infection thread; ic, infected cells; nc, noninfected cells; ac, apoplastic colonies of rhizobia; cw, cell wall; nc, nucleus; vc, vacuoles. Nodules were isolated at 4 wk post‐inoculation with Mesorhizobium plurifarium BOR2.
PanNF‐YA3 and PanNF‐YA6 are expressed during nodule formation
The nf‐ya1 mutants in M. truncatula and L. japonicus are clearly affected in nodule development (Combier et al., 2006; Soyano et al., 2013; Laporte et al., 2014; Xiao et al., 2014). By contrast, no such phenotype was observed in P. andersonii nf‐ya1‐1 mutants. Therefore, we questioned whether additional NF‐YA‐encoding genes perform a symbiotic function in Parasponia.
To determine whether close paralogs of PanNF‐YA1 exist in P. andersonii, as has been reported for the model legumes M. truncatula and L. japonicus (Laloum et al., 2013; Soyano et al., 2013), we reconstructed the phylogeny of the NF‐YA clade. This revealed that P. andersonii possesses seven NF‐YA genes that are divided over seven orthogroups (Figs 4a, S7). We noted that legumes experienced gene duplication events in five orthogroups, including the NF‐YA1 lineage (Fig. 4a). In line with this, we conclude that PanNF‐YA1 is the sole orthologue of two legume genes represented by MtNF‐YA1/LjNF‐YA1 and MtNF‐YA2/LjNF‐YA4 in M. truncatula and L. japonicus. Additionally, we noted that legumes have genes only in six orthogroups, lacking an orthologue of PanNF‐YA7. To determine whether gene duplications are specific to legumes, we reconstructed the phylogeny also including the NF‐YA protein family of the actinorhizal plant species Casuarina glauca (Fagales) and Datisca glomerata (Cucurbitales), and the legume Arachis duranensis. This showed that C. glauca and D. glomerata generally possess a single gene in each of the seven orthogroups, similar to what was observed for P. andersonii, supporting the conclusion that duplication of NF‐YA genes in legumes is the result of a lineage‐specific event (Fig. S7).
Figure 4.
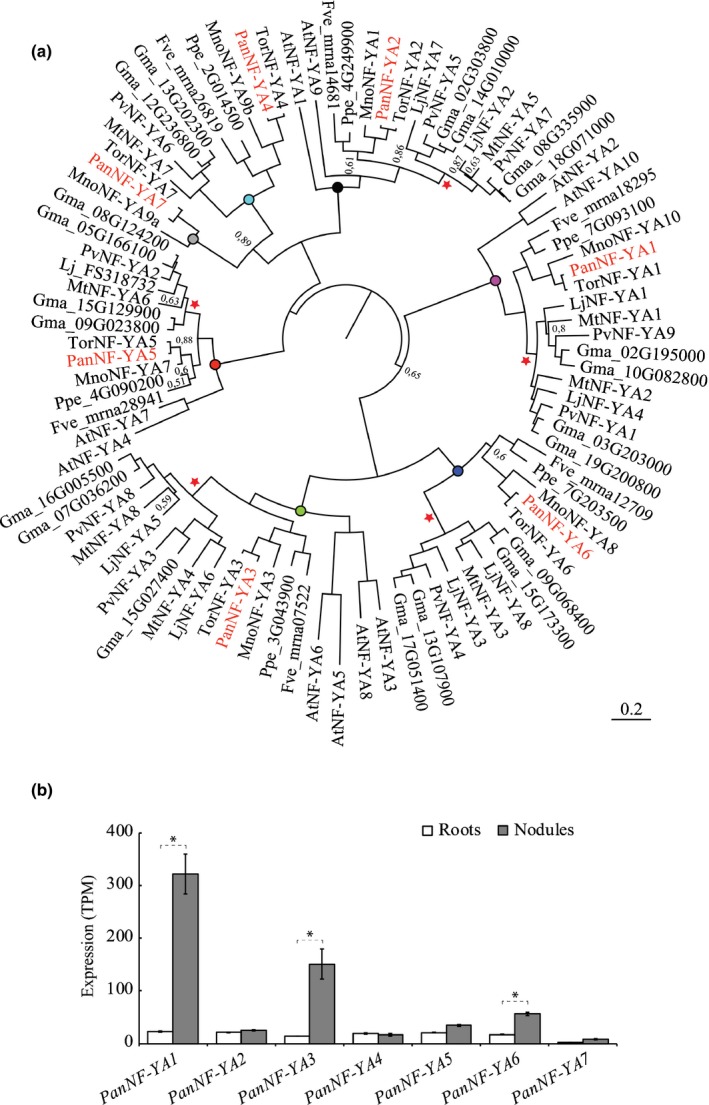
Phylogenetic relation and symbiotic expression of Parasponia andersonii NF‐YA genes. (a) Bayesian phylogeny of NF‐YA proteins reconstructed based on an alignment of protein sequences from the following species: Parasponia andersonii (Pan), Trema orientalis (Tor), Arabidopsis thaliana (At), Medicago truncatula (Mt), Lotus japonicus (Lj), Glycine max (Gma), Phaseolus vulgaris (Pv), Morus notabilis (Mno), Prunus persica (Ppe), Fragaria vesca (Fve). Parasponia andersonii NF‐YA proteins are marked in red. Red pentagrams mark duplication events within the legume family. Orthogroups are indicated by a coloured circle. Node labels indicate posterior probability, Node labels with a value > 0.9 are not shown. (b) Expression level of PanNF‐YA genes in roots and mature nodules. Expression was determined by quantification of RNAseq reads. Data represent average expression in transcripts per million (TPM) (n = 3) ± SD, which were obtained from van Velzen et al. (2018). Nodules were isolated 4 wk post‐inoculation with Mesorhizobium plurifarium BOR2. *, P < 0.01 adjusted for multiple testing based on false discovery rate estimated for two‐fold change in mature nodule vs root sample as described by van Velzen et al. (2018).
To study whether other PanNF‐YA genes might function in rhizobium symbiosis, we determined their expression in nodules using published transcriptome data (van Velzen et al., 2018). This revealed that six PanNF‐YA genes are expressed in nodules (transcripts per million > 10), three of which show a nodule‐enhanced expression profile, namely PanNF‐YA1, PanNF‐YA3 and PanNF‐YA6, respectively (Fig. 4b). To study the symbiotic expression of PanNF‐YA3 and PanNF‐YA6 in more detail, we created promoter‐reporter GUS constructs for both genes. These constructs contain 3.5 and 4.9 kb upstream of the translational start sites of PanNF‐YA3 and PanNF‐YA6, respectively.
Transgenic P. andersonii lines harbouring these constructs revealed that the PanNF‐YA3pro:GUS construct is active in the root apical meristem (Fig. S8a). In the case of PanNF‐YA6, the promoter‐reporter construct is expressed in young parts of the roots, including the meristem (Fig. S8e). Next, we studied their expression patterns following inoculation with rhizobium. In nodule primordia, PanNF‐YA3pro:GUS is active in the dividing epidermal, cortical and pericycle cells, mimicking activity of the PanNF‐YA1pro:GUS reporter (Figs 5a, S8b,c). In young nodules, PanNF‐YA3pro:GUS is expressed in the central region of the nodule lobes where rhizobium infection occurs and in the vascular bundle (Fig. 5b). In mature nodules, PanNF‐YA3pro:GUS activity is observed in the infection zone and nodule vasculature (Figs 5c, S8d). Activity of the PanNF‐YA6pro:GUS reporter is restricted to the nodule vascular meristem (Figs 5d, S8f). ISH confirmed the expression patterns of PanNF‐YA3 and PanNF‐YA6 in young nodules (Fig. 5e,f). Additionally, it showed that PanNF‐YA3 is coexpressed with PanNIN in the lobes of young nodules (Figs 1f, 5e). Therefore, we questioned whether symbiotic PanNF‐YA3 and/or PanNF‐YA6 expression requires a functional PanNIN gene. qRT‐PCR experiments on the same samples used for studying PanNF‐YA1 expression revealed that neither PanNF‐YA3 nor PanNF‐YA6 expression is enhanced within 24 h after inoculation (Fig. S8g,h).
Figure 5.
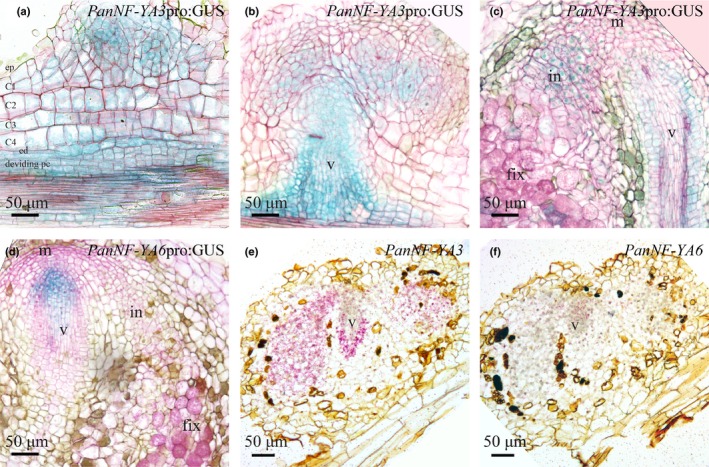
Spatiotemporal expression pattern of PanNF‐YA3 and PanNF‐YA6 in Parasponia andersonii root nodules. (a, c) Spatiotemporal expression pattern of PanNF‐YA3pro:GUS in nodules of different developmental stages. (a) PanNF‐YA3pro:GUS activity is observed in dividing epidermal, cortical, endodermal cells of a nodule primordium as well as the root vasculature. (b) In a young nodule, PanNF‐YA3pro:GUS activity is confined to the nodule lobes that will become intracellularly infected and the nodule vasculature. (c) In a mature nodule PanNF‐YA3pro:GUS is active in the infection zone and the nodule vasculature (v). (d) PanNFYA6pro:GUS is active at the nodule vascular meristem. (e, f) Spatiotemporal expression pattern of PanNFYA3 and PanNF‐YA6 visualized by in situ hybridization on consecutive sections of a young P. andersonii nodule primordium. ep, epidermis; C1–C4, first to fourth cortical cell layer; ed, endodermis; pc, pericycle; m, nodule meristem; in, infection zone; fix, fixation zone; v, nodule vasculature. In (a)–(d), sections (7 µm) were counterstained with Ruthenium Red. Nodules were isolated at 4 wk post‐inoculation with Mesorhizobium plurifarium BOR2.
Taken together, these data suggest a possible symbiotic role for PanNF‐YA3 and, to a lesser extent, PanNF‐YA6, although in roots both genes are not responsive to rhizobium inoculation (1 dpi).
PanNF‐YA1, PanNF‐YA3 and PanNF‐YA6 act redundantly in nodule development
To determine the role of PanNF‐YA3 and PanNF‐YA6 during Parasponia nodule formation, we created CRISPR/Cas9 mutants for both genes. Pannf‐ya3 and Pannf‐ya6 knockout mutant lines were created using three sgRNAs targeting the second and third exons, respectively (Fig. S9a–d). Inoculation with M. plurifarium BOR2 showed that Pannf‐ya3 and Pannf‐ya6 mutants developed a similar number of nodules as transgenic control plants (Fig. S9e). These mutant nodules were able to fix N2, as determined by ARA (Fig. S9f), and display a wild‐type cytoarchitecture (Fig. S9g,h). This indicates that neither PanNF‐YA3 nor PanNF‐YA6 is essential for Parasponia nodule formation.
As we cannot rule out the possibility that PanNF‐YA1, PanNF‐YA3 and/or PanNF‐YA6 function redundantly in nodule organogenesis, we decided to create three double mutants (Pannf‐ya1;Pannf‐ya3, Pannf‐ya1;Pannf‐ya6 and Pannf‐ya3;Pannf‐ya6), and higher‐order triple mutants (Pannf‐ya1;Pannf‐ya3;Pannf‐ya6; Fig. S10). When inoculated with M. plurifarium BOR2, all three double mutant combinations formed nodules (Fig. S11a). Consistent with the phenotype of Pannf‐ya1 single mutant nodules, Pannf‐ya1;Pannf‐ya3‐1 and Pannf‐ya1;Pannf‐ya6‐6 double mutant nodules are devoid of intracellular infection structures (Fig. S12a,b,d,e). Intracellular infection in Pannf‐ya3;Pannf‐ya6‐5 double mutant nodules was not affected (Fig. S12c,f), indicating that intracellular rhizobium infection of P. andersonii nodules is specifically controlled by PanNF‐YA1.
Next, we analysed the nodulation phenotype of three independent Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutant lines. All three lines showed initiation of nodule organogenesis upon rhizobium inoculation with similar efficiency when compared to the transgenic control (Fig. S11a). However, Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutants nodules were irregular in shape and remain substantially smaller than nodules formed on the control (Figs 6a–c, S11b). Approximately half of the Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutant nodules do not develop beyond the primordial stage (Fig. 6a). These nodule‐like structures originated from multiple rounds of cell divisions in the epidermis and outer cortex, but did not develop a vascular bundle (Fig. 6d). By contrast, the somewhat larger nodules formed on the Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutant developed a nodule vascular bundle, but were disturbed in growth (Figs 6b,e, S11b). In M. truncatula, it was shown that the casparian strip was absent from the nodule endodermis in the region close to the meristem (Xiao et al., 2014). We used this criterion to determine whether or not the nodule meristem of the P. andersonii Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutants remained active. Nodule sections were examined under UV light to detect light emitted by the casparian strips. This showed that the meristematic region in P. andersonii triple mutant nodules is fully surrounded by casparian strips, which was not observed in wild‐type nodules of a similar age (Fig. S13). This result indicates that meristematic activity ceased early in the development of Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutant nodules. Like Pannf‐ya1 single mutant nodules, Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutant nodules contain large apoplastic colonies of rhizobium, but are devoid of intracellular infection structures (Fig. 6f). Taken together, these data demonstrate that rhizobium intracellular infection is specifically controlled by PanNF‐YA1, and that PanNF‐YA1, PanNF‐YA3 and PanNF‐YA6 function redundantly to control nodule growth and development.
Figure 6.
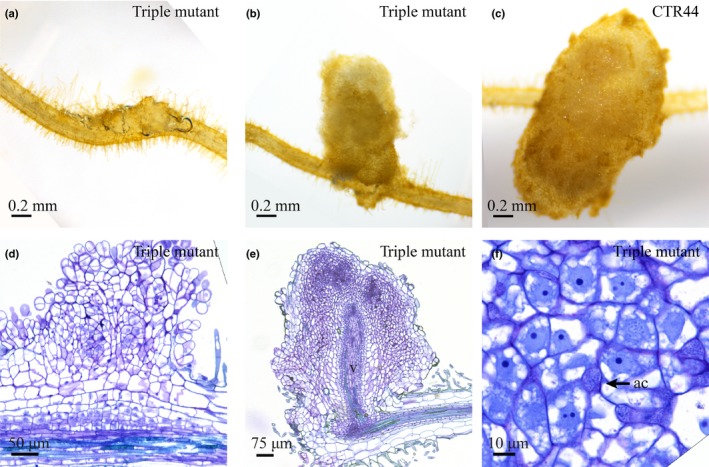
The Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutant is affected in nodule development. (a, b). Nodule‐like structures formed on a Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 mutant. (c) Nodule formed on a transgenic control line (CTR44). (d, e) Sections of the nodule‐like structure shown in (a) and (b). (f) Apoplastic rhizobia (arrow) in a Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 mutant nodule, whereas intracellular infection is absent. v, nodule vasculature; ac, apoplastic colonies of rhizobia. Sections were stained using Toluidine Blue. Nodules were isolated at 4 wk post‐inoculation with Mesorhizobium plurifarium BOR2.
As P. andersonii nf‐ya1 mutant nodules are devoid of intracellular infection, we questioned whether this is specific for rhizobium or, alternatively, whether NF‐YA genes may also function in intracellular colonization by arbuscular mycorrhizal fungi. To test this, control plants, the Pannf‐ya1, Pannf‐ya3 and Pannf‐ya6 single mutants, and the Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 triple mutant were grown under phosphate‐poor conditions and inoculated with 250 spores of the Rhizophagus irregularis strain DOAM197198. The average colonization and arbuscule formation frequency were scored at 6 wk post‐inoculation. This showed that all mutants were equally well mycorrhized when compared with control plants (Fig. S14). Therefore, we conclude that PanNF‐YA1 has a specific role in rhizobium intracellular infection.
Discussion
The transcription factors NIN and NF‐YA1 are essential components in a transcriptional network controlling rhizobium‐induced nodule formation in legumes (Soyano & Hayashi, 2014). Here, we showed that the orthologous genes – PanNIN and PanNF‐YA1 – are essential for the formation of functional root nodules in the nonlegume P. andersonii. Earlier studies, using transient RNA interference‐mediated knockdown, indicated that CgNIN also has a symbiotic function in the nodulating actinorhizal species Casuarina glauca (Clavijo et al., 2015). The Parasponia (Rosales), Casuarina (Fagales) and legume (Fabales) lineages diverged c. 110 Ma, soon after an assumed shared evolutionary event that gave birth to the nodulation trait (Soltis et al., 1995; Wang et al., 2009; van Velzen et al., 2019). As NIN and NF‐YA1 are indispensable for the formation of functional N2‐fixing nodules in distinct taxonomic lineages, we conclude that these transcription factors represent core genes in the nodulation trait. Furthermore, we hypothesize that this recruitment into the nodulation trait has occurred in a species ancestral to the Fabales, Fagales, Cucurbitales and Rosales split.
In L. japonicus, LjNF‐YA1 is a direct transcriptional target of LjNIN (Soyano et al., 2013, 2015). Direct evidence of a similar relationship has not been provided in any other species. Experiments presented here showed that in P. andersonii, rhizobium‐induced PanNF‐YA1 expression is PanNIN‐dependent and that both genes are coexpressed in nodule primordia. In line with the hypothesis that both genes have been recruited in nodulation in a common ancestor of legumes and Parasponia, it is likely that the direct transcriptional regulation of the NF‐YA1 gene by NIN is conserved in nodulating species. This hypothesis is supported by the occurrence of putative NIN‐binding sites in the promoter region of PanNF‐YA1 (Fig. S15). In case these bindings sites find experimental support, the question remains whether the NIN‐NF‐YA1 transcription factor module is ancestral to the N2‐fixing clade, or whether it has evolved in concurrence with the nodulation trait.
Parasponia andersonii NF‐YA1 controls intracellular rhizobium infection, and knockout mutants of this gene are specifically blocked in infection thread formation. This mutant phenotype is different from the phenotypes reported for legume nf‐ya1 knockout and/or knockdown lines. In L. japonicus and M. truncatula, nf‐ya1 mutants and RNAi knockdown lines form smaller nodules (Combier et al., 2006; Soyano et al., 2013; Laporte et al., 2014; Hossain et al., 2016). In M. truncatula, this developmental phenotype is a result of absence or reduced activity of the nodule meristem (Xiao et al., 2014), whereas in L. japonicus LjNF‐YA1 is indispensable for nodule differentiation, including vascular bundle formation (Hossain et al., 2016). Absence of a functional Mtnf‐ya1 gene in M. truncatula also affects rhizobium infection, resulting in an increased number of infection threads that are arrested in the epidermis, and often have a swollen, more bulbous appearance (Laloum et al., 2014; Laporte et al., 2014). In P. andersonii nf‐ya1 knockout mutants are not affected in nodule development. This divergence in phenotype between P. andersonii and legumes is most probably the result of adaptive evolution and subsequent divergence of the nodulation trait in both lineages. For example, intracellular rhizobium infection in M. truncatula and L. japonicus is initiated in curled root hairs, whereas in P. andersonii only nodule cells become invaded. Consequently, infection phenotypes may be observed in different cell types.
Papilionoideae legumes (e.g. L. japonicus, M. truncatula, soybean (Glycine max), and common bean (Phaseolus vulgaris)) experienced gene duplication events in five NF‐YA orthogroups, including NF‐YA1, which is most probably the result of whole‐genome duplication in a common ancestor (Cannon et al., 2006; Young et al., 2011). Subsequent gene redundancy may have allowed subneofunctionalization of NF‐YA1 and its closest paralogue in legumes. Phenotypic analyses of mutant plants where both NF‐YA1 paralogues are targeted simultaneously support the idea that controlling rhizobium intracellular infection is an ancestral symbiotic function of NF‐YA1 and its closest paralogue. For example, by committing MtNF‐YA2 RNAi in a M. truncatula nf‐ya1 mutant background, a more severe rhizobium infection phenotype can be observed (Laloum et al., 2014). Also, in common bean, a strong infection phenotype is observed after silencing of both PvNF‐YA9 and PvNF‐YA1 in A. rhizogenes‐transformed roots (Rípodas et al., 2019). However, in this study, single gene targets have not been analysed. Such gene duplications, which are common in Papilionoideae legumes, complicate reverse genetic studies. P. andersonii did not experience any duplication events in any of the seven NF‐YA orthogroups (Fig. 4a). In line with this, we argue that this species may be more suited to uncover the functioning of NF‐YA genes by reverse genetics.
We also studied the function of two additional NF‐YA genes (PanNF‐YA3 and PanNF‐YA6) in P. andersonii, as both these genes have a nodule‐enhanced expression profile (Fig. 4b). Such a nodule‐enhanced expression profile has also been reported for the M. truncatula orthologues MtNF‐YA8 (orthologous to PanNF‐YA3) and MtNF‐YA3 (orthologous to PanNF‐YA6; Baudin et al., 2015). However, no apparent nodulation phenotype could be observed in P. andersonii single and double mutants. Only upon creating a higher‐order Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 mutant was an effect on nodule organogenesis observed. This suggests that all three PanNF‐YA genes act redundantly in controlling nodule development.
Recent phylogenomic analyses revealed that, within the N2‐fixing clade, absence of the nodulation trait is associated with pseudogenization of the NIN gene (Griesmann et al., 2018; van Velzen et al., 2018). This shows that within the N2‐fixing clade the functioning of this gene correlates with the nodulation trait. In contrast to NIN, no such correlation has been reported between the presence of NF‐YA1 orthologues and the nodulation trait (Griesmann et al., 2018; van Velzen et al., 2018), suggesting that these genes also have nonsymbiotic functions. Arabidopsis thaliana has two orthologues of LjNF‐YA1, MtNF‐YA1 and PanNF‐YA1, named AtNF‐YA2 and AtNF‐YA10 (Fig. 4a). Mutant analysis of these genes has been hampered by the sterility phenotype of Atnf‐ya2 insertion and RNAi lines (Pagnussat et al., 2005; Sorin et al., 2014). Misexpression studies of either gene revealed a function in leaf and root growth and lateral root initiation as well as increased tolerance to several types of abiotic stresses (Leyva‐González et al., 2012; Sorin et al., 2014; Zhang et al., 2017; Soyano et al., 2019). Furthermore, it was shown that, in L. japonicus, ectopic expression of LjNF‐YA1 results in lateral roots with malformed tips (Soyano et al., 2013; Sorin et al., 2014) We observed a mild, though consistent, decrease in lateral roots formed in plantlets containing a mutation in Pannf‐ya1. This supports the findings that NF‐YA1 orthologous genes have a nonsymbiotic function in root development, and may explain why NF‐YA1 is not pseudogenized in species that have lost the nodulation trait (Soyano et al., 2013; Griesmann et al., 2018; van Velzen et al., 2018). As the P. andersonii nf‐ya1 knockout mutants are not affected in the symbiosis with arbuscular mycorrhiza, it suggests that NF‐YA1 symbiotic functioning is exclusively required to allow entry of symbiotic bacteria. As the bacterial infectability of cells is a key characteristic of the nodulation trait, it will be an important future scientific objective to determine the core transcriptional network regulated by NF‐YA1 and its interacting partners. Having a P. andersonii nf‐ya1 mutant available with a strict infection phenotype as a comparative system to legumes where infection and organogenesis phenotypes are intertwined will be instrumental to achieving this objective.
Author contributions
FB, LR and RG planned and designed the research; FB, LR, MR‐F, OK and YPR performed the experiments; FB, AvZ, LR, RG, TB, TO and YPR analysed the data; and FB, AvZ and RG wrote the manuscript.
Supporting information
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Spatiotemporal expression pattern of PanNF‐YA1pro:GUS in Parasponia andersonii roots.
Fig. S2 Structure and expression of the P. andersonii NIN gene and the genotype of CRISPR‐Cas9 Pannin mutants.
Fig. S3 Rhizobium tropici CIAT899.pMP604 constitutively expresses the LCO biosynthesis gene nodC.
Fig. S4 Structure of the P. andersonii NF‐YA1 gene and genotype of CRISPR‐Cas9 Panf‐ya1 mutants.
Fig. S5 Lateral root formation is affected in the P. andersonii nf‐ya1 mutant.
Fig. S6 Phenotyping of P. andersonii nf‐ya1 knockout mutants.
Fig. S7 Phylogenetic analysis of NF‐YA in the nitrogen‐fixing clade.
Fig. S8 Expression of PanNF‐YA3 and PanNF‐YA6 in P. andersonii roots and nodules.
Fig. S9 Gene structure of P. andersonii NF‐YA3 and NF‐YA6, genotype of CRISPR‐Cas9 mutants, and nodulation phenotypes.
Fig. S10 Genotypes of Pannf‐ya1, Pannf‐ya3 and Pannf‐ya6 CRISPR‐Cas9 double and triple mutants.
Fig. S11 Nodulation efficiency and nodule size of Parasponia andersonii nf‐ya single, double and triple knockout mutants.
Fig. S12 Nodule cytoarchitecture of Parasponia andersonii nf‐ya double knockout mutants.
Fig. S13 Casparian strips in the vascular endodermis next to the nodule meristem in Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 mutant plants.
Fig. S14 Parasponia andersonii nf‐ya1, nf‐ya3, nf‐ya6 and Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 mutants can form arbuscular mycorrhiza. Parasponia andersonii nf‐ya1, nf‐ya3 and nf‐ya6 knockout mutants can form arbuscular mycorrhiza.
Fig. S15 Putative NIN‐binding sites in the PanNF‐YA1 promoter region.
Table S1 Sequences of sgRNAs used for creating single, double and triple knockout mutants.
Table S2 Primers used in this work.
Table S3 Putative promoter sequences used for promoter‐reporter GUS assays.
Table S4 Gene identifiers for NF‐YA proteins used to build the phylogenetic tree depicted in Figs 4, S7.
Acknowledgements
This work was supported by an NWO‐VICI grant (865.13.001) to RG, the ENSA project funded by the Bill & Melinda Gates Foundation to the University of Cambridge to RG and TO, a CSC Scholarship (201303250067) to FB, and Ministry of Research, Technology and Higher Education of the Republic of Indonesia (RISET‐PRO grant 8245‐ID) to YPR.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- Baudin M, Laloum T, Lepage A, Ripodas C, Ariel F, Frances L, Crespi M, Gamas PC, Blanco FA, Zanetti ME et al 2015. A phylogenetically conserved group of NF‐Y transcription factors interact to control nodulation in legumes. Plant Physiology 169: 2761–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli DJ, Cannon SB, Froenicke L, Huang G, Farmer AD, Cannon EKS, Liu X, Gao D, Clevenger J, Dash S et al 2016. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nature Genetics 48: 438–446. [DOI] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N et al 2003. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus . Plant Physiology 131: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T et al 2006. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proceedings of the National Academy of Sciences, USA 103: 14959–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet‐Mercey S, Taconnat L, Renou J‐P, Daniel‐Vedele F, Fernandez E et al 2009. The nodule inception‐like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. The Plant Journal 57: 426–435. [DOI] [PubMed] [Google Scholar]
- Clavijo F, Diedhiou I, Vaissayre V, Brottier L, Acolatse J, Moukouanga D, Crabos A, Auguy F, Franche C, Gherbi H et al 2015. The Casuarina NIN gene is transcriptionally activated throughout Frankia root infection as well as in response to bacterial diffusible signals. New Phytologist 208: 887–903. [DOI] [PubMed] [Google Scholar]
- Combier J‐P, Frugier F, de Billy F, Boualem A, El‐Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M et al 2006. MtHAP2‐1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes & Development 20: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Promé JC. 1996. Rhizobium lipo‐chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annual Review of Biochemistry 65: 503–535. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. 2011. Phylogenetic perspectives on the origins of nodulation. Molecular Plant–Microbe Interactions 24: 1289–1295. [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T‐M, Werner S, Jones JDG, Patron NJ, Marillonnet S. 2014. A golden gate modular cloning toolbox for plants. ACS Synthetic Biology 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Geurts R, Lillo A, Bisseling T. 2012. Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Current Opinion in Plant Biology 15: 438–443. [DOI] [PubMed] [Google Scholar]
- Griesmann M, Chang Y, Liu X, Song Y, Haberer G, Crook MB, Billault‐Penneteau B, Lauressergues D, Keller J, Imanishi L et al. 2018. Phylogenomics reveals multiple losses of nitrogen‐fixing root nodule symbiosis. Science 361: eaat1743. [DOI] [PubMed] [Google Scholar]
- He N, Zhang C, Qi X, Zhao S, Tao Y, Yang G, Lee T‐H, Wang X, Cai Q, Li D et al 2013. Draft genome sequence of the mulberry tree Morus notabilis . Nature Communications 4: 2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, Madsen LH, Jurkiewicz A, Nielsen MW, Tirichine L, Stougaard J. 2011. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Molecular Plant–Microbe Interactions 24: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Holmer R, van Velzen R, Geurts R, Bisseling T, de Ridder D, Smit S. 2019. GeneNoteBook, a collaborative notebook for comparative genomics. Bioinformatics 35: 4779–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Shrestha A, Zhong S, Miri M, Austin RS, Sato S, Ross L, Huebert T, Tromas A, Torres‐Jerez I et al 2016. Lotus japonicus NF‐YA1 plays an essential role during nodule differentiation and targets members of the SHI/STY gene family. Molecular Plant–Microbe Interactions 29: 950–964. [DOI] [PubMed] [Google Scholar]
- International Peach Genome Initiative , Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J et al 2013. The high‐quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nature Genetics 45: 487–494. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K‐I, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. 2013. Arabidopsis NIN‐like transcription factors have a central role in nitrate signalling. Nature Communications 4: 1617. [DOI] [PubMed] [Google Scholar]
- Kosuta S, Held M, Hossain MS, Morieri G, Macgillivary A, Johansen C, Antolín‐Llovera M, Parniske M, Oldroyd GED, Downie AJ et al 2011. Lotus japonicus symRK‐14 uncouples the cortical and epidermal symbiotic program. The Plant Journal 67: 929–940. [DOI] [PubMed] [Google Scholar]
- Kulikova O, Franken C, Bisseling T. 2018. In situ hybridization method for localization of mRNA molecules in medicago tissue sections. Methods in Molecular Biology 1822: 145–159. [DOI] [PubMed] [Google Scholar]
- Laloum T, Baudin M, Frances L, Lepage A, Billault‐Penneteau B, Cerri MR, Ariel F, Jardinaud M‐F, Gamas P, de Carvalho‐Niebel F et al 2014. Two CCAAT‐box‐binding transcription factors redundantly regulate early steps of the legume‐rhizobia endosymbiosis. The Plant Journal 79: 757–768. [DOI] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. 2013. CCAAT‐box binding transcription factors in plants: Y so many? Trends in Plant Science 18: 157–166. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia‐Hernandez M et al. 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancelle SA, Torrey JG. 1984. Early development of Rhizobium‐induced root nodules of Parasponia rigida. I. Infection and early nodule initiation. Protoplasma 123: 26–37. [Google Scholar]
- Lancelle SA, Torrey JG. 1985. Early development of Rhizobium‐induced root nodules of Parasponia rigida. II. Nodule morphogenesis and symbiotic development. Canadian journal of botany. Journal Canadien De Botanique 63: 25–35. [Google Scholar]
- Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud M‐F, Mun J‐H, Larrainzar E, Cook DR, Gamas P et al 2014. The CCAAT box‐binding transcription factor NF‐YA1 controls rhizobial infection. Journal of Experimental Botany 65: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. 1990. Symbiotic host‐specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784. [DOI] [PubMed] [Google Scholar]
- Leyva‐González MA, Ibarra‐Laclette E, Cruz‐Ramírez A, Herrera‐Estrella L. 2012. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF‐YA family members. PLoS ONE 7: e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. 2003. LysM domain receptor kinases regulating rhizobial Nod factor‐induced infection. Science 302: 630–633. [DOI] [PubMed] [Google Scholar]
- Liu J, Rutten L, Limpens E, van der Molen T, van Velzen R, Chen R, Chen Y, Geurts R, Kohlen W, Kulikova O et al 2019. A remote cis‐regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula . Plant Cell 31: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N et al 2003. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GED. 2007. Medicago truncatula NIN is essential for rhizobial‐independent nodule organogenesis induced by autoactive Calcium/Calmodulin‐Dependent Protein Kinase. Plant Physiology 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez E, Pardo MA, Palacios R, Miguel AC. 1985. Reiteration of nitrogen fixation gene sequences and specificity of rhizobium in nodulation and nitrogen fixation in Phaseolus vulgaris . Microbiology 131: 1779–1786. [Google Scholar]
- Mun T, Bachmann A, Gupta V, Stougaard J, Andersen SU. 2016. Lotus base: an integrated information portal for the model legume Lotus japonicus . Scientific Reports 6: 39447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers ZA, Holt BF 3rd. 2018. NUCLEAR FACTOR‐Y: still complex after all these years? Current Opinion in Plant Biology 45: 96–102. [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. 2013. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA‐guided endonuclease. Nature Biotechnology 31: 691–693. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews. Microbiology 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JSS, Kudrna D, Wing R, Untergasser A et al 2011. LysM‐type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia . Science 331: 909–912. [DOI] [PubMed] [Google Scholar]
- Op den Camp RHM, Polone E, Fedorova E, Roelofsen W, Squartini A, Op den Camp HJM, Bisseling T, Geurts R. 2012. Nonlegume Parasponia andersonii deploys a broad rhizobium host range strategy resulting in largely variable symbiotic effectiveness. Molecular Plant–Microbe Interactions 25: 954–963. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H‐J, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L‐F, Ye D, Sundaresan V. 2005. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis . Development 132: 603–614. [DOI] [PubMed] [Google Scholar]
- Pasternak T, Tietz O, Rapp K, Begheldo M, Nitschke R, Ruperti B, Palme K. 2015. Protocol: an improved and universal procedure for whole‐mount immunolocalization in plants. Plant Methods 11: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Ott T. 2011. Regulation of signal transduction and bacterial infection during root nodule symbiosis. Current Opinion in Plant Biology 14: 458–467. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N et al 2003. Plant recognition of symbiotic bacteria requires two LysM receptor‐like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Rípodas C, Castaingts M, Clúa J, Villafañe J, Blanco FA, Zanetti ME. 2019. The PvNF‐YA1 and PvNF‐YB7 Subunits of the heterotrimeric NF‐Y transcription factor influence strain preference in the Phaseolus vulgaris–Rhizobium etli symbiosis. Frontiers in Plant Science 10: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rípodas C, Clúa J, Battaglia M, Baudin M, Niebel A, Zanetti ME, Blanco F. 2014. Transcriptional regulators of legume‐rhizobia symbiosis: nuclear factors Ys and GRAS are two for tango. Plant Signaling & Behavior 9: e28847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K et al 2008. Genome structure of the legume, Lotus japonicus . DNA Research 15: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. 1999. A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Schauser L, Wieloch W, Stougaard J. 2005. Evolution of NIN‐like proteins in Arabidopsis, rice, and Lotus japonicus . Journal of Molecular Evolution 60: 229–237. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J et al 2010. Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C et al 2014. A reference genome for common bean and genome‐wide analysis of dual domestications. Nature Genetics 46: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP et al 2011. The genome of woodland strawberry (Fragaria vesca). Nature Genetics 43: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG. 1995. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proceedings of the National Academy of Sciences, USA 92: 2647–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais‐Brière C, Njo MF, Beeckman T, Crespi M, Hartmann C. 2014. A miR169 isoform regulates specific NF‐YA targets and root architecture in Arabidopsis. New Phytologist 202: 1197–1211. [DOI] [PubMed] [Google Scholar]
- Soyano T, Hayashi M. 2014. Transcriptional networks leading to symbiotic nodule organogenesis. Current Opinion in Plant Biology 20: 146–154. [DOI] [PubMed] [Google Scholar]
- Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M. 2014. Nodule Inception creates a long‐distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proceedings of the National Academy of Sciences, USA 111: 14607–14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M. 2013. Nodule inception directly targets NF‐Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus . PLoS Genetics 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Hayashi M. 2015. NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus . Plant & Cell Physiology 56: 368–376. [DOI] [PubMed] [Google Scholar]
- Soyano T, Shimoda Y, Kawaguchi M, Hayashi M. 2019. A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus . Science 366: 1021–1023. [DOI] [PubMed] [Google Scholar]
- Spaink HP, Okker RJ, Wijffelman CA, Tak T, Goosen‐de Roo L, Pees E, van Brussel AA, Lugtenberg BJ. 1989. Symbiotic properties of rhizobia containing a flavonoid‐independent hybrid nodD product. Journal of Bacteriology 171: 4045–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H et al 2014. An improved genome release (version Mt4.0) for the model legume Medicago truncatula . BMC Genomics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick MJ. 1979. Structure of nitrogen‐fixing nodules formed by Rhizobium on roots of Parasponia andersonii Planch. Canadian Journal of Microbiology 25: 565–578. [DOI] [PubMed] [Google Scholar]
- van Velzen R, Doyle JJ, Geurts R. 2019. A resurrected scenario: single gain and massive loss of nitrogen‐fixing nodulation. Trends in Plant Science 24: 49–57. [DOI] [PubMed] [Google Scholar]
- van Velzen R, Holmer R, Bu F, Rutten L, van Zeijl A, Liu W, Santuari L, Cao Q, Sharma T, Shen D et al 2018. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen‐fixing rhizobium symbioses. Proceedings of the National Academy of Sciences, USA 115: E4700–E4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl A, Wardhani TAK, Seifi Kalhor M, Rutten L, Bu F, Hartog M, Linders S, Fedorova EE, Bisseling T, Kohlen W et al 2018. CRISPR/Cas9‐mediated mutagenesis of four putative symbiosis genes of the tropical tree Parasponia andersonii reveals novel phenotypes. Frontiers in Plant Science 9: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho‐Niebel F, Oldroyd GED. 2015. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27: 3410–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE. 2009. Rosid radiation and the rapid rise of angiosperm‐dominated forests. Proceedings of the National Academy of Sciences, USA 106: 3853–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardhani TAK, Roswanjaya YP, Dupin S, Li H, Linders S, Hartog M, Geurts R, Van Zeijl A. 2019. Transforming, genome editing and phenotyping the nitrogen‐fixing tropical Cannabaceae tree Parasponia andersonii . Journal of Visualized Experiments: (150) e59971. [DOI] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T. 2014. Fate map of Medicago truncatula root nodules. Development 141: 3517–3528. [DOI] [PubMed] [Google Scholar]
- Yoro E, Suzaki T, Toyokura K, Miyazawa H, Fukaki H, Kawaguchi M. 2014. A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus . Plant Physiology 165: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H et al 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Hu X, Zhu M, Xu M, Wang L. 2017. Transcription factors NF‐YA2 and NF‐YA10 regulate leaf growth via auxin signaling in Arabidopsis . Scientific Reports 7: 1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Spatiotemporal expression pattern of PanNF‐YA1pro:GUS in Parasponia andersonii roots.
Fig. S2 Structure and expression of the P. andersonii NIN gene and the genotype of CRISPR‐Cas9 Pannin mutants.
Fig. S3 Rhizobium tropici CIAT899.pMP604 constitutively expresses the LCO biosynthesis gene nodC.
Fig. S4 Structure of the P. andersonii NF‐YA1 gene and genotype of CRISPR‐Cas9 Panf‐ya1 mutants.
Fig. S5 Lateral root formation is affected in the P. andersonii nf‐ya1 mutant.
Fig. S6 Phenotyping of P. andersonii nf‐ya1 knockout mutants.
Fig. S7 Phylogenetic analysis of NF‐YA in the nitrogen‐fixing clade.
Fig. S8 Expression of PanNF‐YA3 and PanNF‐YA6 in P. andersonii roots and nodules.
Fig. S9 Gene structure of P. andersonii NF‐YA3 and NF‐YA6, genotype of CRISPR‐Cas9 mutants, and nodulation phenotypes.
Fig. S10 Genotypes of Pannf‐ya1, Pannf‐ya3 and Pannf‐ya6 CRISPR‐Cas9 double and triple mutants.
Fig. S11 Nodulation efficiency and nodule size of Parasponia andersonii nf‐ya single, double and triple knockout mutants.
Fig. S12 Nodule cytoarchitecture of Parasponia andersonii nf‐ya double knockout mutants.
Fig. S13 Casparian strips in the vascular endodermis next to the nodule meristem in Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 mutant plants.
Fig. S14 Parasponia andersonii nf‐ya1, nf‐ya3, nf‐ya6 and Pannf‐ya1;Pannf‐ya3;Pannf‐ya6 mutants can form arbuscular mycorrhiza. Parasponia andersonii nf‐ya1, nf‐ya3 and nf‐ya6 knockout mutants can form arbuscular mycorrhiza.
Fig. S15 Putative NIN‐binding sites in the PanNF‐YA1 promoter region.
Table S1 Sequences of sgRNAs used for creating single, double and triple knockout mutants.
Table S2 Primers used in this work.
Table S3 Putative promoter sequences used for promoter‐reporter GUS assays.
Table S4 Gene identifiers for NF‐YA proteins used to build the phylogenetic tree depicted in Figs 4, S7.


