Abstract
Background
Vitamin C (also known as L‐ascorbic acid) plays a critical role in reactive oxygen species (ROS) reduction and cell regeneration by protecting cell from oxidative stress. Although vitamin C is widely used in cosmetic and therapeutic markets, there is considerable evidence that vitamin C easily undergoes oxidation by air, pH, temperature, and UV light upon storage. This deficiency of vitamin C decreases its potency as an antioxidant and reduces the shelf‐life of products containing vitamin C as its ingredient. To overcome the deficiency of vitamin C, we have developed Aptamin C, an innovative DNA aptamer maximizing the antioxidant efficacy of vitamin C by binding to the reduced form of vitamin C and delaying its oxidation.
Methods
Binding of Aptamin C with vitamin C was determined using ITC analysis. ITC experiment was performed 0.2 mmol/L vitamin C that was injected 25 times in 2 µL aliquots into the 1.8 mL sample cell containing the Aptamin C at a concentration of 0.02 mmol/L. The data were fitted to a one‐site binding isotherm using with origin program for ITC v.5.0.
Results
To investigate the effect of Aptamin C and vitamin C complex in human skins, both in vitro and clinical tests were performed. We observed that the complex of Aptamin C and vitamin C was significantly effective in wrinkle improvement, whitening effect, and hydration increase. In the clinical test, subjects treated with the complex showed dramatic improvement in skin irritation and itching. No adverse reaction was presented by Aptamin C complex in the test.
Conclusion
Taken together, these results showed that Aptamin C, an innovative novel compound, should potentially be served as a key cosmeceutical ingredient for a range of skin conditions.
Keywords: antioxidation, Aptamin C (vitamin C binding Aptamer), oxidation, oxidative stress, vitamin C (L‐ascorbic acid)
1. INTRODUCTION
Reactive oxygen species (ROS) are chemically reactive chemical species containing oxygen, which plays important role in cell signaling and homeostasis.1, 2, 3 These include not only positive effects such as the induction of host defense genes and mobilization of ion transport systems but also roles in apoptosis (programmed cell death).4, 5 ROS level can be increased by environmental stress, leading damage to the cell structures.3 This phenomenon is called “oxidative stress.” Oxidative stress is one of the leading causes of various diseases including neurodegenerative diseases, Lou Gehrig's disease, autism, multiple sclerosis, and skin diseases.6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Furthermore, oxidative stress has direct influence in the skin aging process16; proteins, lipids, and DNA are sensitively reacting to oxidative stress which is caused by ROS.17 Antioxidants are highly effective in skin protection as they react directly to ROS preventing them from reaching the biological target molecules.18, 19 Antioxidants such as vitamin C, vitamin E, coenzyme Q10, and polyphenolic compounds protect skin from damage caused by ROS. Antioxidants thereby help prevent and treat various skin diseases and slow down the skin aging process.20 The use of vitamin C in industries is primarily because of its antioxidizing properties, which are the result of vitamin C's ability to neutralize free oxygen radicals, through a process called radical scavenging.21, 22 However, because of these same antioxidant properties, the molecule itself is inherently susceptible to degradation via oxidation. To solve this problem, we developed Aptamin C, a DNA aptamer that specifically binds to vitamin C and inhibits oxidation of vitamin C. Aptamers are single‐stranded DNA‐ or RNA‐based oligonucleotides capable of selectively binding a wide range of molecules. Aptamers are commonly identified by an in vitro method of selection referred to as Systematic Evolution of Ligands by EXponential enrichment or “SELEX.” We identified that Aptamin C inhibits the oxidation of vitamin C from several oxidizing agents and maintains the antioxidant activity during long‐term storage. To confirm the safety assessment of Aptamin C, experiments were conducted at the cellular level and directly applied to humans and no toxicity was observed. Skin disease such as atopic dermatitis, psoriasis, and acne are related by the immune response and ROS.23 Vitamin C has the ability to remove the ROS and has anti‐inflammation effect.24 Aptamin C prevents oxidation of vitamin c and maximizes its efficacy through slow release. We, therefore, expect expected that Aptamin C‐vitamin C complex will show the synergetic effect.
2. MATERIALS AND METHODS
2.1. Aptamer screening against vitamin C with reduced graphene‐oxide
This method has been carried out by modifying the method of the previous research.25 The ssDNA candidates which can bind specifically to vitamin C were developed from a random ssDNA library consisting of about 1X1018 different sequences. For the ssDNA library, we used custom‐made sequence which size is 60mer and containing 30 randomly generated nucleotide sequences and primer site for amplification (5′‐ATGCGGATCCCGCGC‐(N)30‐GCGCGAAGCTTGCGC‐3′). We performed total five rounds while changing the experimental condition to select more specific sequence. The total volume for reaction was 200 μL. About 20 μL of 10× binding buffer (10× PBS added 10 mmol/L MgCl2), 80 μL of rGO (5 mg/mL, diluted in water), and 200 picomoles of ssDNA library (20 μL of 100 μmol/L stock) were added and filed dH2O to 200 μL. The reaction proceeded for 30 minutes to bind the ssDNA library onto rGO, and then, the mixture was centrifuged at 20 000 g for 20 minutes to eliminate the supernatant. The rGO pellet was washed 1 time with 200 μL of binding buffer which is same concentration with target binding condition of each round. To elute the candidates, 200 nanomoles of vitamin C diluted in 200 μL of binding buffer was added to the rGO pellet, and the elution step did for 1 hour. Eluted ssDNA was divided by centrifugation it was done at 20 000 g for 20 minutes and amplified. We did EtOH precipitation to eliminate impurities except ssDNA before amplification. We did asymmetric PCR to obtain amplified ssDNA. The ratio of forward primer to reverse primer of asymmetric PCR was 10:1. We did electrophoresis on 2.5% agarose gel to confirm the PCR product with a few microliters of it. Asymmetric PCR cannot produce only ssDNA. So, we did crush and soak method to isolate the ssDNA candidates. For the method, we did electrophoresis on a 12% polyacrylamide native gel and staining the gel with ethidium bromide (EtBr). To separate double‐stranded DNA (dsDNA) and ssDNA, the part of gel stained by ssDNA was cut out, pulverized, and extracted ssDNA with crush and soak buffer (500 mmol/L NH4OAc, 0.1% SDS, 0.1 mmol/L EDTA) for overnight. The pulverized gel was separated by centrifugation. The supernatant which contain ssDNA is concentrated and purified by doing EtOH precipitation. Dried ssDNA was gathered with sterilized dH2O and this was used for the next round as library.
2.2. Isothermal titration calorimetry (ITC) experiment
Isothermal titration calorimetry experiment was performed using a VP‐ITC system (MicroCal Inc Northampton) at 25°C in a phosphate buffer saline composed 1 mmol/L magnesium chloride (pH 7.4). Before each titration experiment, Aptamin C 2 mL and vitamin C 600 µL samples were degassed for 30 minutes under vacuum, without stirring, at a temperature a few degrees below that of the experiment. We prepared 0.2 mmol/L vitamin C that was injected 25 times in 2 µL aliquots into the 1.8 mL sample cell containing the Aptamin C at a concentration of 0.02 mmol/L. The data were fitted to a one‐site binding isotherm using with origin program for ITC v.5.0 (MicroCal Inc).
2.3. Fluorescence‐based microplate assays for vitamin C oxidation
Oxidation of vitamin C was measured by detecting the oxidized product dehydroascorbate (DHA) using a modified version of the method described by Vislisel et al.7 In this method, DHA is detected by reaction with o‐phenylenediamine (OPDA) to form the fluorescent condensation product 3‐(dihydroxyethyl)furo[3,4‐b] quinoxaline‐1‐one. The assay was performed as follows in black 384‐well plates (Greiner Bio‐One). Aptamers were first dissolved in phosphate‐buffered saline, pH 7.2, containing 1mM MgCl2, at a concentration of 200 µmol/L then folded by heating to 95°C and allowed to cool slowly to room temperature for 15 minutes. The folded aptamers were then diluted 1:1 (v:v) into a freshly prepared solution of 5 mmol/L vitamin C in assay buffer [50 mmol/L sodium acetate, 1% (w/v) BSA, 0.05% (v/v) Tween 20, 1mM MgCl2 (Sigma, all components) adjusted to pH 5.5], and the mixture was incubated for 30 minutes at room temperature, to allow aptamers to bind prior to addition of oxidizer. Oxidizers were then added to the vitamin C/aptamer solutions at concentrations of (EM) H2O2 (Sigma). Oxidizers were pre‐diluted to their working concentrations in assay buffer. Samples were then incubated at room temperature for 10 minutes before the addition of OPDA (Sigma) at a concentration of 5.5 mmol/L in assay buffer. Immediately following the addition of OPDA, the fluorescence of the samples at 425 nm was determined using a SpectraMax® i3X plate reader (Molecular Devices) with excitation at 345 nm, over a 45‐minutes time course with measurements made every 60 seconds. All sample and reagent containing vessels were wrapped in foil to protect them from light during all incubations performed for the fluorescence assays.
2.4. Measurement of vitamin C reduction using DCPIP (2,6‐Dichlorophenolindophenol)
To determine the reduction of vitamin C, we conducted the experiments using the DCPIP reaction. Vitamin C reacts with DCPIP, changing the color from blue to colorless. The prepared vitamin C was treated with 5% Aptamin CTM, and the untreated vitamin C was incubated at room temperature for 8 weeks. The sample was measured after 2, 4, and 8 weeks. About 2 mL of DCPIP was added to the conical flask using a pipette, and the first untreated vitamin C was added until the solution became colorless. The amount of vitamin C added was measured and repeated with other samples. The degree of reduction of each sample was calculated according to this data.
2.5. In vitro study of anti‐wrinkle effect in human dermal fibroblasts
To evaluate the cell viability in human dermal fibroblasts, cells were treated with different final concentrations of Aptamin C with vitamin C (Aptamin C μg + vitamin C μg/mL) 0.01 + 0.5 μg/mL, 0.1 + 5 μg/mL, 0.5 + 25 μg/mL, 1 + 50 μg/mL, and 2 + 100 μg/mL. And then, cells were treated at concentrations of Aptamin C with vitamin C to detect intracellular collagen, intracellular collagenase (MMP‐1), and elastase activity.
Human dermal fibroblasts (HDFs) were selected on the basis of “Guidelines for efficacy evaluation of functional cosmetics (ΙI)” of MFDS. HDFs were cultured in DMEM/F12 3:1 mixture high glucose supplemented with 10% FBS and 1% Antibiotic‐Antimycotic in a humidified 5% CO2 atmosphere at 37˚C.
The HDFs (5 × 104 cells/well) were seeded into 24‐well plates and incubated for 24 hours, and were treated with various concentrations of Aptamin C with vitamin C and incubated for 24 hours. After 24 hours, the supernatant was collected, and the amount of procollagen liberated into the medium was measured at 450 nm using Procollagen Type I C‐Peptide (PIP) ELISA Kit. The degree of collagen production was calibrated by total protein content and compared with TGF‐β1 as a positive control. Media without test materials was used as a solvent control.
The HDFs (5 × 104 cells/well) were seeded into 24‐well plates and incubated for 24 hours, and were treated with various concentrations of Aptamin C with vitamin C and incubated for 48 hours. After 48 hours, the activity of collagenase was measured at 450 nm using MMP‐1 Human ELISA Kit. MMP‐1 activity was assessed by total protein content and compared with TGF‐β1 as a positive control. Media without test materials was used as a solvent control.
All data were expressed as mean ± standard deviation and came from 3 independent experiments. Statistical analysis was conducted by independent samples t test using the SPSS® software program (IBM) at the P < .05 significance level.
2.6. Measurement of skin wrinkle by 3D image analyzing system
Twenty‐two female subjects (average age: 50.05 ± 2.94 years) participated in this study. Skin wrinkle of the crow's feet was evaluated by 3D image analyzing system at baseline, 4 and 8 weeks. Skin hydration was evaluated by capacitance method, and TEWL by skin surface water diffusion method and skin elasticity by suction method were evaluated at baseline, 2, 4, and 8 weeks after treatment. Also, self‐questionnaires concerning efficacy was filled out by subjects at 2 and 4 and 8 weeks, and that of usability was filled out by subjects at 8 weeks after treatment. All obtained data were statistically analyzed by SPSS® software. The wrinkle parameters of crow's feet were evaluated using PRIMOS® Premium (GFMesstechnik GmbH). This system allowed the quantitative analysis of the roughness, depth, area, and volume of protruding on the skin wrinkle. Image was analyzed same area in terms of skin wrinkle parameters (1, Average depth of wrinkles; 2, Mean depth biggest wrinkle; 3, Max. depth biggest wrinkle; 4, Total wrinkle area; 5, Total wrinkle volume; 6, Total form factor wrinkles; 7, Total length of wrinkles; 8, Ra; 9, Ry; and 10, Rz) at baseline, 4 and 8 weeks after treatment by Primos 5.8 E ver. Software.
3. RESULTS
3.1. A rigorous approach to buffer preparation was required to successfully perform rGO‐SELEX with an unstable target
The ssDNA library, which was bound to rGO via π‐π stacking interactions between the aromatic rings of the graphene surface and the DNA bases 26, 27 and non‐bound ssDNAs on rGO, was separated and removed by centrifugation. ssDNAs that were adsorbed on the rGO surface were eluted through treatment with the target compound. According to literature reports, the conformation of the aptamer changes after binding with target and by weakening the π‐π stacking interactions with rGO.28, 29, 30, 31 This process was repeated for five rounds. Each subsequent round proceeded with harsher buffer conditions and shorter elution times. This performance allowed us to maintain ssDNAs with better specificity to target compound.
The NGS data of the enriched library produced by the rGO‐SELEX process yielded 404071 sequences. We selected 119 sequences from this data, sorted these into 11 groups based on structure similarity, and selected representative sequences for aptamer candidates (Figure 1A).
Figure 1.
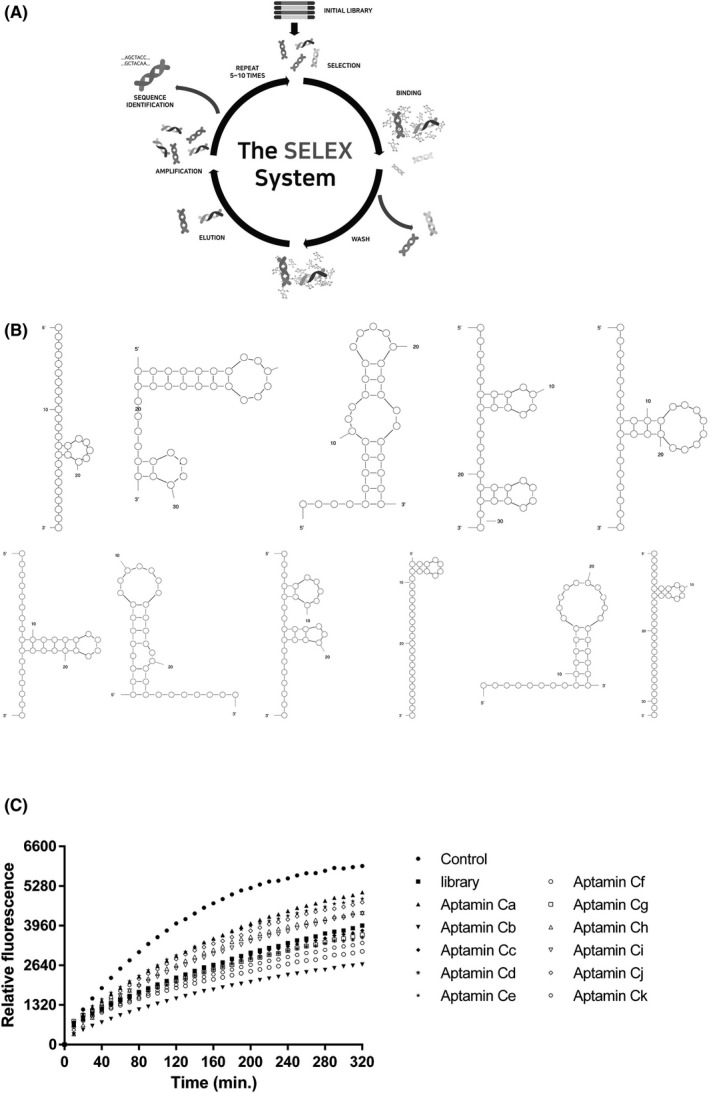
Selection of aptamers capable of binding vitamin C. A, The general scheme of the SELEX method using DNA. B, Representative image of each aptamer group. The secondary structures of the Aptamin C were predicted using the Mfold program (http://mfold.rna.albany.edu/?=mfold). C, Antioxidation (Inhibition) test of representative sequence of each group. Aptamin Cb, Cf, and Ck, these three Aptamin C are the best form to protect vitamin C from oxidation
3.2. Aptamin C selecting
The secondary structures of Aptamin C candidates were predicted by using M‐Fold free software.32, 33 The candidates were grouped by their position, length, shape, and number of stem and loop of the highest potential structure (Figure 1B). DHA, oxide of vitamin C, is detected by its reaction with o‐phenylenediamine (OPDA) which plays the role as an indicator.34 If aptamer binds and prevents its oxidation, to vitamin C and prevent oxidation of it, the fluorescence signal from 3‐(dihydroxyethyl)‐furo‐[3,4‐b] quinoxaline‐1‐one, the condensation product of vitamin C and OPDA, will be lower than that of no‐aptamer control. A plot of each candidate aptamer mixed with vitamin C and oxidizer, with the positive controls, vitamin C plus oxidizer and scramble sequences mixed with vitamin C and oxidizer (Figure 1C). Five aptamers, Aptamin Cb, Cc, Cf, Cg, and Ck, were shown to have effects of antioxidation.
3.3. Inhibition of vitamin C oxidation by Aptamin C
Aptamin C has high binding affinity for vitamin C. Binding of Aptamin C with vitamin C was determined using ITC analysis. From binding isotherm, the enthalpy (ΔH), entropy (ΔS), and stoichiometry (n) of the binding reaction can be obtained. Aptamin Cb exothermic binding was detected from ITC measurement, and the enthalpy (ΔH) was calculated as 302.5 ± 3.788, entropy (ΔS) was calculated as 18.9, stoichiometry(n) was calculated as 56.7 ± 0.426, and the dissociation constants (Kd) were calculated as 2.13uM for vitamin C. Aptamin Cf exothermic binding was detected from ITC measurement, and the enthalpy (ΔH) was calculated as 279.2 ± 2.992, entropy (ΔS) was calculated as 18.4, stoichiometry(n) was calculated as 167 ± 1.15, and the dissociation constants (Kd) were calculated as 0.89uM for vitamin C. Aptamin Ck exothermic binding was detected from ITC measurement, and the enthalpy (ΔH) was calculated as 250.4 ± 3.742, entropy (ΔS) was calculated as 19.8, stoichiometry(n) was calculated as 82.2 ± 0.732, and the dissociation constants (Kd) were calculated as 0.90 µmol/L for vitamin C (Figure 2A). The ITC measurements reveal that the change in entropy upon association with the Aptamin C is a major driving force for vitamin C. To prevent oxidation of vitamin C in the liquid state, all solutions were prepared with deionized water treated with nitrogen. The fluorescence was expressed and quantitatively analyzed when OPDA (o‐phenylenediamine) bound to DHA generated by oxidation of vitamin C. The degree of oxidation of vitamin C according to the concentration of Aptamin C (125, 250, 500 and 1000 nmol/L) was compared and confirmed by OPDA assay. The results demonstrated that the vitamin C conversion to dehydroascorbic acid was slower when the concentration of Aptamin C was higher. (Figure 2B). These data prove that Aptamin C is a dose‐dependent inhibitor of vitamin C oxidation.
Figure 2.
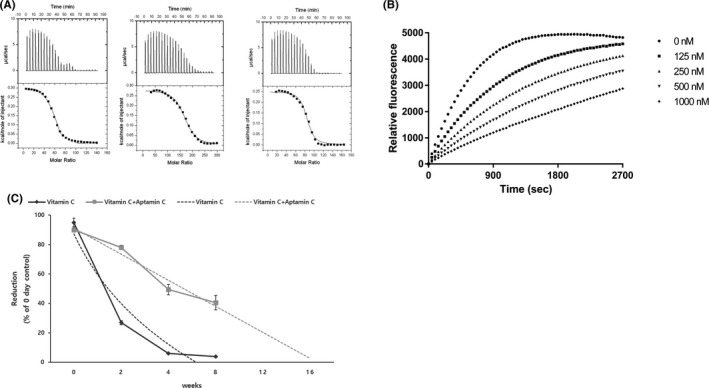
Inhibition of vitamin C oxidation by Aptamin C. A, Isothermal calorimetry. ITC of Aptamin Cb, Cf, and Ck binding to vitamin C. The binding affinity of Aptamin Cb is 2.13 µmol/L, Aptamin Cf is 0.89 µmol/L, and Aptamin Ck is 0.90 µmol/L. B, Comparison of OPDA assay according to the concentration of Aptamin C. Aptamin C concentrations are represented as ● (0 nmol/L), ■ (125 nmol/L), ▲ (250 nmol/L), ▼ (500 nmol/L), or ◆ (1000 nmol/L). C, Aptamin C prevents vitamin C oxidation for a long period of time
We conducted experiments to determine whether Aptamin C could prevent the oxidation of vitamin C over a long period of time. Aptamin C co‐treated vitamin C, and untreated vitamin C was exposed to light and allowed to stand at room temperature for 8 weeks. As a result, when untreated vitamin C was left alone, the degree of reduction was decreased to less than half in 2 weeks, and almost all of them were oxidized after 4 weeks. In the presence of Aptamin C, vitamin C was reduced to about half as much as 8 weeks (Figure 2C). This suggests that Aptamin C prevents oxidation of vitamin C over a long period of time.
3.4. Analysis of intracellular collagenase (MMP‐1) activity and collagen
On analysis of collagenase activity, matrix metalloproteinase‐1 (MMP‐1) production was significantly reduced in a dose‐dependent manner, with 7.06%, 9.29%, and 31.81% at concentrations of 0.01 + 0.5 μg/mL, 0.1 + 5 μg/mL, and 0.5 + 25 μg/mL, respectively (Figure 3A). On analysis of collagen synthesis, procollagen type Ι carboxy‐terminal peptide (PIP) was significantly increased in a dose‐dependent manner, with 25.00% increase at 0.01 + 0.5 μg/mL, 41.99% at 0.1 + 5 μg/mL, and 71.18% at 0.5 + 25 μg/mL (Figure 3B).
Figure 3.
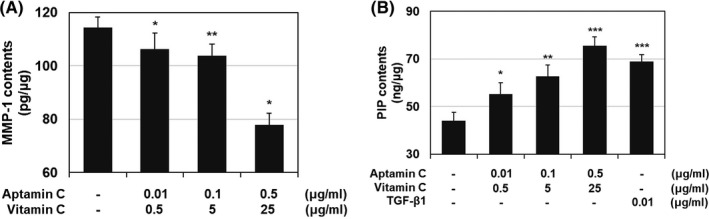
In vitro study of anti‐wrinkle effect in human dermal fibroblasts. A, MMP‐1 contents in human dermal fibroblasts after treatment. The data are expressed as % of solvent control (Mean ± SD, *P < .05, Aptamin C with vitamin C vs solvent control). B, PIP contents in human dermal fibroblasts after treatment. The data are expressed as % of solvent control (Mean ± SD, *P < .05, Aptamin C with vitamin C vs solvent control)
3.5. A clinical study of skin wrinkle improvement effects on human skin
Statistical analysis of wrinkle parameters was performed by 3D image analyzing system, to evaluate the skin wrinkle improvement effects of the test product on human skin. Compared to control group, “Average depth of wrinkles” parameter was decreased 4.77% at 4 weeks and 4.25% at 8 weeks, “Mean depth biggest wrinkle” parameter was decreased 3.37% at 4 weeks and 4.53% at 8 weeks, “Max. depth biggest wrinkle” parameter was decreased 4.36% at 4 weeks and 7.19% at 8 weeks, “Total length of wrinkles” parameter was decreased 1.61% at 4 weeks and 2.67% at 8 weeks, “Ra” parameter was decreased 4.22% at 4 weeks and 3.90% at 8 weeks, “Ry” parameter was decreased 2.66% at 4 weeks and 7.11% at 8 weeks, “Rz” parameter was decreased 3.69% at 4 weeks and 6.22% at 8 weeks, the decrement was 3.69%‐7.11% (Figure 4).
Figure 4.
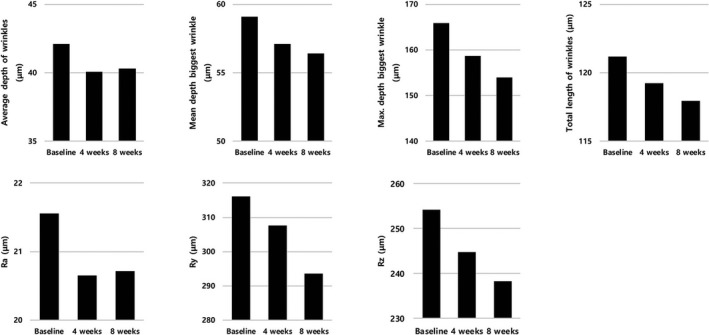
Changes of wrinkle parameters following five consecutive weeks application of the Aptamin C. Compared to baseline, “Average depth of wrinkles,” “Mean depth biggest wrinkle,” “Max. depth biggest wrinkle,” “Total length of wrinkles,” “Ra,” and “Rz” parameters were significantly decreased at 4 and 8 wk, and “Ry” parameter was significantly decreased at 8 wk in test group
4. CONCLUSION
Vitamin C is commercially important yet an unstable molecule, so improving its stability is of interest to various market sectors. Our work demonstrates that Aptamin C, DNA aptamers, potentially extends the shelf‐life and increase the potency of such products by delaying vitamin C oxidation in a solution. We exhibited clinical efficacy of Aptamin C complex in wrinkle improvement and skin hydration. Aptamin C complex could also help to relieve skin in harsh conditions. To the best of our knowledge, this is the first report successfully demonstrates that DNA aptamers prevent oxidation of vitamin C, dramatically enhancing its efficacy.
Choi S, Han J, Kim JH, et al. Advances in dermatology using DNA aptamer “Aptamin C” innovation: Oxidative stress prevention and effect maximization of vitamin C through antioxidation. J Cosmet Dermatol. 2020;19:970–976. 10.1111/jocd.13081
REFERENCES
- 1. Guaiquil VH, Vera JC, Golde DW. Mechanism of vitamin C inhibition of cell death induced by oxidative stress in glutathione‐depleted HL‐60 cells. J Biol Chem. 2001;276(44):40955‐40961. [DOI] [PubMed] [Google Scholar]
- 2. Pullar J, Carr A, Vissers M. The roles of vitamin C in skin health. Nutrients. 2017;9(8):866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Devasagayam T, Tilak JC, Boloor KK, Sane KS, Ghaskadbi SS, Lele RD. Free radicals and antioxidants in human health: current status and future prospects. J Assoc Physicians India. 2004;52:794‐804. [PubMed] [Google Scholar]
- 4. Rada B, Leto TL. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases In: A Egesten, A Schmidt, H Herwald. eds. Trends in Innate Immunity (Vol. 15). Basel, Switzerland: Karger Publishers; 2008;164‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conner GE, Salathe M, Forteza R. Lactoperoxidase and hydrogen peroxide metabolism in the airway. Am J Respir Crit Care Med. 2002;166(supplement_1):S57‐S61. [DOI] [PubMed] [Google Scholar]
- 6. Hollis F, Kanellopoulos AK, Bagni C. Mitochondrial dysfunction in autism spectrum disorder: clinical features and perspectives. Curr Opin Neurobiol. 2017;45:178‐187. [DOI] [PubMed] [Google Scholar]
- 7. Haider L, Fischer MT, Frischer JM, et al. Oxidative damage in multiple sclerosis lesions. Brain. 2011;134(7):1914‐1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel VP, Chu CT. Nuclear transport, oxidative stress, and neurodegeneration. Int J Clin Exp Patho. 2011;4(3):215. [PMC free article] [PubMed] [Google Scholar]
- 9. Nunomura A, Castellani RJ, Zhu X, Moreira PI, Perry G, Smith MA. Involvement of oxidative stress in Alzheimer disease. J Neuropathol Exp Neurol. 2006;65(7):631‐641. [DOI] [PubMed] [Google Scholar]
- 10. Grabnar I, Vovk T, Kores Plesnicar B, Boskovic M. Oxidative stress in schizophrenia. Curr Neuropharmacol. 2011;9(2):301‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramalingam M, Kim S‐J. Reactive oxygen/nitrogen species and their functional correlations in neurodegenerative diseases. J Neural Transm. 2012;119(8):891‐910. [DOI] [PubMed] [Google Scholar]
- 12. Nijs J, Meeus M, DeMeirleir K. Chronic musculoskeletal pain in chronic fatigue syndrome: recent developments and therapeutic implications. Manual therapy. 2006;11(3):187‐191. [DOI] [PubMed] [Google Scholar]
- 13. Handa O, Naito Y, Yoshikawa T. Redox biology and gastric carcinogenesis: the role of Helicobacter pylori. Redox Rep. 2011;16(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trouba KJ, Hamadeh HK, Amin RP, Germolec DR. Oxidative stress and its role in skin disease. Antioxid Redox Signal. 2002;4(4):665‐673. [DOI] [PubMed] [Google Scholar]
- 15. Bickers DR, Athar M. Oxidative stress in the pathogenesis of skin disease. J Invest Dermatol. 2006;126(12):2565‐2575. [DOI] [PubMed] [Google Scholar]
- 16. Rinnerthaler M, Bischof J, Streubel M, Trost A, Richter K. Oxidative stress in aging human skin. Biomolecules. 2015;5(2):545‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stadtman ER. Protein oxidation and aging. Science. 1992;257(5074):1220‐1224. [DOI] [PubMed] [Google Scholar]
- 18. Shindo Y, Witt E, Packer L. Antioxidant defense mechanisms in murine epidermis and dermis and their responses to ultraviolet light. J Invest Dermatol. 1993;100(3):260‐265. [DOI] [PubMed] [Google Scholar]
- 19. Kohen R. Skin antioxidants: their role in aging and in oxidative stress—new approaches for their evaluation. Biomed Pharmacother. 1999;53(4):181‐192. [DOI] [PubMed] [Google Scholar]
- 20. Stojiljković D, Pavlović D, Arsić I. Oxidative stress, skin aging and antioxidant therapy/oksidacioni stres, starenje kože i antioksidaciona terapija. Acta Facultatis Medicae Naissensis. 2014;31(4):207‐217. [Google Scholar]
- 21. Nimse SB, Pal D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Advances. 2015;5(35):27986‐28006. [Google Scholar]
- 22. Padh H. Vitamin C: newer insights into its biochemical functions. Nutr Rev. 1991;49(3):65‐70. [DOI] [PubMed] [Google Scholar]
- 23. Leveque N, Robin S, Muret P, Mac‐Mary S, Makki S, Humbert P. High iron and low ascorbic acid concentrations in the dermis of atopic dermatitis patients. Dermatology. 2003;207(3):261‐264. [DOI] [PubMed] [Google Scholar]
- 24. Sorice A, Guerriero E, Capone F, Colonna G, Castello G, Costantini S. Ascorbic acid: its role in immune system and chronic inflammation diseases. Mini reviews in medicinal. Chemistry. 2014;14(5):444‐452. [DOI] [PubMed] [Google Scholar]
- 25. Kim A‐R, Ha N‐R, Jung I‐P, Kim S‐H, Yoon M‐Y. Development of a ssDNA aptamer system with reduced graphene oxide (rGO) to detect nonylphenol ethoxylate in domestic detergent. J Mol Recognit. 2019;32(3):e2764. [DOI] [PubMed] [Google Scholar]
- 26. Park J‐W, Tatavarty R, Kim DW, Jung H‐T, Gu MB. Immobilization‐free screening of aptamers assisted by graphene oxide. Chem Commun. 2012;48(15):2071‐2073. [DOI] [PubMed] [Google Scholar]
- 27. Wu M, Kempaiah R, Huang P‐J, Maheshwari V, Liu J. Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir. 2011;27(6):2731‐2738. [DOI] [PubMed] [Google Scholar]
- 28. Liu Z, Liu B, Ding J, Liu J. Fluorescent sensors using DNA‐functionalized graphene oxide. Anal Bioanal Chem. 2014;406(27):6885‐6902. [DOI] [PubMed] [Google Scholar]
- 29. Chang H, Tang L, Wang Y, Jiang J, Li J. Graphene fluorescence resonance energy transfer aptasensor for the thrombin detection. Anal Chem. 2010;82(6):2341‐2346. [DOI] [PubMed] [Google Scholar]
- 30. Li M, Zhou X, Guo S, Wu N. Detection of lead (II) with a “turn‐on” fluorescent biosensor based on energy transfer from CdSe/ZnS quantum dots to graphene oxide. Biosens Bioelectron. 2013;43:69‐74. [DOI] [PubMed] [Google Scholar]
- 31. Ahmad Raston NH, Gu MB. Highly amplified detection of visceral adipose tissue‐derived serpin (vaspin) using a cognate aptamer duo. Biosens Bioelectron. 2015;70:261‐267. [DOI] [PubMed] [Google Scholar]
- 32. Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244(4900):48‐52. [DOI] [PubMed] [Google Scholar]
- 33. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406‐3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vislisel JM, Schafer FQ, Buettner GR. A simple and sensitive assay for ascorbate using a plate reader. Anal Biochem. 2007;365(1):31‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]


