Summary
RIPENING INHIBITOR (RIN)‐deficient fruits generated by CRISPR/Cas9 initiated partial ripening at a similar time to wild‐type (WT) fruits but only 10% WT concentrations of carotenoids and ethylene (ET) were synthesized. RIN‐deficient fruit never ripened completely, even when supplied with exogenous ET. The low amount of endogenous ET that they did produce was sufficient to enable ripening initiation and this could be suppressed by the ET perception inhibitor 1‐MCP.
The reduced ET production by RIN‐deficient tomatoes was due to an inability to induce autocatalytic system‐2 ET synthesis, a characteristic feature of climacteric ripening. Production of volatiles and transcripts of key volatile biosynthetic genes also were greatly reduced in the absence of RIN.
By contrast, the initial extent and rates of softening in the absence of RIN were similar to WT fruits, although detailed analysis showed that the expression of some cell wall‐modifying enzymes was delayed and others increased in the absence of RIN.
These results support a model where RIN and ET, via ERFs, are required for full expression of ripening genes. Ethylene initiates ripening of mature green fruit, upregulates RIN expression and other changes, including system‐2 ET production. RIN, ET and other factors are required for completion of the full fruit‐ripening programme.
Keywords: carotenoid, cell wall, MADS‐RIN, post‐harvest softening, ripening initiation, Solanum lycopersicum (tomato), system‐2 ethylene (ET), volatile
Introduction
Fleshy fruits evolved both to protect developing seeds and aid seed dispersal. They have traditionally been classified into climacteric (e.g. apples, pears, bananas, melons and tomato) and nonclimacteric (e.g. pineapple, strawberry, citrus) types. Climacteric fruits such as tomato show a characteristic rise in respiration, the respiratory climacteric, and a marked rise in ethylene (ET) production at the onset of ripening. The tomato (Solanum lycopersicum) has been a model for understanding the molecular basis of ripening for more than 40 yr because of its commercial importance but also because it is well characterized genetically, several crops can be grown year‐round in controlled environments, it can be relatively easily transformed, and several important natural mutations have been identified that affect different aspect of ripening (Giovannoni, 2007; Klee and Giovannoni, 2011). During fruit growth, the cells enlarge and accumulate reserves and at maturity multiple changes are initiated in a coordinated manner which alter colour, flavour, texture and aroma. This ripening process converts a hard, unappealing fruit into a colourful nutritious product that attracts birds, animals and humans to aid in seed dispersal (Giovannoni, 2004).
The transcription factor MADS‐RIN (RIN, RIPENING INHIBITOR) has long been viewed as a master regulator of ripening. RIN affects accumulation of many gene transcripts (Giovannoni, 2004), proteins and their post‐translational degradation (Qin et al., 2012; Wang et al., 2014). Recent advances in knowledge concerning the transcription factors that regulate climacteric fruits ripening (Ito et al., 2017; Gao et al., 2018; Li et al., 2018; Wang et al., 2019), including discrepancies between the phenotypes of natural mutants and CRISPR/Cas9‐induced mutations of ripening inhibitor (rin), non‐ripening (nor) and colourless non‐ripening (Cnr), have called into question the precise roles of these regulatory genes in the ripening control network (Ito et al., 2017; Gao et al., 2018; Li et al., 2019; Wang et al., 2019). The rin mutation was originally thought to correspond to a loss‐of‐function event (Vrebalov et al., 2002) but is now known to generate an active hybrid transcription factor, RIN‐MC, which has a repressor activity (Ito et al., 2017; Li et al., 2018). RIN is still critical for progression of ripening but is not required for ripening initiation (Ito et al., 2017; Li et al., 2018), and the factor actually responsible for ripening initiation in the absence of RIN has not been identified.
Ethylene has long been implicated in ripening control. All plants produce some ET, but this can increase 100‐fold as climacteric fruit transition from low concentration (system‐1) ET, which is self‐inhibitory, to a major burst of autocatalytic ET synthesis, called system‐2 ET (McMurchie et al., 1972; Grierson, 2013, 2014) during ripening. Nonclimacteric fruits also respire and evolve low concentrations of ET, but they do not show a climacteric burst. Analysis of natural mutants which have pleiotropic effects on multiple genes has identified ripening regulators such as MADS‐RIN, NAC‐NOR and the squamosa‐box binding protein CNR, some of which act at least partly independently of ET (Martel et al., 2011; Kumar et al., 2012; Giovannoni et al., 2017). Activator‐ and repressor‐ET response factors (ERFs) and auxin response factors (ARFs) operate downstream of their respective hormone signalling pathways to regulate gene expression and hormone cross‐talk (Liu et al., 2015, 2018). Ethylene promotes ripening (Tucker et al., 2017), abscisic acid also may play a part (Chen et al., 2016; Mou et al., 2016), whereas auxin tends to inhibit ripening (Shin et al., 2019). The way in which this complex network of transcription factors and hormones operates to control the expression of many ripening genes is still being actively investigated (Li et al., 2019), but it is known that together they control production of enzymes for biosynthesis of coloured pigments such as carotenoids in the chromoplasts (or anthocyanins in the vacuoles in other fruits), multiple cell wall softening enzymes, metabolism of acids and sugars affecting taste, and production of multiple aroma volatiles (Giovannoni, 2004; Grierson, 2013; Klee and Tieman, 2018; Li et al., 2019).
Recent work describing the removal of RIN by CRISPR/Cas9 or RNAi strategies (Ito et al., 2017; Li et al., 2018) has shown that the precise role of RIN in climacteric ripening needs further clarification. Here we characterize new RIN‐deficient Ailsa Craig tomato fruits obtained by CRISPR/Cas9 technology and show that they produce sufficient endogenous ET to induce ripening and this RIN‐independent initiation of ripening is inhibited by the ET perception inhibitor 1‐methylcyclopropene (1‐MCP). Ethylene production is low because RIN‐deficient fruits are unable to induce autocatalytic system‐2 ET production and they also are deficient in volatiles and carotenoids and transcripts associated with these pathways. Strikingly, extensive softening occurs independently of RIN, which contrasts strongly with the original rin mutant phenotype. Moreover, late softening of RIN‐deficient fruits coincides with the delayed accumulation of several cell wall enzymes including MAN1, Mside7, MAN4a, TBG4, PG and PME2.1. These results support a model in which ET is required for the initiation of ripening and the combined action of RIN and ET is required for the progression and completion of different facets of the ripening process.
Materials and Methods
Plasmid construction
The target site for CRISPR/Cas9‐mediated RIPENING INHIBITOR (RIN) mutagenesis was selected using the CRISPR‐P program (http://cbi.hzau.edu.cn/cgi-bin/CRISPR) (Supporting Information, Fig. S1a). The 20‐bp oligos were cloned into AtU3d and AtU3b vectors and the sgRNA expression cassettes assembled into pYLCRISPR/Cas9‐Ubi‐H binary plasmid by Golden Gate ligation (Ma et al., 2015). Agrobacterium tumefaciens‐mediated transfer of T‐DNA was used for stable transformation of tomato (Sun et al., 2006; Kimura and Sinha, 2008). For the mutation analysis, genomic DNA was extracted from young tomato leaves using a Plant Genomic DNA Kit (Tiangen, China) and used as a template to amplify the RIN fragment using PCR and the fragments sent for sequencing. The primer pairs used for vector construction and mutation analyses are listed in Table S1.
Plant material and growth conditions
Wild‐type (WT) tomato (Solanum lycopersicum Alisa Craig, AC) and RIN‐CRISPR seedlings were grown in a glasshouse under long‐day conditions (16 h : 8 h, light : dark photperiod) at a temperature of 26°C. For gene expression analysis, organs were collected, frozen in liquid N2, and stored at −80°C until RNA extraction. Three independent samplings were performed for each measurement.
Tomato fruit nuclei isolation and Western blotting
Nuclei were isolated from tomato fruits picked at B + 5 stage and assayed for RIN protein. Fruit samples were ground into a powder under liquid N2 and the mixture was extracted with buffer (0.25 M sucrose, 10 mM Tris‐HCl pH7.5, 1 mM MgCl2, 0.5% PVP, 0.5% Triton X‐100, Roche protease inhibitor tablet) and the suspension filtered using miracloth (475855; Millipore, Pittsburgh, PA, USA). After centrifugation at 10 000 g for 10 min, the precipitate was washed with extraction buffer and centrifuged again at 10 000 g for 10 min, and the pellet was resuspended in percoll buffer (0.25 M sucrose, 95% Percoll, 10 mM Tris‐HCl pH7.5, Roche protease inhibitor tablet). The floating layer was collected after centrifugation at 10 000 g for 10 min, diluted to 30% with extraction buffer, centrifuged at 10 000 g for 10 min, to pellet the nuclei and stored at −80°C or used for SDS‐PAGE assay.
Western blotting was carried out as described (Li et al., 2018). Briefly, protein extracts were separated on 10% SDS‐PAGE gels and transferred to a polyvinylidene fluoride (PVDF) membrane blocked in 5% nonfat milk for 2 h at room temperature. A specific polyclonal antibody produced in rabbit raised against the C‐terminal end of RIN (amino acids 75–242) was added in a ratio of 1 : 1000 and incubated for 2 h at room temperature. Membranes were washed with Tris‐buffered saline plus Tween‐20 three times, 15 min each time. The anti‐rabbit horseradish peroxidase secondary antibody was added at a ratio of 1 : 10 000 and incubated for 2 h at room temperature. After three washes with Tris‐buffered saline plus Tween‐20, the membranes were visualized using a horseradish peroxidase‐enhanced chemiluminescence system.
Ethylene production measurement
For the measurement of ethylene (ET) production, each fruit was placed in a sealed gas‐tight 300 ml container at 25°C for 1 h, and a 1 ml headspace gas sample was analyzed using GC (6890N GC system; Agilent, Folsom, CA, USA) equipped with a flame ionization detector (Ma et al., 2016).
Colour measurement
A Hunter Lab Mini Scan XE Plus colorimeter (Hunter Associates Laboratory Inc., Reston, VA, USA) with the CIE L*a*b colour system was chosen for pericarp colour assay (Komatsu et al., 2016). At least six biological replicates were used for each assay.
Carotenoid content assay
Carotenoid extraction followed the methods reported by Xu et al. (2006); 100 mg tomato fruit samples were ground to a powder and frozen at −80°C, 250 μl methanol was added, vortexed to mix, followed by 500 μl chloroform, vortexed again and 250 μl 50 mM Tris buffer (pH 7.5, containing 1 M NaCl) was added, followed by vortexing. After centrifugation (15 000 g for 10 min at 4°C), the lower chloroform phase was collected. The chloroform extraction was repeated two or three times and the chloroform phases combined and dried under flowing N2. The residue was dissolved in 100 μl ethyl acetate (HPLC grade), and 50 μl transferred to HPLC sample analysis tubes. Carotenoid content was assayed according to the methods reported by Zheng et al. (2015): A volume of 20 μl for each sample was absorbed for HPLC analysis, carried out using a Waters liquid chromatography system (e2695) equipped with a photodiode array (PDA) detector (2998). A C30 carotenoid column (250 mm × 4.6 mm; YMC, Japan) was used to elute the carotenoids with a methanol: H2O (9 : 1, v/v, eluent A) solution and methyl tert‐butyl ether (MTBE) (100%, eluent B) solution containing 0.01% (w/v) butylated hydroxytoluene (BHT). The linear gradient program was performed as follows: 8% B to 25% B for 30 min, 25% B to 70% B for 5 min, 70% B for 5 min, and back to the initial 8% B for re‐equilibration for 10 min. The flow rate was 1 ml min−1. To avoid light degradation of carotenoids the extraction and analysis were performed under subdued light.
Firmness measurement
The firmness of the pericarp was assayed using a penetrometer (TA‐XT2i texture analyzer; Stable Micro Systems, Stable Micro Systems Ltd, Surrey, UK) according to the manufacturer's instructions. At least six biological replicates were used for each assay.
Volatiles assays
Measurements of volatiles were carried out according to Zhang et al. (2010), with modifications. First, 5 g of frozen flesh tissue was ground in liquid N2 and transferred to a 15‐ml vial containing 5 ml of saturated sodium chloride solution. Before vials were sealed, 20 μl of 2‐octanol (0.8 mg ml−1) was added as an internal standard and vortexed for 10 s.
For solid‐phase microextraction (SPME), samples then were equilibrated at 40°C for 30 min before being exposed to a fiber coated with 50/30 μm DVB/CAR/PDMS (Supelco Co., Bellefonte, PA, USA). Volatiles were subsequently desorbed over 5 min at 230°C into the splitless injection port of the GC‐flame ionization detector (FID). An Agilent 7890A GC equipped with an FID and a DB‐WAX column (30 m × 0.32 mm, 0.25 μm internal diameter; J&W Scientific, Folsom, CA, USA) was used for volatile analysis. Chromatography conditions were as follows: injector, 230°C; initial oven temperature, 34°C held for 2 min, increased by 2°C min−1 to 60°C, then increased by 5°C min−1 to 220°C, and held for 2 min. Nitrogen was used as carrier gas at 1.0 ml min−1. Volatiles were identified by comparison with retention times of authentic standards. Further identification of volatile compounds was by capillary gas chromatography‐mass spectrometry (GC‐MS) (7890A‐5975C) performed using an HP‐5 MS column (30 m × 0.25 mm, 0.25 μm; J&W Scientific, Folsom, CA). Injection port temperature was 240°C, with a split ratio of 5 : 1. Helium was used as the carrier gas at a rate of 1.0 ml min−1. The column temperature was held at 40°C for 2 min, increased by 5°C min−1 to 60°C, then increased by 10°C min−1 to 250°C, and held for 5 min. MS conditions were as follows: ion source, 230°C; electron energy, 70 eV; multiplier voltage, 1247 V; GC‐MS interface zone, 280°C; and a scan range, 30–250 mass units. Volatiles were identified on the basis of a comparison of their electron ionization (EI) mass spectra to published data and data from authentic standards. Quantitative determination of compounds was carried out using the peak of the internal standard as a reference value and calculated on the basis of standard curves constructed with authentic compounds.
Ethylene, 1‐methylcyclopropene (1‐MCP) and propylene treatment
Tomato fruits at the mature green (MG) stage, before any sign of colour change, were placed in an air‐tight 1‐l plastic container with 100 ppm ET, 1000 ppm propylene or 10 ppm 1‐MCP. 1000 ppm propylene is equivalent to 10 ppm ET treatment (McMurchie et al., 1972) and is used in order to distinguish it from endogenous ET production by GC equipment. The treatment was conducted continually in an incubator under a 16 h : 8 h, light : dark photoperiod at 25°C, with at least three biological replicates for each treatment. RIN‐CRISPR tomato fruits treated with ET for 48 h, and control WT and RIN‐CRISPR treated with air, were chosen for gene expression assay using qRT‐PCR. The gas environments (air, ET, propylene, 1‐MCP) were replenished every 24 h.
RNA isolation and quantitative reverse transcription (qRT)‐PCR
Isolation of RNA from tomato fruit pericarp at different ripening stages was as described previously (Zhu et al., 2015). Total RNA extraction from tomato fruit pericarp was carried out using Trizol reagent, and RNA integrity was verified by 1.5% (v/v) agar gel electrophoresis. Genomic DNA was removed from RNA preparations by digestion with DNase I (Invitrogen, cat. no. AM1907), and RNA quality and quantity were confirmed by spectrophotometry (Thermo Scientific, Waltham, MA, USA; NanoDrop 1000). RNA was reverse‐transcribed into cDNA using cDNA synthesis kit (Bio‐RAD, cat. no. 1708890) according to the manufacturer's instructions. qRT‐PCR was conducted using FastStart Essential DNA Green Master (Roche, cat. no. 06402712001) with a LightCycler480 (Roche). Relative gene expression values were calculated using the 2‐ΔΔCt method (Livak and Schmittgen, 2001). The tomato ACTIN gene (Solyc03g078400) was used as an internal reference gene. At least three biological replicates were included for each point, and each replicate was from independent sampling. The primer pairs used in qRT‐PCR analyses are listed in Table S2.
Water loss
The water lost by tomato fruits was calculated as FW (%) = fruit weight (g) – fresh fruit weight (g)/ fresh fruit weight (g) × 100%. More than ten biological replicates were used for each assay.
Promoter sequence and motif assay
Promoter sequences 2.0 kb in length were downloaded from Sol Genomics Network (https://solgenomics.net/), various CArG‐box elements were from Fujisawa et al. (2013). The GCC‐box, a characteristic cis‐element binding site for ERFs, was from Licausi et al. (2013). An AP2/ERF binding motif, ATCTA was from Welsch et al. (2007).
Statistical analysis
Microsoft excel 2010 and Spss (IBM SPSS Statistics, v.22; SPSS Inc., Chicago, IL, USA) were used for statistical analyses. Duncan's multiple range test was used (P < 0.05).
Results
Generation of RIN mutants using the CRISPR/Cas9 gene‐editing system
Recent studies using CRISPR/Cas9 or RNAi have shown that ripening initiation is not completely inhibited in RIN‐deficient tomato fruit (Ito et al., 2017; Li et al., 2018), however, the possible role of ET in ripening initiation in the absence of RIN has not been investigated. To clarify the precise role of RIN and other factors controlling the different aspects of climacteric ripening, we generated new RIN loss‐of‐function tomato lines, using the CRISPR/Cas9 gene editing system. Two genomic sites were targeted for cleavage (Fig. S1a), and multiple independent transgenic plants were genotyped by the direct sequencing of PCR products from genomic DNA flanking the target sites. After two generations of screening, ten lines were identified carrying homozygous mutations in exon 2: deletion of 432 bp corresponding to the entire exon 2 (RIN‐CRISPR‐1); deletion of 11 bp (RIN‐CRISPR‐2); insertion of 1 bp (RIN‐CRISPR‐3); deletion of 1 bp (RIN‐CRISPR‐4); deletion of 2 bp (RIN‐CRISPR‐5); insertion of 1bp (RIN‐CRISPR‐6); insertion of 1 bp (RIN‐CRISPR‐7, RIN‐CRISPR‐8); deletion of 2 bp (RIN‐CRISPR‐9); and changes in 4 bp (RIN‐CRISPR‐10) (Fig. S1b). Hereafter, these 10 are referred to as RIN‐deficient lines. The manipulations introduced premature stop codons in the RIN coding sequence, which led to predicted truncated RIN polypeptides of 68, 74, 78, 94, 77, 97, 78, 78, 96 and 242 (with 21 changes) amino acids of the MADS domain (Fig. S1b), compared to the 242 amino acids of WT RIN native protein.
In order to assess the presence of RIN protein in the edited tomato lines, we used antibodies specific to the C‐terminal region (sites 158–242 AAs), which encodes the activation domain of the RIN protein (Qin et al., 2012; Martel et al., 2011; Ito et al., 2008) as the N‐terminal MADS‐box domain is highly conserved among MADS‐box proteins. Western blot analysis revealed that full‐length RIN protein could only be detected in WT and was absent from RIN‐CRISPR fruits (Fig. 1a). All of the lines bearing premature stop codons in the RIN gene displayed similar altered ripening phenotypes. Three of these were studied more closely and two representative lines (named RIN‐CRISPR‐1, ‐2) used for detailed physiological analyses and molecular studies (Fig. S2).
Figure 1.
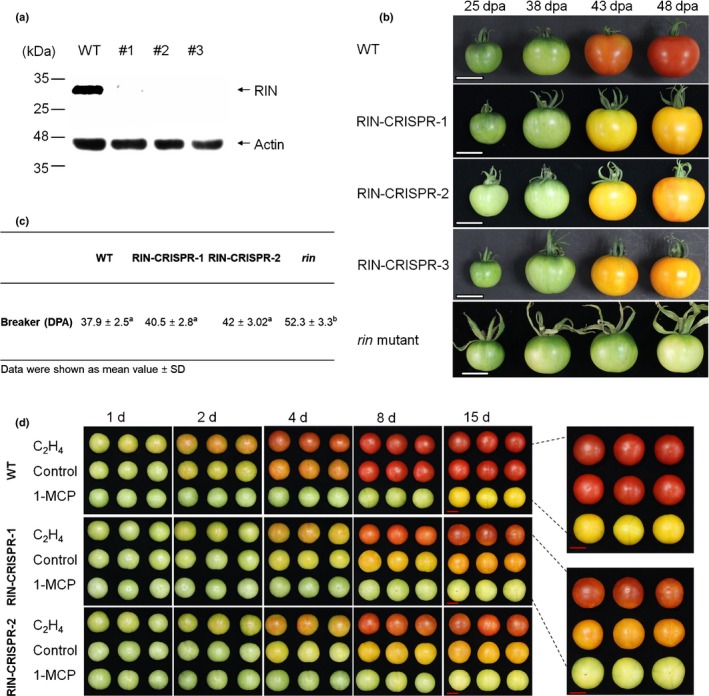
Phenotypes, RIPENING INHIBITOR (RIN) protein content and ethylene (ET) response of wild‐type (WT) and RIN‐deficient tomato (Solanum lycopersicum) fruits. (a) The detection of RIN protein by Western blotting in WT but not RIN‐CRISPR fruits, using RIN‐poly‐antibody. Actin was used as internal control. (b) Tomato fruits from the RIN‐MC mutant (rin mutant) and three RIN‐CRISPR lines (RIN‐CRISPR‐1‐3) generated by CRISPR/Cas9‐mediated mutagenesis of the RIN gene are shown at 25 d after anthesis (dpa), 38 dpa, 43 dpa and 48 dpa, and compared to WT fruit. Bar, 2 cm. (c) Number of days from anthesis to ripening of S. lycopersicum Alisa Craig (AC), RIN‐CRISPR and rin mutant tomato fruits. (d) Effect of exogenous ET and ET perception inhibitor 1‐MCP treatment on ripening progression of RIN‐CRISPR tomato fruit. WT and RIN‐CRISPR tomato fruits were picked at MG stages and treated and. replenished daily with ET (100 ppm) and 1‐MCP (10 ppm) or air continually for up to 15 d. Fruits in horizontal rows are biological replicates. Enlarged photos of representative samples are shown compared to WT fruits on the right. Bar, 2 cm.
RIN‐deficient fruit reveal that ripening can be initiated independently from RIN
Assessing the ripening behaviour of RIN‐deficient fruits revealed that all three independent mutant lines (RIN‐CRISPR‐1, ‐2, ‐3) initiated ripening at approximately the same time as WT fruits and there was no obvious delay in ripening onset (Fig. 1b,c). This contrasted with the situation for rin mutant fruits, where initiation of ripening was delayed by 14 d or more (Fig. 1c) and fruits only ever turned a pale yellow colour and did not soften. The RIN‐deficient fruits not only ripened earlier than rin mutant fruit but became a deep yellow/orange colour and softened, the rin mutant fruit slowly became pale yellow but did not soften, whereas WT fruits softened and turned red (Fig. 1b).
When they were allowed to ripen for longer, RIN‐CRISPR fruits became orange. This appeared to vary slightly in different fruits, according to age and position on the plant, suggesting that they might be sensitive to some other factor(s). The ability of RIN‐deficient fruit to undergo ripening in response to ET was investigated. Detached mature green (MG) WT and RIN‐CRISPR fruits were treated with either external ET (100 ppm) or its competitive inhibitor 1‐methylcyclopropene (1‐MCP) (10 ppm). External ET accelerated red colour development in WT fruit with a visible difference after 2 d treatment, whereas 1‐MCP inhibited this process very significantly for ≥ 15 d (Fig. 1d). Ethylene also accelerated colour change in RIN‐CRISPR fruits, which became faintly orange‐red, whereas 1‐MCP significantly inhibited the transition from green to yellow for many days (Fig. 1d). Although supplying ET externally to picked fruit induced slight red coloration in RIN‐deficient fruits, the response was weaker than in WT fruits and occurred more slowly (Fig. 1d). These results indicated that RIN‐CRISPR fruits are able to respond to ET by enhancing pigment accumulation during ripening, but without approaching the concentrations displayed by WT fruits.
RIN‐CRISPR fruits are deficient in climacteric ET production and unable to induce system‐2 ET synthesis
In order to address whether the reduced ET production of RIN‐deficient tomatoes is due to their inability to induce autocatalytic system‐2 ET synthesis, a characteristic feature of normal climacteric fruit ripening, we treated RIN‐deficient fruit with propylene, an analogue which mimics the hormone effect of ET. In the absence of treatment, WT fruits showed a characteristic burst of ET synthesis after the onset of ripening, whereas ET production by RIN‐CRISPR fruits was substantially inhibited compared to WT (Fig. 2a). Thereafter, detached WT and RIN‐CRISPR MG fruits were challenged by continuous treatment with 1000 ppm propylene which is equivalent to 10 ppm ET treatment (McMurchie et al., 1972). An obvious burst of ET production was detected after 24 h with WT fruits, which reached a peak at 48 h (Fig. 2b). No increase in ET production was detected, however, in RIN‐CRISPR fruits even after 72 h treatment (Fig. 2b), indicating that the autocatalytic system‐2 ET burst could not be induced by propylene treatment in the absence of RIN.
Figure 2.
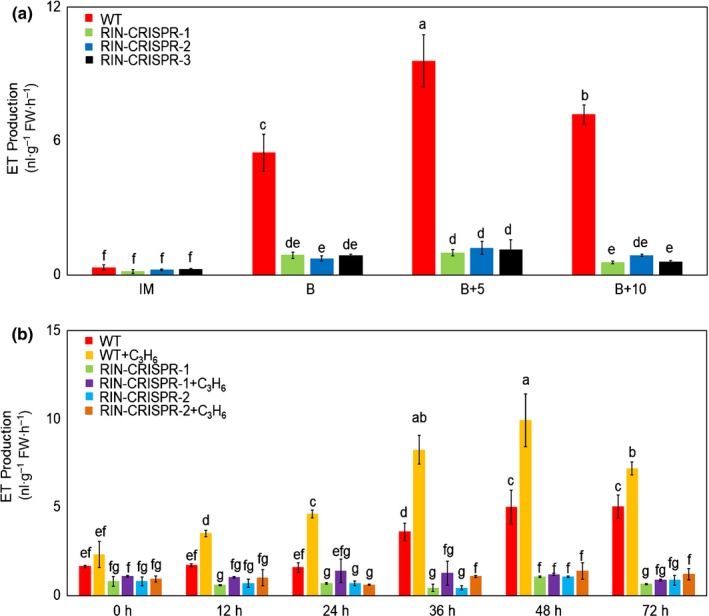
Ethylene (ET) production and response of wild‐type (WT) and RIPENING INHIBITOR (RIN)‐deficient tomato fruits to propylene. (a) Ethylene production in WT and RIN‐CRISPR tomato fruits at immature (IM), breaker (B), breaker + 5 (B + 5) and breaker + 10 (B + 10) stages, measured by GC (average of three biological replicates). (b) Ethylene production of tomato WT and RIN‐CRISPR fruits treated with propylene. Fruit were picked at MG stages and treated with propylene (1000 ppm) (equivalent to 10 ppm ET treatment) (McMurchie et al., 1972) or air (control) continually for 12, 24, 36, 48 and 72 h to test for induction of endogenous system‐2. Propylene was replenished every 12 h. The error bars represent mean ± SD, the lowercase letters indicate significant difference at P = 0.05.
Several differences between RIN‐CRISPR and WT fruits were found in the expression of genes related to ET biosynthesis, perception and signalling (Fig. S3). Transcripts of ACC synthase (ACS) and ACC oxidase (ACO), the ET biosynthesis genes, all were greatly reduced in RIN‐deficient fruits, compared to WT fruits. ACS2 and ACS4 transcripts, the main ACS genes that increase during ripening, accumulated to < 20% of their concentrations in WT respectively, whereas ACO1 and ETR3/NR transcripts reached about 40% and 50% of WT concentrations, respectively. This is consistent with the suggestion that there is a dual control of these genes (Yokotani et al., 2009) and it is noteworthy that they are known to be specifically involved in system‐2 ET synthesis. Moreover, the ACS genes affected in RIN‐deficient fruits are those identified previously among the RIN targets (Fujisawa et al., 2012; Zhong et al., 2013). The classic ET‐induced tomato genes E4 and E8 transcripts also were greatly reduced (Fig. S3; Lincoln and Fischer, 1988).
Pigment accumulation is impaired in RIN‐deficient fruit
Pigment accumulation is a major fruit ripening trait and colour change depends on the degradation of chlorophyll, and the synthesis and accumulation of the coloured carotenoids lycopene and β‐carotene. The tomato fruits pericarp colours were measured with a colorimeter using the CIE L*a*b colour system (Komatsu et al., 2016). The a* value refers to the degree of red to green, determined by the degradation of chlorophyll and the accumulation of carotenoids, such as β‐carotene and lycopene, which produce the characteristic yellow, orange and red coloration (Luo et al., 2013; Fig. S4). At B + 5 and B + 10 stages WT fruits turned red gradually. By contrast, the three RIN‐CRISPR mutants turned yellow and slowly developed an orange tinge 1–2 wk after ripening onset (Fig. 1). The carotenoid content and constituents were determined quantitatively by HPLC in WT and RIN‐CRISPR fruits at B, B + 5 and B + 10 stages. WT fruits accumulated more than a 10‐fold higher concentration of lycopene, the main red colour than RIN‐CRISPR fruits, which, in addition to having low lycopene, accumulated no phytofluene, and very low amounts of phytoene and alpha‐carotene (Fig. 3a). This indicated how severely carotenoid accumulation is inhibited in RIN‐CRISPR fruits. By contrast, the rin mutant fruit remained yellow until B + 10 stage, with almost no detectable lycopene (Li et al., 2018). Transcripts for the key enzymes phytoene synthase1 (PSY1) (Bird et al., 1991; Fray and Grierson, 1993) and non‐heme hydroxylases (CHY/SlBCH2) and carotene isomerase (CRTISO) were inhibited by 90%, 80% and 75%, respectively, in RIN‐deficient fruits and other transcripts of genes in the pathway (1‐d‐deoxyxylulose 5‐phosphate synthase (DXS1), phytoene desaturase (PDS), zeta‐carotene desaturase (ZDS)) were significantly reduced, whereas geranylgeranyl pyrophosphate synthases (GGPS2) was not affected and zeaxanthin epoxidase (ZEP) transcripts actually accumulated to a higher level in RIN‐deficient fruit. In agreement with the positive response to ET of RIN‐deficient fruits, transcripts of both PSY1 (3.0‐ and 3.8‐fold increase in RIN‐CRISPR‐1 and RIN‐CRISPR‐2, respectively) and CHY (3.0‐ and 2.0‐fold increase in RIN‐CRISPR‐1 and RIN‐CRISPR‐2, respectively) accumulated to higher levels in response to ET treatment (Fig. 3b). These data indicate that colour change, a major ripening‐associated trait in tomato fruit, can be induced by ET in a RIN‐independent manner but high‐level accumulation of carotenoids requires RIN.
Figure 3.
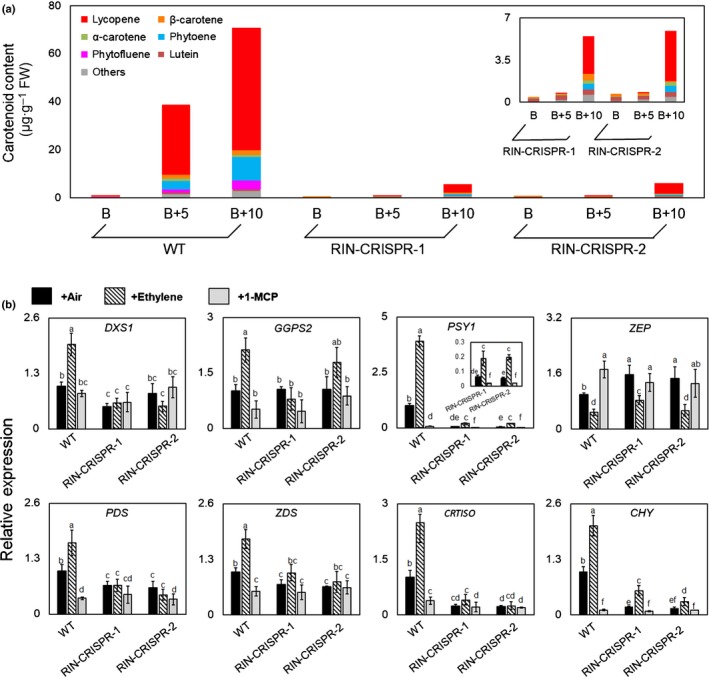
Comparison of changes in pigment production and quantitative reverse transcription (qRT)‐PCR assay of genes involved in pigment accumulation in wild‐type (WT) and RIPENING INHIBITOR (RIN)‐deficient tomato fruits. (a) Carotenoid content of WT and RIN‐CRISPR measured by HPLC (average of three biological replicates). (b) qRT‐PCR of gene transcripts involved in carotenoids biosynthesis (average of three biological replicates). Relative transcript levels are expressed relative to the tomato ACTIN gene internal control, expressed as 2‐ΔΔCt (Livak and Schmittgen, 2001). 1‐d‐deoxyxylulose 5‐phosphate synthase (DXS1); geranylgeranyl pyrophosphate synthases (GGPS2); phytoene synthases 1 (PSY1); phytoene desaturase (PDS); zeta‐carotene desaturase (ZDS); carotene isomerase (CRTISO); non‐heme hydroxylases (SlBCH2/CHY); zeaxanthin epoxidase (ZEP). The error bars represent mean ± SD, the lowercase letters indicate significant difference at P = 0.05. Genes identified as having low level of transcripts by RNA‐Seq (Li et al., 2018) were not measured.
Volatiles accumulation is impaired in RIN‐deficient fruit
Production of volatile compounds is a major trait of ripe fruit that is highly appreciated by consumers. At the B + 10 stage, volatiles derived from different pathways (Fig. S5a) were measured by GC and GC‐MS and were significantly reduced in RIN‐deficient fruits compared to WT (Fig. S5b; Table S3). These included volatiles generated from amino acids (phenylethyl alcohol, 2‐hydroxy‐benzaldehyde, 2‐methoxy‐phenol, benzeneacetaldehyde, benzaldehyde, 2‐Isobutylthiazole), carotenoids (6‐methyl‐5‐hepten‐2‐one, (E)‐3‐Buten‐2‐one), lipids (hexanal, heptanal, (E)‐2‐heptenal, (E)‐2‐Pentenal) (Fig. 4a). Of the volatile compounds shown to be reduced in RIN‐deficient fruits, nine (phenylethyl alcohol, 2‐hydroxy‐benzaldehyde, benzaldehyde, benzeneacetaldehyde, 2‐isobutylthiazole, 6‐methyl‐5‐hepten‐2‐one, heptanal, (E)‐2‐heptenal, (E)‐2‐pentenal) are significantly positively correlated with consumer preferences (Zhang et al., 2016; Klee and Tieman, 2018). Analysis of transcripts of key genes from the different volatile pathways showed that they were greatly reduced (Fig. 4b), including genes encoding branched‐chain aminotransferases (BCAT1) (7.9% and 9.0% of the WT level), lipoxygenase C (LoxC) (5.4%, 3.5%), LoxB (6.9%, 8.0%), hydroperoxide lyase (HPL) (41.4%, 48.5%), alcohol dehydrogenase 2 (ADH2) (69.5%, 72.5%), alcohol acetyltransferase 1 (AAT1) (10.4%, 6.6%) and l‐phenylalanine ammonia lyase (PAL3) (43.8%, 33.9%). Several of these genes have been identified previously as direct targets of RIN including LoxC, HPL, AAT1 and BCAT1, which also could be induced by ET in RIN‐CRISPR fruits (Fujisawa et al., 2012; Zhong et al., 2013; Fig. 4b).
Figure 4.
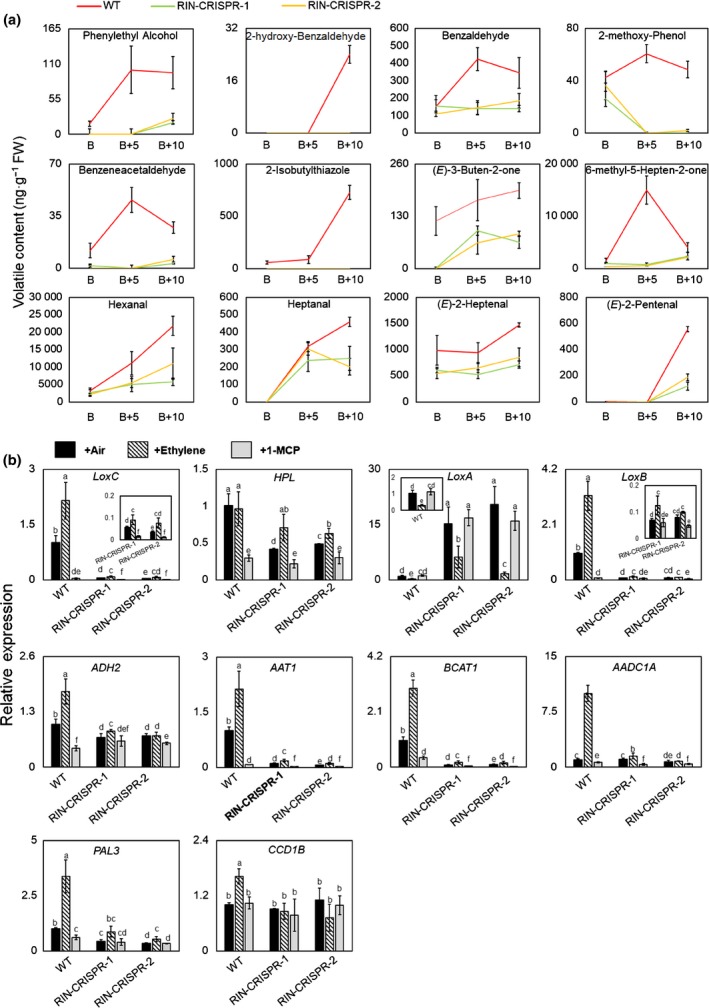
Volatile content and quantitative reverse transcription (qRT)‐PCR assay of genes involved in volatile formation during ripening of wild‐type (WT) and RIPENING INHIBITOR (RIN)‐deficient tomato fruits. (a) Aroma volatiles produced from different pathways including amino acid (phenylethyl alcohol, 2‐hydroxy‐benzaldehyde, 2‐methoxy‐phenol, benzeneacetaldehyde, benzaldehyde, 2‐isobutylthiazole), carotenoid (6‐methyl‐5‐hepten‐2‐one, (E)‐3‐buten‐2‐one), lipid (hexanal, heptanal, (E)‐2‐heptenal, (E)‐2‐pentenal). Volatiles were measured by GC or GC‐MS. (b) qRT‐PCR of gene transcripts involved in major tomato fruit pathways forming volatiles, relative to the expression of the tomato ACTIN gene as internal control, expressed as 2‐ΔΔCt (Livak and Schmittgen, 2001). Lipoxygenase (LoxA, LoxB, LoxC); hydroperoxide lyase (HPL); alcohol dehydrogenase 2 (ADH2); alcohol acetyltransferase 1 (AAT1); branched‐chain aminotransferases (BCAT1); l‐phenylalanine ammonia lyase (PAL3); aromatic amino acid decarboxylase (AADC1A); carotenoid cleavage dioxygenase 1 (CCD1B). RNA‐Seq has shown previously that genes such as carboxymethylesterase (carboxylesterase 1 (CXE1)); PAL2,4,6 have only low transcript levels and PAL1,5 are unaffected if RIN is silenced (Li et al., 2018). The error bars represent Mean ± SD, the lowercase letters indicate significant difference at P = 0.05.
RIN‐deficient fruits soften extensively
Softening is a major component of fleshy fruit ripening. At early ripening stages, WT and RIN‐CRISPR tomato fruits softened at a similar rate and pericarp firmness of mutant lines, measured by penetrometer, was not significantly different from that of the WT fruit from breaker (B) to breaker + 10 (B + 10) (Fig. 5a). Up to 35 d post‐harvest there was no significant difference in water loss between RIN‐deficient and WT fruits during prolonged post‐harvest storage (Fig. 5b). Transcripts of cellulase (CEL2) were greatly reduced in RIN‐deficient fruits (< 1% WT) and polygalacturonase (PG) transcripts also were initially strongly reduced but reached higher levels when fruits were picked at late stages (Fig. 6). Transcripts of other cell wall‐related genes such as expansin 1 (EXP1), pectate lyase (PL), β‐d‐xylosidase (XYL1), β‐mannosidase (Mside1) and pectinmethylesterase (PE1/PME1.9) also were decreased but still accumulated to relatively high levels. CEL2, EXP1, PL and XYL1 have all been reported to be directly targeted by RIN (Fujisawa et al., 2013; Zhong et al., 2013). Of particular note, transcripts of endotransglucosylase/hydrolase (XTH5 and XTH8), which have not been identified among RIN targets (Fujisawa et al., 2013; Zhong et al., 2013), were significantly higher in RIN‐deficient fruits compared to WT. Over longer timescales the RIN‐deficient fruits stored less well than WT fruit (Fig. 5c). The internal structure visibly showed greater disruption and loss of integrity than WT fruits (Fig. 5d). Transcripts of other genes, such as α‐mannosidase (MAN1), endo‐1,4‐β‐mannosidase 7 (Mside7), endo‐1,4‐β‐mannanase (MAN4a), tomato β‐galactosidase (TBG4), polygalacturonase (PG) and pectinmethylesterase (PME2.1) were present at higher levels at later ripening stages in RIN‐deficient fruits compared to WT (Fig. 6). Of these, PG, TBG4, Mside7 and MAN4a have been reported to be directly targeted by RIN (Fujisawa et al., 2013; Zhong et al., 2013).
Figure 5.
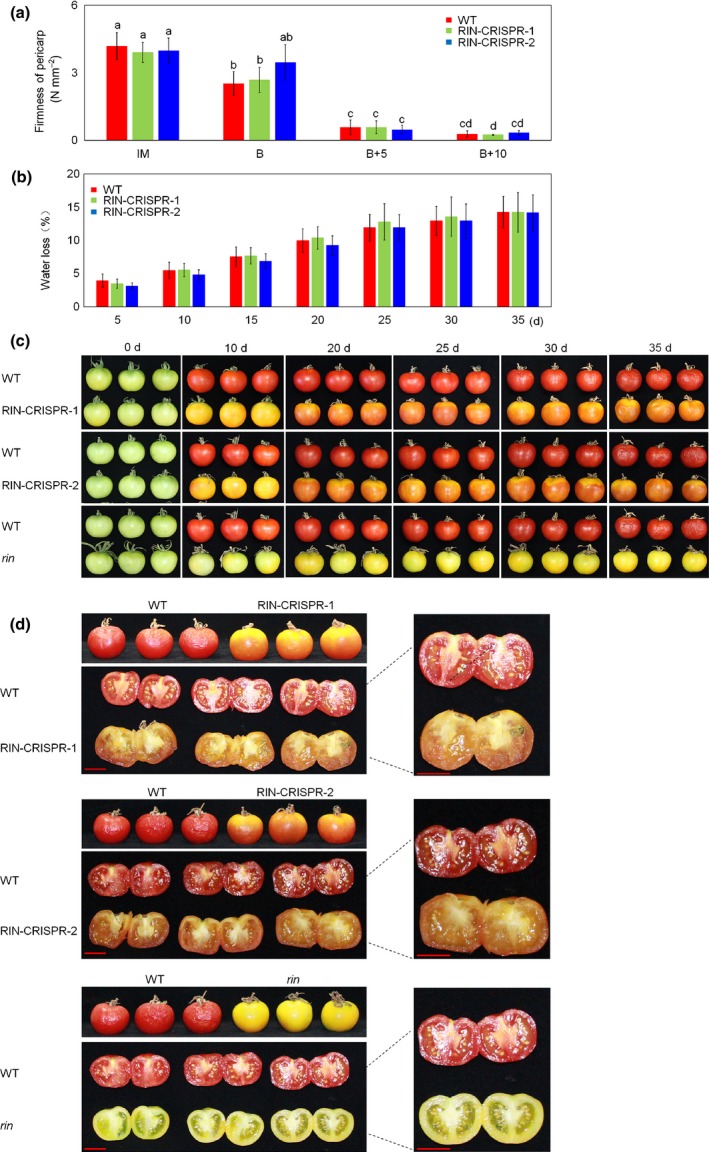
Texture change, water loss and phenotype during post‐harvest stages. (a) Fruit firmness in wild‐type (WT) and RIPENING INHIBITOR (RIN)‐CRISPR tomato measured by penetrometer. Tomato fruits from two RIN‐CRISPR homozygous mutant lines (RIN‐CRISPR‐1, RIN‐CRISPR‐2) and WT were picked at four different ripening stages, including immature (IM), breaker (B), breaker + 5 (B + 5) and breaker + 10 (B + 10). (b) Water loss by WT and RIN‐CRISPR tomato fruits during post‐harvest storage. Tomato fruits from RIN‐CRISPR‐1, RIN‐CRISPR‐2 and WT were picked at early breaker (B), and stored for 0 days (d), 10 d, 20 d, 25 d, 30 d and 35 d. Measurement of water loss was as described in Materials and Methods. The error bars represent mean ± SD. (c) Phenotype of both WT, RIN‐CRISPR and rin mutant tomato fruits during post‐harvest ripening and storage. Tomato fruits from RIN‐CRISPR‐1, RIN‐CRISPR‐2, rin mutant and WT were picked at early B stage and stored for 0 d (d), 10 d, 20 d, 25 d, 30 d and 35 d. (d) Tomato fruits from RIN‐CRISPR‐1, RIN‐CRISPR‐2, rin mutant and WT were picked at early B stage and stored for 35 d and photographed. Enlarged photos of representative samples are shown compared to WT fruits on the right. Bar, 2 cm.
Figure 6.

Transcripts of cell wall‐modifying enzymes in RIPENING INHIBITOR (RIN)‐deficient and wild‐type (WT) fruits measured by quantitative reverse transcription (qRT)‐PCR. (a) Genes whose transcripts are higher in RIN‐deficient fruits compared to WT fruits during at least one ripening stage. (b) Genes whose transcripts are lower in RIN‐deficient fruits compared to WT. Expression of genes was measured in fruits selected at the breaker (B), breaker + 5 (B + 5) and breaker + 10 (B + 10) stages. Transcript levels were determined by qRT‐PCR, relative to the expression of the tomato ACTIN gene internal control, expressed as 2‐ΔΔCt (Livak & Schmittgen, 2001): polygalacturonase (PG), expansin 1 (EXP1), cellulase (CEL2), pectinesterase (PE1/PME1.9, PME2.1), pectate lyase (PL), α‐mannosidase (MAN1), endo‐1,4‐β‐mannanase (MAN4a), β‐d‐xylosidase (XYL1), β‐mannosidase (Mside1), endo‐1,4‐β‐mannosidase 7 (Mside7), tomato β‐galactosidase (TBG4), xyloglucan endotransglucosylase/hydrolase (XTH5 and XTH8). The error bars represent mean ± SD, the lowercase letters indicate significant difference at P = 0.05.
Exogenous ET treatment enhanced the expression of several cell wall‐modifying genes in WT fruit, whereas the same treatment of RIN‐deficient fruit resulted in a much smaller upregulation of some genes, including PG, XYL1 and TBG4 (Fig. S6a), whereas Mside7 and PME1.9 transcripts were slightly reduced by ET treatment in RIN‐deficient fruits (Fig. S6b). By contrast, transcripts of CEL2, EXP1, PL, Mside1, MAN4a and PME2.1 were unaffected by supplying ET externally (Fig. S6c). Furthermore, transcripts for genes such as XTH5, XTH8 and MAN1 displayed no obvious response to ET treatment in RIN‐deficient fruits and were present at similar or higher levels than in WT (Fig. S6d) and XTH1‐4, 6, 7 were undetectable.
Discussion
Ethylene is sufficient to initiate ripening in mature RIN‐deficient fruit
Ethylene (ET) has long been implicated in the control of ripening. Ethylene action inhibitors silver (Ag+) and later 1‐methylcyclopropene (1‐MCP) slow or inhibit ripening and there is a requirement for ET for the stable and continuous expression of ripening genes in fruit expressing RIPENING INHIBITOR (RIN) (Davies et al., 1988; Grierson, 2013). Reducing expression of ACC synthase (ACS) and ACC oxidase (ACO) ET biosynthesis genes by antisense/RNAi also slowed or prevented ripening (Hamilton et al., 1990; Oeller et al., 1991; Picton et al., 1993). Kevany et al. (2007) concluded that degradation of ET receptors, which are negative regulators, could influence the timing of ripening but this analysis was performed with plants expressing an intact functional RIN gene. A striking feature revealed by the present study is that the reduced amount of endogenous ET generated by RIN‐deficient fruits is sufficient to intitiate ripening in the absence of RIN (Figs 1, 2) as proposed by Ito et al. (2017). The inhibition of colour change by 1‐MCP and the responsiveness of carotenoid biosynthesis genes such as phytoene synthase1 (PSY1) and others upon ET treatment (Figs 1, 3) confirms this directly and indicates that this is a genuine ET response. However, ET treatment of RIN‐deficient lines did not restore full ripening and, unlike the situation in wild‐type (WT) fruits, system‐2 ET production could not be induced by propylene in RIN‐CRISPR fruits (Fig. 2). The response to the ET analogue propylene is a classic test for initiation of autocatalytic ET synthesis during ripening (McMurchie et al., 1972) and the lack of a response in RIN‐deficient tomatoes indicates that RIN is required for the induction of system‐2 ET production (Fig. 2). These results are all consistent with the suggestion that the observed RIN‐independent initiation of ripening in mature RIN‐deficient fruits is actually caused by the low endogenous ET that they produce at maturity. This does not solve the ripening initiation problem completely, however, because it is known that an increase in synthesis and responsiveness to ET only occurs in mature green (MG) fruit and does not occur in immature fruit.
Discrepancies between fruits of the rin mutant and RIN‐deficient fruit
The original rin mutation was known to involve a deletion and fusion of parts of two adjacent genes, called RIN and MC (Vrebalov et al., 2002); rin was thought to correspond to a loss‐of‐function mutation and the analysis of the phenotype of rin mutant fruits was influential in shaping the conclusions about RIN function. Recent studies, however, have shown that RIN‐MC protein is an active transcription factor (TF) with a repressor function (Ito et al., 2017; Li et al., 2018).
Our data show that RIN‐deficient fruits are able to initiate ripening at a similar time to WT fruit (Fig. 1), supporting the conclusion that RIN is not required for ripening initiation, although it is needed for the progression and completion of ripening (Ito et al., 2017; Li et al., 2018). This poses the question: what initiates ripening and what is the role of RIN in ripening progression? RIN‐deficient (RIN‐CRISPR) fruits can respond to ET treatment, as indicated by changes in transcript levels of carotenoid pathway genes, required for pigment accumulation, and the accumulation of only a very limited quantity of carotenoids, especially lycopene (Fig. 3). The involvement of ET is confirmed by the demonstration that it is inhibited by the ET perception inhibitor 1‐MCP (Fig. 1d). Our study clarifies the role of RIN in the control of climacteric ripening and highlights the importance of autocatalytic system‐2 ET production (Fig. 2) for the subsequent progression of ripening and development of quality attributes such as coloured carotenoids and volatiles. The observed differences between the original rin mutants and the RIN‐CRISPR lines described in the present study relate to the strong transcription repressor activity of the chimeric RIN‐MC TF generated by the mutation (Ito et al., 2017; Li et al., 2018). Key differences are that RIN is required for volatiles production but not softening, whereas RIN‐MC, generated by the rin mutation, inhibits both.
Post‐harvest RIN‐deficient fruit soften extensively
Breeders have used the rin mutation in hybrids to reduce tomato fruit softening and prolong shelf life. In the present experiments, removal of RIN greatly reduced the production of ET, carotenoids and volatiles but extensive softening still occurred (Figs 5, 6). This is inconsistent with RIN being a ‘master regulator’ of all aspects of ripening. The accumulation of cell wall‐modifying enzymes is greatly reduced in rin mutant fruit (Knapp et al., 1989; Tucker et al., 2017) and this may be related to the repressor function of the RIN‐MC protein generated by the rin mutation (Li et al., 2018). By contrast, however, in RIN‐deficient fruits we found that transcripts of some softening enzyme genes including XTH5 and XTH8 were expressed at higher levels compared to WT at most stages of ripening. This might explain why RIN‐deficient fruits softened in a similar way to WT fruits at early ripening stages (Fig. 5). Surprisingly, early softening occurred in the almost complete absence of CEL2 and reduced levels of EXP1, PL, XYL1, Mside1 and PME1.9 transcripts (Fig. 6), although at later stages MAN1, Mside7, MAN4a, TBG4, PG and PME2.1 transcripts were significantly more abundant than in WT (Fig. 6). The XTH genes, which encode xyloglucanendo‐transglucosylase/hydrolases (XTHs: EC2.4.1.207 and/or EC3.2.1.151) have been proposed to have a dual role integrating newly secreted xyloglucan chains into an existing wall‐bound xyloglucan and restructuring the existing cell wall material by catalyzing transglucosylation between previously wall‐bound xyloglucan molecules (Miedes and Lorences, 2009; Muñoz‐Bertomeu et al., 2013). XTH5 and XTH8 are the only XTHs reported to be expressed in fruits (Miedes and Lorences, 2009) and the higher levels of XTH5 and XTH8 transcripts that we measured (Fig. 6a) might cause cell wall structural changes affecting texture or softening of RIN‐deficient fruits. The fact that these genes are expressed more highly in RIN‐deficient lines may indicate that, when present, RIN can actually repress their expression, but this needs experimental confirmation. RIN‐deficient tomato fruits showed further abnormal softening late in the ripening process (Fig. 5). This is likely to be related to the late accumulation of abnormally high levels of transcripts for several wall‐modifying enzymes, including Mside7, MAN1, MAN4a, TBG4, PG and PME2.1 transcripts (Fig. 6). Mside7 and MAN1, encoding endo‐1,4‐β‐mannosidase 7 and α‐mannosidase, respectively, are likely to affect cell wall structure, because fruits of transgenic α‐Man (MAN1) tomato RNAi lines were approx. 2.5‐fold firmer with c. 30 d extended shelf life, compared to WT, whereas overexpression of α‐Man resulted in excessive fruit softening (Meli et al., 2010).
RIN, ET acting via ET response factors (ERFs) and other TFs are involved in regulating fruit softening
Early studies showed that application of ET action inhibitors to tomatoes inhibited accumulation of PG and other gene transcripts (Davies et al., 1988; 1990) and the present findings support the conclusion that ET enhances accumulation of PG transcripts (Fig. S6). This is in disagreement with results of Oeller et al., (1991) but supports the conclusion of Sitrit & Bennet (1998). An ET response element (GCC‐box) is present in the PG gene promoter (Fig. S7). Transcripts of cell wall‐modifying genes were much lower in response to ET in RIN‐deficient fruits compared to ET‐treated WT fruits (Fig. S6). Without RIN, the fold‐increase in response to ET was similar to WT, although the actual value was much lower (Fig. S6). This may indicate that RIN and ET, acting via ERFs and possibly other TFs, are both required for maximum expression of these cell wall genes. These results support a model where RIN and ET are both required for normal softening during ripening. Direct enhancement of transcription would be expected to involve an ERF activator, but the absence of the classical ERF GCC‐box binding site in the promoters of all cell wall genes except PG has raised doubts about this possibility. There is, however, evidence for involvement of ERFs in recognizing alternative promoter motifs. Tomato MADS protein FUL1 has been shown to interact with ERF8 in vivo (José Ripoll et al., 2015), and in Arabidopsis it has been shown that the AtRAP2.2 ERF binds an ATCTA motif and not the classical GCC‐box. We note that ATCTA motifs are present in all the promoters of the cell wall genes examined (Fig. S7). Thus, full expression of cell wall enzymes may require RIN and ERFs.
Evidence from other recent work (Gao et al., 2018) has confirmed that other TFs, including NACs, also modulate expression of genes involved in cell wall metabolism. There is some evidence that RIN interacts with a NAC protein, NAC4 (Zhu et al., 2014), and may also form heterodimers with other MADS TFs, such as TAGL1, AGL11 and FUL1/2 (reviewed in Li et al., 2019). Cel2, EXP1 and MAN4a gene promoters also can be direct targets of both FUL1 and FUL2, whereas PL and XTH5 are targeted only by FUL1 (Fujisawa et al., 2014). NOR‐like1, another NAC TF, has recently been shown to directly regulate PG, PL, CEL1 and EXP1 genes and also enhances ET production (Gao et al., 2018). Thus, RIN, NAC(s) and an ERF that recognizes the ATCTA motif may all contribute to the regulation of the expression of tomato cell wall‐modifying genes.
Effect of manipulation of RIN on tomato fruit volatiles
The major reduction in aroma volatiles (Figs 4, S5) in RIN‐deficient fruits was related to the reduction in transcripts of key genes that operate in three different biosynthetic pathways to generate compounds that contribute to aroma and flavour (Fig. 4b). These include BCAT1, which is important in the production of branched‐chain amino acids (Maloney et al., 2010;Kochevenko et al., 2012; Zhang et al., 2016), transcripts of LoxC, LoxB, HPL and ADH2 in the lipid pathway, where production of the C5 volatile 1‐penten‐3‐ol is dependent upon LoxC action (Shen et al., 2014) and the C6 volatiles require sequential activity of LoxC and HPL (Chen et al., 2004). AAT1 is important in volatile esters synthesis (Yahyaoui et al., 2002; Goulet et al., 2015) and the final step in the pathway, conversion of aldehydes to alcohols, requires ADH2 activity (Speirs et al., 1998). A major group of amino acid‐derived flavour and aroma compounds, benzenoids (C6‐C1), are synthesized from l‐phenylalanine, including benzaldehyde and benzyl alcohol (Fig. S5). The first step in the biosynthesis of these compounds is catalyzed by l‐phenylalanine ammonia lyase (PAL), which converts phenylalanine to E‐cinnamic acid (Aragüez and Valpuesta, 2013) and the reduction in PAL3 transcripts in RIN‐deficient fruits is consistent with the reduced products from this pathway (Fig. 4b). AADC1A mediates the first step in the production of phenylalanine‐derived volatiles in tomato fruits (Tieman et al., 2006). These transcripts were reduced in RIN‐deficient fruits (Fig. 4b) and this is consistent with lower concentrations of aroma volatiles from the amino acid pathway (Figs 4a, S5). Although transcripts of the CCD1B gene were not significantly different in RIN‐deficient fruits, the accumulation of volatiles such as (E)‐3‐buten‐2‐one, 6‐methyl‐5‐hepten‐2‐one were reduced (Fig. 4). These compounds are carotenoid derivatives and the severely reduced carotenoid content of RIN‐deficient fruits (Ito et al., 2017; Li et al., 2018) probably explains the reduced concentrations of these volatiles.
The use of the rin mutation by tomato breeders to generate hybrids with improved shelf‐life has an adverse effect on tomato flavour (Osorio et al., 2019). Our results indicate that the normal RIN protein in WT fruit makes a major contribution to volatile formation, which suggests that if other RIN mutations were used for breeding, they also could have an adverse effect on flavour.
Models for initiation and progression of ripening and generation of quality attributes
The physiological and molecular analysis of RIN‐deficient tomato fruits sheds new light on the mechanisms underlying initiation and progression of fruit ripening. First, there is a requirement for ET to initiate ripening via a RIN‐independent route (Fig. 1). Second, this leads to the induction of RIN (Giovannoni et al., 2017; Li et al., 2018; Lü et al., 2018). The effect of ET on RIN transcripts was quantified in our previous experiments (Li et al., 2018) and we found that RIN mRNA was reduced 4‐5‐fold by treating MG WT fruit with 1‐MCP compared to the control. RIN and TAGL1 also may be involved in a positive feedback loop leading to system‐2 ET production, because ET biosynthesis genes have been reported to be directly targeted by a RIN‐TAGL1 complex (Lü et al., 2018). We propose a working model (Fig. 7a) where ET is required to initiate ripening and activate expression of a subset of ripening genes. Among these genes is the regulator MADS‐RIN, which activates a further subset of ripening genes, including those required for system‐2 autocatalytic ET synthesis, such as ACS2, ACS4 and ACO1 (Fig. S3; Liu et al., 2015). However, the inability of RIN‐deficient fruits to produce system‐2 ET is insufficient to explain their failure to ripen, because these fruit do not ripen completely, even after 15 d exposure to 100 ppm ET (Fig. 1d). This suggests that other factors also may contribute to the regulation of this process, including other phytohormones such as auxin (Shin et al., 2019). RIN promotes transcript accumulation from some genes involved in cell wall metabolism (Figs 6, S6) and ET stimulates the accumulation of others, discussed in the legend to Fig. 7(b). On the one hand, increases in PG, TBG4, PME2.1, Mside7 and MAN4a transcripts are delayed but they eventually reach very high levels in RIN‐deficient fruits (Fig. 6). XTH5 and XTH8 transcripts, on the other hand, accumulate to higher levels in the absence of RIN, suggesting that when present RIN may cause a significant reduction in their expression. It is important to test these various possibilities in order to clarify the mechanism whereby ET stimulates expression of cell wall‐modifying and other ripening‐related genes.
Figure 7.
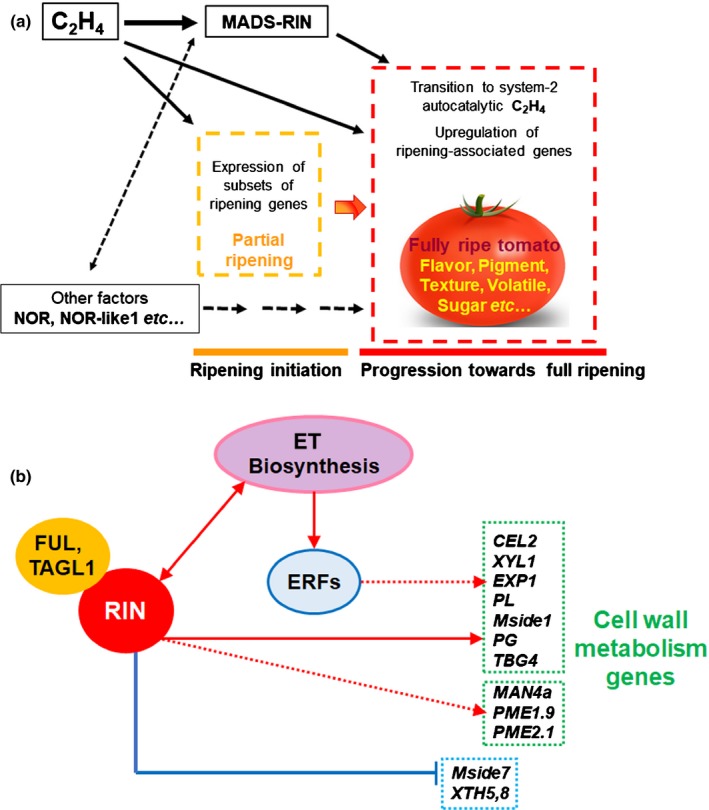
Model outlining the role of ethylene (ET) and RIPENING INHIBITOR (RIN) in initiation and progression of climacteric ripening in tomato fruit. (a) Ethylene can initiate ripening in a RIN‐independent way leading to partial ripening (data provided herein). However, RIN is required for autocatalytic system‐2 of ET production and subsequent full ripening. RIN expression is enhanced by ethylene (Giovannoni et al., 2017; Li et al., 2018; Lü et al., 2018). Other factors such as NOR, NOR‐like1, ERFs, ARFs and TAGL1, not mentioned in this model, also are involved in the ripening genetic programme (Li et al., 2019; Gao et al., 2018). (b) A model of the role of RIN and ET in regulating tomato fruit cell wall changes and softening. Accumulation of CEL2, XYL1, EXP1, PL, Mside1, PG and TBG4 transcripts is stimulated by ethylene (Fig. S6). RIN also regulates the transcription of genes involved in cell wall metabolism, such as CEL2, XYL1, EXP1, PL, Mside1 and PME1.9 (Fig. 6; S6), whereas genes such as XTH5 and XTH8 are expressed at a higher level in the absence of RIN. By contrast, increases in transcripts of PG, TBG4, PME2.1, Mside7 and MAN4a are delayed but eventually reach very high levels in the absence of RIN (Fig. 6).
Author contributions
SL performed the experiments; DG and SL designed the experiments, wrote the article, analyzed the data, selected, designed and generated the figures; BZZ provided the transgenic and mutant tomato materials and participated in design of the study; CX and BZ assisted with the carotenoid and volatile measurements; MB contributed to the design of some experiments and to the analysis and discussion and presentation of the data; JP, MB and DG developed the model and edited the final document; KC participated in design of the study and provided support for the project and the tomato ripening program. All authors approved the article.
Supporting information
Fig. S1 Construction of RIN‐CRISPR tomato mutants.
Fig. S2 Phenotype of RIN‐CRISPR tomato fruits.
Fig. S3 qRT‐PCR assay of genes involved in ET biosynthesis, perception and signaling.
Fig. S4 Colour development in WT and RIN‐CRISPR tomato fruits.
Fig. S5 Biochemical origin of tomato fruit volatiles and the content in WT, RIN‐CRISPR‐1 and RIN‐CRISPR‐2 tomato fruits.
Fig. S6 Transcripts of cell wall‐modifying enzymes in RIN‐deficient and WT fruits treated with ET and 1‐MCP measured by qRT‐PCR.
Fig. S7 RIN and ERF binding motifs in promoters of cell wall‐metabolizing genes.
Table S1 Primer pairs for vector construction and target site mutation analysis.
Table S2 Primer pairs for qRT‐PCR assay.
Table S3 Volatile components of AC, RIN‐CRISPR‐1 and RIN‐CRISPR‐2 tomato fruits at B, B+5 and B+10 stages.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank the Chinese National Key Research and Development Program (2016YFD0400102) for support to KC and the China Postdoctoral Science Foundation (2018M632479) for support to SL. The study also was supported by the 111 project (B17039). We thank Rong Jin (Agricultural Experiment Station, Zhejiang University) for support with growing facilities.
Contributor Information
Mondher Bouzayen, Email: bouzayen@ensat.fr.
Kunsong Chen, Email: akun@zju.edu.cn.
Donald Grierson, Email: donald.grierson@nottingham.ac.uk.
References
- Aragüez I, Valpuesta V. 2013. Metabolic engineering of aroma components in fruits. Biotechnology Journal 8: 1144–1158. [DOI] [PubMed] [Google Scholar]
- Bird CR, Ray JA, Fletcher JD, Boniwell JM, Bird AS, Teulieres C, Blain I, Bramley PM, Schuch W. 1991. Using antisense RNA to study gene function: inhibition of carotenoid biosynthesis in transgenic tomatoes. Nature Biotechnology 9: 635–639. [Google Scholar]
- Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D. 2004. Identification of a specific isoform of tomato lipoxygenase. TomloxC involved in the generation of fatty acid‐derived flavor compounds. Plant Physiology 136: 2641–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Sun YF, Kai WB, Liang B, Zhang YS, Zhai XW, Jiang L, Du YW, Leng P. 2016. Interactions of ABA signaling core components. SlPYLs, SlPP2Cs, and SlSnPK2s. in tomato Solanum lycopersicon . Journal of Plant Physiology 205: 67–74. [DOI] [PubMed] [Google Scholar]
- Davies KM, Hobson GE, Grierson D. 1988. Silver ions inhibit the ethylene‐stimulated production of ripening‐related mRNAs in tomato. Plant, Cell & Environment 11: 729–738. [Google Scholar]
- Davies KM, Hobson GE, Grierson D. 1990. Differential effect of silver ions on the accumulation of ripening related mRNAs in tomato. Journal of Plant Physiology 135: 708–713. [Google Scholar]
- Fray RG, Grierson D. 1993. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co‐suppression. Plant Molecular Biology 22: 589–602. [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y. 2013. A large‐scale identification of direct targets of the tomato MADS‐Box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Higuchi N, Nakano T, Koyama Y, Kasumi T, Ito Y. 2012. Direct targets of the tomato‐ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235: 1107–1122. [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito Y. 2014. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wei W, Zhao X, Tan X, Fan Z, Zhang Y, Jing Y, Meng L, Zhu B, Zhu H et al. 2018. A NAC transcription factor, NOR‐like 1, is a new positive regulator of tomato fruit ripening. Horticulture Research 5: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J, Nguyen C, Ampofo B, Zhong S, Fei Z. 2017. The epigenome and transcriptional dynamics of fruit ripening. Annual Review of Plant Biology 68: 61–84. [DOI] [PubMed] [Google Scholar]
- Giovannoni J. 2007. Fruit ripening mutants yield insights into ripening control. Current Opinion in Plant Biology 10: 283–289. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. 2004. Genetic regulation of fruit development and ripening. Plant Cell 14: S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet C, Kamiyoshihara Y, Lam N, Richard T, Taylor M, Tieman D, Klee H. 2015. Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Molecular Plant 8: 153–162. [DOI] [PubMed] [Google Scholar]
- Grierson D. (2013) Ethylene and the control of fruit ripening In: Seymour GB, Poole M, Giovannoni JJ, Tucker GA, eds. The molecular biology and biochemistry of fruit ripening. Oxford, UK: John Wiley and Sons; (CAB International, 2014), 43–73. Published Online: 5 Apr 2013 doi: https://10.1002/9781118593714.ch3. [Google Scholar]
- Grierson D. 2014. Ethylene biosynthesis In: Nath P, Bouzayen M, Mattoo AK, Pech JC, eds. Fruit ripening physiology, signalling and genomics. Oxfordshire, UK: CAB International, 178–192. [Google Scholar]
- Hamilton A, Lycett GW, Grierson D. 1990. Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346: 284–287. [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. 2008. DNA‐binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit‐ripening regulator RIN. The Plant Journal 55: 212–223. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nishizawa‐Yokoi A, Endo M, Mikami M, Shima Y, Nakamura N, Kotake‐Nara E, Kawasaki S, Toki S. 2017. Re‐evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nature Plants 3: 866–874. [DOI] [PubMed] [Google Scholar]
- José Ripoll J, Bailey LJ, Mai QA, Wu SL, Hon CT, Chapman EJ, Ditta GS, Estelle M, Yanofsky MF. 2015. microRNA regulation of fruit growth. Nature Plants 1: 15036. [DOI] [PubMed] [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ. 2007. Ethylene receptor degradation controls the timing of ripening in tomato fruit: Tomato ethylene receptor degradation. The Plant Journal 51: 458–467. [DOI] [PubMed] [Google Scholar]
- Kimura S, Sinha N. 2008. Tomato Transformation. Cold Spring Harbor Protocols 2008: t5084. [Google Scholar]
- Klee H, Giovannoni J. 2011. Genetics and control of tomato fruit ripening and quality attributes. Annual Review of Genetics 45: 41–59. [DOI] [PubMed] [Google Scholar]
- Klee HJ, Tieman DM. 2018. The genetics of fruit flavour preferences. Nature Reviews Genetics 19: 347–356. [DOI] [PubMed] [Google Scholar]
- Knapp J, Moureau P, Schuch W, Grierson D. 1989. Organisation and expression of polygalacturonase and other ripening related genes in Ailsa Craig, ‘Neverripe’ and ‘Ripening Inhibitor’ tomato mutants. Plant Molecular Biology 12: 105–116. [DOI] [PubMed] [Google Scholar]
- Kochevenko A, Klee HJ, Fernie AR, Araújo WL. 2012. Molecular identification of a further branched‐chain aminotransferase 7 (BCAT7) in tomato plants. Journal of Plant Physiology 169: 437–443. [DOI] [PubMed] [Google Scholar]
- Komatsu T, Mohammadi S, Busa LS, Maeki M, Ishida A, Tani H, Tokeshi M. 2016. Image analysis for a microfluidic paper‐based analytical device using the CIE L*a*b* color system. The Analyst 141: 6507–6509. [DOI] [PubMed] [Google Scholar]
- Kumar R, Sharma MK, Kapoor S, Tyagi AK, Sharma AK. 2012. Transcriptome analysis of rin mutant fruit and in silico analysis of promoters of differentially regulated genes provides insight into LeMADS‐RIN‐regulated ethylene‐dependent as well as ethylene‐independent aspects of ripening in tomato. Molecular Genetics and Genomics 287: 189–203. [DOI] [PubMed] [Google Scholar]
- Li S, Chen K, Grierson D. 2019. A critical evaluation of the role of ethylene and MADS transcription factors in the network. New Phytologist 221: 1724–1741. [DOI] [PubMed] [Google Scholar]
- Li S, Xu H, Ju Z, Cao D, Zhu H, Fu D, Grierson D, Qin G, Luo Y, Zhu B. 2018. The RIN‐MC fusion of MADS‐Box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiology 176: 891–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme‐Takagi M, Perata P. 2013. APETALA2/Ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist 199: 639–649. [DOI] [PubMed] [Google Scholar]
- Lincoln J, Fischer R. 1988. Regulation of gene expression by ethylene in wild‐type and rin tomato Lycopersicon esculentum fruit. Plant Physiology 88: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MC, Chen Y, Chen Y, Shin JH, Mila I, Audran C, Zouine M, Pirrello J, Bouzayen M. 2018. The tomato ethylene response factor Sl‐ERF.B3 integrates ethylene and auxin signalling via direct regulation of Sl‐Aux/IAA27. New Phytologist 219: 631–640. [DOI] [PubMed] [Google Scholar]
- Liu MC, Pirrello J, Chervin C, Roustan JP, Bouzayen M. 2015. Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiology 169: 2380–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lü PT, Yu S, Zhu N, Chen YR, Zhou BY, Pan Y, Tzeng D, Fabi JP, Argyris J, Garcia‐Mas J et al. 2018. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nature Plants 4: 784–791. [DOI] [PubMed] [Google Scholar]
- Luo Z, Zhang J, Li J, Yang C, Wang T, Ouyang B, Li H, Giovannoni J, Ye Z. 2013. A STAY‐GREEN protein SlSGR1 regulates lycopene and b‐carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytologist 198: 442–452. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y et al. 2015. A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Ma Y, Zhou L, Wang Z, Chen J, Qu G. 2016. Oligogalacturonic acids promote tomato fruit ripening through the regulation of 1‐aminocyclopropane‐1‐carboxylic acid synthesis at the transcriptional and post‐translational levels. BMC Plant Biology 16: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney GS, Kochevenko A, Tieman DM, Tohge T, Krieger U, Zamir D, Taylor MG, Fernie AR, Klee HJ. 2010. Characterization of the branched‐chain amino acid aminotransferase enzyme family in tomato. Plant Physiology 153: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni J. 2011. The tomato MADS‐Box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING‐dependent manner. Plant Physiology 157: 1568–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchie EJ, McGlasson WB, Eaks IL. 1972. Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237: 235–236. [DOI] [PubMed] [Google Scholar]
- Meli VS, Ghosh S, Prabha TN, Chakraborty N, Chakraborty S, Datta A. 2010. Enhancement of fruit shelf life by suppressing N‐glycan processing enzymes. Proceedings of the National Academy of Sciences, USA 107: 2413–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E, Lorences EP. 2009. Xyloglucan endotransglucosylase/hydrolases (XTHs) during tomato fruit growth and ripening. Journal of Plant Physiology 166: 489–498. [DOI] [PubMed] [Google Scholar]
- Mou W, Li D, Bu J, Jiang Y, Khan ZU, Luo Z, Mao L, Ying T. 2016. Comprehensive analysis of ABA effects on ethylene biosynthesis and signalling during tomato fruit ripening. PLoS ONE 11: e0154072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz‐Bertomeu J, Miedes E, Lorences EP. 2013. Expression of xyloglucan endotransglucosylase/hydrolase (XTH) genes and XET activity in ethylene treated apple and tomato fruits. Journal of Plant Physiology 170: 1194–1201. [DOI] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A. 1991. Reversible inhibition of tomato fruit senescence by antisence RNA. Science 254: 437–439. [DOI] [PubMed] [Google Scholar]
- Osorio S, Carneiro RT, Lytovchenko A, McQuinn R, Sørensen I, Vallarino JG, Giovannoni JJ, Fernie AR, Rose JKC. 2019. Genetic and metabolic effects of ripening mutations and vine detachment on tomato fruit quality. Plant Biotechnology Journal. doi: 10.1111/pbi.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton S, Barton SL, Bouzayen M, Hamilton AJ, Grierson D. 1993. Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene‐forming enzyme transgene. The Plant Journal 3: 469–481. [Google Scholar]
- Qin GZ, Wang YY, Cao BH, Wang WH, Tian SP. 2012. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. The Plant Journal 70: 243–255. [DOI] [PubMed] [Google Scholar]
- Shen J, Tieman D, Jones JB, Taylor MG, Schmelz E, Huffaker A, Bies D, Chen K, Klee HJ. 2014. A 13‐lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. Journal of Experimental Botany 65: 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Mila I, Liu M, Rodrigues M, Vernoux T, Pirrello J, Bouzayen M. 2019. The RIN‐regulated small auxin‐up RNA SAUR69 is involved in the unripe‐to‐ripe phase transition of tomato fruit via enhancing sensitivity to ethylene. New Phytologist 222: 820–836. [DOI] [PubMed] [Google Scholar]
- Sitrit Y, Bennet AB. 1998. Regulation of tomato fruit polygalacturonase mRNA accumulation by ethylene: a re‐examination. Plant Physiology 116: 1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs J, Lee E, Holt K, Yong‐Duk K, Steele Scott N, Loveys B, Schuch W. 1998. Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiology 117: 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H. 2006. A highly efficient transformation protocol for Micro‐Tom, a model cultivar for tomato functional genomics. Plant and Cell Physiology 47: 426–431. [DOI] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ. 2006. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2‐phenylethanol and 2‐phenylacetaldehyde. Proceedings of the National Academy of Sciences, USA 103: 8287–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker G, Yin X, Zhang A, Wang M, Zhu G, Liu X, Xie X, Chen K, Grierson D. 2017. Ethylene and fruit softening. Food Quality and Safety 1: 253–267. [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. 2002. A MADS‐box gene necessary for fruit ripening at the tomato ripening‐inhibitor (rin) locus. Science 296: 343–346. [DOI] [PubMed] [Google Scholar]
- Wang R, Martinelli E, Angenent G, Lammers M, de Maagd A. 2019. Re‐evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9‐mutagenesis. Scientific Reports 9: 1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang W, Cai J, Zhang Y, Qin G, Tian S. 2014. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin‐conjugating enzymes in fruit ripening. Genome Biology 15: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Maass D, Voegel T, DellaPenna D, Beyer P. 2007. Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiolog 145: 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fraser P, Wang W, Bramley P. 2006. Difference in the carotenoid content of ordinary citrus and lycopene‐accumulating mutants. Journal of Agricultural and Food Chemistry 54: 5474–5481. [DOI] [PubMed] [Google Scholar]
- Yahyaoui FE, Wongs‐Aree C, Latché A, Hackett R, Grierson D, Pech JC. 2002. Molecular and biochemical characteristics of a gene encoding an alcohol acyl‐transferase involved in the generation of aroma volatile esters during melon ripening. European Journal of Biochemistry 269: 2359–2366. [DOI] [PubMed] [Google Scholar]
- Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y. 2009. Ripening‐associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. Journal of Experimental Botany 60: 3433–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Shen JY, Wei WW, Xi WP, Xu CJ, Ferguson I, Chen KS. 2010. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. Journal of Agricultural and Food Chemistry 58: 6157–6165. [DOI] [PubMed] [Google Scholar]
- Zhang B, Tieman DM, Jiao C, Xu Y, Chen K, Fei Z, Giovannoni JJ, Klee HJ. 2016. Chilling‐induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proceedings of the National Academy of Sciences, USA 113: 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Xie Z, Zhu K, Xu Q, Deng X, Pan Z. 2015. Isolation and characterization of carotenoid cleavage dioxygenase 4 genes from different citrus species. Molecular Genetics and Genomics 290: 1589–1603. [DOI] [PubMed] [Google Scholar]
- Zhong SL, Fei ZJ, Chen YR, Zheng Y, Huang MY, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J et al. 2013. Single‐base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nature Biotechnology 31: 154–159. [DOI] [PubMed] [Google Scholar]
- Zhu B, Yang Y, Li R, Fu D, Wen L, Luo Y, Zhu H. 2015. RNA sequencing and functional analysis implicate the regulatory role of long non‐coding RNAs in tomato fruit ripening. Journal of Experimental Botany 66: 4483–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen G, Zhou S, Tu Y, Wang Y, Dong T, Hu Z. 2014. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant and Cell Physiology 55: 119–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Construction of RIN‐CRISPR tomato mutants.
Fig. S2 Phenotype of RIN‐CRISPR tomato fruits.
Fig. S3 qRT‐PCR assay of genes involved in ET biosynthesis, perception and signaling.
Fig. S4 Colour development in WT and RIN‐CRISPR tomato fruits.
Fig. S5 Biochemical origin of tomato fruit volatiles and the content in WT, RIN‐CRISPR‐1 and RIN‐CRISPR‐2 tomato fruits.
Fig. S6 Transcripts of cell wall‐modifying enzymes in RIN‐deficient and WT fruits treated with ET and 1‐MCP measured by qRT‐PCR.
Fig. S7 RIN and ERF binding motifs in promoters of cell wall‐metabolizing genes.
Table S1 Primer pairs for vector construction and target site mutation analysis.
Table S2 Primer pairs for qRT‐PCR assay.
Table S3 Volatile components of AC, RIN‐CRISPR‐1 and RIN‐CRISPR‐2 tomato fruits at B, B+5 and B+10 stages.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


