Abstract
Circular RNAs (circRNAs) represent a new class of usually noncoding transcripts with largely unknown functions. Their research is hampered not least by the inapplicability of traditional analytical methods. Herein we describe a rapid and easy assay for the detection of natural circRNA, based on rolling‐circle amplification (RCA). This technique does not require the use of fluorescently labeled RNA or DNA and can specifically detect circular RNA in the presence of a 1000‐fold excess of the same linear RNA. Only standard devices such as (quantitative) PCR cyclers and gel electrophoresis are used.
Keywords: circRNA, polymerase chain reaction, quantitative PCR, RNA analysis, rolling-circle amplification
Let it roll: Circular RNAs (circRNAs) represent a new class of usually noncoding transcripts with largely unknown functions. Their research is hampered not least by the inapplicability of traditional analytical methods. Herein we describe a rapid and easy assay for the detection of natural circRNA, based on rolling‐circle amplification (RCA) and which uses only standard methods such as qPCR and gel electrophoresis.

Circular RNAs (circRNAs) are a huge class of transcripts with unique structure and largely unknown functions. CircRNAs are formed when the head and the tail of linear RNA are covalently linked together. The first circRNAs were identified in 1976 in an electron microscopy‐based study of RNA viruses,1 but the relevance of those molecules was recognized much later with the proof of circRNAs in eukaryotic organisms.2 They are often expressed in cytoplasm with a size ranging from a few dozen to several thousands of nucleotides2c, 3 and they are naturally generated by a back‐splicing reaction, which contains a joining of a 5′ splice site with a 3′ splice side of an upstream intron.4 CircRNAs have the capability to interact with RNA binding proteins or to bind other nucleic acids, like linear RNA. The key difference between linear and circular RNAs is their life time in living cells. CircRNAs can have lifetimes of more than 24 hours due to the lack of 5′ and 3′ ends that would define them as the substrate of exonucleases.3 In contrast, the half‐life of linear RNAs in the cytoplasm is usually less than 30 minutes.5 Only a few members of the circRNA families have already been characterized, for example CDR1 or ciRS‐7 which acts as a microRNA sponge with over 70 specific binding sites for human miRNA miR‐7. This suggests that CDR1/ciRS 7 acts as a storage repository and regulator of miR‐7 activity.6 This mechanism implies a role of CDR1/ciRS 7 in neurological disorders such as Parkinson's disease and brain tumor development.6 Many other circRNAs are involved in various diseases, like hsa_circ_0002062 and hsa_circ_0022342 in chronic thromboembolic pulmonary hypertension7 or circPVT1 in gastric cancer.8
Given their broad prevalence, role in human disorders and unique properties, further characterization of circRNAs is particularly interesting. However, some of the properties of circRNAs make their further investigation difficult, especially because many classical methods are not applicable. Unlike miRNAs or other siRNAs, circular RNAs are not easily isolated from other RNAs by size or electrophoretic mobility. Also, common techniques like poly(A) enrichment for RNA‐seq studies or rapid amplification of cDNA ends (RACE), which are based on polyadenylated free RNA ends, are not applicable for circRNAs due to the lack of 5′ and 3′ ends. Therefore, most techniques for the investigation of circRNAs are based on sequencing methods, although they are time‐consuming, intricate and laborious. Another problem of many techniques is the need for RNase R treatments to differentiate the circRNA from its linear counterpart. RNase R is an exonuclease capable of degrading linear RNAs, but not circular RNAs. However, this treatment has the possibility of incomplete degradation, which would be associated with false‐positive readout due to linear RNA. Recently, a chemical biology approach to circRNA detection based on specifically tailored fluorescent probes has been reported.9
Herein we present a fast and simple method for the specific detection of natural circular RNA by means of rolling‐circle amplification (RCA) without the need for fluorescent probes or specific equipment. RCA is an established method for isothermal DNA/RNA amplification usually based on a cyclic DNA template and a complementary primer that triggers the DNA polymerase‐driven formation of a linear single‐stranded (ss) DNA product. The DNA strand is continuously elongated by the displacement of its own more in 5′ direction located areas from the template. A key step for this technique is the formation of the circular DNA probe, which can be done prior to RCA or in situ in order to increase specificity when using RCA as an analytical method for the detection of nucleic acids. We have previously used this principle in our studies with noncoding RNA and established assays for microRNA maturation10 and AGO2‐mediated nuclease activity.11 In the course of our investigations on the circularization of (specifically labeled) RNA, we were looking for a method with which circularization could be detected easily, reliably and sensitively. We therefore considered whether circular RNA in combination with reverse transcriptase could be used for RCA (Figure 1). In fact, other groups have already reported the occurrence of so‐called concatemers, which are an indication of an RCA mechanism, after RT PCR.12 For a specific analysis of circularization, the concatemer formed would have to be significantly larger than any copies of the single‐stranded RNA formed. To achieve the most effective differentiation possible, we opted for the branched rolling‐circle amplification (BRCA) format, in which the initially formed cDNA is again amplified by a specific set of primers.13 We chose CRKL as an exemplary and—due to its length of 466 nucleotides—also challenging template. CRKL14 (v‐crk sarcoma virus CT10 oncogene homologue (avian)‐like) is a protein with important stimulatory roles in cancer progression and tumor metastasis.15 Circular CRKL is formed by backsplicing of an exon of the CRKL pre‐mRNA in competition to splicing leading to linear CRKL mRNA. The role of the circular CRKL, also designated as hsa_circRNA_062400 is currently unknown, but it has been shown recently, that it is significantly downregulated in patients with tuberculosis infection.15
Figure 1.
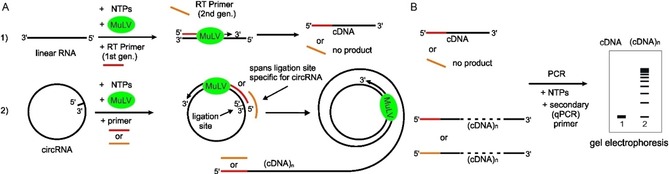
Schematic illustration of the rolling‐circle amplification assay with circular RNA in comparison with linear RNA. A) Reverse transcription of circRNA amplifies a multimeric cDNA by rolling‐circle mechanism but only a monomeric cDNA with linear RNA template by normal amplification, if first‐generation primers (red) are used. The second‐generation RT primer (orange) spans the ligation site of circRNA and is specific to circRNA. B) Gel electrophoresis shows multimeric cDNA (concatemer) only for the circRNA template, confirming RCA mechanism. MuLV (green)=reverse transcriptase.
During our investigations in synthetic circular RNA,16 we optimized the preparation protocol. The sequence of interest was the natural human circular RNA CRKL (466‐mer). First, CRKL was produced by in vitro transcription using a respective DNA plasmid. The linear RNA with 5′‐triphosphate end was dephosphorylated by alkaline phosphatase CIP and purified by PAGE in order to remove CIP. Next, the linear 5′‐hydroxy‐CRKL was divided into two aliquots, with one serving as linear control.
The other portion was used for the production of circular CRKL. To this end, the RNA was phosphorylated by T4 polynucleotide kinase (NEB) and the 5′‐monophosphorylated linear CRKL was then circularized by using T4 RNA Ligase 2 (NEB). The final circular product was purified by gel isolation after denaturing PAGE. Optical analysis by gel staining showed specific ligation and about 80 % circularization of CRKL. The RNA formed showed a migration in the gel corresponding to a linear transcript of more than 1000 nucleotides in length (Figure S1).
To test whether the circCRKL would be suitable for amplification using reverse transcriptase according to the mechanism of an RCA, the purified linear and circular versions of CRKL were used as templates. A first‐generation RT primer (Table S1) and the MuLV reverse transcriptase, included in high‐capacity cDNA reverse transcription kit (Thermo Fisher), present in our laboratory were used to start the synthesis of corresponding cDNA (Figure 1 A). The first‐generation RT primer was designed in such a way that it could bind equally well to linear or circular version of CRKL. Circular or linear CRKL were used at a concentration of 17 fmol, treated with MuLV and then the raw reactions were amplified by real‐time PCR using a second qPCR primer (Table S1, Figure 1 B).
The qPCR showed a cT value of 16.2 for circular and 27.9 for linear CRKL, or a difference of 12 cycles (Figure 2 A), although the sequence and concentration of the template was the same for both reactions. This difference can only be explained by a rolling‐circle amplification mechanism and the formation of a concatemer (Figure 1 A). To directly detect the formation of a concatemer, the various reactions after qPCR were applied to an agarose gel (Figure 2 C). While the linear CRKL provided only one 500 nt product and some small PCR products, the circCRKL template provided a variety of high molecular weight products, which is in line with our hypothesis and explain the better amplification in qPCR. Finally, the primary reaction mixture with circCRKL was treated with RNAse H, which should lead to a cleavage of the circRNA in the presence of the DNA primer. Indeed, the high molecular weight products were always present in the untreated reaction, but disappeared upon RNAse H treatment (Figure S2). Finally, different concentrations of circCRKL were used under these assay conditions and a concentration dependence could be determined (Figure 2 B). These results prove that circRNA is indeed amplified by an RCA mechanism. Next, we sought to achieve an even greater differentiation between circRNA and linear RNA. To this end, we redesigned the primer set. The second‐generation RT primer was designed as to hybridize with the 5′ region and the 3′ region of the RNA by bridging the ligation site (Figure 1). Thereby a sequence was exploited which only occurs in the circRNA and the second‐generation RT primer should therefore be unable to bind to the linear RNA and be specific to the circRNA. Indeed, after reverse transcription and subsequent PCR, the gel showed a “smear” of high molecular weight products that could clearly be attributed to concatemer formation (Figure 3 B). For the linear CRKL, however, no amplification product could be detected on the gel, so that cyclization of CRKL could be confirmed even without expensive equipment. A corresponding qPCR showed a consistent result in the form of a clear fluorescence signal for the circular CRKL or absence of such for the linear CRKL (Figure 3 A).
Figure 2.

PCR analysis of the RT rolling‐circle amplification assay. A) Control without RNA template (blue), circCRKL (green) and linear CRKL (red) 17 fmol each. B) Dose dependence circCRKL (20: blue, 10: orange, 3: grey, 1 fmol: yellow). C) Analysis of real‐time PCR samples by 1.5 % agarose gel electrophoresis: 1) DNA ladder, 2) control, 3) linear CRKL, 4) circular CRKL.
Figure 3.

A) RCA assay with redesigned (second‐generation primer) circular CRKL (green), linear CRKL (red) 4 fmol each. Control without RNA template (blue). B) 1.5 % agarorse gel of qPCR: 1) 50 bp DNA ladder, 2) negative control, 3) linear CRKL, 4) circular CRKL.
Having established reaction conditions that could clearly differentiate between circular and linear CRKL, we wanted to test, the potential of differentiation at higher RNA concentrations. At high concentrations, it should be possible for the primer to partially hybridize to the 3′ RNA segment and initiate reverse transcription or alternatively, the primer could bridge the unlinked 5′ and 3′ ends and form a padlock‐like complex, giving rise to reverse transcription. To this end, we initiated the reaction using 250 fmol linear CRKL, which was 20–50 times higher than in previous reactions, and compared this with a 1000‐fold lower concentration (250 amol) of circCRKL.
Although qPCR still showed higher sensitivity for circCRKL, the response for the 1000‐fold concentrated linear CRKL was similar and clearly showed the limitations of the method (Figure 4 A). Interestingly, this reaction mix on a 1.5 % agarose gel showed only a band of about 500 nt and some shorter by‐products (Figure 4 B). Larger transcripts were however not visible and an RCA reaction could thus be clearly ruled out with the help of the gel. Another method to eliminate larger amounts of linear RNA as an interfering factor is their degradation by RNAse R. In fact, pretreatment of linear RNA with RNAse R led to a complete loss of the qPCR signal, while the qPCR of circCRKL was not altered by this treatment, as expected (Figure 4 C). Another conclusion from this experiment was that RCA amplified the signal about 1000 times and that our assay should be more sensitive by this factor than methods based on qPCR alone.
Figure 4.

A) qPCR with 250 fmol linear CRKL (grey) and 250 amol circular CRKL (orange) and negative control (blue). B) 1.5 % agarose gel of qPCR: 1) DNA ladder, 2) linear CRKL. C) qPCR: control (blue), linear CRKL (orange), linear CRKL+RNaseR (grey), circCRKL (green), circCRKL+RNaseR (dark blue).
To detect circCRKL from cell samples, the total RNA was isolated from different cell lines (HEK293, HeLa, A549, CHO). After RNAse R treatment the samples were incubated with the primary primer and MuLV. This led in all cases to the formation of concatemers that could be easily detected by PAGE (Figure 5 A) or quantified by qPCR. Due to the presence of two independent amplification steps and a non‐homogenous template for the secondary qPCR, a calibration curve recorded under the same condition is mandatory (Figure S3). To ensure the specificity of the method also under these conditions, the RNase R treated total RNA was incubated with MuLV and two random primers, which did not result in any amplification signal (Figure 5 B).
Figure 5.

Agarose gel (1 %) of RCA assay with total RNAs from cell lysates after RNAseR treatment. A) 1) Ladder, 2) circCRKL, 3) linear CRKL, 4) HEK293, 5) HeLa, 6) A549, 7) CHO. B) 1 % agarose gel: 1) 10 kb DNA ladder, 2) circCRKL, 3) RNase R treated total RNA from HEK293 with random DNA primers.
The circular nature of for example, in vitro synthesized RNA is usually assumed simply due to a shift in gel electrophoresis and its resistance to RNAse R treatment. However, partial or full inactivity of the enzyme can lead to fundamentally false assumptions. Here we present a method for the specific detection of circular transcripts that does not require complex probes, special equipment or RNase R treatment. A rolling‐circle transcription, catalyzed by reverse transcriptase was systematically developed for this purpose. Here, the circular nature of RNA is specifically exploited in an RCA. In addition to the specific detection of cellular circRNAs without sequencing, the method is particularly suited to prove the circular nature of chemo‐enzymatically produced RNA. Although it is possible, quantitation of circRNAs is somewhat complicated, as the assay contains two independent amplification steps. This makes the use of calibration curves for each experiment mandatory. Last but not least, the extremely simple method can be used to screen for the presence of a specific circRNA from a large number of different cell lysates. By designing primers that bridge both the 5′ and 3′ ends of a transcript, the assay should be easily extendable to other target RNAs.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
The plasmid containing the CRKL sequence was kindly provided by Nikolaus Rajewsky (Berlin).
M. Boss, C. Arenz, ChemBioChem 2020, 21, 793.
References
- 1. Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K., Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., Marzluff W. F., Sharpless N. E., RNA 2013, 19, 141–157; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Nigro J. M., Cho K. R., Fearon E. R., Kern S. E., Ruppert J. M., Oliner J. D., Kinzler K. W., Vogelstein B., Cell 1991, 64, 607–613; [DOI] [PubMed] [Google Scholar]
- 2c. Salzman J., Gawad C., Wang P. L., Lacayo N., Brown P. O., PLoS One 2012, 7, e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S. D., Gregersen L. H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N., Nature 2013, 495, 333–338. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S., Mol. Cell 2014, 56, 55–66; [DOI] [PubMed] [Google Scholar]
- 4b. Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L. H., Bindereif A., Cell Rep. 2015, 10, 103–111. [DOI] [PubMed] [Google Scholar]
- 5. Shyu A. B., Greenberg M. E., Belasco J. G., Genes Dev. 1989, 3, 60–72. [DOI] [PubMed] [Google Scholar]
- 6. Hansen T. B., Wiklund E. D., Bramsen J. B., Villadsen S. B., Statham A. L., Clark S. J., Kjems J., EMBO J. 2011, 30, 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu N., Jin L., Cai J., Clin. Exp. Hypertens 2017, 39, 454–459. [DOI] [PubMed] [Google Scholar]
- 8. Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., Lyu D., Zheng B., Xu Y., Long Z., Zhou Y., Zhu H., Wang Y., He X., Shi Y., Huang S., Cancer Lett. 2017, 388, 208–219. [DOI] [PubMed] [Google Scholar]
- 9. Jiao J., Gao T., Shi H., Sheng A., Xiang Y., Shu Y., Li G., Chem. Commun. 2018, 54, 13451–13454. [DOI] [PubMed] [Google Scholar]
- 10. Neubacher S., Dojahn C. M., Arenz C., ChemBioChem 2011, 12, 2302–2305. [DOI] [PubMed] [Google Scholar]
- 11. Hesse M., Arenz C., ChemBioChem 2016, 17, 304–307. [DOI] [PubMed] [Google Scholar]
- 12. Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., Laneve P., Rajewsky N., Bozzoni I., Mol. Cell 2017, 66, 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neubacher S., Arenz C., ChemBioChem 2009, 10, 1289–1291. [DOI] [PubMed] [Google Scholar]
- 14. ten Hoeve J., Morris C., Heisterkamp N., Groffen J., Oncogene 1993, 8, 2469–2474. [PubMed] [Google Scholar]
- 15. Fu L., Dong Q., Xie C., Wang Y., Li Q., Tumour Biol. 2015, 36, 1015–1022. [DOI] [PubMed] [Google Scholar]
- 16. Müller S., Appel B., RNA Biol. 2017, 14, 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


