Abstract
Enteric pathogen–host interactions occur at multiple interfaces, including the intestinal epithelium and deeper organs of the immune system. Microbial ligands and activities are detected by host sensors that elicit a range of immune responses. Membrane‐bound toll‐like receptors and cytosolic inflammasome pathways are key signal transducers that trigger the production of pro‐inflammatory molecules, such as cytokines and chemokines, and regulate cell death in response to infection. In recent years, the inflammasomes have emerged as a key frontier in the tussle between bacterial pathogens and the host. Inflammasomes are complexes that activate caspase‐1 and are regulated by related caspases, such as caspase‐11, ‐4, ‐5 and ‐8. Importantly, enteric bacterial pathogens can actively engage or evade inflammasome signalling systems. Extracellular, vacuolar and cytosolic bacteria have developed divergent strategies to subvert inflammasomes. While some pathogens take advantage of inflammasome activation (e.g. Listeria monocytogenes, Helicobacter pylori), others (e.g. E. coli, Salmonella, Shigella, Yersinia sp.) deploy a range of virulence factors, mainly type 3 secretion system effectors, that subvert or inhibit inflammasomes. In this review we focus on inflammasome pathways and their immune functions, and discuss how enteric bacterial pathogens interact with them. These studies have not only shed light on inflammasome‐mediated immunity, but also the exciting area of mammalian cytosolic immune surveillance.
Keywords: E. coli, inflammasomes, Listeria monocytogenes, Salmonella, Shigella, Yersinia
Abbreviations
- A/E lesion
attachment and effacement lesion
- ALRs
AIM2 (absent in melanoma 2)‐like receptors
- ASC
apoptotic speck associated protein with a CARD
- BIR
baculovirus inhibitor of apoptosis protein repeat
- CARD
caspase activation and recruitment domain
- DC
dendritic cell
- GBPs
guanylate binding proteins
- GSDMD
gasdermin D
- LEE
locus for enterocyte effacement
- NAIP
nucleotide‐binding domain and leucine rich repeat containing (NLR)‐containing apoptosis‐inhibitory protein
- NF‐κB
nuclear factor kappa‐light‐chain‐enhancer of activated B cells
- NLR
nucleotide‐binding domain and leucine rich repeat containing (NLR)
- NLRCs
nucleotide‐binding domain and leucine rich repeat containing (NLR) family CARD domain‐containing proteins
- NLRPs
nucleotide‐binding domain and leucine rich repeat containing (NLR) family PYD domain‐containing proteins
- SPI
Salmonella pathogenicity island
- T3SS
type 3 secretion system
- TLRs
toll‐like receptors
- TRIM
tripartite motif
1. INTRODUCTION
Acute gastroenteritis is caused by infection of the stomach or intestinal mucosa with enteric pathogens, which are typically transmitted via contaminated food or water. It is characterised by damage to the mucosa and loss of mucosal barrier integrity, leading to malabsorption, diarrhoea and consequent dehydration (DuPont, 2009). Infectious gastroenteritis can affect people of all ages, thus having a substantial economic impact worldwide. In the USA, 179 million people suffer from acute gastroenteritis per year, leading to over 5,000 deaths (DuPont, 2009; Shane et al., 2017); in the UK the estimate is of 17 million cases per year (Tam et al., 2012). The most common causes of bacterial intestinal infections include the Gram‐negative pathogens Salmonella enterica, Shigella sp., Escherichia coli and Yersinia and the Gram‐positive pathogen Listeria monocytogenes (Kirk et al., 2015; Shane et al., 2017). While its mode of transmission is not currently established, Helicobacter pylori is the most prevalent aetiological agent of bacterial gastritis and is a major risk factor for the development of gastric malignancies (Chey, Leontiadis, Howden, & Moss, 2017).
The gastrointestinal immune system, which encompasses both immune and intestinal epithelial cells (IECs) lining the mucosa, must recognise and be activated by pathogenic insults, while remaining anergic to the presence of the endogenous microbiota. One of the mechanisms involved in this distinction is the multiprotein cytosolic complex known as the inflammasome. The inflammasome acts as a molecular platform for caspase‐1 activation and has been shown to have an increasingly important role in innate immunity since it was described in 2002 (Martinon, Burns, & Tschopp, 2002). It assembles in response to microbial or danger signals, triggering downstream signalling cascades that give rise to the release of pro‐inflammatory factors, including cytokines (e.g. interleukin‐1β [IL‐1β] and IL‐18) and alarmins (such as IL‐1α and HMGB1), as well as pyroptotic cell death (Broz & Dixit, 2016; Hayward, Mathur, Ngo, & Man, 2018). In the intestine, inflammasome signalling is functional within myeloid cells, such as macrophages, dendritic cells (DCs) and neutrophils, as well as epithelial cells, pointing to their pivotal role in the early response to pathogens.
2. THE INFLAMMASOME COMPONENTS
The assembly of inflammasomes is triggered by the recognition of a signal by a cytosolic sensor, which can be a member of the NLR family (e.g. NLRP3 or NLRC4), an ALR, or the TRIM protein PYRIN (Broz & Dixit, 2016) (Figure 1). The NLRs are further divided based on their N‐terminal protein–protein interaction domains, for example NAIPs (also known as NLRBs) contain BIR, NLRCs contain CARD and NLRPs contain pyrin motifs (except NLRP1, which contains a CARD). The pyrin domain is also present in ALRs and PYRIN. The pyrin domain mediates interactions with the adaptor ASC, a small protein that itself consists of a pyrin domain and a CARD, which promotes the recruitment of pro‐caspase‐1 to oligomerised inflammasomes. This leads to caspase‐1 oligomerisation and proximity‐induced activation via autoproteolysis (Figure 1). While non‐CARD‐containing sensors require ASC to recruit pro‐caspase‐1, NLRCs can interact via CARD and directly activate full‐length pro‐caspase‐1. This leads to ASC‐independent pyroptosis, but ASC is still required for caspase‐1 autoproteolysis and cytokine processing (Broz, von Moltke, Jones, Vance, & Monack, 2010). NAIPs can only activate inflammasomes by stimulating NLRC4, which in turn activates pro‐caspase‐1. Proteolytically activated caspase‐1 cleaves pro‐IL‐1β and pro‐IL‐18 into their bioactive forms.
Figure 1.
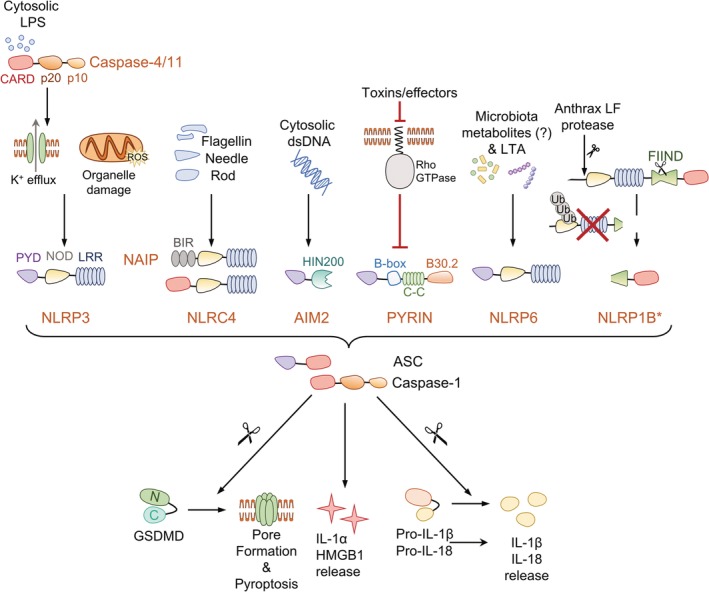
Inflammasome‐forming sensors and their known activators. Inflammasomes are multiprotein complexes that function as platforms to activate caspase‐1. Some inflammasome sensors, such as NLRP3, PYRIN and NLRP1B, are activated following perturbations of cellular homeostasis triggered by damage or microbial associated molecular patterns. For example, mitochondrial or lysosomal disruption will lead to NLRP3 activation, while inhibition of host Rho‐GTPases will allow PYRIN inflammasome assembly and degradation of the NLRP1B N‐terminal will lead to nucleation of the free CARD‐containing NLRP1B C‐terminus. Other inflammasome sensors, exemplified by AIM2, NAIP‐NLRC4 and caspase‐11 (caspase‐4 and 5 in humans), are activated in response to direct detection of their ligands: DNA is recognised by the AIM2 HIN200 domain, NAIP proteins bind flagellin and type 3 secretion system (T3SS) needles and rods, and the caspase‐11 CARD domain interacts with LPS. Active caspase‐11/4/5 cleaves Gasdermin D (GSDMD), leading to pore formation and subsequent potassium efflux, which can trigger non‐canonical activation of the NLRP3 inflammasome, and pyroptosis. NLRP6 functions as a direct sensor of lipoteichoic acid (LTA), but can also be activated by changes in the microbiota and has additionally been shown to perform inflammasome‐independent functions. *Human NLRP1 has an N‐terminal PYD domain. Domain compositions are colour coded and abbreviated as follows: CARD, caspase‐activation and recruitment domain; p20 and p10, large and small catalytic subunits; PYD, pyrin domain; NOD, nucleotide binding and oligomerisation domain; LRR, leucine rich repeat; BIR, baculovirus inhibitor of apoptosis domain; HIN200, haematopoietic expression, interferon inducible, nuclear localised (HIN) DNA binding domain of ∼200 residues; C‐C, coiled‐coil; FIIND, function to find domain
Caspase‐mediated cleavage of GSDMD liberates its N‐terminal fragment that oligomerises, binds to lipids (e.g. phosphatidylinositol phosphates, phosphatidylserine and cardiolipin) and forms large pores in the plasma membrane and mitochondria (Broz, Pelegrín, & Shao, 2019; Ding et al., 2016; Liu et al., 2016). GSDMD pores can then facilitate the release of mature cytokines and eventually lead to osmotic swelling and a form of lytic cell death called pyroptosis (Kayagaki et al., 2015; Shi et al., 2015; Evavold et al., 2018; Orning, Lien, & Fitzgerald, 2019). Not all cells with active inflammasomes undergo pyroptosis, as the endosomal sorting complexes required for transport machinery can promote repair of GSDMD‐mediate plasma membrane damage, promoting cellular survival and reducing cytokine release (Rühl et al., 2018). Notably, pyroptosis‐independent release of inflammasome‐derived cytokines has also been shown, for example from monocytes exposed to LPS (Gaidt et al., 2016) and neutrophils infected with Salmonella (Chen et al., 2014). Other mechanisms of cytokine release during inflammasome activation have been proposed, including autophagy‐associated unconventional secretion (Dupont et al., 2011; Kimura et al., 2017), exosome release from multivesicular bodies (MacKenzie et al., 2001; Qu, Franchi, Nunez, & Dubyak, 2007) and passive release after cell lysis (Cullen, Kearney, Clancy, & Martin, 2015).
3. SIGNALS FOR INFLAMMASOME ACTIVATION
It is commonly accepted that inflammasome activation in myeloid cells and the resultant IL‐1β release require two signals. The ‘priming’ signals (signal 1) activate the transcription and/or post‐translational regulation of inflammasome‐associated genes/proteins and are best understood for the activation of the NLRP3 inflammasome in macrophages. These include: (i) the transcription of pro‐IL‐1β, (ii) the transcriptional and translational licensing of NLRP3 via toll‐like receptors (TLRs) and NF‐κB (Bauernfeind et al., 2009; Fernandes‐Alnemri et al., 2013; Lin et al., 2014), (iii) interferon‐dependent up‐regulation of murine caspase‐11 (Rathinam et al., 2012; Benaoudia et al., 2019) and mouse and human GBPs (Kim et al., 2016) and (iv) IRF2‐driven expression of human caspase‐4 (Benaoudia et al., 2019) and mouse GSDMD (Kayagaki et al., 2019). Importantly, bacterial ligands such as cell wall components and nucleic acids can serve as signal 1, and pathogen‐associated virulence factors and/or activities, as discussed below, serve as a second signal specific to the NLR/ALR/PYRIN inflammasome sensors. Thus, during infection by bacterial pathogens, inflammasome signalling is a fast, inflammatory process that cooperates with other innate immune signalling pathways such as TLRs and interferons.
Several bacterial molecules can serve as signal 2 during activation of NAIPs (Figure 1) (Hayward et al., 2018; Evavold & Kagan, 2019). Bacterial flagellin and evolutionarily conserved rod and needle proteins of the type 3 secretion system (T3SS) injectisome of some Gram‐negative bacteria are recognised by the NAIPs, leading to the assembly of the NLRC4 inflammasome in macrophages (Miao, Mao, et al., 2010; Zhao et al., 2011). There are seven mouse Naip genes, four of which recognise different ligands (Kofoed & Vance, 2011; Zhao et al., 2016), while humans only encode one NAIP gene that can recognise a similar repertoire of ligands (Yang, Zhao, Shi, & Shao, 2013; Kortmann, Brubaker, & Monack, 2015; Grandjean et al., 2017; Reyes Ruiz et al., 2017).
Cytosolic double‐stranded DNA (dsDNA) is recognised by AIM2 via its HIN domain (Hornung et al., 2009). While PYRIN is autoinhibited through interactions with 14‐3‐3 proteins, it can be activated upon the inactivation of cellular RhoA GTPases by various bacterial toxins (Xu et al., 2014; Masters et al., 2016). The mouse NLRP1B sensor is also basally autoinhibited through its N‐terminal regions, which can undergo proteasomal degradation, for example in response to anthrax lethal toxin, leading to the release of the CARD‐containing C‐terminal fragment that oligomerises to recruit caspase‐1 and activate the NLRP1 inflammasome (Chui et al., 2019; Sandstrom et al., 2019; Xu et al., 2019). Other than the inhibitors of the serine proteases DPP8/9 (Okondo et al., 2017; Zhong et al., 2018), physiological activators of human NLRP1 inflammasome remain to be discovered.
The NLRP3 inflammasome responds to the widest range of stimuli, including potassium efflux, extracellular ATP, increased intracellular calcium, mitochondrial DNA and reactive oxygen species (ROS) and liberation of cathepsins from the lysosome (Swanson, Deng, & Ting, 2019). During infection by Gram‐negative bacteria, the NLRP3 inflammasome can be indirectly activated by murine caspase‐11 (represented by caspase‐4 and 5 in humans), through what is now known as the ‘non‐canonical’ inflammasome signalling pathway. In this pathway, cytosolic LPS is recognised by caspase‐4/5 or caspase‐11, which oligomerises and becomes active (Kayagaki et al., 2013; Shi et al., 2014). Activated caspase‐4/5 or caspase‐11 cleaves GSDMD, leading to pyroptosis and/or to potassium efflux and activation of the NLRP3 inflammasome in macrophages (Kayagaki et al., 2015; Shi et al., 2015). Activation of caspase‐11 has also been linked to modulation of the actin cytoskeleton, phagosome maturation and leukocyte migration (Li et al., 2007; Akhter et al., 2012), showing that this caspase also has non‐inflammatory roles.
Other less well‐studied NLRs include NLRP6, NLRP12, NLRC3 and NLRC5. In particular, NLRP6 is important in the gastrointestinal tract as it is expressed in myeloid cells, IECs and goblet cells, where it contributes to intestinal homeostasis (Elinav et al., 2011; Wlodarska et al., 2014). Notably, NLRC3, NLRP6 and NLRP12 are implicated in the suppression of the inflammatory response, and deficiency of these proteins can confer resistance to infection in vivo (Anand et al., 2012; Zaki, Man, Vogel, Lamkanfi, & Kanneganti, 2014; Zhang et al., 2014).
Inflammasomes play an important role in innate immunity, where IL‐1β, IL‐18 and alarmins (e.g. IL‐1α and HMGB1) help promote inflammation, neutrophil recruitment and an adequate adaptive immune response (Mantovani, Dinarello, Molgora, & Garlanda, 2019), while pyroptosis eliminates intracellular bacterial replicative niches (Miao, Leaf, et al., 2010). The antibacterial function of inflammasomes is illustrated by the ability of various inflammasome sensor proteins to recognise and respond to bacterial ligands (Eldridge & Shenoy, 2015; Hayward et al., 2018), and the increased susceptibility to infection shown by mice with deletions in inflammasome components (Man, Karki, & Kanneganti, 2017; Lacey & Miao, 2019). Although inflammasomes have been best studied in myeloid cells, several inflammasome components, such as caspase‐4/11 and pro‐IL‐18, are also expressed in IECs (Man, 2018; Winsor, Krustev, Bruce, Philpott, & Girardin, 2019). The constitutive expression of caspase‐4/11 and NAIPs in IECs (Knodler et al., 2014; Sellin et al., 2014) might allow rapid sensing of Gram‐negative pathogens at mucosal sites. The importance of inflammasomes in innate immune signalling in the gut in response to microbial and sterile insults, as well as the contribution of inflammasomes to diseases such as IBD, cancer and obesity and their role in the maintenance of the intestinal microbiota have been reviewed elsewhere (Levy, Kolodziejczyk, Thaiss, & Elinav, 2017; Man, 2018; Winsor et al., 2019). This review focuses on the host's detection of enteric pathogens via inflammasomes, and the mechanisms used by these pathogens to subvert inflammasome functions to promote their survival and sustain infection.
4. SUBVERSION OF INFLAMMASOMES BY ATTACHING AND EFFACING (A/E) PATHOGENS
Bacteria that cause attaching and effacing (A/E) lesions include the diarrhoeagenic human pathogens enterohaemorrhagic Escherichia coli (EHEC) and enteropathogenic E. coli (EPEC) and the rodent pathogens Citrobacter rodentium and rabbit EPEC (Frankel et al., 1998). While EPEC mainly affects young children in low‐ and middle‐income countries (Hu & Torres, 2015), EHEC infection is prevalent in industrial countries and is associated with haemorrhagic colitis and hemolytic uremic syndrome (HUS) (Kampmeier, Berger, Mellmann, Karch, & Berger, 2018).
A/E lesions are characterised by effacement of the brush border microvilli and intimate bacterial attachment to the apical membrane of IECs. Intimate attachment is mediated by strong interactions between intimin on the bacterial surface and Tir (translocated intimin receptor), which is injected into IECs by the T3SS (Frankel & Phillips, 2008; Slater, Sågfors, Pollard, Ruano‐Gallego, & Frankel, 2018), all of which are encoded within the locus for enterocyte effacement (LEE) common to A/E pathogens (Frankel et al., 1998). Intimin:Tir interactions lead to Tir clustering and actin polymerisation underneath attached bacteria, which facilitates delivery of other T3SS effectors that subvert mammalian cell processes (Shenoy, Furniss, Goddard, & Clements, 2018). Strain to strain variability in both the repertoire of T3SS effectors and other virulence factors affects the outcome of pathogen–host interactions; haemolysins (such as EhxA) and the Shiga toxins (Stx1 and 2) mentioned herein are exclusively expressed by EHEC.
While the multiplicity of virulence mechanisms employed by A/E pathogens underpins their success, it also amplifies the host's ability to sense and respond to their threat. Activation of the NLRP3 inflammasome by EPEC‐ and EHEC‐derived signals is well documented. For example, the EHEC haemolysin EhxA can cause membrane pores, potassium efflux and NLRP3‐caspase‐1 activation in human macrophages (Cheng et al., 2015). Stx also induces pyroptosis in macrophages via canonical NLRP3 activation and IL‐1β release, in addition to apoptosis via caspase‐3/8 activation (Lee et al., 2016). Like other AB5 toxins of its kind (e.g. cholera toxin) (Kayagaki et al., 2013), Stx alone can activate the inflammasome by facilitating the delivery of LPS to the macrophage cytosol, leading to activation of caspase‐4, mitochondrial ROS production and subsequent non‐canonical activation of the NLRP3 inflammasome (Platnich et al., 2018). In contrast, Vanaja et al. found only marginal NLRP3 induction by EhxA or Stx, but identified RNA:DNA hybrids as the microbial molecules that stimulate NLRP3 in murine macrophages (Vanaja et al., 2014). The different findings in these studies could be explained by different bacterial growth conditions, which can affect the levels of EhxA or Stx expression, or the use of macrophages of human vs. mouse origin. Similar responses involving mouse caspase‐11 and NLRP3 were observed with enterotoxigenic E. coli and non‐pathogenic E. coli strains, suggesting this is a broad host‐driven response to cytosolic bacterial LPS (Figure 2a; Table 1).
Figure 2.
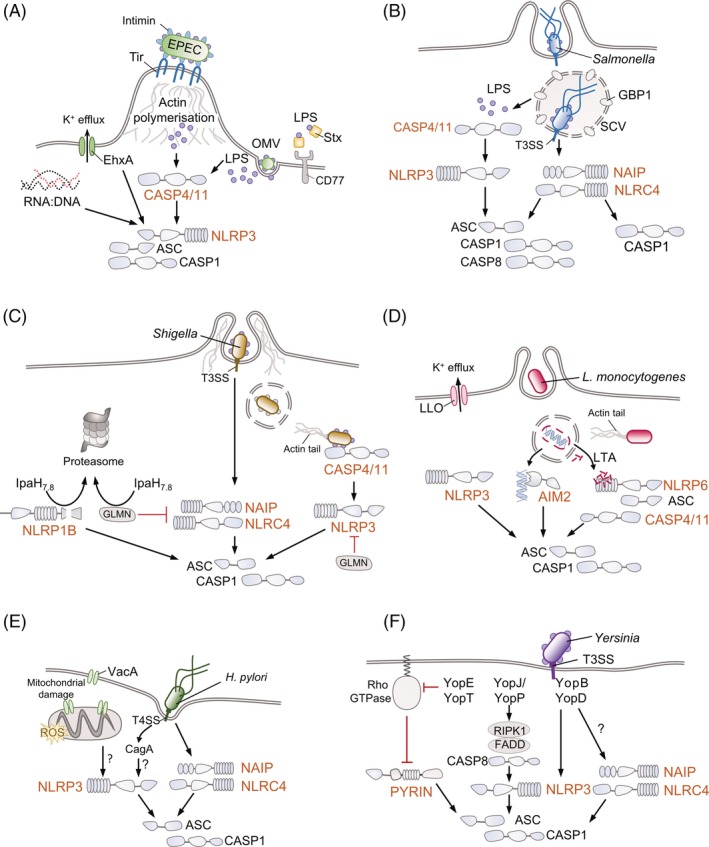
Enteric pathogens can activate multiple inflammasome pathways. Schematics from (a) to (f) show how various enteric pathogens stimulate the assembly and activation of different inflammasomes, focussing on the activating signal (signal 2). Downstream consequences of inflammasome activation shown in Figure 1, that is Gasdermin D cleavage, pore formation and pyroptosis, caspase‐1‐mediated cleavage of pro‐IL‐1β and pro‐IL‐18 into their active forms and the release of these pro‐inflammatory cytokines together with alarmins, are not depicted for simplicity. A/E pathogens (such as Enteropathogenic and Enterohaemorrhagic E. coli, EPEC and EHEC) (a), Helicobacter pylori (e) and Yersinia (f) are mainly extracellular pathogens, while Salmonella survives intracellularly in Salmonella containing vacuoles (SCVs) (b) and Shigella (c) and Listeria monocytogenes (d) are cytosolic bacteria that escape the vacuoles and can move from cell to cell via manipulation of the host actin cytoskeleton. Host proteins are featured in greyscale to emphasise the role of bacterial factors. See text for further information on the mechanisms employed by these pathogens to evade or subvert inflammasomes. OMV, outer membrane vesicle; Stx, Shiga toxin; Tir, translocated intimin receptor; GBP1, guanylate binding protein 1; GLMN, glomulin; LLO, listeriolysin; LTA, lipoteichoic acid; VacA, vacuolating cytotoxin A; CagA, cytotoxin‐associated gene A; ROS, reactive oxygen species; KD, kinase domain; RHIM, RIP (receptor‐interacting serine/threonine‐protein) homotypic interaction motif; DD, death domain; Yop, Yersinia outer protein
Table 1.
Enteric pathogens and the inflammasomes: activation and inhibition
| Pathogen | Inflammasome | Virulence factor | Effect | References |
|---|---|---|---|---|
| A/E pathogens | CASP4/Casp11 | LPS (T3SS‐independent) (EPEC/EHEC) | Activator | (Knodler et al., 2014; Vanaja et al., 2016; Goddard et al., 2019) |
| CASP4 → NLRP3 | Stx (EHEC) | (Lee et al., 2016; Platnich et al., 2018) | ||
| Tir and LPS (T3SS‐dependent) (EPEC) | (Goddard et al., 2019) | |||
| NLRP3 | Haemolysin EhxA (EHEC) | (Zhang et al., 2012; Cheng et al., 2015) | ||
| Nlrp3 | RNA:DNA (EHEC) | (Vanaja et al., 2014) | ||
| Naip2‐Nlrc4 | EscI (T3SS rod; EPEC/EHEC), EprJ (ETT2 rod; EHEC) | (Miao, Mao, et al., 2010; Zhao et al., 2011; Wu et al., 2019) | ||
| NAIP‐NLRC4 OR Naip1‐Nlrc4 | EprI (ETT2 needle; EHEC) | (Yang et al., 2013; Zhao et al., 2016; Wu et al., 2019) | ||
| NLRP3 | NleA/EspI (EPEC) | Inhibitor | (Yen et al., 2015) | |
| CASP4/Casp11 | NleF (EPEC) | (Blasche et al., 2013; Pallett et al., 2017; Pollock et al., 2017) | ||
| Salmonella Typhimurium | NAIP‐NLRC4 OR Naip5/6‐Nlrc4 | Flagellin | Activator | (Broz, Newton, et al., 2010; Miao, Leaf, et al., 2010; Kofoed & Vance, 2011; Zhao et al., 2011; Franchi et al., 2012; Kortmann et al., 2015; Reyes Ruiz et al., 2017) |
| NAIP‐NLRC4 OR Naip2‐Nlrc4 | PrgJ (inner SPI‐1 T3SS rod) | (Kofoed & Vance, 2011; Zhao et al., 2011; Franchi et al., 2012; Tenthorey, Kofoed, Daugherty, Malik, & Vance, 2014; Reyes Ruiz et al., 2017) | ||
| NAIP‐NLRC4 OR Naip1‐Nlrc4 | PrgI (SPI‐1 T3SS needle) | (Zhao et al., 2011; Yang et al., 2013) | ||
| CASP4/Casp11 → NLRP3/Nlrp3 | LPS | (Shenoy et al., 2012; Meunier et al., 2014; Fisch et al., 2019) | ||
| CASP4/Casp11 | LPS | (Knodler et al., 2010; Broz et al., 2012; Knodler et al., 2014) | ||
| Nlrp3 | Curli fibres | (Rapsinski et al., 2015) | ||
| ? (↓NADH, ↑mitoROS)* | (Sanman et al., 2016) | |||
| Nlrp3?* | SlrP | Inhibitor | (Rao et al., 2017) | |
| Casp11 → Nlrp3 | Aconitase | (Wynosky‐Dolfi et al., 2014) | ||
| Shigella | CASP4/Casp11 | LPS | Activator | (Kobayashi et al., 2013; Shi et al., 2014; Watson et al., 2019) |
| Naip2‐Nlrc4 | MxiI (T3SS rod) | (Miao, Mao, et al., 2010; Yang et al., 2013; Suzuki, Franchi, et al., 2014; Zhao et al., 2016) | ||
| Naip‐NLRC4 OR Naip1‐Nlrc4 | MxiH (T3SS needle) | (Yang et al., 2013; Zhao et al., 2016) | ||
| NLRC4/Nlrc4? | IpaB | (Senerovic et al., 2012) | ||
| NLRP3/Nlrp3 & NLRC4/Nlrc4 | IpaH7.8 | (Suzuki, Mimuro, et al., 2014; Suzuki et al., 2018) | ||
| Nlrp1b | IpaH7.8 | (Neiman‐Zenevich et al., 2017; Sandstrom et al., 2019) | ||
| CASP4 | OspC3 | Inhibitor | (Kobayashi et al., 2013) | |
| Listeria monocytogenes | NLRP3/Nlrp3 | Listeriolysin (LLO) | Activator | (Zwaferink, Stockinger, Hazemi, Lemmens‐Gruber, & Decker, 2008; Meixenberger et al., 2010; Sauer et al., 2011; Eldridge et al., 2017) |
| Nlrp3 | p60 | (Schmidt & Lenz, 2012) | ||
| RNA | (Kanneganti et al., 2006) | |||
| Aim2 | dsDNA | (Kim et al., 2010; Rathinam et al., 2010; Tsuchiya et al., 2010; Warren et al., 2010; Wu et al., 2010) | ||
| Naip‐Nlrc4 | Flagellin | (Warren, Mao, Rodriguez, Miao, & Aderem, 2008; Sauer et al., 2010; Tsuchiya et al., 2010; Warren et al., 2010) | ||
| Nlrp6‐Casp4 | Lipoteichoic acid (LTA) | (Meixenberger et al., 2010; Hara et al., 2018) | ||
| CASP4 | ?* | (Hara et al., 2018) | ||
| Nlrp1b | Host cell energy stress?* | (Neiman‐Zenevich et al., 2017) | ||
| Yersinia | Nlrc4?* (in vivo) | YopB, YopD (Yps) | Activator | (Brodsky et al., 2010) |
| Nlrp3 | YopB, YopD (Yps) | (Brodsky et al., 2010; Zwack et al., 2015) | ||
| CASP4/Casp11 | LPS (Yps) | (Casson et al., 2013; Casson et al., 2015) | ||
| Rip1‐Casp8‐Casp1 | YopJ (Yps) | (Philip et al., 2014; Weng et al., 2014) | ||
| Pyrin | YopE, YopT (Yps) | (Chung et al., 2016; Ratner et al., 2016) | ||
| Naip5‐Nlrc4 | Flagellin (Ye) | (Matusiak et al., 2015) | ||
| Nlrp3 | YopK (Yps) | Inhibitor | (Brodsky et al., 2010, Zwack et al., 2015) | |
| Pyrin/ Casp1 | YopM (Yps) | (LaRock & Cookson, 2012; Chung et al., 2014; Chung et al., 2016; Ratner et al., 2016) | ||
| NLRP3/Nlrp3 | YopH, YopE (Ye) | (Schotte et al., 2004; Thinwa et al., 2014) |
Notes: Bacterial components and infection‐mediated alterations of host cells that lead to inflammasome activation are shaded in orange; inflammasome inhibitors are shown shaded in blue. Indirect inhibitors of NLRP3, for example via inhibition of NF‐κB signalling, are not featured in the table (refer to text). H. pylori‐encoded direct activators/inhibitors of inflammasome activation not well defined, and thus the pathogen is not included. When the human inflammasome genes have been involved, these are shown in uppercase (e.g. NLRP3); mouse genes are shown in lowercase except the first letter (e.g. Nlrp3). * indicates when the study does not define which inflammasome is involved or what the bacterial activating signal/factor is.
Abbreviations: mitoROS, mitochondrial reactive oxygen species; Ye, Yersinia enterocolitica; Yps, Yersinia pseudotuberculosis.
Recently, EPEC expressing the T3SS has been shown to activate caspase‐4 and NLRP3 in human macrophages. Induction of cell death was dependent on Tir‐mediated actin polymerisation and binding of LPS to caspase‐4, leading to activation of an atypical, non‐canonical signalling pathway via GSDMD to stimulate the NLRP3 inflammasome (Goddard et al., 2019). Surprisingly, EPEC‐induced caspase‐4 activation was not sufficient to induce pyroptosis; induction of pyroptosis, as well as cytokine processing, required the NLRP3‐ASC‐caspase‐1 inflammasome. LPS stimulation by non‐pathogenic E. coli (including EPEC grown in conditions that do not stimulate virulence gene expression) takes 16–18 h to evoke a non‐canonical inflammasome response (Vanaja et al., 2016), but the translocation of a functional Tir drives non‐canonical NLRP3 inflammasome activation and IL‐1β release within 4 h (Goddard et al., 2019). It is plausible that Tir‐induced actin polymerisation during EPEC infection facilitates entry of LPS and activation of the atypical inflammasome signalling pathway. Notably, EHEC Tir, which alone does not induce actin polymerisation, can only trigger pyroptotic signalling when provided with its cognate actin polymerising effector TccP (Garmendia et al., 2004; Goddard et al., 2019). Consistent with a role for Tir‐driven actin polymerisation in inflammasome activation, over‐expression of EspJ, an ADP ribosyl transferase (Young et al., 2014) targeting multiple non‐receptor tyrosine kinases (Pollard et al., 2018) that phosphorylate and activate EPEC Tir (Swimm et al., 2004), inhibits EPEC‐induced cell death (Goddard et al., 2019) (Figure 2a). Future studies are needed to investigate the interplay between the cytoskeleton and caspase‐4/11, which has been shown to modulate actin polymerisation at the leading edge (Li, Yin, & Yuan, 2008) and cell migration (Li et al., 2007).
In addition to T3SS effectors, structural T3SS components also provoke inflammasome activation. The inner rod protein EscI of the LEE‐encoded T3SS, the inner rod protein EprJ and the needle protein EprI of the E. coli type III secretion system 2 (ETT2) activate murine NLRC4 inflammasomes in vitro when introduced directly into the macrophage cytosol (Miao, Mao, et al., 2010; Zhao et al., 2011; Yang et al., 2013; Wu et al., 2019); however, the ETT2 has accumulated considerable mutational attrition and is not believed to form a functional T3SS (Ren et al., 2004). Critically, the EPEC T3SS needle protein EscF is one of the needle proteins that are not detected in human THP‐1 macrophages (Yang et al., 2013). Similarly, the EPEC/EHEC flagellin (FliC) is not recognised by murine NAIP5/human NAIP (Zhao et al., 2011), whereas its E. coli K12 counterpart is readily detected (Yang et al., 2014). Taken together, murine NLRC4 may detect EPEC/EHEC expressing the LEE T3SS however, these bacteria likely evade this inflammasome in human macrophages.
Inflammasomes also contribute to A/E pathogen–host interactions in vivo. These studies were performed using the extracellular mouse‐specific pathogen C. rodentium (Collins et al., 2014; Mullineaux‐Sanders et al., 2019). Mice lacking NLRP3, caspase‐1, IL‐18 and IL‐1β or NLRC4 exhibit exaggerated colonic pathology and an increased bacterial burden in response to C. rodentium infection (Liu et al., 2012; Nordlander, Pott, & Maloy, 2013). Interestingly, bone‐marrow transplant experiments revealed that NLRP3 and NLRC4 activity in non‐myeloid cells is sufficient for protection against C. rodentium infection (Liu et al., 2012; Nordlander et al., 2013; Song‐Zhao et al., 2013). NLRP6, highly expressed in the colonic epithelial layer, is a major regulator of mucin secretion together with ASC and caspase‐1, and also offers protection against C. rodentium (Wlodarska et al., 2014).
Conversely, A/E pathogens encode T3SS effectors which specifically target inflammasome components. NleA (also named EspI), interacts with the pyrin and leucine‐rich repeat domains of ubiquitylated NLRP3, impairing its deubiquitylation and effectively inhibiting recruitment of caspase‐1 to the inflammasome (Yen, Sugimoto, & Tobe, 2015). Importantly, deletion of the gene encoding NleA/EspI results in severe attenuation of C. rodentium in vivo (Mundy et al., 2004). The T3SS effector NleF inhibits caspase‐4, as well as caspase‐8 and 9 (Blasche et al., 2013; Pallett et al., 2017). Inhibition of caspase‐4 by NleF blocks secretion of processed IL‐18 following infection of Caco‐2 human IEC‐like cells with EPEC at 4 days post‐infection with C. rodentium in mice, which results in reduced neutrophil recruitment (Pallett et al., 2017) (Table 1). Notably, inflammasome responses to the colonic, non‐invasive, pathogen C. rodentium, differ significantly from those triggered by Salmonella, an invasive pathogen that causes inflammation of the small intestine, as discussed next.
5. THE INFLAMMASOME AND SALMONELLA INFECTION
Salmonella enterica subsp. enterica is divided into typhoidal (e.g. S. Typhi) and non‐typhoidal (e.g. S. Typhimurium) serovars, which cause enteric (typhoid) fever and gastroenteritis, respectively (Johnson, Mylona, & Frankel, 2018). S. enterica encodes two T3SSs within pathogenicity islands called SPI‐1 and SPI‐2. The T3SS effectors injected into the host during infection target and manipulate many host pathways, allowing invasion and persistence of Salmonella in the host (Jennings, Thurston, & Holden, 2017; Pinaud, Sansonetti, & Phalipon, 2018). In particular, SPI‐1 effectors are essential for the gastrointestinal stage of the infection, generally involved in invasion of IECs and localised inflammation in the small intestine. In contrast, SPI‐2 effectors are required for the systemic spread of the infection, as they allow survival and replication of the bacteria inside host cells (McGhie, Brawn, Hume, Humphreys, & Koronakis, 2009). Salmonella therefore encounters intestinal myeloid cells, such as microfold cells (M cells), DCs and macrophages, and neutrophils and macrophages at other systemic sites.
The interaction of S. Typhimurium with inflammasomes in vitro using murine bone marrow‐derived macrophages has led to important discoveries, such as the specific and rapid activation of NLRC4 by SPI‐1‐expressing Salmonella (Mariathasan et al., 2004). We briefly summarise a large body of work in this area and refer readers to recent reviews (Crowley, Knodler, & Vallance, 2016; Wemyss & Pearson, 2019). In mouse macrophages S. Typhimurium flagellins and SPI‐1 T3SS needle and rod are strong activators of NAIP‐NLRC4 inflammasome (Franchi et al., 2012; Yang et al., 2013); reduced SPI‐1 T3SS expression results in both late and weak activation of NLRP3 and NLRC4. NLRP3 is activated through the detection of cytosolic LPS by caspase‐4/11 (Broz et al., 2012; Knodler et al., 2014) and through interferon‐inducible guanylate‐binding proteins (GBPs) (Shenoy et al., 2012; Meunier et al., 2014; Pilla et al., 2014; Santos et al., 2018). The mechanism for canonical NLRP3 activation during S. Typhimurium infection has yet to be fully elucidated, although reduced glycolytic flux caused by infection in macrophages has been proposed as a trigger (Sanman et al., 2016). In agreement with their dual roles, both NLRC4 and NLRP3 are recruited to inflammasome foci, along with caspase‐1 and caspase‐8 (Broz, Newton, et al., 2010; Man et al., 2013; Man et al., 2014; Qu et al., 2016; Bierschenk et al., 2019) (Figure 2b). Additionally, CARD9, an adaptor protein involved in immune signalling, inhibits pro‐IL‐1β expression and NLRP3 inflammasomes during S. Typhimurium infection. CARD9 acts via the spleen tyrosine kinase, which blocks ASC speck formation and caspase‐8 recruitment to inflammasome foci (Pereira, Tourlomousis, Wright, Monie, & Bryant, 2016). Macrophages are a heterogeneous cell type and tissue‐resident macrophages differ from the bone marrow‐derived macrophages often used for in vitro studies. Indeed, intestinal mononuclear phagocytes can stay unresponsive to commensal bacteria but activate the NLRC4 inflammasome in response to Salmonella and trigger neutrophil recruitment (Franchi et al., 2012).
As compared to mouse macrophages, the response of human macrophages to Salmonella infection is poorly understood. While human NAIP‐NLRC4 can detect purified S. Typhimurium needle, rod and flagellin components, the importance of NAIP during infection is less well‐defined (Kortmann et al., 2015, Reyes Ruiz et al., 2017). Infection of monocytes with S. Typhimurium activates the NLRP3 inflammasome (Diamond et al., 2017), whereas in macrophages both NLRP3 and NLRC4 are activated (Yang et al., 2013; Baker et al., 2015; Bierschenk et al., 2019). Although S. Typhimurium does not activate caspase‐4‐dependent pyroptosis in naive macrophages, in IFNγ‐activated macrophages GBP1 recruits caspase‐4 directly to S. Typhimurium leading to its activation and enhanced pyroptosis (Fisch et al., 2019). S. enterica serovar Typhi can activate inflammasomes in a flagellin‐dependent manner in human primary monocyte‐derived macrophages (Kortmann et al., 2015), while in the monocytic THP‐1 cell line, which expresses lower levels of NAIP (Kortmann et al., 2015), IL‐1β release in response to S. Typhi infection is flagellin‐independent and caspase‐dependent (Winter et al., 2015).
The role of mouse inflammasomes in the defence against S. Typhimurium in vivo has been extensively studied, which we discuss here briefly. Salmonella invades and replicates within non‐phagocytic cells such as IECs and triggers local inflammation and epithelial barrier breakdown. Inflammasomes are also activated within IECs infected with Salmonella. The NAIP‐NLRC4 inflammasome and caspase‐11 promote cell death and expulsion of infected IECs from the intestinal mucosa, which reduces the available replicative niches for S. Typhimurium (Knodler et al., 2010; Knodler et al., 2014; Sellin et al., 2014; Rauch et al., 2017). However, during later stages of infection, Salmonella dampen NAIP‐NLRC4 inflammasome activation by reducing SPI‐1 expression and up‐regulating the more immunologically silent SPI‐2 T3SS (Miao, Mao, et al., 2010; Zhao et al., 2016; Pérez‐Morales et al., 2017; Reyes Ruiz et al., 2017) and down‐regulating flagellin expression (Cummings, Wilkerson, Bergsbaken, & Cookson, 2006; Ilyas et al., 2018). Notably, NLRC4‐driven pyroptosis of macrophages infected with S. Typhimurium results in bacterial liberation in vivo and subsequent phagocytosis and killing by neutrophils, which are resistant to NLRC4‐mediated pyroptosis (Miao, Leaf, et al., 2010; Chen et al., 2014). Loss of Casp1 increases susceptibility of mice to S. Typhimurium (Lara‐Tejero et al., 2006; Raupach, Peuschel, Monack, & Zychlinsky, 2006). In addition to reduced IL‐1β and IL‐18 that are required for an optimal Th1 immune response for bacterial clearance, the loss of Casp1 also reduces bacterial uptake by neutrophils. As a result, in Casp1 −/− mice macrophage pyroptosis through caspase‐11 leads to higher extracellular bacterial load, resulting in greater susceptibility to infection (Broz et al., 2012). Casp1/Casp11‐double knockout mice, whose cells are resistant to pyroptotic lysis, have fewer extracellular bacteria and are therefore less susceptible than Casp1 −/− single knockouts (Broz et al., 2012). These findings point towards a role of inflammasomes and caspase‐1/11 in pyroptosis, neutrophil function and adaptive immune responses during S. Typhimurium infection in vivo. Furthermore, the effector SlrP (Salmonella leucine‐rich repeat protein), secreted through both T3SSs, inhibits inflammasome activation and IL‐1β release in the small intestine; higher IL‐1β levels during infection with a ΔslrP strain promote anorexia and increases disease severity (Rao et al., 2017) (Table 1).
Additionally, AIM2 has been reported to have a role in preserving epithelial integrity of the intestinal barrier during Salmonella infection, but it is not clear whether this defence mechanism involves inflammasome or non‐inflammasome‐dependent functions of AIM2 (Hu et al., 2016). Conversely, NLRP12 plays an undefined role in dampening the immune response to S. Typhimurium, and deficiency of this NLR leads to resistance to infection (Vladimer et al., 2012; Zaki et al., 2014), independently of caspase‐1 (Zaki et al., 2014). In summary, tissue‐ and cell type‐specific inflammasome signalling is protective against Salmonella infection.
6. SHIGELLA – INFLAMMASOME INTERACTIONS ARE CELL‐TYPE SPECIFIC
Infection by Shigella is a leading cause of diarrhoea in children in low‐ and middle‐income countries, where it is associated with high mortality rates (Tickell et al., 2017). There are four Shigella species, two of which (S. flexneri and S. sonnei) cause 90% of all infections. Shigella infection can cause shigellosis or bacterial dysentery, a severe diarrhoea containing blood and/or mucous which can be quickly spread in the population due to its low infective dose (Shane et al., 2017). The vast majority of research has been conducted on the most prevalent species, S. flexneri, which relies on its ability to utilise a T3SS to invade and replicate within the intestinal mucosal epithelium, causing inflammation and cell death. Adult mice are colonised poorly by Shigella in vivo; however the zebrafish model of infection has been useful in better dissecting host–pathogen interactions (Mostowy et al., 2013). In both epithelial cells and macrophages S. flexneri rapidly escapes the endocytic vacuole and accesses the cytosol where it is vulnerable to cellular defence mechanisms. Despite the cytosolic localisation of S. flexneri in both cell types, the outcome of infection is different; in macrophages cytosolic S. flexneri activates inflammasomes and induces rapid pyroptosis (Hilbi et al., 1998), whereas in epithelial cells S. flexneri avoids inflammasome activation and pyroptosis and instead triggers a delayed calpain‐dependent necrotic cell death mediated by the effector VirA (Bergounioux et al., 2012).
Compared to studies with Salmonella, much less is currently known about the roles inflammasomes play during Shigella infection. S. flexneri LPS can be detected by caspase‐4 in vitro and in epithelial cells (Kobayashi et al., 2013; Shi et al., 2014) (Figure 2c; Table 1). To counteract this and prolong epithelial cell survival, S. flexneri delivers the T3SS effector OspC3. OspC3 interacts with the cleaved caspase‐4 subunit p19 and inhibits its activation by preventing heterodimerisation of the caspase‐4 p19 and p10 subunits (Kobayashi et al., 2013). The importance of OspC3 was confirmed in a synthetic ‘bottom‐up’ approach to identify effectors that inhibit epithelial cell death. This study also identified the effectors OspD2 and IpaH1.4 (Mou, Souter, Du, Reeves, & Lesser, 2018). OspD2 controls the amount of VirA translocated into the host cell, thus regulating VirA‐mediated necrosis (Mou et al., 2018). IpaH1.4 has previously been demonstrated to suppress NF‐κB signalling by acting as an E3 ubiquitin ligase that ubiquitinates and degrades HOIP (HOIL‐1 interacting protein), a component of the linear ubiquitin chain assembly complex (de Jong, Liu, Chen, & Alto, 2016).
In macrophages, S. flexneri activates a number of inflammasome pathways leading to rapid cell death. The T3SS needle and rod proteins (MxiH and MxiI) are recognised by human NAIP (and mouse Naip1 and 2 respectively) to induce NLRC4 inflammasome activation (Kofoed & Vance, 2011; Yang et al., 2013; Suzuki, Franchi, et al., 2014; Zhao et al., 2016). Cellular damage resulting from pore formation by the T3SS effector IpaB may also contribute to NLRC4 inflammasome activation (Senerovic et al., 2012). In addition, IpaH7.8 promotes NLRC4 and NLRP3 inflammasome activation by ubiquitinating glomulin (GLMN) and eliciting its degradation (Suzuki, Mimuro, et al., 2014). GLMN is an inflammasome repressor which specifically targets cellular inhibitor of apoptosis proteins 1 and 2 (cIAP1 and cIAP2), members of the inhibitor of apoptosis family of RING‐E3 ligases, resulting in reduced cIAP E3 ligase activity and consequently diminished cIAP‐mediated inflammasome activation. IpaH7.8‐mediated GLMN depletion therefore results in increased inflammasome‐mediated death of macrophages (Suzuki, Suzuki, Mimuro, Mizushima, & Sasakawa, 2018). IpaH7.8 can also activate murine NLRP1B through ubiquitination and functional degradation of NLRP1B, whereby degradation of the amino‐terminal of NLRP1B releases a carboxyl terminal fragment able to activate caspase‐1 (Neiman‐Zenevich, Stuart, Abdel‐Nour, Girardin, & Mogridge, 2017; Sandstrom et al., 2019). Interestingly, IpaH7.8 does not activate human NLRP1 and, therefore, this pathway would not contribute to inflammasome activation during human Shigella infection. This may play a role in the human specificity displayed by S. flexneri, which does not naturally infect mice (Sandstrom et al., 2019) (Figure 2c). Conversely, during proliferation within epithelial cells, S. flexneri produces LPS with hypoacylated lipid A which, when used to stimulate macrophages prior to S. flexneri infection, results in reduced caspase‐1 activation and IL‐1β production (Paciello et al., 2013). Such LPS modification may also serve to reduce inflammasome activation in macrophages in vivo. Accordingly, S. flexneri employs diverse strategies to fine‐tune inflammasome activation.
Similarly to S. flexneri, S. sonnei also induces caspase‐1 dependent pyroptosis of macrophages. However, the level of cell death induced by S. sonnei is lower, as internalisation and vacuole escape are less efficient, resulting in fewer cytosolic bacteria. This is due to shielding of the T3SS by the O‐antigen, which is present on the lipid A‐core as LPS and as a group 4 capsule (Watson et al., 2019). The inflammatory environment caused by Shigella‐induced macrophage pyroptosis recruits neutrophils to the infection site. While this initially aids infection by destabilising the epithelial barrier (Perdomo, Gounon, & Sansonetti, 1994), it ultimately leads to bacterial killing and the resolution of infection. Interestingly, recent evidence suggests the O‐antigen of S. sonnei enables the bacteria to resist neutrophil killing in zebrafish (Torraca et al., 2019), highlighting the need for further work to characterise this species. Altogether, the presence of effectors and strategies that help evade inflammasomes in Shigella spp. suggests that the host deploys inflammasome pathways for self‐defence.
7. L. MONOCYTOGENES BENEFITS FROM INFLAMMASOME ACTIVATION
L. monocytogenes is a Gram‐positive food‐borne pathogen (Allerberger & Wagner, 2010). Seminal work from Pascale Cossart's lab and others implicated two proteins in the virulence of L. monocytogenes: listeriolysin O (LLO; encoded by the hlyA gene), a cholesterol‐dependent pore‐forming haemolysin, and ActA, a surface bacterial protein that mediates actin‐based motility within host cells and cell‐to‐cell spreading of the bacterium (Hamon, Ribet, Stavru, & Cossart, 2012; Pizarro‐Cerdá, Kühbacher, & Cossart, 2012). After invasion of IECs and myeloid cells, Listeria spreads to deeper tissues such as the liver and spleen, and can also cross the blood–brain barrier and the placenta in pregnant women. Although Listeria resides and replicates in the cytosol of different cell types, its interactions with inflammasomes are best understood in macrophages.
Upon infection of mouse or human macrophages, L. monocytogenes can be detected by multiple inflammasomes, including AIM2, NLRC4, NLRP3, NLRP6, caspase‐11 and NLRP1B. On the bacterial side, LLO (Meixenberger et al., 2010; Hamon & Cossart, 2011; Eldridge, Sanchez‐Garrido, Hoben, Goddard, & Shenoy, 2017), bacterial RNA (Kanneganti et al., 2006), bacterial DNA (Sauer et al., 2010) and the secreted protein p60 (Schmidt & Lenz, 2012) have been implicated in inflammasome activation (Table 1). Notably, conflicting findings have occasionally been reported, which may have arisen from the use of different L. monocytogenes strains, cell‐types (monocytes vs. macrophages), species (human vs. mouse) and/or pre‐treatment with TLR ligands (e.g. LPS) or type I or type II interferons (IFNα/β or IFNγ respectively). Here we summarise key findings on inflammasome activation by L. monocytogenes and refer readers to a detailed discussion elsewhere (Theisen & Sauer, 2016).
In mouse macrophages, AIM2 is the major inflammasome sensor of L. monocytogenes (Sauer et al., 2010; Warren et al., 2010; Wu, Fernandes‐Alnemri, & Alnemri, 2010). The commonly used L. monocytogenes strain 10403S undergoes occasional lysis in the cytosol, leading to the release of its DNA and direct activation of the AIM2 inflammasome. A mutant lacking the lmo2473 gene lyses more in the cytosol of infected macrophages and hyperactivates the AIM2‐ASC‐caspase‐1 inflammasome pathway (Sauer et al., 2010). AIM2 expression requires type I IFN signalling, consistent with which, L. monocytogenes‐induced inflammasome activation is severely reduced in type I IFN‐receptor deficient (Ifnar −/−) macrophages. L. monocytogenes is a poor activator of NLRC4 (Sauer et al., 2010; Warren et al., 2010; Sauer et al., 2011), but can activate NLRP6, caspase‐11 (Hara et al., 2018) and NLRP1B (Neiman‐Zenevich et al., 2017) in mouse macrophages, and NLRP3 in mouse bone‐marrow derived DCs (Clark, Schmidt, McDermott, & Lenz, 2018). Most L. monocytogenes strains turn down flagellin expression at 37°C and thus evade detection by TLR5 and the NAIP5‐NLRC4 pathway (Theisen & Sauer, 2016). However, due to a mutation in MogR (lmo0674), 10403S exhibits low basal flagellin expression which weakly activates the NLRC4 inflammasome (Gründling, Burrack, Bouwer, & Higgins, 2004). L. monocytogenes engineered to overexpress flagellins results in severe attenuation in vivo in an NLRC4‐dependent manner and these strains were also found to be poor vaccine candidates (Sauer et al., 2011). Surprisingly, bacterial lipoteichoic acid activates NLRP6 and caspase‐11 in mouse macrophages infected with L. monocytogenes strain EGD (Hara et al., 2018) (Table 1). NLRP6‐ASC complexes recruited both caspase‐1 and caspase‐11, where the role of active caspase‐11 was to promote caspase‐1 activation, which in turn processed IL‐18 and IL‐1β cytokines (Figure 2d). Importantly, in human cells, NLRP6‐silencing does not affect caspase‐1 activation by L. monocytogenes EGD (Meixenberger et al., 2010), which points towards species‐specific differences. Furthermore, genome sequencing studies performed by P. Cossart have confirmed that the EGD strain is markedly different from 10403S and EGDe strains and has a mutation in the master transcriptional regulator PrfA (PrfA*) which results in constitutive expression of various virulence genes (Bécavin et al., 2014). It is plausible that these differences also contribute to strain‐specific responses in host cells.
Studies in human macrophages showed that NLRP3 is involved in detection of L. monocytogenes; however, this is context‐dependent and has been observed in some (Kanneganti et al., 2006; Meixenberger et al., 2010; Wu et al., 2010; Shenoy et al., 2012; Fernandes‐Alnemri et al., 2013), but not other (Franchi, Kanneganti, Dubyak, & Núñez, 2007; Sauer et al., 2010; Warren et al., 2010), studies. While silencing of NLRP3 in human peripheral blood mononuclear cells reduces inflammasome activation by L. monocytogenes strain EGD, silencing of NLRP1, NLRC4, NLRP12, AIM2, RIP2 or NOD2 has no effect, ruling out their involvement (Meixenberger et al., 2010). Listeria‐mediated NLRP3 activation is mediated by LLO (Meixenberger et al., 2010; Eldridge et al., 2017) (Table 1). Moreover, recombinant LLO can trigger potassium efflux (Hamon & Cossart, 2011), suggesting that extracellular LLO may also trigger NLRP3 activation. Mechanistically, NLRP3 activation during Listeria infection requires lysosomal acidification, rupture and the release of cathepsin B (Meixenberger et al., 2010). Infection of IFNγ‐primed human THP1 cells with L. monocytogenes strain 10403S results in enhanced NLRP3 activation assisted by the IFNγ‐inducible GBP5 (Shenoy et al., 2012). Altogether, these studies are consistent with the recently identified AIM2‐independent, NLRP3‐dependent detection of cytosolic DNA in human, but not mouse, myeloid cells (Gaidt et al., 2017). This new DNA‐sensing pathway in human cells also requires lysosomal damage (Gaidt et al., 2017). Therefore, L. monocytogenes, plausibly via both LLO and release of DNA in the cytosol, activates the human NLRP3 inflammasome and the murine AIM2 inflammasome (Figure 2d).
Early in vivo studies on L. monocytogenes pointed towards a protective role of inflammasome‐related genes (Hirsch, Irikura, Paul, & Hirsh, 1996; Labow et al., 1997; Tsuji et al., 2004); however, these studies were carried out in mice with a mixed C57BL/6 and 129/S background which also carry an inactivating passenger mutation in Casp11. More recent studies suggest that loss of Il18, Nlrp3 or Asc increases resistance to L. monocytogenes, and loss of Casp1/11 −/− does not markedly affect survival or adaptive immune responses (Lochner et al., 2008; Sauer et al., 2011; Tsuchiya et al., 2014; Clark et al., 2018). Intriguingly, Nlrp6 −/− and Casp11 −/− mice, which would not recognise Listeria LTA, are also more resistant to L. monocytogenes infection (Hara et al., 2018). However, increased resistance of Nlrp6 −/− mice to L. monocytogenes has also been attributed to increased canonical NF‐κB signalling and myeloid cell responses that better restrict bacterial growth (Anand et al., 2012). As inflammasome‐driven inflammation increases neutrophil influx, these findings suggest that a heightened neutrophil‐response is detrimental in the defence against L. monocytogenes in vivo. Taken together, most in vivo studies in mice suggest that inflammasome activation benefits Listeria and is detrimental to the host.
8. HELICOBACTER PYLORI, INFLAMMASOME STIMULATION AND PERSISTENT INFECTION
H. pylori is found in the gastric mucosa of around 50% of the world population but only a minority of infected individuals will develop long‐term clinical symptoms (Abadi & Kusters, 2016). Individuals with an active chronic infection can develop gastric ulcers, chronic gastritis, gastric mucosa‐associated lymphoid tissue (MALT) lymphoma and gastric cancers (Chey et al., 2017). Inflammasome‐mediated recognition of H. pylori in myeloid cells is mainly attributed to NLRP3 through a not well‐defined mechanism requiring expression of the bacterial vacuolating cytotoxin A (VacA) and the cytotoxin‐associated genes pathogenicity island, which encodes the cytotoxin associated gene A (CagA) and the type IV secretion system (T4SS) (Kim, Park, Franchi, Backert, & Núñez, 2013; Semper et al., 2014; Kameoka et al., 2016). It is tempting to speculate that VacA, which forms pores in the plasma membrane, causes mitochondrial damage, increases ROS levels and induces cell apoptosis, may likewise contribute to NLRP3 activation. Caspase‐1, IL‐1β and IL‐18 play an important role in H. pylori pathogenesis in vivo, but whether inflammasome activation benefits the host or the pathogen remains to be defined (Hitzler et al., 2012; Kim et al., 2013). The role of NLRP3 in promoting (Hitzler et al., 2012; Koch et al., 2015; Arnold et al., 2017) versus restricting (Semper et al., 2014) H. pylori infection also remains unclear. These differences may be due to the use of mice of different ages, differences in the microbiota or the H. pylori strains, or to variations in the methodologies used in the month‐long infection model. However, supporting the role of inflammasomes in promoting H. pylori pathogenesis, the protective role of Mucin1 expression in the gastric mucosa has been linked to the down‐regulation of NLRP3 expression and consequent IL‐1β release (Ng et al., 2016). Moreover, the NLRC4 inflammasome is activated during H. pylori infection of gastric epithelial cells in a T4SS‐dependent manner, leading to IL‐18 release and robust neutrophil recruitment. NLRC4 activation promotes H. pylori survival as the resulting IL‐18 dampens the IL‐17 response, reducing antimicrobial peptide production and favouring tissue damage and inflammation (Semper, Vieth, Gerhard, & Mejías‐Luque, 2019) (Figure 2e). While inflammasome activation facilitates H. pylori infection, exacerbated inflammation and loss of the replicative niche may prove to be detrimental for both the host and the pathogen. Thus, it is not surprising that H. pylori also employs immune evasion strategies that include the expression of a modified LPS that is less pyrogenic than that of other Gram‐negative enteric pathogens (Moran, 2007) and a distinct flagellin that fails to activate TLR5 and the NLRC4 inflammasome (Andersen‐Nissen et al., 2005; Matusiak et al., 2015). In summary, inflammasome activation in response to H. pylori infection is detrimental for the host, as it promotes H. pylori survival in the gastric mucosa.
9. YERSINIA SPP. AND INFLAMMASOME MANIPULATION
Yersinia enterocolitica and Yersinia pseudotuberculosis are food‐borne Gram‐negative enteric pathogens that cause a self‐limiting enterocolitis, often presented as terminal ileitis in humans, but can also lead to invasive disease and septicaemia (Galindo, Rosenzweig, Kirtley, & Chopra, 2011). Yersinia spp. invade and survive in IECs (Ligeon et al., 2014; Valencia Lopez et al., 2019), cross the intestinal epithelium and target mesenteric lymph nodes (Galindo et al., 2011). Inflammasome activation and evasion by Yersinia spp. is best studied in murine macrophages. The NLRP3 and NLRC4 inflammasomes are activated by the detection of bacterial T3SS and the translocation of the pore‐forming translocon components Yersinia outer protein B (YopB) and YopD (Brodsky et al., 2010; Zwack et al., 2015). The PYRIN inflammasome is activated through the RhoA GTPase‐inhibiting activity of YopE and YopT (Chung et al., 2016; Ratner et al., 2016) (Figure 2f). To counteract detection by the inflammasomes, Yersinia encodes the effector YopK, which interacts with the T3SS and regulates translocation of effectors (Brodsky et al., 2010; Zwack et al., 2015), and YopM, a homologue of the Salmonella SlrP which inhibits caspase‐1 (LaRock & Cookson, 2012) and manipulates host kinases to inhibit PYRIN (Chung et al., 2016; Ratner et al., 2016) (Table 1). Both YopK and YopM are thus essential for Yersinia virulence. Furthermore, during infection with Y. enterocolitica, β1‐integrin signalling triggered by the adhesin invasin increases pro‐IL‐18 levels in IECs; notably, these outcomes are counteracted by YopH and YopE (Thinwa, Segovia, Bose, & Dube, 2014).
The acetyltransferase YopJ (YopP in Y. enterocolitica) is a homologue of Salmonella AvrA that inhibits transforming growth factor beta‐activated kinase 1 (TAK1), IκB kinase β (IKKβ) and mitogen‐activated protein kinase (MAPK) kinases, inhibiting pro‐inflammatory signalling in response to TLR and/or TNF signalling (Peterson et al., 2016; Pinaud et al., 2018). TAK1 inhibition promotes non‐canonical RIPK1‐FADD‐caspase‐8‐dependent GSDMD cleavage, pore formation and cell death in mouse macrophages. The resulting potassium efflux triggers NLRP3 and caspase‐1 activation, and release of IL‐1β (Philip et al., 2014; Orning et al., 2018) (Figure 2f). In addition, blockade of TNF‐mediated pro‐survival NF‐κB and MAPK signalling by YopJ can also lead to RIPK1‐dependent apoptosis (Weng et al., 2014; Peterson et al., 2016; Peterson et al., 2017). Further, YopJ‐mediated inhibition of the Nod2 pathway has also been linked to Nod2‐dependent activation of caspase‐1 and IL‐1β release from Peyer's patches, and loss of intestinal barrier function in mice (Meinzer et al., 2012). During Y. pseudotuberculosis infection in vivo, YopJ‐driven cell death pathways promote bacterial clearance and host survival, seemingly counteracting the pathogen‐mediated blockade of inflammatory signalling (Meinzer et al., 2012; Philip et al., 2014; Peterson et al., 2017).
In addition to secreted effectors, Yersinia LPS can also activate inflammasomes. However, Yersinia evades detection by mouse caspase‐11 in vivo by deacylating its lipid A to four lipid chains at 37°C (Hagar, Powell, Aachoui, Ernst, & Miao, 2013). It is possible that human caspase‐4, which can recognise tetra‐acylated LPS from Francisella novicida, might also recognise tetra‐acylated Yersinia lipid A (Lagrange et al., 2018). While its role during infection with enteric Yersinia species in vivo is yet to be defined, mouse NLRP12, whose activation mechanisms remain poorly understood, also contributes to IL‐1β release from bone marrow‐derived macrophages infected with Y. pseudotuberculosis or Y. enterocolitica (Vladimer et al., 2012). Taken together, inflammasome activation during Yersinia infection is beneficial for the host as it promotes bacterial clearance (Brodsky et al., 2010; Meinzer et al., 2012; Philip et al., 2014; Peterson et al., 2017).
10. CONCLUSIONS
Inflammasomes are key soluble cytosolic surveillance devices that recognise and respond to multiple virulence determinants of both Gram‐negative and Gram‐positive enteric pathogens. Moreover, they operate in multiple cell types that come into early contact with enteric pathogens. Activation of the inflammasomes can trigger the release of pro‐inflammatory cytokines or expel host cells containing intracellular pathogens via cell death. For this reason, it is not surprising that pathogenic bacteria have acquired survival mechanisms to mitigate these host responses. On the other hand, an exuberant inflammasome‐driven inflammatory response results in heightened neutrophil influx and tissue damage, which can promote the systemic spread of invasive pathogens. The pathogens covered in this review selectively infect the gastric mucosa (Helicobacter), invade through the small intestine (Salmonella, Listeria, Yersinia) or colon (Shigella), or are restricted to the colonic mucosa (C. rodentium). Multiple tissues can be colonised upon invasion, including the lymph nodes, blood, spleen, liver, brain or the foetus, and localised cell‐type specific inflammasome responses remain to be better understood. Although the causal role of caspase‐4 and pyroptosis in murine endotoxic shock is well established, more work is required to clarify the role of inflammasomes in infectious sepsis in humans. Future work should also explore whether immunomodulation of inflammasomes with small molecules (e.g. MCC950) could promote microbial clearance by reducing deregulated inflammation.
Pascale Cossart was among the pioneers who championed studies on the interactions of intracellular bacteria with various host cell machineries, including membrane‐bound surface receptors activated during bacterial entry, manipulation of the cytoskeletal and endocytic trafficking systems (Cossart, Boquet, Normark, & Rappuoli, 1996). Studies from her group and others also ushered the phase of discovery of cytosolic immune surveillance pathways aided by the use of various bacterial pathogens as invaluable cell biological tools. Studies have now revealed the benefits and disadvantages of inflammasome signalling in host defence. No doubt, the near future will reveal yet new inflammasome functions and other subversion mechanisms, writing new a chapter of the never‐ending story of an intimate relationship between bacteria and the host.
CONFLICT OF INTEREST
The authors have no interests to declare.
AUTHOR CONTRIBUTIONS
J.S.G., S.L.S., A.C., A.R.S. and G.F. wrote the review. J.S.G. and A.R.S. created the figures.
ACKNOWLEDGEMENTS
The authors would like to acknowledge funding from the MRC (MR/P022138/1 to AS; MR/R020671/1 to GF) and the Wellcome Trust 107057/Z/15/X to GF).
Sanchez‐Garrido J, Slater SL, Clements A, Shenoy AR, Frankel G. Vying for the control of inflammasomes: The cytosolic frontier of enteric bacterial pathogen–host interactions. Cellular Microbiology. 2020;22:e13184 10.1111/cmi.13184
Funding information Medical Research Council, Grant/Award Numbers: MR/P022138/1, MR/R020671/1; Wellcome Trust, Grant/Award Number: 107057/Z/15/X
REFERENCES
- Abadi, A. T. B. , & Kusters, J. G. (2016). Management of Helicobacter pylori infections. BMC Gastroenterology, 16(1), 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter, A. , Caution, K. , Abu Khweek, A. , Tazi, M. , Abdulrahman, B. A. , Abdelaziz, D. H. A. , … Amer, A. O. (2012). Caspase‐11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity, 37(1), 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerberger, F. , & Wagner, M. (2010). Listeriosis: A resurgent foodborne infection. Clinical Microbiology and Infection, 16(1), 16–23. [DOI] [PubMed] [Google Scholar]
- Anand, P. K. , Malireddi, R. K. S. , Lukens, J. R. , Vogel, P. , Bertin, J. , Lamkanfi, M. , & Kanneganti, T. D. (2012). NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature, 488(7411), 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen‐Nissen, E. , Smith, K. D. , Strobe, K. L. , Barrett, S. L. R. , Cookson, B. T. , Logan, S. M. , & Aderem, A. (2005). Evasion of toll‐like receptor 5 by flagellated bacteria. Proceedings of the National Academy of Sciences of the United States of America, 102(26), 9247–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, I. C. , Zhang, X. , Urban, S. , Artola‐Borán, M. , Manz, M. G. , Ottemann, K. M. , & Müller, A. (2017). NLRP3 controls the development of gastrointestinal CD11b+ dendritic cells in the steady state and during chronic bacterial infection. Cell Reports, 21(13), 3860–3872. [DOI] [PubMed] [Google Scholar]
- Baker, P. J. , Boucher, D. , Bierschenk, D. , Tebartz, C. , Whitney, P. G. , D'Silva, D. B. , … Masters, S. L. (2015). NLRP3 inflammasome activation downstream of cytoplasmic LPS recognition by both caspase‐4 and caspase‐5. European Journal of Immunology, 45(10), 2918–2926. [DOI] [PubMed] [Google Scholar]
- Bauernfeind, F. , Horvath, G. , Stutz, A. , Alnemri, E. S. , MacDonald, K. , Speert, D. , … Latz, E. (2009). NF‐κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. Journal of Immunology, 183(2), 787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécavin, C. , Bouchier, C. , Lechat, P. , Archambaud, C. , Creno, S. , Gouin, E. , … Cossart, P. (2014). Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD‐e highlights genomic variations underlying differences in pathogenicity. mBio, 5(2), e00969‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaoudia, S. , Martin, A. , Puig Gamez, M. , Gay, G. , Lagrange, B. , Cornut, M. , … Henry, T. (2019). A genome‐wide screen identifies IRF2 as a key regulator of caspase‐4 in human cells. EMBO Reports, 0(0), e48235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergounioux, J. , Elisee, R. , Prunier, A.‐L. , Donnadieu, F. , Sperandio, B. , Sansonetti, P. , & Arbibe, L. (2012). Calpain activation by the Shigella flexneri effector VirA regulates key steps in the formation and life of the Bacterium's epithelial niche. Cell Host & Microbe, 11(3), 240–252. [DOI] [PubMed] [Google Scholar]
- Bierschenk, D. , Monteleone, M. , Moghaddas, F. , Baker, P. J. , Masters, S. L. , Boucher, D. , & Schroder, K. (2019). The Salmonella pathogenicity Island‐2 subverts human NLRP3 and NLRC4 inflammasome responses. Journal of Leukocyte Biology, 105(2), 401–410. [DOI] [PubMed] [Google Scholar]
- Blasche, S. , Mörtl, M. , Steuber, H. , Siszler, G. , Nisa, S. , Schwarz, F. , … Kögl, M. (2013). The E. coli effector protein NleF is a Caspase inhibitor. PLoS One, 8(3), e58937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, I. E. , Palm, N. W. , Sadanand, S. , Ryndak, M. B. , Sutterwala, F. S. , Flavell, R. A. , … Medzhitov, R. (2010). A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host & Microbe, 7(5), 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, P. , & Dixit, V. M. (2016). Inflammasomes: Mechanism of assembly, regulation and signalling. Nature Reviews. Immunology, 16(7), 407–420. [DOI] [PubMed] [Google Scholar]
- Broz, P. , Newton, K. , Lamkanfi, M. , Mariathasan, S. , Dixit, V. M. , & Monack, D. M. (2010). Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. The Journal of Experimental Medicine, 207(8), 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, P. , Pelegrín, P. , & Shao, F. (2019). The gasdermins, a protein family executing cell death and inflammation. Nature Reviews Immunology. 10.1038/s41577-019-0228-2 [DOI] [PubMed] [Google Scholar]
- Broz, P. , Ruby, T. , Belhocine, K. , Bouley, D. M. , Kayagaki, N. , Dixit, V. M. , & Monack, D. M. (2012). Caspase‐11 increases susceptibility to Salmonella infection in the absence of caspase‐1. Nature, 490, 288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, P. , von Moltke, J. , Jones, J. W. , Vance, R. E. , & Monack, D. M. (2010). Differential requirement for Caspase‐1 autoproteolysis in pathogen‐induced cell death and cytokine processing. Cell Host & Microbe, 8(6), 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson, C. N. , Copenhaver, A. M. , Zwack, E. E. , Nguyen, H. T. , Strowig, T. , Javdan, B. , … Shin, S. (2013). Caspase‐11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathogens, 9(6), e1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson, C. N. , Yu, J. , Reyes, V. M. , Taschuk, F. O. , Yadav, A. , Copenhaver, A. M. , … Shin, S. (2015). Human caspase‐4 mediates noncanonical inflammasome activation against gram‐negative bacterial pathogens. Proceedings of the National Academy of Sciences of the United States of America, 112(21), 6688–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Groß, C. , Sotomayor, F. , Stacey, K. , Tschopp, J. , Sweet, M. , & Schroder, K. (2014). The neutrophil NLRC4 inflammasome selectively promotes IL‐1β maturation without pyroptosis during acute Salmonella challenge. Cell Reports, 8(2), 570–582. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.‐L. , Song, L.‐Q., Huang, Y. M., Xiong, Y. W., Zhang, X. A., Sun, H., … Ren, Z. H. (2015). Effect of enterohaemorrhagic Escherichia coli O157:H7‐specific enterohaemolysin on interleukin‐1β production differs between human and mouse macrophages due to the different sensitivity of NLRP3 activation. Immunology, 145(2), 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chey, W. D. , Leontiadis, G. I. , Howden, C. W. & Moss, S. F. (2017). ACG clinical guideline: Treatment of Helicobacter pylori infection. American Journal of Gastroenterology 112(2): 212–239. [DOI] [PubMed] [Google Scholar]
- Chui, A. J. , Okondo, M. C. , Rao, S. D. , Gai, K. , Griswold, A. R. , Johnson, D. C. , … Bachovchin, D. A. (2019). N‐terminal degradation activates the NLRP1B inflammasome. Science, 364(6435), 82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, L. K. , Park, Y. H. , Zheng, Y. , Brodsky, I. E. , Hearing, P. , Kastner, D. L. , … Bliska, J. B. (2016). The Yersinia virulence factor YopM hijacks host kinases to inhibit type III effector‐triggered activation of the pyrin inflammasome. Cell Host & Microbe, 20(3), 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, L. K. , Philip, N. H. , Schmidt, V. A. , Koller, A. , Strowig, T. , Flavell, R. A. , … Bliska, J. B. (2014). IQGAP1 is important for activation of caspase‐1 in macrophages and is targeted by Yersinia pestis type III effector YopM. mBio 5(4): e01402‐e01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. E. , Schmidt, R. L. , McDermott, D. S. , & Lenz, L. L. (2018). A Batf3/Nlrp3/IL‐18 axis promotes natural killer cell IL‐10 production during Listeria monocytogenes infection. Cell Reports, 23(9), 2582–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, J. W. , Keeney, K. M. , Crepin, V. F. , Rathinam, V. A. K. , Fitzgerald, K. A. , Finlay, B. B. , & Frankel, G. (2014). Citrobacter rodentium: Infection, inflammation and the microbiota. Nature Reviews Microbiology, 12, 612–623. [DOI] [PubMed] [Google Scholar]
- Cossart, P. , Boquet, P. , Normark, S. , & Rappuoli, R. (1996). Cellular microbiology emerging. Science, 271(5247), 315–316. [DOI] [PubMed] [Google Scholar]
- Crowley, S. M. , Knodler, L. A. , & Vallance, B. A. (2016). Salmonella and the inflammasome: Battle for intracellular dominance In Backert S. (Ed.), Inflammasome signaling and bacterial infections (pp. 43–67). Cham: Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- Cullen, S. P. , Kearney, C. J. , Clancy, D. M. , & Martin, S. J. (2015). Diverse activators of the NLRP3 inflammasome promote IL‐1β secretion by triggering necrosis. Cell Reports, 11(10), 1535–1548. [DOI] [PubMed] [Google Scholar]
- Cummings, L. A. , Wilkerson, W. D. , Bergsbaken, T. , & Cookson, B. T. (2006). In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Molecular Microbiology, 61(3), 795–809. [DOI] [PubMed] [Google Scholar]
- de Jong, M. F. , Liu, Z. , Chen, D. , & Alto, N. M. (2016). Shigella flexneri suppresses NF‐κB activation by inhibiting linear ubiquitin chain ligation. Nature Microbiology, 1(7), 16084–16084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, C. E. , Leong, K. W. K. , Vacca, M. , Rivers‐Auty, J. , Brough, D. , & Mortellaro, A. (2017). Salmonella typhimurium‐induced IL‐1 release from primary human monocytes requires NLRP3 and can occur in the absence of pyroptosis. Scientific Reports, 7(1), 6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J. , Wang, K. , Liu, W. , She, Y. , Sun, Q. , Shi, J. , … Shao, F. (2016). Pore‐forming activity and structural autoinhibition of the gasdermin family. Nature, 535, 111–116. [DOI] [PubMed] [Google Scholar]
- DuPont, H. L. (2009). Bacterial diarrhea. New England Journal of Medicine, 361(16), 1560–1569. [DOI] [PubMed] [Google Scholar]
- Dupont, N. , Jiang, S. , Pilli, M. , Ornatowski, W. , Bhattacharya, D. , & Deretic, V. (2011). Autophagy‐based unconventional secretory pathway for extracellular delivery of IL‐1β. The EMBO Journal, 30, 4701–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge, M. J. G. , Sanchez‐Garrido, J. , Hoben, G. F. , Goddard, P. J. , & Shenoy, A. R. (2017). The atypical ubiquitin E2 Conjugase UBE2L3 is an indirect Caspase‐1 target and controls IL‐1β secretion by inflammasomes. Cell Reports, 18(5), 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge, M. J. G. and Shenoy, A. R. (2015). Antimicrobial inflammasomes: Unified signalling against diverse bacterial pathogens. Current Opinion in Microbiology 23(0): 32–41. [DOI] [PubMed] [Google Scholar]
- Elinav, E. , Strowig, T. , Kau, A. L. , Henao‐Mejia, J. , Thaiss, C. A. , Booth, C. J. , … Flavell, R. A. (2011). NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell, 145(5), 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold, C. L. , & Kagan, J. C. (2019). Inflammasomes: Threat‐assessment organelles of the innate immune system. Immunity, 51, 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold, C. L. , Ruan, J. , Tan, Y. , Xia, S. , Wu, H. and Kagan, J. C. (2018). The pore‐forming protein Gasdermin D regulates Interleukin‐1 secretion from living macrophages. Immunity 48(1): 35‐44.e36, 44.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes‐Alnemri, T. , Kang, S. , Anderson, C. , Sagara, J. , Fitzgerald, K. A. , & Alnemri, E. S. (2013). Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. The Journal of Immunology, 191(8), 3995–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch, D. , Bando, H. , Clough, B. , Hornung, V. , Yamamoto, M. , Shenoy, A. R. , & Frickel, E. M. (2019). Human GBP1 is a microbe‐specific gatekeeper of macrophage apoptosis and pyroptosis. The EMBO Journal, 38(13), e100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi, L. , Kamada, N. , Nakamura, Y. , Burberry, A. , Kuffa, P. , Suzuki, S. , … Núñez, G. (2012). NLRC4‐driven production of IL‐1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nature Immunology, 13, 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi, L. , Kanneganti, T.‐D. , Dubyak, G. R. , & Núñez, G. (2007). Differential requirement of P2X7 receptor and intracellular K+ for Caspase‐1 activation induced by intracellular and extracellular bacteria. Journal of Biological Chemistry, 282(26), 18810–18818. [DOI] [PubMed] [Google Scholar]
- Frankel, G. , & Phillips, A. D. (2008). Attaching effacing Escherichia coli and paradigms of Tir‐triggered actin polymerization: Getting off the pedestal. Cellular Microbiology, 10(3), 549–556. [DOI] [PubMed] [Google Scholar]
- Frankel, G. , Phillips, A. D. , Rosenshine, I. , Dougan, G. , Kaper, J. B. , & Knutton, S. (1998). Enteropathogenic and enterohaemorrhagic Escherichia coli: More subversive elements. Molecular Microbiology, 30(5), 911–921. [DOI] [PubMed] [Google Scholar]
- Gaidt, M. M. , Ebert, T. S. , Chauhan, D. , Ramshorn, K. , Pinci, F. , Zuber, S. , O'Duill F., Schmid‐Burgk J. L., Hoss F., Buhmann R., Wittmann G., Latz E., Subklewe M., Hornung V. (2017). The DNA inflammasome in human myeloid cells is initiated by a STING‐cell death program upstream of NLRP3. Cell 171(5): 1110–1124.e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt, M. M. , Ebert, T. S. , Chauhan, D. , Schmidt, T. , Schmid‐Burgk, J. L. , Rapino, F. , … Hornung, V. (2016). Human monocytes engage an alternative inflammasome pathway. Immunity, 44(4), 833–846. [DOI] [PubMed] [Google Scholar]
- Galindo, C. L. , Rosenzweig, J. A. , Kirtley, M. L. , & Chopra, A. K. (2011). Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in human Yersiniosis. Journal of Pathogens, 2011, 182051–182051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia, J. , Phillips, A. D. , Carlier, M.‐F. , Chong, Y. , Schüller, S. , Marches, O. , … Frankel, G. (2004). TccP is an enterohaemorrhagic Escherichia coli O157:H7 Type III effector protein that couples Tir to the actin‐cytoskeleton. Cellular Microbiology, 6(12), 1167–1183. [DOI] [PubMed] [Google Scholar]
- Goddard, P. J. , Sanchez‐Garrido, J. , Slater, S. L. , Kalyan, M. , Ruano‐Gallego, D. , Marchès, O. , Fernández L. Á., Frankel G., Shenoy A. R. (2019). Enteropathogenic Escherichia coli stimulates effector‐driven rapid Caspase‐4 activation in human macrophages. Cell Reports 27(4): 1008–1017.e1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean, T. , Boucher, A. , Thepaut, M. , Monlezun, L. , Guery, B. , Faudry, E. , … Dessein, R. (2017). The human NAIP‐NLRC4‐inflammasome senses the Pseudomonas aeruginosa T3SS inner‐rod protein. International Immunology, 29(8), 377–384. [DOI] [PubMed] [Google Scholar]
- Gründling, A. , Burrack, L. S. , Bouwer, H. G. A. , & Higgins, D. E. (2004). Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proceedings of the National Academy of Sciences of the United States of America, 101(33), 12318–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar, J. A. , Powell, D. A. , Aachoui, Y. , Ernst, R. K. , & Miao, E. A. (2013). Cytoplasmic LPS activates Caspase‐11: Implications in TLR4‐independent endotoxic shock. Science, 341(6151), 1250–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon, M. A. , & Cossart, P. (2011). K+ efflux is required for histone H3 dephosphorylation by Listeria monocytogenes listeriolysin O and other pore‐forming toxins. Infection and Immunity, 79(7), 2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon, M. A. , Ribet, D. , Stavru, F. , & Cossart, P. (2012). Listeriolysin O: The Swiss army knife of Listeria. Trends in Microbiology, 20(8), 360–368. [DOI] [PubMed] [Google Scholar]
- Hara, H. , Seregin, S. S. , Yang, D. , Fukase, K. , Chamaillard, M. , Alnemri, E. S. , Inohara N., Chen G. Y., Núñez G. (2018). The NLRP6 inflammasome recognizes Lipoteichoic acid and regulates gram‐positive pathogen infection. Cell 175(6): 1651–1664.e1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward, J. A. , Mathur, A. , Ngo, C. , & Man, S. M. (2018). Cytosolic recognition of microbes and pathogens: Inflammasomes in action. Microbiology and Molecular Biology Reviews, 82(4), e00015–e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbi, H. , Moss, J. E. , Hersh, D. , Chen, Y. , Arondel, J. , Banerjee, S. , … Zychlinsky, A. (1998). Shigella‐induced apoptosis is dependent on Caspase‐1 which binds to IpaB. Journal of Biological Chemistry, 273(49), 32895–32900. [DOI] [PubMed] [Google Scholar]
- Hirsch, E. , Irikura, V. M. , Paul, S. M. , & Hirsh, D. (1996). Functions of interleukin 1 receptor antagonist in gene knockout and overproducing mice. Proceedings of the National Academy of Sciences of the United States of America, 93(20), 11008–11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzler, I. , Sayi, A. , Kohler, E. , Engler, D. B. , Koch, K. N. , Hardt, W.‐D. , & Müller, A. (2012). Caspase‐1 has both proinflammatory and regulatory properties in Helicobacter infections, which are differentially mediated by its substrates IL‐1β and IL‐18. The Journal of Immunology, 188(8), 3594–3602. [DOI] [PubMed] [Google Scholar]
- Hornung, V. , Ablasser, A. , Charrel‐Dennis, M. , Bauernfeind, F. , Horvath, G. , Caffrey, D. R. , … Fitzgerald, K. A. (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase‐1‐activating inflammasome with ASC. Nature, 458(7237), 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G. Q. , Song, P. X. , Li, N. , Chen, W. , Lei, Q. Q. , Yu, S. X. , … Yang, Y. J. (2016). AIM2 contributes to the maintenance of intestinal integrity via Akt and protects against Salmonella mucosal infection. Mucosal Immunology, 9, 1330–1339. [DOI] [PubMed] [Google Scholar]
- Hu, J. , & Torres, A. G. (2015). Enteropathogenic Escherichia coli: Foe or innocent bystander? Clinical Microbiology and Infection, 21(8), 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas, B. , Mulder, D. T. , Little, D. J. , Elhenawy, W. , Banda, M. M. , Pérez‐Morales, D. , … Coombes, B. K. (2018). Regulatory evolution drives evasion of host inflammasomes by Salmonella Typhimurium. Cell Reports 25(4): 825‐832.e825. [DOI] [PubMed]
- Jennings, E. , Thurston, T. L. M. , & Holden, D. W. (2017). Salmonella SPI‐2 type III secretion system effectors: Molecular mechanisms and physiological consequences. Cell Host & Microbe, 22(2), 217–231. [DOI] [PubMed] [Google Scholar]
- Johnson, R. , Mylona, E. , & Frankel, G. (2018). Typhoidal Salmonella: Distinctive virulence factors and pathogenesis. Cellular Microbiology, 20(9), e12939. [DOI] [PubMed] [Google Scholar]
- Kameoka, S. , Kameyama, T. , Hayashi, T. , Sato, S. , Ohnishi, N. , Hayashi, T. , … Takaoka, A. (2016). Helicobacter pylori induces IL‐1β protein through the inflammasome activation in differentiated macrophagic cells. Biomedical Research, 37(1), 21–27. [DOI] [PubMed] [Google Scholar]
- Kampmeier, S. , Berger, M. , Mellmann, A. , Karch, H. , & Berger, P. (2018). The 2011 German enterohemorrhagic Escherichia coli O104:H4 outbreak—The danger is still out there In Frankel G. and Ron E. Z. (Eds.), Escherichia coli, a versatile pathogen (pp. 117–148). Cham: Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- Kanneganti, T.‐D. , Özören, N. , Body‐Malapel, M. , Amer, A. , Park, J.‐H. , Franchi, L. , … Núñez, G. (2006). Bacterial RNA and small antiviral compounds activate caspase‐1 through cryopyrin/Nalp3. Nature, 440(7081), 233–236. [DOI] [PubMed] [Google Scholar]
- Kayagaki, N. , Lee, B. L. , Stowe, I. B. , Kornfeld, O. S. , Rourke, K. , Mirrashidi, K. M. , … Dixit , V. M. (2019). IRF2 transcriptionally induces GSDMD expression for pyroptosis. Science Signaling, 12(582), eaax4917. [DOI] [PubMed] [Google Scholar]
- Kayagaki, N. , Stowe, I. B. , Lee, B. L. , O/'Rourke, K., Anderson, K., Warming, S., … Dixit, V. M. (2015). Caspase‐11 cleaves gasdermin D for non‐canonical inflammasome signalling. Nature, 526(7575), 666–671. [DOI] [PubMed] [Google Scholar]
- Kayagaki, N. , Wong, M. T. , Stowe, I. B. , Ramani, S. R. , Gonzalez, L. C. , Akashi‐Takamura, S. , … Dixit, V. M. (2013). Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science, 341(6151), 1246–1249. [DOI] [PubMed] [Google Scholar]
- Kim, B.‐H. , Chee, J. D. , Bradfield, C. J. , Park, E.‐S. , Kumar, P. , & MacMicking, J. D. (2016). Interferon‐induced guanylate‐binding proteins in inflammasome activation and host defense. Nature Immunology, 17(5), 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.‐J. , Park, J.‐H. , Franchi, L. , Backert, S. , & Núñez, G. (2013). The cag pathogenicity Island and interaction between TLR2/NOD2 and NLRP3 regulate IL‐1β production in Helicobacter pylori infected dendritic cells. European Journal of Immunology, 43(10), 2650–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Bauernfeind, F. , Ablasser, A. , Hartmann, G. , Fitzgerald, K. A. , Latz, E. , & Hornung, V. (2010). Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. European Journal of Immunology, 40(6), 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, T. , Jia, J. , Kumar, S. , Choi, S. W. , Gu, Y. , Mudd, M. , … Deretic, V. (2017). Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. The EMBO Journal, 36(1), 42–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, M. D. , Pires, S. M. , Black, R. E. , Caipo, M. , Crump, J. A. , Devleesschauwer, B. , … Angulo, F. J. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Medicine, 12(12), e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler, L. A. , Crowley, S. M. , Sham, H. P. , Yang, H. , Wrande, M. , Ma, C. , … Vallance, B. A. (2014). Noncanonical inflammasome activation of Caspase‐4/Caspase‐11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host & Microbe, 16(2), 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler, L. A. , Vallance, B. A. , Celli, J. , Winfree, S. , Hansen, B. , Montero, M. , & Steele‐Mortimer, O. (2010). Dissemination of invasive Salmonella via bacterial‐induced extrusion of mucosal epithelia. Proceedings of the National Academy of Sciences of the United States of America, 107(41), 17733–17738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T. , Ogawa, M. , Sanada, T. , Mimuro, H. , Kim, M. , Ashida, H. , … Sasakawa, C. (2013). The Shigella OspC3 effector inhibits Caspase‐4, antagonizes inflammatory cell death, and promotes epithelial infection. Cell Host & Microbe, 13(5), 570–583. [DOI] [PubMed] [Google Scholar]
- Koch, K. N. , Hartung, M. L. , Urban, S. , Kyburz, A. , Bahlmann, A. S. , Lind, J. , … Müller, A. (2015). Helicobacter urease‐induced activation of the TLR2/NLRP3/IL‐18 axis protects against asthma. The Journal of Clinical Investigation, 125(8), 3297–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed, E. M. , & Vance, R. E. (2011). Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature, 477(7366), 592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann, J. , Brubaker, S. W. , & Monack, D. M. (2015). Cutting edge: Inflammasome activation in primary human macrophages is dependent on Flagellin. The Journal of Immunology, 195(3), 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labow, M. , Shuster, D. , Zetterstrom, M. , Nunes, P. , Terry, R. , Cullinan, E. B. , … McIntyre, K. (1997). Absence of IL‐1 signaling and reduced inflammatory response in IL‐1 type I receptor‐deficient mice. The Journal of Immunology, 159(5), 2452–2461. [PubMed] [Google Scholar]
- Lacey, C. A. , & Miao, E. A. (2019). Programmed cell death in the evolutionary race against bacterial virulence factors. Cold Spring Harbor Perspectives in Biology, 2020; 12:a036459. 10.1101/cshperspect.a036459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange, B. , Benaoudia, S. , Wallet, P. , Magnotti, F. , Provost, A. , Michal, F. , … Henry, T. (2018). Human caspase‐4 detects tetra‐acylated LPS and cytosolic Francisella and functions differently from murine caspase‐11. Nature Communications, 9(1), 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara‐Tejero, M. , Sutterwala, F. S. , Ogura, Y. , Grant, E. P. , Bertin, J. , Coyle, A. J. , … Galán, J. E. (2006). Role of the caspase‐1 inflammasome in Salmonella typhimurium pathogenesis. The Journal of Experimental Medicine, 203(6), 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock, C. N. , & Cookson, B. T. (2012). The Yersinia virulence effector YopM binds caspase‐1 to arrest inflammasome assembly and processing. Cell Host & Microbe, 12(6), 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.‐S. , Kwon, H. , Lee, E.‐Y. , Kim, D.‐J. , Park, J.‐H. , Tesh, V. L. , … Kim, M. H. (2016). Shiga toxins activate the NLRP3 inflammasome pathway to promote both production of the Proinflammatory cytokine interleukin‐1β and apoptotic cell death. Infection and Immunity, 84(1), 172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, M. , Kolodziejczyk, A. A. , Thaiss, C. A. , & Elinav, E. (2017). Dysbiosis and the immune system. Nature Reviews Immunology, 17, 219–232. [DOI] [PubMed] [Google Scholar]
- Li, J. , Brieher, W. M. , Scimone, M. L. , Kang, S. J. , Zhu, H. , Yin, H. , … Yuan, J. (2007). Caspase‐11 regulates cell migration by promoting Aip1–Cofilin‐mediated Actin depolymerization. Nature Cell Biology, 9, 276–286. [DOI] [PubMed] [Google Scholar]
- Li, J. , Yin, H. L. , & Yuan, J. (2008). Flightless‐I regulates proinflammatory caspases by selectively modulating intracellular localization and caspase activity. The Journal of Cell Biology, 181(2), 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligeon, L.‐A. , Moreau, K. , Barois, N. , Bongiovanni, A. , Lacorre, D.‐A. , Werkmeister, E. , … Lafont, F. (2014). Role of VAMP3 and VAMP7 in the commitment of Yersinia pseudotuberculosis to LC3‐associated pathways involving single‐ or double‐membrane vacuoles. Autophagy, 10(9), 1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K.‐M. , Hu, W. , Troutman, T. D. , Jennings, M. , Brewer, T. , Li, X. , … Pasare, C. (2014). Irak‐1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proceedings of the National Academy of Sciences of the United States of America, 111(2), 775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhang, Z. , Ruan, J. , Pan, Y. , Magupalli, V. G. , Wu, H. , & Lieberman, J. (2016). Inflammasome‐activated gasdermin D causes pyroptosis by forming membrane pores. Nature, 535(7610), 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Zaki, M. H. , Vogel, P. , Gurung, P. , Finlay, B. B. , Deng, W. , … Kanneganti, T. D. (2012). Role of inflammasomes in host defense against Citrobacter rodentium infection. Journal of Biological Chemistry, 287(20), 16955–16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner, M. , Kastenmüller, K. , Neuenhahn, M. , Weighardt, H. , Busch, D. H. , Reindl, W. , & Förster, I. (2008). Decreased susceptibility of mice to infection with Listeria monocytogenes in the absence of interleukin‐18. Infection and Immunity, 76(9), 3881–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie, A. , Wilson, H. L. , Kiss‐Toth, E. , Dower, S. K. , North, R. A. , & Surprenant, A. (2001). Rapid secretion of interleukin‐1β by microvesicle shedding. Immunity, 15(5), 825–835. [DOI] [PubMed] [Google Scholar]
- Man, S. M. (2018). Inflammasomes in the gastrointestinal tract: Infection, cancer and gut microbiota homeostasis. Nature Reviews Gastroenterology & Hepatology, 15(12), 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, S. M. , Hopkins, L. J. , Nugent, E. , Cox, S. , Glück, I. M. , Tourlomousis, P. , … Bryant, C. E. (2014). Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proceedings of the National Academy of Sciences of the United States of America, 111(20), 7403–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, S. M. , Karki, R. , & Kanneganti, T. D. (2017). Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunological Reviews, 277(1), 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, S. M. , Tourlomousis, P. , Hopkins, L. , Monie, T. P. , Fitzgerald, K. A. , & Bryant, C. E. (2013). Salmonella infection induces recruitment of Caspase‐8 to the inflammasome to modulate IL‐1β production. The Journal of Immunology, 191(10), 5239–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani, A. , Dinarello, C. A. , Molgora, M. and Garlanda, C. (2019). Interleukin‐1 and related cytokines in the regulation of inflammation and immunity. Immunity 50(4): 778–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan, S. , Newton, K. , Monack, D. M. , Vucic, D. , French, D. M. , Lee, W. P. , … Dixit, V. M. (2004). Differential activation of the inflammasome by caspase‐1 adaptors ASC and Ipaf. Nature, 430, 213–218. [DOI] [PubMed] [Google Scholar]
- Martinon, F. , Burns, K. , & Tschopp, J. (2002). The inflammasome: A molecular platform triggering activation of inflammatory Caspases and processing of proIL‐β. Molecular Cell, 10(2), 417–426. [DOI] [PubMed] [Google Scholar]
- Masters, S. L. , Lagou, V. , Jéru, I. , Baker, P. J. , Van Eyck, L. , Parry, D. A. , … Liston, A. (2016). Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Science Translational Medicine 8(332): 332ra345. [DOI] [PubMed]
- Matusiak, M. , Van Opdenbosch, N. , Vande Walle, L. , Sirard, J.‐C. , Kanneganti, T.‐D. , & Lamkanfi, M. (2015). Flagellin‐induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proceedings of the National Academy of Sciences of the United States of America, 112(5), 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie, E. J. , Brawn, L. C. , Hume, P. J. , Humphreys, D. , & Koronakis, V. (2009). Salmonella takes control: Effector‐driven manipulation of the host. Current Opinion in Microbiology, 12(1), 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinzer, U. , Barreau, F. , Esmiol‐Welterlin, S. , Jung, C. , Villard, C. , Léger, T. , … Hugot, J. P. (2012). Yersinia pseudotuberculosis effector YopJ subverts the Nod2/RICK/TAK1 pathway and activates Caspase‐1 to induce intestinal barrier dysfunction. Cell Host & Microbe, 11(4), 337–351. [DOI] [PubMed] [Google Scholar]
- Meixenberger, K. , Pache, F. , Eitel, J. , Schmeck, B. , Hippenstiel, S. , Slevogt, H. , … Opitz, B. (2010). Listeria monocytogenes‐infected human peripheral blood mononuclear cells produce IL‐1β, depending on Listeriolysin O and NLRP3. The Journal of Immunology, 184(2), 922–930. [DOI] [PubMed] [Google Scholar]
- Meunier, E. , Dick, M. S. , Dreier, R. F. , Schurmann, N. , Broz, D. K. , Warming, S. , … Broz, P. (2014). Caspase‐11 activation requires lysis of pathogen‐containing vacuoles by IFN‐induced GTPases. Nature, 509(7500), 366–370. [DOI] [PubMed] [Google Scholar]
- Miao, E. A. , Leaf, I. A. , Treuting, P. M. , Mao, D. P. , Dors, M. , Sarkar, A. , … Aderem, A. (2010). Caspase‐1‐induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nature Immunology, 11(12), 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, E. A. , Mao, D. P. , Yudkovsky, N. , Bonneau, R. , Lorang, C. G. , Warren, S. E. , … Aderem, A. (2010). Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proceedings of the National Academy of Sciences of the United States of America, 107(7), 3076–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, A. P. (2007). Lipopolysaccharide in bacterial chronic infection: Insights from Helicobacter pylori lipopolysaccharide and lipid A. International Journal of Medical Microbiology, 297(5), 307–319. [DOI] [PubMed] [Google Scholar]
- Mostowy, S. , Boucontet, L. , Mazon Moya, M. J. , Sirianni, A. , Boudinot, P. , Hollinshead, M. , … Colucci‐Guyon, E. (2013). The Zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathogens, 9(9), e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, X. , Souter, S. , Du, J. , Reeves, A. Z. , & Lesser, C. F. (2018). Synthetic bottom‐up approach reveals the complex interplay of Shigella effectors in regulation of epithelial cell death. Proceedings of the National Academy of Sciences of the United States of America, 115(25), 6452–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux‐Sanders, C. , Sanchez‐Garrido, J. , Hopkins, E. G. D. , Shenoy, A. R. , Barry, R. , & Frankel, G. (2019). Citrobacter rodentium–host–microbiota interactions: Immunity, bioenergetics and metabolism. Nature Reviews Microbiology, 17(11), 701–715. [DOI] [PubMed] [Google Scholar]
- Mundy, R. , Petrovska, L. , Smollett, K. , Simpson, N. , Wilson, R. K. , Yu, J. , … Frankel, G. (2004). Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infection and Immunity, 72(4), 2288–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman‐Zenevich, J. , Stuart, S. , Abdel‐Nour, M. , Girardin, S. E. , & Mogridge, J. (2017). Listeria monocytogenes and Shigella flexneri activate the NLRP1B inflammasome. Infection and Immunity, 85(11), e00338–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, G. Z. , Menheniott, T. R. , Every, A. L. , Stent, A. , Judd, L. M. , Chionh, Y. T. , … Sutton, P. (2016). The MUC1 mucin protects against Helicobacter pylori pathogenesis in mice by regulation of the NLRP3 inflammasome. Gut, 65(7), 1087–1099. [DOI] [PubMed] [Google Scholar]
- Nordlander, S. , Pott, J. , & Maloy, K. J. (2013). NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal Immunology, 7, 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo, M. C. , Johnson, D. C. , Sridharan, R. , Go, E. B. , Chui, A. J. , Wang, M. S. , … Bachovchin, D. A. (2017). DPP8 and DPP9 inhibition induces pro‐caspase‐1‐dependent monocyte and macrophage pyroptosis. Nature Chemical Biology, 13(1), 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orning, P. , Lien, E. and Fitzgerald, K. A. (2019). Gasdermins and their role in immunity and inflammation. The Journal of Experimental Medicine 216 (11), 2453–2465. [DOI] [PMC free article] [PubMed]
- Orning, P. , Weng, D. , Starheim, K. , Ratner, D. , Best, Z. , Lee, B. , … Lien, E. (2018). Pathogen blockade of TAK1 triggers caspase‐8‐dependent cleavage of gasdermin D and cell death. Science, 362(6418), 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciello, I. , Silipo, A. , Lembo‐Fazio, L. , Curcurù, L. , Zumsteg, A. , Noël, G. , … Bernardini, M. L. (2013). Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proceedings of the National Academy of Sciences of the United States of America, 110(46), E4345–E4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallett, M. A. , Crepin, V. F. , Serafini, N. , Habibzay, M. , Kotik, O. , Sanchez‐Garrido, J. , … Frankel, G. (2017). Bacterial virulence factor inhibits Caspase‐4/11 activation in intestinal epithelial cells. Mucosal Immunology, 10(3), 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo, J. J. , Gounon, P. , & Sansonetti, P. J. (1994). Polymorphonuclear leukocyte transmigration promotes invasion of colonic epithelial monolayer by Shigella flexneri . The Journal of Clinical Investigation, 93(2), 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, M. , Tourlomousis, P. , Wright, J. , Monie, T. P. , & Bryant, C. E. (2016). CARD9 negatively regulates NLRP3‐induced IL‐1β production on Salmonella infection of macrophages. Nature Communications, 7(1), 12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Morales, D. , Banda, M. M. , Chau, N. Y. E. , Salgado, H. , Martínez‐Flores, I. , Ibarra, J. A. , … Bustamante, V. H. (2017). The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PLoS Pathogens, 13(7), e1006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, L. W. , Philip, N. H. , DeLaney, A. , Wynosky‐Dolfi, M. A. , Asklof, K. , Gray, F. , … Brodsky, I. E. (2017). RIPK1‐dependent apoptosis bypasses pathogen blockade of innate signaling to promote immune defense. The Journal of Experimental Medicine, 214(11), 3171–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, L. W. , Philip, N. H. , Dillon, C. P. , Bertin, J. , Gough, P. J. , Green, D. R. , Brodsky I. E. (2016). Cell‐extrinsic TNF collaborates with TRIF signaling to promote Yersinia‐induced apoptosis. The Journal of Immunology: 1601294, 197, 4110, 4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, N. H. , Dillon, C. P. , Snyder, A. G. , Fitzgerald, P. , Wynosky‐Dolfi, M. A. , Zwack, E. E. , … Brodsky, I. E. (2014). Caspase‐8 mediates caspase‐1 processing and innate immune defense in response to bacterial blockade of NF‐κB and MAPK signaling. Proceedings of the National Academy of Sciences of the United States of America, 111(20), 7385–7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla, D. M. , Hagar, J. A. , Haldar, A. K. , Mason, A. K. , Degrandi, D. , Pfeffer, K. , … Coers, J. (2014). Guanylate binding proteins promote caspase‐11–dependent pyroptosis in response to cytoplasmic LPS. Proceedings of the National Academy of Sciences of the United States of America, 111(16), 6046–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud, L. , Sansonetti, P. J. , & Phalipon, A. (2018). Host cell targeting by enteropathogenic bacteria T3SS effectors. Trends in Microbiology, 26(4), 266–283. [DOI] [PubMed] [Google Scholar]
- Pizarro‐Cerdá, J. , Kühbacher, A. , & Cossart, P. (2012). Entry of Listeria monocytogenes in mammalian epithelial cells: An updated view. Cold Spring Harbor Perspectives in Medicine, 2(11), a010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platnich, J. M. , Chung, H. , Lau, A. , Sandall, C. F. , Bondzi‐Simpson, A. , Chen, H.‐M. , Komada T., Trotman‐Grant A. C., Brandelli J. R., Chun J., Beck P. L., Philpott D. J., Girardin S. E., Ho M., Johnson R. P., MacDonald J. A., Armstrong G. D., Muruve D. A. (2018). Shiga toxin/lipopolysaccharide activates Caspase‐4 and Gasdermin D to trigger mitochondrial reactive oxygen species upstream of the NLRP3 inflammasome. Cell Reports 25(6): 1525‐1536.e1527, 1536.e7. [DOI] [PubMed] [Google Scholar]
- Pollard, D. J. , Berger, C. N. , So, E. C. , Yu, L. , Hadavizadeh, K. , Jennings, P. , … Frankel, G. (2018). Broad‐Spectrum regulation of nonreceptor tyrosine kinases by the bacterial ADP‐Ribosyltransferase EspJ. mBio, 9:e00170‐18. https://doi.org/10.1128/mBio.00170‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, G. L. , Oates, C. V. L. , Giogha, C. , Wong Fok Lung, T. , Ong, S. Y. , Pearson, J. S. , & Hartland, E. L. (2017). Distinct roles of the antiapoptotic effectors NleB and NleF from enteropathogenic Escherichia coli . Infection and Immunity, 85(4), e01071–e01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Y. , Franchi, L. , Nunez, G. , & Dubyak, G. R. (2007). Nonclassical IL‐1β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. The Journal of Immunology, 179(3), 1913–1925. [DOI] [PubMed] [Google Scholar]
- Qu, Y. , Misaghi, S. , Newton, K. , Maltzman, A. , Izrael‐Tomasevic, A. , Arnott, D. , & Dixit, V. M. (2016). NLRP3 recruitment by NLRC4 during Salmonella infection. The Journal of Experimental Medicine, 213(6), 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, S. , Schieber, A. M. P. , O'Connor, C. P. , Leblanc, M. , Michel, D. and Ayres, J. S. (2017). Pathogen‐mediated inhibition of anorexia promotes host survival and transmission. Cell 168(3): 503–516.e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapsinski, G. J. , Wynosky‐Dolfi, M. A. , Oppong, G. O. , Tursi, S. A. , Wilson, R. P. , Brodsky, I. E. , & Tükel, Ç. (2015). Toll‐like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, Curli. Infection and Immunity, 83(2), 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam, V. A. K. , Jiang, Z. , Waggoner, S. N. , Sharma, S. , Cole, L. E. , Waggoner, L. , … Fitzgerald, K. A. (2010). The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature Immunology, 11(5), 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam, V. A. K. , Vanaja, S. K. , Waggoner, L. , Sokolovska, A. , Becker, C. , Stuart, L. M. , … Fitzgerald, K. A. (2012). TRIF licenses Caspase‐11‐dependent NLRP3 inflammasome activation by gram‐negative bacteria. Cell, 150(3), 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner, D. , Orning, M. P. A. , Proulx, M. K. , Wang, D. , Gavrilin, M. A. , Wewers, M. D. , … Lien, E. (2016). The Yersinia pestis effector YopM inhibits pyrin inflammasome activation. PLoS Pathogens, 12(12), e1006035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch, I. , Deets, K. A. , Ji, D. X. , von Moltke, J. , Tenthorey, J. L. , Lee, A. Y. , … Vance, R. E. (2017). NAIP‐NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL‐18 release via activation of Caspase‐1 and ‐8. Immunity, 46(4), 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raupach, B. , Peuschel, S.‐K. , Monack, D. M. , & Zychlinsky, A. (2006). Caspase‐1‐mediated activation of interleukin‐1β (IL‐1β) and IL‐18 contributes to innate immune defenses against Salmonella enterica Serovar Typhimurium infection. Infection and Immunity, 74(8), 4922–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, C.‐P. , Chaudhuri, R. R. , Fivian, A. , Bailey, C. M. , Antonio, M. , Barnes, W. M. , & Pallen, M. J. (2004). The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. Journal of Bacteriology, 186(11), 3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes Ruiz, V. M. , Ramirez, J. , Naseer, N. , Palacio, N. M. , Siddarthan, I. J. , Yan, B. M. , … Shin, S. (2017). Broad detection of bacterial type III secretion system and flagellin proteins by the human NAIP/NLRC4 inflammasome. Proceedings of the National Academy of Sciences of the United States of America, 114(50), 13242–13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl, S. , Shkarina, K. , Demarco, B. , Heilig, R. , Santos, J. C. , & Broz, P. (2018). ESCRT‐dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science, 362(6417), 956–960. [DOI] [PubMed] [Google Scholar]
- Sandstrom, A. , Mitchell, P. S. , Goers, L. , Mu, E. W. , Lesser, C. F. , & Vance, R. E. (2019). Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science, 364(6435), eaau1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanman, L. E. , Qian, Y. , Eisele, N. A. , Ng, T. M. , van der Linden, W. A. , Monack, D. M. , … Bogyo, M. (2016). Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. eLife, 5, e13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, J. C. , Dick, M. S. , Lagrange, B. , Degrandi, D. , Pfeffer, K. , Yamamoto, M. , … Broz, P. (2018). LPS targets host guanylate‐binding proteins to the bacterial outer membrane for non‐canonical inflammasome activation. The EMBO Journal, 37(6), e98089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, J.‐D. , Pereyre, S. , Archer, K. A. , Burke, T. P. , Hanson, B. , Lauer, P. , & Portnoy, D. A. (2011). Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proceedings of the National Academy of Sciences of the United States of America, 108(30), 12419–12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, J.‐D. , Witte, C. E. , Zemansky, J. , Hanson, B. , Lauer, P. , & Portnoy, D. A. (2010). Listeria monocytogenes triggers AIM2‐mediated Pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host & Microbe, 7(5), 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R. L. , & Lenz, L. L. (2012). Distinct licensing of IL‐18 and IL‐1β secretion in response to NLRP3 inflammasome activation. PLoS One, 7(9), e45186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte, P. , Denecker, G. , Van Den Broeke, A. , Vandenabeele, P. , Cornelis, G. R. , & Beyaert, R. (2004). Targeting Rac1 by the Yersinia effector protein YopE inhibits Caspase‐1‐mediated maturation and release of interleukin‐1β. Journal of Biological Chemistry, 279(24), 25134–25142. [DOI] [PubMed] [Google Scholar]
- Sellin, M. E. , Müller, A. A. , Felmy, B. , Dolowschiak, T. , Diard, M. , Tardivel, A. , … Hardt, W. D. (2014). Epithelium‐intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host & Microbe, 16(2), 237–248. [DOI] [PubMed] [Google Scholar]
- Semper, R. P. , Mejías‐Luque, R. , Groß, C. , Anderl, F. , Müller, A. , Vieth, M. , … Gerhard, M. (2014). Helicobacter pylori‐induced IL‐1β secretion in innate immune cells is regulated by the NLRP3 inflammasome and requires the cag Pathogenicity Island. The Journal of Immunology, 193(7), 3566–3576. [DOI] [PubMed] [Google Scholar]
- Semper, R. P. , Vieth, M. , Gerhard, M. and Mejías‐Luque, R. (2019). Helicobacter pylori exploits the NLRC4 inflammasome to dampen host defenses. The Journal of Immunology: ji1900351, 203, 2183, 2193. [DOI] [PubMed] [Google Scholar]
- Senerovic, L. , Tsunoda, S. P. , Goosmann, C. , Brinkmann, V. , Zychlinsky, A. , Meissner, F. , & Kolbe, M. (2012). Spontaneous formation of IpaB ion channels in host cell membranes reveals how Shigella induces pyroptosis in macrophages. Cell Death & Disease, 3(9), e384–e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane, A. L. , Mody, R. K. , Crump, J. A. , Tarr, P. I. , Steiner, T. S. , Kotloff, K. , … Pickering, L. K. (2017). 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and Management of Infectious Diarrhea. Clinical Infectious Diseases, 65(12), e45–e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy, A. R. , Furniss, R. C. D. , Goddard, P. J. , & Clements, A. (2018). Modulation of host cell processes by T3SS effectors In Frankel G. and Ron E. Z. (Eds.), Escherichia coli, a versatile pathogen (pp. 73–115). Cham: Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- Shenoy, A. R. , Wellington, D. A. , Kumar, P. , Kassa, H. , Booth, C. J. , Cresswell, P. , & MacMicking, J. D. (2012). GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science, 336(6080), 481–485. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Zhao, Y. , Wang, K. , Shi, X. , Wang, Y. , Huang, H. , … Shao, F. (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature, 526(7575), 660–665. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Zhao, Y. , Wang, Y. , Gao, W. , Ding, J. , Li, P. , … Shao, F. (2014). Inflammatory caspases are innate immune receptors for intracellular LPS. Nature, 514(7521), 187–192. [DOI] [PubMed] [Google Scholar]
- Slater, S. L. , Sågfors, A. M. , Pollard, D. J. , Ruano‐Gallego, D. , & Frankel, G. (2018). The type III secretion system of pathogenic Escherichia coli In Frankel G. and Ron E. Z. (Eds.), Escherichia coli, a versatile pathogen (pp. 51–72). Cham: Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- Song‐Zhao, G. X. , Srinivasan, N. , Pott, J. , Baban, D. , Frankel, G. , & Maloy, K. J. (2013). Nlrp3 activation in the intestinal epithelium protects against a mucosal pathogen. Mucosal Immunology, 7, 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, S. , Franchi, L. , He, Y. , Muñoz‐Planillo, R. , Mimuro, H. , Suzuki, T. , … Núñez, G. (2014). Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcδ. PLoS Pathogens, 10(2), e1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, S. , Mimuro, H. , Kim, M. , Ogawa, M. , Ashida, H. , Toyotome, T. , … Sasakawa, C. (2014). Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proceedings of the National Academy of Sciences of the United States of America, 111(40), E4254–E4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, S. , Suzuki, T. , Mimuro, H. , Mizushima, T. , & Sasakawa, C. (2018). Shigella hijacks the glomulin–cIAPs–inflammasome axis to promote inflammation. EMBO Reports, 19(1), 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, K. V. , Deng, M. , & Ting, J. P. Y. (2019). The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nature Reviews Immunology, 19(8), 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swimm, A. , Bommarius, B. , Li, Y. , Cheng, D. , Reeves, P. , Sherman, M. , … Kalman, D. (2004). Enteropathogenic Escherichia coli use redundant tyrosine kinases to form Actin pedestals. Molecular Biology of the Cell, 15(8), 3520–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, C. C. , Rodrigues, L. C. , Viviani, L. , Dodds, J. P. , Evans, M. R. , Hunter, P. R. , … IID2 Study Executive Committee . (2012). Longitudinal study of infectious intestinal disease in the UK (IID2 study): Incidence in the community and presenting to general practice. Gut, 61(1), 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenthorey, J. L. , Kofoed, E. M. , Daugherty, M. D. , Malik, H. S. , & Vance, R. E. (2014). Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Molecular Cell, 54(1), 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen, E. , & Sauer, J.‐D. (2016). Listeria monocytogenes and the inflammasome: From cytosolic bacteriolysis to tumor immunotherapy. Current Topics in Microbiology and Immunology, 397, 133–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinwa, J. , Segovia, J. A. , Bose, S. , & Dube, P. H. (2014). Integrin‐mediated first signal for inflammasome activation in intestinal epithelial cells. The Journal of Immunology, 193(3), 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickell, K. D. , Brander, R. L. , Atlas, H. E. , Pernica, J. M. , Walson, J. L. , & Pavlinac, P. B. (2017). Identification and management of Shigella infection in children with diarrhoea: A systematic review and meta‐analysis. The Lancet Global Health, 5(12), e1235–e1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torraca, V. , Kaforou, M. , Watson, J. , Duggan, G. M. , Guerrero‐Gutierrez, H. , Krokowski, S. , … Mostowy, S. (2019). Shigella sonnei infection of zebrafish reveals that O‐antigen mediates neutrophil tolerance and dysentery incidence. PLoS Pathogens, 15(12), e1008006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, K. , Hara, H. , Fang, R. , Hernandez‐Cuellar, E. , Sakai, S. , Daim, S. , … Kawamura, I. (2014). The adaptor ASC exacerbates lethal Listeria monocytogenes infection by mediating IL‐18 production in an inflammasome‐dependent and ‐independent manner. European Journal of Immunology, 44(12), 3696–3707. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, K. , Hara, H. , Kawamura, I. , Nomura, T. , Yamamoto, T. , Daim, S. , … Mitsuyama, M. (2010). Involvement of absent in melanoma 2 in inflammasome activation in macrophages infected with Listeria monocytogenes . The Journal of Immunology, 185(2), 1186–1195. [DOI] [PubMed] [Google Scholar]
- Tsuji, N. M. , Tsutsui, H. , Seki, E. , Kuida, K. , Okamura, H. , Nakanishi, K. , & Flavell, R. A. (2004). Roles of caspase‐1 in listeria infection in mice. International Immunology, 16(2), 335–343. [DOI] [PubMed] [Google Scholar]
- Valencia Lopez, M. J. , Schimmeck, H. , Gropengießer, J. , Middendorf, L. , Quitmann, M. , Schneider, C. , … Ruckdeschel, K. (2019). Activation of the macroautophagy pathway by Yersinia enterocolitica promotes intracellular multiplication and egress of yersiniae from epithelial cells. Cellular Microbiology, 0(0), e13046. [DOI] [PubMed] [Google Scholar]
- Vanaja, S. K. , Rathinam, V. A. K. , Atianand, M. K. , Kalantari, P. , Skehan, B. , Fitzgerald, K. A. , & Leong, J. M. (2014). Bacterial RNA:DNA hybrids are activators of the NLRP3 inflammasome. Proceedings of the National Academy of Sciences of the United States of America, 111(21), 7765–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaja, S. K. , Russo, A. J. , Behl, B. , Banerjee, I. , Yankova, M. , Deshmukh, S. D. , & Rathinam, V. A. K. (2016). Bacterial outer membrane vesicles mediate cytosolic localization of LPS and Caspase‐11 activation. Cell, 165(5), 1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimer, G. I. , Weng, D. , Paquette, S. W. M. , Vanaja, S. K. , Rathinam, V. A. K. , Aune, M. H. , … Lien, E. (2012). The NLRP12 inflammasome recognizes Yersinia pestis . Immunity, 37(1), 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, S. E. , Armstrong, A. , Hamilton, M. K. , Mao, D. P. , Leaf, I. A. , Miao, E. A. , & Aderem, A. (2010). Cutting edge: Cytosolic bacterial DNA activates the inflammasome via Aim2. Journal of Immunology, 185(2), 818–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, S. E. , Mao, D. P. , Rodriguez, A. E. , Miao, E. A. , & Aderem, A. (2008). Multiple nod‐like receptors activate caspase 1 during Listeria monocytogenes infection. Journal of Immunology, 180(11), 7558–7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, J. L. , Sanchez‐Garrido, J. , Goddard, P. J. , Torraca, V. , Mostowy, S. , Shenoy, A. R. , & Clements, A. (2019). Shigella sonnei O‐antigen inhibits internalization, vacuole escape, and inflammasome activation. mBio, 10(6), e02654–e02619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemyss, M. A. , & Pearson, J. S. (2019). Host cell death responses to non‐typhoidal Salmonella infection. Frontiers in Immunology, 10(1758). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, D. , Marty‐Roix, R. , Ganesan, S. , Proulx, M. K. , Vladimer, G. I. , Kaiser, W. J. , … Lien, E. (2014). Caspase‐8 and RIP kinases regulate bacteria‐induced innate immune responses and cell death. Proceedings of the National Academy of Sciences of the United States of America, 111(20), 7391–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor, N. , Krustev, C. , Bruce, J. , Philpott, D. J. , & Girardin, S. E. (2019). Canonical and noncanonical inflammasomes in intestinal epithelial cells. Cellular Microbiology, 0(0), e13079. [DOI] [PubMed] [Google Scholar]
- Winter, S. E. , Winter, M. G. , Atluri, V. , Poon, V. , Romão, E. L. , Tsolis, R. M. , & Bäumler, A. J. (2015). The flagellar regulator TviA reduces pyroptosis by Salmonella enterica Serovar Typhi. Infection and Immunity, 83(4), 1546–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska, M. , Thaiss, C. A. , Nowarski, R. , Henao‐Mejia, J. , Zhang, J.‐P. , Brown, E. M. , … Flavell, R. A. (2014). NLRP6 inflammasome orchestrates the colonic host‐microbial interface by regulating goblet cell mucus secretion. Cell, 156(5), 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Lu, W. , Zhang, Y. , Zhang, G. , Shi, X. , Hisada, Y. , Grover S. P., Zhang X., Li L., Xiang B., Shi J., Li X.A., Daugherty A., Smyth S. S., Kirchhofer D., Shiroishi T., Shao F., Mackman N., Wei Y., Li Z. (2019). Inflammasome activation triggers blood clotting and host death through pyroptosis. Immunity 50(6): 1401–1411.e1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Fernandes‐Alnemri, T. , & Alnemri, E. S. (2010). Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in Caspase‐1 activation by Listeria monocytogenes . Journal of Clinical Immunology, 30(5), 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynosky‐Dolfi, M. A. , Snyder, A. G. , Philip, N. H. , Doonan, P. J. , Poffenberger, M. C. , Avizonis, D. , … Brodsky, I. E. (2014). Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. The Journal of Experimental Medicine, 211(4), 653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Shi, J. , Gao, H. , Liu, Y. , Yang, Z. , Shao, F. , & Dong, N. (2019). The N‐end rule ubiquitin ligase UBR2 mediates NLRP1B inflammasome activation by anthrax lethal toxin. The EMBO Journal, 38(13), e101996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Yang, J. , Gao, W. , Li, L. , Li, P. , Zhang, L. , … Shao, F. (2014). Innate immune sensing of bacterial modifications of rho GTPases by the pyrin inflammasome. Nature, 513, 237–241. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Zhang, E. , Liu, F. , Zhang, Y. , Zhong, M. , Li, Y. , … Yan, H. (2014). Flagellins of Salmonella Typhi and nonpathogenic Escherichia coli are differentially recognized through the NLRC4 pathway in macrophages. Journal of Innate Immunity, 6(1), 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Zhao, Y. , Shi, J. , & Shao, F. (2013). Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proceedings of the National Academy of Sciences of the United States of America, 110(35), 14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, H. , Sugimoto, N. , & Tobe, T. (2015). Enteropathogenic Escherichia coli uses NleA to inhibit NLRP3 inflammasome activation. PLoS Pathogens, 11(9), e1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. C. , Clements, A. , Lang, A. E. , Garnett, J. A. , Munera, D. , Arbeloa, A. , … Frankel, G. (2014). The Escherichia coli effector EspJ blocks Src kinase activity via amidation and ADP ribosylation. Nature Communications, 5, 5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, M. H. , Man, S. M. , Vogel, P. , Lamkanfi, M. , & Kanneganti, T.‐D. (2014). Salmonella exploits NLRP12‐dependent innate immune signaling to suppress host defenses during infection. Proceedings of the National Academy of Sciences of the United States of America, 111(1), 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Mo, J. , Swanson, K. V. , Wen, H. , Petrucelli, A. , Gregory, S. M. , … Ting, J. P.‐Y. (2014). NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity, 40(3), 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Cheng, Y. , Xiong, Y. , Ye, C. , Zheng, H. , Sun, H. , … Xu, J. (2012). Enterohemorrhagic Escherichia coli specific enterohemolysin induced IL‐1β in human macrophages and EHEC‐induced IL‐1β required activation of NLRP3 inflammasome. PLoS One, 7(11), e50288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Shi, J. , Shi, X. , Wang, Y. , Wang, F. , & Shao, F. (2016). Genetic functions of the NAIP family of inflammasome receptors for bacterial ligands in mice. The Journal of Experimental Medicine, 213(5), 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Yang, J. , Shi, J. , Gong, Y.‐N. , Lu, Q. , Xu, H. , … Shao, F. (2011). The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature, 477(7366), 596–600. [DOI] [PubMed] [Google Scholar]
- Zhong, F. L. , Robinson, K. , Teo, D. E. T. , Tan, K.‐Y. , Lim, C. , Harapas, C. R. , … Reversade, B. (2018). Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. The Journal of Biological Chemistry, 293(49), 18864–18878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack, E. E. , Snyder, A. G. , Wynosky‐Dolfi, M. A. , Ruthel, G. , Philip, N. H. , Marketon, M. M. , … Brodsky, I. E. (2015). Inflammasome activation in response to the Yersinia type III secretion system requires hyperinjection of translocon proteins YopB and YopD. mBio 6(1): e02095‐02014. [DOI] [PMC free article] [PubMed]
- Zwaferink, H. , Stockinger, S. , Hazemi, P. , Lemmens‐Gruber, R. , & Decker, T. (2008). IFN‐β increases listeriolysin O‐induced membrane permeabilization and death of macrophages. The Journal of Immunology, 180(6), 4116–4123. [DOI] [PubMed] [Google Scholar]


