Abstract
Allele-specific polymerase chain reaction (PCR) (amplification-refractory mutation system, ARMS) is one of the most commonly used methods for mutation detection. However, a main limitation of ARMS-PCR is the false positive results obtained due to nonspecific priming that can take place with wild-type (WT) DNA, which often precludes detection of low-level mutations. To improve the analytical specificity of ARMS, we present here a new technology, NAPA: NaME-PrO-assisted ARMS, that overcomes the ARMS deficiency by adding a brief enzymatic step that reduces wild-type alleles just prior to ARMS. We performed this technology for the simultaneous detection of two hot-spot PIK3CA mutations (E545 K and H1047R) in circulating tumor cells (CTCs) and cell free DNA (cfDNA). The developed protocol could simultaneously detect mutation-allelic-frequency of 0.5% for PIK3CA exon 9 (E545 K) and 0.1% for PIK3CA exon 20 (H1047R) with high specificity. We further compared the developed NAPA assay with (a) ddPCR considered as the gold standard and (b) our previous assay based on the combination of allele-specific, asymmetric rapid PCR, and melting analysis. Our data show that the newly developed NAPA assay gives consistent results with both these assays (p = 0.001). The developed assay resolves the false positive signals issue derived through classic ARMS-PCR and provides an ideal combination of speed, accuracy, and versatility and should be easily applicable in routine diagnostic laboratories.
Liquid biopsy is considered today as one of the most promising approaches for the management of cancer patients; it is mainly based on the analysis of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating miRNAs, and exosomes.1 Robust methods with high sensitivity is necessary in order to evaluate the mutational status of specific genes in liquid biopsy material. Detection of DNA mutations in liquid biopsy samples provides a powerful tool for the management of cancer patients.2-4 However, the limited amount of circulating cell free DNA (cfDNA) and the excess amount of circulating wild-type (WT) DNA obtained through a standard blood draw are persistent issues that often compromise the diagnostic results.
The amplification-refractory mutation system (ARMS) is a simple approach for detecting any mutation involving single base changes or small deletions, since Taq DNA polymerase is very effective at distinguishing between a match and a mismatch at the 3′-end of a PCR primer. When the primer is fully matched to the template, the amplification proceeds with full efficiency, but when the 3′-base is mismatched, amplification is inefficient. Hence primers matched to the mutated allele can amplify the mutant alleles at the expense of WT alleles. ARMS is probably the most commonly used method for low-level mutation detection5 and has been used clinically for many years as an FDA approved assay for detecting KRAS mutations.6 However, when using ARMS primers in most cases, inefficient priming still occurs on the WT sample, yielding a late “background Cq”, originating from WT DNA, i.e., false positive results. It is highly desirable to correct for this well-known ARMS defect especially in liquid biopsy applications where a high sensitivity is required.
Nuclease-assisted minor allele enrichment using overlapping probes (NaME-PrO),7 is an enzymatic approach to remove WT DNA from multiple DNA targets selected at will, prior to DNA amplification, after which current genomic analysis processes remain substantially unchanged. In NaME-PrO, after DNA denaturation, the temperature is reduced to allow addition of a thermostable double-stranded DNA Duplex specific nuclease (DSN) and mutation-overlapping oligonucleotide probes that guide nuclease digestion to the selected WT DNA sequences.
Here we present a new, combined approach, NAPA: NaME-PrO-assisted ARMS for bypassing the known ARMS defect (i.e., generation of false positives) to enable an assay with high specificity by adding a brief enzymatic step just prior to ARMS. This novel methodology is based on a combination of NAME-PrO, multiplex ARMS-PCR, and melting analysis. By using NAME-PrO, we remove WT DNA from DNA targets and we avoid the inefficient priming of ARMS primers and the false positive signal that results from this false priming. Moreover, due to the different size of the designed PCR products, we managed to discriminate and identify each of two specific PIK3CA hotspot mutations simultaneously, by melting analysis.
We selected as a novel pilot application system the detection of PIK3CA mutations in CTCs and ctDNA. PIK3CA somatic mutations play an important role in target therapies and are the most common mutations in several types of human cancers.8 Nowadays, novel PI3K/mTOR/AKT inhibitors used as “personalized” treatment due to the importance of individual PIK3CA mutations.9,10 In breast cancer (BrCa) patients PIK3CA mutations have been detected in 18–40%;11 up to 80% of all PIK3CA mutations are localized in exons 9 and 20 (E542 K, E545 K, and H1047R). PIK3CA mutational status has already been studied in CTCs13-16 and plasma-ctDNA.17,18 Based on these studies, 30–40% of PIK3CA hotspot mutations were detected in CTC and plasma-ctDNA. Here we show that NAPA represents a time- and labor-saving as well as cost-efficient method that is readily and broadly applicable in clinical molecular diagnostic laboratories.
EXPERIMENTAL SECTION
Cell Lines.
Genomic DNA from MCF-7 cell line (c.1633G > A: E545 K; heterozygous) and T47D cell line (c.3140A > G: H1047R; heterozygous) were used as positive PIK3CA mutation controls.
Clinical Samples.
We analyzed a total of 61 samples: 24 DNAs isolated from epithelial cell adhesion molecule (EpCAM)-positive CTC fractions and 17 ctDNA samples isolated from plasma of breast cancer patients with clinically confirmed metastasis. Peripheral blood from 10 healthy female volunteers, collected for specificity studies, was processed by using the same procedure as used for patient’s samples. Thus, we analyzed 10 DNAs isolated from EpCAM-positive fractions and 10 ctDNA samples isolated from plasma of healthy volunteers.
DNA isolation from EpCAM-positive CTCs. CTCs were enriched from 20 mL of peripheral blood, using anti-EpCAM-coated immunomagnetic beads as previously described.19 gDNA was extracted from the EpCAM+ CTC-fraction and cell lines using Trizol as previously described.20,21 Isolated gDNA was dissolved in 50 μL of 8 mmol/L NaOH. The concentration of gDNA was quantified by the Nanodrop ND-1000 spectrophotometer. The quality of DNA was checked by RT-qPCR with specific primers for a WT region of the PIK3CA gene as previously described.
ctDNA Isolation from Plasma.
Plasma samples were obtained from blood collected into EDTA tubes before 2–4 h after blood draw by centrifugation at 530g for 10 min at room temperature. Plasma isolated samples were centrifuged again at 2000g for 10 min, transferred into clean 2 mL tubes, and stored at −70 °C. cfDNA was isolated from plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Germany).
Primer and Probe Designs.
For each DNA target interrogated for mutations, a distinct pair of oligonucleotide probes overlapping the target region was designed. Probes were designed to be comprised of 20–25 bp oligonucleotides in a way that they have a Tm of an average 65 °C and a range of 63–67 °C when they are fully matched to WT DNA. NAME-PRO probes are designed to bind, respectively, to the top and bottom DNA template strands with an overlap “target” region of about 10–15 bp. There probes can optionally contain a polymerase block on their 3′ end to prevent polymerase extension in subsequent amplification reactions. Probe to probe interactions including those from top and bottom strand overlap were designed to have a Tm of <50 °C. Integrated DNA Technologies OligoAnalyzer 3.1 software was used to calculate the probe Tm. A list of overlapping probes used for NaME-PrO reaction are listed in Table S1.
For each of PIK3CA exons 9 and 20, we designed in silico novel primer pairs using the Primer Premier 5.00 software (Premier Biosoft, CA). An allele-specific primer was designed for each mutation, for exon 9 was the reverse primer while for exon 20 forward primer was designed as an allele specific primer. Moreover, the other primer was designed to match with the WT region. The ARMS primers contain an additional mismatch at one of the last five nucleotides. Primer set 1 for exon 9 was designed to amplify the region (70 bp) that includes the hotspot mutation of exon 9 and primer set 2 was designed to amplify a region (104 bp) that includes the hotspot mutation of exon 20. All primers and probes were designed with attention to avoiding amplification of a pseudogene on chromosome 22 that has >95% homology to exon 9 of PIK3CA. All primers and probes sequences are given in detail in Table S1.
NAME-PRO Assay.
The NAME-PRO reaction was performed as previously described.6 A total of at least 10 ng of genomic DNA for each sample and overlapping oligonucleotides (probes) were mixed in 1× DSN (Evrogen) buffer to a final 10 μL volume, probe (concentration, 100 nM), and then the sample was placed in a Thermal Cycler (MJ Cycler) for denaturation at 98 °C for 2 min. The temperature was then reduced to 63 °C and 0.5 units of DSN (Evrogen) was added into the mixture followed by 20 min incubation at 63 °C and 2 min at 95 °C for DSN inactivation as previously described.6
Multiplex-PCR and Melting Analysis.
A multiplex real-time PCR was designed for both PIK3CA mutations. Real-time and melting curves were obtained using the Cobas 480 LightCycler (Roche). The LC-Green Plus fluorescent dye (Idaho Technology) was used for fluorescence measurements. PCR conditions and melting analysis protocols for the detection of both exons are outlined in Tables S2 and S3.
Droplet Digital PCR (ddPCR) for the Detection of PIK3CA E545 K Hotspot Mutation.
PIK3CA E545 K hotspot mutation (in exon 9) detection was also performed by using ddPCR in 41/61 of DNA samples analyzed by NAPA. These samples were analyzed with the QX200 Droplet Digital PCR System (Bio-Rad Laboratories) using TaqMan hydrolysis probes for detecting and quantifying wild-type PIK3CA, as well as PIK3CA E545 K in exon 9. One probe was specific for the wild-type sequence (wild-type reference assay/HEX, Channel 2), and the other was specific for each PIK3CA E545 K hotspot mutation (mutation assay/FAM, Channel 1). Briefly, a 20 μL reaction mixture containing 10 μL of 2× ddPCR Supermix for probes (No dUTP) (Bio-Rad Laboratories), 1 μL of 20× target primers/probe (FAM), 1 μL of 20× reference primers/probe (HEX), and 2 μL of the extracted DNA in a reaction volume of 20 μL, adjusted with sterile water, according to the manufacturer’s instructions. After droplet formation using 70 μL of generation oil (Bio-Rad) and the entire volume of the PCR mix, 40 μL of droplets was transferred for thermal cycling as follows: 95 °C for 10 min (1 cycle), 94 °C for 30 s, and 55 °C for 60 s (55 cycles) and an infinite hold at 12 °C. For each series of ddPCR assays, a nontemplate control was carried out and a positive control (MCF-7 cell line), which contained enough mutant and wild type DNA to create a double-positive cluster of greater than 100 droplets. Data analysis was performed using the QuantaSoft v1.7.4 software.
RESULTS
Development of the NAPA Assay for Simultaneous Detection of Two PIK3CA Hotspot Mutations.
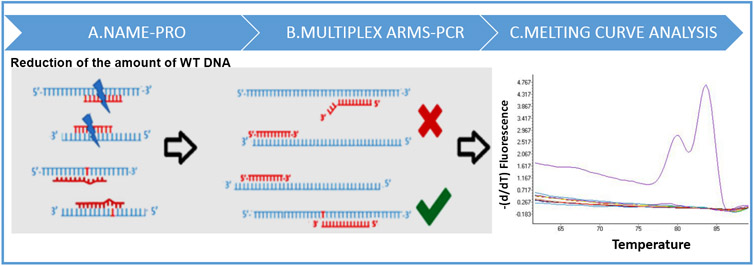
The NAPA assay is based on the combination of (a) elimination of WT DNA by multiplex NAME-PrO assay, (b) multiplex ARMS-PCR for both PIK3CA mutations, and (c) melting curve analysis (Figure 1). In this assay, we aim to enhance the specificity of ARMS-PCR by including a two-target NAME-PrO step to digest enzymatically WT DNA from both PIK3CA hotspot mutations before DNA amplification. NAME-PRO creates a strand break on both sense and antisense WT DNA strands and removes the ability of WT templates to amplify. Detection of both mutations was performed by melting curve analysis, which was based on the identification of specific peaks for each ARMS-PCR product that indicated the presence of PIK3CA mutations in both exons simultaneously as the Tm of each product was designed to be significantly different (79 and 83 °C for E545 K and H1047L, respectively). In this way, each hotspot mutation is specifically detected by the derivative melting of the mutant PIK3CA sequence for each PCR product, after ARMS-PCR amplification using the mutant allele-specific primer.
Figure 1.
Schematic representation of the experimental procedure of the NAPA assay.
Protocol Optimization.
We optimized the NAPA PIK3CA hotspot mutation assay for both E545 K and H1047L, using gDNA samples as a positive control from the cancer cell lines (MCF-7 and T47D) and as negative control wild-type from gDNA isolated from healthy donors. Analytical parameters like PCR annealing temperature, Mg2+ concentration, primers concentration, duration of each PCR step, and melting analysis conditions were optimized in detail (data not shown). The final PCR conditions and mix are outlined in Table S2. All experiments were performed in triplicate.
Analytical Sensitivity.
The analytical sensitivity of the NAPA PIK3CA assay was evaluated by mixing known concentrations of mutated gDNA (isolated from cell lines) with WT gDNA at ratios of 50%, 10%, 5%, 1%, 0.5%, 0.1%, and 0%. Melting curves were generated, and the ability to discriminate melting transitions of the cell line dilutions from that of WT sample was assessed. For PIK3CA exon 9 (E545 K), we could clearly discriminate a mutant allelic frequency of 0.5% (MCF-7 DNA), whereas for PIK3CA exon 20 (H1047R), a mutant allelic frequency of 0.1% (T47D DNA) was detectable (Figure 2).
Figure 2.
Analytical sensitivity of the NAPA assay in samples prepared by mixing known concentrations of mutated gDNA (isolated from cell lines), with WT gDNA at ratios of 50%, 10%, 5%, 1%, 0.5%, 0.1%, and 0%.
Analytical Specificity.
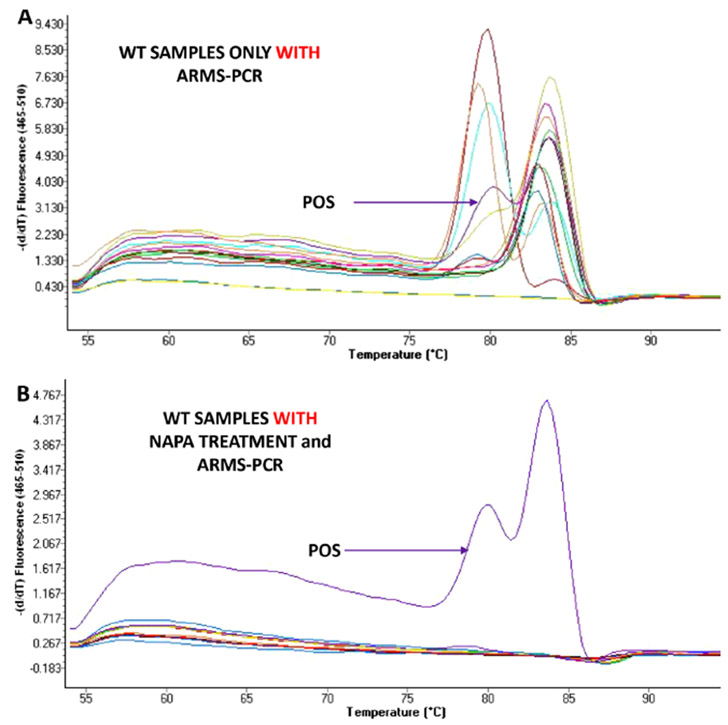
We evaluated the NAPA PIK3CA assay specificity by analyzing gDNA isolated from 10 EPCAM+ CTC fraction of healthy female volunteers and from 10 plasma ctDNA of healthy female volunteers. It is important to mention that false positive detection was observed in all cases of healthy female volunteer samples if we did not perform NAME-PrO assay prior to ARMS (Figure 3A). This observation could be explained by the fact that even by using the PIK3CA hotspot mutation-specific primers, a low amount of the wild-type sequence that is present at very high concentrations could be nonspecifically amplified. On the contrary, the developed methodology is highly specific, as we did not detect these PIK3CA mutations in any of these same samples and we did not detect false positive signals when NAME-PrO assay was performed prior to ARMS (Figure 3B).
Figure 3.
Analytical specificity of the NAPA assay by analyzing gDNA isolated from healthy female volunteers: (A) without NAME-PrO assay treatment prior to ARMS-PCR and (B) with NAME-PrO assay treatment prior to ARMS-PCR.
Detection of PIK3CA Mutations in Peripheral Blood of Breast Cancer Patients by Using the NAPA PIK3CA Assay.
EpCAM-positive CTC fraction. We applied the developed NAPA PIK3CA assay to analyze 24 peripheral blood samples from patients with clinically confirmed metastasis for both hotspot PIK3CA mutations. We detected, in total, PIK3CA mutations in 4/24 (16.7%) patients with metastatic breast cancer, 2/24 (8.3%) in exon 9, and 2/24 (8.3%) in exon 20. No-treatment control samples were also run in parallel throughout the process by omitting NaME-PrO to ensure the importance of this assay in ARMS-PCR. If the NAME-PrO step was not included, false positive signals were detected in most of these samples (20/24).
Plasma-ctDNA.
We further applied the developed NAPA PIK3CA assay to analyze 17 plasma-ctDNA samples from breast cancer patients with clinically confirmed metastasis. We detected, in total, PIK3CA mutations in 10/17 (58.8%) patients with metastatic breast cancer, 6/17 (35.3%) in exon 9, and 4/17 (23.5%) in exon 20. All healthy donor samples were negative 0/10 (0%). However, when healthy control samples were also run in parallel throughout the process, by omitting the NaME-PrO step, false positive signals were detected in all samples 10/10 (100%).
PIK3CA Mutational Status in CTCs and Corresponding Primary Tumors.
In our previous study, we have already shown that PIK3CA mutational status in CTCs could change during disease recurrence or progression in patients with breast cancer.13 In the present study, the PIK3CA mutational status in CTCs or plasma and corresponding primary tumors could only be compared in nine paired patient samples that were available. Six out these samples were found negative for both PIK3CA mutations both in the primary tumors and CTCs. For one patient that was carrying the 1633G > A PIK3CA hotspot mutation in the primary tumor, the identical mutation was also detected in the CTCs. In one patient, we identified this mutation in the primary tumor but not in CTCs, while in another patient the mutation was detected in the CTCs but not in the primary tumor. The 3140 A > G mutation was observed in 2/9 (22.2%) of primary tumors but not in CTCs and in 1/9 (11.1%) CTC samples but not in primary tumors. Six patient samples were found negative for this mutation both in the primary tumor and in CTCs.
Comparison Study between NAPA PIK3CA Assay and ddPCR for the Detection of E545 K PIK3CA Hotspot Mutation.
The developed NAPA assay gave comparable results with ddPCR (p = 0.001) (Table 1). More specifically, we analyzed for the presence of PIK3CA E545 K mutation by ddPCR in 41 samples of breast cancer patients with verified metastasis: 24 genomic DNA isolated from EpCAM-positive CTC fraction and 17 plasma-ctDNA samples (Figure 4). Moreover, 20 samples from healthy volunteers were also analyzed in order to define the cutoff of the droplets. In total, the concordance between the novel NAPA PIK3CA assay and ddPCR for PIK3CA E545 K hotspot mutation was 37/41 (90.2%) (Table 1). More specifically, 30 samples were found negative by both assays, 7 samples were found positive by the developed NAPA assay. There were only two samples found positive by the developed NAPA assay and negative by ddPCR, while two samples were found positive by ddPCR and negative by NAPA. In ddPCR, the results were based on the definition of the threshold line for each subpopulation of mutant or wild type droplets. All droplets above the threshold line of 3 500 droplets were scored as positive, whereas the negative droplets (those below the threshold line of 3 500) were scored as negative (background). Clinical samples, were considered as positive for PIK3CA E545 K hotspot mutation by ddPCR, when at least three droplets of PIK3CA E545 K DNA mutated sequences were detected.
Table 1.
Direct Comparison between the Developed NAPA Assay and ddPCR for Detecting PIK3CA E545 K Hotspot Mutation
| ddPCR | ||||
|---|---|---|---|---|
| NAPA PIK3CA assay | EpCAM-positive CTC fraction | |||
| + | + | − | total | |
| 2 | 0 | 2 | ||
| − | 2 | 20 | 22 | |
| total | 4 | 20 | 24 | |
| concordance: (22/24) 91.7% (p = 0.022 chi-square test) | ||||
| cfDNA | ||||
| + | 5 | 2 | 7 | |
| − | 0 | 10 | 10 | |
| total | 5 | 12 | 17 | |
| concordance: (15/17) 88.2% (p = 0.003, chi-square test) | ||||
Figure 4.
Representative graphs of the analysis of PIK3CA hotspot mutations by (a) droplet digital PCR: Each sample has two plots one representing the WT DNA (FAM channel, blue) and the other the mutated DNA (HEX channel, green). Each point represents a single droplet, which is scored as positive (colored) or negative (blank) depending on the fluorescent amplitude. (b) NAPA assay: Detection of both mutations by melting curve analysis as the Tm of each product was designed to be significantly different (79 and 83 °C for E545 K and H1047L, respectively). The red lines represent clinical samples, while the green lines are the positive controls of the assays.
Comparison between ddPCR and Allele-Specific/Asymmetric-Rapid PCR/Melting Analysis.
We further compared the established ddPCR with our previously developed ultrasensitive assay, based on the combination of allele-specific priming, competitive probe blocking of wild-type amplification, asymmetric PCR, and probe melting analysis.12 The concordance between ddPCR and our previously developed ultrasensitive assay for DNA samples isolated from EpCAM-positive CTC fractions was 22/24 (91.7%) for PIK3CA E545 K DNA mutation. More specifically, in EpCAM-positive CTC fractions, 19 samples were found negative by both methods. There was one sample that was found positive by our previous ultrasensitive assay and negative by ddPCR, and one that was negative by the ultrasensitive assay and positive by ddPCR (Table 2). The corresponding concordance for cfDNA samples was 12/17 (70.6%) (Table 2). Nine samples were found negative by both assays, and three samples were found positive by both assays (Table 2).
Table 2.
Direct Comparison of Allele-Specific, Asymmetric Rapid PCR and Melting Analysis Assay with ddPCR for E545 K Mutation
| ddPCR | ||||
|---|---|---|---|---|
| allele-specific, asymmetric rapid PCR and melting analysis assay | EpCAM-positive CTC fraction | |||
| + | + | − | total | |
| 3 | 1 | 4 | ||
| − | 1 | 19 | 20 | |
| total | 4 | 20 | 24 | |
| concordance: (22/24)91.7% (p = 0.008 chi-square test) | ||||
| cfDNA | ||||
| + | 3 | 2 | 5 | |
| − | 3 | 9 | 12 | |
| total | 6 | 11 | 17 | |
| concordance:(12/17)70.6% (p = 0.205, chi-square test) | ||||
DISCUSSION
While alternative mutation enrichment methods like COLD-PCR22,23 can also be used to enrich mutation-containing alleles prior to performing ARMS, a concern is that polymerase errors can occur during the initial COLD-PCR step, thus introducing false positives. Accordingly, to improve the analytical specificity of ARMS-PCR assays here we applied a new approach, NAPA: NaME-PrO-assisted ARMS, where WT alleles are removed enzymatically at the genomic DNA level, prior to amplification. Enzymatic approaches using restriction endonucleases to improve ARMS have been reported. Zhang et al. applied restriction fragment length polymorphism (RFLP) cleavage prior to ARMS TaqMan quantitative (qPCR) genotyping to screen clinical melanoma samples for the BRAF V600E mutation in a single reaction tube. To assess the specificity, the method was performed using wild-type genomic DNA and a cutoff ΔCq value was determined to exclude the nonspecific signals.24 In another approach, namely, ARMS-Plus, Zhang et al. performed the detection of epidermal growth factor receptor (EGFR) mutations in plasma samples with high specificity (97.2%), but background EGFR mutations in circulating cell-free DNA of healthy individuals were still present, thus a cutoff values of EGFR L858R and 19del as 5 copies/mL and 2 copies/mL, respectively, was used.25 Nevertheless, restriction enzyme based ARMS requires an enzyme with a recognition site at the position of the mutation, which is not always available.
In contrast, NAPA enables enzymatic removal of WT alleles at all sequence positions and hence comprises a generalized approach to improve ARMS. The assay design overcomes the known ARMS defect by adding a brief enzymatic step just prior to ARMS. Restriction enzymes specifically digest wild-type alleles, leaving the mutant alleles available for analysis and overcome false positive signals. Development of multiplex ARMS tests is more complex than for single tests, but once optimized the use of these tests is straightforward and their performance is reliable.26,27
In this assay, we added the enzymatic digestion step in order to improve the analytical specificity of ARMS-PCR. This is clearly achieved, since the oligonucleotide-probes enhance double-stranded-DNA-specific nuclease digestion at the selected targets, with high preference toward WT over mutant DNA. Our results clearly indicate that after the addition of this enzymatic step, the analytical specificity is highly improved, while the analytical sensitivity of the assay is not affected.
We performed this technology for the simultaneous detection of two hot-spot PIK3CA mutations (E545 K and H1047R) in liquid biopsy samples, CTCs and cfDNA. The developed protocol could simultaneously detect 0.5% of mutated dsDNA for PIK3CA exon 9 (E545 K) and 0.1% of mutated dsDNA for PIK3CA exon 20 (H1047R) and was highly specific. When we performed our developed NAPA assay in clinical samples of metastatic breast cancer patients, we detected PIK3CA mutations in 16.7% of EpCAM positive CTC-fraction and in 58.8% in ctDNA. The comparison of our developed assay with ddPCR and our previous ultrasensitive assay, which was based on the combination of an allele-specific, asymmetric rapid PCR and a melting analysis assay, gave comparable results with both these assays (p = 0.001).
Although, ddPCR is an emerging technology to detect a low abundance of plasma ctDNA with ultrasensitivity and absolute quantification, in many cases ddPCR instruments are not available, hence NAPA provides a simple, low cost solution for clinically relevant mutations. Not only instrumentation but the cost of reagents and consumables in the NAPA approach is much lower than ddPCR, while it is also faster to perform. In addition, the high multiplexity afforded by NAME-PRO combines well with multiplexed ARMS. On the contrary, for multiplex application, ddPCR requires multicolor instruments, otherwise each mutation should be analyzed in a separate reaction, increasing not only the cost but mainly the precious and usually limited available material in liquid biopsy samples.
Very recently, the U.S. Food and Drug Administration approved Piqray (alpelisib) tablets to be used in combination with the FDA-approved endocrine therapy fulvestrant to treat postmenopausal women, and men, with hormone receptor (HR)-positive, human epidermal growth factor receptor 2 (HER2)-negative, PIK3CA-mutated, advanced, or metastatic breast cancer following progression on or after an endocrine-based regimen.28,29 Therefore, the detection of PIK3CA mutations is of high clinical importance, and the present application to such mutations especially in liquid biopsy samples for the follow-up of patients over time provides an immediate “payoff”. The developed assay eliminates the false positive signals derived through classic ARMS-PCR and provides an ideal combination of speed, accuracy, and versatility. Moreover, it is easily applicable in many routine diagnostic laboratories and assays.
Supplementary Material
ACKNOWLEDGMENTS
This project has received funding from the Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Technology (GSRT), under Grant Agreement No. 1964. G.M.M. and I.L. were funded by U.S. National Institutes of Health Grants R33 CA217652 and R01 CA221874 to G.M.M.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b03325.
SI Tables: All primers and probes sequences and PCR conditions (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Lianidou E; Pantel K Genes, Chromosomes Cancer 2019, 58, 219–232. [DOI] [PubMed] [Google Scholar]
- (2).Bettegowda C; Sausen M; Leary RJ; Kinde I; Wang Y; Agrawal N Sci. Transl. Med 2014, 6, 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Diehl F; Li M; Dressman D; He Y; Shen D; Szabo S; et al. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 16368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Thierry AR; Mouliere F; El Messaoudi S; Mollevi C; Lopez-Crapez E; et al. Nat. Med 2014, 20, 430–5. [DOI] [PubMed] [Google Scholar]
- (5).Milbury CA; Li J; Makrigiorgos GM Clin. Chem 2009, 55, 632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bando H; Yoshino T; Tsuchihara K; Ogasawara N; Fuse N; Kojima T; et al. Br. J. Cancer 2011, 105, 403–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Song C; Liu Y; Fontana R; Makrigiorgos A; Mamon H; Kulke MH; Makrigiorgos GM Nucleic Acids Res. 2016, 44, e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Cizkova M; Susini A; Vacher S; Cizeron-Clairac G; Andrieu C; et al. Breast Cancer Res. 2012, 14, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Martini M; Ciraolo E; Gulluni F; Hirsch E Front. Oncol 2013, 3, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Andersen JN; Sathyanarayanan S; Di Bacco A; Chi A; Zhang T Sci. Transl. Med 2010, 2, 43ra55. [DOI] [PubMed] [Google Scholar]
- (11).Barbareschi M; Buttitta F; Felicioni L; Cotrupi S; Barassi F; et al. Clin. Cancer Res 2007, 13, 6064–9. [DOI] [PubMed] [Google Scholar]
- (12).Campbell IG; Russell SE; Choong DY; Montgomery KG; Ciavarella ML; et al. Cancer Res. 2004, 64, 7678–81. [DOI] [PubMed] [Google Scholar]
- (13).Markou A; Farkona S; Schiza C; Efstathiou T; Kounelis S; et al. Clin. Cancer Res 2014, 20, 5823–34. [DOI] [PubMed] [Google Scholar]
- (14).Bingham C; Fernandez SV; Fittipaldi P; Dempsey PW; Ruth KJ; et al. Breast Cancer Res. Treat 2017, 163, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tzanikou E; Markou A; Politaki E; Koutsopoulos A; Psyrri A; et al. Mol. Oncol 2019, DOI: 10.1002/1878-0261.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gasch C; Oldopp T; Mauermann O; Gorges TM; Andreas A; et al. Mol. Oncol 2016, 10, 1330–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Moynahan ME; Chen D; He W; Sung P; Samoila A; et al. Br. J. Cancer 2017, 116, 726–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Takeshita T; Yamamoto Y; Yamamoto-Ibusuki M; Tomiguchi M; Sueta A; et al. Mol. Cancer 2018, 17, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Strati A; Markou A; Parisi C; Politaki E; Mavroudis D; Georgoulias V; Lianidou E BMC Cancer 2011, 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Chimonidou M; Strati A; Tzitzira A; Sotiropoulou G; Malamos N; Georgoulias V; et al. Clin. Chem 2011, 57, 1169–1177. [DOI] [PubMed] [Google Scholar]
- (21).Mastoraki S; Strati A; Tzanikou E; Chimonidou M; Politaki E; et al. Clin. Cancer Res 2018, 24, 1500–1510. [DOI] [PubMed] [Google Scholar]
- (22).Li J; Wang L; Mamon H; Kulke MH; Berbeco R; Makrigiorgos GM Nat. Med 2008, 14, 579–84. [DOI] [PubMed] [Google Scholar]
- (23).Galbiati S; Brisci A; Lalatta F; Seia M; Makrigiorgos GM; Ferrari M; Cremonesi L Clin. Chem 2011, 57, 136–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhang Y; Qu S; Zhao J; Yu T; Guo L; Yin S; Hu X; Chen W; Lai W; Huang J Oncol. Lett 2018, 16, 1615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhang X; Chang N; Yang G; Zhang Y; Ye M; Cao J; et al. Oncotarget. 2017, 8, 112014–112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ferrie RM; Schwarz MJ; Robertson NH; Vaudin S; Super M; et al. Am. J. Hum. Genet 1992, 51, 251–262. [PMC free article] [PubMed] [Google Scholar]
- (27).Fortina P; Conant R; Monokian G; Dotti G; Parrella T; et al. Hum. Genet 1992, 90, 375–378. [DOI] [PubMed] [Google Scholar]
- (28).André F; Ciruelos E; Rubovszky G; Campone M; Loibl S; et al. N. Engl. J. Med 2019, 380, 1929–1940. [DOI] [PubMed] [Google Scholar]
- (29).Di Leo A; Johnston S; Lee KS; Ciruelos E; Lønning PE; et al. Lancet Oncol. 2018, 19, 87–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.