Abstract
Background
The sensitive detection of recurrent colorectal cancer (CRC) by the measurement of circulating tumor DNA (ctDNA) might improve the chance of a cure. This study compared a quantitative methylated ctDNA test with carcinoembryonic antigen (CEA) in the setting of surveillance for recurrence.
Methods
Blood samples collected either during surveillance or within 12 months of the confirmation of recurrence were assayed for ctDNA (methylated branched‐chain amino acid transaminase 1 [BCAT1]/Ikaros family zinc‐finger 1 protein [IKZF1]) and CEA. The optimal ctDNA threshold was determined by receiver operating characteristic analysis, and the test performance for the detection of recurrence was compared with CEA (5 ng/mL threshold).
Results
The study cohort comprised 144 eligible patients and included 50 recurrence events. The sensitivity of the methylated ctDNA test for recurrence was 66.0% (95% confidence interval [CI], 57.1%‐69.3%), which was significantly higher than the sensitivity of CEA (31.9%; 95% CI, 22.8%‐36.6%; P < .001). The sensitivity for resectable recurrence (n = 20) was also higher (ctDNA, 60.0%; CEA, 20.0%; P = .01). The specificity did not differ between the tests (ctDNA, 97.9%; 95% CI, 93.2%‐99.6%; CEA, 96.4%; 95% CI, 91.4%‐99.0%). When adjustments were made for other predictors of the presence of recurrence, a positive ctDNA test was an independent predictor (odds ratio, 155.7; 95% CI, 17.9‐1360.6; P < .001), whereas CEA was not (odds ratio, 2.5; 95% CI, 0.3‐20.6; P = .407).
Conclusions
The quantitative ctDNA test showed superior sensitivity in comparison with CEA without a difference in the specificity for detecting recurrent CRC. Longitudinal studies are warranted to further assess the utility (specifically the survival benefit) of methylated BCAT1/IKZF1 ctDNA in the surveillance of patients with CRC.
Keywords: blood test, branched-chain amino acid transaminase 1 (BCAT1), circulating tumor DNA (ctDNA), colorectal cancer, Ikaros family zinc‐finger 1 protein (IKZF1), methylation, recurrence
Short abstract
An optimal positivity threshold has been determined for an epigenetic circulating tumor DNA panel of biomarkers (methylated BCAT1 and IKZF1), and it has been applied to investigating the panel's utility in the detection of colorectal cancer recurrence. The sensitivity of the circulating tumor DNA test is superior to that of the clinically used carcinoembryonic antigen test for all recurrences (66% vs 32%) and those considered curable (60% vs 20%), with both tests having a very high specificity (98% vs 96%).
Introduction
Randomized trials have demonstrated a significant benefit of routine follow‐up investigations in identifying resectable (ie, potentially curable) metastatic disease after the resection of colorectal cancer (CRC). For this reason, surveillance is provided for those deemed to be at higher risk (stage III or high‐risk stage II). Despite this, multiple unresectable metastases are still the predominant finding at the time of detection of relapse, and this makes subsequent therapy palliative with a median survival under 3 years. This may be due to the poor sensitivity of the currently applied surveillance tools, with guidelines focused on radiological imaging (mostly yearly computed tomography [CT] scans) and regular blood tests for carcinoembryonic antigen (CEA).1 In addition to its limited sensitivity, CEA has been reported to have suboptimal specificity; the threshold value triggering a radiological assessment is not universally established; and the role of CEA, independent of imaging in clinical trials, has not been determined. There is a need to improve the timely detection of metastatic disease while it is still confined to a resectable state.2, 3 Noninvasive blood biomarkers, such as cell‐free circulating tumor DNA (ctDNA),4, 5 that are more sensitive than CEA are likely to improve survival after recurrence because they would allow earlier detection of presymptomatic recurrence as well as the detection of lower volume distant metastases.
A number of studies have personalized ctDNA biomarkers to patient‐specific mutation profiles by first detecting which somatic alterations are present in the primary tumor and then applying either 1 biomarker or a panel of the biomarkers for measurement in blood.6, 7 This technique, however, has limitations because the heterogeneity of tumor tissue and the development of subclones, which may further change during therapy, can result in a blood ctDNA mutation profile different than what was found in the primary tumor with the original testing.8
Aberrantly methylated genes are common to CRC,9 and these methylation changes can also be detected in ctDNA.10, 11 One such ctDNA test is COLVERA, which detects hypermethylated regions within branched‐chain amino acid transaminase 1 (BCAT1) and Ikaros family zinc‐finger 1 protein (IKZF1).12, 13 Furthermore, an initial evaluation of these biomarkers when they were assessed in a qualitative manner showed that this methylated ctDNA test could be up to twice as sensitive for detecting recurrent CRC in comparison with CEA.14
Because the quantitative level of methylated BCAT1/IKZF1 in the blood reflects a response to CRC resection,13 we investigated whether quantitative reporting would further improve the utility of COLVERA in a CRC monitoring setting. A cross‐sectional observational study was conducted to determine the optimal threshold of COLVERA for the detection of recurrence in patients with CRC undergoing surveillance. The performance of the ctDNA test was then compared with CEA measurements.
Materials and Methods
Adults (≥18 years old) undergoing surveillance for CRC recurrence at Flinders Medical Centre (Bedford Park, South Australia, Australia) were invited to the study (July 2014 to October 2018). Radiological examinations (CT of the chest, abdomen, and pelvis) were planned at 12‐month intervals at the discretion of the clinician in accordance with national guidelines. Consenting patients with adenocarcinoma were followed longitudinally after their cancer diagnosis, and blood samples were collected at the time of regular clinic visits, at approximately 6‐month intervals, or at the time of confirmed recurrence and were assayed for levels of ctDNA and CEA. The blood test results were not disclosed to the managing physician.
Enrolled patients were excluded if their initial treatment had not been completed, if residual disease was evident, if radiological imaging had not been performed for surveillance, if no blood samples were available for analysis that had been collected within 12 months of the determination of the recurrence status, if blood had been collected within 6 weeks of chemotherapy, or if imaging findings were indeterminate for recurrence. Patients who developed another cancer (cancer of any organ, including metachronous CRC) during the period of surveillance were also excluded.
The patients included in the analysis were those who had undergone either surgical resection or neoadjuvant therapy only (for rectal cancer) and had no evidence of residual disease on either a CT scan or rectal magnetic resonance imaging. Only 1 blood result was used per patient in this analysis: either that collected at the time of recurrence or the sample collected closest to the time of imaging (the proximate sample) as long as it had been collected within 12 months (either side) of imaging and no adjuvant therapy or surgery had been undertaken between the date of blood sampling and the date of imaging.
The study was approved by the Southern Adelaide Clinical Human Research Ethics Committee. Written informed consent was obtained before any procedures. The trial is registered with the Australian New Zealand Clinical Trials Registry (No. 12611000318987).
Clinical Procedures
Venous blood was collected (2 × 9‐mL K3‐EDTA vacutainers), prepared, and stored as previously described.14 All procedures were performed by hospital‐accredited specialists and met site‐specific standards. All pathology was reviewed by one of the authors (P.R.).
A combination of analytical and clinical information for establishing the recurrence status was applied as previously detailed.14 Recurrence was defined as locoregional when it was present at the site of anastomosis or in draining lymph nodes in patients with rectal cancer. Evidence of recurrence in the liver, lung, or other distant organs was defined as distant recurrence, with this being the principal diagnosis when locoregional recurrence was also present.
To ensure rigor in determining those with no apparent evidence of recurrence, final estimates of test accuracy were made with just patients for whom 2 sequential imaging examinations were clear (approximately 12 months apart) because CT has limited sensitivity for small lesions (<1 cm in diameter).15
Blood Testing
Each plasma sample was assayed for both ctDNA and CEA. The analysis for ctDNA used the COLVERA test (Clinical Genomics Pty, Ltd, North Ryde, New South Wales, Australia) and involved DNA extraction from plasma and bisulfite conversion, which was followed by triplex assays with real‐time polymerase chain reaction using primers for methylated BCAT1 and IKZF1 as well as β‐actin for the internal control as previously described12 but with some postanalytical changes (described in the Supporting Methods). The level of methylation was expressed as the total sum of methylated BCAT1 and IKZF1 per processed specimen. The plasma concentration of CEA was determined with the LIAISON CEA test as recommended by the manufacturer (DiaSorin SpA, Saluggia, Italy). A CEA result ≥5 ng/mL was deemed positive. All samples were analyzed without knowledge of the clinical status.
Statistical Analyses
We previously reported a difference of 31% in sensitivity with an indeterminate result for proportion discordance between the qualitative methylated BCAT1/IKZF1 ctDNA and CEA blood tests.14 Under the assumption of a 31% difference and a 55% discordant proportion, a sample size of 46 events of recurrence was estimated to provide 80% power to compare the 2 tests with the McNemar test for difference of proportions.
The principal outcome measures were true‐ and false‐positive rates of the methylated BCAT1/IKZF1 ctDNA and CEA tests according to recurrence status. A receiver operating characteristic analysis was applied with Youden's index to determine the optimal threshold for the ctDNA test. A 2‐sided McNemar test was used for paired concordance analysis. Discrete and continuous data were compared with a Z‐score 2‐population proportion test and a Wilcoxon rank‐sum test, respectively. A multivariate logistic regression analysis was performed to determine independent predictors of recurrence (sex, age, tumor location, lesion size, differentiation, apical lymph node involvement, and lymphatic and perineural invasion). A backward selection method was used to remove variables that did not provide evidence of an association with recurrence to create a parsimonious model. All analyses were completed in Stata version 13.1 with the exception of the comparison of predictive values, which used the R package DTComPair. P values <.05 were considered statistically significant.
Results
Characteristics of the Patients
A total of 548 volunteers were enrolled into the study (61.1% male; median age at diagnosis, 66.3 years; range, 23.9‐85.9 years; Fig. 1). The main reason for the exclusion of patients from the analysis was that imaging had not yet been undertaken. In 340 patients under surveillance (Table 1), 67 were diagnosed with a recurrence (19.7%), and 50 had a blood sample collected within 12 months of the recurrence confirmation (of the 17 not eligible for analysis, 1 provided insufficient plasma; 16 did not have blood collected within 12 months of the recurrence status assessment, or blood was not collected before the treatment of recurrence). Among the eligible 177 patients without clinically apparent recurrences, 94 (53.1%) had 2 clear consecutive radiological assessments (minimizing the chance of a small lesion being missed), with the second clear image taken a median of 12.6 months after the first image (interquartile range [IQR], 11.1‐17.4 months). Among the 96 patients not eligible for analysis, 9 provided insufficient plasma for the assay; 87 did not have blood collected within 12 months of the recurrence status assessment, or blood was not collected before treatment. The timing between the blood collection and the imaging event is shown in Supporting Figure 1.
Figure 1.
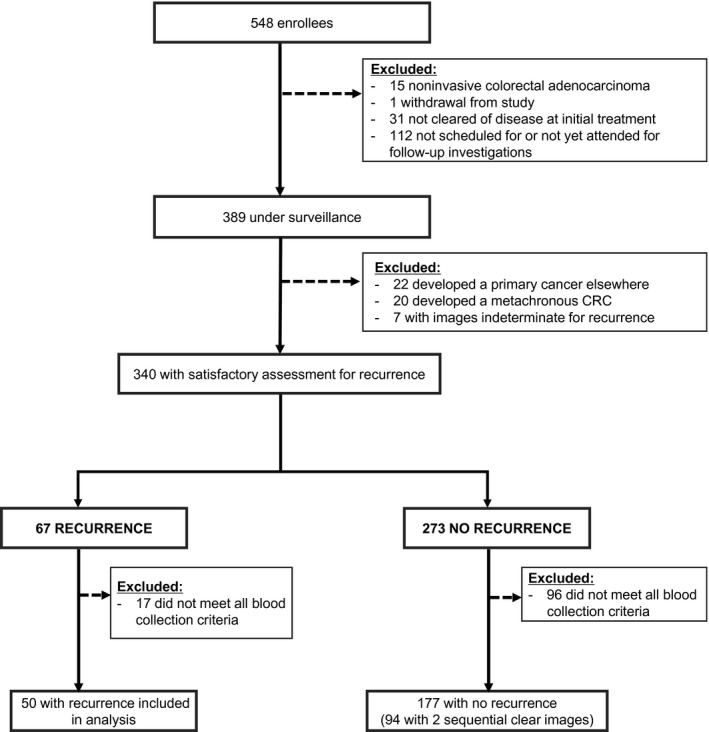
Disposition of the volunteers. CRC indicates colorectal cancer.
Table 1.
Patient Characteristics
| Characteristic | Recurrence (n = 50) | No Recurrence (n = 94) | P |
|---|---|---|---|
| Age at diagnosis, median (IQR), y | 65.1 (55.7‐73.7) | 62.5 (51.1‐72.3) | .284 |
| Sex: male, No. (%) | 34 (68.0) | 58 (61.7) | .454 |
| Primary cancer, No. (%) | |||
| Stage | |||
| I | 0 (0) | 21 (22.3) | <.001 |
| II | 13 (26.0) | 37 (39.4) | .109 |
| III | 30 (60.0) | 32 (34.8) | .004 |
| IV | 7 (14.0) | 4 (4.4) | .040 |
| Location | |||
| Right colona | 12 (24.0) | 37 (39.4) | .064 |
| Left colon | 16 (32.0) | 30 (31.9) | 1.000 |
| Rectum | 22 (44.0) | 27 (28.7) | .065 |
| T stage | |||
| T1 | 0 (0) | 12 (12.8) | .008 |
| T2 | 2 (4.0) | 14 (14.9) | .048 |
| T3 | 26 (52.0) | 51 (54.3) | .798 |
| T4 | 20 (40.0) | 17 (18.1) | 0004 |
| Tx | 2 (4.0) | 0 (0) | .515 |
| Nature of treatment | |||
| Resection ± adjuvant therapy | 41 (82.0) | 93 (98.9) | <.001 |
| Neoadjuvant therapy only | 9 (18.0) | 1 (1.1) | <.001 |
| Location of recurrence, No. (%) | |||
| Locoregionalb | 13 (26.0) | N/A | N/A |
| Distant | 37 (74.0) | N/A | N/A |
| Time between diagnosis and imaging status, median (IQR), mo | 24.2 (16.1‐35.0) | 18.2 (13.0‐28.9) | .010 |
| Time between proximate blood and imaging status, median (IQR), mo | 2.6 (0.8‐5.5) | 1.4 (0.3‐4.7) | .258 |
| Time between CRC surgery (or rectal neoadjuvant therapy) and blood collection, median (IQR), mo | 18.8 (9.6‐28.0) | 17.3 (11.0‐28.3) | .528 |
| Follow‐up since diagnosis, median (IQR), mo | 47.8 ( 33.7‐61.9) | 46.3 (31.7‐53.3) | .205 |
| Follow‐up after imaging status, median (IQR), mo | 17.1 (9.1‐29.1) | 12.6 (10.9‐17.9) | .102 |
| Follow‐up after blood collection, median (IQR), mo | 18.2 (9.1‐31.5) | 23.7 (12.3‐34.1) | .182 |
Abbreviations: CRC, colorectal cancer; IQR, interquartile range; N/A, not applicable.
The right colon included the colon proximal to the splenic flexure.
All were rectal cancers clear of residual disease on imaging at the conclusion of neoadjuvant therapy.
The median time from the cancer diagnosis to the latest CT scan or death for all analyzed patients was 46.3 months (IQR, 32.7‐57.2 months), with follow‐up times for nonrecurrence and recurrence patients given in Table 1. The time between the primary diagnosis and the imaging event used in the analysis was 6 months shorter in the nonrecurrence group versus the recurrence group (Table 1); nonetheless, the median follow‐up times for both groups were long at 46.3 months (IQR, 31.7‐53.3 months) for nonrecurrence patients and 47.8 months (IQR, 33.7‐61.9 months) for patients diagnosed with recurrence. Similar features were observed in patients who were classed as being without recurrence on the basis of a single image (Supporting Table 1).
Patients with recurrence were more likely to have had the primary tumor diagnosed at a later stage (III or IV) or to have deeper invasion (T3 or T4). The majority of recurrences were distant in location (37 of 50 [74.0%]; Table 1), with 43.2% of these patients having metastases found in the lungs, 29.7% having metastases found in the liver, and 43.2% having metastases found in other locations (including distant nodes [para‐aortic, mediastinal, neck, and peripancreatic nodes], the pancreas, and the peritoneum). Overall, 20 of the 50 patients (40%) with recurrence underwent surgery with curative intent.
Although all patients had COLVERA testing performed, CEA results were available for only 47 of the 50 recurrence patients and for 158 of the 177 nonrecurrence patients.
Test Detection of Recurrence
The optimal positivity threshold of the ctDNA test for recurrence detection was 12.8 pg per sample (Fig. 2). The area under the curve was 0.819 (95% confidence interval [CI], 0.744‐0.894; P < .001). Comparing the qualitative and quantitative thresholds, we found that the specificity significantly improved for the quantitative test from 90.4% to 97.9% (P = .023) without a reduction in sensitivity (66%; Table 2). The quantitative values for each patient are displayed in Figure 3.
Figure 2.
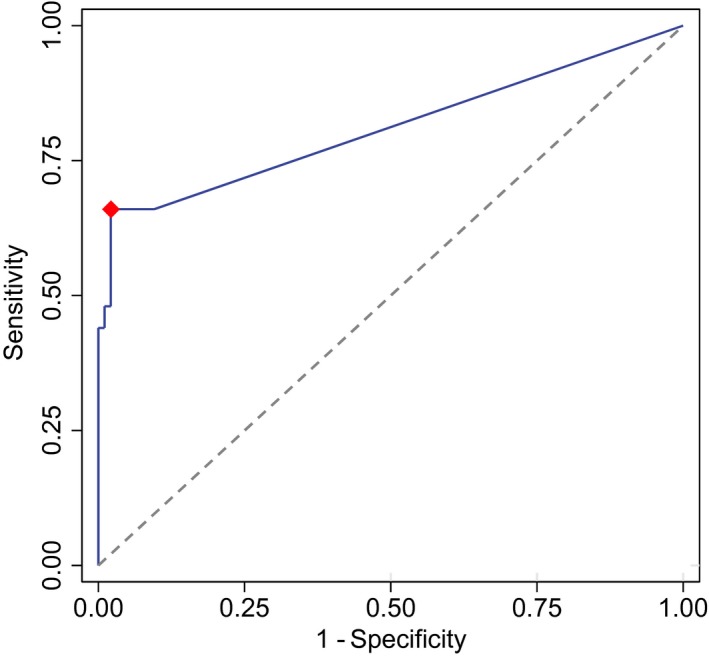
Receiver operating characteristic analysis of quantitative BCAT1/IKZF1 circulating tumor DNA (n = 144). The red dot is the optimal point (Youden's index). BCAT1 indicates branched‐chain amino acid transaminase 1; IKZF1, Ikaros family zinc‐finger 1 protein.
Table 2.
Performance Statistics for CEA and Methylated BCAT1/IKZF1 ctDNA
| CEA | Qualitative ctDNAa | Quantitative ctDNAb | |
|---|---|---|---|
| No. | 131 (47 with recurrence) | 144 (50 with recurrence) | 144 (50 with recurrence) |
| Sensitivity, % | 31.9 (22.8‐36.6) | 66.0 (55.3‐74.0) | 66.0 (57.1‐69.3) |
| Specificity, % | 96.4 (91.4‐99.0) | 90.4 (84.7‐94.7) | 97.9 (93.2‐99.6) |
| Likelihood ratio (+) | 8.94 (2.64‐38.04) | 6.89 (3.62‐13.89) | 31.0 (8.35‐183.81) |
| Likelihood ratio (–) | 0.706 (0.640‐0.845) | 0.376 (0.275‐0.528) | 0.347 (0.308‐0.460) |
| Odds ratio | 12.7 (3.43‐46.69) | 18.3 (7.44‐45.20) | 89.3 (19.56‐407.53) |
Abbreviations: BCAT1, branched‐chain amino acid transaminase 1; CEA, carcinoembryonic antigen; ctDNA, circulating tumor DNA; IKZF1, Ikaros family zinc‐finger 1 protein.
All patients with recurrences (n = 50) and all patients without recurrences on the basis of 2 serial images (n = 94) were included. The values within parentheses are 95% confidence intervals.
The threshold was 0 pg per sample.
The threshold was 12.8 pg per sample.
Figure 3.
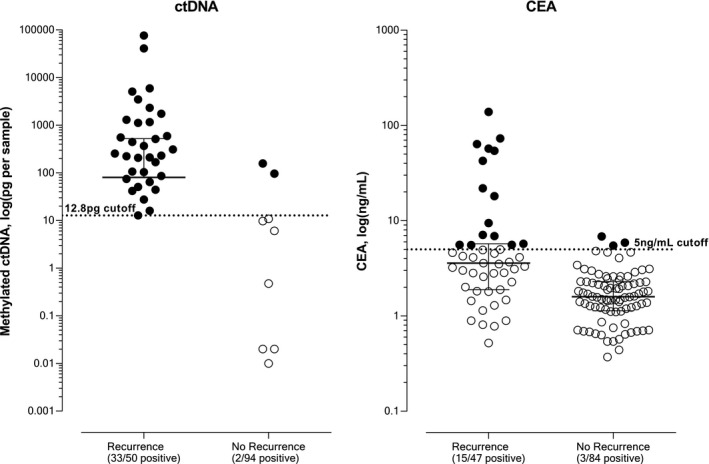
Quantitative levels of total methylation and CEA for patients with and without recurrence. Test positivity thresholds are shown with the broken lines. Closed markers represent results considered positive, and open markers represent results considered negative. CEA indicates carcinoembryonic antigen; ctDNA, circulating tumor DNA.
According to samples with paired CEA and ctDNA testing (n = 131 [47 with recurrence]), the sensitivity of the quantitative ctDNA test for the detection of recurrence was significantly greater than that of CEA (32 of 47 [68.1%] vs 15 of 47 [31.9%]; P = .0002 [McNemar test]), whereas there was no statistically significant difference between the specificities of ctDNA and CEA (82 of 84 [97.6%] vs 81 of 84 [96.4%]; P = 1.000). The respective test performance characteristics for the full data set are provided in Table 2. An analysis of the 83 additional patients who had only a single radiological investigation to exclude recurrence revealed no change in the specificity for either the quantitative ctDNA test (96.8%) or the CEA test (94.9%; P = .579). The superior sensitivity of the ctDNA test remained when patients with rectal cancer given only neoadjuvant therapy were excluded from the analysis (61.0% vs 31.6%; Table 3).
Table 3.
Sensitivity of Quantitative Methylated BCAT1/IKZF1 ctDNA and CEA for Recurrence According to Cancer Features and Treatment
| Test Sensitivity for Recurrence, n/N (%) | P | ||
|---|---|---|---|
| ctDNA | CEA | ||
| Stage at primary diagnosis | |||
| I | N/A | N/A | N/A |
| II | 9/13 (69.2) | 4/13 (30.8) | .0499 |
| III | 21/30 (70.0) | 11/29 (37.9) | .013 |
| IV | 3/7 (42.9) | 0/5 (0) | .310 |
| Treatment of primary tumor | |||
| Resection ± adjuvant therapy | 25/41 (61.0) | 12/38 (31.6) | .009 |
| Neoadjuvant therapy only (low rectal cancers) | 8/9 (88.9) | 3/9 (33.3) | .016 |
| Nature of recurrence | |||
| Locoregional | 10/13 (76.9) | 2/13 (15.4) | .006 |
| Distant | 23/37 (62.1) | 13/34 (38.2) | .044 |
| Treatment of recurrence | |||
| Treatment with curative intent | 12/20 (60.0) | 4/20 (20.0) | .010 |
Abbreviations: BCAT1, branched‐chain amino acid transaminase 1; CEA, carcinoembryonic antigen; ctDNA, circulating tumor DNA; IKZF1, Ikaros family zinc‐finger 1 protein; N/A, not applicable.
The difference between the positive predictive values of the quantitative ctDNA test and CEA (94.3% vs 83.3%; P = .262) was not significant, but the negative predictive value was significantly greater for ctDNA (84.4% vs 71.7%; P < .001). Because predictive values are dependent on the prevalence of recurrence in a population, the positive and negative predictive values across a broad range of prevalences for each test are shown in Figure 4. The predictive values were consistently higher for the methylated ctDNA test, especially within the expected range for the prevalence of recurrence of 0.1 to 0.3.5
Figure 4.
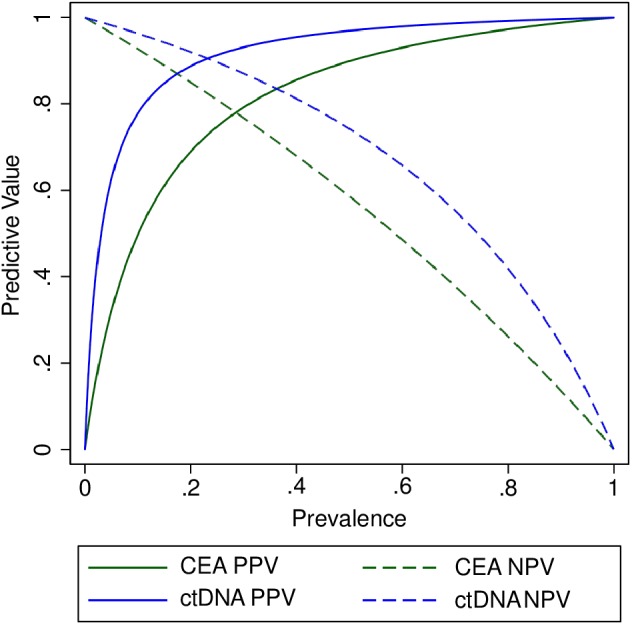
Predictive values according to prevalence based on results from matched blood samples for methylated BCAT1/IKZF1 ctDNA and CEA (n = 131). The expected range for the prevalence of recurrence is 0.1 to 0.3.5 The thresholds are 5 ng/mL for CEA and 12.8 pg per sample for ctDNA. BCAT1 indicates branched‐chain amino acid transaminase 1; CEA, carcinoembryonic antigen; ctDNA, circulating tumor DNA; IKZF1, Ikaros family zinc‐finger 1 protein; NPV, negative predictive value; PPV, positive predictive value.
The odds ratio for recurrence for a positive CEA test versus a negative CEA test was 12.7 (95% CI, 3.4‐46.7), whereas the odds ratio for recurrence for a positive quantitative methylated ctDNA test versus a negative methylated ctDNA test was 89.3 (95% CI, 19.6‐407.5; Table 4). When adjustments were made for other predictors of recurrence, including the stage at primary diagnosis, a positive CEA result was not an independent predictor, but a positive methylated ctDNA result remained independently associated with recurrence (odds ratio, 155.7; 95% CI, 17.9‐1360.6).
Table 4.
ORs for the Presence of Recurrence Associated With a Positive Blood Test (Quantitative Methylated BCAT1/IKZF1 ctDNA or CEA) or Late‐Stage CRC
| Univariate OR for Recurrence (95% CI) | P | Adjusted OR for Recurrence (95% CI) | P | |
|---|---|---|---|---|
| Positive ctDNA (vs negative ctDNA) | 89.3 (19.6‐407.5) | <.001 | 155.7 (17.9‐1360.6) | <.001 |
| Positive CEA (vs negative CEA) | 12.7 (3.4‐46.7) | <.001 | 2.5 (0.3‐20.6) | .407 |
| Late‐stage (stage III or IV) CRC (vs stage I or II) | 4.3 (2.1‐9.4) | <.001 | 4.6 (1.4‐15.5) | .013 |
Abbreviations: BCAT1, branched‐chain amino acid transaminase 1; CEA, carcinoembryonic antigen; CI, confidence interval; CRC, colorectal cancer; ctDNA, circulating tumor DNA; IKZF1, Ikaros family zinc‐finger 1 protein; OR, odds ratio.
Nature of Recurrence
Table 3 shows the test sensitivity stratified by diagnostic details and recurrence outcomes. There were no differences in the sensitivity of the methylated ctDNA test between locoregional (n = 13) and distant recurrences (n = 37; 76.9% and 62.1%, respectively; P = .53), but both values were higher than those for CEA (15.4% [P = .006] and 38.2% (P = .044], respectively; Table 3). For recurrences considered amendable to surgery with curative intent (n = 20), the quantitative methylated ctDNA test was positive in 60.0% of patients, whereas the CEA test was positive in only 20.0% (P = .010).
As for recurrences with a detectable methylated ctDNA signal (>0 pg per sample), those with metastases found in distant locations (n = 23) had a significantly higher total mass of methylated ctDNA than those with recurrences located in the bowel or a regional location (n = 10; median, 446.0 vs 69.2 pg per sample; P = .002). There were no significant differences seen in the positive CEA concentrations for distant (21.9 ng/mL) and locoregional recurrences (7.5 ng/mL; P = .219), but the sample sizes were small (13 and 2, respectively).
Test Concordance
The concordance between the 2 tests is shown in Supporting Table 2. Fourteen (29.8%) were positive by both tests, whereas methylated ctDNA detected an additional 18 patients (38%) missed by CEA (P < .001). Only 1 case (2.1%) with recurrence was positive for CEA but negative for ctDNA. In patients without clinically detectable recurrence, no specimen was positive for both tests; 2 (2.4%) were positive by methylated ctDNA only, and 3 (3.6%) were positive by CEA only (P = .655). Clinical details for the nonconcordant cases are summarized in Supporting Table 3; however, because just 1 case was positive for CEA and negative for ctDNA, it was not possible with this limited data set to ascertain whether recurrences differed in their characteristics when they were detected only by CEA or by methylated ctDNA.
When either the ctDNA or CEA test was positive, the sensitivity for the detection of recurrence was 70.2% (33 of 47) with a corresponding specificity of 94.0% (79 of 84). This was similar to the results with methylated ctDNA alone and indicated little incremental utility from combining the 2 tests.
Test Results Over Time
The timing of blood collection with respect to recurrence could be divided into the following: 1) at the time of recurrence, 2) up to 6 months before the confirmation of recurrence, and 3) 6 to 12 months before the confirmation of recurrence. At the time of recurrence, the methylated ctDNA test was positive in 78.3% (18 of 23), whereas CEA was positive in 34.8% (8 of 23; P = .004 [McNemar test]). Up to 6 months before knowledge of recurrence, the ctDNA test was positive for 65.0% of the patients (13 of 20), whereas the CEA test was positive for 35% (7 of 20; P = .0771). No CEA test was positive in blood samples collected more than 6 months before the confirmation of recurrence, whereas 40% of ctDNA tests were positive (2 of 5) during this time period, with 1 blood test positive for ctDNA 11.6 months before the confirmation of recurrence in the lungs.
Discussion
This cross‐sectional, observational study set out to determine whether a quantitative methylated BCAT1/IKZF1 ctDNA assay was a better test for detecting recurrent CRC than CEA. This study, conducted in the context of usual care, showed that the ctDNA test facilitated more accurate and earlier detection of recurrent CRC than CEA, regardless of whether the recurrence was locoregional or distant. The quantitative methylated ctDNA test was twice as sensitive as CEA but with similar specificity. The odds for recurrence given a positive test were significantly higher for methylated ctDNA than CEA. Furthermore, CEA was not a significant identifier of recurrence in a multivariate analysis including the methylated ctDNA test. Because of the better detection of recurrence (including the detection of recurrences treated with curative intent), if this test were to be used to trigger earlier‐than‐scheduled radiological imaging, a survival benefit might be achieved.
It is important that any tool for surveillance monitoring for the recurrence of cancer be highly sensitive and specific to detect disease early (while it is curable) while also preventing unnecessary clinical procedures in the case of a false‐positive. CEA is the only blood test currently recommended for use in the surveillance of patients with CRC to aid in the detection of recurrence.1, 16, 17, 18, 19 However, studies have questioned the suitability of CEA for surveillance because of its low sensitivity and specificity.20 In the current study, we used a commonly applied cutoff value of 5 ng/mL, and although specificity estimates were high, the sensitivity of CEA for recurrence was only 32%. This sensitivity falls within the lower range that has been reported.21 The reasons are unclear, but this is similar to what we have previously observed.14 It is possible that serial changes in the CEA concentration might be more informative than a single test, but this could also be true for the methylated ctDNA test and warrants future evaluation.
In contrast to causes for false positivity in CEA,22 previous studies have shown that smoking, age, and sex are not associated with higher positivity rates for methylated BCAT1/IKZF1 ctDNA.14, 23 Several studies have reported that ctDNA levels significantly correlate with tumor volume for both mutation24 and methylation biomarkers.25, 26 We, therefore, investigated whether applying a threshold level based on total methylation could improve test performance. Using the quantitative assay, we now demonstrate that it is possible to improve specificity from 90.4% to 97.9% without compromising sensitivity, and this results in the high odds ratio for recurrence of a positive methylated ctDNA result. Because imaging requires a lesion to be at least 5 to 10 mm to be detectable, a clinically undetectable recurrence of smaller size might account for a positive ctDNA test. Consequently, we also determined specificity only in those patients for whom 2 serial imaging investigations were negative. This minimized the possibility of apparent false‐positive results based on a single imaging event.
The overall sensitivity of the quantitative methylated ctDNA assay of 66% was twice the sensitivity of CEA (32%). The methylated ctDNA test detected 18 additional cases of recurrence (38%) in comparison with CEA. If one were to use both tests together, the sensitivity for recurrence if either were positive rose a little, but specificity fell in this data set. However, in the multivariate analysis, CEA was not a significant predictor for the presence of recurrence when the ctDNA result was included. In other words, there may be little to gain by complementary use of these 2 markers.
We have previously demonstrated that significant methylation of either BCAT1 or IKZF1 is observed in more than 95% of CRC tissue.13 This observation contrasts with ctDNA tests based on DNA mutations because few specific gene mutations are present in more than 5% to 10% of cancers and mutation patterns are highly variable on account of tumor heterogeneity; this means that mutation profiles of distant recurrences may not match that of the primary tumor.8, 27 Use of a mutation panel improves test performance and may identify patients at high risk for recurrence7; however, there are challenges in identifying individual‐specific mutations.7, 28 Patient‐specific somatic mutation tests for recurrence require tissue genotyping followed by interrogation of large proportions of the genome provided that the DNA is reliably released into the circulation. In contrast to mutation detection using a large panel of biomarkers, a small panel of epigenetic ctDNA biomarkers may provide a more cost‐effective and technologically simpler approach, although the utility of either assay in patient monitoring is subject to the same limitations of release of ctDNA into the circulation.7, 14, 29
Studies addressing biomarker accuracy for the detection of recurrence require a rigorous “diagnostic gold standard” as well as sampling for the biomarker at a time when recurrence is known to be present without the external influence of other variables such as treatment. We chose a window within 12 months of either side of the confirmation of recurrence status to allow for variability in the timing of the collection of samples, which were not similarly timed in all patients. A limitation of the study is that the analysis was performed in a test set without a separate validation set, and it focused on the detection of recurrence rather than the potential survival value of detection by the ctDNA. Such a study, in which a positive ctDNA test triggers earlier imaging than would otherwise occur, is now ethically justified but would require a prospective study with a different design.
Our data were collected from patients undergoing post‐CRC surveillance, with this study cohort having a recurrence rate of 20%, which is similar to that reported in the literature.5 Therefore, although the data collected allowed a focus on test accuracy for the detection of recurrence when present, a positive methylated BCAT1/IKZF1 ctDNA test did not trigger earlier‐than‐scheduled imaging. Thus, the true value of the ctDNA test for the detection of actionable recurrences might have been underestimated in cases with a positive ctDNA test before imaging because the lesion might have progressed in the interim. On the other hand, because the ctDNA result was not used to trigger imaging, we were able to observe that this blood test was positive up to 11.6 months before the confirmation of recurrence, whereas no CEA test was positive more than 6 months before the confirmation of recurrence. The clinical utility for methylated BCAT1/IKZF1 ctDNA testing for the surveillance of recurrence will be more conclusively determined with a randomized controlled trial. A trial should commence at the end of the initial CRC treatment with survival as the endpoint, and separate arms should use frequent CEA or methylated BCAT1/IKZF1 ctDNA testing to trigger earlier‐than‐scheduled imaging. This would determine whether surveillance that includes frequent methylated BCAT1/IKZF1 ctDNA testing has an incremental benefit over current guidelines in which regular imaging is scheduled.
A further limitation of the study is that the treatment protocols did not always include surgery for patients with rectal cancer. A small proportion of patients did not undergo resection of the primary rectal cancer after neoadjuvant therapy, although they were included in the analysis only if there was no evidence of residual disease after such treatment. This watch‐and‐wait approach is being trialed.30 Nonetheless, we showed that the ctDNA test had better sensitivity than CEA for recurrence whether these patients were considered or not (Table 3), and the ctDNA test was positive in 8 of the 9 patients (88.9%) with rectal cancer with only neoadjuvant therapy who had a recurrence. This highlights the ability of the methylated ctDNA test to be applied after different treatment protocols.
The strengths of the current study are as follows: 1) test results could be paired in almost all individuals, and when they were paired, plasma was collected at the same time; 2) the study population was typical of current clinical practice; 3) apparent false‐positives were verified by the determination of specificity primarily in those patients without recurrence for whom 2 serial images were taken; and 4) because the methylated ctDNA test did not trigger an early CT scan, we were able to identify the potential for a shorter lead time to a recurrence diagnosis in comparison with CEA.
Although recurrence occurs less frequently in patients with stage I or II CRC and adjuvant therapy is less likely to be used in these patients, methylated BCAT1/IKZF1 was present above threshold levels in 69% of the recurrences occurring in patients initially diagnosed with stage II CRC. This observation suggests that the methylated ctDNA blood test could be usefully applied in monitoring these patients. These findings prompt the need to perform interventional studies to assess where a positive methylated BCAT1/IKZF1 test result might inform the recommendation of administering adjuvant therapy to stage II patients, such as that being investigated in clinical trials with panels of personalized somatic mutations.7, 31
In conclusion, the methylated BCAT1/IKZF1 ctDNA test is twice as sensitive as CEA for detecting recurrent CRC during the monitoring of patients after their initial treatment. We have shown that the quantitative assay allows a substantial improvement in specificity, while maintaining high sensitivity, in comparison with a qualitative determination. The adjusted odds ratio of a positive methylated ctDNA test for recurrent disease being present was 156, whereas it was 2.5 for CEA. Moreover, if methylated BCAT1/IKZF1 were to be used to monitor patients with CRC for recurrence after initial successful treatment and lead to earlier scheduled radiological imaging when positive, we predict that recurrence would be likely to be detected at a time when it is more amenable to curative therapy. Consequently, these findings justify a prospective randomized controlled trial comparing the methylated BCAT1/IKZF1 ctDNA test with CEA, including serial changes in both markers during longitudinal surveillance, to determine whether the better sensitivity of the ctDNA assay translates into improvements in survival.
Funding Support
This study was funded in part by the National Health and Medical Research Council (APP1006242 and APP1017083) and Clinical Genomics Pty, Ltd. Graeme P. Young and Christos Karapetis are recipients of a grant funded by the financial support of Cancer Council SA's Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health together with the support of the Flinders Medical Centre Foundation, its donors, and partners.
Conflict of Interest Disclosures
Erin L. Symonds reports grants and nonfinancial support from Clinical Genomics Pty, Ltd, and Eiken Chemical Company outside the submitted work. Susanne K. Pedersen, David Murray, Frederick S. Jones, and Lawrence LaPointe are paid employees of Clinical Genomics. Pedersen also reports 4 patents co‐owned by Clinical Genomics and the Commonwealth Scientific and Industrial Research Organisation, and LaPointe is a shareholder of Clinical Genomics, which owns multiple patents for biomarkers presented in this article. Eva Segelov reports grants from Clinical Genomics outside the submitted work. Graeme P. Young is a paid consultant for Clinical Genomics and reports that his family hold shares in the company. The other authors made no disclosures.
Author Contributions
Erin L. Symonds: Study design, data curation, analysis, and writing. Susanne K. Pedersen: Study design, analysis, and writing. David Murray: Data curation, statistical analysis, and writing. Susan E. Byrne: Methodology, project administration, data curation, clinical analysis, and critical revisions. Amitesh Roy: Advisor for study design, pathology review, clinical analysis, and critical revisions. Christos Karapetis: Advisor for study design, clinical analysis, and critical revisions. Paul Hollington: Methodology, clinical analysis, and critical revisions. Philippa Rabbitt: Methodology, pathology review, clinical analysis, and critical revisions. Frederick S. Jones: Study design and critical revisions. Lawrence LaPointe: Study design and critical revisions. Eva Segelov: Advisor for study design, clinical analysis, and critical revisions. Graeme P. Young: Study design, data curation, analysis, and writing.
Supporting information
We acknowledge the contributions of Lorraine Sheehan‐Hennessy and Kathryn Cornthwaite to patient enrollment, and we thank the patients for their involvement.
References
- 1. Meyerhardt JA, Mangu PB, Flynn PJ, et al. Follow‐up care, surveillance protocol, and secondary prevention measures for survivors of colorectal cancer: American Society of Clinical Oncology clinical practice guideline endorsement. J Clin Oncol. 2013;31:4465‐4470. [DOI] [PubMed] [Google Scholar]
- 2. Tsang ME, Jayaraman S, Karanicolas PJ, Wei AC. Colorectal liver metastases In: Wright FC, Escallon J, Cukier M, Tsang ME, Hameed U, eds. Surgical Oncology Manual. Springer; 2016:101‐113. [Google Scholar]
- 3. Neo EL, Beeke C, Price T, et al. South Australian clinical registry for metastatic colorectal cancer. ANZ J Surg. 2011;81:352‐357. [DOI] [PubMed] [Google Scholar]
- 4. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early‐ and late‐stage human malignancies. Sci Transl Med. 2014;6:224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saluja H, Karapetis CS, Pedersen SK, Young GP, Symonds EL. The use of circulating tumor DNA for prognosis of gastrointestinal cancers. Front Oncol. 2018;8:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell‐free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124‐1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883‐892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Behrouz Sharif S, Hashemzadeh S, Mousavi Ardehaie R, et al. Detection of aberrant methylated SEPT9 and NTRK3 genes in sporadic colorectal cancer patients as a potential diagnostic biomarker. Oncol Lett. 2016;12:5335‐5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Symonds EL, Pedersen SK, Baker RT, et al. A blood test for methylated BCAT1 and IKZF1 vs. a fecal immunochemical test for detection of colorectal neoplasia. Clin Transl Gastroenterol. 2016;7:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Symonds EL, Pedersen SK, Murray DH, et al. Circulating tumour DNA for monitoring colorectal cancer—a prospective cohort study to assess relationship to tissue methylation, cancer characteristics and surgical resection. Clin Epigenetics. 2018;10:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Young GP, Pedersen SK, Mansfield S, et al. A cross‐sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor‐derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med. 2016;5:2763‐2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Metser U, You J, McSweeney S, Freeman M, Hendler A. Assessment of tumor recurrence in patients with colorectal cancer and elevated carcinoembryonic antigen level: FDG PET/CT versus contrast‐enhanced 64‐MDCT of the chest and abdomen. AJR Am J Roentgenol. 2010;194:766‐771. [DOI] [PubMed] [Google Scholar]
- 16. Labianca R, Nordlinger B, Beretta GD, Brouquet A, Cervantes A; ESMO Guidelines Working Group . Primary colon cancer: ESMO clinical practice guidelines for diagnosis, adjuvant treatment and follow‐up. Ann Oncol. 2010;21(suppl 5):v70‐v77. [DOI] [PubMed] [Google Scholar]
- 17. Poston GJ, Tait D, O’Connell S, Bennett A, Berendse S; Guideline Development Group . Diagnosis and management of colorectal cancer: summary of NICE guidance. BMJ. 2011;343:d6751. [DOI] [PubMed] [Google Scholar]
- 18. Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313‐5327. [DOI] [PubMed] [Google Scholar]
- 19. Duffy MJ, van Dalen A, Haglund C, et al. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348‐1360. [DOI] [PubMed] [Google Scholar]
- 20. Chao M, Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow‐up for patients with early‐stage colon cancer. J Clin Oncol. 2009;27:e279‐e280. [DOI] [PubMed] [Google Scholar]
- 21. Nicholson BD, Shinkins B, Pathiraja I, et al. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev. 2015;12:CD011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruibal Morell A. CEA serum levels in non‐neoplastic disease. Int J Biol Markers. 1992;7:160‐166. [DOI] [PubMed] [Google Scholar]
- 23. Pedersen SK, Symonds EL, Baker RT, et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer. 2015;15:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26:1715‐1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhangu JS, Beer A, Mittlbock M, et al. Circulating free methylated tumor DNA markers for sensitive assessment of tumor burden and early response monitoring in patients receiving systemic chemotherapy for colorectal cancer liver metastasis. Ann Surg. 2018;268:894‐902. [DOI] [PubMed] [Google Scholar]
- 26. Overman MJ, Morris V, Moinova H, et al. Phase I/II study of azacitidine and capecitabine/oxaliplatin (CAPOX) in refractory CIMP‐high metastatic colorectal cancer: evaluation of circulating methylated vimentin. Oncotarget. 2016;7:67495‐67506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer‐associated genes. Nature. 2013;499:214‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu B, Yan P, Zhang S, et al. Cell‐free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Markers. 2018;2018:6437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sposato LA, Lam Y, Karapetis C, et al. Observation of “complete clinical response” in rectal cancer after neoadjuvant chemoradiation: the Flinders experience. Asia Pac J Clin Oncol. 2018;14:439‐445. [DOI] [PubMed] [Google Scholar]
- 31. Day D, Frentzas S, Naidu CA, Segelov E, Green M. Current utility and future applications of ctDNA in colorectal cancer In: Segelov E, ed. Advances in the Molecular Understanding of Colorectal Cancer. IntechOpen; 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


