Abstract
Increasing evidence suggests that the process of alpha‐synuclein (α‐syn) aggregation from monomers into amyloid fibrils and Lewy bodies, via oligomeric intermediates plays an essential role in the pathogenesis of different synucleinopathies, including Parkinson's disease (PD), multiple system atrophy and dementia with Lewy bodies (DLB). However, the nature of the toxic species and the mechanisms by which they contribute to neurotoxicity and disease progression remain elusive. Over the past two decades, significant efforts and resources have been invested in studies aimed at identifying and targeting toxic species along the pathway of α‐syn fibrillization. Although this approach has helped to advance the field and provide insights into the biological properties and toxicity of different α‐syn species, many of the fundamental questions regarding the role of α‐syn aggregation in PD remain unanswered, and no therapeutic compounds targeting α‐syn aggregates have passed clinical trials. Several factors have contributed to this slow progress, including the complexity of the aggregation pathways and the heterogeneity and dynamic nature of α‐syn aggregates. In the majority of experiment, the α‐syn samples used contain mixtures of α‐syn species that exist in equilibrium and their ratio changes upon modifying experimental conditions. The failure to quantitatively account for the distribution of different α‐syn species in different studies has contributed not only to experimental irreproducibility but also to misinterpretation of results and misdirection of valuable resources. Towards addressing these challenges and improving experimental reproducibility in Parkinson's research, we describe here a simple centrifugation‐based filtration protocol for the isolation, quantification and assessment of the distribution of α‐syn monomers, oligomers and fibrils, in heterogeneous α‐syn samples of increasing complexity. The protocol is simple, does not require any special instrumentation and can be performed rapidly on multiple samples using small volumes. Here, we present and discuss several examples that illustrate the applications of this protocol and how it could contribute to improving the reproducibility of experiments aimed at elucidating the structural basis of α‐syn aggregation, seeding activity, toxicity and pathology spreading. This protocol is applicable, with slight modifications, to other amyloid‐forming proteins.

Keywords: alpha‐synuclein, amyloid fibrils, oligomers and monomers, Parkinson's disease
We describe here a simple protocol for the isolation, quantification and assessment of the distribution of different types of α‐syn species, such as monomers, oligomers and fibrils, in α‐syn samples. The protocol is simple, does not require any special instrumentation and can be performed rapidly on multiple samples using small volumes. We believe that the application of this protocol by the α‐syn research community could contribute significantly to improving the reproducibility of experiments aimed at elucidating the structural basis of α‐syn aggregation, seeding activity, toxicity and pathology spreading, and facilitate the development of α‐synuclein‐based biomarkers and therapeutics.

Read the Editorial Highlight for this article on page https://doi.org/10.1111/jnc.14973.
Abbreviations
- AFM
atomic force microscopy
- BCA
bicinchoninic acid
- CD
circular dichroism spectroscopy
- CSF
cerebrospinal fluid
- DLB
dementia with Lewy bodies
- EM
electron microscopy
- kDa
kilodalton
- LBs
Lewy bodies
- MDa
megadalton
- MSA
multiple system atrophy
- MWCO
molecular weight cut‐off
- nm
nanometer
- PBS
phosphate‐buffered saline
- PD
Parkinson's disease
- PFF
pre‐formed fibril
- SDS‐PAGE
sodium dodecyl sulphate–polyacrylamide gel electrophoresis
- SEC
size exclusion chromatography
- TBS
tris‐buffered saline
- TFA
trifluoroacetic acid
- UV
ultra‐violet
- WT
wild‐type
- α‐syn
Alpha‐Synuclein
1. INTRODUCTION
Alpha‐synuclein (α‐syn) is a 14 kDa protein that is found abundantly in the pre‐synaptic terminals of neurons, but is also expressed in the heart, muscles and other tissues (Iwai et al., 1995; Jakes, Spillantini, & Goedert, 1994). While the precise function of α‐syn is not yet well understood, accumulating evidence from genetic and neuropathological studies point to a central role of α‐syn in several neurodegenerative disorders that are collectively referred as synucleinopathies (Spillantini & Goedert, 2000). These disorders include Parkinson's disease (PD), dementia with Lewy bodies (DLB), the Lewy body variant of Alzheimer's disease, multiple system atrophy (MSA) and neurodegeneration with brain iron accumulation (Galvin, Lee, & Trojanowski, 2001; Jellinger, 2003). A common pathological link connecting these disorders is the intracytoplasmic accumulation of misfolded and aggregated forms of α‐syn in neurons and in glial cells (Baba et al., 1998; Spillantini & Goedert, 2000; Spillantini et al., 1997; Takeda et al., 1998). Despite differences in the cell types and affected brain regions, the fibrillar structures of α‐syn enriched in the neuronal inclusions is a salient feature of these diseases (Goedert, 2015). α‐syn aggregates (oligomers and fibrils) are released by neurons and, upon internalization by the neighbouring cell, induce the misfolding and/or seed the aggregation of endogenous α‐syn in the host cells (Danzer, Krebs, Wolff, Birk, & Hengerer, 2009; Mahul‐Mellier, Altay, & Burtscher, 2018). This process is thought to be the underlying mechanism for the cell‐to‐cell spreading and propagation of α‐syn to different brain regions during the different stages of disease progression (Braak et al., 2003; Hansen, Angot, & Bergstrom, 2011; Li et al., 2008). Although there is a consensus that the process of α‐syn fibril formation plays a central role in the initiation and progression of PD and other synucleinopathies, the nature of the toxic species and how they contribute to the different putative disease mechanisms underlying the pathogenesis of PD and other synucleinopathies remain unknown.
Several studies have shown that the presence and/or abundance of Lewy bodies (LBs) do not always correlate with the development of PD symptoms (e.g. the presence of LBs in healthy brains) (Duffy & Tennyson, 1965; Forno, 1969; Gibb & Lees, 1988; Lipkin, 1959; Mikolaenko et al., 2005; Schmidt et al., 1991) or disease severity. These observations, combined with the absence of a direct correlation between α‐syn fibril formation and neurodegeneration in different models of PD and the detection of soluble oligomeric forms of α‐syn in the cerebrospinal fluid, have led to a shift of interest from fibrils towards prefibrillar aggregates (oligomers) (Park, Cheon, Bae, Kim, & Kim, 2011). Therefore, significant efforts and resources have been devoted to dissecting the different steps of α‐syn fibril formation in vitro and isolating individual species along the pathway of α‐syn fibrillization. These efforts led to the development of several protocols for the generation and isolation of α‐syn oligomeric preparations with a distinct secondary structure, size and morphological distributions (Table S2).
Despite many attempts over the past two decades, it has been possible to generate homogeneous preparations that comprise a single type of oligomer of a specific size or morphology (Alam, Bousset, Melki, & Otzen, 2019; Lansbury & Lashuel, 2006; Lashuel, 2005; Lashuel & Lansbury, 2006). Most of the existing methods and protocols led to the generation of a population of oligomers with variable size and morphological distribution that is often in equilibrium with monomers. Depending on the ratio of oligomers to monomers, the propensity for fibril formation in these preparations varies. Very often, chemical cross‐linking is used to stabilize oligomers, but this significantly alters their dynamic properties (Nasstrom, Fagerqvist, & Barbu, 2011) and the ability to undergo conformational changes that may be required for exerting their biological activity. Therefore, the vast majority of α‐syn oligomeric preparations used in research today are heterogeneous in terms of the size distribution of the oligomers and the ratio of monomers, oligomers and fibrils.
During the past 6–8 years, α‐syn fibrils have made a comeback, mainly because of the emergence of the prion‐like hypothesis for the pathological spreading during the progression of PD. This hypothesis suggests that α‐syn aggregates (fibrils or oligomers) are secreted by neurons (Desplats et al., 2009; Lee, Patel, & Lee, 2005; Lee et al., 2008), and exhibit a potent seeding capacity that allows them to induce aggregation and inclusion formation upon internalization into neighbouring neurons and other cell types (Karpowicz, Trojanowski, & Lee, 2019; Luk et al., 2009; Luna, Decker, & Riddle, 2018; Volpicelli‐Daley et al., 2011). Consistent with this hypothesis, inoculation of fibrils produced in vitro (Abdelmotilib, Maltbie, & Delic, 2017; Luk et al., 2012; Shimozawa, Ono, & Takahara, 2017) or α‐syn aggregate‐enriched extracts from diseased brains is sufficient to induce α‐syn fibrillization in neurons, rodent models and non‐human primates (Masuda‐Suzukake et al., 2013; Peng, Gathagan, & Covell, 2018; Recasens & Dehay, 2014; Recasens, Dehay, & Bove, 2014; Tu, Galvin, & Baba, 1998; Watts et al., 2013). This has led to a resurgence in the number of studies focusing on fibrils and strategies to interfere with their seeding capacity and role in pathology spreading. The vast majority of this research relies on the ability to reproducibly generate fibril preparations of the desired properties. While incubating concentrated samples of α‐syn (3–5 mg/ml) at 37°C with agitation reproducibly leads to the formation of fibrils, very often these preparations contain different amounts of monomers and oligomers. Given that most of these samples are analysed using electron microscopy (EM) or atomic force microscopy (AFM), which are not quantitative imaging techniques, the presence of the monomers and oligomers is either not detectable (monomers) or under‐appreciated. Because of the differences in the dynamic and structural properties of each of the three species, the nature of the samples could change dramatically from one preparation to another and even for the same preparation when exposed to different conditions. Furthermore, all fibril preparations that are used to investigate cellular toxicity, α‐syn seeding mechanisms and pathology spreading are usually subjected to sonication or mechanical disruption procedures to generate short fibril seeds with a similar size distribution (50–200 nm). This procedure leads to fibril fragmentation resulting in the generation of more fibril ends and an increase in the amounts of monomers/oligomers released from the fibrils. The levels of monomers/oligomers in fibril seed preparation have been shown to be a critical determinant of their dynamics, growth and toxicity (Mahul‐Mellier et al., 2015), and if not account for, could contribute to significant experimental variability.
The increasing reliance on α‐syn aggregate preparations (fibrils and oligomers) in the development of PD models and their use to investigate the mechanisms linked to α‐syn dysfunction and toxicity underscore the critical importance of using high quality and reproducible preparations in such studies. However, experience teaches us that it is nearly impossible to generate oligomer and fibril preparations of identical species, size and morphology distribution. This challenge could be easily addressed by (1) developing protocols and methods that allow the separation and proper quantification of the amount of each of the three species, and (2) providing basic morphological and structural characterizations of the crude mixtures and isolated species by imaging techniques (EM and AFM) and circular dichroism spectroscopy (CD) respectively. This level of characterization would at least enable better interpretation of the structure‐activity studies and improve the reproducibility by allowing accurate comparison of data and results across different laboratories.
Towards achieving these goals, we present here a straightforward protocol based on centrifugation and filtration procedures, which allows rapid quantitative and qualitative assessment of the different types of α‐syn species such as monomers, oligomers and fibrils in samples of different complexities. This protocol is based on years of experience in our group (Fauvet, Mbefo, & Fares, 2012; Khalaf, Fauvet, & Oueslati, 2014) and the evidence that it has been successfully applied in α‐syn research by other groups (Ghosh et al., 2015; Kumar, Das, & Mohite, 2018; Zhang, Griggs, Rochet, & Stanciu, 2013). The flexibility of this method relies on the fact that it can be used to characterize purified α‐syn samples at different time points during α‐syn aggregation. The protocol only requires a minimum sample volume (100 μL) and can be implemented using basic laboratory tools. The total running time of the protocol is 3–4 hr, including (1) isolation of all three species (fibrils, monomers and oligomers, (2) assessment of the distribution of three species by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS‐PAGE) and (3) concentration determination using ultra‐violet (UV) absorbance and/or bicinchoninic acid (BCA) assay. Biophysical characterization of the α‐syn species using CD and EM is necessary to allow batch to batch comparison and to improve experimental reproducibility. Below, we present and discuss several examples that illustrate the potential applications of this protocol, how it could contribute to improving the reproducibility of experiments aimed at elucidating the structural basis of α‐syn aggregation, seeding activity, toxicity and pathological spreading.
2. MATERIALS AND METHODS
2.1. Key equipment and materials
| Equipment/material | Company | Catalogue |
|---|---|---|
|
Centrifuges Benchtop Ultracentrifuge |
Eppendorf 5417R Beckmann Coulter Optima Max XD |
|
|
Spin filters 30 kDa 50 kDa 100 kDa |
Microcon−30 Ultracel YM 30 Amicon Ultra−2 ml, Ultracel−50K Microcon DNA fast flow Ultracel Vivaspin 500 (GE healthcare) |
MRCF0R030 UFC205024 MRCF0R100 28–9322–37 |
| Sonicator | Sonics Vibra‐Cell™ |
VCX 130 630–0422 (microtip) |
2.2. Recombinant over‐expression and purification of wild‐type (WT) α‐syn and cysteine variants
BL21 (DE3) cells transformed with the pT7‐7 plasmid encoding the WT α‐syn or M1C or A140C α‐syn mutants were grown in LB medium at 37°C and induced with 1 mM 1‐thio‐β‐d‐galactopyranoside (AppliChem) at an O.D. between 0.4–0.6 and continued to grow for 4 hr at 37°C. Induced bacterial cultures were pelleted and sonicated for the lysis of the cells. After centrifugation at 18,000 g for 20 min, the supernatant was boiled for 5 min and centrifuged again for 20 min. The supernatant was purified by anion exchange chromatography (HiPrep 16/10 Q FF, GE Healthcare Life Sciences), followed by reverse‐phase HPLC (Jupiter 300 C4, 20 mm I.D. × 250 mm, 10 µm average bead diameter, Phenomenex) and lyophilized. The M1C α‐syn mutant was purifed as thiazolidine adducts from bacterial expression. To make cysteine residue available for the formation of disulphide‐linked homodimer, purified M1C α‐syn was dissolved in reaction buffer (0.1% trifluoroacetic acid, 5% acetonitrile, 5% acetic acid in water) to a final concentration of 0.5 mM. To this solution 100 eq of Silver trifluoromethanesulphonate (Sigma) from 1 M solution in 0.1% trifluoroacetic acid H2O was added. The solution was then incubated at RT without shaking. After 30 min, the reaction was analysed by electron spray ionization mass spectrometry which showed quantitative deprotection of thiazolidine adducts. The reaction mixture was desalted using a PD‐10 column (GE Healthcare) and lyophilized.
2.3. Preparation WT α‐syn oligomers
To prepare WT α‐syn oligomers, 60 mg of lyophilized protein was dissolved in 5 ml of phosphate‐buffered saline (10 mM disodium hydrogenphosphate, 2 mM potassium dihydrogen phosphate (pH 7.4), 137 mM NaCl and 2.7 mM potassium chloride) and followed by incubation for 5 hr at 900 rpm constant shaking at 37°C. Then, the solution was centrifuged to remove any insoluble particles at 12,000 g for 10 min at 4°C. 5 ml of supernatant was loaded onto a HiLoad® 26/600 Superdex® 200 preparation grade (GE lifesciences) XK pre‐packed column equilibrated with phosphate‐buffered saline, and protein was eluted as 2.5 ml fractions at a flow‐rate of 1 ml/min. Fractions corresponding to the void volume peak (oligomers) were split to 500 μL aliquots, snap‐frozen and stored at −20°C.
2.4. Step‐by‐step explanation of the centrifugation‐based filtration protocol
This protocol can be applied to the characterization of α‐syn preparations containing variable amounts of α‐syn monomers, oligomers or fibrils, such as the freshly dissolved purified protein, oligomers or fibrils. The protocol has been divided into three steps: (1) a centrifugation step for the isolation of the fibrils from the soluble species; (2) a filtration step for the separation of monomers and oligomers and (3) recovery of the soluble oligomers. A standard procedure for the application of this protocol is described below:
A total sample volume between 50 μL and 500 μL is subjected to ultracentrifugation at 100,000 g for 30 min at 4°C.
After centrifugation, a careful pipetting of the supernatant from the pellet is carried out to isolate the soluble species. The isolated pellets can be used for further analysis after resuspension in the buffer of interest.
Next, the supernatant is transferred into the filtration membrane unit (100 kDa), placed into a new collection tube and centrifuged at 15,300 g for 20 min at 4°C.
After centrifugation, the membrane unit is separated from the collection tube. The sample which passes through the membrane and enters the new tube, is named as filtrate sample and contains predominantly monomeric or dimeric α‐syn species.
To collect the oligomeric species that were retained on the membrane unit, named retentate, nine parts of the buffer from the initial volume of the sample is added to the membrane (e.g. use 450 μL of buffer if 500 μL was the initial volume of the sample), pipette gently 2–3 times and place the filtration membrane unit into a new collection tube by carefully turning it upside‐down. Next, pipette one part of the buffer (e.g. 50 μL) into the inverted bottom and centrifuge at low speed at 425 g for 4 min to recover the retentate sample.
In all cases, the spin filters were equilibrated with the sample buffer and subjected to a short centrifuge run prior to applying the samples to filtration.
The different α‐syn species separated during the centrifugation‐based filtration protocol are obtained in standard buffer solutions and thus can be characterized, without the need to exchange buffer, using common protein concentration determination methods, SDS‐PAGE analysis, CD spectroscopy, light scattering, sedimentation velocity or electron microscopy, among other techniques.
2.5. Filtration analysis of WT α‐syn monomers using different MWCO filter membranes
WT α‐syn was dissolved in 600 μL of Tris‐buffered saline (TBS) pH 7.5 to give a final concentration of 35 μM while placing it on ice cold condition. The solution was ultracentrifuged for 30 min at 100,000 g at 4°C to remove any preformed insoluble aggregates (both fibrillar and amorphous aggregates). The supernatant was collected and filtered through different MWCO centrifugal filters (30 kDa, 50 kDa or 100 kDa). 100 µl of the supernatant solution was used for each filtration. The sources of the centrifugal filter membranes used are described in the table above.
For 30 kDa and 100 kDa filtration, 100 μL of the supernatant solution was centrifuged at 15,300 g for 20 min and the filtrate was collected. The retentate was collected by inverted centrifugation using 40 µl of TBS (pH 7.5) at 425 g for 4 min at 4°C. The volume is diluted to 100 µl using TBS. For the 50 kDa filter, 100 μL of the supernatant was centrifuged at 4000 g for 20 min at 4°C in a swinging bucket. The filtrate was collected and diluted to 100 μL using TBS. Next, 100 μL TBS was added to the membrane, pippeted up and down for few times and collected back into a new Eppendorf tube using pippete as the retentate.
Twenty microlitre of each filtrate and retentate for each membrane was mixed with 20 μL of 2 × Laemmli buffer for SDS‐PAGE analysis and heated at 95°C for 5 min. The samples were analysed by SDS‐PAGE (1.5 mm thickness) immediately or snap frozen in liquid nitrogen and stored at −80°C until further use. 10 μL of the SDS‐PAGE samples were loaded onto a gel. The remaining filtrate and retentate for each membrane were snap‐frozen in liquid nitrogen and stored at −80°C.
2.6. Filtration analysis of WT α‐syn monomers using 100 kDa membranes of different material
WT α‐syn was dissolved in 600 μL of TBS (pH 7.5) to a final concentration of 35 μM while placing it on ice‐cold condition. The solution was ultracentrifuged for 30 min at 100,000 g at 4°C to remove any insoluble aggregates (both fibrillar and amorphous aggregates). The supernatant was collected and filtered through a 100 kDa centrifugal filter composed of a Microcon 100 kDa membrane made of regenerated cellulose or a Vivaspin 500 (100 kDa) made up of polyethersulphone membrane. As stated earlier in the methods section, 100 μL of WT α‐syn supernatant was centrifuged at 15,300 g for 20 min at 4°C. The filtrate was collected and diluted to 100 μL using TBS. The retentate from the Microcon membrane was recovered following the usual procedure. However, for Vivaspin, the retentate was collected by addition of 100 μL TBS to the membrane, pippeted up and down for 3‐4 times and collected back into a new Eppendorf tube. The SDS‐PAGE samples were prepared and analysed as described earlier. SDS‐PAGE analysis was performed on a 1‐mm‐thick gel.
2.7. Filtration analysis of M1C and A140C α‐sSyn mutants (dimers) using 100 kDa filter membrane
M1C α‐syn was dissolved in 270 μL of TBS, and A140C α‐syn was dissolved in 560 μL of TBS (pH 7.5) to reach a final concentration of 35 μM. Both solutions were kept at RT for 3 day without shaking and analysed using mass spectrometry for validation of dimer formation. After 3 day, the solutions were centrifuged at 100,000 g for 30 min at 4°C to remove any insoluble aggregates (both fibrillar and amorphous aggregates). The supernatants were collected and subjected to the usual filtration analysis using Microcon 100 kDa as follows. 100 µl of supernatant was centrifuged at 15,300 g for 20 min at 4°C. The filtrate was collected, and the retentate was collected by inverted centrifugation at 425 g for 4 min at 4°C using 40 μL of TBS and the volume diluted to 100 μL using TBS. Fractions were collected at each step and equal volumes of samples were used for SDS‐PAGE analysis.
2.8. Transmission electron microscopy
Prior to the application of the sample, Formvar and carbon‐coated 200 mesh‐containing copper EM grids (Electron Microscopy Sciences) were glow‐discharged for 30 s at 20 mA using a PELCO easiGlow™ Glow Discharge Cleaning System (TED PELLA, Inc). Subsequently, 5 μL of the sample was placed onto the EM grid for a minute. Then, the sample was carefully blotted using filter paper and air‐dried for 30 s. Then, the grids were washed three times with ultrapure water and stained with 0.7% (w/v) uranyl formate solution. Grids were examined using a Tecnai Spirit BioTWIN electron microscope. The microscope was equipped with a LaB6 gun operated at an acceleration voltage of 80 kV, and images were captured using a 4K × 4K charge‐coupled device camera (FEI Eagle). The length (ln) or diameter (d) of respective α‐syn oligomeric fractions was measured using ImageJ software (NIH).
2.9. SDS‐PAGE analysis
Samples for SDS‐PAGE were mixed with 2 × Laemmli buffer (4% SDS, 20% glycerol, 0.004% bromphenol blue, 0.125 M Tris‐Cl, 10% 2‐mercaptoethanol pH 6.8) and loaded onto 15% polyacrylamide gels. The gel was run at 180 V for 1 hr in running buffer (25 mM Tris, 192 mM Glycine, 0.1% SDS, pH 8.3), followed by staining with a solution of 25% (v/v) isopropanol, 10% acetic acid (v/v) and 0.05% (w/v) Coomassie brilliant blue R (Applichem) and destaining with boiling distilled water.
2.10. Protein concentration estimation
Concentration of α‐syn samples such as monomers, oligomers and fibrils are estimated using BCA assay and amino acid analysis. For BCA assay, microplate measurements were carried out using BCA protein assay reagents (Pierce, catalog number: 23227). Briefly, triplicates of known concentrations (from 10 μg/mL to 1000 μ g/mL) of bovine serum albumin (concentration standard) and equal volume of α‐syn samples were pipetted into microplate wells. To which, 200 μL of BCA working reagent was added and incubated at 37 C for 30 min. Absorbance at 562 nm was measured using Tecan plate reader. Using BCA assay‐based concentration estimation as standards, known concentrations (2–3 μg) of α‐syn samples were pipetted into a conical insert, flash‐frozen and lyophilised. The dried form of α‐syn samples were shipped to Functional Genomic Center Zurich for subjecting for amino acid analysis (AAA) for absolute quantification of α‐syn samples concentrations. Destained gels were imaged and analysed (only in the case of figure 2) using ImageJ software (U.S. National Institutes of Health, Maryland, USA; RRID:SCR_001935) and GraphPad Prism was used to present statistical results.
Figure 2.
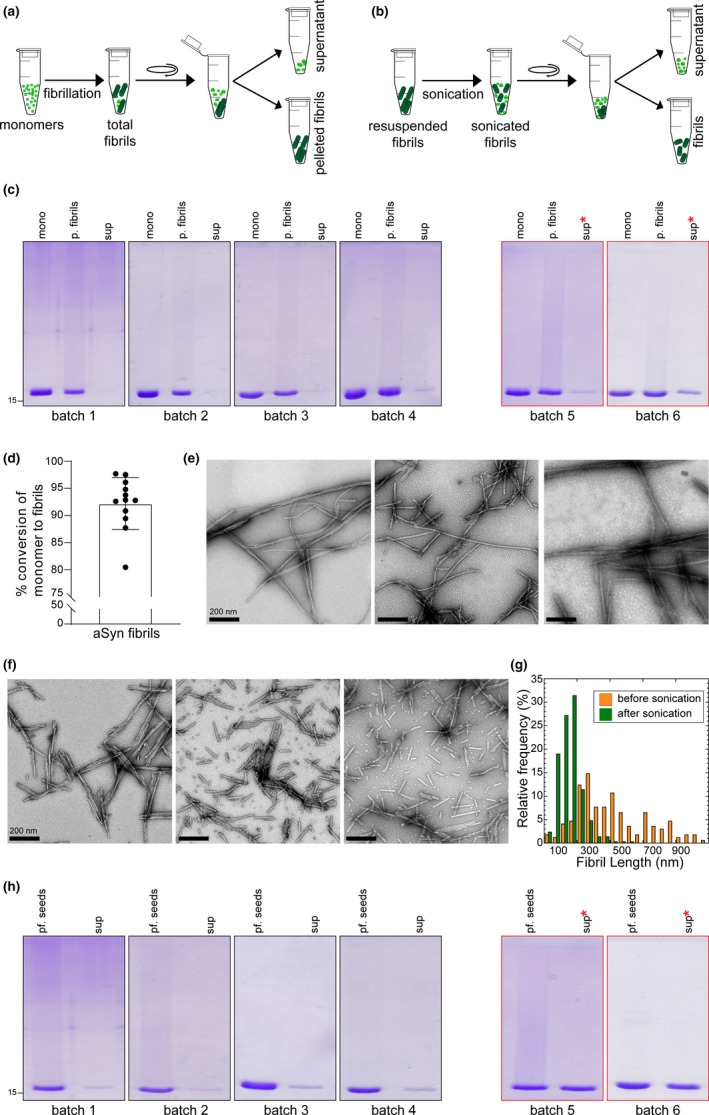
Schematic illustrations of (a) the separation of α‐syn fibrils from a crude mixture of total α‐syn fibrils following fibrillization and (b) following sonication (20% amplitude, one‐second ON/OFF pulse cycle for 20 s), to assess the amounts of monomers/oligomers present in the supernatant fraction. (c) sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the different batches of α‐syn fibrils during fibrillization; mono: α‐syn monomers prior to initiating fibrillization; p. fibrils: pelleted fibrils after resuspension; and sup: supernatant α‐syn species following centrifugation at the end of fibrillization. (d) Bar chart displaying the percentage of α‐syn monomers converted to fibrils in different batches of fibril preparation. Bullet points represent the different batches of fibril preparation (n = 15). (e) EM images of total α‐syn fibrils at the end of fibrillization showing three representative images from three random batches of fibril preparations. (f) EM images of sonicated α‐syn fibrils showing three representative images from three random batches. (g) Differences in the lengths of the fibrils before and after sonication. (h) SDS‐PAGE analysis of different batches of α‐syn fibrils following sonication; pf. seeds: the fibril seeds sedimented as pellets after sonication; sup: the soluble supernatant α‐syn species following centrifugation. * in (c) and (h) denotes the increase in the amount of soluble α‐syn species in the supernatant fractions in some non‐reproducible fibril batches
2.11. Mass spectrometry analysis
To perform MALDI‐TOF (matrix‐assisted laser desorption/ionization‐ time‐of‐flight) mass spectroscopy analysis, 5 μL of the protein was mixed with 5 μL of sinapinic acid (100 mM) and 2 μL from the reaction mixture was directly spotted on a opti‐TOF MALDI plate (384 spots) and dried under vacuum. Then the intact mass of the proteins was measured on a 4,800 MALDI TOF/TOF™ Analyzer (from Applied Biosystems).
2.12. Far‐UV circular dichroism spectroscopy
CD spectra of α‐syn were obtained by loading 150 microlitres of samples into a quartz cuvette with a 1‐mm path length and collected using a Jasco J‐815 CD spectrophotometer or Chirascan spectropolarimeter (Applied Photophysics) operated at 20°C within the range 198–250 nm. When employing the Jasco J‐815 CD spectrophotometer, data acquisition used the following parameters: data pitch, 0.2 nm; bandwidth, 1 nm; scanning speed, 50 nm/min; digital integration time, 2 s. A spectrum of each sample is the average of 10 repeats followed by a binomial approximation.
3. RESULTS
3.1. A simple centrifugation‐based filtration protocol for the separation and characterization of distinct α‐syn species (fibrils, monomers and oligomers)
Although different methods have been shown to be effective in separating insoluble α‐syn aggregates (both fibrillar and amorphous aggregates) from soluble α‐syn species and monomers from oligomers, (Conway et al., 2000; Cremades, Cohen, & Deas, 2012; Gao, Carroni, & Nussbaum‐Krammer, 2015; Lashuel et al., 2002), these methods suffer from the requirement of large sample volumes and/or lead to significant dilution of the sample of interest. Furthermore, many of these methods require at least 1–2 hr of processing time per sample and are thus not amenable to rapid analysis of large number of samples. They also require access to specialized instruments (e.g. fast protein liquid chromatography (FPLC)) and expertise that may not be available in all laboratories.
The protocol described here addresses these limitations and takes advantage of (1) the large differences in the size of the three α‐syn species (monomers at 14 kDa, oligomers between 28 kDa–1 MDa and fibrils > 2 MDa), and (2) the difference in solubility of the fibrils compared to the monomers and oligomers. The fibrils, but not the oligomers and monomers, can be sedimented by centrifugation. For samples containing the three forms of α‐syn, the first step involves the sedimentation and separation of the fibrils from the monomers and oligomers by ultracentrifugation of the sample at 100,000 g for 30 min (scheme 1 (1)). This allows for the separation of the sedimentable fibrils as a pellet from the soluble supernatant, which contains both monomers and oligomers of different sizes (scheme 1 (2)). The supernatant (scheme 1 (3)) is then separated into monomers and mixtures of oligomers by filtration through a 100 kDa molecular weight cut‐off (MWCO) filter, which allows only the monomers to pass through (scheme 1 (4)). The oligomers are retained on the filter membrane are then recovered, as shown in scheme 1 (5). Determination of the concentration of each species after isolation, using UV absorbance and/or BCA assay, allows for the robust estimation of the distribution of the different species in the original sample. For more accurate determination of the concentration of each species, we recommend using amino acid analysis (AAA). The requirements for the implementation of this protocol are having access only to: (1) the spin filters (Microcon (100 kDa MWCO, Ireland)); (2) a benchtop centrifuge or an ultracentrifuge; (3) a UV absorbance‐based spectrometer or a plate reader; and (4) a gel electrophoresis setup. These are standard pieces of equipment to which research laboratories have easy access. Most importantly, this procedure could be performed on sample volumes ranging from 50 to 500 μL.
The protocol described in Scheme 1 can be used to: (1) prepare α‐syn monomeric solutions that are free of any pre‐formed aggregates which may have formed during the process of storage, lyophilization or resolubilization of α‐syn samples, (2) separate the non‐fibrillized α‐syn species (monomers and oligomers) from fibrils and quantify both species and (3) assess the amount of released monomers or the formation of oligomers because of sonication of the fibrils or treatment with chaperones or small molecule disaggregases (Bieschke et al., 2010; DeSantis et al., 2012; Gao et al., 2015) and (4) enable more comprehensive profiling of the complexity of α‐syn samples. These capabilities should enable the assessment of batch to batch variations of α‐syn sample preparation, comparison of data from different research groups and improving experimental reproducibility across different laboratories. However, for accurate interpretation of the results using this protocol, it is crucial to verify by EM that this separation protocol works for the sample/protein of interest of under the experimental conditions used.
3.2. Separation of fibrils from soluble α‐syn species (oligomers and monomers)
Here, we provide an overview of the standard methods for the generation and characterization of α‐syn fibril preparations and discuss some of the factors that could influence batch‐to‐batch variations. Because of their simplicity, centrifugation‐based sedimentation protocols have emerged as a substitute for SEC‐based methods, and are commonly used to study protein aggregation by monitoring and quantifying the amount of sedimentable aggregates and the remaining soluble proteins in the supernatant (Fauvet et al., 2012; Khalaf et al., 2014). These protocols have proven to be reliable and effective in separating fibrils or sedimentable aggregates from the soluble protein species. However, it is crucial to validate such protocols (through analysis of the supernatant) to ensure that fibrils formed by the protein of interest are sedimentable. In most studies, a relatively short and low centrifugal force of approximately 14,000–16,000 g is sufficient to sediment mature, fully‐grown insoluble α‐syn fibrils and other large amorphous aggregates (Table S1), but not small fibrils that are < 150 nm in length. For example, the pre‐formed fibril (PFF) seed preparations that are commonly used in cellular seeding assays and animal models of pathology spreading are usually subjected to sonication prior to use. This procedure is used to induce fibril fragmentation with the aim of obtaining a fibril preparation with an average length of 50–150 nm (Mahul‐Mellier et al., 2018, 2015; Tarutani et al., 2016). Once fragmented to this size, the fibrils are non‐sedimentable at this centrifugal force (Table S1). For such preparations, high centrifugal force at 100,000 g is generally applied to sediment and separate the shortened/fragmented fibril seeds from the soluble α‐syn species.
Figure 1 illustrates how the protocol described above can be used to ensure reproducible preparations of PFF seeds for use in seeding‐based aggregation assays (Polinski et al., 2018; Tarutani et al., 2016) or in cellular and animal studies to investigate aSyn pathology formation and spreading (Abdelmotilib et al., 2017; Luk et al., 2012, 2009; Shimozawa et al., 2017; Volpicelli‐Daley et al., 2011). This is achieved by assessing the following parameters for each PFF preparation: (1) the percentage of fibril formation; (2) the percentage of remaining soluble α‐syn species in each preparation and (3) the relative stability of the fibrils as determined by the amount of soluble α‐syn species released after sonication of the fibrils. The protocol can be also modified to assess the presence and amounts of oligomers in such preparations (see Figures 4 and 5, Figure S3).
Figure 1.
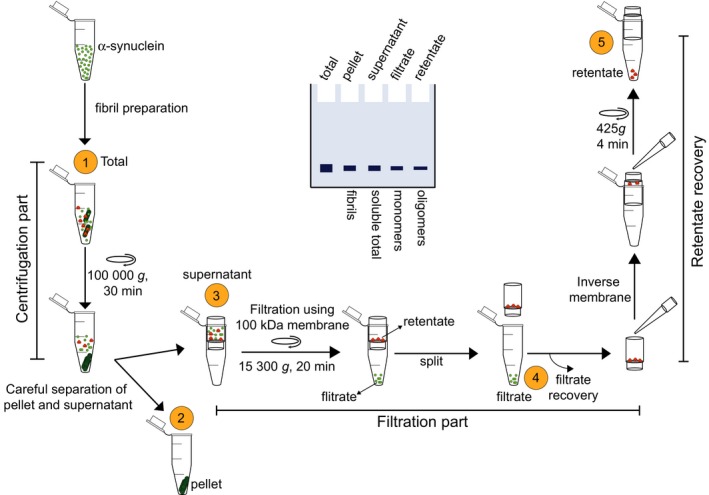
Schematic depiction of the centrifugation‐based filtration protocol. Briefly, the protocol includes a centrifugation step for the isolation of the fibril (pellet) from soluble α‐syn (monomers and oligomers), a filtration step (through 100 kDa MWCO) for the separation of the monomers from the oligomers, and a final step for the recovery of the oligomers from the spin filters. A depiction of the sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the samples at each step is shown in the middle. Quantification of band intensity should allow for a quick assessment of the distribution of coexisting α‐syn species
Figure 4.
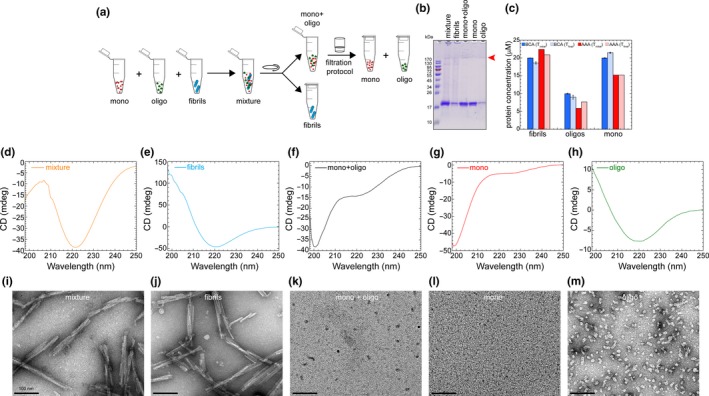
(a) A scheme illustrating the experimental setup for assessing efficiency to separate α‐syn species from a prepared mixture containing known concentration of α‐syn monomers, oligomers and fibril. (b) sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the samples collected at each step of the protocol. Red arrow head points the band of SDS‐resistant oligomers. (c) Comparison of protein concentration estimation (μM; based on BCA assay and AAA) of α‐syn samples (fibrils, oligomers and monomers) from the initial concentrations, that is before the preparation of α‐syn mixture versus the obtained concentrations of α‐syn fractions isolated from the α‐syn mixture during the steps of the protocol. (d–h) CD spectra of the total α‐syn mixture (d), fibrils (e), monomers and oligomers (mono + oligo) in the soluble α‐syn supernatant (f), monomers (mono) in the filtrate (g) and oligomers (oligo) in the retentate (h) samples. (i–m) Representative electron micrograph images of the α‐syn mixture (i), fibrils (j), monomers and oligomers (mono + oligo) in the soluble α‐syn supernatant (k), monomers (mono) in the filtrate (l) and oligomers (oligo) in the retentate (m) samples isolated from the different steps of the protocol
Figure 5.
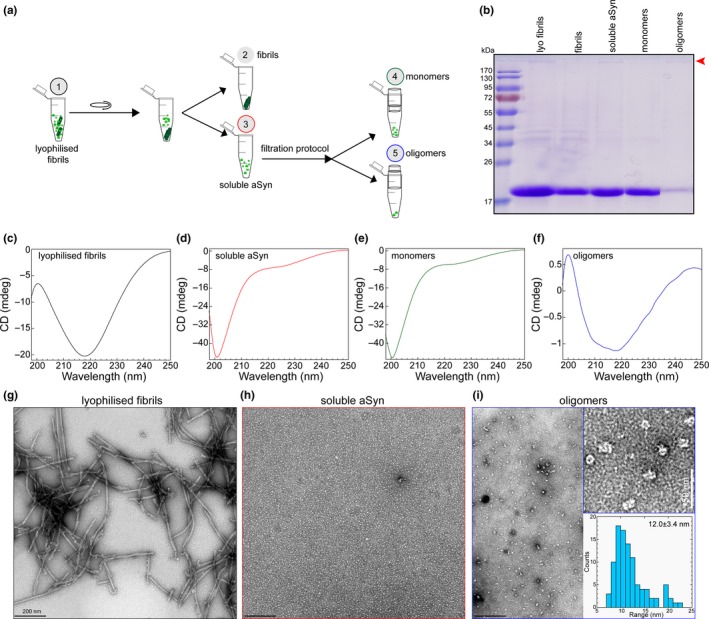
(a) Schematic illustration showing the experimental setup of the centrifugation‐based filtration analysis of lyophilized and resuspended α‐syn fibrils (lyophilized fibrils). (b) sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of α‐syn samples isolated from the different steps of the protocol. Red arrow head points the band of SDS‐resistant oligomers. (c–f) CD spectra of lyophilized fibrils (c), soluble α‐syn (d), monomers (filtrate, e) and oligomers (retentate, f) in the samples. (g–i) Representative EM images of lyophilized fibrils (g), soluble α‐syn (h), and oligomers (retentate, i) in the samples from the different steps of the protocol. The top insert on (i) is the zoom of the EM images of the oligomer sample. The bottom insert of I shows the distribution of the diameter of oligomeric particles based on the quantification from EM images
An important criterion for improving the reproducibility of PFF‐based experiments is to ensure that the PFF batches are well characterized. The major source of variation between different PFF preparations is differences in the stability of the fibrils and distribution of α‐syn species in the PFF preparations. However, a large concern with the commonly used methods to characterize PFFs is the focus on assessing the morphology of the fibrils and their size distribution rather than focusing on the sample heterogeneity, that is the presence of other α‐syn species (e.g. monomers and oligomers). Below, we present a series of examples that illustrate how the protocol described here allows for a more in‐depth characterization of the PFF preparations and enables the assessment of the levels of the different α‐syn species (monomers and oligomers) aSyn PFF preparations.
For the generation of different batches of α‐syn fibril preparations, we follow the standard protocol (Mahul‐Mellier et al., 2018). Our experience shows that despite following the same method carefully, it is not unusual to observe variations between different fibrils batches with respect to the extent of the amount of monomers converted to fibrils (Figure 2a, c and d) and/or the stability of the fibrils. This is indeed the case for all α‐syn fibrillization protocols. Thus, we strongly recommend quantitative assessment of the amount of both fibrils and soluble α‐syn species in each PFF preparation. This can be achieved by quantifying (1) the amount of monomers before initiating the fibrillization reaction; (2) the amount of α‐syn converted to fibrils (Figure 2a scheme, pellet) and the remaining aSyn soluble species at the end of fibrillization (Figure 2a scheme, supernatant). After 5 days (Figure 2c), we typically see >90%–95% conversion of α‐syn monomers to fibrils, as evidenced by the almost total absence of an α‐syn band in the supernatant lane for the fibril batches 1–4. Figure 2d shows the quantitative analysis of the extent of fibril formation from 12 independent fibril preparation experiments, estimated by quantifying the amounts of remaining soluble α‐syn species by SDS‐PAGE. Two of the fibril preparations, batch 5 and 6, show higher levels of soluble α‐syn species (batch 5:12.3% and batch 6:19.5%), which could indicate either incomplete fibrillization or the formation of less stable (SDS‐sensitive) fibrils. Importantly, such variations are not captured by the standard qualitative analysis of fibril preparation using techniques such as EM and CD. Figure 2e shows the EM images three different fibril batches which reveal predominantly straight and long fibrils and the absence of any amorphous structures. α‐syn monomers are not detectable by EM and when present as minor species, their existence and levels cannot be easily discerned by CD. Finally, it is important to emphasize that the fibrils exists in equilibrium with monomers and further manipulation of the fibrils could influence this equilibrium. For example prior to their use in seeding‐based assay, PFFs are subjected to sonication to induce the fibril fragmentation and to generate homogeneous (in terms of their average length, 50–150 nm) PFF resuspensions that are easily taken up by cells (Figure 2b). Figure 2f and g show an example of the average fibril lengths before and after sonication for a typical PFF preparation based on EM analysis. For the great majority of seeding‐studies, only the average length and morphology of the fibrils are usually assessed. We strongly recommend that the amount of soluble α‐syn species should also be assessed prior to and after sonication and preferably on the day the PFF preparations are used.
To illustrate how this could be done, the sonicated fibril sample shown in Figure 2b was subjected to ultracentrifugation at 100,000 g for 30 min, as described above. SDS‐PAGE analysis showed the differences between the levels of sedimented fibril seeds and soluble α‐syn species released after sonication for different PFF batches (Figure 2h). For stable fibril preparations, we typically see ~5%–10% soluble α‐syn after sonication (Figure 2h, batches 1–4). The stability of fibrils here is defined by their propensity to disassociate and the amount of soluble α‐syn species detected after sonication. Depending on the stability of the fibrils, the amount of soluble α‐syn released after sonication could vary significantly. For instance, batches 5 and 6 show approximately more than 30% soluble α‐syn species in the supernatant after sonication.
Modifying the sonication parameters such as the length of sonication time has been shown to strongly influence the length of the fibrils in the PFF preparations (Tarutani et al., 2016). We have also observed that variations in these parameters strongly influence the extent of soluble aSyn species released from the fibrils. For example, when using handheld sonicators, the position of the sonicator with respect to both the sample and the wall of the tube were found to influence the release of soluble α‐syn species from the fibrils. Therefore, we highly recommend exercising extreme care when performing this procedure and assessing fibril morphology, size and the ratio of soluble‐to‐fibrillar α‐syn species in all aSyn fibril preparations.
3.3. Detection and quantification of α‐syn oligomers in PFF preparations
While the protocol described above could be used to quantify the levels of α‐syn monomers and fibrils before and after sonication of each preparation, it is not suitable for quantifying oligomers in these PFF preparations. Given the potent toxicity of α‐syn oligomers (Danzer et al., 2009; Fusco, Chen, & Williamson, 2017; Mahul‐Mellier et al., 2015), the presence of small amounts of oligomers could be a major contributor to experimental variability. In the following sections, we describe the extension of the protocol explained above to allow for the separation and quantification of oligomers from different α‐syn samples, including PFF preparations.
Several protocols for the preparation of α‐syn oligomers have been developed (Table S2). Analysis of the size distribution of the oligomers obtained using these preparations suggests that it is still possible to develop a single simple protocol that allows for the separation of monomers from oligomers (greater than a dimer). Based on the method of preparation, oligomers generally show different shapes (such as spherical, chain‐like and annular structures), a wide range of sizes (usually ranging from 2 to 60 nm in diameter), and a molecular weight distribution ranging from 88 to 5,800 kDa (Table S2). The large difference in the size and molecular weight between monomers and oligomers suggests that spin filters with a MWCO of 30 kDa or greater should retain the oligomers on the membrane while allowing the monomers to pass through. However, when dealing with natively unfolded proteins such as α‐syn, one must consider that they exhibit a higher hydrodynamic radius, which makes aSyn behave as a globular protein of a larger size. For example the α‐syn monomer, which has a theoretical molecular weight of 14.46 kDa, behaves like a 57–60 kDa protein in size exclusion chromatography (Fauvet et al., 2012; Weinreb, Zhen, Poon, Conway, & Lansbury, 1996).
To determine which MWCO and membrane material are best for separating aSyn monomers from oligomers, we screened membranes with different MWCO (30 kDa, 50 kDa and 100 kDa) and material properties (Figure S1). We found that 100 kDa MWCO cellulose membranes (Microcon) enabled the complete separation of oligomers and recovery of the monomers into the filtrate fraction. Next, we sought to determine if oligomers of different size distribution can be separated from α‐syn monomers. We used a Superose 6 column to subfractionate our α‐syn oligomer preparations into fractions of different size distribution. As shown in Figure 3a (total chromatogram) and b (oligomer peak), the oligomeric peak was fractionated into four oligomeric fractions (Figure 3b). Using EM analysis, we measured the average size of the oligomers in each fraction (Figure 3c). Despite the heterogeneity of each oligomer fraction, they exhibited different size distribution ranging from 33 to 3 nm. Fraction 4 showed predominantly spherical oligomers as small as 3–4 nm. Next, we applied the filtration protocol on all these different oligomeric fractions separately. Figure 3d–g show the EM images (total and filtrate) and SDS‐PAGE analysis of total, filtrate and retentate samples of each oligomeric fraction. We did not observe any significant release of monomers from all the oligomeric fractions. In addition, we did not detect any oligomeric band in the filtrate samples as evidenced by SDS‐PAGE gel and EM analyses (Figure 3d–g). These results show that filtration through 100 kDa MWCO filters allows for the efficient separation of aSyn monomers and recovery of α‐syn oligomers of different sizes greater than a dimer. Similar result was obtained using unfractionated oligomers using Superdex 200 SEC column, which is commonly used to purify α‐syn (Figure S2).
Figure 3.
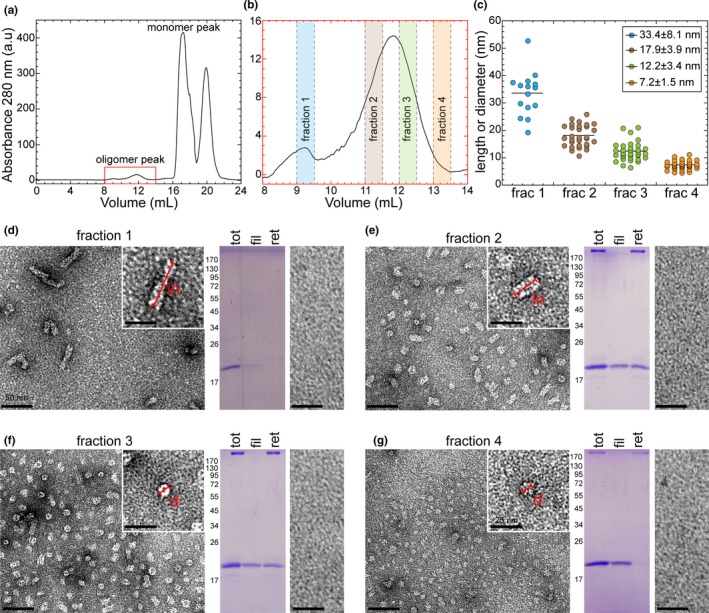
Size‐based separation of α‐syn oligomers using Superose 6 column and filtration analysis. (a) Superose 6 based purification of α‐syn oligomers. Oligomeric fractions (highlighted in red box) eluted from 8 ml to 14 ml and monomeric fractions from 16 ml to 19 ml. (b) Highlighted oligomeric peak from (a) showing the elution of different fractions (fraction 1, fraction 2, fraction 3 and fraction 4 in varying colours) of oligomers collected for filtration analysis. (c) EM images based size distribution analysis of different fractions of oligomeric populations. (d, e, f and g) EM analysis of total (left) and filtrate (right) of oligomeric fraction 1 (d), fraction 2 (e), fraction 3 (f) and fraction 4 (g). Scale bars of total and filtrate samples are 50 nm. Zoomed EM images (scale bars: 25 nm) on the top right of each fraction shows the length (ln) or diameter (d) of differently sized oligomers measured in (c). Coomassie‐stained sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS‐PAGE) gel in the middle of each oligomeric fraction shows the total (tot) oligomeric fraction used for filtration analysis using 100 kDa MWCO membrane, filtrate (fil) and retentate (ret) samples following the filtration protocol
Several studies have shown that using monomeric aSyn preparations that are free of any preformed fibrils or oligomers are essential to obtain reproducible and reliable aggregation kinetic profiles and results (Buell et al., 2014; Fredenburg et al., 2007; Singh et al., 2013). This is commonly achieved using SEC. However, this procedure requires at least 2–5 hr and is not suitable for experiments that involve comparative analysis of multiple samples or the use of highly precious aSyn samples, for example site‐specifically modified synthetic or semi‐synthetic proteins. Here, we show that a simple protocol based on filtration through a 100 kDa MWCO membrane provides an efficient method for generating an aggregate‐free aSyn preparation and the rapid removal or isolation of aSyn oligomers (>aSyn dimers).
3.4. Analysis and separation of α‐syn mixtures containing monomers, oligomers and fibrils
To demonstrate the robustness of the protocol, we tested it on an α‐syn sample mixture composed of known concentrations (determined by BCA assay and AAA) of monomers, oligomers and fibrils (referred to as the aSyn mixture), as illustrated in Figure 4a. This mixture was then subjected to a centrifugation‐based filtration protocol, and the α‐syn species isolated in each step of the protocol were characterized using SDS‐PAGE analysis, amino acid analysis, CD and EM. Figure 4b shows the SDS‐PAGE analysis of the mixture as well as aSyn samples recovered from each step of the protocol, that is the pellet (fibrils), supernatant (monomers and oligomers), filtrate (monomers) and retentate (oligomers). This analysis shows that we are able to recover each species. Figure 4c shows the initial concentrations of individual α‐syn samples (fibrils, oligomers and monomers) used in the α‐syn mixture, and the concentrations of the recovered α‐syn species isolated during the different steps of the protocol (determined by BCA and AAA). Comparison of the initial protein concentration estimation between BCA assay and AAA reveals that there is not any significant difference on the concentrations except for oligomers. However, comparing the concentrations of initial versus recovered α‐syn species shows the almost total recovery of the oligomers (10 μM was the initial concentration and 9.0 μM was the recovered concentration from BCA assay, and with AAA 5.9 μM was the initial and 7.7 μM was the recovered) and monomers (20 μM was the initial and 21.4 μM was the recovered concentration with BCA assay, and with AAA both initial and recovered has 15.2 μM concentration), confirming that there was no significant reduction in the concentrations of soluble α‐syn species during the recovery. Using AAA, the concentration of fibrils recovered (20.9 µM) was also close to that of initial concentration (22.4 μM) added to the mixture.
The importance of this protocol is emphasized by the results obtained from CD and EM analyses. Figure 4d and I show the CD and EM analyses of the total mixture of α‐syn before applying our protocol (Figure 4d and i) and the fibrils (Figure 4e and j), monomers (Figure 4g and l) and oligomers (Figure 4h and m) after separation after applying the protocol. The EM image of the aSyn mixture is dominated by the fibrils (Figure 4i). The oligomers are seen in the background and are often underestimated or overlooked in samples that are rich in fibrils, whereas the monomers cannot be detected by EM. The CD spectra are characterized by a minimum at 221 nm, which is consistent with a secondary structure that is dominated by β‐sheets (Figure 4d). This signal comes primarily from the oligomers and fibrils, both of which are rich in β‐sheet structures which have been previously shown in several studies (Iyer et al., 2017; Khalaf et al., 2014; Del Mar, Greenbaum, Mayne, Englander, & Woods, 2005; Skamris, Marasini, Madsen, Foderà, & Vestergaard, 2019). The relative contribution of the monomers to the CD spectra is not easy to delineate. This illustrates the difficulties in assessing the complexity and heterogeneity of α‐syn samples containing multiple species by EM or CD, and underscores the critical importance of using protocols that provide a quantitative assessment of the distribution of the different α‐syn species. The data in Figure 4e–h and j–m show how this is achievable using the protocol described here. Figure 4e and j show the CD spectra and EM images of the α‐syn fibrils separated by ultracentrifugation of the α‐syn mixture at 100,000 g for 30 min at 4°C (Figure 4a). As expected, and because of their enriched cross‐β structure, the CD spectra of the sedimented and resuspended fibrils show the classical signal for the β‐sheet structure (minimum at 221 nm, Figure 4e), and the EM image (Figure 4j) shows the appearance of straight and twisted fibrils. Analysis of the supernatant fraction containing soluble α‐syn species (monomers and oligomers) by CD spectroscopy revealed a spectrum (Figure 4f) that is dominated by a minimum at 200 nm and a shoulder minima band at 223 nm, consistent with the presence of both disordered (monomers) and structured (oligomers) α‐syn species. Analysis of this sample by EM shows the presence of oligomers, but their morphological and size diversity is challenging to discern (Figure 4k). Upon separation of the oligomers from the monomers by filtration through a 100 kDa MWCO filter, the CD spectra for the isolated monomer shows a single with minimum at 199 nm (without a shoulder), indicating the effective removal of all oligomers, which was also further confirmed by the absence of any oligomers in the sample (Figure 4g and l). The recovered oligomers were resuspended in the same volume as the supernatant. They exhibited a CD spectrum with a broad minimum centred at 219 nm (Figure 4h), consistent with the presence of mixed secondary structure contents dominated by β‐sheet structures. EM analysis of the recovered oligomers allowed more easy assessment of their morphological diversity and revealed the presence of spherical, annular and tubular‐shaped structures (Figure 4m). Taken together, these data show that applying our protocol to α‐syn samples could help unmask their complexity, and that relying solely on CD or EM analyses may lead to incorrect conclusions about the heterogeneity of the sample. This could partially explain some of the variations and/or irreproducibility of experimental observations in the field.
3.5. Assessing the effect of PFF lyophilization on distribution of α‐syn species
Very often, fibril and oligomeric preparations of α‐syn are shared and exchanged between different research groups. The samples are usually frozen and shipped, or as more recently, done by Proteos (https://www.michaeljfox.org/grant/external-validation-proteos-alpha-synuclein-preformed-fibrils) stored and shipped in freeze‐dried form (lyophilized powder). Previous studies have shown that subjecting α‐syn samples to freeze/thaw cycles induces/increases oligomer/soluble α‐syn species formation (Mollenhauer, Batrla, & El‐Agnaf, 2017; Polinski et al., 2018; Stephens et al., 2018). The extent to which these storage procedures alter the biophysical properties of the fibrils will most likely depend on the type of fibrils present and the number of freeze/thaw cycles. Therefore, it is crucial to reassess the properties of the samples after thawing or resolubilization.
Here, we applied the centrifugation‐based filtration protocol to an α‐syn fibril sample, which was lyophilized and resolubilized in water (referred to as lyophilized fibrils). Figure 5a illustrates the steps and analysis we performed on this sample. SDS‐PAGE analysis (Figure 5b) revealed that only half of the α‐syn protein is present as the mature and sedimentable fully grown fibrils as identified in the pellet fraction. The CD and EM analyses of the lyophilized fibrils show the presence of β‐sheet secondary structures (Figure 5c) and long and straight α‐syn fibrils (Figure 5g). Similar to Figure 4, α‐syn monomers and oligomers are not easily detectable or quantifiable by these techniques. However, once we applied our protocol, one can clearly see that this sample of fibrils indeed contained a large fraction of monomers and a very small percentage of α‐syn oligomers (Figure 5b, e, f and i). SDS‐PAGE analysis showed that approximately 5% of the soluble α‐syn sample was retained in the spin filter and recovered as the oligomer fraction. Filtered α‐syn monomers show the typical CD spectrum and characteristics of the unstructured α‐syn species with a minimum signal at 200 nm (Figure 5e). Again, CD and EM analyses of the soluble α‐syn failed to detect the small percentage of existing oligomers, showing an absence of any oligomers‐like structures by EM (Figure 5h) and a CD spectrum that reflects a predominantly disordered conformation. However, analysis of the recovered oligomers clearly showed the presence of small spherical shaped α‐syn oligomers with an average diameter of approximately 12 nm (Figure 5i, inserts) and the CD spectra (Figure 5f) showed a peak minimum at 218 nm, revealing the structured oligomeric α‐syn species. Taken together, this example illustrated how applying the centrifugation‐based filtration protocol enabled the separation of complex mixtures of α‐syn aggregate, and facilitated the detection and characterization of low abundance species which cannot be easily recognized in the EM images and contribute minimially to the CD spectra of such complex mixtures. We have also used the same protocol on the same lyophilized fibrils sample to assess the presence and level of α‐syn oligomers after sonication as done in Figure 5 (Figure S3). These results were then used to help improve the procedure for generating stable and reproducible fibril preparations.
In summary, the centrifugation‐based protocol presented here is flexible, that is it can be easily applied to the same α‐syn fibrils resuspended after lyophilization and following sonication. Very importantly, the protocol is robust and shows that it is possible to separate the different α‐syn species from the same α‐syn sample, such as fibrils, monomers and oligomers, even when present at levels that are not detected by CD and EM analyses. Furthermore, the protocol captures changes in the concentration of α‐syn oligomeric species following manipulation of the samples, for example PFF sonication. Such careful characterizations of the samples will improve the reproducibility of PFF preparations, which will lead to more reliable and reproducible results from experiments aimed at assessing the structural properties, interactome, toxicity, seeding capacity and pathology spreading of α‐syn fibrils.
4. DISCUSSION
There is a consensus that the heterogeneity and variation in the distribution of different species (monomers, oligomers and fibrils) in α‐syn aggregate preparations are major factors contributing to the experimental differences and irreproducibility in α‐syn research today. Previous efforts to address these experimental differences have focused on comparing the polymorphisms and structural properties of α‐syn fibrils or oligomers as determined by TEM or AFM which do not allow for quantitative determination of the relative ratio of the different species in the sample. We believe that advancing our understanding on the role of α‐syn aggregation in the pathogenesis of PD requires the development of reliable methods that not only enable the preparation of different species of α‐syn or isolation of distinct α‐syn species from complex sample mixtures but also quantitative assessment of the distribution of different α‐syn species in such preparations. This is crucial since most α‐syn oligomeric and fibrillar species are in dynamic equilibrium with monomers and could undergo conformational and quaternary structure changes in response to changes in their environment. Such methods should be accessible to most research laboratories and be suitable for the analysis of small amounts/volumes of samples. While several biophysical techniques have been widely used to characterize the size, morphology and structural properties of α‐syn aggregates, those methods usually require high quantities of protein and special instrumentation and/or do not allow for the analysis of multiple samples within a short period of time (1–2 hr). These limitations make it very difficult to achieve detailed analysis of the distribution of α‐syn samples prior to performing biophysical or biological studies and/or during these studies.
Herein, we describe a simple centrifugation‐based filtration protocol that overcomes the aforementioned limitations. The protocol is based on multiple separations and analytical procedures that have been validated by multiple research groups. This protocol offers several advantages: (1) it requires only basic laboratory tools and equipment, which makes it easy to implement in most research laboratories, (2) it can be applied to very low sample volumes (50–500 μL); (3) it allows for isolation and characterization of each species at submicromolar concentrations and (4) it can be performed in a very short time (3–4 hr). To illustrate the utility of this protocol, we presented several examples that demonstrate how it can be used to: (1) assess the distribution of α‐syn monomers, oligomers and fibrils in α‐syn preparations; (2) isolate small amount of each of these species from complex mixtures; (3) investigate the extent of monomer release from α‐syn fibrils; (4) capture the batch‐to‐batch variability in α‐syn PFF preparations and changes in α‐syn species upon manipulations of these preparations (e.g. lyophilization and sonication). We also showed that it enables the detection of small amounts of oligomers in α‐syn fibril preparations, the presence of which is usually undetectable or unappreciated by standard techniques used to characterize fibril preparations (e.g. CD and EM). These capabilities will contribute significantly to improve sample‐to‐sample variations and improve the level of characterization of monomeric, oligomeric and fibrillar α‐syn samples used in structure‐function/dysfunction relationship studies. This is crucial to allow accurate interpretation of experiments and comparison of results obtained using different preparations.
This simple protocol has wide‐ranging applications beyond what is described above. For example, it can be used to assess the disaggregase activity of small molecules and chaperones, allow determination of amounts of fibrils remaining or monomers release upon co‐incubation with chaperones or drugs, and allow insight into the mechanism by which these disaggregases disassemble the fibrils, that is through fibril fragmentation, oligomer formation or monomer release. It is worth reminding that this protocol has been developed and validated for the characterization of α‐syn samples in simple buffer solutions. We plan to explore the possibilities to modify it and extend its application to the characterization of α‐syn species in complex samples derived from culture media, cell extracts, biological fluids or brain homogenates.
Supporting information
ACKNOWLEDGEMENTS AND CONFLICT OF INTEREST DISCLOSURE
We thank Somanath Jagannath for his assistance during the preparation of WT α‐syn oligomers and Melek Firat Altay for her critical review of the manuscript. This project was funded by the École Polytechnique Fédérale de Lausanne and the Michael J Fox Foundation. We also acknowledge the EPFL CIME central facility for their assistance during the EM imaging. The preprint version of this article, prior to peer review, was published on bioRxiv (Kumar et al. 2019; https://www.biorxiv.org/content/10.1101/772160v1). The authors do not have any conflict of interest to disclose. Prof. Hilal A. Lashuel is the founder and chief scientific officer at ND BioSciences, Epalinges, Switzerland.
Kumar ST, Donzelli S, Chiki A, Muazzam Kamil Syed M, Lashuel HA. A simple, versatile and robust centrifugation‐based filtration protocol for the isolation and quantification of α‐synuclein monomers, oligomers and fibrils: Towards improving experimental reproducibility in α‐synuclein research. J. Neurochem.. 2020;153:103–119. 10.1111/jnc.14955
Read the Editorial Highlight for this article on page https://doi.org/10.1111/jnc.14973.
REFERENCES
- Abdelmotilib, H. , Maltbie, T. , Delic, V. , Liu, Z., Hu, X. , Fraser, K. B. … West, A. (2017). Alpha‐Synuclein fibril‐induced inclusion spread in rats and mice correlates with dopaminergic Neurodegeneration. Neurobiology of Diseases, 105, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam, P. , Bousset, L. , Melki, R. , & Otzen, D. E. (2019). Alpha‐synuclein oligomers and fibrils: A spectrum of species, a spectrum of toxicities. Journal of Neurochemistry, 150, 522–534. [DOI] [PubMed] [Google Scholar]
- Baba, M. , Nakajo, S. , Tu, P. H. , Tomita, T. , Nakaya, K. , Lee, V. M. , … Iwatsubo, T. (1998). Aggregation of alpha‐synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. American Journal of Pathology, 152, 879–884. [PMC free article] [PubMed] [Google Scholar]
- Bieschke, J. , Russ, J. , Friedrich, R. P. , Ehrnhoefer, D. E. , Wobst, H. , Neugebauer, K. , & Wanker, E. E. (2010). EGCG remodels mature alpha‐synuclein and amyloid‐beta fibrils and reduces cellular toxicity. Proceedings of the National Academy of Sciences of the United States of America, 107, 7710–7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak, H. , Del Tredici, K. , Rub, U. , de Vos, R. A. , Jansen Steur, E. N. , & Braak, E. (2003). Staging of brain pathology related to sporadic Parkinson's disease. Neurobiology of Aging, 24, 197–211. 10.1016/S0197-4580(02)00065-9 [DOI] [PubMed] [Google Scholar]
- Buell, A. K. , Galvagnion, C. , Gaspar, R. , Sparr, E. , Vendruscolo, M. , Knowles, T. P. , … Dobson, C. M. (2014). Solution conditions determine the relative importance of nucleation and growth processes in alpha‐synuclein aggregation. Proceedings of the National Academy of Sciences of the United States of America, 111, 7671–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway, K. A. , Lee, S. J. , Rochet, J. C. , Ding, T. T. , Williamson, R. E. , & Lansbury, P. T. Jr (2000). Acceleration of oligomerization, not fibrillization, is a shared property of both alpha‐synuclein mutations linked to early‐onset Parkinson's disease: Implications for pathogenesis and therapy. Proceedings of the National Academy of Sciences of the United States of America, 97, 571–576. 10.1073/pnas.97.2.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremades, N. , Cohen, S. I. , Deas, E. , Abramov, A. Y. , Chen, A. Y. , Orte, A. , … Klenerman, D. (2012). Direct observation of the interconversion of normal and toxic forms of alpha‐synuclein. Cell, 149, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer, K. M. , Krebs, S. K. , Wolff, M. , Birk, G. , & Hengerer, B. (2009). Seeding induced by alpha‐synuclein oligomers provides evidence for spreading of alpha‐synuclein pathology. Journal of Neurochemistry, 111, 192–203. [DOI] [PubMed] [Google Scholar]
- Del Mar, C. , Greenbaum, E. A. , Mayne, L. , Englander, S. W. , & Woods, V. L. Jr (2005). Structure and properties of alpha‐synuclein and other amyloids determined at the amino acid level. Proceedings of the National Academy of Sciences of the United States of America, 102, 15477–15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis, M. E. , Leung, E. H. , Sweeny, E. A. , Jackrel, M. E. , Cushman‐Nick, M. , Neuhaus‐Follini, A. , … Shorter, J. (2012). Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell, 151, 778–793. 10.1016/j.cell.2012.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats, P. , Lee, H. J. , Bae, E. J. , Patrick, C. , Rockenstein, E. , Crews, L. , … Lee, S. J. (2009). Inclusion formation and neuronal cell death through neuron‐to‐neuron transmission of alpha‐synuclein. Proceedings of the National Academy of Sciences of the United States of America, 106, 13010–13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, P. E. , & Tennyson, V. M. (1965). Phase and electron microscopic observations of Lewy bodies and melanin granules in the substantia nigra and locus Caeruleus in Parkinson's disease. Journal of Neuropathology & Experimental Neurology, 24, 398–414. [Google Scholar]
- Fauvet, B. , Mbefo, M. K. , Fares, M. B. , Desobry, C. , Michael, S. , Ardah, M. T. , … Lashuel, H. A. (2012). Alpha‐Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. Journal of Biological Chemistry, 287, 15345–15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forno, L. S. (1969). Concentric hyalin intraneuronal inclusions of Lewy type in the brains of elderly persons (50 incidental cases): Relationship to parkinsonism. Journal of the American Geriatrics Society, 17, 557–575. 10.1111/j.1532-5415.1969.tb01316.x [DOI] [PubMed] [Google Scholar]
- Fredenburg, R. A. , Rospigliosi, C. , Meray, R. K. , Kessler, J. C. , Lashuel, H. A. , Eliezer, D. , & Lansbury, P. T. Jr (2007). The impact of the E46K mutation on the properties of alpha‐synuclein in its monomeric and oligomeric states. Biochemistry, 46, 7107–7118. [DOI] [PubMed] [Google Scholar]
- Fusco, G. , Chen, S. W. , Williamson, P. T. F. , Cascella, R. , Perni, M. , Jarvis, J. A. , … Simone, A. D. (2017). Structural basis of membrane disruption and cellular toxicity by alpha‐synuclein oligomers. Science, 358, 1440–1443. [DOI] [PubMed] [Google Scholar]
- Galvin, J. E. , Lee, V. M. , & Trojanowski, J. Q. (2001). Synucleinopathies: Clinical and pathological implications. Archives of Neurology, 58, 186–190. 10.1001/archneur.58.2.186 [DOI] [PubMed] [Google Scholar]
- Gao, X. , Carroni, M. , Nussbaum‐Krammer, C. , Mogk, A. , Nillegoda, N. B. , Szlachcic, A. , … Bukau, B. (2015). Human Hsp70 disaggregase reverses Parkinson's‐linked alpha‐synuclein amyloid fibrils. Molecular Cell, 59, 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, D. , Singh, P. K. , Sahay, S. , Jha, N. N. , Jacob, R. S. , Sen, S. , … Maji, S. K. (2015). Structure based aggregation studies reveal the presence of helix‐rich intermediate during alpha‐Synuclein aggregation. Scientific Reports, 5, 9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb, W. R. , & Lees, A. J. (1988). The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. Journal of Neurology, Neurosurgery and Psychiatry, 51, 745–752. 10.1136/jnnp.51.6.745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert, M. (2015). NEURODEGENERATION. Alzheimer's and Parkinson's diseases: The prion concept in relation to assembled Abeta, tau, and alpha‐synuclein. Science, 349, 1255555. [DOI] [PubMed] [Google Scholar]
- Hansen, C. , Angot, E. , Bergstrom, A. L. , Steiner, J. A. , Pieri, L. , Paul, G. … Brundin, P. (2011). Alpha‐Synuclein propagates from mouse brain to grafted dopaminergic neurons and seeds aggregation in cultured human cells. Journal of Clinical Investigation, 121, 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai, A. , Masliah, E. , Yoshimoto, M. , Ge, N. , Flanagan, L. , de Silva, H. A. , … Saitoh, T. (1995). The precursor protein of non‐A beta component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron, 14, 467–475. [DOI] [PubMed] [Google Scholar]
- Iyer, A. , Roeters, S. J. , Kogan, V. , Woutersen, S. , Claessens, M. M. A. E. , & Subramaniam, V. (2017). C‐Terminal truncated α‐synuclein fibrils contain strongly twisted β‐sheets. Journal of the American Chemical Society, 139, 15392–15400. 10.1021/jacs.7b07403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakes, R. , Spillantini, M. G. , & Goedert, M. (1994). Identification of two distinct synucleins from human brain. FEBS Letters, 345, 27–32. 10.1016/0014-5793(94)00395-5 [DOI] [PubMed] [Google Scholar]
- Jellinger, K. A. (2003). Neuropathological spectrum of synucleinopathies. Movement Disorders, 18(Suppl 6), S2–12. 10.1002/mds.10557 [DOI] [PubMed] [Google Scholar]
- Karpowicz, R. J. Jr , Trojanowski, J. Q. , & Lee, V. M. (2019). Transmission of alpha‐synuclein seeds in neurodegenerative disease: Recent developments. Laboratory Investigation, 99, 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf, O. , Fauvet, B. , Oueslati, A. , Dikiy, I. , Mahul‐Mellier, N.‐L. , Ruggeri, F. S. … Lashuel, H. A. (2014). The H50Q mutation enhances alpha‐synuclein aggregation, secretion, and toxicity. Journal of Biological Chemistry, 289, 21856–21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R. , Das, S. , Mohite, G. M. , Rout, S. K. , Halder, S. , Jha, N. N. , … Maji, S. K. (2018). Cytotoxic oligomers and fibrils trapped in a gel‐like state of alpha‐synuclein assemblies. Angewandte Chemie (International Ed. in English), 57, 5262–5266. [DOI] [PubMed] [Google Scholar]
- Kumar, S. T. , Donzelli, S. , Chiki, A. , Syed, M. M. K. , & Lashuel, H. A. (2019) A simple, versatile and robust centrifugation‐based filtration protocol for the isolation and quantification of α‐synuclein monomers, oligomers and fibrils: towards improving experimental reproducibility in α‐synuclein research. bioRxiv 772160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury, P. T. , & Lashuel, H. A. (2006). A century‐old debate on protein aggregation and neurodegeneration enters the clinic. Nature, 443, 774–779. 10.1038/nature05290 [DOI] [PubMed] [Google Scholar]
- Lashuel, H. A. (2005) Membrane permeabilization: A common mechanism in protein‐misfolding diseases. Science of Aging Knowledge Environment, 2005, pe28–pe28. [DOI] [PubMed] [Google Scholar]
- Lashuel, H. A. , & Lansbury, P. T. Jr (2006). Are amyloid diseases caused by protein aggregates that mimic bacterial pore‐forming toxins? Quarterly Reviews of Biophysics, 39, 167–201. 10.1017/S0033583506004422 [DOI] [PubMed] [Google Scholar]
- Lashuel, H. A. , Petre, B. M. , Wall, J. , Simon, M. , Nowak, R. J. , Walz, T. , & Lansbury, P. T. Jr (2002). Alpha‐synuclein, especially the Parkinson's disease‐associated mutants, forms pore‐like annular and tubular protofibrils. Journal of Molecular Biology, 322, 1089–1102. [DOI] [PubMed] [Google Scholar]
- Lee, H. J. , Patel, S. , & Lee, S. J. (2005). Intravesicular localization and exocytosis of alpha‐synuclein and its aggregates. Journal of Neuroscience, 25, 6016–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. J. , Suk, J. E. , Bae, E. J. , Lee, J. H. , Paik, S. R. , & Lee, S. J. (2008). Assembly‐dependent endocytosis and clearance of extracellular alpha‐synuclein. International Journal of Biochemistry & Cell Biology, 40, 1835–1849. [DOI] [PubMed] [Google Scholar]
- Li, J.‐Y. , Englund, E. , Holton, J. L. , Soulet, D. , Hagell, P. , Lees, A. J. , … Brundin, P. (2008). Lewy bodies in grafted neurons in subjects with Parkinson's disease suggest host‐to‐graft disease propagation. Nature Medicine, 14, 501–503. 10.1038/nm1746 [DOI] [PubMed] [Google Scholar]
- Lipkin, L. E. (1959). Cytoplasmic inclusions in ganglion cells associated with parkinsonian states: A neurocellular change studied in 53 cases and 206 controls. American Journal of Pathology, 35, 1117–1133. [PMC free article] [PubMed] [Google Scholar]
- Luk, K. C. , Kehm, V. , Carroll, J. , Zhang, B. , O'Brien, P. , Trojanowski, J. Q. , & Lee, V. M. (2012). Pathological alpha‐synuclein transmission initiates Parkinson‐like neurodegeneration in nontransgenic mice. Science, 338, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk, K. C. , Song, C. , O'Brien, P. , Stieber, A. , Branch, J. R. , Brunden, K. R. , … Lee, V. M. (2009). Exogenous alpha‐synuclein fibrils seed the formation of Lewy body‐like intracellular inclusions in cultured cells. Proceedings of the National Academy of Sciences of the United States of America, 106, 20051–20056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, E. , Decker, S. C. , Riddle, D. M. , Caputo, A. , Zhang, B. , Cole, T. … Luk, K. C. (2018). Differential alpha‐synuclein expression contributes to selective vulnerability of hippocampal neuron subpopulations to fibril‐induced toxicity. Acta Neuropathologica, 135, 855–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahul‐Mellier, A.‐L. , Altay, M. F. , Burtscher, J. et al (2018). The making of a lewy body: The role of α‐synuclein post‐fibrillization modifications in regulating the formation and the maturation of pathological inclusions. bioRxiv, 500058. [Google Scholar]
- Mahul‐Mellier, A. L. , Vercruysse, F. , Maco, B. , Ait‐Bouziad, N. , De Roo, M. , Muller, D. , & Lashuel, H. A. (2015). Fibril growth and seeding capacity play key roles in alpha‐synuclein‐mediated apoptotic cell death. Cell Death and Differentiation, 22, 2107–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda‐Suzukake, M. , Nonaka, T. , Hosokawa, M. , Oikawa, T. , Arai, T. , Akiyama, H. , … Hasegawa, M. (2013). Prion‐like spreading of pathological alpha‐synuclein in brain. Brain, 136, 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolaenko, I. , Pletnikova, O. , Kawas, C. H. , O'Brien, R. , Resnick, S. M. , Crain, B. , & Troncoso, J. C. (2005). Alpha‐synuclein lesions in normal aging, Parkinson disease, and Alzheimer disease: Evidence from the Baltimore Longitudinal Study of Aging (BLSA). Journal of Neuropathology and Experimental Neurology, 64, 156–162. 10.1093/jnen/64.2.156 [DOI] [PubMed] [Google Scholar]
- Mollenhauer, B. , Batrla, R. , El‐Agnaf, O. et al (2017). A user's guide for alpha‐synuclein biomarker studies in biological fluids: Perianalytical considerations. Movement Disorders, 32, 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasstrom, T. , Fagerqvist, T. , Barbu, M. , Karlsson, M. , Nikolajeff, F. , Kasrayan, A. , …, Bergström, J. (2011). The lipid peroxidation products 4‐oxo‐2‐nonenal and 4‐hydroxy‐2‐nonenal promote the formation of alpha‐synuclein oligomers with distinct biochemical, morphological, and functional properties. Free Radical Biology and Medicine, 50, 428–437. [DOI] [PubMed] [Google Scholar]
- Park, M. J. , Cheon, S. M. , Bae, H. R. , Kim, S. H. , & Kim, J. W. (2011). Elevated levels of alpha‐synuclein oligomer in the cerebrospinal fluid of drug‐naive patients with Parkinson's disease. Clinical Neurology and Neurosurgery, 7, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C. , Gathagan, R. J. , Covell, D. J. , Medellin, C. , Stieber, A. , Robinson, J. L. , … Lee, V. M.‐Y. (2018). Cellular milieu imparts distinct pathological alpha‐synuclein strains in alpha‐synucleinopathies. Nature, 557, 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polinski, N. K. , Volpicelli‐Daley, L. A. , Sortwell, C. E. , Luk, K. C. , Cremades, N. , Gottler, L. M. , … Dave, K. D. (2018). Best practices for generating and using alpha‐synuclein pre‐formed fibrils to model Parkinson's disease in rodents. Journal of Parkinson's Disease, 8, 303–322. 10.3233/JPD-171248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recasens, A. , & Dehay, B. (2014). Alpha‐synuclein spreading in Parkinson's disease. Frontiers in Neuroanatomy, 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recasens, A. , Dehay, B. , Bove, J. , Carballo‐Carbajal, I. , Dovero, S. , Pérez‐Villalba, A. … Vila, M. (2014). Lewy body extracts from Parkinson disease brains trigger alpha‐synuclein pathology and neurodegeneration in mice and monkeys. Annals of Neurology, 75, 351–362. [DOI] [PubMed] [Google Scholar]
- Schmidt, M. L. , Murray, J. , Lee, V. M. , Hill, W. D. , Wertkin, A. , & Trojanowski, J. Q. (1991). Epitope map of neurofilament protein domains in cortical and peripheral nervous system Lewy bodies. American Journal of Pathology, 139, 53–65. [PMC free article] [PubMed] [Google Scholar]
- Shimozawa, A. , Ono, M. , Takahara, D. , Tarutani, A. , Imura, S. , Masuda‐Suzukake, M. , … Hasegawa, M. (2017). Propagation of pathological alpha‐synuclein in marmoset brain. Acta Neuropathol Commun, 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. K. , Kotia, V. , Ghosh, D. , Mohite, G. M. , Kumar, A. , & Maji, S. K. (2013). Curcumin modulates alpha‐synuclein aggregation and toxicity. ACS Chemical Neuroscience, 4, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamris, T. , Marasini, C. , Madsen, K. L. , Foderà, V. , & Vestergaard, B. (2019). Early stage alpha‐synuclein amyloid fibrils are reservoirs of membrane‐binding species. Scientific Reports, 9, 1733 10.1038/s41598-018-38271-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini, M. G. , & Goedert, M. (2000). The alpha‐synucleinopathies: Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy. Annals of the New York Academy of Sciences, 920, 16–27. [DOI] [PubMed] [Google Scholar]
- Spillantini, M. G. , Schmidt, M. L. , Lee, V. M. , Trojanowski, J. Q. , Jakes, R. , & Goedert, M. (1997). Alpha‐synuclein in Lewy bodies. Nature, 388, 839–840. [DOI] [PubMed] [Google Scholar]
- Stephens, A. D. , Nespovitaya, N. , Zacharopoulou, M. , Kaminski, C. F. , Phillips, J. J. , & Kaminski Schierle, G. S. (2018). Different structural conformers of monomeric alpha‐synuclein identified after lyophilizing and freezing. Analytical Chemistry, 90, 6975–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, A. , Mallory, M. , Sundsmo, M. , Honer, W. , Hansen, L. , & Masliah, E. (1998). Abnormal accumulation of NACP/alpha‐synuclein in neurodegenerative disorders. American Journal of Pathology, 152, 367–372. [PMC free article] [PubMed] [Google Scholar]
- Tarutani, A. , Suzuki, G. , Shimozawa, A. , Nonaka, T. , Akiyama, H. , Hisanaga, S. , & Hasegawa, M. (2016). The effect of fragmented pathogenic alpha‐synuclein seeds on prion‐like propagation. Journal of Biological Chemistry, 291, 18675–18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, P. H. , Galvin, J. E. , Baba, M. , Giasson, B. , Tomita, T. , Leight, S. , … Lee, V. M. (1998). Glial cytoplasmic inclusions in white matter oligodendrocytes of multiple system atrophy brains contain insoluble alpha‐synuclein. Annals of Neurology, 44, 415–422. [DOI] [PubMed] [Google Scholar]
- Volpicelli‐Daley, L. A. , Luk, K. C. , Patel, T. P. , Tanik, S. A. , Riddle, D. M. , Stieber, A. , … Lee, V. M. (2011). Exogenous alpha‐synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron, 72, 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, J. C. , Giles, K. , Oehler, A. , Middleton, L. , Dexter, D. T. , Gentleman, S. M. , … Prusiner, S. B. (2013). Transmission of multiple system atrophy prions to transgenic mice. Proceedings of the National Academy of Sciences of the United States of America, 110, 19555–19560. 10.1073/pnas.1318268110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb, P. H. , Zhen, W. , Poon, A. W. , Conway, K. A. , & Lansbury, P. T. Jr (1996). NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry, 35, 13709–13715. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Griggs, A. , Rochet, J. C. , & Stanciu, L. A. (2013). In vitro study of alpha‐synuclein protofibrils by cryo‐EM suggests a Cu(2+)‐dependent aggregation pathway. Biophysical Journal, 104, 2706–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


