SUMMARY
Deficiency of ubiquitin-specific peptidase 18 (USP18) is a severe type I interferonopathy. USP18 down-regulates type I interferon signaling by blocking the access of Janus-associated kinase 1 (JAK1) to the type I interferon receptor. The absence of USP18 results in unmitigated interferon-mediated inflammation and is lethal during the perinatal period. We describe a neonate who presented with hydrocephalus, necrotizing cellulitis, systemic inflammation, and respiratory failure. Exome sequencing identified a homozygous mutation at an essential splice site on USP18. The encoded protein was expressed but devoid of negative regulatory ability. Treatment with ruxolitinib was followed by a prompt and sustained recovery. (Funded by King Saud University and others.)
TYPE I INTERFERONOPATHIES ARE MONOGENIC AUTOINFLAMMATORY DIS-orders1–3 that are characterized by the overproduction of, or an enhanced response to, type I interferons.1–4 USP18 down-regulates signaling by type I interferons by blocking the interaction between JAK1 and the type I interferon receptor 2 (IFNAR2) subunit of the type I interferon receptor complex5–7 (Fig. 1). USP18 also has enzymatic activity in removing the covalently conjugated 15kDa protein, encoded by interferon-stimulated gene 15 (ISG15), from its targets in a process called de-ISGylation.8 Finally, independent of its affinity for ISGylated proteins, USP18 also binds free ISG15, which protects USP18 against proteasomal degradation, thereby enhancing its negative regulatory capacity.9
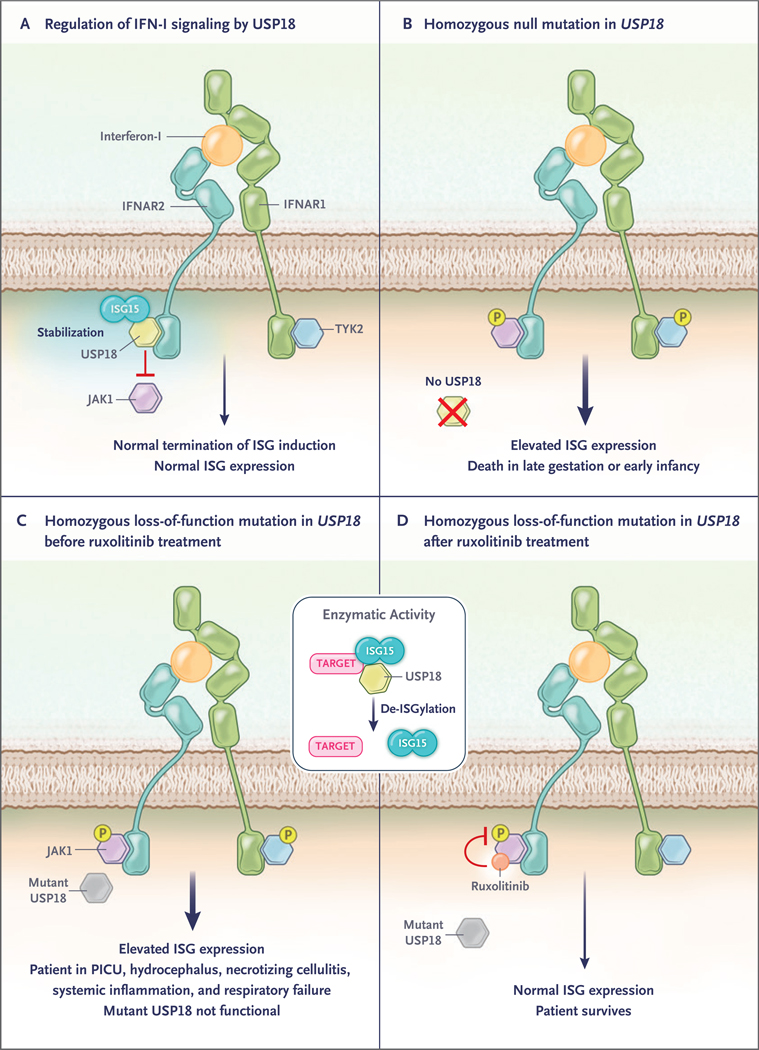
Figure 1 (facing page). Molecular Functions of USP18.
Type I interferon (IFN–I) binds IFN–I receptor, consisting of two subunits, IFN–I receptor 1 (IFNAR1) and IFN–I receptor 2 (IFNAR2). It signals through activation of two receptor–associated protein tyrosine kinases, Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), which phosphorylate (P) signal transducer and activator of transcription 1 (STAT1) and 2 (STAT2) to induce the transcription of hundreds of interferon–stimulated genes (ISGs). Panel A shows the activity of USP18 (an ISG) as a negative–feedback regulator of IFN–I signaling through the inhibition of JAK1 recruitment to IFNAR2. ISG15 stabilizes USP18 and protects it against proteasomal degradation, resulting in normal ISG expression. Panel B shows that homozygous mutations resulting in no synthesis of USP18 cause unrestrained IFN–I signaling and are lethal. Panels C and D show the outcome in the patient in the case report before and after the initiation of ruxolitinib, a JAK1 inhibitor. The box in the middle of the figure shows that in addition to inhibiting JAK1, USP18 has catalytic activity: it cleaves the ubiquitin–like ISG15 protein from conjugated proteins in a process called de–ISGylation. PICU denotes pediatric intensive care unit.
We previously described five patients from two unrelated families with homozygous or compound heterozygous USP18 mutations resulting in a complete lack of USP18 expression.10 This deficiency was characterized by perinatal-onset intracranial hemorrhage, calcifications, brain malformations, liver dysfunction, septic-shock–like presentations, and thrombocytopenia. All the patients had respiratory insufficiency and seizures, which resulted in death in late gestation or early infancy.10 The clinical, pathological, and radiologic features of these patients resembled those of congenital intrauterine infections characteristic of TORCH (toxoplasmosis, other agents [including human immunodeficiency virus, syphilis, varicella, and fifth disease], rubella, cytomegalovirus, and herpes simplex virus) but without detectable infection, which resulted in a diagnosis of pseudo-TORCH. Complete USP18 deficiency is currently the most severe genetic cause of the rapidly expanding group of type I interferonopathies.2,11 Here, we report the case of a Saudi Arabian boy who had an inherited complete USP18 deficiency and survived well beyond the perinatal period owing to supportive care, rapid genetic diagnosis by whole-exome sequencing, and prompt treatment with the JAK inhibitor ruxolitinib (Fig. 1).
CASE REPORT
The patient was born to first-cousin parents from Saudi Arabia after in vitro fertilization. At 38 weeks of gestation, emergency cesarean section was performed because of cardiotocographic findings indicating fetal distress. At birth, the patient was vaccinated with bacille Calmette– Guérin, with no adverse effects. At the age of 13 days, he presented with septic shock and multiple organ failure, including severe acute respiratory distress syndrome (ARDS) (Fig. 2A, subpanel a) and disseminated intravascular coagulopathy without organomegaly. He was admitted to the pediatric intensive care unit (PICU). A nasopharyngeal swab was positive for Bordetella pertussis on polymerase chain reaction (PCR). All cultures from blood, cerebrospinal fluid, and urine were negative. He was treated with supportive care and antibiotics and was discharged after 17 days of hospitalization with grade II intraventricular hemorrhage, as evidenced on transfontanelle ultrasonography and computed tomography (CT) of the head (Fig. 2A, subpanel b; and Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).
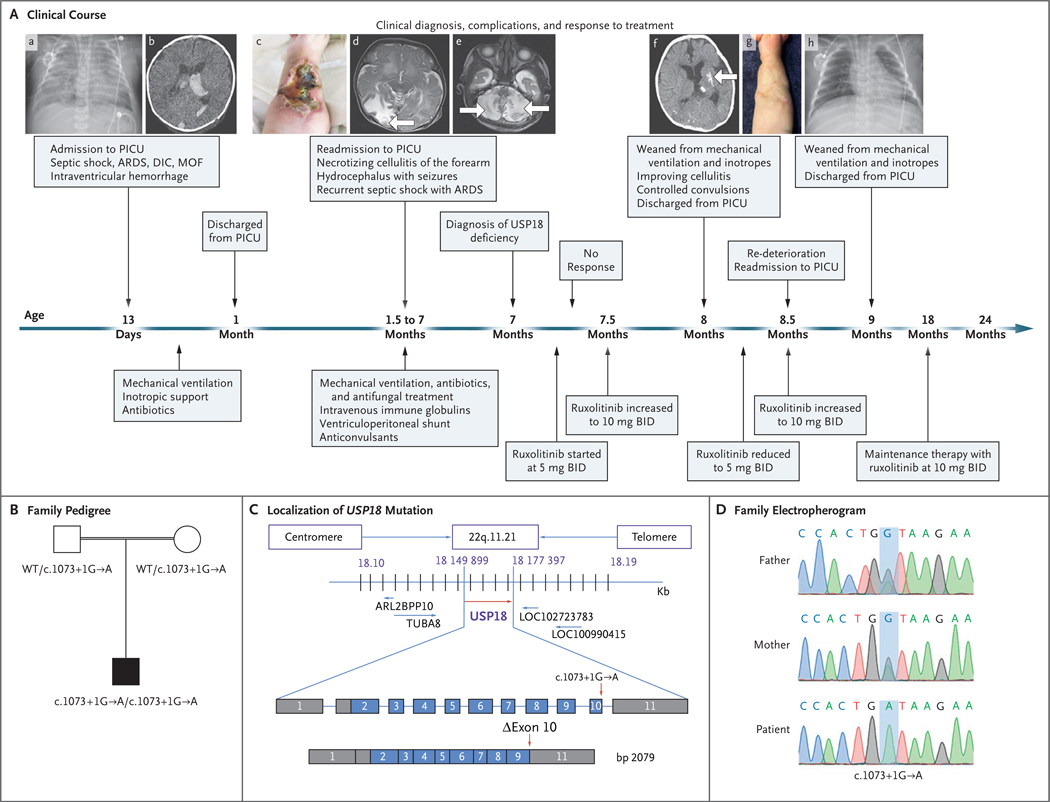
Figure 2 (facing page). Clinical Course of the Patient and Identification of a USP18 Mutation.
In Panel A, at the age of 13 days, the patient was admitted to the pediatric intensive care unit (PICU), where he underwent chest radiography, which showed patches of space opacification with an air bronchogram of both lungs characteristic of severe acute respiratory distress syndrome (ARDS) (subpanel a). At that time, multiple organ failure (MOF) and disseminated intravascular coagulopathy (DIC) had also developed, along with septic shock. Computed tomography (CT) of the brain at the age of 23 days showed mildly dilated lateral ventricles with intraventricular hyperdense acute hemorrhage more marked on the left, corresponding to grade II intraventricular hemorrhage (subpanel b). At the age of 45 days, the patient was readmitted with necrotizing cellulitis at the site of a peripheral venous catheter in the right forearm (subpanel c). Magnetic resonance imaging (MRI) of the brain at the age of 6 weeks showed a well–defined high–intensity signal in the right occipital region (subpanel d, arrow). MRI of the brain at the age of 11 weeks showed a well–defined high–intensity signal in the bilateral cerebellum owing to subacute infarction (subpanel e, arrows). One month after the diagnosis of USP18 deficiency and the initiation of ruxolitinib (at a dose of 5 mg twice daily [BID]) at the age of 7 months, CT of the brain showed resolution of hydrocephalus, hemorrhage, and ischemia with only small areas of residual calcifications in the left putamen (subpanel f, arrow) and the formation of scar tissue on the forearm after healing of the necrotizing cellulitis (subpanel g). Chest radiography after 2 months of ruxolitinib therapy showed sufficient improvement for weaning from mechanical ventilation (subpanel h). Panel B shows familial segregation of the c.1073+1G→A USP18 allele. The unaffected parents, who were first cousins (as indicated by the double horizontal line), were heterozygous for the mutation, and the patient was homozygous. Panel C shows the localization of the USP18 mutation in the genomic DNA and messenger RNA, with coding regions in blue, untranslated regions in gray, and the mutation indicated by red arrows, with the predicted transcript skipping exon 10. Panel D shows the electro-pherographic results of the homozygous c.1073+1G→A USP18 mutation in the patient and the heterozygous mutation in the unaffected parents.
At the age of 45 days, the patient was readmitted with cellulitis, which rapidly progressed to necrotizing cellulitis at the site of a peripheral venous catheter in the right forearm (Fig. 2A, subpanel c). A culture of fluid from the lesion was positive for Enterobacter cloacae. The development of seizures resulted in the administration of two antiepileptic drugs (phenytoin and levetiracetam). CT and magnetic resonance imaging (MRI) of the brain provided evidence of acute hydrocephalus for which a ventriculoperitoneal shunt was placed. Follow-up brain imaging revealed multiple areas of mixed hemorrhagic and ischemic damage in both cerebellar hemispheres, the right parietal and occipital lobes, and the left basal ganglia and external capsule (Fig. 2A, subpanels d and e), and there was evidence of calcification in multiple regions (Fig. S2), none of which were present on previous imaging. The patient progressed to a state of shock with the development of ARDS, resulting in the initiation of ventilatory support, which lasted for 6 months. A culture of tracheal aspirate was positive for mixed organisms on different occasions (Pseudomonas aeruginosa, Haemophilus parainfluenzae, and Candida parapsilosis), whereas blood, cerebrospinal fluid, and urine cultures were negative. On the basis of the culture and sensitivity results, the patient was treated with supportive care together with antibiotics and antifungal therapy. The patient did not show features of classic immunodeficiency, except for low levels of IgG; intravenous infusion of immune globulin was not followed by clinical improvement (Table S1). We detected no likely causative autoantibodies (Fig. S3).
METHODS
Whole-Exome Sequencing and Variant Analysis
Whole-exome sequencing and mitochondrial genome sequencing were performed at the Centogene genetic diagnosis laboratory (Rostock, Germany), as described previously.12 Briefly, a preparation enriched in approximately 37 Mb (214,405 exons) of the consensus coding sequences was obtained from fragmented genomic DNA with 340,000 probes directed against the human genome (Nextera Rapid Capture Exome, Illumina). The library that was generated was sequenced on an Illumina platform to a mean coverage depth of 100 to 130×. An end-to-end in-house bioinformatics pipeline — including base calling, the primary filtering of low-quality reads and probable artifacts, and the annotation of variants — was applied. All disease-causing variants that were reported in the Human Gene Mutation Database, ClinVar database, or CentoMD (class 1) and all variants with a minor allele frequency below 1% in the ExAC database were considered. The evaluation focused on exons and intron boundaries (20 residues on either side).
Cells Obtained from the Patient and Controls
An institutional review board at the College of Medicine, King Saud University, in Saudi Arabia approved the study, and the child’s parents provided written informed consent. Dermal fibroblasts obtained from the patient and controls were immortalized by stable transduction with a pBABE construct (a retroviral vector for cloning and expressing a gene of interest) containing human telomerase reverse transcriptase (hTERT). Fibroblasts obtained from the patient and the controls were stably complemented with wild-type USP18 or luc (encoding the luciferase protein) by lentiviral transduction with a construct encoding an internal ribosomal entry site and red fluorescent protein (RFP). USP18-deficient fibroblasts were transduced with wild-type USP18, with ΔEx10 USP18 (USP18 complementary DNA [cDNA] lacking exon 10), or with a luciferase-expressing construct. RFP-expressing fibroblasts were sorted (FACSAria II, BD Biosciences) and cultured for further analysis.
3′ RACE PCR
Total RNA was extracted from the patient’s hTERT-immortalized fibroblasts. We performed 3′ RACE (rapid amplification of cDNA ends) PCR (Invitrogen) according to the manufacturer’s instructions. PCR products were cloned by TOPO-TA cloning (Thermo Fisher Scientific). Twenty-six colonies were grown; we performed Sanger sequencing on plasmid purified from each colony.
PLASMIDS AND TRANSFECTION
HEK293T cells were transiently transfected with the aid of Lipofectamine 2000 (Invitrogen). The following constructs were used: pCAGGS-Ube1L (encoding ubiquitin-activating enzyme E1-like protein), pCS2+–Herc5, pFlagCMV2-UbcH8 (encoding E2 ubiquitin-conjugating enzyme L6), pTRIP-USP18 (wild-type or ΔEx10 USP18), and pTRIP-ISG15.
EXPRESSION ASSAYS
RNA was extracted from hTERT-immortalized fibroblasts with the RNeasy Mini Kit (Qiagen) or whole blood (PAXgene Blood RNA Kit) and reverse-transcribed (ABI High Capacity Reverse Transcriptase). The expression of interferon-stimulated genes (IFIT1, MX1, RSAD2, IFI27, IFI44L, and ISG15), relative to the 18S housekeeper gene, was analyzed by TaqMan quantitative real-time PCR with the use of Roche LightCycler 480 II.
PROTEIN ANALYSES
Cells were lysed in radioimmunoprecipitation-assay buffer, and protein extract was used for Western blotting. The antibodies that were used were directed against STAT1 (Santa Cruz Biotechnology), STAT2 (Millipore), phospho-Tyr701 STAT1 (Cell Signaling Technology), phosphor-Tyr689 STAT2 (Millipore), USP18 (Cell Signaling Technology), ISG15 (Santa Cruz Biotechnology), β-actin (Cell Signaling Technology), and IFIT1 (Cell Signaling Technology).
AUTOANTIBODY DETECTION
We queried the presence of autoantibodies using IgG autoantibody arrays (RayBiotech). Plasma and serum samples were diluted at a ratio of 1:200. Genepix Pro 7.0 software was used to analyze the images.
Results
WHOLE-EXOME SEQUENCING
In samples obtained from the patient at the age of 7 months, whole-exome sequencing revealed a private (i.e., not listed in a public database) homozygous variant in an essential donor splice site after exon 10 in USP18 (c.1073+1G→A). This variant was confirmed on Sanger sequencing; analysis of samples obtained from the parents yielded results consistent with an autosomal-recessive mode of inheritance (Fig. 2B, 2C, and 2D). No other relevant rare nonsynonymous or splice variants were identified in protein-coding genes associated with, or known to cause, pediatric inflammatory disorders.13
The combined annotation-dependent depletion score of 23.70 for the c.1073+1G→A allele was well above the cutoff score of 2.31,14 indicating mutational significance. The variant lies in an essential donor splice site. We predicted that the variant would force the skipping of exon 10 and cause a frameshift mutation; we therefore considered it to be damaging. We observed no representation of persons who were homozygous for damaging USP18 variants in public genetic databases. On the basis of these results and the clinical symptoms, we reached a tentative diagnosis of autosomal recessive USP18 deficiency.10 Given the established lethality of this condition, we promptly introduced treatment with a JAK inhibitor (a pharmacologic inhibitor of type I interferon signaling), despite having yet to confirm the damaging effects of this novel variant. We reasoned that the genetic results, previous studies supporting the lethality of USP18 deficiency in neonates, and the deteriorating clinical status of the patient justified this intervention.
TREATMENT
We treated the patient with ruxolitinib, a JAK1/2 inhibitor, at a dose of 5 mg administered by mouth twice daily for 1 week, with continuation of the other supportive measures (Fig. 2). Because we observed no improvement, we subsequently increased the dose to 10 mg twice daily. The child’s symptoms rapidly improved, which resulted in weaning from ventilatory support. CT and MRI of the head performed after shunt placement revealed a resolution of hydrocephalus, hemorrhage, and ischemia (Fig. S4), with only residual calcifications and a small residual area of signal intensity in the right occipital region (Fig. 2A, subpanel f). In addition, the necrotizing cellulitis of the right forearm completely healed with residual scarring (Fig. 2A, subpanel g). We tapered the dose of ruxolitinib to 5 mg twice daily with the aim of establishing a minimal dose that would maintain remission, but respiratory failure developed, so the dose was again increased to 10 mg twice daily and recovery ensued. After weaning from mechanical ventilation, the patient was discharged from the PICU (Fig. 2A, subpanel h). At the time, he had hemiparesis on the right side of his body related to associated neurologic complications. A nasogastric tube had been placed for feeding, and the patient was being treated with anticonvulsant medications, physiotherapy, and ruxolitinib at a dose of 10 mg twice daily.
At 24 months, Denver II testing for developmental assessment15 showed abilities consistent with a developmental age of 9 to 10 months. Motor activity on the right side continued to improve, as evidenced by the child’s ability to stand up with support and to hold a bottle with the right hand. He had a score of 4/5 in motorpower assessment in both upper and lower right limbs. The patient’s respiratory status remained stable, except for mild upper respiratory tract infections, which were easily managed.
MOLECULAR PATHOGENESIS
In parallel, we isolated and reverse-transcribed RNA from the peripheral blood of the patient and a healthy control and amplified the region encompassing exon 10 on PCR. We detected a PCR product of lower molecular weight in the patient’s sample that reflected the loss of USP18 exon 10 (Fig. S5A). Using RACE PCR, we observed two different transcripts, resulting from the c.1073+1G→A mutation: a transcript with exon 10 omitted (ΔEx10), which resulted in a frameshift mutation and a putative addition of nine amino acids translated from the 3′ untranslated region, and a transcript with the insertion of part of the intervening intron, which resulted in a premature stop codon at the end of exon 10 (frequencies of 92% and 8%, respectively). We detected no wild-type transcripts (Fig. S5B, S5C, and S5D).
Patients with type I interferonopathy have high levels of expression of interferon-stimulated genes in bulk peripheral-blood mononuclear cells (PBMCs). We thus assessed the messenger RNA (mRNA) levels of six interferon-stimulated genes (IFIT1, MX1, IFI27, RSAD2, IFIT2, and ISG15) in PBMCs collected after the initiation of ruxolitinib treatment. Unfortunately, no blood sample obtained before the initiation of treatment was available. Levels of mRNA transcribed from five of these genes (IFIT1, MX1, RSAD2, IFIT2, and ISG15) in the patient’s PBMCs were not higher than in those obtained from healthy controls, a finding that was consistent with the improvement in the patient’s health status (Fig. S6A through S6F).
We then analyzed the molecular basis of the disease in detail. We transiently transfected HEK293T cells with wild-type or ΔEx10 USP18 and assayed mRNA and protein levels. The absence of exon 10 affected neither mRNA nor protein levels (Fig. 3A and 3B). We then evaluated the different molecular functions of the mutant protein. We first investigated whether its levels were sustained by ISG15. The transient expression of ΔEx10 USP18 with increasing amounts of ISG15 did not lead to its stabilization, as occurs with wild-type USP18 (Fig. 3C). We next analyzed its ability to suppress signaling by type I interferon by transiently expressing wild-type or ΔEx10 USP18 in HEK293T cells and assaying the extent of STAT activation in response to a 30-minute pulse with interferon-α. Unlike the wild-type protein, the mutant protein did not suppress the phosphorylation of STAT1 and STAT2 induced by type I interferon (Fig. 3D). Finally, we assessed the catalytic activity by transiently cotransfecting HEK293T cells with wild-type or ΔEx10 USP18, together with constructs encoding each of the components of the ISGylation machinery (Ube1L, UbcH8, Herc5, and ISG15). The levels of ISG15 conjugates were mildly underrepresented in the HEK293T cells transfected with ΔEx10 USP18 (Fig. 3E), which supports the conclusion that ΔEx10 USP18 has largely intact catalytic activity and so can remove covalently bound ISG15 from protein conjugate, despite its inability to be stabilized by ISG15 or to suppress type I interferon signaling.
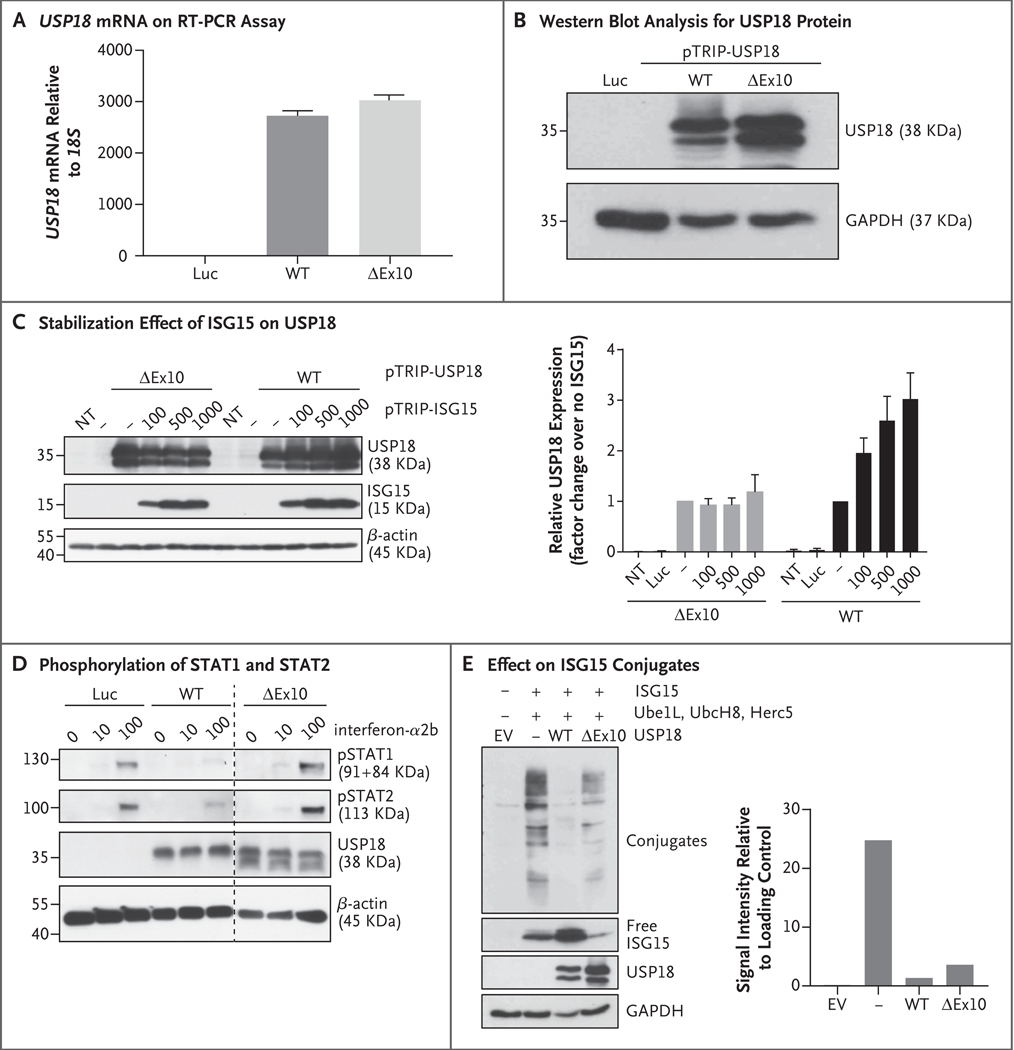
Figure 3. Characterization of the USP18 Allele.
Panel A shows the relative levels of messenger RNA (mRNA), as normalized with the 18S housekeeper gene, in wild-type (WT) USP18 or the c.1073+1G→A (ΔEx10) variant and in a control with lentiviral particles expressing luciferase (Luc), as assayed on quantitative reverse–transcriptase–polymerase chain reaction (RT–PCR) in three independent experiments, each with technical triplicates; the results of a representative experiment are shown. Panel B shows a representative experiment involving cell lysates that were analyzed by Western blotting for the USP18 protein. Panel C shows HEK293T cells that were transfected with wild–type USP18 or the ΔEx10 variant, together with various amounts of pTRIP–ISG15 (graph at the left). Cell lysates were analyzed by Western blotting with the antibodies indicated. The graph at the right shows the densitometric analysis of USP18 protein expression levels normalized over an endogenous control. T bars represent the standard error of three independent experiments. NT denotes not transfected. Panel D shows HEK293T cells that were transduced with wild–type USP18, the ΔEx10 variant, or Luc. Twenty–four hours later, the cells were treated with the indicated concentrations of interferon–α2b for 20 minutes. Cell lysates were analyzed by Western blotting with the antibodies indicated. Samples were run on the same gel, and extra lanes were deleted to remove irrelevant data, as indicated by the vertical dashed line. Panel E shows HEK293T cells that were cotransfected with ISG15 and E1, E2, and E3 enzymes for ISG15 (Ube1L, UbcH8, and Herc5) in combination with the different variants of USP18. The graph at the right shows the densitometric analysis of ISGylated proteins normalized over an endogenous control. Cell lysates were analyzed by Western blotting with the antibodies indicated. The results of a representative experiment are shown. EV denotes empty vector.
We monitored the capacity of the patient’s derived hTERT-immortalized fibroblasts to induce expression of interferon-stimulated genes. We found that the patient’s fibroblasts robustly expressed mRNA from interferon-stimulated genes and protein (Fig. S7A, S7B, and S7C), similar to fibroblasts from a patient with complete USP18 deficiency10 (Fig. S8). For further confirmation that c.1073+1G→A USP18 is the causal variant of type I interferon dysregulation, we transduced fibroblasts from the patient and from a control with wild-type USP18 or used luciferase as a control. Transduction with wild-type USP18 rescued the patient’s cellular phenotype, which resulted in normal levels of MX1 mRNA levels encoding ISG15 (Fig. S7D). Finally, we also transduced USP18-deficient fibroblasts with wild-type or ΔEx10 USP18 lentiviral constructs. Complementation with the ΔEx10 USP18 plasmid not only did not rescue complete USP18 deficiency but also phenocopied the type I interferon response, as reflected by MX1 mRNA levels of complete USP18 deficiency (Fig. S7E).
DISCUSSION
We report here a kindred with inherited complete USP18 deficiency that defines a second form of this disorder: the protein is produced and retains most of its catalytic activity while lacking the ability to be stabilized by ISG15 and to suppress type I interferon signaling. The clinical features of the patients from each of the three kindreds reported so far are similar. All previously reported patients died perinatally.10 To our knowledge, the child with USP18 deficiency in this study is the only reported patient who survived until at least 7 months of age on supportive therapy and then, after genetic diagnosis and ruxolitinib therapy, had resolution of the associated complications and survived the acute critical illness. At the time of this report, the child was 3 years of age and was continuing to receive oral ruxolitinib. He continues to grow normally but with slower progress in his developmental milestones.
The response to ruxolitinib was observed 2 weeks after an increase in the dose from 5 mg twice daily to 10 mg twice daily. The child remained in full remission of the clinical manifestations, with 2 years of follow-up in the outpatient clinic. JAK inhibitors have been reported to improve signs and symptoms and control disease activity in patients with other monogenic type I interferonopathies.16–21 Although long-term use of JAK inhibitors may predispose patients to mycobacterial and viral infections, along with limiting vaccine responses and otherwise influencing immune homeostasis, the benefit of ruxolitinib in this patient would seem to outweigh the potential for adverse effects. This study supports rapid genetic diagnosis of inherited disorders as a means of identifying existing drugs for experimental treatment.
Supplementary Material
Acknowledgments
Supported by a grant (RGP-190, to Dr. Alangari) from the Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia; by grants (R01AI127372, R21AI134366, and R21AI129827, to Dr. Bogunovic) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health; and by the Hirschl Scholar Award and the March of Dimes (to Dr. Bogunovic). The Laboratory of Human Genetics of Infectious Diseases is supported by grants from the St. Giles Foundation, the Jeffrey Modell Foundation, and the Rockefeller University Center for Clinical and Translational Science; by a grant (8UL1TR000043) from the National Center for Research Resources and the National Center for Advancing Sciences; by grants (5R01AI089970–02 and 5R37AI095983) from the National Institute of Allergy and Infectious Diseases; and by the French National Research Agency through a grant (ANR-10-LABX-62-IBEID) from the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence, a grant (ANR-10-IAHU-01) from the “Investments for the Future” program, and a grant (ANR-16-CE17–0005-01, to Dr. Bustamante) from GENMSMD (Human Genetic Dissection of Mendelian Susceptibility to Mycobacterial Disease).
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank the patient and his family for their participation in this study.
References
- 1.Rodero MP, Decalf J, Bondet V, et al. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J Exp Med 2017; 214: 1547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodero MP, Crow YJ. Type I interferon-mediated monogenic autoinflammation: the type I interferonopathies, a conceptual overview. J Exp Med 2016; 213: 2527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crow YJ. Type I interferonopathies: mendelian type I interferon up-regulation. Curr Opin Immunol 2015; 32: 7–12. [DOI] [PubMed] [Google Scholar]
- 4.Hermann M, Bogunovic D. ISG15: in sickness and in health. Trends Immunol 2017; 38: 79–93. [DOI] [PubMed] [Google Scholar]
- 5.Malakhova OA, Kim KI, Luo JK, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J 2006; 25: 2358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francois-Newton V, Livingstone M, Payelle-Brogard B, Uzé G, Pellegrini S. USP18 establishes the transcriptional and anti-proliferative interferon α/β differential. Biochem J 2012; 446: 509–16. [DOI] [PubMed] [Google Scholar]
- 7.François-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, et al. USP18based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS One 2011; 6(7): e22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem 2002; 277: 9976–81. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Bogunovic D, Payelle-Brogard B, et al. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 2015; 517: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meuwissen ME, Schot R, Buta S, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J Exp Med 2016; 213: 1163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boisson-Dupuis S, Kong XF, Okada S, et al. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr Opin Immunol 2012; 24: 364–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, et al. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet 2017; 25: 176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bousfiha A, Jeddane L, Picard C, et al. The 2017 IUIS phenotypic classification for primary immunodeficiencies. J Clin Immunol 2018; 38: 129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itan Y, Shang L, Boisson B, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods 2016; 13: 109–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankenburg WK, Dodds J, Archer P, Shapiro H, Bresnick B. The Denver II: a major revision and restandardization of the Denver Developmental Screening Test. Pediatrics 1992; 89: 91–7. [PubMed] [Google Scholar]
- 16.Frémond ML, Rodero MP, Jeremiah N, et al. Efficacy of the Janus kinase 1/2 inhibitor ruxolitinib in the treatment of vasculopathy associated with TMEM173-activating mutations in 3 children. J Allergy Clin Immunol 2016; 138: 1752–5. [DOI] [PubMed] [Google Scholar]
- 17.Higgins E, Al Shehri T, McAleer MA, et al. Use of ruxolitinib to successfully treat chronic mucocutaneous candidiasis caused by gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation. J Allergy Clin Immunol 2015; 135: 551–3. [DOI] [PubMed] [Google Scholar]
- 18.König N, Fiehn C, Wolf C, et al. Familial chilblain lupus due to a gain-offunction mutation in STING. Ann Rheum Dis 2017; 76: 468–72. [DOI] [PubMed] [Google Scholar]
- 19.Seo J, Kang JA, Suh DI, et al. Tofacitinib relieves symptoms of stimulator of interferon genes (STING)-associated vasculopathy with onset in infancy caused by 2 de novo variants in TMEM173. J Allergy Clin Immunol 2017; 139(4): 1396.e121399.e12. [DOI] [PubMed] [Google Scholar]
- 20.Vargas-Hernández A, Mace EM, Zimmerman O, et al. Ruxolitinib partially reverses functional natural killer cell deficiency in patients with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations. J Allergy Clin Immunol 2018; 141(6): 2142.e52155.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinacht KG, Charbonnier LM, Alroqi F, et al. Ruxolitinib reverses dysregulated T helper cell responses and controls autoimmunity caused by a novel signal transducer and activator of transcription 1 (STAT1) gain-of-function mutation. J Allergy Clin Immunol 2017; 139(5): 1629.e21640.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.