Abstract
Equid herpesviruses (EHVs) threaten equine health and can cause significant economic losses to the equine industry worldwide. Different equid herpesviruses, EHV‐1, EHV‐2, EHV‐4 and EHV5 are regularly detected among horse populations. In Egypt, monitoring is sporadic but EHV‐1 or EHV‐4 have been reported to circulate in the horse population. However, there is a lack of reports related to infection and health status of horses, likely due to the absence of regular diagnostic procedures. In the current study, the circulation of four infectious equid herpesviruses (EHV‐1, EHV‐2, EHV‐4 and EHV‐5) among different Arabian horse populations and donkeys residing the same farm was monitored. Different samples were collected and DNA was extracted and subjected to quantitative (q)‐PCR to detect the four equid herpesviruses using specific primers and probes. Antibody titres against EHV‐1 and EHV‐4 were tested using virus neutralization test and type‐specific ELISA. The results showed that EHV‐1, EHV‐2, EHV‐4 and EHV‐5 are endemic and can be a continuous threat for horses in the absence of vaccination programs and frequent virus reactivation. There is an urgent need for introduction of active regular surveillance measures to investigate the presence of different equid herpesviruses, and other equine viral pathogens, in various horse populations around Egypt and to establish a standardized cataloguing of equine health status.
Keywords: alphaherpesviruses, gammaherpesviruses, arabian horses, donkeys, co‐infection
Equid herpesviruses are endemic in horse population in Egypt and can be a continuous threat for horses in the absence of vaccination programs and frequent virus reactivation. There is an urgent need for introduction of active regular surveillance measures to investigate the presence of different equid herpesviruses, and other equine viral pathogens, in various horse populations around the country and to establish a standardized cataloguing of equine health status.

Introduction
Horses are constantly exposed to equid herpesviruses (EHVs), because the viruses circulate in horse populations in all countries and regions of the world. Among the Equidae, nine herpesviruses have been identified to date. The viruses belong to the Alphaherpesvirinae subfamily [six viruses: equid herpesviruses type 1 (EHV‐1), EHV‐3, EHV‐4, EHV‐6, EHV‐8 and EHV‐9] or the Gammaherpesvirinae subfamily [three viruses; EHV‐2, EHV‐5 and EHV‐7] (Davison et al. 2009). Horses are the natural host to EHV‐1, EHV‐2, EHV‐3, EHV‐4 and EHV‐5, while donkeys are the primary host of EHV‐6 (also referred to as asinine herpesvirus 1, AsHV‐1), EHV‐7 (AsHV‐2), and EHV‐8 (AsHV‐3). EHV‐9, the newest member of the equid herpesviruses, was first isolated from an infected Thomson gazelle; however, recent studies suggested that zebras or other perissodactyls such as the rhinoceros could be the definitive host for this virus (Fukushi et al. 1997; Abdelgawad et al. 2015, 2016).
EHV‐1 and its close relative, EHV‐4, cause significant economic losses to the equine industry due to clinical illnesses, which are associated with lost time for training and performance (Allen & Bryans 1986; Patel & Heldens 2005). Although both viruses cause respiratory disease, only infection with EHV‐1 may result in abortion, perinatal mortality and neurological disorders with clinical signs that vary in severity but can result in complete paralysis (Patel & Heldens 2005). Infection of horses with either of the two viruses is clinically and serologically difficult to distinguish because of their high genetic and antigenic similarity. Primary infection with either EHV‐1 or EHV‐4 occurs mainly via direct horse‐to‐horse contact. Direct contact with EHV‐1‐infected aborted fetuses, placental tissues or fomites contaminated by respiratory secretions may serve as a source of infection although the viruses are reported to be relatively short‐lived in the environment (Reed & Toribio 2004; Harless & Pusterla 2006). However, it was shown that EHV‐1 can remain stable and infectious for over a week and up to 3 weeks in water under different conditions of salinity, pH, temperature and turbidity in controlled in vitro experiments (Dayaram et al. 2017).
EHV‐2 and EHV‐5 are slow growing cell‐associated gammaherpesviruses. Both viruses are widespread in the equine population worldwide (Borchers et al. 2006; Hue et al. 2014). EHV‐2 has been reported to be associated with upper respiratory tract disease, lymphadenopathy, immunosuppression, keratoconjunctivitis, general malaise and poor performance (Borchers et al. 2006). EHV‐5 in contrast has been reported to be associated with pulmonary fibrosis in horses (Williams et al. 2007). Equine herpesviruses, like all herpesviruses, enter a latent state but reactivation from latency may result in recurring disease, which is accompanied by virus shedding and transmission to other horses (Allen & Bryans 1986; Edington et al. 1994; Crabb & Studdert 1995).
Standard diagnostic methods for EHV‐1 and related viruses are well‐established, including virus isolation and serological assays, particularly virus neutralization tests (VNT) and type‐specific ELISA assays (Crabb & Studdert 1993; Crabb et al. 1995; Lang et al. 2013; OIE, 2018). Quantitative PCR (qPCR) methods offer alternatives to virus isolation and have been proven sensitive and time‐effective (Pusterla et al. 2005).
Currently there are different inactivated or modified live EHV‐1 and EHV‐4 vaccines commercially available. However, they cannot completely block virus infection and most vaccines only induce reliable protection against respiratory disease (Heldens et al. 2001; Patel & Heldens 2005; Ma et al. 2013). Possible protection against neurological disease under experimental conditions (Goodman et al. 2006) and abortion under field conditions using EHV‐1‐based vaccines (Bresgen et al. 2012) was reported.
In Egypt, there are more than 1.5 million equids, with horses representing around 40% of the total equine population (Animal Wealth development sector, Egyptian Ministry of Agriculture, 2010 http://www.agr-egypt.gov.e.g/Uploads/Studies/d14ee0cb-c15d-4fb9-9f2d-008f5b7ef873.pdf). These horses include pure‐bred registered Egyptian Arabian horses and are mainly used for showcasing, semen collection, and export. Although there are no definitive statistics, horses play an important role in Egyptian economics. This includes the direct economic impact of the horse industry (breeding, employment and education) and the indirect impact (related to horse activities like organization of social events). Currently, there is no accurate or official documentation on the health status of horses or vaccination programs. Recently, one study in Egypt detected EHV‐1, EHV‐2 and EHV‐4 DNA in clinical samples collected from horses from 2005 to 2006 (Amer et al. 2011). Furthermore, isolation of EHV‐1 from aborted fetuses in Egypt was documented (Soliman et al. 2008; Abd El‐Hafeiz et al. 2010). In general, lack of proper monitoring of equine infectious diseases in Egypt could threaten the stability of the horse industry in Egypt and beyond due to international trade in horses. The aim of the current study was to investigate the circulation of different equid herpesviruses, particularly EHV‐1, EHV‐4, EHV‐2 and EHV‐5, in selected Arabian horse and donkey populations in Egypt.
Materials and methods
Sample collection
Samples were collected from Arabian horses (n = 176) and donkeys (n = 16) from different parts of Egypt, including the main provinces of the North Egypt (Cairo, Alexandria, Giza, Sharkia, Gharbia and Monufia; Table 1 and Data S1). The Arabian horses were used mainly for show purposes and donkeys were used as work and draft animals. The samples were collected either at horse farms or from horses admitted to horse clinics. Tissue samples and placentae (placenta: n = 23 and tissues from aborted fetuses: n = 17; total cases of abortion = 40) were collected from aborted fetuses and mares immediately after abortion. Thirteen nasal swabs (Egyptian Company for Medical Equipment) were collected from horses suffering from fever and respiratory disorders using sterile swabs. The swabs were placed directly in virus transport medium (serum‐free MEM with 1% penicillin streptomycin, 1% gentamicin, and 0.1% fungizone). Cerebrospinal fluids (CSF; n = 4) were collected from stallions suffering from neurological disorders (Table 1 and Data S1). Whole blood collected into tubes with EDTA anticoagulant (n = 135) and without anticoagulant, to obtain serum, (n = 110) was obtained from apparently healthy horses and with no history of previous vaccination. After collection, all samples were packed in coolers with ice packs and transported immediately to the laboratory for analysis.
Table 1.
List of samples used in this study
| Samples* | Number of samples | History | Sex | Age range/median (year) | Location |
|---|---|---|---|---|---|
| Animals with clinical problems | |||||
| Aborted fetus tissues and placenta | 40† | Abortion and still birth | F: 40‡ | 5–12/7 | Cairo, Alexandria, Giza, Sharkia, Monufia |
| Nasal swabs | 13 | Fever, nasal discharge, and respiratory disease |
M: 1 F: 12 |
2–12/5 | Sharkia, Giza, Gharbia |
| Cerebrospinal fluid | 4 | Ataxia and hind limb paralysis | M: 4 | 3–8/4.5 | Giza, Alexandria |
| Total | 57 | ||||
| Apparently healthy animals | |||||
| Whole blood | 135 | No obvious clinical signs |
M: 67 F: 68 |
2–12/7 | Gharbia, Giza, Alexandria, Sharkia |
Total
|
192 | ||||
| Serology | |||||
| Serum | 110 | No obvious clinical signs |
M: 53 F: 57 |
2–12/7 | Gharbia, Giza, Alexandria, Sharkia |
Samples were collected in 2015 and 2016.
Placenta: n = 23; tissues from aborted fetuses: n = 17.
Numbers indicate the number of tested animals of each sex.
DNA extraction and PCR
Viral DNA was extracted from collected samples and infected cell cultures using DNA/RNA Virus, Tissue or Blood Mini Kits (Stratec Biomedical, Birkenfeld, Germany) according to the manufacturer's instructions.
PCR reactions were performed using the extracted DNA as a template. Nested PCR amplifications using degenerate PCR primers (Table 2) that specifically amplify a 250‐bp fragment of herpesvirus DNA polymerase gene were performed on all samples as described previously (VanDevanter et al. 1996). Fourteen randomly selected amplicons were purified and directly sequenced by Sanger sequencing (LGC Genomics) to confirm that the amplified fragments contain herpesvirus DNA. All DNA samples were re‐analysed by qPCR with the Applied Biosystems 7500 FAST (ABI, Foster City, CA) using specific primers and probes targeting the highly conserved gB gene (Table 2) (Dynon et al. 2001; Pusterla et al. 2005; Hussey et al. 2006; Dunowska et al. 2011). Positive (virus‐infected cell culture) and negative (water) controls were included from the beginning of the extraction procedure until the reading of the results.
Table 2.
Primers and probes used in the study
| Product | Primer | Sequence | Fragment | References |
|---|---|---|---|---|
| All herpesviruses | ||||
| Pan herpesvirus | DFA (For) | GAYTTYGCNAGYYTNTAYCC* | 700 bp | VanDevanter et al. (1996) |
| ILK (For) | TCCTGGACAAGCAGCARNYSGCNMTNAA* | |||
| KG1 (Rev) | GTCTTGCTCACCAGNTCNACNCCYTT* | |||
| Pan herpesvirus (nested PCR) | TGV (For) | TGTAACTCGGTGTAYGGNTTYACNGGNGT* | 250 bp | |
| IYG (Rev) | CACAGAGTCCGTRTCNCCRTADAT* | |||
| Equid herpesvirus 1 (EHV‐1)† | ||||
| glycoprotein B (gB) | For | 61722‐CACTTCCATGTCAACGCACT‐61741 | 869 bp | Designed for this study |
| Rev | 62591‐TCGACTTTCTTCTCGGTCCA‐62572 | |||
| DNA polymerase (POL) | For | 54885‐ACCTCCGGAGGCAAAGTTCA‐54904 | 709 bp | |
| Rev | 54195‐TTCGCCCCGTTGAGCGACAC‐54214 | |||
| EHV‐4† | ||||
| gB | For | 61314‐CATGTCTAAAGACTCGACAT‐61333 | 1369 bp | Designed for this study |
| Rev | 62664‐GATTGGTATTATGGTTTGCG‐62683 | |||
| POL | For | 54660‐CATCACAGTACACTTTTGGG‐54679 | 843 bp | |
| Rev | 53836‐ACTATAAGCTACTGTGTTTT‐53855 | |||
| EHV‐2† | ||||
| POL | For | 36400‐GCGCGTGTTGCGCGAGTACT‐36419 | 1270 bp | Designed for this study |
| Rev | 37651‐GGTGCAGGCACAGCCTGTCT‐37670 | |||
| EHV‐5† | ||||
| POL | For | 32972‐AAGGGTTTTGAAACAATACA‐32991 | 1115 bp | Designed for this study |
| Rev | 34068‐GAACTTTCCTTGTTGCCCGA‐34087 | |||
| qPCR | ||||
| EHV‐1 gB | For | CATACGTCCCTGTCCGACAGAT | Hussey et al. (2006) | |
| Rev | GGTACTCGGCCTTTGACGAA | |||
| Probe | 6FAM‐TGAGACCGAAGATCTCCTCCACCGA‐BHQ1 | |||
| EHV‐4 gB | For | CGCAGAGGATGGAGACTTTTACA | Pusterla et al. (2005) | |
| Rev | CATGACCGTGGGGGTTCAA | |||
| Probe | 6FAM‐CTGCCCGCCGCCTACTGGATC‐TAMRA | |||
| EHV‐2 gB | For | AGGACTACTACTATGTCAG | Dunowska et al. (2011) | |
| Rev | ATGGTCTCGATGTCAAACAC | |||
| Probe | 6FAM‐TGACATACCCACCCTACACACCATGA‐BHQ1 | |||
| EHV‐5 gB | For | ATGAACCTGACAGATGTGCC | Dynon et al. (2001) | |
| Rev | CACGTTCACTATCACGTCGC | |||
| Probe | 6FAM‐TCCATCCACGATGGCAGGGA‐BHQ1 | |||
Bold letters indicate amino acid residues included in the primers.
Numbers before and after primer sequences represent their positions in the genome.
EHV‐specific primers (Table 2) that target various regions of two genes, gB (glycoprotein B; ORF33), and/or POL (DNA polymerase; ORF30), were designed and employed to amplify the corresponding fragments. The amplified products were purified, sequenced by Sanger sequencing (LGC Genomics), and submitted to GenBank (accession numbers: MG732975‐ MG732978).
Cells
Equine dermal (ED) cells were grown in DMEM medium (PAN Biotech, Aidenbach, Germany) supplemented with 10% fetal bovine serum (FBS; PAN Biotech) and 100 U ml−1 penicillin (Roth, Karlsruhe, Germany) and 100 μg ml−1 streptomycin (Alfa Aesar, Kandel, Germany) in a 37°C incubator with 5% CO2 atmosphere. Cells were grown to confluency in a 100 mm tissue culture dish, washed with phosphate buffer saline (PBS), trypsinized with 0.25% trypsin supplemented with 2.5 μmol L−1 EDTA and counted in a Neubauer counting chamber.
Virus neutralization test
Virus neutralization test (VNT) was performed according to the OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (OIE, 2018). Briefly, serum samples were inactivated at 56°C for 30 min. In 96‐well plates, serial twofold dilutions of inactivated serum were incubated with 100 plaque forming units (PFU)/100 μl of either EHV‐1 or EHV‐4 at 37°C. After 1 h, a total of 5 × 105 equine dermal cells (ED) were added and plates were incubated for 2 h at 37°C. The monolayers were overlaid with 1.6% methylcellulose medium and plates were incubated at 37°C for 3 days. The reaction was stopped with 3% formalin and the plaques were stained with Giemsa. EHV‐1‐ and EHV‐4‐positive horse serum and fetal calf serum were included as positive and negative controls, respectively. Antibody titres were expressed as the reciprocal of the highest serum dilution that protected the cells from infection in each well. A reciprocal titre of greater than or equal to 4 was considered positive (Lang et al. 2013). Each test was validated with the positive and negative sera controls.
Peptide‐based ELISA
The ELISA test was carried out as described before (Lang et al. 2013) with few modifications. Briefly, 96‐well plates were coated with 1 μg ml−1 streptavidin (100 μl well−1 dissolved in 50 nmol L−1 carbonate‐biocarbonate buffer; pH 9.6) overnight at 4°C. The wells were washed three times with PBS containing 0.1% Tween 20 (PBST). After coating with 100 μl well−1 of the respective biotinylated peptide [EHV‐1 E (E1) peptide: KQPQPRLRVKTPPPVTVP and EHV‐4G (G4) peptide: TEGMKNNPVYSESLMLNV; 2 μg ml−1 in 50 nmol L−1 carbonate‐biocarbonate buffer], the plates were incubated at 37°C for 2 h. The plates were then blocked with 1% goat serum diluted in PBST and incubated for 1 h at 37°C. After washing, serum samples (100 μl well−1 in 1:400 dilution) were added and plates were incubated for 1 h at 37°C. Purified goat anti‐horse IgG conjugated with horseradish peroxidase (1:20 000; Dianova, Hamburg, Germany) was added and plates were incubated for 1 h. After washing, binding was detected by addition of 100 μl well−1 TMB [3,3,5,5‐tetramethylbenzidine, dissolved in 42 μg ml−1 citric acid, 0.01% H2O2 (pH 3.95)]. The reaction was stopped after 10 min with 100 μl ml−1 of 1 mol L−1 sulphuric acid and the plates were read at a wavelength of 450 nm on a spectrophotometer (TriStar LB 941, Berthold Technologies, Bad Wildbad, Germany). Based on our previous publication (Lang et al. 2013), we have set a cut‐off value (OD value = 0.118) above which a sample was considered positive. Samples that produced OD values between 0.118 and 0.100 were considered questionable, and samples with OD values of <0.100 were considered negative. The same cut‐off values were used for both EHV‐1 and EHV‐4 ELISA. Each serum sample was tested for EHV‐1 and EHV‐4 antibody two independent times. Negative and positive serum controls were included in each plate.
Statistical analysis
Statistical analyses and graphs of the serology data were performed using GraphPad Prism version 5.0 software (GraphPad Software, San Diego, CA). Fisher's exact test was used to compare the frequencies or proportions of EHV‐1 and EHV‐4 positive samples. Differences were considered statistically significant when the P value was less than 0.05.
Results
Detection of different equid herpesviruses
Using consensus herpesvirus PCR, a 250‐bp fragment was amplified in 110/192 (57%) samples. Using virus‐specific qPCR, 133/192 (69%) samples were positive for at least one of the four EHVs. The number of samples detected by both assays was n = 64 for EHV‐1, n = 5 for EHV‐4, n = 63 for EHV‐2 and n = 37 for EHV‐5. The number of samples detected only by virus‐specific qPCR was EHV‐1 (n = 21), EHV‐4 (n = 4), EHV‐2 (n = 10) and EHV‐5 (n = 8). Four samples were detected only by consensus herpesvirus PCR (Data S1). The 250‐bp fragment of 14 amplicons were sequenced and the data revealed 95‐100% identity at the nucleotide level to previously reported EHV‐1 (n = 5), EHV‐4 (n = 1), EHV‐2 (n = 7) and EHV‐5 (n = 1) DNA polymerase sequences.
Using specific qPCR protocols, we found that the most prevalent virus was EHV‐1 (85/192; 44%), followed by EHV‐2 (73/192; 38%), EHV‐5 (45/192; 23%) and EHV‐4 (9/192; 4%). Among the 40 tested samples collected from aborted cases, EHV‐1 and EHV‐2 were detected in 29/40 (72%) and 16/40 (40%) samples, respectively (Table 3). EHV‐4 (4/40; 10%) and EHV‐5 (1/40; 2.5%) were also detected. All four herpesviruses (mainly EHV‐1 and EHV‐5) were detected in donkeys that were present at the same farm as positive horses (Table 3). No herpesviruses were detected in the CSF samples. Co‐infections with different herpesviruses were common, especially for EHV‐1 and EHV‐2 or EHV‐2 and EHV‐5 (Table 4). Triple co‐infections were also detected for all viruses; however, we did not identify coinfection with all four viruses in a single animal (Table 4).
Table 3.
qPCR results from different samples
Table 4.
Incidence of co‐infection as detected by qPCR
| Classification | Virus* | No. detected in horses | No. detected in donkeys |
|---|---|---|---|
| Unique detection | EHV‐1 only | 32 | 3 |
| EHV‐4 only | 0 | 0 | |
| EHV‐2 only | 21 | 0 | |
| EHV‐5 only | 9 | 3 | |
| Double detection | EHV‐1 and EHV‐4 | 4 | 0 |
| EHV‐1 and EHV‐2 | 24 | 2 | |
| EHV‐1 and EHV‐5 | 4 | 2 | |
| EHV‐2 and EHV‐5 | 15 | 0 | |
| Triple detection | EHV‐1, EHV‐4, EHV‐2 | 2 | 0 |
| EHV‐1, EHV‐4, EHV‐5 | 2 | 1 | |
| EHV‐1, EHV‐2, EHV‐5 | 9 | 0 | |
| Quadrupel detection | EHV‐1, EHV‐4, EHV‐2, EHV‐5 | 0 | 0 |
This includes virus detection in samples collected from clinically affected and apparently healthy animals.
Four sequences, Egy‐01 (EHV‐1 gB sequence; GenBank: MG732975), Egy‐02 (EHV‐1 POL sequence; GenBank: MG732976), Egy‐03 (EHV‐2 gB; GenBank: MG732977), and Egy‐04 (EHV‐5 gB; GenBank: MG732978), were obtained from the isolated DNA and blasted against available EHV‐sequences from GenBank.
Detection of EHV‐1 and EHV‐4 neutralizing antibodies
Of 110 tested serum samples, 59 (54%) and 78 (71%) were seropositive by VNT for EHV‐1 and EHV‐4 antibodies, respectively (Fig. 1a and Data S1). In horse serum samples (n = 103), 54 (52%) and 75 (73%) tested positive for EHV‐1 and EHV‐4 antibodies, respectively. In samples collected from donkeys living in the same farm (n = 7), 5 (71%) and 3 (43%) tested positive for EHV‐1 and EHV‐4 antibodies, respectively (Data S1). The highest neutralizing titre was 128 (Data S1). It is worth mentioning that the horse with this high titre was apparently healthy and positive only for EHV‐5 DNA by qPCR. The prevalence of EHV‐4 antibodies in the tested serum samples was significantly higher than that of EHV‐1 (P = 0.01; Fisher's exact test).
Figure 1.
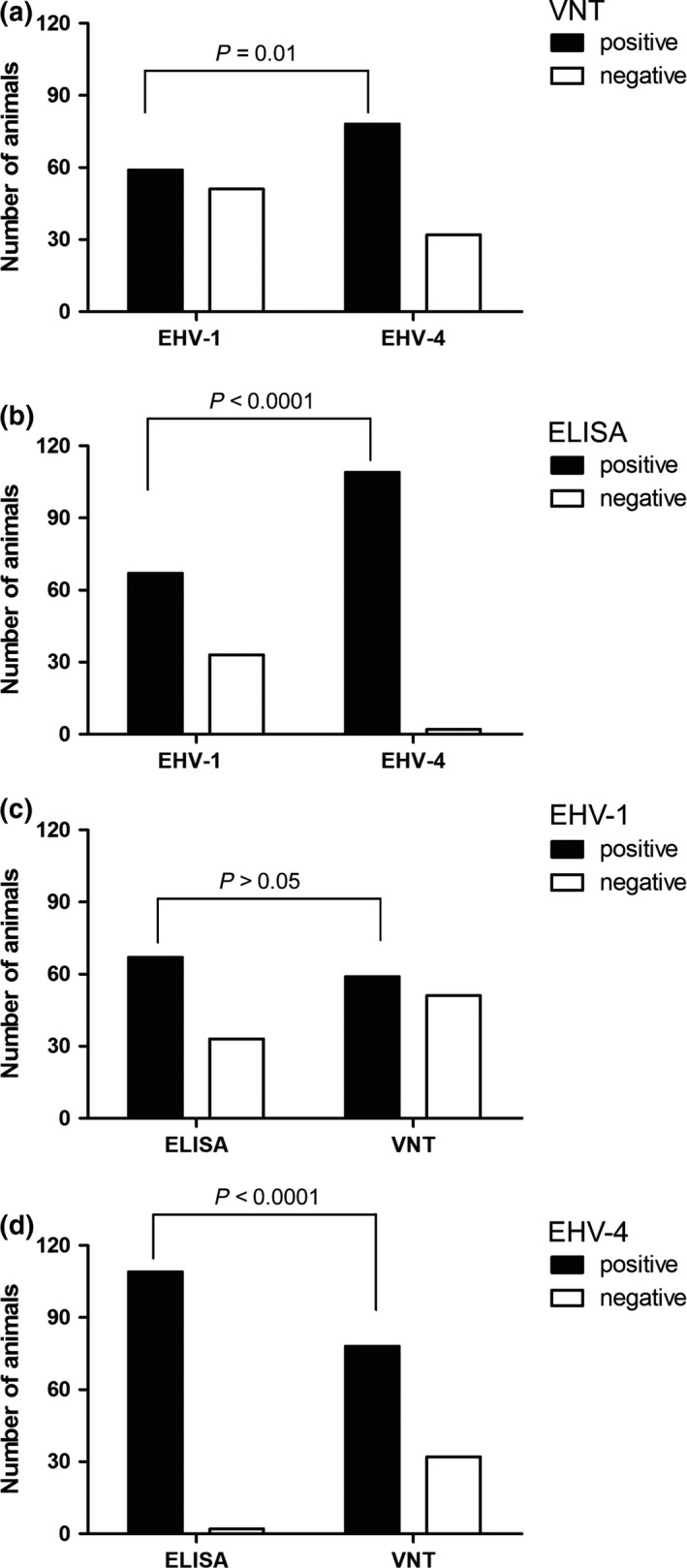
Detection of EHV‐1 and EHV‐4 antibodies. Number of animals (n = 110) positive or negative for the presence of specific antibodies either by virus neutralization test (a) or peptide‐specific ELISA (b). The prevalence of EHV‐4 antibodies was significantly higher in the serum samples tested by virus neutralization test (VNT) or ELISA (Fisher's exact test). Number of animals positive or negative for the presence of EHV‐1‐specific (c) or EHV‐4‐specific (d) antibodies using ELISA and VNT. The number of animals tested positive for EHV‐4 antibodies by ELISA was significantly (Fisher's exact test) higher than this tested by VNT.
Discrimination between EHV‐1‐ and EHV‐4‐specific antibodies by peptide‐based ELISA
EHV‐1 gE (EHV‐1_E)‐ and EHV‐4 gG (EHV‐4_G)‐specific peptides were used to differentiate between EHV‐1‐ and EHV‐4‐specific antibodies as described before (Lang et al. 2013). In total, 67/110 (61%) samples tested positive for EHV‐1 antibodies and 109/110 (99%) samples for EHV‐4 antibodies, including seven donkeys that were all positive for both EHV‐1 and EHV‐4 antibodies (Fig. 1b and Data S1). Ten additional samples were classified as questionable for EHV‐1 antibodies (Data S1). The data showed that the prevalence of EHV‐4 antibodies was significantly higher than that of EHV‐1 (P < 0.0001; Fisher's exact test).
There were 14/59 (24%) horse serum samples that tested positive for EHV‐1 antibodies by VNT, were negative by EHV‐1 ELISA (Data S1). In addition, there were 8 and 31 samples that were negative by VNT and tested positive with EHV‐1 and EHV‐4 ELISA, respectively (Data S1). The data showed that ELISA was significantly more sensitive for EHV‐4, but not EHV‐1, antibodies (Fig. 1c,d; P < 0.0001; Fisher's exact test).
Discussion
The loss of valuable horses and revenue from major horse events has negative consequences for the horse breeding and sports industry. In Egypt, few monitoring studies have been performed but the existence of equine herpesviruses, particularly EHV‐1 and EHV‐4, in the horse population has been documented (Abd El‐Hafeiz et al. 2010; Amer et al. 2011; Al‐Shammari et al. 2016; Fararh et al. 2016). Egyptian Arabian horses are an important part of the country's economy due to global trade linked to the international reputation of the Egyptian breeding lines. In the current study, we investigated the circulation of equine alpha‐ and gammaherpesviruses among Arabian horses and donkeys in Egypt.
A total of 192 samples were collected from apparently healthy animals or horses with clinical signs of abortion, respiratory disease or neurological disorders. EHV‐1 and EHV‐2, respectively, were the main viruses detected in clinically affected horses; however, EHV‐4 and EHV‐5 were also detected to a lesser extent. It is difficult to state whether any of these viruses were responsible for the clinical diseases among horses sampled due to the absence of clear case history and proper diagnosis at the time of disease onset. On the other hand, EHV‐2, EHV‐1 and EHV‐5, respectively, were mostly detected in the apparently healthy horses. Interestingly, all viruses, but predominantly EHV‐1 and EHV‐5, were detected in the tested donkey samples. Our data as well as previous studies clearly indicate that these equine herpesviruses are circulating in equid populations in Egypt (Abd El‐Hafeiz et al. 2010; Amer et al. 2011; Al‐Shammari et al. 2016; Mohamed et al. 2017). The frequency of detection of each virus among equids varied between studies. This can be explained by the age, health status and breed of horses as well as geographical variability and environmental factors. Detection of EHV in donkeys (in this study) and mules (Mohamed et al. 2017) is interesting but it is not clear if the donkeys contracted the infection from horses or whether they can play a role in further spreading of the infection.
Co‐infections with up to three viruses were detected in both clinically infected and apparently healthy horses and donkeys. This dual infection was reported before in horses but also in wild equids (Amer et al. 2011; Back et al. 2015; Abdelgawad et al. 2016; Laabassi et al. 2017; Negussie et al. 2017). However, their potential synergistic effect on the disease outcome remains to be elucidated. The detection of EHV‐1, associated with other EHVs in donkeys, provides hallmarks on the potential source of infection. Furthermore, donkeys in Egypt are always stressed due to the assigned hard work, which might increase the probability of EHV shedding after reactivation.
It was clear that both EHV‐2‐ and EHV‐5 are circulating among apparently healthy horses (Table 3). This high prevalence may increase the risk of other possible infections due to compromising horse immunity (Nordengrahn et al. 1996; Negussie et al. 2017). Further studies are needed to better understand the clinical outcomes of EHV‐2 and EHV‐5. The role of EHV‐4 in disease outcome cannot be ideally assessed due to the low number of detected positive samples.
Serological analysis was conducted for all collected serum samples. Although VNT is known to be an efficient and robust test, it cannot accurately discriminate between antigenically similar viruses, like EHV‐1 and EHV‐4 (Crabb et al. 1995; Lang et al. 2013); this was further confirmed in our current data. We found that 14 of 59 horse serum samples, which tested positive for EHV‐1 antibodies by VNT, were negative by EHV‐1 ELISA but positive by EHV‐4 ELISA. This result indicates that the detected EHV‐1 neutralizing antibodies were a result of cross‐reactivity with EHV‐4. In addition, there was a clear difference in sensitivity when comparing VNT with ELISA (Fig. 1c,d). In general, our data clearly support the previous data indicating that peptide‐ELISA is considered a specific and sensitive serological test for detection of EHV‐1 and EHV‐4 seroprevalence (Lang et al. 2013; Damiani et al. 2014). VNT and specific peptide‐based ELISA demonstrated that the prevalence of EHV‐4 antibodies among sampled horses was significantly higher than that of EHV‐1. These results strongly suggest that EHV‐4 is omnipresent in horse population in Egypt and that most of the horses probably contracted the infection during their early life.
Equid herpesviruses EHV‐1, EHV‐4, EHV‐2 and EHV‐5 circulate among various Arabian horse populations in Egypt. In the present study, all viruses were detected in apparently healthy and clinically ill equids. Although Arabian horses are considered one of the most proficient and valuable breeds used in breeding, show competitions and exportation, no regular monitoring programs are performed to track the health status of these animals, including EHV infections. In addition, vaccination programs against equid herpesviruses, that might reduce the risk of infection, are not routinely performed by veterinary practitioners in Egypt. In the current study, we shed light on the frequency of detection of various equine herpesviruses among sampled population(s) of Arabian horses in Egypt. These data will help to guide efficient diagnosis and preventive measures against these viruses through establishing regular testing (molecular and serological) and vaccination programs.
Source of funding
This study was supported by grant from the Alexander von Humboldt‐Research Group Linkage Program to Walid Azab.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributions
Walid Azab and Sameh Bedair have contributed equally to this work by designing and carrying out the experiments, analyssing and interpreting the data. Azza Abdelgawad did peptide‐specific ELISA, data analysis and helped with manuscript writing. Kathrin Eschke helped with PCR, sequencing and data analysis. Gemelat K. Farag helped with sample preparation and data analysis. Ali Abdel‐Rahiem, Alex D. Greenwood, Nikolaus Osterrieder and Ahmed A. H. Ali contributed to the drafting of the manuscript, discussion and writing the manuscript.
Ethical approval
Sample collection was done according to the Ministry of Health and Population (General Administration of Medical Licenses; approval number 36490).
Supporting information
Data S1. Detailed descriptions of collected samples and presentation of molecular and serological results of each tested sample.
Acknowledgements
We thank Sebastian Bischofberger, for technical assistance.
References
- Abd El‐Hafeiz Y.G.M., Abu Maaty A. & Darwish S. (2010) Isolation of equine herpesvirus‐1 (EHV‐1) as a cause of reproductive disorders with emphasis on antigenic and genetic identifications. International Journal of Microbiological Research 1, 26–32. [Google Scholar]
- Abdelgawad A., Hermes R., Damiani A., Lamglait B., Czirják G.Á., East M. et al (2015) Comprehensive serology based on a peptide ELISA to assess the prevalence of closely related equine herpesviruses in zoo and wild animals. PLoS ONE 10, e0138370 10.1371/journal.pone.0138370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelgawad A., Damiani A., Ho S., Strauss G., Szentiks C., East M. et al (2016) Zebra alphaherpesviruses (EHV‐1 and EHV‐9): genetic diversity, latency and co‐infections. Viruses 8, 262 10.3390/v8090262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.P. & Bryans J.T. (1986) Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus‐1 infections. Progress in Veterinary Microbiology and Immunology 2, 78–144. [PubMed] [Google Scholar]
- Al‐Shammari Z., Ahmed B., Haroun M., Afify A., Elsanousi A.A. & Shalaby M. (2016) a first molecular phylogeny of an egyptian equine herpesvirus‐4 strain derived from a fetal Arabian horse. Journal of Veterinary Science & Medical Diagnosis 5 10.4172/2325-9590.1000186. [DOI] [Google Scholar]
- Amer H., Shaltout A., El‐Sabagh I., El‐Sanousi A. & Shalaby M. (2011) Prevalence of equine herpes viruses 1, 2 and 4 in Arabian horse population in Egypt. African Journal of Microbiology Research 5, 4805–4811. [Google Scholar]
- Back H., Ullman K., Treiberg Berndtsson L., Riihimaki M., Penell J., Stahl K. & Pringle J. (2015) Viral load of equine herpesviruses 2 and 5 in nasal swabs of actively racing Standardbred trotters: temporal relationship of shedding to clinical findings and poor performance. Veterinary Microbiology 179, 142–148. 10.1016/j.vetmic.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Borchers K., Ebert M., Fetsch A., Hammond T. & Sterner‐Kock A. (2006) Prevalence of equine herpesvirus type 2 (EHV‐2) DNA in ocular swabs and its cell tropism in equine conjunctiva. Veterinary Microbiology 118, 260–266. 10.1016/j.vetmic.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Bresgen C., Lammer M., Wagner B., Osterrieder N. & Damiani A.M. (2012) Serological responses and clinical outcome after vaccination of mares and foals with equine herpesvirus type 1 and 4 (EHV‐1 and EHV‐4) vaccines. Veterinary Microbiology 160, 9–16. 10.1016/j.vetmic.2012.04.042. [DOI] [PubMed] [Google Scholar]
- Crabb B.S. & Studdert M.J. (1993) Epitopes of glycoprotein G of equine herpesviruses 4 and 1 located near the C termini elicit type‐specific antibody responses in the natural host. Journal of Virology 67, 6332–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb B.S. & Studdert M.J. (1995) Equine herpesviruses 4 (equine rhinopneumonitis virus) and 1 (equine abortion virus). Advances in Virus Research 45, 153–190. [DOI] [PubMed] [Google Scholar]
- Crabb B.S., MacPherson C.M., Reubel G.H., Browning G.F., Studdert M.J. & Drummer H.E. (1995) A type‐specific serological test to distinguish antibodies to equine herpesviruses 4 and 1. Archives of Virology 140, 245–258. [DOI] [PubMed] [Google Scholar]
- Damiani A.M., de Vries M., Reimers G., Winkler S. & Osterrieder N. (2014) A severe equine herpesvirus type 1 (EHV‐1) abortion outbreak caused by a neuropathogenic strain at a breeding farm in northern Germany. Veterinary Microbiology 172, 555–562. 10.1016/j.vetmic.2014.06.023. [DOI] [PubMed] [Google Scholar]
- Davison A.J., Eberle R., Ehlers B., Hayward G.S., McGeoch D.J., Minson A.C. & Thiry E. (2009) The order Herpesvirales. Archives of Virology 154, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaram A., Franz M., Schattschneider A., Damiani A.M., Bischofberger S., Osterrieder N. & Greenwood A.D. (2017) Long term stability and infectivity of herpesviruses in water. Scientific Reports 7, 46559 10.1038/srep46559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunowska M., Howe L., Hanlon D. & Stevenson M. (2011) Kinetics of Equid herpesvirus type 2 infections in a group of Thoroughbred foals. Veterinary Microbiology 152, 176–180. 10.1016/j.vetmic.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Dynon K., Varrasso A., Ficorilli N., Holloway S., Reubel G., Li F. & Drummer H. (2001) Identification of equine herpesvirus 3 (equine coital exanthema virus), equine gammaherpesviruses 2 and 5, equine adenoviruses 1 and 2, equine arteritis virus and equine rhinitis A virus by polymerase chain reaction. Australian Veterinary Journal 79, 695–702. [DOI] [PubMed] [Google Scholar]
- Edington N., Welch H.M. & Griffiths L. (1994) The prevalence of latent Equid herpesviruses in the tissues of 40 abattoir horses. Equine Veterinary Journal 26, 140–142. [DOI] [PubMed] [Google Scholar]
- Fararh K.M., Kandil O.M., Abd‐Allah O.A. & Thabet N.F. (2016) Clinicopathological changes in equine herpes virus type 1 (EHV‐1) infection in Arabian foals. International Journal of PharmTech Research 9, 138–149. [Google Scholar]
- Fukushi H., Tomita T., Taniguchi A., Ochiai Y., Kirisawa R., Matsumura T. & Hirai K. (1997) Gazelle herpesvirus 1: a new neurotropic herpesvirus immunologically related to equine herpesvirus 1. Virology 227, 34–44. [DOI] [PubMed] [Google Scholar]
- Goodman L.B., Wagner B., Flaminio M.J., Sussman K.H., Metzger S.M., Holland R. & Osterrieder N. (2006) Comparison of the efficacy of inactivated combination and modified‐live virus vaccines against challenge infection with neuropathogenic equine herpesvirus type 1 (EHV‐1). Vaccine 24, 3636–3645. 10.1016/j.vaccine.2006.01.062. [DOI] [PubMed] [Google Scholar]
- Harless W. & Pusterla N. (2006) Equine herpesvirus 1 and 4 respiratory disease in the horse. Clinical Techniques in Equine Practice 5, 197–202. [Google Scholar]
- Heldens J.G., Hannant D., Cullinane A.A., Prendergast M.J., Mumford J.A., Nelly M. et al (2001) Clinical and virological evaluation of the efficacy of an inactivated EHV1 and EHV4 whole virus vaccine (Duvaxyn EHV1, 4). Vaccination/challenge experiments in foals and pregnant mares. Vaccine 19, 4307–4317. [DOI] [PubMed] [Google Scholar]
- Hue E.S., Fortier G.D., Fortier C.I., Leon A.M., Richard E.A., Legrand L.J. & Pronost S.L. (2014) Detection and quantitation of equid gammaherpesviruses (EHV‐2, EHV‐5) in nasal swabs using an accredited standardised quantitative PCR method. Journal of Virological Methods 198, 18–25. 10.1016/j.jviromet.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Hussey S.B., Clark R., Lunn K.F., Breathnach C., Soboll G., Whalley J.M. & Lunn D.P. (2006) Detection and quantification of equine herpesvirus‐1 viremia and nasal shedding by real‐time polymerase chain reaction. Journal of Veterinary Diagnostic Investigation 18, 335–342. 10.1177/104063870601800403. [DOI] [PubMed] [Google Scholar]
- Laabassi F., Hue E., Fortier C., Morilland E., Legrand L., Hans A. & Pronost S. (2017) Epidemiology and molecular detection of equine herpesviruses in western Algeria in 2011. Veterinary Microbiology 207, 205–209. 10.1016/j.vetmic.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Lang A., de Vries M., Feineis S., Muller E., Osterrieder N. & Damiani A.M. (2013) Development of a peptide ELISA for discrimination between serological responses to equine herpesvirus type 1 and 4. Journal of Virological Methods 193, 667–673. 10.1016/j.jviromet.2013.07.044. [DOI] [PubMed] [Google Scholar]
- Ma G., Azab W. & Osterrieder N. (2013) Equine herpesviruses type 1 (EHV‐1) and 4 (EHV‐4)–masters of co‐evolution and a constant threat to equids and beyond. Veterinary Microbiology 167, 123–134. 10.1016/j.vetmic.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Mohamed S.I., Shalaby M.A., El Deeb A.H. & Salem S.A. (2017) Prevalence of equine herpes viruses 1 and 4 in Arabian horses population in Egypt. Journal of Virological Sciences 2, 1–9. [Google Scholar]
- Negussie H., Gizaw D., Tesfaw L., Li Y., Oguma K., Sentsui H. et al (2017) Detection of equine herpesvirus (EHV)‐1,‐2,‐4 and‐5 in ethiopian equids with and without respiratory problems and genetic characterization of EHV‐2 and EHV‐5 strains. Transboundary and Emerging Diseases 64, 1970–1978. 10.1111/tbed.12601. [DOI] [PubMed] [Google Scholar]
- Nordengrahn A., Rusvai M., Merza M., Ekstrom J., Morein B. & Belak S. (1996) Equine herpesvirus type 2 (EHV‐2) as a predisposing factor for Rhodococcus equi pneumonia in foals: prevention of the bifactorial disease with EHV‐2 immunostimulating complexes. Veterinary Microbiology 51, 55–68. 10.1016/0378-1135(96)00032-6. [DOI] [PubMed] [Google Scholar]
- OIE . (2018) OIE manual for terrestial animals, 2018. Equine rhinopneumonitis, http://www.oie.int/standard-setting/terrestrial-manual/access-online/2.05.09_EQUINE_RHINO.pdf (Chapter 2.5.9).
- Patel J.R. & Heldens J. (2005) Equine herpesviruses 1 (EHV‐1) and 4 (EHV‐4)–epidemiology, disease and immunoprophylaxis: a brief review. Veterinary Journal 170, 14–23. [DOI] [PubMed] [Google Scholar]
- Pusterla N., Leutenegger C.M., Wilson W.D., Watson J.L., Ferraro G.L. & Madigan J.E. (2005) Equine herpesvirus‐4 kinetics in peripheral blood leukocytes and nasopharyngeal secretions in foals using quantitative real‐time TaqMan PCR. Journal of Veterinary Diagnostic Investigation 17, 578–581. [DOI] [PubMed] [Google Scholar]
- Reed S.M. & Toribio R.E. (2004) Equine herpesvirus 1 and 4. Veterinary Clinics of North America: Equine Practice 20, 631–642. [DOI] [PubMed] [Google Scholar]
- Soliman E., Kalad M., Elkbany M., Ebied E., Warda S., Abdelhamid N., Mohamed N. (2008) Recent isolation and identification of equine Herpes Viral abortion (EHV‐1) in Egypt ‐2007 9th Vet.Med.Zag.Conference.
- VanDevanter D.R., Warrener P., Bennett L., Schultz E.R., Coulter S., Garber R.L. & Rose T.M. (1996) Detection and analysis of diverse herpesviral species by consensus primer PCR. Journal of Clinical Microbiology 34, 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.J., Maes R., Del Piero F., Lim A., Wise A., Bolin D.C. & Bolin S.R. (2007) Equine multinodular pulmonary fibrosis: a newly recognized herpesvirus‐associated fibrotic lung disease. Veterinary Pathology 44, 849–862. 10.1354/vp.44-6-849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Detailed descriptions of collected samples and presentation of molecular and serological results of each tested sample.


