Abstract
Proteasomes are a class of protease that carry out the degradation of a specific set of cellular proteins. While essential for eukaryotic life, proteasomes are found only in a small subset of bacterial species. In this chapter, we present the current knowledge of bacterial proteasomes, detailing the structural features and catalytic activities required to achieve proteasomal proteolysis. We describe the known mechanisms by which substrates are doomed for degradation, and highlight potential non-degradative roles for components of bacterial proteasome systems. Additionally, we highlight several pathways of microbial physiology that rely on proteasome activity. Lastly, we explain the various gaps in our understanding of bacterial proteasome function and emphasize several opportunities for further study.
INTRODUCTION
Proteasomes are a class of protease found in all three domains of life. A minimum proteasome complex is a barrel-shaped, 28-subunit structure known as the 20S core particle (CP, Fig. 1a), named in accordance with its sedimentation coefficient. 20S CPs are gated to prevent uncontrolled proteolysis, and associate with proteasomal activator proteins in order to degrade specific sets of cellular substrates.
Fig. 1. The M. tuberculosis Pup-proteasome system.
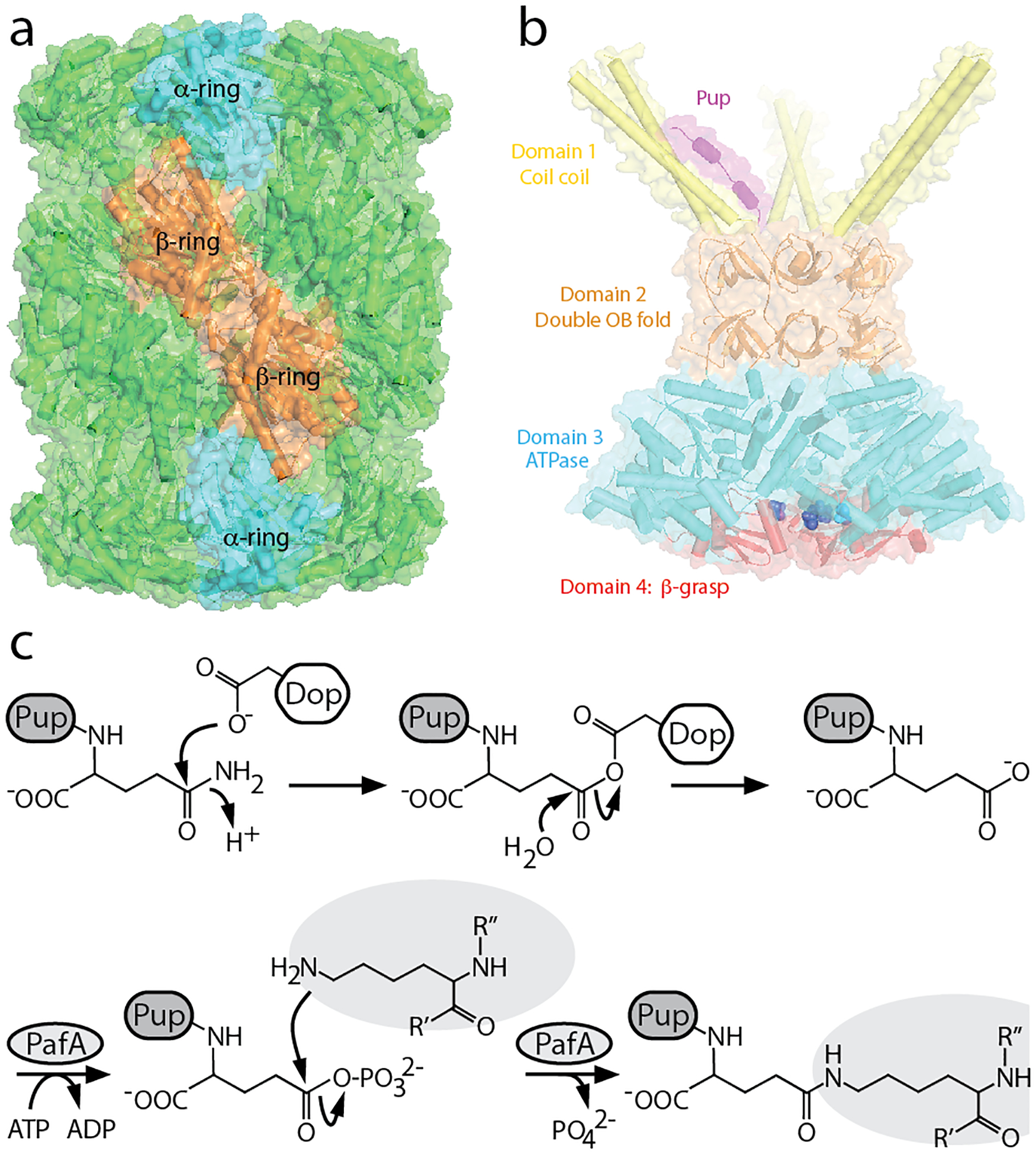
(a) Crystal structure of the M. tuberculosis 20S core particle (20S CP) (PBD entry: 2FHG). Individual subunits of PrcA (blue) and PrcB (orange) are highlighted within each α- and β-ring. (b) Model of an Mpa hexamer bound to the N-terminal region of Pup, indicating the four structural domains of Mpa (PDB entries: 3M9D and 5KZF). Note that the last resolved residue Thr-601, shown in sphers, is receded inside the axial chamber. (c) Deamidation of Pup and ligation to a target protein. An aspartate on Dop acts either as a direct nucleophile as shown (Burns et al. 2012) or activates a water molecule (Ozcelik et al. 2012) to attack the C-terminal Gln of Pup, generating a Pup-Dop intermediate; Dop then catalyzes the nucleophilic attack by a water molecule to generate PupGlu. PafA subsequently uses ATP hydrolysis to phosphorylate PupGlu, rendering Pup suitable for nucleophilic attack by the side chain of a substrate lysine residue (Guth et al. 2011).
Proteasomes are essential in all eukaryotes and archaea. Among the bacteria, however, 20S CP genes are not ubiquitous, and are found almost exclusively in the Actinomycetales and Nitrospirales orders (Darwin et al. 2003; De Mot 2007; Lehmann et al. 2017; Lupas et al. 1994; Nagy et al. 1998; Pouch et al. 2000; Tamura et al. 1995). Among the Actinomycetales are the mycobacteria, including Mycobacterium tuberculosis (M. tuberculosis), a major human pathogen. In eukaryotes, proteins are destined for proteasomal degradation primarily through their ligation to a small protein called ubiquitin (Ub) [reviewed in (Streich and Lima 2014)]. Proteasome-bearing bacteria do not have Ub, but instead can modify proteins with Pup (prokaryotic ubiquitin-like protein), after which they are recognized by proteasomal activators for delivery into 20S CPs (Burns et al. 2009; Burns et al. 2010b; Pearce et al. 2008; Striebel et al. 2010). More recent studies have shown that proteins can be sent to bacterial proteasomes independently of pupylation (Delley et al. 2014; Jastrab et al. 2015). Furthermore, there are now data showing that Pup performs degradation-independent functions in several bacterial species (Elharar et al. 2014; Kuberl et al. 2016). Here, we describe the current understanding of bacterial proteasomes and their accessory factors, focusing on both the mechanisms of targeted proteolysis as well as the aspects of bacterial physiology that depend on these macromolecular machines.
20S PROTEASOME CORE PARTICLES
In all domains of life, 20S CPs are composed of four stacked, heptameric rings: two outer rings, composed of α-subunits, act as gated antechambers controlling entry of substrates into the proteasome core, and two inner rings, made up of β-subunits, harbor the active sites that carry out proteolysis (Fig. 1a) (Choi et al. 2016; Groll et al. 2000; Groll et al. 1997; Heinemeyer et al. 1997; Hu et al. 2006; Lin et al. 2006; Lowe et al. 1995; Tamura et al. 1995). All α- and β-subunits belong to the amino (N)-terminal nucleophilic hydrolase protein family (Brannigan et al. 1995). While eukaryotes encode fourteen different α- and β-subunit genes (Groll et al. 1997), most bacteria and archaea have single genes for α- and β-subunits, respectively (Lowe et al. 1995; Tamura et al. 1995).
In contrast to the assembly of eukaryotic and archaeal proteasomes, which requires accessory chaperones (Le Tallec et al. 2007; Ramos et al. 1998), bacterial proteasomes studied to date self-assemble. Bacterial 20S CP assembly initiates with the formation of a half-proteasome consisting of an α- and a β-ring (Groll et al. 2003; Zuhl et al. 1997). Bacterial β-subunits are translated with an N-terminal propeptide that prevents catalytic activity; the association of two half-proteasomes at the β- to β-ring interface initiates autocatalytic cleavage of the propeptide, yielding an N-terminal threonine (Thr1) residue (Li et al. 2010; Pouch et al. 2000; Zuhl et al. 1997; Zwickl et al. 1994). The role of the propeptide appears to differ between bacterial species. In Rhodococcus erythropolis (R. erythropolis), propeptides promote 20S CP assembly (Zuhl et al. 1997); in contrast, the propeptide of M. tuberculosis must be cleaved before half-proteasomes can fully associate (Li et al. 2010; Lin et al. 2006).
The mature β-subunit active site harbors a set of conserved catalytic residues consisting of Thr1 as well as a lysine (Lys) and either an aspartate (Asp) or a glutamate (Glu), a configuration that is present in 20 CPs across all domains of life (Huber et al. 2016). The Lys deprotonates the terminal amine of Thr1, which in turn carries out the nucleophilic attack of an amide bond in a peptide substrate. Meanwhile, an Asp or Glu serves to orient Thr1 within the active site (Hu et al. 2006; Huber et al. 2016; Mc Cormack et al. 1997). Cleavage of the propeptide occurs in an identical manner, where Thr1 attacks the amide bond linking it to the residue in the −1 position (Huber et al. 2016).
Despite a unified catalytic mechanism in 20S CPs, the residues lining β-subunit active sites are variable, and ultimately determine the specificity of proteolytic activity. Archaeal 20S CPs studied to date appear to preferentially cleave following hydrophobic residues within peptides (Hu et al. 2006; Tamura et al. 1995). The three catalytically active β-subunits in eukaryotic 20S CPs have distinct active sites, each with specificity for hydrophobic, basic, or acidic peptide residues (Heinemeyer et al. 1997; Hu et al. 2006). Interestingly, despite only having a single type of β-subunit (PrcB), the M. tuberculosis 20S CP can degrade all three peptide classes due to the presence of both hydrophobic and hydrophilic residues lining the active site (Hu et al. 2006; Lin et al. 2006).
To prevent uncontrolled proteolysis, α-subunits have N-termini that gate 20S CPs to hinder proteins or peptides from entering the central channel. In turn, the 20S CP gate must be displaced by a proteasomal activator in order for substrates to be degraded (Groll et al. 2000; Li et al. 2010; Lin et al. 2006). For the M. tuberculosis α-subunit PrcA, removal of the N-terminal gating residues (“20S open gate CP”) is sufficient to allow for the in vitro degradation of peptide or unfolded protein substrates in the absence of an activator (Lin et al. 2006; Striebel et al. 2010).
ATP-DEPENDENT PROTEASOME ACTIVATION
In order for proteins to be degraded by a proteasome, they must be specifically targeted for unfolding and delivery into the 20S CP. In eukaryotes, the 19S regulatory particle is a heteromeric complex that contains functionally distinct subunits for substrate recognition and unfolding [reviewed in (Finley et al. 2016)]. In contrast, archaeal and bacterial proteasome activators consist of a single gene product that simultaneously recognizes and unfolds substrates. Mpa (mycobacterial proteasome ATPase) in mycobacteria and ARC (AAA ATPase forming ring-shaped complexes) in R. erythropolis are currently the most well-characterized bacterial proteasome activators (Darwin et al. 2005; Wolf et al. 1998). Mpa/ARC belongs to the AAA ATPase (ATPases Associated with diverse cellular Activities) family, which contains a variety of proteasomal and non-proteasomal protease activators (Djuranovic et al. 2009; Erzberger and Berger 2006; Striebel et al. 2009b; Wang et al. 2009).
Mpa/ARC monomers consist of four domains, arranged from the N- to carboxyl (C)-terminus: the coiled-coil domain, the interdomain, the ATPase domain, and the β-grasp domain (Djuranovic et al. 2009; Wang et al. 2009; Wu et al. 2017) (Fig. 1b). At the N-termini of Mpa/ARC hexamers, helices from each subunit intercalate to form three coiled-coils, each of which can act as a substrate-binding site (Djuranovic et al. 2009; Wang et al. 2009).
The interdomains of Mpa/ARC proteins contain tandem regions that adopt an oligosaccharide-binding (OB) fold (Murzin 1993). Compared to the highly flexible coiled-coil domain, the series of β-barrels within the OB folds gives the interdomain a rigid structure that is capable of self-assembly in vitro (Wang et al. 2009). While OB folds are a conserved feature of all proteasomal AAA ATPases, the specific function of the interdomain is unknown, though it has been proposed to promote the stability of the mature hexamer while structural movement is generated during substrate binding and ATPase activity (Djuranovic et al. 2009; Wang et al. 2009).
The ATPase domain is required for protein unfolding using a mechanism conserved across protease activators. The defining characteristic of AAA ATPase family proteins is an aromatic-hydrophobic-glycine motif, also known as the pore loop, which makes direct contact with substrates as they translocate through the ATPase channel (Martin et al. 2008; Striebel et al. 2010; Wang et al. 2009). Studies of AAA ATPases associated with non-proteasomal proteases, as well as the archaeal proteasome activator PAN and the eukaryotic 19S subunit Rpt1, have demonstrated a mechanism by which successive rounds of ATP hydrolysis induce movement of pore loops to pull the substrate through the ATPase channel (Kim et al. 2015; Martin et al. 2008).
The C-termini of Mpa/ARC proteins include a disordered region with the sequence glycine-glutamine-tyrosine-leucine, also known as the GQYL motif, which is essential for interaction with 20S CPs (Darwin et al. 2003; Hu et al. 2018; Jastrab et al. 2015; Pearce et al. 2006). A conserved feature of bacterial proteasome activators, the GQYL motif is thought to act similarly to the C-terminal HbYX (hydrophobic-tyrosine-any amino acid) motif on PAN as well as the 19S RP ATPases (Delley et al. 2014; Jastrab et al. 2015; Smith et al. 2007). The HbYX motif inserts into a pocket in between α-subunits, inducing a conformational change that opens the 20S CP gate (Rabl et al. 2008; Smith et al. 2007; Yu et al. 2010). Notably, a peptide of just the GQYL sequence is sufficient to activate M. tuberculosis 20S CPs for peptide degradation in vitro (Jastrab et al. 2015).
It has long been observed that Mpa associates weakly with 20S CPs in vitro, preventing the robust degradation of substrates (Striebel et al. 2010; Wang et al. 2009). The observation that Mpa and 20S CPs interact poorly could be explained by the discovery of a new domain in M. tuberculosis Mpa, between the ATPase domain and the C-terminus, forming a β-grasp fold (Burroughs et al. 2007; Wu et al. 2017). This feature so far appears to be unique to bacterial proteasome activators and prevents the GQYL motif from interacting with PrcA subunits of a 20S CP. Artificial extension of the Mpa C-terminus (“MpaC-ext”) allows for increased binding between Mpa and 20S CPs. While MpaC-ext could not activate proteolysis by wild type 20S CPs, the use of “open gate” 20S CPs, in which the N-termini of the PrcA subunits are truncated, allows for the in vitro degradation of a model proteasome substrate, Pup~FabD (Wu et al. 2017). It is therefore likely that other factors regulate the ability of Mpa to activate proteasomal degradation in vivo.
Recently, another bacterial proteasome interactor was identified. Rv0435c, or cpa (Cdc48-like protein of actinobacteria), genetically co-occurs with bacterial 20S CP genes, and encodes a protein with structural homologs in eukaryotes and prokaryotes. M. tuberculosis Cpa forms a hexamer that has ATPase activity in vitro and that can associate with the R. erythropolis 20S CPs. While disruption of cpa in M. smegmatis leads to broad changes in the proteome, it remains to be determined if Cpa associates with 20S CPs in vivo and what its role is in the physiology of bacteria (Ziemski et al. 2018).
PUP: A DEGRADATION SIGNAL, AND MORE
Pupylation
One mechanism by which bacterial proteins are targeted for proteasomal degradation is through post-translational modification with Pup. Pup is a small protein that is covalently linked by its C-terminus to the side chain of a lysine on target proteins (Fig. 1c) (Burns et al. 2009; Pearce et al. 2008). In the Pup-proteasome system (PPS), pupylated proteins are specifically recognized by Mpa/ARC, which unfolds and delivers them into 20S CPs (Burns et al. 2010b; Striebel et al. 2010; Wang et al. 2010). While functionally similar to Ub, Pup is mostly disordered, unlike the highly structured Ub (Chen et al. 2009; Sutter et al. 2009).
Mycobacterium Pup is translated with a C-terminal glutamine (Gln) residue. However, initial characterizations of pupylated proteins in M. tuberculosis and M. smegmatis found that lysine-bound Pup contains a C-terminal Glu (PupGlu) (Burns et al. 2009; Pearce et al. 2008). It was found that, following translation, PupGln must be deamidated by Dop (deamidase of Pup) to generate PupGlu in a reaction that is dependent on the presence, but not hydrolysis of ATP (Fig. 1c) (Imkamp et al. 2010a; Striebel et al. 2009a). Covalent linkage of PupGlu to a lysine on a target protein is achieved by a second enzyme, PafA (proteasome accessory factor A) (Striebel et al. 2009a). Upon binding to PafA, Pup adopts a helical conformation, positioning the Pup C-terminus within the PafA active site (Barandun et al. 2013). Upon ATP hydrolysis, PafA phosphorylates PupGlu, preparing it for nucleophilic attack by the ε-amino group of a lysine in a target protein (Guth et al. 2011) (Fig. 1c). Dop, PafA, and PupGln together are necessary and sufficient to pupylate proteins in vitro, and the need for Dop can be bypassed if PupGlu is used (Striebel et al. 2009a).
In eukaryotes, chains of Ub (polyubiquitylation) on protein substrates are generally required for recognition and degradation by proteasomes (Thrower et al. 2000). While polypupylation has been observed in vitro and upon overproduction of Pup in bacteria (Chen et al. 2016; Elharar et al. 2014; Regev et al. 2015), it has never been observed under physiologic conditions. Although a role for polypupylation may exist in certain circumstances, it appears that only a single pupylation event is sufficient to target a substrate for degradation (Burns et al. 2010b). The majority of natively pupylated proteins are modified at a single, specific lysine, although some proteins have up to four different lysines that can be targeted (Fascellaro et al. 2016; Festa et al. 2010; Kuberl et al. 2014; Poulsen et al. 2010; Watrous et al. 2010). There are little reliable data to suggest substrates are pupylated at more than one lysine on the same molecule; i.e., a single polypeptide can only be pupylated once. However, an oligomeric complex can have more than one Pup attached: Mycobacterium Ino1 (inositol 1-phosphate synthetase) forms tetramers and can be pupylated on two monomers within the same complex (Burns et al. 2010a). These observations suggest that the Pup ligase PafA is hindered from interacting with a polypeptide or complex once it is pupylated.
In Pup-bearing bacteria, specific proteins are pupylated at steady state, and these proteins are likely to vary under different laboratory conditions (Fascellaro et al. 2016; Festa et al. 2010; Kuberl et al. 2014; Kuberl et al. 2016; Pearce et al. 2008; Poulsen et al. 2010; Samanovic et al. 2015; Watrous et al. 2010). While numerous targets of pupylation have been identified, there is no unifying model to explain how proteins are selected for pupylation. Furthermore, there is no known consensus sequence that is targeted for pupylation. This observation may not be surprising in light of data showing that free lysine can be pupylated in vitro (Guth et al. 2011) and that the production of the M. tuberculosis pupylation system genes in E. coli, which does not have a PPS, results in the pupylation of over 50 different proteins (Cerda-Maira et al. 2011). While it may seem that PafA is highly promiscuous, it has a higher affinity for at least one native pupylation substrate than for free lysine (Guth et al. 2011). Furthermore, the presence of surface-exposed lysines is insufficient to target a protein for pupylation, suggesting that there are intrinsic features of specific proteins that make them suitable pupylation targets. Pertinent to this observation, a study has identified a loop region of PafA near its active site that appears to be important for the efficient binding of PafA to pupylation substrates (Regev et al. 2016).
Delivery of pupylated proteins to the proteasome
In mycobacteria, pupylated proteins interact with Mpa in a manner that facilitates their unfolding and entry into a 20S CP. Upon binding to the coiled-coil domain of Mpa, the N-terminal region of Pup adopts a helical structure similar to that observed in Pup-PafA interactions (Barandun et al. 2013; Striebel et al. 2010; Wang et al. 2010) (Fig. 1b). The flexible N-terminal region of Pup is required to achieve unfolding by Mpa, leading to a model by which the N-terminus of Pup is delivered “headfirst” into the Mpa ATPase domain (Maldonado et al. 2013; Striebel et al. 2010; Wang et al. 2009). The precise manner by which the pupylated proteins themselves are translocated through Mpa is unknown. In the case of other protease activator ATPases, this is activity is enhanced by adaptor proteins that interact with both the ATPase and the substrate [reviewed in (Sauer and Baker 2011)], though no such adaptors have been identified for bacterial proteasomes.
Fate of Pup: Depupylation and transpupylation
Besides deamidating Pup, Dop also functions to remove Pup from substrates (Burns et al. 2010a; Imkamp et al. 2010b). There is substantial evidence that depupylation is required for the normal function of the PPS. Notably, many bacterial species, including Streptomyces coelicolor (S. coelicolor), encode PupGlu, which suggested Dop has functions beyond the deamidation of PupGln (Compton et al. 2015; Striebel et al. 2009a). Although the production of PupGlu can bypass the requirement of Dop in M. smegmatis, robust pupylation in M. tuberculosis requires Dop activity, even if PupGlu is ectopically over-produced (Cerda-Maira et al. 2011; Cerda-Maira et al. 2010; Imkamp et al. 2010a). These observations led to the hypothesis that a depupylase activity by Dop is required to maintain steady-state levels of Pup (Cerda-Maira et al. 2010). Depupylation by Dop has since been directly demonstrated using a mechanism that appears similar to Pup deamidation: a conserved aspartate residue is proposed to act as a nucleophile that attacks either the Gln side-chain amide to release ammonium (for deamidation) or the isopeptide bond between Pup and a substrate lysine (for depupylation) (Burns et al. 2012; Ozcelik et al. 2012).
The observation that some bacterial species encode PupGlu instead of PupGln suggests that depupylation is a major function of Dop. Furthermore, depupylation likely contributes to the cellular levels of proteasome substrates by rescuing them from degradation. Along these lines, it is unclear whether Pup is recycled by Dop following engagement of a pupylated substrate with Mpa, or is instead degraded along with a doomed protein. In in vitro experiments, Mpa and 20S CPs can degrade a model pupylated substrate where Pup is also degraded (Striebel et al. 2010), but it remains to be determined if Pup is degraded with a linked substrate in vivo.
Recently, a second function of PafA was identified in its ability to transfer Pup from one substrate to another. The “transpupylation” activity of PafA proceeds first through the depupylation of a donor substrate, followed by the ligation of the freed Pup to a new, recipient protein (Zhang et al. 2017). Importantly, transpupylation may help to explain why some pupylated proteins are not conspicuously degraded in vivo under several conditions tested (Fascellaro et al. 2016; Festa et al. 2010). It is possible that some proteins function as Pup “donors” from which Pup may be transferred to other proteins that must be degraded, rather than as proteasome substrates themselves. While this is a provocative idea, it will be challenging to determine if transpupylation occurs in vivo and has physiologic consequences.
Degradation-independent functions of pupylation
Some bacterial species encode components of the PPS, but are missing genes for a 20S CP, suggesting roles for pupylation that do not involve proteolysis. In Corynebacterium glutamicum (C. glutamicum), which encodes PupGlu, PafA, Dop, and ARC, disruption of pup or arc renders this species sensitive to iron starvation. It was shown that ferritin, a protein that oligomerizes to sequester intracellular iron stores, is pupylated. A model was thus proposed whereby dissociation of pupylated ferritin complexes by ARC allows for the release of iron stores necessary during iron-depleted conditions (Kuberl et al. 2016).
There is also evidence that Pup has degradation-independent functions in bacteria that have 20S CPs. In S. coelicolor, mutant strains that are unable to pupylate proteins but still have 20S CPs are defective in sporulation and have altered levels of several metabolites (Boubakri et al. 2015). In another example, an M. smegmatis prcSBA (pup is also known as prcS in M. smegmatis) mutant is deficient in survival during nitrogen starvation; this phenotype is almost fully complemented by pup alone, suggesting that degradation by proteasomes is either not involved or plays a minor role in the ability of this species to withstand nitrogen starvation (Elharar et al. 2014). It is possible that Pup functions in the proteolysis of proteins by another protease in M. smegmatis. Alternatively, there may be non-degradative functions of Pup that facilitate survival during nitrogen starvation. For example, several M. smegmatis proteins involved in nitrogen metabolism are targets of pupylation, which perhaps may affect their activities or interactions (Fascellaro et al. 2016). Thus it remains to be determined how pupylation affects nitrogen metabolism in M. smegmatis.
ATP-INDEPENDENT PROTEASOME ACTIVATION
An M. tuberculosis mutant lacking 20S CP genes has a severe growth defect compared to wild type, dop, pafA or mpa strains, an observation that suggested substrate recognition by Mpa is not the only route for proteasomal degradation in this species (Cerda-Maira et al. 2010; Darwin et al. 2003; Gandotra et al. 2010). Studies seeking novel interaction partners with M. tuberculosis 20S CPs identified a second proteasomal activator, PafE (proteasome accessory factor E), also known as Bpa (bacterial proteasome activator) (Delley et al. 2014; Jastrab et al. 2015). PafE operates separately from the PPS, and is required for the proteasomal degradation of an apparently independent set of substrates (Jastrab et al. 2015).
PafE is unique among protease activators as it assembles into dodecameric rings (Bai et al. 2016; Jastrab et al. 2015) (Fig. 2a). Furthermore, like Mpa, PafE has a canonical C-terminal GQYL motif required for its ability to open the 20S CP gate. Unlike the subunits of an Mpa hexamer, the C-termini of PafE subunits extend outwards from the main structure of the dodecamer (Bai et al. 2016; Bolten et al. 2016) (Fig. 2b). This structure is in direct contrast to Mpa, which has buried C-termini (Wu et al. 2017). This major difference likely explains why PafE, but not Mpa, can activate wild type 20S CPs in vitro (Bolten et al. 2016; Delley et al. 2014; Hu et al. 2018; Jastrab et al. 2015).
Fig. 2. Structural features of the proteasome activator PafE.
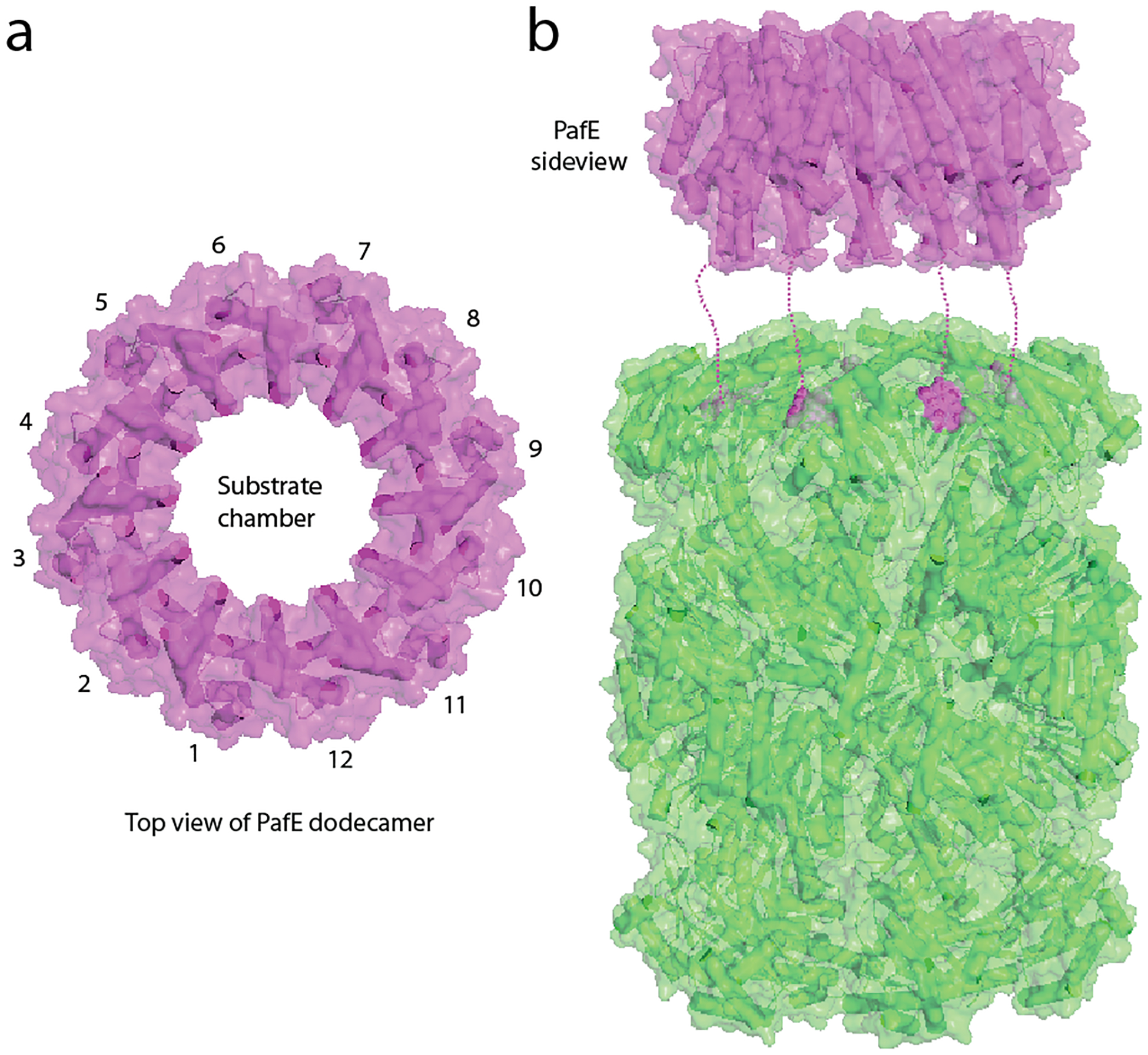
(a) Crystal structure of M. tuberculosis PafE (top view) (PDB entry: 5IET). (b) Model of PafE association with a 20S CP (side view). The GQYL motif of PafE inside the top alpha-ring of the 20S core particle is shown as purple spheres. Dashed lines approximate the disordered C-terminal peptide connecting the last α-helix and the terminal GQYL of PafE.
In contrast to Mpa/ARC, PafE does not hydrolyze ATP. Rather than functioning as an unfoldase, PafE appears to facilitate the translocation of proteins into 20S CPs by acting simply as a gate-opener. Remarkably and consistent with this model, PafE-proteasomes can robustly degrade a model unfolded protein (β-casein) as well as native, folded M. tuberculosis proteins without any apparent post-translational modifications in vitro (Delley et al. 2014; Hu et al. 2018; Jastrab et al. 2015). There are many questions about how specific substrates are degraded by PafE-proteasomes, including how native proteins are translocated into the proteasome in the absence of ATP. It also remains to be determined how PafE-dependent proteolysis is regulated in vivo in the absence of post-translational modifiers.
ROLES OF THE PROTEASOME IN BACTERIAL PHYSIOLOGY
Nitric oxide resistance
Significant attention was given to the bacterial proteasome upon the recognition of its essentiality for M. tuberculosis to resist nitric oxide (NO) toxicity and cause lethal infections in animals. NO is an antimicrobial molecule produced by macrophages upon infection with M. tuberculosis; mice unable to produce NO rapidly succumb to M. tuberculosis infections (MacMicking et al. 1997). However, in wild type mice, M. tuberculosis is able to avoid complete killing by NO, persisting in a host for years after initial infection. M. tuberculosis strains with mutations in mpa, pafA, dop, and prcBA are hypersensitive to NO in vitro and are highly attenuated in mice (Cerda-Maira et al. 2010; Darwin et al. 2003; Gandotra et al. 2010). It was since discovered that the failure to degrade a single PPS substrate, Log (lonely guy), renders M. tuberculosis more susceptible to NO-mediated killing in vitro (Fig. 3a) (Samanovic et al. 2015).
Fig. 3. Regulatory roles of the M. tuberculosis proteasome.
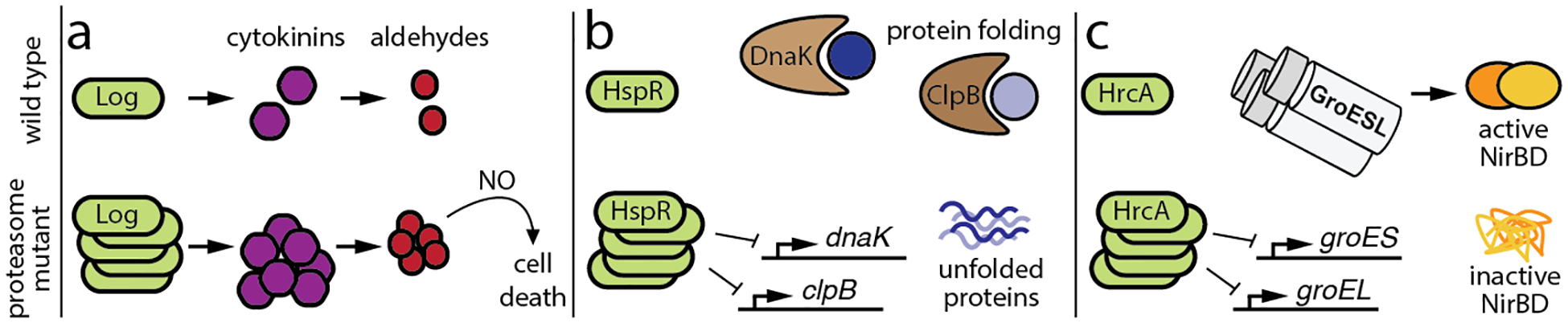
(a) Log is a pupylated proteasome substrate that synthesizes cytokinins, which break down into aldehydes. In the absence of proteasomal degradation, accumulation of Log leads to the overproduction of cytokinins, and the ensuing accumulation of aldehydes synergizes with nitric oxide (NO) to kill M. tuberculosis (Samanovic et al. 2015). (b) HspR, a repressor of M. tuberculosis genes encoding the protein-folding chaperones DnaK and ClpB, is normally degraded by the PafE-proteasome. In the absence of PafE, accumulation of HspR leads to the repression of chaperone genes, which is associated with defective growth and an inability to fully withstand heat shock (Jastrab et al. 2017; Jastrab et al. 2015). (c) M. tuberculosis HrcA represses the genes encoding the GroES/GroEL chaperonins, which are required for the full activity of the nitrite reductase NirBD. It is proposed that failure to pupylate and degrade HrcA prevents full chaperonin expression, leading to a loss of NirBD activity; this prohibits M. tuberculosis from using nitrate as a nitrogen source (Becker et al. 2019).
Log is an orthologue of LONELY GUY, a phosphoribohydrolase that was first identified in plants as an enzyme required for the biosynthesis of a class of adenine-based hormone molecules called cytokinins (Kurakawa et al. 2007). M. tuberculosis Log synthesizes cytokinins, and an M. tuberculosis mpa mutant makes more cytokinins than a wild type strain due to a failure to degrade Log. In turn, aldehydes derived from cytokinins synergize with NO to kill M. tuberculosis through an unknown mechanism (Fig. 3a). Importantly, while disruption of log fully restored NO resistance to an M. tuberculosis mpa mutant, it did not fully rescue the virulence defect of this strain in mice (Samanovic et al. 2015). This result suggests that there are additional proteins whose degradation by the PPS is required for the full pathogenicity of M. tuberculosis.
Copper homeostasis
An early attempt to characterize PPS-dependent gene expression in M. tuberculosis identified a previously unrecognized transcriptional repressor, RicR (regulated in copper repressor, Rv0190). RicR represses M. tuberculosis genes including the metallothionein-encoding mymT as well as mmcO, which encodes a copper-detoxifying enzyme (Festa et al. 2011). mymT and mmcO are each required for the resistance of M. tuberculosis to copper in vitro (Shi et al. 2014). While growth of wild type bacteria in copper is associated with de-repression of the RicR regulon, RicR-regulated genes are constitutively repressed in PPS mutants. These observations suggested that RicR itself is a PPS substrate; however, there is currently no evidence that RicR is pupylated or that its levels increase in the absence of proteasomal degradation (Festa et al. 2011). An alternative explanation is that RicR acutely senses changes in the intracellular availability of copper: the accumulation of one or more copper-binding proteins or molecules in a PPS mutant could titrate copper away from RicR, preventing its dissociation from RicR-repressed promoters.
Deletion and disruption of individual RicR-regulated genes does not affect the ability of M. tuberculosis to cause lethal infections; however, an M. tuberculosis strain with a constitutively active RicR variant is attenuated in mice (Shi et al. 2014). These observations suggest that M. tuberculosis copper homeostasis is maintained by a combination of proteins whose levels are controlled by RicR, and that PPS-dependent de-repression of the RicR regulon may be important for the full virulence of this pathogen.
Protein quality control
Initial efforts to characterize the function of PafE in M. tuberculosis identified HspR (heat shock protein repressor) as a PafE-proteasome substrate (Jastrab et al. 2015). HspR is a transcriptional repressor conserved in many bacteria as a regulator of genes encoding molecular chaperones and proteases (Narberhaus 1999; Roncarati et al. 2007). In M. tuberculosis, HspR represses the expression of dnaKJ, clpB and hsp, which encode chaperones (Stewart et al. 2001; Stewart et al. 2002). ClpB, DnaKJ, and Hsp are highly conserved proteins that help disaggregate and refold misfolded proteins that form under conditions such as heat shock (reviewed in (Kim et al. 2013)). The accumulation of HspR in the absence of PafE-mediated degradation results in reduced expression of dnaK and clpB in M. tuberculosis; accordingly, a pafE mutant has a growth defect and is sensitive to heat shock compared to a parental strain (Fig. 3b) (Jastrab et al. 2015). Perhaps unsurprisingly, point mutations in hspR that abolish repressor activity are frequently acquired in a pafE strain; analysis of these hspR loss-of-function mutations showed that they suppressed the growth defect and heat shock sensitivity caused by disruption of pafE (Jastrab et al. 2017).
The observations that the PafE-proteasome can degrade an unfolded protein in vitro (Delley et al. 2014; Jastrab et al. 2015) and that PafE-dependent degradation of HspR is required for the robust production of an essential protein quality control system (Jastrab and Darwin 2015; Jastrab et al. 2017) suggest that the M. tuberculosis proteasome has a regulatory function in mitigating proteotoxic stress. This activity may be critical in M. tuberculosis, which lacks Lon protease that is important for the degradation of misfolded proteins in E. coli (Gottesman and Zipser 1978). It is possible that the PafE-proteasome fulfills some functions of Lon in M. tuberculosis. Because PafE-dependent proteolysis does not require ATP, unlike the ClpXP, HslUV, and Lon proteases, the PafE-proteasome may be particularly useful when bacteria are metabolically stressed, a condition that could lead to both ATP depletion and the accumulation of improperly folded proteins.
Nitrogen metabolism
In eukaryotic cells, amino acid recycling through proteasomal degradation of proteins is essential for life (Suraweera et al. 2012; Vabulas and Hartl 2005). In Mycobacteria, amino acids are an optimal source of nitrogen (Gouzy et al. 2014a; Gouzy et al. 2013; Gouzy et al. 2014b; Song and Niederweis 2012), and it was thus proposed that bacterial proteasomes are important for nutrient homeostasis by turning over proteins into amino acids. This hypothesis was tested in M. smegmatis, where a prcSBA strain was found to be deficient in survival during carbon or nitrogen starvation compared to a wild type strain. Importantly, sensitivity of the prcSBA mutant to nitrogen limitation is almost fully rescued by the re-introduction of prcS into this strain (Elharar et al. 2014), suggesting that while the PPS is important for maintaining nitrogen homeostasis, its role is primarily in the regulation of individual substrates through pupylation, rather than amino acid turnover by proteasomal proteolysis. Alternatively, pupylation may be targeting proteins to another protease as suggested above. In an important distinction, pupylation does not support M. tuberculosis survival during nitrogen starvation (Becker et al. 2019), suggesting that M. smegmatis and M. tuberculosis have different nutrient requirements and may differ in the proteins that are targeted for pupylation or proteasomal degradation.
Supporting the hypothesis that the PPS regulates nitrogen metabolism in M. smegmatis, a second study observed that M. smegmatis proteins involved in nitrogen transport and assimilation are pupylated during nitrogen starvation. Interestingly, only a subset of these proteins accumulated in a Pup-deficient strain, further supporting a model whereby pupylation provides both degradation-dependent and -independent functions in M. smegmatis (Fascellaro et al. 2016).
Finally, recent work has identified a novel role for the PPS in M. tuberculosis nitrate metabolism. M. tuberculosis mutants lacking an intact PPS cannot use nitrate as a nitrogen source for in vitro growth. A screen to identify proteins whose pupylation and degradation is required for M. tuberculosis nitrate metabolism identified the transcriptional repressor HrcA as a putative proteasome substrate (Becker et al. 2019). In M. tuberculosis, HrcA represses genes encoding GroES/GroEL chaperonins (Stewart et al. 2002), a class of essential molecular chaperones that facilitate the folding of many bacterial proteins [reviewed in (Hayer-Hartl et al. 2016)]. It was determined that chaperonin production promotes the activity of the nitrite reductase NirBD, an enzyme that is essential for the assimilation of nitrogen from nitrate in M. tuberculosis. Chaperonin gene expression is impaired in strains that are unable to degrade pupylated proteins, and a model was proposed that pupylation and degradation of HrcA, leading to de-repression of the chaperonin genes, is required for the full activity of NirBD (Fig. 3c) (Becker et al. 2019). It is unclear whether this novel mechanism of control over M. tuberculosis chaperonin production is required during conditions other than growth in nitrate, or if there are additional M. tuberculosis proteins whose function strictly depends on robust chaperonin expression.
REMAINING QUESTIONS
While many aspects of bacterial proteasome systems have been uncovered over the past several decades, important questions remain to be answered. Structural studies of purified proteasome components suggest that in vivo association of activators with 20S CPs may require additional factors. The C-terminus of PafE is twice as long as those found in ATP-independent proteasome activators from eukaryotes, leading to a very weak interaction between PafE and 20S CPs (Bai et al. 2017). Meanwhile, in the case of Mpa/ARC, the C-terminal 20S-interacting peptide appears to be receded inside the Mpa/ARC central chamber, due to the presence of a β-grasp domain that is absent from eukaryotic or archaeal proteasomal ATPase activators (Wu et al. 2017). Accordingly, PafE and Mpa/ARC cannot efficiently activate protein degradation in vitro (Bai et al. 2016; Hu et al. 2018; Wang et al. 2009; Wu et al. 2017). It is possible that post-translational modifications or interactions with additional proteins alter the structure of bacterial proteasome activators or 20S CPs to make them more conducive to binding in vivo.
Another poorly-understood aspect of bacterial proteasomes is the manner in which substrates are selected for pupylation, depupylation, and transpupylation. Because abundant pupylation occurs upon production of PafA and Pup in E. coli (Cerda-Maira et al. 2011), it may be that the structures of individual proteins simply makes them inherently more or less prone to pupylation. However, it is possible that bacteria that have pupylation may harbor additional mechanisms of substrate selectivity. Additionally, the breadth of substrate selection by the PafE-proteasome system is currently unknown. Because PafE can facilitate the degradation of unfolded proteins (Delley et al. 2014; Jastrab et al. 2015), the PafE-proteasome may be important for the clearance of un- or mis-folded proteins in bacteria under certain proteotoxic stress conditions.
Lastly, repeated observations that bacteria with an intact PPS only appear to degrade a subset of pupylated proteins under routine culture conditions suggest that there are yet-to-be-discovered mechanisms of control over proteasomal proteolysis. In M. smegmatis, along with the observation that the PPS is important for survival during nitrogen starvation (Elharar et al. 2014), it was also shown that the number of proteins with altered abundance in a pupylation mutant compared to a wild type strain was much greater upon total nitrogen starvation than during nitrogen-replete conditions (Fascellaro et al. 2016). Along similar lines in M. tuberculosis, the steady-state levels of pupylated proteins is lower when bacteria are grown in nitrate, a relatively poor source of nitrogen for M. tuberculosis, than in a more ideal nitrogen source (Becker et al. 2019). These observations together offer an intriguing hypothesis that for M. smegmatis, M. tuberculosis and possibly other bacteria, the capacity of the PPS to degrade proteins is determined in some manner by nutrient availability. Therefore, obtaining a better understanding of the precise roles of pupylation and degradation in maintaining nitrogen homeostasis may lead the way towards new regulatory functions mediating proteasome activity.
ACKNOWLEDGEMENTS
Proteasome research in the Darwin lab is supported by NIH grants HL92774 and AI088075 awarded to K.H.D. NIH grant T32AI007180 supported S.H.B. HL is supported by AI070285.
REFERENCES
- Bai L, Hu K, Wang T, Jastrab JM, Darwin KH, Li H (2016) Structural analysis of the dodecameric proteasome activator PafE in Mycobacterium tuberculosis Proc Natl Acad Sci U S A doi: 10.1073/pnas.1512094113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L et al. (2017) Structural Analysis of Mycobacterium tuberculosis Homologues of the Eukaryotic Proteasome Assembly Chaperone 2 (PAC2) J Bacteriol 199 doi: 10.1128/JB.00846-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barandun J, Delley CL, Ban N, Weber-Ban E (2013) Crystal structure of the complex between prokaryotic ubiquitin-like protein and its ligase PafA J Am Chem Soc 135:6794–6797 doi: 10.1021/ja4024012 [DOI] [PubMed] [Google Scholar]
- Becker SH et al. (2019) The Mycobacterium tuberculosis Pup-proteasome system regulates nitrate metabolism through an essential protein quality control pathway Proc Natl Acad Sci U S A (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolten M, Delley CL, Leibundgut M, Boehringer D, Ban N, Weber-Ban E (2016) Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome Structure 24:2138–2151 doi: 10.1016/j.str.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Boubakri H et al. (2015) The Absence of Pupylation (Prokaryotic Ubiquitin-Like Protein Modification) Affects Morphological and Physiological Differentiation in Streptomyces coelicolor J Bacteriol 197:3388–3399 doi: 10.1128/JB.00591-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation Nature 378:416–419 doi: 10.1038/378416a0 [DOI] [PubMed] [Google Scholar]
- Burns KE, Cerda-Maira FA, Wang T, Li H, Bishai WR, Darwin KH (2010a) “Depupylation” of prokaryotic ubiquitin-like protein from mycobacterial proteasome substrates Mol Cell 39:821–827 doi: 10.1016/j.molcel.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE 3rd (2009) Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein J Biol Chem 284:3069–3075 doi: 10.1074/jbc.M808032200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE et al. (2012) Mycobacterium tuberculosis prokaryotic ubiquitin-like protein-deconjugating enzyme is an unusual aspartate amidase J Biol Chem 287:37522–37529 doi: 10.1074/jbc.M112.384784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Pearce MJ, Darwin KH (2010b) Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates J Bacteriol 192:2933–2935 doi: 10.1128/JB.01639-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Balaji S, Iyer LM, Aravind L (2007) Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold Biol Direct 2:18 doi: 10.1186/1745-6150-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira FA, McAllister F, Bode NJ, Burns KE, Gygi SP, Darwin KH (2011) Reconstitution of the Mycobacterium tuberculosis pupylation pathway in Escherichia coli EMBO Rep 12:863–870 doi: 10.1038/embor.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira FA, Pearce MJ, Fuortes M, Bishai WR, Hubbard SR, Darwin KH (2010) Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis Mol Microbiol 77:1123–1135 doi: 10.1111/j.1365-2958.2010.07276.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li C, Wang L, Liu Y, Li C, Zhang J (2016) The Mechanism of Mycobacterium smegmatis PafA Self-Pupylation PLoS One 11:e0151021 doi: 10.1371/journal.pone.0151021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ (2009) Prokaryotic ubiquitin-like protein pup is intrinsically disordered J Mol Biol 392:208–217 doi: 10.1016/j.jmb.2009.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WH et al. (2016) Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation Nat Commun 7:10963 doi: 10.1038/ncomms10963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton CL, Fernandopulle MS, Nagari RT, Sello JK (2015) Genetic and Proteomic Analyses of Pupylation in Streptomyces coelicolor J Bacteriol 197:2747–2753 doi: 10.1128/JB.00302-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF (2003) The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide Science 302:1963–1966 doi: 10.1126/science.1091176 [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF (2005) Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue Mol Microbiol 55:561–571 doi: 10.1111/j.1365-2958.2004.04403.x [DOI] [PubMed] [Google Scholar]
- De Mot R (2007) Actinomycete-like proteasomes in a Gram-negative bacterium Trends Microbiol 15:335–338 doi: 10.1016/j.tim.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Delley CL, Laederach J, Ziemski M, Bolten M, Boehringer D, Weber-Ban E (2014) Bacterial proteasome activator bpa (rv3780) is a novel ring-shaped interactor of the mycobacterial proteasome PLoS One 9:e114348 doi: 10.1371/journal.pone.0114348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S et al. (2009) Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases Mol Cell 34:580–590 doi: 10.1016/j.molcel.2009.04.030 [DOI] [PubMed] [Google Scholar]
- Elharar Y et al. (2014) Survival of mycobacteria depends on proteasome-mediated amino acid recycling under nutrient limitation EMBO J 33:1802–1814 doi: 10.15252/embj.201387076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM (2006) Evolutionary relationships and structural mechanisms of AAA+ proteins Annu Rev Biophys Biomol Struct 35:93–114 doi: 10.1146/annurev.biophys.35.040405.101933 [DOI] [PubMed] [Google Scholar]
- Fascellaro G et al. (2016) Comprehensive Proteomic Analysis of Nitrogen-Starved Mycobacterium smegmatis Deltapup Reveals the Impact of Pupylation on Nitrogen Stress Response J Proteome Res 15:2812–2825 doi: 10.1021/acs.jproteome.6b00378 [DOI] [PubMed] [Google Scholar]
- Festa RA et al. (2011) A novel copper-responsive regulon in Mycobacterium tuberculosis Mol Microbiol 79:133–148 doi: 10.1111/j.1365-2958.2010.07431.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH (2010) Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis [corrected] PLoS One 5:e8589 doi: 10.1371/journal.pone.0008589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Chen X, Walters KJ (2016) Gates, Channels, and Switches: Elements of the Proteasome Machine Trends Biochem Sci 41:77–93 doi: 10.1016/j.tibs.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra S, Lebron MB, Ehrt S (2010) The Mycobacterium tuberculosis proteasome active site threonine is essential for persistence yet dispensable for replication and resistance to nitric oxide PLoS Pathog 6:e1001040 doi: 10.1371/journal.ppat.1001040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Zipser D (1978) Deg phenotype of Escherichia coli lon mutants J Bacteriol 133:844–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A et al. (2014a) Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection PLoS Pathog 10:e1003928 doi: 10.1371/journal.ppat.1003928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A et al. (2013) Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate Nat Chem Biol 9:674–676 doi: 10.1038/nchembio.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A, Poquet Y, Neyrolles O (2014b) Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence vol 12. doi: 10.1038/nrmicro3349 [DOI] [PubMed] [Google Scholar]
- Groll M et al. (2000) A gated channel into the proteasome core particle Nat Struct Biol 7:1062–1067 doi: 10.1038/80992 [DOI] [PubMed] [Google Scholar]
- Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R (2003) Investigations on the maturation and regulation of archaebacterial proteasomes J Mol Biol 327:75–83 [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 A resolution Nature 386:463–471 doi: 10.1038/386463a0 [DOI] [PubMed] [Google Scholar]
- Guth E, Thommen M, Weber-Ban E (2011) Mycobacterial ubiquitin-like protein ligase PafA follows a two-step reaction pathway with a phosphorylated pup intermediate J Biol Chem 286:4412–4419 doi: 10.1074/jbc.M110.189282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer-Hartl M, Bracher A, Hartl FU (2016) The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding Trends Biochem Sci 41:62–76 doi: 10.1016/j.tibs.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH (1997) The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing J Biol Chem 272:25200–25209 [DOI] [PubMed] [Google Scholar]
- Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H (2006) Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate Mol Microbiol 59:1417–1428 doi: 10.1111/j.1365-2958.2005.05036.x [DOI] [PubMed] [Google Scholar]
- Hu K, Jastrab JB, Zhang S, Kovach A, Zhao G, Darwin KH, Li H (2018) Proteasome substrate capture and gate opening by the accessory factor PafE from Mycobacterium tuberculosis J Biol Chem 293:4713–4723 doi: 10.1074/jbc.RA117.001471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber EM, Heinemeyer W, Li X, Arendt CS, Hochstrasser M, Groll M (2016) A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome Nat Commun 7:10900 doi: 10.1038/ncomms10900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E (2010a) Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo Mol Microbiol 75:744–754 doi: 10.1111/j.1365-2958.2009.07013.x [DOI] [PubMed] [Google Scholar]
- Imkamp F, Striebel F, Sutter M, Ozcelik D, Zimmermann N, Sander P, Weber-Ban E (2010b) Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway EMBO Rep 11:791–797 doi: 10.1038/embor.2010.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrab JB, Darwin KH (2015) Bacterial Proteasomes Annu Rev Microbiol 69:109–127 doi: 10.1146/annurev-micro-091014-104201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrab JB, Samanovic MI, Copin R, Shopsin B, Darwin KH (2017) Loss-of-Function Mutations in HspR Rescue the Growth Defect of a Mycobacterium tuberculosis Proteasome Accessory Factor E (pafE) Mutant J Bacteriol 199 doi: 10.1128/JB.00850-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrab JB et al. (2015) An adenosine triphosphate-independent proteasome activator contributes to the virulence of Mycobacterium tuberculosis Proc Natl Acad Sci U S A 112:E1763–1772 doi: 10.1073/pnas.1423319112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Snoberger A, Schupp J, Smith DM (2015) ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function Nat Commun 6:8520 doi: 10.1038/ncomms9520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis Annu Rev Biochem 82:323–355 doi: 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Kuberl A, Franzel B, Eggeling L, Polen T, Wolters DA, Bott M (2014) Pupylated proteins in Corynebacterium glutamicum revealed by MudPIT analysis Proteomics 14:1531–1542 doi: 10.1002/pmic.201300531 [DOI] [PubMed] [Google Scholar]
- Kuberl A, Polen T, Bott M (2016) The pupylation machinery is involved in iron homeostasis by targeting the iron storage protein ferritin Proc Natl Acad Sci U S A 113:4806–4811 doi: 10.1073/pnas.1514529113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T et al. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme Nature 445:652–655 doi: 10.1038/nature05504 [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A (2007) 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals Mol Cell 27:660–674 doi: 10.1016/j.molcel.2007.06.025 [DOI] [PubMed] [Google Scholar]
- Lehmann G, Udasin RG, Livneh I, Ciechanover A (2017) Identification of UBact, a ubiquitin-like protein, along with other homologous components of a conjugation system and the proteasome in different gram-negative bacteria Biochem Biophys Res Commun 483:946–950 doi: 10.1016/j.bbrc.2017.01.037 [DOI] [PubMed] [Google Scholar]
- Li D, Li H, Wang T, Pan H, Lin G, Li H (2010) Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome EMBO J 29:2037–2047 doi: 10.1038/emboj.2010.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G et al. (2006) Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity Mol Microbiol 59:1405–1416 doi: 10.1111/j.1365-2958.2005.05035.x [DOI] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution Science 268:533–539 [DOI] [PubMed] [Google Scholar]
- Lupas A, Zwickl P, Baumeister W (1994) Proteasome sequences in eubacteria Trends Biochem Sci 19:533–534 [DOI] [PubMed] [Google Scholar]
- MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF (1997) Identification of nitric oxide synthase as a protective locus against tuberculosis Proc Natl Acad Sci U S A 94:5243–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AY, Burz DS, Reverdatto S, Shekhtman A (2013) Fate of pup inside the Mycobacterium proteasome studied by in-cell NMR PLoS One 8:e74576 doi: 10.1371/journal.pone.0074576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT (2008) Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding Nat Struct Mol Biol 15:1147–1151 doi: 10.1038/nsmb.1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Cormack T et al. (1997) Active site-directed inhibitors of Rhodococcus 20 S proteasome. Kinetics and mechanism J Biol Chem 272:26103–26109 [DOI] [PubMed] [Google Scholar]
- Murzin AG (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences EMBO J 12:861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R (1998) The 20S proteasome of Streptomyces coelicolor J Bacteriol 180:5448–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F (1999) Negative regulation of bacterial heat shock genes Mol Microbiol 31:1–8 [DOI] [PubMed] [Google Scholar]
- Ozcelik D et al. (2012) Structures of Pup ligase PafA and depupylase Dop from the prokaryotic ubiquitin-like modification pathway Nat Commun 3:1014 doi: 10.1038/ncomms2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH (2006) Identification of substrates of the Mycobacterium tuberculosis proteasome EMBO J 25:5423–5432 doi: 10.1038/sj.emboj.7601405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis Science 322:1104–1107 doi: 10.1126/science.1163885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouch MN, Cournoyer B, Baumeister W (2000) Characterization of the 20S proteasome from the actinomycete Frankia Mol Microbiol 35:368–377 [DOI] [PubMed] [Google Scholar]
- Poulsen C et al. (2010) Proteome-wide identification of mycobacterial pupylation targets Mol Syst Biol 6:386 doi: 10.1038/msb.2010.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases Mol Cell 30:360–368 doi: 10.1016/j.molcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ (1998) Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly Cell 92:489–499 [DOI] [PubMed] [Google Scholar]
- Regev O, Korman M, Hecht N, Roth Z, Forer N, Zarivach R, Gur E (2016) An Extended Loop of the Pup Ligase, PafA, Mediates Interaction with Protein Targets J Mol Biol 428:4143–4153 doi: 10.1016/j.jmb.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Regev O, Roth Z, Korman M, Khalaila I, Gur E (2015) A kinetic model for the prevalence of mono- over poly-pupylation FEBS J 282:4176–4186 doi: 10.1111/febs.13413 [DOI] [PubMed] [Google Scholar]
- Roncarati D, Danielli A, Spohn G, Delany I, Scarlato V (2007) Transcriptional regulation of stress response and motility functions in Helicobacter pylori is mediated by HspR and HrcA J Bacteriol 189:7234–7243 doi: 10.1128/JB.00626-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanovic MI et al. (2015) Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide Mol Cell 57:984–994 doi: 10.1016/j.molcel.2015.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA (2011) AAA+ proteases: ATP-fueled machines of protein destruction Annu Rev Biochem 80:587–612 doi: 10.1146/annurev-biochem-060408-172623 [DOI] [PubMed] [Google Scholar]
- Shi X, Festa RA, Ioerger TR, Butler-Wu S, Sacchettini JC, Darwin KH, Samanovic MI (2014) The copper-responsive RicR regulon contributes to Mycobacterium tuberculosis virulence MBio 5 doi: 10.1128/mBio.00876-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL (2007) Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry Mol Cell 27:731–744 doi: 10.1016/j.molcel.2007.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Niederweis M (2012) Uptake of sulfate but not phosphate by Mycobacterium tuberculosis is slower than that for Mycobacterium smegmatis J Bacteriol 194:956–964 doi: 10.1128/JB.06132-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GR et al. (2001) Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection Nat Med 7:732–737 doi: 10.1038/89113 [DOI] [PubMed] [Google Scholar]
- Stewart GR et al. (2002) Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays Microbiology 148:3129–3138 doi: 10.1099/00221287-148-10-3129 [DOI] [PubMed] [Google Scholar]
- Streich FC Jr., Lima CD (2014) Structural and functional insights to ubiquitin-like protein conjugation Annu Rev Biophys 43:357–379 doi: 10.1146/annurev-biophys-051013-022958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Hunkeler M, Summer H, Weber-Ban E (2010) The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup’s N-terminus EMBO J 29:1262–1271 doi: 10.1038/emboj.2010.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E (2009a) Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes Nat Struct Mol Biol 16:647–651 doi: 10.1038/nsmb.1597 [DOI] [PubMed] [Google Scholar]
- Striebel F, Kress W, Weber-Ban E (2009b) Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes Curr Opin Struct Biol 19:209–217 doi: 10.1016/j.sbi.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Suraweera A, Munch C, Hanssum A, Bertolotti A (2012) Failure of amino acid homeostasis causes cell death following proteasome inhibition Mol Cell 48:242–253 doi: 10.1016/j.molcel.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Striebel F, Damberger FF, Allain FH, Weber-Ban E (2009) A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa FEBS Lett 583:3151–3157 doi: 10.1016/j.febslet.2009.09.020 [DOI] [PubMed] [Google Scholar]
- Tamura T et al. (1995) The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus Curr Biol 5:766–774 [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal EMBO J 19:94–102 doi: 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU (2005) Protein synthesis upon acute nutrient restriction relies on proteasome function Science 310:1960–1963 doi: 10.1126/science.1121925 [DOI] [PubMed] [Google Scholar]
- Wang T, Darwin KH, Li H (2010) Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation Nat Struct Mol Biol 17:1352–1357 doi: 10.1038/nsmb.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T et al. (2009) Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa Structure 17:1377–1385 doi: 10.1016/j.str.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J et al. (2010) Expansion of the mycobacterial “PUPylome” Mol Biosyst 6:376–385 doi: 10.1039/b916104j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S et al. (1998) Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis J Mol Biol 277:13–25 doi: 10.1006/jmbi.1997.1589 [DOI] [PubMed] [Google Scholar]
- Wu Y et al. (2017) Mycobacterium tuberculosis proteasomal ATPase Mpa has a beta-grasp domain that hinders docking with the proteasome core protease Mol Microbiol 105:227–241 doi: 10.1111/mmi.13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y (2010) Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions EMBO J 29:692–702 doi: 10.1038/emboj.2009.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Burns-Huang KE, Janssen GV, Li H, Ovaa H, Hedstrom L, Darwin KH (2017) Mycobacterium tuberculosis Proteasome Accessory Factor A (PafA) Can Transfer Prokaryotic Ubiquitin-Like Protein (Pup) between Substrates MBio 8 doi: 10.1128/mBio.00122-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemski M, Jomaa A, Mayer D, Rutz S, Giese C, Veprintsev D, Weber-Ban E (2018) Cdc48-like protein of actinobacteria (Cpa) is a novel proteasome interactor in mycobacteria and related organisms Elife 7 doi: 10.7554/eLife.34055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhl F, Seemuller E, Golbik R, Baumeister W (1997) Dissecting the assembly pathway of the 20S proteasome FEBS Lett 418:189–194 [DOI] [PubMed] [Google Scholar]
- Zwickl P, Kleinz J, Baumeister W (1994) Critical elements in proteasome assembly Nat Struct Biol 1:765–770 [DOI] [PubMed] [Google Scholar]


