Abstract
Objective:
To review the most recent advances in human and bacterial genomics as applied to pathogenesis and clinical management of otitis media.
Data Sources:
PubMed articles published since the last meeting in June 2015 up to June 2019.
Review Methods:
A panel of experts in human and bacterial genomics of otitis media was formed. Each panel member reviewed the literature in their respective fields and wrote draft reviews. The reviews were shared with all panel members, and a merged draft was created. The panel met at the 20th International Symposium on Recent Advances in Otitis Media in June 2019, discussed the review and refined the content. A final draft was made, circulated, and approved by the panel members.
Conclusion:
Trans-disciplinary approaches applying pan-omic technologies to identify human susceptibility to otitis media and to understand microbial population dynamics, patho-adaptation and virulence mechanisms are crucial to the development of novel, personalized therapeutics and prevention strategies for otitis media.
Implications for Practice:
In the future otitis media prevention strategies may be augmented by mucosal immunization, combination vaccines targeting multiple pathogens, and modulation of the middle ear microbiome. Both treatment and vaccination may be tailored to an individual’s otitis media phenotype as defined by molecular profiles obtained by using rapidly developing techniques in microbial and host genomics.
Keywords: otitis media, otitis-prone, microbiome, metagenomics, genome-wide association study, precision medicine
1. Introduction
Despite significant decreases in incidence and prevalence worldwide, otitis media (OM) remains a very common diagnosis particularly among children and the elderly. OM continues to impose a major public health burden across human populations and disproportionately affects socially and disadvantaged persons, e.g. indigenous communities. Part of the reason why OM burden cannot be easily eliminated is the incomplete understanding of the OM phenotype and its underlying disease mechanisms.
Human studies of genetic predisposition to experience OM rely on the supposition that the history of diagnosis of OM is accurate. Contamination of the defined susceptible population by individuals who have not experienced OM complicates analysis. The extent of contamination of the susceptible population by “non-susceptibles” currently cannot be known and may vary widely. Precision in diagnosis varies over time for several reasons. The definitions of OM, acute (A)OM and OM with effusion (OME) have changed as recently as 2004 following guidelines from the American Academy of Pediatrics (AAP), then revised again by the AAP in 2013 [1–2]. Moreover, the clinical diagnosis is challenging especially in young children and as such estimates of misdiagnosis can range as high as 50% [3–9]. Clinicians that are specifically trained and tested on their accuracy of diagnosis (proven by tympanocentesis after clinical diagnosis) are termed “validated otoscopists” but they represent <1% of those making OM diagnoses. Tympanocentesis-proven OM is the gold standard of diagnosis but very few clinicians practice this procedure, mostly due to the required skill set for performing tympanocentesis. From a prospective, longitudinal study of children from Rochester, NY, 27% of the children studied met the criteria of otitis-prone (3 AOM infections within 6 months or 4 AOM infections within a year) by age 30 months old if children were diagnosed with AOM by primary care clinicians [10]. In contrast, if the diagnosis was made by validated otoscopists, 14% met the otitis-prone definition, while if diagnosis was made by tympanocentesis, 6% met the otitis-prone definition [10]. That study gives a perspective on the magnitude of the problem of misclassification of OM. Note that such misclassification may occur not only for AOM but also for chronic (C)OM with effusion especially if with an intact eardrum.
At this time of rapid development in molecular -omic technologies, OM clinicians and scientists must take advantage of these advances to improve understanding and classification of the OM phenotype by integrating knowledge of host, pathogen, and microbiome interactions with overall health status and to enable improvements in OM management and reduction of OM incidence, with the ultimate goal of personalizing treatment and prevention. Here we review the most recent discoveries in human and microbial genomics as applied to OM pathogenesis and precision medicine.
2. Methods
Published literature in PubMed was searched since the last symposium. Search terms included “otitis media”, “acute otitis media”, “recurrent acute otitis media”, “chronic otitis media”, “chronic otitis media with effusion”, “gene”, “genome-wide association studies”, “GWAS”, “loci”, “mouse model”, “animal model”, “microbiome”, “metagenomics”, “epigenetic”, “personalized medicine”, “precision medicine”. The search included all published literature with available English abstracts. No further exclusion or selection criteria were employed. Additional inputs were also taken from each panel member on their specific areas of OM expertise.
3. Discussion
3.1. Discovery of OM Susceptibility and Protective Loci in Humans
Twenty-one significant loci have been identified in five genome-wide association studies (GWAS; Table 1). Of these variants, 15 were identified in a 23andMe study that included >120,000 European-descent individuals [12,13]. Additionally four of the loci were either coding variants or in linkage disequilibrium (LD) with coding variants (Table 1): (A) The PLG c.112A>G (p. Lys38Glu; rs73015965) variant is pathogenic for autosomal recessive type I plasminogen deficiency that includes COM as a clinical manifestation [16]. P/g-knockout mice are also known to spontaneously develop COM [17]. In the GWAS, the heterozygous genotype for the rs73015965 is a risk factor for childhood ear infections (OR=1.43; 95%CI: 1.26, 1.63; p=3.8×10−8) [12]. (B) A synonymous variant within FUT2, c.249C>T (p.Tyr83=; rs681343), was initially identified as a protective variant in homozygous individuals with OM but was later reported as a risk allele [12,13,18]. The rs681343 variant is in almost complete LD with FUT2 c.461G>A (p.Trp154*; rs601338) [12,13,18] which has in the genome Aggregation Database (gnomAD.broadinstitute.org) a minor allele frequency (MAF) 0.26-0.50 in various populations except East Asians (MAF=0.002). This stop variant has been well-studied as a protective factor against multiple non-OM infections [18,19]. It also results in higher plasma vitamin B12 levels [20]. Conversely rs601338 increases risk for autoimmune disorders such as Behcet’s, celiac and inflammatory bowel diseases [18,21]. In multi-ethnic families and probands with OM, several coding variants within FUT2 were identified to confer OM susceptibility [18]. This is further supported by the transient expression of Fut2 in non-typeable Haemophilus influenzae (NTHi)- inoculated mouse middle ears and by decreased epithelial A antigen levels due to FUT2 variants [18]. (C) The intronic TBX1 rs1978060 variant is in almost complete LD with TBX1 c.1189A>C (p.Asn397His; rs72646967) [13]. T6×1-knockout mice have COM that is characteristic of 22q11.2 deletion syndrome in humans [22,23]. In the 23andMe GWAS the TBX1 rs1978060 variant was identified as a risk factor for both childhood ear infections and myringotomy [12,13]. (D) The intronic variant CDHR3 rs114947103 is in almost complete LD with CDHR3 c.1586G>A (p.Cys529Tyr; rs6967330) [13]. Previously CDHR3 rs6967330 was implicated in a GWAS for asthma, particularly for wheezing after rhinovirus infection [24–26]. On the contrary for childhood ear infections, the CDHR3 rs114947103 variant was identified as a protective factor (OR=0.94; 95%CI: 0.91, 0.96; p=5.4×10−9) [12].
Table 1.
Genome-wide significanta loci from genome-wide association studies on otitis media
| Ref | dbSNP ID | Gene | Variant | All MAF | NFE MAF | Lat MAF | Afr MAF |
|---|---|---|---|---|---|---|---|
| 11 | rs2406176 | TMPRSS15 | c.1172-3049A>C | 0.72 | 0.75 | 0.80 | 0.61 |
| 12,13 | rs681343 | FUT2c | c.249C>T (p.Tyr83=) | 0.39 | 0.47 | 0.27 | 0.50 |
| 11 | rs4825724 | C1GALT1C1—[]—CT47B1 | Intergenic | 0.47 | 0.40 | 0.45 | 0.63 |
| 12,13 | rs1978060c | TBX1 | c.410+722A>G | 0.64 | 0.61 | 0.46 | 0.75 |
| 12,13 | rs2808290 | RAB18--[]—MKX | Intergenic | 0.38 | 0.49 | 0.40 | 0.18 |
| 12 | rs7174062 | SPATA8--[]—ARRDC4 | Intergenic | 0.78 | 0.72 | 0.81 | 0.89 |
| 12,13 | rs4329147d | HLA-DRB5--[]--HLA-DQA1 | Intergenic | 0.84 | 0.85 | 0.88 | 0.83 |
| 12,13 | rs8176643 | ABO | c.28+869delG | 0.14 | 0.20 | 0.13 | 0.04 |
| 12,13 | rs1802575 | EFEMP1 | c.*1004C>G | 0.07 | 0.11 | 0.05 | 0.03 |
| 12,13 | rs5829676 | NT5C1B-RDH14--[]--OSR1 | Intergenic | 0.31 | 0.40 | 0.39 | 0.19 |
| 11 | rs885932 | HLA-G—[]—HLA-H | Intergenic | 0.11 | 0.15 | 0.08 | 0.05 |
| 11 | rs3821170 | ADAM23 | c.1852+432C>T | 0.14 | 0.10 | 0.14 | 0.23 |
| 12,13 | rs72931768 | FGF3p--[]--ANO1 | Intergenic | 0.10 | 0.12 | 0.11 | 0.05 |
| 12,13 | rs35213789 | AUTS2 | c.310−96260C>T | 0.25 | 0.26 | 0.19 | 0.23 |
| 12,13 | rs114947103 | CDHR3 | c.1653+409T>C | 0.21 | 0.17 | 0.17 | 0.27 |
| 12,13 | rs13281988 | NIPAL2--[]--KCNS2 | Intergenic | 0.31 | 0.29 | 0.33 | 0.32 |
| 14 | rs10497394c | CDCA7--[]—SP3 | Intergenic | 0.23 | 0.26 | 0.34 | 0.12 |
| 12,13 | rs67035515 | BSN | c.225–5877_225–5874delTGAA | 0.86 | 0.83 | 0.89 | 0.87 |
| 15 | rs2932989c | FNDC1 | ~6kb from 3’UTR | 0.88 | 0.87 | 0.91 | 0.87 |
| 12,13 | rs73015965 | PLG | c.112A>G (p.Lys38Glu) | 0.003 | 0.005 | 0.003 | 0.001 |
| 12 | rs151208372 | DCBLD2--[]--COL8A1 | Intergenic | 0.09 | 0.08 | 0.15 | 0.13 |
Abbreviations: Ref, reference; MAF, minor allele frequencies from gnomAD database; NFE, non-Finnish European; Lat, Latino, Afr, African.
p<5×10−8; Variants are listed in order of increasing p-value.
RefSeq accession numbers for coding and intronic variants: TMPRSS15, NM_002772.2; FUT2, NM_000511.5; TBX1, NM_005992.1; ABO, NM_020469.2; EFEMP1, NM_001039348.2; ADAM23, NM_003812.3; AUTS2, NM_015570.3; CDHR3, NM_152750.4; BSN, NM_003458.3; PLG, NM_000301.3.
Only these variants/genes have been replicated in human subjects with otitis media using an independent dataset.
This variant was associated with childhood ear infections in the initial GWAS; however reanalysis with conditioning for the HLA-DRB1 Gln96 allele resulted in loss of association with the rs4329147 variant.
From these GWAS only five genes or variants were replicated in an independent OM dataset (Table 1): (a) FUT2 [12,13,18]; (b) TBX1 variant rs1978060 [12,13]; (c) ABO [12,27,28]; (d) intergenic variant rs10497394 on chromosome 2q31.1 which lies between genes CDCA7 and SP3 [14]; and (e) a 3’UTR variant in FNDC1 [15]. ABO blood types were previously known to be associated with different otitis media types, such that type O is protective while type A increases risk for OM [27,28]. The initial GWAS for the rs10497394 and FNDC1-3’UTR variants reported independent replication, indicating that they are credibly associated with OM [14,15]. Moreover FNDC1 variants were positively correlated with FNDC1 expression levels but negatively correlated with methylation status of FNDC1 [15]. To date, the FNDC1 variant rs2932989 is the only significant epigenetic signal identified for OM in humans.
All of the five replicated GWAS loci were identified as expression quantitative trait loci (eQTL) for various mucosal and epithelial tissues in the Genotype-Tissue Expression (GTEx) database (gtexportal.org, accessed 21 August 2019), suggesting a regulatory function for these variants (Table 2). Unfortunately middle ear mucosal epithelium is not represented in GTEx. While the majority of these eQTL variants affect expression levels of genes where they lie, some of the variants have more distant effects (e.g. variant rs885932 at 6p22.1 is 0.87Mb proximal to target gene IER3 at 6p21.33). Of note, four of the ten eQTL-GWAS loci affect expression of multiple genes in various tissues (Table 2). The rs4329147 variant not only primarily affects HLA-DRB5 expression but also influences the expression of six other HLA genes and four additional non-HLA genes, including complement gene C4A (Table 2). While the initial 23andMe study identified the rs4329147 variant as the most significant locus among HLA alleles (Table 1), a follow-up study on the same dataset demonstrated that this association is lost when conditioning for the HLA-DRB1 Gln96 allele is performed, implying that the association is largely driven by the amino acid variant within HLA-DRB1 [12,13].
Table 2.
Single-tissue expression quantitative trait loci (eQTL) in the Genotype-Tissue Expression (GTEx) database among genome-wide significant loci for otitis media
| Variant (Chromosomal Band) | Gene | Tissue | eQTL p-value |
|---|---|---|---|
| rs681343 (19q13.33) | FUT2 | Esophageal mucosa | 1.4×10−102 |
| Unexposed skin | 5.0×10−30 | ||
| Exposed skin | 1.5×10−25 | ||
| Transverse colon | 2.7×10−15 | ||
| Small intestine (ileum) | 2.9×10−10 | ||
| Lung | 7.4×10−06 | ||
| NTN5 | Transverse colon | 2.7×10−15 | |
| Unexposed skin | 2.0×10−13 | ||
| RASIP1 | Esophageal mucosa | 4.6×10−16 | |
| Exposed skin | 4.9×10−14 | ||
| Unexposed skin | 3.1×10−10 | ||
| Sigmoid colon | 2.0×10−07 | ||
| IZUMO1 | Lung | 1.2×10−06 | |
| FAM83E | Exposed skin | 3.8×10−06 | |
| rs1978060 (22q11.21) | TBX1 | Prostateb | 5.2×10−12 |
| rs2808290 (10p12.1) | PTCHD3 | Testisb | 9.7×10−06 |
| rs4329147 (6p21.32) | HLA-DRB5 | Lung | 6.4×10−86 |
| Exposed skin | 7.0×10−76 | ||
| Esophageal mucosa | 3.8×10−73 | ||
| Unexposed skin | 1.2×10−64 | ||
| Stomach | 5.8×10−43 | ||
| Sigmoid colon | 2.6×10−33 | ||
| Vagina | 1.1×10−21 | ||
| Uterus | 1.8×10−19 | ||
| Small intestine (ileum) | 2.3×10−18 | ||
| HLA-DRB6 | Exposed skin | 2.6×10−45 | |
| Lung | 2.8×10−42 | ||
| Esophageal mucosa | 3.7×10−39 | ||
| Unexposed skin | 3.5×10−32 | ||
| Transverse colon | 6.8×10−28 | ||
| Stomach | 6.0×10−27 | ||
| Small intestine (ileum) | 9.9×10−16 | ||
| Sigmoid colon | 4.8×10−15 | ||
| Uterus | 1.2×10−10 | ||
| Vagina | 3.2×10−10 | ||
| HLA-DQB1 | Lung | 1.2×10−22 | |
| Esophageal mucosa | 1.1×10−20 | ||
| Exposed skin | 7.2×10−20 | ||
| Stomach | 6.5×10−15 | ||
| Transverse colon | 4.9×10−13 | ||
| Sigmoid colon | 2.0×10−11 | ||
| Small intestine (ileum) | 8.8×10−07 | ||
| HLA-DRB9 | Lung | 1.0×10−18 | |
| Small intestine (ileum) | 9.1×10−10 | ||
| Unexposed skin | 1.6×10−08 | ||
| Vagina | 1.3×10−05 | ||
| Exposed skin | 3.1×10−05 | ||
| HLA-DRB1 | Lung | 9.6×10−14 | |
| Exposed skin | 1.9×10−12 | ||
| Esophageal mucosa | 2.2×10−09 | ||
| Unexposed skin | 2.9×10−07 | ||
| Transverse colon | 2.4×10−05 | ||
| HLA-DQA2 | Esophageal mucosa | 2.4×10−13 | |
| Lung | 5.7×10−12 | ||
| Exposed skin | 5.0×10−11 | ||
| Transverse colon | 1.1×10−09 | ||
| Sigmoid colon | 2.8×10−06 | ||
| Unexposed skin | 4.8×10−06 | ||
| Stomach | 5.5×10−06 | ||
| PRRT1 | Esophageal mucosa | 6.1×10−11 | |
| Exposed skin | 2.8×10−06 | ||
| Transverse colon | 2.1×10−05 | ||
| HLA-DQB2 | Lung | 5.1×10−10 | |
| Transverse colon | 4.4×10−09 | ||
| Sigmoid colon | 4.8×10−07 | ||
| Stomach | 1.8×10−06 | ||
| HLA-DQA1 | Lung | 2.8×10−09 | |
| Exposed skin | 7.3×10−06 | ||
| Esophageal mucosa | 2.5×10−05 | ||
| NOTCH4 | Unexposed skin | 3.1×10−09 | |
| CYP21A2 | Exposed skin | 4.3×10−05 | |
| Unexposed skin | 6.6×10−05 | ||
| C4A | Exposed skin | 5.1×10−05 | |
| rs8176643 (9q34.2) | ABO | Esophageal mucosa | 1.2×10−06 |
| SURF1 | Exposed skin | 2.4×10−06 | |
| Transverse colon | 2.8×10−06 | ||
| Lung | 1.1×10−05 | ||
| rs1802575 (2p16.1) | EFEMP1 | Unexposed skin | 8.0×10−07 |
| Exposed skin | 2.0×10−06 | ||
| rs885932 (6p22.1) | ZFP57 | Exposed skin | 6.0×10−35 |
| Esophageal mucosa | 1.6×10−32 | ||
| Lung | 1.4×10−29 | ||
| Unexposed skin | 1.2×10−28 | ||
| Transverse colon | 4.0×10−24 | ||
| Small intestine (ileum) | 5.7×10−13 | ||
| Sigmoid colon | 5.7×10−11 | ||
| Vagina | 2.4×10−08 | ||
| HLA-K | Unexposed skin | 2.4×10−15 | |
| Transverse colon | 3.0×10−13 | ||
| Esophageal mucosa | 6.4×10−13 | ||
| Lung | 5.0×10−12 | ||
| Small intestine (ileum) | 2.0×10−06 | ||
| Sigmoid colon | 1.1×10−05 | ||
| HLA-A | Exposed skin | 1.4×10−13 | |
| Unexposed skin | 3.4×10−12 | ||
| Esophageal mucosa | 6.0×10−11 | ||
| TRIM31 | Unexposed skin | 8.2×10−12 | |
| Lung | 1.1×10−06 | ||
| Exposed skin | 4.5×10−06 | ||
| HLA-J | Lung | 5.4×10−11 | |
| Exposed skin | 3.9×10−09 | ||
| Transverse colon | 5.6×10−09 | ||
| Unexposed skin | 2.0×10−07 | ||
| Esophageal mucosa | 7.0×10−07 | ||
| Sigmoid colon | 1.4×10−06 | ||
| HLA-V | Lung | 2.7×10−08 | |
| Esophageal mucosa | 7.4×10−05 | ||
| HLA-T | Lung | 9.0×10−05 | |
| HLA-U | Unexposed skin | 2.1×10−05 | |
| Transverse colon | 4.9×10−05 | ||
| Esophageal mucosa | 1.1×10−04 | ||
| IER3 | Esophageal mucosa | 3.5×10−05 | |
| TRIM27 | Exposed skin | 1.1×10−04 | |
| RNF39 | Exposed skin | 1.2×10−04 | |
| rs3821170 (2q33.3) | LOC200726 | Heart (left ventricle)b | 1.2×10−07 |
| rs10497394 (2q31.1) | CDCA7 | Lung | 9.0×10−05 |
| rs2932989 (6q25.3) | FNDC1 | Esophageal muscle | 1.6×10−09 |
No significant single-tissue eQTLs in the GTEx database were identified for variants rs2406176 (21q21.1), rs4825724 (Xq24), rs7174062 (15q26.2), rs72931768 (11q13.3), rs35213789 (7q11.22), rs114947103 (7q22.3), rs13281988 (8q22.2), rs67035515 (3p21.31), rs73015965 (6q26) and rs151208372 (3q12.1). Variant rs5829676 (2p24.1) was not in the GTEx database.
No other mucosal or epithelial tissue identified for single-tissue eQTLs.
When the genes associated with eQTL variants are analyzed in NetworkAnalyst [29], significant pathways based on protein-protein interactions included the following: protein binding, MHC class II receptor activity, antigen processing and presentation, endocytosis, immune response, regulation of binding, regulation of TGF-β receptor signaling, pattern specification process, endoderm development, collagen and chromatin binding, chromatin remodeling, and regulation of Ras GTPase activity and of catabolic process. These results provide a glimpse of which pathways are important in the development of OM based on the genes that have been identified so far by GWAS.
3.2. Host Genomics in Animal Models for OM
Several animal models of OM have been developed over the years (Table 3). The chinchilla, Chinchilla lanigera, has long been used as model of OM because it recapitulates the human condition very faithfully with respect to pathogen detection [30] and the severity of disease induced by different strains of the same bacterial species [31]. Based on the demonstrated usefulness of this model, the ISOM community convinced the National Human Genome Research Institute to conduct a chinchilla genome project which culminated in the publication of a comprehensive chinchilla genomic resource [32] which is being exploited for host-pathogen interactions. The mouse has also become one of the favored models for genetic studies because of its lifespan, easy breeding and well-established methods for introducing genetic modifications. In addition, the similarities in auditory structure between mouse and human and the close evolutionary relationship of these two genomes, make the mouse a valuable model to study the genetics of hearing [52], although there are also limitations in inter-species comparisons, and comparison of syndromic to non-syndromic disease. In the past four years, mouse studies have identified several genes and genetic pathways involved in the predisposition to OME including the following examples.
Table 3.
Otitis media (OM)-related genes: animal studies 2014-2019
| Gene | Protein product | Known role in OM | Ref. |
|---|---|---|---|
| Bpifal | Antibacterial | Deletion increases OM susceptibility | 30 |
| Casp4 | Inflammasome effector | Inflammasome deficiency enhances OM | 31 |
| Ccl3 | Chemotactic chemokine | Deletion enhances OM | 32 |
| Ccr2 | Chemokine receptor | Deletion enhances OM | U |
| Cd44 | Hyaluronin receptor, multifunctional | Deletion alters leukocyte recruitment in OM | 33 |
| Celsr1 | Intercellular organization | Mutation causes OM | J |
| Coro1a | Lytic granule secretion | Mutation causes immunodeficiency and chronic OM | 34 |
| Dusp1 | MAP kinase phosphatase | Inhibits MUC5AC production in response to NTHi | 35 |
| Enpp1 | Transmembrane glycoprotein | Mutations cause OM with ectopic bone formation | 36 |
| Entpd1 | ATP hydrolase | Predicted to increase chronic OM | A |
| Fbxo11 | Transcription factor | Mutation causes a middle ear cavitation | 37 |
| Fli1 | Transcription factor | Haploinsufficiency causes OM | 38 |
| Grn | Granulin growth factor precursor | Deletion reduces bacterial clearance in OM | 39 |
| Hbegf | Epithelial growth factor | Stimulates middle ear epithelial growth | 40 |
| Il1rn | IL1 receptor antagonist | Deletion exacerbates OM | U |
| Mapk9 | JNK isoform 2, gene regulation | JNK2 mutation exacerbates OM | 41 |
| Mif | Pro-inflammatory mediator | Blocking MIF alleviates OM | 42 |
| Mkp1 | MAPK phosphatase | Upregulation reduces MUC5AB production in OM | 35 |
| Ncf2 | Superoxide source in neutrophils | Predicted to increase chronic OM | A |
| Nfkb1 | Immune, growth signaling | Deletion enhances OM | U |
| Nisch | Cell signaling | Mutation causes OM | 43 |
| Nlrp3 | Pathogen receptor, inflammasome | Inflammasome deficiency enhances OM | 31 |
| Nod1 | Pathogen receptor | Mutation exacerbates OM | U |
| Nod2 | Pathogen receptor | Mutation reduces bacterial clearance in OM | 44 |
| Pai1 | Plasminogen inhibitor | Deletion exacerbates OM | 45 |
| Pax9 | Growth-related transcription factor | Down-regulation leads to OM | 46 |
| Pycard | ASC inflammasome component | Deletion enhances OM | 31 |
| Ripk2 | Pathogen receptor signaling | Deletion disables OM recovery | U |
| Spag6 | Cilia gene | Deletion causes OM | 47 |
| Tbx1 | Transcription factor | Mutation alters facial morphology, increases OM incidence | 22 |
| Tlr2 | Pathogen receptor | Deletion exacerbates OM | 48 |
Abbreviations: U, unpublished observations from D.G. Hur, B. Nuyen and A. Kurabi; J, phenotype from Jackson Labs; NTHi, non-typeable Haemophilus influenzae; A, ARCHS4 database
FLI1 and ETS1 are transcription factors from the ETS (E26 transformation-specific) family, known to be hemizygous in Jacobsen syndrome. It has been demonstrated that these genes have a role in the development of the nose, middle ear cavity and ossicles. The Fli1+/− and Ets1+/−/Fli1+/− mice exhibit hearing impairment associated with COM, inflamed middle ear epithelium, abnormally small middle ear cavity, fusion of ossicles to the middle ear wall, and deformed stapes [41].
Mouse mutants called asj (ages with stiffened joints), which have a point mutation in the Enppl gene, were also reported to have conductive hearing loss. At six weeks of age, the mutants exhibited effusion in the middle ear, thickened middle ear epithelium, impaired Eustachian tube function due to epithelia proliferation, fusion of the malleus and incus, and calcification of middle ear structures [39].
Mutations in EDA, EDAR and EDARADD genes have been described as triggering HED (hypohidrotic ectodermal dysplasia). Eda- (Ectodysplasin a) and Edar- (Ectodysplasin a receptor) mutant mice (EdaTa and EdardlJ/dlJ) were reported to develop OM, rhinitis and nasopharyngitis. They also display reduced mucociliary clearance and the loss of glandular secretions [53].
Studying the ENU mutant edison revealed that a point mutation in the Nisch (Nischarin) gene results in the development of conductive hearing loss due to COM. Homozygous edison mice spontaneously develop COM as early as three weeks of age. The mutants also demonstrate mild craniofacial defects, middle ear cavities filled with fluid and lined with thickened mucoperiosteum, polypoid growths into the middle ear cavity, and in the more severely affected mice inflamed tympanic membranes. Furthermore, an investigation of the impact of the edison mutation on pathways in which Nischarin is involved in has implicated LIMK1 and NF-κB pathways in the development of COM [46].
BPIFA1 is a member of the bacterial permeability-increasing (BPI) fold containing family of putative innate defense proteins and is one of the most abundant secretory proteins in the upper respiratory tract. Bpifa1-knockout mice do not develop spontaneous OM up to six months although BPIFA1 is highly expressed in the middle ear epithelium. However, deletion of Bpifa1 in Junbo mice, one of the first models of COM, results in significant exacerbation of the phenotype including thickening of the middle ear mucosa and increased collagen deposition. This finding indicates a role for BPIFA1 in mucosal protection [54].
Down syndrome is caused by an extra copy of some or all of the genes of human chromosome 21 (Hsa21). The orthologs to the genes from Hsa21 are spread among three regions on the mouse genome located on chromosomes 10 (Mmu10), 16 (Mmu16) and 17 (Mmu17). Ongoing study at the MRC Harwell Institute on the phenotype of the DpTyb mice, which are mouse models of Down syndrome, revealed that mice with full duplication of the genes from Mmu16 have conductive hearing loss due to the development of middle ear inflammation. A detailed study on the OM phenotype of mouse strains with full duplication of the genes from Mmu10 and 17 and with duplication of smaller segments of the Dp1Tyb region will provide a better understanding of the genes and genetic pathways involved in the development of OM in Down syndrome.
3.3. Advances in Microbiome Analyses
Although viruses are known to play a role in OM, genomic studies on viruses in the middle ear were not conducted during the study period. Thus this section concentrates on bacterial genomics.
3.3.1. New technologies for 16S rRNA sequencing for OM bacterial pathogens
Recently an improved pan-bacterial domain molecular diagnostic (MDx) that uses PCR and circular consensus reads (CCS) of the entire bacterial 16S rRNA (FL16S) gene on Pacific Bioscience (PacBio) 3rd-4th generation sequencers has been developed [55,56]. The process incorporates both laboratory and informatic improvements over extant systems. Briefly, CCS provides for multiple polymerase-driven sequencing-read-passes of a single DNA molecule, where each ‘pass’ of the molecule corresponds to the sequence of sense or antisense strand. Each pass is used towards the creation of a consensus sequence, generating ultra-high-quality sequences (as individual PacBio errors are not sequence context dependent) providing for unambiguous species calls when paired with the companion informatics system, called MCSMRT. This pipeline for quantitatively and specifically profiling microbiota includes a custom full-length 16S gene database. Beginning with PCR of the entire 16S rRNA gene the resultant amplimers are then sequenced on a PacBio sequencer. CCS read sequences are calculated based on average >15 passes resulting in expected error rates of ≤1 incorrectly called base per FL16S gene read in over half the data. MCSMRT has extended and improved upon previous applications of PacBio 16S sequencing [57–64], including: (A) incorporation of stringent filters eliminating sources of sequencing artifacts, effectively eliminating inflated operational taxonomic unit (OTU) counts, a pervasive problem in short read microbiome protocols [65]; (B) assignment of taxonomy and confidence values to OTUs using the custom-designed full-length 16S database which provides a uniform Linnaean hierarchy for each read. The combination of these factors resulted in a pan-domain MDx that is highly quantitative, even for complex microbiota. Analysis of the Critical Assessment of Metagenome Interpretation (CAMI) community (>250 species) demonstrated its ability to identify and determine abundances of hundreds of species over >3 logs of variance (Fig. 1) [55].
Fig. 1.
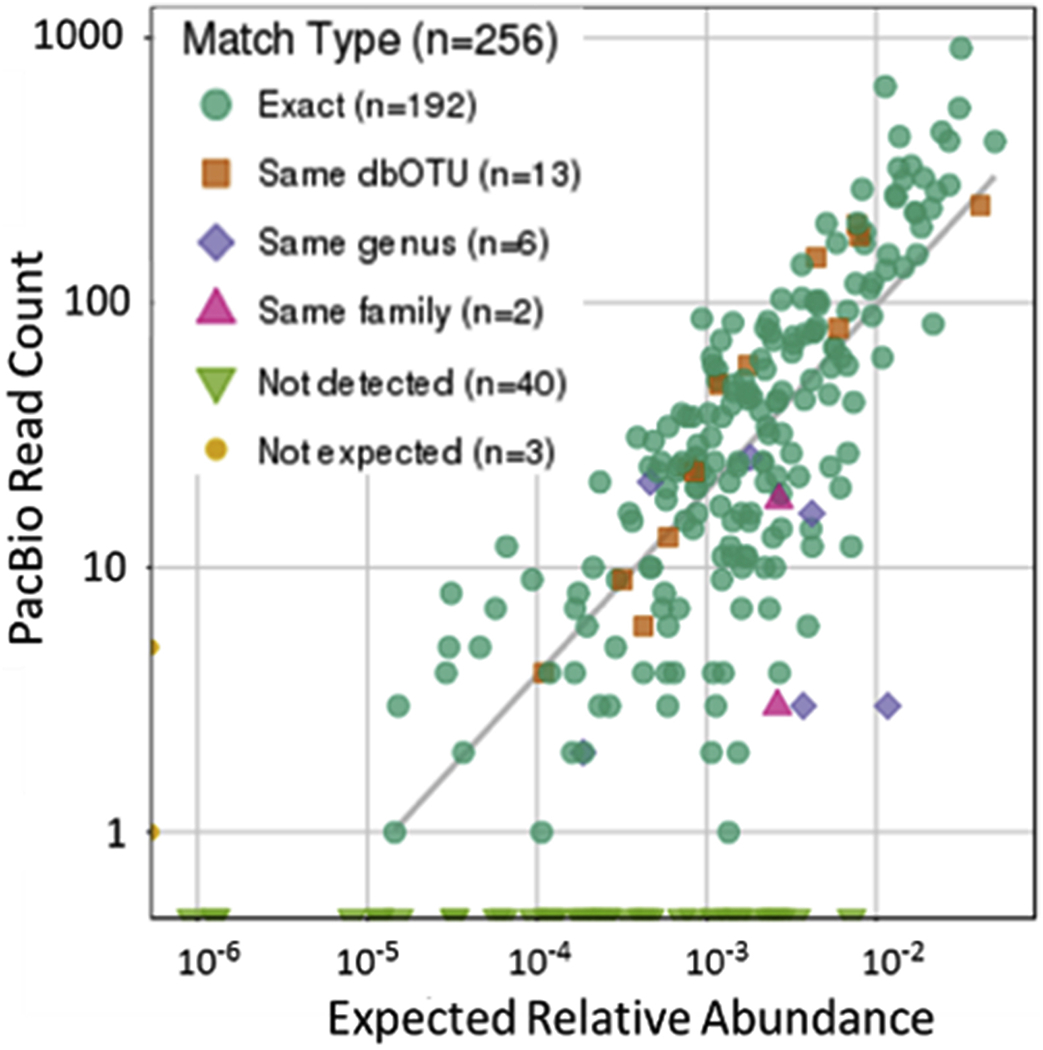
Analysis of the Critical Assessment of Metagenome Interpretation community (>250 species) demonstrated the ability of MDx to identify and determine abundances of hundreds of species over >3 logs of variance. Reproduced from reference 52 which is an author owned paper
Two recent innovations that improve the accuracy and ability to discriminate among closely related species and strains have been added to MCSMRT: (A) upon ascertaining that ~5% of the 16S amplimers were hybrids of mismatched, closely related single-stranded DNAs (from closely related species, or different intragenomic copies), the ability to produce CCS reads derived from each strand was added as an option; and (B) a new, more accurate algorithm for calling sequences based on graph deconvolution, as opposed to a multiple sequence alignment strategy was added. This latter technique builds on the algorithm introduced in 2002 based on a directed acyclic graph [66]. Using these algorithmic improvements, the quality of even the unfiltered CCS reads substantially improved from an average of 8.0 to 2.2 expected errors, and resulting in improved post-filter yields, improving from 86% to 98% recovery.
Informatic analyses have seen additional recent advances. Significant challenges in assessing differential abundance of microbial taxa exist due to unbalanced sampling, and artifacts from both PCR and sequencing. Traditional methods (such as rarefaction) have been shown to be untenable for accurate statistical comparisons within microbiome data [67]. New analysis software has been designed that properly normalize OTU counts across samples using Gaussian mixture models, and have been added to popular R microbiome analysis library frameworks, such as phyloseq [68–70]. OTU representation of the microbiome (where sequences are usually clustered together based on an identity threshold of 97%) have also been recently challenged as inadequate to properly account for all microbial diversity within a sample [71]. Recent programmatic packages now exist that can be applied after OTU analysis (such as that done in MCSMRT) to further examine diversity based on single nucleotide variants within the OTU sequences [72,73].
Another recent advancement with strong implications for a FL16S microbiome pipeline is the development of a kit by the company Shoreline Biome (Farmington, CT, USA), created specifically to go from sample to prepared PacBio sequencing libraries in a series of predefined laboratory steps. The advantages of this pipeline are a pre-designed system for cell lysis, PCR amplification of FL16S, and sample pooling of up to 100 samples per run. This kit streamlines the process resulting in a significant reduction of man hours required for sample library preparation (estimated reduction from 16-25 hours to ~2 hours). Taken together, these exciting new developments in microbial sequencing technologies are expected to have a major impact in scientific discoveries and novel clinical applications in otopathogen detection and characterization.
3.3.2. Application of Novel Techniques in Microbiome Studies on COM with Effusion
The above-mentioned microbiome MDx was used to characterize 20 bilateral COM with effusion cases to investigate their polymicrobial nature (Fig. 2). Right and left middle ears typically had similar communities though exceptions existed (patients 12, 27 and 32 in Fig. 2). The major otopathogens causing AOM are NTHi, Streptococcus pneumoniae (Spn) and Moraxella catarrhalis (Mcat). These OM pathogens were prevalent among the chronic OM patients (6 Mcat, 4 NTHi, 3 Spn: 55% of cases with 1+), but under-recognized species were more prevalent: 40% Alloiococcus otitis, 40% Staphylococcus auricularis, 30% Turicella otitidis, 30% Staphylococcus caprae, and 10% Aggregatibacter aphrophilus [74]. The extremely high abundances of these “atypical” species in all cases suggest that they are likely OM etiological agents, and not simply contaminants from the external auditory canal.
Fig. 2.
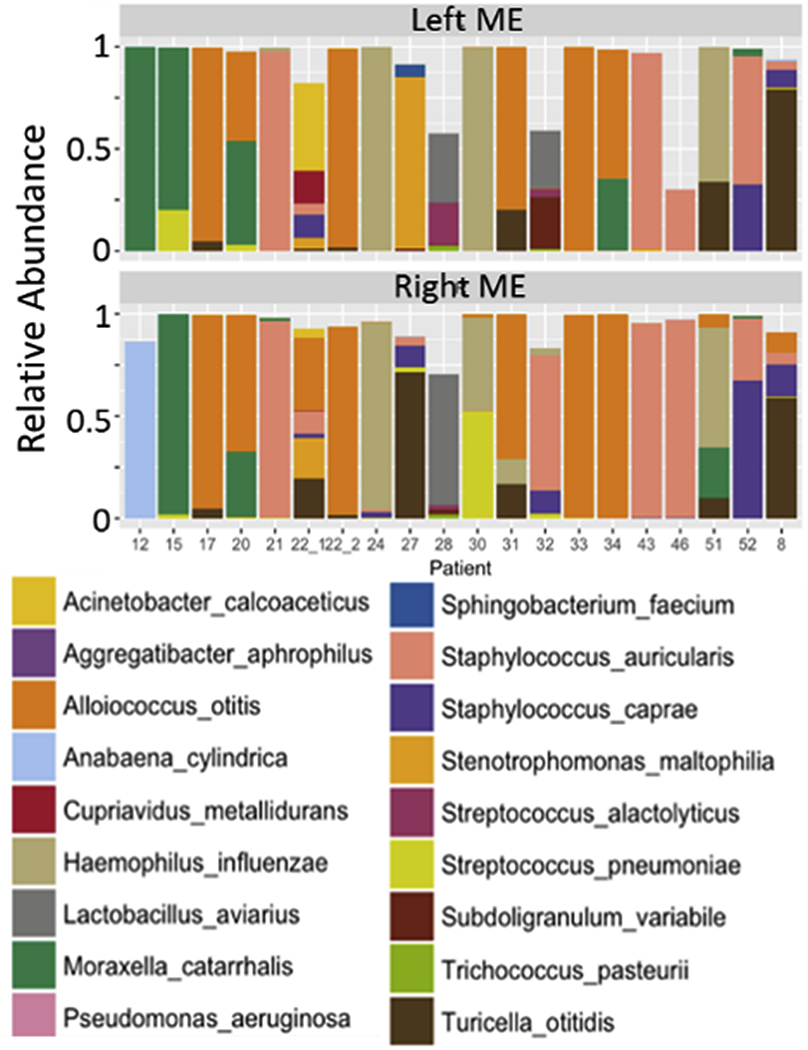
Twenty bilateral microbiomes for chronic otitis media with effusion. The top 18 most prevalent species are shown as colored bars. Reproduced from reference 52 which is an author owned paper
3.4. Bacterial population genomics to dissect otopathogen-specific virulence genes and mechanisms
Whole-genome shotgun (WGS) sequencing of clinical bacterial isolates is rapidly revolutionizing clinical microbiology, not only by increasing the throughput and resolution of strain typing but also enabling detection of associations between bacterial genetic variation and clinical pathogenesis phenotypes [75–78]. Bacterial supra-GWAS (SGWAS) presents special challenges [79–81], including inadequate or biased sampling (especially of carriage isolates), limited clinical metadata availability, strong phylogenetic substructure and genomic (gene content) diversity among strains, and a lack of consensus among varied bioinformatic analysis pipelines. Nevertheless, increasingly large datasets and sophisticated tools [82–84] are now detecting signatures of in-host bacterial evolution by identifying recurrent and parallel genomic changes arising over the course of infections or between healthy and diseased states. Isolating these in situ genomic signatures of bacterial transitions to pathogenesis (“pathoadaptations”) in turn informs our understanding of pathogenic mechanisms in the human host [85–87]. Combining comparative genomic analysis with transcriptomic and other -omic technologies to investigate in vivo and in situ host-pathogen interactions will allow the integration of clinical and molecular mechanistic studies of bacterial pathogenesis.
This emerging discipline of “genomic epidemiology” has been broadly applied to bacterial pathogens, but has only begun to be used to dissect OM pathogenesis. The three main bacterial otopathogens NTHi, Spn and Mcat are all amenable to large-scale SGWAS [88–105], a technique that takes into account that the majority of bacterial species’ genes are not found in all strains of the species, but are rather distributed among the many strains. Indeed, all three organisms have many publicly available WGS assemblies including complete reference genomes at NCBI (as of June 2019: Sp=10,462 assemblies, NTHi=735, Mcat=114), and additional genomes are continuing to accrue from diverse sources and disease states. In contrast to studies of environmental opportunistic pathogens that typically colonize subjects as single clones that subsequently evolve and diversify (e.g. Pseudomonas and Burkholderia cenocepacea in cystic fibrosis infections) [106,107], population-level genomic studies in human-restricted normal commensals—like the three main otopathogens—are complicated by the high diversity and polyclonality of these species when isolated from diverse health and disease states. Below, we summarize recent WGS surveys and association studies from NTHi. Although none have yet been specifically focused on OM diseases, the findings suggest that our understanding of OM pathogenesis could be greatly expanded by GWAS and other -omics approaches.
H. influenzae became an early genomic model system when the lab strain Rd became the first cellular organism to have its complete genome sequenced in 1995 [108], and the pace of WGS has continued to increase, with 735 assemblies publically available at NCBI (as of June 7, 2019) and >1,000 more expected in short order. Early and continuing comparative analysis of NTHi genomes were among the first to recognize the immense diversity in genic content seen in many bacterial species [88,89,99,100]. More recently, SGWAS studies identified gene presence/absence differences strongly associated with strains isolated from carriage or disease [98], and follow-up studies identified a novel multi-gene family associated with virulence, the vSLR genes, one of which was shown to increase survival in macrophages (msfl) and also enhancing the ability of OM that increases the severity of disease in the chinchilla model of OM [102]. Recent studies of NTHi in chronic obstructive pulmonary disease (COPD) have revealed recurrent genomic changes accumulating over time in different patients, particularly at phase variable genes encoding in lipooligosaccharide synthesis enzymes and outer membrane proteins [103,104]. This also identified recurrent loss-of-function mutations in the bifunctional ompPl/fadL gene as a specific NTHi patho-adaptation to the COPD lung, and also showed that this is a likely evolutionary trade-off between ompP1 mutants’ losing their ability to adhere and invade airway epithelial cells, but gaining resistance to arachidonic acid, an abundant inflammatory mediator in the COPD lung [103]. Similar work with a small number of strains in pediatric pulmonary infections found limited evidence for parallel evolution but nasal-lung isolate pairs also showed convergent transcriptional signatures by in vitro RNA-seq [105]. Additional SGWAS has been been performed for population-level and epidemiological analysis of the NTHi, including new invasive isolates from the US [106]and Portugal [107], as well as a detailed analysis of 265 strains collected in the UK that identified lineage-specific associations with pneumococcus and shifts in NTHi population structure in carriage upon introduction of the PCV13 vaccine [108]. These results all show the power of comparative analysis of bacterial genomes.
Excitingly, recent technical and informatic advances in metagenomic sequencing together with novel cultural protocols provide for the systematic capture of much deeper and more complete views of highly complex polyclonal and polymicrobial populations while providing for the near complete genome sequencing and assembly of both very rare strains and strains that are recalcitrant to culture. These advances provide for deep, strain-traceable, metagenomic/WGS characterization of polyclonal populations in situ – when combined with a multipartite specimen collection approach to provide comprehensive strain-, gene-, and pan(supra)genome-based analyses of any clinical sample -- and particularly useful for following over time the evolution of clonal lineages and inter-strain population dynamics. A comprehensive approach to understanding these issues with respect to the evolution of otopathogenesis at the population-level, particularly with regard to horizontal gene transfer includes: (A) WGS of large numbers of individual isolates recovered simultaneously from middle ear effusions and the nasopharynx; (B) the metagenomic, and ligation-proximity-metagenomic sequencing of all recovered colonies from each specimen that have been pooled to form a ‘gimish’ [109,110] to construct genomes/clonal lineages of lower prevalence strains; (C) metagenomic sequencing of uncultured clinical specimen DNA that has been selected using a species-specific capture reagent [98] to provide coverage for unculturable strains; and (D) the use of gel microdroplets (GMD) to encapsulate single cells for the production of sequestered clonal microcolonies grown in coculture which can provide sufficient genomic DNA for whole genome amplification (MDA) and subsequent genomic assemblies [111,112]. Selection of individual GMD microcolonies for sequencing is achieved via FACS using labeled probes made from metagenomically identified genes that are not present in the strains which underwent WGS.
3.5. Epigenetic regulation in bacterial otopathogens
Many bacterial pathogens that are adapted to the human host display random, high-frequency on/off switching of gene expression, called phase variation [113]. This gene regulation strategy generates a diverse bacterial population that provides many possible solutions to the challenges posed by distinct immunological memories. In most cases, the phase variably expressed genes encode surface exposed antigens such as outer membrane proteins and variable oligosaccharide structures. Two of the major otopathogens H. influenzae [114] and Mcat [115] are archetypal examples of bacterial pathogens that exhibit phase variable gene expression and have many phase variable genes.
In 2005 a new type of epigenetic regulation system called a phase variable regulon (phasevarion), was described in H. influenzae [116]. Phasevarions are controlled by DNA methyltransferases that randomly switch on and off, leading to changes in DNA methylation at thousands of sites in the genome and consequent changes in global gene expression [117]. Both H. influenzae [118] and Mcat [119] contain multiple phasevarion systems that control virulence and immune evasion. In the case of H. influenzae, the impact of phasevarion switching in OM has been confirmed in the chinchilla model [118]. Spn also has a DNA methyltransferase that randomly switches between six different DNA methylation patterns by recombination that regulates virulence in a murine model system and immune evasion [120].
Studies in in vitro model systems indicate the potential importance of phasevarions in the pathogenesis of all major bacterial otopathogens causing OM [118–120]. Further studies on human clinical samples are required to assess the impact of these systems in regulating the adaptation of the pathogen to the human host in distinct phases of disease and distinct host niches, e.g. to address the question of a potential role for phasevarion-mediated regulation of the transition from nasopharyngeal colonization to active OM infection.
The epigenetic regulation of phasevarions is mediated by changes in methylation at thousands of sites in the genome. The vast majority of these changes are neutral with respect to impact on gene expression. Thus, it is not possible at this time to conduct in silico prediction of which genes are subject to expression changes upon phasevarion switching. The inability to predict which genes will be subject to phasevarion regulation presents particular problems for genomics-based vaccine development [121] for these pathogens. Detailed studies of phasevarion-mediated gene expression are required in all three common AOM pathogens to define the repertoire of stably expressed immunological targets.
3.6. Precision Medicine for OM
Precision Medicine (PM), also called personalized medicine, is designed to treat the right patient, at the correct time, with the right treatment strategy. This takes into consideration the patients’ genetic and environmental factors, including the molecular basis of disease. This strategy has been implemented in cancer treatment for some time [122–125]. Studies on primary immune deficiency have revealed several hundred genetic defects associated with various disorders, helped understand immunobiology and resulted in targeted new therapies [125]. For example, cystic fibrosis is a monogenic disease, but more than 2000 different mutations in the CFTR gene have been reported. Targeted therapies for some of these mutations, e.g. for carriers of the CFTR p.Phe508del variant, are already on the market [126].
Application of PM in infectious diseases is much more complex than in many other medical fields and, combined with the lack of new efficacious antibiotics to deal with established bacterial resistance to traditional antibiotics [127–130], makes for a very challenging environment for the adoption of PM. For OM, the recognition that the expression of particular allelic forms of the gel-forming mucins and aquaporins are specifically linked to pediatric hearing loss [131] provides the potential for targeted therapeutic interventions. Similarly,the recent identification of specific host microRNAs that are associated with OM [132] (mir-223) and the hyperplastic response associated with COME [133] (mir-146) provide high value targets for future interventional strategies Treatment strategies for PM in infectious diseases include pathogen-targeted antibiotic treatment, adjuvant therapy to antibiotics, precision antimicrobial therapies, and subclassification of patient groups according to intrinsic and extrinsic factors.
Bacterial culture and antimicrobial sensitivity tests are available but are slow to perform and therefore microbial WGS to find resistant strains is evolving [130,134]. WGS can be used to select an appropriate and efficacious medication, reducing side effects and excluding toxic drugs. PM based on the bacterial strain is possible in OM with suppuration from tympanic membrane perforation or tympanostomy tube discharge.
PM in infectious diseases also includes developing non-antibiotic strategies. Precision antimicrobial treatment can target certain virulence factors in pathogens, whether a gene, a cellular process, or a specific microbe. These strategies also leave the rest of the human microbiota intact. An unbalanced microbiota is associated with a wide range of disorders, from autoimmune to psychiatric diseases. Precision antibiotic treatments include lysins, nucleic acid-based systems, synthetic peptides, and mannosides. One strategy in OM would be to identify genes that favor colonization of OM pathogens in the middle ear and to develop small antagonists against molecules produced by bacteria [127,128,135].
The term -omics comprises genomics, transcriptomics, proteomics, metabolomics, and microbiomes, while translational -omics refers to clinical use of multi-omic data to treat disease in a precise way. It enables the definition of different endotypes of a phenotype or trait. In a Cochrane review of antibiotic treatment in OM, an example of an endotype that benefits most from antibiotic treatment is defined as “children under two years of age with bilateral AOM, or with both AOM and otorrhea” [136]. Such endotypes are used to identify patients who will benefit from different treatment strategies with fewer side effects [127,137].
3.7. OM Therapeutics
Understandably translation of -omics findings to therapeutic use takes a long time. Recent studies on OM therapeutics have focused on steroids and antibiotics, which are not –omics-guided and mostly with negative results. Francis et al. (2018) performed a randomized trial of oral steroids for treatment of hearing loss in OM, and found no effect [138]. Ranakusuma et al. (2018) reviewed studies of oral corticosteroids for AOM, and found no evidence of effectiveness in reducing duration of symptoms [139]. Lewnard et al. (2018) found that long-term amoxicillin treatment did not increase the carriage of resistant Spn strains [140]. Ruohola et al. (2018) noted that immediate amoxicillin treatment for AOM did not enhance clearance of later middle ear effusion [141]. Hoberman et al. (2016) found that a shortened antibiotic regimen was not as effective as a full course for the resolution of OM [142]. Te Molder et al. (2016) found that antibiotic treatment for a first OM episode does not affect the probability of future recurrences [143]. While these studies were negative, two other studies that had positive findings showed that for children less than two years old, immediate amoxicillin was the most cost-effective treatment for AOM [144], while immediate amoxicillin treatment was modestly more effective in treating OM than delayed antibiotics [145]. Outside of steroids and antibiotics, only one study by Kondura et al. (2016) found that circumin reduced inflammation in an animal model of OM [38].
Regarding delivery of drugs to the middle ear, Yang et al. (2016) found that delivery of ciprofloxacin and penetrants in a hydrogel to the external surface of the intact tympanic membrane resulted in delivery of therapeutic amounts of drug to the middle ear [146]. Kurabi et al. (2017) discovered rare peptides that support active transport of large particles across the intact eardrum, and enhanced transport by lengthening the peptides [147,148]. They also found that the peptides can transit the intact human tympanic membrane [149].
3.8. Vaccination for OM towards PM
3.8.1. Pneumococcal vaccination
At onset of AOM, Spn (58%) was the predominant organism in the nasopharynx followed by Mcat (55.1%) and then H. influenzae (38.2%) [150], which shows low prediction value of nasopharyngeal cultures to determine the etiology of middle ear bacterial pathogens and the importance of collection of middle ear fluid to identify vaccines for AOM prevention. The only current licensed vaccines targeting Spn are pneumococcal conjugate vaccines (PCVs). These vaccines contain 10 (PCV10) or 13 (PCV13) polysaccharide Spn serotypes conjugated to a protein carrier. PCVs are effective in reducing AOM caused by the serotypes contained within the vaccines [151–154]. In addition, administration of PCVs during infancy may reduce the risk of recurrent AOM and progression to more complex disease caused by non-vaccine serotypes [155,156]. However, increases in both nasopharyngeal carriage and diseases caused by non-vaccine Spn capsule types are on the rise [157]. This has resulted in vaccine companies developing newer PCVs such as PCV15 (Merck) [158–160] and PCV20 (Pfizer). PCV15 contains the same 13 serotypes as in PCV13 plus 22F and 33F. PCV20 contains same serotypes as in PCV15 plus 8, 10A, 11A, 12F and 15B/C. Both PCV15 and PCV20 are in Phase 3 clinical studies in adults.
Although these newer PCVs will help to reduce the incidence of diseases caused by the additional vaccine serotypes, history tells us that non-vaccine serotype replacements will occur over time. Hence, there is a need to develop non-serotype dependent pneumococcal vaccines. Efforts have been made formulating pneumococcal protein-based vaccines either alone or in combination with PCVs [160,161]. However, none of these vaccines moved to Phase 3 clinical trials because they failed to reduce nasopharyngeal colonization similar to PCVs and the indirect herd immunity effect of PCVs is sought for overall public health benefit. Another approach being tested is a whole cell vaccine (WCV) that generates both antibody and Th1/Th17 cellular immunity thereby offering the potential to prevent Spn carriage [162,163]. A genetically engineered unencapsulated killed strain of Spn that elicits IL-17A production is in clinical trials [164,165]. Additional preclinical WCVs using different engineered strains and different routes of immunization (intraperitoneal or intranasal) are being studied [166,167].
3.8.2. Vaccination against H. influenzae
The only licensed vaccine is the H. influenzae serotype b (PRP) polysaccharide conjugate vaccine. This vaccine is not effective against other H. influenzae capsule types that cause invasive disease nor NTHi which, aside from being a major AOM pathogen, is a major pathogen causing COPD and invasive diseases in infants and young children [168–170]. PCV10 vaccine contains protein D from H. influenzae conjugated to eight of the pneumococcal polysaccharides. In a pediatric clinical trial using an earlier version of PCV10 and PCV11 where all 11 polysaccharides were conjugated to protein D, a significant reduction in NTHi-caused AOM was measured [171]. Subsequent clinical and observational studies using PCV10 showed marginal efficacy against NTHi-caused AOM and had no impact on nasopharyngeal carriage of H. influenzae [172,173].
Current strategies for new vaccines to prevent H. influenzae infections are being explored and are focused on relatively conserved proteins expressed by the bacteria [174–176]. Two phase 1 clinical trials investigated a multi-component H. influenzae protein-based vaccine and a multi-component combination Spn and H. influenzae protein-based vaccine [177,178]. Another Phase 1 trial investigated a multi-component combination H. influenzae and Mcat protein-based vaccine to target subjects with acute exacerbations of COPD [179]. Oral vaccines using inactivated NTHi to prevent acute exacerbations of COPD have been in clinical studies [180–182]. A novel preclinical study using a transcutaneous immunization of a chimeric protein with a band aid prevented OM in chinchillas [183]. Animal studies mimicking the natural human mode of AOM as a result of nasal coinfection of virus and NTHi show promise in identifying vaccine candidates [184]. An outer membrane vesicle of NTHi has been shown to be protective against OM in a chinchilla model [185].
3.8.3. Vaccination for Meat
There are no licensed vaccines against Meat. One of the major issues in developing a vaccine against OM is lack of an animal model due to Meat being a human-restricted pathogen [186]. Mouse pulmonary clearance and chinchilla nasopharyngeal colonization models have been used for testing Mcat vaccine candidates. A number of protein and lipooligosaccharide candidates have been studied preclinically [186,187]. Only one investigational clinical study of a vaccine containing three NTHi proteins and one Mcat protein has been reported targeting acute exacerbations of COPD [179].
3.8.3. Vaccination against Otopathogens within the Context of PM
From a PM perspective pneumococcal vaccines should be effective in the population which experience the greatest disease burden, e.g. otitis-prone children. Otitis-prone children have multiple dysfunctions in adaptive immunity including responses to vaccines [188–194]. In particular, otitis-prone children respond with significantly lower antibody levels and lower generation of memory B cells following PCV13 vaccination [188,194]. They respond to natural immunization induced by nasopharyngeal colonization and AOM infections with less antibody against pneumococcal proteins which are considered as vaccine candidates [192,195,196]. Otitis-prone children also have diminished Th1 and Th17 immune responses [193] and generally have poor response to most routine vaccination [188]. Similar to the response to pneumococcal vaccines, otitis-prone children respond less well to H. influenzae type b vaccine [188], as well as to several proteins expressed by H. influenzae and Mcat that are vaccine candidates [195,197,198].
4. Implications for Practice
Despite the considerable morbidity and health care costs for OM treatment, a barrier that prevents development of new vaccines and treatments for OM including those arising from knowledge brought forth by new genomic technologies is the lack of appreciation of OM as a major public health threat, particularly by industry. Genetic diversity among OM pathogens is another major challenge that must be addressed. From a public health perspective vaccines targeting otopathogens would be most cost-effective if they not only reduced OM but also reduced nasopharyngeal carriage to produce an indirect herd immunity effect. However, for any vaccine, is total elimination of nasopharyngeal carriage risky? Might elimination of potential otopathogens open a niche for other pathogenic organisms? Some scientists recommend that elimination of nasopharyngeal colonization is not desirable, and rather enhancing the host immunity to prevent bacteria from reaching a pathogenic threshold should be the goal [160]. Eliciting mucosal responses by formulating vaccines to enhance Th17 responses or by mucosal immunization is a strategy under investigation. Combination vaccines targeting Spn, NTHi and Mcat would be the preferred formulation to target OM and other diseases caused by these pathogens. Modulating the microbiome by monitoring bacterial abundances not just of OM pathogens but also of the commensal middle ear bacteria is another potential goal for OM prevention.
The increasing pace of large-scale WGS of clinical bacterial isolates and novel insights into bacterial population dynamics and pathogenesis offer great promise to dissect how bacteria adapt during pathogenesis, pointing to the molecular mechanisms important within the human host. The methods and tools are now available to gain direct insights into the specific bacterial genomic variation that impacts OM. In combination with carefully collected and curated clinical isolate collections, applying these analyses to bacterial otopathogens will not only improve epidemiological tracking but also dissecting bacterial pathoadaptation and virulence mechanisms. Increasing use of functional approaches, like transcriptome and metabolome analyses [199,200] —applied in a comparative context—will help inform new disease management strategies, identify new diagnostic biomarkers, and suggest new routes for therapeutic intervention.
In the near future, human GWAS using next-generation sequence data will further illuminate how coding and non-coding variants with a wide MAF spectrum, i.e. both common and rare variants, play a role in OM susceptibility. Further studies in non-European populations are also needed to elucidate OM susceptibility variants that are important in various human ethnic groups. In addition there continue to be many advances in the identification of genes that play a role in OM from animal studies, virtually all in the mouse. These include natural and ENU-induced mutations, as well as studies of knockout and other gene-modified mice. The categories that influence OM include genes related to immunity, inflammation, secretory activity, morphology, and tissue growth. This diversity of OM-related pathways suggests that many more such OM-related genes will be discovered in human and mouse studies, which are important to understand mechanism of disease in OM and to determine pathways that may be targeted for treatment and prevention. Collaboration between disciplines and across populations is mandatory in the development of PM, where research and development in translational -omics will be the cornerstone.
Acknowledgments
Funding: Funding for the generation and publication of this panel report was made possible in part by 1 R13 DC017389-01 from the National Institute on Deafness and Other Communication Disorders.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: Dr. Allen F. Ryan is a Co-founder of and equity holder in Otonomy, Inc., a company that develops slow-release compounds for local delivery of medications to the middle and inner ear. The authors declare no competing interests.
References
- 1.Subcommittee on Management of Acute Otitis Media, Diagnosis and Management of Acute Otitis Media, Pediatrics 113 (2004) 1451–1465. [DOI] [PubMed] [Google Scholar]
- 2.Lieberthal AS, Carroll AE, Chonmaitree T, Ganiats TG, Hoberman A, Jackson MA, Joffe MD, Miller DT, Rosenfeld RM, Sevilla XD, Schwartz RH, Thomas PA, Tunkel DE, The diagnosis and management of acute otitis media, Pediatrics 131 (2013) e964–e999. [DOI] [PubMed] [Google Scholar]
- 3.Laine MK, Tahtinen PA, Ruuskanen O, Huovinen P, Ruohola A, Symptoms or symptom-based scores cannot predict acute otitis media at otitis-prone age, Pediatrics 125 (2010) e1154–e1161. [DOI] [PubMed] [Google Scholar]
- 4.Pichichero ME, Poole MD, Assessing diagnostic accuracy and tympanocentesis skills in the management of otitis media, Arch. Pediatr. Adolesc. Med 155 (2001) 1137–1142. [DOI] [PubMed] [Google Scholar]
- 5.Balasundaram N, Phan D, Mazzoni D, Duong E, Sweeny A, Del Mar C, Keijzers G, Acute otitis media in children presenting to the emergency department: Is it diagnosed and managed appropriately? J. Paediatr. Child Health (2019). doi: 10.1111/jpc.14414. [DOI] [PubMed] [Google Scholar]
- 6.Pichichero ME, Poole MD, Comparison of performance by otolaryngologists, pediatricians, and general practioners on an otoendoscopic diagnostic video examination, Int. J. Pediatr. Otorhinolaryngol 69 (2005) 361–366. [DOI] [PubMed] [Google Scholar]
- 7.Paul CR, Gjerde CL, McIntosh G, Weber LS, Teaching the pediatric ear exam and diagnosis of Acute Otitis Media: a teaching and assessment model in three groups, BMC Med. Educ 17 (2017) 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pichichero ME, Diagnostic accuracy of otitis media and tympanocentesis skills assessment among pediatricians, Eur. J. Clin. Microbiol. Infect. Dis 22 (2003) 519–524. [DOI] [PubMed] [Google Scholar]
- 9.Legros JM, Hitoto H, Garnier F, Dagorne C, Parot-Schinkel E, Fanello S, Clinical qualitative evaluation of the diagnosis of acute otitis media in general practice, Int. J. Pediatr. Otorhinolaryngol 72 (2008) 23–30. [DOI] [PubMed] [Google Scholar]
- 10.Pichichero ME, Casey JR, Almudevar A, Reducing the Frequency of Acute Otitis Media by Individualized Care, Pediatr. Infect. Dis. J 32 (2013) 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einarsdottir E, Hafren L, Leinonen E, Bhutta MF, Kentala E, Kere J, Mattila PS, Genome-wide association analysis reveals variants on chromosome 19 that contribute to childhood risk of chronic otitis media with effusion, Sci. Rep 6 (2016) 33240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA, Detection and interpretation of shared genetic influences on 42 human traits, Nat. Genet 48 (2016) 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian C, Hromatka BS, Kiefer AK, Eriksson N, Noble SM, Tung JY, Hinds DA, Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections, Nat. Commun 8 (2017) 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen EK, Chen WM, Weeks DE, Chen F, Hou X, Mattos JL, Mychaleckyj JC, Segade F, Casselbrant ML, Mandel EM, Ferrell RE, Rich SS, Daly KA, Sale MM, A genome-wide association study of chronic otitis media with effusion and recurrent otitis media identified a novel susceptibility locus on chromosome 2, J. Assoc. Res. Otolaryngol 14 (2013) 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Ingen G, Li J, Goedegebure A, Pandey R, Li YR, March ME, Jaddoe VW, Bakay M, Mentch FD, Thomas K, Wei Z, Chang X, Uitterlinden AG, Moll HA, van Duijn CM, Rivadeneira F, Raat H, Baatenburg de Jong RJ, Sleiman PM, van der Schroeff MP, Hakonarson H, Genome-wide association study for acute otitis media in children identifies FNDC1 as disease contributing gene, Nat. Commun 7 (2016) 12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tefs K, Gueorguieva M, Klammt J, Allen CM, Aktas D, Anlar FY, Aydogdu SD, Brown D, Ciftci E, Contarini P, Dempfle CE, Dostalek M, Eisert S, Gokbuget A, Gunhan O, Hidayat AA, Hugle B, Isikoglu M, Irkec M, Joss SK, Klebe S, Kneppo C, Kurtulus I, Mehta RP, Ornek K, Schneppenheim R, Seregard S, Sweeney E, Turtschi S, Veres G, Zeitler P, Ziegler M, Schuster V, Molecular and clinical spectrum of type I plasminogen deficiency: a series of 50 patients, Blood 108 (2006) 3021–3026. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson PO, Li J, Ny T, Hellstrom S S, Spontaneous development of otitis media in plasminogen-deficient mice, Int. J. Med. Microbiol 296 (2006) 501–509. [DOI] [PubMed] [Google Scholar]
- 18.Santos-Cortez RLP, Chiong CM, Frank DN, Ryan AF, Giese APJ, Bootpetch Roberts TC, Daly KA, Steritz MJ, Szeremeta W, Pedro M, Pine H, Yarza TKL, Scholes MA, Llanes E.G.d.V, Yousaf S, Friedman N, Tantoco MLC, Wine TM, Labra PJ, Benoit J, Ruiz AG, de la Cruz RAR, Greenlee C, Yousaf A, Cardwell J, Nonato RMA, Ray DC, Ong KMC, So E, Robertson CE, Dinwiddie J, Lagrana-Villagracia SM, University of Washington Center for Mendelian Genomics, Gubbels SP, Shaikh RS, Cass SP, Einarsdottir E, Lee NR, Schwartz DA, Gloria-Cruz TLI, Bamshad MJ, Yang IV, Kere J, Abes GT, Prager JD, Riazuddin S, Chan AL, Yoon PJ, Nickerson DA, Cutiongco-de la Paz EMC, Streubel SO, Reyes-Quintos MRT, Jenkins HA, Mattila PS, Chan KH, Mohlke KL, Leal SM, Hafren L, Chonmaitree T, Sale MM, Ahmed ZM, FUT2 variants confer susceptibility to familial otitis media, Am. J. Hum. Genet 103 (2018) 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorven M, Grahn A, Hedlund KO, Johansson H, Wahlfrid C, Larson G, Svensson L, A homozygous nonsense mutation (428G->A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections, J. Virol 79 (2005) 15351–15355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazra A, Kraft P, Selhub J, Giovannucci EL, Thomas G, Hoover RN, Chanock SJ, Hunter DJ, Common variants of FUT2 are associated with plasma vitamin B12 levels, Nat. Genet 40 (2008) 1160–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi M, Mizuki N, Meguro A, Ombrello MJ, Kirino Y, Satorius C, Le J, Blake M, Erer B, Kawagoe T, Ustek D, Tugal-Tutkun I, Seyahi E, Ozyazgan Y, Sousa I, Davatchi F, Francisco V, Shahram F, Abdollahi BS, Nadji A, Shafiee NM, Ghaderibarmi F, Ohno S, Ueda A, Ishigatsubo Y, Gadina M, Oliveira SA, Gul A, Kastner DL, Remmers EF, Dense genotyping of immune-related loci implicates host responses to microbial exposure in Behcet’s disease susceptibility, Nat. Genet 449 (2017) 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Zhang X, Li J, Song C, Jia Y, Xiong W, Identification of a Novel ENU-Induced Mutation in Mouse Tbx1 Linked to Human DiGeorge Syndrome, Neural Plast. (2016)5836143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs JC, Linden JF, Baldini A, Tucker AS AS, A defect in early myogenesis causes Otitis media in two mouse models of 22q11.2 Deletion Syndrome, Hum. Mol. Genet 24 (2015) 1869–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bønnelykke K, Sleiman P, Nielsen K, Kreiner-Møller E, Mercader JM, Belgrave D, den Dekker HT, Husby A, Sevelsted A, Faura-Tellez G, Mortensen LJ, Paternoster L, Flaaten R, Mølgaard A, Smart DE, Thomsen PF, Rasmussen MA, Bonàs-Guarch S, Holst C, Nohr EA, Yadav R, March ME, Blicher T, Lackie PM, Jaddoe VW, Simpson A, Holloway JW, Duijts L, Custovic A, Davies DE, Torrents D, Gupta R, Hollegaard MV, Hougaard DM, Hakonarson H, Bisgaard H, A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations, Nat. Genet 46 (2014) 51–55. [DOI] [PubMed] [Google Scholar]
- 25.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, Palmenberg AC, Gern JE, Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication, Proc. Natl. Acad. Sci. U S A 112 (2015) 5485–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bønnelykke K, Coleman AT, Evans MD, Thorsen J, Waage J, Vissing NH, Carlsson CJ, Stokholm J, Chawes BL, Jessen LE, Fischer TK, Bochkov YA, Ober C, Lemanske RF Jr, Jackson DJ, Gern JE, Bisgaard H, CDHR3 Genetics and Rhinovirus C Respiratory Illnesses, Am. J. Respir. Crit. Care Med 197 (2018) 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortensen EH, Lildholdt T, Gammelgard NP, Christensen PH, Distribution of ABO blood groups in secretory otitis media and cholesteatoma, Clin. Otolaryngol. Allied Sci 8 (1983) 263–265. [DOI] [PubMed] [Google Scholar]
- 28.Apostopoulos K, Labropoulou E, Konstantinos B, Rhageed S, Ferekidis E, Blood group in otitis media with effusion, ORL J. Otorhinolaryngol. Relat. Spec 64 (2002) 433–435. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, Soufan O, Ewald J, Hancock REW, Basu N, Xia J, Network Analyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis, Nucleic Acids Res. 2019. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aul JJ, Anderson KW, Wadowsky R, Doyle WJ, Kingsley LA, Post JC, Ehrlich GD Comparative evaluation of culture and PCR for the detection and determination of persistence of bacterial strains and DNAs in the Chinchilla laniger model of otitis media. Annals of Otology, Rhinology and Laryngology 107:508–513, 1998. [DOI] [PubMed] [Google Scholar]
- 31.Buchinsky FJ, Forbes M, Hayes J, Hu FZ, Greenberg P, Post JC, and Ehrlich GD Phenotypic Plurality among Clinical Strains of nontypeable Haemophilus influenzae determined by symptom severity in the Chinchilla model of Otitis Media. BMC Microbiology 14;7:56, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimoyama M, Hong W, Khampang P, Ehrlich GD, Bakaletz L, Kerschner JE. The Chinchilla Research Resource Database: resource for an otolaryngology disease model. Database – The Journal of Biological Databases and Curation 2016. May 12 Epub pii: baw073 doi:10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartlett JA, Meyerholz DK, Wohlford-Lenane CL, Naumann PW, Salzman NH, McCray PB, Increased susceptibility to otitis media in a Splunc1-deficient mouse model, Dis. Model Mech 8 (2015) 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurabi A, Lee J, Wong C, Pak K, Hoffman H, Ryan AF, Wasserman S, The inflammasome adaptor ASC contributes to multiple innate immune processes in the resolution of otitis media, Innate Immun. 21 (2015) 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deniffel D, Nuyen B, Pak K, Suzukawa K, Hung J, Kurabi A, Wasserman SI, Ryan AF, Otitis Media and Nasopharyngeal Colonization in ccl3(−/−) Mice, Infect Immun. 85 (2017). doi: 10.1128/IAI.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim HW, Pak K, Kurabi A, Ryan AF, Lack of the hyaluronan receptor CD44 affects the course of bacterial otitis media and reduces leukocyte recruitment to the middle ear, BMC Immunol. 20 (2019) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Punwani D, Pelz B, Yu J, Arva NC, Schafernak K, Kondratowicz K, Makhija M, Puck JM, Coronin-1A: immune deficiency in humans and mice, J. Clin. Immunol 35 (2015) 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konduru AS, Matsuyama S, Lee BC, Komatsu K, Li JD, Curcumin Inhibits NTHi-Induced MUC5AC Mucin Overproduction in Otitis Media via Upregulation of MAPK Phosphatase MKP-1, Int. J. Inflam 2017 (2017) 4525309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian C, Harris B, Johnson K, Ectopic mineralization and conductive hearing loss in enpp1/asj mutant mice, a new model for otitis media and tympanosclerosis, PLoS One 11 (2016) e0168159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del-Pozo J, MacIntyre N, Azar A, Glover J, Milne E, Cheeseman M, Chronic otitis media is initiated by a bulla cavitation defect in the FBXO11 mouse model, Dis. Model Mech 12 (2019). doi: 10.1242/dmm.037804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpinelli MR, Kruse EA, Arhatari BD, Debrincat MA, Ogier JM, Bories JC, Kile BT, Burt RA, Mice haploinsufficient for Ets1 and Fli1 display middle ear abnormalities and model aspects of Jacobsen syndrome, Am. J. Pathol 185 (2015) 1867–1876. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, He Q, Zhang X, Ma Y, Fan F, Dong Y, Xu W, Yin Y, He Y, Innate antimicrobial and anti-chemotaxis properties of progranulin in an acute otitis media mouse model, Front. Immunol 9 (2018) 2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzukawa K, Tomlin J, Pak K, Chavez E, Kurabi A, Baird A, Wasserman SI, Ryan AF, A mouse model of otitis media identifies HB-EGF as a mediator of inflammation-induced mucosal proliferation, PLoS One 9 (2014) e102739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao W, Frei M, Pan J, Pak K, Webster N, Wasserman SI, Ryan AF, C-Jun N-terminal Kinase (JNK) isoforms play differing roles in otitis media, BMC Immunol. 15 (2014) 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Xu M, Zheng Q, Zhang Y, Ma W, Zhang Z, Blocking macrophage migration inhibitory factor activity alleviates mouse acute otitis media in vivo, Immunol. Lett 162 (2014) 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crompton M, Purnell T, Tyrer HE, Parker A, Ball G, Hardisty-Hughes RE, Gale R, Williams D, Dean CH, Simon MM, Mallon AM, Wells S, Bhutta MF, Burton MJ, Tateossian H, Brown SDM, A mutation in Nischarin causes otitis media via LIMK1 and NF-κB pathways, PLoS Genet. 13 (2017) e1006969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo JI, Oh S, Webster P, Lee YJ, Lim DJ, Moon SK, NOD2/RICK-dependent β-defensin 2 regulation is protective for nontypeable Haemophilus influenzae-induced middle ear infection, PLoS One 9 (2014) e90933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin SG, Koh SH, Woo CH, Lim JH, PAI-1 inhibits development of chronic otitis media and tympanosclerosis in a mouse model of otitis media, Acta Otolaryngol. 134 (2014) 1231–1238. [DOI] [PubMed] [Google Scholar]
- 49.Maguire S, Estabel J, Ingham N, Pearson S, Ryder E, Carragher DM, Walker N, Sanger MGP Slc25a21 Project Team, Bussell J, Chan WI, Keane TM, Adams DJ, Scudamore CL, Lelliott CJ, Ramírez-Solis R, Karp NA, Steel KP, White JK, Gerdin AK, Targeting of Slc25a21 is associated with orofacial defects and otitis media due to disrupted expression of a neighbouring gene, PLoS One 9 (2014) e91807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Xu L, Li J, Li B, Bai X, Strauss JF 3rd, Zhang Z Z, Wang H, Otitis media in sperm-associated antigen 6 (Spag6)-deficient mice, PLoS One 9 (2014) e112879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y, Wang Z, Jin C, Wang L, Zhang X, Xu W, Xiang Y, Wang W, He X, Yin Y, He Y, TLR2 promotes macrophage recruitment and Streptococcus pneumoniae clearance during mouse otitis media, Pediatr Res. 80 (2016) 886–893. [DOI] [PubMed] [Google Scholar]
- 52.Brown SD, Hardisty-Hughes RE, Mburu P P, Quiet as a mouse: dissecting the molecular and genetic basis of hearing, Nat. Rev. Genet 9 (2008) 277–290. [DOI] [PubMed] [Google Scholar]
- 53.Azar A, Piccinelli C, Brown H, Headon D, Cheeseman M, Ectodysplasin signalling deficiency in mouse models of hypohidrotic ectodermal dysplasia leads to middle ear and nasal pathology, Hum. Mol. Genet 25 (2016) 3564–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulay A, Hood DW, Williams D, Russell C, Brown SDM, Bingle L, Cheeseman M, Bingle CD, Loss of the homeostatic protein BPIFA1, leads to exacerbation of otitis media severity in the Junbo mouse model, Sci Rep. 8 (2018) 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Earl JP, Adappa ND, Krol J, Bhat AS, Balashov S, Ehrlich RL, Palmer JN, Workman AD, Blasetti M, Sen B, Hammond J, Cohen NA, Ehrlich GD, Mell JC, Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16A rRNA genes, Microbiome 23 (2018) 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, Polley EC, Bowman ED, Khan MA, Robles AI, Cooks T, Ryan BM, Padgett N, Dzutsev AH, Trinchieri G, Pineda MA, Bilke S, Meltzer PS, Hokenstad AN, Stickrod TM, Walther-Antonio MR, Earl JP, Mell JC, Krol JE, Balashov SV, Bhat AS, Ehrlich GD, Valm A, Deming C, Conlan S, Oh J, Segre JA, Harris CC, Interaction between the microbiome and TP53 in human lung cancer, Genome Biol. 19 (2018) 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babauta JT, Atci E, Ha PT, Lindemann SR, Ewing T, Call DR, Frederickson JK, Beyenal H, Localized electron transfer rates and microelectrode-based enrichment of microbial communities within a phototrophic microbial mat, Front. Microbiol 5 (2014) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Amore R, Ijaz UZ, Schirmer M, Kenny JG, Gregory R, Darby AC, Shakya M, Podar M, Quince C, Hall N, A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling, BMC Genomics 17 (2016) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fichot EB, Norman RS, Microbial phylogenetic profiling with the Pacific Biosciences sequencing platform, Microbiome 1 (2013) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mosher JJ, Bernberg EL, Shevchenko O, Kan J, Kaplan LA, Efficacy of a 3rd generation high-throughput sequencing platform for analyses of 16S rRNA genes from environmental samples, J. Microbiol. Methods 95 (2013) 175–181. [DOI] [PubMed] [Google Scholar]
- 61.Mosher JJ, Bowman B, Bernberg EL, Shevchenko O, Kan J, Korlach J, Kaplan LA, Improved performance of the PacBio SMRT technology for 16S rDNA sequencing, J. Microbiol. Methods 104 (2014) 59–60. [DOI] [PubMed] [Google Scholar]
- 62.Schloss PD, Jenior ML, Koumpouras CC, Westcott SL, Highlander SK, Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system, PeerJ 4 (2016) e1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singer E, Bushnell B, Coleman-Derr D, Bowman B, Bowers RM, Levy A, Gies EA, Cheng JF, Copeland A, Klenk HP, Hallam SJ, Hugenholtz P, Tringe SG, Woyke T, High-resolution phylogenetic microbial community profiling, ISME J. 10 (2016) 2020–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner J, Coupland P, Browne HP, Lawley TD, Francis SC, Parkhill J, Evaluation of PacBio sequencing for full-length bacterial 16S rRNA gene classification, BMC Microbiol. 16 (2016) 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P, Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates, Environ. Microbiol 12 (2010) 118–123. [DOI] [PubMed] [Google Scholar]
- 66.Lee C, Grasso C, Sharlow MF, Multiple sequence alignment using partial order graphs, Bioinformatics 18 (2002) 452–464. [DOI] [PubMed] [Google Scholar]
- 67.McMurdie PJ, Holmes S, Waste not, want not: why rarefying microbiome data is inadmissible, PLoS Comput. Biol 10 (2014) e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho NT, Li F, Wang S, Kuhn L, metamicrobiomeR: an R package for analysis of microbiome relative abundance data using zero-inflated beta GAMLSS and meta-analysis across studies using random effects models, BMC Bioinformatics 20 (2019) 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMurdie PJ, Holmes S, phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, PLoS One 8 (2013) e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McMurdie PJ, Holmes S, Shiny-phyloseq: Web application for interactive microbiome analysis with provenance tracking, Bioinformatics 31 (2015) 282–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML, Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data, Methods Ecol. Evol 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods 13 (2016) 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO, Anvi’o: an advanced analysis and visualization platform for ‘omics data, PeerJ 3 (2015) e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Norskov-Lauritsen N, Kilian M, Reclassification of actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates, Int. J. Syst. Evol. Microbiol 56 (2006) 2135–2146. [DOI] [PubMed] [Google Scholar]
- 75.Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW, Transforming clinical microbiology with bacterial genome sequencing, Nat. Rev. Genet 13 (2012) 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ, Within-host evolution of bacterial pathogens, Nat. Rev. Microbiol 14 (2016) 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klemm E, Dougan G, Advances in Understanding Bacterial Pathogenesis Gained from Whole-Genome Sequencing and Phylogenetics, Cell Host Microbe 19 (2016) 599–610. [DOI] [PubMed] [Google Scholar]
- 78.Sheppard SK, Guttman DS, Fitzgerald JR, Population genomics of bacterial host adaptation, Nat. Rev. Genet 19 (2018) 549–565. [DOI] [PubMed] [Google Scholar]
- 79.Chen PE, Shapiro BJ, The advent of genome-wide association studies for bacteria, Curr. Opin. Microbiol 25 (2015) 17–24. [DOI] [PubMed] [Google Scholar]
- 80.Lees JA, Bentley SD, Bacterial GWAS: not just gilding the lily, Nat. Rev. Microbiol 14 (2016) 406. [DOI] [PubMed] [Google Scholar]
- 81.Power RA, Parkhill J, de Oliveira T, Microbial genome-wide association studies: lessons from human GWAS, Nat. Rev. Genet 18 (2017) 41. [DOI] [PubMed] [Google Scholar]
- 82.Aun E, Brauer A, Kisand V, Tenson T, Remm M, A k-mer-based method for the identification of phenotype-associated genomic biomarkers and predicting phenotypes of sequenced bacteria, PLoS Comput. Biol 14 (2018) e1006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Collins C, Didelot X, A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination, PLoS Comput. Biol 14 (2018) e1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaillard M, Lima L, Tournoud M, Mahé P, Van Belkum A, Lacroix V, Jacob L, A fast and agnostic method for bacterial genome-wide association studies: Bridging the gap between k-mers and genetic events, PLoS Genet. 14 (2018) e1007758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caballero JD, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YC, Waters VJ, Hwang DM, Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung, MBio 6 (2015) e00981–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sokurenko EV, Hasty DL, Dykhuizen DE, Pathoadaptive mutations: gene loss and variation in bacterial pathogens, Trends Microbiol 7 (1999) 191–195. [DOI] [PubMed] [Google Scholar]
- 87.Young BC, Wu CH, Gordon NC, Cole K, Price JR, Liu E, Sheppard AE, Perera S, Charlesworth J, Golubchik T, Severe infections emerge from commensal bacteria by adaptive evolution, eLife 6 (2017). doi: 10.7554/eLife.30637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen K, Antalis P, Gladitz J, Sayeed S, Ahmed A, Yu S, Hayes J, Johnson S, Dice B, Dopico R, Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae, Infect. Immun 73 (2005) 3479–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hogg JS, Hu FZ, Janto B, Boissy R, Hayes J, Keefe R, Post JC, Ehrlich GD, Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains, Genome Biol. 8 (2007) R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen K, Antalis P, Gladitz J, Dice B, Janto B, Keefe R, Hayes J, Ahmed A, Dopico R, Ehrlich N, Jocz J, Kropp J, Yu S, Nistico L, Greenberg DP, Barbadora K, Post JC, Ehrlich GD, Hu FZ, Characterization, Distribution and Expression of Novel Genes Among Eight Clinical Isolates of Streptococcus pneumoniae, Infect. Immun 74 (2006) 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ, Comparative Genomic Analyses of Seventeen Streptococcus pneumoniae Strains: Insights into the Pneumococcal Supragenome, J. Bacteriol 189 (2007) 8186–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Riley D, Covacci A, Bentley SD, Kilian M, Ehrlich GD, Hu FZ, Rappuoli R, Moxon ER, Masignani V, Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species, Genome Biol. 11 (2010) R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boissy R, Ahmed A, Janto B, Earl J, Hall BJ, Hogg J, Pusch GD, Hiller NL, Powell E, Hayes J, Yu S, Kathju S, Stoodley P, Post JC, Ehrlich GD, Hu FZ, Comparative Supragenomic Analyses among the Pathogens Staphylococcus aureus, Streptococcus pneumoniae, and Haemophilus influenzae Using a Modification of the Finite Supragenome Model, BMC Genomics 12 (2011) 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davie JJ, Earl J, de Vries SPW, Ahmed A, Hu FZ, Bootsma HJ, Stol K, Hermans PWM, Wadowsky RM, Ehrlich GD, Hays J, Campagnari AA, Comparative Analysis and Supragenome Modeling of Twelve Moraxella catarrhalis Clinical Isolates, BMC Genomics 12 (2011) 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Earl JP, de Vries SPW, Ahmed A, Powell E, Schultz MP, Hermans PWM, Hill DJ, Constantinidou CI, Hu FZ, Bootsma HJ, Ehrlich GD, Comparative Genomic Analyses of the Moraxella Catarrhalis Serosensitive and Seroresistant Lineages Demonstrates Their Independent Evolution, Genome Biol. Evol 8 (2016) 955–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rau RH, Marvig RL, Ehrlich GD, Molin S, Jelsbak L, Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment, Environ. Microbiol 14 (2012) 2200–2211. [DOI] [PubMed] [Google Scholar]
- 97.Lee AHY, Flibotte S, Sinha S, Paiero A, Ehrlich RL, Balashov S, Ehrlich GD, Zlosnik JEA, Mell JC, Nislow C, Phenotypic diversity and genotypic flexibility of Burkholderia cenocepacia during long-term infection of cystic fibrosis lungs, Genome Res. 27 (2017) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, Whole-genome random sequencing and assembly of Haemophilus influenzae Rd, Science 269 (1995) 496–512. [DOI] [PubMed] [Google Scholar]
- 99.Harrison A, Dyer DW, Gillaspy A, Ray WC, Mungur R, Carson MB, Zhong H, Gipson J, Gipson M, Johnson LS, Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20, J. Bacteriol 187 (2005) 4627–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Chiara M, Hood D, Muzzi A, Pickard DJ, Perkins T, Pizza M, Dougan G, Rappuoli R, Moxon ER, Soriani M, Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure, Proc. Natl. Acad. Sci. U S A 111 (2014) 5439–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eutsey RA, Hiller NL, Earl JP, Janto BA, Dahlgren ME, Ahmed A, Powell E, Schultz MP, Gilsdorf JR, Zhang L, Design and validation of a supragenome array for determination of the genomic content of Haemophilus influenzae isolates, BMC Genomics 14 (2013) 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kress-Bennett JM, Hiller NL, Eutsey RA, Powell E, Longwell MJ, Hillman T, Blackwell T, Byers B, Mell JC, Post JC, Identification and characterization of msf, a novel virulence factor in Haemophilus influenzae, PloS One 11 (2016) e0149891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moleres J, Fernández-Calvet A, Ehrlich RL, Martí S, Pérez-Regidor L, Euba B, Rodríguez-Arce I, Balashov S, Cuevas E, Liñares J, Antagonistic Pleiotropy in the Bifunctional Surface Protein FadL (OmpP1) during Adaptation of Haemophilus influenzae to Chronic Lung Infection Associated with Chronic Obstructive Pulmonary Disease, mBio 9 (2018) e01176–01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pettigrew MM, Ahearn CP, Gent JF, Kong Y, Gallo MC, Munro JB, D’Mello A, Sethi S, Tettelin H, Murphy TF, Haemophilus influenzae genome evolution during persistence in the human airways in chronic obstructive pulmonary disease, Proc. Natl. Acad. Sci. U S A 115 (2018) E3256–E3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aziz A, Sarovich DS, Nosworthy E, Beissbarth J, Chang AB, Smith-Vaughan H, Price EP, Harris TM, Molecular signatures of non-typeable Haemophilus influenzae lung adaptation in paediatric chronic lung disease, bioRxiv (2019) 614214 10.1101/614214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Topaz N, Boxrud D, Retchless AC, Nichols M, Chang HY, Hu F, Wang X, BMScan: using whole genome similarity to rapidly and accurately identify bacterial meningitis causing species, BMC Infect. Dis 18 (2018) 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pinto M, González-Díaz A, Machado M, Duarte S, Vieira L, Carriço J, Marti S, Bajanca-Lavado M, Gomes J, Insights into the population structure and pan-genome of Haemophilus influenzae, Infect. Genet. Evol 67 (2019) 126–135. [DOI] [PubMed] [Google Scholar]
- 108.Cleary D, Devine V, Morris D, Osman K, Gladstone R, Bentley S, Faust S, Clarke S, Pneumococcal vaccine impacts on the population genomics of non-typeable Haemophilus influenzae, Microb. Genom 4 (2018). doi: 10.1099/mgen.0.000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stewart RD, Auffret MD, Warr A, Wiser AH, Press MO, Langford KW, Liachko I, Snelling TJ, Dewhurst RJ, Walker AW, Roehe R, Watson M, Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen, Nat. Commun 9 (2018) 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bickhart DM, Watson M, Koren S, Panke-Buisse K, Cersosimo LM, Press MO, Van Tassell CP, Van Kessel JAS, Haley BJ, Kim SW, Heiner C, Suen G, Bakshee K, Liachko I, Sullivan ST, Myer PR, Ghurye J. Pop M, Weimer PJ, Phillippy AM, Smith TPL, Assignment of virus and antimicrobial resistance genes to microbial hosts in a complex microbial community by combined long-read assembly and proximity ligation, Genome Biol. 153 (2019). doi: 10.1101/491175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dichosa AEK, Daughton AR, Reitenga KG, Fitzsimons MS, Han CS, Capturing and cultivating single bacterial cells in gel microdroplets to obtain near-complete genomes, Nat. Protoc 9 (2014) 610. [DOI] [PubMed] [Google Scholar]
- 112.Dichosa AEK, Davenport DW, Li PE, Ahmed SA, Daligault H, Gleasner CD, Kunde Y, McMurry K, Lo CC, Reitenga KG, Daughton AR, Shen C, Frietze S, Wang D, Johnson SL, Drautz-Moses DI, Schuster S, Chain PS, Han C, Draft Genome Sequence of Thauera sp. Strain SWB20, Isolated from a Singapore Wastewater Treatment Facility Using Gel Microdroplets, Genome Announc, 3 (2015) e00132–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moxon R, Bayliss C, Hood D, Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation, Annu. Rev. Genet 40 (2006) 307–333. [DOI] [PubMed] [Google Scholar]
- 114.Power PM, Sweetman WA, Gallacher NJ, Woodhall MR, Kumar GA, Moxon ER, Hood DW, Simple sequence repeats in Haemophilus influenzae, Infect. Genet. Evol 9 (2009) 216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blakeway LV, Tan A, Peak IRA, Seib KL, Virulence determinants of Moraxella catarrhalis: distribution and considerations for vaccine development, Microbiology 163 (2017) 1371–1384. [DOI] [PubMed] [Google Scholar]
- 116.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP, The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes, Proc. Natl. Acad. Sci. U S A 102 (2005) 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Srikhanta YN, Fox KL, Jennings MP, The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes, Nat. Rev. Microbiol 8 (2010) 196–206. [DOI] [PubMed] [Google Scholar]
- 118.Atack JM, Srikhanta YN, Fox KL, Jurcisek JA, Brockman KL, Clark TA, Boitano M, Power PM, Jen FE, McEwan AG, Grimmond SM, Smith AL, Barenkamp SJ, Korlach J, Bakaletz LO, Jennings MP, A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae, Nat. Commun 6 (2015) 7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blakeway LV, Power PM, Jen FE, Worboys SR, Boitano M, Clark TA, Korlach J, Bakaletz LO, Jennings MP, Peak IR, Seib KL, ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media, FASEB J. 28 (2014) 5197–5207. [DOI] [PubMed] [Google Scholar]
- 120.Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR, A random six-phase switch regulates pneumococcal virulence via global epigenetic changes, Nat. Commun 5 (2014) 5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Capecchi B, Serruto D, Adu-Bobie J, Rappuoli R, Pizza M, The genome revolution in vaccine research, Curr. Issues Mol. Biol 6 (2004) 17–27. [PubMed] [Google Scholar]
- 122.Filipp FV, Precision medicine driven by cancer systems biology, Cancer Metastasis Rev. 36 (2017) 91–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rossi G, Ignatiadis M, Promises and pitfalls of using liquid biopsy for precision medicine, Cancer Res. 79 (2019) 2798–2804. [DOI] [PubMed] [Google Scholar]
- 124.Morganti S, Tarantino P, Ferraro E, D’Amico P, Viale G, Trapani D, Duso BA, Curigliano G, Complexity of genome sequencing and reporting: Next generation sequencing (NGS) technologies and implementation of precision medicine in real life, Crit. Rev. Oncol. Hematol 133 (2019) 171–182. [DOI] [PubMed] [Google Scholar]
- 125.Leiding JW, Ballow M, Precision medicine in the treatment of primary immunodeficiency diseases, Curr. Opin. Allergy Clin. Immunol 18 (2018) 159–166. [DOI] [PubMed] [Google Scholar]
- 126.Skov M, Hansen CR, Pressler T, Cystic fibrosis - an example of personalized and precision medicine, APMIS 127 (2019) 352–360. [DOI] [PubMed] [Google Scholar]
- 127.Pletz MW, Bauer M, Brakhage AA, One step closer to precision medicine for infectious diseases, Lancet Infect. Dis 19 (2019) 564–565. [DOI] [PubMed] [Google Scholar]
- 128.Spaulding CN, Klein RD, Schreiber HL 4th, Janetka JW, Hultgren SJ, Precision antimicrobial therapeutics: The path of least resistance? NPJ Biofilms Microbiomes 4 (2018) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Atreya MR, Wong HR, Precision medicine in pediatric sepsis, Curr. Opin. Pediatr 31 (2019) 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mahomed S, Padayatchi N, Singh J, Naidoo K, Precision medicine in resistant tuberculosis: Treat the correct patient, at the correct time, with the correct drug, J. Infect 78 (2019) 261–268. [DOI] [PubMed] [Google Scholar]
- 131.Samuels TL, Yan JC, Khampang P, Dettmar PW, MacKinnon A, Hong W, Johnston N, Papsin BC, Chun RH, McCormick ME, Kerschner JE. Association of gel-forming mucins and aquaporin gene expression with hearing loss, effusion viscosity and inflammation in otitis media with effusion. JAMA Otolaryngology-Head & Neck Surgery 2017;143(8):810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Val S, Jeong S, Poley M, Krueger A, Nino G, Brown K, Preciado D. Purification and characterization of microRNAs within middle ear fluid exosomes: implication in otitis media pathophysiology. Pediatric Research 2017;81(6):911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Samuels TL, Yan J, Khampang P, MacKinnon A, Hong W, Johnston N, Kerschner JE. Association of microRNA 146 with middle ear hyperplasia in pediatric otitis media. International Journal of Pediatric Otorhinolaryngology 2016;88(9);104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park CG, Kim S, Lee EJ, Jeon HS, Han S, Clinical relevance of point mutations in the 23S rRNA gene in helicobacter pylori eradication: A prospective, observational study, Medicine (Baltimore) 97 (2018) e11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de la Fuente-Nunez C, Torres MD, Mojica FJ, Lu TK, Next-generation precision antimicrobials: Towards personalized treatment of infectious diseases, Curr. Opin. Microbiol 37 (2017) 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Venekamp RP, Sanders SL, Glasziou PP PP, Del Mar CC, Rovers MM, Antibiotics for acute otitis media in children, Cochrane Database Syst. Rev 6 (2015) CD000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wafi A, Mirnezami R, Translational -omics: Future potential and current challenges in precision medicine, Methods 151 (2018) 3–11. [DOI] [PubMed] [Google Scholar]
- 138.Francis NA, Waldron CA, Cannings-John R, Thomas-Jones E, Winfield T, Shepherd V, Harris D, Hood K, Fitzsimmons D, Roberts A, Powell CV, Gal M, Jones S, Butler CC, Oral steroids for hearing loss associated with otitis media with effusion in children aged 2-8 years: the OSTRICH RCT, Health Technol. Assess 22 (2018) 1–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ranakusuma RW, Pitoyo Y, Safitri ED, Thorning S, Beller EM, Sastroasmoro S, Del Mar CB, Systemic corticosteroids for acute otitis media in children, Cochrane Database Syst. Rev 3 (2018) CD012289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lewnard JA, Tähtinen PA, Laine MK, Lindholm L, Jalava J, Huovinen P, Lipsitch M, Ruohola A, Impact of Antimicrobial Treatment for Acute Otitis Media on Carriage Dynamics of Penicillin-Susceptible and Penicillin-Nonsusceptible Streptococcus pneumoniae, J. Infect. Dis 218 (2018) 1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ruohola A, Laine MK, Tähtinen PA, Effect of Antimicrobial Treatment on the Resolution of Middle-Ear Effusion After Acute Otitis Media, J. Pediatric Infect. Dis. Soc 7 (2018) 64–70. [DOI] [PubMed] [Google Scholar]
- 142.Hoberman A, Paradise JL, Rockette HE, Kearney DH, Bhatnagar S, Shope TR, Martin JM, Kurs-Lasky M, Copelli SJ, Colborn DK, Block SL, Labella JJ, Lynch TG, Cohen NL, Haralam M, Pope MA, Nagg JP, Green MD, Shaikh N, Shortened Antimicrobial Treatment for Acute Otitis Media in Young Children, N. Engl. J. Med 375 (2016) 2446–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Te Molder M, de Hoog ML, Uiterwaal CS, van der Ent CK, Smit HA, Schilder AG, Damoiseaux RA, Venekamp RP, Antibiotic Treatment for First Episode of Acute Otitis Media Is Not Associated with Future Recurrences, PLoS One 11 (2016) e0160560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shaikh N, Dando EE, Dunleavy ML, Curran DL, Martin JM, Hoberman A, Smith KJ, A Cost-Utility Analysis of 5 Strategies for the Management of Acute Otitis Media in Children, J. Pediatr 189 (2017) 54–60.e3. [DOI] [PubMed] [Google Scholar]
- 145.Spurling GK, Del Mar CB, Dooley L, Foxlee R, Farley R, Delayed antibiotic prescriptions for respiratory infections, Cochrane Database Syst. Rev 9 (2017) CD004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang R, Sabharwal V, Okonkwo OS, Shlykova N, Tong R, Lin LY, Wang W, Guo S, Rosowski JJ, Pelton SI, Kohane DS, Treatment of otitis media by transtympanic delivery of antibiotics, Sci. Transl. Med 8 (2016) 356ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kurabi A, Beasley KA, Chang L, McCann J, Pak K, Ryan AF, Peptides actively transported across the tympanic membrane: Functional and structural properties, PLoS One 12 (2017) e0172158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kurabi A, Schaerer D, Chang L, Pak K, Ryan AF, Optimisation of peptides that actively cross the tympanic membrane by random amino acid extension: a phage display study, J. Drug Target 26 (2018) 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kurabi A, Schaerer D, Noack V, Bernhardt M, Pak K, Alexander T, Husseman J, Nguyen Q, Harris JP, Ryan AF, Active Transport of Peptides Across the Intact Human Tympanic Membrane, Sci. Rep 8 (2018) 11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu Q, Gill S, Xu L, Gonzalez E, Pichichero ME. Comparative Analysis of Microbiome in Nasopharynx and Middle Ear in Young Children With Acute otitis Media. Front Genet. 2019. November 19;10:1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mrkvan T, Pelton SI, Ruiz-Guinazu J, Palmu AA, Borys D, Effectiveness and impact of the 10-valent pneumococcal conjugate vaccine, PHiD-CV: review of clinical trials and post-marketing experience, Expert Rev. Vaccines 17 (2018) 797–818. [DOI] [PubMed] [Google Scholar]
- 152.Zhou X, de Luise C, Gaffney M, Burt CW, Scott DA, Gatto N, Center KJ, National impact of 13-valent pneumococcal conjugate vaccine on ambulatory care visits for otitis media in children under 5 years in the United States, Int. J. Pediatr. Otorhinolaryngol 119 (2019) 96–102. [DOI] [PubMed] [Google Scholar]
- 153.Fortanier AC, Venekamp RP, Boonacker CW, hak E, Schilder AG, Sanders EA, Damoiseaux RA, Pneumococcal conjugate vaccines for preventing acute otitis media in children, Cochrane Database Syst. Rev 5 (2019) CD001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pichichero M, Kaur R, Scott DA, Gruber WC, Trammel J, Almudevar A, Center KJ, Effectiveness of 13-valent pneumococcal conjugate vaccination for protection against acute otitis media caused by Streptococcus pneumoniae in healthy young children: a prospective observational study, Lancet Child Adolesc. Health 2 (2018) 561–568. [DOI] [PubMed] [Google Scholar]
- 155.Dagan R, Pelton S, Bakaletz L, Cohen R, Prevention of early episodes of otitis media by pneumococcal vaccines might reduce progression to complex disease, Lancet Infect. Dis 16 (2016) 480–492. [DOI] [PubMed] [Google Scholar]
- 156.Dagan R, Pelton S, Bakaletz L, Cohen R, First Otitis Media and Pneumococcal Conjugate Vaccine Serotypes in Infants, Pediatr. Infect. Dis. J 37 (2018) e351–e352. [DOI] [PubMed] [Google Scholar]
- 157.Kaur R, Casey JR, Pichichero ME, Emerging Streptococcus pneumoniae Strains Colonizing the Nasopharynx in Children After 13-valent Pneumococcal Conjugate Vaccination in Comparison to the 7-valent Era, 2006-2015, Pediatr. Infect. Dis. J 35 (2016) 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Greenberg D, Hoover PA, Vesikari T, Peltier C, Hurley DC, McFetridge RD, Dallas M, Hartzel J, Marchese RD, Coller BG, Stek JE, Abeygunawardana C, Winters MA, MacNair JE, Pujar NS, Musey L, Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine (PCV15) in healthy infants, Vaccine 36 (2018) 6883–6891. [DOI] [PubMed] [Google Scholar]
- 159.Stacey HL, Rosen J, Peterson JT, Williams-Diaz A, Gakhar V, Sterling TM, Acosta CJ, Nolan KM, Li J, Pedley A, Benner P, Abeygunawardana C, Kosinski M, Smith WJ, Pujar H, Musey LK, Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine (PCV-15) compared to PCV-13 in healthy older adults. Hum. Vaccin. Immunother 15 (2019) 530–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Peterson JT, Stacey HL, MacNair JE, Li J, Hartzel JS, Sterling TM, Benner P, Tamms GM, Musey LK, Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine compared to 13-valent pneumococcal conjugate vaccine in adults ≥65 years of age previously vaccinated with 23-valent pneumococcal polysaccharide vaccine, Hum. Vaccin. Immunother 15 (2019) 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pichichero ME, Pneumococcal whole-cell and protein-based vaccines: changing the paradigm, Expert Rev. Vaccines 16 (2017) 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alderson MR, Status of research and development of pediatric vaccines for Streptococcus pneumoniae, Vaccine 34 (2016) 2959–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gabutti G, Azzari C, Bonanni P, Prato R, Tozzi AE, Zanetti A, Zuccotti G, Pertussis. Hum. Vaccin. Immunother 11 (2015) 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hoe E, Boelsen LK, Toh ZQ, Sun GW, Koo GC, Balloch A, Marimla R, Dunne EM, Tikoduadua L, Russell FM, Satzke C, Mulholland EK, Licciardi PV, Reduced IL-17A Secretion Is Associated with High Levels of Pneumococcal Nasopharyngeal Carriage in Fijian Children, PLoS One 10 (2015) e0129199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Campo JJ, Le TQ, Pablo JV, Hung C, Teng AA, Tettelin H, Tate A, Hanage WP, Alderson MR, Liang X, Malley R, Lipsitch M, Croucher NJ, Panproteome-wide analysis of antibody responses to whole cell pneumococcal vaccination, Elife (2018) 7. doi: 10.7554/eLife.37015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Moffitt KL, Yadav P, Weinberger DM, Anderson PW, Malley R, Broad antibody and T cell reactivity induced by a pneumococcal whole-cell vaccine, Vaccine 30 (2012) 4316–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.David SC, Laan Z, Minhas V, Chen AY, Davies J, Hirst TR, McColl SR, Alsharifi M, Paton JC, Enhanced safety and immunogenicity of a pneumococcal surface antigen A mutant whole-cell inactivated pneumococcal vaccine, Immunol. Cell Biol (2019). doi: 10.1111/imcb.12257. [DOI] [PubMed] [Google Scholar]
- 168.Giufre M, Fabiani M, Cardines R, Riccardo F, Caporali MG, D’Ancona F, Pezzotti P, Cerquetti M, Increasing trend in invasive non-typeable Haemophilus influenzae disease and molecular characterization of the isolates, Italy, 2012-2016, Vaccine 36 (2018) 6615–6622. [DOI] [PubMed] [Google Scholar]
- 169.Naito S, Takeuchi N, Ohkusu M, Takahashi-Nakaguchi A, Takahashi H, Imuta N, Nishi J, Shibayama K, Matsuoka M, Sasaki Y, Ishiwada N, Clinical and Bacteriologic Analysis of Nontypeable Haemophilus influenzae Strains Isolated from Children with Invasive Diseases in Japan from 2008 to 2015, J. Clin. Microbiol 56 (2018). doi: 10.1128/JCM.00141-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pettigrew MM, Ahearn CP, Gent JF, Kong Y, Gallo MC, Munro JB, D’Mello A, Sethi S, Tettelin H, Murphy TF, Haemophilus influenzae genome evolution during persistence in the human airways in chronic obstructive pulmonary disease, Proc. Natl. Acad. Sci. U S A 115 (2018) E3256–E3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Prymula R, Peeters P, Chrobok V, Kriz P, Novakova E, Kaliskova E, Kohl I, Lommel P, Poolman J, Prieels JP, Schuerman L, Pneumococcal capsular polysaccharides conjugated to protein D for prevention of acute otitis media caused by both Streptococcus pneumoniae and non-typable Haemophilus influenzae: a randomised double-blind efficacy study, Lancet 367 (2006) 740–748. [DOI] [PubMed] [Google Scholar]
- 172.Clarke C, Bakaletz LO, Ruiz-Guinazu J, Borys D, Mrkvan T, Impact of protein D-containing pneumococcal conjugate vaccines on non-typeable Haemophilus influenzae acute otitis media and carriage, Expert Rev. Vaccines 16 (2017) 1–14. [DOI] [PubMed] [Google Scholar]
- 173.van den Bergh MR, Spijkerman J, Swinnen KM, Francois NA, Pascal TG, Borys D, Schuerman L, Ijzerman EP, Bruin JP, van der Ende A, Veenhoven RH, Sanders EA, Effects of the 10-valent pneumococcal nontypeable Haemophilus influenzae protein D-conjugate vaccine on nasopharyngeal bacterial colonization in young children: a randomized controlled trial, Clin. Infect. Dis 56 (2013) e30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jalalvand F, Riesbeck K, Update on non-typeable Haemophilus influenzae-mediated disease and vaccine development, Expert Rev. Vaccines 17 (2018) 503–512. [DOI] [PubMed] [Google Scholar]
- 175.Novotny LA, Brockman KL, Mokrzan EM, Jurcisek JA, Bakaletz LO, Biofilm biology and vaccine strategies for otitis media due to nontypeable Haemophilus influenzae, J. Pediatr. Infect. Dis 14 (2019) 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Khan MN, Ren D, Kaur R, Basha S, Zagursky R, Pichichero ME, Developing a vaccine to prevent otitis media caused by nontypeable Haemophilus influenzae, Expert Rev. Vaccines 15 (2016) 863–878. [DOI] [PubMed] [Google Scholar]
- 177.Leroux-Roels G, Van Damme P, Haazen W, Shakib S, Caubet M, Aris E, Devaster JM, Peeters M, Phase I, randomized, observer-blind, placebo-controlled studies to evaluate the safety, reactogenicity and immunogenicity of an investigational non-typeable Haemophilus influenzae (NTHi) protein vaccine in adults, Vaccine 34 (2016) 3156–3163. [DOI] [PubMed] [Google Scholar]
- 178.Berglund J, Vink P, Tavares Da Silva F, Lestrate P, Boutriau D, Safety, immunogenicity, and antibody persistence following an investigational Streptococcus pneumoniae and Haemophilus influenzae triple-protein vaccine in a phase 1 randomized controlled study in healthy adults, Clin. Vaccine Immunol 21 (2014) 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van Damme P, Leroux-Roels G, Vandermeulen C, De Ryck I, Tasciotti A, Dozot M, Moraschini L, Testa M, Arora AK, Safety and immunogenicity of non-typeable Haemophilus influenzae-Moraxella catarrhalis vaccine, Vaccine 37 (2019) 3113–3122. [DOI] [PubMed] [Google Scholar]
- 180.Tandon MK, Phillips M, Waterer G, Dunkley M, Comans P, Clancy R, Oral immunotherapy with inactivated nontypeable Haemophilus influenzae reduces severity of acute exacerbations in severe COPD, Chest 137 (2010) 805–811. [DOI] [PubMed] [Google Scholar]
- 181.Clancy RL, Dunkley ML, Sockler J, McDonald CF, Multi-site placebo-controlled randomised clinical trial to assess protection following oral immunisation with inactivated non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease, Intern. Med. J 46 (2016) 684–693. [DOI] [PubMed] [Google Scholar]
- 182.Teo E, Lockhart K, Purchuri SN, Pushparajah J, Cripps AW, van Driel ML, Haemophilus influenzae oral vaccination for preventing acute exacerbations of chronic bronchitis and chronic obstructive pulmonary disease, Cochrane Database Syst. Rev 6 (2017) CD010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Novotny LA, Clements JD, Goodman SD, Bakaletz LO, Transcutaneous Immunization with a Band-Aid Prevents Experimental Otitis Media in a Polymicrobial Model, Clin. Vaccine Immunol 24 (2017) e00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Michel LV, Kaur R, Zavorin M, Pryharski K, Khan MN, LaClair C, O’Neil M, Xu Q, Pichichero ME, Intranasal coinfection model allows for assessment of protein vaccines against nontypeable Haemophilus influenzae in mice, J. Med. Microbiol 67 (2018) 1527–1532. [DOI] [PubMed] [Google Scholar]
- 185.Winter LE, Barenkamp SJ, Immunogenicity of Nontypeable Haemophilus influenzae Outer Membrane Vesicles and Protective Ability in the Chinchilla Model of Otitis Media, Clin. Vaccine Immunol 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Perez AC, Murphy TF, A Moraxella catarrhalis vaccine to protect against otitis media and exacerbations of COPD: An update on current progress and challenges, Hum. Vaccin. Immunother 13 (2017) 2322–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ren D, Pichichero ME, Vaccine targets against Moraxella catarrhalis, Expert Opin. Ther. Targets 20 (2016) 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pichichero ME, Casey JR, Almudevar A, Nonprotective responses to pediatric vaccines occur in children who are otitis prone, Pediatr. Infect. Dis. J 32 (2013) 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pichichero ME, Ten-Year Study of the Stringently Defined Otitis-prone Child in Rochester, NY, Pediatr. Infect. Dis. J 35 (2016) 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Basha S, Pichichero ME, Poor memory B cell generation contributes to non-protective responses to DTaP vaccine antigens in otitis-prone children, Clin. Exp. Immunol 182 (2015) 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sharma SK, Casey JR, Pichichero ME, Reduced memory CD4+ T-cell generation in the circulation of young children may contribute to the otitis-prone condition, J. Infect. Dis 204 (2011) 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sharma SK, Casey JR, Pichichero ME, Reduced serum IgG responses to pneumococcal antigens in otitis-prone children may be due to poor memory B-cell generation, J. Infect. Dis 205 (2012) 1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Basha S, Kaur R, Mosmann TR, Pichichero ME, Reduced T-Helper 17 Responses to Streptococcus pneumoniae in Infection-Prone Children Can Be Rescued by Addition of Innate Cytokines, J. Infect. Dis 215 (2017) 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Basha S, Pichichero ME, Decreased TNF family receptor expression on B-cells is associated with reduced humoral responses to Streptococcus pneumoniae infections in young children, Cell Immunol. 320 (2017) 11–19. [DOI] [PubMed] [Google Scholar]
- 195.Kaur R, Casey JR, Pichichero ME, Serum antibody response to five Streptococcus pneumoniae proteins during acute otitis media in otitis-prone and non-otitis-prone children, Pediatr. Infect. Dis. J 30 (2011) 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xu Q, Casey JR, Newman E, Pichichero ME, Otitis-Prone Children Have Immunologic Deficiencies in Naturally Acquired Nasopharyngeal Mucosal Antibody Response after Streptococcus pneumoniae Colonization, Pediatr. Infect. Dis. J 35 (2016) 54–60. [DOI] [PubMed] [Google Scholar]
- 197.Ren D, Murphy TF, Lafontaine ER, Pichichero ME, Stringently Defined Otitis Prone Children Demonstrate Deficient Naturally Induced Mucosal Antibody Response to Moraxella catarrhalis Proteins, Front. Immunol 8 (2017) 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ren D, Almudevar AL, Murphy TF, Lafontaine ER, Campagnari AA, Luke-Marshall N, Pichichero ME, Serum antibody response to Moraxella catarrhalis proteins in stringently defined otitis prone children, Vaccine 37 (2017) 4637–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Harrison A, Dubois LG, John-Williams LS, Moseley MA, Hardison RL, Heimlich DR, Stoddard A, Kerschner JE, Justice SS, Thompson JW, Comprehensive proteomic and metabolomic signatures of nontypeable Haemophilus influenzae-induced acute otitis media reveal bacterial aerobic respiration in an immunosuppressed environment, Mol. Cell. Proteomics, 15 (2016) 1117–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Baddal B, Muzzi A, Censini S, Calogero RA, Torricelli G, Guidotti S, Taddei AR, Covacci A, Pizza M, Rappuoli R, Dual RNA-seq of nontypeable Haemophilus influenzae and host cell transcriptomes reveals novel insights into host-pathogen cross talk, MBio 6 (2015) e01765–15. [DOI] [PMC free article] [PubMed] [Google Scholar]


