Abstract
Objective:
To identify differences in the transcriptomic profiles during placentation from pregnancies conceived spontaneously to those with infertility, using non-in vitro fertilization (NIFT) or in vitro fertilization (IVF).
Design:
Cohort study.
Setting:
Academic Medical Center.
Patient(s):
Women undergoing chorionic villus sampling at gestational age 11–13 weeks (n=141), with pregnancies that were conceived spontaneously (n=74), with NIFT (n=33), or IVF (n=34) resulting in the delivery of viable offspring.
Intervention(s):
Collection of chorionic villus samples from women who conceived spontaneously, with NIFT or IVF for gene expression analysis using RNA sequencing.
Main Outcome Measure(s):
Baseline maternal, paternal and fetal demographics, maternal medical conditions, pregnancy complications and outcomes. Differential gene expression of first trimester placenta.
Result(s):
There were few differences in the transcriptome of first trimester placenta from NIFT, IVF, and spontaneous pregnancies. There was one protein-coding differentially expressed gene (DEG) between the spontaneous and infertility groups, CACNA1I, one protein-coding DEG between the spontaneous and IVF groups, CACNA1I, and five protein-coding DEGs between the NIFT and IVF groups, SLC18A2, CCL21, FXYD2, PAEP, and DNER.
Conclusion(s):
This is the first and largest study looking at transcriptomic profiles of first trimester placenta demonstrating similar transcriptomic profiles in pregnancies conceived using NIFT or IVF and spontaneous conceptions. Gene expression differences found to be highest in the NIFT group suggest that the underlying infertility, in addition to treatmentrelated factors, may contribute to the observed gene expression profiles.
Keywords: Non-IVF fertility treatment (NIFT), in vitro fertilization (IVF), placentation, RNA Sequencing, Transcriptome
Capsule
The transcriptome during placentation of pregnancies conceived using infertility treatment are similar to that of spontaneously conceived pregnancies with gene expression differences found mainly in the non-IVF fertility treatment pregnancies.
Introduction
Infertility affects about 6.1 million people in the U.S., equivalent to ten percent of the reproductive age population (1). The use of assisted reproductive technologies (ART), including in vitro fertilization (IVF) contributes to 1.5% of live births in the United States and other non-IVF fertility treatments (NIFT) contribute to 4.6% (2, 3). Adverse pregnancy outcomes have been associated with ART, including low birth weight and small for gestational age babies, preeclampsia, retained placenta, placental abruption, placenta previa, preterm labor and delivery, and birth defects compared to pregnancies conceived spontaneously (2, 4–11). However, it is unclear whether these adverse outcomes are the result of the ART procedures, such as IVF or the underlying infertility, as pregnancies conceived by couples utilizing other types of fertility treatments, NIFT, are also at increased risk of adverse outcomes, including placental abruption, fetal loss, and gestational diabetes (12). Furthermore, pregnancies conceived in couples with infertility regardless of treatment are at increased risk of adverse outcomes, including placenta accreta (13), earlier gestational age at delivery, late preterm birth, greater neonatal intensive care unit admissions (11, 14) as well as birth defects (15–17).
As many of these adverse outcomes are related to placentation, a better understanding of placental function in pregnancies conceived in couples with infertility, utilizing fertility treatments, may uncover the underlying pathology leading to adverse outcomes. Previous studies examining the transcriptome of human placenta have been limited to term placentas with specific placenta pathologies, predominantly preeclampsia (18). Yet it is during the first trimester of a pregnancy, during placentation when trophoblast proliferation, differentiation, and invasion, as well as angiogenesis and vasculogenesis, crucial for laying the groundwork for successful placental function, are taking place and a necessary time point to study placental development (19). It is also the closest timepoint to conception, either spontaneously or through fertility treatments, that can be studied, in order to minimize placental changes due to placental pathology and not the mode of conception. This is the first and largest study looking at the first trimester placenta (chorionic villi) transcriptome in our SMAART (Spontaneously/Medically Assisted/ART) study cohort consisting of pregnancies that delivered from couples either with infertility, utilizing NIFT or IVF, and pregnancies conceived spontaneously.
Materials and Methods
Patient Population
Our SMAART Study cohort consisted of pregnancies conceived either spontaneously or in couples with infertility who conceived either through NIFT or IVF, that are in the late first trimester of pregnancy at the time of Chorionic Villus Sampling (CVS) and followed until delivery. NIFT was defined as treatment using either medications for ovulation induction or controlled ovarian stimulation and intrauterine insemination (IUI). IVF pregnancies included fresh or frozen embryo transfers using either cleavage stage or blastocyst stage embryos. Our SMAART Study cohort consisted of 409 singleton pregnancies, of which 208 were spontaneous conceptions and 201 pregnancies conceived with a history of infertility. Of the infertility group, 90 were conceived with NIFT and 111 were conceived with IVF. All pregnancies had a normal karyotype and delivered (20).
Transcriptomic profiling was performed on a subset of 141 subjects that had chorionic villi available from the first trimester. The SMAART Transcriptome cohort consisted of 74 spontaneous conceptions and 67 pregnancies conceived with a history of infertility. Of the infertility group, 33 were conceived with NIFT and 34 were conceived with IVF.
Demographics Statistical Analysis
T-test and Analysis of Variance (ANOVA) were used for the baseline and pregnancy outcome demographics.
CVS collection
Chorionic villus samples were collected at 11–13 weeks gestation at the Cedars-Sinai Prenatal Diagnostic Center as previously described (21, 22). Left over tissue after clinical genetic testing was collected from consenting patients per institutional review board (IRB)-approved protocol and placed in RNAlater RNA Stabilization Reagent (Qiagen, Valencia, CA) and stored at −80°C in Cedars-Sinai Prenatal Biorepository until processing.
RNA Extraction for RNA-sequencing
CVS tissues were thawed on ice in 1% β-mercaptoethanol-containing lysis buffer using AllPrep DNA/RNA Extraction Mini kit (Qiagen). Cells were homogenized by repeatedly passing through single-use needles with decreasing gauge (22G, 25G, then 27G) attached to a 1ml sterile, RNase-free syringe before being loaded onto AllPrep spin columns (21, 22). DNA/RNA extraction was performed according to manufacturer’s instructions and previously described. Purity of the extracted total RNA was evaluated using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE) and the concentration of the total RNA measured using the Qubit Fluorometer (Thermo Fisher Scientific, Carlsbad, CA). RNA integrity was assessed using the Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA) that analyzes the integrity of the total RNA by measuring the ratio between the 18S and 28S ribosomal peaks.
RNA-Sequencing and Statistical Analysis
RNA-Seq libraries were constructed from 200 ng of total RNA using Illumina TruSeq Stranded Total RNA with Ribo-Zero Gold sample prep kits (Illumina, Carlsbad, CA). Constructed libraries contained RNAs >200 nt and were depleted of cytoplasmic and mitochondrial ribosomal RNAs. RNA-seq reads were assessed for quality using FastQC (23). Transcript abundances were then quantified against the human reference genome, (Ensembl build GRCh38) using Kallisto (24), and read into the R statistical computing environment (25) as gene-level counts using the tximport package (26). We then used the DESeq2 Bioconductor package (27, 28) to normalize for differences in sequencing depth between samples (using the default median-of-ratios method), estimate dispersion, and fit a negative binomial model for each gene. Datasets were adjusted for fetal sex and sequencing runs. The Benjamini-Hochberg False Discovery Rate (FDR) procedure (29) was then used to reestimate the adjusted p-values.
Pathway Analysis
To investigate biological significance of expressed transcripts, Ingenuity Pathway Analysis (IPA) software (Qiagen, Redwood City, CA, http://www.qiagen.com/ingenuity) was used to analyze enriched pathways. Differentially expressed genes were first filtered as significant at P-value < 0.05 and uploaded to IPA platform to perform Core Analyses, which rank pathways represented by the dataset and upstream regulators according to P-values calculated using Fisher’s exact test.
Results
Clinical characteristics of SMAART Transcriptome cohort
From the SMAART cohort which consisted of 409 singleton pregnancies, all subjects underwent chorionic villus sampling for diagnostic testing. All subjects had a fetus with normal karyotype analysis and birth outcomes (20). Extra chorionic villi from the first trimester was available after tissue was removed for genetic testing for 141 subjects that was available for transcriptome profiling and analysis. This SMAART Transcriptome cohort, which was matched for maternal age, race, ethnicity and fetal sex, consisted of 74 spontaneous conceptions and 67 pregnancies conceived with a history of infertility. Of the infertility group, 33 were conceived with NIFT and 34 were conceived with IVF. When comparing pregnancies conceived spontaneously versus those from couples with infertility, there was no significant difference in maternal and paternal age and race. There was no significant difference in fetal sex distribution or fetal race. There was no difference in maternal underlying conditions but a significant difference in maternal BMI (22 ± 3.2 and 24 ± 5.5, respectively). When pregnancy outcomes were evaluated, there was a higher rate of Cesarean section and pregnancy-related diabetes in the infertility group (Table 1). When birth outcomes were examined, there was no difference in gestational age at delivery but a significant difference in birthweight (3430 ± 590 grams for spontaneous and 3192 ± 623 grams for infertility). When the infertility group was divided into pregnancies conceived with NIFT or IVF, the significant difference in maternal BMI was found to be between spontaneous and NIFT pregnancies (22 ± 3.2 versus 25 ± 6.3, respectively), rather than between spontaneous and IVF pregnancies. There was a higher rate of Cesarean section in the NIFT and IVF pregnancies, as was observed in the SMAART cohort. The difference in birthweight was found to be between spontaneous and IVF pregnancies (3430 ± 590 grams in spontaneous versus 3082 ± 586 grams in IVF) (Table 1).
Table 1.
Baseline demographics and pregnancy outcomes for spontaneous versus infertility, and spontaneous versus NIFT versus IVF SMAART Transcriptome cohort.
| SPONT (N=74) |
INFERT (N=67) |
P-value SPONT vs INFERT |
NIFT (N=33) |
IVF (N=34) |
P-value SPONT vs NIFT vs IVF |
P-value Differences among groups |
|
|---|---|---|---|---|---|---|---|
| BASELINE DEMOGRAPHICS | |||||||
| Maternal age (years) | 39 ± 2.2 | 40 ± 3.0 | 0.2483 | 40 ± 2.7 | 40 ± 3.2 | 0.4985 | |
| Paternal age (years) | 41 ± 4.9 | 41 ± 6.9 | 0.6589 | 40 ± 6.3 | 43 ± 7.1 | 0.1092 | |
| Maternal race | 0.275 | 0.326 | |||||
| Caucasian | 71, 96% | 60, 90% | 30, 91% | 30, 88% | |||
| Asian | 3, 4.1% | 6, 9.0% | 3, 9.1% | 3, 8.8 | |||
| African American | |||||||
| Biracial (Caucasian/Asian) | 0 | 1, 1.5% | 0 | 1, 2.9% | |||
| Biracial (Other) | |||||||
| Paternal race | 0.308 | 0.546 | |||||
| Sex of fetus | 0.455 | 0.416 | |||||
| Male | 40, 54% | 32, 48% | 18, 55% | 14, 41% | |||
| Female | 34, 46% | 35, 52% | 15, 45% | 20, 59% | |||
| Fetal race | 0.130 | 0.181 | |||||
| Caucasian | 68, 92% | 56, 84% | 29, 88% | 27, 79% | |||
| Asian | |||||||
| African American | |||||||
| Biracial (Caucasian/Asian) | 6, 8.1% | 11, 16% | 4, 12% | 7, 21% | |||
| Biracial (Other) | |||||||
| Multiracial | |||||||
| Maternal BMI | 22 ± 3.2 | 24 ± 5.5 | 0.0078 | 25 ± 6.3 | 23 ± 4.5 | 0.0108 | 0.009 (Spont vs NIFT) |
| Hypertension | 2, 2.7% | 2, 3.0% | 0.92 | 1, 3.0% | 1, 2.9% | 0.995 | |
| Diabetes | 0 | 3, 4.5% | 0.066 | 1, 3.0% | 2, 5.9% | 0.133 | |
| Thyroid disease | 16, 22% | 14, 21% | 0.916 | 7, 21% | 7, 21% | 0.993 | |
| Other medical conditions | 1, 1.4% | 2, 3.0% | 0.502 | 1, 3.0% | 1, 2.9% | 0.798 | |
| Gestational age at CVS (days) | 83 ± 6.1 | 82 ± 6.4 | 0.4572 | 82 ± 6.6 | 82 ± 6.4 | 0.7324 | |
| Crown rump length (mm) | 56 ± 11 | 53 ± 11 | 0.1701 | 54 ± 11 | 53 ± 12 | 0.3504 | |
| PREGNANCY OUTCOMES | |||||||
| Gestational age at delivery (days) | 273 ± 13 | 270 ± 14 | 0.2004 | 272 ± 13 | 268 ± 15 | 0.1399 | |
| Birthweight (grams) | 3430 ± 590 | 3192 ± 623 | 0.0317 | 3294 ± 647 | 3082 ± 586 | 0.04 | 0.035 (Spont vs IVF) |
| Mode of delivery | 0.007 | ||||||
| Vaginal, Spontaneous | 44, 64% | 23, 37% | 12, 39% | 11, 34% | |||
| Cesarean Section | 25, 36% | 40, 63% | 0.002 | 19, 61% | 21, 66% | ||
| Pregnancy complications | 10, 14% | 18, 27% | 0.047 | 7, 21% | 11, 32% | 0.073 | |
| Hypertension | 3, 4.1% | 4, 6.0% | 0.601 | 1, 3.0% | 3, 8.8% | 0.481 | |
| Diabetes | 5, 6.8% | 13, 19% | 0.025 | 6, 18% | 7, 21% | 0.077 | |
| Coagulation disorders | 0 | 2, 3.0% | 0.134 | 1, 3.0% | 1, 2.9% | 0.326 | |
| Placenta previa | 2, 2.7% | 2, 3.0% | 0.920 | 0 | 2, 5.9% | 0.348 | |
| Placental abruption | 1, 1.4% | 0 | 0.340 | 0 | 0 | 0.634 | |
| Placenta other | 0 | 0 | |||||
P-values < 0.05 are in bold. (SPONT: spontaneous, INFERT: infertility)
Transcriptome
Two-Dimensional Principal component analysis (2D-PCA), which projects the RNA-seq dataset onto two dimensions to identify principal causes of variation in the dataset as represented by clustering of samples as data points (30), revealed that RNA sequencing samples clustered together with similar data points regardless of pre-pregnancy maternal health conditions and medications, pregnancy complications, or in cases of IVF pregnancies, whether embryo transfer was performed with fresh versus frozen embryos or at cleavage versus blastocyst stages (data not shown) and therefore did not contribute to differences in outcomes. As there was clustering based on fetal sex, also demonstrated by other studies showing differences in the first trimester placenta due to sex (31), we corrected the dataset for fetal sex. In addition, there was clustering due to fetal race so we selected to use our largest populations that clustered together which included Caucasian and Caucasian-Asian fetuses.
All samples had at least 10 million uniquely mapped reads with an average of 20.8 million uniquely mapped reads per sample. All samples had at least 67% reads uniquely mapped with an average of 82% reads uniquely mapped per sample.
Overall, there were small differences in the transcriptome of first trimester placenta from spontaneous, NIFT, and IVF pregnancies (Figure 1) as the PCA plots of spontaneous versus infertility (Figure 1A) and spontaneous versus NIFT versus IVF pregnancies (Figure 1B) illustrate. Samples clustered according to sample subset (shown on the PC1 axis) and fetal sex (shown on the PC2 axis) (Figure 1), both of which were corrected in our analysis. When the transcriptome of spontaneous pregnancies was compared to that of pregnancies conceived with infertility treatment, of the 61799 genes examined, following adjustment for multiple comparisons, one protein-coding gene, CACNA1I was significantly differentially expressed at FDR of 5%, with 6.82-fold higher expression in the spontaneous group than in the infertility group (Table 2).
Figure 1.
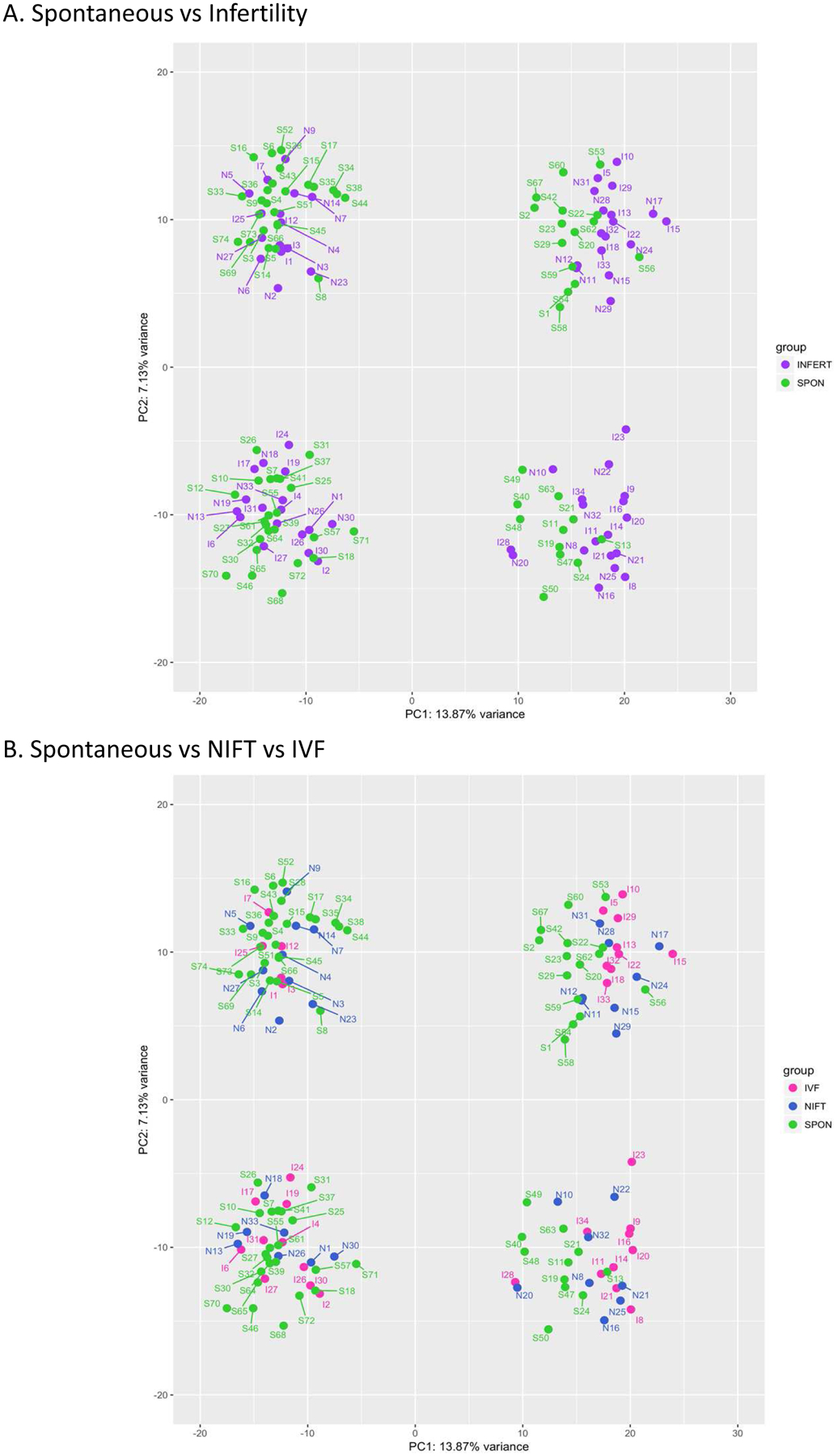
Principal component analysis (PCA) plots of (A) spontaneous versus infertility pregnancies, (B) spontaneous versus NIFT versus IVF pregnancies. Samples clustered according to sample subset (shown on the PC1 axis) and fetal sex (shown on the PC2 axis).
(SPON: spontaneous, INFERT: infertility, Green: spontaneous, Purple: infertility, Blue: NIFT, Pink: IVF)
Table 2.
Differentially expressed genes in spontaneous versus infertility, spontaneous versus IVF, and NIFT versus IVF pregnancies.
| Spontaneous vs Infertility | |||
|---|---|---|---|
| SYMBOL | DESCRIPTION | FC | P adj |
| CACNA1I | Calcium voltage-gated channel subunit alphal | 6.82 | 0.000779 |
| Spontaneous vs IVF | |||
| SYMBOL | DESCRIPTION | FC | P adj |
| CACNA1I | Calcium voltage-gated channel subunit alpha1 | 9.85 | 0.000568 |
| NIFT vs IVF | |||
| SYMBOL | DESCRIPTION | FC | P adj |
| SLC18A2 | Solute carrier family 18 member A2 | 6.41 | 0.000283 |
| CCL21 | C-C motif chemokine ligand 21 | 6.02 | 0.0118 |
| FXYD2 | FXYD domain containing ion transport regulator 2 | 3.39 | 0.0235 |
| PAEP | Progestagen associated endometrial protein | 3.94 | 0.0137 |
| DNER | Delta/notch like EGF repeat containing | 3.73 | 0.0291 |
(FC: fold change, P adj: adjusted P-value)
When we compared the transcriptome profiles among spontaneous, NIFT and IVF conceptions, CACNA1I was found to be the only significantly differentially expressed gene between spontaneous and IVF pregnancies again, with its expression 9.85-fold higher in the spontaneous group (Table 2). There were no protein-coding genes significantly differentially expressed between spontaneous and NIFT pregnancies at FDR of 5%. Comparing the transcriptome of NIFT versus IVF pregnancies, five protein-coding genes (SLC18A2, CCL21, FXYD2, PAEP and DNER) were significantly upregulated in NIFT pregnancies compared to IVF pregnancies, with fold changes ranging from 3.39 to 6.41 (Table 2). When expression levels of these DEGs represented by fragments per kilobase transcript per million (FPKM), a normalized version of RNA expression which accounts for gene length and sequencing depth (32), among the three groups were plotted and examined, the NIFT group had the highest expression of SLC18A2, CCL21, FXYD2, PAEP and DNER, with IVF pregnancies clustering with spontaneous pregnancies (Figure 2C).
Figure 2.
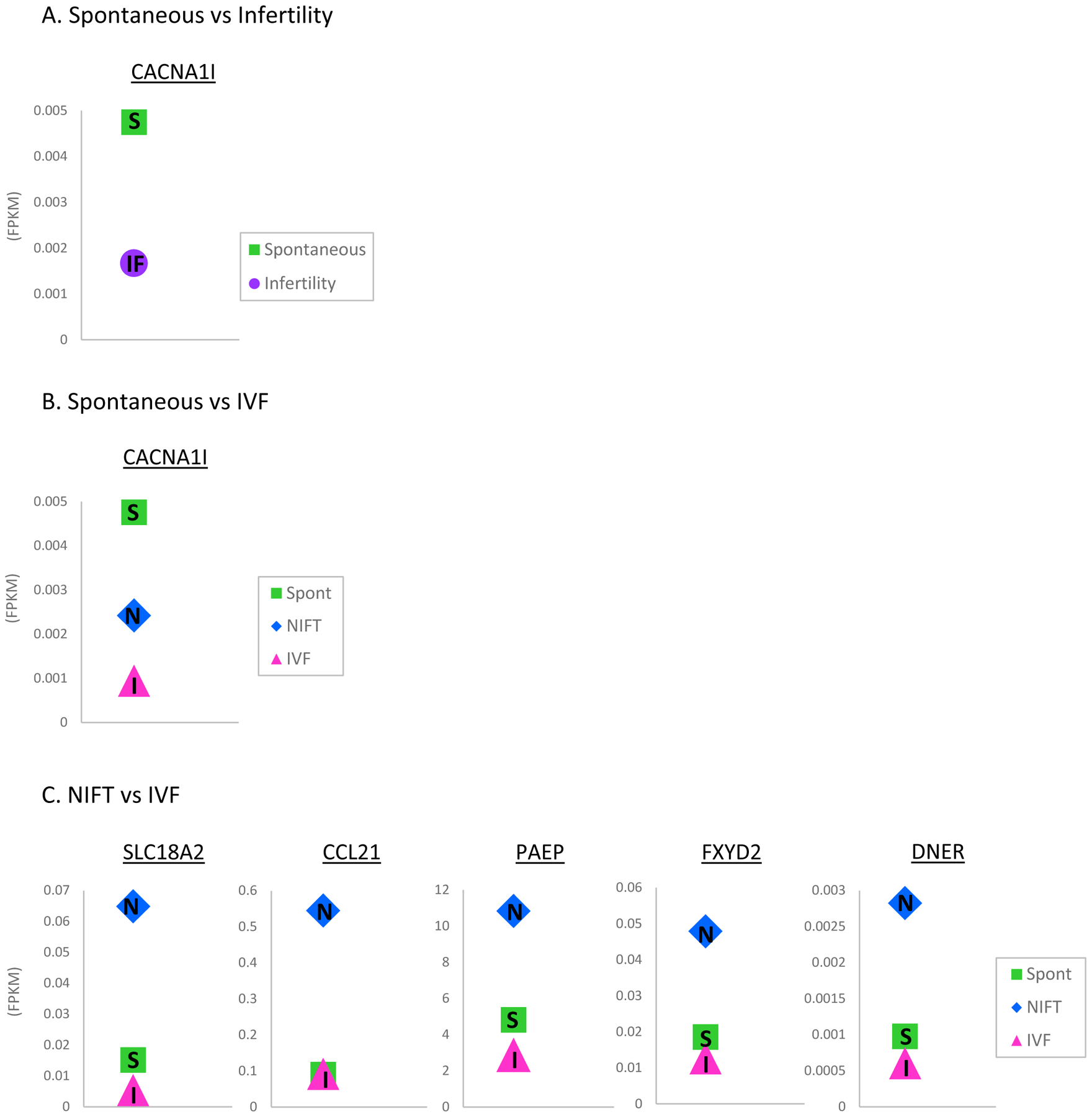
Gene expression levels of differentially expressed genes in (A) CACNA1I is differentially expressed in spontaneous versus infertility pregnancies,(B) CACNA1I is differentially expressed in spontaneous versus IVF, and (C) SLC18A2, CCL21, FXYD2, PAEP and DNER are significantly upregulated in NIFT pregnancies compared to IVF pregnancies with spontaneous pregnancies segregating with IVF pregnancies.
(S: spontaneous, IF: infertility, N: NIFT, I: IVF, Green: spontaneous, Purple: infertility, Blue: NIFT, Pink: IVF, FPKM: fragments per kilobase of transcript per million mapped reads)
Pathway Analysis
Due to the small number of differentially expressed genes, gene ontology studies were not possible for the DEGs that were identified following adjustment for multiple comparisons. However, in order to identify enriched pathways, differentially expressed genes with P-value < 0.05 were analyzed with IPA. When we used the p-value cutoff of 0.05 there were 966 genes that were significantly different between spontaneous and infertility, 729 genes between spontaneous and NIFT, 893 genes between spontaneous and IVF, and 482 between NIFT and IVF pregnancies. IPA of these DEGs revealed similar canonical pathways in all comparisons with IVF subjects, including spontaneous versus infertility, spontaneous versus IVF, and NIFT versus IVF pregnancies (Supplemental Table 1). Pathogenesis of Multiple Sclerosis was the most represented in those comparison datasets (Supplemental Table 1), but this pathway was not represented in spontaneous versus NIFT pregnancies. Cytokines and cytokine receptors involved in Pathogenesis of Multiple Sclerosis were down-regulated in infertility and IVF pregnancies compared to spontaneous pregnancies; CCR5, CXCL9, and CXCL10 were down-regulated in infertility compared to spontaneous pregnancies, CCR5, CXCL9, CXCL10, and CXCL11 down-regulated in IVF compared to spontaneous pregnancies, and CXCL9, CXCL10, and CXCL11 were down-regulated in IVF compared to NIFT pregnancies. None of these genes were differentially expressed in spontaneous versus NIFT (P-values ranged from 0.305 to 0.928). Another enriched canonical pathway, Neuroinflammatory Signaling Pathway, was significantly enriched in all three comparisons to spontaneous pregnancies, (Supplemental Table 1), but not significantly enriched in NIFT versus IVF (P-value = 0.11). Neuroinflammatory Signaling Pathway genes were more represented in spontaneous versus IVF pregnancies than other comparisons, and nine pathway genes overlapped in all comparisons to spontaneous pregnancies, five consistently downregulated in spontaneous pregnancies (CALB1, GABRR1, HLA-A, IKBKG, and TNF) and four consistently upregulated (GABRA2, GABRG3, HLA-A, PIK3CA, and PPP3CB). Antigen Presentation Pathway was significantly enriched in spontaneous versus IVF (P-value = 5.85 × 10−4) and NIFT versus IVF (P-value = 1.07 × 10−3) (Supplemental Table 1), and less significant in spontaneous versus NIFT (P-value = 0.0309). In general, all enriched canonical pathways were immune related, but spontaneous versus NIFT pregnancies were represented by a more diverse set of canonical pathways which included Crosstalk between Dendritic Cells and Natural Killer Cells and Production of Nitric Oxide and Reactive Oxygen Species in Macrophages (Supplemental Table 1).
Upstream analysis was performed to identify master regulators of DEGs with P-value < 0.05. In spontaneous versus IVF pregnancies, cytokines were abundantly represented among the most significant upstream regulators of DEGs, including interleukins such as IL27, various interferon-alpha subtypes, chemokine ligands such as CCL11, and other cytokine families (Supplemental Table 2). Notably, cytokines were rarely found among upstream regulators of spontaneous versus NIFT pregnancy DEGs (Supplemental Table 3).
Discussion
Transcriptomic profiling of the first trimester placenta (chorionic villi) from spontaneous, NIFT, and IVF pregnancies demonstrated little differences in global gene expression among the three groups. One protein-coding gene (CACNA1I) was significantly differentially expressed in infertility pregnancies when compared to spontaneous pregnancies and this differential expression of CACNA1I was significant between spontaneous and IVF pregnancies as well. There were no DEGs between spontaneous versus NIFT pregnancies. Five protein-coding genes (SLC18A2, CCL21, FXYD2, PAEP, and DNER) were significantly differentially expressed in NIFT versus IVF pregnancies.
A number of DEGs had higher expression levels in NIFT pregnancies with IVF pregnancies segregating more with spontaneous pregnancies (Figure 2C). This result complements our previous finding in a pilot study demonstrating differential methylation between NIFT and IVF pregnancies at select loci, with global methylation remaining unchanged among spontaneous, NIFT and IVF pregnancies (33). In addition, it may reflect the differences in the underlying cause of infertility, and not the IVF itself, where fertilization occurs in the laboratory and has been a source implicated in adverse outcomes. This is also consistent with characteristics of our cohort, where the NIFT group had a higher BMI, which may indicate a different etiology for infertility, including polycystic ovary syndrome (PCOS). As adverse outcomes exist, some likely due to an underlying placental pathology, it also remains to be determined if adverse outcomes are due to the underlying genetics of infertility or the intrauterine environment throughout gestation, as differences in methylation status in placenta at delivery have been reported in pregnancies conceived spontaneously versus those conceived using ART (33–37).
Since race did play a potential role in our transcriptome study, we opted to look at the Caucasian and Caucasian-Asian cohort as this group had the most similar global transcriptomic profile and would limit variability based on race, since race has been reported to play a role in reproductive and birth outcomes (38, 39).
Although global transcriptomic profiles were not found to differ, differences in specific gene expression and their potential roles in placental development may provide insight into long term regulation of placental function, as we did find differences in fetal growth with birth weight, with a lower birth weight in the IVF group among our transcriptomic cohort (Table 1).
The one gene found to be differentially expressed between pregnancies conceived spontaneously and those conceived with infertility treatment, specifically with IVF was CACNA1I. CACNA1I encodes the pore-forming alpha subunit of a voltage-gated calcium channel (VGCC) belonging to the low voltage-activated, T-type calcium channel subfamily. VGCCs mediate calcium (Ca2+) signaling upon depolarization of membrane potential created by the Ca2+ gradient, and activates a variety of downstream events, including muscle contraction, excitation of neurons, regulation of gene expression, and release of hormones or neurotransmitters (40–42). In the placenta, Ca2+ signaling has been reported to be primarily mediated by the high voltage-activated, L-type VGCCs and transient receptor potential (TRP) channels (43, 44).
While the role of T-type VGCC in placentation remains to be elucidated, the presence of other calcium channels including L-type VGCCs has been identified in syncytiotrophoblasts (43, 44) and Ca2+ plays a role in hormone secretion regulation, including human chorionic gonadotropin (hCG) and human placental lactogen, which is important in fetal growth and development (45–52). Calcium channel-mediated Ca2+ influx regulates vasoconstriction of uterine radial arteries through L- and T-type VGCCs (53). Remodeling of uterine vasculature is critical for development of a healthy placenta to ensure normal fetal development. Impaired vascular remodeling has been associated with spontaneous abortion, intrauterine growth restriction, preeclampsia, and lower birth weights (54–59). Thus, decreased expression of CACNA1I in IVF pregnancies may be impacting regulation of Ca2+ homeostasis in the placenta and leading to impaired vascular remodeling and altered hormone secretion necessary for normal placentation and fetal development.
Among the genes significantly differentially expressed between NIFT versus IVF pregnancies are SLC18A2, CCL21, FXYD2 and PAEP, which may be involved in regulation of immune responses and transport of signaling molecules at the maternal-fetal interface. SLC18A2 encodes a vesicular monoamine transporter which acts to accumulate cytosolic monoamines, such as norepinephrine, serotonin, dopamine and histamine, into synaptic vesicles. It has been reported that histamine plays a role in decidualization, implantation, as well as immune and blood flow regulation (60–63). There are many transporter proteins expressed in the placenta which normally keep extracellular monoamine concentrations low (64, 65). During decidualization, however, the accumulated histamine in intracellular vesicles is released and stimulates initial decidual formation (66, 67), as monoamines are potent vasoactive agents. In fact, an elevated concentration of monoamines was reported in preeclamptic pregnancies (68), possibly through monoamine transporters. Thus, SLC18A2 may be important in regulating monoamines necessary for regulation of uterine and placental blood flow at the maternal fetal interface.
CCL21 is a small cytokine belonging to the CC chemokine family and is a ligand for chemokine receptor 7 (CCR7). As their function is to recruit immune cells in response to inflammation or homeostatic conditions, cytokines play an active role at the maternal-fetal interface by modulating interactions between the decidua and trophoblasts through its selective recruitment of leukocytes (69–71). Trophoblasts have been shown to actively recruit immune cells through chemokine production and expression of different chemokine-receptor profiles. For example, it has been shown that CXCL12 secreted by human first trimester trophoblast cells stimulates the decidualization process through increased production of CXCR4 (72). In other cell types, CCL21/CCR7 pair has been shown to promote growth and metastasis in tumors by stimulating angiogenesis and lymphangiogenesis (73, 74). Thus, CCL21 may be acting as a chemokine promoting decidualization at the maternal-fetal interface by stimulating angiogenesis/lymphangiogenesis necessary for proper placentation and placental function throughout gestation as it remains expressed in term placental mesenchymal stem cells (75). FXYD2 encodes the sodium/potassium (Na+/K+)-transporting ATPase subunit gamma of the FXYD family of transmembrane protein. In the placenta, Na+/K+ ATPases are found in both the microvillous and basal membranes of syncytiotrophoblast (76), and play an important role in transport of nutrients. Inhibition of Na+/K+ ATPase, impairs first trimester cytotrophoblast cell proliferation, migration, and invasion (77) and reductions in Na+/K+ ATPase activity were seen in syncytiotrophoblast plasma membranes from intrauterine growth restricted placentas (76). Thus, misregulation of FXYD2 may affect function of Na+/K+ transporters impairing nutrient transport to the fetus and resulting in growth restricted phenotypes.
Glycodelin-A, the protein encoded by PAEP is a glycoprotein found abundantly in the endometrium, decidua, and amniotic fluid and has been identified as a marker of endometrial receptivity (78). It plays an important role in regulation of immune responses at the maternal-fetal interface to help prevent immune rejection of the fetus. Specifically, glycodelin-A facilitates the type 1 T helper cell (Th1)-type 2 T helper cell (Th2) shift of T lymphocytes in the decidua during pregnancy to increase Th2 and decrease Th1, balancing activity and cytokine secretion by the different T lymphocyte populations. Additionally, glycodelin-A regulates cytokine production by natural killer cells and dendritic cells to induce a tolerogenic phenotype (78–80). Glycodelin-A has also been suggested to induce differentiation of cytotrophoblast, as evidenced by an increase in the extravillous trophoblast (EVT) lineage population and stimulation of hCG production (81, 82). Thus, altered expression of PAEP may lead to impaired immune regulation at the maternal-fetal interface as well as a disrupted trophoblast differentiation, resulting in placental dysfunction.
DNER encodes a transmembrane protein that binds Notch1 (83). In early pregnancy, the Notch signaling pathway functions through cell-cell interactions to regulate stem cell renewal and differentiation, decidualization, implantation, placentation, and angiogenesis (84). Notch1 protein expression is higher in the mid-secretory (receptive phase) endometrium of fertile women, compared to infertile women (85). Thus, DNER may be important for maternal embryonic/fetal communication leading to appropriate placentation and downregulation of DNER in IVF pregnancies may impact this communication. Further functional studies are necessary to understand differences at the maternal- embryonic/fetal interface.
Enrichment analysis of top canonical pathways suggests that differences in fertility are largely linked to differences in inflammatory pathways during the late first trimester. Specifically, the C-X-C motif chemokine ligand family (including CXCL9, CXCL10, CXCL11) associated with autoimmunity (e.g. Multiple Sclerosis) seems to be lower in IVF pregnancies, driving differences in the infertility group. These C-X-C motif cytokines are important in immune cell migration at the maternal-fetal interface and increase implantation success (86, 87). Further, in upstream analysis, multiple cytokine families were among the top upstream regulators that predicted transcript differences in spontaneous versus IVF pregnancies, but not spontaneous versus NIFT pregnancies. These results suggest that IVF pregnancies may have slightly altered regulation of cytokine signaling pathways in late first trimester, compared to other pregnancies, which may be altering normal placentation. By contrast, spontaneous versus NIFT pregnancies had more diverse top canonical pathways and upstream regulators, which may reflect different etiology of infertility between NIFT and IVF pregnancies.
Limitations in our study include the racial distribution of our study population as the majority of our cohort was Caucasian. Future studies looking at the impact of race on transcriptomic differences of placenta will be important. Additionally, as CVS is more commonly performed in women of advanced reproductive age, we were limited by advanced maternal age in our cohort. However, multiple studies, including our own have identified increased maternal age with infertility (10, 11, 15). Although maternal age in our infertility cohort was greater, it was only slightly increased which may not be clinically significant and the spontaneous conceptions of advanced maternal age minimize differences that may be due to maternal age. Another limitation was that chorionic villi consist of multiple cell types. Future studies looking at specific cell types within the chorionic villus tree will be necessary to determine their functional activity in overall fetal outcomes and how they relate to the underlying infertility or treatments utilized.
Conclusions
In conclusion, this is the first and largest study to demonstrate that the transcriptomic profiles of first trimester placenta, during placentation, in pregnancies conceived with infertility using NIFT or IVF and spontaneous conceptions are similar, which is reassuring, that overall pregnancies conceived as a result of infertility treatments are similar to spontaneous conceptions. However, there is a select group of genes that are differentially expressed, with the greatest differences found in the NIFT cohort, suggesting that it may be the underlying infertility, which may account for differences in overall outcomes. Further studies are needed to look at the functional role of these genes in placental development and function.
Supplementary Material
Supplemental Table 1. Enriched canonical pathways from differentially expressed genes (P-values < 0.05) in spontaneous versus infertility, spontaneous versus NIFT, spontaneous versus IVF, and NIFT versus IVF pregnancies.
Supplemental Table 2. Upstream regulators of differentially expressed genes (P-values < 0.05), spontaneous versus IVF pregnancies.
Supplemental Table 3. Upstream regulators of differentially expressed genes (P-values < 0.05), spontaneous versus NIFT pregnancies.
Grant Support:
The research was supported by the NICHD of the National Institutes of Health under award number R01HD074368. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conference presentation: Presented at the 65th Annual Meeting of the Society for Reproductive Investigation, March 6–10, 2018, San Diego, CA.
Conflicts of Interest: None
References
- 1.Faddy MJ, Gosden MD, Gosden RG. A demographic projection of the contribution of assisted reproductive technologies to world population growth. Reprod Biomed Online 2018;36:455–8. [DOI] [PubMed] [Google Scholar]
- 2.Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS. Low and very low birth weight in infants conceived with use of assisted reproductive technology. The New England journal of medicine 2002;346:731–7. [DOI] [PubMed] [Google Scholar]
- 3.Sunderam S, Kissin DM, Crawford SB, Folger SG, Boulet SL, Warner L et al. Assisted Reproductive Technology Surveillance - United States, 2015. MMWR Surveill Summ 2018;67:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson RA, Gibson KA, Wu YW, Croughan MS. Perinatal outcomes in singletons following in vitro fertilization: a meta-analysis. Obstetrics and gynecology 2004;103:551–63. [DOI] [PubMed] [Google Scholar]
- 5.Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. The New England journal of medicine 2002;346:725–30. [DOI] [PubMed] [Google Scholar]
- 6.Verlaenen H, Cammu H, Derde MP, Amy JJ. Singleton pregnancy after in vitro fertilization: expectations and outcome. Obstetrics and gynecology 1995;86:906–10. [DOI] [PubMed] [Google Scholar]
- 7.Klemetti R, Gissler M, Sevon T, Koivurova S, Ritvanen A, Hemminki E. Children born after assisted fertilization have an increased rate of major congenital anomalies. Fertility and sterility 2005;84:1300–7. [DOI] [PubMed] [Google Scholar]
- 8.Rimm AA, Katayama AC, Diaz M, Katayama KP. A meta-analysis of controlled studies comparing major malformation rates in IVF and ICSI infants with naturally conceived children. J Assist Reprod Genet 2004;21:437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stromberg B, Dahlquist G, Ericson A, Finnstrom O, Koster M, Stjernqvist K. Neurological sequelae in children born after in-vitro fertilisation: a population-based study. Lancet 2002;359:461–5. [DOI] [PubMed] [Google Scholar]
- 10.Jackson S, Hong C, Wang ET, Alexander C, Gregory KD, Pisarska MD. Pregnancy outcomes in very advanced maternal age pregnancies: the impact of assisted reproductive technology. Fertil Steril 2015;103:76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ET, Ramos L, Vyas N, Bhasin G, Simmons CF, Pisarska MD. Maternal and neonatal outcomes associated with infertility. J Matern Fetal Neonatal Med 2018:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shevell T, Malone FD, Vidaver J, Porter TF, Luthy DA, Comstock CH et al. Assisted reproductive technology and pregnancy outcome. Obstetrics and gynecology 2005;106:1039–45. [DOI] [PubMed] [Google Scholar]
- 13.LW S, JL C, R B, R D, O M, K C et al. Mode of conception does not affect fetal or placental growth parameters or ratios in early gestation or at delivery. In. Vol. Accepted. Journal of Assited Reproduction and Genetics, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang ET, Sundheimer LW, Spades C, Quant C, Simmons CF, Pisarska MD. Fertility Treatment Is Associated with Stay in the Neonatal Intensive Care Unit and Respiratory Support in Late Preterm Infants. J Pediatr 2017;187:309–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies MJ, Moore VM, Willson KJ, Van Essen P, Priest K, Scott H et al. Reproductive technologies and the risk of birth defects. N Engl J Med 2012;366:1803–13. [DOI] [PubMed] [Google Scholar]
- 16.Merritt TA, Goldstein M, Philips R, Peverini R, Iwakoshi J, Rodriguez A et al. Impact of ART on pregnancies in California: an analysis of maternity outcomes and insights into the added burden of neonatal intensive care. J Perinatol 2014;34:345–50. [DOI] [PubMed] [Google Scholar]
- 17.Boulet SL, Kirby RS, Reefhuis J, Zhang Y, Sunderam S, Cohen B et al. Assisted Reproductive Technology and Birth Defects Among Liveborn Infants in Florida, Massachusetts, and Michigan, 2000–2010. JAMA pediatrics 2016;170:e154934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sõber S, Reiman M, Kikas T, Rull K, Inno R, Vaas P et al. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci Rep 2015;5:13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton GJ, Jauniaux E. What is the placenta? American journal of obstetrics and gynecology 2015;213:S6 e1, S6–8. [DOI] [PubMed] [Google Scholar]
- 20.Sun T, Lee B, Kinchen J, Wang ET, Gonzalez TL, Rotter JI et al. The First Trimester Maternal Metabolomic Profiles in Pregnancies Conceived with In Vitro Fertilization (IVF). J Clin Endocrinol Metab 2018;Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pisarska MD, Akhlaghpour M, Lee B, Barlow GM, Xu N, Wang ET et al. Optimization of techniques for multiple platform testing in small, precious samples such as Human Chorionic Villus Sampling. Prenatal diagnosis 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee B, Kroener LL, Xu N, Wang ET, Banks A, Williams J et al. Function and Hormonal Regulation of GATA3 in Human First Trimester Placentation. Biol Reprod 2016;95:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S A. FastQC A Quality Control tool for High Throughput Sequence Data. In. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, 2010.
- 24.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 25.RDC TR: A Language and Environment for Statistical Computing. In: R Foundation for Statistical Computing, 2010. [Google Scholar]
- 26.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S A, W H. Differential expression analysis for sequence count data. In. Vol. 11: Genome Biology, 2010:R106–R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Y B, Y H. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. In. Vol. 57: Journal of the Royal Statistical Society. Series B (Methodological), 1995:289–300. [Google Scholar]
- 30.Ringner M What is principal component analysis? Nat Biotechnol 2008;26:303–4. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez TL, Sun T, Koeppel AF, Lee B, Wang ET, Farber CR et al. Sex differences in the late first trimester human placenta transcriptome. Biol Sex Differ 2018;9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nature reviews Genetics 2014;15:121–32. [DOI] [PubMed] [Google Scholar]
- 33.Xu N, Barlow GM, Cui J, Wang ET, Lee B, Akhlaghpour M et al. Comparison of Genome-Wide and Gene-Specific DNA Methylation Profiling in First-Trimester Chorionic Villi From Pregnancies Conceived With Infertility Treatments. Reprod Sci 2017;24:996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 2009;18:3769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP et al. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet 2010;6:e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng HY, Tang Y, Niu J, Li P, Ye DS, Chen X et al. Aberrant DNA methylation of imprinted loci in human spontaneous abortions after assisted reproduction techniques and natural conception. Hum Reprod 2013;28:265–73. [DOI] [PubMed] [Google Scholar]
- 37.Song S, Ghosh J, Mainigi M, Turan N, Weinerman R, Truongcao M et al. DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clinical epigenetics 2015;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buescher PA, Mittal M. Racial disparities in birth outcomes increase with maternal age: recent data from North Carolina. N C Med J 2006;67:16–20. [PubMed] [Google Scholar]
- 39.Shiao SY, Andrews CM, Helmreich RJ. Maternal race/ethnicity and predictors of pregnancy and infant outcomes. Biol Res Nurs 2005;7:55–66. [DOI] [PubMed] [Google Scholar]
- 40.Dolphin AC. Voltage-gated calcium channels and their auxiliary subunits: physiology and pathophysiology and pharmacology. The Journal of physiology 2016;594:5369–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangoni ME, Couette B, Bourinet E, Platzer J, Reimer D, Striessnig J et al. Functional role of L-type Ca(v)13Ca(2+) channels in cardiac pacemaker activity. P Natl Acad Sci USA 2003;100:5543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putzier I, Kullmann PH, Horn JP, Levitan ES. Cav1.3 channel voltage dependence, not Ca2+ selectivity, drives pacemaker activity and amplifies bursts in nigral dopamine neurons. J Neurosci 2009;29:15414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreau R, Hamel A, Daoud G, Simoneau L, Lafond J. Expression of calcium channels along the differentiation of cultured trophoblast cells from human term placenta. Biol Reprod 2002;67:1473–9. [DOI] [PubMed] [Google Scholar]
- 44.Bernucci L, Henriquez M, Diaz P, Riquelme G. Diverse calcium channel types are present in the human placental syncytiotrophoblast basal membrane. Placenta 2006;27:1082–95. [DOI] [PubMed] [Google Scholar]
- 45.Hochberg Z, Bick T, Perlman R, Lahav M, Barzilai D. The modulation of placental lactogen secretion by calcium: studies with cultured human term trophoblast. Mol Cell Endocrinol 1984;37:359–62. [DOI] [PubMed] [Google Scholar]
- 46.Hochberg Z, Perlman R, Bick T. Interrelated calcium ion and cyclic AMP inhibition of placental lactogen secretion by cultured human term trophoblast. Acta Endocrinol (Copenh) 1987;114. [DOI] [PubMed] [Google Scholar]
- 47.Meuris S, Polliotti B, Robyn C, Lebrun P. Ca2þ entry through L-type voltage-sensitive Ca2þ channels stimulates the release of human chorionic gonadotrophin and placental lactogen by placental explants. Biochim Biophys Acta 1994;1220. [DOI] [PubMed] [Google Scholar]
- 48.Robidoux J, Simoneau L, Masse A, Lafond J. Activation of L-type calcium channels induces corticotropin-releasing factor secretion from human placental trophoblasts. . J Clin Endocrinol Metab 2000;85:3356e64. [DOI] [PubMed] [Google Scholar]
- 49.Mathialagan N, Rao AJ. A role for calcium in gonadotrophin-releasing hormone (GnRH) stimulated secretion of chorionic gonadotrophin by first trimester human placental minces in vitro. Placenta 1989;10:61–70. [DOI] [PubMed] [Google Scholar]
- 50.Sharma SC, Rao AJ. Role of calcium in secretion of chorionic gonadotropin by first trimester human placenta. Indian journal of experimental biology 1992;30:1105–10. [PubMed] [Google Scholar]
- 51.Sharma SC, Rao AJ. Effect of calcium depletion on the secretion of newly synthesised human chorionic gonadotropin by first trimester human placenta. Cell Calcium 1993;14:601–7. [DOI] [PubMed] [Google Scholar]
- 52.Handwerger S, Freemark M. The roles of placental growth hormone and placental lactogen in the regulation of human fetal growth and development. Journal of pediatric endocrinology & metabolism : JPEM 2000;13:343–56. [DOI] [PubMed] [Google Scholar]
- 53.Senadheera S, Bertrand PP, Grayson TH, Leader L, Tare M, Murphy TV et al. Enhanced contractility in pregnancy is associated with augmented TRPC3, L-type, and T-type voltage-dependent calcium channel function in rat uterine radial artery. American journal of physiology Regulatory, integrative and comparative physiology 2013;305:R917–26. [DOI] [PubMed] [Google Scholar]
- 54.Meekins JW, Pijnenborg R, Hanssens M, McFadyen IR, van Asshe A. A study of placental bed spiral arteries and trophoblast invasion in normal and severe pre-eclamptic pregnancies. British journal of obstetrics and gynaecology 1994;101:669–74. [DOI] [PubMed] [Google Scholar]
- 55.Vailhe B, Dietl J, Kapp M, Toth B, Arck P. Increased blood vessel density in decidua parietalis is associated with spontaneous human first trimester abortion. Hum Reprod 1999;14:1628–34. [DOI] [PubMed] [Google Scholar]
- 56.Aardema MW, Oosterhof H, Timmer A, van Rooy I, Aarnoudse JG. Uterine artery Doppler flow and uteroplacental vascular pathology in normal pregnancies and pregnancies complicated by pre-eclampsia and small for gestational age fetuses. Placenta 2001;22:405–11. [DOI] [PubMed] [Google Scholar]
- 57.Lisman BA, Boer K, Bleker OP, van Wely M, van Groningen K, Exalto N. Abnormal development of the vasculosyncytial membrane in early pregnancy failure. Fertil Steril 2004;82:654–60. [DOI] [PubMed] [Google Scholar]
- 58.Prefumo F, Güven M, Ganapathy R, Thilaganathan B. The longitudinal variation in uterine artery blood flow pattern in relation to birth weight. Obstet Gynecol 2004;103:764–8. [DOI] [PubMed] [Google Scholar]
- 59.Prefumo F, Sebire NJ, Thilaganathan B. Decreased endovascular trophoblast invasion in first trimester pregnancies with high-resistance uterine artery Doppler indices. Hum Reprod 2004;19:206–9. [DOI] [PubMed] [Google Scholar]
- 60.Hatanaka K, Kitamura Y, Maeyama K, Watanabe T, Matsumoto K. Deciduoma formation in uterus of genetically mast cell-deficient W/Wv mice. Biol Reprod 1982;27:25–8. [DOI] [PubMed] [Google Scholar]
- 61.Dey SK. Role of histamine in implantation: inhibition of histidine decarboxylase induces delayed implantation in the rabbit. Biol Reprod 1981;24:867–9. [DOI] [PubMed] [Google Scholar]
- 62.Cocchiara R, Di Trapani G, Azzolina A, Albeggiani G, Geraci D. Early embryonic histamine-releasing factor: a new model for human implantation. Hum Reprod 1986;1:445–7. [DOI] [PubMed] [Google Scholar]
- 63.Barkai U, Kraicer PF. Intrauterine signaling and embryonic implantation. Biol Signals 1996;5:111–21. [DOI] [PubMed] [Google Scholar]
- 64.Hansson SR, Bottalico B, Noskova V, Casslén B. Monoamine transporters in human endometrium and decidua. Hum Reprod Update 2009;15:249–60. [DOI] [PubMed] [Google Scholar]
- 65.Myllynen P, Immonen E, Kummu M, Vähäkangas K. Developmental expression of drug metabolizing enzymes and transporter proteins in human placenta and fetal tissues. Expert Opin Drug Metab Toxicol 2009;5:1483–99. [DOI] [PubMed] [Google Scholar]
- 66.Bottalico B, Pilka R, Larsson I, Casslen B, Marsal K, Hansson SR. Plasma membrane and vesicular monoamine transporters in normal endometrium and early pregnancy decidua. Mol Hum Reprod 2003;9:389–94. [DOI] [PubMed] [Google Scholar]
- 67.Bottalico B, Larsson I, Brodszki J, Hernandez-Andrade E, Casslén B, Marsál K et al. Norepinephrine transporter (NET), serotonin transporter (SERT), vesicular monoamine transporter (VMAT2) and organic cation transporters (OCT1, 2 and EMT) in human placenta from pre-eclamptic and normotensive pregnancies. Placenta 2004;25:518–29. [DOI] [PubMed] [Google Scholar]
- 68.Carrasco G, Cruz MA, Dominguez A, Gallardo V, Miguel P, González C. The expression and activity of monoamine oxidase A, but not of the serotonin transporter, is decreased in human placenta from pre-eclamptic pregnancies. Life Sci 2000;67:2961–9. [DOI] [PubMed] [Google Scholar]
- 69.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 2011;11:762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robertson SA, Mau VJ, Hudson SN, Tremellen KP. Cytokine-leukocyte networks and the establishment of pregnancy. Am J Reprod Immunol 1997;37:438–42. [DOI] [PubMed] [Google Scholar]
- 71.Gouwy M, Struyf S, Proost P, Van Damme J. Synergy in cytokine and chemokine networks amplifies the inflammatory response. Cytokine Growth Factor Rev 2005;16:561–80. [DOI] [PubMed] [Google Scholar]
- 72.Ramhorst R, Grasso E, Paparini D, Hauk V, Gallino L, Calo G et al. Decoding the chemokine network that links leukocytes with decidual cells and the trophoblast during early implantation. Cell Adh Migr 2016;10:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tutunea-Fatan E, Majumder M, Xin X, Lala PK. The role of CCL21/CCR7 chemokine axis in breast cancer-induced lymphangiogenesis. Mol Cancer 2015;14:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pickens SR, Chamberlain ND, Volin MV, Pope RM, Talarico NE, Mandelin AM et al. Role of the CCL21 and CCR7 pathways in rheumatoid arthritis angiogenesis. Arthritis Rheum 2012;64:2471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abumaree MH, Al Jumah MA, Kalionis B, Jawdat D, Al Khaldi A, AlTalabani AA et al. Phenotypic and functional characterization of mesenchymal stem cells from chorionic villi of human term placenta. Stem cell reviews 2013;9:16–31. [DOI] [PubMed] [Google Scholar]
- 76.Johansson M, Karlsson L, Wennergren M, Jansson T, Powell TL. Activity and protein expression of Na+/K+ ATPase are reduced in microvillous syncytiotrophoblast plasma membranes isolated from pregnancies complicated by intrauterine growth restriction. J Clin Endocrinol Metab 2003;88:2831–7. [DOI] [PubMed] [Google Scholar]
- 77.Uddin MN, Horvat D, Glaser SS, Mitchell BM, Puschett JB. Examination of the cellular mechanisms by which marinobufagenin inhibits cytotrophoblast function. J Biol Chem 2008;283:17946–53. [DOI] [PubMed] [Google Scholar]
- 78.Lee CL, Lam KK, Vijayan M, Koistinen H, Seppala M, Ng EH et al. The Pleiotropic Effect of Glycodelin-A in Early Pregnancy. Am J Reprod Immunol 2016;75:290–7. [DOI] [PubMed] [Google Scholar]
- 79.Lee CL, Lam KK, Koistinen H, Seppala M, Kurpisz M, Fernandez N et al. Glycodelin-A as a paracrine regulator in early pregnancy. Journal of reproductive immunology 2011;90:29–34. [DOI] [PubMed] [Google Scholar]
- 80.Alok A, Karande AA. The role of glycodelin as an immune-modulating agent at the fetomaternal interface. Journal of reproductive immunology 2009;83:124–7. [DOI] [PubMed] [Google Scholar]
- 81.Lam KK, Chiu PC, Chung MK, Lee CL, Lee KF, Koistinen R et al. Glycodelin-A as a modulator of trophoblast invasion. Hum Reprod 2009;24:2093–103. [DOI] [PubMed] [Google Scholar]
- 82.Jeschke U, Richter DU, Reimer T, Bergemann C, Briese V, Karsten U et al. Glycodelin A and differentiation of first trimester trophoblast cells in vitro. Archives of gynecology and obstetrics 2005;272:151–9. [DOI] [PubMed] [Google Scholar]
- 83.Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T et al. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nature neuroscience 2005;8:873–80. [DOI] [PubMed] [Google Scholar]
- 84.Haider S, Pollheimer J, Knofler M. Notch signalling in placental development and gestational diseases. Placenta 2017;56:65–72. [DOI] [PubMed] [Google Scholar]
- 85.Van Sinderen M, Cuman C, Gamage T, Rainczuk K, Osianlis T, Rombauts L et al. Localisation of the Notch family in the human endometrium of fertile and infertile women. Journal of molecular histology 2014;45:697–706. [DOI] [PubMed] [Google Scholar]
- 86.Han J, Gu MJ, Yoo I, Choi Y, Jang H, Kim M et al. Analysis of cysteine-X-cysteine motif chemokine ligands 9, 10, and 11, their receptor CXCR3, and their possible role on the recruitment of immune cells at the maternal-conceptus interface in pigs. Biology of reproduction 2017;97:69–80. [DOI] [PubMed] [Google Scholar]
- 87.Imakawa K, Imai M, Sakai A, Suzuki M, Nagaoka K, Sakai S et al. Regulation of conceptus adhesion by endometrial CXC chemokines during the implantation period in sheep. Molecular reproduction and development 2006;73:850–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Enriched canonical pathways from differentially expressed genes (P-values < 0.05) in spontaneous versus infertility, spontaneous versus NIFT, spontaneous versus IVF, and NIFT versus IVF pregnancies.
Supplemental Table 2. Upstream regulators of differentially expressed genes (P-values < 0.05), spontaneous versus IVF pregnancies.
Supplemental Table 3. Upstream regulators of differentially expressed genes (P-values < 0.05), spontaneous versus NIFT pregnancies.


