Abstract
Non-coding RNAs (ncRNAs) participate in regulation of gene expression, and are highly relevant to pathological development. They are found to be stably present in diverse body fluids, including those in the circulatory system, which can be sampled non-invasively for clinical tests. Thus, circulating ncRNAs have great potential to be disease biomarkers. However, tremendous efforts are desired to discover and utilize ncRNAs as biomarkers in clinical diagnosis, calling for technological advancement in analysis of circulating ncRNAs in biospecimens. Hence, this review summarizes the recent developments in this area, highlighting the works devoted to cancer diagnosis and prognosis. Three main directions are focused: 1) Extraction and purification of ncRNAs from body fluids; 2) Quantification of the purified circulating ncRNAs; and 3) Microfluidic platforms for integration of both steps to enable point-of-care diagnostics. These technologies have laid a solid foundation to move forward the applications of circulating ncRNAs in disease diagnosis and cure.
Keywords: Disease biomarker, Non-coding RNA, Liquid biopsy, Extraction, Purification, Detection, Quantification, Point-of-care diagnostics
1. Introduction
Cancer has unquestionably been a great health challenge to the entire society due to its destructive effects and very poor survival rates [1]. Early detection is one of the key factors to improve cancer survival; and prompt monitoring of cancer progression and response to therapy are also imperative to achieve promising therapeutic outcomes [2]. The conventional approaches in cancer diagnosis and prognosis rely on biopsy procedures to collect tissues and analyze the pathological signals displayed by the samples. However, biopsy is invasive to patients, and some tumors are not easily accessible, not to mention the long and complex protocols needed to analyze the solid tissues [3]. Hence, patients will be greatly benefited if tumors can be located at an early stage with minimal invasion and monitored in a timely manner during disease progression and treatment. Such benefits are likely obtained by the promising approach termed liquid biopsy.
Liquid biopsy collects biofluids, including blood, urine, saliva, breast milk, cerebrospinal, synovial, serous and amniotic fluids, which are relatively easy to be sampled compared to solid tissues in a cost-effective and non-invasive manner [4]. These biofluids have been proven to contain promising markers for cancer diagnosis, prognosis and treatment supervision, such as circulating tumor cells (CTC), extracellular vesicles (EVs), proteins, and cell-free nucleic acids (cfDNA, miRNA, long non-coding RNA) (Figure 1) [5–8]. In addition, biofluids could carry molecules related to cancer metastasis, one of the major causes tied to cancer-related deaths and its progression, which could be secreted by tumor cells to help with cancer dissemination and vascular invasion; and participate in extravasation into a secondary site, to finally induce metastatic colonization [9, 10]. Identification of such molecules by liquid biopsy could help recognize the potential of metastasis for applications of early intervention.
Figure 1.
Circulating biomarkers in patients with cancer. Reprinted with permission from Ref. 8. Copyright 2018, The Authors.
Among all molecules identified by liquid biopsy, non-coding RNAs, including microRNAs, PIWI-interacting RNAs, small nucleolar RNAs, long non-coding RNAs, etc., have attracted tremendous attention. Although with limited or no protein-coding potential, non-coding RNAs (ncRNAs) are transcribed from an overwhelming majority of the mammalian genome. ncRNAs that can be found in biofluids such as plasma and urine are defined as circulating ncRNAs, which are processed by parent cells, released into the extracellular space and then circulated into biofluids (Figure 2) [11–13]. Despite the ubiquitous presence of ribonucleases (RNases) in circulation, circulating ncRNAs display remarkable resistance to degradation and are present at consistent concentrations with sufficient integrity. Several models for robust stability and biogenesis have been proposed. Attachment to RNA-binding proteins or high-density lipoproteins and encapsulation within EVs are considered three important ways that safeguard ncRNAs from degradation [14–16].
Figure 2.
(A) Schematic diagram of biogenesis of circulating lncRNAs. Reprinted with permission from Ref. 11. Copyright 2016, The Authors. (B) Schematic diagram of biogenesis of circulating miRNAs. Reprinted with permission from Ref. 12. Copyright 2014, Wiley.
ncRNAs are of crucial functional importance and contribute to the development of various human disorders and diseases, including cancers [17, 18]. For example, two ncRNA categories have been widely studied for their functions in cancer diagnosis and prognosis: long non-coding RNAs (lncRNA) and microRNAs (miRNA) have greatly inspired scientific interest in cancer research. lncRNAs are RNA transcripts of 200 nucleotides or greater in length and play fundamental roles in all levels of gene expression, including epigenetic, transcriptional and post-transcriptional regulation [19–21]. Analogous to protein-coding oncogenes, various lncRNAs are differentially expressed in various types of cancer and can activate cellular pathways that lead to tumorigenesis, tumor progression, and metastasis, if their expression is dysregulated. The dysregulation can be observed not only at the intracellular/tissue levels but also at the levels in extracellular body fluids. The potential usage of lncRNAs as circulating biomarkers for cancer diagnosis, prognosis and therapy have recently been demonstrated [11, 22–25].
miRNAs are a class of endogenous, non-coding, single-stranded, small RNAs (< 200 nt) of 19–25 nucleotides. Different from lncRNAs, miRNAs regulate gene expression just at the post-transcriptional level by guiding the RNA-induced silencing complex to the miRNA target sites in the 3’ untranslated region of mRNA, which leads to mRNA degradation or translation inhibition [26]. Increasing evidence has been found about their key roles in controlling a variety of biological processes, such as cell cycle and differentiation, development and metabolism [27–30]. The abnormal expression profiles of miRNAs are highly associated with human disease progression, including cancer [31–34]. There are research findings that suggest uptake of circulating miRNAs could lead to modulation of cell proliferation/apoptosis, angiogenesis, tumor cell invasion and cell-cell communication, indicating tight association with various pathological conditions [35–38].
Both their biological functions and stable presence in body fluids make circulating ncRNAs promising biomarkers for early diagnosis, prognosis, and prediction of cancer [39–47]. In recent years, rapid technological advances have been obtained in biofluid sampling and ncRNA profiling. These advancements have led to the discovery of more ncRNA-based markers, facilitating understanding of their functions in cancer development and progression [48–50]. Still, to eventually improve cancer diagnosis and thus patient survival rates, significant technological advancements are needed to overcome the challenges in analysis of circulating ncRNAs. These challenges are mainly stemmed from the diverse forms of body fluids and the highly diluted concentrations of ncRNAs in the fluids. For clinical applications, the detection methods of ncRNAs should also provide high sensitivity, accuracy and precision; and be rapid and deployable under typical clinical settings with low sample consumption. Herein, this work reviews recent advancements in isolation and detection of circulating ncRNAs (mainly lncRNAs and miRNAs) present in body fluids; and highlight the developments of point-of-care diagnostics based on ncRNAs. By illuminating the current state-of-the-art technologies available in the field, we hope to identify the emerging needs to promote further development and utilization of circulating RNAs as disease markers.
2. Extraction and purification of circulating ncRNAs from biological samples
The first barrier to overcome in utilizing circulating ncRNAs as biomarkers is their direct sampling from body fluids, which are sophisticated in nature and inundated with large numbers of biomolecules and cells. What further exacerbates this issue is the fact that circulating ncRNAs tend to be encapsulated in EVs, or attached to lipids and proteins [51]. Therefore, prior to analysis, an extraction step is usually required to release the ncRNAs from their carriers and remove the impurities, so that the detection assays can be carried out with reduced interferences, enhanced performance, and improved reproducibility. This step is also needed to collect adequate amounts of ncRNAs for downstream profiling, which would require a large sample volume to afford an appreciable yield, making this matter even more complicated.
To date, several strategies to extract and purify circulating ncRNAs have been formulated to overcome the aforementioned challenges. In this section, we review the common approaches of liquid-liquid extraction (LLE) and solid phase extraction (SPE) employed for extraction of circulating ncRNAs, and the recent progress in development of new materials to address the challenges in the field. Both matured (i.e. methods using commercial kits) and contemporary extraction methods (i.e. those reported by individual research labs) are included. Each technique has its advantages and disadvantages that need to be weighed in consideration. In the end, the favored sample preparation method depends largely on the amount of precious sample on hand, the achievable recovery desired, and the project budget.
2.1. Liquid-liquid extractions (LLE)
Liquid-liquid extractions (LLE) have been widely employed to isolate RNAs, because the procedure of LLE is straightforward, it does not require sophisticated equipment to perform, operates at low expense with low technical demand, and a large number of solvents can be selected. Moreover, the solvents employed can disrupt the structures of ncRNA carriers like EVs and lipoproteins for recovery of total ncRNAs. By far, two main categories of solvents have been employed for extraction of nucleic acids: organic solvents and ionic liquids.
2.1.1. LLE with conventional organic solvents
Organic LLE relies on phase separation to achieve RNA isolation from a dirty sample matrix. The chosen solvents should simultaneously separate and release nucleic acids from hydrophobic cellular content by virtue of polarity. The most well-known solvent system is the mixture of phenol-chloroform. Chloroform is immiscible with water, and the phenol-chloroform phase is easily separated from the aqueous phase. The negatively charged backbone of nucleic acids partitions them into the aqueous phase, while hydrophobic contaminants, such as proteins and lipids, partition into the organic phase. Phenol and chloroform can respectively denature proteins and dissolve lipids, liberating any protein- or lipid-bound nucleic acids into the aqueous phase [52, 53]. The DNAs and RNAs can be further sorted by adjusting the pH – both DNAs and RNAs will remain in the aqueous layer under neutral conditions, but DNAs will partition into the organic layer at acidic pH (4–6) [52, 54–56], allowing collection of the total RNAs from the aqueous layer by alcohol precipitation.
To improve the consistency in nucleic acid extraction, the commercialized version of phenol-chloroform extraction [57], called the TRIzol reagent [58] has been developed. In addition to phenol and chloroform, TRIzol contains guanidinium thiocyanate, a common chaotropic agent, to further increase the denaturing strength on the more tightly protein-bound and EV-encapsulated RNAs [52]. It is one of the most common approaches taken by research groups to obtain circulating ncRNAs for genomic profiling. For example, Kim et al. [59] isolated miR-16 and miR-223 from healthy human plasma and quantified their recoveries by qPCR. It was found that their recovery depended on the sample matrix and anticoagulant used. In the case of plasma, post-treatment with NaF/KOx further increased the recovery of miR-16, but did not work well for miR-223, the recovery of which was 500 times lower and much less reproducible.
Sourvinou and colleagues [60] tested the recovery of the endogenous miR-16, miR-21, and the spiked cel-miR-39 from the plasma samples taken from healthy individuals and carcinoma patients. They compared the recovery obtained by TRIzol LS Reagent and several commercial extraction kits that combine the advantages of organic and solid-phase extractions; and found that the tested kits gave higher percentage yields and reproducibility than a standard LLE protocol with TRIzol LS Reagent. It was also noted in their work that the method of blood collection and storage temperatures were critical factors to RNA recovery.
More recently, colorectal cancer biomarkers were investigated by Dong et al. [61] to help unlock their diagnostic value. Particularly, they extracted total RNAs from several types of circulating extracellular cargo, including apoptotic bodies, microvesicles, and exosomes, in both healthy and patient sera using TRIzol and examined the differential expression of interesting lncRNAs and mRNAs by qPCR. They reported was that, of the RNA carriers studied, exosomes were the most abundant source of all measured lncRNAs.
2.1.2. LLE with ionic liquids
Ionic liquids (ILs) are an emerging solvent system used to improve LLE of nucleic acids while also being less toxic than most organic solvents [62]. ILs are organic molten salts with melting points of 100 °C or less. They have versatile physical properties, including tunable viscosity, thermal and chemical stability, and near all-purpose solvation power [62]. The physical properties of ILs can also be altered based on the identity of the cation and anion [63]. The anion generally characterizes the melting point of the solvent system, and both cation and anion determine the miscibility in aqueous systems [64]. More importantly, the selected cation is the essential driving force in nucleic acid extractions since it electrostatically interacts with the negatively charged phosphate backbone. Further optimization of the cation structure can lead to an IL of high extraction efficiency and specific for ssDNA [65].
Anderson and his research group have been pioneering applications of ILs in nucleic acid extraction. They initially investigated the factors that enable optimal IL performance in LLE (Figure 3A) [62]. They synthesized and tested six ILs, holding the organic anion constant while changing the identity of the cation. The DNA solution and prepared IL were essentially mixed together and a metathesis reagent was added to induce phase separation. It was found that extending the alkyl chain length of the cation and adding hydroxyl handles enhanced the recovery, with extraction efficiencies higher than 97%, even in mock biological samples spiked with albumin and metal ions. More importantly, this preconcentration method is also rapid in demanding only 5 minutes of extraction under favorable conditions.
Figure 3.
(A) Assay schematic of in situ dispersive liquid-liquid microextraction using an ionic liquid (IL). Reprinted with permission from Ref. 62. Copyright 2013, Elsevier B.V. (B) Magnetic ionic liquid (MIL) extraction of DNA. The MIL is recovered using a magnetic rod (B = 0.66 T). Reprinted with permission from Ref. 66. Copyright 2015, American Chemical Society. (C) Illustration of solid-phase microextraction (SPME) for RNA purification. Polyacrylate sorbent fibers are either coated with ionic liquid or cross-linked to dT oligos to enhance mRNA capture. The extraction was evaluated by reverse transcription quantitative polymerase chain reaction (RT-qPCR). Reprinted with permission from Ref. 73. Copyright 2017, American Chemical Society.
In order to reduce liquid handling, this group continued to test nucleic acid extraction with magnetic ILs (MIL) for nucleic acid extraction (Figure 3B) [66]. The magnetic property stems from the ferric-centered complex anions, and allows direct user control of material transfer with an external magnet. Their initial attempt has showed that the MIL approach is capable of recovering PCR-amplifiable plasmid DNA from processed cell lysates with no apparent wash steps required. Recently, the group has developed a new series of MILs focused around the trihexyl(tetradecyl)phosphonium cation, [P6,6,6,14+], with different metal-ligand anions. One Ni-centered MIL, [P6,6,6,14+][Ni(hfacac)3”], was found to be qPCR-friendly and can be used directly in the amplification mixture without inhibiting the reaction [65]. The extraction itself requires only 0.3-6 μL of MIL (compared to 20 μL in earlier stages of MIL development) and 10 minutes of sample binding to achieve an amplification efficiency of 91.1%. The other metal centers of the anion component studied were Co, Mn, and Dy, and each required additives, such as EDTA, b, Mg2+, or extra SYBR Green I, to be suitable for qPCR use.
In general, the downside of conventional LLE methods is that they require a rather large sample volume input, ranging anywhere from 0.2 to 1 mL, in order to provide enough detectable analyte. The toxicity of organic solvents may also compromise the quality of extracted products. Moreover, LLE can be very tedious and irreproducible, and is generally coupled with commercial kits to maximize the yield. IL-based extractions partially amend these problems and have proven to be much more functional in their use. However, most of the IL systems reviewed have yet to be demonstrated on RNA extraction from body fluids like serum and plasma. As ILs await successful application on circulating ncRNAs, they have thus far paved the groundwork for tougher challenges and are currently regarded as a developing trend in this field.
2.2. Solid phase-based extractions (SPE)
Compared to LLE, solid phase extraction (SPE) provides several advantages like the simplified procedure, reduced labor, and decreased sample and solvent consumption. In this type of extraction, nucleic acids are concentrated onto a solid phase, typically a silica support, by using a binding buffer that provides the optimal pH and salt conditions for silica-analyte interaction. Under an acidic pH [67], nucleic acids will bind tightly to silica through proton or salt bridges; and an alkaline pH [68] will release them because of electrostatic repulsion of the ionized silanol groups and the phosphate backbone. Water can also be the elution solvent because it contains no salt to bridge the nucleic acid-silica binding. This sort of sorption/desorption mechanism is the most established mode of action for many spin column-based extraction kits. After nucleic acid adsorption, the column can be rinsed with a wash buffer to remove the impurities in samples. Because of their good performance and easy handling, many commercial kits have been developed based on SPE, while progress continues to be made in research labs for improved performance.
2.2.1. Commercial SPE kits
Commercialized kits are thoroughly optimized products that boast a level of guaranteed quality and safe sample preparation, and make nucleic acid isolation common practice for many labs. Most kits utilize spin columns packed with sorbent, typically silica powder or glass beads, to purify nucleic acids.
The quality of extracted products and recovery of commercial kits, like the miRNeasy® and the miRCURY™ kit, were tested by many groups. El-Khoury et al. [69] focused on how influential the isolation method and volume input were in recovering several select miRNAs: miR-106a, miR-222, miR-16 and miR-223. In both human plasma and circulating exosomes, the miRNeasy® kit recovered consistently more than miRCURY™ across all tested miRNAs. In particular, miRNeasy® recovered significantly better (P < 0.001) than miRCURY™ with the input plasma volume between 50–200 μL, but experienced some column saturation when tending towards 200 μL.
miRNeasy® was also employed by Rubio et al. [70] to extract RNAs from the plasma samples of recent postpartum mothers. An average of 81.14 ng/mL total RNAs were recovered from 3 mL of plasma, which was 50 times less than the amount found in 1 mL of processed breast milk with the same kit and protocol. Various RNA subclasses, like miRNA, snoRNA, snRNA, lncRNA, and piRNA were found in the total recovered plasma RNAs, and potential biomarkers indicative of postpartum diseases may be found from all these different groups of RNAs.
Extraction of RNA from blood, a more complicated matrix than plasma, using various commercial kits was compared by Häntzsch et al. [71]. The purpose of the work was to examine both mRNA and miRNA signatures of acute myocardial infarction patients enrolled in the small scale LIFE study. Blood collection was performed via either PAXgene® Blood RNA Tubes or Tempus™ Blood RNA Tubes, with the former processing lasting for 2 hours at room temperature and the latter in 30 minutes on ice.
Extraction processing was compared among several manual (the Norgen Preserved Blood RNA Purification Kit II) or semi-automatic kits (QIAsymphony® PAXgene® Blood RNA Kit, or MagMAX™ for Stabilized Blood Tubes RNA Isolation Kit). While the PAXgene® and Norgen kits are spin column-based extractions, the MagMAX™ kit’s working principle is through magnetic separation. The total assay time ranges from 2.5 to 3.5 hours for 24 samples.
Taken together, the combination of time, blood collection and assay processing has a profound effect on the RNA yield. For example, total RNA yields were found to be 30% higher, on average, using Tempus™ Blood RNA Tubes than PAXgene® Blood RNA Tubes for blood collection [71]. It was also reported that blood collection with the PAXgene® tubes in conjunction with the Norgen II Kit for extraction resulted in higher mRNA recoveries but lower for miRNA. Intriguingly, the study found no significant differences between manual and semi-automated processing, although the obvious advantage of semi-automation is the elimination of sample bias and reduced bench labor.
The Maxwell® RSC miRNA Tissue Kit from Promega is another semi-automated kit that isolates nucleic acids through proprietary buffers and paramagnetic particles. Despite its name, the kit can interestingly be employed to extract ncRNAs from biofluids. Mensah and Borzi [72] established a consistent protocol using this kit to collect, extract and quantify total plasma RNAs from smokers of 50–75 years old. Specific miRNA markers for lung cancer were detected using homemade qPCR microfluidic cards.
Overall, the work of these authors demonstrated that the recovery and quality of circulating RNA extracted from biofluids can still vary depending on the kit used. These innate variances could be attributed to difference in extraction materials and buffer composition which are proprietary information not available to users, and also to personal adjustments or modifications to the recommended protocol. The degree to which these kits differ can apparently have very consequential impacts on RNA recovery and quality, eventually affecting the results of profiling circulating ncRNAs.
2.2.2. Ionic liquid-based solid-phase microextraction (SPME)
In the consideration of minimizing sample consumption and streamlining the work flow from RNA extraction to qPCR analysis, solid-phase microextraction (SPME) using the polymeric ILs (PIL) as the coating of a polyacrylate sorbent fiber has been explored by the Anderson group [73, 74]. In principle, the binding mechanism is still based off backbone-cation attractions, except that the cations here are immobilized on the fiber substrate, which provides a concentrated locale for nucleic acid recruitment (Figure 3C). To increase specific recovery of mRNA, the surface of the fiber was modified with dT20 to allow mRNA docking through the poly-A tail [73]. Desorption via ion-exchange was easily induced by immersion in 500 mM to 1 M NaCl for 30 minutes. The extracted RNA was suitable for downstream RT-qPCR analysis, showing amplification efficiencies of 100% or higher [74].
The PIL-coated fibers offer some very attractive features: they are portable, renewable, and can be chemically restored by immersion in 2 M NaCl for 20 minutes. It was demonstrated that such fibers were capable of handling extractions in crude cell lysates and mock food samples containing FeCl3 and CaCl2. It would be of great interest to see if further development and optimization can lead to direct sampling of circulating ncRNAs followed by RT-qPCR quantification. Still, the ultralow concentration of the circulating ncRNAs could present a big challenge in extracting a quantifiable amount by this SPME method and in how trace amounts of metal ions and the IL matrix would affect the stability of short ncRNAs, like miRNAs.
2.3. Nanomaterial-assisted extractions
Despite of the wide usage of commercial SPE kits, new materials need to be developed to further improve extraction recovery, reduce sample consumption, and shorten sample processing. Such developments are extremely important in order to meet the challenges faced by analysis of circulating ncRNAs: low concentrations and highly complex matrices of biofluids. Nanomaterials, because of their ultra-high surface area-to-volume ratios that greatly increase adsorption efficiency, have been widely explored for their utility in extraction of nucleic acids [75]. Their unique physiochemical properties could also be important factors for enhancement of nucleic acid recovery. For example, transition metal-based nanomaterials have repeatedly been demonstrated to bind prodigiously to both DNA and RNA species with high affinity under favorable conditions created by pH or salt concentrations [76]. The metals exposed on the surface of such nanomaterials could coordinate with the phosphate backbone; or attract nucleic acids via electrostatic interaction [77]. Additionally, with proper selection of the buffer conditions, nanomaterials allow controllable release of bound ligands [78], and even permit discrimination of dsDNA from ssDNA [79]. These outstanding characteristics render nanomaterials highly valuable for extraction of low abundance ncRNAs from biofluids.
2.3.1. Iron oxide nanoparticles
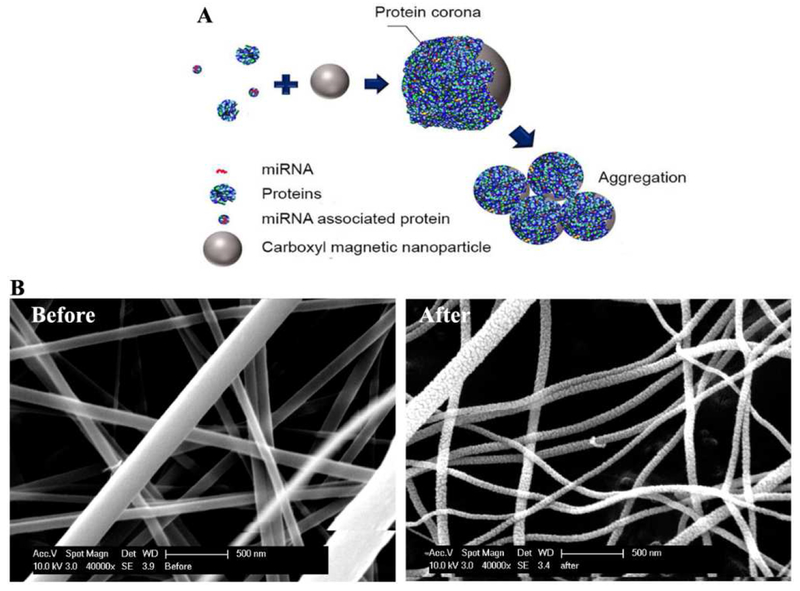
Iron oxide nanoparticles are among the most commonly explored nanomaterials for biomolecule enrichment, including nucleic acids, because of their magnetic properties, easy preparation, and wide availability. The iron oxide nanoparticles used in nucleic acid extraction are typically formed by coating an iron core surrounded with a silica shell, which could be used directly for nucleic acid binding, or be modified to facilitate extraction via other mechanisms. For instance, Xu et al. functionalized their magnetic nanoparticles with carboxylic acids, which can form protein coronas through simple adsorption and aggregate in biofluids (Figure 4A) [80]. The aggregation enabled quick enrichment of circulating protein-bound miRNA and exosomes, both of which can be readily isolated and quantified. Our lab has also employed silica magnetic particles for on-chip extraction and amplification of small RNAs [81]. In our case, the silica magnetic particles served as a solid support for direct nucleic acid binding, after which they were magnetically maneuvered through a series of wells on a homemade chip containing wash buffers, eluate, and RT-PCR amplification mix.
Figure 4.
(A) Graphic depicting carboxylated magnetic nanoparticles adsorbing proteins from urine or cell culture medium and forming protein corona upon aggregation. Reprinted with permission from Ref. 80. Copyright 2018, American Chemical Society. (B) Transmission electron microscopy (TEM) image of TiO2 nanofibers before and after calcination. Calcination caused TiO2 nanofibers to appear rougher and more brittle than non-calcinated ones. Scale bars represent 500 nm. Reprinted with permission from Ref. 83. Copyright 2018, Springer-Verlag GmbH.
2.3.2. Titania nanofibers
Titanium is another transition metal that has previously shown great potential in isolating phosphorylated proteins and nucleic acids. Zhang et al. [82] demonstrated that TiO2 nanoparticles could tightly bind to DNA under very acidic conditions (pH 2). This is a result of titania’s pKa being ~ 4.2. At pH 2, the metal surface is positively charged and can interact electrostatically with the negative phosphate backbone.
Fibers, however, possess a long morphology that endows a larger surface area than particles for nucleic acid adsorption (Figure 4B). Our lab developed the TiO2 nanofibers [83], which were fabricated by electrospinning [84], for the extraction of miRNAs from biological samples, like serum and cell lysates. miRNAs are more challenging to extract than long mRNAs and lncRNAs due to their short lengths. Using a judiciously constructed binding buffer comprised mainly of guanidine and a series of ethanol-water washing steps, the nanofibers outperformed the PureLink™ commercial column extraction kit, recovering 100-folds more endogenous miRNA (miR-21 and miR-191) from MDA-MB-231 breast cancer cells. As a result of high recovery, sample consumption can be reduced.
2.3.3. Other nanomaterials with potential for RNA extraction
There are many other nanomaterials as well that have been discovered with untapped potential for nucleic acid extraction. In many cases, researchers so far have mostly used these nanomaterials for signal enhancement or as a tool for detection. For instance, graphene [85–87], Au nanoparticles [87], carbon nanotubes [87, 88], MoS2 nanosheets [89], ZnO nanowires [90], and Cu-TCPP metal-organic framework nanosheets [91] all have been shown to exhibit strong binding affinities to nucleic acids. However, none of them have been used directly in extraction applications, but instead have played roles as signal quenchers or substrates for surface modifications. Cobalt oxyhydroxide (CoOOH) nanoflake [79] is one example of the nanomaterial that has not yet been exploited for circulating miRNA recovery but has shown high affinity for ssDNAs. Perhaps this property could be further explored and unlocked to enrich shorter ssRNA species that are biologically interesting.
2.4. EV isolation to obtain EV-enclosed ncRNAs
More and more works have revealed that a good portion of circulating ncRNAs is encapsulated in diverse sub-types of EVs, including exosomes, microvesicles, apoptotic bodies, and oncosomes that shield them from RNase degradation [92–94]. The protection is also considered improving sample quality because the ncRNA contents would be less affected by handling and storage conditions. Thus, another direction in extraction and purification of circulating ncRNAs is to isolate the EVs and obtain the EV-enclosed ncRNA fraction.
Conventional strategies to isolate EVs from biological fluids are by targeting their differences in physical properties, such as size, density, and surface hydrophobicity, or by molecular signatures (Table 1). Among the aforementioned techniques, density gradient, centrifugation, and ultracentrifugation are considered gold standards [95–97], but they are tedious with low yield. To speed up EV isolation, commercial kits [98] that rely on polymers to aggregate and precipitate lipid structures have been developed to simplify the process and improve the throughput. However, non-EV lipid impurities could co-precipitate and interfere with downstream analyses. The polymer-based methods also introduce polymer impurities that may compromise the structural integrity of the EV and further impact ncRNAs analyses as well.
Table 1.
Comparison of commonly used exosomes isolation methods.
| Property to be targeted | Method | EV yield | Purity | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|---|
| Density | Ultracentrifugation | Medium | Low | Compatible with large sample volumes; possible for parallel processing of multiple samples | Time consuming; high equipment cost; inducing exosome damage | 95, 96 |
| Density Gradient Centrifugation | Medium | High | Capable of removing lipids and separating EVs by density | Technically demanding | 97 | |
| Size | Ultrafiltration | Low | High | Capable of removing big particles from EVs | Low recovery, causing exosome damage and deformation | 99 |
| Size Exclusion Chromatography | Medium | Medium | Capable of removing small proteins and lipids without causing EV aggregation; fast speed | Large dilution of the collected EVs that require re-concentration | 100, 101 | |
| Surface hydrophobicity | Polymer-based precipitation | High | Low | Easy to use, high sample capacity | Co-precipitation of non-exosomal contaminants | 98 |
| Surface markers | Immunological separation | Medium | High | Rapid, high purity and efficiency | Low sample throughput; high reagent cost; non-inclusive | 106 |
Size-based separation technologies, like ultrafiltration, size exclusion chromatography and flow field flow fractionation [99–104] can separate EVs from proteins and lipids; but they still suffer from low sample throughput and labour-intensive processing. Recently, a filtration-based EV isolation platform, ExoTIC (exosome total isolation chip), was developed by Demirci and his co-workers [105] via the layer-by-layer assembly of a polycarbonate track-etched nanoporous filter membrane, a poly(ether sulfone) (PES) layer, and a cellulose pad. This platform rapidly isolates EVs within the size range of 30–200 nm with high simplicity and flushes out small biomolecules, such as free nucleic acids, proteins, lipids, and other tissue/cell debris in biological samples prior to EV enrichment.
Although convenient and inclusive to all EVs, physical separation does not provide selectivity over EV sub-groups which may contain ncRNAs more relevant to disease development and progression. Particular EV sub-groups could be obtained through immuno-precipitation targeting specific surface markers by antibodies, aptamers, or protein receptors. With more and more knowledge gained on EV biogenesis, EV subgroups, like exosomes and microvesicles, can be obtained by targeting their corresponding surface proteins with satisfactory yield, purity, and integrity [106]. A brief summary and comparison of these conventional EV isolation methods can be found in Table 1.
With regard to analysis of the EV-encapsulated RNAs, EVs isolated by centrifugation-based approaches could be contaminated by the non-EV nucleic acids which could be adsorbed onto the EVs and co-precipitated [107]. Filtration- or column-based isolation may lead to loss of EVs due to irreversible attachment to the membrane or columns or damaged while passing through the separation device [108–109]. These flaws would induce fatal errors in quantification of the enclosed ncRNAs. EV characterization and cargo quantification should be conducted before analysis of the EV-encapsulated ncRNAs to assess EV integrity, recovery, and purity. In particular, protein-to-particle, protein-to-lipid, and RNA-to-particle ratios should be evaluated when comparing the ncRNA contents among EV samples, as suggested by the Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018) summarized by the International Society of EV [110].
Still, for analysis of circulating ncRNAs enclosed in EVs, there are significant needs for more efficient EV isolation followed by more stream-lined ncRNA extraction and quantification. Several microchip platforms thus have been developed for such a purpose. Some of them provide the possibility of direct EV inspection on the chip after isolation using microscopic techniques; and others are able to release the captured EVs for off-chip analysis of the EV cargos including ncRNAs. These novel microchip nanoplatforms are discussed in the following sections.
2.4.1. Acoustofluidics for exosome isolation
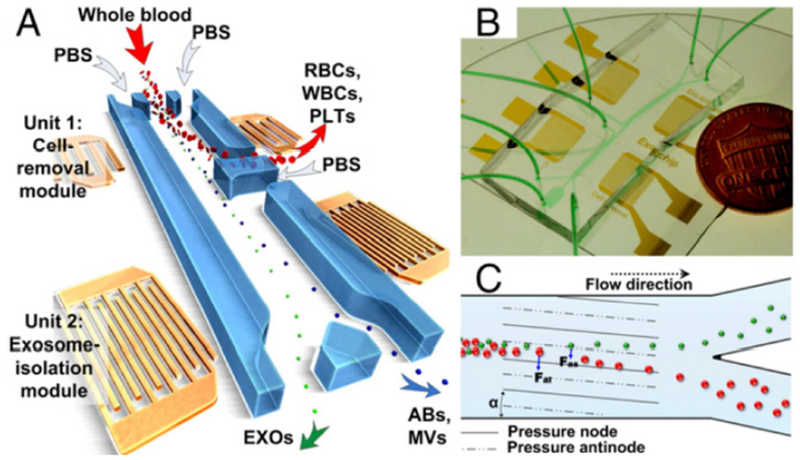
An acoustofluidic chip has been employed by Wu et al. [111] to extract and enrich exosomes in many major biological fluids. The operating principle underlying the innovation utilizes acoustofluidics, which is a combination of surface acoustic wave (SAW) and microfluidics. By using interdigital transducers, the platform employs two modules of separation: 1) a lower frequency scan on whole blood to segregate components greater than 1 μm in diameter (RBCs, WBCs, PLTs), and 2) a higher frequency scan on the resulting plasma to further remove submicron particles (microvesicles and apoptotic bodies). The chip is constructed in such a way that the first SAW module directs larger cells into a separate channel; then the second module diverts the exosomes away into a streamline different from the remaining particulates (Figure 5). Overall, the mechanism is superior to differential centrifugation techniques in affording intact biologically preserved exosomes.
Figure 5.
Overall schematic for acoustofluidic enrichment of exosomes. (A) RBCs, WBCs, and PLTs are depleted from whole blood through Unit 1 to provide cell-free plasma in downstream analysis. The filtrate containing total extracellular vesicles is further filtered through Unit 2, where exosomes are segregated from ABs and MVs. (B) Optical image of fully integrated acoustofluidic chip. (C) Size-based separation occurs in each module due to the lateral deflection induced by a taSSAW field. The periodic distribution of pressure nodes and antinodes generates an acoustic radiation force to push large particles toward node planes. Abbreviations: RBC = red blood cell, WBC = white blood cell, PLT = platelet, AB = apoptotic body, MV = microvesicle, taSSAW = tilted-angle standing SAW. Reprinted with permission from Ref. 111. Copyright 2017, National Academy of Sciences.
This integrated platform satisfies several of the needs for a useful POCT device. Being partly microfluidic-based, it offers automation, minimal operator training and handling, and concomitant reproducibility. Results can be obtained rapidly within 25 minutes with excellent yield (82%) and purity (98%). The chip is patterned by photolithography and molded from PDMS, both of which are inexpensive and accessible fabrication methods. More importantly, the design can accommodate direct processing of biological fluids, which is a key feature sought in any decent sample preparation protocol.
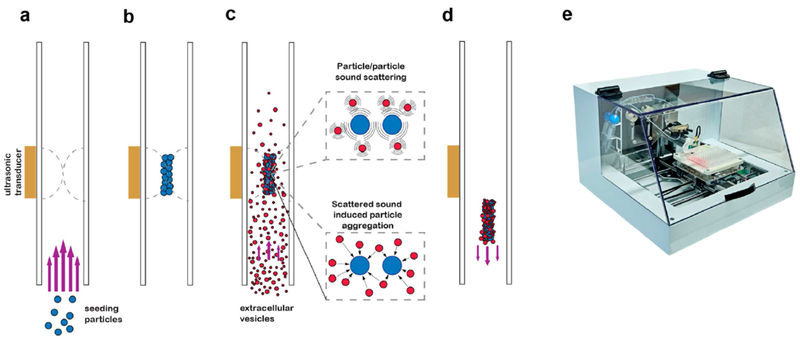
The AcouSort Laboratory in Lund, Sweden, developed the AcouTrap 2.1, a fully automated research table top platform designed specifically for acoustic seed particle trapping. It was successfully applied by Ku and coauthors [112] to isolate EVs directly from human urine and plasma (Figure 6). In comparison to ultracentrifugation, the AcouTrap had a predilection for enriching vesicles closer to the size range of exosomes (93–100 nm) in both urine and plasma. However, acoustic trapping performed worse than ultracentrifugation in urine enrichment. It was also found that acoustic trapping preserved the morphology of the enriched exosomes and eluded the co-precipitation of Tamm Horsfall fibers that tend to manifest from UC-treated urine.
Figure 6.
Mechanistic scheme of acoustic trapping for extracellular vesicle isolation. (a) Seeding particles are introduced into the fluidic channel and (b) are packed together by the acoustic standing wave generated by the ultrasonic transducer. Excess seeding particles are washed away. (c) Total extracellular vesicles (EV’s) are introduced into the fluidic channel and are trapped by the seed cluster via secondary acoustic forces and particle-particle interactions. (d) After cleaning, the seed cluster-EV aggregate is released upon terminating the acoustic wave. (e) Photograph of the automated trapping device, AcouTrap. Reprinted with permission from Ref. 112. Copyright 2018, American Chemical Society.
The group also extracted the corresponding total exRNA using mirVana Paris RNA and Protein isolation kit and analyzed the miR-16, miR-21, and miR-24 content via RT-qPCR. Since they observed comparable levels between UC and acoustic trapping for each miRNA, it can be said that the two enrichment methods are complementary and divulge similar information on ncRNAs. However, the AcouTrap clearly wields winning advantages over UC, including rapid assay time, smaller volume consumption, high throughput yields, and very robust performance. These qualities are ideal for diagnostic applications and, with further development, the AcouTrap may very well find itself in POC settings soon.
2.4.2. Patterned surface for EV trapping
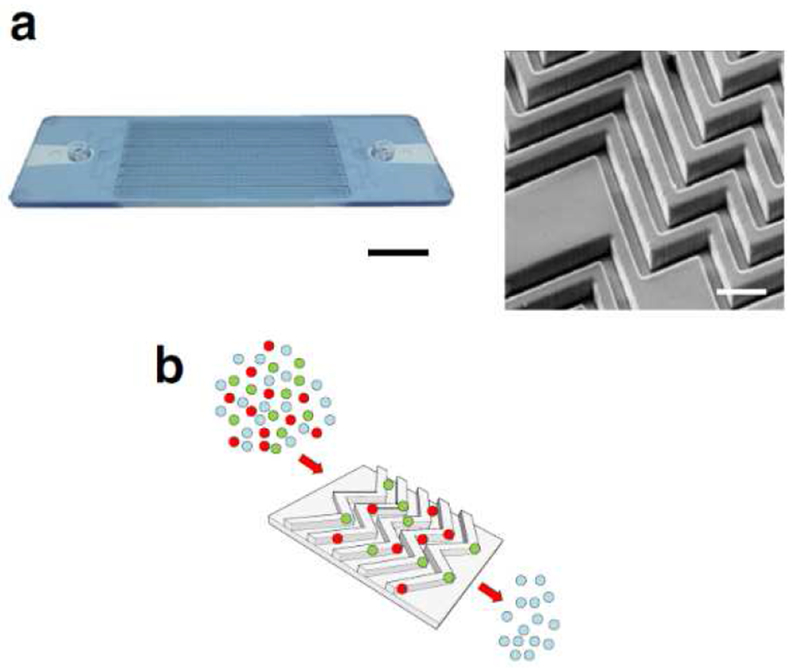
Microfluidic chips with patterned surface have also been developed to trap EVs. Reátegui and co-authors [113] constructed a microfluidic chip patterned with herringbone grooves (EVHB-Chip) to improve chaotic mixing, and applied it to isolate tumor-derived EVs (exosomes, microvesicles and oncosomes) from glioblastoma multiforme patients (Figure 7). They novelized the design by adding a nanostructured thermo-active gelatin substrate raised with PEG spacers and streptavidin-coated nanoparticles that maximized EV capture by tumor-specific antibodies on the channel surface. Controlled release of the isolated EVs is triggered by dissolving the gelatin at 37 °C. The device can process 100 μL to 5 mL of filtered plasma or serum in 3 hours and isolate EVs, which are washed from cell-free DNA, lysed, and profiled for RNA via Qiagen miRNeasy Mini Kit and RNA sequencing. Compared to ultracentrifugation and bead-based separation, this technology afforded 10-fold higher mRNA yield from EVs.
Figure 7.
EVHB-Chip design. a) Image of the operating COC EVHB-Chip. Pressure-driven flow-pumped serum or plasma through an inlet at 1mL/h. Waste serum or plasma is collected in a single outlet (scale bar 1 cm). On the right, an SEM micrograph of the 3D herringbone features of the microfluidic device (scale bar 100 μm). b) Serum or plasma from healthy donors with spiked-in fluorescent EVs (in red and green) was run through the microfluidic device coated with tumor-specific antibodies to capture tumor EVs. Abbreviations: EV = extracellular vesicle, HB = herringbone. Reprinted with permission from Ref. 113. Copyright 2018, The Authors.
2.4.3. Dielectrophoretic separation of EVs
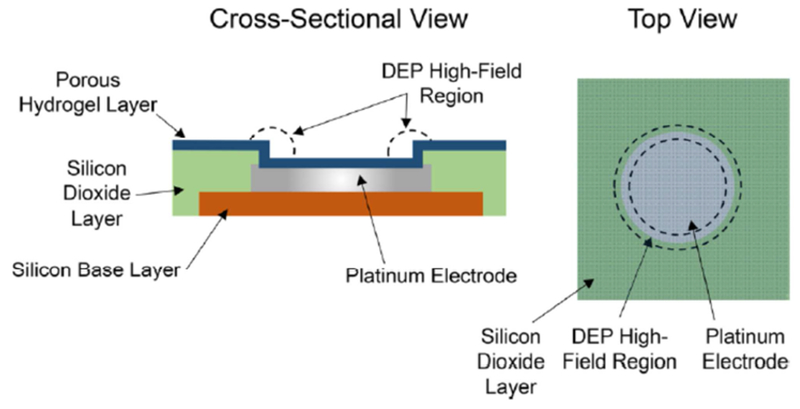
Ibsen and colleagues [114] fabricated an alternating current electrokinetic (ACE) microarray chip on an impetus to recover glioblastoma-derived EVs. Controlled by platinum microelectrodes, the emitted alternating current produces dielectrophoretic forces: a high-field region surrounds the border of the microelectrodes and attract nanoparticles while a low-field region exists between the microelectrodes that draw in larger scale particles (Figure 8). Gentle conditions are ensured by coating the microelectrodes with a porous hydrogel to prevent plasma redox and electrolytic degradation of the EVs. Collected EVs could either be detected directly on-chip via immunostaining or eluted by reversal of the dielectrophoretic forces and verified by TEM. The ACE chip provides several advantages compared to conventional methods, including low sample consumption of 30-50 μL, rapid assay time, simple workflow, tolerance for raw samples. However, while the authors intended the ACE chip for wholesome exosome isolation, they did not formally report a quantitative yield or recovery. Like other platforms, the ACE chip is also limited in EV resolution and cannot segregate exosomes from other EVs.
Figure 8.
Schematic of the microelectrode array chip showing the cross-sectional and top views of a single electrode. Over 1000 electrodes can be in a single device. The DEP high-field regions where particles are collected are shown within the dotted lines. The darker color of the silicon dioxide layer and electrode in the top view represents the overlying transparent porous hydrogel layer. DEP stands for dielectrophoretic. Reprinted with permission from Ref. 114. Copyright 2017, American Chemical Society.
2.4.4. EV isolation with nanomaterials fabricated on chips
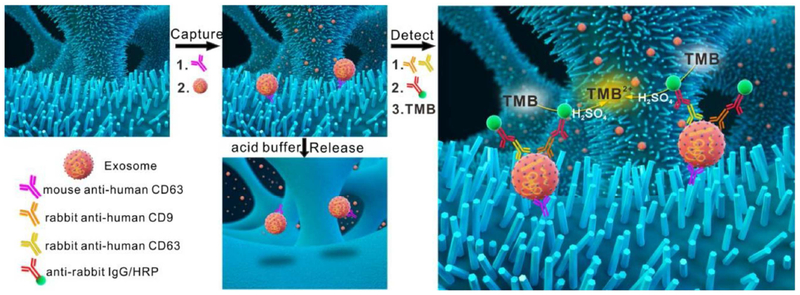
Chen and co-authors [115] recently discovered a method to fabricate a ZnO nanowire chip that is capable of both detecting and isolating intact exosomes from serum and plasma. The fabrication method allows ZnO nanowires to be directly embedded within the PDMS matrix. The ZnO surface was moderately functionalized with a series of organic coupling steps to introduce streptavidin, which was joined to biotinylated anti-CD63 for EV immobilization. EV release was controlled by flushing in citric acid buffer due to the nanowires being pH sensitive (Figure 9). The clear advantage of the ZnO nanowire chip is compliance with raw samples. This not only preserves the integrity of the exosomes, but also simplifies the assay’s workflow. The morphology and large surface area of the nanowires also provide a size-exclusion effect to further improve the EV capture efficiency. The EVs isolated by the ZnO nanowire chip can be recovered and destroyed for downstream total ncRNA analysis.
Figure 9.
Schematic representation of the capture, detection, and release of exosomes by the ZnO-chip device. Prior to the aforementioned stages, ZnO nanowires are grown on a 3D porous PDMS scaffold and cross-linked to capture and detection antibodies. Reprinted with permission from Ref. 115. Copyright 2018, Elsevier B.V.
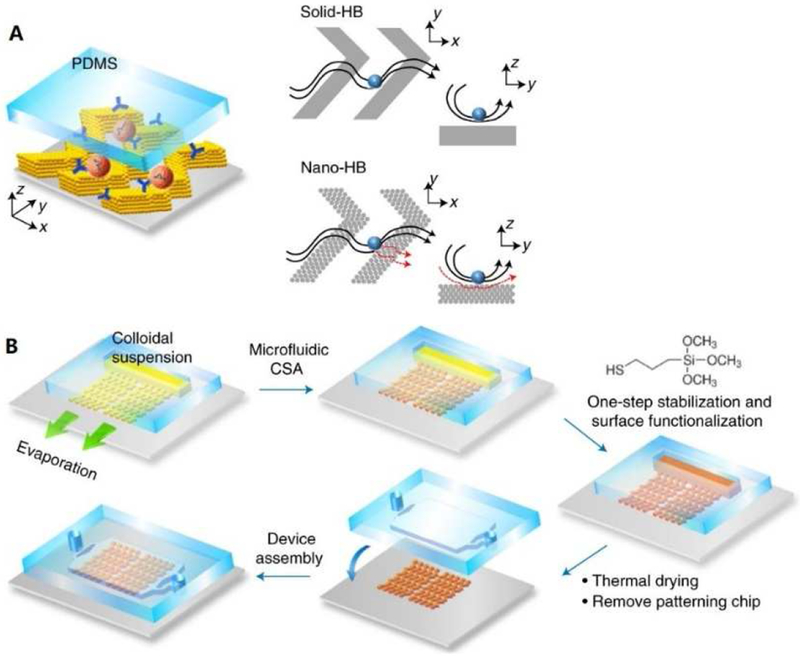
Very recently, Zeng’s group reported a 3D-nanopatterned microfluidic chip for isolation of circulating EVs [116]. Microfluidically engineered colloidal self-assembly (CSA) was utilized in multiscale integration by designed self-assembly (MINDS), with flow manipulation and molecular recognition handled by a functional microelement herringbone (HB) mixer (Figure 10). Compared with current microfluidic methods, this novel microfluidic design addressed the fundamental limitations of mass transfer, surface reaction and boundary effects. Consequently, the nano-HB chip not only shows extremely high sensitivity for detection of EVs spiked into samples (LOD is 10 exosomes/mL), but also realizes quantitative determination and surface protein biomarker (CD24) profiling of the low-abundant circulating exosome in plasma collected from ovarian cancer patients and healthy controls.
Figure 10.
(A) Schematic of the MINDS strategy that improves biosensing by 3D nanostructuring of microfluidic elements, such as the herringbone mixer. The conventional, solid-HB mixer creates microvortices to promote mass transfer of targets. A particle will experience hydrodynamic resistance near a solid surface that reduces direct surface contact. In a 3D nano-HB chip, fluid near the surface can be drained through the porous structure (red dashed lines) to increase the probability of particle-surface collisions. (B) Workflow for fabricating a 3D nano-HB chip by MINDS. Reprinted with permission from Ref. 116. Copyright 2019, Springer Nature.
3. Characterization, detection and quantification of circulating non-coding RNA
Upon purification from biofluids, the circulating RNAs are to be characterized and quantified for marker discovery and detection. The main focus in this step is to improve detection specificity and sensitivity. To date, microarrays and next-generation sequencing (NGS) are the most advanced technologies applied for studying circulating RNAs in the high-throughput manner. Microarrays offer the remarkable advantages of comprehensive coverage, adaptable to microfluidic devices, and ability to be customized towards different RNA categories. In contrast, NGS offers the advantages of extreme sensitivity and high throughput, and more importantly, it does not require knowledge of target RNAs. However, both suffer from high cost, and NGS is highly sophisticated in its implementation. They are mostly accessible in core laboratories and their availability to general clinical or biochemical labs is very limited. Consequently, simpler techniques that permit rapid amplification of the signals from target nucleic acids have been developed. There are two main approaches to realize sensitive and selective detection of circulating RNAs: amplifying the nucleic acid sequences, or amplifying the signal without generating more strands.
3.1. Nucleic acid amplification
In terms of whether DNA/RNA enzymes are employed, nucleic acid amplification can be classified into two categories: enzyme-assisted or enzyme-free amplification. Among the enzyme-assisted amplification, polymerase chain reaction (PCR) is the most popular owing to its high sensitivity, simplicity, and cost-effectiveness. To further improve amplification simplicity, sensor systems that do not rely on temperature cycling have also been actively developed, such as rolling circle amplification, duplex-specific nuclease signal amplification, and so on. Moreover, enzyme-free amplification strategies, like hybridization chain reaction and self-assembled DNA cascades, have also been developed and applied for detection of circulating RNAs.
3.1.1. Polymerase chain reaction (PCR)
PCR remains the gold standard for nucleic acid identification and quantification because of its matured protocols and high sensitivity. It employs a heat-stable polymerase to amplify a specific region of a nucleic acid strand. After three reaction steps (denaturing, annealing and extension) controlled by varied temperatures, thousands to millions of copies of a particular sequence can be generated. During the past years, abundant PCR-based strategies for measuring and quantifying circulating lncRNA had been developed by researchers [117–123]. For example, Arita et al. analyzed the levels of lncRNA H19, the HOX antisense intergenic RNA, in the plasma of gastric cancer patients and healthy controls through reverse transcription polymerase chain reaction (RT-PCR) assay [117]. Tests results clearly indicate that the concentration of lnc-RNA H19 was much higher in patients, showing the potential as a cancer marker.
In 2016, by using quantitative reverse transcription polymerase chain reactions (qRT-PCR), Huang et al. reported that the expression level of lncRNA MALAT1 in serum samples collected from patients with non-small cell lung cancer (NSCLC) was different from that in healthy controls [118]. The authors have proved that, the customized PCR method not only has high sensitivity and specificity, but also has satisfactory discriminative accuracy for the diagnosis of NSCLC. Another circulating lncRNA biomarker for NSCLC was reported by Shi et al. who employed qRT-PCR to detect significantly higher expression level of AFAP1-AS1 in cancer patients [119]. Both works discovered the circulating lncRNAs with promising potential in clinical diagnosis of NSCLC.
Circulating miRNA as cancer markers were also discovered in works employing PCR, prompted by their high resistance to endogenous RNase and excellent stability in blood samples [124–129]. Abnormal levels of circulating miR-141 in serum were found to be able to distinguish prostate cancer patients from healthy controls by Mitchell and co-workers, with the purified miR-141 amplified by RT-qPCR [124]. Their results established a very useful and practical approach for liquid biopsy-based detection of human prostate cancer. In 2014, Lu et al. developed a universal and time-saving RT-qPCR strategy for measurement of circulating miRNA in plasma with high sensitivity, high efficiency and low cost [125]. In this method, a random pre-adenylated DNA oligonucleotide was first adopted to ligate miRNAs with the T4 RNA ligase, and then bound to a universal tailing primer for reverse transcription with a reverse transcriptase. Finally, the traditional real-time qPCR was conducted to quantify the amount of miRNAs. To verify the practicability of this system, circulating miRNAs from lung cancer patients was tested. It showed that this ligation-based method may be a convenient tool for identifying tissue-specific and disease-specific circulating miRNA biomarkers.
Determination of circulating exosome-enclosed RNA (namely exosomal RNA) species were reported as well in numerous works with the assistance of PCR amplification technology during the past few years [130–143]. For instance, Isin et al. purified circulating exosome in urine samples from prostate cancer (PCa) and benign prostatic hyperplasia (BPH) patients, and quantified the exosomal lncRNAs by real-time PCR, revealing the significant difference in the exosomal lncRNA-p21 level between these patients [130]. Similarly, after the isolation of exosomes in the plasma collected from PCa, BPH patients and healthy donors, Wang and his co-worker investigated the expression levels of two tumor-derived exosomal lncRNAs SAP30L-AS1 and SChLAP1 by using RT-qPCR [131]. The level of lncRNA SAP30L-AS1 were found to be upregulated in BPH, while that of lncRNA SChLAP1 was significantly higher in PCa than that in BPH and healthy donors. Both work testify that exosomal lncRNAs could act as potential biomarkers in clinical diagnosis to distinguish cancer patients from healthy controls, and even predict the malignant states.
Potential exosomal miRNA markers have also been reported. Zhao et al. evaluated the levels of miR-21, miR-133a, and miR-181b obtained from the serum of colon cancer patients using RT-PCR [143], and reported a higher level of exosomal miR-21 in colon cancer patients than that in normal individuals. Besides, the levels of exosomal miR-574–3p, miR-141–5p, and miR-21–5p in urine samples from prostate cancer patients were found to be significantly upregulated that those in the samples from healthy donors by Samsonov and his co-workers using RT-qPCR [142].
3.1.2. Other enzyme-assisted nucleic acid amplification
Although powerful, PCR requires temperature cycling and instruments with sophisticated thermal sensors need to be employed. Thus, tremendous research efforts have been devoted to develop isothermal amplification strategies that amplify nucleic acids at a single temperature and permit usage of much simpler and cheaper heating instruments for easy implementation in the ill-equipped labs. As an additional benefit, isothermal amplification has been reported to be less hindered by analytes found in whole blood that would otherwise interfere with PCR [144, 145].
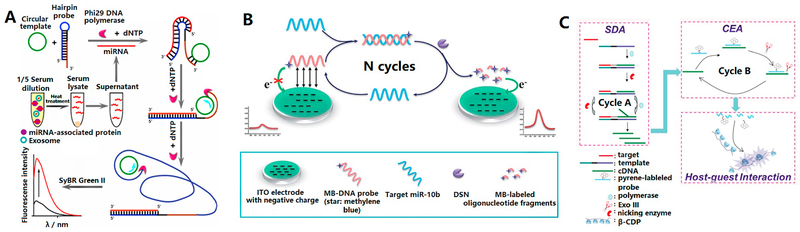
As a highly efficient isothermal amplification process, rolling circle amplification (RCA) can produce thousands of tandem repeats using a polymerase with strand-displacement capability, a circle template and a primer. In 2013, Zhang and his co-workers proposed a simple, highly sensitive and selective RCA method, so called hairpin probe (HP)-based RCA, for detection of lung cancer-related circulating miR-486–5p [146]. In this study, a stable hairpin probe which integrated the recognition parts for the target miRNA and the RCA primer was designed. Its binding to the cancer biomarker of miR-486–5p exposed the blocked primer which would then hybridize with the circular template and triggered RCA (Figure 11A). As a result, quantitative analysis of miR-486–5p with a detection limit as low as 10 fM was realized with SYBR Green II staining. Furthermore, this method was applied to accurately discriminate the difference in the expression level of miR-486–5p in the serum samples that collected from lung cancer patients and healthy individuals.
Figure 11.
(A) Schematic illustration of circulating miR-486–5p detection based on the hairpin probe-assisted rolling circle amplification (RCA). Reprinted with permission from Ref. 146. Copyright 2013, American Chemical Society. (B) Schematic diagram of the designed electrochemical biosensing strategy for specific miRNA detection with DSN-based target recycling amplification. Reprinted with permission from Ref. 149. Copyright 2016, The Royal Society of Chemistry. (C). Schematic of the enzyme-assisted multiple amplification technology based on SDA (strand displacement amplification) and CEA (cyclic enzymatic amplification). Reprinted with permission from Ref. 152. Copyright 2015, The Royal Society of Chemistry.
Similarly, based on the excellent selectivity of the p19 RNA binding protein towards double-stranded RNAs, direct and simple detection of the circulating miR-21 in human serum was realized by Yang’s group [147]. A specially designed Janus probe recognized the target miR-21, causing its enrichment by the magnetic beads modified with the p19 protein. Following RCA, the enriched miR-21 was detected by SYBR Green I. This strategy showed very high sensitivity and specificity in detecting the target miRNA in clinical samples. Additionally, combining solid-phase RCA and chronocoulometry, Yao et al. prepared a novel microchip to detect miR-143 in human blood, achieving a detection limit of 1 pM [148].
Duplex-specific nuclease signal amplification (DSNSA) is another isothermal amplification approach that employs the duplex-specific nuclease (DSN), an enzyme selectively cleaving DNA in DNA-RNA hybrids while showing little activity towards ssDNA or RNA-RNA duplexes. The DSN assisted target recycling amplification was coupled with electrochemistry by Fu et al. to build a sensor for miRNA detection (Figure 11B) [149]. Owing to the high efficiency of the DSN-based target-recycling, this electrochemical biosensor showed high sensitivity with a limit of detection reaching 0.1 pM. Its immobilization-free feature also simplified the assay. The effectiveness of this method in clinical applications was demonstrated by discriminating the expression differences of the lung cancer biomarker of miR-10b between lung cancer patients and healthy controls.
In addition to electrochemical biosensor, colorimetric assays utilizing DSN for detection of miRNA in human serum was developed as well. In 2013, Shen et al. demonstrated a simple and label-free strategy by exploiting the AuNP networks and DSN [150]. The AuNP networks, which was designed to bind to the target miRNA complementarily, was fabricated with the help of a bifunctional DNA capture probe (CP) with the (T)10 and (CH2)6 spacer at each end. Generation of AuNP networks from crosslinking of the two types of AuNPs can be conveniently achieved by the well-known 1-ethyl-3-(3-dimethylpropyl)-carboiimide chemistry. When DSN was present, the CP within the miRNA-CP duplex was specifically digested, releasing the miRNA. The intact miRNA was then constantly hybridized with another CP to trigger its digestion by DSN. As a result, free AuNPs were generated from the AuNP networks and quantification of target miRNA was realized by monitoring the change of absorbance at λ = 530 nm. A limit of detection of 0.1 fM was achieved; and circulating miRNAs in blood was successfully analyzed. Likewise, Guo et al. reported another colorimetric method for detection of circulating microRNAs [151]. In this work, the DNA probe 1 (P1) and probe 2 (P2) modified AuNPs were employed and the amount of dimers was decreased with the increase of target miRNA concentration. The obvious color change from blue to red can be easily discerned even with the naked eyes. Serum samples collected from cancer patients and healthy donors were measured by the proposed method, and showed significantly different colors.
Besides the enzyme-assisted methods mentioned above, the Wang group developed a detection system for assaying miRNA in human serum by utilizing isothermal strand displacement amplification (SDA) coupled with exonuclease III-aided cyclic enzymatic amplification (CEA) [152]. As illustrated in Figure 11C, the target miR-21 in complex biological samples initiates SDA and generates a good number of substrates to bind to the pyrene conjugated strands and trigger CEA. CEA results in cleavage of pyrene from the DNA and generates strong fluorescence signal based on the host-guest interaction between β-CDP and pyrene.
3.1.3. Enzyme-free nucleic acid amplification
All of the aforementioned enzyme-assisted isothermal amplification methods exhibit great potential in clinical diagnosis using circulating RNAs. Their success can be attributed to their high simplicity, high amplification efficiency, and low limits of detection. Further simplification of such strategies focuses on the complete removal of biological enzymes to reduce the cost and complexity of the assay. Enzyme removal can also avoid problems of enzyme denaturation induced under non-optimal transportation or storage conditions.
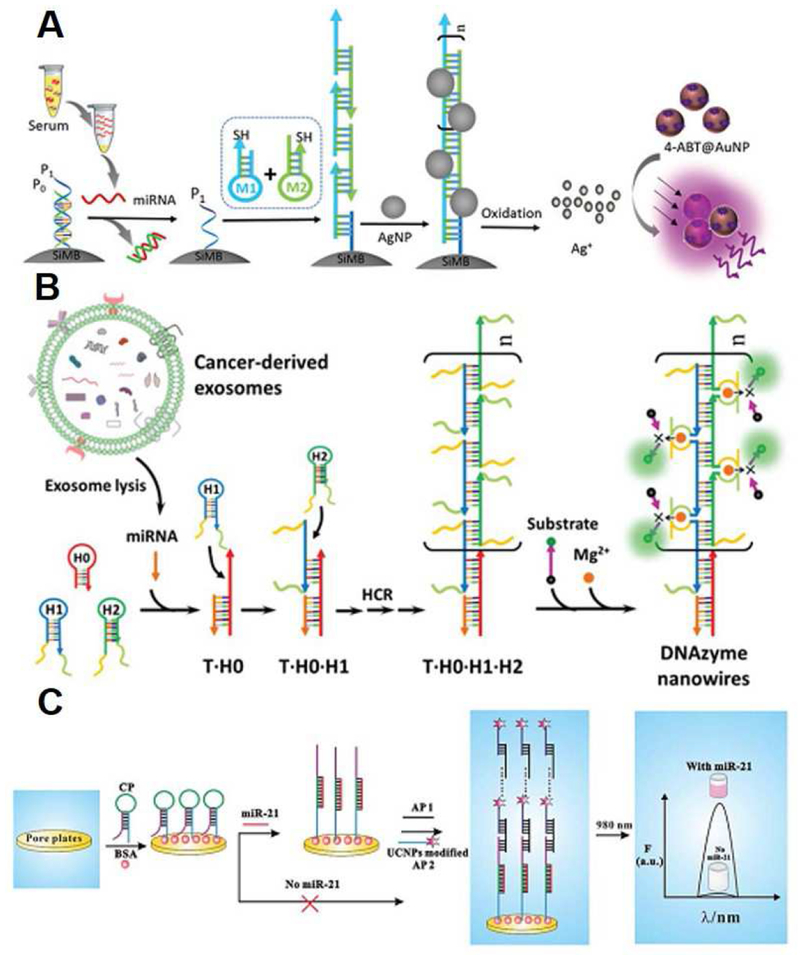
Hybridization chain reaction (HCR) is one of the most popular enzyme-free isothermal amplification strategy widely employed in diverse bioassays [153]. In this approach, two metastable DNA hairpins coexist in solution and will assemble into a long dsDNA-like nicked duplex triggered by an initiator that binds to one of the hairpins. In 2015, Zheng et al. applied HCR for detection of miR-21 in human serum (Figure 12A) [154]. In their strategy, when the target circulating miRNA was present, the sealed initiator on the surface of silica microbeads (SiMBs) was exposed and further triggered HCR to generate long self-assembled and thiol-contained DNA polymer. Subsequently, multiple AgNPs were conjugated to the DNA polymer, which can be dissolved into numerous silver ions (Ag+). Ag+ can control the gaps between neighboring 4-aminobenzenethiol (4-ABT) modified gold nanoparticles (AuNP@4-ABT) to form a ‘‘hot-spot’’ and surface-enhanced Raman spectroscopy (SERS) signal were greatly improved. Therefore, quadratic cascade amplification was achieved and this biosensing platform allows a limit of detection of miR-21 down to 0.3 fM. In addition, it can recognize the expression level differences of miR-21 in the serum samples collected from healthy donors and patients with different stages of chronic lymphocytic leukemia (CLL). Similarly, by using HCR and AuNPs, Rana and co-workers enabled multiplexed detection of circulating oncomiRNAs with the detection signal observable with the naked eyes [155]. In this method, HCR took place on the surface of AuNP only when the specific miRNA marker was present. Long HCR hybridization products prevented the AuNPs from ion-induced aggregation. Even without the assistance of any analytical equipment, the limit of detection of the proposed biosensor was as low as 20 fM, much more sensitive than those reported previously.
Figure 12.
(A) Schematic illustration of enzyme-free quadratic amplification for detecting circulating miRNA based on hybridization chain reaction (HCR) and Ag+-mediated cascade SERS signal amplification. Ref. 154. Copyright 2015, The Royal Society of Chemistry. (B) Schematic of enzyme-free amplified detection of circulating exosomal miRNA with the assistance of the target-initiated HCR and DNAzyme. Ref. 156. Copyright 2018, The Royal Society of Chemistry. (C) Design scheme of photoluminescent biosensor for circulating microRNA detection based on self-assembled DNA cascades. Ref. 157. Copyright 2015, The Royal Society of Chemistry.
HCR has also been coupled with DNAzyme to further amplify the detection signal. [156] Li et al. rationally designed three hairpin DNA probes, which included H0, H1 and H2 with the split Mg2+-dependent DNAzyme sequences at their 5’ and 3’ ends, so that miR-21 can open the initially enclosed initiator in the stem region of H0 to allow continuous hybridization of H1 and H2 and thus generation of the long DNA duplex nanostructure consisting of numerous adjacent DNAzyme units (Figure 12B). Once hybridized with their specific single-stranded DNA substrate modified with quencher and fluorophore, the ribonucleobase (rA) within the substrate was cleaved in the presence of Mg2+ ions to release the fluorophore from the quencher, resulting in bright fluorescence signal to realize ultrasensitive detection of tumor-specific exosomal miRNAs in the serum samples from cancer patients.
Self-assembled DNA cascades, another enzyme-free signal amplification method, was also exploited to design various sensing platforms for circulating RNAs. Unlike HCR, linear auxiliary probe1 (AP1) and auxiliary probe2 (AP2) participate in the formation of duplex assemblies upon initiation, which subsequently is labeled by upconversional nanoparticles to produce a photoluminescent biosensor (Figure 12C) [157]. This sensing platform was found to provide extremely high sensitivity for miRNA quantification in biological samples, offering a very simple and convenient tool for early cancer diagnosis.
3.2. Nanomaterials-based signal amplification
Signal enhancement can also be achieved without generating more strands but relying on nanomaterials with unique chemical or optical properties. With a large number of nanomaterials being developed for biomolecular detection, our review only focuses on those applied for detection of circulating RNAs in biospecimens that are published in the past 5 years. In these applications, nanomaterials are typically employed to enhance electrochemical signals or to quench fluorophores for them to be turned back on by the target strands.
Gold nanoparticles (AuNPs) are one of the most common nanomaterials employed in sensors for biomolecules because of their unique electrochemical and optical properties. For instance, a nanogenosensor based on cysteamine-capped gold nanoparticles (Cys-AuNPs) for electrochemical detection of miR-25, a marker for lung cancer, was proposed by Zare et al. [158]. In this sensor, a single stranded DNA (ssDNA) probe was firstly covalently immobilized on the surface of a Cys-AuNPs/glassy carbon electrode (GCE), establishing the sensing surface for analysis of the target miRNA (Figure 13A). Once the probe was hybridized with the lung cancer-specific miR-25, an electrochemical signal was generated and the concentration of miR-25 was quantified by electrochemical impedance spectroscopy (EIS). Experimental results showed that this nanogenosensor had high sensitivity, with an LOD of 2.5 × 10”13 M, and good selectivity towards the target miRNA. Later, it was successfully applied for detection of miR-25 expression in human plasma without requiring any sample extraction and/or amplification.
Figure 13.
(A) Schematic illustration of the fabrication of electrochemical nanogenosensor for the detection of circulating miR-25 with the assistance of cysteamine-capped gold nanoparticles (Cys-AuNPs). Reprinted with permission from Ref. 158. Copyright 2017, The Royal Society of Chemistry. (B) Schematic presentation of the biosensing mechanism for detecting miR-1246 in circulating exosomes collected from human plasma based on Au nanoflare-based probe. Ref. 159. Copyright 2018, American Chemical Society. (C) Schematic representation of fluorophore labeled probes/nanographene oxide (nGO) assembly for detecting of multiple target circulating miRNAs. Reprinted with permission from Ref. 160. Copyright 2014, American Chemical Society. (D). Principle of general biosensing method for probing circulating microRNA biomarkers based on oligonucleotide-templated reaction (OTR). Ref. 162. Copyright 2016, American Chemical Society.
The optical property of AuNPs was taken advantage by Duan et al. to develop a spherical Au nanoflare probe, formed by modifying the bare AuNP with a DNA duplex (Figure 13B) [159]. The DNA duplex was composed of the sulfhydryl-labeled antisense oligonucleotide sequence of the target, miR-1246, i.e. the capture DNA, and a shorter complementary single-strand DNA consisting of a Cy5 fluorophore (the reporter DNA). Because of fluorescence resonance energy transfer (FRET), the fluorescence of Cy5 was efficiently quenched by the AuNPs, which would be recovered once the reporter DNA is displaced by miR-1246. Interestingly, the authors found that Au nanoflares were able to directly enter exosomes extracted from breast cancer cells with simple incubation without any transfection agents. It is thus possible to eliminate the time-consuming step of exosome isolation from the plasma samples. Moreover, differentiation of breast cancer patients from healthy controls with high sensitivity and specificity was realized, proving its potential as a cancer diagnosis tool.
Graphene is a two-dimension nanomaterial that also possesses excellent physical and chemical properties; and thus its applications in detection of circulating ncRNAs have been exploited as well. By using graphene oxide, Yigit et al. designed a nanoplatform which enables simultaneous detection of the exogenous oncomiRs: miR-21 and miR-141, from different kinds of human body fluids including blood, urine and saliva (Figure 13C) [160]. miR-141 is an oncomiR which is substantially elevated in advanced prostate cancer, but its expression level is normal in the early cancer development stage. On contrary, miR-21 is overexpressed in the early stage of prostate cancer but not in the advanced stage. In this strategy, two DNA probes respectively labelled with the fluorophores of FAM and Cy5 were tightly adsorbed on the surface of graphene oxide through π-π stacking and the fluorescence was dramatically quenched. When two miRNA targets were introduced, the formation of the DNA/miRNA hybrid structure led to desorption of the two probes, recovering the fluorescence. The content and ratio of the miR-21 and miR-141 in different samples were successfully determined and the status of the disease was easily distinguished with the assistance of the nanoplatform. It provides a non- or minimally invasive approach for evaluating early, advanced, metastatic, or nonmetastatic stages of prostate cancer.
Interestingly, Au NPs and graphene can be combined and form excellent sensors for detection of circulating RNAs. For instance, a label-free colorimetric sensor based on the graphene/Au-NP hybrids was established to detect miR-21. Instead of taking advantage of the outstanding quenching performance of graphene, this work exploited the intrinsic peroxidase-like activity of the graphene/Au NP hybrid [161]. The ssPNA-21 probe was strongly adsorbed to the surface of the graphene/Au NP hybrid via π-π stacking between the ssPNA and the graphene. The adsorption almost completely inhibited the peroxidase-like activity of the hybrid, leading to no color change when the colorimetric peroxidase substrate was added. Upon the addition of miR-21, hybridization between the PNA probe (ssPNA-21) and miR-21 released the ssPNA from the graphene/Au NP hybrid, restoring its catalytic activity. The LOD of this sensor was determined to be 3.2 nM with a linear range spanning from 10 to 980 nM. It was also successfully applied to detect miR-21 in human serum.
3.3. Amplification-free techniques
To simplify the detection scheme, elegant work that does not rely on nucleic acid amplification nor nanoparticles for signal enhancement has been developed for analysis of circulating RNAs. Peptide nucleic acids (PNAs) are engineered uncharged NDA or RNA analogues that can hybridize to complementary nucleic acids with high affinity and high selectivity. Ladame et al. developed a PNA-based fluorogenic biosensor for detection of circulating miRNAs with minimal sample processing in blood [162]. As illustrated in Figure 13D, two non- or weakly fluorescent molecules respectively labelling two PNA probes were brought to the close vicinity by the target miRNA, which triggered the reaction between these two molecules and produced a fluorescent molecule. Compared to the gold standard of RT-qPCR, this versatile design provides the advantages of simple, low-cost, isothermal, and highly specific. It also has the potential to be incorporated in portable devices for public screening.
With the help of an anthraquinone compound, Oracet Blue (OB), Manesh et al. proposed an electrochemical biosensor for miRNA quantification [163]. In this sensor, a thiol modified ssDNA probe was immobilized on the surface of the Au electrode and hybridized with the target miRNA. OB intercalated with the hybridized duplex and generated electrochemical signals in the presence of the target miRNA. The sensor could discriminate between the target miRNA, the single-base mismatch, and the non-complementary strand. The sensor was applied to detect miR-155 in human serum; and demonstrated a wide sensing range from 50 pM to 15 nM and an LOD of 13.5 pM.
4. Integration for Point-of-care (POC) testing of circulating ncRNAs
Sensitive and selective nucleic acid signal amplification methods permit detection of circulating ncRNAs at trace levels; and the advanced extraction techniques reduce interferences from sample matrices. Combination of both on POC devices broadens the biomedical applications of circulating ncRNAs outside of laboratory or clinical settings, and enables reliable analysis to take place near or at the patient’s location. The field of POC diagnostics mainly focuses on developing and miniaturizing laboratory-based procedures into user-friendly platforms that are portable, simple, and disposable [164–166]. Ideally, true integration would offer direct sampling on a breadth of unprocessed clinical samples, including blood, plasma, serum, sweat, urine, and saliva. This would be an improvement to laboratory procedures that require very pure samples to attain high sensitivity, specificity and accuracy in diagnosis. The amount of sample needed from the end-user is also an important consideration. A platform that can manage an accurate readout while demanding very little input [167] is a boon to quantifying low-abundance biomarkers in complex matrices, such as circulating miRNAs in serum [168–170].
Still, it is an ongoing challenge to integrate all these beneficial elements into a singular, convenient and foolproof platform. Innovating how sample preparation, amplification and detection are handled and tying them into a simple device that is easy enough to use with minimal instruction are the present motivations [171–174]. In this section, several platforms recently developed for POC applications are discussed, with some of them focusing to detect circulating RNAs for disease diagnosis, and the others being integrated platforms that enable both sample processing and in situ ncRNA analysis.
4.1. Commercial point-of-care testing technology
Several POC testing devices have been on the market that simplify ncRNA detection. For example, Gene-Z [175–176] is a POC device that is based on loop-mediated isothermal amplification (LAMP) and fluorescence detection. It has been applied to detect several miRNAs (miR-92, miR-141) from mouse blood, plasma, and even sputum with good specificity [175]. The platform itself is a disposable microfluidic chip containing wells where the reaction mixture is injected. Gene-Z houses the chip, automates the analysis and is operated by an iPod touch that receives and emails data to a local PC.
Digital PCR (dPCR) is a variant of real-time qPCR that offers absolute quantification of nucleic acids. By splitting the prepared sample into many small reaction volumes, the amplification is accomplished through numerous parallel reactions that collectively result in higher reproducibility than traditional qPCR. Moreover, the number of targets managed per reaction in dPCR can be very few and concentrated in an extremely small volume, leading to higher sensitivity in detecting small-fold changes. This is genuinely fitting for profiling miRNA dysregulation in cancers [177–179] and exosomal miRNA signatures in other diseases [180–182]. The QuantStudio™ 3D Digital PCR System marketed by ThermoFisher Scientific has been an invaluable asset in detecting ncRNAs of serum/plasma [183–186] and exosome origin [187–189]. At the heart of the device is a chip housing 20,000 60 μm-wide wells that can each hold 14.5 μL of volume. Detection is endpoint-based and can utilize several channels for FAM™/SYBR Green, VIC™, and ROX™. As a dependable resource, this platform is relatively cheap, portable, high throughput and rapid as one sample takes only 30 seconds of assay time.
POC platforms that integrate ncRNA extraction and analysis have been commercialized as well. One good example is GeneXpert, retailed by Cepheid Inc. [190–193]. This diagnostic system boasts a single-use disposable cartridge that consolidates direct sampling of raw specimen, RT-PCR, and fluorescence detection of RNA in plasma samples. The GeneXpert instrument automatically coordinates these assay steps and culminates them into printable test results. In terms of the analytical specifications, GeneXpert can reach ultrasensitivity (4.0 IU/mL) and calls for an assay time of 105 minutes, but requires 1–3 mL of sample volume.
Another platform that integrates nucleic acid extraction, purification, and reverse transcription is called the BioFire FilmArray BioThreat-E [194, 195]. It is currently used to detect a panel of viral RNA in blood, but it should be useful for analysis circulating ncRNAs. The elegance of the platform lies in its simplicity for automating the entire assay and accepting whole blood rather than serum, so minimal personnel training is needed for sample preparation. Setup takes about 2 minutes, in which the raw patient sample is assembled in a special pouch containing manufacture reagents [194] and placed in the FilmArray machine. The blisters of the pouch act as reaction chambers that house nucleic acid extraction, purification, and reverse transcription, respectively. The resulting RT products are transferred to a PCR array for amplification and detection, which is analyzed and engendered as a straightforward readout via the FilmArray Software.
4.2. Microfluidic platforms for ncRNA detection
Many microfluidic platforms that have been developed in research laboratories and not yet brought to the market are only for on-chip ncRNA detection when loaded with purified samples. Although they suffer from necessitating a separate sample preparation step, which complicates the workflow to get urgent results, they provide the capability to profile multiple ncRNAs simultaneously.
For example, an isothermal microarray platform was developed by Roy et al. [196] for miRNA detection in both cardiac tissue and plasma. Because these samples contain relatively complex matrices with thousands of proteins [197–200], to prevent nonspecific surface adsorption, the authors covalently capped the glass surface with PEG to 1) minimize background and 2) enhance target capture on surface (Figure 14A). Target capture immobilized AuNPs on the surface, which were quantified via imaging. The counts of AuNPs were then converted to miRNA quantity. The assay was tested by adding 20 μL unprocessed human serum spiked with miR-208b, which has been found to be upregulated and can serve as a biomarker for damaged cardiac muscle cells [197, 198], and results were obtained in 4 hours to afford a detection limit of 10 fM. Roy et al. [196] also demonstrated successful duplex detection using Cy3-labeled miR-208b and FAM-labeled miR-335. Selectivity was challenged using different members of the let-7 family and showed good discrimination.
Figure 14.
(A) Cartoon illustration of a custom microarray fabricated from a glass slide substrate that is functionalized with carboxyl-PEG. A1: Hairpin CP’s cross-linked to glass surface and awaiting target. A2: Target-induced stem separation of hairpin CP leaving SP sequence exposed. A3: Hybridization of AuNP-conjugated SP to exposed SP sequence marking target in nucleic acid duplex for DIC microscopy. Abbreviations: CP = capture probe, SP = signaling probe, AuNP = gold nanoparticle, DIC = differential interference contrast. Reprinted with permission from Ref. 196. Copyright 2016, Elsevier B.V. (B) Schematic illustration delineating morpholino nanochannel functionalization with PMOs and miRNA sensing. The blue spheres represent negative charges along the miRNA phosphate backbone. PMO stands for phosphorodiamidate morpholino oligos. Reprinted with permission from Ref. 201. Copyright 2017, American Chemical Society. (C) Overview of TORNADO platform. C1: Detailed view of TORNADO disc assembly. The grey layers are PMMA, the green layers are PSA. Abbreviations: PMMA = poly(methyl methacrylate) and PSA = pressure sensitive adhesive. C2: Image of fully assembled disc with integrated electrodes (cropped to view one section). Chambers labeled (i)-(x) are preloaded with assay reagents. C3: Hybridization of miRNA strands on surface of working electrode inside chamber (v). Reprinted with permission from Ref. 213. Copyright 2017, The Authors.
Another novel isothermal detection platform was recently reported by Liao and colleagues [201] involving morpholino-modified nanochannels. Structurally, phosphorodiamidate morpholino oligos (PMO) have a neutral backbone, where the – OH groups are replaced with -NH2 groups [202]. This confers greater aqueous solubility, flexibility, stability and specificity [202–205] and makes PMOs excellent probes for miRNA detection. Furthermore, PMOs also stabilize their RNA targets upon binding by blocking other biomolecules via steric hindrance [206]. The platform support is a 12 μm thick PET membrane. Conical nanochannels were chemically etched onto the surface with dimensions measuring about 1.1 μm in diameter for the large opening and 51 nm in diameter for the small hole. After etching, the nanochannels were functionalized with PMOs and incubated with the appropriate miRNA target spiked in serum. Since the PMOs are neutral, detection was carried out by ion current measurements which reflect the change in surface charge density upon miRNA binding (Figure 14B). In serum, the platform showed good response in the dynamic range of 100 aM to 1 nM when tested with let-7b, with the limit of detection being10 fM. Specificity was evaluated in 0.5* PBS, however, and showed good distinguishability among let-7b, let-7c and miR-21. As a potential POC device, this platform also demonstrated reusability after several assays. Although this strategy is sound, it suffers from long incubation periods that last over 12 hours for surface modification and 4 hours for target capture. This would not be practical in acute care settings that demand high throughput results. Additionally, many groups outside a laboratory may not have access to the tools or technology needed to fabricate the platform features, thus limiting its use in clinical diagnosis.
4.3. Integrated system for on-chip sample processing and ncRNA detection
Several microfluidic systems have been reported with the capability of providing the detection results with input of raw, unprocessed samples, taking advantage of the nice features of microfluidics in miniaturization, portability, ease of fabrication and small volume consumption.
4.3.1. Integrated platform for ncRNA isolation and detection
One excellent example of the integrated systems is the centrifugal microfluidic platforms that have been studied extensively to further decrease the sample volume requirement and limit of detection in the past decades [207, 208]. Such systems have recently been sought to detect miRNA like miR-134, which is a brain-derived biomarker detectable in plasma and is indicative of several brain illnesses [209–212], by McArdle and her colleagues [213]. They designed a unique microfluidic disc, termed Theranostic One-Step RNA Detector (TORNADO), that was assembled from 4 layers of poly(methyl acrylate) and 5 layers of pressure sensitive adhesive. These discs were etched with ten chambers that served as sites for miR-134 capture, reagent storage, or waste disposal (Figure 14C). Together, the discs arranged a valve network connected by pneumatic channels that allow the flow of one liquid in one chamber to trigger the flow of the next liquid reservoir from another chamber. Centrifugal microfluidic flow was controlled and directed by a computer motor to allow release of the chamber contents at will and at a predetermined rate. Detection of miR-134 was directly conducted on either unprocessed plasma or cerebrospinal fluid in a sandwich hybridization assay. Successful target capture left an exposed overhang region of miR-134 to hybridize with a secondary probe tagged (also by thiol interaction) with electrocatalytic 50–70 nm platinum nanoparticles. When H2O2 was added, electrocatalytic reduction of H2O2 occurred on the PtNP’s and the current difference was measured by an external potentiostat. A linear range between 1 pM to 1 μM and an LOD of 1 pM were achieved for detection of miR-134 with excellent sensitivity to discriminate between strands with one-base difference in sequence.
TORNADO can be theoretically tailored for any miRNA of interest. The assay itself is also relatively quick: the total turnaround time was approximately 1 hour and 45 minutes, including all hybridization and handling steps. The computer controls the centrifugal spin frequency to properly dispense and manage the liquid contents from one chamber to the next without a pump, cutting back on personnel handling and thereby increasing reproducibility. However, integration can be pressed further to include automated plasma isolation and an internal detection unit for current measurements. The need for the platform to be capable of multiplexing is also a very important consideration since insightful correlations can often be drawn among several disease biomarkers.
In 2019, the Microfluidic Exponential Rolling Circle Amplification (MERCA) platform was developed by Zeng and his co-workers [214]. This system integrates and streamlines solid-phase miRNA isolation, miRNA-adapter ligation, and dual-phase exponential rolling circle amplification (eRCA) assays all in one analytical workflow. In their design, the three-layer device integrates seven parallel units and each unit combines a three-valve pump and several pneumatically addressable assay chambers. Flow channels were fabricated, assembled with a thick PDMS slab, and patterned with a pneumatic control circuit to open the assay chambers for flow-through miRNA capture and close the chambers during isothermal amplification and fluorescence detection. Then, a glass slide patterned with miRNA capture probe was bound to the PDMS assembly. Target miRNAs present in raw cell lysate and cell-derived exosomes were captured, amplified by eRCA, and detected by SYBR Green, proving the high value of such a platform in improving circulating ncRNA analysis in biological and clinical applications.
4.3.2. Integrated system for EV isolation and ncRNA detection
While microchip-based EV isolation has been reviewed in the early section, this part of the discussion focuses on those that integrate EV isolation, lysis and detection of EV-enclosed RNAs. Taller and cohorts [215] demonstrated that this type of accommodation is possible through two separate chip devices. Their lysis platform is SAW-driven while their exRNA detection platform uses an ion-exchange nanomembrane sensor infused with complementary oligos (Figure 16). The dual-chip assay performs both tasks in about 1.5 hours using a minimum of 100 pL raw cell medium. It was also shown to have 2 pM sensitivity and a dynamic linear region spanning from pM to nM. This detection limit is toward the higher end of circulating ncRNA concentrations.
Figure 16.
(a) Schematic of surface acoustic wave (SAW) device (side view) and SAW-induced lysing of exosomes to release RNA for detection. SAWs generated at the transducer refract into the liquid bulk, inducing fluid motion, and electromechanical coupling also generates a complimentary electric wave at the surface of the substrate. (b) Schematic of ion-exchange nanomembrane sensor consisting of two reservoirs separated by the membrane. RNA in the sensing reservoir hybridize to complimentary oligos immobilized on the surface of the membrane. The inset shows the ion transport through the device to generate current and the right image is a characteristic current-voltage curve illustrating the under-limiting, limiting, and over-limiting regimes. Reprinted with permission from Ref. 215. Copyright 2015, The Royal Society of Chemistry.
Hu, Kwak and co-authors [216] developed an in situ method for detecting RNAs stored in EVs, thus eliminating the obligation to first release the encapsulated contents and preserving the exosomes. Their strategy relies on the coalescence of negatively charged EVs and cationic lipoplex nanoparticles (CLNs), which are immobilized onto a Neutravidin-coated gold surface biochip and enclose molecular beacons (MBs) that are complementary to the target RNA (Figure 17A). They designed the MB with a toehold domain that, upon hybridization with the target, facilitates stem separation. MBs with this toehold overhang were termed Oh-MBs and exhibited remarkable stability over extended periods of time and at elevated temperatures. Strategic placement of the dye and quencher pair at the end of the stem provided low background and appropriate signal for total internal reflection fluorescence (TIRF) microscopy, which has excellent sensitivity for single molecules < 300 nm from the glass-liquid interface. This method is sensitive down to 40 amol, has a linear range spanning 2.42–77.3 fmol, and is able to discriminate point mutations, owing to the structural stability of the Oh-MBs against nonspecific sequences. The CLN-Oh-MB biochip is also capable of detection of multiple strands of EV RNA, while preserving the structural integrity of EVs. But it is nontrivial to use TIRF to perform single vesicle counting. Suitability in a low-resource setting is a hurdle that many specialized platforms find difficult to surmount.
Figure 17.
(A) Principle of CLN-Oh-MB biochip. A1: Illustration of CLN-TIRF technology. A2: Schematic illustration of Oh-MB for hybridization and signal gain with target RNA in CLN-EV complex. A3: Structure comparison of Co-MB, Ohe-MB and Ohi-MB. Abbreviations: Co-MB = conventional molecular beacon, Ohe-MB = overhang molecular beacon with external dye, Ohi-MB = overhang molecular beacon with internal dye. Reprinted with permission from Ref. 216. Copyright 2018, Elsevier Ltd. (B) Overview of integrated chip for detection of diseased exosomal miR-21 and miR-126. B1: A schematic illustration of the integrated microfluidic chip for detecting miRNA biomarkers for cardiovascular disease. B2: Exposed view of the integrated microfluidic chip. B3: A photograph of the integrated microfluidic chip. The chip was comprised of four layers: an air control layer, a liquid channel layer, a double-sided tape layer, and an epoxy substrate equipped with an FET detection sensor. FET stands for field-effect transistor. Reprinted with permission from Ref. 217. Copyright 2018, The Royal Society of Chemistry.
An integrated platform that streamlines three modules of EV extraction, miRNA recovery and detection was developed by Cheng et al. to target exosomal miR-21 and miR-126, which are respectively upregulated and downregulated in heart disease [217]. The chip was designed to perform extraction, recovery and detection in a linear concatenate (Figure 17B). The EV extraction module accomplished vesicular capture through magnetic beads cross-linked to antibodies. An external magnet anchored the beads while micropumps were used to wash away unwanted materials. The EVs was destructed by lysis buffer to release exosomal miRNAs. In the miRNA recovery module, the exosomal miRNAs were treated with fresh magnetic beads conjugated to capture probes. While the magnetic beads are anchored, the annealed exosomal miRNAs are washed, eluted and transported to the detection module, where they hybridize to complementary probes immobilized on the Au-coated field-effect transistor (FET) sensor surface. The limit of detection of this system towards the target miRNAs was reported to be 1 fM, appropriate for physiological detection in patient samples. Still, fabrication of the chip is nontrivial, requiring an assembly of four layers with meticulous channeling. On-chip EV capture efficiency was 54.3%, and to offset this loss and provide enough analyte, 2–5 mL of plasma were required for sufficient detection signal, which was quite large sample demand. The total assay time of 5 hours is relatively longer than other systems reviewed above.
5. Conclusion and future perspectives
Cancer has become one of the most devastating epidemics that mankind has faced with few satisfying remedies. In cancer diagnosis and therapy, early detection of cancer is most conducive to cancer survival. Compared to traditional tissue biopsy in clinical diagnosis of cancer, liquid biopsy is a non-invasive and inexpensive alternative to collecting and profiling cancer biomarkers. More importantly, acquiring liquid biopsies is much more tolerated by patients than obtaining solid tumor tissues. An important class of cancer biomarkers found in biofluids is circulating ncRNAs, which play key roles in cancer genesis and progression and show great promise in cancer theranostics.
Up until now, salient achievements and accomplishments in the study of circulating ncRNAs have already been established that encourage further progress in this field. Hence, in this review, some prominent research works focusing on analysis of circulating ncRNAs have been systemically discussed, covering various aspects of the analysis. ncRNA extraction and purification from various biological samples were first summarized, including liquid-liquid, solid phase-based and nanomaterial-assisted extractions. Characterization, detection and quantification of circulating ncRNAs via nucleic acid amplification technologies and nanomaterial-based signal enhancement were discussed next. Finally, point-of-care testing technologies that integrated sample preparation and detection into a fully unified system for circulating ncRNAs analysis were presented.
For analysis of circulating ncRNAs, information of their carriers cannot be omitted which may enhance their efficacy as the potential biomarkers. In addition, ncRNA carriers like EVs and lipoproteins are different in their physiological properties, which could influence ncRNA recovery during purification. As found by Lasser [218] and Murillo et al. [219], RNA isolation kits can exert bias towards certain carrier types. However, our understanding on carrier-bound RNAs remains limited due to technical difficulties in carrier differentiation and isolation. The most focused carrier group is EVs, but all cells secrete EVs. Therefore, recent developments on EV isolation followed by ncRNA analysis were included in this review as well. While the current focus is mainly on isolation of total EVs, future studies should be devoted more towards specific EV subsets. For example, the ncRNAs enclosed in the EV subsets secreted from specific cell groups, such as those belonging to distant organs and those undergoing unique pathological development, will be highly beneficial markers because the nature of their specific cell of origin can provide more information to guide accurate disease diagnosis and proper treatment. Isolation of EV subsets then requires more knowledge about the properties of such EVs, like their specific surface proteins, and typically relies on immuno-precipitation, the capability of which in targeting multiple markers simultaneously remains to be greatly improved.
Another challenge in studying ncRNAs is their implementation in bioinformatics, such as the two RNA-seq pipelines, exceRpt and Atlas. Such pipelines are essential for mapping the infrastructure of extracellular RNA communication within humans and facilitate exchange of metadata among users. However, the quality of RNA-seq data heavily depends on RNA isolation, which is bottlenecked by contamination issues. exceRpt and Atlas combat this by aligning and filtering RNA-seq reads with known potential contaminants, although this data treatment cannot cover more ground than preventative measures during sample preparation. Thus, a strong emphasis should be placed on accurately fractionating biofluids into carrier subtypes in order to progress forward with our knowledge of extracellular RNA communication and utilization of exRNA as clinical biomarkers.
Significant technical improvements and continuous evolution of our repertoire and knowledge are crucial in the perennial battle against cancer. We hope that this review can help deepen readers’ insight on the diagnostic potential of circulating ncRNAs, facilitate the discovery of new circulating ncRNA biomarkers, improve their applications in the biomedical field, and incentivize the development of circulating ncRNA-based technologies for diagnosis and treatment of cancer and other diseases in the near future.
Figure 15.
Schematic illustration of the MERCA principle for miRNA detection. The on-chip chamber is patterned with probes to capture target miRNAs. An adapter hybridizes with the miRNA and is ligated with the capture probe by T4 DNA ligase. Reprinted with permission from Ref. 214. Copyright 2019, Elsevier B.V
Highlights:
Circulating non-coding RNAs are promising biomarkers for disease diagnosis.
These RNAs need to be extracted and enriched from the complex biofluid.
Trace amounts of non-coding RNAs prepared are detected via signal amplification.
Microfluidic devices enables integrated, point-of-care analysis of non-coding RNAs.
Acknowledgements
W. Zhong is thankful to the National Cancer Institute of the National Institutes of Health for the R01CA188991 award that supports the work on circulating biomarkers in her group.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interest Statement
The authors have no competing interests to declare.
References
- [1].Wu L, Qu X, Chem. Soc. Rev. 44 (2015) 2963. [DOI] [PubMed] [Google Scholar]
- [2].Kim BY, Rutka JT, Chan WC, Engl N. J. Med. 363(2010) 2434. [DOI] [PubMed] [Google Scholar]
- [3].Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A, Nat. Rev. Clin. Oncol. 10 (2013) 472. [DOI] [PubMed] [Google Scholar]
- [4].Trecate G, Sinues PM, Orlandi R, Future Oncol. 12 (2016) 1395. [DOI] [PubMed] [Google Scholar]
- [5].Tong YK, Lo YM, Clin. Chim. Acta. 363 (2006) 187. [DOI] [PubMed] [Google Scholar]
- [6].Yoruker EE, Holdenrieder S, Gezer U. Clin. Chim. Acta. 455 (2016) 26. [DOI] [PubMed] [Google Scholar]
- [7].Xu MJ, Dorsey JF, Amaravadi R, Karakousis G, Simone CB II, Xu X, Xu W, Carpenter EL, Schuchter L Kao GD, Oncologist 21 (2016) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lim SY, Lee JH, Diefenbach RJ, Kefford RF, Rizos H. Mol. Cancer 17 (2018) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Weigelt B, Peterse JL, van ‘t Veer LJ, Nat. Rev. Cancer 5 (2005) 591. [DOI] [PubMed] [Google Scholar]
- [10].Mehlen P, Puisieux A, Nat. Rev. Cancer 6 (2006) 449. [DOI] [PubMed] [Google Scholar]
- [11].Qi P, Zhou XY, Du X, Mol. Cancer 15 (2016) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liang H, Gong F, Zhang S, Zhang CY, Zen K, Chen X, Wiley Interdiscip Rev. RNA 5 (2014) 285. [DOI] [PubMed] [Google Scholar]
- [13].Arrese M, Eguchi A, Feldstein AE, Semin.Liver Dis. 35 (2015) 43. [DOI] [PubMed] [Google Scholar]
- [14].Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Proc. Natl. Acad. Sci. USA 108 (2011) 5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT, Nat. Cell Biol. 13 (2011) 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO, Nat. Cell Biol. 9 (2007) 654. [DOI] [PubMed] [Google Scholar]
- [17].Esteller M, Nat. Rev. Genet. 12 (2011) 861. [DOI] [PubMed] [Google Scholar]
- [18].Mercer TR, Dinger ME, Mattick JS, Nat. Rev. Genet. 10 (2009) 155. [DOI] [PubMed] [Google Scholar]
- [19].Gardini A, Shiekhattar R, FEBS J 282 (2015) 1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mercer TR, Mattick JS, Nat. Struct. Mol. Biol. 20 (2013) 300. [DOI] [PubMed] [Google Scholar]
- [21].Fang Y, Fullwood MJ, Genomics Proteomics Bioinformatics 14 (2016) 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Reis EM, Verjovski-Almeida S, Front. Genet. 3 (2012) 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jiang X, Lei R, Ning Q, Biomark. Med. 10 (2016) 757. [DOI] [PubMed] [Google Scholar]
- [24].Silva A, Bullock M, Calin G, Cancers 7 (2015) 2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ellinger J, Gevensleben H, Muller SC, Dietrich D, Expert Rev. Mol. Diagn. 16 (2016) 1059. [DOI] [PubMed] [Google Scholar]
- [26].Bartel DP, Cell 116 (2004) 281. [DOI] [PubMed] [Google Scholar]
- [27].Garzon R, Calin GA, Croce CM, Annu. Rev. Med. 60(2009) 167. [DOI] [PubMed] [Google Scholar]
- [28].Carleton M, Cleary MA, Linsley PS, Cell Cycle 6 (2007) 2127. [DOI] [PubMed] [Google Scholar]
- [29].Harfe BD, Curr. Opin. Genet. Dev. 15 (2005) 410. [DOI] [PubMed] [Google Scholar]
- [30].Boehm M, Slack FJ, Cell Cycle 5(2006) 837. [DOI] [PubMed] [Google Scholar]
- [31].Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M, Nature 432 (2014) 226. [DOI] [PubMed] [Google Scholar]
- [32].Jin P, Alisch RS, Warren ST, Nat. Cell Biol. 6 (2004) 1048. [DOI] [PubMed] [Google Scholar]
- [33].Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR, Nature 435 (2005) 834. [DOI] [PubMed] [Google Scholar]
- [34].Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM, Proc. Natl. Acad. Sci. USA 103 (2006) 2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fritz JV, Heintz-Buschart A, Ghosal A, Wampach L, Etheridge A, Galas D , Wilmes P, Annu Rev. Nutr. 36 (2016) 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K, Clin. Chem. 56 (2010) 1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sohel MH, Achv. Life Sci. 10 (2016) 175. [Google Scholar]
- [38].Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T, J. Biol. Chem. 285 (2010)17442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kosaka N, Iguchi H, Ochiya T, Cancer Sci. 101 (2010) 2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kawaguchi T, Komatsu S, Ichikawa D, Tsujiura M, Takeshita H, Hirajima S, Miyamae M, Okajima W, Ohashi T, Imamura T, Kiuchi J, Konishi H, Shiozaki A, Okamoto K, Otsuji E, Int J Mol Sci. 17 (2016) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Huang YK, Yu JC, World J. Gastroenterol. 21 (2015) 9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sita-Lumsden A, Dart DA, Waxman J, Bevan CL, Br. J. Cancer 108 (2013) 1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C, Int. J. Oncol. 41 (2012) 1897. [DOI] [PubMed] [Google Scholar]
- [44].Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K, World J. Gastroenterol. 21 (2015) 8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].He Y, Lin J, Kong D, Huang M, Xu C, Kim TK, Etheridge A, Ding YY, Wang K, Clin Chem. 61 (2015) 1138. [DOI] [PubMed] [Google Scholar]
- [46].Thind A, Wilson C, J. Extracell. Vesicles. 5 (2016) 31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wong DT, Clin. Ther. 37 (2015) 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jackson JM, Witek MA, Kamande JW, Soper SA, Chem. Soc. Rev. 46 (2017) 4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Schwarzenbach H, Hoon DS, Pantel K, Nat. Rev. Cancer 11 (2011) 426. [DOI] [PubMed] [Google Scholar]
- [50].Hanash SM, Baik CS, Kallioniemi O, Nat. Rev. Clin. Oncol. 8 (2011) 142. [DOI] [PubMed] [Google Scholar]
- [51].Penfornis P, Vallabhaneni KC, Whitt J, Pochampally R, Int. J. Cancer. 138 (2016) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chomczynski P, Sacchi N, Anal. Biochem. 162 (1987) 156. [DOI] [PubMed] [Google Scholar]
- [53].Bligh EG, Dyer WJ, Can. J. Biochem. Phys. 37 (1959) 911. [DOI] [PubMed] [Google Scholar]
- [54].Brawerman G, Mendecki J, Lee S, Biochemistry 11 (1972) 637. [DOI] [PubMed] [Google Scholar]
- [55].Perry RP, Kelley DE, J. Mol. Biol. 70 (1972) 265. [DOI] [PubMed] [Google Scholar]
- [56].Puissant C, Houdebine LM, BioTechniques. 8 (1990) 148. [PubMed] [Google Scholar]
- [57].Rio DC, Ares M Jr., Hannon GJ, Nilsen TW, Cold Spring Harb. Protoc. 6 (2010) pdb.prot5438. [DOI] [PubMed] [Google Scholar]
- [58].Rio DC, Ares M Jr., Hannon GJ, Nilsen TW, Cold Spring Harb. Protoc. 6 (2010) pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- [59].Kim D, Linnstaedt S, Palma J, Park JC, Ntrivalas E, Kwak-Kim JYH, Gilman-Sachs A, Beaman K, Hastings ML, Martin JN, Duelli DM, J. Mol. Diagn. 14 (2012) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sourvinou IS, Markou A, Lianidou ES, J. Mol. Diagn. 15 (2013) 827. [DOI] [PubMed] [Google Scholar]
- [61].Dong L, Lin W, Qi P, Xu M, Wu X, Ni S, Huang D, Weng W, Tan C, Sheng W, Zhou X, Du X, Cancer Epidemiol. Biomarkers Prev. 25 (2016) 1158. [DOI] [PubMed] [Google Scholar]
- [62].Li T, Joshi MD, Ronning DR, Anderson JL, J. Chromatogr. A. 1272 (2013) 8. [DOI] [PubMed] [Google Scholar]
- [63].Hallett JP, Welton T, Chem. Rev. 111 (2011) 3508. [DOI] [PubMed] [Google Scholar]
- [64].Gale RJ, Osteryoung RA, Inorg. Chem. 18 (1979) 1603. [Google Scholar]
- [65].Emaus MN, Clark KD, Hinners P, Anderson JL, Anal. Bioanal. Chem. 410 (2018) 4135. [DOI] [PubMed] [Google Scholar]
- [66].Clark KD, Nacham O, Yu H, Li T, Yamsek MM, Ronning DR, Anderson JL, Anal. Chem. 87 (2015) 1552. [DOI] [PubMed] [Google Scholar]
- [67].Hourfar MK, Michelsen U, Schmidt M, Berger A, Seifried E, Roth WK, Clin. Chem. 51 (2005)1217. [DOI] [PubMed] [Google Scholar]
- [68].Marko MA, Chipperfield R, Birnboim HC, Anal. Biochem. 121 (1982) 382. [DOI] [PubMed] [Google Scholar]
- [69].El-Khoury V, Pierson S, Kaoma T, Bernardin F, Berchem G, Sci. Rep. 6 (2016) 19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Rubio M, Bustamante M, Hernandez-Ferrer C, Fernandez-Orth D, Pantano L, Sarria Y, Pique-Borras M, Vellve K, Agramunt S, Carreras R, Estivill X, Gonzalez JR, Mayor A, PLoS ONE 13 (2018) e0193527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hantzsch M, Tolios A, Beutner F, Nagel D, Thiery J, Teupser D, Holdt LM, PLoS ONE 9 (2014)e113298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mensah M, Borzi C, Verri C, Suatoni P, Conte D, Pastorino U, Orazio F, Sozzi G, Boeri M, J. Vis. Exp. 128 (2017) e56326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nacham O, Clark KD, Varona M, Anderson JL, Anal. Chem. 89 (2017) 10661. [DOI] [PubMed] [Google Scholar]
- [74].Nacham O, Clark KD, Anderson JL, Anal. Chem. 88 (2016) 7813. [DOI] [PubMed] [Google Scholar]
- [75].Wang H, Yang R, Yang L and Tan W, ACS Nano. 3 (2009) 2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Berg JM, Romoser A, Banerjee N, Zebda R, Sayes CM, Nanotoxicology. 3 (2009) 276. [Google Scholar]
- [77].Dausend J, Musyanovych A, Dass M, Walther P, Schrezenmeir H, Landfester K, Mailander V, Macromol. Biosci. 8 (2008) 1135. [DOI] [PubMed] [Google Scholar]
- [78].Feng Q, Zhang Z, Ma Y, He X, Zhao Y, Chai Z, Nanoscale Res. Lett. 7 (2012) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chang Y, Zhang Z, Liu H, Wang N, Tang J, Analyst 141 (2016) 4719. [DOI] [PubMed] [Google Scholar]
- [80].Xu S, Nasr SH, Chen D, Zhang X, Sun L, Huang X, Qian C, ACS Biomater. Sci. Eng. 4 (2018) 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhong R, Flack K, Zhong W, Analyst 137 (2012) 5546. [DOI] [PubMed] [Google Scholar]
- [82].Zhang X, Wang F, Liu B, Kelly EY, Servos MR, Liu J, Langmuir. 30 (2014) 839. [DOI] [PubMed] [Google Scholar]
- [83].Jimenez LA, Gionet-Gonzales MA, Sedano S, Carballo JG, Mendez Y, Zhong W, Anal. Bioanal. Chem. 410 (2018) 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Madhugiri S, Sun B, Smirniotis PG, Ferraris JP, Balkus KJ Jr., Microporous Mesoporous Mater. 69 (2004) 77. [Google Scholar]
- [85].Hashemi E, Akhavan O, Shamsara M, Valimehra S, Rahighi R, RSC Adv. 4 (2014) 60720. [Google Scholar]
- [86].Park JS, Goo N, Kim D, Langmuir. 30 (2014) 12587. [DOI] [PubMed] [Google Scholar]
- [87].Li F, Pei H, Wang L, Lu J, Gao J, Jiang B, Zhao X, Fan C, Adv. Funct. Mater. 23 (2013) 4140. [Google Scholar]
- [88].Zhao X, Johnson JK, J. Am. Chem. Soc. 129 (2007) 10438. [DOI] [PubMed] [Google Scholar]
- [89].Zhu C, Zeng Z, Li H, Li F, Fan C, Zhang H, J. Am. Chem. Soc. 135 (2013) 5998. [DOI] [PubMed] [Google Scholar]
- [90].Saha S, Sarkar P, Phys. Chem. Chem. Phys. 16 (2014) 15355. [DOI] [PubMed] [Google Scholar]
- [91].Zhao M, Wang Y, Ma Q, Huang Y, Zhang X, Ping J, Zhang Z, Lu Q, Yu Y, Xu H, Zhao Y, Zhang H, Adv. Mater. 27 (2015) 7372. [DOI] [PubMed] [Google Scholar]
- [92].Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, Matsumura Y, J. Gastrointest. Oncol. 2 (2011) 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cheng L, Sharples RA, Scicluna BJ, Hill AF, Extracell J. Vesicles 3 (2014) 23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wang K, Zhang S, Weber J, Baxter D, Galas DJ, Nucleic Acid Res. 38 (2010) 7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, Generozov EV, Govorun VM, Sci. Rep. 5 (2015) 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cvjetkovic A, Lotvall J, Lasser C, Extracell J. Vesicles 3 (2014) 23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Salih M, Zietse R, Hoorn EJ, Am. J. Physiol. Renal. Physiol. 306 (2014) F1251. [DOI] [PubMed] [Google Scholar]
- [98].Abdalla M, Haj-Ahmad T, Lam B, Kim W, Simkin M, Rghei N, Haj-Ahmad Y, Norgen Biotek Corp. 2012. [Google Scholar]
- [99].Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK, Kidney Int. 82 (2012) 1024. [DOI] [PubMed] [Google Scholar]
- [100].Nordin JZ, Lee Y, Vader P, Mager I, Johansson HJ, Heusermann W, Wiklander OP, Hallbrink M, Seow Y, Bultema JJ, Gilthorpe J, Davies T, Fairchild PJ, Gabrielsson S, Meisner-Kober NC, Lehtio J, Smith CI, Wood MJ, EL Andaloussi S, Nanomedicine 11 (2015) 879. [DOI] [PubMed] [Google Scholar]
- [101].Boing AN, van der Pol E, Grootemaat AE, Coumans FAW, Sturk A, Nieuwland R, J Extracell. Vesicles 3 (2014) 23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ashby J, Flack K, Jimenez LA, Duan Y, Khatib AK, omlo GS, Wang SE, Cui X, Zhong W, Anal. Chem. 86 (2014) 9343. [DOI] [PubMed] [Google Scholar]
- [103].Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, Chow A, O’Connor ST, Li S, Chin AR, Somlo G, Palomares M, Li Z, Tremblay JR, Tsuyada A, Sun G, Reid MA, Wu X, Swiderski P, Ren X, Shi Y, Kong M, Zhong W, Chen Y, Wang SE, Nat. Cell Biol. 17 (2015) 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhaes A, Ferreira JA, Osorio H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA, Lyden D, Nat. Cell Biol. 20 (2018) 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, Hsu EC, Gowrishankar G, Kanada M, Jokerst JV, Sierra RG, Chang E, Lau K, Sridhar K, Bermudez A, Pitteri SJ, Stoyanova T, Sinclair R, Nair VS, Gambhir SS, Demirci U, ACS Nano 11 (2017) 10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yoo CE, Kim G, Kim M, Park D, Kang HJ, Lee M, Huh N, Anal. Biochem. 431 (2012) 96. [DOI] [PubMed] [Google Scholar]
- [107].Szatanek R, Baran J, Siedlar M, Baj-Krzyworzeka M, Int. J. Mo.l Med. 36 (2015) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Taylor DD, Shah S, Methods 87 (2015) 3. [DOI] [PubMed] [Google Scholar]
- [109].Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Hoen ENN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F, J. Extracell. Vesicles. 2 (2013) 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Thery C, Witwer KW et al. , J. Extracell. Vesicles. 7 (2018) 1535750.30637094 [Google Scholar]
- [111].Wu M, Ouyang Y, Wang Z, Zhang R, Huang P, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y, Huang TJ, Proc. Natl. Acad. Sci. USA 114 (2017) 10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ku A, Lim HC, Evander M, Lilja H, Laurell T, Scheding S, Ceder Y, Anal. Chem. 90 (2018)8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Reategui E, van der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B, Floyd FP Jr., Khankhel AH, Thapar V, Hochberg FH, Sequist LV, Nahed BV, Carter BS, Toner M, Balaj L, Ting DT, Breakefield XO, Stott SL, Nat. Commun. 9 (2018) 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ibsen SD, Wright J, Lewis JM, Kim S, Ko S, Ong J, Manouchehri S, Vyas A, Akers J, Chen CC, Carter BS, Esener SC, Heller MJ, ACS Nano 11 (2017) 6641. [DOI] [PubMed] [Google Scholar]
- [115].Chen Z, Cheng S, Cao P, Qiu Q, Chen Y, Xie M, Xu Y, Huang W, Biosens. Bioelectron. 122 (2018)211. [DOI] [PubMed] [Google Scholar]
- [116].Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK, Zeng Y, Nat. Biomed. Eng. 3 (2019) 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, Fujiwara H, Okamoto K, Otsuji E, Anticancer Res. 33 (2013) 3185. [PubMed] [Google Scholar]
- [118].Peng H, Wang J, Li J, Zhao M, Huang SK, Gu YY, Li Y, Sun XJ, Yang L, Luo Q, Huang CZ, Life Sci. 151 (2016) 235. [DOI] [PubMed] [Google Scholar]
- [119].Li W, Li N, Kang X, Shi K, Clin. Chim. Acta. 475 (2017) 152. [DOI] [PubMed] [Google Scholar]
- [120].Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L, Qian X, Zhang F, Sun B, Cell Death Dis. 5 (2014) e1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Zhang K, Luo Z, Zhang Y, Wang Y, Cui M, Liu L, Zhang L, Liu J, Thorac. Cancer 9 (2018) 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C, Oncotarget 7 (2016) 85551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M, Tu J, Technol, Cancer Res. Treat. 16 (2017) 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M, Proc. Natl. Acad. Sci. USA 105 (2008) 10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Ge Q, Tian F, Zhou Y, Zhu Y, Lu J, Bai Y, Lu Z, Anal. Methods 6 (2014) 9101. [Google Scholar]
- [126].Tomasetti M, Staffolani S, Nocchi L, Neuzil J, Strafella E, Manzella N, Mariotti L, Bracci M, Valentino M, Amati M, Santarelli L, Clin. Biochem. 45 (2012) 575. [DOI] [PubMed] [Google Scholar]
- [127].Liu X, Luo HN, Tian WD, Lu J, Li G , Wang L, Zhang B, Liang BJ, Peng XH, Lin SX, Peng Y, Li XP, Cancer Biol. Ther. 14 (2013) 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H, Respir. Res. 15 (2014) 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, Aldahmash A, Alajez NM, Sci. Rep. 6 (2016) 25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Isin M, Uysaler E, Ozgur E, Koseoglu H, Sanli O, Yucel OB, Gezer U, Dalay N, Front. Genet. 6 (2015) 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wang YH, Jia J, Wang BC, Chen H, Yang ZH, Wang K, Luo CL, Zhang WW, Wang FB, Zhang XL, Cell Physiol. Biochem. 46 (2018) 532. [DOI] [PubMed] [Google Scholar]
- [132].Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H, Cancer 119 (2013) 1159. [DOI] [PubMed] [Google Scholar]
- [133].Que R, Ding G, Chen J, Cao L, World J Surg. Oncol. 11 (2013) 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Cazzoli R, Buttitta F, Nicola MD, Malatesta S, Marchetti A, Rom WN, Pass HI, J. Thorac Oncol. 8 (2013) 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Liu J, Sun H, Wang X, Yu Q, Li S, Yu X, Gong W, Int. J. Mol. Sci. 15 (2014) 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F, Tan W, Nandy D, Bevan GH, Longenbach S, Sun Z, Lu Y, Wang T, Thibodeau SN, Boardman L, Kohli M, Wang L, Eur. Urol. 67 (2015) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Alegre E, Sanmamed MF, Rodriguez C, Carranza O, Martin-Algarra S, Gonzalez A, Arch. Pathol. Lab Med. 138 (2014) 828. [DOI] [PubMed] [Google Scholar]
- [138].Eichelser C, Stuckrath1 I, Muller V, Milde-Langosch K, Wikman H, Pantel K, Schwarzenbach H, Oncotarget 5 (2014) 9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, Shim SG, Paik YH, Exp. Mol. Med. 47 (2015) e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Samsonov R, Burdakov V, Shtam T, Radzhabova Z, Vasilyev D, Tsyrlina E, Titov S, Ivanov M, Berstein L, Filatov M, Kolesnikov N, Gil-Henn H, Malek A, Tumor Biol. 37 (2016) 12011. [DOI] [PubMed] [Google Scholar]
- [141].Hannafon BN, Trigoso1 YD , Calloway CL, Zhao YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC, Ding WQ, Breast Cancer Res. 18 (2016) 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Samsonov R, Shtam T, Burdakov V, Glotov A, Tsyrlina E, Berstein L, Nosov A, Evtushenko V, Filatov M, Malek A, The Prostate 76 (2016) 68. [DOI] [PubMed] [Google Scholar]
- [143].Zhao L, Yu J, Wang J, Li H, Che J, Cao B, Cancer J. 8 (2017) 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Craw P, Balachandran W, Lab Chip 12 (2012) 2469. [DOI] [PubMed] [Google Scholar]
- [145].Ramalingam N, San TC, Kai TJ, Mak MYM, Gong HQ, Microfluid. Nanofluid. 7 (2009) 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Li Y, Liang L, Zhang CY, Anal. Chem. 85 (2013) 11174. [DOI] [PubMed] [Google Scholar]
- [147].Hong CY, Chen X, Li J, Chen JH, Chen G, Yang HH, Chem. Commun. 50 (2014) 3292. [DOI] [PubMed] [Google Scholar]
- [148].Yao B, Liu Y, Tabata M, Zhu H, Miyahara Y, Chem. Commun. 50 (2014) 9704. [DOI] [PubMed] [Google Scholar]
- [149].Fu C, Liu C, Wang S, Luo F, Lin Z, Chen G, Anal. Methods 8 (2016) 7034. [Google Scholar]
- [150].Shen W, Deng H, Ren Y, Gao Z, Chem. Commun. 49 (2013) 4959. [DOI] [PubMed] [Google Scholar]
- [151].Guo L, Lin Y, Chen C, Qiu B, Lin Z, Chen G, Chem. Commun. 52 (2016) 11347. [DOI] [PubMed] [Google Scholar]
- [152].Guo X, Liu P, Yang X, Wang K, Wang Q, Guo Q, Huang J, Liu J, Song C, Li W, Analyst 140 (2015)2016. [DOI] [PubMed] [Google Scholar]
- [153].Dirks RM, Pierce NA, Proc. Natl. Acad. Sci. USA 101 (2004) 15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Zheng J, Ma D, Shi M, Bai J, Li Y, Yang J, Yang R, Chem. Commun. 51 (2015) 16271. [DOI] [PubMed] [Google Scholar]
- [155].Rana M, Balcioglu M, Kovach M, Hizir MS, Robertson NM, Khan I, Yigit MV, Chem. Commun. 52 (2016) 3524. [DOI] [PubMed] [Google Scholar]
- [156].He D, Hai L, Wang H, Wu R, Li HW, Analyst 143 (2018) 813. [DOI] [PubMed] [Google Scholar]
- [157].Lan J, Wen F, Fu F, Zhang X, Cai S, Liu Z, Wu D, Li C, Chen J, Wang C, RSC Adv. 5 (2015)18008. [Google Scholar]
- [158].Asadzadeh-Firouzabadi A, Zare HR, Anal. Methods 9 (2017) 3852. [Google Scholar]
- [159].Zhai LY, Li MX, Pan WL, Chen Y, Li MM, Pang JX, Zheng L, Chen JX, Duan WJ, ACS Appl. Mater. Interfaces 10 (2018) 39478. [DOI] [PubMed] [Google Scholar]
- [160].Hizir MS, Balcioglu M, Rana M, Robertson NM, Yigit MV, ACS Appl. Mater. Interfaces 6(2014)14772. [DOI] [PubMed] [Google Scholar]
- [161].Zhao H, Qu Y, Yuan F, Quan X, Anal. Methods 8 (2016) 2005. [Google Scholar]
- [162].Metcalf GA, Shibakawa A, Patel H, Sita-Lumsden A, Zivi A, Rama N, Bevan CL, Ladame S, Anal. Chem. 88 (2016) 8091. [DOI] [PubMed] [Google Scholar]
- [163].Azimzadeh M, Rahaie M, Nasirizadeh N, Naderi-Manesh H, Anal. Methods 7 (2015) 9495. [Google Scholar]
- [164].Mabey D, Peeling RW, Ustianowski A, Perkins MD, Nat. Rev. Microbiol. 2 (2004) 231. [DOI] [PubMed] [Google Scholar]
- [165].Nichols JH, Clin. Lab. Med. 27 (2007) 893. [DOI] [PubMed] [Google Scholar]
- [166].Tudos AJ, Besselink GAJ, Schasfoort RBM, Lab Chip 1 (2001) 83. [DOI] [PubMed] [Google Scholar]
- [167].Sista R, Hua Z, Thwar P, Sudarsan A, Srinivasan V, Eckhardt A, Pollack M, Pamula V, Lab Chip 8 (2008) 2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Bhomia M, Balakathiresan NS, Wang KK, Papa L, Maheshwari RK, Sci. Rep. 6 (2016) 28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Mar-Aguilar F, Mendoza-Ramirez JA, Malagon-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodriguez-Padilla C, Resendez-Perez D, Dis. Markers 34 (2013) 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Wang K, Yuan Y, Cho J, McClarty S, Baxter D, Galas DJ, PLoS ONE 7 (2012) e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Niemz A, Ferguson TM, Boyle DS, Trends Biotechnol. 29 (2011) 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Hanafiah KM, Garcia M, Anderson D, Biomark. Med. 7 (2013) 333. [DOI] [PubMed] [Google Scholar]
- [173].John AS, Price CP, Clin. Biochem. Rev. 35 (2014) 155. [PMC free article] [PubMed] [Google Scholar]
- [174].Luppa PB, Muller C, Schlichtiger A, Schlebusch H, Trends Anal. Chem. 30 (2011) 887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Williams MR, Stedtfeld RD, Stedtfeld TM, Tiedje JM, Hashsham SA, Biomed. Microdevices 19 (2017) 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Stedtfeld RD, Tourlousse DM, Seyrig G, Stedtfeld TM, Kronlein M, Price S, Ahmad F, Gulari E, Tiedje JM, Hashsham SA, Lab Chip 12 (2012) 1454. [DOI] [PubMed] [Google Scholar]
- [177].Croce CM, Nat. Rev. Genet. 10 (2009) 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Palanichamy JK, Rao DS, Front. Genet. 5 (2014) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Iorio MV, Croce CM, EMBO Mol. Med. 4 (2012) 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO, Nat. Cell. Biol. 9 (2007) 654. [DOI] [PubMed] [Google Scholar]
- [181].Kosaka N, Iguchi H, Ochiya T, Cancer Sci. 10 (2010) 2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I, Oral Dis. 16 (2010) 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Conte D, Verri C, Borzi C, Suatoni P, Pastorino U, Sozzi G, Fortunato O, BMC Genomics 16(2015)849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Borzi C, Calzolari L, Conte D, Sozzi G, Fortunato O, Methods Mol. Biol. 1580 (2017) 239. [DOI] [PubMed] [Google Scholar]
- [185].Suzuki R, Asama H, Waragai Y, Takagi T, Hikichi T, Sugimoto M, Konno N, Watanabe K, Nakamura J, Kikuchi H, Sato Y, Marubashi S, Masamune A, Ohira H, Oncotarget 9 (2018) 4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Gasparello J, Allegretti M, Tremante E, Fabbri E, Amoreo CA, Romania P, Melucci E, Messana K, Borgatti M, Giacomini P, Gambari R, Finotti A, J. Exp. Clin. Cancer Res. 37 (2018) 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Bellingham SA, Shambrook M, Hill AF, Methods Mol. Biol. 1545 (2017) 55. [DOI] [PubMed] [Google Scholar]
- [188].Yoshikawa M, Iinuma H, Umemoto Y, Yanagisawa T, Matsumoto A, Jinno H, Oncol. Lett. 15 (2018)9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Perge P, Decmann A, Pezzani R, Bancos I, Fassina A, Luconi M, Canu L, Toth M, Boscaro M, Patocs A, Igaz P, Endocrine 59 (2018) 280. [DOI] [PubMed] [Google Scholar]
- [190].Gupta E, Agarwala P, Kumar G, Maiwall R, Sarin SK, J. Clin Virol. 88 (2017) 46. [DOI] [PubMed] [Google Scholar]
- [191].Hillemann D, Rusch-Gerdes S, Boehme C, Richter E, J. Clin. Microbiol. 49 (2011) 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Rossney AS, Herra CM, Brennan GI, Morgan PM, O’Connell B, J. Clin. Microbiol. 46 (2008)3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Zeka AN, Tasbakan S, Cavusoglu C, J. Clin. Microbiol. 49 (2011) 4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Gay-Andrieu F, Magassouba N, Picot V, Phillips CL, Peyrefitte CN, Dacosta B, Dore A, Kourouma F, Ligeon-Ligeonnet V, Gauby C, Longuet C, Scullion M, Faye O, Machuron JL, Miller M, J. Clin. Virol. 92 (2017) 20. [DOI] [PubMed] [Google Scholar]
- [195].Weller SA, Bailey D, Matthews S, Lumley S, Sweed A, Ready D, Eltringham G, Richards J, Vipond R, Lukaszewski R, Payne PM, Aarons E, Simpson AJ, Hutley EJ, Brooks T, J. Clin. Microbiol. 54 (2016) 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Roy S, Soh JH, Ying JY, Biosens. Bioelectron. 75 (2016) 238. [DOI] [PubMed] [Google Scholar]
- [197].Cai BZ, Pan ZW, Lu YJ, Curr. Med. Chem. 17 (2010) 407. [DOI] [PubMed] [Google Scholar]
- [198].Han M, Toli J, Abdellatif M, Curr. Opin. Cardiol. 26 (2011) 181. [DOI] [PubMed] [Google Scholar]
- [199].Stern E, Vacic A, Rajan NK, Criscione JM, Park J, Ilic BR, Mooney DJ, Reed MA, Fahmy TM, Nat. Nanotechnol. 5 (2010) 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Nissum M, Foucher AL, Expert Rev. Proteomics 5 (2008) 571. [DOI] [PubMed] [Google Scholar]
- [201].Liao T, Li X, Tong Q, Zou K, Zhang H, Tang L, Sun Z, Zhang G, Anal. Chem. 89 (2017) 5511. [DOI] [PubMed] [Google Scholar]
- [202].Summerton J, Weller D, Antisense Nucleic Acid Drug Dev. 7 (1997) 187. [DOI] [PubMed] [Google Scholar]
- [203].Bill BR, Petzold AM, Clar KJ, Schimmenti LA, Ekker SC, Zebrafish 6 (2009) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Youngblood DS, Hatlevig SA, Hassinger JN, Iversen PL, Moulton HM, Bioconjugate Chem. 18 (2007) 50. [DOI] [PubMed] [Google Scholar]
- [205].Hudziak RM, Barofsky E, Barofsky DF, Weller DL, Huang SB, Weller DD, Antisense Nucleic Acid Drug Dev. 6 (2009) 267. [DOI] [PubMed] [Google Scholar]
- [206].Summerton J, Biochim. Biophys. Acta 1489 (1999) 141. [DOI] [PubMed] [Google Scholar]
- [207].Ducree J, Haeberle S, Lutz S, Pausch S, von Stetten F, Zengerle R, Micromech J. Microengineering 17 (2007) S103. [Google Scholar]
- [208].Gorkin R, Park J, Siegrist J, Amasia M, Lee BS, Park JM, Kim J, Kim H, Madou M, Cho YK, Lab Chip 10 (2010) 1758. [DOI] [PubMed] [Google Scholar]
- [209].Wang S, Du D, Sensors 2 (2002) 41. [Google Scholar]
- [210].Roncon P, Soukupova M, Binaschi A, Falcicchia C, Zucchini S, Ferracin M, Langley SR, Petretto E, Johnson MR, Marucci G, Michelucci R, Rubboli G, Simonato M, Sci. Rep. 5 (2015)14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ, Biol. Psychiatry 69 (2011) 180. [DOI] [PubMed] [Google Scholar]
- [212].Rong H, Liu TB, Yang KJ, Yang HC, Wu DH, Liao CP, Hong F, Yang HZ, Wan F, Ye XY, Xu D, Zhang X, Chao CA, Shen QJ, J. Psychiatr. Res. 45 (2011) 92. [DOI] [PubMed] [Google Scholar]
- [213].McArdle H, Jimenez-Mateos EM, Raoof R, Carthy E, Boyle D, ElNaggar H, Delanty N, Hamer H, Dogan M, Huchtemann T, Kortvelyessy P, Rosenow F, Forster RJ, Henshall DC, Spain E, Sci. Rep. 7 (2017) 1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Cao H, Zhou X, Zeng Y, Sens. Actuators. B Chem. 279 (2019) 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Taller D, Richards K, Slouka Z, Senapati S, Hill R, Go DB, Chang H, Lab Chip 15 (2015) 1656. [DOI] [PubMed] [Google Scholar]
- [216].Hu J, Kwak KJ, Shi J, Yu B, Sheng Y, Lee LJ, Biomaterials 183 (2018) 20. [DOI] [PubMed] [Google Scholar]
- [217].Cheng H, Fu C, Kuo W, Chen Y, Chen Y, Lee Y, Li K, Chen C, Ma H, Huang P, Wang Y, Lee G, Lab Chip 18 (2018) 2917. [DOI] [PubMed] [Google Scholar]
- [218].Lasser C, Cell 177 (2019) 228. [DOI] [PubMed] [Google Scholar]
- [219].Murillo OD, Thistlethwaite W, Rozowsky J, Subramanian SL, Lucero R, Shah N, Jackson AR, Srinivasan S, Chung A, Laurent CD, Kitchen RR, Galeev T, Warrell J, Diao JA, Welsh JA, Hanspers K, Riutta A, Burgstaller-Muehlbacher S, Shah RV, Yeri A, Jenkins LM, Ahsen ME, Cordon-Cardo C, Dogra N, Gifford SM, Smith JT, Stolovitzky G, Tewari AK, Wunsch BH, Yadav KK, Danielson KM, Cell 177 (2019) 463. [DOI] [PMC free article] [PubMed] [Google Scholar]