Abstract
Several research lines are currently ongoing to address the multitude of facets of the pandemic COVID-19. In line with the One-Health concept, extending the target of the studies to the animals which humans are continuously interacting with may favor a better understanding of the SARS-CoV-2 biology and pathogenetic mechanisms; thus, helping to adopt the most suitable containment measures. The last two decades have already faced severe manifestations of the coronavirus infection in both humans and animals, thus, circulating epitopes from previous outbreaks might confer partial protection from SARS-CoV-2 infections. In the present study, we provide an in-silico survey of the major nucleocapsid protein epitopes and compare them with the homologues of taxonomically-related coronaviruses with tropism for animal species that are closely inter-related with the human beings population all over the world. Protein sequence alignment provides evidence of high sequence homology for some of the investigated proteins. Moreover, structural epitope mapping by homology modelling revealed a potential immunogenic value also for specific sequences scoring a lower identity with SARS-CoV-2 nucleocapsid proteins. These evidence provide a molecular structural rationale for a potential role in conferring protection from SARS-CoV-2 infection and identifying potential candidates for the development of diagnostic tools and prophylactic-oriented strategies.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Personalized medicine, Diagnosis
As of 30 March 2020, SARS-CoV-2 diffusion becomes a world-wide issue with nearly all countries of the globe being interested in a steadily growing number of infected subjects and the tendentially increasing mortality rate, by means of the COVID-19, in the newly involved countries. On the other hand, the SARS-CoV-2 diffusion seems to be approaching an end in Wuhan, China, its original epicentre. Here, the registration of new COVID-19 cases is currently near zero, enabling the development of fair and unbiased statistics on the COVID-19 data in the Hubei region [1,2]. The thorough review of the patient cases describes, besides the common pulmonary issues, a wide variety of sequelae reported by diverse patients from diverse hospitals following the viral infection. These range from hearth injury until hepatic and kidney failure, hindering a clear depiction of the pathogenetic mechanisms of the novel virus and the definition of informative clinical signs pathognomonic of SARS-CoV-2 infection [3,4]. Although the appropriate consideration of the patient’s health status is of crucial importance while assessing COVID-19 impact, it is believed that the severity of COVID-19 depends on a plurality of factors that include, among others, the load of viral particles transmitted between individuals, the transmission route and the individual immune system efficiency [5,6].
The causal agent of the current pandemic has been timely classified as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [7]. In humans, coronavirus infections are commonly manifested through weak clinical signs attributable to the seasonal flu-like symptoms. Nevertheless, the last two decades have already seen a couple of severe manifestations of the coronavirus infection. The first outbreak was described as the SARS epidemy registered in 2003 in China [8]; whereas a second coronavirus outbreak, known as Middle East Respiratory Syndrome (MERS), dates back to 2012 in Saudi Arabia. Besides humans [9], SARS viruses are able to infecting a wide plethora of animals such as birds and mammals, including synanthropic birds, dogs, camels, dromedaries, and pangolins [10]. SARS-CoV-2 is a member of the genus Betacoronavirus, featured by a positive-sense, single-stranded RNA genome of approximately 30 Kbs. The viral genome encodes for a relatively low number of proteins classified as either structural or nonstructural ones. Among structural proteins, the spike glycoprotein (S), envelope protein (E), membrane protein (M) and the nucleocapsid protein (NC) are the major ones [7].
Nucleocapsid protein is a 50 KDa protein commonly involved in the replication, transcription and packaging of the viral genome, other than hindering the reproductive cycle of the host cell [11]. Also, NC is the most abundant protein in coronaviruses, is highly immunogenic and its amino acid sequence is normally conserved, making of this protein a suitable candidate target for both vaccine formulations and diagnostic assays [12,13]. Previous studies on SARS-CoV reported NC protein epitopes as capable of eliciting a massive production of antibodies in infected subjects while its prevalence was significantly reduced in convalescent patients, suggesting such epitopes as candidates for the design of diagnostic tools. Contrarywise, epitopes triggering T-cell response might confer prolonged protection, in some cases estimated up to 11 years; thus, representing a valid alternative for the design of prophylactic measures [14].
Acknowledged the recent identification of SARS-CoV-2, several research lines are currently ongoing to address the multitude of facets of this pandemic issue. Among these, defining the immunological features of the SARS-CoV-2 might provide crucial knowledge for the design of efficient diagnostic tools and/or vaccine strategy. In addition, comparing epitopes of taxonomically related viruses can help to track an eventual cross-protection occurring between humans and the domestic animals, meant as the consequence of the continuous interrelation that, for various reasons, occur among the diverse animal species.
In the present study, we provide an in-silico survey of the major NC epitopes and compare them with the NC immunological domains of taxonomically related coronaviruses with tropism for synanthropic animal species that are closely inter-related with the human beings population all over the world. Given the ongoing status of the pandemic, both epitopes eliciting antibodies production and T-cell responses will be taken into account, as even the short-term antibody production can be protective for both human and animals; thus, help to weaken the clinical manifestation of the SARS-CoV-2 infection.
2. Material and methods
2.1. Data collection
The virus species and accession number of the proteins employed in this survey are provided in Table 1 . Nucleocapsid protein sequences were downloaded from the NCBI Protein repository (https://www.ncbi.nlm.nih.gov/protein/) except for the Pangolin nucleocapsid protein. The latter was obtained from the recent publication of Zhang et al. (supplementary material: [13], https://doi.org/10.1016/j.cub.2020.03.022) as no protein sequence for the pangolin coronavirus was available at the time of the study.
Table 1.
Selected virus overview. The table lists the viral specimens employed in this study along with the relative details.
| Virus | NCBI TaxID | NCBI Genome | Protein GI |
|---|---|---|---|
| Bat CoV RaTG13 | 693998 | MN996532 | QHR63308.1 |
| SARS-CoV | 694009 | NC_004718.3 | AAX16200.1 |
| Pangolin CoV | 2708335 | MT084071 | NC_PangolinCoV from [13] |
| Camel CoV | 1335626 | MK967708 | QGV13487.1 |
| MERS-CoV | 1335626 | NC_019843.3 | YP_009047211.1 |
| Dromedarius CoV | 1335626 | MH259486 | QCI31487.1 |
| H-Enteric CoV | 166124 | FJ415324 | ACJ35489.1 |
| Canine CoV | 215681 | KX432213 | AQT26504.1 |
| Bovine CoV | 11128 | NC_003045 | NP_150083.1 |
| Avian CoV | 11120 | NC_001451 | NP_040838.1 |
2.2. Computational processing of the protein sequences
The selected protein sequences were analyzed by using the Basic Local Alignment Search Tool for protein sequences (pBLAST) [15]. This tool implements the BLOSUM62 algorithm to compare protein sequences and calculates the statistical significance of matches as means of e-values. Multiple sequence alignment was performed by keeping default settings. Only the best hit for each coronavirus was considered and hits with alignment length less than or equal to 4 amino acids were manually removed. Statistically significant matches were employed for calculating the similarity tree of the taxonomically-related coronaviruses on the basis of their relative NC protein sequences. Fast minimum evolution algorithm was employed to produce a tree from given distances between sequences by setting a maximum fraction of mismatch between pairs of sequences of 0.85.
2.3. Structural mapping of epitopes to PDB structures
The mapped protein epitopes to SARS-CoV-2 NC protein (YP_009724397.2 |P0DTC9) were also mapped on two available PDB structures using the PyMOL Molecular Graphics System, Version 2.3.4 (Schrödinger, LLC.). Briefly, two NC 3D structure available in the RCSB PDB database (6M3M and 2JW8, https://www.rcsb.org/) covering respectively residues number 47–173 and 247–364 were downloaded and mapping epitopes were visualized through PyMOL. Distances between residues representing different sequences located on the surface of the 3D model were measured using the wizard measurement to identify possible areas of cross-reactivity and antigen-binding.
3. Results
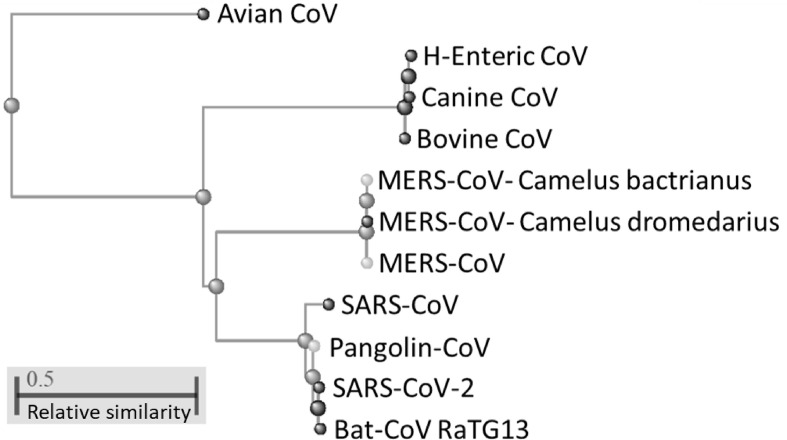
The whole aminoacidic sequences of the nucleocapsid protein from the selected coronavirus representatives were compared to assess the similarity level existing between the conserved protein sequences of the taxonomically related viruses. Fig. 1 depicts the phylogenetic classification of the coronaviruses on the basis of their NC protein. In this view, bat coronavirus RaTG13 is classified as the most similar to the circulating SARS-CoV-2, followed by SARS-CoV and the pangolin NC protein sequence. A separate clade is reserved to MERS-CoV and the NC protein sequences of the coronaviruses with tropism for camels (Camelus bactrianus) and dromedaries (Camelus dromedarius). Nucleocapsid proteins of bovine, canine and human enteric coronaviruses are depicted as sister groups of a third clade with lower similarity in relation to SARS-CoV-2; whereas, the avian coronavirus cluster apart since featured by the lowest similarity with the circulating SARS-CoV-2. Phylogenetic classification of the viruses on the basis of the NC protein sequence is further supported by the multiple sequence alignment performed on the whole protein sequence. Here, NC from bat coronavirus RaTG13 shows a similarity with the SARS-CoV-2 homologue slightly above 99%. NC from SARS-CoV, instead, is approximately 90.3% similar with the causal agent of COVID-19, followed by the NC sequence from pangolin CoV with almost 88% sequence similarity. Other selected coronavirus representatives have minor sequence similarity as reported in Table 2 .
Fig. 1.
Phylogenetic classification of the selected coronaviruses according to the whole nucleocapsid protein sequence homology.
Table 2.
Multiple sequence alignment of the Nucleocapside protein from taxonomically-related coronaviruses.
| Virus | Protein GI | Identity (%) | Alignment length | Mismatches | e-value |
|---|---|---|---|---|---|
| Bat CoV | QHR63308.1 | 99.045 | 419 | 4 | – |
| SARS-CoV | AAX16200.1 | 90.284 | 422 | 38 | – |
| Pangolin CoV | NC_PangolinCoV | 87.857 | 420 | 8 | – |
| Camel CoV | QGV13487.1 | 48.492 | 398 | 179 | 7.14e-93 |
| MERS-CoV | YP_009047211.1 | 48.492 | 398 | 179 | 7.14e-93 |
| Dromedarius CoV | QCI31487.1 | 48.492 | 398 | 179 | 9.55e-93 |
| Bovine CoV | NP_150083.1 | 38.938 | 339 | 158 | 5.97e-54 |
| H-Enteric CoV | ACJ35489.1 | 38.348 | 339 | 160 | 1.98e-54 |
| Canine CoV | AQT26504.1 | 38.348 | 339 | 160 | 9.97e-54 |
| Avian CoV | NP_040838.1 | 29.664 | 327 | 210 | 2.00e-31 |
Nucleocapsid protein sequences were further investigated for the presence of major immunogenic sequences capable of eliciting either antibodies production or eliciting T-cell response (Table 3 ) according to the evidence reported in Ref. [14].
Table 3.
Selected nucleocapsid protein epitopes.
| Epitope | Virus | % Identity | Length | Alignment length | Position | e-value |
|---|---|---|---|---|---|---|
| KHWPQIAQ FAPSASAFF |
SARS-CoV | 100 | 17 | 17 | 300–316 | 2.46e-16 |
| Bat CoV | 100 | 17 | 299–315 | 2.46e-16 | ||
| Dromedarius CoV | 78.571 | 14 | 293–306 | 7.55e-09 | ||
| Camel CoV | 78.571 | 14 | 293–306 | 7.55e-09 | ||
| MERS-CoV | 78.571 | 14 | 293–306 | 7.55e-09 | ||
| H-Enteric CoV | 52.941 | 17 | 308–324 | 7.05e-04 | ||
| Canine CoV | 52.941 | 17 | 308–324 | 7.05e-04 | ||
| Bovine CoV | 52.941 | 17 | 308–324 | 8.30e-05 | ||
| Pangolin CoV | 100 | 16 | 290–305 | 2.72e-16 | ||
| AQFAPSA SAFFGMSR |
SARS-CoV | 100 | 15 | 15 | 306–320 | 3.34e-13 |
| Bat CoV | 100 | 15 | 305–319 | 3.34e-13 | ||
| Dromedarius CoV | 71.429 | 14 | 297–310 | 2.75e-06 | ||
| Camel CoV | 71.429 | 14 | 297–310 | 2.75e-06 | ||
| MERS-CoV | 71.429 | 14 | 297–310 | 2.75e-06 | ||
| H-Enteric CoV | 63.636 | 11 | 314–324 | 0.001 | ||
| Canine CoV | 63.636 | 11 | 314–324 | 0.001 | ||
| Avian CoV | 58.333 | 12 | 276–286 | 13 | ||
| Bovine CoV | 63.636 | 11 | 314–324 | 1.73e-04 | ||
| Pangolin CoV | 100 | 15 | 295–309 | 3.33e-14 | ||
| PKGFYAEG SRGGSQASSR |
SARS-CoV | 100 | 18 | 18 | 169–186 | 2.32e-15 |
| Bat CoV | 100 | 18 | 168–185 | 2.32e-15 | ||
| Dromedarius CoV | 61.111 | 18 | 157–174 | 1.02e-04 | ||
| Camel CoV | 61.111 | 18 | 157–174 | 1.02e-04 | ||
| MERS-CoV | 61.111 | 18 | 157–174 | 1.02e-04 | ||
| H-Enteric CoV | 48 | 25 | 183–207 | 2.88e-04 | ||
| Canine CoV | 48 | 25 | 183–207 | 2.88e-04 | ||
| Avian CoV | 100 | 5 | 183–187 | 0.15 | ||
| Bovine CoV | 48 | 25 | 183–207 | 3.40e-05 | ||
| Pangolin CoV | 100 | 18 | 168–185 | 2.30e-16 | ||
| QFAPSASAF FGMSRIGM |
SARS-CoV | 100 | 17 | 17 | 307–323 | 6.91e-16 |
| Bat CoV | 100 | 17 | 306–322 | 6.91e-16 | ||
| Dromedarius CoV | 81.818 | 11 | 300–310 | 1.54e-05 | ||
| Camel CoV | 81.818 | 11 | 300–310 | 1.54e-05 | ||
| MERS-CoV | 81.818 | 11 | 300–310 | 1.54e-05 | ||
| H-Enteric CoV | 50 | 16 | 309–324 | 0.004 | ||
| Canine CoV | 50 | 16 | 309–324 | 0.004 | ||
| Avian CoV | 53.846 | 13 | 276–287 | 7.8 | ||
| Bovine CoV | 50 | 16 | 309–324 | 4.73e-04 | ||
| Pangolin CoV | 100 | 17 | 296–312 | 6.88e-17 | ||
| QLPQGTTLPKGF YAEGSRGGSQ |
SARS-CoV | 100 | 22 | 22 | 161–182 | 7.79e-20 |
| Bat CoV | 100 | 22 | 160–181 | 7.79e-20 | ||
| H-Enteric CoV | 66.667 | 15 | 177–191 | 3.16e-05 | ||
| Canine CoV | 66.667 | 15 | 177–191 | 3.16e-05 | ||
| Dromedarius CoV | 61.111 | 18 | 153–170 | 4.88e-04 | ||
| Camel CoV | 61.111 | 18 | 153–170 | 4.88e-04 | ||
| MERS-CoV | 61.111 | 18 | 153–170 | 4.88e-04 | ||
| Avian CoV | 100 | 5 | 183–187 | 0.22 | ||
| Bovine CoV | 66.667 | 15 | 177–191 | 3.72e-06 | ||
| Pangolin CoV | 100 | 22 | 160–181 | 7.75e-21 | ||
| YNVTQAFGR RGPEQTQGNF |
SARS-CoV | 100 | 19 | 19 | 269–287 | 4.02e-18 |
| Bat CoV | 100 | 19 | 268–286 | 4.02e-18 | ||
| Dromedarius CoV | 63.158 | 19 | 260–278 | 1.86e-06 | ||
| Camel CoV | 63.158 | 19 | 260–278 | 1.86e-06 | ||
| MERS-CoV | 63.158 | 19 | 260–278 | 1.86e-06 | ||
| H-Enteric CoV | 58.824 | 17 | 282–295 | 3.32e-04 | ||
| Canine CoV | 58.824 | 17 | 282–295 | 3.32e-04 | ||
| Bovine CoV | 58.824 | 17 | 282–295 | 3.91e-05 | ||
| Pangolin CoV | 100 | 15 | 261–275 | 2.40e-14 |
Epitopes falling within protein sequence domains for which homology modelling based on the experimental crystallographic structure is available are mapped and provided in Fig. 2 and Fig. 3 ; whereas, the full list of the tested epitopes against the selected viral nucleocapsid protein sequences is provided as Supplementary material (S1).
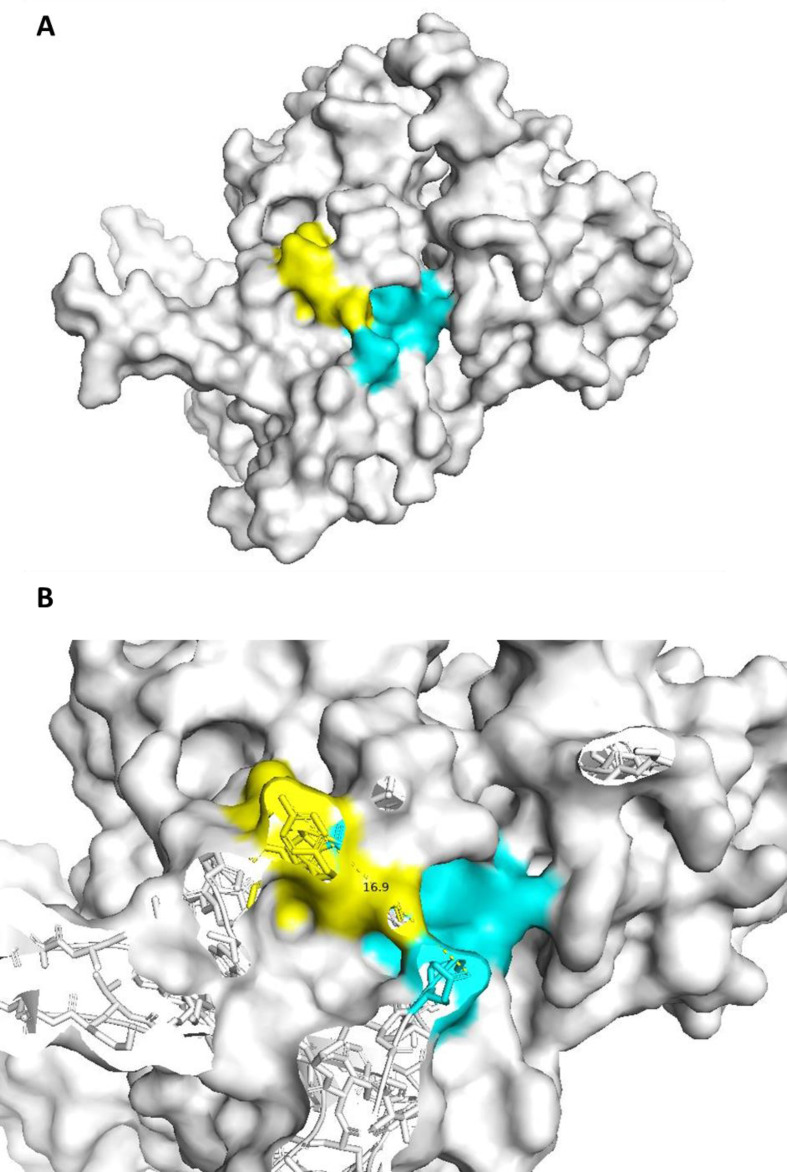
Fig. 2.
Epitope mapping into 6M3M structural model. The figure depicts mapping of the epitope sequence AQFAPSASAFFGMSR (yellow) and QFAPSASAFFGMSRIGM (cyan). A-panel shows the surface mapping of the epitopes in the D structure of the nucleocapsid protein domain. B-panel highlights distances occurring between the mapped epitopes.
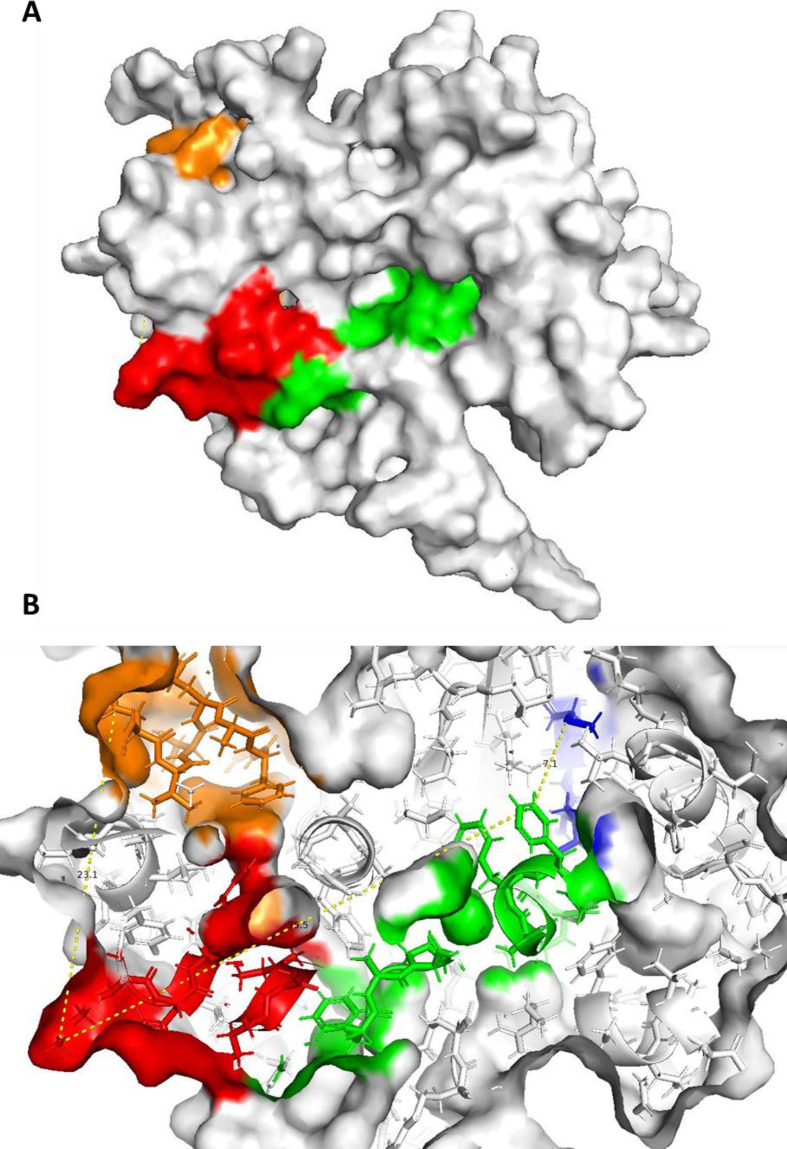
Fig. 3.
Epitope mapping into 2JW8 structural model. The figure depicts mapping of the epitope sequence PKGFYAEGSRGGSQASSR (blue), YNVTQAFGRRGPEQTQGNF (orange), AQFAPSASAFFGMSR (green) and KHWPQIAQFAPSASAFF (red). A-panel shows the surface mapping of the epitopes in the D structure of the nucleocapsid protein domain. B- panel highlights distances occurring between mapped epitopes.
Some of the mapped epitopes are “conserved” between the circulating SARS-CoV-2, the SARS-CoV, bat RaTG13 CoV and the pangolin CoV NC protein, i.e. the clade including the most similar specimens; whereas, other epitopes are “shared” with a multitude of coronavirus, including those from other animals that clustered as dissimilar from the circulating SARS-CoV-2 (Fig. 1, Table 3 and Table S1). With regard to interspecies distribution, all epitopes are identified in the bat RaTG13 CoV and SARS-CoV, whilst the pangolin NC sequence fail to significatively align two of the epitope sequences classified as B-cells activating. In addition, epitopes sequence comparison highlights that both bat RaTG13 coronavirus and SARS-CoV scored 100% sequence identity with SARS-CoV-2 epitopes, whereas NC protein from pangolin results in a handful of epitopes sharing a lower percentage of identity with the novel 2019 virus (Table 3 and Table S1).
Besides the most similar virus specimens, some epitope sequences are also identified in the viruses responsible for previous coronavirus outbreaks and coronaviruses with tropism for synanthropic and domestic animals. Although identity percentage with SARS-CoV-2 is below 100%, high homology levels have been scored for MERS-CoV, camel-CoV, dromedary-CoV. Interestingly, bovine CoV, canine CoV, human enteric CoV and avian-CoV have also shared epitope sequences, despite the dissimilarity highlighted on the basis of the whole NC sequence (Fig. 1, Table 3).
Structural mapping of the epitopes listed in Table 3 into two high-fidelity 3D models of the NC protein domains (6M3M and 2JW8 entries of PDB database) shows that a plurality of epitopes is mapped adjacent each other in a well define structural domain which is defined by a radius of approximately 15 Å (Fig. 2, Fig. 3). Interestingly, these epitopes are coded in distal primary sequence region of the NC protein which forms a conformational immunogenic domain (Fig. 2, Fig. 3). Unfortunately, not all the investigated epitopes can be mapped in the tridimensional model of the NC protein due to the lack of confident model structure for the whole protein length.
4. Discussion
The current health emergency raised by the COVID-19 aroused the interest of the world-wide scientific community that actively operates under diverse research lines on the attempt to address the multiple facets of this pandemic issue, including the elucidation of the immunological peculiarities of the novel virus.
The present study provides an in-silico survey of the major epitope sequences of the nucleocapsid protein from taxonomically related coronaviruses. Coronavirus infections and outbreaks were already registered in the past decades in both humans and animals [8,9]. Thus, investigating whether immunological features of taxonomically related viruses are shared with the SARS-CoV-2 might be helpful for planning innovative diagnostic and prophylactic interventions. According to the One-Health approach, defining the distribution of epitopes among viruses capable of infecting synanthropic and/or domestic animals might represent a key strategy for assessing the plausible occurrence of cross-protection elicited by the continuous inter-relation existing between human and animal population. In this view, exposure to epitopes from synanthropic animals might confer partial protection from SARS-CoV-2 infection. This would explain, at least in part, the early detection of class G immunoglobulins in newly infected subjects and the big divergence occurring in the clinical manifestation of the SARS-CoV-2 infections, other than preventing epitopes-exposed subjects from the severe form of COVID-19 [16].
Nucleocapsid protein is a highly conserved protein involved in the active phase of the viral infection. Being highly immunogenic is commonly targeted in studies aimed at developing alternative diagnostic tools and prophylactic strategies [11,14,[17], [18], [19]]. Multiple alignments of the whole protein sequences highlight a very high homology between the NC sequence of SARS-CoV-2 and bat RaTG13 CoV.
This evidence does not reflect the overall genome alignments recently published where the draft whole-genome sequence of the pangolin coronavirus is clustered as the closest relative to SARS-CoV-2 [13]. Our outcomes, instead, highlight a closer functional relationship between the bat RaTG13 and SARS-CoV-2 than the pair pangolin CoV and SARS-CoV-2 when restricting the investigation to a key structural protein. Also, NC protein of pangolin CoV fails to align two epitope sequences, suggesting a separation between the functional adaptation and molecular evolution. Moreover, acknowledged the overall co-share of epitope sequences, the lack of epitope alignments might be of importance for an accurate elucidation of the role of pangolin as the intermediate host in the virus life cycle.
With regard to epitopes distribution across viruses with tropism for synanthropic and/or domestic animals, the survey highlights that some epitope sequences are more “conserved” among the most related specimens, while other epitopes are shared among a wider ensemble of coronaviruses. Similar outcomes were also described for the spike proteins epitopes [20]. In this view, a detailed description of the epitopes distribution over the viral population might provide valuable information driving future researches aimed at setting efficient prophylactic strategies and/or the design of tool capable of differential diagnosis on the basis of serological tests.
The adjacent mapping of the tested epitopes in the tridimensional models of the NC protein domains suggest the mapped area as being highly immunogenic, exposed to the solvent and, consequently, to molecular immune system effectors. Furthermore, this supports the immunogenicity of the selected epitope sequences which is of particular interest for that sequences scoring a lower level of identity in relation to SARS-CoV-2 such as the canine CoV, bovine CoV and the human enteric CoV. Although featured by a lower level of homology with SARS-CoV-2 NC protein and the presence of some mismatches in the epitope sequence, these sequences are mapping in the same structural domains as SARS-CoV-2 epitopes and are likely to be recognized by the host immune system; thus, potentially involved in conferring partial protection against SARS-CoV-2 infection other being considered good candidates for the development of diagnostic tools and prophylactic-oriented strategies.
To conclude, COVID-19 pandemic is a global issue that stimulates the synergistic cooperation between research groups all over the world. At the same manner, planning studies that seek to address multiple aspects of COVID-19 enable a fast and effective control of the SARS-CoV-2 infections. In line with the One-Health concept, extending the target of the studies to the environment and the animals which humans are continuously interacting with favor a better understanding of this complex phenomenon and help to adopt the most suitable containment measures. Continuing mapping the major epitopes known for the viral proteins is, in our view, a promising strategy. In this light, defining the map of the circulating epitopes among diverse geographical areas and considering the diverse people habits and animal population they are interacting with might also provide unevaluable information for the design of personalized prophylactic and diagnostic strategies, shaped in dependence of the geographical context and the subject’s habits. Thus, an incisive focus on these aspects can contribute to a relatively fast control of this pandemic issue.
Conflict of interest
The authors declare no conflict on interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2020.04.002.
Contributor Information
Andrea Urbani, Email: andrea.urbani@unicatt.it.
Paola Roncada, Email: roncada@unicz.it.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhou X., Wu Z., Yu R., Cao S., Fang W., Jiang Z. Modelling-based evaluation of the effect of quarantine control by the Chinese government in the coronavirus disease 2019 outbreak. MedRxiv. 2020 doi: 10.1101/2020.03.03.20030445. https://ssrn.com/abstract=3551346 [Online ahead of print]. Available at SSRN: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H., Wang Z., Dong Y., Chang R., Xu C., Yu X. Phase-adjusted estimation of the number of coronavirus disease 2019 cases in wuhan, China. Cell Discov. 2020;6:10. doi: 10.1038/s41421-020-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi Q., Wu Y., Mei S., Ye C., Zou X., Zhang Z. Epidemiology and Transmission of COVID-19 in Shenzhen China: analysis of 391 cases and 1,286 of their close contacts. MedRxiv. 2020 doi: 10.1101/2020.03.03.20028423. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiao R., Tran N.H., Shan B., Ghodsi A., Li M. 2020. Personalized workflow to identify optimal T-cell epitopes for peptide-based vaccines against COVID-19. arXiv:2003.10650 [Online ahead of print] [Google Scholar]
- 7.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;3:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang M.S., Lu Y.T., Ho S.T., Wu C.C., Wei T.Y., Chen C.J. Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem Biophys Res Commun. 2004;314:931–936. doi: 10.1016/j.bbrc.2003.12.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020:ciaa310. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1–6. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S.F., Wootton J.C., Gertz E.M., Agarwala R., Morgulis A., Schäffer A.A. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P., Gao Q., Wang T., Ke Y., Mo F., Jia R. Evaluation of recombinant nucleocapsid and spike proteins for serological diagnosis of novel coronavirus disease 2019 (COVID-19) MedRxiv. 2020 doi: 10.1101/2020.03.17.20036954. [Online ahead of print]. [DOI] [Google Scholar]
- 18.He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem Biophys Res Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. 2020. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell host microbe. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilocca B., Soggiu A., Musella V., Britti D., Sanguinetti M., Urbani A. Molecular basis of COVID-19 relationships in different species: a one health perspective. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.03.002. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.