Abstract
Small-molecule aggregates are a leading cause of artifacts in early drug discovery, but little is known about their interactions with proteins, nor why some proteins are more susceptible to inhibition than others. A possible reason for this apparent selectivity is that aggregation-based inhibition, as a stoichiometric process, is sensitive to protein concentration, which varies across assays. Alternatively, local protein unfolding by aggregates may lead to selectivity since stability varies among proteins. To deconvolute these effects, we used differentially stable point mutants of a single protein, TEM-1 β-lactamase. Broadly, destabilized mutants had higher affinities for and were more potently inhibited by aggregates versus more stable variants. The addition of the irreversible inhibitor moxalactam-destabilized several mutants, and these typically bound tighter to a colloidal particle, while the only mutant it stabilized bound weaker. These results suggest that less-stable enzymes are more easily sequestered and inhibited by colloidal aggregates.
Graphical Abstract

■ INTRODUCTION
Many organic small molecules aggregate in a concentration-dependent manner in aqueous buffer.1,2 These densely packed particles have long been known to promiscuously inhibit proteins in vitro and in vivo3,4 by sequestration and partial unfolding5-7 and are a leading cause of artifacts in early drug discovery.8-11 Intriguingly, the same properties that enable them to sequester proteins also make them interesting candidates as self-assembling drug nanoparticles. Proteins that bind to the surface of aggregates can stabilize them, increasing their longevity in buffered solutions.12,13 Additionally, although aggregated drug particles have difficulty diffusing through the cell membrane,14,15 surface-adsorbed proteins can enable uptake of drug aggregates through receptor-mediated endocytosis and can even target them to specific cell populations.12 This has made colloidal aggregates, long considered problematic in early discovery, targets for formulation as drug-rich nanoparticles.12,16
To understand their role as artifactual inhibitors in early discovery and to optimize them for nanoparticle drug delivery, it would be useful to understand the properties of proteins, if any, that drive protein–aggregate interactions. Several studies have noted that aggregates composed of a given molecule can have different half maximal inhibitory concentration (IC50) values against different proteins, an observation that has been attributed to differences in protein concentration in the relevant assays.17,18 However, unusually stable and well-behaved co-formulated colloidal aggregates have been shown to have higher affinities for some proteins than others in the same protein concentration range, suggesting that aggregate-based inhibition may have the potential for selectivity.5 This, along with the observation that binding to colloidal aggregates leads to local protein unfolding, suggests that the apparent selectivity may partly reflect differential protein stability. A challenge to investigating this hypothesis is that binding to colloidal aggregates is effectively a stoichiometric event. In such a domain, proteins that are assayed at higher concentrations will always seem less sensitive to inhibition by a colloidal aggregate because there is more protein to bind, and hence more aggregator is needed.
To deconvolute protein stability from protein concentration effects and other potential gross variations among proteins (e.g., exposed hydrophobic surface area, isoelectric point, and protein size), we engineered five point mutants of TEM-1 β-lactamase, each of which occurs in clinical isolates. Mostly, these substitutions are distributed around the overall ligand-binding site of the TEM enzymes. Only one (G238S) is near the catalytic center, while M182T is far from the active site. This particular mutation is functionally silent (it is a stability mutant, occurring naturally in the evolution of TEM resistance, whose major role appears to be restabilizing gain-of-function mutants that have lost stability, consistent with a trade-off between stability and activity, Figure 1A).19-21 These mutants, all previously characterized,20 provide a quantitative range of stability relative to the native enzyme. As point mutants of the same enzyme, any effect of concentration can be controlled for, whereas other changes in gross physical properties in the enzymes can be essentially discounted. This allows us to isolate any possible effect of protein stability in aggregate-based enzyme inhibition. The implications of these results for understanding protein–aggregate interactions, both for artifactual inhibition and for nanoparticle design, will be discussed.
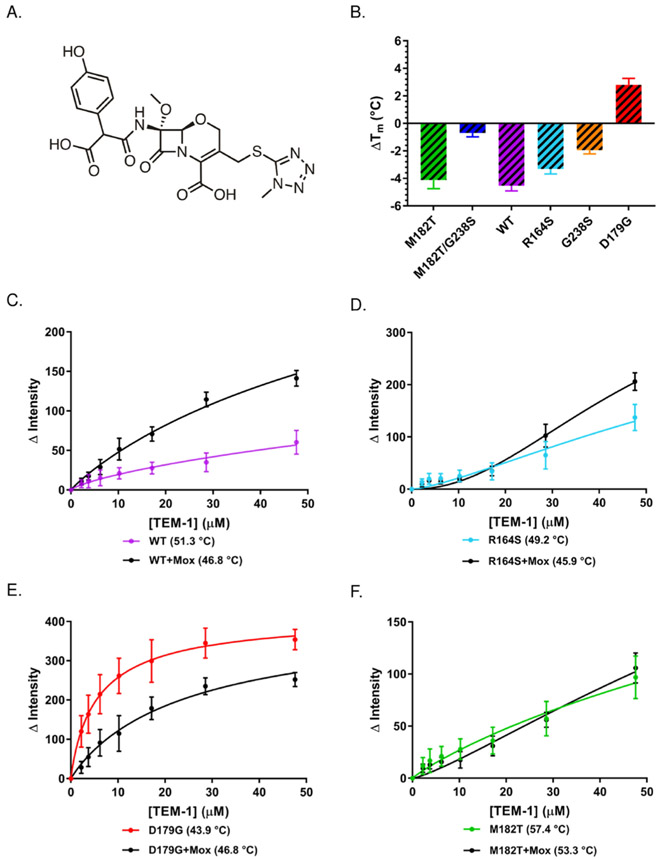
Figure 1.
Thermodynamic stabilities and binding affinities of TEM-1 mutants. (A) Location of TEM-1 mutations M182T (green), R164S (cyan), G238S (orange), and D179G (red) in TEM-1 M182T mutant PDB structure 1JWP relative to catalytic residues S70, K73, S130, and E166 (pink). (B) Melting temperatures of TEM-1 mutants determined by differential scanning fluorimetry (DSF) (see Table 1 for Tm and ΔΔG values). (C) The competitive displacement of 100 nM 5-maleimido-fluorescein-ribosomal protein L2 globular domain (5-MF-L2gd) bound to 20 μM Sor/CR colloids by TEM-1 stability mutants.
■ RESULTS
To investigate whether stability effects are present in aggregate-based enzyme inhibition, we studied TEM-1 β-lactamase point mutants with different thermodynamic stabilities. In addition to the wild-type (WT) enzyme, these included three destabilized mutants, R164S, G238S, and D179G, and two stabilized mutants, M182T/G238S and M182T. The stabilities of these enzymes were previously characterized by reversible, equilibrium thermal denaturation using circular dichroism,20 which we verified here by differential scanning fluorimetry (DSF; Table S1; the two sets of measurements are consistent). Differences in melting temperatures and ΔΔG values between the least and most stable mutants spanned 13.5 °C and 6.7 kcal/mol, with the least stable mutant, D179G, having a Tm of 43.9 °C and the most stable mutant, M182T, having a Tm of 57.4 °C (Table 1 and Figure 1B). We determined binding affinities for all six TEM-1 variants against co-formulated Sorafenib/Congo red (Sor/CR) colloids5,22 by a fluorescence-based competitive binding assay. 5-MF-L2gd, the globular domain of the ribosomal protein L2 labeled with fluorescein-5-maleimide loses fluorescence upon aggregate adsorption. When a competing protein, here TEM-1 or its mutants, is introduced fluorescence recovers as the TEM-1 outcompetes 5-MF-L2gd for colloid binding.5 By monitoring fluorescence recovery upon displacement of 5-MF-L2gd by the competing TEM-1 enzymes, we can determine apparent binding constants to the Sor/CR colloids (expressed here as EC50 values). Destabilized mutants R164S, G238S, and D179G had lower (more potent) apparent EC50 values against Sor/CR colloid (85.8, 38.2, and 6.0 μM, respectively) versus the more stabl WT and M182T enzymes (577 and 198.5 μM, respectively, (Table 1 and Figure 1C). The highly destabilized D179G had a 33-fold lower EC50 value than the stabilized M182T enzyme and bound 96-fold better than WT. There was a 15-fold and 5-fold decrease (improvement) in EC50 of the destabilized G238S versus the WT and M182T enzymes, respectively, while the destabilized R164S showed a modest decrease (improvement) in EC50 versus both WT and M182T. Admittedly, not all of the mutants had a monotonic relationship between stability and colloid binding. Indeed, one of the more stable mutants, M182T/G238S, with a ΔTm of 3.2 °C and a ΔΔG of 1.08 kcal/mol versus WT, was the third best binder, after D179G and G238S, the two most destabilized mutants in the series, with an EC50 of 58.5 μM (Table 1 and Figure 1C). We note that M182T/G238S has an unusually low van’t Hoff enthalpy of unfolding, lower even than the WT enzyme (Table S1), reflecting a broader temperature range over which this mutant unfolds; this may partly explain this unexpected result, a point to which we will return.
Table 1.
Thermodynamic Stabilities and Binding Affinities of TEM-1 mutants to Sor/CR Colloidsa
| mutant | Tm (°C) | ΔΔG (kcal mol−1) | EC50 (μM) |
|---|---|---|---|
| M182T | 57.4 ± 0.3 | 2.67 ± 0.13 | 198.5 ± 47.5 |
| M182T/G238S | 54.5 ± 0.08 | 1.08 ± 0.107 | 58.5 ± 7.5 |
| WT | 51.3 ± 0.3 | 577 ± 303 | |
| R164S | 49.2 ± 0.2 | −0.73 ± 0.05 | 85.8 ± 9.1 |
| G238S | 46.7 ± 0.07 | −1.94 ± 0.12 | 38.2 ± 4.5 |
| D179G | 43.9 ± 0.2 | −4.04 ± 0.21 | 6.0 ± 0.9 |
EC50 is the concentration of TEM-1 mutant at which 50% of fluorescent L2gd is displaced from the colloid.
To further explore the possibility of protein stability as a driving force in protein–aggregate binding, we perturbed the TEM-1 point mutants using a well-studied covalent inhibitor, moxalactam (Figure 2A). Unlike noncovalent ligands, which stabilize their respective enzymes upon binding, covalent inhibitors can either destabilize or stabilize an enzyme. This depends on the noncovalent interactions between the inhibitor and the enzyme. Although binding is enforced by the covalent bond, the noncovalent interactions can either be favorable or unfavorable.23-26 Based on a crystallographic complex (PDB ID: 1FCO), mutant analysis, and protein stability measurements,26 moxalactam is thought to destabilize another β-lactamase, AmpC, via an unfavorable interaction imposed by its 6(7)-methoxy group in the covalent enzyme adduct. A similar effect is observed in a TEM-1 covalent complex with imipenem (PDB ID: 1BT5), where the methyl in the 6α-1R-hydroxyethyl group is thought to destabilize via a steric clash in the covalent adduct.26,27 Substantial changes in the overall enzyme structure are not observed in either case versus the apo structures. Accordingly, we investigated shifts in Tm after incubation of the enzymes with moxalactam using DSF. As previously reported, the irreversible inhibitor destabilized the native enzyme, lowering its Tm by 4.5 °C.26 Moxalactam also destabilized M182T, R163S, and G238S by 4.1, 3.3, and 1.9 °C, respectively, while having no substantial effect on the M182T/G238S mutant. Unexpectedly, moxalactam actually stabilized D179G, increasing its Tm by 2.9 °C, reflecting favorable noncovalent interactions within the covalent complex, between the inhibitor and this particular mutant (Table 2 and Figure 2B). We determined binding affinities to the Sor/CR colloids for the native enzyme, M182T, R164S, and D179G, those most significantly impacted by moxalactam. Consistent with the idea that protein stability affects colloid binding, moxalactam-destabilized WT and R164S enzymes experienced a 6- and 1.8-fold decrease (improvement) in EC50 values, respectively (Table 2 and Figure 2C,D). Conversely, D179G, the only enzyme to be stabilized by moxalactam, experienced a 4.3-fold increase (worsening) in its EC50 against the Sor/CR colloids (Table 2 and Figure 2B,E). An enzyme that was inconsistent with this trend was M182T. This mutant was substantially destabilized by moxalactam, and curve fitting suggested a 1.7-fold improvement in its EC50. However, little discernable binding change was observed in the binding isotherm (Table 2 and Figure 2F).
Figure 2.
Thermodynamic stabilities and binding affinities of TEM-1 mutants in complex with moxalactam. (A) The chemical structure of moxalactam. (B) Change in melting temperatures of TEM-1 mutants incubated with 100-fold molar excess of β-lactamase inhibitor moxalactam relative to respective apo-enzymes, determined by DSF (see Table 2). The competitive displacement of 100 nM 5-MF-L2gd bound to 20 μM Sor/CR colloids by TEM-1 stability mutants, (C) WT and WT-inhibitor complex, (D) R164S and R164S-inhibitor complex, (E) D179G and D179G-inhibitor complex, and (F) M182T and M182T-inhibitor complex (see Tables 1 and 2 for EC50 values).
Table 2.
Thermodynamic Stabilities and Binding Affinities of TEM-1-moxalactam Complexes to Sor/CR Colloidsa
| mutant | Tm (°C) + Mox | ΔTm (°C) | EC50 (μM) + Mox |
|---|---|---|---|
| M182T | 53.3 ± 0.4 | −4.1 ± 0.6 | 118 ± 9.3 |
| M182T/G238S | 53.8 ± 0.2 | −0.7 ± 0.3 | |
| WT | 46.8 ± 0.1 | −4.5 ± 0.4 | 93.8 ± 5.5 |
| R164S | 45.9 ± 0.2 | −3.3 ± 0.4 | 48.8 ± 1.1 |
| G238S | 44.8 ± 0.2 | −1.9 ± 0.3 | |
| D179G | 46.8 ± 0.3 | 2.9 ± 0.4 | 25.9 ± 1.5 |
Change in melting temperature of TEM-1 enzymes in complex with moxalactam is relative to respective apo-enzymes. EC50 is the concentration of the TEM-1 mutant or mutant–inhibitor complex at which 50% of fluorescent L2gd is displaced from the colloid.
To investigate whether the stability effects observed in enzyme-aggregate binding translated to enzyme inhibition, we turned to enzyme kinetics assays. We determined IC50 values for the Sor/CR colloids against the native enzyme and all TEM-1 point mutants, excluding D179G. The D179G mutation causes the enzyme to lose kinetic activity against penicillins, including the widely used substrate CENTA, which we use in our activity assays.20 The IC50 values reflect the ability of Sor/CR colloids to inhibit enzyme-catalyzed hydrolysis of a β-lactam substrate. The Sor/CR colloids inhibited the relatively stable WT and M182T mutants with IC50 values of 629 and 710 μM, respectively (Figure 3A and Table 3). Against their less-stable counterparts R164S and G238S, IC50 values dropped to 141 and 18.8 μM. G238S, with a ΔTm of −10.7 °C and ΔΔG of −4.6 kcal/mol compared to M182T, had a 38-fold more potent IC50. Likewise, R164S, with a ΔTm of −2.1 °C and ΔΔG of −0.73 kcal/mol versus WT, was inhibited by Sor/CR 4.5-fold more potently (Figure 3A and Table 3). Once again, M182T/G238S, one of the more stable enzymes in the mutant panel, had the second-lowest IC50 at 21.4 μM, just after G238S, in alignment with its high binding affinity to the Sor/CR colloids. We also determined the ability of three other well-characterized colloid-forming compounds (Fulvestrant, 3′,3″,5′,5″-tetraiodophenolphthalein (TIPT), and Miconazole) to inhibit TEM-1 and its mutants. Like Sor/CR, all three aggregators inhibited the G238S mutant most potently, followed by M182T/G238S, R164S, WT, and M182T (Figure 3B-D and Table 3). Thus, the stability trends seem to hold irrespective of the aggregating molecule.
Figure 3.
TEM-1 inhibition dose–response curves against (A) Sor/CR, (B) Fulvestrant, (C) TIPT, and (D) Miconazole colloids (see Table 3 for IC50 values).
Table 3.
Half Maximal Inhibitory Concentrations (IC50) of Sor/CR, Fulvestrant, TIPT, and Miconazole Colloids Against TEM-1 Stability Mutants
| mutant | Sor/CR (μM) | Fulvestrant (μM) | TIPT (μM) | Miconazole (μM) |
|---|---|---|---|---|
| M182T | 710 ± 37 | 78.4 ± 14.3 | 4.7 ± 0.2 | 130 ± 9.3 |
| M182T/G238S | 21.4 ± 0.3 | 17.6 ± 1.9 | 0.92 ± 0.1 | 58.2 ± 17.6 |
| WT | 629 ± 29 | 55.9 ± 12.3 | 3.4 ± 0.3 | 125 ± 7.9 |
| R164S | 141 ± 13 | 35.8 ± 2.5 | 1.7 ± 0.2 | 95.3 ± 6.9 |
| G238S | 18.8 ± 1.6 | 13.1 ± 0.7 | 0.88 ± 0.1 | 52.8 ± 7.9 |
| D179G | ||||
■ DISCUSSION AND CONCLUSIONS
The key observation from this study is that destabilized enzymes bind colloidal aggregates with higher affinities compared to their stable counterparts, all other things being equal. Among the six variants studied here, the most destabilized, D179G, binds the colloids most tightly, with an EC50 of 6.0 μM, whereas the WT and M182T proteins, which have temperatures of melting that are 7.4–13.5° higher than D179G, exhibit EC50 values that are 96- to 33-fold worse, respectively. Admittedly, the increase is not fully monotonic, with the more stable M182T mutant binding slightly better than WT, and M182T/G238S binding far better to the colloids than its stability would suggest. Still, the EC50 values of the intermediate stability mutants G238S and R164S do follow stability rank order. Moreover, the effects of moxalactam on stability and binding are consistent with the overall trend: this covalent inhibitor stabilizes D179G by 2.9° and its EC50 for aggregate binding worsens by 4.3 fold. Conversely, moxalactam destabilizes WT and R164S enzymes, improving their apparent EC50 values. Crucially, these stability effects are not only observed in direct binding experiments, which measure the displacement of the fluorescently labeled L2gd protein from the surface of the colloidal particles by the β-lactamase mutants, but also in the functional assay, which measures the susceptibility of these β-lactamase mutants to inhibition by the same colloids. This remains true for three additional aggregators tested against TEM-1 and its mutants.
In examining the correlations between enzyme stability and affinity to small-molecule aggregates, we expected to observe one of two things: a monotonic relationship between enzyme stability and susceptibility to inhibition by small-molecule colloids or no relation at all. The idea that aggregate binding would be sensitive to protein stability is supported by previous experiments, namely, hydrogen–deuterium exchange mass spectrometry and protease sensitivity assays, which suggested that enzymes are locally unfolded upon colloid binding.6 This idea is further supported by fluorescence binding experiments that showed that different enzymes bind more or less tightly to the same colloidal particles, at least to the long-lived and stable co-formulated ones investigated here.5 Conversely, it remains true that protein concentration plays an important role in nonspecific inhibition by colloidal aggregates. Because these particles are present at mid-femtomolar to low picomolar concentrations and bind their protein targets tightly, they act as stoichiometric inhibitors for which increased protein concentration linearly decreases measured inhibition.28,29 Thus, both mechanisms are likely at play in protein binding to colloidal aggregates. By studying a point mutant stability series of a single enzyme, the two may be deconvoluted, revealing a role for protein stability in protein–aggregate association for the first time.
It is important to note that stability did not correlate quantitatively or even strictly monotonically with colloid binding. The most egregious example was the M182T/G238S mutant enzyme, which had a far lower (better) EC50 value than expected given its high stability. This may be partly explained by the relatively low van’t Hoff enthalpy of M182T/G238S (126.5 kcal/mol, Table S1). With its increased stability relative to WT, one would typically expect this double mutant to have a van’t Hoff enthalpy higher than that of WT (139.5 kcal/mol, Table S1), part way to that of the further stabilized M182T itself (160.3 kcal/mol, Table S1). The lower van’t Hoff enthalpy of M182T/G238S reflects a broader unfolding transition for this mutant, beginning to unfold at a lower temperature than would be expected. This potentially makes it more sensitive to colloid binding, an event likely driven by preferential sequestration of hydrophobic residues exposed upon unfolding. Still, this can only be a partial explanation, and it remains true that although there is a good rank order correlation between stability and colloid binding, it is an imperfect one.
Notwithstanding this caveat, the key results from this study remain clear: the less stable a protein is, the more likely it is to bind to and be inhibited by a colloidal aggregate. Although protein concentration will always play a role, along with variations in other physical properties among entirely different proteins, stability will also determine colloid binding. This shapes our understanding of apparent selectivity effects in early screening for ligand discovery and of protein–nanoparticle design (e.g., antibody–colloid conjugates12). In screening, it is sometimes thought that if a molecule is selective for one target over another, then it is unlikely to be behaving as a colloidal aggregator. This study suggests caution in drawing this conclusion: selectivity must control for variations in protein concentration, buffer conditions, and protein stabilities in the two assays. Pragmatically, this will be difficult, and direct interrogation of colloidal aggregation, for instance through dynamic light scattering or detergent-dependent inhibition,30 remains the best way to check for this effect. For nanoparticle design, this study suggests, nonintuitively, that less-stable proteins are better suited for loading onto colloids as targeting agents since they exhibit higher affinities for these drug-rich aggregates and can readily fold into their functional forms upon colloid disruption.22 This has implications for the design and use of drug-rich colloidal aggregates as delivery vehicles.12,31
■ EXPERIMENTAL SECTION
Cloning and Site-Directed Mutagenesis.
A TEM-1 WT gBlock (Integrated DNA Technologies) was cloned into the pET24a (+) vector DNA (Millipore Sigma, 69749) via Gibson assembly.32 Mutagenesis was performed on the WT vector using a modified version of Gibson cloning to include site-directed mutagenesis.32,33 For each mutant, a set of primers (MutF and MutR) were designed within the TEM-1 gene containing the point mutation and another set (PairF and PairR) opposite the target gene. Two polymerase chain reaction (PCR) reactions were carried out, using a combination of the MutF and PairR or MutR and PairF primer pairs. Equal molar ratios of the purified PCR products were used in a standard Gibson assembly reaction (New England Biolabs, E5510S). Mutations were confirmed by sequencing of the gene.
Periplasmic Expression and Purification of TEM-1 via Osmotic Shock.
Mutant genes were transformed into BL21(DE3) chemically competent cells (Thermo Fisher, C600003) and grown in the 2XYT medium. TEM-1 was expressed and purified with a modified procedure from Wang et al.20 The cells were induced with 0.5 mM IPTG (Fisher Scientific, 42032210GM) and expressed for 22 h at 16 °C. After expression, cells were centrifuged at 8000 rpm and resuspended in 100 mL of 5 mM Tris/HCl, pH 8.0, 1 mM EDTA, 20% sucrose, followed by centrifugation at 16 000 rcf and resuspension in 25 mL of 5 mM MgCl2. The MgCl2 cell slurry was incubated on ice for 10 min and centrifuged at 16 000 rcf. The supernatants containing M182T, M182T/G238S, and WT proteins were filtered, concentrated, and purified via get filtration using a Superdex 75 10/300 column (GE Life Sciences, 17-5174-01) with 50 mM Tris/HCl, 500mM NaCl, pH 8.0 running buffer. Filtered supernatants of destabilized mutants R164S, G238S, and D179G were dialyzed into 50 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid, 50 mM NaCl, pH 8.0 and purified via anion exchange using a Hi-Trap Q-Sepharose Fast Flow column (GE Life Sciences, 17515601), followed by gel filtration. All six TEM-1 variants were dialyzed into 50 mM potassium phosphate buffer (KPi), pH 7.0. All steps were carried out at 4 °C, and the homogeneity of TEM-1 mutants was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Differential Scanning Fluorimetry (DSF).
TEM-1 mutants and mutant–moxalactam complexes were incubated with SYPRO Orange dye (Thermo Fisher, S6650) in a 384-well PCR microplate (VWR, 10011-194), with a final volume of 15 μL/well, 2 μM TEM-1, and 2.5× dye in 50 mM KPi, pH 7.0.34 The temperature was ramped from 30 to 95 °C at a rate of 1 °C/min and fluorescence of the dye monitored by qPCR machine. Melting temperatures were determined by the Life Cycler Thermal Shift Analysis software. To initiate mutant–inhibitor complexes, TEM-1 variants were incubated with 100 times molar excess of moxalactam sodium salt (Sigma-Aldrich, M8158-1G) for 1 h at room temperature prior to DSF.
Fluorescence Spectroscopy.
Competitive binding of TEM-1 mutants to Sor/CR colloids was determined by the displacement of 5-MF-L2gd.5 Sor/CR colloids were formulated as described by McLaughlin et al. at a 25:1 ratio in 50 mM KPi, pH 7.0.22 The colloid (160 μM) was mixed with 800 nM L2gd and TEM-1 mutants varying in concentration from 0 to 63.5 μM, rendering final concentrations of 20 μM, 100 nM, and 0–47.5 μM. For mutant–inhibitor complex binding experiments, mutants were incubated with 100 times molar excess of moxalactam for 1 h at room temperature. Fluorescence recovery measurements were taken with a SpectraMax M5 Microplate Reader, with wavelengths of excitation and emission set to 485 and 538 nm, respectively, with a 530 nm cutoff filter in 384-well black clear bottom plates (Sigma-Aldrich, CLS3540-10EA), 20 μL/well, at room temperature, by monitoring fluorescence recovery as a function of TEM-1 concentration. Data was analyzed, and EC50 values determined with the GraphPad Prism version 7.03 for Windows (GraphPad Software) by fitting to [Agonist] versus response-Variable slope while constraining the maximum fluorescence value to be the same for all TEM-1 variants.
Enzyme Inhibition.
TEM-1 enzyme activity assays were performed in methacrylate cuvettes (Fisher Scientific, 14955128) with CENTA, a chromogenic β-lactamase substrate (Millipore Sigma, 219475-25MG) with a final volume of 1 mL for all mutants except D179G.35,36 Incubation was performed for 20 nM TEM-1 and its mutants with varying concentrations of Sor/CR, Fulvestrant, TIPT, and Miconazole (MedChemExpress, HY-10201; Millipore Sigma, C6277; MedChemExpress, HY-13636; Spectrum Chemical, T0126; Santa Cruz Biotechnology, sc-204806) for 5 min at room temperature in 50 mM KPi, pH 7.0. The reaction was initiated by 138 μM CENTA, and the change in absorbance was monitored at 405 nm over 150 s by an HP8453a spectrophotometer in the kinetic mode using the UV–Vis Chemstation software (Agilent Technologies). IC50 values were determined by fitting to the log [inhibitor] versus normalized response-Variable slope using the GraphPad Prism version 7.03. Sorafenib, Congo red, Fulvestrant, TIPT, and Miconazole were tested by liquid chromatography–mass spectrometry or high performance liquid chromatography and have the following purities: 99.92%, 85% dye content, 99.99%, 95%, and 97%.
Supplementary Material
■ ACKNOWLEDGMENTS
This work was supported by NIH grant R35GM122481. We thank P. Lak for experimental assistance and P. Lak, I. Singh, and J. Pottel for reading this manuscript. We also thank M. Korczynska for thoughtful discussions on assay trouble-shooting and D. Agard for providing the labeled L2gd protein for the binding assays.
■ ABBREVIATIONS
- IC50
half maximal inhibitory concentration
- WT
wild-type
- DSF
differential scanning fluorimetry
- Tm
melting temperature
- Sor/CR
sorafenib/Congo red
- 5-MF-L2gd
5-maleimido-fluorescein-ribosomal protein L2 globular domain
- EC50
half maximal effective concentration
- TIPT
3′,3″,5′,5″-tetraiodophenolphthalein
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.9b01019.
TEM-1 melting temperatures determined by circular dichroism; differential scanning fluorimetry (PDF)
The authors declare no competing financial interest.
■ REFERENCES
- (1).McGovern SL; Caselli E; Grigorieff N; Shoichet BK A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem 2002, 45, 1712–1722. [DOI] [PubMed] [Google Scholar]
- (2).Seidler J; McGovern SL; Doman TN; Shoichet BK Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem 2003, 46, 4477–4486. [DOI] [PubMed] [Google Scholar]
- (3).Frenkel YV; Clark AD Jr.; Das K; Wang YH; Lewi PJ; Janssen PA; Arnold E Concentration and pH dependent aggregation of hydrophobic drug molecules and relevance to oral bioavailability. J. Med. Chem 2005, 48, 1974–1983. [DOI] [PubMed] [Google Scholar]
- (4).Boulton S; Selvaratnam R; Ahmed R; Van K; Cheng X; Melacini G Mechanisms of specific versus nonspecific interactions of aggregation-prone inhibitors and attenuators. J. Med. Chem 2019, 62, 5063–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Duan D; Torosyan H; Elnatan D; McLaughlin CK; Logie J; Shoichet MS; Agard DA; Shoichet BK Internal structure and preferential protein binding of colloidal aggregates. ACS Chem. Biol 2017, 12, 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Coan KE; Maltby DA; Burlingame AL; Shoichet BK Promiscuous aggregate-based inhibitors promote enzyme unfolding. J. Med. Chem 2009, 52, 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).McGovern SL; Helfand BT; Feng B; Shoichet BK A specific mechanism of nonspecific inhibition. J. Med. Chem 2003, 46, 4265–4272. [DOI] [PubMed] [Google Scholar]
- (8).Jadhav A; Ferreira RS; Klumpp C; Mott BT; Austin CP; Inglese J; Thomas CJ; Maloney DJ; Shoichet BK; Simeonov A Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J. Med. Chem 2010, 53, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Aldrich C; Bertozzi C; Georg GI; Kiessling L; Lindsley C; Liotta D; Merz KM Jr.; Schepartz A; Wang S The ecstasy and agony of assay interference compounds. J. Med. Chem 2017, 60, 2165–2168. [DOI] [PubMed] [Google Scholar]
- (10).Feng BY; Shelat A; Doman TN; Guy RK; Shoichet, B. K. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol 2005, 1, 146–148. [DOI] [PubMed] [Google Scholar]
- (11).Feng BY; Simeonov A; Jadhav A; Babaoglu K; Inglese J; Shoichet BK; Austin CP A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem 2007, 50, 2385–2390. [DOI] [PubMed] [Google Scholar]
- (12).Ganesh AN; McLaughlin CK; Duan D; Shoichet BK; Shoichet MS A new spin on antibody-drug conjugates: Trastuzumab-fulvestrant colloidal drug aggregates target her2-positive cells. ACS Appl. Mater. Interfaces 2017, 9, 12195–12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ganesh AN; Aman A; Logie J; Barthel BL; Cogan P; Al-Awar R; Koch TH; Shoichet BK; Shoichet MS Colloidal drug aggregate stability in high serum conditions and pharmacokinetic consequence. ACS Chem. Biol 2019, 14, 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Owen SC; Doak AK; Wassam P; Shoichet MS; Shoichet BK Colloidal aggregation affects the efficacy of anticancer drugs in cell culture. ACS Chem. Biol 2012, 7, 1429–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Raina SA; Zhang GGZ; Alonzo DE; Wu J; Zhu D; Catron ND; Gao Y; Taylor LS Enhancements and limits in drug membrane transport using supersaturated solutions of poorly water soluble drugs. J. Pharm. Sci 2014, 103, 2736–2748. [DOI] [PubMed] [Google Scholar]
- (16).Shamay Y; Shah J; Isik M; Mizrachi A; Leibold J; Tschaharganeh DF; Roxbury D; Budhathoki-Uprety J; Nawaly K; Sugarman JL; Baut E; Neiman MR; Dacek M; Ganesh KS; Johnson DC; Sridharan R; Chu KL; Rajasekhar VK; Lowe SW; Chodera JD; Heller DA Quantitative self-assembly prediction yields targeted nanomedicines. Nat. Mater 2018, 17, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sassano MF; Doak AK; Roth BL; Shoichet BK Colloidal aggregation causes inhibition of g protein-coupled receptors. J. Med. Chem 2013, 56, 2406–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Duan D; Doak AK; Nedyalkova L; Shoichet BK Colloidal aggregation and the in vitro activity of traditional chinese medicines. ACS Chem. Biol 2015, 10, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shoichet BK; Baase WA; Kuroki R; Matthews BW A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. U.S.A 1995, 92, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wang X; Minasov G; Shoichet BK Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol 2002, 320, 85–95. [DOI] [PubMed] [Google Scholar]
- (21).Beadle BM; Shoichet BK Structural bases of stability-function tradeoffs in enzymes. J. Mol. Biol 2002, 321, 285–296. [DOI] [PubMed] [Google Scholar]
- (22).McLaughlin CK; Duan D; Ganesh AN; Torosyan H; Shoichet BK; Shoichet MS Stable colloidal drug aggregates catch and release active enzymes. ACS Chem. Biol 2016, 11, 992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Beadle BM; McGovern SL; Patera A; Shoichet BK Functional analyses of ampc beta-lactamase through differential stability. Protein Sci. 1999, 8, 1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Trehan I; Beadle BM; Shoichet BK Inhibition of ampc beta-lactamase through a destabilizing interaction in the active site. Biochemistry 2001, 40, 7992–7999. [DOI] [PubMed] [Google Scholar]
- (25).Beadle BM; Nicholas RA; Shoichet BK Interaction energies between beta-lactam antibiotics and E. coli penicillin-binding protein 5 by reversible thermal denaturation. Protein Sci. 2001, 10, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wang X; Minasov G; Shoichet BK Noncovalent interaction energies in covalent complexes: Tem-1 beta-lactamase and beta-lactams. Proteins 2002, 47, 86–96. [PubMed] [Google Scholar]
- (27).Maveyraud L; Mourey L; Kotra L; Pedelacq J; Guillet V; Mobashery S; Samama J Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class a β-lactamases from the antibiotic-resistant bacteria. J. Am. Chem. Soc 1998, 120, 9748–9752. [Google Scholar]
- (28).Coan KE; Shoichet BK Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J. Am. Chem. Soc 2008, 130, 9606–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Giannetti AM; Koch BD; Browner MF Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J. Med. Chem 2008, 51, 574–580. [DOI] [PubMed] [Google Scholar]
- (30).Irwin JJ; Duan D; Torosyan H; Doak AK; Ziebart KT; Sterling T; Tumanian G; Shoichet BK An aggregation advisor for ligand discovery. J. Med. Chem 2015, 58, 7076–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ganesh AN; Donders EN; Shoichet BK; Shoichet MS Colloidal aggregation: From screening nuisance to formulation nuance. Nano Today 2018, 19, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gibson DG; Young L; Chuang RY; Venter JC; Hutchison CA III; Smith HO Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- (33).Kunkel TA Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A 1985, 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Huynh K; Partch CL Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci 2015, 79, 28.9.1–28.9.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bebrone C; Moali C; Mahy F; Rival S; Docquier JD; Rossolini GM; Fastrez J; Pratt RF; Frere JM; Galleni M Centa as a chromogenic substrate for studying beta-lactamases. Antimicrob. Agents Chemother 2001, 45, 1868–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Feng BY; Shoichet BK A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc 2006, 1, 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.