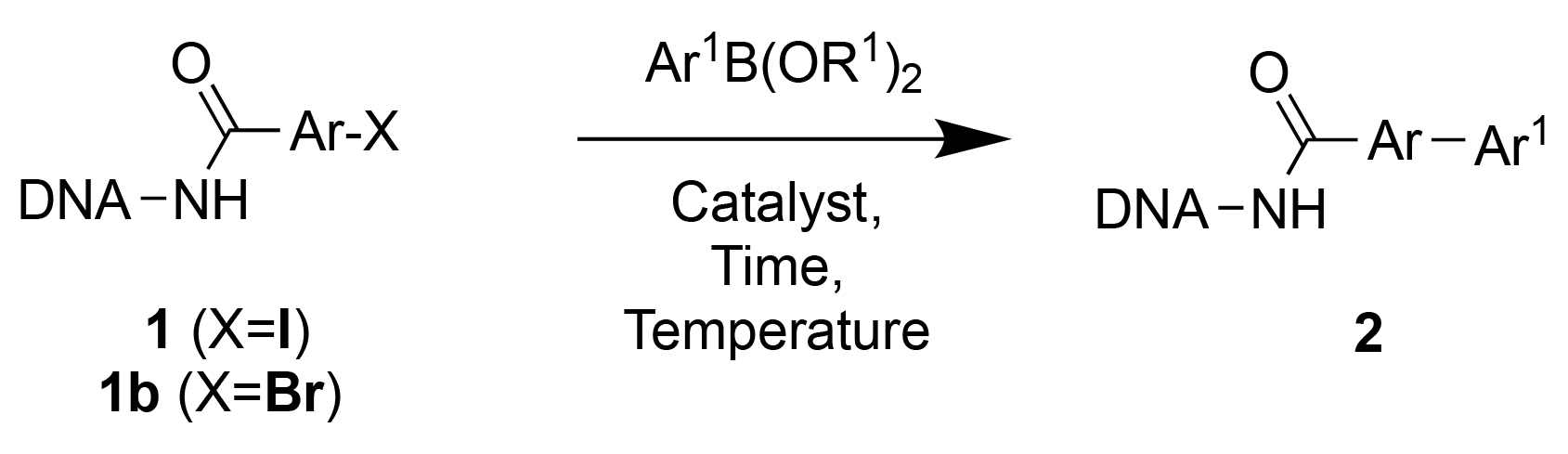
Abstract
The construction of DNA-encoded chemical libraries (DECLs) crucially relies on the availability of chemical reactions, which are DNA-compatible and which exhibit high conversion rates for a large number of diverse substrates. In this work, we present our optimization and validation procedures for three copper and palladium-catalyzed reactions (Suzuki cross-coupling, Sonogashira cross-coupling and copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC)), which have been successfully used by our group for the construction of large encoded libraries.
Keywords: DNA-encoded chemical libraries, Suzuki coupling, Sonogashira coupling, CuAAC reaction, DNA-compatible reactions
Introduction
The encoding of individual small organic molecules with distinctive DNA fragments, serving as amplifiable identification barcodes, allows the construction and screening of chemical libraries of unprecedented size.[1–11] Libraries containing billions of compounds from a relatively small number of building blocks (e.g., few thousand chemical compounds) can be constructed using a variety of different experimental approaches, such as pool-and-split methodologies,[12] DNA-templated chemical reactions with preformed oligonucleotide derivatives[13–15] or the use of encoded self-assembling strategies.[16–21] The attractiveness of encoding chemical compounds with DNA fragments relate to the possibility of constructing and storing large libraries as a mixture, that can be interrogated by affinity capture procedures on immobilized proteins of interest, followed by decoding, using high-throughput DNA sequencing.[12, 22, 23] Indeed, the identity and relative frequency of each compound in the library (e.g., before and after a screening experiment) can be directly retrieved from the results of a DNA sequencing experiment, since each library member is encoded by a distinctive DNA fragment.[12, 22, 23] The construction of large DNA-encoded chemical libraries crucially relies on the availability of “robust” reactions, which can be performed in water, are DNA-compatible[24–26] and which accept a large number of structurally diverse substrates. One of the most widely used reactions for the construction of DNA-encoded chemical libraries is the formation of amide bonds, since excellent procedures are available and since thousands of building blocks (e.g., amines and carboxylic acids) can be purchased from commercial sources at moderate costs. However, even a robust reaction such as amide bond formation with DMT-MM methodology displays acceptable conversion yields only for approximately 44% of carboxylic acids. For these reasons improved methodologies are constantly being explored.[27]
In this work, we aimed at implementing and optimizing copper and palladium-catalyzed reactions, which could be readily used for the construction of DNA-encoded chemical libraries. We focused on Suzuki cross-coupling, Sonogashira cross-coupling and copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC). There have been recent investigations on the implementation of these reactions on substrates coupled to double-stranded DNA fragments.[24, 28–34] However, it is often convenient to construct libraries using single-stranded DNA fragments, as these molecular assemblies can be expanded using encoding self-assembling chemistry (ESAC)[16–21] or used for innovative screening techniques (e.g., interaction-dependent PCR[35] or the hybridization with complementary oligonucleotide-photocrosslinker conjugates.[36, 37] In principle, the DNA-compatibility of a reaction could be different for single- and double-stranded DNA derivatives, due to the different shielding of the bases.
Results
Suzuki cross-coupling
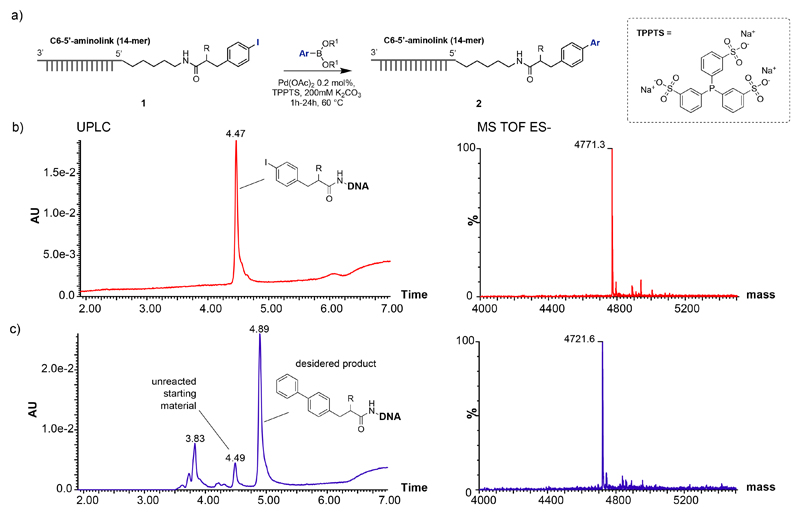
We first focused on Suzuki coupling procedures, since these transformations are broadly used in Medicinal Chemical research for the formation of C-C bonds. Figure 1 illustrates the reaction of a p-iodophenyl derivatives, coupled to an amino-tagged single-stranded oligonucleotide comprising 14 bases, with phenyl boronic acid. The purity and identity of the DNA conjugates is typically monitored both by HPLC and by LC/MS, thus allowing an experimental determination of the performance of the reaction. Throughout the paper, this methodology has been used for the calculation of conversion yields and for the characterization of optimal reaction conditions. As a first step, we explored conversion yields for the reaction of various boronic acids with iodophenyl or bromophenyl propionic acid derivatives on single-stranded DNA, using Pd(OAc)2 as catalyst [Table 1]. Overall, conversion yields were better for the iodophenyl derivative, which convinced us to focus on iodinated aromatic compounds for the rest of our investigations.
Figure 1.
a) Palladium-catalyzed Suzuki cross-coupling reactions between aryl halides (Iodide and bromide) - DNA conjugates and boronic acids or esters. b) UPLC chromatograms registered at 260 nM of starting material (1) and the deconvoluted TOF MS ES(-) spectrum of the corresponding peak (4.47). c) Chromatogram of crude Suzuki reaction between 1 and phenlyboronic acid and the deconvolution of TOF MS ES(-) spectrum of the product (4.89).
Table 1.
Suzuki cross-coupling performed on aryl iodide and aryl bromide DNA conjugates. The reactions were carried out at 60 °C for 3 hours using 0.4 eq. of Pd(OAc)2 - TPPTS complex as catalyst. The reported conversions were calculated by integrating UPLC chromatograms of crude reactions.
| Reactants vs. Substrate |
|
|
|---|---|---|
|
0% | 0% |
|
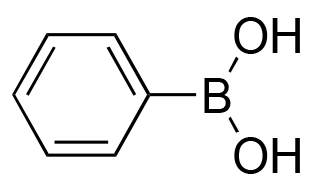
88% | 0% |
|
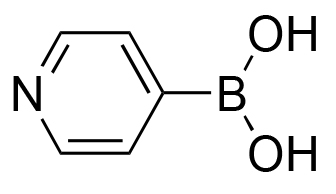
>98% | 60% |
|
>98% | 40% |
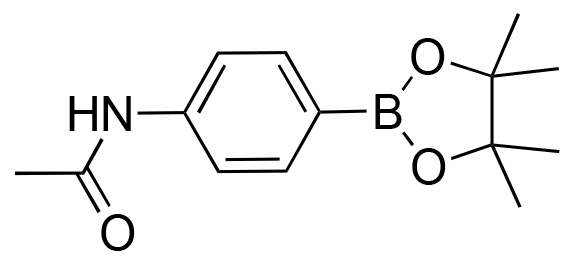
|
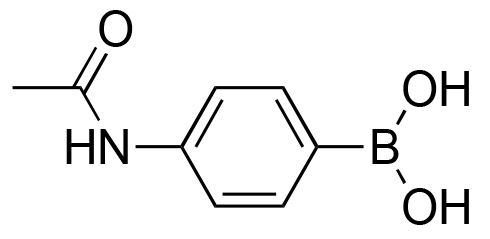
>98% | 40% |
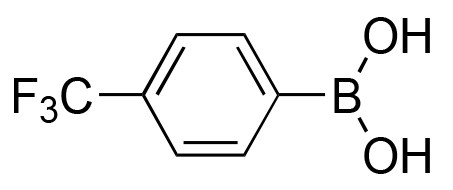
|
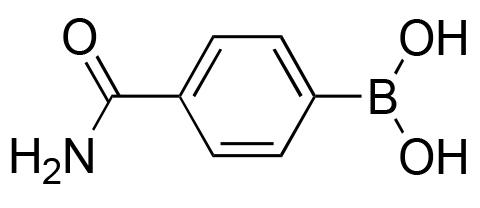
>98% | 50% |
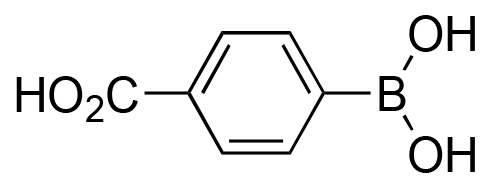
|
70% | 0% |
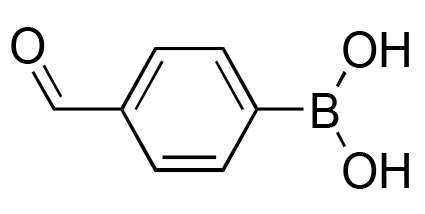
|
70% | 0% |
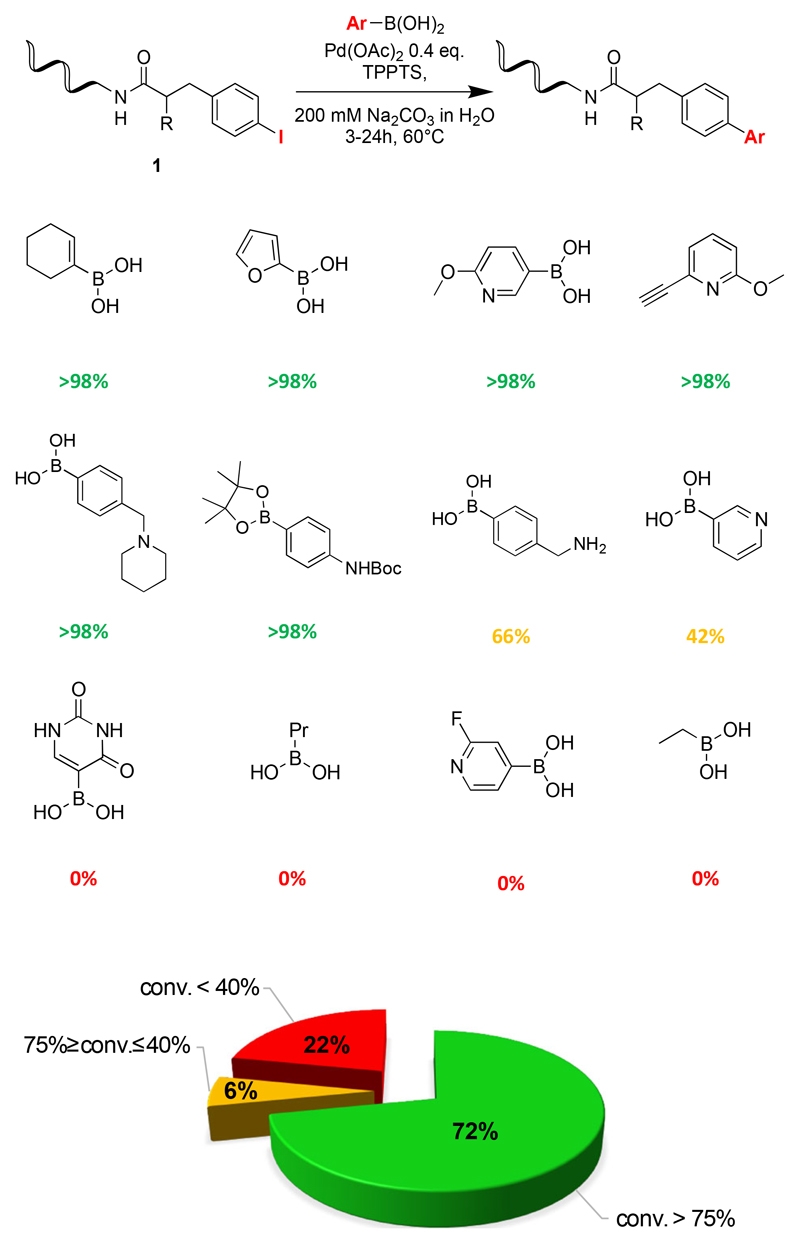
We then explored different sources and equivalents of palladium complexes, as well as temperature conditions [Table 2]. We obtained the best results using Pd(OAc)2, using at least 0.4 equivalents of palladium complexes. Reactions were often satisfactory with as little as 0.2 equivalents. The use of >0.4 equivalents did not result in further improvements. The optimal reaction condition was tested on 32 boronic acids [Figure 2, Supplementary Table 1]. In 72% of the cases, conversion yields were >75%, while 6% of the boronic acids exhibited conversion yields between 40 and 75%. Importantly, 22% of the substrates exhibited conversions lower than 40%. These boronic acids were typically sp3 boronates, acrylboronates, p-pyridinlboronic acid derivatives and compounds which are not soluble in DMA/water mixture. Indeed, methylester derivatives were deprotected yielding the corresponding carboxylic acids [Supplementary Table 1].
Table 2. Screening of different catalysts, substrates and reaction conditions for Suzuki “on-DNA” cross-couplings.
| ||||
|---|---|---|---|---|
|
| ||||
| Pd(II) Source |
Monitored after | Temp. | Aryl halides | Eq. of Pd [a] |
| Pd(Oac)2 | 10 mins | 50 °C | Ar-I | 0.2 |
| PdCl2 | 1 hour | 60 °C | Ar-Br | 0.4 |
| PdCl2(PPh3)2 | 2 hours | 65 °C | 1.0 | |
| [PdCl(allyl)]2 | 4 hours | 70 °C | 1.5 | |
| 24 hours | 2.0 | |||
| 4.0 | ||||
Equivalents with respect to DNA – aryl halide conjugate (1).
Figure 2.
Screening results of Suzuki cross coupling reactions between 32 boronic acids and a modified 14-mer oligonucleotide with a p-I-phenylalanine scaffold. Red, yellow and green colors correspond to reactions with poor, intermediate or good conversions.
In conclusion, we found that Suzuki reaction could reliably be implemented with iodophenyl DNA derivatives, using Pd(OAc)2 as catalyst (0.4 equivalents, 60°C, 3-24 hours) with aromatic, heteroaromatic and vinylboronic acids.
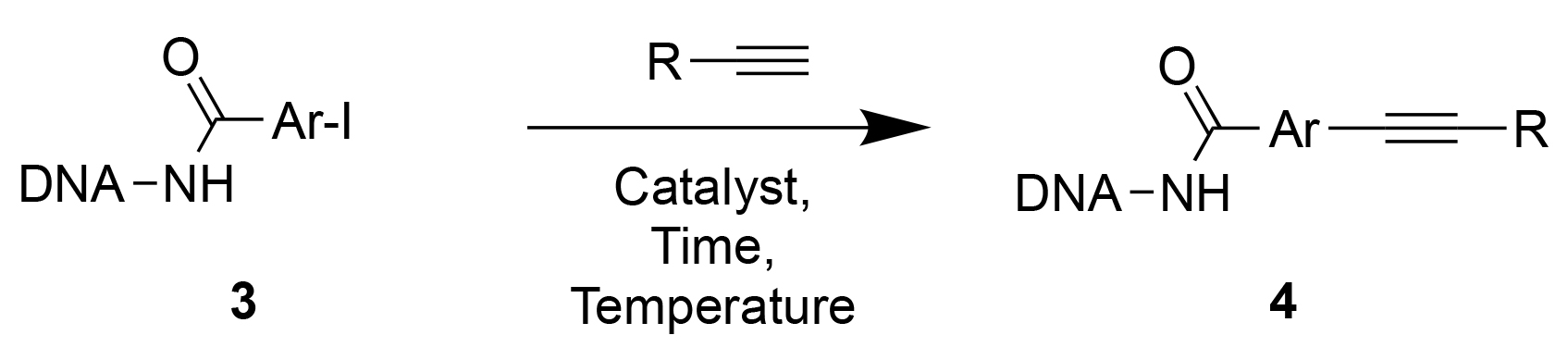
Sonogashira cross-coupling
In full analogy to the optimization of Suzuki reaction, we first studied the choice of catalyst (type and equivalents) and of temperature conditions [Table 3]. Various palladium and copper complexes were tested, alone and in combination. The best conversions were obtained using 1 equivalent of [PdCl(allyl)]2 at 70 °C for 3 hours.
Table 3. Screening of different catalysts, substrates and reaction conditions for Suzuki and Sonogashira “on-DNA” cross-couplings.
| ||||
|---|---|---|---|---|
|
| ||||
| Pd(II) Source |
Cu(II) Source |
Monitored after | Temp. | Eq. of Pd [a] |
| Pd(Oac)2 | Cu(Oac)2 | 10 mins | 50 °C | 0.2 |
| PdCl2 | Cu(SO4)2 | 1 hour | 60 °C | 0.5 |
| PdCl2(PPh3)2 | Cu(Otf)2 | 2 hours | 65 °C | 1.0 |
| [PdCl(allyl)]2 | Cu(Oac)2L | 4 hours | 70 °C | 1.5 |
| Copper-free | 24 hours | 2.0 | ||
| 4.0 | ||||
Equivalents with respect to DNA - aryl halide conjugate (3).
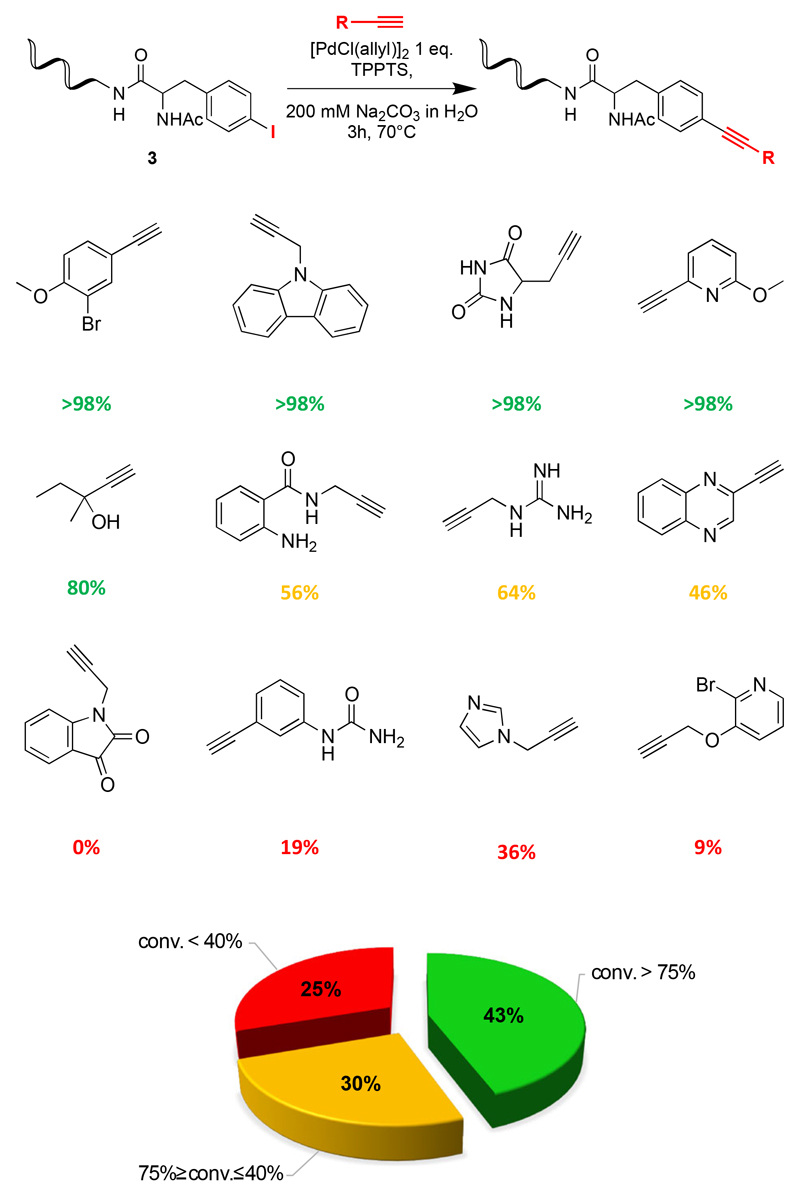
Figure 3 shows representative coupling results for some of the 44 alkynes that were tested. For 43% of the reagents, >75% conversion was achieved. A conversion between 40% and 75% was observed for 30% of the alkynes, whereas 25% of the compounds exhibited a conversion < 40%. “Difficult” alkynes included structures that were more prone to decomposition [Figure 3, Supplementary Table 2].
Figure 3.
Screening results of Sonogashira cross coupling reactions between 44 alkynes and a modified 14-mer oligonucleotide with a p-I-phenyl moiety. Red, yellow and green colors correspond to reactions with poor, intermediate or good conversions.
Copper-catalyzed azido-alkyne cycloaddition (CuAAC)
CuAAC is a frequently used reaction for the construction of DNA-encoded chemical libraries,[33, 38, 39] because of its robustness and of the broad availability of azides and terminal alkynes.
Previous activities of our laboratory featured the use of pseudo-solid phase "on-DNA CuAAC" chemistry. The reactions were performed using organic solvents (DMSO and tBuOH mixtures) and the conventional TBTA (1-(1-benzyltriazol-4-yl)-N,N-bis[(1-benzyltriazol-4-yl)methyl]methanamine) as copper (I) ligand.[40]
In order to perform reactions in aqueous solution, with a considerable saving in the amounts of reagents to be used, we synthesised a water-soluble pre-catalyst obtained by mixing Copper (II) acetate with a potassium salt of 8 (potassium 4,4',4''-(((nitrilotris(methylene))tris(1H-1,2,3-triazole-4,1-diyl))tris(methylene))tribenzoate). The complex was subsequently reduced in situ by sodium ascorbate obtaining the Cu(I) catalyst which promoted efficient conversions, with a broad scope of substrates [Table 4]. The importance of keeping Cu(I) in stable complexes has previously been recognized,[41] as Cu(I) ions may promote DNA cleavage.[41–43]
Table 4. A comparison between pseudo-solid phase CuAAC and the optimized water-compatible CuAAC conditions.
| Reaction Conditions |
Pseudo-solid phase CuAAC |
Solution-phase CuAAC |
|---|---|---|
| Pre-catalyst | CuSO4 + TBTA | Cu(OAc)2 + 8 |
| reducing agent | sodium ascorbate | sodium ascorbate |
| Eq. of Cu [a] | >10 | 0.1 |
| Solvent(s) | DMSO:tBuOH:H2O mixtures | H2O |
| Base | triethylamine | K2CO3 |
| temperature | r.t. | 35 °C |
| Eq. of Alkyne [a] | 400 | 40 |
| Reaction time | 24 hours | 3 hours |
Equivalents with respect to azido-DNA conjugate (1).
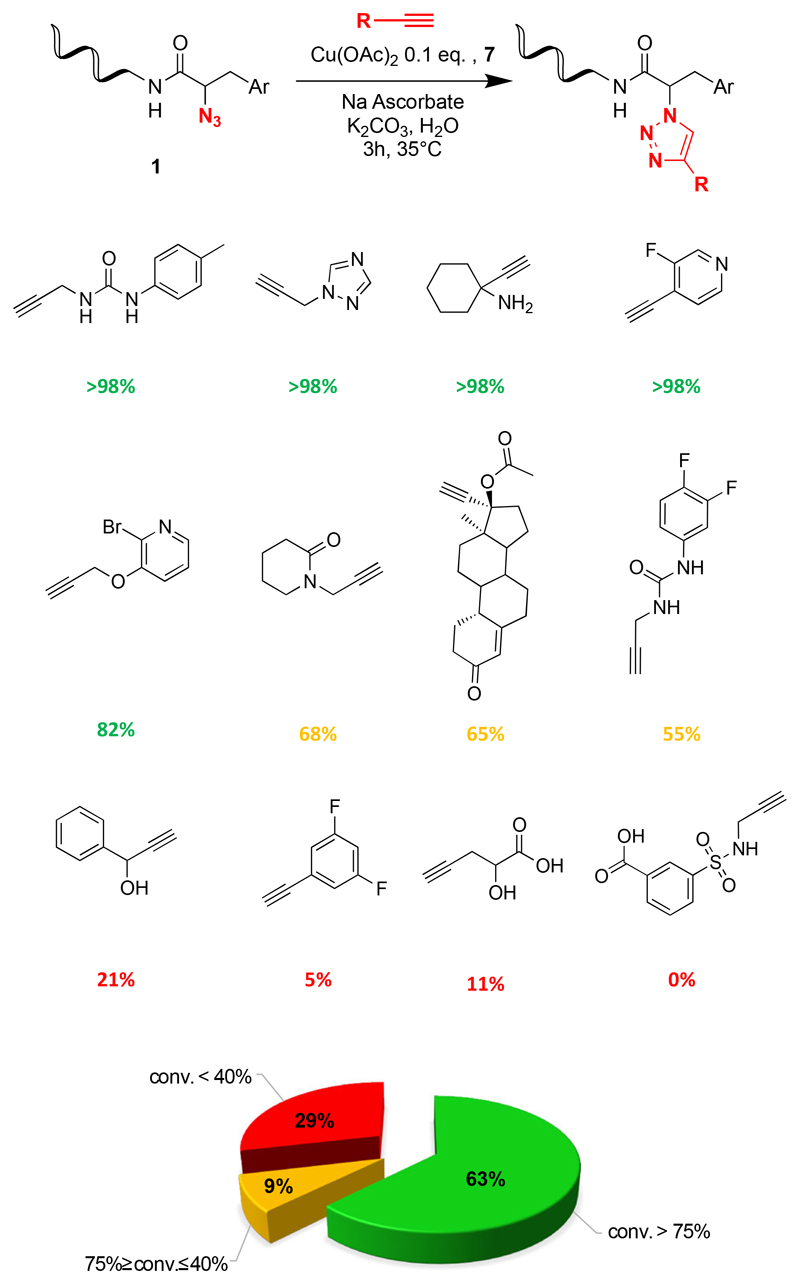
Figure 5 shows representative results for the reaction of an azide derivative on single-stranded DNA with 115 different terminal alkynes. For 65% of the reagents, a conversion >75% was observed. Only 9% of the alkynes exhibited a conversion between 40% and 75%, while 29% of the reagents showed a worse performance [Figure 4, Supplementary Table 3]. Conversions lower than 40% were often observed for hydrophobic compounds which do not readily dissolve in water and for structures (e.g., methyl esters or carboxylic acids), which may compromise the integrity of the catalyst.
Figure 5.
Screening results of CuAAC coupling reactions between 115 alkynes and a modified 14-mer oligonucleotide with a phenylalanine based-scaffold bearing an azido group. Red, yellow and green colors correspond to reactions with poor, intermediate or good conversions.
Figure 4.
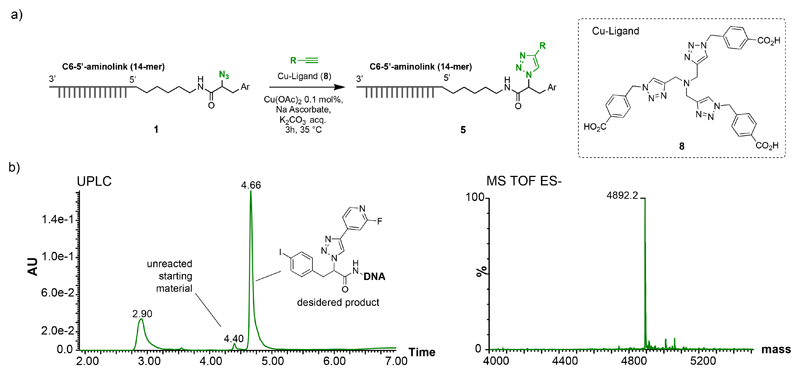
a) Copper-catalyzed azido-alkyne cycloaddition (CuAAC) reactions between an azido modified oligonucleotide and alkynes. b) UPLC Chromatogram of crude CuAAC reaction between 1 and 4-ethynyl-2-fluoropyridine and the deconvolution of TOF MS ES- spectra of the product (4.66). UPLC Chromatogram and MS spectra of starting material 1 is reported in figure 1b.
Discussion
Suzuki coupling, Sonogashira coupling and copper-catalyzed alkyne-azide cycloaddition reactions were extensively tested with ~200 substrates. We established conditions for all three reactions, which worked with the majority of the building blocks.
Similar to previous studies with amide bond forming reactions, it is extremely difficult to identify experimental procedures, which yield conversions >75% for all possible substrates. The construction of DNA-encoded chemical libraries can often afford to be performed using lower conversion rates (e.g., >40%), since bioactive compounds are readily identified in screening procedures also when products are not completely pure.[12, 15, 44–46] Nonetheless, scientists aim at constantly improve library purity, in order to have consistent correlations between enrichment factors in screening experiments and affinity measurements at the hit validation stage.[39, 47]
The pie charts presented in the article, describing the cumulative results of conversion rates for Suzuki coupling, Sonogashira coupling and copper-catalyzed alkyne-azide cycloaddition reactions, provide a strong rationale for the quantitative evalution of model reactions prior to library construction. Building blocks which show poor conversions may be replaced by other molecules, which give better yields. The data contained in Supplementary Tables 1-3 may serve as reference data base for other groups, who may wish to use the same experimental conditions for library construction.
It has often been assumed that double-stranded DNA may be more suitable for library construction, as the heteroduplex could potentially contribute to the integrity of the DNA bases. In our experience, however, single-stranded DNA structures preserve their integrity in the reactions described in this paper and in other reactions [data not shown]. It is particularly convenient to use single-stranded DNA for library construction, since n oligonucleotides are required for the encoding of n building blocks. The use of double-stranded DNA fragments typically requires the use of twice the number of oligonucleotides, as well as the need for a linker that bridges and stabilizes the heteroduplex.[48] Single-stranded DNA libraries are conveniently encoded using splint ligation procedures, with short oligonucleotides that can easily be removed at the end of the encoding step.[17, 38]
Single-stranded DNA-encoded chemical libraries containing millions of compounds can be easily synthesized.[33, 38, 39, 49] They can be converted into double-stranded DNA using Klenow polymerization procedures.[12, 38, 39, 45, 47] Alternatively, the pairing of two single-stranded DNA-encoded chemical libraries leads to ESAC libraries of very large size.[16, 17, 19–21] The total number of compound combinations in ESAC libraries corresponds to the product of the sizes of the individual libraries.
While efficient reaction conditions could be found for Suzuki coupling, Sonogashira coupling and copper-catalyzed alkyne-azide cycloadditions, the field of DNA-encoded chemistry will crucially rely on similar studies on other reactions, in order to expand the chemical diversity that can be used for library screens. While dozens of DNA-compatible reactions have been proposed,[24] the scope of those reactions with a sufficiently broad set of building blocks is often missing.
Experimental Section
General Experimental Information
All reagents and solvents used were of analytical grade. Buffers were prepared with ultrapure water (Merk Millipore Quantum® TEX). All chemicals were purchased from Acros, Alfa Aeaser, Sigma-Aldrich or TCI. The 5'-aminomodified-14mer oligonucleotide (5′-C6-amino-GGAGCTTCTGAATT) used for tests was purchased by LGC Biosearch Technologies. All boronic acid solutions were purchased from Apollo Scientific and alkynes solutions were purchased from Enamine. Purifications were carried out on a semi-preparative HPLC with XTerra® Shield RP18 (125 Å, 5 μm) using 100 mM triethylammonium acetate (TEAA pH=7.0) : acetonitrile (MeCN) as mobile phase (flow = 4.0 mL/min). Databases [Supplementary Table 1-3] were managed with Instantjchem 18.
Synthesis of 1, 1b (general procedure for "on-DNA" amidic coupling formation)
200 mM 2-azido-3-(4-iodophenyl)propanoic acid (for the synthesis of 1) and 2-azido-3-(4-ibromophenyl)propanoic acid (for the synthesis of 1b) were activated by adding 100 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) in DMSO (0.95 eq.) and 100 mM N-hydroxysuccinimide (S-NHS) in DMSO: water = 2:1 (1.5 eq.). The resulting mixtures were agitated for 20 minutes at 30 °C. The 5'-aminomodified oligonucleotide (5′-C6-amino-GGAGCTTCTGAATT) in 50 mM TEA-HCl buffer (pH=10) was subsequently added to 100 equivalents of the corresponding activated S-NHS ester. The reactions were heated at 37 °C for 12 hours and the products were precipitated with EtOH and purified by HPLC.
Synthesis of 2 (optimized procedure for "on-DNA" Suzuki cross-coupling)
The catalyst solution was prepared by mixing 20 μL of 10 mM palladium (II) acetate in dimethylacetamide (DMA), 100 μL of 100 mM Trisodium 3,3',3-phosphinetriyltribenzenesulfonate (TPPTS) in water and 480 μL of water, resulting in a 0.33 mM solution of Pd(0) - TPPTS complex. To a 100 mM solution of oligonucleotide 1 (2 nmol, 20 μL) in water were subsequently added 60 μL of 200 mM Na2CO3 in water, 2.5 μL of catalyst solution (0.8 nmol in Pd) and 10 μL of 200 mM ArB(OH)2 [Supplementary Table 1] in DMA. The resulting mixture was agitated overnight at 60 °C.
Synthesis of 3
The azido oligonucleotide conjugate 1 was treated with 30 mM tris(2-carboxyethyl)phosphine (TCEP) in 500 mM Tris×HCl buffer (pH=7.4) for 3 hours at 35 °C yielding the corresponding primary amine. The obtained product was acetylated by adding activated acetic acid S-NHS ester (procedure 1). The product was precipitated and purified by HPLC.
Synthesis of 4 (optimized procedure for "on-DNA" Sonogashira cross coupling)
The catalyst solution was prepared by mixing 100 μL of 10 mM allylpalladium (II) chloride dimer in DMA, 100 μL of 100 mM TPPTS in water and 800 μL of water, resulting in a 1 mM solution of Pd(0) - TPPTS complex. To a 100 mM solution of oligonucleotide 3 (2 nmol, 20 μL) in water were subsequently added 50 μL of 200 mM Na2CO3 in water, 2 μL of catalyst solution (2 nmol in Pd) and 5 μL of 100 mM alkyne [Supplementary Table 2] in DMSO. The resulting mixture was agitated for 3 hours at 70 °C.
Synthesis of 5 (optimized procedure for "on-DNA" CuAAC)
All solvents were degassed in argon atmosphere. The pre-catalyst solution was prepared by mixing 25 μL of 10 mM Cu(OAC)2, 100 μL of 10 mM solution of 8 in 200 mM K2CO3 and 2'325 μL of water, resulting in a 100 μM solution of Cu(II) - 8 complex. To a 100 mM solution of oligonucleotide 1 (10 nmol in 100 μL) in water were subsequently added 100 μL of 50 mM K2CO3, 10 μL of pre-catalyst solution (1 nmol) and 40 μL of 10 mM alkyne [Supplementary Table 3] in DMSO. The solution was mixed and the catalyst was activated by adding 10μL of 10 mM L-sodium-ascorbate in water. The reaction mixture was agitated at 35 °C for 3 hours.
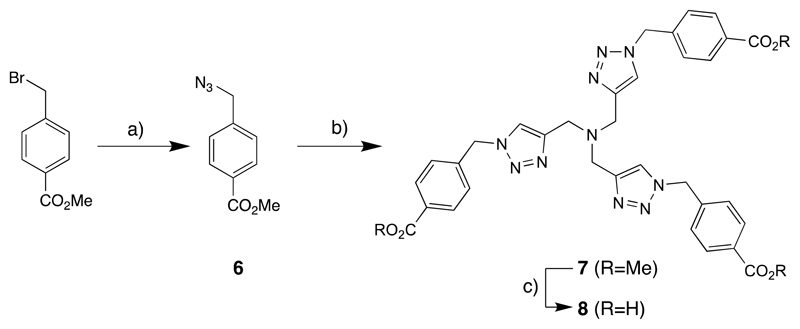
Synthesis of Copper(I) ligand (8)
Synthesis of 6
The commercially available methyl 4-(bromomethyl)benzoate (3.0 g, 13.1 mmol) was dissolved in DMF (20 mL) and then NaN3 (8.5 g, 130.8 mmol) and water (2 mL) were added. The resulting mixture was heated at 80 °C for 4 hours then concentrated under reduced pressure. The crude product was diluted with CH2Cl2 and washed with water. The organic phases were collected and concentrated. The pure product 6 was obtained by silica gel chromatography with hexane:EtOAc = 8:2 as an eluent with 96% of yield. 1H NMR (400 MHz, CDCl3) δ 8.09 – 7.99 (m, 2H), 7.40 – 7.34 (m, 2H), 4.39 (s, 2H), 3.90 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 166.59, 140.42, 130.10, 127.91, 119.99, 54.26, 52.18.
Synthesis of 7
All solvents were degassed in argon atmosphere. Compound 6 (2.4 g, 12.6 mmol) was dissolved in DMF (10 mL) and tripropargylamine (4.1 mmol, 590 μL), CuI (48 mg, 0.25 mmol) and 100 mg of tris(benzyltriazolylmethyl)amine (TBTA, 132 mg, 0.25 mmol) were added. The reaction was stirred at room temperature for 48 hours. The solvent was evaporated under reduce pressure and the pure product 7 was obtained by silica gel chromatography with CH2Cl2:MeOH = 9:1 as an eluent with >98% of yield. 1H NMR (400 MHz, DMSO-d6) δ 8.17 (s, 3H), 7.93 (d, J = 8.2 Hz, 5H), 7.37 (d, J = 8.3 Hz, 6H), 5.70 (s, 6H), 3.83 (s, 9H), 2.54 (s, 6H). 13C NMR (101 MHz, DMSO-d6) δ 166.27, 141.79, 130.05, 129.71, 128.35, 52.66, 40.91. m/z (TOF MS ES+): 623 [M-3N2], 651 [M-2N2], 679 [M-N2], 706 [M+H+], 707 [M+2H+], 727 [M+Na+]1410 [2M].
Synthesis of 8
The methyl ester 7 (1.0 g, 1.41 mmol) was dissolved in MeOH (10 mL) and 3M LiOH water solution (30 mmol, 10 mL) was added. The resulting suspension was stirred at 80 °C for 1 hour. The reaction was cooled and quenched with 3M HCl. The product was extracted with CH2Cl2 and purified by RP-HPLC with 0.1% trifluoroacetic acid (TFA) in water:acetonitrile as mobile phase and obtained with 50% of yield. 1H NMR (400 MHz, DMSO-d6) δ 8.13 (s, 3H), 7.94 – 7.87 (m, 6H), 7.38 – 7.29 (m, 6H), 5.67 (s, 6H), 3.64 (s, 6H). 13C NMR (101 MHz, DMSO-d6) δ 167.90, 144.21, 140.63, 132.68, 130.09, 127.98, 124.92, 52.80, 47.43. m/z (TOF MS ES-): 661.226 [M-H+], 662.220 [M], 1323.356 [2M-H+], 1324.356 [2M], 1986.485 [3M]. m/z (TOF MS ES+): 635.230 [M-N2]. 663.207 [M+H+], 664.236 [M+2H+], 665.238 [M+3H+], 1325.370 [2M+H+].
Supplementary Material
Scheme 1.
Synthesis of 8. a) NaN3 (10 eq.), DMF:H2O = 9:1, 80 °C, 4h. b) tripropargylamine, CuI (2%), TBTA (2%), DMF, TEA, r.t., 48h. c) 3M LiOH, H2O:MeOH = 1:1, 80 °C, 1h.
Acknowledgements
Financial support from the ETH Zürich, the Swiss National Science Foundation (Grants 310030B_163479), Sinergia (CRS112_160699) and from the ERC Advanced Grant “ZAUBERKUGEL” (Grant agreement 670603) is gratefully acknowledged.
Biographies
Dario Neri was born in Rome (Italy) in 1963, obtained a Master Degree in Chemistry from the Scuola Normale Superiore in Pisa (Italy) and performed a PhD in Chemistry at ETH Zürich (Switzerland), under the supervision of Prof. Dr. Kurt Wüthrich. After a 4-year post-doctoral research activity in Cambridge (UK) under the supervision of Sir Gregory Winter, Dario Neri joined the Faculty at ETH Zürich (first in the Department of Biology, later in the Department of Chemistry and Applied Biosciences) in 1996. He is currently full professor of Biomacromolecules at ETH Zürich. His research interests include the development of antibody therapeutics and the exploitation of DNA-encoded chemical libraries for disease-targeting applications. 
Nicholas Favalli was born in Verona (Italy) in 1990, obtained a Master Degree in Chemistry at Università degli Studi di Milano in Milan (Italy). He is currently doing a PhD in Chemistry and applied Biosciences at ETH Zürich (Switzerland), under the supervision of Prof. Dr. Dario Neri. 
Gabriele Bassi was born in Voghera (Italy) in 1992. He obtained his master degree in Chemistry at Università degli studi di Pavia (Italy). He is currently doing a PhD in chemistry and applied biosciences at ETH Zürich (Switzerland), under the supervision of Prof. Dr. Dario Neri. 
Tania Zanetti was born in Poschiavo (Switzerland) in 1993 and is a Master student in Pharmacy at ETH Zürich (Switzerland). This work represented part of her Master thesis. 
Jörg Scheuermann was born in Mannheim (Germany) and studied Chemistry at the Ruprecht-Karls-University Heidelberg (Germany) and ETH Zurich (Switzerland). Together with Prof. Dario Neri he developed DNA-Encoded Library technology, e.g., DNA-Encoded Self-Assembling Chemical (ESAC) libraries. He habilitated at ETH Zürich on DNA-Encoded Chemical Library Technology and currently works as Privatdozent at the Institute of Pharmaceutical Sciences at the ETH Zürich. 
References
- [1].Brenner S, Lerner RA. Encoded combinatorial chemistry. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Melkko S, Dumelin CE, Scheuermann J, Neri D. Lead discovery by DNA-encoded chemical libraries. Drug discovery today. 2007;12:465–471. doi: 10.1016/j.drudis.2007.04.007. [DOI] [PubMed] [Google Scholar]
- [3].Wrenn SJ, Harbury PB. Chemical evolution as a tool for molecular discovery. Annual review of biochemistry. 2007;76:331–349. doi: 10.1146/annurev.biochem.76.062205.122741. [DOI] [PubMed] [Google Scholar]
- [4].Clark MA. Selecting chemicals: the emerging utility of DNA-encoded libraries. Current opinion in chemical biology. 2010;14:396–403. doi: 10.1016/j.cbpa.2010.02.017. [DOI] [PubMed] [Google Scholar]
- [5].Kleiner RE, Dumelin CE, Liu DR. Small-molecule discovery from DNA-encoded chemical libraries. Chemical Society reviews. 2011;40:5707–5717. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sadhu KK, Winssinger N. Nucleic acid-tagged peptides: encoding libraries and controlling dimerization and conformation. Chimia. 2013;67:905–909. doi: 10.2533/chimia.2013.905. [DOI] [PubMed] [Google Scholar]
- [7].Chan AI, McGregor LM, Liu DR. Novel selection methods for DNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:55–61. doi: 10.1016/j.cbpa.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yuen LH, Franzini RM. Achievements, Challenges, and Opportunities in DNA-Encoded Library Research: An Academic Point of View. Chembiochem : a European journal of chemical biology. 2017;18:829–836. doi: 10.1002/cbic.201600567. [DOI] [PubMed] [Google Scholar]
- [9].Shi B, Zhou Y, Huang Y, Zhang J, Li X. Recent advances on the encoding and selection methods of DNA-encoded chemical library. Bioorganic & medicinal chemistry letters. 2017;27:361–369. doi: 10.1016/j.bmcl.2016.12.025. [DOI] [PubMed] [Google Scholar]
- [10].Favalli N, Bassi G, Scheuermann J, Neri D. DNA-encoded chemical libraries - achievements and remaining challenges. FEBS letters. 2018;592:2168–2180. doi: 10.1002/1873-3468.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Neri D, Lerner R. DNA-Encoded Chemical Libraries: A Selection System Based On Endowing Organic Compounds with Amplifiable Information. Annual Review of Biochemistry. 2018;87:24. doi: 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mannocci L, Zhang Y, Scheuermann J, Leimbacher M, De Bellis G, Rizzi E, Dumelin C, Melkko S, Neri D. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proceedings of the National Academy of Sciences. 2008;105:17670–17675. doi: 10.1073/pnas.0805130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-Templated Organic Synthesis and Selection of a Library of Macrocycles. Science. 2004;305:1601. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li X, Liu DR. DNA-templated organic synthesis: nature's strategy for controlling chemical reactivity applied to synthetic molecules. Angew Chem Int Ed Engl. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- [15].Blakskjaer P, Heitner T, Hansen NJ. Fidelity by design: Yoctoreactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery. Curr Opin Chem Biol. 2015;26:62–71. doi: 10.1016/j.cbpa.2015.02.003. [DOI] [PubMed] [Google Scholar]
- [16].Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded self-assembling chemical libraries. Nat Biotechnol. 2004;22:568–574. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- [17].Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J. Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat Chem. 2015;7:241–249. doi: 10.1038/nchem.2158. [DOI] [PubMed] [Google Scholar]
- [18].Winssinger N. Nucleic acid-programmed assemblies: translating instruction into function in chemical biology. Chimia (Aarau) 2013;67:340–348. doi: 10.2533/chimia.2013.340. [DOI] [PubMed] [Google Scholar]
- [19].Scheuermann J, Dumelin CE, Melkko S, Zhang Y, Mannocci L, Jaggi M, Sobek J, Neri D. DNA-Encoded Chemical Libraries for the Discovery of MMP-3 Inhibitors. Bioconjugate Chem. 2008;19:7. doi: 10.1021/bc7004347. [DOI] [PubMed] [Google Scholar]
- [20].Reddavide FV, Lin W, Lehnert S, Zhang Y. DNA-Encoded Dynamic Combinatorial Chemical Libraries. Angew Chem Int Ed Engl. 2015;54:7924–7928. doi: 10.1002/anie.201501775. [DOI] [PubMed] [Google Scholar]
- [21].Zhou Y, Li C, Peng J, Xie L, Meng L, Li Q, Zhang J, Li XD, Li X, Huang X, Li X. DNA-Encoded Dynamic Chemical Library and Its Applications in Ligand Discovery. J Am Chem Soc. 2018;140:15859–15867. doi: 10.1021/jacs.8b09277. [DOI] [PubMed] [Google Scholar]
- [22].Buller F, Steiner M, Scheuermann J, Mannocci L, Nissen I, Kohler M, Beisel C, Neri D. High-throughput sequencing for the identification of binding molecules from DNA-encoded chemical libraries. Bioorg Med Chem Lett. 2010;20:4188–4192. doi: 10.1016/j.bmcl.2010.05.053. [DOI] [PubMed] [Google Scholar]
- [23].Zhu Z, Cuozzo J. Review article: high-throughput affinity-based technologies for small-molecule drug discovery. J Biomol Screen. 2009;14:1157–1164. doi: 10.1177/1087057109350114. [DOI] [PubMed] [Google Scholar]
- [24].Satz AL, Cai J, Chen Y, Goodnow R, Gruber F, Kowalczyk A, Petersen A, Naderi-Oboodi G, Orzechowski L, Strebel Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjug Chem. 2015;26:1623–1632. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- [25].Malone ML, Paegel BM. What is a "DNA-Compatible" Reaction? ACS Comb Sci. 2016;18:182–187. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li H, Sun Z, Wu W, Wang X, Zhang M, Lu X, Zhong W, Dai D. Inverse-Electron-Demand Diels-Alder Reactions for the Synthesis of Pyridazines on DNA. Org Lett. 2018;20:7186–7191. doi: 10.1021/acs.orglett.8b03114. [DOI] [PubMed] [Google Scholar]
- [27].Li Y, Gabriele E, Samain F, Favalli N, Sladojevich F, Scheuermann J, Neri D. Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb Sci. 2016;18:438–443. doi: 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].de Pedro Beato E, Priego J, Gironda-Martinez A, Gonzalez F, Benavides J, Blas J, Martin-Ortega MD, Toledo MA, Ezquerra J, Torrado A. Mild and Efficient Palladium-mediated C-N Cross-Coupling Reaction between DNA-conjugated Aryl Bromides and Aromatic Amines. ACS Comb Sci. 2019 doi: 10.1021/acscombsci.8b00142. [DOI] [PubMed] [Google Scholar]
- [29].Li JY, Huang H. Development of DNA-Compatible Suzuki-Miyaura Reaction in Aqueous Media. Bioconjug Chem. 2018;29:3841–3846. doi: 10.1021/acs.bioconjchem.8b00676. [DOI] [PubMed] [Google Scholar]
- [30].El-Sagheer AH, Brown T. Click chemistry with DNA. Chem Soc Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- [31].Hervé G, Len C. Heck and Sonogashira couplings in aqueous media – application to unprotected nucleosides and nucleotides. Sustainable Chemical Processes. 2015:3. [Google Scholar]
- [32].Fan L, Davie CP. Zirconium(IV)-Catalyzed Ring Opening of on-DNA Epoxides in Water. chembiochem. 2017;18:4. doi: 10.1002/cbic.201600563. [DOI] [PubMed] [Google Scholar]
- [33].Litovchick A, Dumelin CE, Habeshian S, Gikunju D, Guie MA, Centrella P, Zhang Y, Sigel EA, Cuozzo JW, Keefe AD, Clark MA. Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors from a 334-Million Member Library. Sci Rep. 2015;5 doi: 10.1038/srep10916. 10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ding Y, Franklin GJ, DeLorey JL, Centrella PA, Mataruse S, Clark MA, Skinner SR, Belyanskaya S. Design and Synthesis of Biaryl DNA-Encoded Libraries. ACS Comb Sci. 2016;18:625–629. doi: 10.1021/acscombsci.6b00078. [DOI] [PubMed] [Google Scholar]
- [35].McGregor LM, Gorin DJ, Dumelin CE, Liu DR. Interaction-Dependent PCR: Identification of Ligand–Target Pairs from Libraries of Ligands and Libraries of Targets in a Single Solution-Phase Experiment. J Am Chem Soc. 2010;132:3. doi: 10.1021/ja107677q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li G, Liu Y, Liu Y, Chen L, Wu S, Liu Y, Li X. Photoaffinity labeling of small-molecule-binding proteins by DNA-templated chemistry. Angew Chem Int Ed Engl. 2013;52:9544–9549. doi: 10.1002/anie.201302161. [DOI] [PubMed] [Google Scholar]
- [37].Wang D-Y, Cao Y, Zheng L-Y, Chen L-D, Chen X-F, Hong Z-Y, Zhu Z-Y, Li X, Chai Y-F. Target Identification of Kinase Inhibitor Alisertib (MLN8237) by Using DNA-Programmed Affinity Labeling. Chemistry a European Journal. 2017;23:9. doi: 10.1002/chem.201702033. [DOI] [PubMed] [Google Scholar]
- [38].Li Y, De Luca R, Cazzamalli S, Pretto F, Bajic D, Scheuermann J, Neri D. Versatile protein recognition by the encoded display of multiple chemical elements on a constant macrocyclic scaffold. Nature Chemistry. 2018 doi: 10.1038/s41557-018-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Favalli N, Biendl S, Hartmann M, Piazzi J, Sladojevich F, Gräslund S, Brown PJ, Näreoja K, Schüler H, Scheuermann J, Franzini R, et al. A DNA-encoded library of chemical compounds based on common scaffolding structures reveals the impact of ligand geometry on protein recognition. ChemMedChem. 2018 Jul 6;13(13):1303–1307. doi: 10.1002/cmdc.201800193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as Copper(I)-Stabilizing Ligands in Catalysis. ORGANIC LETTERS. 2004;6:3. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- [41].Li DD, Tian JL, Gu W, Liu X, Yan SP. A novel 1,2,4-triazole-based copper(II) complex: synthesis, characterization, magnetic property and nuclease activity. J Inorg Biochem. 2010;104:171–179. doi: 10.1016/j.jinorgbio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- [42].Basile LA, Raphael AL, Barton JK. Metal-Activated Hydrolytic Cleavage of DNA. J Am Chem Soc. 1987;109:2. [Google Scholar]
- [43].J DCA, Douglas aKT. A common chemical mechanism used for DNA cleavage by copper(II) activated by thiols and ascorbate is distinct from that for copper(II): hydrogen peroxide cleavage. Transition Met Chem. 1996;21:3. [Google Scholar]
- [44].Decurtins W, Wichert M, Franzini RM, Buller F, Stravs MA, Zhang Y, Neri D, Scheuermann J. Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat Protoc. 2016;11:764–780. doi: 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Leimbacher M, Zhang Y, Mannocci L, Stravs M, Geppert T, Scheuermann J, Schneider G, Neri D. Discovery of small-molecule interleukin-2 inhibitors from a DNA-encoded chemical library. Chemistry. 2012;18:7729–7737. doi: 10.1002/chem.201200952. [DOI] [PubMed] [Google Scholar]
- [46].Zhao P, Chen Z, Li Y, Sun D, Gao Y, Huang Y, Li X. Selection of DNA-encoded small molecule libraries against unmodified and non-immobilized protein targets. Angew Chem Int Ed Engl. 2014;53:10056–10059. doi: 10.1002/anie.201404830. [DOI] [PubMed] [Google Scholar]
- [47].Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, Nauer A, Zimmermann M, Samain F, Scheuermann J, Brown PJ, Hall J, et al. Identification of structure-activity relationships from screening a structurally compact DNA-encoded chemical library. Angew Chem Int Ed Engl. 2015;54:3927–3931. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- [48].Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Belyanskaya SL, Ding Y, Callahan JF, Lazaar AL, Israel DI. Discovering Drugs with DNA-Encoded Library Technology: From Concept to Clinic with an Inhibitor of Soluble Epoxide Hydrolase. Chembiochem. 2017;18:837–842. doi: 10.1002/cbic.201700014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.