Abstract
The 26S proteasome is the essential compartmental protease in eukaryotic cells required for the ubiquitin-dependent clearance of damaged polypeptides and obsolete regulatory proteins. Recently, a combination of high-resolution structural, biochemical, and biophysical studies has provided critical new insights into the mechanisms of this fascinating molecular machine. A multitude of new cryo-electron microscopy structures provided snapshots of the proteasome during ATP-hydrolysis-driven substrate translocation, and detailed biochemical studies revealed the timing of individual degradation steps, elucidating the mechanisms for substrate selection and the commitment to degradation through conformational transitions. It was uncovered how ubiquitin removal from substrates is mechanically coupled to degradation, and cryo-electron tomography studies gave a glimpse of active proteasomes inside the cell, their sub-cellular localization, and interactions with protein aggregates. Here, we summarize these advances in our mechanistic understanding of the proteasome, with a particular focus on how its structural features and conformational transitions enable the multi-step degradation process.
Architecture and function of the 26S proteasome.
Targeted degradation of eukaryotic proteins occurs primarily through the ubiquitin-proteasome system (UPS), whose executor, the 26S proteasome, must process hundreds of protein substrates with vastly different chemical and structural properties in a highly selective manner. Damaged or no longer needed regulatory proteins are targeted for proteasomal degradation through the attachment of either several single ubiquitins [1] or poly-ubiquitin chains [2–4] by the E1/E2/E3 ubiquitination machinery [5]. The proteasome’s ability to balance promiscuity with selectivity is accomplished in part by its architecture. Central to the degradative function is the 20S core particle (CP), composed of seven different alpha and seven different beta subunits that are each present in two copies and form a compartmental, double-stacked barrel structure [6]. Three of the seven beta subunits contain proteolytic active sites that have trypsin-like, chymotrypsin-like, and caspase-like cleavage specificities, and are sequestered within the CP’s internal degradation chamber. Narrow axial pores restrict access to this chamber, with gates formed by the flexible N-termini of the CP’s alpha subunits and controlled by the 19S regulatory particle (RP) [7]. The RP caps one or both ends of the CP, and is responsible for the recognition of ubiquitinated substrates, their deubiquitination, and ATP-hydrolysis-dependent delivery into the CP for proteolysis. The RP can be further subdivided into the base and lid subcomplexes [8,9]. The 10-subunit base includes the ring-shaped AAA+ (ATPases Associated with various cellular Activities) ATPase motor of the proteasome, as well as three intrinsic ubiquitin receptors, Rpn1, Rpn10, and Rpn13. Detailed biochemical and structural studies have previously revealed how these receptors interact with ubiquitin in isolation, showing their general preference for Lys48-linked chains despite vastly distinct ubiquitin-binding motifs [9–12]. Interestingly, deletion of these three intrinsic receptors in yeast is not lethal [10], suggesting that additional receptors or ubiquitin-binding sites may exist on the proteasome. Rpn1, Rpn10, and Rpn13 occupy key positions on the RP near the central processing channel to position substrates for entering the AAA+ motor (Figure 1A). This ring-shaped motor consists of six distinct ATPase subunits, Rpt1 -Rpt6, that drive the mechanical unfolding and translocation of protein substrates. Once a ubiquitin modification is recognized by a receptor, an obligate unstructured region of the substrate [2,13,14] has to enter the central pore of the ATPase motor, where it is engaged through interactions with conserved pore loops of the six Rpts and mechanically pulled on for unfolding and translocation into CP (Figure 1D, E). Five of the Rpts contain conserved Hydrophobic-Tyrosine-X (HbYX) or related motifs at their C-terminus to dock into hydrophobic pockets of the CP’s alpha subunits and trigger opening of the access gate for substrate passage [15] (Figure 1G).
Figure 1:
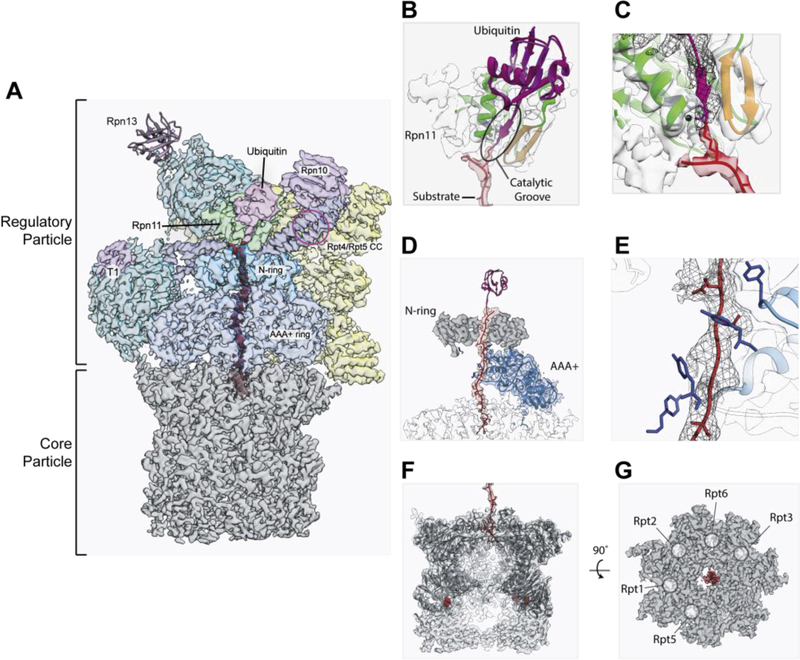
High-resolution substrate-bound structures of the 26S proteasome reveal mechanistic and regulatory details of substrate processing. (A) Architecture of the substrate-bound proteasome (EMD: 9045), displaying the core particle in gray and regulatory particle in multi-color, with substrate in red and ubiquitin in magenta. Ubiquitin receptors Rpn13 (PDB: 6FVW), Rpn10, and the T1 site on Rpn1, as well as a potential binding site on the Rpt4/Rpt5 coiled-coil (CC) are presented in purple. The hexameric AAA+ ATPase motor consists of three N-terminal coiled-coils (navy blue) and a ring formed by the six N-domains (N-ring, sky blue) atop the ring of AAA+ domains (cornflower blue). Rpn1 and Rpn2 are presented in dark cyan, and the Rpn11 deubiquitinase in green. Lid subunits are shown in yellow. (B) X-ray structure of Rpn11-bound ubiquitin (PDB: 5U4P; Rpn11 in green, Ins-1 loop in orange, active-site residues and catalytic zinc in dark gray, ubiquitin in magenta) docked into the cryo-EM density of substrate-bound proteasome (EMD: 9045; regulatory particle in gray, substrate in red) and overlaid with the atomic model for the substrate-bound proteasome (PDB: 6EF3; ubiquitin in violet red, substrate in red). The catalytic groove formed by active Rpn11 is highlighted. (C) Zoomed-in representation of (B), omitting the ubiquitin model from the crystal structure and including density for ubiquitin from the cryo-EM reconstruction (EMD: 9045), displayed in dark gray mesh. (D) Cut-away view of the cryo-EM density (EMD: 9045) and atomic model (PDB: 6EF3) for the AAA+-motor engaged substrate, with substrate in red, ubiquitin in violet, Rpt1 in blue, and the pore-loop tyrosine residues of all six Rpts shown in dark blue. The substrate traverses through the N-domain ring (dark gray), makes contact with five of the six Rpts (Rpt1 density and atomic mode shown in cornflower blue), and reaches into the core particle (light gray). (E) Zoomed-in representation of (D) with substrate density (EMD: 9045) shown dark gray mesh and four of the five engaged pore loops in dark blue. (F) Cut-away density (EMD: 9045; PDB: 6EF3) of the substrate (red) entering through the gate of the core particle (gray). The atomic model (PDB: 6EF3) for the core particle is displayed in dark gray, with the proteolytic active site residues highlighted as red spheres and the atomic model for substrate shown in red. (G) Representation of core-particle density (EMD: 9045) from the top-down, with the gate shown fully open and occupied with substrate (red; PDB: 6EF3). The docking sites for Rpts’ C-terminal HbYX motifs are circled and labeled with the respective Rpt binding partner.
The 9-subunit lid subcomplex of the RP is bound to one side of the base and makes additional contacts with alpha subunits of the CP (Figure 1A). It includes the essential deubiquitinase (DUB) Rpn11 (or Poh1 in humans) that is positioned near the entrance to the central pore of the AAA+ motor and responsible for the en-bloc removal of ubiquitin modifications from substrates prior to their translocation into the CP [16,17]. The proteasome can harbor additional DUBs, such as Uch37 and Ubp6/Usp14, whose activity is strongly dependent on proteasome association to edit a substrate’s ubiquitin signal [18–20]. Furthermore, Ubp6 modulates the DUB activity of Rpn11 [21] and allosterically influences the conformational state of the proteasome [22–24].
Previous electron microscopy (EM) studies indicated that the RP undergoes major conformational changes upon substrate engagement, leading to a coaxial alignment of the base and the CP, the formation of a more planar ATPase ring and a continuous channel for substrate processing, and the positioning of Rpn11 directly above the central pore [25]. Several additional sub-states had been identified and proposed to represent alternative substrate-free conformations (often termed s1 and s2) and substrate-processing conformations (s3, s4, and s6) [25–28], providing a basic structural framework for understanding the mechanisms of the proteasome. However, the functional roles of these snapshots and how they relate to each other within RP’s conformational landscape or in terms of the degradation process remained elusive. New high-resolution cryogenic electron microscopy (cryo-EM) and crystal structures, along with complex biochemical and biophysical analyses, have now illuminated how the proteasome components are functionally orchestrated to catalyze ubiquitin-dependent substrate degradation, and how the RP transitions through distinct conformational states for substrate recognition, engagement, and processing. In addition, advances in cryogenic electron tomography (cryo-ET) have captured spatial information about the localization of 26S proteasomes within live cells. Here we summarize the significance of these findings and outline our current understanding of how the conformational dynamics of the RP allow the proteasome to select and degrade protein substrates.
Degradation-coupled deubiquitination by Rpn11.
Recent biochemical and structural data revealed the detailed mechanism of ubiquitin removal from translocating substrates by the deubiquitinase Rpn11 as an integral step of ubiquitin-dependent degradation. Rpn11 is a member of the JAB1/MPN/Mov34 metalloenzyme (JAMM) DUB family and utilizes a catalytic Zinc (II) ion to hydrolyze the isopeptide bond between the ε-amino group of a modified lysine side chain and the C-terminus of ubiquitin. In isolation, Rpn11 is highly promiscuous and able to cleave between ubiquitin moieties linked through any of ubiquitin’s seven lysines, albeit with relatively low rates and affinities [29]. To prevent this promiscuous DUB activity before full proteasome assembly, Rpn11 in the isolated lid subcomplex is inhibited through steric occlusion and coordination of its catalytic Zinc by the neighboring lid subunit Rpn5 [30]. Upon lid incorporation into the 26S holoenzyme, this inhibition is released, and Rpn11 is positioned in close proximity to the AAA+ motor, which sterically restricts the space on the proximal side of the isopeptide bond, prevents cleavage between folded ubiquitin moieties, and favors the en-bloc removal of ubiquitin from unstructured substrate polypeptides at the entrance of the ATPase motor [29,31].
Premature deubiquitination prior to substrate engagement with the AAA+ motor would lead to substrate escape from the proteasome. The appropriate timing of ubiquitin removal only after a substrate polypeptide is grabbed by the Rpt pore loops and committed to degradation is therefore critical for efficient turnover. An important prerequisite for this coupling of degradation and deubiquitination is the conformation of the Insert-1 region in Rpn11, which adopts an occluding loop structure over the catalytic active site [29,32] until ubiquitin binding induces the transition to a beta hairpin that forms a 3-stranded beta sheet with ubiquitin’s flexible C-terminus and stabilizes it in Rpn11’s active site for cleavage (highlighted in orange in Figure 1B,C) [33–35]. Importantly, this loop-to-hairpin transition is rate-limiting for Rpn11 cleavage and accelerated by almost an order of magnitude when the AAA+ motor pulls an ubiquitinated substrate into the Rpn11 active site [33,36]. Mechanical substrate translocation thus has a dual effect on deubiquitination: it decreases KM through the vectorial delivery of ubiquitin to the Rpn11 active site and increases kcat by accelerating the Insert-1 conformational switch from an inhibitory loop to a ubiquitin-stabilizing beta hairpin. This mechano-chemical coupling is enabled by the perfect alignment of Rpn11’s catalytic groove with the trajectory of the translocating polypeptide through the AAA+ ATPase motor (Figure 1A, B, C) [34,35]. Fast ubiquitin removal is therefore limited to motor-engaged, translocating substrates that are committed to degradation. Deubiquitination of these substrates is consequently not rate-limiting, even when multiple ubiquitin modifications are present [36]. Consistently, a mutation that disfavors the inhibitory loop in Rpn11 leads to accelerated, premature deubiquitination of un-engaged substrates and their escape from degradation [33], which highlights the importance of degradation-coupled deubiquitination for efficient protein turnover. On the other hand, inhibiting Rpn11-mediated deubiquitination, for instance by targeting its catalytic Zinc ion, stalls substrates on the proteasome and is deleterious to degradation. Rpn11 has therefore been identified as a potent target for small-molecule inhibitors to treat cancers refractory to CP-proteolysis inhibitors, like bortezomib [37–40].
Cryo-EM reveals the proteasome conformational landscape during substrate processing.
Several critical cryo-EM studies were instrumental in defining RP’s subunit architecture [31,41] and various conformations of the 26S proteasome [25–28]. Together, they laid out a landscape of RP conformational states whose distribution can be influenced by numerous factors [42]. In its substrate-free ground state, the proteasome shows an equilibrium primarily between two conformations, s1 and s2 [26,27,43] (Figure 2B). Rpn11, the AAA+ motor, and CP are offset from each other in the s1 conformation, leading to a discontinuous processing channel, but a central pore that is openly accessible and therefore facilitates initial entry of a substrate polypeptide (Figure 2B). In contrast, the s2 conformation as well as the proposed substrate-processing states s3, s4, and s6 are characterized by a coaxial alignment of Rpn11, the AAA+ motor, and CP, which establishes a wider, continuous central channel whose entrance is occluded by Rpn11. These non-s1 states thus seem well suited for processive substrate translocation into CP and translocation-coupled deubiquitination by Rpn11, which acts as a gatekeeper at the motor entrance. Factors that shift the conformational equilibrium away from the s1 state include substrate engagement by the AAA+ motor [25,34,35], binding of free K48-linked ubiquitin chains [44], binding of non-or slowly-hydrolyzable ATP analogs to Rpt subunits [26–28,45–48], the inactivation of ATP hydrolysis in individual Rpts through Walker-B mutations [28], the disruption of lid-base contacts [43], binding of CP inhibitors [49], and interactions of the ATPase ring with Ubp6 in its ubiquitin-free or ubiquitin-bound state [22,23,26].
Figure 2:
Substrate degradation by the 26S proteasome depends on large conformational changes that can be observed in vitro and in situ. (A) Cartoon representation of the substrate-degradation pathway with time constants derived from a recent study [36]. A substrate (red) containing an unstructured initiation region and an ubiquitin modification (light purple) is recruited to the proteasome via intrinsic proteasome receptors (dark purple). The substrate’s unstructured region can passively diffuse into the central channel before being gripped by the pore loops of the AAA+ ATPase (blue). This substrate engagement drives a major conformational switch, committing a substrate to degradation via a kinetic gateway and placing Rpn11 into a coaxially aligned position with the AAA+ motor for co-translocational deubiquitination. Substrates are then unfolded and translocated into the core particle for proteolytic cleavage. Unfolding appears to be the rate-limiting step of degradation, with time constants depending on a substrate’s thermodynamic stability. (B) Top: EM densities of the 26S proteasome in the s1 (EMD: 3534, left) and s2 (EMD:3535, right) conformations, with Rpn11 (green) offset or coaxially aligned with the pore of the AAA+ motor (dark blue). Ubiquitin receptors are in purple, lid subunits in yellow, and Rpn1 and Rpn2 are in dark blue. Red and purple stars represent locations on the 26S proteasome where fluorescent dyes were incorporated to track conformational switching by FRET [36]. Bottom: cryo-EM densities of the 26S proteasome in the s1 (EMD: 3534, left) and s3 (EMD: 3536, right) conformations, highlighting the s1-specific contacts (orange) between the lid subunit Rpn5 (yellow) the AAA+ motor (cornflower blue), and the coaxial alignment of the AAA+ motor with the core particle (gray) in the s3 conformation. Other lid subunits are in light gray. (C) Bar graph of the fraction of proteasomes in s1 (green) or non-s1 (blue) conformations observed in in vitro cryo-EM and in situ cryo-ET studies. Gray represents unknown functional states. For in vitro studies, experimental conditions or mutations, and the observed degradation activities are noted. For in situ studies, subcellular localization and experimental conditions and noted where applicable. (D) Cartoon representation of the cell, showing proteasomes (core particle in dark gray, regulatory particle in light gray) localized near the nuclear pore complexes and/or protein aggregates (red). 3D representations of the substrate-processing proteasome conformations SPS1 (EMD: 3914) and SPS2 (EMD: 3915) from Guo et al. [53] are shown with the core particle in gray, the AAA+ motor in cornflower blue, Rpn1 and Rpn2 in dark blue, lid subunits in yellow, and potential density for bound substrate in red.
While previous EM reconstructions revealed the major conformational changes that occur upon substrate engagement with the AAA+ motor, two recent studies have captured details of the substrate-bound and actively processing proteasome. In one study conducted by our group, substrate translocation was stalled by inhibiting Rpn11’s deubiquitination activity with a Zinc chelator, which trapped the ATP-hydrolyzing proteasome at the stage of ubiquitin-chain removal and revealed a ubiquitin-bound state of Rpn11 that was identical to the previously described crystal structure of the isolated ubiquitin-bound DUB [34](Figure 1B, C). This approach captured four distinct motor states, with three of them likely representing consecutive stages of the ATP-hydrolysis and substrate-translocation cycles, and thus providing critical new insights into the mechano-chemical coupling of the AAA+ motor. Furthermore, the defined stall of a specific model substrate led to clearly resolved density for the substrate polypeptide, spanning from the ubiquitin-modified lysine at Rpn11’s active site, through the AAA+ motor, and into the proteolytic core (Figure 1F). The polypeptide was observed stably engaged with the pore loops of five Rpt subunits (Figure 1D, E), which appear to undergo hand-over-hand substrate translocation, driven by sequential ATP hydrolysis around the Rpt hexameric ring. In addition, the motor communicates with the CP through the docking of five Rpt C-terminal tails, which induce complete gate opening for substrate passage into the CP’s degradation chamber (Figure 1G). The requirement of five docking sites on CP’s alpha ring to be filled with Rpt tails for productive gate opening was also uncovered through genetics and cryo-EM studies of various proteasome mutants [28]. In a separate study, Mao and colleagues trapped the proteasome with bound substrate by adding a non-hydrolyzable ATP analog, which allowed capturing of six distinct motor conformations [35]. In addition to a state with ubiquitin-bound Rpn11, this study revealed proteasome states in which ubiquitin appears to interact with Rpn1 or the N-terminal coiled coil of Rpt4 and Rpt5, which had previously been suggested as a ubiquitin-binding site based on crosslinking experiments [50]. Furthermore, this study proposed three different modes of ATP hydrolysis corresponding to substrate deubiquitination, initiation, and translocation, which is in contrast to the single-mode hydrolysis model derived from the structures for Rpn11-inhibited, actively ATP-hydrolyzing proteasomes [34] and highlights an aspect of AAA+ motor research requiring further experimental inquiry.
Biochemical and biophysical assays reveal proteasome kinetics and conformational dynamics.
While extensive structural studies of the 26S holoenzyme and its constituents have provided snapshots of the proteasome in action, more details on the timing and dynamics of this molecular machine have been contributed by a combination of classical biochemical experiments with bulk and single-molecule fluorescence approaches.
Several recent studies aimed to dissect the kinetics and coordination of individual degradation steps, as well as the significance of the conformational changes observed upon substrate engagement. A single-molecule TIRF-microscopy-based approach was employed to investigate initial substrate binding to ubiquitin receptors on the proteasome [51]. This study revealed a direct correlation between the number of ubiquitin modifications and a substrate’s residence time on the proteasome, and suggested that the linkage type of ubiquitin chains affects productive degradation. Monitoring the co-localization of substrate and proteasome did, however, not allow capturing the entry of a substrate into the proteasome pore and its engagement by the AAA+ motor, which is considered the commitment step of degradation [25,34,35]. A comprehensive kinetic map of proteasomal substrate processing was generated using a variety of fluorescence assays and the site-specific labeling of proteasomes on incorporated unnatural amino acids [36](Figure 2A). Two FRET based probes were designed to monitor the insertion of a substrate’s unstructured initiation region into the AAA+ motor and the global conformational changes of the RP when switching from a substrate-free to substrate-processing states (Fig 2B). These studies revealed that substrate engagement by ATPase subunits in the central pore induces the major conformational switch, providing both, an important selectivity filter for appropriate substrates with sufficiently long initiation regions and a kinetic trap for committing substrates to processive degradation. This mechanism restricts fast deubiquitination to actively translocating substrates and allows the proteasome to reject ubiquitinated proteins that lack the necessary unstructured regions. These studies also demonstrated that unfolding represents the rate-limiting step of degradation, and they constitute an important platform for future mechanistic studies into substrate prioritization and the regulation of proteasomal turnover.
To address how perturbations to the RP’s conformational landscape affect proteasome activities, the Tomko group developed a method that allows the readout of s1 versus non-s1 states through disulfide crosslinking [28]. Using this approach combined with genetics and cryo-EM, they investigated how inactivating Walker-B mutations in individual Rpt subunits affect the RP’s conformational landscape (Figure 2C) and cause certain degradation defects, though the exact mechanisms remain elusive. Focusing on the interface between the lid and base subcomplexes, we recently found that s1-state-specific interactions of the lid subunit Rpn5 with Rpt3 in the ATPase ring influence the RP’s conformational landscape (Figure 2C), likely by affecting the dwell time in the s1 conformation and thereby regulating the proteasome’s ability to engage a substrate [43](Figure 2B). Taken together, these studies highlight the critical dependence of proteasomal degradation on the RP’s conformational switch between the engagement-competent s1 state and the non-s1 processing states that commit substrates to degradation and translocation-coupled deubiquitination (Figure 2B, C).
Cryo-Electron Tomography shows the proteasome in action within the cell.
While cryo-EM and biochemistry have provided a structural, kinetic, and mechanistic understanding of the proteasome, cryo-ET contributes important spatial information about proteasome localization and distribution in the cell. Using cryo-ET, the Baumeister group had first located 26S proteasomes within intact neurons and confirmed that the conformations observed by cryo-EM in vitro also exist in vivo, with the majority of complexes apparently in a resting, unengaged state [52] (Figure 2C). A subsequent study overexpressing aggregation-prone and ALS-relevant poly-Gly-Ala repeat proteins revealed that proteasomes cluster on aggregates and get trapped in a substrate-processing state, likely due to stalled degradation [53]. This is especially thought-provoking given reports that the proteasome can fragment tau and alpha-synuclein fibrils into smaller aggregates with increased cellular toxicity [54]. Another study investigating proteasomes near the nuclear pore complex (NPC) found that they attach to both the membrane surrounding the NPC and the basket structure of the NPC itself [55]. The proximity to the NPC suggests that proteasomes may play a key role in regulating nuclear transport of proteins. Cryo-ET also provided important new insight into processes or pathways that lie upstream or downstream of the UPS and rely on physical proximity. For instance, proteasomes were localized near the tripeptidyl peptidase II (TPPII), a large protease complex proposed to function in the exopeptidolytic cleavage of small peptides into amino acids downstream of the proteasome [56]. Furthermore, the proteasome appears to co-localize with the chaperonin TRiC under proteostatic stress [53], suggesting that protein unfolding, degradation, and peptide clearance could be coupled through subcellular localization.
In the future, cryo-ET may provide more information about how the 26S proteasome is organized around other molecular machines, such as Cdc48/p97 that is critically involved in ER-associated degradation and retro-translocation, and acts immediately upstream of the proteasome in the UPS [57,58].
Conclusions and future directions.
The past few years have led to significant advancements in the understanding of how the various activities of the 26S proteasome are coordinated and coupled with conformational changes to enable efficient ubiquitin-dependent degradation. A complete kinetic map of proteasomal degradation, together with atomic-resolution structural information, allows us now to perturb the system and identify the allosteric networks responsible for the regulation and fine-tuning of protein turnover, and investigate the effects of transiently bound effectors or posttranslational modifications on proteasomal activities and conformations. We look forward to elucidating how the ubiquitin code of a substrate, in combination with multivalent receptor interactions or ubiquitin editing on the proteasome surface, influences degradation kinetics and substrate prioritization, and how other proteostasis pathways connect to the UPS.
Highlights.
New structures show the 26S proteasome with engaged substrate during ATP hydrolysis.
The proteasomal Rpn11 is an essential, AAA+ motor-assisted deubiquitinase.
Detailed biochemical assays now outline the kinetic landscape of the 26S proteasome.
Proteasome conformational switching allows regulation of substrate commitment.
Cryo-electron tomography shows proteasome conformations and distributions in cells.
Acknowledgements:
A.M. acknowledges support by the Howard Hughes Medical Institute and the US National Institutes of Health.
Funding sources:
A.M. is an investigator of the Howard Hughes Medical Institute. This work was funded by the US National Institutes of Health (R01-GM094497 to A.M.) and the Howard Hughes Medical Institute (K.C.D. and A.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as: * of special interest
** of outstanding interest
References
- 1.Braten O, Livneh I, Ziv T, Admon A, Kehat I, Caspi LH, Gonen H, Bercovich B, Godzik A, Jahandideh S, et al. : Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci U S A 2016, 113:E4639–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H, Matouschek A: Recognition of Client Proteins by the Proteasome. Annu Rev Biophys 2017, 46:149–173. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Rape M: The ubiquitin code. Annual Review of Biochemistry 2012, 81:203–229. [DOI] [PubMed] [Google Scholar]
- 4.Oh E, Akopian D, Rape M: Principles of Ubiquitin-Dependent Signaling. Annu Rev Cell Dev Biol 2018, 34:137–162. [DOI] [PubMed] [Google Scholar]
- 5.Mevissen TET, Komander D: Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem 2017, 86:159–192. [DOI] [PubMed] [Google Scholar]
- 6.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R: Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386:463–471. [DOI] [PubMed] [Google Scholar]
- 7.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D: A gated channel into the proteasome core particle. Nat Struct Biol 2000, 7:1062–1067. [DOI] [PubMed] [Google Scholar]
- 8.Glickman MH, Rubin DM, Fried VA, Finley D: The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol 1998, 18:3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D: A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94:615–623. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee B-H, Shi Y, Zhang N, Poot SAH, Tuebing F, et al. : Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351:aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I: Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsasser S, Chandler-Militello D, Müller B, Hanna J, Finley D: Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. The Journal of Biological Chemistry 2004, 279:26817–26822. [DOI] [PubMed] [Google Scholar]
- 13.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A: An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 2004, 11:830–837. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi J, Chen H, Coffino P: Proteasome substrate degradation requires association plus extended peptide. The EMBO Journal 2007, 26:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y: Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell 2008, 30:360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao T, Cohen RE: A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419:403–407. [DOI] [PubMed] [Google Scholar]
- 17.Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, Deshaies RJ: Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298:611–615. [DOI] [PubMed] [Google Scholar]
- 18.Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK: Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell 2015, 57:887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vander Linden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T, Hill CP: Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol Cell 2015, 57:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D: Multiple Associated Proteins Regulate Proteasome Structure and Function. Molecular Cell 2002, 10:495–507. [DOI] [PubMed] [Google Scholar]
- 21.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D: Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006, 127:99–111. [DOI] [PubMed] [Google Scholar]
- 22.Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W, Förster F: Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proceedings of the National Academy of Sciences 2015, 112:8626–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A: Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nature structural & molecular biology 2015, 22:712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HT, Goldberg AL: UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proc Natl Acad Sci U S A 2018, 115:E11642–E11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matyskiela ME, Lander GC, Martin A: Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol 2013, 20:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Forster F, Schulten K, Baumeister W, Sakata E: Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci U S A 2017, 114:1305–1310.* Using different non-hydrolyzable ATP analogs, the authors resolved an s4 conformation of the proteasome with a fully open CP gate that closely resembled substrate-bound structures to follow and also had resolvable density for the deubiquitinase Ubp6. The authors revealed distributions of proteasome conformational states that provided a framework for conformational landscapes.
- 27.Unverdorben P, Beck F, Sledz P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W, Forster F: Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci U S A 2014, 111:5544–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisele MR, Reed RG, Rudack T, Schweitzer A, Beck F, Nagy I, Pfeifer G, Plitzko JM, Baumeister W, Tomko RJ Jr., et al. : Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep 2018, 24:1301–1315 e1305.** Using a combination of yeast genetics and cryo-EM, the authors demonstrated how site-specific ATP hydrolysis can affect proteasome conformational states with in vivo consequences. The authors also provide robust evidence for the docking of five Rpt subunits being required for full CP gate opening.
- 29.Worden EJ, Padovani C, Martin A: Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nature Structural & Molecular Biology 2014, 21:220–227. [DOI] [PubMed] [Google Scholar]
- 30.Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC: Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. Elife 2016, 5:e13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A: Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathare GR, Nagy I, Sledz P, Anderson DJ, Zhou HJ, Pardon E, Steyaert J, Forster F, Bracher A, Baumeister W: Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci U S A 2014, 111:2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worden EJ, Dong KC, Martin A: An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Molecular Cell 2017, 67:799–811.e798.** Using X-ray crystallography and biochemical assays, the authors established the active conformation of the Rpn11 deubiquitinase with the Insert-1 loop in an open beta-hairpin structure. The authors also identified the Insert-1 conformational switch as the rate-limiting step for ubiquitin cleavage, which is strongly accelerated when the AAA+ motor pulls the substrate-attached ubiquitin into the DUB active site for translocation-coupled deubiquitination.
- 34.de la Pena AH, Goodall EA, Gates SN, Lander GC, Martin A: Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 2018, 362.** Cryo-EM structures of Rpn11-inhibited, translocation-stalled proteasomes reveal the mechanisms of coordinated ATP hydrolysis and substrate translocation by the AAA+ motor, and confirm that full CP gate opening depends on docking of five Rpt C-terminal tails, as identified by Eisele et al.[28]. Additionally, these structures show the Rpn11 Insert-1 loop in an open, active beta-hairpin conformation bound to the C-terminus of ubiquitin, in close agreement with Worden et al. [33].
- 35.Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D, Mao Y: Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2018.** Cryo-EM structures of the human proteasome during substrate degradation and bound to a non-hydrolyzable ATP analog, indicating a different ATP-hydrolysis mode during substrate commitment in addition to the processive hydrolysis during translocation. The authors also observed fully active Rpn11 with its Insert-1 loop in the open, active beta-hairpin conformation, and a potential ubiquitin-binding site at the Rpt4/Rpt5 coiled-coil region of the proteasome.
- 36.Bard JAM, Bashore C, Dong KC, Martin A: The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 2019, 177:286–298 e215.** Using novel fluorescence-and FRET-based assays to monitor individual steps of proteasomal degradation, the authors revealed the complete kinetic picture of substrate processing and found that insertion of the flexible initiation region into the AAA+ motor triggers the proteasome conformational switch that commits a substrate to degradation. In addition, authors identified substrate unfolding as the rate-limiting step for degradation.
- 37.Song Y, Li S, Ray A, Das DS, Qi J, Samur MK, Tai YT, Munshi N, Carrasco RD, Chauhan D, et al. : Blockade of deubiquitylating enzyme Rpn11 triggers apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Oncogene 2017, 36:5631–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Yakushi T, Parlati F, Mackinnon AL, Perez C, Ma Y, Carter KP, Colayco S, Magnuson G, Brown B, et al. : Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat Chem Biol 2017, 13:486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Zhang Y, Da Silva Sil Dos Santos B, Wang F, Ma Y, Perez C, Yang Y, Peng J, Cohen SM, Chou TF, et al. : Epidithiodiketopiperazines Inhibit Protein Degradation by Targeting Proteasome Deubiquitinase Rpn11. Cell Chem Biol 2018, 25:1350–1358 e1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauinger L, Li J, Shostak A, Cemel IA, Ha N, Zhang Y, Merkl PE, Obermeyer S, Stankovic-Valentin N, Schafmeier T, et al. : Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat Chem Biol 2017, 13:709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. : Near-atomic resolution structural model of the yeast 26S proteasome. Proceedings of the National Academy of Sciences of the United States of America 2012, 109:14870–14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A: Structure and Function of the 26S Proteasome. Annu Rev Biochem 2018, 87:697–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greene ER, Goodall EA; de la Pena AH; Matyskiela ME; Lander GC; Martin A: Specific lid-base contacts in the 26S proteasome control the conformational switching required for substrate degradation. BioRxiv 2019, 10.1101/687921** The authors found that specific contacts between the lid and base subcomplexes stabilize the s1 conformation and coordinate the overall conformational transitions of the regulatory particle. The study uncovers some of the mechanisms underlying the degradation defects of proteasome mutants with perturbed conformational landscapes, suggesting that proteasome activity in vivo could be tuned through conformational biasing by binding partners or post-translational modifications.
- 44.Ding Z, Xu C, Sahu I, Wang Y, Fu Z, Huang M, Wong CCL, Glickman MH, Cong Y: Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations. Mol Cell 2019, 73:1150–1161 e1156.* The authors found through cryo-EM studies that K48-linked tetra-ubiquitin, the commonly accepted degradation signal appended to condemned substrates, can influence the conformational landscape of the proteasome through binding to Rpn1.
- 45.Ding Z, Fu Z, Xu C, Wang Y, Wang Y, Li J, Kong L, Chen J, Li N, Zhang R, et al. : High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx. Cell Res 2017, 27:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sledz P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Forster F, Baumeister W: Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A 2013, 110:7264–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S, Wu J, Lu Y, Ma YB, Lee BH, Yu Z, Ouyang Q, Finley DJ, Kirschner MW, Mao Y: Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci U S A 2016, 113:12991–12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y, Wang WL, Yu D, Ouyang Q, Lu Y, Mao Y: Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome. Nat Commun 2018, 9:1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haselbach D, Schrader J, Lambrecht F, Henneberg F, Chari A, Stark H: Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs. Nat Commun 2017, 8:15578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM: A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 2002, 416:763–767. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y, Lee BH, King RW, Finley D, Kirschner MW: Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science 2015, 348:1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, Baumeister W: A molecular census of 26S proteasomes in intact neurons. Science 2015, 347:439–442. [DOI] [PubMed] [Google Scholar]
- 53.Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU, et al. : In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell 2018, 172:696–705 e612.** Authors used cryo-ET of neuronal cells to investigate how cells respond to severe proteotoxic stress from C9orf72 poly-GA transfection. A high density of proteasomes around aggregates was found and two resolvable, substrate-bound 3D classes were elucidated, suggesting that these proteasomes were stalled on substrate.
- 54.Cliffe R, Sang JC, Kundel F, Finley D, Klenerman D, Ye Y: Filamentous Aggregates Are Fragmented by the Proteasome Holoenzyme. Cell Rep 2019, 26:2140–2149 e2143.* The authors found that the proteasome can cleave aggregate fibrils of both Tau and alpha-synuclein into smaller fragments with increased cytotoxicity.
- 55.Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko JM, Beck M, Baumeister W, Engel BD: Proteasomes tether to two distinct sites at the nuclear pore complex. Proc Natl Acad Sci U S A 2017, 114:13726–13731.* The authors used in situ cryo-ET to show that proteasomes cluster near nuclear pore complexes through a linkage potentially mediated by the proteasome subunit Rpn9. It is postulated that this localization enables regulation of transcriptional processes and protein quality control.
- 56.Fukuda Y, Beck F, Plitzko JM, Baumeister W: In situ structural studies of tripeptidyl peptidase II (TPPII) reveal spatial association with proteasomes. Proc Natl Acad Sci U S A 2017, 114:4412–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye Y, Tang WK, Zhang T, Xia D: A Mighty “Protein Extractor” of the Cell: Structure and Function of the p97/CDC48 ATPase. Front Mol Biosci 2017, 4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van den Boom J, Meyer H: VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol Cell 2018, 69:182–194. [DOI] [PubMed] [Google Scholar]
- 59.Beckwith R, Estrin E, Worden EJ, Martin A: Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol 2013, 20:1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]