Abstract
Many studies have highlighted the importance of the tight regulation of mRNA stability in the control of gene expression. mRNA stability largely depends on the mRNA nucleotide sequence, which affects the secondary and tertiary structures of the mRNAs, and the accessibility of various RNA-binding proteins to the mRNAs. Recent advances in high-throughput RNA-sequencing techniques have resulted in the elucidation of the important roles played by mRNA modifications and mRNA nucleotide sequences in regulating mRNA stability. To date, hundreds of different RNA modifications have been characterized. Among them, several RNA modifications, including N6-methyladenosine (m6A), N6,2′-O-dimethyladenosine (m6Am), 8-oxo-7,8-dihydroguanosine (8-oxoG), pseudouridine (Ψ), 5-methylcytidine (m5C), and N4-acetylcytidine (ac4C), have been shown to regulate mRNA stability, consequently affecting diverse cellular and biological processes. In this review, we discuss our current understanding of the molecular mechanisms underlying the regulation of mammalian mRNA stability by various RNA modifications.
Subject terms: RNA modification, RNA decay
Gene activity: the significance of RNA modification
Messenger RNA molecules play their key role in directing the manufacture of proteins via translating the genetic information in DNA. Sung Ho Boo and Yoon Ki Kim at Korea University in Seoul review current understanding of the significance of modifications to messenger RNA with respect to RNA stability in mammalian cells. Recent advances in RNA sequencing technology have revealed hundreds of different modifications which, by influencing RNA stability, can control gene expression. The modifications largely involve small chemical groups added to any of the four nucleotide groups that make up an RNA molecule. This can affect all aspects of mature RNA biogenesis, including transcription, pre-mRNA splicing, RNA export form the nucleus to the cytoplasm, translation, and stability.
Introduction
Many recent studies have demonstrated that RNA undergoes various modifications in a manner similar to DNA. These RNA modifications play a role in many cellular and biological processes, thereby opening up an emerging research field known as epitranscriptomics1–7. According to the MODOMICS database, ~170 different RNA modifications have been identified in coding and noncoding RNAs5,8,9. In certain types of modifications, the specific nucleotide sequences and positions targeted for RNA modification have been well characterized due to recent advances in specialized high-throughput RNA-sequencing technologies10.
Generally, the fate of a modified transcript is determined by the coordinated actions of the following three effector proteins (Fig. 1)1,3–7: (i) writer proteins (RNA-modifying enzymes), which transfer a specific chemical group to a target position on an RNA molecule; (ii) RNA-binding proteins (RBPs), which specifically recognize the modified nucleotides (reader proteins); and (iii) eraser proteins, which remove specific chemical groups from the modified nucleotides, converting them back into unmodified nucleotides. In certain cases, endogenous or exogenous chemical damage can also generate RNA modifications without the involvement of writer proteins11,12. In addition, some modifications are reversible, while others are irreversible.
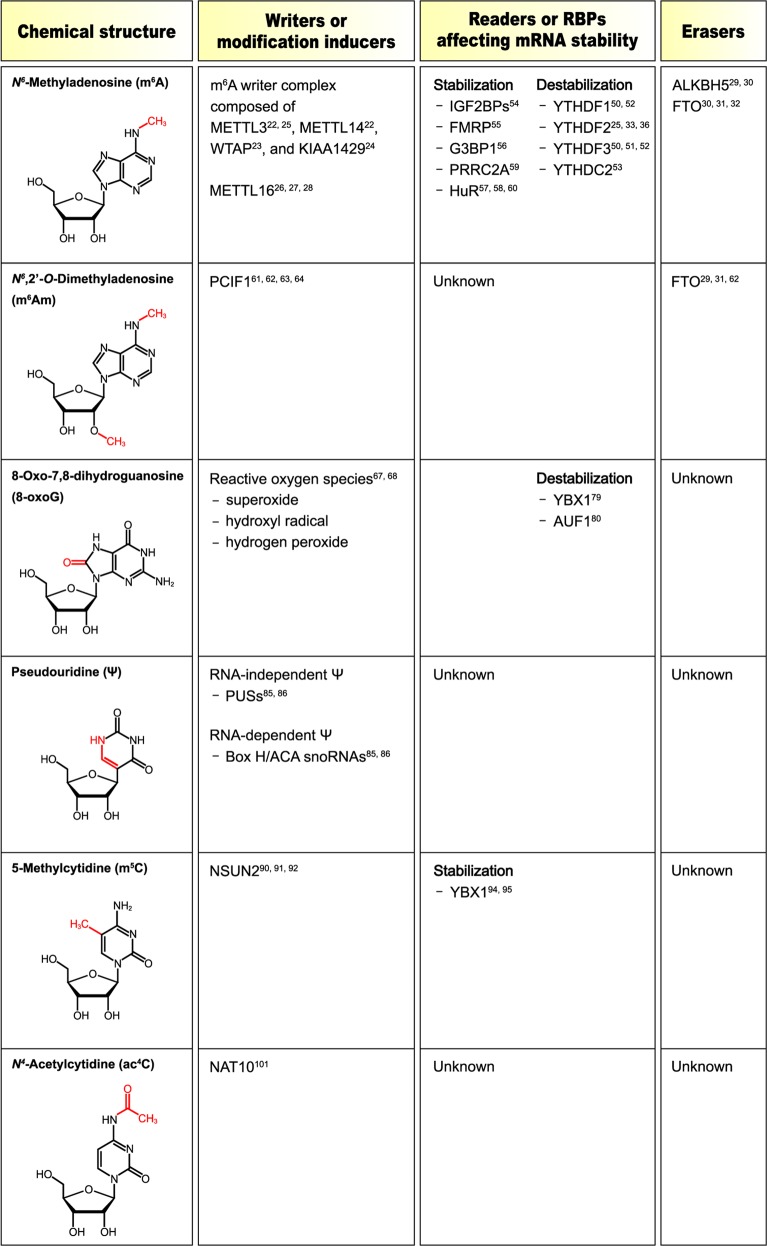
Fig. 1. Chemical structures of RNA modifications affecting mRNA stability.
The chemical structures of the six RNA modifications discussed in this review are shown. The modified chemical groups are depicted in red. The known writers (or modification inducers), readers (or RBPs) involved in mRNA stability, and erasers for each RNA modification are also summarized.
These RNA modifications can affect a variety of molecular processes, such as transcription, pre-mRNA splicing, RNA export, mRNA translation, and RNA degradation1,3–7. All of these molecular events contribute to shaping the cellular transcriptome and proteome1–7,13. In particular, recent reports have posited that the regulation of mRNA stability through RNA modification is a crucial step for the tight regulation of gene expression1–7,13. Therefore, in this review, we aim to highlight recent progress made in our understanding of the molecular mechanisms underlying the regulation of mRNA stability through various mRNA modifications, including N6-methyladenosine (m6A), N6,2′-O-dimethyladenosine (m6Am), 8-oxo-7,8-dihydroguanosine (8-oxoG), pseudouridine (Ψ), 5-methylcytidine (m5C), and N4-acetylcytidine (ac4C).
N6-methyladenosine
m6A is the most abundant internal mRNA modification, and it affects various cellular and physiological processes, such as maternal-to-zygotic transition (MZT), cortical neurogenesis, and the regulation of cancer stem cells in acute myeloid leukemia7,14–19. A transcriptome-wide analysis for identifying the consensus sequence motifs for the m6A modification in the human transcriptome revealed that m6A sites (Gm6AC or Am6AC) are found in noncoding RNAs and mRNAs, with a greater number within long exons and adjacent to stop codons20,21.
The m6A modification is cotranscriptionally generated in nascent transcripts by a methyltransferase complex comprising methyltransferase like 3 (METTL3), METTL14, WTAP, and KIAA1429 (Fig. 1)7,17–19,22–25. The methyltransferase complex transfers a methyl group to the N-6 position of the adenosine base. The second m6A writer protein, METTL16 also contributes to m6A modification of both coding and noncoding RNAs26–28. However, only a handful of mRNAs, such as MAT2A mRNA, have been identified as substrates of METTL16. m6A is reversibly converted into adenosine by m6A erasers (demethylases), which remove the methyl group from m6A. The α-ketoglutarate-dependent dioxygenase alkB homolog 5 protein (ALKBH5) is known to be the primary and specific m6A demethylase29,30. In addition, fat mass and obesity-associated protein (FTO) demethylase is known to have a weak preference for m6A30–32.
The m6A modification plays a regulatory role in diverse molecular processes, such as transcription, pre-mRNA splicing, mRNA export, mRNA stability, and translation7,17–19. The molecular events that occur through the m6A modification are guided by various m6A-recognizing reader proteins, such as YT521-B homology (YTH) domain-containing proteins. The YTHDF2 protein is the most representative m6A reader protein involved in the decay of m6A-containing RNA (Fig. 2)25. YTHDF2 contains a P/Q/N-rich unstructured region in its N-terminal half, which is critical for YTHDF2 interactions with other cellular factors, and an RNA-binding domain in the C-terminal half, which is crucial for binding m6A-containing transcripts25,33–35. When an m6A-containing mRNA is recognized by YTHDF2, rapid degradation of mRNA is initiated in one of two distinct pathways: the deadenylation pathway or the endoribonucleolytic pathway19,33,36. Rapid deadenylation of m6A-containing mRNA is accelerated by the recruitment of a deadenylase complex (CCR4–NOT complex)37,38 to the m6A-containing mRNA via a direct interaction with the N-terminus of YTHDF2 and the SH domain of CNOT1, a component of the CCR4–NOT complex33. The resulting deadenylated m6A-containing mRNA would be more vulnerable to 3′–5′ exoribonucleolytic cleavage by the exosome complex or DIS3-like enzymes39–43.
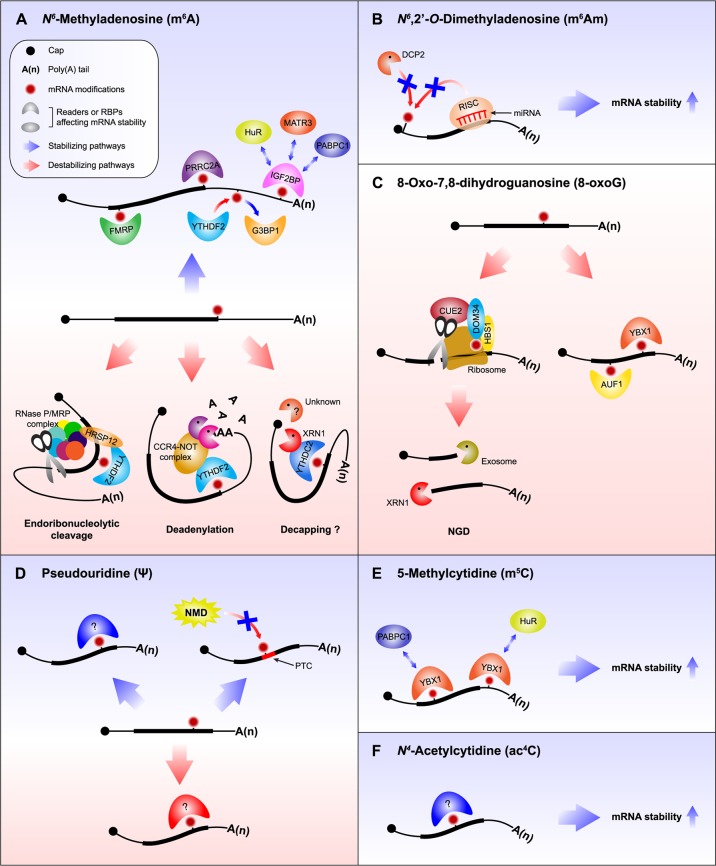
Fig. 2. Molecular mechanisms underlying the regulation of mRNA stability through diverse RNA modifications.
a N6-methyladenosine (m6A): in general, YTH domain-containing proteins destabilize m6A-containing mRNAs. When m6A is recognized by YTHDF2, the degradation of m6A-containing mRNAs is initiated by deadenylation through the CCR4–NOT complex. If the YTHDF2-bound m6A-containing mRNA harbors an HRSP12-binding site, the degradation of the mRNA is preferentially initiated through an endoribonucleolytic cleavage reaction mediated by the RNase P/MRP complex. YTHDC2 binds to m6A and recruits XRN1, thereby triggering 5′–3′ exoribonucleolytic cleavage. In contrast, m6A-containing mRNA can be stabilized by other m6A reader proteins or RBPs, including IGF2BP, FMRP, G3BP1, PRRC2A, and HuR. b N6,2′-O-dimethyladenosine (m6Am): the presence of m6Am at the 5′-end of mRNA blocks its accessibility to DCP2, thus stabilizing the mRNA. m6Am also enables mRNA to become more resistant to microRNA-mediated mRNA degradation. c 8-Oxo-7,8-dihydroguanosine (8-oxoG): the presence of 8-oxoG in mRNA causes ribosome stalling, thereby triggering NGD. Alternatively, 8-oxoG-containing mRNAs are degraded through 8-oxoG reader proteins, such as YBX1 and AUF1. d Pseudouridine (Ψ): Ψ can stabilize or destabilize mRNA. In particular, Ψ on PTCs results in the inhibition of NMD. As a consequence, the mRNA is stabilized. e 5-Methylcytidine (m5C): YBX1 specifically recognizes m5C on mRNA and recruits either PABPC1 or HuR, thereby stabilizing the mRNA. f N4-acetylcytidine (ac4C). The presence of ac4C stabilizes mRNA by unknown mechanisms.
As an alternative to deadenylation followed by 3′–5′ exoribonucleolytic cleavage, endoribonucleolytic cleavage of m6A-containing mRNAs can be initiated by the interplay among YTHDF2, heat-responsive protein 12 (HRSP12, also known as reactive intermediate imine deaminase A homolog), and an endoribonuclease RNase P/MRP complex (Fig. 2)36. HRSP12 has been identified as a cellular factor involved in glucocorticoid receptor-mediated mRNA decay43–46. The RNase P/MRP complex has been characterized as an endoribonuclease that cleaves long noncoding RNAs and mRNAs, as well as precursor forms of 5.8 S rRNA and tRNA47–49. When YTHDF2 binds to an m6A-containing mRNA, it recruits HRSP12, which functions as an adaptor protein that links YTHDF2 and POP1, a component of the RNase P/MRP complex36. Results from transcriptome analyses have shown that HRSP12 preferentially binds to the GGUUC motif, typically located upstream of YTHDF2-binding sites36. Intriguingly, HRSP12 and YTHDF2 bind to mRNA in a cooperative manner36. This cooperative interaction facilitates the efficient recruitment of the RNase P/MRP complex to the m6A-containing mRNA. Consequently, the recruited RNase P/MRP complex triggers endoribonucleolytic cleavage, mostly downstream of the YTHDF2-binding site in the mRNA36. The two resulting products, 5′ and 3′ fragments, are degraded by 3′–5′ exoribonucleolytic cleavage and 5′–3′ exoribonucleolytic cleavage, respectively39–43.
It should be noted that both HRSP12 and CNOT1 bind to the unstructured N-terminal region of YTHDF2, but at different residues33,36. Amino acids 101‒200 in the N-terminal half of YTHDF2 are required for binding to CNOT133. In contrast, HRSP12 efficiently interacts with amino acids 1‒100 as well as a truncated N-terminal YTHDF2 lacking amino acids 101‒20036. Therefore, the discrete binding residues in YTHDF2 can activate two distinct pathways for the decay of m6A-containing mRNA, either deadenylation by the YTHDF2–CCR4–NOT complex or endoribonucleolytic cleavage by the YTHDF2–HRSP12–RNase P/MRP complex, depending on whether HRSP12-binding sites are present in the m6A-containing mRNA. Currently, it is unknown whether components in these two pathways communicate with each other to regulate the destabilization of m6A-containing mRNA, and which cellular environments are responsible for the preferential activation of each decay pathway.
Recent studies have shown that, in addition to YTHDF2, other YTH domain-containing proteins are also engaged in mRNA degradation. For instance, YTHDF1, YTHDF2, and YTHDF3 share a subset of target transcripts that they destabilize50–52. In addition, the N-terminal half of YTHDC2, another YTH domain-containing protein, interacts with XRN1, a cytoplasmic 5′–3′ exoribonuclease53, suggesting that YTHDC2 recruits XRN1 and triggers rapid degradation of m6A-containing mRNA.
m6A-modified mRNA can also be targeted toward an opposite fate, depending on the m6A reader proteins and other RBPs (Fig. 2). For instance, a recently identified m6A reader protein, insulin-like growth factor 2 mRBP (IGF2BP), binds to the UGGAC motif54, which overlaps with the m6A motif, and increases the half-life of m6A-containing mRNA20,25,54. In addition to IGF2BP, other m6A reader proteins or RBPs, such as fragile X mental retardation protein (FMRP), Ras-GTPase-activating protein SH3 domain-binding protein (G3BP1), proline-rich coiled-coil 2 A (PRRC2A), and human antigen R (HuR; also known as ELAVL1), have been shown to stabilize m6A-containing mRNA at the transcriptome or gene-specific level54–60.
N6,2′-O-dimethyladenosine
The first transcribed nucleotide next to the 5′ m7G cap structure in mRNA is generally methylated on the ribose ring at the 2′-OH position3–7. In particular, when the first nucleotide is adenosine, the methylated adenosine at the 2′-OH position, known as 2′-O-methyladenosine (Am), is further methylated at the N-6 position of Am, generating m6Am (Fig. 1)3–7. Therefore, m6Am and m6A are generated by very similar chemical reactions: methylation at the N-6 position of adenosine, and Am generates m6A and m6Am, respectively. However, m6Am has several properties that distinguish it from m6A. First, m6Am is generated by the methylation of Am, which is primarily located in the first nucleotide position adjacent to the m7G cap structure of mRNA3–7. Second, a unique writer protein, phosphorylated CTD-interacting factor 1, is responsible for generating the m6Am modification61–64, whereas a methyltransferase complex comprising METTL3, METTL14, WTAP, and KIAA1429 is involved in the generation of the m6A modification22–25. Finally, whereas m6A is largely demethylated by ALKBH5 but also by FTO, with a weak preference29–32, m6Am is preferentially and specifically demethylated by FTO29,31,62.
Although controversial63,64, findings from recent studies have revealed that m6Am-initiated mRNAs are in greater abundance and have longer half-lives than mRNAs with Am, Gm, Cm, or Um31,62. In vitro decapping experiments have shown that m6Am-initiated mRNAs are more resistant to decapping by decapping mRNA 2 (DCP2)65, resulting in the increased mRNA stability (Fig. 2)31. Furthermore, m6Am-initiated mRNAs are more resistant to microRNA-mediated mRNA degradation31, which also involves decapping (Fig. 2)66. Further investigation is needed to understand the molecular mechanism underlying the stabilization of m6Am-initiated mRNAs.
8-Oxo-7,8-dihydroguanosine
The bases in RNA are vulnerable to various forms of chemical damage, such as those induced by reactive oxygen species (ROS), ultraviolet light, and alkylating agents11. In particular, ROS—including superoxide, hydroxyl radicals, and hydrogen peroxide—are produced as byproducts of normal oxygen metabolism (e.g., cellular respiration in the mitochondria) and are also generated by various environmental stresses, such as ultraviolet irradiation and heat shock67. It should be noted that ROS oxidize RNA bases and generate numerous forms of oxidized RNAs11,67. These oxidized bases include 8-oxoG, 8-oxo-7,8-dihydroadenosine, 5-hydroxyuridine, 5-hydroxycytidine, and cytosine glycol (Fig. 1). Among these forms, 8-oxoG (an oxidized form of the guanine base) is the most abundant within mammalian cells, and its accumulation is associated with many neurodegenerative diseases68,69.
The oxidation of mRNA affects multiple steps of mRNA fate determination, including mRNA stability and translation12,70–73. For instance, the oxidation of mRNA (typically 8-oxoG) inhibits the efficiency of peptide bond formation by >1000-fold, regardless of the codon position71. This inhibition at the elongation step of translation causes the accumulation of stalled ribosomes71, which triggers the rapid mRNA degradation via the no-go decay (NGD) pathway, one of the mRNA surveillance pathways in eukaryotes (Fig. 2)41,74–76. The NGD pathway identifies stalled ribosomes caused by various impediments to translation elongation, such as robust secondary structures or stretches of rare codons in the mRNA77. The stalled ribosome is then disassembled from the mRNA and recycled. Concomitantly, the mRNA is rapidly degraded by the NGD pathway, with the coordinated action of HBS1 and DOM3477. Recently, CUE2 was identified as an endoribonuclease that initiates the internal cleavage of NGD targets upstream of the stalled ribosome78. The resulting 5′ and 3′ fragments generated by endoribonucleolytic cleavage are degraded via the exosome complex and XRN1, respectively. In this way, NGD minimizes the production of truncated polypeptides (which are potentially detrimental to cells) from the ribosomes stalled because of the mRNA oxidation.
In addition to NGD, 8-oxoG–containing mRNAs are degraded by 8-oxoG reader proteins via unknown mechanisms (Fig. 2). Y-box binding protein 1 (YBX1) and AU-rich element RBP 1 (AUF1; also known as hnRNP D) have been shown to preferentially bind to 8-oxoG, thereby triggering the rapid degradation of 8-oxoG–containing mRNAs79,80. A recent study also identified poly(rC)-binding protein 1 (PCBP1) as an 8-oxoG reader protein. Notably, unlike AUF1, which recognizes a single 8-oxoG, the binding of PCBP1 to RNA requires two 8-oxoGs located in close proximity. In addition, the binding of PCBP1 to oxidized RNA is associated with apoptosis—under conditions of oxidative stress81—rather than with the rapid degradation of 8-oxoG–containing mRNAs, as has been observed in the case of AUF180. Several important questions regarding the molecular mechanisms of mRNA decay remain unanswered. First, how do 8-oxoG reader proteins trigger the rapid mRNA degradation? Second, how do the 8-oxoG reader proteins recruit general RNA-degrading enzymes? Third, is 8-oxoG reversibly converted into a normal guanosine base by a specific enzyme, as observed in the case of many other RNA modifications?
Pseudouridine
Ψ is generated by the C–C glycosidic isomerization of a uridine base (Fig. 1). Although Ψ was first discovered in rRNA, tRNA, and small nuclear RNAs, evidence from recent transcriptome-wide analysis of Ψ profiles in humans and yeast revealed that hundreds of human and yeast mRNAs contain Ψ82,83. More recently, another transcriptome-wide profiling study identified thousands of Ψ sites in human mRNAs84. The conversion of uridine into Ψ is catalyzed by either RNA-independent or RNA-dependent mechanisms85,86. In the RNA-independent mechanism, Ψ is deposited by various Ψ synthases (PUSs) with different substrate specificities, different chemical reactions, and different subcellular localizations. In contrast, the RNA-dependent mechanism is guided and catalyzed by Box H/ACA small nucleolar RNAs (snoRNAs).
The chemical properties of Ψ differ from those of uridine85,86. For instance, Ψ makes the phosphodiester backbone more rigid, and the base pairing between Ψ and adenine is stronger than that between uridine and adenine. Because of these properties, the presence of Ψ in mRNAs can affect the local secondary structures and the protein-coding potential of the mRNA. Therefore, despite the lack of sufficient experimental evidence, it is plausible that Ψ may directly or indirectly influence pre-mRNA splicing, mRNA translation, mRNA localization, and/or mRNA stability. Indeed, the artificially targeted conversion of U-to-Ψ in translation termination codons (UAA, UGA, and UAG) turns them into missense codons87. In particular, the U-to-Ψ change at premature termination codons (PTCs) can inhibit the rapid mRNA degradation triggered by nonsense-mediated mRNA decay (NMD)87, an mRNA surveillance mechanism by which faulty (e.g., PTC containing) mRNAs are specifically recognized and removed before the production of truncated (and potentially toxic) polypeptides (Fig. 2)41–43.
Several lines of evidence support the hypothesis that Ψ affects mRNA stability (Fig. 2). PUS7 deletion in yeast causes a reduction in the amount of Ψ-containing mRNAs83, suggesting that Ψ stabilizes mRNA. In agreement with this finding, in vitro-synthesized Ψ-containing mRNAs are more stable than unmodified mRNAs with identical nucleotide sequences in mammalian cells88. In contrast, another study showed that, in the eukaryotic parasite Toxoplasma gondii, the half-life of the mRNAs pseudouridylated by PUS1 is significantly increased in the PUS1 mutant89, suggesting that Ψ destabilizes mRNA. Therefore, future studies should focus on elucidating the molecular mechanism underlying Ψ-mediated regulation of mRNA stability. In addition, it should be determined whether certain RBPs have the ability to directly recognize Ψ, thereby affecting the stability of Ψ-containing mRNAs, as observed in the case of other mRNA modifications.
5-Methylcytidine
m5C is generated in transcripts by NOP2/Sun RNA methyltransferase 2 (NSUN2), which catalyzes the deposition of a methyl group at the 5 position of cytosine (Fig. 1)90–92. m5C is recognized by m5C reader proteins, such as ALYREF93 or YBX194,95. It remains unknown whether m5C is a reversible process because m5C erasers have not yet been identified. Similar to other RNA modifications, this modification is also present in mRNAs96. Bisulfite-sequencing analysis used to determine the m5C landscape in the human transcriptome has revealed that m5C sites are highly enriched in the 3′-UTR of mRNAs or near the translation initiation codon93,97–99.
A possible role of m5C in the regulation of mRNA stability has been previously implied (Fig. 2)98,100. Downregulation of NSUN2 causes a decrease in the amount and the half-life of p16INK4 mRNA, suggesting that NSUN2 functions as a stabilizer of p16INK4 mRNA100. Furthermore, two recent studies have shown that YBX1 preferentially binds to m5C-containing RNA through a π–π interaction between the target RNA and two tryptophan residues (Trp45 and Trp65) in the cold-shock domain of YBX194,95. This interaction contributes to the stabilization of m5C-containing RNA, consequently affecting physiological events, such as the MZT (a reprogramming process during which maternal transcripts are eliminated and embryonic identity is established) and oncogene activation in human urothelial carcinoma of the bladder (UCB)94,95. During early MZT in zebrafish, the interaction between m5C and YBX1 stabilizes a subset of maternal mRNAs by recruiting poly(A)-binding protein cytoplasmic 1 (PABPC1)95. Failure of this stabilization leads to early gastrulation defects in zebrafish embryos95. Another recent report also showed that, in human UCB, a subset of oncogenic mRNAs have hypermethylated m5C sites and that the levels of these mRNAs are upregulated in an NSUN2-dependent manner94. In addition, the levels of NSUN2 and YBX1 proteins are higher in UCB than those in normal cells94. Mechanistically, YBX1 binds to and stabilizes oncogenic mRNAs with hypermethylated m5C sites (e.g., heparin-binding growth factor mRNA, which is critical for UCB progression and pathogenesis) by recruiting HuR, thus indicating an essential oncogenic role of m5C in UCB94.
N4-acetylcytidine
Recent transcriptome-wide profiling of another cytidine modification, ac4C, in human cells showed that ac4C is widely distributed within noncoding RNAs and coding RNAs, with greater abundance near the translation initiation codon in mRNA (Fig. 1)101. mRNAs modified by ac4C are known to have increased half-lives and promoted translation (Fig. 2)101. Knocking out N-acetyltransferase 10 (NAT10) reduces the level of ac4C modification on RNA, indicating that NAT10 is a primary ac4C writer protein (RNA cytosine acetyltransferase)101. In yeast, orphan box C/D snoRNAs specifically guide Kre33 (a yeast homolog of human NAT10) to ac4C target sites in rRNA102, similar to the way that Ψ is guided by the H/ACA snoRNAs85,86. However, it remains unknown whether human NAT10 also uses box C/D snoRNAs to generate ac4C on its target RNAs. To date, neither ac4C reader protein nor an active deacetylation process has been reported. It is also unknown whether ac4C modification is a reversible process. Therefore, future studies should address the molecular mechanism underlying the stabilization of ac4C-containing mRNAs.
Concluding remarks
Cellular mRNA levels are determined by various quantity control pathways (such as transcription, capping, splicing, and 3′-end formation) and quality control pathways (such as NMD and NGD). All of these molecular events are mediated by diverse cis-acting elements (e.g., nucleotide sequences and secondary structures) and trans-acting factors (e.g., RBPs and noncoding RNAs). Furthermore, recent advances have been made toward understanding the roles of RNA modifications in regulating mRNA stability. Although only certain types of modifications are discussed in this review, recent studies imply that several other RNA modifications might have the ability to influence mRNA stability. For instance, 2′-O-methylation (Nm)—in which a methyl group is added to the 2′-OH of the ribose ring—is highly enriched in the first and second transcribed nucleotides next to the cap structure2. The presence of Nm is known to increase the levels of peroxidasin mRNA103. Furthermore, in vitro decapping experiments have shown that a capped RNA with an Nm modification is resistant to hydrolysis by the decapping exoribonuclease DXO, which specifically recognizes and removes pre-mRNAs harboring a defective cap structure104. Therefore, these two recent reports suggest that Nm functions as an mRNA stabilizer. As they are oxidized, RNAs can also be alkylated upon exposure to alkylating agents that are either endogenously produced during normal metabolic processes or exogenously provided in the environment11. Bases, riboses, and the phosphate backbone of RNA are all vulnerable to alkylation because they contain oxygen and nitrogen atoms. As a result, numerous alkylated nucleosides can be generated in RNA, possibly affecting the RNA structure and/or the protein-coding potential of mRNA. A recent study even showed that alkylated mRNA is subject to rapid degradation via NGD12. Finally, the presence of internal N7-methylguanosine (m7G) in mRNA is known to promote translation. The positive charge of internal m7G may affect the RNA secondary structure and thereby affect the mRNA stability105,106.
Future investigations should aim to determine the transcriptome profiles of all RNA modifications, extend a list of RNA modifications that affect mRNA stability, and elucidate the underlying molecular mechanisms. In addition, considering that several mRNA modifications, such as Ψ, oxidation, and alkylation, are associated with mRNA surveillance pathways, it will be interesting to investigate whether mRNA surveillance pathways (NMD, NGD, and no-stop decay)41–43,74 are associated with the mRNA degradation caused by other types of RNA modifications.
Acknowledgements
This work was supported by a National Research Foundation (NRF) of Korea grant funded by the Korean government (Ministry of Science, ICT and Future Planning; NRF-2015R1A3A2033665 and NRF-2018R1A5A1024261).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019;21:552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 2.Dimitrova DG, Teysset L, Carre C. RNA 2’-O-methylation (Nm) modification in human diseases. Genes (Basel) 2019;10:117. doi: 10.3390/genes10020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonkhout N, et al. The RNA modification landscape in human disease. RNA. 2017;23:1754–1769. doi: 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadumuri RV, Janga SC. Epitranscriptomic code and its alterations in human disease. Trends Mol. Med. 2018;24:886–903. doi: 10.1016/j.molmed.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nachtergaele S, He C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 2018;52:349–372. doi: 10.1146/annurev-genet-120417-031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helm M, Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017;18:275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
- 9.Boccaletto P, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods. 2016;14:23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- 11.Yan LL, Zaher HS. How do cells cope with RNA damage and its consequences? J. Biol. Chem. 2019;294:15158–15171. doi: 10.1074/jbc.REV119.006513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan LL, Simms CL, McLoughlin F, Vierstra RD, Zaher HS. Oxidation and alkylation stresses activate ribosome-quality control. Nat. Commun. 2019;10:5611. doi: 10.1038/s41467-019-13579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh G, Pratt G, Yeo GW, Moore MJ. The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao BS, et al. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon KJ, et al. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell. 2017;171:877–889 e817. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paris J, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–148 e136. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Weng H, Chen J. The biogenesis and precise control of RNA m(6)A methylation. Trends Genet. 2020;36:44–52. doi: 10.1016/j.tig.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, Y., Choe, J., Park, O. H. & Kim, Y. K. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet.36, 177–188 (2020). [DOI] [PubMed]
- 20.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 21.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendel M, et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 2018;71:986–1000 e1011. doi: 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nance DJ, et al. Characterization of METTL16 as a cytoplasmic RNA binding protein. PLoS ONE. 2020;15:e0227647. doi: 10.1371/journal.pone.0227647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pendleton KE, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835 e814. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mauer J, et al. Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei J, et al. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018;71:973–985 e975. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du H, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park OH, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74:494–507 e498. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 37.Collart MA. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA. 2016;7:438–454. doi: 10.1002/wrna.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ukleja M, Valpuesta JM, Dziembowski A, Cuellar J. Beyond the known functions of the CCR4-NOT complex in gene expression regulatory mechanisms: new structural insights to unravel CCR4-NOT mRNA processing machinery. Bioessays. 2016;38:1048–1058. doi: 10.1002/bies.201600092. [DOI] [PubMed] [Google Scholar]
- 39.Schmid M, Jensen TH. The nuclear RNA exosome and its cofactors. Adv. Exp. Med. Biol. 2019;1203:113–132. doi: 10.1007/978-3-030-31434-7_4. [DOI] [PubMed] [Google Scholar]
- 40.Saramago M, da Costa PJ, Viegas SC, Arraiano CM. The implication of mRNA degradation disorders on human disease: focus on DIS3 and DIS3-like enzymes. Adv. Exp. Med. Biol. 2019;1157:85–98. doi: 10.1007/978-3-030-19966-1_4. [DOI] [PubMed] [Google Scholar]
- 41.Wolin SL, Maquat LE. Cellular RNA surveillance in health and disease. Science. 2019;366:822–827. doi: 10.1126/science.aax2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019;20:406–420. doi: 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YK, Maquat LE. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA. 2019;25:407–422. doi: 10.1261/rna.070136.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park OH, et al. Identification and molecular characterization of cellular factors required for glucocorticoid receptor-mediated mRNA decay. Genes Dev. 2016;30:2093–2105. doi: 10.1101/gad.286484.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park OH, Do E, Kim YK. A new function of glucocorticoid receptor: regulation of mRNA stability. BMB Rep. 2015;48:367–368. doi: 10.5483/BMBRep.2015.48.7.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho H, et al. Glucocorticoid receptor interacts with PNRC2 in a ligand-dependent manner to recruit UPF1 for rapid mRNA degradation. Proc. Natl Acad. Sci. USA. 2015;112:E1540–E1549. doi: 10.1073/pnas.1409612112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarrous N. Roles of RNase P and its subunits. Trends Genet. 2017;33:594–603. doi: 10.1016/j.tig.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Mattijssen S, et al. Viperin mRNA is a novel target for the human RNase MRP/RNase P endoribonuclease. Cell Mol. Life Sci. 2011;68:2469–2480. doi: 10.1007/s00018-010-0568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jarrous N, Gopalan V. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res. 2010;38:7885–7894. doi: 10.1093/nar/gkq701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu W, et al. N(6)-Methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J. Biol. Chem. 2018;293:12992–13005. doi: 10.1074/jbc.RA118.004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi H, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirumuru N, et al. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife. 2016;5:e15528. doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kretschmer J, et al. The m(6)A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1. RNA. 2018;24:1339–1350. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang F, et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 2018;27:3936–3950. doi: 10.1093/hmg/ddy292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edupuganti RR, et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017;24:870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visvanathan A, et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu R, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41. doi: 10.1038/s41422-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun H, Zhang M, Li K, Bai D, Yi C. Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res. 2019;29:80–82. doi: 10.1038/s41422-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boulias K, et al. Identification of the m(6)Am methyltransferase PCIF1 reveals the location and functions of m(6)Am in the transcriptome. Mol. Cell. 2019;75:631–643.e638. doi: 10.1016/j.molcel.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sendinc E, et al. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell. 2019;75:620–630.e629. doi: 10.1016/j.molcel.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akichika, S. et al. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science363, eaav0080. 10.1126/science.aav0080 (2019). [DOI] [PubMed]
- 65.Mugridge JS, Coller J, Gross JD. Structural and molecular mechanisms for the control of eukaryotic 5’-3’ mRNA decay. Nat. Struct. Mol. Biol. 2018;25:1077–1085. doi: 10.1038/s41594-018-0164-z. [DOI] [PubMed] [Google Scholar]
- 66.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013;5:a012559. doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nunomura A, Lee HG, Zhu X, Perry G. Consequences of RNA oxidation on protein synthesis rate and fidelity: implications for the pathophysiology of neuropsychiatric disorders. Biochem Soc. Trans. 2017;45:1053–1066. doi: 10.1042/BST20160433. [DOI] [PubMed] [Google Scholar]
- 69.Poulsen HE, et al. RNA modifications by oxidation: a novel disease mechanism? Free Radic. Biol. Med. 2012;52:1353–1361. doi: 10.1016/j.freeradbiomed.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 70.Jamar NH, Kritsiligkou P, Grant CM. The non-stop decay mRNA surveillance pathway is required for oxidative stress tolerance. Nucleic Acids Res. 2017;45:6881–6893. doi: 10.1093/nar/gkx306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014;9:1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKinlay A, Gerard W, Fields S. Global analysis of RNA oxidation in Saccharomyces cerevisiae. Biotechniques. 2012;52:109–111. doi: 10.2144/000113801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc. Natl Acad. Sci. USA. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ikeuchi K, Izawa T, Inada T. Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front. Genet. 2018;9:743. doi: 10.3389/fgene.2018.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simms CL, Thomas EN, Zaher HS. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip. Rev. RNA. 2017;8:e1366. doi: 10.1002/wrna.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Graille M, Seraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat. Rev. Mol. Cell Biol. 2012;13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- 77.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D’Orazio KN, et al. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. Elife. 2019;8:e49117. doi: 10.7554/eLife.49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hayakawa H, et al. Binding capacity of human YB-1 protein for RNA containing 8-oxoguanine. Biochemistry. 2002;41:12739–12744. doi: 10.1021/bi0201872. [DOI] [PubMed] [Google Scholar]
- 80.Ishii T, Hayakawa H, Sekiguchi T, Adachi N, Sekiguchi M. Role of Auf1 in elimination of oxidatively damaged messenger RNA in human cells. Free Radic. Biol. Med. 2015;79:109–116. doi: 10.1016/j.freeradbiomed.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 81.Ishii T, Hayakawa H, Igawa T, Sekiguchi T, Sekiguchi M. Specific binding of PCBP1 to heavily oxidized RNA to induce cell death. Proc. Natl Acad. Sci. USA. 2018;115:6715–6720. doi: 10.1073/pnas.1806912115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 85.Adachi H, De Zoysa MD, Yu YT. Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:230–239. doi: 10.1016/j.bbagrm.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Penzo M, Guerrieri AN, Zacchini F, Trere D, Montanaro L. RNA pseudouridylation in physiology and medicine: for better and for worse. Genes (Basel) 2017;8:301. doi: 10.3390/genes8110301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kariko K, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakamoto MA, Lovejoy AF, Cygan AM, Boothroyd JC. mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii. RNA. 2017;23:1834–1849. doi: 10.1261/rna.062794.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hussain S, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013;31:458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang X, et al. 5-Methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen X, et al. 5-Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019;21:978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 95.Yang Y, et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell. 2019;75:1188–1202 e1111. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 96.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Amort T, et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18:1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X, et al. The tRNA methyltransferase NSun2 stabilizes p16INK(4) mRNA by methylating the 3’-untranslated region of p16. Nat. Commun. 2012;3:712. doi: 10.1038/ncomms1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arango D, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–1886. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sharma S, et al. Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 2017;13:e1006804. doi: 10.1371/journal.pgen.1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elliott BA, et al. Modification of messenger RNA by 2’-O-methylation regulates gene expression in vivo. Nat. Commun. 2019;10:3401. doi: 10.1038/s41467-019-11375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Picard-Jean F, et al. 2’-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO. PLoS ONE. 2018;13:e0193804. doi: 10.1371/journal.pone.0202308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang LS, et al. Transcriptome-wide mapping of internal N(7)-methylguanosine methylome in mammalian mRNA. Mol. Cell. 2019;74:1304–1316. e1308. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malbec L, et al. Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Res. 2019;29:927–941. doi: 10.1038/s41422-019-0230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]