Dear Editor,
Multiple myeloma (MM) is a clonal plasma cell disorder whose prognosis is driven by the presence or absence of a wide gamut of primary (trisomies and translocations) and secondary (monosomies/deletions and amplifications) genetic abnormalities. Translocations involving the immunoglobulin heavy chain (IgH) region on chromosome 14 and one of its partners on chromosomes 4, 6, 11, 16 and 20 occur in approximately 40% of MM patients. Among the IgH translocations, t(11;14) is the most commonly observed lesion (occurring in 16–24% of MM patients), while t(14;16) is rather uncommon (<5% of patients, in most reported series)1–3. The International Myeloma Working Group (IMWG)4,5 and the m-SMART6 risk-stratification currently consider t(11;14) as a standard-risk chromosomal abnormality7, hile t(4;14), t(14;16) and t(14;20) have been associated with poor survival, although the data on the prognostic impact of t(14;16) in the era of newer anti-myeloma drugs have not been extensively reported and few retrospective studies have reported conflicting results on the prognostic significance of this abnormality1,8,9. t(14;16)(q32;q23) involves the IgH locus and the c-musculoaponeurotic fibrosarcoma (c-MAF) oncogene locus, the latter likely being responsible, at least partially, of the MAF-mediated innate resistance to proteasome inhibition, a backbone treatment in current myeloma regimens10.
Having considered the availability of newer combination regimens and the recent outcomes in the treatment of MM, we conducted a multi-institutional collaborative study among patients harboring t(14;16) diagnosed at different European and American centers, with the aim of describing their clinical features and investigating the prognostic value of t(14;16).
Newly diagnosed (ND)MM patients meeting the IMWG criteria for symptomatic MM were included in the analysis. Patients were diagnosed between December 2006 and March 201711,12 and registered in databases of: clinical trials coordinated by the Italian group and with centralized fluorescent in situ hybridization (FISH) at the Torino site (TO, Italy; see Table S1 for the list of source trials with centralized FISH at TO); the Winship Cancer Institute (WCI, Emory University, Atlanta US); and the Department of Clinical Therapeutics, National and Kapodistrian University of Athens (NKUA, Greece). Data regarding demographic and baseline characteristics, treatments administered, responses to treatment and survival outcomes were collected from the institutional review board databases approved by the respective centers. Response to treatment and disease progression were assessed using the IMWG uniform response criteria13,14.
FISH analysis was performed on CD138+ enriched bone marrow plasma cells; the cut-off levels ranged from 10 to 20% for numerical aberrations and from 5 to 15% for IgH translocations.
Progression-free survival (PFS) was calculated from the date of initial therapy to either the date of the first relapse or death due to any cause; overall survival (OS) was calculated from the date of initial therapy to the date of death. PFS and OS were estimated using the Kaplan–Meier method and compared between groups using the log-rank test. We used the Cox proportional hazards model to identify factors affecting PFS and OS. A two-tailed P-value < 0.05 was considered significant for all statistical tests. Data were analyzed as of April 2018 using R (v3.1.1).
We identified 123 NDMM patients with t(14;16)-positive FISH at diagnosis (TO, n = 76/1678; WCI, n = 29/926; NKUA, n = 18/571) with a median age at diagnosis of 66 years (range, 38–87 years; see Table S2. Patient characteristics at diagnosis). Twenty-three percent of patients had lactate dehydrogenase (LDH) levels above the upper limit of normal; creatinine ≥2 mg/dl and hypercalcemia were observed in 16% of patients. Most of the patients presented with International Staging System (ISS) stages 2 (34%) or 3 (43%) and with concomitant chromosomal abnormalities, including del(13q) (71%), gain(1q) (51%), and del(17p) (23%).
All but one patient received novel agents as part of induction therapy, including immunomodulatory drugs (42%), proteasome inhibitors (29%) or a combination of both (28%) (Table S2). Autologous stem-cell transplantation (ASCT) was performed in 42% of patients, 28% upfront and 14% at relapse; 41% of patients received maintenance therapy.
The objective response rate to first-line treatment was 85% (partial response [PR]: 26%; very-good PR [VGPR]: 29%; at least complete response [≥CR]: 28%); 5% of patients were primary refractory to induction therapy.
After a median follow-up of 53 months (95% CI 35–63), median PFS and OS for the entire cohort were, respectively, 19 months (95% CI 16–30) and 53 months (95% CI 36–63), with 39% of patients being alive at 5 years.
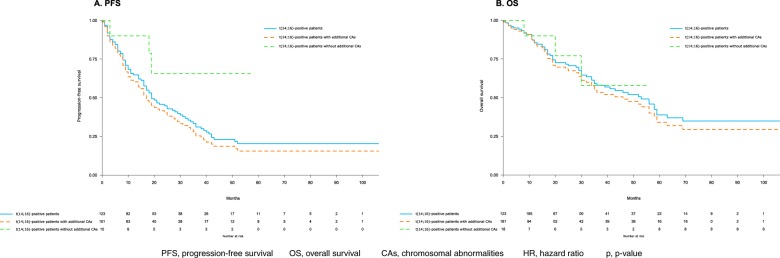
t(14;16)-positive patients with del(17p), del(13q), or amp(1q) (n = 101) had significantly shorter median PFS (17 months vs. NR; HR: 3.33, p = 0.04) and inferior, although not statistically significant, OS (median, 46 months vs. NR; HR: 1.54, p = 0.47), as compared to those without additional chromosomal abnormalities (n = 10) (Fig. 1). More specifically, the risk of progression or death of t(14;16) patients increased by 1.35 fold in patients also harboring del(17p) (p = 0.23), by 1.64 in presence of del(13q) (p = 0.05) and by 2.2 in patients carrying amp(1q) (p = 0.02); a shorter OS, although not statistically significant, was observed in presence of del(17p) (HR: 1.3; p = 0.47) and amp(1q) (HR: 1.7; p = 0.16), whereas a significantly shorter OS was observed with del(13q) (HR: 1.96; p = 0.04).
Fig. 1. Time-to-event analysis.
Kaplan–Meier progression-free survival (a PFS progression-free survival) and overall survival (b OS overall survival) curves in t(14;16)-positive patients. Blue curve: t(14;16)-positive patients overall; orange curve: t(14;16)-positive patients with additional CAs chromosomal abnormalities [del(17p) and/or del(13q) and/or amp(1q)]; green curve: t(14;16)-positive patients without additional CAs.
Among younger, ASCT-eligible patients, median PFS and OS were, respectively, 24 (95% CI 17–38) and 52 months (95% CI 34–69); ASCT upfront prolonged median PFS (36 vs. 17 months; HR: 0.5, p = 0.036), as compared to standard-dose chemotherapy without ASCT. Maintenance therapy significantly prolonged median PFS, as compared to no maintenance (36 vs. 19 months, HR: 0.59; p = 0.045). The achievement of ≥CR as best response to treatment was related to prolonged PFS (HR 0.29, p < 0.001), as compared to the achievement of PR/VGPR.
In a multivariate analysis, ISS-3 (HR: 1.79; p = 0.014) and the presence of an additional chromosomal abnormality (del(17p), del(13q) or amp(1q); HR: 3.24; p = 0.049) were associated to a shorter PFS, while ISS-3 (HR: 1.6; p = 0.09), hypercalcemia (HR: 2.4; p = 0.043) and elevated LDH (HR: 2.13; p = 0.026) were associated with a significantly shorter OS (see Table S3. Univariate and multivariate analyses).
To our knowledge, this is one of the largest studies on NDMM patients harboring t(14;16) describing their clinical features and survival outcomes in the era of novel agents.
In a previous report from the Mayo Clinic (2003), the presence of t(14;16) was associated with shorter PFS (median, 9 vs. 30 months) and OS (median, 16 vs. 41 months), as compared to t(14;16)-negative MM (see Table S4. Previous reports of t(14;16))1. However, that analysis included a limited number of patients (n = 15) before the introduction of novel agents. These results were not confirmed in a larger cohort of patients (n = 1003) treated by the Intergroupe Francophone du Myélome (IFM): here, t(14;16) was detected in 32 patients and was not prognostically significant9. Nonetheless, the IMWG listed t(14;16) among the unfavorable high-risk chromosomal abnormalities7. Narita et al. showed that both PFS (median, 0.6 vs. 1.2 years) and OS (median, 3.06 vs. 4.40 years) were significantly shorter among t(14;16)-positive patients (N = 35) than among t(14;16)-negative patients (N = 124)8.
According to our analysis, the majority of t(14;16) patients presented at diagnosis with at least one other high-risk feature, such as additional chromosomal abnormality (81%), ISS-3 (43%) and elevated LDH (23%), which were all significantly associated with inferior survival (Table S3).
Median PFS and OS for the entire cohort were 19 and 53 months. Although this study does not include a control population, the median OS of t(14;16) patients was shorter than that observed in a cohort of patients treated with novel agents (median, 72 months)15. Interestingly, t(14;16)-positive patients who harbored additional chromosomal abnormalities [del(17p), del(13q), or amp(1q)] displayed worse PFS (HR: 3.33) and OS (HR: 1.54), as compared to t(14;16) patients without further chromosomal abnormalities (Table S3). Despite the limited number of patients, this observation casts doubt on the unfavorable prognostic significance of isolated t(14;16), which seems to occur infrequently, and raises the question of whether the poor prognosis of these patients is related to t(14;16) per se rather than to the presence of additional genetic lesions.
Our analysis also confirmed the role of upfront ASCT in prolonging PFS and OS and the role of maintenance treatment in deepening the quality of response and prolonging PFS as compared to fixed-duration therapy.
This study has some limitations. First, the absence of a control population limits our ability to precisely estimate the risk conferred by the presence of t(14;16) in the era of novel agents. Secondly, the heterogeneity of treatment therapies does not allow us to speculate on the efficacy of specific regimens. Finally, since 62% of patients in this cohort were enrolled in clinical trials, the rates of patients with renal failure or aggressive disease requiring immediate treatment were underestimated and consequently affected the results. Despite these caveats, PFS and OS of t(14;16) patients in the era of novel agents seem to be shorter than those of standard-risk patients15. Whether the poor prognosis of t(14;16) patients is associated with t(14;16) per se or with the frequent co-existence of other high-risk features is an issue that needs to be addressed.
Supplementary information
Author contributions
R.M., F.G., A.K.N. and M.A.D. conceived and designed the work that led to the submission. All the authors acquired the data, and interpreted the results. R.M., F.G., A.K.N. and E.K. drafted the first version of the manuscript. All the authors revised the manuscript and approved the final version. All the authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of interest
R.M. has received honoraria from Amgen, Celgene, Takeda, and Janssen; has served on the advisory boards for Janssen. F.G. has received honoraria from Amgen, Celgene, Janssen, Takeda, and Bristol-Myers Squibb; has served on the advisory boards for Amgen, Celgene, Janssen, Takeda, Bristol-Myers Squibb, Roche, AbbVie, Adaptive, and Seattle Genetics. E.K. has received honoraria by Amgen, Genesis Pharma, Janssen, Takeda, and research grants from Amgen and Janssen. M.T.P. has received honoraria from and served on the advisory boards for Amgen, Bristol-Myers Squibb, Celgene, Janssen, and Takeda. J.L.K. has consulted for Roche, AbbVie, Janssen, BMS, Takeda, and Karyopharm. V.M. has received honoraria from Amgen, Celgene, Bristol-Myers-Squibb, and Janssen. F.P.: advisory role: Janssen, Celgene, MSD Italy; travel, accomodations, expenses: Celgene, Jazz, Medac. P.O. has served on the advisory board for Janssen. L.H.B. has received honoraria from AstraZeneca and undertaken consultancy for AbbVie and AstraZeneca. N.G. has received honoraria from Bristol-Myers Squibb, Celgene, Janssen; has served on the advisory boards for Amgen, Celgene, Takeda, Janssen; has received research funding from Celgene, Janssen; has received sponsorship for clinical trials from GlaxoSmithKline, Janssen, and Takeda. S.O. has received honoraria from Amgen, Celgene, and Janssen; has served on the advisory boards for Adaptive Biotechnologies, Janssen, Amgen, and Takeda. M.O. has received honoraria from Amgen, BMS, Celgene, Janssen, Takeda, and served on the advisory boards for Amgen, BMS, Celgene, Janssen, and Takeda. A.B. has served on the advisory boards for Amgen, Celgene, and Janssen. L.D.P. has received honoraria from and served on the advisory boards for Amgen, Janssen, Celgene, and Shire. E.T. has received honoraria from Amgen, Celgene, Janssen, Takeda, Bristol-Myers Squibb; has received research support from Amgen, Celgene, and Janssen. S.L. has consulted for Takeda, Celgene, Novartis, Janssen, GSK, Bristol-Myers Squibb, and Merck. M.B. has received honoraria from Sanofi, Celgene, Amgen, Janssen, Novartis, AbbVie, and Bristol-Myers Squibb; has received research funding from Celgene, Janssen, Amgen, Bristol-Myers Squibb, Mundipharma, Novartis, and Sanofi. A.K.N. has served on the advisory board for GSK, Spectrum, Celgene, Amgen, Novartis Pharmaceuticals, Adaptive Biotechnologies; has received honoraria from Bristol-Myers Squibb and Janssen Pharmaceuticals. M.A.D. has received honoraria from participation in advisory boards for Amgen, Celgene, Janssen, Bristol-Myers Squibb, and Takeda. The remaining authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41408-020-0307-4).
References
- 1.Fonseca R, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 2.Boyd KD, et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2012;26:349–355. doi: 10.1038/leu.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakshman A, et al. Natural history of t(11;14) multiple myeloma. Leukemia. 2018;32:131–138. doi: 10.1038/leu.2017.204. [DOI] [PubMed] [Google Scholar]
- 4.Chng WJ, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28:269–277. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo A, et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J. Clin. Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikhael JR, et al. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin. Proc. 2013;88:360–376. doi: 10.1016/j.mayocp.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Sonneveld P, et al. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127:2955–2962. doi: 10.1182/blood-2016-01-631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita T, et al. t(14;16)-positive multiple myeloma shows negativity for CD56 expression and unfavorable outcome even in the era of novel drugs. Blood Cancer J. 2015;5:e285–e285. doi: 10.1038/bcj.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avet-Loiseau H, et al. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood. 2011;117:2009–2011. doi: 10.1182/blood-2010-07-295105. [DOI] [PubMed] [Google Scholar]
- 10.Qiang Y-W, et al. MAF protein mediates innate resistance to proteasome inhibition therapy in multiple myeloma. Blood. 2016;128:2919–2930. doi: 10.1182/blood-2016-03-706077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyle RA, et al. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br. J. Haematol. 2003;121:749–757. doi: 10.1046/j.1365-2141.2003.04355.x. [DOI] [PubMed] [Google Scholar]
- 12.Rajkumar SV, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 13.Durie B, et al. International uniform response criteria for multiple myeloma. Leukemia. 2006;20:1467–1473. doi: 10.1038/sj.leu.2404284. [DOI] [PubMed] [Google Scholar]
- 14.Kyle RA, Rajkumar SV. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia. 2009;23:3–9. doi: 10.1038/leu.2008.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar SK, et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28:1122–1128. doi: 10.1038/leu.2013.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.