Abstract
Irradiation, or chemoradiotherapy, is a curative treatment for oropharyngeal squamous cell carcinoma (OPSCC). Its invasiveness, however, can often negate its efficacy. Therefore, developing methods to predict which patients would benefit from irradiation is urgent. Promoter DNA hypermethylation was recently reported to correlate with favorable OPSCC prognosis. It is still unclear, however, whether there is an association between promoter DNA methylation and response to irradiation. In this study, we analyzed DNA methylation in the specimens from 40 OPSCC patients who had undergone irradiation, using the Infinium assay. Our results showed significant correlation between high levels of promoter DNA methylation and better response to treatment (P < 0.01). We used the 10 most differentially‐methylated genes between responders and non–responders to develop a panel of predictive markers for efficacy. Our panel had high sensitivity, specificity and accuracy (92%, 93% and 93%, respectively). We conducted pyrosequencing to quantitatively validate the methylation levels of 8 of the 10 marker genes (ROBO1, ULK4P3, MYOD1, LBX1, CACNA1A, IRX4, DPYSL3 and ELAVL2) obtained by Infinium. The validation by pyrosequencing showed that these 8 genes had a high prediction performance for the training set of 40 specimens and for a validation set of 35 OPSCC specimens, showing 96% sensitivity, 89% specificity and 94% accuracy. Methylation of these markers correlated significantly with better progression‐free and overall survival rates, regardless of human papillomavirus status. These results indicate that increased DNA methylation is associated with better responses to irradiation therapy and that DNA methylation can help establish efficacy prediction markers in OPSCC.
Keywords: biomarker, DNA methylation, irradiation, oropharyngeal squamous cell carcinoma, precision medicine
We investigated the association between promoter DNA methylation and irradiation efficacy against oropharyngeal squamous cell carcinoma (OPSCC), so that an appropriate stratification by establishing molecular classifier markers could help therapeutic optimization and de–escalation of OPSCC treatment. A methylation marker panel was developed to act as an efficacy predictor with high sensitivity, specificity and accuracy in the training samples. It is noteworthy that the utility of the established prediction marker panel was consistent regardless of human papillomavirus status, and that multivariate analysis verified the methylation status of the marker panel as the only independent prognostic factor.

1. INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide. Human papillomavirus (HPV) infection is a risk factor for HNSCC, especially for oropharyngeal squamous cell carcinoma (OPSCC). The increase in HPV infections has led to a significant escalation in HNSCC cases.1, 2, 3 Irradiation, or chemoradiotherapy, is a potential treatment for HNSCC,4, 5 but its invasiveness often leads patients to stop treatment, causing a severe decline in quality of life.6, 7 Therefore, the optimization of irradiation therapy is an urgent issue.
Ang et al8 reported significantly better prognosis of HPV‐positive than HPV‐negative OPSCC patients, but they also suggested the prevalence of a different prognostic factor. Completed clinical trials have evaluated the therapeutic de–escalation of OPSCC therapy, but they are yet to obtain promising results.9, 10 These trials stratified OPSCC patients into two groups according to HPV status but did not consider any other molecular factors that might be expected to affect mortality or irradiation efficacy.11, 12 Therefore, an appropriate stratification of OPSCC patients and the development of classifier markers based on molecular biology could help to personalize OPSCC treatment.
Foy et al13 predicted the efficacy of radiation therapy using the 13‐gene expression‐based radioresistant score. They established the efficacy prediction score through in vitro experiments and information extracted from The Cancer Genome Atlas (TCGA) database but did not validate the predictive performance using clinical samples. Li et al14 successfully extracted candidate genes to predict the efficacy of radiotherapy using a comprehensive expression analysis of long non–coding RNA (lncRNA). The prediction performance of these genes, however, has not been verified. Although there have been attempts to predict therapeutic efficacy using approaches based on molecular biology, the stratification of OPSCC patients using molecular markers aimed at therapeutic de–escalation has not been established.
Promoter DNA methylation is a critical epigenetic mechanism for regulation of gene expression.15, 16, 17, 18 We previously conducted a comprehensive DNA methylation analysis using Infinium 450K and revealed differential methylation levels in HNSCC cases. In that report, we also showed a significant association between DNA methylation of gene promoters (eg RXRG) and better prognosis in HNSCC.19 Shen et al20 proposed a prognosis prediction algorithm based on methylome and transcriptome data from clinical specimens of oral squamous cell carcinoma. The results suggest that aberrant DNA methylation could help to predict HNSCC prognosis or the efficacy of the therapies against this disease.
In this study, we thus conducted a genome‐wide analysis of promoter DNA methylation in samples of OPSCC patients who had undergone irradiation using Infinium BeadArray. We established a panel of methylation marker genes to predict the efficacy of irradiation therapy with high accuracy. The panel was further validated using independent samples and pyrosequencing analysis to confirm the impact of DNA methylation on progression‐free survival (PFS) and overall survival (OS) after irradiation.
2. MATERIALS AND METHODS
Full information is provided in the Supporting Materials and Methods (Data S1).
2.1. Clinical samples
In this study, we analyzed 75 clinical OPSCC specimens (40 for the training set and 35 for the validation set).
For the training sample set, we enrolled 57 untreated OPSCC patients who visited Chiba University Hospital and received irradiation therapy (Figure 1). A total of 17 patients were excluded from this study because they were transferred to other hospitals, had undergone radical surgery before irradiation therapy, had interrupted treatment due to adverse events, or withdrew before treatment (Figure 1A). The specimens of the remaining 40 patients formed the training set. For the validation set, we enrolled 35 OPSCC patients who received irradiation therapy as first‐line treatment at Chiba University Hospital or Hamamatsu University Hospital (Figure 1B). The enrolled patients were all Asian.
Figure 1.
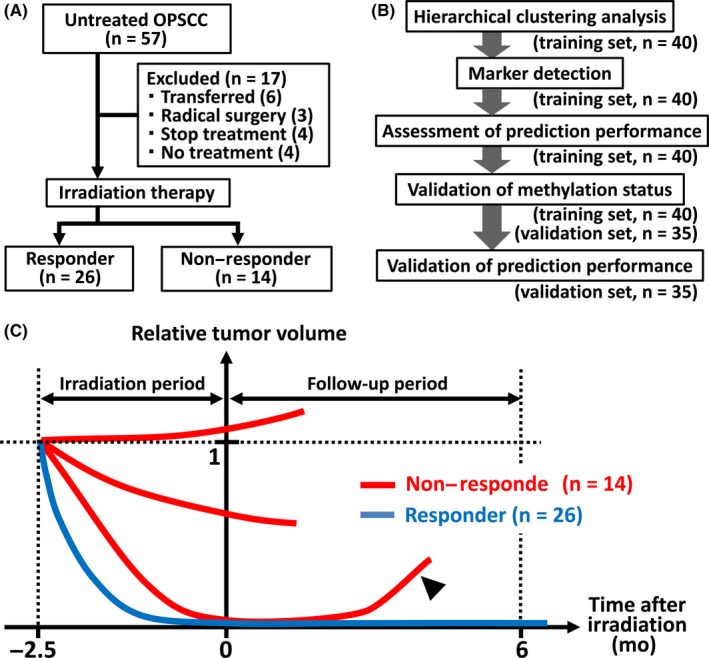
Study participants and design. A, Diagram of participant inclusion and exclusion criteria in the training sample set (n = 40). B, Flowchart of the study. C, Overview of the definition of therapeutic efficacy. The vertical axis represents total tumor volume and the horizontal axis represents the time lapse from the end of the irradiation period. Red and blue lines, non–responder and responder cases, respectively. Arrowhead, relapse including local or locoregional relapse and distant metastasis
We collected the following clinicopathological factors for all the patients enrolled in this study: age, gender, tumor site, HPV‐L1 status, clinical stage (T, N, stage based on UICC 8th edition21, 22), Brinkman index of 200 as smoking behavior,23, 24 irradiation dose and combination chemotherapy (Table S1). The 40 specimens of the training set were collected during biopsies performed before treatment, immediately frozen in liquid nitrogen and stored at −80°C. Among the 35 validation samples, 24 were frozen specimens and 11 were formalin‐fixed paraffin‐embedded (FFPE) specimens.
All the participants were classified as responders or non–responders. We defined responders as patients who reached complete remission (CR) status and maintained it for >6 months after irradiation therapy or those for whom we did not find viable cancer cells in the specimen collected during rescue surgery after irradiation. We defined non–responders as patients who did not reach CR, those who had viable cancer cells in the specimen collected during rescue surgery after irradiation or those who temporarily achieved CR but relapsed locally or by distant metastasis within 6 months after irradiation (Figure 1C).
Two independent pathologists microscopically examined the tumor cell content on the frozen or FFPE clinical specimens (Table S2). The specimens were dissected to enrich the tumor cells when necessary. We used 75 samples with >50% tumor cell content for subsequent molecular analyses. DNA was extracted using a QIA Quick DNA Mini Kit (Qiagen) for frozen material and a QIAamp DNA FFPE Tissue Kit (Qiagen) for FFPE material. The institutional review boards at Chiba University and Hamamatsu University approved the study protocol.
2.2. Amplification of L1 DNA region in high‐risk human papillomavirus
We evaluated HPV infection by amplifying a portion of the L1 region of high‐risk HPV, including HPV 16, 18, 31 and 33 in the 40 training and 35 validation samples, using GP5 and GP6 primers as previously reported.25 Positive PCR amplification was designated as HPV‐L1(+).
2.3. Infinium assay
The Infinium assay of the training sample set was previously performed and the data were registered at GEO [accession number GSE124633]. The Infinium HumanMethylation450 BeadChip (Illumina) contains approximately 485 577 individual CpG sites. The β‐value, ranging from 0.00 to 1.00, was measured by a methylated probe relative to the sum of the methylated and unmethylated probes. The CpG score for each probe was calculated based on previous reports.26, 27 Before analyzing the Infinium data, we extracted the upstream probe nearest to the transcription start site (TSS) of each gene (excluding sex chromosome genes) (n = 15 212) among 485 577 probes designed on the human genome. Among 15 212 probes, 10 048 (66%) were located in CpG islands and 2852 (19%) in CpG island shores, according to annotation by Illumina. Probes showing standard deviation (SD) >0.11 among the training samples (n = 2112) were extracted and used for the unsupervised two‐way hierarchical clustering analysis.
2.4. Gene Ontology analysis
The Gene Ontology (GO) enrichment analysis was conducted based on GO terms (biological process, cellular component and molecular function) using the Gene Annotation and Analysis Resource at Metascape (http://metascape.org/gp/index.html#/main/step1).
2.5. Pyrosequencing analysis
The quantitative validation of methylation level in the 40 training and 35 validation samples was performed by pyrosequencing using the PyroMark Q96 (Qiagen) as previously reported.27 Bisulfite conversion was performed using the Zymo EZ DNA Methylation Kit (Zymo Research) and 500 ng of genomic DNA for each sample. We amplified the promoter region covering the interested Infinium probe site and several surrounding CpG sites using bisulfite‐treated DNA as a template. The amplification primers were designed by Pyro Q‐CpG Software (Qiagen) and are shown in the Supporting Information (Table S3). We calculated the average methylation value of the analyzed CpG sites as the representative methylation level of the gene. We prepared methylation control samples (0%, 25%, 50%, 75% and 100%) as described previously,28, 29 and analyzed them to confirm the quantification quality of the pyrosequencing assays (Figure S1).
2.6. Statistical analysis
The association between clinicopathological factors and HPV‐L1 status or DNA methylation was analyzed using Fisher's exact test, the χ2 test and the Student's t test. Unsupervised two‐way hierarchical clustering was performed using Cluster 3.0 with the C Clustering Library version 1.59 software. We conducted the receiver operating characteristic (ROC) analysis in the training and validation sample sets to calculate the area under the curve (AUC) value. Progression‐free survival (PFS) was measured from the completion date of irradiation therapy until the date of any relapse (local, locoregional or distant metastasis) or death. OS was measured from the date of biopsy until the date of death. We estimated the PFS and OS distribution using the Kaplan‐Meier method, and the difference between groups was determined by a log‐rank test using R software (http://www.r-project.org/). The univariate and multivariate analyses were performed using the Cox proportional hazard model for PFS P < 0.05 was considered statistically significant.
3. RESULTS
3.1. Patient characteristics
Human papillomavirus status, clinical stage and combination chemotherapy were significantly different between the responder and non–responder groups in all 75 samples of the training and validation sets. HPV infection was significantly more frequent in the responder group (73%) than in the non–responder group (35%, P = 0.003). Lymph node metastasis was significantly less common in the responder group (67%) than the non–responder group (91%, P = 0.04). Significantly fewer patients in the responder group (12%) had clinical Stage IVA than in the non–responder group (48%, P = 0.002). Age and gender were not significantly associated with irradiation response. Smokers were frequently observed in both groups: 81% in the responder group and 74% in the non–responder group (P ≥ 0.5).
3.2. DNA methylation and Gene Ontology analyses
The unsupervised two‐way hierarchical clustering analysis of the 40 training samples classified them into two clusters: high‐methylation and low‐methylation groups. Responder samples were significantly more enriched in the high‐methylation group than in the low‐methylation group (23/27 vs 6/13, P = 0.02).
The two‐way hierarchical clustering divided the 2112 genes into three clusters (Figure 2):
A cluster of genes hypermethylated (mean β‐value >0.2) in both the high‐methylation and low‐methylation groups. This cluster included 722 genes, which showed considerably higher methylation levels in the high‐methylation group (Figure 2). The Infinium probes representing these genes were significantly enriched in either CpG islands or CpG island shores (651 of 722, 90%, P = 1 × 10−4). GO terms such as “neuron fate commitment,” “brain development” and “diencephalon development” were enriched in this cluster.
A cluster of genes hypermethylated in the high‐methylation group but not in the low‐methylation group. This cluster included 756 genes, and the Infinium probes representing these genes were significantly enriched in either CpG islands or CpG island shores (694 of 756, 92%, P = 2 × 10−7). They showed significant enrichment of GO terms, such as cell “morphogenesis,” “neuron differentiation” and “renal system development.”
A cluster of genes showing very high methylation levels in the high methylation and low‐methylation groups. This cluster included 634 genes, which did not show considerably higher methylation levels in the high‐methylation group. The Infinium probes representing these genes were less frequently located in either CpG islands or CpG island shores (446 of 634, 70%, P = 2 × 10−16). GO terms such as “cell adhesion,” “regulation of appetite” and “gene silencing by RNA,” were significantly enriched in this cluster.
Figure 2.
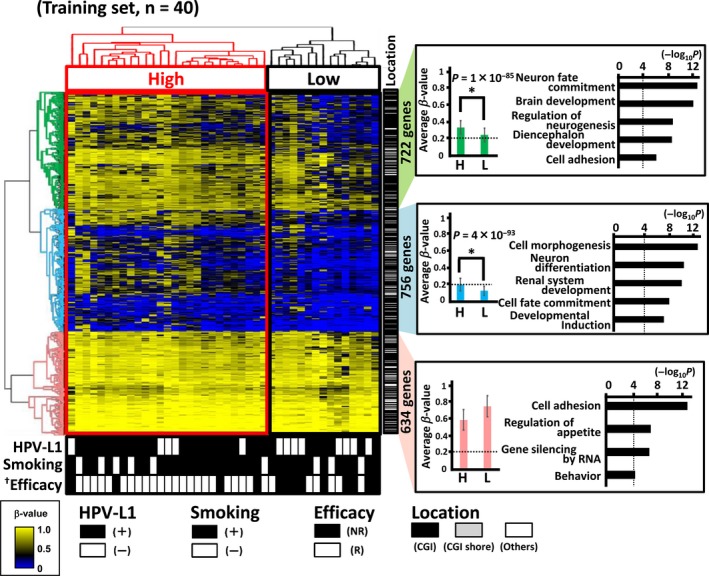
Infinium 450K analysis of oropharyngeal squamous cell carcinoma (OPSCC). The patients underwent curative irradiation therapy/concurrent chemoradiotherapy (including cetuximab‐based bioradiotherapy) as first‐line treatment. Unsupervised two‐way hierarchical clustering was performed based on Infinium 450K data, using 2112 genes with β‐values that highly deviated between the analyzed samples (SD > 0.11) (top left). The high methylated group were significantly associated with good response (P = 0.02). † P < 0.05 (Fisher's exact test). Closed box, HPV‐L1(+), smoking(+) (Brinkman index ≥200), and responder for irradiation therapy; open box, HPV‐L1(−), smoking(−) (Brinkman index <00) and non–responder for irradiation therapy (bottom). Location of Infinium probes is shown at the center: closed bar, CpG island (CGI); grey bar, CGI shore; open bar, other region. Three gene clusters showing different methylation levels between high‐methylation and low‐methylation groups (right). *Significantly hypermethylated in the high‐methylation group. The terms of significantly enriched categories in the GO analysis are shown. High or H, high‐methylation group; Low or L, low‐methylation group; NR, non–responder; R, responder
3.3. Extraction of prediction markers for irradiation response
Because the hierarchical clustering revealed a significant correlation between promoter DNA methylation and irradiation efficacy, we tried to extract prediction markers of therapeutic efficacy by comparing the methylation status of each gene in the responder and non–responder groups. A total of 92 genes had a significantly different methylation status in the two groups (P < 0.01). Overall, 87 of the 92 genes (95%) showed significantly higher methylation levels in the responder group. The residual 5 genes (5%) showed higher methylation levels in the non–responder group, presumably regarded as noise in the genome‐wide analysis (Figure 3A). GO terms such as “cell adhesion” and “regulation of cell proliferation” were significantly enriched in the 87 genes with higher methylation levels in the responder group. Genes associated with these GO terms included ROBO1, FOXA2, LBX1 and SHISA6 (Figure 3A). We extracted the 10 genes (ROBO1, FOXA2, ULK4P3, MYOD1, LBX1, CACNA1A, SHISA6, IRX4, DPYSL3 and ELAVL2), with the most significant P‐values between the responder and non–responder groups to serve as our candidate prediction markers of therapeutic efficacy (Figure 3A,B). All the Infinium probes representing these 10 marker genes were located within either CpG islands or CpG island shores, and at 0‐200 base pairs upstream from the TSS.
Figure 3.
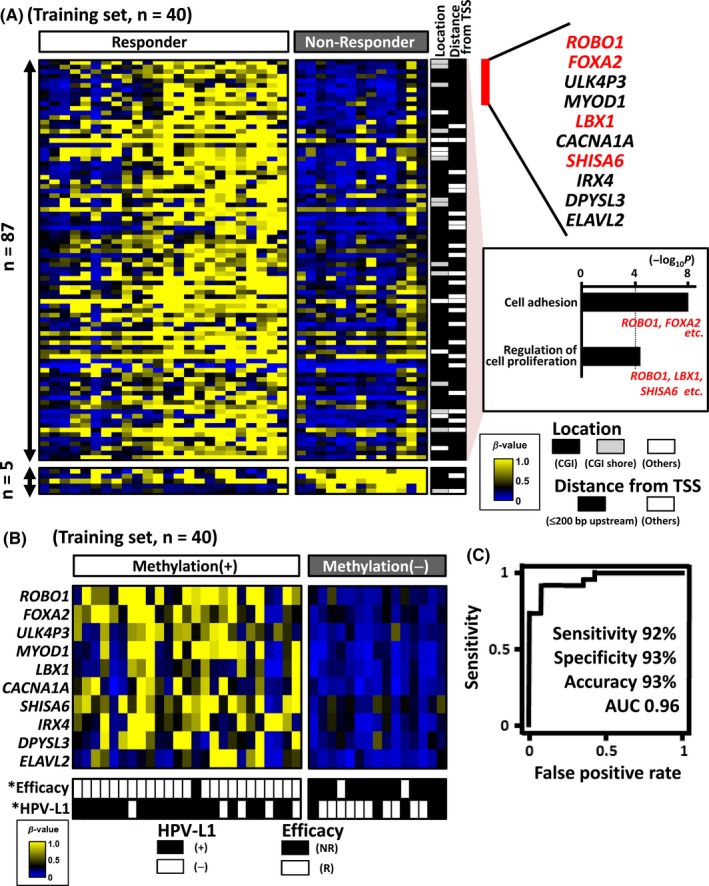
Extraction of prediction markers for irradiation response using 40 training samples. A, Comparison of methylation status between responders and non–responders in the training sample set (n = 40). Genes with significantly different methylation status (P < 0.01) were extracted (n = 92) (left). Location of Infinium probes is shown at the center: closed bar, CpG island (CGI); grey bar, CGI shore; open bar, other region. Distance from the TSS is also shown: closed bar, 0‐200 base pairs upstream from the TSS; open bar, other region. The GO analysis identified 87 significantly hypermethylated genes in the responder group; GO terms such as “cell adhesion” and “regulation of cell proliferation” were significantly enriched (right). Among the genes significantly methylated in the responder group, we extracted the most significant 10 genes as candidate efficacy prediction markers. B, Efficacy prediction performance of irradiation therapy based on extracted marker candidates. The methylation(+) group was significantly associated with the responder (P = 7 × 10−7) and HPV status (P = 0.006). *P < 0.01 (Fisher's exact test). C, A receiver operating characteristic (ROC) curve was drawn using β‐values and therapeutic efficacy of the training samples. The sensitivity, specificity and accuracy were 92%, 93% and 93%, respectively, where β‐value = .28 maximized the accuracy of the efficacy prediction
Among the 40 samples of the training set, 25 showed hypermethylation (β‐value >0.2) in at least 1 of the 10 candidate marker genes, and significantly correlated with responder cases (24/25 vs 2/15, P = 7 × 10−7). To assess the efficacy prediction performance of the 10 markers for the training samples, we performed ROC analysis using the irradiation efficacy as an objective variable and the minimum β‐value of the 10 genes in each sample as an explanatory variable. The AUC value was 0.96, and the sensitivity, specificity and accuracy were 92%, 93% and 93%, respectively, with a β‐value threshold of 0.28 maximizing the accuracy of the efficacy prediction.
3.4. Validation of promoter DNA methylation status by pyrosequencing
To quantitatively verify the methylation status observed by the Infinium analysis, we conducted a pyrosequencing analysis for the candidate markers using the 40 training samples. Pyrosequencing primers were successfully generated for 8 genes (ROBO1, ULK4P3, MYOD1, LBX1, CACNA1A, IRX4, DPYSL3 and ELAVL2), for which methylation control samples were quantified with a correlation coefficient R 2 > .98 (Figure S1). We could not generate appropriate primers for the remaining two genes. To confirm prediction performance using these 8 successful markers only, ROC analysis was performed with Infinium data of these genes. The AUC value reached 0.97, and the sensitivity, specificity and accuracy were 92%, 92% and 92%, respectively, indicating that this 8‐gene panel is comparable in prediction performance to the 10‐gene panel (Figures 3 and 4).
Figure 4.
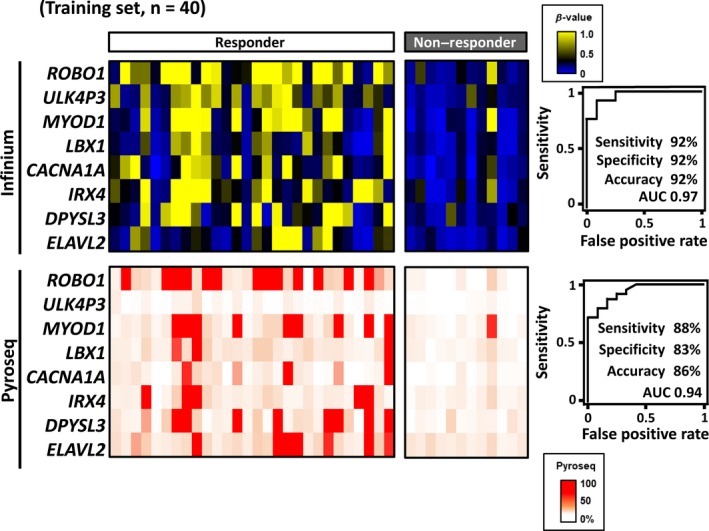
Validation of promoter DNA methylation status by pyrosequencing. The methylation levels of candidate marker genes analyzed by Infinium for the 40 training samples are shown on the color scale (top left). Validation by quantitative methylation analysis was performed using pyrosequencing. Pyrosequencing primers for 8 of the 10 markers could be generated (bottom left). Receiver operating characteristic curves were drawn using Infinium and pyrosequencing data of the 8 markers (right)
Then, we validated the methylation levels of these 8 genes against the training sample set quantitatively by pyrosequencing. The methylation levels by pyrosequencing were confirmed to be similar to those by Infinium, and the ROC analysis resulted in an AUC value of 0.94, and sensitivity, specificity and accuracy of 88%, 83% and 86%, respectively (Figure 4).
3.5. Validation of the prediction performance
We next performed pyrosequencing to detect the methylation status of the 8 markers using 35 additional samples, which were independent of the training set. We also analyzed the correlation between methylation status and irradiation efficacy in the validation sample set (Figure 5). The threshold methylation level was set at 28%, which was indicated as best in the training sample set. The 35 validation samples were classified into 26 methylation(+) and 9 methylation(−) samples, and there was a significant correlation between the methylation(+) and responder groups (25/26 vs 1/9, P = 3 × 10−6). When we performed the ROC analysis for the 35 validation samples using the 8 marker genes, the AUC value reached 0.98, and the sensitivity, specificity and accuracy were 96%, 89% and 94%, respectively (Figure 5A). We further analyzed the prediction performances by adjusting the methylation level using the tumor cell content of each sample (Table S2 and Figure S2). When adjusted, the accuracy for the efficacy prediction was slightly elevated. Adjustment using tumor cell content might improve the prediction performance, although it could be too complicated in clinical settings and accuracy was as high as 94% without adjustment (Figure 5A).
Figure 5.
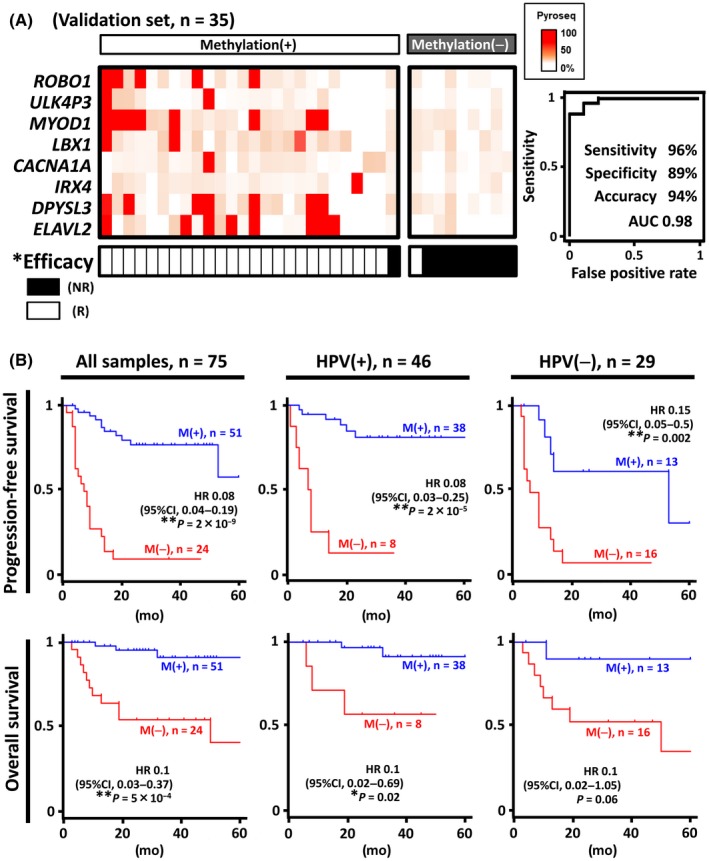
Validation of prediction performance using 35 validation samples. A, The significant correlation between the marker panel status and the therapeutic efficacy with the high‐accuracy prediction performance was verified by pyrosequencing of the 8 markers in the validation sample set. B, Kaplan‐Meier curves of progression‐free survival (PFS) and overall survival (OS). PFS and OS were analyzed using 75 oropharyngeal squamous cell carcinoma (OPSCC) samples, showing a markedly favorable outcome in the methylation(+) group compared to the methylation(−) group. Even when HPV(+) and HPV(−) patients were analyzed separately, the methylation(+) group showed significantly better outcomes, regardless of human papillomavirus (HPV) status (see also Figure S3). **P < 0.01 (Fisher's exact test); *P < 0.05 (Fisher's exact test); NR, non–responder; R, responder; M(+), the methylation(+) group; M(−), the methylation(−) group; HR, hazard ratio
Finally, we compared PFS between the methylation(+) and methylation(−) groups defined by the panel of 8 marker genes. We analyzed the 75 samples of both sets and found that the hazard ratio (HR) (95% confidence interval [CI]) of the methylation(+) group relative to the methylation(−) group was 0.08 (0.04, 0.19). PFS was significantly higher in the methylation(+) group (P = 2 × 10−9, log‐rank test) (Figure 5B). As HPV(+) OPSCC is known to correlate with better prognosis, HPV(+) samples in our cohort showed significantly favorable PFS than HPV(−) samples (P = 0.002) (Figure S3). When the 46 HPV(+) and 29 HPV(−) samples were analyzed separately, the HR of the methylation(+) group remained significantly lower than that of the methylation(−) group in both HPV(+) patients (P = 2 × 10−5) and HPV(−) patients (P = 0.002) (Figure 5B). The univariate analysis using the Cox proportional hazard model showed some clinicopathological factors such as HPV status (P = 0.002), clinical stage ⅣA and ⅣB (P = 0.002, P = 0.007, respectively), and the panel marker status (≥1 marker with methylation level >28%, P = 5 × 10−13) as prognostic factors for PFS (Table 1). We then performed the multivariate analysis using the Cox proportional hazard model, and the adjusted HR for critical clinicopathological factors showed that the panel marker status (P = 1 × 10−5) was the only independent prognostic factor for favorable PFS (Table 1, Figure S3). In addition, the Kaplan‐Meier curve of OS showed significantly better prognosis of the methylation(+) group comparted to the methylation(−) group (HR 0.1 [95%CI: 0.03‐0.4], P = 2 × 10−5) (Figure 5B, Figure S3).
Table 1.
Analysis of prognostic factors using Cox proportional hazard model
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P‐value | HR | 95%CI | P‐value | |
| Age (>60) | 1.1 | 0.4‐2.6 | 2.6 | |||
| Gender (male) | 0.9 | 0.2‐4.5 | 0.9 | |||
| Tumor site | ||||||
| Tonsils | 0.7 | 0.3‐1.6 | 0.4 | |||
| Base of tongue | 0.3 | 0.08‐1.4 | 0.1 | |||
| Soft palate | 5 | 1.8‐14 | 6 × 10−4* | 0.3 | 0.01‐9.3 | 0.5 |
| Posterior wall | 7.2 | 0.7‐72 | 0.1 | |||
| HPV | 0.3 | 0.2‐0.7 | 0.002 | 1 × 10−9 | 0‐∞ | 1.0 |
| Clinical N stage | ||||||
| N0 | 0.3 | 0.1‐1.1 | 0.1 | |||
| N1 | 0.3 | 0.1‐2 | 0.3 | |||
| N2 | 1.4 | 0.7‐3.3 | 0.3 | |||
| N3 | 2.7 | 0.9‐8.4 | 0.1 | |||
| Clinical stage | ||||||
| I | 0.1 | 0.02‐1.3 | 0.1 | |||
| II | 0.4 | 0.2‐0.9 | 0.03* | 34 | 0‐∞ | 1.0 |
| III | 1.2 | 0.4‐3 | 0.7 | |||
| IVA | 3.1 | 1.5‐6.7 | 0.002* | 5.9 | 0.5‐7 × 103 | 0.2 |
| IVB | 5.1 | 1.4‐20 | 0.007* | N/A | N/A | N/A |
| Smoking behaviour | ||||||
| BI ≥ 200 | Reference | |||||
| BI < 200 | 0.5 | 0.3‐1.3 | 0.2 | |||
| N/A | ||||||
| Irradiation dose (>60 Gy) | 0.9 | 0.1‐7 | 0.9 | |||
| Combination chemotherapy | ||||||
| None | 0.6 | 0.2‐2 | 0.4 | |||
| CDDP | 0.5 | 0.2‐1 | 0.1 | |||
| Cetuximab | 1.5 | 0.6‐3 | 0.3 | |||
| CDDP + 5‐Fluorouracil | 2.9 | 1.2‐6.7 | 0.008* | N/A | N/A | N/A |
| Marker status | 0.08 | 0.03‐0.2 | 5 × 10−13* | 0.1 | 0.03‐0.3 | 1 × 10−5* |
BI, Brinkman index; CDDP, cisplatin; CI, confidence interval; HPV, human papillomavirus; HR, hazard ratio; N/A, not applicable.
*P < 0.05.
4. DISCUSSION
In irradiation therapy against OPSCC, its invasiveness is considerable, while ongoing stratification by HPV status does not necessarily predict which patients the irradiation would benefit. Therefore, we investigated the association between promoter DNA methylation and irradiation efficacy in this study, so that an appropriate stratification by establishing molecular classifier markers could help therapeutic optimization and de–escalation of OPSCC treatment. A methylation marker panel was successfully developed to act as an efficacy predictor with high accuracy. It is noteworthy that the utility of the established prediction marker panel was consistent regardless of HPV status.
Gene ontology terms enriched in the 87 genes highly methylated in the responder samples included “cell adhesion” and “Ras signaling.” Notably, 3 of 8 marker genes (ROBO1, LBX1 and SHISA6) were associated with these GO terms. Zhou et al30 previously reported that ROBO1 expression is involved in liver metastasis and proliferation of colorectal carcinoma. Another study reported that inhibition of ROBO1 signaling promotes metastatic invasion in pancreatic ductal adenocarcinoma.31 Although the function of ROBO1 differs depending on the organ, this gene is involved in important mechanisms related to tumor progression. Previous experimental studies showed that LBX1 was involved in breast cancer metastasis.32 Another marker gene, MYOD1, was reportedly indirectly involved in MMP‐mediated cancer metastasis.33 Although the functions of ROBO1, LBX1 and MYOD1 in head and neck cancer have not been reported, it has been suggested that some marker genes might be involved in cancer pathologies such as metastasis and proliferation in this type of cancer. Therefore, some of the marker genes might be involved in metastasis or proliferation of cancer, leading to better prognosis after irradiation therapy, because irradiation therapy is presumably a curative treatment for the local area.
HPV(+) OPSCC is distinct, with p53 degradation and retinoblastoma pathway inactivation contributing to carcinogenesis.34, 35, 36 Recently, HPV was described as an activator of DNA methyltransferase (DNMT), which might play a role in aberrant DNA methylation in HPV‐associated cancers.37 This may explain in part why HPV‐positive samples correlated with the methylation(+) OPSCC (Figure 3B). HPV(+) OPSCC is reportedly more sensitive to irradiation therapy than HPV(−) OPSCC.8, 35, 38, 39 Therefore, we hypothesized that the significant correlation between the responder samples and the high methylation shown in this study might be due to HPV. However, the results from the PFS analysis for all samples showed a better PFS in methylation(+) samples than in methylation(−) samples, regardless of HPV status. In addition, the univariate analysis found that HPV status, marker status and clinical stage were prognostic factors for PFS. Surprisingly, multivariate analysis revealed the methylation status of the marker panel as the only independent prognostic factor for PFS. The established marker panel also distinguished the OS regardless of HPV status. These results suggested clinical importance of aberrant methylation status as well as a biological tumorigenic role.
This study has some limitations. First, the irradiation conducted in our cohort was all curative irradiation therapy, and the established marker panel exhibited high prediction performance. The performance against irradiation other than curative therapy (eg adjuvant or neo‐adjuvant irradiation), however, is yet to be evaluated. Second, we defined the non–responder patients as those whose cancer relapsed within 6 months after receiving irradiation therapy, as previously defined in the clinical trials that enrolled HNSCC patients with radio‐resistance or chemo‐resistance.40, 41 The marker panel established in this study could successfully predict patients with favorable outcomes for long periods (Figure 5B). If more favorable or unfavorable subgroups are desired for predictions for precision medicine, it may be necessary to screen larger cohorts using various definitions of non–responder patients, such as relapsing within longer or shorter periods. Further study is necessary to elucidate why DNA methylation markers exhibit the high prediction performance demonstrated in this study, as well as how high the performance could be improved.
In summary, we established a panel of gene markers to predict irradiation efficacy against OPSCC based on methylation status.
DISCLOSURE
The authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
We thank Eriko Ikeda, Haruka Maruyama and Tsubasa Matsusaka for technical assistance. We also thank Editage (http://www.editage.jp) for English language editing. This study was funded by the Japan Agency for Medical Research and Development (AMED; Practical Research for Innovative Cancer Control 17ck0106263h0001 to AK), the Japan Society for the Promotion of Science (JSPS; 19K18722 to TK), and a grant from Global and Prominent Research, Chiba University to AK.
Kurokawa T, Nakagawa T, Matsusaka K, et al. Establishment of epigenetic markers to predict irradiation efficacy against oropharyngeal cancer. Cancer Sci. 2020;111:1407–1416. 10.1111/cas.14338
REFERENCES
- 1. Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294‐4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mehanna H, Beech T, Nicholson T, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta‐analysis of trends by time and region. Head Neck. 2013;35:747‐755. [DOI] [PubMed] [Google Scholar]
- 3. Windon MJ, D'Souza G, Rettig EM, et al. Increasing prevalence of human papillomavirus‐positive oropharyngeal cancers among older adults. Cancer. 2018;124:2993‐2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous‐cell carcinoma of the head and neck. N Engl J Med. 2006;354:567‐578. [DOI] [PubMed] [Google Scholar]
- 5. Lefebvre JL, Pointreau Y, Rolland F, et al. Induction chemotherapy followed by either chemoradiotherapy or bioradiotherapy for larynx preservation: the TREMPLIN randomized phase II study. J Clin Oncol. 2013;31:853‐859. [DOI] [PubMed] [Google Scholar]
- 6. Wee JT, Anderson BO, Corry J, et al. Management of the neck after chemoradiotherapy for head and neck cancers in Asia: consensus statement from the Asian Oncology Summit 2009. Lancet Oncology. 2009;10:1086‐1092. [DOI] [PubMed] [Google Scholar]
- 7. Markert A, Thierry V, Kleber M, Behrens M, Engelhardt M. Chemotherapy safety and severe adverse events in cancer patients: strategies to efficiently avoid chemotherapy errors in in‐ and outpatient treatment. Int J Cancer. 2009;124:722‐728. [DOI] [PubMed] [Google Scholar]
- 8. Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mehanna H, Robinson M, Hartley A, et al. Radiotherapy plus cisplatin or cetuximab in low‐risk human papillomavirus‐positive oropharyngeal cancer (De‐ESCALaTE HPV): an open‐label randomised controlled phase 3 trial. Lancet. 2019;393:51‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gillison ML, Trotti AM, Harris J, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus‐positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non–inferiority trial. Lancet. 2019;393:40‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mirghani H, Blanchard P. Treatment de‐escalation for HPV‐driven oropharyngeal cancer: Where do we stand? Clin Transl Radiat Oncol. 2018;8:4‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wirth LJ, Burtness B, Nathan CO, Gregoire V, Richmon J. Point/counterpoint: Do we de–escalate treatment of HPV‐associated oropharynx cancer now? And how? Am Soc Clin Oncol Educ Book. 2019;39:364‐372. [DOI] [PubMed] [Google Scholar]
- 13. Foy JP, Bazire L, Ortiz‐Cuaran S, et al. A 13‐gene expression‐based radioresistance score highlights the heterogeneity in the response to radiation therapy across HPV‐negative HNSCC molecular subtypes. BMC Med. 2017;15:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li G, Liu Y, Liu C, et al. Genome‐wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next‐generation deep sequencing. BMC Cancer. 2016;16:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777‐786. [DOI] [PubMed] [Google Scholar]
- 16. Boscolo‐Rizzo P, Furlan C, Lupato V, Polesel J, Fratta E. Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: role of HPV and lifestyle factors. Clin Epigenetics. 2017;9:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20:590‐607. [DOI] [PubMed] [Google Scholar]
- 18. Zilberman D. The human promoter methylome. Nat Genet. 2007;39:442. [DOI] [PubMed] [Google Scholar]
- 19. Nakagawa T, Matsusaka K, Misawa K, et al. Frequent promoter hypermethylation associated with human papillomavirus infection in pharyngeal cancer. Cancer Lett. 2017;407:21‐31. [DOI] [PubMed] [Google Scholar]
- 20. Shen S, Wang G, Shi Q, et al. Seven‐CpG‐based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin Epigenetics. 2017;9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. James D, Brierley MKG, Wittekind C. Classification of Malignant Tumours , 8th edn Hoboken, NJ: John Wiley and Sons; 2017. [Google Scholar]
- 22. Lydiatt WM, Patel SG, O'Sullivan B, et al. Head and Neck cancers‐major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:122–137. [DOI] [PubMed] [Google Scholar]
- 23. Oba S, Inaba Y, Shibuya T, et al. Changes in oxidative stress levels during two weeks of smoking cessation treatment and their association with nutritional characteristics in Japanese smokers. Exp Ther Med. 2019;17:2757‐2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Umeda A, Kato T, Yamane T, et al. Does smoking cessation with varenicline worsen vascular endothelial function? BMJ Open. 2013;3:e003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mehlhorn G, Obermann E, Negri G, et al. HPV L1 detection discriminates cervical precancer from transient HPV infection: a prospective international multicenter study. Mod Pathol. 2013;26:967‐974. [DOI] [PubMed] [Google Scholar]
- 26. Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457‐466. [DOI] [PubMed] [Google Scholar]
- 27. Matsusaka K, Kaneda A, Nagae G, et al. Classification of Epstein‐Barr virus‐positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187‐7197. [DOI] [PubMed] [Google Scholar]
- 28. Florea AM. DNA methylation pyrosequencing assay is applicable for the assessment of epigenetic active environmental or clinical relevant chemicals. Biomed Res Int. 2013;2013:486072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Delaney C, Garg SK, Yung R. Analysis of DNA methylation by pyrosequencing. Methods Mol Biol. 2015;1343:249‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou WJ, Geng ZH, Chi S, et al. Slit‐Robo signaling induces malignant transformation through Hakai‐mediated E‐cadherin degradation during colorectal epithelial cell carcinogenesis. Cell Res. 2011;21:609‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gohrig A, Detjen KM, Hilfenhaus G, et al. Axon guidance factor SLIT2 inhibits neural invasion and metastasis in pancreatic cancer. Cancer Res. 2014;74:1529‐1540. [DOI] [PubMed] [Google Scholar]
- 32. Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial‐mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead‐like‐2. Cancer Res. 2013;73:6299‐6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Echizenya M, Kondo S, Takahashi R, et al. The membrane‐anchored MMP‐regulator RECK is a target of myogenic regulatory factors. Oncogene. 2005;24:5850‐5857. [DOI] [PubMed] [Google Scholar]
- 34. Narisawa‐Saito M, Kiyono T. Basic mechanisms of high‐risk human papillomavirus‐induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505‐1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marur S, D'Souza G, Westra WH, Forastiere AA. HPV‐associated head and neck cancer: a virus‐related cancer epidemic. Lancet Oncol. 2010;11:781‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taberna M, Mena M, Pavon MA, Alemany L, Gillison ML, Mesia R. Human papillomavirus‐related oropharyngeal cancer. Ann Oncol. 2017;28:2386‐2398. [DOI] [PubMed] [Google Scholar]
- 37. Duenas‐Gonzalez A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer. 2005;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gubanova E, Brown B, Ivanov SV, et al. Downregulation of SMG‐1 in HPV‐positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012;18:1257‐1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang C, Marsit CJ, McClean MD, et al. Biomarkers of HPV in head and neck squamous cell carcinoma. Cancer Res. 2012;72:5004‐5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferris RL, Blumenschein G Jr, Fayette J, et al. Nivolumab for recurrent squamous‐cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856‐1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bauml J, Seiwert TY, Pfister DG, et al. Pembrolizumab for platinum‐ and cetuximab‐refractory head and neck cancer: results from a single‐arm. Phase II Study. J Clin Oncol. 2017;35:1542‐1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


