Abstract
Cancer is a leading cause of death worldwide, and the incidence continues to increase. Despite major research aimed at discovering and developing novel and effective anticancer drugs, oncology drug development is a lengthy and costly process, with high attrition rates. Drug repositioning (DR, also referred to as drug repurposing), the process of finding new uses for approved noncancer drugs, has been gaining popularity in the past decade. DR has become a powerful alternative strategy for discovering and developing novel anticancer drug candidates from the existing approved drug space. Indeed, the availability of several large established libraries of clinical drugs and rapid advances in disease biology, genomics/transcriptomics/proteomics and bioinformatics has accelerated the pace of activity‐based, literature‐based and in silico DR, thereby improving safety and reducing costs. However, DR still faces financial obstacles in clinical trials, which could limit its practical use in the clinic. Here, we provide a brief review of DR in cancer and discuss difficulties in the development of DR for clinical use. Furthermore, we introduce some promising DR candidates for anticancer therapy in Japan.
Keywords: cancer, clinical trial, drug repositioning, intellectual property, Japan
We provide a brief review of drug repositioning (DR) in cancer and discuss difficulties in the development of DR for clinical use. Furthermore, we introduce some promising DR candidates for anticancer therapy in Japan.
1. INTRODUCTION
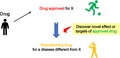
Drug repositioning (DR) is a strategy for drug development that seeks to establish new drugs from existing approved drugs by discovering novel effects or targets of the approved drugs.1 DR can be likened to finding a baseball player’s talent in an active soccer player (Figure 1). DR has generated increasing interest in the past decade owing to its use as an explicit development strategy and substantial benefits over the traditional development of new drugs (de novo drugs) by reducing development risks and facilitating the development process.1, 2, 3, 4, 5 Indeed, the number of publications related to DR in PubMed has increased rapidly since 2004.6 Moreover, the number of registrations of clinical trials for DR has increased in Japan (Figure 2).
Figure 1.
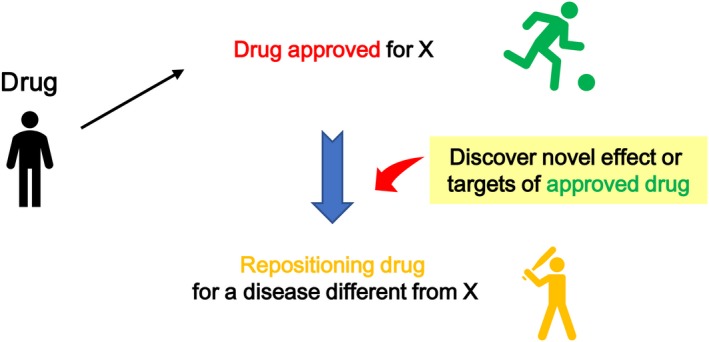
Drug repositioning: Discovery strategy of new drugs (baseball player) from existing approved drugs (soccer player)
Figure 2.
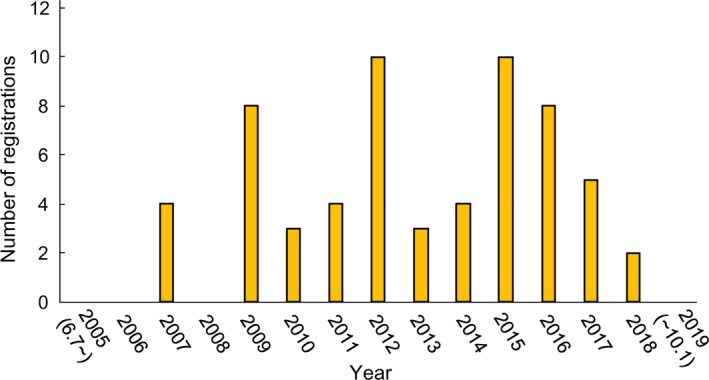
The number of registrations of clinical trials for drug repositioning in University Hospital Medical Information Network Clinical Trials Registry (UMIN‐CTR). UMIN‐CTR is one of the major registry systems of clinical trials in Japan
The discovery process for de novo drugs is costly and time‐consuming.7, 8, 9 Currently, on average, the cost for conventional de novo discovery and development of a drug is approximately US$2.5bn and requires around 10‐15 years for the drug to reach the market.7, 9 In Japan, the average time and costs to launch a drug de novo are 9.2 years and 55 billion yen, respectively.10 Moreover, only approximately 1 in 10 drugs entering phase I clinical trials is finally approved by the US Food and Drug Administration (FDA), and the proportion is decreased to 1 in 15 for drugs with an oncology indication in the USA.8
Repositioned drugs have already been used in humans and tested at various stages of drug development; therefore, such drugs have information available regarding safety, pharmacology and toxicology. In addition, later stages of the process, such as manufacturing and formulation, can be reused for the new drug product.5 In oncology, the demand for new therapies continues to increase, and DR could offer a faster and economically more attractive way of fighting malignancies.11 For example, the drug thalidomide, which was initially used to treat morning sickness but was found to cause malformations in newborns, was successfully repositioned for use in anticancer therapy owing to its anti–angiogenic and anti–inflammatory effects. Thus, thalidomide was approved by the FDA for combination treatment with dexamethasone in patients with multiple myeloma in the USA.11 This drug was also re–approved for use in patients with multiple myeloma and erythema nodosum leprosum in 2008 in Japan.
In this current report, we provide a brief review of DR in cancer and discuss difficulties in the development of DR for clinical use. In addition, we introduce some promising examples of DR for anticancer therapy in Japan.
2. APPROACH FOR DRUG REPOSITIONING
To efficiently reposition drugs that are already approved for human use, careful selection is required, followed by a thorough demonstration of the effectiveness of the drugs in other biological contexts. Here, we provide an overview of three representative methods used for the selection of effective testing and DR in cancer therapy: activity‐based, literature‐based and in silico DR.
2.1. Activity‐based drug repositioning
Activity‐based DR involves testing of actual drugs using in vitro or in vivo assays.12 Two public comprehensive drug libraries (ie, Johns Hopkins Clinical Compound Library (JHCCL) and NCGC Pharmaceutical Collection [NPC]) are often used for drug screening in these assays. JHCCL is the largest publicly accessible collection of existing drugs, with approximately 3100 available compounds, many of which are approved by the FDA or its foreign counterparts.11 NPC was built by the NIH Chemical Genomics Center (NCGC) and possesses 2500 small compounds that have been approved for clinical use by US (FDA), European (European Medicines Agency [EMA]), Japanese (National Health Insurance) and Canadian (Health Canada) authorities.13 Activity‐based DR can employ both protein target‐based and cell‐based screenings, which shed light on clinical trial outcomes. The antifungal agent itraconazole is a promising anticancer drug that has been explored through this approach using JHCCL.14
2.2. Literature‐based drug repositioning
This approach is used to identify licensed noncancer drugs with published evidence of anticancer activity using PubMed, MEDLINE and databases of clinical trials, such as ClinicalTrials.gov or University Hospital Medical Information Network Clinical Trials Registry (UMIN‐CTR), which is used mainly in Japan, chemical databases (DrugQuest) and other large‐scale databases.15 One example of successful use of literature‐based DR in cancer is the Repurposing Drugs in Oncology (ReDO) project, which was designed to rapidly identify new and effective cancer treatments characterized by low toxicity and cost‐effectiveness.16 Researchers successfully applied a literature‐based approach using all forms of published data to identify compounds to repurpose.16, 17
2.3. In silico drug repositioning
In silico DR utilizes public databases and bioinformatics tools to systematically identify interaction networks between drugs and protein targets.18 This approach has become successful with the large amounts of information on the structure of proteins (structural biology), genomics/transcriptomics/proteomics (omics) and pharmacophores that have accumulated in recent decades, along with the advancements in bioinformatics and computational science. Most pharmaceutical companies have already adopted in silico models for drug discovery from diverse chemical spaces.11 Using artificial intelligence algorithms and other bioinformatics tools, researchers have attempted to systematically identify interaction networks between drugs and protein targets.19 Clearly, in silico DR is a powerful technology with significant advantages, including speed and reduced costs.20, 21 Indeed, this approach has three main advantages: (a) robustness of algorithms for gene expression data analysis; (b) cost‐effectiveness; and (c) availability of freely accessible resources.22, 23 However, there are some limitations to this approach because it requires high‐resolution structural information for the targets. Moreover, information on the disease phenotype or gene expression profiles of drugs is necessary when a screen does not involve protein targets.11 In silico DR has been recently reviewed elsewhere in detail.24, 25, 26, 27
The main idea underlying several current methods, including in silico DR, is to identify genes whose expression levels are inversely correlated in the context of disease and drug treatment.28, 29, 30, 31, 32 These three approaches represent alternative and complementary approaches to the discovery of repositioned drugs. Notably, before conceiving any clinical use, in vitro and in vivo studies are essential to confirm predictions and to design clinical trials.
3. ADVANTAGES OF DRUG REPOSITIONING
Drugs still under patent but shelved after unsuccessful or partially successful clinical trials could have applications in DR. Indeed, for such drugs, many resources have already been spent to develop these drugs, and their use in similar or novel indications could lead to cost savings and avoidance of risks related to drug development. Furthermore, prior extensive investigations of the pharmacokinetic and pharmacodynamic characteristics of drugs can allow researchers to avoid studies assessing drug dosage, safety and toxicity.19 In a previous report, Pantziarka et al15 summarized the advantages of DR as follows: availability of pharmacokinetic, pharmacodynamics and posology data; knowledge of safety, toxicity and adverse events; clinical experience derived from the original indications; widespread availability worldwide; understanding of the mechanisms of action and molecular targets of the drug; and low cost. In most cases, DR can allow researchers to avoid carrying out preclinical tests and phase I trials, thereby reducing the cost of the drug development process (Figure 3). Some estimates have suggested that DR requires an average of 6.5 years and US$300m dollars to approve and launch a drug.10
Figure 3.
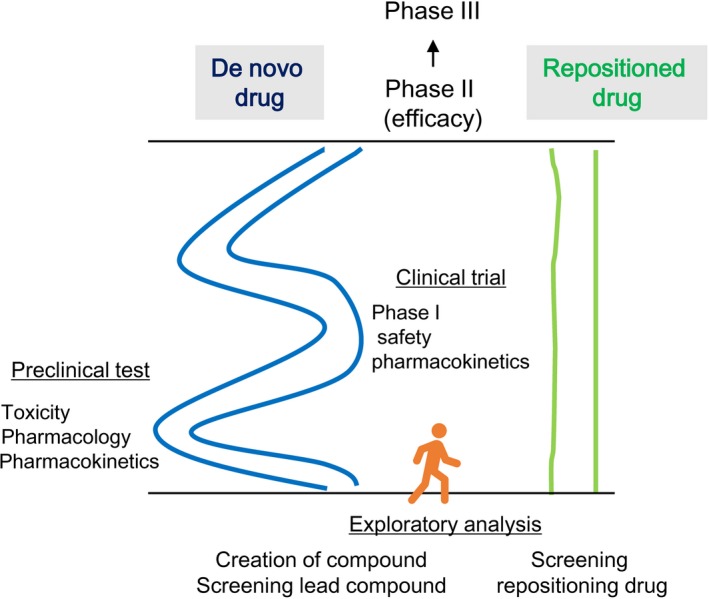
Advantage of drug repositioning (DR) over the development of de novo drugs. DR can be used to skip preclinical tests and phase I trials, thereby reducing costs throughout drug development
In addition to these advantages of repurposed drugs, repositioned drugs should be considered to treat patients when there are no approved medications or when a patient has exhausted all available treatment options.
4. LIMITATIONS
Despite the multiple advantages of DR, this approach still has some limitations. First, DR faces financial obstacles in that there are costs associated with licensing a drug for a new indication. The DR process involves intellectual property protection of the repositioned drugs, particularly for drugs that are off patents. Repositioned drugs are currently no longer patentable, resulting in low commercial interest from pharmaceutical companies. DR trials, particularly phase III efficacy trials, are, therefore, at a disadvantage in that they must rely on philanthropic sources for funding.
Next, in general, repurposed drugs are rarely effective in monotherapy, with anticancer activity more frequently observed in association with other repurposed drugs or known cytotoxic drugs.33 Furthermore, for novel applications, achieving sufficient efficacy by repositioned drugs sometimes requires treatment at higher doses, for an extended treatment period, or with a different formulation compared with the conventional indication. This may result in unexpected side effects, jeopardizing the idea of the “fast‐tracking” of the compound due to the necessity for full‐scale preclinical and clinical investigations.34
5. CLINICAL TRIALS OF DRUG REPOSITIONING IN JAPAN
For searching clinical trials of DR in Japan, we used the following search terms on 1 October 2019: “malignancy” AND “Japan” AND “interventional” using the website of UMIN‐CTR (https://www.umin.ac.jp/ctr/index-j.htm), which is a major registry system in Japan that has been recognized by the ICMJE as an “acceptable registry.” We then extracted clinical trials related to DR. Among all 7999 trials from June 7, 2005, to October 1, 2019, 63 trials were associated with DR. As shown in Figure 2, the number of registered clinical trials for DR has increased in Japan. We found 29 candidates of repositioned drugs in these 63 trials. These drugs are listed in Table 1. We checked the number of registrations of candidate repositioned drugs around the world using ClinicalTrials.gov (https://clinicaltrials.gov/ct2/home), which is a database of publicly and privately supported clinical studies of human participants conducted worldwide (Table 1). This database is provided by the US National Library of Medicine. To our knowledge, none of the 29 candidates have been clinically approved as repositioned drugs in the world (as of 1 October 2019). Among them, aspirin, zoledronic acid, metformin, atrial natriuretic polypeptide (ANP) and propagermanium (PG) are often subject to clinical trials as repositioned drugs in Japan. We briefly review these five drugs below.
Table 1.
Candidate repositioned drugs with anticancer effect in Japan in UMIN‐CTR
| Candidate drug | Effect as approved drug | Disease targeted by DR | Number of registrations | UMIN ID | Number of registrations (overseas)a |
|---|---|---|---|---|---|
| Aspirin | NSAID | CRC, FAP | 5 | 000031532, 000018736, 000018734, 000000698, 000000697 | 84 |
| Zoledronic acid | Osteoclastic inhibitor | BC, MPM, ATL, PrC, AML | 8 | 000008701, 000008093, 000007569, 000006736, 000003493, 000003261, 000002577, 000000796 | 62 |
| Nitroglycerin | Angina | NSCLC | 2 | 000001820, 000000813 | 7 |
| Menatetrenone | Hemostatic agent | HCC | 1 | 000001815 | 0 |
| Pipsissewa extract | Prostatic hyperplasia | Patients without PrC with high PSA | 1 | 000002126 | 0 |
| Candesartan cilexetil | Hypertension | HCC, PaC | 2 | 000002435, 000002152 | 0 |
| Metformin | Diabetes mellitus | Solid cancer, UC, PaC, LC | 7 | 000028405, 000020856, 000020681, 000020306, 000017694, 000004852, 000002210 | 163 |
| Branched‐chain amino acid | Hepatic encephalopathy | HCC | 1 | 000002330 | 0 |
| Celecoxib | NSAID (COX‐2 inhibitor) | HCC | 1 | 000002435 | 175 |
| Sodium valproate | Seizures | ESCC, PaC, BtC | 2 | 000010817, 000004525 | 63 |
| Atrial natriuretic polypeptide | Natriuretic peptide hormone | CRC,NSCLC, LC | 6 | 000025877, 000024302, 000019627, 000018851, 000018480, 000004880 | 0 |
| Irsogladine Maleate | Gastric mucosal protectant | NSCLC | 1 | 000005901 | 0 |
| Pioglitazone | Diabetes mellitus | HCC | 1 | 000007344 | 24 |
| Rifampicin | Antibiotic | HCC | 2 | 000007668, 000007667 | 29 |
| Mefenamic acid | NSAID | Head and neck cancer | 1 | 000007871 | 0 |
| Cilostazol | Antithrombotic agent | Head and neck cancer | 1 | 000007871 | 0 |
| Tranilast | Anti–allergy drug | Lymphatic tumor | 1 | 000008210 | 0 |
| Disulfiram | Alcoholism | HCC | 1 | 000008529 | 18 |
| Deferoxamine mesilate | Iron chelator | PaC | 1 | 000009054 | 0 |
| Sulfasalazine | Rheumatoid arthritis | NSCLC (non–SCC), GC | 2 | 000017854, 000010254 | 5 |
| Ribavirin | Antiviral agent | PrC | 1 | 000012521 | 21 |
| Azilsartan | Hypertension | PrC | 1 | 000012656 | 0 |
| Retinoic acid | Vitamin A | Neuroblastoma | 1 | 000013672 | 72 |
| Itraconazole | Antifungal medication | PaC, solid cancer | 3 | 000025398, 000018388, 000013951 | 49 |
| Propagermanium | Chronic hepatitis type B | PaC, BC, CRC, GC | 6 | 000027441, 000022494, 000022440, 000022129, 000017123, 000014689 | 0 |
| Mesalazine | Inflammatory bowel disease | FAP | 1 | 000018736 | 3 |
| Naftopidil | Prostatic hyperplasia | PrC | 1 | 000022862 | 0 |
| Cefadroxil | Bactericidal antibiotic | CML | 1 | 000027904 | 0 |
| Bezafibrate | Hyperlipidemia | NSCLC | 1 | 000029854 | 0 |
Abbreviations: AML, acute myeloid leukemia; ATL, adult T‐cell leukemia; BC, breast cancer; BtC, biliary tract cancer; CML, chronic myeloid leukemia; CRC, colorectal cancer; ESCC, esophageal squamous cell cancer; FAP, familial adenomatous polyposis; GC, gastric cancer; GyC, gynecologic cancer; HCC, hepatocellular cancer; LC, lung cancer; MPM, malignant pleural mesothelioma; NSAID, nonsteroidal anti–inflammatory drug; NSCLC, non–small cell lung cancer; PaC, pancreatic cancer; PrC, prostate cancer; UC, urological cancer.
The numbers are obtained using ClinicalTrials.gov.
5.1. Aspirin
Aspirin is the most widely used nonsteroidal anti–inflammatory drug worldwide and functions as a cyclooxygenase (COX)‐1 and COX‐2 inhibitor to suppress the conversion of arachidonic acid to prostaglandins and thromboxanes.35 COX‐2 promotes inflammation, pain and fever, and the development of colorectal cancer is often promoted by chronic inflammation caused by COX‐2 overexpression.36 Aspirin induces apoptosis via both COX‐dependent and COX‐independent mechanisms to combat malignancies.37 The details of clinical trials for aspirin are reviewed elsewhere.38
5.2. Zoledronic acid
Zoledronic acid, a bisphosphonate, is clinically used to treat osteoporosis and cancer‐related hypercalcemia and prevents skeletal complications due to bone metastases.39 Moreover, zoledronic acid inhibits osteoclastic bone resorption through negative regulation of the action of the farnesyl pyrophosphate synthase enzyme in the mevalonate pathway.12 Zoledronic acid has anticancer effects40, 41 by inhibiting cell proliferation, adhesion, invasion, angiogenesis and bone metastases, and shows immunomodulatory effects, possibly through inhibition of both the Ras/mitogen‐activated protein kinase and Akt/mammalian target of rapamycin (mTOR) pathways.42
5.3. Metformin
Metformin, an antidiabetic drug that is used for type 2 diabetes, lowers plasma glucose by suppressing hepatic gluconeogenesis, improving insulin sensitivity, and enhancing peripheral glucose uptake and utilization through activation of AMP‐activated protein kinase (AMPK). In the past decade, metformin has also been reported to exhibit anticancer activities. These anticancer effects are mainly mediated by activation of AMPK, which inhibits mTOR, a key mediator of the phosphatidylinositol 3‐kinase/Akt signaling pathway in tumor cells that activates the tumor suppressor p53, leading to cancer cell apoptosis.43, 44 Details of clinical trials of metformin are reviewed elsewhere.38, 45
5.4. Atrial natriuretic polypeptide
Atrial natriuretic polypeptide is a heart peptide hormone that belongs to a family of cardiac‐derived and vascular‐derived hormones with crucial roles in cardiovascular homeostasis (ie, regulation of blood volume and pressure).46, 47 In addition to its role in cardiovascular homeostasis, ANP has recently been shown to inhibit tumor growth in vitro and in vivo.48, 49, 50, 51, 52, 53, 54 Furthermore, Nojiri et al reported that mice pretreated with ANP exhibit dramatic reductions in lipopolysaccharide‐induced pulmonary metastasis in inoculated cancer cells, suggesting that ANP prevents cancer metastasis by suppressing the inflammatory reaction of endothelial cells, thereby inhibiting cancer cell adhesion to vascular endothelial cells.55 Thus, ANP may be a promising candidate for innovative anticancer therapy.56, 57 The anti–proliferative efficacy of ANP has been demonstrated in: pancreatic adenocarcinoma; breast, prostate, colon, renal, ovarian, small cell and squamous cell lung cancers; and other tumors.48, 49, 50, 51, 52, 53, 54 The JANP phase III clinical study is ongoing in Japan.58
5.5. Propagermanium
Propagermanium is an organic germanium compound that has been used clinically in Japan since 1994. PG is a safe therapeutic agent against hepatitis B virus e antigen (HBe)‐positive chronic hepatitis type B. Administration of PG decreases or eliminates HBe antigen and increases anti–HBe antibodies, resulting in seroconversion. Moreover, decreases in HBV‐DNA polymerase activity and HBV‐DNA content in blood have been reported.59 PG targets glycosylphosphatidylinositol‐anchored proteins that are closely associated with C‐C motif chemokine receptors (CCR) and selectively inhibit C‐C motif chemokine ligand (CCL) 2/CCR2 signaling.60 We recently showed that the CCL2/CCR2 signaling pathway in bone marrow‐derived stromal host cells promotes cancer metastasis via the formation of premetastatic niches.61 Furthermore, we showed that the administration of a CCL2 inhibitor, PG, blocked the enhancement of metastasis by inhibiting the formation of premetastatic niches in a mouse model.61 Based on these findings, we conducted a phase I dose‐escalation study of PG administration as an antimetastatic drug for perioperative patients with primary breast cancer and showed that PG could be given safely (UMIN000022494).62 In addition, Kikuchi et al demonstrated that PG has antitumor activity due to its effects on promoting natural killer cell maturation for patients with metastatic gastric or oral cancer.63 Details of PG are reviewed elsewhere.64
6. CONCLUSIONS
Drug repositioning is a growing endeavor supported by both the pharmaceutical industry and academia due to its advantages of cost savings and time savings relative to the traditional de novo drug development process. The EMA in Europe and the NIH and the FDA in the USA have already launched DR programs to identify new uses for existing pipeline medications developed by the pharmaceutical industry.17, 65 Furthermore, the ReDO project is an ongoing collaborative project that has focused exclusively on the potential use of approved noncancer medications as sources of new anticancer therapeutics.15, 66 An open‐access version is available online via http://www.redo-project.org/db. This website will be periodically updated to make information available to oncologists as new drugs are added to the database or as new data become available for drugs listed in the database.
Few bioventures support DR in Japan, although there are some DR candidates currently being evaluated in clinical trials in Japan, as described here. However, to promote DR in Japan, three Japanese pharmaceutical companies have started joint implementation of a drug discovery program, Joint Open INnovation of drUg repositioning (JOINUS), using a drug repositioning compound library constructed by these companies in 2017 (https://open-innovation-joinus.jp/joinus/). Nishimura et al described in detail the ongoing efforts to stimulate DR in Japan.10 Furthermore, the re–establishment of intellectual property for repositioned drugs may solve major problems of DR for practical use in patients.
Drug repositioning provides a rational, evidence‐based and cost‐effective approach for cancer treatment, although there are some practical limitations in clinical use. In Japan, the Clinical Trials Act was enacted in 2017 to establish procedures for the conduct of clinical trials, which may affect the number of new clinical trial registrations for DR candidates. We believe that DR is valuable and may result in the addition of new tools for use by oncologists.
DISCLOSURE
The authors have no conflicts of interest to declare.
ACKNOWLEDGEMENTS
We thank Dr Tyler Lahusen for helpful comments and English proofreading. This work was supported in part by the following grants and foundations: the Japan Agency for Medical Research and Development (18ae0101016, 18cm0106504, 18kk0205003 and 18ck0106259); the Japan Society for the Promotion of Science (JSPS) Grant‐in‐Aid for Science Research (19H03715, 19K09176, 19K18057, 18K08649 and 18K15323); JST AIP‐PRISM (JPMJCR18Y5); OITA Cancer Research Foundation; and Priority Issue on Post‐K computer (hp170227).
Masuda T, Tsuruda Y, Matsumoto Y, Uchida H, Nakayama KI, Mimori K. Drug repositioning in cancer: The current situation in Japan. Cancer Sci. 2020;111:1039–1046. 10.1111/cas.14318
REFERENCES
- 1. Langedijk J, Mantel‐Teeuwisse AK, Slijkerman DS, Schutjens M‐HDB. Drug repositioning and repurposing: terminology and definitions in literature. Drug Discov Today. 2015;20:1027‐1034. [DOI] [PubMed] [Google Scholar]
- 2. Sivapalarajah S, Krishnakumar M, Bickerstaffe H, et al. The prescribable drugs with efficacy in experimental epilepsies (PDE3) database for drug repurposing research in epilepsy. Epilepsia. 2018;59:492‐501. [DOI] [PubMed] [Google Scholar]
- 3. Mercorelli B, Palu G, Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol. 2018;26:865‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kakkar AK, Singh H, Medhi B. Old wines in new bottles: repurposing opportunities for Parkinson’s disease. Eur J Pharmacol. 2018;830:115‐127. [DOI] [PubMed] [Google Scholar]
- 5. Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673‐683. [DOI] [PubMed] [Google Scholar]
- 6. Shah RR, Stonier PD. Repurposing old drugs in oncology: opportunities with clinical and regulatory challenges ahead. J Clin Pharm Ther. 2019;44:6‐22. [DOI] [PubMed] [Google Scholar]
- 7. Paul SM, Mytelka DS, Dunwiddie CT, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203‐214. [DOI] [PubMed] [Google Scholar]
- 8. Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32:40‐51. [DOI] [PubMed] [Google Scholar]
- 9. Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534:314‐316. [DOI] [PubMed] [Google Scholar]
- 10. Nishimura Y, Tagawa M, Ito H, Tsuruma K, Hara H. Overcoming obstacles to drug repositioning in Japan. Front Pharmacol. 2017;8:729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shim JS, Liu JO. Recent advances in drug repositioning for the discovery of new anticancer drugs. Int J Biol Sci. 2014;10:654‐663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turanli B, Grøtli M, Boren J, et al. Drug repositioning for effective prostate cancer treatment. Front Physiol. 2018;9:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang R, Southall N, Wang Y, et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med. 2011;3:80ps16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chong CR, Sullivan DJ Jr. New uses for old drugs. Nature. 2007;448:645‐646. [DOI] [PubMed] [Google Scholar]
- 15. Pantziarka P, Verbaanderd C, Sukhatme V, et al. ReDO_DB: the repurposing drugs in oncology database. Ecancermedicalscience. 2018;12:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pantziarka P, Sukhatme V, Bouche G, Meheus L, Sukhatme VP. Repurposing drugs in oncology (ReDO)‐diclofenac as an anti–cancer agent. Ecancermedicalscience. 2016;10:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nowak‐Sliwinska P, Scapozza L, Altaba ARI. Drug repurposing in oncology: compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim Biophys Acta Rev Cancer. 2019;1871:434‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frattini V, Trifonov V, Chan JM, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141‐1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abbruzzese C, Matteoni S, Signore M, et al. Drug repurposing for the treatment of glioblastoma multiforme. J Exp Clin Cancer Res. 2017;36:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Z, Fang H, Reagan K, et al. In silico drug repositioning: what we need to know. Drug Discov Today. 2013;18:110‐115. [DOI] [PubMed] [Google Scholar]
- 21. Patel MN, Halling‐Brown MD, Tym JE, Workman P, Al‐Lazikani B. Objective assessment of cancer genes for drug discovery. Nat Rev Drug Discov. 2013;12:35‐50. [DOI] [PubMed] [Google Scholar]
- 22. Iorio F, Rittman T, Ge H, Menden M, Saez‐Rodriguez J. Transcriptional data: a new gateway to drug repositioning? Drug Discov Today. 2013;18:350‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cardone L. Biocomputing drug repurposing toward targeted therapies. Aging (Albany NY). 2016;8:2609‐2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng F. In silico oncology drug repositioning and polypharmacology. Methods Mol Biol. 2019;1878:243‐261. [DOI] [PubMed] [Google Scholar]
- 25. Vanhaelen Q, Mamoshina P, Aliper AM, et al. Design of efficient computational workflows for in silico drug repurposing. Drug Discov Today. 2017;22:210‐222. [DOI] [PubMed] [Google Scholar]
- 26. Turanli B, Altay O, Borén J, et al. Systems biology based drug repositioning for development of cancer therapy. Semin Cancer Biol. 2019. 10.1016/j.semcancer.2019.09.020 [DOI] [PubMed] [Google Scholar]
- 27. Mottini C, Napolitano F, Li Z, Gao X, Cardone L. Computer‐aided drug repurposing for cancer therapy: approaches and opportunities to challenge anticancer targets. Semin Cancer Biol. 2019. 10.1016/j.semcancer.2019.09.023 [DOI] [PubMed] [Google Scholar]
- 28. Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene‐expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929‐1935. [DOI] [PubMed] [Google Scholar]
- 29. Yamanishi Y, Kotera M, Kanehisa M, Goto S. Drug‐target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics. 2010;26:i246‐i254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dudley JT, Sirota M, Shenoy M, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwata M, Sawada R, Iwata H, Kotera M, Yamanishi Y. Elucidating the modes of action for bioactive compounds in a cell‐specific manner by large‐scale chemically‐induced transcriptomics. Sci Rep. 2017;7:40164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boyer A, Pasquier E, Tomasini P, et al. Drug repurposing in malignant pleural mesothelioma: a breath of fresh air? Eur Respir Rev. 2018;27:170098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahmed K, Shaw H, Koval A, Katanaev V. A second WNT for old drugs: drug repositioning against WNT‐dependent cancers. Cancers. 2016;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patrono C, García Rodríguez LA, Landolfi R, Baigent C. Low‐dose aspirin for the prevention of atherothrombosis. N Engl J Med. 2005;353:2373‐2383. [DOI] [PubMed] [Google Scholar]
- 36. Fong W, To KKW. Drug repurposing to overcome resistance to various therapies for colorectal cancer. Cell Mol Life Sci. 2019;76:3383‐3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schror K. Pharmacology and cellular/molecular mechanisms of action of aspirin and non–aspirin NSAIDs in colorectal cancer. Best Pract Res Clin Gastroenterol. 2011;25:473‐484. [DOI] [PubMed] [Google Scholar]
- 38. Umezawa S, Higurashi T, Komiya Y, et al. Chemoprevention of colorectal cancer: past, present, and future. Cancer Sci. 2019;110:3018‐3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perry CM, Figgitt DP. Zoledronic acid: a review of its use in patients with advanced cancer. Drugs. 2004;64:1197‐1211. [DOI] [PubMed] [Google Scholar]
- 40. Rogers MJ. New insights into the molecular mechanisms of action of bisphosphonates. Curr Pharm Des. 2003;9:2643‐2658. [DOI] [PubMed] [Google Scholar]
- 41. Green JR. Antitumor effects of bisphosphonates. Cancer. 2003;97:840‐847. [DOI] [PubMed] [Google Scholar]
- 42. Kato J, Futamura M, Kanematsu M, et al. Combination therapy with zoledronic acid and cetuximab effectively suppresses growth of colorectal cancer cells regardless of KRAS status. Int J Cancer. 2016;138:1516‐1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alimova IN, Liu B, Fan Z, et al. Metformin inhibits breast cancer cell growth, colony formation and induces cell cycle arrest in vitro. Cell Cycle. 2009;8:909‐915. [DOI] [PubMed] [Google Scholar]
- 44. Kalender A, Selvaraj A, Kim SY, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase‐dependent manner. Cell Metab. 2010;11:390‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deng J, Peng M, Wang Z, et al. Novel application of metformin combined with targeted drugs on anticancer treatment. Cancer Sci. 2019;110:23‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Epstein FH, Levin ER, Gardner DG, Samson WK. Natriuretic peptides. N Engl J Med. 1998;339:321‐328. [DOI] [PubMed] [Google Scholar]
- 47. Wilkins MR, Redondo J, Brown LA. The natriuretic‐peptide family. Lancet. 1997;349:1307‐1310. [DOI] [PubMed] [Google Scholar]
- 48. Vesely BA, McAfee Q, Gower WR, Vesely DL. Four peptides decrease the number of human pancreatic adenocarcinoma cells. Eur J Clin Invest. 2003;33:998‐1005. [DOI] [PubMed] [Google Scholar]
- 49. Vesely BA, Song S, Sanchez‐Ramos J, et al. Four peptide hormones decrease the number of human breast adenocarcinoma cells. Eur J Clin Invest. 2005;35:60‐69. [DOI] [PubMed] [Google Scholar]
- 50. Vesely BA, Fitz SR, Gower WR, Vesely DL. Vessel dilator: most potent of the atrial natriuretic peptides in decreasing the number and DNA synthesis of human squamous lung cancer cells. Cancer Lett. 2006;233:226‐231. [DOI] [PubMed] [Google Scholar]
- 51. Vesely BA, Song S, Sanchez‐Ramos J, et al. Five cardiac hormones decrease the number of human small‐cell lung cancer cells. Eur J Clin Invest. 2005;35:388‐398. [DOI] [PubMed] [Google Scholar]
- 52. Gower WR, Vesely BA, Alli AA, Vesely DL. Four peptides decrease human colon adenocarcinoma cell number and DNA synthesis via cyclic GMP. Int J Gastrointest Cancer. 2005;36:77‐87. [DOI] [PubMed] [Google Scholar]
- 53. Vesely BA, Eichelbaum EJ, Alli AA, Sun Y, Gower WR, Vesely DL. Urodilatin and four cardiac hormones decrease human renal carcinoma cell numbers. Eur J Clin Invest. 2006;36:810‐819. [DOI] [PubMed] [Google Scholar]
- 54. Vesely DL. Cardiac and renal hormones: anticancer effects in vitro and in vivo. J Investig Med. 2009;57:22‐28. [DOI] [PubMed] [Google Scholar]
- 55. Nojiri T, Hosoda H, Tokudome T, et al. Atrial natriuretic peptide prevents cancer metastasis through vascular endothelial cells. Proc Natl Acad Sci USA. 2015;112:4086‐4091. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Vesely DL, Clark LC, Garces AH, McAfee QW, Soto J, Gower WR. Novel therapeutic approach for cancer using four cardiovascular hormones. Eur J Clin Invest. 2004;34:674‐682. [DOI] [PubMed] [Google Scholar]
- 57. Vesely DL. New anticancer agents: hormones made within the heart. Anticancer Res. 2012;32:2515‐2521. [PubMed] [Google Scholar]
- 58. Nojiri T, Yamamoto H, Hamasaki T, et al. A multicenter randomized controlled trial of surgery alone or surgery with atrial natriuretic peptide in lung cancer surgery: study protocol for a randomized controlled trial. Trials. 2017;18:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hirayama C, Suzuki H, Ito M, Okumura M, Oda T. Propagermanium: a nonspecific immune modulator for chronic hepatitis B. J Gastroenterol. 2003;38:525‐532. [DOI] [PubMed] [Google Scholar]
- 60. Yokochi S, Hashimoto H, Ishiwata Y, et al. An anti–inflammatory drug, propagermanium, may target GPI‐anchored proteins associated with an MCP‐1 receptor, CCR2. J Interferon Cytokine Res. 2001;21:389‐398. [DOI] [PubMed] [Google Scholar]
- 61. Yumimoto K, Akiyoshi S, Ueo H, et al. F‐box protein FBXW7 inhibits cancer metastasis in a non–cell‐autonomous manner. J Clin Invest. 2015;125:621‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Masuda T, Noda M, Kogawa T, et al. Phase I dose‐escalation trial to repurpose propagermanium, an oral CCL2 Inhibitor, in patients with breast cancer. Cancer Sci. 2020. 10.1111/cas.14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kikuchi S, Noguchi K, Wakai K, et al. Propagermanium induces NK cell maturation and tends to prolong overall survival of patients with refractory cancer. Anticancer Res. 2019;39:4687‐4698. [DOI] [PubMed] [Google Scholar]
- 64. Yumimoto K, Sugiyama S, Mimori K, Nakayama KI. The potentials of CCL2‐CCR2 blockers including propagermanium as anticancer agents. Cancer Sci. 2019;110:2090‐2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Allison M. NCATS launches drug repurposing program. Nat Biotechnol. 2012;30:571‐572. [DOI] [PubMed] [Google Scholar]
- 66. Pantziarka P, Bouche G, Meheus L, Sukhatme V, Sukhatme VP, Vikas P. The repurposing drugs in oncology (ReDO) project. Ecancermedicalscience. 2014;8:442. [DOI] [PMC free article] [PubMed] [Google Scholar]


