Abstract
Background
Genetic testing has been conducted in patients with prostate cancer (PCa) using multigene panels, but no centralized guidelines for genetic testing exist. To overcome this limitation, we investigated the demographic and clinical characteristics of patients with pathogenic variants.
Methods
We sequenced eight genes associated with hereditary PCa in 7636 unselected Japanese patients with PCa and 12 366 male, cancer-free control individuals. We assigned clinical significance for all 1456 variants using the American College of Medical Genetics and Genomics guidelines and ClinVar. We compared the frequency of carriers bearing pathogenic variants between cases and control participants with calculated PCa risk in each gene and documented the demographic and clinical characteristics of patients bearing pathogenic variants. All statistical tests were two-sided.
Results
We identified 136 pathogenic variants, and 2.9% of patients and 0.8% of control individuals had a pathogenic variant. Association with PCa risk was statistically significant for variants in BRCA2 (P < .001, odds ratio [OR] = 5.65, 95% confidence interval [CI] = 3.55 to 9.32), HOXB13 (P < .001, OR = 4.73, 95% CI = 2.84 to 8.19), and ATM (P < .001, OR = 2.86, 95% CI = 1.63 to 5.15). We detected recurrent new pathogenic variants such as p.Gly132Glu of HOXB13. Patients with pathogenic variants were 2.0 years younger at diagnosis and more often had smoking and alcohol drinking histories as well as family histories of breast, pancreatic, lung, and liver cancers.
Conclusions
This largest sequencing study of PCa heredity provides additional evidence supporting the latest consensus among clinicians for developing genetic testing guidelines for PCa.
Prostate cancer (PCa) is the second most common cancer in men worldwide and has the highest incidence rate in developed countries (1). Among common cancers at 11 anatomical sites, PCa was found to be the most heritable (2), and genome-wide association studies have identified more than 150 variants associated with PCa (3). However, the identified variants were common and had low penetrance, limiting the clinical utility of genetic risk scores (4,5). Familial clustering of PCa has been reported (6), and around 5% of PCa cases could be primarily attributable to rare, highly penetrant mutations in genes such as BRCA1, BRCA2, and HOXB13 (7).
Genetic testing using multigene panels has the potential to guide PCa screening, targeted treatment, and surveillance for patients and their relatives (8). Because variants in genes such as BRCA1 and BRCA2 are associated with increased risk of multiple cancer types including PCa (9), identifying pathogenic variants in patients with PCa has implications for surveillance of various cancer types in relatives. Patients with a pathogenic variant in ATM, BRCA2, and CHEK2 are reported to have a higher risk for metastatic PCa (10). Patients with metastatic PCa who have germline or somatic mutations in DNA-repair machinery have sustained responses to poly–adenosine diphosphate ribose polymerase inhibitors (11) and platinum-based chemotherapy (12). The ongoing IMPACT study (NCT00261456) is evaluating the use of targeted PCa screening in men with BRCA1/2 mutations (13). However, no centralized guidelines for PCa genetic testing exist.
Guidelines for genetic testing serve as a resource to identify individuals who may benefit from cancer-risk assessment and genetic counseling, to guide decisions related to genetic testing, and to facilitate a multidisciplinary approach in managing individuals at increased risk (9). In contrast to breast, ovarian (9), and colon cancers (14), studies on PCa have been limited, and information regarding the clinical significance of genetic variants in ClinVar is much sparser than information available for other cancers. In this setting, a large-scale, case-control study could provide important information on classifications of individual germline variants, PCa disease risk for each gene, and demographic and clinical characteristics of patients bearing pathogenic variants. Although most studies have analyzed only patients with PCa (8), our previous study on breast cancer (15) showed that population-matched control individuals were indispensable because 5% or fewer variants found in Japanese patients were registered in the most closely matched population in ExAC (16). Therefore, various types of information from a large-scale, case-control study would help in developing guidelines for genetic testing in PCa.
In this study, we performed the largest case-control sequencing study on PCa heredity (to the best of our knowledge), involving 7636 unselected Japanese patients with PCa and 12 366 control participants. We sequenced coding regions of eight genes (7), assigned clinical significance for all variants detected, and calculated PCa risk estimates for presumed pathogenic variants in each gene. We investigated the demographic and clinical characteristics of patients with pathogenic variants.
Methods
Study Population
We obtained all study samples from BioBank Japan (17,18), which is a multi-institutional, hospital-based registry that collects DNA and clinical information from patients with various common diseases, including PCa (19), from all over Japan between 2003 and 2018. Clinical characteristics of cases and control individuals were collected by interview or medical record survey using a standard questionnaire at the point of entry to Biobank Japan. These PCa samples are considered likely to be representative of Japanese patients because the age-specific distribution of PCa patients in BioBank Japan was similar to that described in the Japanese Ministry of Health, Labour, and Welfare Patient Survey (19). In this study, we performed a hospital registry-based study in 7744 patients with PCa and 12 520 male controls. Among the 7744 patients with PCa, 7229 individuals were diagnosed before enrollment, and the remaining 515 patients were diagnosed during a follow-up period. We used the same 12 520 male controls age 60 years and older with no personal or family history of cancer from our previous study on breast cancer (15). Owing to this selection criterion, the control group may exhibit a lower frequency of pathogenic variants than that observed in the general controls individuals, and as a result, disease risk may be calculated to be higher. All participants provided their written informed consent. The study was approved by the ethical committees of the Institute of Medical Sciences, the University of Tokyo, and the RIKEN Center for Integrative Medical Sciences.
Sequencing and Bioinformatics Analysis
We selected eight genes (ATM, BRCA1, BRCA2, BRCA1 interacting protein C-terminal helicase 1 [BRIP1], CHEK2, HOXB13, NBN, and PALB2) whose rare germline variants were reported to show high penetrance for PCa in a review article (7) because there were no guidelines about gene selection for genetic testing. We analyzed the complete coding regions and 2-bp flanking intronic sequences of all eight genes (37 982 bp) by a multiplex polymerase chain reaction–based target sequence method (20) (Supplementary Methods, available online). Finally, we identified 1456 genetic variants in 7636 patients and 12 366 control individuals, and 99.98% of the target region was covered by at least 20 sequence reads.
Annotation of Variants
We assigned clinical significance (pathogenic, benign, or uncertain) for all variants as in our previous study (15) (Supplementary Methods, available online). Briefly, we determined clinical significance using the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines (21,22) (Supplementary Tables 1 and 2, available online) as well as pathogenicity assertions registered in ClinVar (23). We used the same procedure for all genes except HOXB13 because a gain-of-function missense variant in HOXB13 was considered pathogenic (24). We considered variants as pathogenic based on classification as pathogenic by the ACMG/AMP guidelines and/or classification as pathogenic in ClinVar. Specific details are described in Supplementary Methods (available online). Variants not registered in ClinVar on August 1, 2017, were considered novel.
Statistical Analysis
Case-control association analysis was performed using Fisher exact test under a dominant model. To investigate the association of pathogenic variants with demographic and clinical characteristics, we used t tests for continuous variables and Fisher exact tests or Cochran-Armitage tests for discrete variables. All statistical tests were two-sided, and P less than .05 was considered statistically significant except when Bonferroni correction was applied for the association analysis between each of the eight genes and PCa (P < .006 = 0.05/8). All analyses were performed using R statistical package (ver. 3.1.3).
Results
Demographic and Clinical Characteristics of Participants
The mean age at PCa diagnosis was 71.0 years (SD = 6.9) (Table 1). Positive smoking history in cases (69.4%) was statistically significantly lower than that in controls (76.1%, P < .001). This reflects the fact that this proportion was the fifth smallest among 42 male diseases registered in the BioBank Japan (17), whereas control participants consisted of patients with complex diseases other than cancer in the same biobank. Family history of prostate, breast, or pancreatic cancers was observed in 6.8%, 4.7%, and 3.3% of patients, respectively. Other clinical characteristics are shown in Table 1.
Table 1.
Demographic and clinical characteristics of study participants
| Variable | PCa patients | Controls |
|---|---|---|
| No. (%) | No. (%) | |
| No. of participants | 7744 (100) | 12 520 (100) |
| Mean age at entry, (SD), y | 72.9 (7.1) | 70.4 (7.0) |
| Mean age at diagnosis, (SD), y* | 71.0 (6.9) | — |
| Smoking history | ||
| Yes | 5263 (69.4) | 9490 (76.1) |
| No | 2322 (30.6) | 2984 (23.9) |
| Missing | 159 | 46 |
| Alcohol drinking history | ||
| Yes | 5307 (70.0) | 8698 (69.9) |
| No | 2270 (30.0) | 3751 (30.1) |
| Missing | 167 | 71 |
| Body mass index, (SD) kg/m2* | 23.4 (2.9) | 23.4 (3.1) |
| Family history of PCa | ||
| Yes | 530 (6.8) | 0† (0) |
| No | 7214 (93.2) | 12 520 (100.0) |
| Family history of breast cancer | ||
| Yes | 362 (4.7) | 0† (0) |
| No | 7382 (95.3) | 12 520 (100.0) |
| Family history of pancreatic cancer | ||
| Yes | 255 (3.3) | 0† (0) |
| No | 7489 (96.7) | 12 520 (100.0) |
| Histological type | ||
| Adenocarcinoma | 7085 (99.5) | — |
| Others | 38 (0.5) | — |
| Missing | 621 | — |
| TNM classification: T | ||
| T4 | 172 (4.5) | — |
| T3 | 805 (20.9) | — |
| T2 | 1794 (46.5) | — |
| T1 | 1080 (28.0) | — |
| T0 | 9 (0.2) | — |
| Missing | 3884 | — |
| TNM classification: N | ||
| N1 | 229 (6.1) | — |
| N0 | 3517 (93.9) | — |
| Missing | 3998 | — |
| TNM classification: M | ||
| M1 | 297 (8.0) | — |
| M0 | 3397 (92.0) | — |
| Missing | 4050 | — |
| Gleason score | ||
| Aggressive (≥8) | 1713 (29.7) | — |
| Indolent (<8) | 4064 (70.3) | — |
| Missing | 1967 | — |
| Maximum value of serum PSA before treatment, ng/mL | ||
| >20 | 1470 (31.0) | — |
| 10–20 | 1117 (23.5) | — |
| 4–10 | 1991 (42.0) | — |
| ≤4 | 166 (3.5) | — |
| Missing | 3000 | — |
| Complicated with benign prostatic hypertrophy | ||
| Yes | 2722 (57.8) | — |
| No | 1985 (42.2) | — |
| Missing | 3037 | — |
The number of missing data is 311 in mean age at diagnosis; 268 in cases and 753 in controls in body mass index. PCa = prostate cancer; PSA = prostate- specific antigen. Em dashes (—) indicate no information for control participants.
Controls with no past history or family history of cancers were selected for this study. TNM = Tumor, Node, Metastasis.
Pathogenic Germline Variants
Sequencing of the eight PCa-relevant genes identified 1456 germline variants in total. We categorized the variants according to the ACMG/AMP guidelines as follows: five pathogenic, 117 likely pathogenic, 49 benign, 90 likely benign, two variants of uncertain significance (VUS) with pathogenic as well as benign evidence, and 1193 VUS with insufficient evidence. When we compared these results with assertions in ClinVar (Supplementary Table 3, available online), there were no apparent discrepancies (variants that were pathogenic in one classification and benign in the other). Finally, we considered 136, 284 and 1036 variants as pathogenic, benign, and VUS, respectively (Supplementary Table 4, available online). Single variant association results are shown in Supplementary Table 5 (available online) for the 136 pathogenic variants and in Supplementary Table 6 (available online) for the 284 benign variants and the 1036 VUS. More than one-half (57.4%) of the pathogenic variants were novel, and 71.4% and 61.5% of pathogenic variants in BRCA1 and BRCA2, respectively, were previously described in ClinVar, whereas more than one-half of pathogenic variants in the other genes were novel (Supplementary Figure 1, available online). We revisited the latest database in ClinVar on February 11, 2019, and other published works to ensure that our data were current. In total, 32 pathogenic variants were reported in four publications or ClinVar. The majority (21 variants) came from our previous large-scale sequencing study on breast cancer (15). Finally, 46 of 136 pathogenic variants (33.8%) are still considered novel.
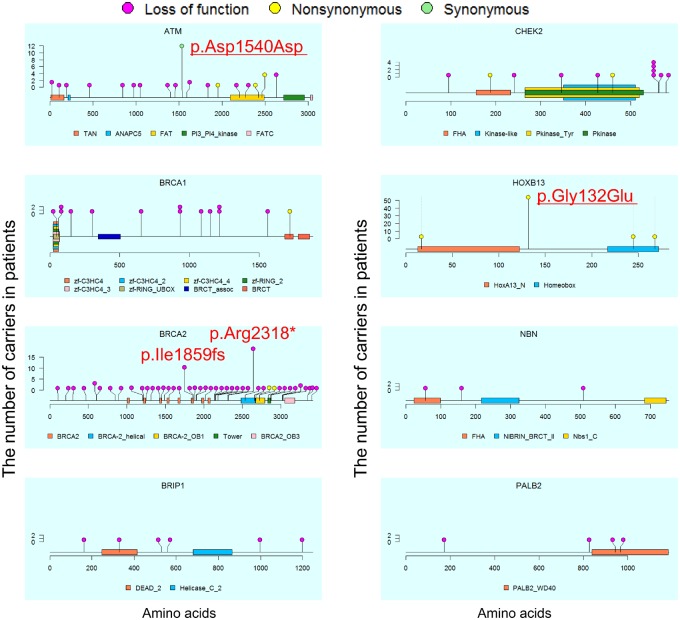
We checked the locations and frequencies of the pathogenic variants (Figure 1). We observed four frequent pathogenic variants shared in five or more patients: p.Asp1540Asp in ATM (n = 12, P < .001, odds ratio [OR] = 19.46, 95% confidence interval [CI] = 2.88 to 828.78), p.Gly132Glu in HOXB13 (n = 56, P < .001, OR = 6.08, 95% CI = 3.39 to 11.59), and p.Ile1859fs (n = 11, P < .001, OR = 8.92, 95% CI = 1.95 to 82.99) and p.Arg2318* (n = 19, P < .001, OR = 15.42, 95% CI = 3.72 to 136.50) in BRCA2. The first two variants were novel. Note that we did not observe any association between the carrier status of these four variants and the 10 principal components calculated using single nucleotide polymorphism array data from 6269 of 7636 patients with PCa (25,26). This finding suggests that these pathogenic variants may be spread across Japanese populations.
Figure 1.
Location and number of pathogenic variants in Japanese patients with prostate cancer. Locations of pathogenic variants found in patients (n = 219) and domains in proteins encoded by the six genes are shown by lollipop structures, with the variant type indicated by color. The x axis reflects the number of amino acid residues, and the y axis shows the total number of patients with each pathogenic variant. HGVS.p of frequent variants with five or more patients are shown, and two variants newly identified as pathogenic variants are underlined. ANAPC5 = anaphase-promoting complex subunit 5; BRCT = BRCA1 C terminus; DEAD_2 = DEAD/DEAH box helicase 2; FAT = FRAP, ATM and TRRAP; FATC = FRAP, ATM, TRRAP C-terminal; FHA = forkhead-associated; HOXA13_N= hox protein A13 N terminal; Nbs1_C = DNA damage repair protein Nbs1; NIBRIN_BRCT_II = second BRCT domain on Nijmegen syndrome breakage protein; PALB2_WD40 = partner and localizer of BRCA2 WD40 domain; PI3_PI4_kinase = phosphatidylinositol 3- and 4-kinase; TAN = telomere-length maintenance and DNA damage repair; zf-C3HC4 = zinc finger, C3HC4 type; zf-RING = zinc finger, C3HC4 type.
We compared the frequency of carriers bearing pathogenic variants between cases and controls (Table 2). In total, 2.9% of patients and 0.8% of control participants carried a pathogenic variant (P < .001, OR = 3.66, 95% CI = 2.87 to 4.69). BRCA2 (P < .001, OR = 5.65, 95% CI = 3.55 to 9.32), HOXB13 (P < .001, OR = 4.73, 95% CI = 2.84 to 8.19), and ATM (P < .001, OR = 2.86, 95% CI = 1.63 to 5.15) were statistically significantly associated with PCa after Bonferroni correction. Only one patient carried two major pathogenic variants (p.Arg2318* in BRCA2 and p.Gly132Glu in HOXB13). He was diagnosed with T2bN0M0 and GS7 PCa at age 61 years and had no previous cancer history but did have a family history of breast cancer.
Table 2.
Results of gene-based association test using pathogenic variants
| Gene | No. of pathogenic variants | Case (n = 7636) |
Control (n = 12 366) |
P * | OR† (95% CI) | ||
|---|---|---|---|---|---|---|---|
| No. of carriers | Carrier frequency (%) | No. of carriers | Carrier frequency (%) | ||||
| BRCA2 | 52 | 83 | 1.1 | 24 | 0.2 | <.001 | 5.65 (3.55 to 9.32) |
| HOXB13 | 5 | 61 | 0.8 | 21 | 0.2 | <.001 | 4.73 (2.84 to 8.19) |
| ATM | 31 | 37 | 0.5 | 21 | 0.2 | <.001 | 2.86 (1.63 to 5.15) |
| BRCA1 | 14 | 14 | 0.2 | 10 | 0.1 | .06 | 2.27 (0.94 to 5.71) |
| CHEK2 | 9 | 12 | 0.2 | 8 | 0.1 | .06 | 2.43 (0.91 to 6.86) |
| PALB2 | 6 | 4 | 0.1 | 4 | 0.0 | .49 | 1.62 (0.30 to 8.70) |
| BRIP1 | 12 | 6 | 0.1 | 7 | 0.1 | .58 | 1.39 (0.39 to 4.83) |
| NBN | 7 | 3 | 0.0 | 4 | 0.0 | 1.00 | 1.21 (0.18 to 7.18) |
| Sum | 136 | 219‡ | 2.9 | 99 | 0.8 | <.001 | 3.66 (2.87 to 4.69) |
Two-sided Fisher exact test was used. BRIP1 = BRCA1 interacting protein C-terminal helicase 1; CI = confidence interval; OR = odds ratio.
Odds ratio for cases bearing pathogenic variants vs controls bearing pathogenic variants.
Because one patient had a pathogenic variant both in BRCA2 and HOXB13, the total number of carriers was one smaller than the sum of carriers in each gene.
Sensitivity Analyses Regarding the Annotation of Variants
Various procedures of the ACMG/AMP guidelines (21) followed in different laboratories have resulted in different interpretations of variants (27). We performed two types of sensitivity analyses. The first sensitivity analysis compared our method to ClinVar. We performed gene-based association analysis using pathogenic variants registered in ClinVar (n = 58, Supplementary Table 7, available online) and determined by the ACMG/AMP guidelines (n = 122, Supplementary Table 8, available online). The odds ratio of all genes was comparable (ClinVar: OR = 3.99, 95% CI = 2.68 to 6.07; ACMG/AMP: OR = 3.66, 95% CI = 2.84 to 4.74). For each gene, similar odds ratios were observed, with the exception of HOXB13–0/58 pathogenic ClinVar records vs 5/122 ACMG/AMP classifications. These data suggest that our interpretation of the ACMG/AMP guidelines would be comparable with that of ClinVar.
The second sensitivity analysis involves gene-based association analysis of rare benign variants exhibiting Minor allele frequency less than 0.01 (n = 248, Supplementary Table 9, available online) and of rare VUS (n = 1036, Supplementary Table 10, available online). The gene-based association test using benign variants actually showed no genes possessing a P less than .05. Conversely, the gene-based association test using VUS indicated that CHEK2 exhibited a P less than .001 and an OR = 1.62 (95% CI = 1.30 to 2.00). Three missense variants possessed a P less than .05, and these included p.Ala496Pro (P = .006, OR = 4.22, 95% CI = 1.41 to 15.11), p.Arg223Cys (P = .03, OR = 1.98, 95% CI = 1.01 to 3.92), and p.His414Tyr (P = .03, OR = 2.25, 95% CI = 1.04 to 4.99). Although p.Arg223Cys possessed sufficient pathogenic (PS3, PM1, and PP3) as well as benign evidence (BS1 and BP5), the others lacked sufficient evidence. Therefore, certain variants within CHEK2 may be pathogenic, and CHEK2 may contribute to the development of PCa, although additional research, including functional tests, is required to clarify this issue. It might also suggest further improvement of the ACMG/AMP guidelines.
Demographic and Clinical Characteristics of Patients , With Pathogenic Variants
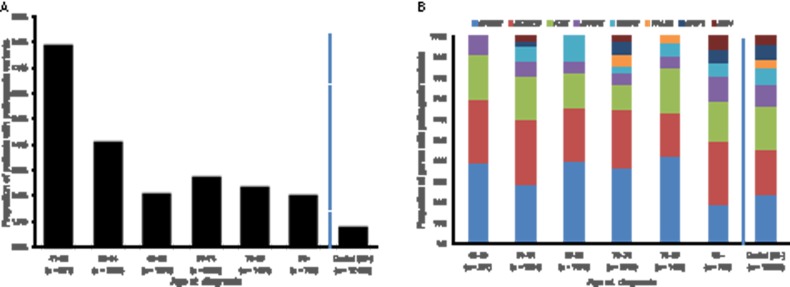
To investigate the association of pathogenic variants with demographic and clinical characteristics of PCa, we compared these between the 219 carrier patients and the 7417 noncarrier patients (Table 3). The carriers were on average 2.0 years younger at PCa diagnosis (P < .001) and more often had histories of smoking (77.0% in carriers vs 69.2% in noncarriers, P = .02) and alcohol drinking (82.6% vs 69.8%, P < .001). Carriers also more often had a family history of breast (11.9% vs 4.5%, P < .001), pancreatic (8.7% vs 3.2%, P < .001), lung (12.3% vs 7.5%, P = .01), or liver (7.8% vs 4.6%, P = .03) cancers. There was no difference between carriers (8.2%) and noncarriers (6.8%) in family history of PCa (P = .41). The carriers also showed worse clinical characteristics in terms of Tumor, Node, Metastasis (TNM) classifications (P = .03 and P = .04 for T and M, respectively), Gleason score of at least 8 (41.0% vs 29.1%, P = .002), and prostate-specific antigen (PSA) greater than 10 (64.0% vs 54.1%, P = .02). We also performed logistic regression analysis with carrier status of a pathogenic variant as a dependent variable against these 11 variables because they may be interrelated. We used 2427 of 7636 patients who did not have missing data. Supplementary Table 11 (available online) indicates that only three variables (age at diagnosis, alcohol drinking history, and family history of breast cancer) showed P less than .05, which would be explained by interrelation between some variables and/or the decreased number of samples in this analysis. The other variables should be carefully considered in further studies. We also examined the impact of pathogenic variants on age at PCa diagnosis (Figure 2, A and B). Pathogenic variants were found in 7.9% of patients diagnosed at younger than 60 years old. The proportion of pathogenic variants statistically significantly decreased with advancing age at diagnosis (P < .001) but was stable between 2% and 3% in individuals age 65 years or older.
Table 3.
Statistically significant differences in demographic and clinical characteristics between patients with PCa with and without pathogenic variants
| Variable | No. of patients with pathogenic variants (%) | No. of patients without pathogenic variants (%) | P * | OR (95% CI) |
|---|---|---|---|---|
| No. of patients | 219 (100) | 7417 (100) | — | — |
| Age at diagnosis, mean (SD), y | 69.0 (7.7) | 71.0 (6.9) | <.001 | — |
| Smoking history | ||||
| Yes | 167 (77.0) | 5046 (69.2) | .02 | 1.48 (1.07 to 2.09) |
| No | 50 (23.0) | 2243 (30.8) | 1.00 (Referent) | |
| Alcohol drinking history | ||||
| Yes | 180 (82.6) | 5078 (69.8) | <.001 | 2.05 (1.44 to 3.01) |
| No | 38 (17.4) | 2202 (30.2) | 1.00 (Referent) | |
| Family history of PCa† | ||||
| Yes | 18 (8.2) | 505 (6.8) | .41 | 1.23 (0.71 to 2.01) |
| No | 201 (91.8) | 6912 (93.2) | 1.00 (Referent) | |
| Family history of breast cancer | ||||
| Yes | 26 (11.9) | 333 (4.5) | <.001 | 2.87 (1.80 to 4.40) |
| No | 193 (88.1) | 7084 (95.5) | 1.00 (Referent) | |
| Family history of pancreatic cancer | ||||
| Yes | 19 (8.7) | 234 (3.2) | <.001 | 2.92 (1.69 to 4.78) |
| No | 200 (91.3) | 7183 (96.8) | 1.00 (Referent) | |
| Family history of lung cancer | ||||
| Yes | 27 (12.3) | 553 (7.5) | .01 | 1.73 (1.10 to 2.64) |
| No | 192 (87.7) | 6864 (92.5) | 1.00 (Referent) | |
| Family history of liver cancer | ||||
| Yes | 17 (7.8) | 340 (4.6) | .03 | 1.75 (0.99 to 2.92) |
| No | 202 (92.2) | 7077 (95.4) | 1.00 (Referent) | |
| TNM classification: T | ||||
| T3/T4 | 37 (33.6) | 902 (24.6) | .03 | 1.56 (1.01 to 2.36) |
| T0 -2 | 73 (66.4) | 2769 (75.4) | 1.00 (Referent) | |
| TNM classification: M | ||||
| M1 | 14 (13.2) | 270 (7.7) | .04 | 1.83 (0.95 to 3.28) |
| M0 | 92 (86.8) | 3244 (92.3) | 1.00 (Referent) | |
| Gleason score | ||||
| Aggressive (≥8) | 66 (41.0) | 1610 (29.1) | .002 | 1.70 (1.21 to 2.36) |
| Indolent (<8) | 95 (59.0) | 3930 (70.9) | 1.00 (Referent) | |
| Maximum value of serum PSA before treatment, ng/mL | ||||
| >10 | 87 (64.0) | 2461 (54.1) | .02 | 1.51 (1.04 to 2.19) |
| ≤10 | 49 (36.0) | 2087 (45.9) | 1.00 (Referent) |
Two-sided Fisher exact test was used for all analyses except for age at diagnosis. Two-sided t test was used for age at diagnosis. CI = confidence interval; OR = odds ratio; PCa = prostate cancer; PSA = prostate-specific antigen; TNM = Tumor, Node, Metastasis.
Family history of PCa was not associated with carrier status but is shown because its association was expected.
Figure 2.
Proportion of patients and genes with pathogenic variants by age groups at prostate cancer diagnosis. A) Proportion of patients with pathogenic variants is shown. Two-sided Cochran-Armitage test was used (P < .001). B) Proportion of genes with pathogenic variants is shown.
We further analyzed these demographic and clinical characteristics in patients with variants in ATM (n = 37), BRCA2 (n = 83), or HOXB13 (n = 61) (Supplementary Table 12, available online). Although some association tests had limited statistical power because of the low number of carriers, associations with age at diagnosis, smoking history, and alcohol drinking history showed similar tendencies among the three genes. Family history of breast cancer was associated with both variants in BRCA2 (15.7% in BRCA2 carriers vs 4.5% in noncarriers, P < .001) and in HOXB13 (16.4%, P < .001). Family history of pancreatic cancer was associated with variants in BRCA2 (16.9% in BRCA2 variant carriers vs 3.2% in noncarriers, P < .001). Patients bearing pathogenic variants of BRCA2 alone showed worse clinical characteristics as shown by TNM classifications (T: P = .001, M: P = .008), Gleason score of at least 8 (51.7% in BRCA2 carriers vs 29.1% in noncarriers, P < .001), and PSA greater than 10 (70.2% vs 54.1%, P = .03).
Discussion
We identified 136 pathogenic variants in eight genes in 7636 patients with PCa and 12 366 control individuals. Finally, 33.8% of the pathogenic variants were newly identified in this study. Pathogenic variants were found in 2.9% of unselected Japanese PCa patients; HOXB13, BRCA2, and ATM were the statistically significant causative genes. Patients with pathogenic variants showed specific demographic and clinical characteristics.
The Philadelphia Prostate Cancer Consensus 2017 (the Consensus) was recently published to establish a genetic evaluation framework for inherited PCa (8). We investigated if our study results could add more evidence to the Consensus. Regarding gene selection, the Consensus considered that both BRCA1 and BRCA2 had high-grade evidence for being related to PCa. However, our results revealed that only BRCA2 showed a statistically significant contribution to PCa, and patients carrying a pathogenic variant in this gene showed worse clinical characteristics. BRCA1 pathogenic variants, altogether or considering only the most frequent pathogenic variant observed in patients with breast cancer (15), p.Leu63*, showed no statistically significant association with PCa risk. PCa disease risks were calculated as 5.65 for BRCA2 and 2.27 for BRCA1. This is consistent with results of previous studies (28) (BRCA2: 4.7–8.6 and BRCA1: 1.1–3.8). Therefore, our results suggest that BRCA1 and BRCA2 should be separately considered in genetic testing among Japanese patients with PCa.
We validated the importance of HOXB13, although its main pathogenic variant in our population, p.Gly132Glu, was different from those in European (p.Gly84Glu) (24) and Chinese (p.Gly135Glu) populations (29). According to the Genome Aggregation Database (16), p.Gly84Glu was also observed in African, Ashkenazi Jewish, and Latino populations, but p.Gly132Glu and p.Gly135Glu were found only in East Asian populations. However, p.Gly132Glu and p.Gly135Glu may be subpopulation specific, given that p.Gly132Glu was not observed in 96 Chinese patients (29) and p.Gly135Glu was not found in our Japanese cohort, suggesting that the existence of frequent pathogenic variants in each subpopulation would strongly affect the importance of HOXB13 in genetic testing for PCa.
We also provided additional support regarding the contribution to PCa of ATM with emerging evidence in the Consensus (8). NBN also had the same level of evidence, but we did not observe any contribution from this gene in this study of Japanese individuals. The primary evidence supporting a role of NBN came from 657del5 (p.Lys219Asnfs, rs587776650) in a Polish population (30), but this variant was not observed in our Japanese population. Therefore, the lack of association in this study indicates less importance of this gene in Japanese PCa rather than evidence against association in other populations. We did not observe the importance of CHEK2 and PALB2 with low or insufficient evidence in the Consensus (8). Taken together, the Japanese data provide evidence about the contribution of each gene to PCa and also highlight the importance of population-specific data for genetic testing.
Regarding the selection of patients for genetic testing in the Consensus, we validated the importance of family histories of breast and pancreatic cancers. Our results also showed weaker associations with family history of lung and liver cancers. Note that we did not detect a statistically significant association with family history of PCa, although it is recognized as an established risk factor (7). Because the proportion of patients with a family history of PCa was lower (6.8%) in Japan than that reported elsewhere at 10–15% (31), this finding might be explained by a lower incidence of PCa in the Japanese population (32). However, because incidence of PCa (32) and life expectancy (33) are rapidly increasing in Japan, family histories of PCa would presumably become useful for the next generation to be aware of. The importance of age at diagnosis was confirmed because patients who were younger at diagnosis were more likely to carry pathogenic variants. We also observed associations between smoking and alcohol drinking histories with carrier status, although its importance was not previously recognized (34). Taken together, our results provide a series of additional and unexpected evidence above and beyond the Consensus (8), which should inform genetic testing in PCa and promote studies about specific questions in other populations internationally.
This study had some limitations. First, risks associated with pathogenic variants might be overestimated because we selected controls age 60 years and older with no individual or family history of cancers, although the disease risk for BRCA1/2 variants was comparable with that found in previous studies. Second, we focused on eight genes based on a review article (7) because the Consensus did not exist when this study started. The Consensus (8) proposed DNA mismatch repair genes for Lynch syndrome, although they have less support than BRCA1/2 and HOXB13. Third, we derived the variant classifications against ACMG/AMP criteria (21) using an automated approach and recognize that additional manual curation is required to ensure “clinical-grade” classifications. Finally, we considered gain-of-function missense variants in HOXB13 as pathogenic based on the previous publications examining European (24) and Chinese (29) populations. Further functional studies are required to confirm that the pathogenic missense variants found in this study are indeed gain-of-function variants (35).
In conclusion, variants predicted to alter function of BRCA2, HOXB13, and ATM were associated with PCa, and patients with a pathogenic variant showed specific demographic and clinical characteristics. The findings reported in this study may inform future genetic testing for PCa.
Funding
This work was conducted as part of the BioBank Japan Project supported by the Japan Agency for Medical Research and Development and by the Ministry of Education, Culture, Sports, Sciences and Technology of the Japanese government. A.B. Spurdle is supported by an Australian National Health and Medical Research Council Senior Research Fellowship.
Notes
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. The authors have no conflicts of interest to disclose.
We thank the individuals who participated in this study. We acknowledge C. Inai, S. Takata, N. Hakozaki, H. Hashinokuchi, M. Endo, and other staff members of the Laboratory for Genotyping Development, RIKEN Center for the Integrative Medical Sciences, and the staff of the BioBank Japan project.
Supplementary Material
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. [DOI] [PubMed] [Google Scholar]
- 2. Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. [DOI] [PubMed] [Google Scholar]
- 3. Schumacher FR, Al Olama AA, Berndt SI, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakagawa H, Akamatsu S, Takata R, et al. Prostate cancer genomics, biology, and risk assessment through genome-wide association studies. Cancer Sci. 2012;103(4):607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eeles R, Goh C, Castro E, et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol. 2014;11(1):18–31. [DOI] [PubMed] [Google Scholar]
- 6. Bratt O, Drevin L, Akre O, et al. Family history and probability of prostate cancer, differentiated by risk category: a nationwide population-based study. J Natl Cancer Inst. 2016;108(10). doi: 10.1093/jnci/djw110. [DOI] [PubMed] [Google Scholar]
- 7. Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet. 2016;387(10013):70–82. [DOI] [PubMed] [Google Scholar]
- 8. Giri VN, Knudsen KE, Kelly WK, et al. Role of genetic testing for inherited prostate cancer risk: Philadelphia Prostate Cancer Consensus Conference 2017. J Clin Oncol. 2018;36(4):414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daly MB, Pilarski R, Berry M, et al. NCCN guidelines insights: genetic/familial high-risk assessment: breast and ovarian, version 2.2017. J Natl Compr Canc Netw. 2017;15(1):9–20. [DOI] [PubMed] [Google Scholar]
- 10. Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng HH, Pritchard CC, Boyd T, et al. Biallelic inactivation of BRCA2 in platinum-sensitive metastatic castration-resistant prostate cancer. Eur Urol. 2016;69(6):992–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bancroft EK, Page EC, Castro E, et al. Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur Urol. 2014;66(3):489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Provenzale D, Gupta S, Ahnen DJ, et al. Genetic/familial high-risk assessment: Colorectal version 1.2016, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2016;14(8):1010–1030. [DOI] [PubMed] [Google Scholar]
- 15. Momozawa Y, Iwasaki Y, Parsons MT, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9(1):4083.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirata M, Kamatani Y, Nagai A, et al. Cross-sectional analysis of BioBank Japan clinical data: a large cohort of 200,000 patients with 47 common diseases. J Epidemiol. 2017;27(3S):S9–S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagai A, Hirata M, Kamatani Y, et al. Overview of the BioBank Japan project: study design and profile. J Epidemiol. 2017;27(3S):S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ukawa S, Nakamura K, Okada E, et al. Clinical and histopathological characteristics of patients with prostate cancer in the BioBank Japan project. J Epidemiol. 2017;27(3S):S65–S70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Momozawa Y, Akiyama M, Kamatani Y, et al. Low-frequency coding variants in CETP and CFB are associated with susceptibility of exudative age-related macular degeneration in the Japanese population. Hum Mol Genet. 2016;25(22):5027–5034. [DOI] [PubMed] [Google Scholar]
- 21. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eilbeck K, Quinlan A, Yandell M.. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet. 2017;18(10):599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ewing CM, Ray AM, Lange EM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366(2):141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takata R, Akamatsu S, Kubo M, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42(9):751–754. [DOI] [PubMed] [Google Scholar]
- 26. Akamatsu S, Takata R, Haiman CA, et al. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44(4):426–429, S1. [DOI] [PubMed] [Google Scholar]
- 27. Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016;98(6):1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng HH, Pritchard CC, Montgomery B, et al. Prostate cancer screening in a new era of genetics. Clin Genitourin Cancer. 2017;15(6):625–628. [DOI] [PubMed] [Google Scholar]
- 29. Lin X, Qu L, Chen Z, et al. A novel germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate. 2013;73(2):169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cybulski C, Wokołorczyk D, Kluźniak W, et al. An inherited NBN mutation is associated with poor prognosis prostate cancer. Br J Cancer. 2013;108(2):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13(suppl 1):R103–R121. [DOI] [PubMed] [Google Scholar]
- 32. Hsing AW, Tsao L, Devesa SS.. International trends and patterns of prostate cancer incidence and mortality. Int J Cancer. 2000;85(1):60–67. [DOI] [PubMed] [Google Scholar]
- 33. Nomura S, Sakamoto H, Glenn S, et al. Population health and regional variations of disease burden in Japan, 1990–2015: a systematic subnational analysis for the Global Burden of Disease Study 2015. Lancet. 2017;390(10101):1521–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Theodoratou E, Timofeeva M, Li X, et al. Nature, nurture, and cancer risks: genetic and nutritional contributions to cancer. Annu Rev Nutr. 2017;37(1):293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cardoso M, Maia S, Paulo P, et al. Oncogenic mechanisms of HOXB13 missense mutations in prostate carcinogenesis. Oncoscience. 2016;3(9–10):288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.