Abstract
Background
Isocitrate dehydrogenase wild-type (WT) glioblastoma (GBM) accounts for 90% of all GBMs, yet only 27% of isocitrate dehydrogenase WT-GBMs have p53 mutations. However, the tumor surveillance function of WT-p53 in GBM is subverted by mechanisms that are not fully understood.
Methods
We investigated the proteolytic inactivation of WT-p53 by asparaginyl endopeptidase (AEP) and its effects on GBM progression in cancer cells, murine models, and patients’ specimens using biochemical and functional assays. The sera of healthy donors (n = 48) and GBM patients (n = 20) were examined by enzyme-linked immunosorbent assay. Furthermore, effects of AEP inhibitors on GBM progression were evaluated in murine models (n = 6–8 per group). The statistical significance between groups was determined using two-tailed Student t tests.
Results
We demonstrate that AEP binds to and directly cleaves WT-p53, resulting in the inhibition of WT-p53-mediated tumor suppressor function in both tumor cells and stromal cells via extracellular vesicle communication. High expression of uncleavable p53-N311A-mutant rescue AEP-induced tumorigenesis, proliferation, and anti-apoptotic abilities. Knock down or pharmacological inhibition of AEP reduced tumorigenesis and prolonged survival in murine models. However, overexpression of AEP promoted tumorigenesis and shortened the survival time. Moreover, high AEP levels in GBM tissues were associated with a poor prognosis of GBM patients (n = 83; hazard ratio = 3.94, 95% confidence interval = 1.87 to 8.28; P < .001). A correlation was found between high plasma AEP levels and a larger tumor size in GBM patients (r = 0.6, P = .03), which decreased dramatically after surgery.
Conclusions
Our results indicate that AEP promotes GBM progression via inactivation of WT-p53 and may serve as a prognostic and therapeutic target for GBM.
Glioblastoma (GBM) is the most common and aggressive central nervous system neoplasm and is divided into two subtypes: isocitrate dehydrogenase (IDH)-wild-type (WT)-GBM and IDH-mutant-GBM. Besides their broadly similar histopathologies, most GBMs appear to be driven by dysregulation of p53, RB transcriptional corepressor 1, and receptor tyrosine kinase Ras signaling (1). Among these regulators, p53 is a master regulator of diverse cellular processes (2–4). Mutations that directly inactivate or delete p53 itself are relatively few in IDH-WT-GBM (5). Although the p53 pathway can be compromised by amplification of MDM2, the mechanisms that suppress p53 functions remain elusive in most IDH-WT-GBMs (6–9). Therefore, understanding the inhibitory mechanisms targeting WT-p53 is extremely important for addressing GBM, especially the IDH-WT form.
In addition to cancer cells, the malignant transformation of host stromal cells within the tumor microenvironment is essential for tumor progression (10). Reactive astrocytes and endothelial cells are stromal cells that constitute a large portion of the tumor mass and are associated with GBM progression. Recently, extracellular vesicles (EVs) have been shown to be involved in cell-cell communication and proposed as tumor biomarkers and novel therapeutic targets for GBM (11–16). The functions and underlying mechanisms of EVs in the malignant transformation of stromal cells in the GBM microenvironment are barely known.
Asparaginyl endopeptidase (AEP), also called legumain, is currently the only known AEP of the mammalian genome that specifically cleaves peptide bonds at asparaginyl residue (17,18). AEP has been found to play important roles in diseases including immunity and neurodegenerative disease through the specific proteolysis of substrates such as Toll-like receptor 9 and Foxp3 (19–29). AEP has been observed to be highly expressed in a variety of tumors with little knowledge of its substrates (18, 25–30).
Here, we sought to determine the functional role of AEP in GBM malignant progression and analyzed the prognostic significance of AEP in GBM patients. Moreover, we explored the underlying mechanisms of AEP, which involve proteolytic inactivation of WT-p53 in GBM cells as well as in noncancerous stromal cells via EVs.
Methods
Co-immunoprecipitation
Cell extracts were incubated with 0.5 μg individual antibody, and 20 μL protein G beads (GE Healthcare, Pittsburgh, PA). After overnight incubation, beads were washed four times with lysis buffer, separated by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis, and analyzed by immunoblotting.
Immunofluorescence
The following primary antibodies were used for immunofluorescence: anti-AEP (R&D Systems, MN, USA, AF2199, 1: 200) and anti-p53 (Santa Cruz Biotechnology, sc-126, 1: 250); donkey anti-goat or anti-mouse Alexa fluor 488 or 568 (Invitrogen, CA, USA, 1: 1000) were used as secondary antibodies.
Cell Functional Assays
For the colony formation assay, GBM cells with or without AEP knockdown or overexpression were examined. For the tube formation assay, GBM cells or human umbilical vein endothelial cells (HUVECs) were added on the top of Matrigel in 96-well plates. For the invasion assay, GBM cells or human normal glial cells (HEBs) were plated in the upper chambers of Matrigel-coated-Transwell inserts (Millipore, Billerica, MA). For details, please see the Supplementary Methods (available online).
In Vivo Analysis of GBM Progression
All mice were maintained and treated in accordance with the guidelines approved by the Institutional Animal Care and Use Committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine. p16Ink4a/p19Arf; K-Rasv12; LucR Cre-LoxP-based conditional mice aged 4 to 6 weeks, which were a kind gift from Professor Olaf van Tellingen, were used in the experimental procedures (31).
Patients
Glioma tissues were collected from patients at Renji Hospital, Shanghai Jiao Tong University School of Medicine, after obtaining the patients’ informed consent. Our study was approved by the Ethics Committee of the Renji Hospital, Shanghai Jiao Tong University School of Medicine. All patients obtained a confirmed diagnosis of glioma after resection. Supplementary Table 2 (available online) describes the glioma patient characteristics by AEP expression level. AEP expression levels were classified as low expression and high expression. The median follow-up time of glioblastoma was 12 (12.19) months. Patients who were alive before April 1, 2019, were designated no death, and patients who died before April 1, 2019, were designated death.
Enzyme-Linked Immunosorbent Assay
AEP concentrations in culture medium or serum were measured as previously described (30). Recombinant human AEP (R&D Systems) was used as the standard to determine the optical density of each well immediately using a microplate reader at 450 nm, with correction at 570 nm.
Statistical Analysis
The statistical significance of the differences between two groups was determined using the Student t test. For multiple group comparisons, analysis of variance was followed by two-tailed Dunn’s post hoc analysis, or the Tukey’s multiple comparisons test was performed. Survival analyses comparing the prognoses of glioblastoma patients with low AEP or high AEP took confounding risk factors into account. Statistical analysis was performed using SPSS 15.0 software (Chicago, IL). All P values are two-sided. P values of less than .05 were considered statistically significant.
Detailed materials and methods are in the Supplementary Materials (available online).
Results
Interaction Between AEP and Wild-Type p53 in GBM
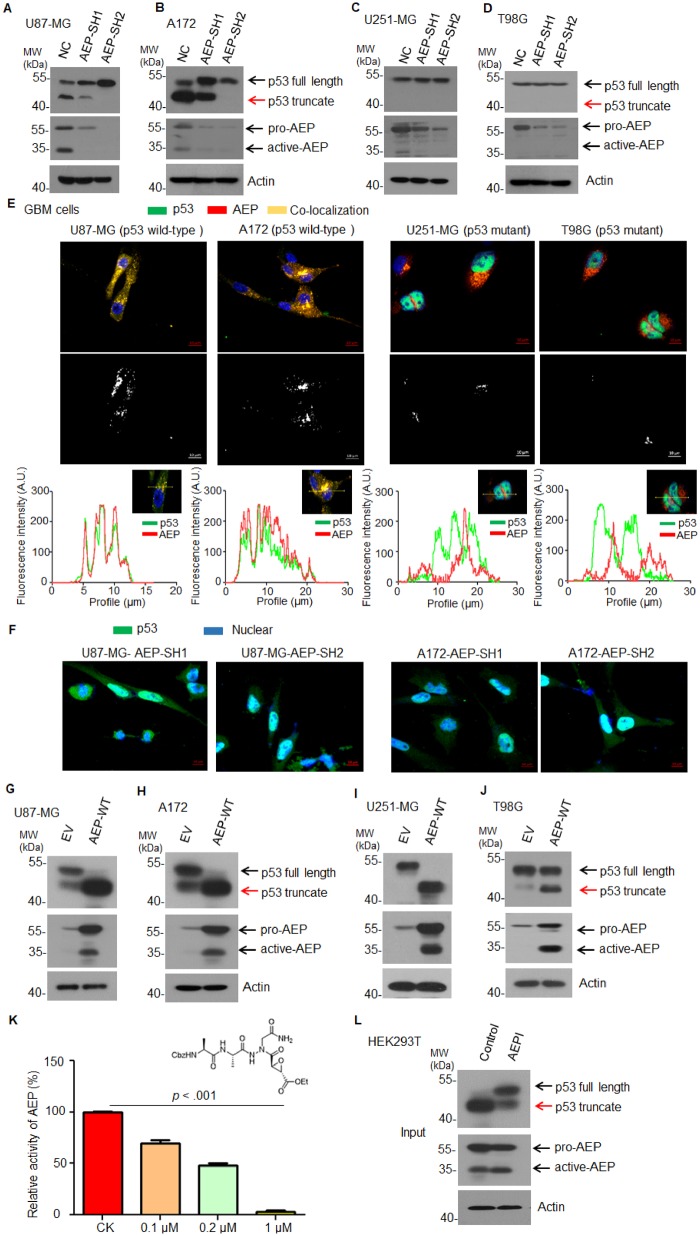
To find the substrates of AEP in GBM, we screened a series of tumor suppressor genes and found that WT-p53 could be cleaved by AEP (Supplementary Figure 1A, available online, antibody epitope mapping between amino acid residues 11–25 of p53). The cysteine 189 (C189) residue is essential for AEP enzymatic activity (18) and inactivation of this critical site (AEP-C189S) failed to cleave p53 (Supplementary Figure 1B, available online). We further knocked down AEP in p53-WT-GBM cells (U87-MG and A172) and found that the silencing of AEP abolished p53 cleavage (Figure 1, A and B). However, the active form of AEP was much lower in U251-MG and T98G (GBM cells with p53 mutations: p53-R273H in U251-MG and p53-M237I in T98G) (Figure 1, A–D). Correspondingly, we did not notice any cleavage of mutant p53 in these cells (Figure 1, C and D). Immunofluorescent confocal microscopy revealed colocalization of p53 and AEP in U87-MG and A172 but not in U251-MG and T98G (Figure 1E). Moreover, silencing of AEP in U87-MG and A172 remarkably elevated the level of nuclear p53 (Figure 1F). To further determine the cellular compartment in which AEP and p53 interaction occurs, we performed co-immunostaining of AEP, p53 with endosomal marker-EEA1, and lysosomal marker-LAMP1 as well as endoplasmic reticulum (ER)- and Golgi-associated marker-ERPp72 in U87-MG and A172. We found that AEP and p53 could only colocalize with endosomal and lysosomal markers, whereas no colocalization occurred with the ER- and Golgi-associated marker (Supplementary Figure 2, available online), indicating that AEP and p53 interaction occurs in the endolysosomal system. Moreover, AEP and p53 were colocalized with endolysosomal markers but not with the ER- and Golgi-associated marker in GFP-P53 and AEP cotransfected 293 T (Supplementary Figure 3, available online). Consistently, overexpressing WT-AEP in U87-MG and A172 increased the endogenous cleaved p53 truncates (Figure 1, G and H). Surprisingly, overexpression of AEP in U251-MG and T98G also induced mutant p53 cleavage (Figure 1, I and J).
Figure 1.
Effect of AEP on p53 in GBM cells. A) Immunoblot analysis of p53, AEP, and actin in U87-MG cells with or without AEP silencing. B) Immunoblot analysis of p53, AEP, and actin in A172 cells with or without AEP silencing. C) Immunoblot analysis of p53, AEP, and actin in U251-MG cells with or without AEP silencing. D) Immunoblot analysis of p53, AEP, and actin in T98G cells with or without AEP silencing. E) Immunofluorescence analysis of p53 and AEP in U87-MG, A172, U251-MG, and T98G cells (magnification: 400×; scale bar = 10 μm). The middle graph shows the colocalized signals between the green signal (p53) and the red signal (AEP). The intensity profile plots also analyzed using image J software. F) Immunofluorescence analysis of p53 in U87-MG and A172 cells with AEP silencing (magnification: 400×; scale bar = 10 μm). G) Immunoblot analysis of p53, AEP, and actin in U87-MG cells with or without AEP overexpression. H) Immunoblot analysis of p53, AEP, and actin in A172 cells with or without AEP overexpression. I) Immunoblot analysis of p53, AEP, and actin in U251-MG cells with or without AEP overexpression. J) Immunoblot analysis of p53, AEP, and actin in T98G cells with or without AEP overexpression. K) Structure and activity analysis of AEPI. Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. L) Immunoblot analysis of p53 and AEP in AEP-overexpressing HEK293T cells with or without AEPI treatment. AEP = asparaginyl endopeptidase; AEPI = AEP inhibitor; EV = empty vector; GBM = glioblastoma; MW = molecular weight; NC = negative control; SH = short hairpin; WT = wild type.
AEP activity is pH dependent, remaining active at pH 6.0 and inactive at pH 7.0. P53 was completely degraded at pH 6.0, whereas it remained intact at pH 7.0 (Supplementary Figure 4A, available online). Consistently, AEP-C189S mutant failed to degrade p53 at pH 6.0 (Supplementary Figure 4B, available online). We also lysed primary GBM tissues at pH 6.0 or pH 7.0 and found that p53 was robustly cleaved at pH 6.0 but remained intact at pH 7.0 (Supplementary Figure 4C, available online). Furthermore, we synthesized aza-Asn epoxide, which is an AEP-specific small inhibitory compound (AEPI). AEPI effectively suppressed the enzymatic activity of AEP at 1 µM (AEPI [SD] = 2.86[2.19]%, control [SD]= 100.18[0.27]%; P < .001) (Figure 1K). AEP-induced cleavage of p53 was inhibited by AEPI (Figure 1L).
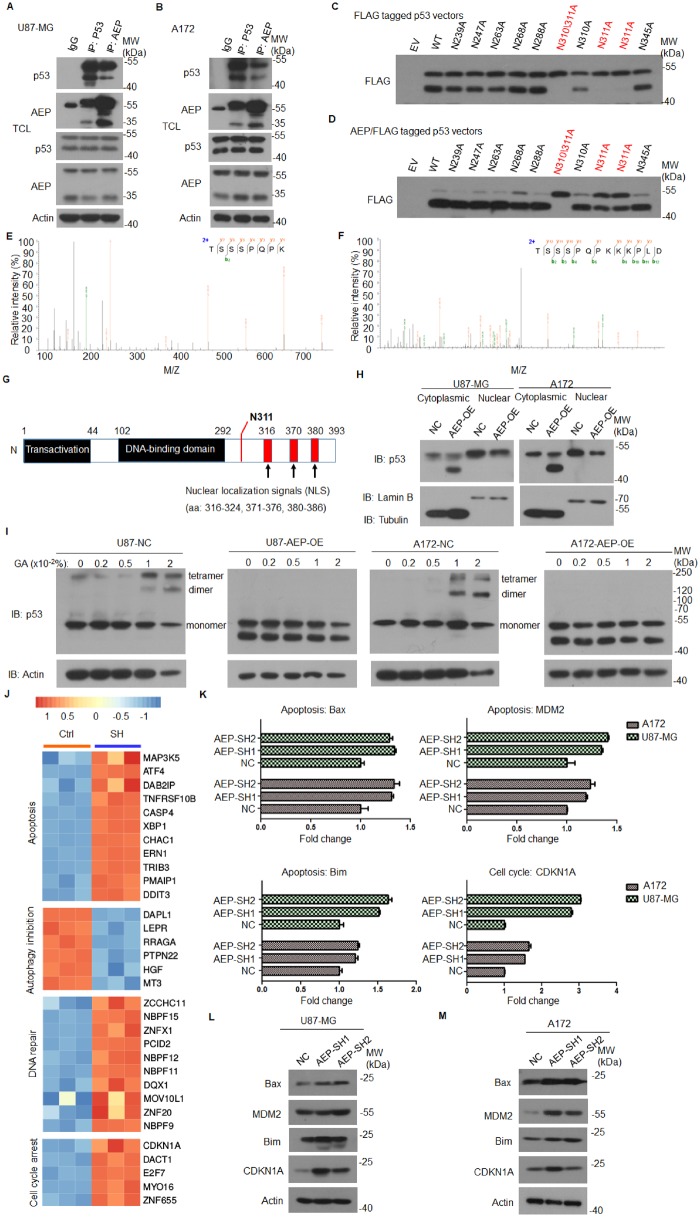
AEP and p53 interacted directly with each other in U87-MG and A172, and p53 can interact with pro-AEP and a smaller amount of p53 could also interact with active AEP (Figure 2, A and B). To determine the precise cleavage site in p53, we mutated each individual asparagine (N) residue of p53 into an alanine (A). We found that a p53-N311A-mutant remained intact, whereas the WT-p53 was cleaved, indicating that p53 was selectively cleaved at N311 (Figure 2, C and D). We further purified the fragmented glutathione S-transferase (GST)-p53 proteins and performed mass spectrometry analysis. The liquid chromatography-tandem mass spectrometry analysis revealed that the p53 peptide fragments cleaved at N311 from the partial digested peptides with trypsin or Glu-C, respectively (Figure 2, E and F).
Figure 2.
Interaction between AEP and WT p53 in GBM. A) Co-immunoprecipitation analysis of AEP and p53 in U87-MG cells. B) Co-immunoprecipitation analysis of AEP and p53 in A172 cells. C) Immunoblot analysis of FLAG-tagged p53 in HEK293T cells transfected with WT p53 and a series of mutations of its asparagine residues. D) Immunoblot analysis of FLAG-tagged p53 in HEK293T cells transfected with AEP and WT p53 or a series of mutations of its asparagine residues. E and F) Mass spectrometry analysis of recombinant p53 fragmented by AEP. The detected Mass Spectrometer/Mass Spectrometer peptides spectra are listed. Protein samples were in-gel digested with trypsin (E) or Glu-C (F), respectively. G) Domain analysis of p53 with nuclear localization signals located at its C-terminal. H) Subcellular fractionation of cytoplasmic and nuclear to gain quantitative insight into the amount of p53 that is relocated to the cytoplasm. I) The oligomeric forms of p53 were analyzed in GBM cells with or without AEP-OE. J) Heatmap of a subset of p53 downstream genes differentially regulated in AEP-silenced cells. K) Reverse transcriptase polymerase chain reaction (RT-PCR) analysis of BAX, MDM2, Bim, and CDKN1A expression in U87-MG and A172 cells with or without AEP knockdown. Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. L and M) Immunoblot analysis of BAX, MDM2, Bim, and CDKN1A in U87-MG and A172 cells with or without AEP silencing. AEP = asparaginyl endopeptidase; EV = empty vector; GBM = glioblastoma; MW = molecular weight; NC = negative control; OE = overexpression; SH = short hairpin; TCL = total cell lysate; WT = wild type.
There are three potential nuclear localization signals located in the C-terminus of p53 after N311. AEP increased cytoplasmic p53 but reduced its nuclear levels. Indeed, the active tetrameric form of p53 in AEP-overexpressing cells was also reduced (Figure 2, G–I; Supplementary Figure 5, A and B, available online).
We then studied the impact of AEP in GBM cells by performing an RNA-array analysis of the control and AEP-silenced U87-MG. More than 3388 transcripts were found to be differentially expressed (fold change ≥ 2) upon AEP silencing (P < .01). Among the p53 targeted genes, those related to cell cycle arrest (37.1% of the p53 downstream effector genes), apoptosis (81.7% of the p53 downstream effector genes), DNA repair (74.3% of the p53 downstream effector genes), and autophagy (47.1% of the p53 downstream effector genes) were suppressed when p53 was cleaved by AEP (Figure 2J;Supplementary Table 1, available online). Transcription of genes such as Bax, MDM2, Bim, and CDKN1A was elevated after AEP silencing (Figure 2K). The protein levels of these genes were also increased in the same cells (Figure 2, L and M). The chromatinic immunoprecipitation of p53 in AEP-overexpressing cells displayed a reduction of gene occupancy (Supplementary Figure 5, C, available online).
Moreover, both the proform and active form of AEP were upregulated by estimated glomerular filtration rate (EGFR) activation, especially in U87-MG and A172 (Supplementary Figure 6, A and B, available online). Overexpression of the EGFRvIII, which is the predominant form in GBMs, consistently upregulated AEP expression (Supplementary Figure 6, C–E, available online). Although it is published that p53 regulates AEP expression (32), knockdown of p53 in GBM cells did not alter the expression of AEP (Supplementary Figure 7, available online).
Altogether, these data suggest that AEP binds to and directly cleaves p53 at N311, resulting in the transcriptional inhibition of WT-p53 downstream effector genes in WT-p53 GBM cells.
Effect of AEP on GBM Cell Malignant Functions
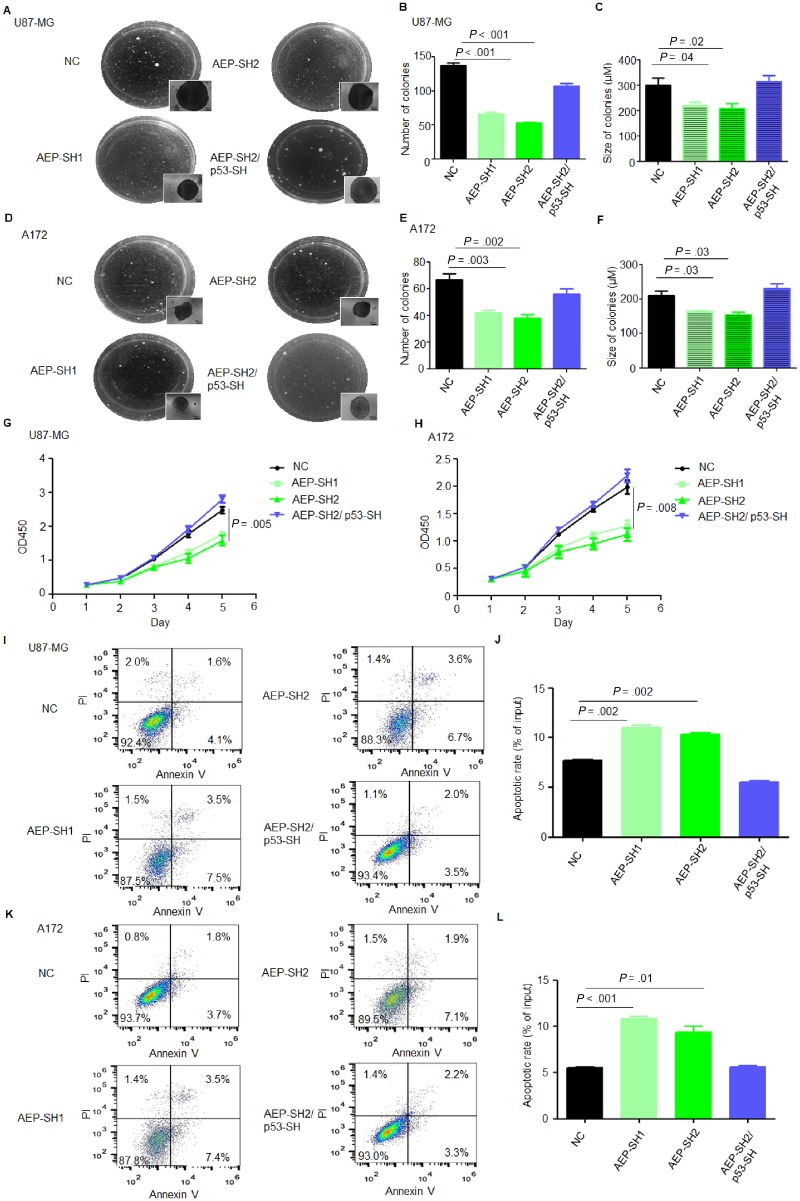
We then investigated the pathological functions of AEP in GBM cells. Colony formation assays showed that AEP silencing reduced colony formation (Figure 3, A–F). Cell growth was slower after AEP silencing in U87-MG and A172 (Figure 3, G and H). Additionally, the percentage of apoptotic cells was increased among AEP-silenced cells (Figure 3, I–L). However, suppression of p53 in AEP-silenced cells rescued the tumorigenesis, proliferation, and anti-apoptotic abilities (Figure 3). The invasive capacity was also reduced in AEP-silenced cells but remained intact in AEP and p53 double-silenced cells (Supplementary Figure 8, available online).
Figure 3.
Effect of AEP inhibition on p53 wild-type GBM cell malignant functions. A) Colony formation assay of U87-MG cells with the negative control (NC), AEP silencing, or AEP plus p53 silencing (magnification: 400×; scale bar = 50 μm). B and C) Statistical analysis of the (B) number and (C) size of the colonies shown in (A). D) Colony formation assay of A172 cells with the NC, AEP silencing, or AEP plus p53 silencing (magnification: 400×; scale bar = 50 μm). E and F) Statistical analysis of the (E) number and (F) size of the colonies shown in (D). G) CCK8 analysis of U87-MG cells with the NC, AEP silencing, or AEP plus p53 silencing. H) CCK8 analysis of A172 cells with the NC, AEP silencing, or AEP plus p53 silencing. I and J) Apoptosis analysis of U87-MG cells with the NC, AEP silencing, or AEP plus p53 silencing. K and L) Apoptosis analysis of A172 cells with the NC, AEP silencing, or AEP plus p53 silencing. Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. AEP = asparaginyl endopeptidase; GBM = glioblastoma; NC = negative control; PI = Propidium Iodide; SH = short hairpin.
AEP overexpression promoted colony formation (Figure 4, A–F). Cell growth was also faster after its overexpression (Figure 4, G and H). The percentage of apoptotic cells among AEP-overexpressing cells was decreased (Figure 4, I–L). However, forcible expression of p53-N311A in AEP-overexpressing cells suppressed the tumorigenesis, proliferation, and anti-apoptotic abilities induced by AEP in U87-MG and A172 (Figure 4). AEP overexpression also suppressed the DNA repair responses of U87-MG after radiation (Supplementary Figure 9, available online). Additionally, autophagy was increased after AEP silencing or overexpression of the mutant-p53-N311A (Supplementary Figure 10, available online). These results indicated that AEP promotes malignant behaviors of GBM cells partially through inactivation of p53.
Figure 4.
Effect of AEP overexpression on p53 wild-type GBM cell malignant functions. A) Colony formation assay of U87-MG cells with AEP overexpression or AEP plus p53 N311A overexpression (magnification: 400×; scale bar = 50 μm). B and C) Statistical analysis of the (B) number and (C) size of colonies shown in (A). D) Colony formation assay of A172 cells with AEP overexpression or AEP plus p53 N311A overexpression (magnification: 400×; scale bar = 50 μm). E and F) Statistical analysis of the (E) number and (F) size of colonies shown in (D). G) CCK8 analysis of U87-MG cells with AEP overexpression or AEP plus p53 N311A overexpression. H) CCK8 analysis of A172 cells with AEP overexpression or AEP plus p53 N311A overexpression. I and J) Apoptosis analysis of U87-MG cells with AEP overexpression or AEP plus p53 N311A overexpression. K and L) Apoptosis analysis of A172 cells with AEP overexpression or AEP plus p53 N311A overexpression. Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. AEP = asparaginyl endopeptidase; GBM = glioblastoma; NC = negative control; OE = overexpression; PI = Propidium Iodide.
Effect of AEP-Containing EVs Derived from GBM Cells on Stromal Cell Functions
It is increasingly clear that the tumor microenvironment can influence tumorigenesis. P53 has been found to exhibit loss of function in the mesenchyme (10). Surprisingly, the culture medium collected from GBM cells contained a large amount of AEP protein, and p53 in HEBs and HUVECs was cleaved after incubation with culture medium derived from GBM cells (Figure 5, A and B). We immunostained AEP in GBM cells and observed by immunoelectron microscopy. AEP existed in the EVs secreted by GBM cells (Figure 5, C and D). Thus, we separated the EVs and incubated them with HUVECs and HEBs and found that the EVs were endocytosed by these cells (Supplementary Figure 11, available online). Consistently, p53 was cleaved in these cells after incubation (Figure 5, E and F). The tube formation activity of HUVECs was increased with GBM-EVs but not with EVs collected from GBM cells in which AEP was silenced (Figure 5, G and H). However, HUVECs with p53 suppression showed increased tube formation, even when the cells were incubated with EVs collected from AEP-silenced GBM cells. Additionally, the invasive ability of HEBs was regulated likewise (Figure 5, I and J). Moreover, overexpression of AEP enhanced HEBs invasive ability and remarkably elevated the tube formation of HUVECs. P53 truncation and target gene repression were increased when AEP were overexpressed in these cells (Supplementary Figure 12, available online). Altogether, these results indicate that AEP derived from the EVs of GBM cells regulates the tumor-promoting functions of stromal cells in the tumor microenvironment.
Figure 5.
Effect of AEP-containing EVs derived from GBM cells on stromal cell functions. A) Immunoblot analysis of p53, AEP, and actin in HEB cells cultured with or without GBM cell-conditioned medium. B) Immunoblot analysis of p53, AEP, and actin in HUVECs cultured with or without GBM cell-conditioned medium. C) Immunoelectron microscopy analysis of AEP in extracellular vesicles released by U87-MG cells (magnification: 33000×; scale bar = 250 nm). D) Immunoelectron microscopy analysis of AEP in extracellular vesicles released by A172 cells (magnification, 46000×; scale bar = 100 nm). E) Immunoblot analysis of p53, AEP, and actin in HEB cells cultured with or without GBM cell-derived extracellular vesicles. F) Immunoblot analysis of p53, AEP, and actin in HUVECs cultured with or without GBM cell-derived extracellular vesicles. G and H) Tube formation assay of HUVECs with or without p53 silencing cultured with conditioned medium derived from wild-type GBM cells or AEP-suppressed GBM cells (magnification: 100×; scale bar = 50 μm). Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. I and J) Transwell assay of HEB cells with or without p53 silencing cultured with conditioned medium derived from wild-type GBM cells or AEP-suppressed GBM cells (magnification: 200×; scale bar = 100 μm). Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. AEP = asparaginyl endopeptidase; CM = culture medium; EVs = extracellular vesicles; GBM = glioblastoma; HEB = human normal glial cells; HUVEC = human umbilical vein endothelial cells; MW = molecular weight; NC = negative control; SH = short hairpin.
Effect of AEP on GBM Progression In Vivo
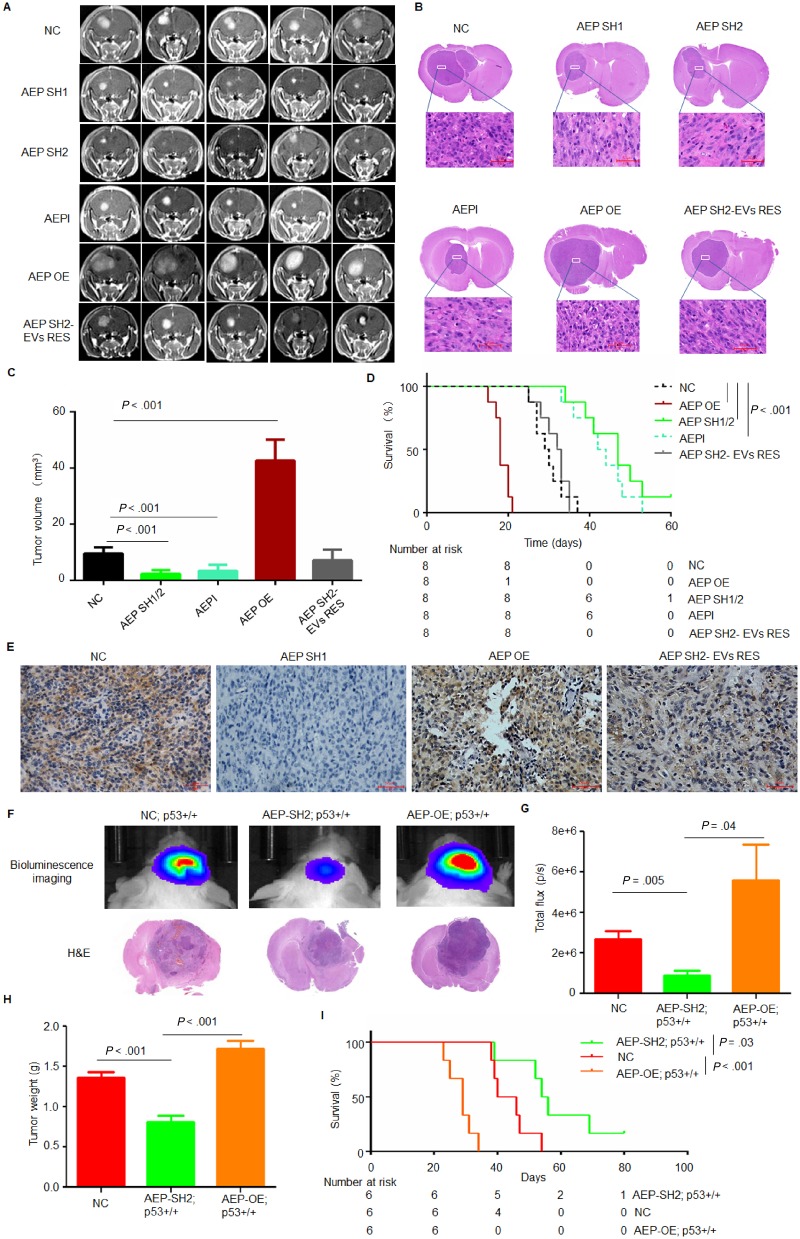
Next, we studied the in vivo function of AEP by implanting a subline of U87-NC, U87-AEP-short hairpin (SH), or U87-AEP-OE cells into the corpus striata of athymic nude mice. After 16 days, at which point a few animals started to show signs of morbidity, mice in each experimental group were assessed by magnetic resonance imaging to confirm intracranial tumor formation. Tumor growth in the AEP-SH group was much slower than that in the control group. Treatment with AEPI statistically significantly suppressed tumor progression (n = 8 per group; AEPI [SD] = 3.42 [2.04] mm3, negative control [NC] [SD] = 9.54 [2.20] mm3; P = .002). In contrast, AEP-overexpression (OE) resulted in a statistically significant increased tumor volume (n = 8 per group; AEP-OE [SD] = 42.64 [7.44] mm3; P < .001). Injection of EVs collected from U87-NC into the mice inoculated with AEP-SH cells restimulated tumor progression (Figure 6, A–C).
Figure 6.
Effect of AEP on GBM progression in vivo. A) Representative axial magnetic resonance images of xenograft GBM tumors orthotopically inoculated with U87-NC, U87-AEP-SH, or U87-AEP-OE cells on day 16 postimplantation. Mice inoculated with U87-NC were treated with AEPI (1 mg/mL) by tail vein injection. Mice inoculated with U87-AEP-SH were treated with EVs collected from U87-NC cells by tail vein injection. B) Representative H&E images of every group are shown (magnification: 400×; scale bar = 50 μm). C) The tumor volume was calculated in every group (n = 8 per group). Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. D) Survival time comparison of all groups of mice. The time of death was recorded as days after the GBM cell implantation. The K-M method was used to estimate survival curves. The K-M survival analysis was performed by SPSS15.0. E) Representative immunohistochemistry images of AEP in tumors collected from every group are shown (magnification: 400×; scale bar = 50 μm). F and G) Representative 2-D luminescence images of gliomas induced in p16Ink4a/p19Arf; K-Rasv12; LucR mice with AEP knockdown or overexpression (n = 6 per group). The data are presented as the average radiance efficiency [(photons/s/cm2/steradian)/(μW/cm2)]. Images are presented with the same scale bar. H) The tumor weight of each group is shown. Data are presented as mean (SD). The two-tailed Student t test was used to analyze the differences between the groups. I) Survival time comparison of all groups of mice. The time of death was recorded as days after the Cytomegalovirus promoter-cyclization recombinase (CMV-CRE) lentivirus injection. The K-M method was used to estimate survival curves. The K-M survival analysis was performed by SPSS15.0. AEP = asparaginyl endopeptidase; AEPI = AEP inhibitor; EVs RES = extracellular vesicles rescue; GBM = glioblastoma; H&E = hematoxylin and eosin; K-M = Kaplan-Meier; NC = negative control; OE = overexpression; SH = short hairpin.
Consequently, mice implanted with AEP-OE cells died within 21 days, whereas 100% of the control mice survived for that duration. Silencing of AEP or AEPI dramatically prolonged the survival of the mice compared with that of the control group (n = 8 per group; median survival: AEP-SH = 47, 95% confidence interval [CI] = 38.95 to 55.05 days; AEPI = 42, 95% CI = 37.84 to 46.16 days; NC = 29, 95% CI = 24.84 to 33.16 days; AEP-OE vs NC: P < .001; AEP-SH vs NC: P < .001; AEPI vs NC: P < .001) (Figure 6D). The expression of AEP in each group of tumors was also examined (Figure 6E). TUNEL assays of sections of inoculated tumors revealed that apoptotic cells were increased in tumors inoculated with AEP-silenced cells and were reduced in the AEP-overexpressed group (Supplementary Figure 13, available online).
We further investigated the tumor-promoting function of AEP by utilizing transgenic high-grade glioma mice, which were established through stereotactic intracranial injections of CMV-Cre lentivirus into LoxP-conditional mice, resulting in K-Rasv12 expression and loss of p16Ink4a and p19Arf (33). Lentiviruses targeting AEP or overexpressing AEP were intracranially injected into the transgenic mice. AEP-SH reduced glioma progression (Figure 6F), and the total flux and the tumor weight were also reduced after AEP-SH (n = 6 per group; total flux: AEP-SH [SD] = 0.89 × 106 [0.58 × 106] p/s; NC [SD] = 2.68 × 106 [0.98 × 106] p/s; P = .005; tumor weight: AEP-SH [SD] = 0.82 [0.20] g; NC [SD] = 1.37 [0.16] g; P < .001) (Figure 6, G and H). Consistently, the survival time of these tumor-bearing mice indicated that AEP suppression statistically significantly prolonged survival (n = 6 per group; median survival: AEP-SH = 54, 95% CI = 49.2 to 58.8 days; NC = 40, 95% CI = 31.6 to 48.4 days; P = .03) (Figure 6I). However, overexpression of AEP enhanced glioma progression and shortened survival (n = 6 per group; median survival: AEP-OE = 29, 95% CI = 24.47 to 33.53 days; NC = 40, 95% CI = 31.6 to 48.4 days; P < .001) (Figure 6, F–I).
The Influence of AEP Expression on Survival in GBM
We collected human low-grade gliomas (LGG), GBMs, and normal tissue specimens to evaluate AEP expression. Immunoblotting results showed that AEP was upregulated in GBM tissues (Figure 7A). By analyzing TCGA data, we found that AEP mRNA levels were not prognostic in GBMs as well as in other solid tumors (data not shown), thus we analyzed the protein levels of AEP in GBM tissues. We performed immunohistochemical analyses to examine AEP in a tissue array of 99 human glioma specimens by using an antibody with validated specificity (Supplementary Table 2, available online). AEP staining was strong in tumor cells as well as in stromal cells (Figure 7B). The survival durations of GBM patients with low and high AEP expression were compared (n = 83; hazard ratio [HR] = 3.94, 95% CI = 1.87 to 8.28; P < .001) (Figure 7, C and D). Patients whose tumors showed low AEP expression had a median survival time of 26 months (95% CI = 8.91 to 43.09), whereas the median survival time of patients decreased to 8 months when their tumors showed high levels of AEP (Figure 7D;Supplementary Table 3, available online). These results demonstrate that AEP is highly expressed in GBM and associated with poor prognosis. The enzyme-linked immunosorbent assay results showed that the AEP concentration in the serum of GBM patients was statistically significantly higher than of LGG patients or healthy donors (GBM n = 20, GBM [SD] = 411.77 [414.20] ng/mL; LGG n = 16, LGG [SD] = 116.13 [165.15] ng/mL; healthy donors n = 48, healthy donors [SD] = 50.37 [66.19] ng/mL; GBM vs LGG: P = .004; GBM vs healthy donors: P = .001) (Figure 7E). Intriguingly, the concentration of AEP in serum was decreased after surgery (n = 15, presurgery [SD] = 203.48 [260.15] ng/mL; postsurgery [SD] = 40.95 [77.56] ng/mL; P = .003) (Figure 7F). Pearson correlation analysis showed a statistically significant positive correlation of the plasma AEP concentration with the tumor size (r = 0.6; P = .03, n = 14) (Figure 7G).
Figure 7.
The influence of AEP expression on survival in GBM. A) Immunoblot analysis of AEP in fresh GBM tissues (n = 4), LGG (n = 4), and normal controls (n = 4). The experiment was repeated three times. B) IHC staining with an antibody to AEP was performed on 83 glioma specimens. Images with representative staining are presented (magnification: 400×; scale bar = 50 μm). C) Representative cases indicating negative, weak, moderate, and strong AEP staining in 67 GBM tissues are shown. The patient tumors were stratified into high and low AEP expression based on median cutoff of continuous H-scoring (cutoff value = 6 score; range = 0–12; magnification: 400×; scale bar = 50 μm). D) K-M curves of AEP expression in GBM tumor tissues in relation to overall survival (n = 83; P < .001). The K-M survival analysis was performed by SPSS13.1. E) ELISA of the AEP concentrations in the sera of GBM patients, low-grade glioma patients, and healthy controls (GBM: n = 20, LGG: n = 16, healthy donors: n = 48). F) ELISA of the AEP concentrations in the sera of GBM patients pre- and postsurgery (n = 15). The two-tailed Student t test was used to analyze the differences between the groups. G) Pearson correlation analysis showed a statistically significant positive correlation between plasma AEP concentration and tumor size (n = 14, r = 0.6; P = .03). H and I) Immunoblot analysis of AEP and p53 in fresh p53-WT/IDH-WT GBM tissues is shown (n = 30). J) The enzymatic activity of AEP in fresh p53-WT/IDH-WT GBM tissues is shown (n = 30). AEP = asparaginyl endopeptidase; dF = differential fluorescence; ELISA = enzyme-linked immunosorbent assay; GBM = glioblastoma; IB = immunoblot; IHC = immunohistochemical; K-M = Kaplan-Meier; LGG = low-grade gliomas; MW = molecular weight; WT = wild type.
We further analyzed WT-p53 cleavage in protein lysates from newly diagnosed human GBMs with WT-p53. We found that 46.7% (14 of 30) of human GBMs with p53-WT and IDH-WT showed cleavage of p53 (Figure 7H). Furthermore, in the p53-WT and IDH-WT-GBMs, the range in terms of the percent of p53 that was cleaved in an individual tumor was 0.04–53.6% (Figure 7I). The AEP activity was higher in samples where p53 cleavage was more profound (Figure 7J). Thus, these data indicate that AEP is highly expressed in GBMs and associated with poor prognosis.
Discussion
Among all cancers, IDH-WT-GBM has the lowest rate of p53 mutations, but the p53 pathway is functionally disabled (34, 35). Our findings suggest a novel mechanism of WT-p53 inactivation in GBMs. Highly expressed AEP cleaves p53 specifically at the asparaginyl bond, which disrupts the p53 tumor-suppressing function and is critical for GBM progression. The strict specificity of AEP for asparagine bonds is striking, which makes the identification of new AEP substrates important. In our study, we found that mutated p53 was barely cleaved by AEP. This might be caused by a different property of mutant p53 proteins that leads to a high accumulation in tumor cells.
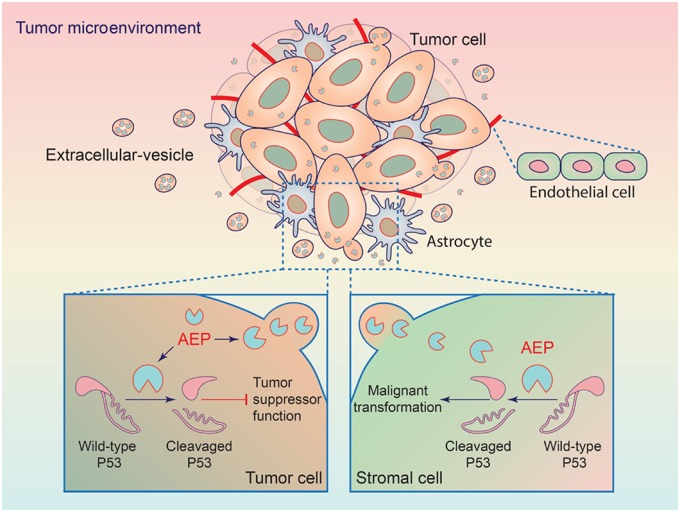
In addition, AEP cleavage of p53 occurs in not only GBM cells but also stromal cells in the tumor microenvironment through EVs. GBM-derived EVs have been found to participate in modifying the phenotype of stromal cells (36, 37). EVs derived from glioma carry a variety of biomolecules, such as oncogenic growth factors, receptors, and enzymes (36, 38, 39). We found that AEP was loaded in EVs and promoted GBM progression (Figure 8). This finding provides more potential and importance of AEP regarding the circumstances in which p53 mutant tumor cells propagate into an environment where stromal cells express WT-p53. Moreover, p53 inhibition may improve the efficiency of the genome editing of untransformed cells (40, 41). AEP proteolysis of p53 may provide an alternative way to achieve p53 inhibition.
Figure 8.
Model for the mechanism of action of AEP in GBM. Wild-type p53 proteolytic inactivation by AEP in GBM tumor cells (bottom left) promotes tumor cell functions. Additionally, AEP is secreted by GBM tumor cells and endocytosed by stromal cells including astrocytes and endothelial cells to promote the malignant transformation of these cells (bottom right). AEP = asparaginyl endopeptidase; GBM = glioblastoma.
As in other cancer types, AEP was highly expressed in GBM and associated with poor prognosis, which indicated that AEP is a potential biomarker and therapeutic target for GBM. A potential limitation of the study concerns the mechanism of AEP upregulation that remains to be uncharacterized in GBM. AEP expression is increased in response to stress stimuli including hepatocyte growth factor stimulation, hypoxia, and oxidative stresses in tumors (30, 42). Our study found that AEP was likely upregulated by EGFR activation in GBM. A second issue that needs to be investigated further is the regulation of enzymatic activity of AEP, which is important for its biological functions. As a protease, AEP requires a multistep activation process to reach its mature form, which is not completely elucidated. Besides its pH dependence, AEP activity is regulated by cystatin C, cystatin E/M (18). Integrin binding to AEP can also prompt AEP activity (43).
Targeting AEP with a vaccine has been found to be effective in the targeted therapy of cancer (30). Individuals who overexpress AEP with their tumor infiltrating lymphocytes may be more responsive to programmed cell death 1– and/or programmed death ligand 1–based immunotherapeutics (22). Moreover, the strict substrate specificity of AEP combined with its overexpression in various tumors has motivated the exploitation of AEP as a prodrug activator in cancer treatment, such as by adding a cleavable peptide chain to doxorubicin or auristatin (25). Finally, we propose the small compound used in our study as a potent inhibitor of AEP (44), which might have therapeutic potential in GBM therapy.
Funding
This work was supported by the National Natural Science Foundation of China (81671203, 81402042, 81772654), Shanghai Science and Technology (14140903400, 14YF1402600, 16140902900), Shanghai Municipal Population and Family Planning Commission (2013SY024), and the training plan for scientific research of Renji Hospital (RJZZ13-021).
Notes
The study sponsors had no role in in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication. The authors have declared that no conflict of interest exists.
YL, ZQ, YM, JJ, and YQ contributed to designing research studies, conducting experiments, acquiring data, analyzing data, providing reagents, and writing the manuscript. KL, ZF, XY, QN, GJ, JL, YY, XZ, and YH contributed to conducting experiments and acquiring data. All authors agreed to submit this manuscript for publication.
We gratefully thank Professor Olaf van Tellingen at the Netherlands Cancer Institute/Antoni van Leeuwenhoek Hospital.
Supplementary Material
References
- 1.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kastenhuber ER, Lowe SW.. Putting p53 in context. Cell. 2017;170(6):1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soragni A, Janzen DM, Johnson LM.. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell. 2016;29(1):90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Comel A, Sorrentino G, Capaci V, et al. The cytoplasmic side of p53’s oncosuppressive activities. FEBS Lett. 2014;588(16):2600–2609. [DOI] [PubMed] [Google Scholar]
- 5. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 6. Biernat W, Kleihues P, Yonekawa Y, et al. Amplification and overexpression of MDM2 in primary (de novo) glioblastomas. J Neuropathol Exp Neurol. 1997;56(2):180–185. [DOI] [PubMed] [Google Scholar]
- 7. Sayan BS, Sayan AE, Knight RA, et al. p53 is cleaved by caspases generating fragments localizing to mitochondria. J Biol Chem. 2006;281(19):13566–13573. [DOI] [PubMed] [Google Scholar]
- 8. Qin Q, Liao G, Baudry M, et al. Role of calpain-mediated p53 truncation in Semaphorin 3A-induced axonal growth regulation. Proc Natl Acad Sci USA. 2010;107(31):13883–13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kubbutat MH, Vousden KH.. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol Cell Biol. 1997;17(1):460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hill R, Song Y, Cardiff RD, et al. Selective evolution of stromal mesenchyme with p53 loss in response to epithelial tumorigenesis. Cell. 2005;123(6):1001–1011. [DOI] [PubMed] [Google Scholar]
- 11. Wendler F, Favicchio R, Simon T, et al. Extracellular vesicles swarm the cancer microenvironment: from tumor-stroma communication to drug intervention. Oncogene. 2017; 36(7):877–884. [DOI] [PubMed] [Google Scholar]
- 12. Glioblastoma produces tumor-promoting microvesicles. Nat Clin Pract Neurol. 2009;5(3):120–121. [PubMed] [Google Scholar]
- 13. Burgess DJ. Glioblastoma: microvesicles as major biomarkers? Nat Rev Cancer. 2013;13(1):8.. [DOI] [PubMed] [Google Scholar]
- 14. Giusti I, Francesco M, Dolo V.. Extracellular vesicles in glioblastoma: role in biological processes and in therapeutic applications. Curr Cancer Drug Targets. 2017;17(3):221–235. [DOI] [PubMed] [Google Scholar]
- 15. Treps L, Edmond S, Harford-Wright E, et al. Extracellular vesicle-transported Semaphorin3A promotes vascular permeability in glioblastoma. Oncogene. 2016;35(20):2615–2623. [DOI] [PubMed] [Google Scholar]
- 16. Erkan EP, Senfter D, Madlener S, et al. Extracellular vesicle-mediated suicide mRNA/protein delivery inhibits glioblastoma tumor growth in vivo. Cancer Gene Ther. 2017;24(1):38–44. [DOI] [PubMed] [Google Scholar]
- 17. Dall E, Brandstetter H.. Structure and function of legumain in health and disease. Biochimie. 2016;122(12):126–150. [DOI] [PubMed] [Google Scholar]
- 18. Chen JM, Dando PM, Rawlings ND, et al. Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J Biol Chem. 1997;272(12):8090–8098. [DOI] [PubMed] [Google Scholar]
- 19. Miller G, Matthews SP, Reinheckel T, et al. Asparagine endopeptidase is required for normal kidney physiology and homeostasis. FASEB J. 2011;25(5):1606–1617. [DOI] [PubMed] [Google Scholar]
- 20. Manoury B, Hewitt EW, Morrice N, et al. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396(6712):695–699. [DOI] [PubMed] [Google Scholar]
- 21. Ye KQ, Liu ZX, Jang SW, et al. Neuroprotective actions of PIKE-L by inhibition of SET proteolytic degradation by asparagine endopeptidase. Mol Cell. 2008;29(6):665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stathopoulou C, Gangaplara A, Mallett G, et al. PD-1 inhibitory receptor downregulates asparaginyl endopeptidase and maintains FOXp3 transcription factor stability in induced regulatory T cells. Immunity. 2018;49(2):247–263.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clerin V, Shih HH, Deng N, et al. Expression of the cysteine protease legumain in vascular lesions and functional implications in atherogenesis. Atherosclerosis. 2008;201(1):53–66. [DOI] [PubMed] [Google Scholar]
- 24. Manoury B, Sepulveda FE, Maschalidi S, et al. Critical role for asparagine endopeptidase in endocytic toll-like receptor signaling in dendritic cells. Immunity. 2009;31(5):737–748. [DOI] [PubMed] [Google Scholar]
- 25. Liu C, Sun C, Huang H, et al. Overexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapy. Cancer Res. 2003;63(11):2957–2964. [PubMed] [Google Scholar]
- 26. Murthy RV, Arbman G, Gao J, et al. Legumain expression in relation to clinicopathologic and biological variables in colorectal cancer. Clin Cancer Res. 2005;11(6):2293–2299. [DOI] [PubMed] [Google Scholar]
- 27. Gawenda J, Traub F, Luck HJ, et al. Legumain expression as a prognostic factor in breast cancer patients. Breast Cancer Res Treat. 2007;102(1):1–6. [DOI] [PubMed] [Google Scholar]
- 28. Patel N, Krishnan S, Offman MN, et al. A dyad of lymphoblastic lysosomal cysteine proteases degrades the antileukemic drug L-asparaginase. J Clin Invest. 2009;119(7):1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andrade VA, Jardim CA, Melo FM, et al. Nucleoplasmic calcium regulates proliferation of hepatocytes through legumain and reticulon4. Gastroenterology. 2009;136(5):A-792. [Google Scholar]
- 30. Lin Y, Qiu Y, Xu C, et al. Functional role of asparaginyl endopeptidase ubiquitination by TRAF6 in tumor invasion and metastasis. J Natl Cancer Inst. 2014;106(4):dju012.. [DOI] [PubMed] [Google Scholar]
- 31. Lan J, Guo P, Lin Y, et al. Role of glycosyltransferase PomGnT1 in glioblastoma progression. Neuro Oncol. 2015;17(2):211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamane T, Murao S, Kato-Ose I, et al. Transcriptional regulation of the legumain gene by p53 in HCT116 cells. Biochem Biophys Res Commun. 2013;438(4):613–618. [DOI] [PubMed] [Google Scholar]
- 33. de Vries NA, Bruggeman SW, Hulsman D, et al. Rapid and robust transgenic high-grade glioma mouse models for therapy intervention studies. Clin Cancer Res. 2010;16(13):3431–3441. [DOI] [PubMed] [Google Scholar]
- 34. Kang YJ, Balter B, Csizmadia E, et al. Contribution of classical end-joining to PTEN inactivation in p53-mediated glioblastoma formation and drug-resistant survival. Nat Commun. 2017;8(1):14013.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leung SY, Yuen ST, Chan TL, et al. Chromosomal instability and p53 inactivation are required for genesis of glioblastoma but not for colorectal cancer in patients with germline mismatch repair gene mutation. Oncogene. 2000;19(35):4079–4083. [DOI] [PubMed] [Google Scholar]
- 36. Bastida E, Ordinas A, Escolar G, et al. Tissue factor in microvesicles shed from U87MG human glioblastoma cells induces coagulation, platelet aggregation, and thrombogenesis. Blood. 1984;64(1):177–184. [PubMed] [Google Scholar]
- 37. Liu S, Sun J, Lan Q.. Glioblastoma microvesicles promote endothelial cell proliferation through Akt/beta-catenin pathway. Int J Clin Exp Pathol. 2014;7(8):4857–4866. [PMC free article] [PubMed] [Google Scholar]
- 38. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koch CJ, Lustig RA, Yang XY, et al. Microvesicles as a biomarker for tumor progression versus treatment effect in radiation/temozolomide-treated glioblastoma patients. Transl Oncol. 2014;7(6):752–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haapaniemi E, Botla S, Persson J, et al. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24(7):927–930. [DOI] [PubMed] [Google Scholar]
- 41. Ihry RJ, Worringer KA, Salick MR, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24(7):939–946. [DOI] [PubMed] [Google Scholar]
- 42. Andrade V, Guerra M, Jardim C, et al. Nucleoplasmic calcium regulates cell proliferation through legumain. J Hepatol. 2011;55(3):626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao L, Hua T, Crowley C, et al. Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage. Cell Res. 2014;24(3):344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Niestroj AJ, Feussner K, Heiser U, et al. Inhibition of mammalian legumain by Michael acceptors and AzaAsn-halomethylketones. Biol Chem. 2002;383(7–8):1205–1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.