Abstract
The C9orf72 repeat expansion causes amyotrophic lateral sclerosis and frontotemporal dementia, but the poor correlation between C9orf72‐specific pathology and TDP‐43 pathology linked to neurodegeneration hinders targeted therapeutic development. Here, we addressed the role of the aggregating dipeptide repeat proteins resulting from unconventional translation of the repeat in all reading frames. Poly‐GA promoted cytoplasmic mislocalization and aggregation of TDP‐43 non‐cell‐autonomously, and anti‐GA antibodies ameliorated TDP‐43 mislocalization in both donor and receiver cells. Cell‐to‐cell transmission of poly‐GA inhibited proteasome function in neighboring cells. Importantly, proteasome inhibition led to the accumulation of TDP‐43 ubiquitinated within the nuclear localization signal (NLS) at lysine 95. Mutagenesis of this ubiquitination site completely blocked poly‐GA‐dependent mislocalization of TDP‐43. Boosting proteasome function with rolipram reduced both poly‐GA and TDP‐43 aggregation. Our data from cell lines, primary neurons, transgenic mice, and patient tissue suggest that poly‐GA promotes TDP‐43 aggregation by inhibiting the proteasome cell‐autonomously and non‐cell‐autonomously, which can be prevented by inhibiting poly‐GA transmission with antibodies or boosting proteasome activity with rolipram.
Keywords: antibody therapy, C9orf72, neurodegeneration, nucleocytoplasmic transport, proteasome
Subject Categories: Molecular Biology of Disease, Neuroscience
Poly‐GA dipeptide‐repeat expansion in C9orf72 inhibits proteasome activity to cause TDP‐43 pathology even in neighboring cells not carrying it, explaining how TDP‐43 aggregation arises in C9orf72 patients.
Introduction
Neuronal cytoplasmic aggregates of the nuclear RNA‐binding protein TDP‐43 are the key feature of sporadic amyotrophic lateral sclerosis (ALS) and define a large subgroup of frontotemporal dementia (FTD) neuropathologically (Geser et al, 2009; Ling et al, 2013; Scotter et al, 2015; Prasad et al, 2019). TDP‐43 undergoes constitutive nucleocytoplasmic shuttling mediated by a bipartite nuclear localization signal (NLS) and diffusion‐driven nuclear egress (Winton et al, 2008; Ederle et al, 2018) and active export (Aksu et al, 2018; Archbold et al, 2018). Normally, TDP‐43 is located predominantly in the nucleus and regulates expression, splicing, and polyadenylation of hundreds of target genes (Polymenidou et al, 2011; Tollervey et al, 2011). In ALS/FTD, cytoplasmic TDP‐43 aggregates are strongly correlated with neurodegeneration and contain ubiquitinated C‐terminal fragments (CTFs) that show characteristic hyperphosphorylation (Neumann et al, 2006; Geser et al, 2009; Igaz et al, 2009; Zhang et al, 2009). Cytoplasmic TDP‐43 aggregation most likely causes neurodegeneration through a combination of direct toxicity and loss of function due to nuclear clearance of TDP‐43 in affected cells (Gendron & Petrucelli, 2011; Walker et al, 2015; Ederle & Dormann, 2017; Prasad et al, 2019). The discovery of genetic mutations that cause familial ALS and/or FTD with TDP‐43 pathology similar to sporadic cases has highlighted the role of the ubiquitin–proteasome system (e.g., UBQLN2, VCP, SQSTM1) and the autophagy pathway (e.g., C9orf72, TBK1, OPTN) in pathogenesis (Ling et al, 2013; Scotter et al, 2015; Gotzl et al, 2016; Gao et al, 2017). However, apart from rare aggregation‐enhancing mutations directly in the TDP‐43 encoding gene, the cause of the pathological TDP‐43 mislocalization and aggregation in familial and sporadic cases remains elusive (Ederle & Dormann, 2017; Prasad et al, 2019).
The most common pathogenic mutation found in about 10% of all ALS/FTD patients is a massive (GGGGCC)n repeat expansion in the first intron of C9orf72 (DeJesus‐Hernandez et al, 2011; Renton et al, 2011). In addition to the typical TDP‐43 inclusion pathology, C9orf72 cases show nuclear foci of sense and antisense repeat RNA transcripts and unique aggregates of dipeptide repeat (DPR) proteins resulting from unconventional non‐ATG translation of the expanded repeat into poly‐GA/‐GP/‐GR/‐PA and poly‐PR (Edbauer & Haass, 2016). Moreover, C9orf72 protein expression from the mutant allele is reduced (Frick et al, 2018; Saberi et al, 2018). Multiple downstream effects of these three proposed pathomechanisms have been reported, but none of the C9orf72‐specific pathologies correlates reproducibly with TDP‐43 pathology and neurodegeneration in end‐stage tissue (Mackenzie et al, 2013, 2015; DeJesus‐Hernandez et al, 2017). Since all of the mutation‐specific effects occur many years or even decades prior to disease onset (Vatsavayai et al, 2016), chronic and possibly non‐cell‐autonomous effects that synergistically trigger disease once compensatory mechanisms fail seem the most likely explanation (Edbauer & Haass, 2016). We and others have reported cell‐to‐cell transmission of DPRs suggesting they may have non‐cell‐autonomous effects (Westergard et al, 2016; Zhou et al, 2017). In C9orf72 animal models, the most robust TDP‐43 pathology has so far been reported upon viral expression of the (GGGGCC)n repeat at high levels (Chew et al, 2015) and to a lesser extent in one of the BAC transgenic C9orf72 mouse lines (Liu et al, 2016) suggesting gain‐of‐function mechanisms are most important. Modest TDP‐43 pathology has been observed in transgenic poly‐GA mouse models (Zhang et al, 2016; Schludi et al, 2017). In cellular systems, poly‐GA expression has been linked to subtle TDP‐43 mislocalization (Khosravi et al, 2017) and phosphorylation (Nonaka et al, 2018). Using cryo‐electron tomography, we have shown that poly‐GA inclusions consist of amyloid‐like twisted ribbons that sequester large amounts of proteasomes stalled in an otherwise rare transition state (Guo et al, 2018b). Proteasome inhibitors promote TDP‐43 pathology in vitro (van Eersel et al, 2011), but DPR and TDP‐43 inclusions rarely occur within the same cell in patients (Mori et al, 2013).
Cell‐to‐cell transmission of cytoplasmic Tau and α‐synuclein aggregates drives stereotypic spreading of these pathologies during the progression of Alzheimer's and Parkinson's disease, respectively (Jucker & Walker, 2018). Therefore, we asked whether non‐cell‐autonomous effects of DPRs could trigger TDP‐43 pathology in neighboring cells focusing on effects on the proteasome and nucleocytoplasmic transport. Using co‐culture assays and antibody treatment to inhibit cell‐to‐cell transmission, we discovered that poly‐GA inhibits the proteasome non‐cell‐autonomously. Although the proteasome shows high constitutive activity that is largely limited by substrate availability in most cell types, activity can be boosted by rolipram through the cAMP/protein kinase A‐depended phosphorylation of PSMD11 (Lokireddy et al, 2015). We show that chemical and genetic proteasomal activation rescues poly‐GA‐induced mislocalization of TDP‐43 caused by ubiquitination within the TDP‐43 NLS.
Results
Cell‐to‐cell transmission of poly‐GA causes cytoplasmic mislocalization of TDP‐43
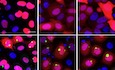
Among the DPR proteins, poly‐GA has been most robustly linked to TDP‐43 aggregation in vitro, although the mechanism is still unknown (Schludi et al, 2015; Khosravi et al, 2017; Lee et al, 2017; Nonaka et al, 2018; Solomon et al, 2018). We co‐expressed all DPRs with the aggregation‐prone CTF of TDP‐43 tagged with RFP. CTFs of TDP‐43 constitute the major aggregating TDP‐43 species in patient tissue and are generated by caspase cleavage (Neumann et al, 2006; Igaz et al, 2009; Zhang et al, 2009). Co‐expression of GA175‐GFP but not the other GFP‐tagged DPR species increased aggregation of TDP‐CTF (Appendix Fig S1A–D) without affecting turn‐over of TDP‐43‐CTF (Appendix Fig S1E and F) similar to the previous reports (Nonaka et al, 2018). In addition, we quantified mislocalization of endogenous TDP‐43 in anterior horn motor neurons of our transgenic mice expressing GA149‐CFP (Fig EV1A and B). Consistent with the increased phosphorylation of TDP‐43 at the disease‐associated residue S409/410 (Schludi et al, 2017), we detected enhanced levels of cytoplasmic TDP‐43 in the spinal cord of poly‐GA transgenic mice without detectable proteolytic cleavage (Fig EV1A–C). In this mouse model, TDP‐43 mislocalization is mostly seen in ChAT‐positive motor neurons where poly‐GA expression is most prominent, while the posterior horn shows no overt changes (Fig EV1D). A fully automated analysis pipeline revealed that poly‐GA‐positive neurons in the frontal cortex of C9orf72 FTLD cases show higher frequency of cytoplasmic mislocalization of TDP‐43 than neurons without poly‐GA aggregates (Fig EV1E and F).
Figure EV1. Poly‐GA induces cytoplasmic TDP‐43 mislocalization.
-
A, BImmunofluorescence analysis of endogenous TDP‐43 in the anterior horn of the spinal cord of GA149‐CFP transgenic mice 8–12 months of age (Schludi et al, 2017). Single confocal sections are shown in (A). Arrow indicates neuron with cytoplasmic TDP‐43 punctae. (B) Manual quantification of neurons with cytoplasmic TDP‐43 in the anterior horn. To allow blinded quantification, poly‐GA expression was not taken into account. Scatter plot with bar graphs of mean ± SD. Statistical analysis using unpaired t‐test and Welch's correction (three wild‐type and six transgenic animals).
-
CImmunoblotting of three wild‐type and three GA149‐CFP transgenic mice spinal cord 8 months of age. Immunoblotting of one wild‐type and one GA149‐CFP transgenic mouse spinal cord is shown. Proteolytic processing of TDP‐43 was not detected in both genotypes.
-
DImmunofluorescence analysis of endogenous TDP‐43 in large ChAT‐positive motoneurons in the anterior and posterior horns of the spinal cord of GA149‐CFP transgenic mice 8–12 months of age (Schludi et al, 2017). Maximum intensity projections are shown. Arrow indicates neurons with cytoplasmic TDP‐43 punctae.
-
E, FAutomated analysis of cytoplasmic mislocalization of TDP‐43 in frontal cortex of C9orf72 FTLD patients. Representative raw image and the resulting CellProfiler mask (see Materials and Methods for details). Poly‐GA‐positive neurons were significantly more likely to have detectable cytoplasmic TDP‐43 than neighboring poly‐GA‐negative neurons (paired t‐test t (7) = 5.58, partial η2 = 0.816, mean ± SD).
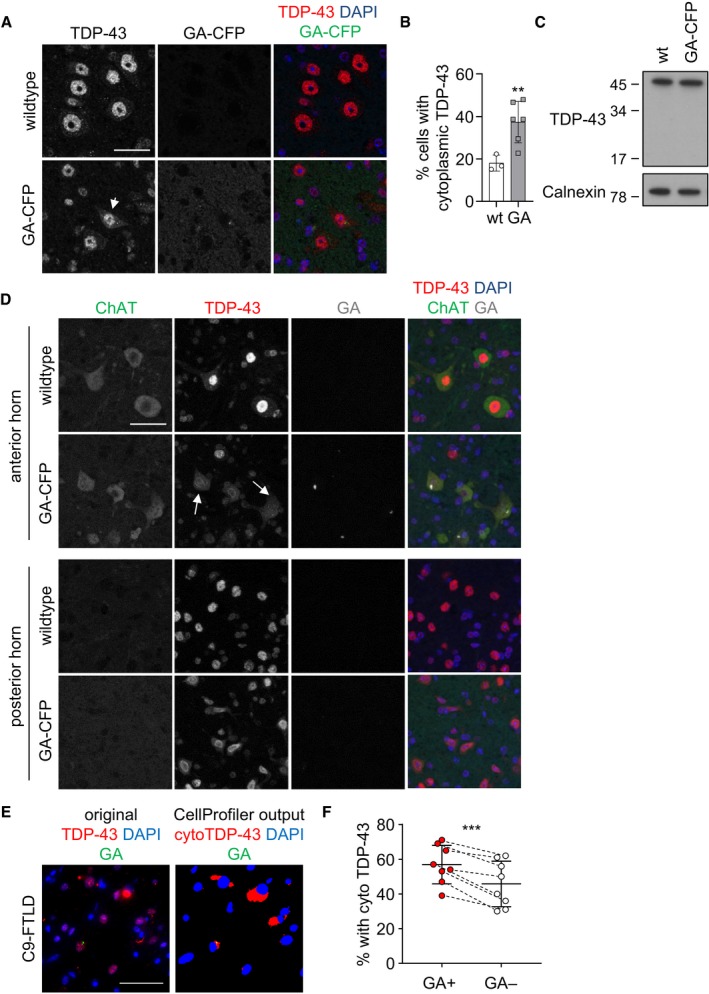
Since TDP‐43 and poly‐GA only occasionally co‐aggregate in patient tissue, we investigated potential non‐cell‐autonomous effects of poly‐GA in a neuronal co‐culture system. We transduced primary rat hippocampal neurons growing on coverslips with either GFP or GA175‐GFP (“donor cells”). Four days later, we transferred the coverslips with extensively washed donor cells into a new well containing untreated primary neurons (“receiver cells”) separated by ~ 1‐mm spacers (Fig 1A) and co‐cultured donor and receiver cells for another 4 days. Immunofluorescence of the donor cells showed enhanced cytoplasmic localization of TDP‐43 in GA175‐GFP‐transduced compared with GFP‐transduced donor cells (Fig 1B and C and Appendix Fig S2A) as we had reported previously (Khosravi et al, 2017). Automated quantification of the number of poly‐GA aggregates per cell (2.23 ± 0.18 [mean ± SD] in the donor compartment vs 0.70 ± 0.22 in the receiver compartment) showed robust transmission of GA175‐GFP aggregates between neurons consistent with previous results (Westergard et al, 2016; Zhou et al, 2017). Using automated image analysis of single confocal sections, we compared cytoplasmic TDP‐43 in GFP‐positive and GFP‐negative cells (Khosravi et al, 2017). Strikingly, cytoplasmic TDP‐43 expression was not only enhanced in cells taking up visible GA175‐GFP aggregates but also enhanced in neurons without obvious GA175‐GFP, both on donor and receiver coverslips (Fig 1B and Appendix Fig S2A red arrows, and Fig 1C). Thus, poly‐GA release from transduced neurons leads to TDP‐43 mislocalization in neighboring neurons presumably even by uptake of small amounts of soluble or aggregated poly‐GA. At the time scale of our experiments, no large TDP‐43 aggregates or proteolytic processing was detected (Fig 1B and Appendix Fig S2B). To exclude unspecific effects due to DPR toxicity, we repeated the experiment with expression of arginine‐rich DPR proteins poly‐GR and poly‐PR that show stronger acute toxicity in most model systems (Wen et al, 2014). However, expression of poly‐GR/PR did not alter TDP‐43 localization in either the donor or receiver compartment (Appendix Fig S2C and D).
Figure 1. Cell‐to‐cell transmission of poly‐GA causes cytoplasmic mislocalization of TDP‐43.
-
A–CPrimary hippocampal neurons were transduced with GFP or GA175‐GFP (DIV4 + 4) and co‐cultured with naïve primary neurons for 4 days. Endogenous TDP‐43 and poly‐GA aggregates in donor and receiver coverslips were analyzed by immunofluorescence. (A) Schematic representation of co‐culture experiments. (B) Cytoplasmic TDP‐43 immunostaining is elevated not only in poly‐GA‐transduced neurons, but also in the non‐transduced receiver cells. White and red arrows indicate cells with cytoplasmic TDP‐43 in GFP‐positive and GFP‐negative cells, respectively. (C) Automated quantification of cells with cytoplasmic TDP‐43 in GFP‐ or GA175‐GFP‐transduced (donor) and non‐transduced (receiver) neurons. Cells with and without GFP signal were counted separately (indicated by +/−). Two groups (GFP‐negative donor and GFP‐positive receiver) were excluded due to very high GFP transduction rate and very low GFP transmission rate. n = 4 biological replicates. In total, 283 donor GFP, 273 donor GA175‐GFP, 284 receiver GFP, and 266 receiver GA175‐GFP cells were analyzed. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. ***P < 0.001.
-
D–GCo‐culture model in HeLa cells transfected with iRFP or GA175‐iRFP in the donor compartment and TDP‐43ΔNLS‐GFP in donor and receiver compartments. (D) Immunofluorescence staining and (E) automatic quantification of TDP‐43ΔNLS aggregate number per cell, (F) in addition to filter trap assay of SDS‐insoluble TDP‐43ΔNLS‐GFP aggregates compared in iRFP‐ or GA175‐iRFP‐transfected cells. In (E) n = 3 biological replicates with 368 donor iRFP, 251 donor GA175‐iRFP, 430 receiver iRFP, and 328 GA175‐iRFP cells were analyzed. Cells with and without GFP signal were analyzed separately (indicated by +/−). White and red arrows indicate cells with cytoplasmic TDP‐43 in GFP‐positive and GFP‐negative cells, respectively. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. (G) GFP mRNA expression levels were measured by qPCR. RNA levels were normalized to GAPDH, β‐actin, and β2‐microglobulin mRNA. Bar graphs of mean ± SD. Unpaired two‐tailed t‐test with Welch's correction. *P < 0.05, and **P < 0.01.
To differentiate the effect of poly‐GA on nucleocytoplasmic transport and aggregation of TDP‐43, we analyzed receiver cells expressing GFP‐tagged TDP‐43 lacking the nuclear localization signal (ΔNLS). Since TDP‐43ΔNLS is highly toxic to primary neurons as shown in mouse models (Walker et al, 2015), we conducted these co‐culture experiments in HeLa cells. Donor cells were co‐transfected with TDP‐43ΔNLS‐GFP and either iRFP or GA175‐iRFP, while receiver cells were transfected only with GFP‐tagged TDP‐43ΔNLS. Twenty‐four hours after separate transfection, the washed coverslips were co‐cultured for another 24 h before analysis of poly‐GA and TDP‐43 fluorescence. Strikingly, both co‐expression of poly‐GA and co‐culture with poly‐GA‐expressing cells resulted in partial cytoplasmic aggregation of TDP‐43ΔNLS‐GFP suggesting poly‐GA has profound cell‐autonomous and non‐cell‐autonomous effects on TDP‐43 solubility even in cells without detectable poly‐GA inclusions (Fig 1D and E, Appendix Fig S2E). About 10–20% of poly‐GA inclusions also contained TDP‐43‐ΔNLS. In addition, we confirmed that poly‐GA induced GFP‐TDP‐43ΔNLS aggregation using a filter trap assay of cell extracts from the donor and receiver cells (Fig 1F). These effects are not caused by enhanced mRNA expression (Fig 1G). Thus, transmission of small amounts of poly‐GA may trigger TDP‐43 mislocalization in cells without obvious DPR pathology.
Anti‐GA antibodies block the non‐cell‐autonomous effects of poly‐GA on TDP‐43
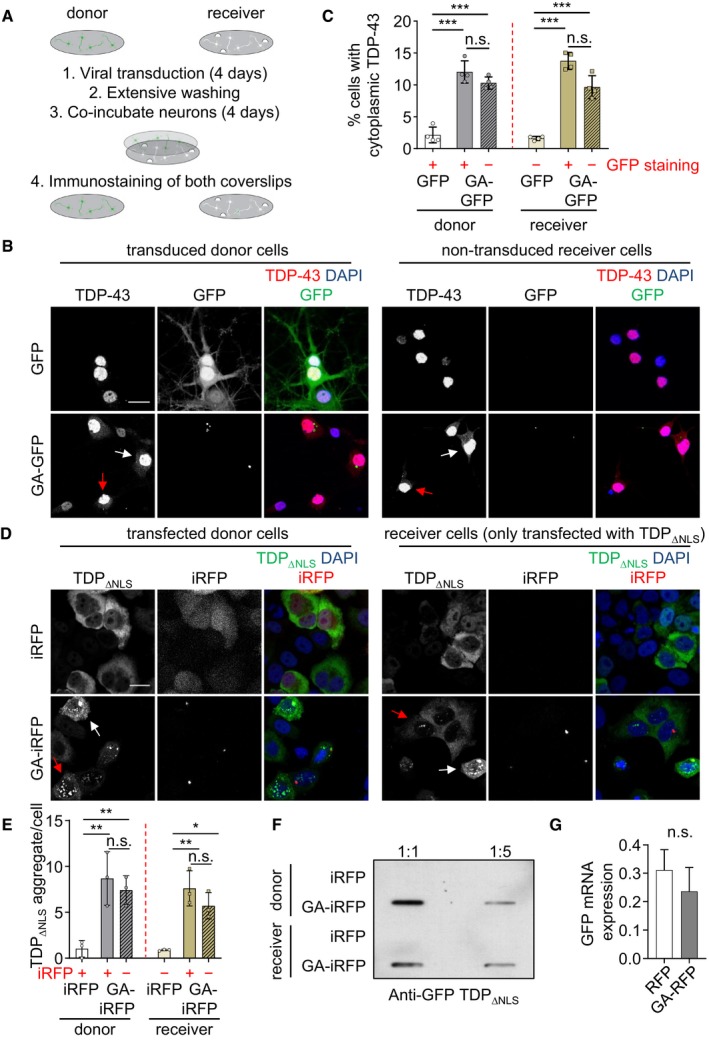
Cell‐to‐cell transmission of aggregating proteins is a potential target for therapeutic antibodies, and we have previously shown that monoclonal antibodies can inhibit transmission of poly‐GA (Zhou et al, 2017). We asked whether anti‐GA antibodies would also inhibit poly‐GA‐dependent TDP‐43 mislocalization. Thus, we added an anti‐GA antibody (clone 5F2) or purified mouse IgG as control to the co‐culture model from Fig 1A and analyzed TDP‐43 localization in the poly‐GA‐transduced donor compartment and the non‐transduced receiver compartment (Fig EV2). Importantly, anti‐GA treatment reduced cytoplasmic TDP‐43 levels in 5F2 treated neurons in both the donor and receiver compartments (Fig EV2A and B). Immunoblotting confirmed reduction in poly‐GA in both compartments (Fig EV2C).
Figure EV2. Anti‐GA antibodies block the non‐cell‐autonomous effects of poly‐GA on TDP‐43 in a co‐culture assay.
Primary hippocampal neurons were transduced with GFP or GA175‐GFP (DIV4 + 4) and treated with IgG control and anti‐GA (5F2) antibody.
- Confocal imaging revealed that anti‐GA antibody treatment reduces Poly‐GA‐induced cytoplasmic mislocalization of TDP‐43 in hippocampal neurons. White and red arrows show cells with cytoplasmic TDP‐43 in GFP‐positive and GFP‐negative cells, respectively. Scale bar denotes 20 μm.
- Automated quantification of cells with cytoplasmic TDP‐43 in GFP or GA175‐GFP‐transduced cells. Cells with and without GFP signal were analyzed separately (indicated by +/−). As in Fig 1C, GFP‐negative donor and GFP‐positive receiver cells were excluded due to high transduction and low transmission rate of GFP. n = 4 biological replicates. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. *P < 0.05, and ***P < 0.001.
- Immunoblotting shows reduced poly‐GA expression upon anti‐GA antibody treatment.
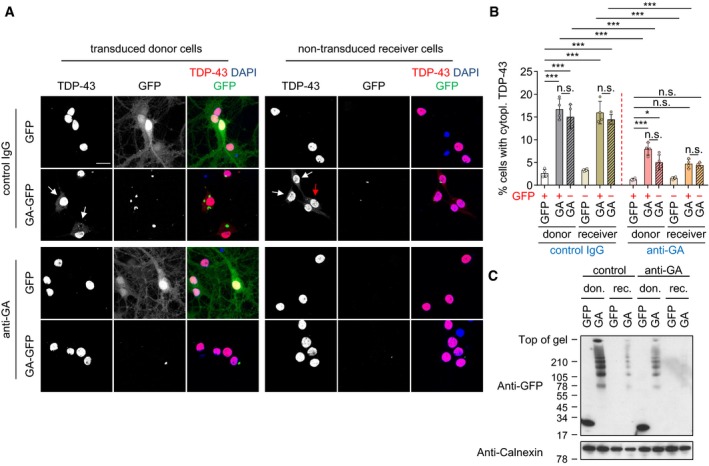
To exclude potential indirect effects due to unknown factors secreted from the donor cells in response to poly‐GA expression beyond mere toxicity (compare Appendix Fig S2C and D), we repeated the experiments using anti‐GA immunodepletion of conditioned medium from poly‐GA expressing neurons (Fig 2). Consistent with the co‐culture experiments, supernatant of GA175‐GFP‐transduced cells induced TDP‐43 mislocalization in receiver cells compared with GFP supernatant (Fig 2A and B). Moreover, immunodepletion with anti‐GA (clone 5F2) prevented poly‐GA uptake in receiver cells and strongly reduced TDP‐43 mislocalization compared to depletion with control IgG, suggesting that the effects on TDP‐43 are directly mediated by poly‐GA released from donor cells into the conditioned media. Poly‐GA immunoblotting and immunoassays confirmed successful precipitation and nearly complete clearance of poly‐GA (and anti‐GA antibodies) from the conditioned supernatant using 5F2 antibody (Fig 2C and D).
Figure 2. Anti‐GA immunodepletion in conditioned media prevents the non‐cell‐autonomous effects of poly‐GA on TDP‐43.
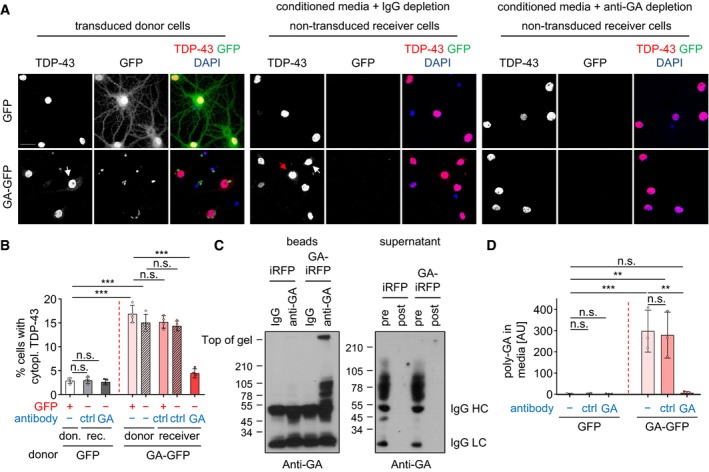
- Confocal imaging showed anti‐GA antibody treatment reduces poly‐GA aggregates and TDP‐43 mislocalization in hippocampal neurons. White and red arrows show cells with cytoplasmic TDP‐43 in GFP‐positive and GFP‐negative cells, respectively. Scale bar denotes 30 μm.
- Automated quantification of cells with cytoplasmic TDP‐43 in GFP‐ or GA175‐GFP‐transduced (donor) and non‐transduced (receiver) neurons. Four groups were excluded due to very high GFP transduction rate (GFP‐negative donor) and very low GFP transmission rate (GFP‐positive receiver with IgG and anti‐GA) and complete prevention of GA‐RFP transmission of anti‐GA immunodepletion (GA‐GFP receiver with anti‐GA). n = 3 biological replicates. In total, 280 donor GFP, 284 receiver GFP with IgG, 317 receiver GFP with anti‐GA, 277 donor GA175‐GFP, 294 receiver GA175‐GFP with IgG, and 311 receiver GA175‐GFP with anti‐GA cells were analyzed. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. ***P < 0.001.
- Immunoblotting of poly‐GA immunoprecipitated from conditioned media using antibody‐coupled beads and antibody leftover pre‐ and post‐immunoprecipitation in the conditioned media.
- Poly‐GA levels in conditioned media before and after immunodepletion with control IgG and anti‐GA antibody were determined by immunoassay. n = 3 biological replicates. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. **P < 0.01, and ***P < 0.001.
Taken together, anti‐GA antibodies reduce poly‐GA aggregation and transmission as well as cytoplasmic mislocalization of TDP‐43. Immunodepletion corroborates the direct effects of transmitted poly‐GA on TDP‐43 in receiver cells.
Poly‐GA inhibits the proteasome cell‐autonomously and non‐cell‐autonomously
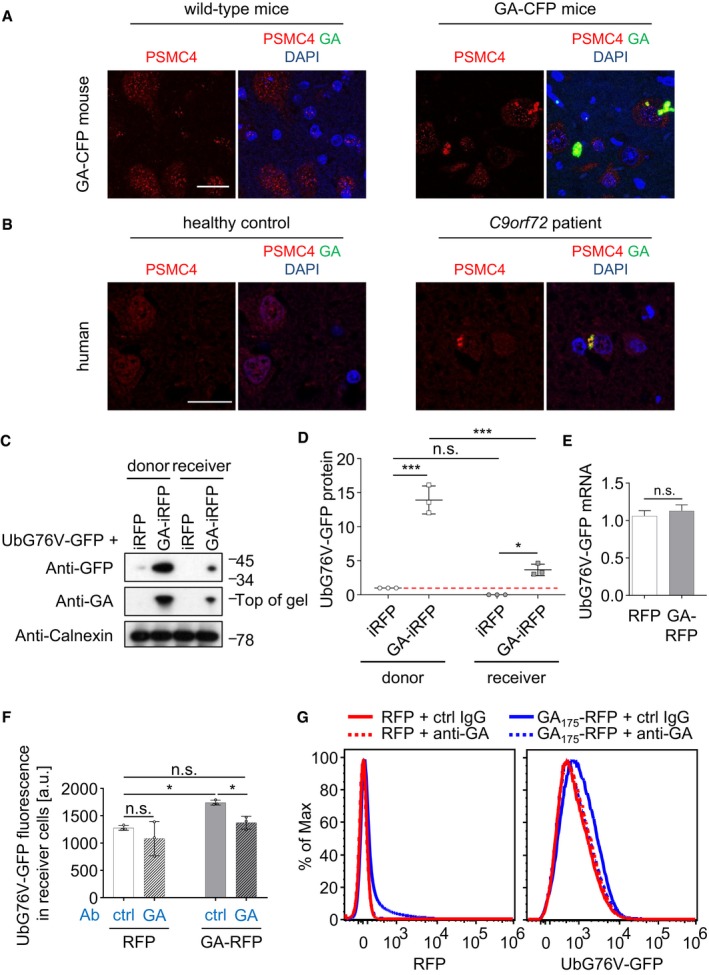
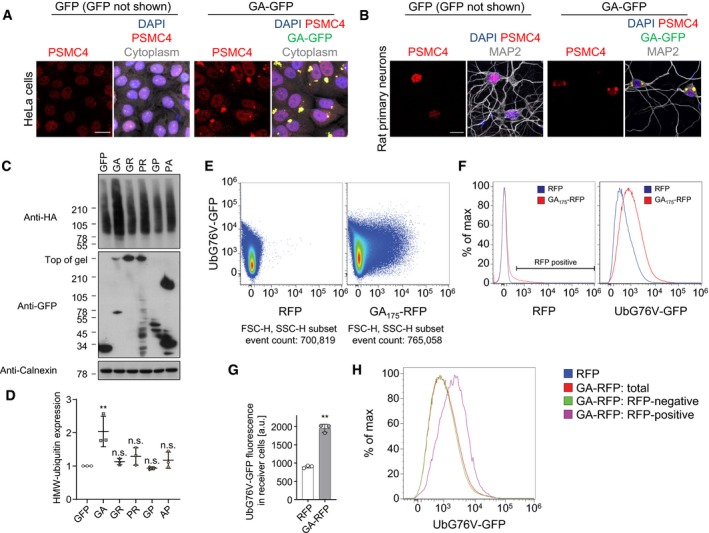
To investigate the mechanism of poly‐GA on TDP‐43 mislocalization and aggregation, we investigated the effects of poly‐GA on the proteasome. Cryo‐electron tomography has revealed that poly‐GA inclusions sequester large amounts of stalled proteasomes (Guo et al, 2018b). Here, we confirmed enrichment of the proteasome subunit PSMC4 by immunofluorescence in GA149‐CFP transgenic mice and C9orf72 ALS/FTD patients (Fig 3A and B) as well as in poly‐GA‐expressing HeLa cells and primary neurons (Fig EV3A and B). Moreover, only expression of poly‐GA, but not the other DPR species, promoted accumulation of high‐molecular weight ubiquitin species in HEK293 cells (Fig EV3C and D). To address non‐cell‐autonomous effects, we interrogated proteasome function in donor and receiver cells using the UbG76V‐GFP reporter, which accumulates upon proteasome inhibition (Dantuma et al, 2000). Thus, we co‐transfected HeLa cells with UbG76V‐GFP and the donor compartment additionally with iRFP or GA175‐iRFP. Strikingly, poly‐GA expression strongly increased UbG76V‐GFP levels in both the GA175‐iRFP‐transduced donor cells and the receiver cells as measured by Western blot without affecting mRNA expression of the reporter (Fig 3C–E). Flow cytometry using a HEK293 reporter line stably expressing UbG76V‐GFP confirmed that the mean UbG76V‐GFP fluorescence was significantly increased in cells co‐cultured with GA175‐RFP‐expressing cells, compared to cells co‐cultured with RFP alone (Fig EV3E–G). Uptake of GA175‐RFP was detectable in over 10% of receiving cells compared to < 1% uptake in receiving cells co‐cultured with RFP (Fig EV3E–G), which is consistent with previous reports (Westergard et al, 2016; Zhou et al, 2017). Differential analysis of GA175‐RFP‐positive vs GA175‐RFP‐negative receiver cells showed that UbG76V‐GFP levels were much higher in the cells with clear GA175‐RFP uptake than RFP‐negative receiver cells (Fig EV3H). Importantly, conditioned media from GA175‐RFP‐transduced cells also mediated poly‐GA transmission and induced UbG76V‐GFP levels in receiver cells, which was completely rescued by immunodepletion of poly‐GA with our monoclonal antibody (Fig 3F and G, compare Fig 2).
Figure 3. Poly‐GA inhibits the proteasome cell‐autonomously and non‐cell‐autonomously.
-
A, BDouble immunofluorescence of the proteasome subunit PSMC4 and poly‐GA inclusions in spinal cord of GA149‐CFP transgenic mouse and cortex of a C9orf72 patient compared with controls. Scale bar denotes 20 μm.
-
C, DCo‐culture model of HeLa cells transfected with iRFP or GA175‐iRFP in the donor compartment and an UbG76V‐GFP proteostasis reporter in donor and receiver compartments (48 h). (C) Separate analysis of both compartments by immunoblot and (D) immunoblot quantification. For quantitative analysis of immunoblots, UbG76V‐GFP was normalized to calnexin. n = 3 biological replicates. Scatter plot with mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. Red dashed line indicates the control's expression level. *P < 0.05, ***P < 0.001.
-
EGFP mRNA expression levels were measured by qPCR. RNA levels were normalized to GAPDH, β‐actin, and β2‐microglobulin mRNA. Bar graphs of mean ± SD. n = 3 biological replicates. Unpaired two‐tailed t‐test with Welch's correction.
-
F, GFlow cytometry analysis of a UbG76V‐GFP reporter cell line incubated 48 h with conditioned media from RFP or GA175‐RFP‐transfected cells upon immunodepletion of poly‐GA or control depletion using unspecific IgG. n = 3 biological replicates. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. *P < 0.05. (G) Comparisons of the corresponding histograms for compensated RFP and UbG76V‐GFP fluorescence from one representative experiment that shows specific transmission of GA175‐RFP associated with accumulation of UbG76V‐GFP in cells incubated with GA175‐RFP conditioned media. For flow cytometry analysis of co‐culture experiments, see Fig EV3E–H.
Source data are available online for this figure.
Figure EV3. Poly‐GA inclusions sequester the proteasome.
-
A, BImmunofluorescence of the proteasome subunit PSMC4 and poly‐GA inclusions in GA175‐GFP‐transfected HeLa cells and GA175‐GFP‐transduced rat primary neurons. To confirm cell viability, the cytoplasm of HeLa cells was stained with HCS CellMask™ Deep Red Stain and neuronal dendrites were labeled with MAP2. Scale bar denotes 20 μm.
-
C, DImmunoblots of HeLa cells that were co‐transfected with HA‐ubiquitin and GFP, GA175‐GFP, GFP‐GR149, PR175‐GFP, GFP‐GP47, and PA175‐GFP and incubated for 48 h and analyzed by densitometry. Scatter plot, mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. **P < 0.01.
-
E–HFlow cytometry analysis of non‐cell‐autonomous proteasome inhibition using HEK293 UbG76V‐GFP reporter cells co‐cultured for 48 h with GA175‐RFP‐ or RFP‐transfected cells. (E) Two‐color scatter plots, presented as pseudo‐color density plots, with compensated RFP fluorescence plotted on the x‐axis and compensated GFP fluorescence on the y‐axis. A representative experiment out of three independent repeats is shown. (F) Comparisons of the corresponding histograms for compensated RFP and UbG76V‐GFP fluorescence from one representative experiment that shows specific transmission of GA175‐RFP associated with accumulation of UbG76V‐GFP in cells co‐cultured with GA175‐RFP. (G) Accumulation of UbG76V‐GFP signal in non‐transfected receiver cells that were co‐incubated for 48 h with GA175‐RFP‐transfected donor cells compared with RFP control. n = 3 biological replicates. Scatter plot with bar graphs of mean ± SD. Unpaired two‐tailed t‐test with Welch's correction. **P < 0.01. (H) Histograms for UbG76V‐GFP intensity showing separate analysis of RFP‐positive and RFP‐negative receiver cells for the GA175‐RFP condition. RFP gating as indicated in (F).
Source data are available online for this figure.
Taken together, this suggests that cell‐to‐cell transmission of small amounts of poly‐GA is sufficient to induce significant proteasome inhibition in neighboring cells within a short time frame.
Rolipram rescues poly‐GA‐dependent TDP‐43 mislocalization and aggregation by boosting proteasome activity
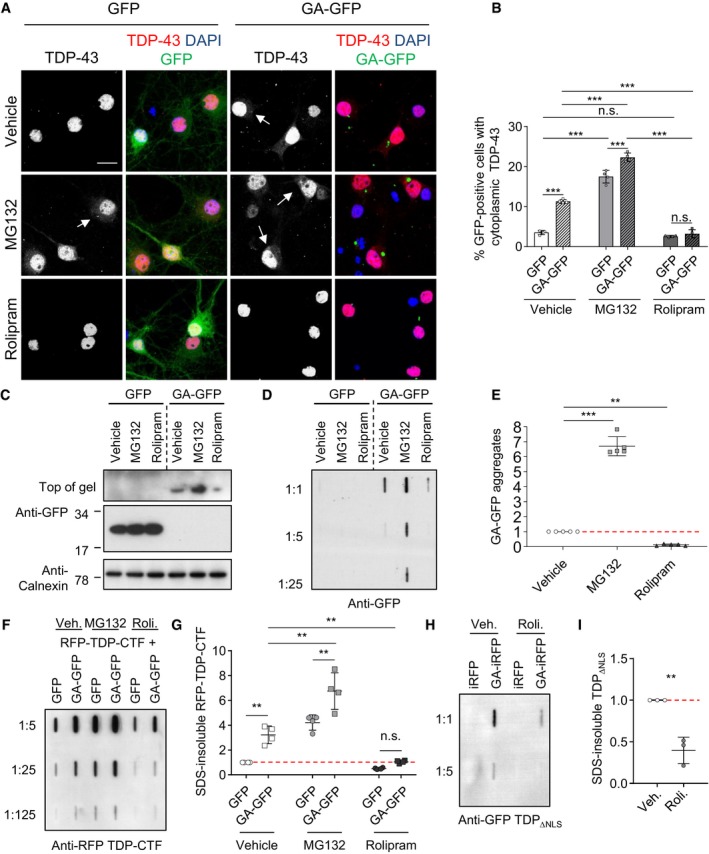
To test whether proteasome inhibition triggers TDP‐43 mislocalization to the cytoplasm upon poly‐GA expression, we transduced rat primary hippocampal neurons with GFP or GA175‐GFP and additionally inhibited the proteasome using MG132 or stimulated proteasomal activity using rolipram (Fig 4A). Automated image analysis revealed that MG132 treatment (10 μM, 16 h) significantly increased cytoplasmic TDP‐43 levels compared with the vehicle control in both GFP and GA175‐GFP expressing neurons. Strikingly, rolipram treatment (30 μM, 16 h) reduced cytoplasmic TDP‐43 levels in the GA175‐GFP‐transduced neurons (Fig 4A and B).
Figure 4. Rolipram rescues poly‐GA‐dependent TDP‐43 mislocalization and aggregation by boosting proteasome activity.
-
A–EPrimary hippocampal neurons were transduced with GFP or GA175‐GFP after 4 days in vitro, incubated for 7 days (DIV 4 + 7), and treated with vehicle (DMSO), MG132 (10 μM), or rolipram (30 μM) for 16 h. (A) Immunofluorescence reveals enhanced cytoplasmic TDP‐43 levels in neurons with poly‐GA aggregates or treated with MG132. Arrows mark punctate TDP‐43 staining. Rolipram treatment reduced cytoplasmic TDP‐43 in GA175‐GFP neurons. Scale bar denotes 20 μm. (B) Automated quantification of cells with cytoplasmic TDP‐43 in GFP‐ or GA175‐GFP‐transduced neurons. n = 4 biological replicates. In total, 462 GFP and 371 GA175‐GFP cells treated with vehicle, and 386 GFP and 529 GA175‐GFP cells treated with MG132, and 513 GFP and 434 GA175‐GFP cells treated with rolipram were analyzed. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. (C) Immunoblot to show effects of MG132 and rolipram on GA175‐GFP and GFP expression. (D and E) Filter trap assay with quantification of SDS‐insoluble aggregated GA175‐GFP. n = 5 biological replicates. Scatter plot with mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test.
-
F, GHeLa cells were co‐transfected with RFP‐TDP‐CTF and GFP or GA175‐GFP for 2 days. For the final 16 h, cells were treated with rolipram (30 μM) or MG132 (10 μM). Filter trap assay of SDS‐insoluble TDP‐CTF aggregates quantified by densitometry. n = 4 biological replicates. Scatter dot plot, mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. See also Appendix Fig S3.
-
H, IHeLa cells were co‐transfected with TDP‐43ΔNLS‐GFP and iRFP or GA175‐iRFP for 2 days. For the final 16 h, cells were treated with either vehicle or rolipram (30 μM). Filter trap assay of SDS‐insoluble TDP‐43ΔNLS‐GFP aggregates quantified by densitometry. n = 3 biological replicates. Scatter dot plot, mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test.
Immunoblotting confirmed that MG132 and rolipram treatment had little effect on the GFP control, but increased or decreased GA175‐GFP levels, respectively (Fig 4C). In addition, we used a filter trap assay to quantify the levels of SDS‐insoluble poly‐GA aggregates upon proteasome manipulation in primary neurons relative to control (Fig 4D and E). While MG132 significantly enhanced GA175‐GFP aggregation, rolipram reduced poly‐GA aggregates. Immunofluorescence confirmed that MG132 and rolipram also affected aggregate number accordingly (Appendix Fig S3A and B). In HeLa cells, poly‐GA expression also increased the levels of high‐molecular weight species of co‐expressed HA‐ubiquitin similar to proteasome inhibition using MG132, which was rescued by rolipram (Appendix Fig S3C and D). Moreover, the UbG76V‐GFP reporter confirmed that overall proteostasis was improved upon rolipram treatment (Appendix Fig S3E and F). To test whether proteasomal activation with rolipram can also rescue poly‐GA‐induced TDP‐43 aggregation, we co‐transfected HeLa cells with RFP‐TDP‐CTF and GFP or GA175‐GFP, treated them with MG132 or rolipram, and analyzed protein aggregation by filter trap (Fig 4F and G). Both proteasome inhibition by MG132 and expression of GA175‐GFP significantly increased the amount of TDP‐CTF inclusions compared with the GFP control (Fig 4G). In contrast, rolipram strongly reduced poly‐GA‐dependent RFP‐TDP‐CTF aggregation. Similarly, rolipram also reduced poly‐GA‐induced aggregation of TDP‐43‐ΔNLS in a filter trap assay (Fig 4H and I). Thus, proteasome activation prevents the formation or promotes the clearance of poly‐GA aggregates and reduces the cytoplasmic mislocalization and aggregation of TDP‐43 in cells with residual poly‐GA aggregates.
Boosting proteasomal activity prevents poly‐GA‐induced cytoplasmic accumulation of TDP‐43
We speculated that poly‐GA‐induced proteasomal inhibition might directly contribute to the cytoplasmic mislocalization of TDP‐43. To test the effects of poly‐GA on TDP‐43 localization independent of aggregation, we used RFP fused to the NLS of TDP‐43 as a nuclear import reporter in HeLa cells (Khosravi et al, 2017). Co‐transfection of the RFP‐TDP‐43‐NLS reporter with GA175‐GFP increased cytoplasmic reporter levels in inclusion‐bearing cells compared with the GFP control as measured by automated quantification (Fig 5A–C). Rolipram (30 μM, 16 h) did not affect localization of the RFP‐TDP‐43 NLS in GFP‐transfected cells, but largely prevented poly‐GA‐induced cytoplasmic mislocalization of the reporter (Fig 5A and B). This effect was phenocopied by overexpression of PSMD11 (Fig 5D–F), which is known to enhance proteasome assembly and activity and is the direct target of rolipram (Vilchez et al, 2012; Lokireddy et al, 2015). Similar to primary neurons (Appendix Fig S3A and B), rolipram also reduced the number of GA175‐GFP inclusions in HeLa cells consistent with stimulated proteasomal degradation of poly‐GA (Fig 5G–I). Neither rolipram treatment nor PSMD11 transfection altered the mRNA levels of the RFP‐TDP‐43‐NLS reporter (Fig 5C, F and I). To test whether the effects of proteasome activation result from improved clearance of cytoplasmic TDP‐43 or improved nuclear import, we added rolipram (30 μM) to reporter cells treated with 10 μM ivermectin, an inhibitor of the importin‐α/β pathway (Wagstaff et al, 2011). Rolipram reduced reporter mislocalization even under these conditions suggesting that it mainly enhances degradation of cytoplasmic TDP‐43 (Appendix Fig S4). Together, these findings suggest that proteasome inhibition by poly‐GA promotes cytoplasmic mislocalization of TDP‐43 by affecting the NLS of TDP‐43, which can be prevented by proteasome activation.
Figure 5. Boosting proteasomal activity prevents poly‐GA‐induced cytoplasmic accumulation of TDP‐43.
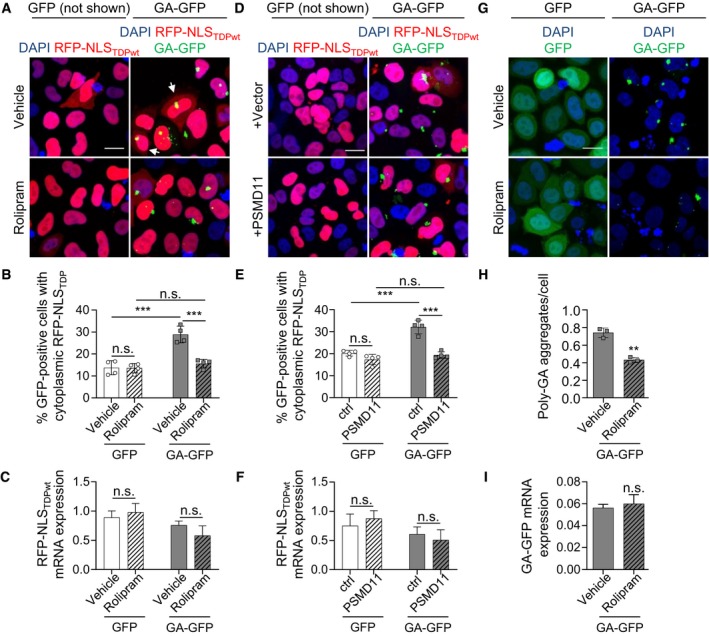
-
A–CHeLa cells were co‐transfected with an RFP‐based TDP‐NLS reporter and GFP or GA175‐GFP. Twenty‐four hours after transfection, cells were treated with rolipram (30 μM) for 16 h. In the immunofluorescence, GFP is not shown because diffuse GFP expression would hide the cytoplasmic RFP reporter. White arrows indicate cells with cytoplasmic TDP‐43. (B) Automated quantification of cells with cytoplasmic TDP‐NLS reporter in GFP‐ and GA175‐GFP‐positive cells. n = 4 biological replicates. In total, 345 GFP and 386 GA175‐GFP cells treated with vehicle, and 371 GFP and 404 GA175‐GFP cells treated with rolipram were analyzed. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. (C) RFP‐NLSTDPwt mRNA expression levels were measured by qPCR. n = 3 biological replicates. Bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test.
-
D–FHeLa cells were co‐transfected with the RFP‐based TDP‐NLS reporter, GFP or GA175‐GFP, and PSMD11 or empty vector. Image analysis as in (A). n = 4 biological replicates. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. 354 GFP and 330 GA175‐GFP cells with vector, and 367 GFP and 369 GA175‐GFP cells with PSMD11 in total were analyzed. (F) RFP‐NLSTDPwt mRNA expression levels were measured by qPCR. n = 3 biological replicates. Bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test.
-
G–IImmunofluorescence of HeLa cells transfected with GFP or GA175‐GFP showing reduced poly‐GA aggregation upon rolipram treatment (30 μM, 16 h). (H) Automated quantification of poly‐GA aggregate number per cell. n = 3 biological replicates. In total, 223 cells treated with vehicle and 286 cells treated with rolipram were analyzed. Scatter plot with bar graphs of mean ± SD. Unpaired two‐tailed t‐test with Welch's correction. (I) GA‐GFP mRNA expression levels were measured by qPCR. n = 3 biological replicates. Bar graphs of mean ± SD. Unpaired two‐tailed t‐test with Welch's correction.
Lysine 95 is critical for the inhibition of nuclear import of TDP‐43 by poly‐GA
Since proteasomal activation rescues nuclear import of the TDP‐43 NLS reporter, we speculated that poly‐GA expression might inhibit import via ubiquitination within the TDP‐43 NLS. Indeed, several previous proteome‐wide mass spectrometry studies had identified ubiquitination sites within the TDP‐43 NLS at lysine 84 and lysine 95 (Fig 6A) (Kim et al, 2011; Lumpkin et al, 2017; Akimov et al, 2018). To test the role of both residues in nuclear import of TDP‐43, we generated RFP‐based reporters containing lysine‐to‐alanine and lysine‐to‐arginine mutations at these sites to block ubiquitination while either removing or maintaining the positive charge. All constructs were expressed at comparable levels (Fig 6B) and showed similar turn‐over in a cycloheximide experiment (Appendix Fig S5A and B). Next, we assessed the nuclear import efficacy of the mutant NLS reporters in HeLa cells co‐transfected with GA175‐GFP or GFP control. Compared to the wild‐type, K84A and K84R mutations largely prevented nuclear import of the RFP reporter even in the absence of poly‐GA indicating K84 is crucial for the function of the TDP‐43 NLS, which precludes the analysis of poly‐GA‐specific effects on K84 in this assay (Fig 6C and D). However, both K95A and K95R mutants were imported to the nucleus as efficiently as the wild‐type NLS in cells co‐transfected with GFP (Fig 6C and D), suggesting a positive charge at this position is not required for NLS activity. In striking contrast to the wild‐type NLS, the K95A and K95R reporters remained largely nuclear even in inclusion‐bearing cells (Fig 6C and D), indicating that lack of this putative ubiquitination site protects the TDP‐43 NLS from the inhibitory effect of poly‐GA. MG132 treatment phenocopied the effects of poly‐GA on wild‐type and mutant reporters, suggesting that proteasome inhibition is a main driver of reporter mislocalization. Importantly, K95 mutations also prevented poly‐GA‐dependent cytoplasmic mislocalization of full‐length TDP‐43 without affecting overall TDP‐43 clearance (Appendix Fig S5C–F).
Figure 6. Lysine 95 is critical for the inhibition of nuclear import of TDP‐43 by poly‐GA .
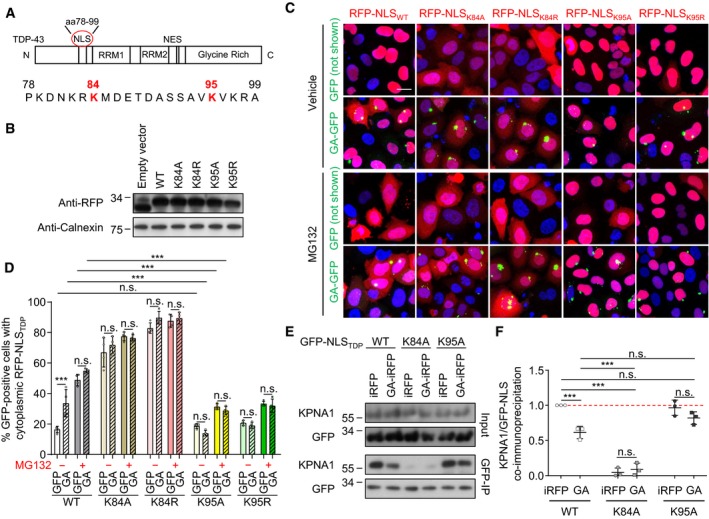
-
ADomain structure of TDP‐43 and location of the bipartite NLS at positions 78–99 (Winton et al, 2008). Known ubiquitination sites listed on http://www.phosphosite.org at K84 and K95 are highlighted.
-
BImmunoblot of HeLa cells transfected with RFP‐based TDP‐NLS wild type (WT) or mutants (K84A, K84R, K95A, K95R).
-
C, DHeLa cells were co‐transfected with the indicated TDP‐NLS reporters as well as GFP or GA175‐GFP, and treated with MG132 (10 μM) or vehicle for 16 h. (D) Automated quantification of RFP‐NLS reporters in GFP‐positive cells. Note that K84A and K84R block overall import, while K95A and K95R allow import but are resistant to inhibition by poly‐GA. n = 4 biological replicates. The total number of cells analyzed per group was (from left to right) 667, 581, 789, 783, 809, 708, 628, 721, 938, 557, 857, 861, 886, 699,789, 539, 636, 577, 638, and 870. Scatter plot with bar graphs of mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. ***P < 0.001. Scale bar denotes 20 μm.
-
E, FHeLa cells were co‐transfected with the indicated GFP‐TDP‐NLS reporters and iRFP or GA175‐iRFP. Cell lysates were immunoprecipitated with anti‐GFP and immunoblotted with indicated antibodies to detect co‐immunoprecipitation of the TDP‐43 NLS with importin‐α5/KPNA1 nuclear import receptor. (F) Quantification of KPNA1 levels normalized to total GFP‐NLSTDP reporter levels in anti‐GFP immunoprecipitates. n = 3 biological replicates. Scatter plot with mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. ***P < 0.001. See also Fig EV4. Red dashed line indicates the control's expression level.
Source data are available online for this figure.
The TDP‐43 NLS acts via the classical nuclear import receptor importin‐α (Winton et al, 2008; Nishimura et al, 2010). To test how poly‐GA interferes with this pathway, we performed co‐immunoprecipitation of the GFP‐NLS reporter constructs with endogenous importin‐α5/KPNA1 in HeLa cells co‐expressing iRFP or GA175‐iRFP (Fig 6E and F). The wild‐type GFP‐NLS and the K95A mutant co‐immunoprecipitated KPNA1 under control conditions, whereas the K84A mutation severely impaired binding to the import receptor independent of poly‐GA consistent with poor nuclear import. Moreover, poly‐GA co‐expression reduced KPNA1 binding to the wild‐type GFP‐NLS but not to the K95A construct that was resistant to poly‐GA induced mislocalization (compare Fig 6C and D). Similarly, poly‐GA reduced binding of full‐length TDP‐43 to importin‐α5/KPNA1, which was blocked by the K95A mutation (Fig EV4). Taken together, poly‐GA‐induced ubiquitination or other post‐translational modifications at K95 are likely inhibiting the nuclear import of TDP‐43.
Figure EV4. Poly‐GA reduces KPNA1 binding of full‐length TDP‐43.
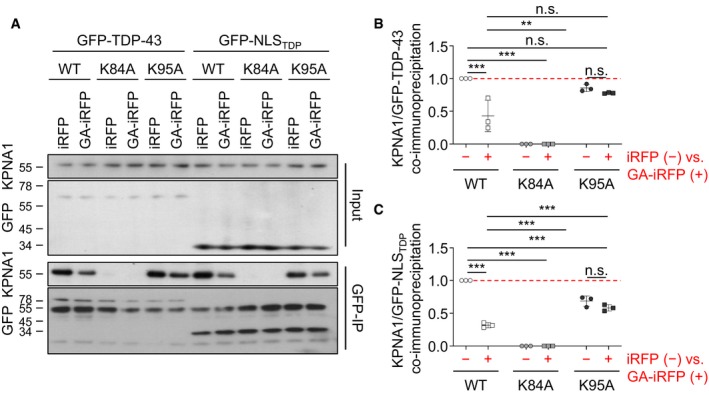
-
AHeLa cells were co‐transfected with either full‐length GFP‐TDP‐43 (wild type, K84A, K95A) or GFP‐NLSTDP (wild type, K84A, K95A) as well as iRFP or GA175‐iRFP and incubated for 48 h. Lysates were immunoprecipitated with anti‐GFP and immunoblotted with an anti‐importin‐α5/KPNA1 antibody to detect binding of the nuclear import receptor. Protein expression in the input is also shown.
-
B, CQuantification of KPNA1 levels normalized to total GFP‐TDP‐43 and GFP‐NLSTDP reporter levels in anti‐GFP immunoprecipitates. n = 3 biological replicates. Scatter plot with mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. **P < 0.01, ***P < 0.001. Red dashed line indicates the control's expression level.
Source data are available online for this figure.
Poly‐GA induced poly‐ubiquitination of TDP‐43 within the NLS at lysine 95
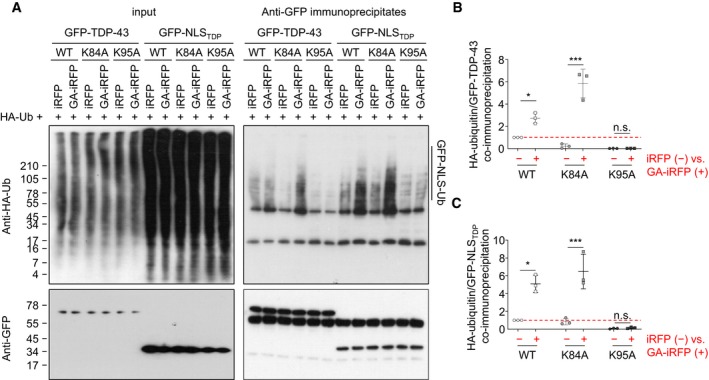
To test whether poly‐GA induced ubiquitination within the TDP‐43 NLS, we co‐transfected HeLa cells with the GFP‐NLSTDP reporters, poly‐GA and HA‐ubiquitin, and analyzed the amount of ubiquitin chains in GFP‐NLSTDP immunoprecipitates (Fig 7). HA immunoblotting clearly showed poly‐ubiquitination of the wild‐type GFP‐NLS reporter compared to control immunoprecipitates from cells without HA‐ubiquitin expression (Fig 7A). Importantly, poly‐ubiquitination of the wild‐type NLS reporter increased upon poly‐GA expression. In contrast, basal ubiquitination of the K95A reporter was much lower than wild‐type and did not increase upon poly‐GA co‐expression suggesting that K95 (and not K84) is the main ubiquitination site within the TDP‐43 NLS. The proteasome inhibitor MG132 induced accumulation of poly‐ubiquitinated wild‐type but not K95A reporter (Fig 7A and B). In contrast, rolipram reduced basal ubiquitination of the wild‐type reporters to the level of the K95A mutant. Finally, introducing the K95A mutation into full‐length TDP‐43 largely prevented the poly‐GA‐induced accumulation of ubiquitinated TDP‐43 (Fig EV5). Together, these data indicate that poly‐GA‐mediated proteasome inhibition leads to the cytoplasmic accumulation of TDP‐43 NLS ubiquitinated predominantly at K95, and this mislocalized TDP‐43 can be effectively cleared by boosting proteasome activity.
Figure 7. Poly‐GA induces poly‐ubiquitination of TDP‐43 within the NLS at lysine 95.
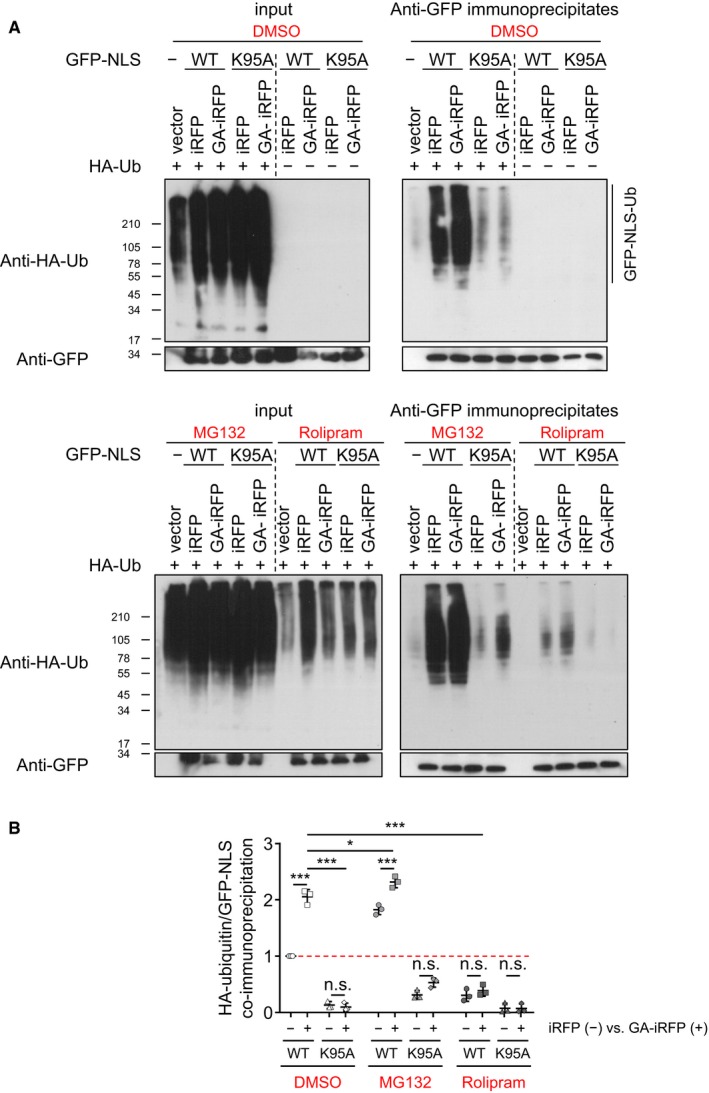
- Immunoblotting of input (left panels) and anti‐GFP immunoprecipitates (right panels) to show GFP reporter levels and poly‐ubiquitination.
- Quantification of HA‐ubiquitin levels normalized to total GFP‐NLSTDP reporter levels in anti‐GFP immunoprecipitates. n = 3 biological replicates. Scatter plot, mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. *P < 0.05, and ***P < 0.001. Red dashed line indicates the control's expression level.
Figure EV5. Poly‐GA induces poly‐ubiquitination of TDP‐43 at lysine 95.
-
A–CHeLa cells were co‐transfected with either full‐length GFP‐TDP‐43 (wild type, K84A, K95A) or GFP‐NLSTDP (wild type, K84A, K95A) as well as HA‐ubiquitin and iRFP or GA175‐iRFP, and incubated for 48 h. Lysates were immunoprecipitated with anti‐GFP antibody. Immunoblotting of input (left panels) and anti‐GFP immunoprecipitates (right panels) to show TDP‐43 bait levels and poly‐ubiquitination. (B, C) Quantification of HA‐ubiquitin levels normalized to total GFP‐TDP‐43 and GFP‐NLSTDP reporter levels in anti‐GFP immunoprecipitates. n = 3 biological replicates. Scatter plot, mean ± SD. One‐way ANOVA with Tukey's multiple comparisons test. *P < 0.05, and ***P < 0.001. Red dashed line indicates the control's expression level.
Source data are available online for this figure.
Discussion
Dysfunction of the ubiquitin–proteasome system has been reported for many neurodegenerative diseases, but actual sequestration and proteasome stalling has so far been detected only for poly‐GA (Guo et al, 2018b). Here, we show in a co‐culture model that poly‐GA inhibits the proteasome and promotes TDP‐43 mislocalization and aggregation even in neighboring cells that uptake only small amounts of poly‐GA. TDP‐43 mislocalization by poly‐GA is mediated by ubiquitination at lysine 95 within the NLS, which inhibits binding to importin‐α. We show that inhibiting poly‐GA transmission with antibodies and chemically activating the proteasome with rolipram ameliorate both poly‐GA and TDP‐43 pathology and may thus break the pathogenic cascade in C9orf72 patients.
Proteasome activation reduces poly‐GA and TDP‐43 aggregate formation
Proteasome inhibition is known to promote TDP‐43 aggregation in vitro (Igaz et al, 2009), but it is unclear whether this mechanism occurs in patients and how it would be triggered only in motoneurons and/or the frontotemporal cortex. Here, we analyzed cell‐autonomous and non‐cell‐autonomous effects of poly‐GA on the proteasome. We show that poly‐GA aggregates partially sequester the proteasome in C9orf72 ALS/FTD patients and a GA175‐CFP expressing mouse model, which confirms our in vitro data (Guo et al, 2018b). Moreover, poly‐GA expression promotes cytoplasmic mislocalization of endogenous TDP‐43 in our mouse model. In primary neurons, MG132 treatment or poly‐GA expression acutely triggers cytoplasmic mislocalization of endogenous TDP‐43 and an RFP‐NLSTDP reporter, suggesting that proteasome impairment is sufficient to inhibit nuclear import of TDP‐43. Consistent with previous reports, only poly‐GA but not the other DPR species promoted aggregation of a C‐terminal TDP‐43 fragment (Khosravi et al, 2017; Nonaka et al, 2018). Recent data from a primate model of TDP‐43 pathology suggest that rodent caspases cleave TDP‐43 less efficiently to generate aggregation‐prone CTFs, which may explain the absence of large TDP‐43 aggregates in mouse models and primary neurons (Yin et al, 2019).
We tested the activation of the proteasome chemically using rolipram or genetically by overexpressing PSMD11 to ameliorate poly‐GA toxicity. The PDE4 inhibitor rolipram leads to PSMD11 activation via serine‐14 phosphorylation by PKA, which boosts proteasome assembly (Lokireddy et al, 2015). In addition, proteasomes from rolipram‐treated cells have a higher ATPase activity, suggesting that substrate processing is enhanced (Lokireddy et al, 2015). The short side chains of poly‐GA may impair translocation into the catalytic subunit of the proteasome as has been shown for glycine/alanine‐rich sequences of EBNA1 (Levitskaya et al, 1997; Kraut, 2013). PSMD11 phosphorylation may promote translocation efficacy to allow degradation of poly‐GA. Moreover, enhanced degradation of soluble poly‐GA forms may reduce aggregate formation. In other disease contexts, activating the proteasome in a transgenic Tau mouse model reduced Tau levels and improved cognition (Myeku et al, 2016). Proteasome activation was shown to promote degradation of full‐length TDP‐43, SOD1 and FUS in vitro (Lokireddy et al, 2015). In our experiments, rolipram treatment had no effect on basal TDP‐43 localization but prevented cytoplasmic mislocalization in poly‐GA‐expressing cells. In addition, rolipram reduced the aggregation of poly‐GA and TDP‐43 CTFs. Therefore, we analyzed the effects of rolipram in all immunofluorescence assays only in cells containing visible poly‐GA inclusions to exclude confounding effects. Our findings are most consistent with the model that rolipram activates both poly‐GA and TDP‐43 degradation and additionally inhibits poly‐GA‐dependent effects on TDP‐43.
Non‐cell‐autonomous effects of poly‐GA on TDP‐43
Most neuropathological studies in end‐stage tissue found no correlation between reduced C9orf72 expression, RNA foci or the five DPR species and neurodegeneration (Mackenzie et al, 2013, 2015; Schludi et al, 2015; DeJesus‐Hernandez et al, 2017). We and others reported cell‐to‐cell transmission of DPRs (Westergard et al, 2016; Zhou et al, 2017) and uptake of synthetic poly‐GA aggregates is neurotoxic (Chang et al, 2016; Flores et al, 2016), but downstream effects were unknown. DPR inclusions were reported to cluster within human tissue which may support paracrine effects (Zu et al, 2013).
Here, we show by using fluorescent reporters that poly‐GA affects proteasome function as well as TDP‐43 localization and aggregation even in neighboring cells that do not contain obvious poly‐GA aggregates, which may explain the poor regional overlap of DPRs and TDP‐43 pathology in patients. We propose that some neuron populations (e.g., in cerebellum) are very efficient at non‐canonical translation of the expanded C9orf72 repeat, without being overly susceptible to their toxicity (e.g., due to higher basal proteasome activity), while motoneurons express only low levels of DPRs, but may be highly susceptible to proteasomal inhibition (Tashiro et al, 2012) by uptake of soluble or aggregated poly‐GA. Our data show that anti‐GA antibodies can break this cascade at least in cultured cells by blocking transmission of DPRs. Immunodepletion of poly‐GA from conditioned media completely prevented mislocalization of TDP‐43 in receiver cells, suggesting that in vitro the effects are mainly driven by released poly‐GA. We cannot exclude that poly‐GA expression triggers additional indirect effects in vivo, for example by directly releasing other molecules that promote TDP‐43 aggregation in neighboring cells or triggering release of such factors from glial cells. Our findings provide mechanistic insights into very recent active and passive antibody therapy approaches in C9orf72 mouse models by us and others (Nguyen et al, 2020; Zhou et al, 2020). Nguyen et al (2020) also reported that anti‐GA antibodies partially restore proteasome function in poly‐GA‐expressing cells and show that antibodies clear poly‐GA via the proteasome and autophagy pathway depending on the intracellular Fc‐receptor TRIM21. Moreover, boosting proteasome function in donor and receiver cells with small molecules such as rolipram may overcome poly‐GA‐induced proteasome impairment and lead to clearance of ubiquitinated substrates such as TDP‐43.
TDP‐43 ubiquitination regulates nuclear import
Driving TDP‐43 to the cytoplasm promotes its aggregation and is highly toxic, potentially through both gain‐ and loss‐of‐function mechanisms (Ederle & Dormann, 2017; Prasad et al, 2019). We provide ample evidence that ubiquitination at K95 inhibits its NLS function, possibly through steric hindrance of importin‐α binding. Surprisingly, mutagenizing K95 of the bipartite NLS to alanine or arginine preserves activity but prevents ubiquitination and poly‐GA‐mediated inhibition of nuclear import. In addition, we confirm reduced binding of wild‐type but not K95A to importin‐α upon poly‐GA expression. Consistent with the data by Hans et al (2018), K84 mutants completely block NLS activity even in the absence of poly‐GA, and it is conceivable that ubiquitination at K84 may also inhibit nuclear import (Kim et al, 2011; Lumpkin et al, 2017; Akimov et al, 2018). However, removing K95 largely prevented ubiquitination in our assays, suggesting that K95 is the main ubiquitination site within the TDP‐43 NLS. Interestingly, a recent study linked K95 ubiquitination to pathological phosphorylation at S409/410 (Hans et al, 2018). Boosting proteasome activity in poly‐GA‐expressing cells may allow more efficient degradation of TDP‐43 ubiquitinated at K95 (and other sites) and thus prevent accumulation of cytoplasmic TDP‐43. A similar inhibition of nuclear transport by ubiquitination within an NLS has been described for p53 (Marchenko et al, 2010) and CCTα (Chen & Mallampalli, 2009), suggesting that this may be a common regulatory mechanism. Since poly‐GA also promotes TDP‐43‐ΔNLS and TDP‐CTF accumulation, ubiquitination or other post‐translational modification at additional sites may also favor aggregation or liquid–liquid phase separation (Ederle & Dormann, 2017; Prasad et al, 2019).
So far, nucleocytoplasmic transport defects in C9orf72 ALS/FTD have been mostly attributed to a direct effect of the repeat RNA and/or poly‐GR/PR on the nuclear pore involving phase separation, but clear or even preferential effects on nuclear import of TDP‐43 have not been reported (Freibaum et al, 2015; Jovicic et al, 2015; Zhang et al, 2015; Boeynaems et al, 2016). Moreover, poly‐PR expression promotes recruitment of TDP‐43 in stress granules upon arsenite treatment, but is ~ 100‐fold less abundant than poly‐GA in patients (Mackenzie et al, 2015; Boeynaems et al, 2017). Cytoplasmic TDP‐43 aggregates further inhibit nucleocytoplasmic transport, which may trigger a vicious cycle (Chou et al, 2018; Solomon et al, 2018). We speculate that the combined effect of proteasome inhibition by poly‐GA specifically on the TDP‐43 NLS and a (subtle) general transport deficit caused by ubiquitous low‐level expression of the repeat RNA and rare poly‐PR cause the preferential mislocalization and aggregation of TDP‐43 in C9orf72 ALS/FTD patients. Recent findings on the role of TNPO1 as a chaperone for FUS (Guo et al, 2018a; Hofweber et al, 2018) suggest that the proteins affected by dysfunction of multiple pathways may be most sensitive to impairment of nucleocytoplasmic transport, when most other cargos are still trafficked normally.
Summary
Together, this work links the UPS dysfunction due to poly‐GA aggregation with the deficits in nucleocytoplasmic transport recently reported in C9orf72 FTD/ALS and other neurodegenerative diseases. Among the DPR proteins, poly‐GA is the key driver of TDP‐43 pathology in C9orf72 disease, although it is not sufficient to trigger full pathology by itself in mouse models, which may be explained by additional impact of other DPR species, the repeat RNA itself, haploinsufficiency, or poor caspase cleavage of TDP‐43 in rodents (Yin et al, 2019). Our work indicates that boosting proteasome activity or targeting poly‐GA with antibodies may be a promising therapeutic strategy because it reduces not only poly‐GA aggregation but also TDP‐43 mislocalization and aggregation.
Materials and Methods
Plasmids, transfection, and viral packaging
Synthetic expression constructs containing an ATG start codon in pEF6 backbone (EF1 promoter) for transient transfection or in FhSynW backbone (human synapsin promoter) for lentiviral expression were described before (May et al, 2014; Schludi et al, 2015). Here, we additionally generated variants tagged with iRFP670 (Shcherbakova & Verkhusha, 2013).
We fused the NLS of human TDP‐43 (PKDNKRKMDETDASSAVKVKRA, position 78–99) to the C‐terminus of GFP or tagRFP‐T2 (abbreviated as RFP throughout the manuscript; gift from Michael Davidson) in FUW2 backbone as described previously (Khosravi et al, 2017). Similar constructs containing mutations of lysine 84 (K84) or lysine 95 (K95) to alanine or arginine were cloned using synthetic oligonucleotides. Human TDP‐43 C‐terminal (amino acids 220–414, CTF) fragments were generated by PCR and fused to the C‐terminus of tagRFP‐T2 in FUW2 backbone.
The UbG76V‐GFP reporter for the ubiquitin–proteasome system (Dantuma et al, 2000) was subcloned into the FUW2 vector. Full‐length TDP‐43 and ΔNLS (K95A/K97A/R98A as described before (Winton et al, 2008)) were fused to the C‐terminus of GFP in the FUW2 vector.
Lentivirus was packaged in HEK293FT cells (Life Technologies) as previously described (Guo et al, 2018b).
Antibodies
TDP‐43 (Cosmo Bio Co, TIP‐TD‐P09), TDP‐43 (Proteintech, 10782‐2‐AP), TDP‐43 phospho‐S409/410 (Cosmo Bio Co., Ltd, TIP‐PTD‐P02), TDP‐43 (C‐terminal; Proteintech, 12892‐1‐AP), ChAT (Merck, AB144P), GFP (UC Davis/NIH NeuroMab Facility, N86/8 and N86/38), PSMC4 (Bethyl Laboratories, A303‐850A and A303‐849A), tagRFP (Thermo Fisher Scientific, R10367), KPNA1 clone 114‐E12 (Thermo Fisher Scientific, 37‐0800), calnexin (Enzo Life Sciences, ADI‐SPA‐860‐F), HA 3F10 (Merck, 11867423001), GA 5F2 (Mackenzie et al, 2013), control IgG from mouse serum (Merck, I5381), and HCS CellMask™ Deep Red Stain (Thermo Fisher Scientific, H32721) were used.
Primary neuron culture and immunofluorescence
Primary hippocampal neuron cultures were prepared from embryonic day 19 rats as described previously (Guo et al, 2018b). Primary neurons were then plated on sterilized poly‐d‐lysine‐coated coverslips. For co‐culture experiments, three 1‐ to 2‐mm dots of melted paraffin were spotted on the coverslips as a spacer. Then, primary neurons transduced on separate coverslips (DIV4 + 4) were extensively washed with media and put face to face for another 4 days of incubation in fresh media. For antibody treatment in neuronal co‐cultures, primary neurons on coverslips were transduced (DIV4 + 4) and washed with media and incubated face to face with non‐transduced cells for 4 days, followed by 7 days of treatment with IgG control and anti‐GA antibody.
HeLa cell culture and transfection
HeLa cells were cultured in DMEM, high glucose, GlutaMAX ™ Supplement containing 10% FCS and 1% penicillin/streptomycin together with MEM Non‐Essential Amino Acids Solution at 37°C with 5% CO2. HeLa cells were transfected using Lipofectamine 2000 (Thermo Scientific) according to the manufacturer's instructions, followed by 24‐ to 48‐h incubation at 37°C with 5% CO2. For co‐culture experiments, HeLa cells were transfected separately on two sets of coverslips with paraffin spacers for 24 h. After extensive washing with media, both coverslips were placed face to face and incubated for another 24 h.
C9orf72 patients
We selected nine C9orf72 cases from the Brain Bank München Regina Feederle and stained frontal cortex sections for GA (Helmholtz Zentrum, 1A12) and TDP‐43 (Proteintech, 10782‐2‐AP). One case was excluded from analysis due to extremely poor DAPI staining that precluded quantification of the frequency of poly‐GA and cytoplasmic TDP‐43.
Transgenic mice
Generation and characterization of Thy1‐GA149‐CFP (abbreviated as GA‐CFP) mice was reported previously (Schludi et al, 2017). Expression of GA149‐CFP was driven by Thy1.2 promoter. GA‐CFP transgenic mice were kept in the C57BL/6N background. Animal handling was performed in accordance with animal law of the Government of Upper Bavaria, Germany. Animals were housed in standard cages with ad libitum access to food and water in pathogen‐free facility on a 12‐h day/night cycle. Six transgenic (four male and two female) mice and three littermates (two male and one female) were analyzed. Manual image analysis was performed blinded to the genotype.
Immunofluorescence and confocal imaging
For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde and 4% sucrose for 10 min at RT and incubated with the indicated antibodies in GDB buffer (0.1% gelatin, 0.3% Triton X‐100, 450 mM NaCl, 16 mM sodium phosphate pH 7.4) and washed with PBS. All antibodies are listed in the key resources table.
For endogenous TDP‐43 staining, primary hippocampal neurons (DIV4 + 7) were fixed with 4% paraformaldehyde, then permeabilized (0.2% Triton X‐100, 50 mM NH4Cl in PBS), and blocked for 30 min (2% fetal bovine serum, 2% serum albumin, 0.2% fish gelatin in PBS) and incubated with antibodies in the same buffer. In both protocols, the primary antibodies were incubated overnight at 4°C and the secondary antibodies for 1 h at room temperature.
For mouse experiments, 8‐ to 12‐month‐old mice were euthanized with CO2 followed by cervical dislocation. Postmortem spinal cord was formalin fixated for 24 h, decalcificated with 5% formic acid for 48 h, and embedded in paraffin. Immunofluorescence staining was performed on 5‐μm‐thick paraffin sections as described previously (Schludi et al, 2017).
LSM710 confocal laser scanning system (Carl Zeiss) with Plan‐APOCHROMAT 10X/NA 0.45 (420640‐9900) or oil immersion 40×/NA 1.4 (420762‐9900) objectives equipped with the ZEN 2011 software package (black edition, Zeiss) was used for acquiring images. For all analyses, at least three images per group were taken blind to the experimental condition at 1,024 × 1,024 pixel resolution. For z‐stacked imaging, images were taken with z‐step size of 0.8 μm at 5–7 μm thickness.
Automated image analysis
To quantify the fraction of cells with cytoplasmic signaling of endogenous TDP‐43 staining in neurons, or RFP‐NLSTDP in HeLa, together with TDP‐CTF intensity in HeLa cells, Columbus Acapella version 2.6.0 (PerkinElmer) was used as described before (Khosravi et al, 2017). Nucleus objects were detected based on DNA staining “Find Nuclei” (area > 30 μm2, common threshold 0.10, split factor 7.0, individual threshold 0.4, contrast 0.45). In order to reject dead and mitotic nuclei, intensity properties were calculated at a standard method (mean and coefficient variance selected at quantile fraction 50%) by linear classification with the “select Population” function. Morphology of the nuclei was calculated by area, roundness, and Haralick features (including Haralick contrast, Haralick correlation, Haralick sum variance, Haralick homogeneity selected). The training set composed of ∼ 60 manually selected nuclei across all populations. For all selected nuclei, cell region was determined by expanding the nucleus region for 6 μm with morphological dilation. We selected the GFP/GFP‐DPR‐positive cells by laying a threshold on the mean intensity in the nucleus region. From this selection, we selected RFP‐positive cells by setting a second threshold based on the RFP channel. We determined different thresholds for HeLa cells and primary neurons, while thresholds were maintained constant for all subpopulations. We analyzed the mean of cytoplasmic and nuclear HA‐TDP‐43/RFP‐TDP‐NLS intensities, and cytoplasmic‐to‐nuclear ratio and finally determined percentage of GFP‐positive cells with cytoplasmic TDP‐43/RFP‐TDP‐NLS. Average results from two tile images per experiment were treated as n = 1 for the statistical analysis.
The aggregate/cell ratio was quantified using Image J (version 1.52i). The Otsu image threshold was determined automatically, followed by binary water shedding. Finally, particles with 2–18 μm diameter were counted. For DAPI channel, particles > 30 μm with circularity factor 0.7–1.00 were identified as nuclei to determine the cell number.
For TDP‐43 analysis from patient brains, we used a Leica fluorescent microscope (LAS X software) and imaged 50 fields of view per case in the gray matter, in manually determined grid patterns separated by 1 mm in each direction. Using CellProfiler (3.0.0), we identified DAPI‐stained nuclei, cytoplasmic TDP‐43 signal, bright TDP‐43 inclusions, and GA aggregates. We removed the majority of glial nuclei from analysis using thresholds that identified the brightest, smallest nuclei. We summed the total nuclei, GA, and TDP‐43 signals for all images per case and calculated the % GA‐positive and GA‐negative cells with cytoplasmic TDP‐43 signal.
Western blot and filter trap assays
For Western blot analysis, cells were lysed on ice in RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% SDS, 1.0% Triton X‐100, 1% deoxycholate, 5 mM EDTA) supplemented with 0.2 mg/ml DNase in PBS and protease and phosphatase inhibitors. Lysates were then centrifuged at 1,000 g for 10 min at 4°C or 15 min at 18,000 g 4°C depending on experiments. Protein concentration was adjusted according to measurements using a BCA assay (Interchim). After adding 4× Laemmli buffer (Bio‐Rad) containing 2‐mercaptoethanol, samples were denatured at 95°C for 10 min and loaded on Novex 10–20% Tris‐Tricine gels (Life Technologies).
For filter tap, cells were lysed on ice in Triton buffer (1% Triton X‐100, 15 mM MgCl2 in PBS) supplemented with 0.2 mg/ml DNase and protease inhibitor (Mori et al, 2013). Lysates were centrifuged at 13,000 rpm 4°C 30 min. Pellets were resuspended in SDS buffer (2% SDS in 100 mM Tris pH 7) and incubated for 2 h at RT. Samples were then filtered through a nitrocellulose membrane (0.2 μm pore). Membranes were then blocked with 2% I‐Block (Thermo Scientific) according to the manufacturer's instructions and detected with antibodies as indicated. Immunoblots were analyzed by using Fiji software. Immunoblot lanes were first detected by rectangle tool and then plotted, followed by peak labeling.
Immunoprecipitation assay
HeLa cells were lysed in 2% Triton X‐100, 0.75 M NaCl, 1 mM KH2PO4, and 3 mM Na2HPO4 supplemented with Benzonase Nuclease (6.7 U/ml) and protease inhibitors. 40 μl Protein G Sepharose beads were coupled with 3.96 mg/ml anti‐GFP antibody for 1 h at 4°C. Lysed samples were cleared by centrifugation (1,000 g for 5 min), and 10% of the supernatant was taken out as input. The remainder was incubated with GFP‐coupled beads overnight at 4°C, followed by extensive washing (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5% glycerol).
Anti‐GA immunodepletion and immunoassay
For immunodepletion experiments, 50 μl Protein G Dynabeads were coupled with 10 μg anti‐GA (5F2) or control IgG antibodies for 1 h at RT followed by three washing steps with PBS. Cell supernatant was incubated with antibody‐coupled beads for 3 h at RT. Supernatant was then collected, equilibrated to 37°C, and added to receiver cells for 96 h. To confirm immunodepletion, washed beads (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5% glycerol) were analyzed by immunoblotting and aliquots of the supernatant were analyzed by immunoassay on the Meso Scale platform (MSD) as described (Zhou et al, 2017). Briefly, streptavidin plates (MSD Gold 96‐well streptavidin) were coated with biotinylated 5F2 antibody (capture antibody, 1:1,000) in PBS overnight. After washing and blocking, the plates were then incubated with media for 2 h at RT on a shaking platform. Plates were washed three times and incubated with MSD sulfo‐tag‐labeled 5F2 antibody (detection antibody, 1:1,000) for 2 h at RT on a shaking platform followed by three final washing steps. The plates were measured shortly after adding 100 μl MSD Read Buffer T. MESO QuickPlex SQ 120 instrument was used to detect the electrochemical signal. Data are shown in arbitrary units after background correction.
Flow cytometry
HEK293 cells stably expressing UbG76V‐GFP (Dantuma et al, 2000; De Smet et al, 2017) were transfected with the indicated constructs for co‐culture assays or incubated with conditioned media from GA175‐RFP‐ or RFP‐expressing cells for 48 h. Subsequently, receiver cells were harvested and analyzed by flow cytometry for GFP and RFP fluorescence using an Attune NxT Cytometric Analyser (Thermo Fisher) at the Imaging Facility of the Max Planck Institute of Biochemistry, Martinsried. Fluorescence was detected using the following settings: GFP Ex 488 nm, Em 530/30 nm, tagRFP Ex 561 nm, and Em 586/15 nm. At least 500,000 cells were analyzed per sample. Fluorescence intensities were corrected for spectral overlap using HEK293 cells expressing single fluorophores, and compensated flow cytometry data were further analyzed using FlowJo software (version 9.9; Tree Star).
RNA isolation and quantitative RT–PCR
HeLa cells were transfected and incubated for 48 h. Next, RNA isolation was performed using the QIAshredder and RNeasy Mini Kit (Qiagen) according to manufacturer's instructions. To generate cDNA, the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) was used with random hexamer primers. CFX384 Touch Real‐Time PCR Detection System (Bio‐Rad Laboratories) was used to perform RT‐qPCR. The following primers were used: EGFP (Mr04097229_mr, Thermo Fisher Scientific), ACTB (Hs01060665_g1, Thermo Fisher Scientific), B2M (4326319E, Thermo Fisher Scientific), GAPDH (Hs02758991_g1, Thermo Fisher Scientific), and tagRFP (PrimerQuest Tool and Supply, Integrated DNA Technologies). Signals were normalized to ACTB, GAPDH, and B2M with the Bio‐Rad CFX Manager Software (Bio‐Rad Laboratories) by using the ΔΔCT method.
Statistical analyses
Statistical analysis was done in GraphPad Prism (version 7.01) using one‐way ANOVA with Tukey's multiple comparisons test. Family‐wise significance and confidence level were set at 0.05 (95% confidence interval). For experiments with only two groups, unpaired two‐tailed t‐test with 95% confidence level was performed. For comparison of cytoplasmic TDP‐43 in poly‐GA‐positive vs poly‐GA‐negative cells within patients, a paired two‐tailed t‐test was used.
Author contributions
BK performed most cell biological and biochemical experiments. KDL, HH, and TA provided and analyzed human samples. QZ, NM, and MM provided and analyzed mouse samples. HR performed qPCR analysis. FF and MSH performed flow cytometry analysis. BK, DF, and HR generated reagents. MC performed immunoassays. FUH and DE acquired funding. FUH and MSH supervised research and contributed to writing. DE designed the study, supervised research, and wrote the manuscript. All authors discussed the data and the manuscript.
Conflict of interest
D.E. holds a patent on “Dipeptide‐repeat proteins as therapeutic target in neurodegenerative diseases with hexanucleotide repeat expansion” (EP2948777 and US10066007).
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Dorothee Dormann, Ruben Fernandez‐Busnadiego, Qiang Guo, Saskia Hutten, and Bettina Schmid for critical comments to the manuscript, and Markus Oster and Martin Spitaler from the MPIB Imaging Facility for assistance with flow cytometry. Christoph Möhl from the DZNE Image and Data Analysis Facility wrote the original script for Columbus analysis. This work was supported by NOMIS Foundation and the Hans und Ilse Breuer Foundation (D.E.), the Munich Cluster of Systems Neurology (SyNergy) (EXC 2145/ID 390857198 to T.A., F.U.H., M.S.H., and D.E.), and the European Community's Health Seventh Framework Programme under grant agreement 617198 [DPR‐MODELS] (D.E.).
The EMBO Journal (2020) 39: e102811
Data availability
Source data are provided with the manuscript.
References
- Akimov V, Barrio‐Hernandez I, Hansen SVF, Hallenborg P, Pedersen AK, Bekker‐Jensen DB, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM et al (2018) UbiSite approach for comprehensive mapping of lysine and N‐terminal ubiquitination sites. Nat Struct Mol Biol 25: 631–640 [DOI] [PubMed] [Google Scholar]
- Aksu M, Pleiner T, Karaca S, Kappert C, Dehne HJ, Seibel K, Urlaub H, Bohnsack MT, Gorlich D (2018) Xpo7 is a broad‐spectrum exportin and a nuclear import receptor. J Cell Biol 217: 2329–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archbold HC, Jackson KL, Arora A, Weskamp K, Tank EM, Li X, Miguez R, Dayton RD, Tamir S, Klein RL et al (2018) TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Sci Rep 8: 4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovicic A, De Baets G, Scheveneels W, Steyaert J, Cuijt I et al (2016) Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep 6: 20877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH et al (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 65: 1044–1055.e1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Jeng US, Chiang YL, Hwang IS, Chen YR (2016) The glycine‐alanine dipeptide repeat from C9orf72 hexanucleotide expansions forms toxic amyloids possessing cell‐to‐cell transmission properties. J Biol Chem 291: 4903–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BB, Mallampalli RK (2009) Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol Cell Biol 29: 3062–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes‐Casey M, Lee CW, Jansen‐West K, Kurti A, Murray ME et al (2015) Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP‐43 pathology, neuronal loss, and behavioral deficits. Science 348: 1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, Sayegh M, Donlin‐Asp PG, Chen YH, Duong DM et al (2018) TDP‐43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci 21: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG (2000) Short‐lived green fluorescent proteins for quantifying ubiquitin/proteasome‐dependent proteolysis in living cells. Nat Biotechnol 18: 538–543 [DOI] [PubMed] [Google Scholar]
- De Smet F, Saiz Rubio M, Hompes D, Naus E, De Baets G, Langenberg T, Hipp MS, Houben B, Claes F, Charbonneau S et al (2017) Nuclear inclusion bodies of mutant and wild‐type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J Pathol 242: 24–38 [DOI] [PubMed] [Google Scholar]
- DeJesus‐Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p‐linked FTD and ALS. Neuron 72: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus‐Hernandez M, Finch NA, Wang X, Gendron TF, Bieniek KF, Heckman MG, Vasilevich A, Murray ME, Rousseau L, Weesner R et al (2017) In‐depth clinico‐pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathol 134: 255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Haass C (2016) An amyloid‐like cascade hypothesis for C9orf72 ALS/FTD. Curr Opin Neurobiol 36: 99–106 [DOI] [PubMed] [Google Scholar]
- Ederle H, Dormann D (2017) TDP‐43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett 591: 1489–1507 [DOI] [PubMed] [Google Scholar]
- Ederle H, Funk C, Abou‐Ajram C, Hutten S, Funk EBE, Kehlenbach RH, Bailer SM, Dormann D (2018) Nuclear egress of TDP‐43 and FUS occurs independently of exportin‐1/CRM1. Sci Rep 8: 7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eersel J, Ke YD, Gladbach A, Bi M, Gotz J, Kril JJ, Ittner LM (2011) Cytoplasmic accumulation and aggregation of TDP‐43 upon proteasome inhibition in cultured neurons. PLoS ONE 6: e22850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores BN, Dulchavsky ME, Krans A, Sawaya MR, Paulson HL, Todd PK, Barmada SJ, Ivanova MI (2016) Distinct C9orf72‐associated dipeptide repeat structures correlate with neuronal toxicity. PLoS ONE 11: e0165084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez‐Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC et al (2015) GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525: 129–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick P, Sellier C, Mackenzie IRA, Cheng CY, Tahraoui‐Bories J, Martinat C, Pasterkamp RJ, Prudlo J, Edbauer D, Oulad‐Abdelghani M et al (2018) Novel antibodies reveal presynaptic localization of C9orf72 protein and reduced protein levels in C9orf72 mutation carriers. Acta Neuropathol Commun 6: 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Almeida S, Lopez‐Gonzalez R (2017) Dysregulated molecular pathways in amyotrophic lateral sclerosis‐frontotemporal dementia spectrum disorder. EMBO J 36: 2931–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Petrucelli L (2011) Rodent models of TDP‐43 proteinopathy: investigating the mechanisms of TDP‐43‐mediated neurodegeneration. J Mol Neurosci 45: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser F, Martinez‐Lage M, Robinson J, Uryu K, Neumann M, Brandmeir NJ, Xie SX, Kwong LK, Elman L, McCluskey L et al (2009) Clinical and pathological continuum of multisystem TDP‐43 proteinopathies. Arch Neurol 66: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotzl JK, Lang CM, Haass C, Capell A (2016) Impaired protein degradation in FTLD and related disorders. Ageing Res Rev 32: 122–139 [DOI] [PubMed] [Google Scholar]
- Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O'Donovan K, Fare CM, Diaz Z, Singh N et al (2018a) Nuclear‐import receptors reverse aberrant phase transitions of RNA‐binding proteins with prion‐like domains. Cell 173: 677–692.e620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Lehmer C, Martinez‐Sanchez A, Rudack T, Beck F, Hartmann H, Perez‐Berlanga M, Frottin F, Hipp MS, Hartl FU et al (2018b) In situ structure of neuronal C9orf72 poly‐GA aggregates reveals proteasome recruitment. Cell 172: 696–705.e612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F, Eckert M, von Zweydorf F, Gloeckner CJ, Kahle PJ (2018) Identification and characterization of ubiquitinylation sites in TAR DNA‐binding protein of 43 kDa (TDP‐43). J Biol Chem 293: 16083–16099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner‐Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T et al (2018) Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173: 706–719.e713 [DOI] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Chen‐Plotkin A, Winton MJ, Unger TL, Xu Y, Neumann M, Trojanowski JQ, Lee VM (2009) Expression of TDP‐43 C‐terminal fragments in vitro recapitulates pathological features of TDP‐43 proteinopathies. J Biol Chem 284: 8516–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18: 1226–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M, Walker LC (2018) Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat Neurosci 21: 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi B, Hartmann H, May S, Mohl C, Ederle H, Michaelsen M, Schludi MH, Dormann D, Edbauer D (2017) Cytoplasmic poly‐GA aggregates impair nuclear import of TDP‐43 in C9orf72 ALS/FTLD. Hum Mol Genet 26: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ et al (2011) Systematic and quantitative assessment of the ubiquitin‐modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut DA (2013) Slippery substrates impair ATP‐dependent protease function by slowing unfolding. J Biol Chem 288: 34729–34735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Baskaran P, Gomez‐Deza J, Chen HJ, Nishimura AL, Smith BN, Troakes C, Adachi Y, Stepto A, Petrucelli L et al (2017) C9orf72 poly GA RAN‐translated protein plays a key role in amyotrophic lateral sclerosis via aggregation and toxicity. Hum Mol Genet 26: 4765–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG (1997) Inhibition of ubiquitin/proteasome‐dependent protein degradation by the Gly‐Ala repeat domain of the Epstein‐Barr virus nuclear antigen 1. Proc Natl Acad Sci USA 94: 12616–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79: 416–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LP (2016) C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90: 521–534 [DOI] [PubMed] [Google Scholar]
- Lokireddy S, Kukushkin NV, Goldberg AL (2015) cAMP‐induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci USA 112: E7176–E7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin RJ, Gu H, Zhu Y, Leonard M, Ahmad AS, Clauser KR, Meyer JG, Bennett EJ, Komives EA (2017) Site‐specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat Commun 8: 1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D et al (2013) Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico‐pathological correlations. Acta Neuropathol 126: 859–879 [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D et al (2015) Quantitative analysis and clinico‐pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130: 845–861 [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Hanel W, Li D, Becker K, Reich N, Moll UM (2010) Stress‐mediated nuclear stabilization of p53 is regulated by ubiquitination and importin‐alpha3 binding. Cell Death Differ 17: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf‐Strathmann J et al (2014) C9orf72 FTLD/ALS‐associated Gly‐Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128: 485–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide‐repeat proteins in FTLD/ALS. Science 339: 1335–1338 [DOI] [PubMed] [Google Scholar]
- Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, Duff KE (2016) Tau‐driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP‐PKA signaling. Nat Med 22: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM et al (2006) Ubiquitinated TDP‐43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133 [DOI] [PubMed] [Google Scholar]
- Nguyen L, Montrasio F, Pattamatta A, Tusi SK, Bardhi O, Meyer KD, Hayes L, Nakamura K, Banez‐Coronel M, Coyne A et al (2020) Antibody therapy targeting RAN proteins rescues C9 ALS/FTD phenotypes in C9orf72 mouse model. Neuron 105: 645–662.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Zupunski V, Troakes C, Kathe C, Fratta P, Howell M, Gallo JM, Hortobagyi T, Shaw CE, Rogelj B (2010) Nuclear import impairment causes cytoplasmic trans‐activation response DNA‐binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133: 1763–1771 [DOI] [PubMed] [Google Scholar]
- Nonaka T, Masuda‐Suzukake M, Hosokawa M, Shimozawa A, Hirai S, Okado H, Hasegawa M (2018) C9ORF72 dipeptide repeat poly‐GA inclusions promote intracellular aggregation of phosphorylated TDP‐43. Hum Mol Genet 27: 2658–2670 [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier‐Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C et al (2011) Long pre‐mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP‐43. Nat Neurosci 14: 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Bharathi V, Sivalingam V, Girdhar A, Patel BK (2019) Molecular mechanisms of TDP‐43 misfolding and pathology in amyotrophic lateral sclerosis. Front Mol Neurosci 12: 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon‐Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21‐linked ALS‐FTD. Neuron 72: 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi S, Stauffer JE, Jiang J, Garcia SD, Taylor AE, Schulte D, Ohkubo T, Schloffman CL, Maldonado M, Baughn M et al (2018) Sense‐encoded poly‐GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co‐localize with TDP‐43 in dendrites of repeat‐expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol 135: 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T; German Consortium for Frontotemporal Lobar Degeneration; Bavarian Brain Banking Alliance , Arzberger T et al (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol 130: 537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, Popper B, Dimou L, Strom TM, Winkelmann J et al (2017) Spinal poly‐GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol 134: 241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotter EL, Chen HJ, Shaw CE (2015) TDP‐43 proteinopathy and ALS: insights into disease mechanisms and therapeutic targets. Neurotherapeutics 12: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Verkhusha VV (2013) Near‐infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods 10: 751–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Stepto A, Au WH, Adachi Y, Diaper DC, Hall R, Rekhi A, Boudi A, Tziortzouda P, Lee YB et al (2018) A feedback loop between dipeptide‐repeat protein, TDP‐43 and karyopherin‐alpha mediates C9orf72‐related neurodegeneration. Brain 141: 2908–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Urushitani M, Inoue H, Koike M, Uchiyama Y, Komatsu M, Tanaka K, Yamazaki M, Abe M, Misawa H et al (2012) Motor neuron‐specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J Biol Chem 287: 42984–42994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V et al (2011) Characterizing the RNA targets and position‐dependent splicing regulation by TDP‐43. Nat Neurosci 14: 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsavayai SC, Yoon SJ, Gardner RC, Gendron TF, Vargas JN, Trujillo A, Pribadi M, Phillips JJ, Gaus SE, Hixson JD et al (2016) Timing and significance of pathological features in C9orf72 expansion‐associated frontotemporal dementia. Brain 139: 3202–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT et al (2012) Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489: 304–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff KM, Rawlinson SM, Hearps AC, Jans DA (2011) An AlphaScreen(R)‐based assay for high‐throughput screening for specific inhibitors of nuclear import. J Biomol Screen 16: 192–200 [DOI] [PubMed] [Google Scholar]
- Walker AK, Spiller KJ, Ge G, Zheng A, Xu Y, Zhou M, Tripathy K, Kwong LK, Trojanowski JQ, Lee VM (2015) Functional recovery in new mouse models of ALS/FTLD after clearance of pathological cytoplasmic TDP‐43. Acta Neuropathol 130: 643–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB et al (2014) Antisense proline‐arginine RAN dipeptides linked to C9ORF72‐ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84: 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard T, Jensen BK, Wen X, Cai J, Kropf E, Iacovitti L, Pasinelli P, Trotti D (2016) Cell‐to‐cell transmission of dipeptide repeat proteins linked to C9orf72‐ALS/FTD. Cell Rep 17: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM (2008) Disturbance of nuclear and cytoplasmic TAR DNA‐binding protein (TDP‐43) induces disease‐like redistribution, sequestration, and aggregate formation. J Biol Chem 283: 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Guo X, Yang W, Yan S, Yang S, Zhao T, Sun Q, Liu Y, Li S, Li XJ (2019) Caspase‐4 mediates cytoplasmic accumulation of TDP‐43 in the primate brains. Acta Neuropathol 137: 919–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Xu YF, Cook C, Gendron TF, Roettges P, Link CD, Lin WL, Tong J, Castanedes‐Casey M, Ash P et al (2009) Aberrant cleavage of TDP‐43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA 106: 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S et al (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525: 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen‐West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M et al (2016) C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci 19: 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Lehmer C, Michaelsen M, Mori K, Alterauge D, Baumjohann D, Schludi MH, Greiling J, Farny D, Flatley A et al (2017) Antibodies inhibit transmission and aggregation of C9orf72 poly‐GA dipeptide repeat proteins. EMBO Mol Med 9: 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Mareljic N, Michaelsen M, Parhizkar S, Heindl S, Nuscher B, Farny D, Czuppa M, Schludi C, Graf A et al (2020) Active poly‐GA vaccination prevents microglia activation and motor deficits in a C9orf72 mouse model. EMBO Mol Med 12: e10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez‐Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH et al (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110: E4968–E4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Data Availability Statement
Source data are provided with the manuscript.


