Abstract
Study Objectives
Sleep problems are common, serving as both a predictor and symptom of posttraumatic stress disorder (PTSD), with these bidirectional relationships well established in the literature. While both sleep phenotypes and PTSD are moderately heritable, there has been a paucity of investigation into potential genetic overlap between sleep and PTSD. Here, we estimate genetic correlations between multiple sleep phenotypes (including insomnia symptoms, sleep duration, daytime sleepiness, and chronotype) and PTSD, using results from the largest genome-wide association study (GWAS) to date of PTSD, as well as publicly available GWAS results for sleep phenotypes within UK Biobank data (23 variations, encompassing four main phenotypes).
Methods
Genetic correlations were estimated utilizing linkage disequilibrium score regression (LDSC), an approach that uses GWAS summary statistics to compute genetic correlations across traits, and Mendelian randomization (MR) analyses were conducted to follow up on significant correlations.
Results
Significant, moderate genetic correlations were found between insomnia symptoms (rg range 0.36–0.49), oversleeping (rg range 0.32–0.44), undersleeping (rg range 0.48–0.49), and PTSD. In contrast, there were mixed results for continuous sleep duration and daytime sleepiness phenotypes, and chronotype was not correlated with PTSD. MR analyses did not provide evidence for casual effects of sleep phenotypes on PTSD.
Conclusion
Sleep phenotypes, particularly insomnia symptoms and extremes of sleep duration, have shared genetic etiology with PTSD, but causal relationships were not identified. This highlights the importance of further investigation into the overlapping influences on these phenotypes as sample sizes increase and new methods to investigate directionality and causality become available.
Keywords: insomnia, genetics, posttraumatic stress disorder, sleep and psychiatric conditions, genetic correlation, LDSC, LDSR, endelian randomization, sleep duration, sleep disorders
Statement of Significance.
This paper provides the first formal investigation into genetic overlap between sleep phenotypes and posttraumatic stress disorder (PTSD), which, despite known epidemiologic relationships, has yet to be studied at the genetic level. We present evidence for shared genetic influences between PTSD and several sleep traits, including insomnia and sleep duration, harnessing the power of the Psychiatric Genomics Consortium for PTSD and incorporating genetic data from the largest publicly available datasets. We also conduct Mendelian randomization analyses to examine potential causal relationships, although we do not find evidence for causality. Despite this, moderate genetic correlations shown here highlight genetic overlap, indicating potential shared pathophysiology. Additional work to further examine the basis of these correlations and test bidirectionality and causality is warranted.
Introduction
Approximately 70% of adults worldwide are exposed to at least one traumatic event in their lifetime [1], with 7%–30% developing post-traumatic stress disorder (PTSD) [2], characterized by symptoms of intrusion, avoidance, negative alterations in mood and cognition, and alterations in arousal and reactivity [3]. Disruptions in sleep are common complaints among individuals with PTSD [4] and, indeed, sleep difficulties are included within the arousal and reactivity (insomnia) and intrusion (trauma-related nightmares) clusters [3]. However, the relationship between sleep disturbance and PTSD is likely greater than symptom overlap, as research supports a complex and bidirectional relationship between sleep and disorders that share internalizing characteristics, such as major depressive disorder (MDD) and PTSD [5, 6]. Longitudinal studies in Veterans demonstrate that insomnia prior to deployment is a strong predictor of future PTSD or MDD diagnosis [7] and that insomnia is associated with PTSD and depressive symptom severity in this population [8]. Often, sleep symptoms are persistent and difficult to treat in the context of psychiatric disorders, serving as global markers of disorder severity and negative sequelae [9–12]. Understanding this comorbidity is important, as sleep problems themselves are also common: one in three adults in the general population report at least one symptom of sleep disturbance and 6%–10% of people meet the DSM diagnostic criteria for insomnia disorder [5].
One approach to further elucidating the nature of these bidirectional relationships is through genetics. Both insomnia and PTSD are moderately heritable, with estimates from the twin literature ranging from approximately 20% to 60% for insomnia [13] and 30% to 70% for PTSD [14]. There is also some evidence for quantitative sex differences across both phenotypes, although notably only sex effects for PTSD have been shown to be consistent within the molecular genetics literature [15–17]. Gene identification efforts have been rapidly growing, encompassing both genome-wide association studies (GWAS) as well as consortia efforts that afford meta-analysis of individual GWAS studies to gain statistical power. At the time of writing, there had been 10 individual GWAS of PTSD [18] and one combined GWAS conducted by the Psychiatric Genomics Consortia (PGC) PTSD workgroup [16], with the second PGC–PTSD GWAS under review (now published) [17]. There were nine GWAS of insomnia-related phenotypes [19], 12 GWAS of sleep duration [20], and four of chronotype [21]. Relationships between sleep and PTSD at the genetic level have yet to be investigated in great detail. However, there is evidence for shared genetic etiology between PTSD and numerous disorders characterized by both internalizing and externalizing symptomatology [22] that also share genetic influences with sleep [23].
For example, longitudinal twin studies demonstrate substantial genetic overlap between insomnia and depression [23, 24], and shared genetic influences between MDD and PTSD have also been documented in the twin literature, with genetic correlations estimated at over 50% [25, 26]. Similarly, there is evidence for this overlap at the molecular genetics level as well, with several recently published GWAS reporting moderate genetic correlations between insomnia and depression phenotypes (range 0.34–0.53) [19, 27, 28], and the most recent PGC–PTSD publication reporting a correlation of 0.62 between PTSD and depression [17]. However, no studies to date have explicitly examined genetic overlap, biometric, or molecular, between sleep phenotypes and PTSD. As large-scale GWAS of insomnia phenotypes increasingly show more robust genetic correlations with psychiatric disorders, as opposed to other sleep phenotypes [29, 30], a better understanding of genetic relationships between sleep and PTSD is clearly warranted.
Statistical genetic methods that utilize genomic data in aggregate allow for the examination of molecular overlap across phenotypes, often without the need for individual-level genetic data. An example of this is Linkage Disequilibrium score regression (LDSR or LDSC) [31], which incorporates data from GWAS summary statistics to calculate genetic correlations (rg) between phenotypes based on available single nucleotide polymorphisms (SNPs) and LD scores. This method has been used to demonstrate genetic correlations between many medical and psychiatric traits [31, 32], including genetic correlations between insomnia and metabolic traits [19] and between PTSD and schizophrenia [16, 17]. While genetic correlations provide information regarding relationships across traits, they cannot identify causal associations or provide information regarding directionality. There are also methods that utilize summary-level data to conduct Mendelian randomization (MR) analyses [33], which allow one to test whether or not a specific exposure is causally related to an outcome variable through genetic polymorphisms [34]. The current study addresses gaps in the literature, outlined above, by examining genetic correlations between PTSD and sleep phenotypes (i.e. insomnia symptoms, sleep duration, daytime sleepiness, chronotype), and then conducting MR analyses on phenotypes with significant correlations to determine whether genetic markers associated with sleep phenotypes are causally linked with PTSD.
Methods
Samples
PTSD
PGC–PTSD meta-analysis summary statistics from Freeze 2 [17], which include 174 659 (European ancestry [EA]) individuals (23 212 cases with PTSD and 151 447 controls; all adults) from 51 studies, were used for genetic correlation analyses. Only EA summary statistics were used given that the current version of LDSC cannot be used on genetic data from individuals with admixed ancestry (e.g. African Americans and many Latinos). For MR analyses, PGC–PTSD meta-analysis summary statistics from Freeze 1.5 (total N = 48 471; 12 823 cases with PTSD and 35 648 controls), which excludes individuals from the UK Biobank dataset, were used. This is necessary, as MR analyses are not robust to sample overlap [33] and including the full results from Freeze 2 would bias results.
Sleep
Sleep phenotypes included those present in a total of 23 publicly available sets of summary statistics. Seventeen sets of summary statistics were chosen from sleep phenotypes available on LD Hub in April 2018; two phenotypes explicitly stated to reflect sleep apnea were excluded [35]. Of note, seven of these phenotypes were taken from the UK Biobank GWAS (run by Neale and colleagues; http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank/, last accessed May 30, 2019, see description below). An additional six sleep phenotypes (results from newly published manuscripts of updated UK Biobank data with larger sample sizes) were downloaded from the Sleep Disorders Knowledge Portal (http://sleepdisordergenetics.org/informational/data/, last accessed May 30, 2019) in May 2019. Although also present in the Sleep Disorders Knowledge Portal, results from two additional publications (one with actigraphy data and another investigating nighttime oxygen) were not included, as these phenotypes are outside the scope of this manuscript.
Phenotypes used were as follows: insomnia symptoms (N = 4), sleep duration (N = 8), daytime sleepiness (N = 4), chronotype (N = 6), and ICD-10 sleep diagnosis (N = 1), which encompasses multiple categories of sleep disorders, including insomnias and hypersomnias. All sleep GWAS utilized the UK Biobank dataset, which is a large, population-based sample of adults aged 40–69 residing in England, Scotland, and Wales [36]. It is important to note that analyses of UK Biobank participant characteristics have demonstrated evidence of selection bias toward healthy individuals [36]. Sleep phenotypes varied in their conceptualization, and all samples included both sexes. Each of the 23 sleep phenotypes does not necessarily represent a unique data point; instead there are multiple conceptualizations of the same category of phenotype analyzed across different subsets of the UK Biobank data. A subset of included studies provided results for phenotypes separated by sex (N = 4; insomnia symptoms [19, 27], daytime sleepiness [19], and sleep duration [19]).
UK Biobank GWAS
The UK Biobank GWAS, accessible at http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank/ (last accessed May 30, 2019), is a genetics resource developed by Neale and colleagues, who conducted a series of genome-wide analyses across thousands of phenotypes available in the UK Biobank dataset (N ~ 337 000). The 2017 release (used in analyses presented here) also includes LDSC heritability estimates. In order to conduct genetic analyses on such a large scale, an automated system was used to create phenotypes, making all statistical distributions normal or binomial, running all models as linear, and modeling all alleles with additive effects. Only individuals of EA who were not related were used in genetic analyses. Furthermore, genetic analyses also had the following restrictions with regard to inclusion criteria: SNPs imputed from the Haplotype Research Consortium (initial analyses), MAF > 0.1, INFO > 0.8, and Hardy–Weinberg equilibrium p > 1E−10. Thus, while this resource provides summary statistics for a wide range of phenotypes with large sample sizes, a limited number of SNPs are included, and phenotypes have not been validated. Consequently, all results should be considered preliminary and may change as new iterations of the data are developed. Detailed information regarding phenotypes, methods, and results can be accessed at the website listed above.
Data analytic methods and plan
Note that this work conforms to the ethical standards of the Declaration of Helsinki.
Genetic correlations
Genetic correlations between PTSD and sleep phenotypes were estimated using LDSC [31] version 1.0.0, downloaded from https://github.com/bulik/ldsc/ (Accessed August 17, 2019). Estimates were also stratified by sex where possible (i.e. for females only, as the heritability estimate for PTSD in males within the PGC–PTSD is not significantly different from zero [17]). Summary statistics files were filtered initially for removal of SNPs with INFO < 0.9 or MAF < 0.01 or > 0.99. Next, all files were run through the munge_sumstats.py script included in the LDSC program and filtered on a list of HapMap 3 SNPs in order to retain only well-imputed SNPs; the major histocompatibility complex was also removed, resulting in 1 215 001 potential SNPs retained for analysis. European ancestry LD scores from 1000 genomes were used (https://data.broadinstitute.org/alkesgroup/LDSCORE/, Acessed August 17, 2019). The intercept was not constrained for LDSC analyses. To correct for multiple testing, a Bonferroni-adjusted p-value of 0.0022 (p = 0.05/23 genetic correlations) was used as the threshold for significance.
Mendelian randomization
For sleep phenotypes found to be significantly correlated with PTSD, we conducted MR analyses to further investigate potential causal and bidirectional relationships between these traits. MR is a method that uses genetic data (i.e. independent genome-wide significant [GWS] SNPs often referred to as “instruments”) to determine if an exposure (e.g. short sleep duration) is causally related to an outcome (e.g. PTSD diagnosis). Analyses can also be run in the reverse direction (i.e. by flipping the exposure and the outcome variables) to examine bidirectionality of causal effects. Note that here, we utilize sleep variables as the exposure; this is in part due to low power for running PTSD as the exposure (discussed in detail elsewhere in the manuscript), but also makes sense based on the literature: Sleep phenotypes are modifiable (e.g. insomnia can be treated with cognitive behavioral therapy [37]), and longitudinal studies clearly demonstrate effects of pre-trauma sleep disturbances on post-trauma psychiatric phenotypes [7, 8]. For analysis, we utilized the R package TwoSampleMR (https://mrcieu.github.io/TwoSampleMR/, last accessed May 30, 2019) [33], which provides a streamlined approach to run MR analyses using summary statistics level data. For MR analyses, we used PTSD summary statistics from Freeze 1.5 (see Table 1 for sample size and heritability).
Table 1.
PTSD and sleep phenotypes utilized in genetic correlation analyses
| Source* | Phenotype | N | SNP-heritability (SE) |
|---|---|---|---|
| PTSD [17] | |||
| PGC, Freeze 1.5 | PTSD, binary | 48 471 (12 823 Ca; 35 648 Co) | 0.05 (0.018) |
| PGC, Freeze 2† | PTSD, binary | 174 659 (23 212 Ca; 151 447 Co) | 0.05 (0.010) |
| Insomnia | |||
| Lane et al. [19] | Insomnia symptoms,‡ binary | 58 702 (31 767 Ca; 26 935 Co) | 0.13 (0.012) |
| Hammerschlag et al. [27] | Insomnia symptoms,§ binary | 113 006 (32 384 Ca; 80 622 Co) | 0.09 (0.008) |
| UK Biobank GWAS | Insomnia symptoms, continuous | 336 965 | 0.06 (0.004) |
| Lane et al. [29] | Insomnia symptoms,‡ binary | 237 622 (129 270 Ca; 108 352 Co) | 0.18 (0.007)|| |
| Sleep duration | |||
| Jones et al. [20] | Sleep duration, continuous | 127 573 | 0.07 (0.007) |
| Lane et al. [19] | Sleep duration, continuous | 111 975 | 0.06 (0.007) |
| UK Biobank GWAS | Sleep duration, continuous | 335 410 | 0.07 (0.005) |
| Dashti et al. [42] | Sleep duration, continuous | 446 118 | 0.07 (0.003)|| |
| Jones et al. [20] | Long sleepers,¶ binary | 91 306 (10 102 Ca; 81 204 Co) | 0.07 (0.016)|| |
| Dashti et al. [42] | Long sleepers,¶ binary | 339 926 (34 184 Ca; 305 742 Co) | 0.08 (0.006)|| |
| Jones et al. [20] | Short sleepers,# binary | 110 184 (28 980 Ca; 81 204 Co) | 0.09 (0.009)|| |
| Dashti et al. [42] | Short sleepers,# binary | 411 934 (106 192 Ca; 305 742 Co) | 0.09 (0.004)|| |
| Daytime sleepiness | |||
| Lane et al. [19] | Excessive daytime sleepiness, continuous | 111 648 | 0.05 (0.005) |
| UK Biobank GWAS | Daytime dozing/sleeping (narcolepsy), continuous | 336 082 | 0.05 (0.003) |
| UK Biobank GWAS | Nap during the day, continuous | 337 074 | 0.08 (0.004) |
| UK Biobank GWAS | Easy to get up in morning, continuous | 336 501 | 0.07 (0.004) |
| Chronotype | |||
| Jones et al. [20] | Chronotype, continuous | 127 898 | 0.12 (0.007) |
| Lane et al. [21] | Chronotype, continuous | 100 420 | 0.12 (0.007)|| |
| UK Biobank GWAS | Chronotype, continuous | 301 143 | 0.12 (0.006) |
| Jones et al. [51] | Chronotype, continuous | 449 734 | 0.11 (0.004)|| |
| Lane et al. [21] | Extreme chronotype [eveningness],** binary | 35 672 (8 724 Ca; 26 948 Co) | 0.40 (0.034)|| |
| Jones et al. [51] | Morningness, binary | 403 195 (252 287 Ca; 150 908 Co) | 0.17 (0.006)|| |
| Other | |||
| UK Biobank GWAS | ICD-10 Sleep Diagnosis,†† binary | 337 199 (2025 Ca; 335 174 Co) | 0.13 (0.051) |
Ca, case; Co, control; GWAS, genome-wide association study; ICD, International Classification of Diseases; PGC, Psychiatric Genomics Consortium; PTSD, posttraumatic stress disorder; SE, standard error; SNP, single nucleotide polymorphism.
*All sources analyzed data from the UK Biobank. The description “UK Biobank GWAS” refers to a series of analyses conducted on UK Biobank data (2017 release), which can be accessed at: http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank/ (last accessed May 30, 2019)
†This is the PTSD phenotype used for genetic correlations presented in this manuscript; both PTSD heritabilities were scaled to a population prevalence of 30%.
‡Insomnia cases were defined as individuals endorsing “Often” on the sleeplessness item, while controls included individuals endorsing “Never/Rarely.” Individuals endorsing “Sometimes” were designated as missing and not used in analysis.
§Insomnia cases were defined as individuals endorsing “Often” on the sleeplessness item, while controls were those endorsing “Never/Rarely” and “Sometimes.”
||Denotes that this heritability estimate was not published; instead it was estimated from downloaded summary statistic data using LDSC and converted to the liability scale (with sample prevalence = population prevalence), if appropriate. Otherwise, the estimate was taken from the original source publication or https://nealelab.github.io/UKBB_ldsc/, (last accessed May 30, 2019) as appropriate.
¶Long sleepers were defined as individuals reporting 9 or more hours of sleep, while controls were those endorsing 7–8 h.
#Short sleepers were defined as individuals reporting 6 or fewer hours of sleep, while controls were those endorsing 7–8 h.
**Extreme chronotype cases were defined as individuals endorsing “Definitely an evening person” or “Maybe an evening person,” while controls were individuals endorsing “Definitely a morning person” or “Maybe a morning person.” All other individuals were defined as missing.
††Note that this phenotype encompasses multiple categories of sleep disorders, including insomnias, hypersomnias, circadian rhythm disorders, sleep apnea, narcolepsy and cataplexy, other sleep disorders, and unspecified sleep disorder.
Summary statistics were formatted per the guidelines specified in TwoSampleMR, beginning with the same files utilized for LDSC (i.e. filtered based on MAF and INFO, as above). The online tutorial (https://mrcieu.github.io/TwoSampleMR/, last accessed May 30, 2019) was used as a guide to conduct the appropriate steps of data management prior to analysis. For each sleep phenotype (“exposure”), SNPs with p < 5 × 10−8 were chosen as potential instruments. The “clump” command, with default settings, was then run to retain only independent GWS SNPs for use in MR analysis. Next, data for each of the chosen SNPs was extracted from summary statistics of the outcome variable (i.e. PTSD). Data harmonization was conducted for each combination of exposure phenotype and PTSD to verify the presence of corresponding effect alleles. Once this was completed, MR analyses were run using the default setting for the methods list, which runs MR using five different methods: MR Egger, weighted median, inverse variance weighted (IVW), simple mode, and weighted mode. The IVW method is the default method and the simplest, but does have assumptions regarding horizontal pleiotropy (i.e. where an instrument [SNP] has an effect on the outcome variable that is independent [not mediated by] of its effect on the exposure variable), whereas these assumptions are less stringent within the MR Egger method [33, 38]. We also conducted MR analyses by single SNP and ran additional tests examining heterogeneity and horizontal pleiotropy. A detailed explanation of each MR method and its assumptions, as well as additional tests, is beyond the scope of this manuscript, but further information can be found in the main methods of the original publication for TwoSampleMR and supporting statistical papers, referenced in the tutorial [33]. A Bonferroni-adjusted p-value of 0.0125 (p = 0.05/4 sleep exposures) was used as the threshold for significance for MR.
Note that several limitations of the data affected MR analysis. First, given that all sleep phenotypes were from the UK Biobank dataset, and there were significant correlations across multiple versions of the same phenotype, we chose a representative phenotype for each significant genetic correlation (i.e. insomnia symptoms, sleep duration, short sleepers, long sleepers). Note that the two significant daytime sleepiness phenotypes from the UK Biobank GWAS were not included in MR analyses, as these correlations are weaker, and the phenotypes have not been validated. Furthermore, we did not specifically test for reverse directionality (PTSD to sleep phenotypes), as the PTSD phenotype is not yet well powered for MR (i.e. there are not sufficient GWS instruments; only two GWS SNPs were identified in EA individuals) [17]. The use of Freeze 1.5 (as opposed to Freeze 2), which has a much smaller sample size, also decreased the power to detect associations through MR.
Results
Univariate SNP-heritability estimates
Briefly, heritability estimates based on additive genetic variance from autosomal SNPs () for PTSD and sleep phenotypes were derived from previous research and are shown in Table 1, including the original GWAS sample size and phenotype description. PTSD (Freeze 2) had a small, but significant, = 0.05 (SE = 0.010), with a similar estimate seen for Freeze 1.5 ( = 0.05 [SE = 0.018]). For sleep phenotypes, estimates were generally small and ranged from 0.06 to 0.18 for insomnia symptoms, 0.06–0.09 for sleep duration, 0.05–0.08 for daytime sleepiness, 0.11–0.40 for chronotype, and 0.13 for ICD-10 sleep diagnosis.
Main genetic correlation analyses
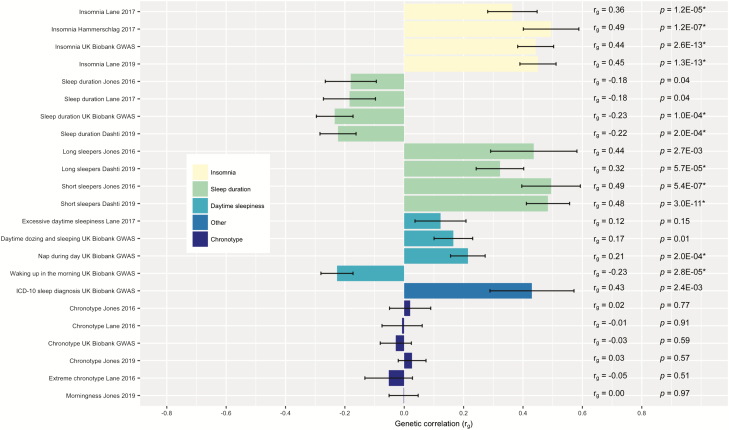
Figure 1 presents genetic correlations and standard errors for analyses that include both sexes. Specific rg estimates and uncorrected p-values are embedded in the right side of the figure. Moderate, positive genetic correlations were found between PTSD and insomnia symptoms (all four definitions of the phenotype; rg range 0.36–0.49), short sleepers (rg range 0.48–0.49), and long sleepers (rg range 0.32–0.44), and all except for the initial estimate for long sleepers (rg = 0.44; p = 0.0027) remained significant following Bonferroni correction (p < 0.0022; see Figure 1 for raw p-values; range 1.3E−13 to 5.7E−05 for these phenotypes). Several daytime sleepiness phenotypes showed small positive correlations with PTSD, although only napping during the day (rg = 0.21) remained statistically significant following correction. In contrast, small, negative genetic correlations were found between PTSD and continuous sleep duration, with the correlations becoming larger in magnitude and significant after correction with increasing sample size (rg range [significant] = −0.22 to −0.23). There was also a significant correlation between waking up in the morning (rg = −0.23; with higher scores indicating that the participant finds it easier to wake up on a typical day) and PTSD. Finally, there were no significant genetic correlations between any variation of chronotype (six different sets of summary statistics) and PTSD.
Figure 1.
Genetic correlations (rg) between sleep phenotypes and posttraumatic stress disorder (PTSD). Genetic correlation estimates obtained through linkage disequilibrium score regression (LDSC) are shown here, grouped by sleep phenotype, with each specific study name on the left. Error bars represent standard error. p-Values that pass multiple testing correction (p < 0.0022 per Bonferroni correction) are indicated by an (*).
Sex-specific analyses (i.e. for females only)
Results of sex-specific analyses are shown in Table 2, along with estimates and other sample information. There were four phenotypes available for analysis (two for insomnia symptoms, one for daytime sleepiness, and one for sleep duration). Both insomnia phenotypes had significant, small to moderate correlations with PTSD in females (rg = 0.26, SE = 0.10, p = 0.0069 for Lane et al. [19]; rg = 0.44, SE = 0.12, p = 0.0002 for Hammerschlag et al. [27]), consistent with analyses within the full sample. However, neither daytime sleepiness (rg = −0.14, SE = 0.10, p = 0.15) nor sleep duration (rg = 0.07, SE = 0.11, p = 0.50) were significantly correlated with PTSD for females, which is also consistent with results presented above.
Table 2.
Genetic correlations between sleep phenotypes and PTSD, females only
| Genetic correlation with PTSD | |||||
|---|---|---|---|---|---|
| Source† | Phenotype | N | SNP- heritability | r g (SE) | p value |
| PTSD [17] | |||||
| PGC, Freeze 1.5 | PTSD, binary | 15 656 (6128 Ca; 9528 Co) | 0.21 (0.05) | — | — |
| PGC, Freeze 2‡ | PTSD, binary | 86 600 (12 973 Ca; 73 627 Co) | 0.10 (0.02) | — | — |
| Sleep phenotypes | |||||
| Lane et al. [19] | Insomnia symptoms,§ binary | 30 445 (NR Ca; NR Co) | 0.14 (0.02)¶ | 0.26 (0.10) | 0.007* |
| Hammerschlag et al. [27] | Insomnia symptoms,# binary | 59 367 (19 521 Ca; 39 846 Co) | 0.08 (0.01) | 0.44 (0.12) | 0.0002* |
| Lane et al. [19] | Excessive daytime sleepiness, continuous | 59 576 | 0.06 (0.01) | −0.14 (0.10)e | 0.148 |
| Lane et al. [19] | Sleep duration, continuous | 59 365 | 0.08 (0.01) | 0.07 (0.11)e | 0.498 |
Ca, cases; Co, controls; NR, not reported; PGC, Psychiatric Genomics Consortium; PTSD, posttraumatic stress disorder; SNP, single nucleotide polymorphism.
†All sources analyzed data from the UK Biobank. The description “UK Biobank GWAS” refers to a series of analyses conducted on UK Biobank data (2017 release), which can be accessed at: http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank/ (last accessed May 30, 2019)
‡This is the PTSD phenotype used for genetic correlations presented in this manuscript; both PTSD heritabilities were scaled to a population prevalence of 30%.
§Insomnia cases were defined as individuals endorsing “Often” on the sleeplessness item, while controls included individuals endorsing “Never/Rarely.” Individuals endorsing “Sometimes” were designated as missing and not used in analysis.
#Insomnia cases were defined as individuals endorsing “Often” on the sleeplessness item, while controls were those endorsing “Never/Rarely” and “Sometimes.”
¶This heritability estimate was not published; instead it was estimated from downloaded summary statistic data using LDSC. Otherwise, the estimate was taken from the original source publication.
*p < 0.05.
Mendelian randomization analyses
Results from TwoSampleMR analyses investigating potential causal relationships between four sleep phenotypes (exposures) and PTSD (outcome) are shown in Table 3. There was no evidence to support causal relationships between any of the four sleep phenotypes (insomnia symptoms, sleep duration, short sleepers, long sleepers) and PTSD utilizing any of the five MR methods. Furthermore, none of the phenotypes showed strong evidence for significant heterogeneity (all p values > 0.05 for both MR Egger and IVW analysis) or horizontal directional pleiotropy (all p values > 0.05) contributing to the results. As there were no significant results within the main MR analyses, we are not presenting results of single SNP analyses. Scatter plots showing MR results for each method, as well as forest plots of each individual SNP, are available upon request. Of note, there were only six SNPs included in analyses of long sleepers following clumping, so results should be interpreted in light of the smaller number of instruments for this phenotype.
Table 3.
Results of two-sample Mendelian randomization [MR] analyses examining causal relationships between four sleep phenotypes (insomnia symptoms, sleep duration, short sleepers, and long sleepers), and PTSD
| Mendelian randomization* | Heterogeneity test† | Pleiotropy test‡ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | nSNP | Beta | SE | P value | Q | Q df | Q p val | Egger Int. | SE | p value |
| Insomnia symptoms (Lane et al. [19]) | ||||||||||
| MR Egger | 38 | −0.563 | 0.986 | 0.571 | 24.732 | 36 | 0.922 | 0.011 | 0.010 | 0.280 |
| Weighted median | 38 | 0.372 | 0.403 | 0.356 | — | — | — | |||
| Inverse variance weighted | 38 | 0.471 | 0.288 | 0.102 | 25.937 | 37 | 0.914 | |||
| Simple mode | 38 | 0.385 | 0.838 | 0.648 | — | — | — | |||
| Weighted mode | 38 | 0.036 | 0.688 | 0.958 | — | — | — | |||
| Sleep duration (Dashti et al. [42] | ||||||||||
| MR Egger | 70 | −0.639 | 0.555 | 0.254 | 86.188 | 68 | 0.067 | 0.011 | 0.009 | 0.240 |
| Weighted median | 70 | −0.136 | 0.214 | 0.525 | — | — | — | |||
| Inverse variance weighted | 70 | −0.006 | 0.153 | 0.966 | 87.967 | 69 | 0.062 | |||
| Simple mode | 70 | 0.236 | 0.482 | 0.626 | — | — | — | |||
| Weighted mode | 70 | −0.136 | 0.363 | 0.708 | — | — | — | |||
| Short sleepers (Dashti et al. [42]) | ||||||||||
| MR Egger | 24 | 4.340 | 2.917 | 0.151 | 28.138 | 22 | 0.171 | −0.025 | 0.020 | 0.224 |
| Weighted median | 24 | 0.293 | 0.796 | 0.713 | — | — | — | |||
| Inverse variance weighted | 24 | 0.771 | 0.635 | 0.225 | 30.144 | 23 | 0.145 | |||
| Simple mode | 24 | −0.551 | 1.457 | 0.709 | — | — | — | |||
| Weighted mode | 24 | −0.218 | 1.347 | 0.873 | — | — | — | |||
| Long sleepers (Dashti et al. 2019) | ||||||||||
| MR Egger | 6 | −1.331 | 5.092 | 0.807 | 1.619 | 4 | 0.805 | 0.009 | 0.029 | 0.772 |
| Weighted median | 6 | 0.160 | 1.852 | 0.931 | — | — | — | |||
| Inverse variance weighted | 6 | 0.178 | 1.521 | 0.907 | 1.716 | 5 | 0.887 | |||
| Simple mode | 6 | 0.454 | 2.594 | 0.868 | — | — | — | |||
| Weighted mode | 6 | 0.076 | 2.721 | 0.979 | — | — | — | |||
Int., intercept; MR, Mendelian randomization; PTSD, posttraumatic stress disorder; SE, standard error; SNP, single nucleotide polymorphism.
*Mendelian randomization was conducted utilizing the TwoSampleMR package in R.
†Heterogeneity tests were conducted on the MR Egger and inverse variance weighted results within the TwoSampleMR package.
‡Horizontal directional pleiotropy was tested using the TwoSampleMR package.
Discussion
This analysis is the first demonstration that PTSD and sleep-related traits have overlapping genetic architecture based on molecular genetic data. We also present the first analyses examining causal relationships between sleep phenotypes and PTSD, although we did not find substantial evidence for causality within this data. While correlations are expected based on both the twin and molecular genetics literatures (PTSD has genetic overlap with symptoms of other psychiatric disorders, particularly depression [22], which themselves overlap with insomnia [23]), genetic overlap between insomnia and PTSD has not been explicitly examined. However, results should be interpreted in the context of low SNP heritability estimates, as shown in Table 1: PTSD itself has a SNP heritability of 5% within the PGC Freeze 2 sample [17] and most sleep phenotypes used within this manuscript have SNP heritabilities under 15%. This means that while significant, correlations represent small portions of shared variance in the traits examined (although note that all phenotypes had the recommended heritability Z-score > 4, which indicates that power is appropriate and estimates are interpretable) [35].
Genetic correlations, mixed sexes
The strongest and most consistent genetic correlations across studies were found for PTSD and insomnia symptoms, which is not surprising given that insomnia symptoms are included within PTSD symptom clusters. Furthermore, published GWAS of insomnia phenotypes support that insomnia symptoms have substantial shared genetics with psychiatric disorders in general and less of a relationship with other sleep phenotypes such as chronotype and sleep duration [29, 30]. Thus, one would expect to see the most robust genetic correlation with PTSD for insomnia symptoms when compared to other phenotypes examined here. Estimates of genetic correlations between insomnia symptoms and PTSD reported here ranged from 0.36 to 0.49. These estimates are similar in magnitude to those reported in other studies estimating molecular genetics overlap between insomnia and depression, where estimates have ranged from 0.34 to 0.51 [19, 27, 28, 39]. The magnitude is also similar to the reported genetic correlations between PTSD and depression (rg = 0.34 in initial PGC–PTSD results [16] and rg = 0.62 in PGC–PTSD Freeze 2 [17]). Given that PTSD and depression are highly comorbid [40] and known to share genetic architecture as demonstrated in twin and molecular studies [16, 25, 26], similarities across genetic correlations are logical. Notably, genetic correlations discussed here are moderate, suggesting that each trait has a considerable degree of unique genetic architecture not accounted for by the insomnia/PTSD overlap.
For other sleep-related traits, the highest correlations were between short sleepers (sleeping 6 or less hours a night), and long sleepers (sleeping 9 or more hours a night), with weaker but significant correlations seen with napping during the day and waking up in the morning. A moderate, positive correlation was seen for any ICD-10 sleep diagnosis (rg = 0.43), but this was only nominally significant, as it did not pass multiple testing correction. Positive genetic correlations between both oversleeping and undersleeping are consistent with the epidemiologic literature demonstrating that both short- and long-sleep increase risk for PTSD in veterans [41] and provide evidence for a relationship between genetic influences on the extremes of sleep duration and PTSD. In contrast, only two of four continuous sleep duration phenotypes survived multiple testing correction, and sleep duration was negatively correlated with PTSD, with the highest correlation at −0.23. With increasing sample sizes, and thus increasing power, we provide evidence that sleep duration is indeed correlated with PTSD, although not at the same magnitude as the extremes of duration (short/long sleepers). Correlations between the extremes of sleep duration and sleep duration itself (estimated using LDSC and summary statistics data from Dashti et al. [42]) are high, with rg = 0.69 for long sleepers and rg = −0.89 short sleepers (this makes sense given the U-shaped curve of sleep duration). There are also published genetic correlations between sleep duration and schizophrenia (0.29) [19] and schizophrenia and PTSD (0.33) [16], which provides some support for a sleep duration and PTSD association.
Large GWAS are beginning to identify a substantial number of loci for sleep duration [42], and combining self-report with actigraphy measures, but it may be that sleep duration is less heritable and/or a more complex phenotype. It does not appear to be strongly correlated with insomnia symptoms [19], and as shown in Table 1, may be less heritable than insomnia symptoms and chronotype. Results from a twin study of sleep duration and depression may help reconcile these findings: Watson and colleagues [43] found that sleep duration moderates genetic influences on depression. Results indicated that, while initial models did not demonstrate genetic overlap, shared genetic influences on the two traits increased as sleep duration became more extreme. Furthermore, the heritability of depression was higher for individuals who were short or long (vs. normal) sleepers within this sample [43]. These results highlight the complexity of genetic influences on sleep duration, and thus it may be that shared genetic influences between sleep duration and PTSD can best be seen at extremes of duration, as opposed to a linear relationship with sleep duration.
Finally, there was no evidence for a significant genetic correlation between PTSD and chronotype. Genetic variants in clock genes have shown association with mood disorders [44, 45] RORA has been implicated in PTSD [18], and a recent meta-analysis of PTSD candidate gene studies implicates a polymorphism in ADCYAP1R1 [46], the gene coding for the pituitary adenylate cyclase-activating polypeptide (PACAP) receptor, which is an important neurotransmitter in circadian regulation [47, 48]. However, this evidence for a role of circadian genes in PTSD is not robust and should be interpreted in light of numerous problems with the candidate gene method (e.g. false positives, assumption of incorrect effect sizes) and the potential for false-positive GWAS results, particularly with smaller sample sizes [49, 50]. Thus, it is plausible that shared genetic influences between circadian rhythms and PTSD are not significant. Consistent with our findings, one GWAS failed to find any genetic overlap between chronotype and insomnia symptoms, despite a known role of circadian rhythms in insomnia [19]. Additional research is needed to further elucidate the role of circadian genes in PTSD, and a better understanding of the role of circadian loci in chronotype, as is emerging from new GWAS [51], will also be useful in this endeavor.
Sex-specific genetic correlations
We also attempted to estimate sex-based genetic correlations where possible, given existing evidence for quantitative sex differences across both phenotypes [15–17]. Note, however, that recent GWAS have not identified substantial sex differences in SNP-heritability estimates at the molecular level for insomnia symptoms [27, 30]. Analyses were unfortunately limited to females only, as the SNP-heritability of PTSD for males in the PGC does not differ from zero [17]. We report similar estimates for females only as in the full samples for two insomnia phenotypes (0.26 and 0.44, as compared to 0.36 and 0.49 in the mixed-sex samples), and the standard errors are larger. This is not surprising given the loss in power due to smaller sample size in sex-specific analyses. It may be that with increasing sample sizes, significant differences in the genetic correlation become observation (i.e. more robust for females than in males), but this will need to be further investigated in future studies.
Mendelian randomization analyses
In order to better elucidate the nature of the relationships between significantly correlated sleep phenotypes and PTSD, we conducted the first set of MR analyses for sleep exposure variables (insomnia symptoms, sleep duration, short sleepers, long sleepers) and PTSD. We did not find substantial evidence for causal relationships between sleep and PTSD, despite running analyses using results from the largest available GWAS of validated sleep phenotypes and PGC–PTSD data. However, given the lack of robust, replicated associations for sleep phenotypes that can be used as instruments, particularly when compared to other psychiatric phenotypes like schizophrenia [52] and low number of GWS hits for PTSD [17], this is not necessarily unexpected. The MR approach itself has limitations, particularly with regard to pleiotropy and weak instrument bias [33]. We were also interested in examining bidirectionality, given complex relationships between sleep and PTSD [6], but were not powered to conduct analyses of causality in the reverse direction, given that there are only two GWS SNPs for PTSD in Freeze 2 [17]. TwoSampleMR does include a test of bidirectionality [33], but this was likely under powered as well. Although these results are largely inconclusive, further examination of the shared genetics between sleep and PTSD with regard to bidirectionality and causality is warranted as methods evolve and more datasets become available.
Limitations
The present study included data from the largest PTSD GWAS meta-analysis to date (N = 174 659 EA individuals) and the largest published sleep-related GWAS results. However, there are a number of limitations to consider. Given that sleep disturbance is part of PTSD diagnostic criteria, this may induce some degree of correlation based on overlapping phenotype definitions. In one twin study, this confound was reduced by removing sleep items from depression operationalization [23] but that was not possible here. The sleep-related phenotypes are mostly limited to single-item self-report items rather than validated questionnaires, clinician diagnoses, or objective data. Furthermore, analyses were only conducted on individuals of EA given limitations of LDSC. As our understanding of how to approach admixed populations in genetic studies increases, future studies that replicate findings across other ancestry groups will be useful. Similarly, all sleep phenotypes used UK Biobank data, which limits the generalizability of results. Finally, summary statistics for many sleep phenotypes came from the UK Biobank GWAS (http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank/, last accessed May 30, 2019) and should be considered preliminary, as phenotypes have not been validated and genetic analyses have specific constraints (see Methods section for more details).
Conclusions
In summary, these results indicate that there are common genetic factors shared between PTSD and sleep-related phenotypes, in particular insomnia symptoms. This has implications for gene finding efforts, as genes that have been identified (or are identified in the future) for PTSD may also be important in sleep phenotypes; the reverse may also be true. Thus, further investigation of specific variants and genes may shed light on the underlying pathophysiology of both disorders. For example, one could hypothesize that stress response pathways may be involved in the development of both sleep problems and PTSD, given relationships between sleep and stress [53]. As sleep problems and PTSD are highly comorbid and the literature regarding the optimal way to treat both is a work in progress [54], a better understanding of the relationships between sleep and PTSD at the genetic level could lead to the development of novel treatment approaches as the field advances. Future studies with more in-depth phenotyping and genotyping, functional studies of identified variants, and analyses that test for causality and bidirectionality are clearly needed to advance this line of research.
Acknowledgements
We would like to thank everyone who contributed to the data utilized within this manuscript, as these analyses would not be possible without their prior work. This includes members of the PGC–PTSD workgroup, who provided summary statistic data for Freeze 2 (Nievergelt et al. 2019) and all contributors to the PGC–PTSD (both researchers and participants), all authors of manuscripts utilizing UK Biobank sleep phenotypes (PMIDs 26955885, 27494321, 27992416, 28604731, 30696823, 30804566, 30846698), Ben Neale and colleagues, who developed the UK Biobank GWAS, which includes GWAS results, as well as heritability estimates, for thousands of phenotypes within the UK Biobank sample (http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank, last accessed May 30, 2019) and was supported by UK Biobank applications 18597 and 11898, as well as the individuals who maintain the Sleep Disorders Knowledge Portal (http://sleepdisordergenetics.org/informational/data/, last accessed May 30, 2019).
Group Information: Members of the Psychiatric Genomics Consortium-Posttraumatic Stress Disorder (PGC–PTSD) are Allison E. Aiello (Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill); Lynn M. Almli (Department of Psychiatry and Behavioral Sciences, Emory University, Atlanta, Georgia); Ananda B. Amstadter (Department of Psychiatry, Virginia Institute for Psychiatric and Behavioral Genetics, Richmond); Søren B. Andersen (Research and Knowledge Centre, The Danish Veteran Centre, Ringsted, Denmark); Ole A. Andreassen (Institute of Clinical Medicine, University of Oslo, Oslo, Norway); Paul A. Arbisi (Mental Health Service Line, Minneapolis Veterans Affairs [VA] Health Care System, Minneapolis, Minnesota); Allison E. Ashley-Koch (Duke Molecular Physiology Institute, Duke University, Durham, North Carolina); Elizabeth G. Atkinson (Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts; Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts); S. Bryn Austin (Department of Pediatrics, Harvard Medical School, Boston, Massachusetts; Department of Social and Behavioral Sciences, Harvard School of Public Health; Division of Adolescent and Young Adult Medicine, Boston Children’s Hospital, Boston, Massachusetts; Channing Division of Network Medicine, Brigham & Women’s Hospital, Boston, Massachusetts); Esmina Avdibegovic (Department of Psychiatry, University Clinical Center of Tuzla, Tuzla, Bosnia and Herzegovina); Dragan Babić (Department of Psychiatry, University Clinical Center of Mostar, Mostar, Bosnia and Herzegovina); Marie Bækvad-Hansen (Department for Congenital Disorders, Statens Serum Institut, Copenhagen, Denmark; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH); Dewleen G. Baker (Department of Psychiatry, University of California, San Diego; Center of Excellence for Stress and Mental Health and Psychiatry Service, VA San Diego Healthcare System); Jean C. Beckham (Department of Psychiatry and Behavioral Sciences, Duke University; Research, Durham VA Medical Center; Genetics Research Laboratory, VA Mid-Atlantic Mental Illness Research, Education, and Clinical Center [MIRECC]); Laura J. Bierut (Department of Psychiatry, Washington University in Saint Louis School of Medicine, St Louis, Missouri); Jonathan I. Bisson (Medical Research Council Centre for Psychiatric Genetics and Genomics, National Centre for Mental Health, Cardiff University, Cardiff, United Kingdom); Marco P. Boks (Department of Translational Neuroscience, Utrecht Brain Center Rudolf Magnus, University Medical Center, Utrecht, the Netherlands); Elizabeth A. Bolger (McLean Hospital, Belmont, Massachusetts; Department of Psychiatry, Harvard Medical School); Anders D. Børglum (Centre for Integrative Sequencing, iSEQ, and Department of Biomedicine—Human Genetics, Aarhus University, Aarhus, Denmark; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH); Bekh Bradley (Mental Health Service Line, Atlanta VA Health Care System, Decatur, Georgia; Department of Psychiatry and Behavioral Sciences, Emory University); Megan Brashear (School of Public Health and Department of Epidemiology, Louisiana State University Health Sciences Center, New Orleans); Gerome Breen (National Institute for Health Research Biomedical Research Centre (NIHR BRC) at the Maudsley and Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, United Kingdom); Richard A. Bryant (Department of Psychology, University of New South Wales, Sydney, Australia); Angela C. Bustamante (Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor); Jonas Bybjerg-Grauholm (Department for Congenital Disorders, Statens Serum Institut; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH); Joseph R. Calabrese (Department of Psychiatry, University Hospitals, Cleveland, Ohio); José M. Caldas-de-Almeida (Lisbon Institute of Global Mental Health, Chronic Diseases Research Centre [CEDOC], Lisbon, Portugal); Chia-Yen Chen (Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard; Analytic and Translational Genetics Unit and Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital); Jonathan R. I. Coleman (Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, NIHR BRC at the Maudsley, King’s College London); Anders M. Dale (Department of Radiology and Department of Neurosciences, University of California, San Diego); Shareefa Dalvie (South African Medical Research Council Unit on Risk and Resilience in Mental Disorders, Department of Psychiatry, University of Cape Town, Cape Town, Western Cape, South Africa); Mark J. Daly (Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital); Nikolaos P. Daskalakis (Department of Psychiatry, Harvard Medical School; Cohen Veterans Bioscience; Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York; McLean Hospital); Jürgen Deckert (Center of Mental Health, Psychiatry, Psychosomatics and Psychotherapy, University Hospital of Würzburg, Würzburg, Germany); Douglas L. Delahanty (Department of Psychological Sciences, Research and Sponsored Programs, Kent State University, Kent, Ohio); Michelle F. Dennis (Department of Psychiatry and Behavioral Sciences, Duke University; Research, Durham VA Medical Center; Genetics Research Laboratory, VA MIRECC); Seth G. Disner (Research Service Line, Minneapolis VA Health Care System); Katharina Domschke (Department of Psychiatry and Psychotherapy, Faculty of Medicine, Medical Center University of Freiburg, Freiburg, Germany; Center for Basics in Neuromodulation, Faculty of Medicine, University of Freiburg, Freiburg, Germany); Laramie E. Duncan (Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, California); Alma Dzubur-Kulenovic (University Clinical Center of Sarajevo, Department of Psychiatry, Sarajevo, Bosnia and Herzegovina); Christopher R. Erbes (Center for Care Delivery and Outcomes Research [CCDOR], Minneapolis VA Health Care System; Department of Psychiatry, University of Minnesota, Minneapolis); Alexandra Evans (Medical Research Council Centre for Psychiatric Genetics and Genomics, National Centre for Mental Health, Cardiff University); Lindsay A. Farrer (Department of Medicine, Boston University School of Medicine); Norah C. Feeny (Department of Psychological Sciences, Case Western Reserve University, Cleveland, Ohio); Janine D. Flory (Department of Psychiatry, Icahn School of Medicine at Mount Sinai); David Forbes (Department of Psychiatry, University of Melbourne, Melbourne, Australia); Carol E. Franz (Department of Psychiatry, University of California, San Diego); Sandro Galea (Department of Psychological and Brain Sciences, Boston University); Melanie E. Garrett (Department of Psychiatry and Behavioral Sciences, Duke University); Bizu Gelaye (Department of Epidemiology, Harvard T.H. Chan School of Public Health, Harvard University); Joel Gelernter (Department of Psychiatry, US Department of Veterans Affairs, West Haven, Connecticut; VA Connecticut Healthcare Center, West Haven; Department of Genetics and Neuroscience, Yale University School of Medicine, New Haven, Connecticut); Elbert Geuze (Brain Research and Innovation Centre, Netherlands Ministry of Defence, Utrecht, the Netherlands; Department of Psychiatry, Utrecht Brain Center Rudolf Magnus, University Medical Center); Charles Gillespie (Department of Psychiatry and Behavioral Sciences, Emory University); Aferdita Goci Uka (Department of Psychiatry, University Clinical Centre of Kosovo, Prishtina, Republic of Kosovo); Scott D. Gordon (Department of Genetics and Computational Biology, Queensland Institute of Medical Research [QIMR] Berghofer Medical Research Institute, Brisbane, Australia); Guia Guffanti (Department of Psychiatry, Harvard Medical School; McLean Hospital); Magali Haas (Cohen Veterans Bioscience); Rasha Hammamieh (US Army Center for Environmental Health Research, US Army Medical Research and Materiel Command, Fort Detrick, Maryland); Michael A. Hauser (Department of Psychiatry and Behavioral Sciences, Duke University); Andrew C. Heath (Department of Genetics, Washington University in Saint Louis School of Medicine); Sian M.J. Hemmings (Department of Psychiatry, Stellenbosch University Faculty of Medicine and Health Sciences, Cape Town, Western Cape, South Africa); David Michael Hougaard (Department for Congenital Disorders, Statens Serum Institut; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH); Miro Jakovljevic (Department of Psychiatry, University Hospital Center of Zagreb, Zagreb, Croatia); Marti Jett (US Army Center for Environmental Health Research, US Army Medical Research and Materiel Command); Eric Otto Johnson (Behavioral Health and Criminal Justice Division, Research Triangle Institute International, Research Triangle Park, North Carolina); Ian Jones (Medical Research Council Centre for Psychiatric Genetics and Genomics, National Centre for Mental Health, Cardiff University); Tanja Jovanovic (Department of Psychiatry and Behavioral Sciences, Emory University); Angela G. Junglen (Department of Psychological Sciences, Kent State University); Karen-Inge Karstoft (Research and Knowledge Centre, the Danish Veteran Center; Department of Psychology, University of Copenhagen, Copenhagen, Denmark); Milissa L. Kaufman (Department of Psychiatry, Harvard Medical School); Ronald C. Kessler (Department of Health Care Policy, Harvard Medical School); Alaptagin Khan (McLean Hospital; Department of Health Care Policy, Harvard Medical School); Nathan A. Kimbrel (Duke Molecular Physiology Institute, Duke University; Research, Durham VA Medical Center; Genetics Research Laboratory, VA Mid-Atlantic MIRECC); Anthony P. King (Department of Psychiatry, University of Michigan Medical School); Nastassja Koen (South Africa Medical Research Council Unit on Risk and Resilience in Mental Disorders, Department of Psychiatry, University of Cape Town); Karestan C. Koenen (Department of Epidemiology, Harvard School of Public Health; Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard); Henry R. Kranzler (Department of Psychiatry, University of Pennsylvania Perelman School of Medicine, Philadelphia; Crescenz VA Medical Center, MIRECC); William S. Kremen (Department of Psychiatry, University of California, San Diego; Center of Excellence for Stress and Mental Health, VA San Diego Healthcare System); Bruce R. Lawford (Institute of Health and Behavioral Innovation, School of Biomedical Sciences, Queensland University of Technology, Brisbane, Australia); Lauren A. M. Lebois (Department of Psychiatry, Harvard Medical School; McLean Hospital); Catrin E. Lewis (National Centre for Mental Health, Medical Research Council Centre for Psychiatric Genetics and Genomics, Cardiff University); Israel Liberzon (Department of Psychiatry, University of Michigan Medical School); Sarah D. Linnstaedt (Department of Anesthesiology, University of North Carolina Institute for Trauma Recovery, Chapel Hill); Mark W. Logue (National Center for PTSD, VA Boston Healthcare System, Boston, Massachusetts); Adriana Lori (Department of Gynecology and Obstetrics, Emory University); Bozo Lugonja (National Centre for Mental Health, Medical Research Council Centre for Psychiatric Genetics and Genomics, Cardiff University); Jurjen J. Luykx (Department of Translational Neuroscience and Department of Psychiatry, University Medical Center, Utrecht Brain Center Rudolf Magnus); Michael J. Lyons (Dean’s Office, Boston University, Boston, Massachusetts); Adam X. Maihofer (Department of Psychiatry, University of California, San Diego; Center of Excellence for Stress and Mental Health, Research Service, VA San Diego Healthcare System); Jessica Maples-Keller (Department of Psychiatry and Behavioral Sciences, Emory University); Charles Marmar (Department of Psychiatry, New York University School of Medicine, New York); Nicholas G. Martin (Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute); Douglas Maurer (Command, US Army, Fort Sill, Oklahoma); Matig R. Mavissakalian (Department of Psychiatry, University Hospitals); Alexander McFarlane (Centre for Traumatic Stress Studies, University of Adelaide, Adelaide, Australia); Regina E. McGlinchey (GRECC/TRACTS, VA Boston Healthcare System, Boston, Massachusetts); Katie A. McLaughlin (Department of Psychology, Harvard University); Samuel A. McLean (Department of Anesthesiology and Department of Emergency Medicine, University of North Carolina Institute for Trauma Recovery); Sarah McLeay (PTSD Initiative, Gallipoli Medical Research Institute, Greenslopes, Australia); Divya Mehta (Institute of Health and Biomedical Innovation, School of Psychology and Counseling, Faculty of Health, Queensland University of Technology); William P. Milberg (GRECC/TRACTS, VA Boston Healthcare System); Mark W. Miller (National Center for PTSD, VA Boston Healthcare System); Rajendra A. Morey (Duke Molecular Physiology Institute, Duke University); Charles Phillip Morris (Institute of Health and Biomedical Innovation, School of Biomedical Sciences, Queensland University of Technology); Ole Mors (Psychosis Research Unit, Aarhus University Hospital, Risskov, Denmark; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH); Preben B. Mortensen (Centre for Integrated Register-Based Research, Centre for Integrative Sequencing, iSEQ, and National Centre for Register-Based Research, Aarhus University, Aarhus, Denmark; The Lundbeck Foundation Initiative for Integrative Psychiatric Research); Elliot C. Nelson (Department of Psychiatry, Washington University in Saint Louis School of Medicine); Caroline M. Nievergelt (Department of Psychiatry, University of California, San Diego; Center of Excellence for Stress and Mental Health, Research Service, VA San Diego Healthcare System); Merete Nordentoft (The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH; Mental Health Services in the Capital Region of Denmark, Mental Health Center Copenhagen, University of Copenhagen, Copenhagen, Denmark); Sonya B. Norman (Executive Division, National Center for Posttraumatic Stress Disorder, White River Junction, Vermont; Department of Psychiatry, University of California, San Diego; Department of Research and Psychiatry, VA San Diego Healthcare System); Meaghan O’Donnell (Department of Psychiatry, University of Melbourne); Holly K. Orcutt (Department of Psychology, Northern Illinois University, DeKalb); Matthew S. Panizzon (Department of Psychiatry, University of California, San Diego); Edward S. Peters (School of Public Health and Department of Epidemiology, Louisiana State University Health Sciences Center); Alan L. Peterson (Department of Psychology, University of Texas Health Science Center at San Antonio); Matthew Peverill (Department of Psychology, University of Washington, Seattle); Robert H. Pietrzak (US Department of Veterans Affairs National Center for Posttraumatic Stress Disorder, West Haven, Connecticut; Department of Psychiatry, Yale University School of Medicine); Melissa A. Polusny (Department of Mental Health and Psychology, Minneapolis VA Health Care System; Department of Psychiatry, University of Minnesota, Minneapolis); Xue-Jun Qin (Duke Molecular Physiology Institute, Duke University); Andrew Ratanatharathorn (Department of Epidemiology, Harvard T.H. Chan School of Public Health, Harvard University); Kerry J. Ressler (Department of Psychiatry, Harvard Medical School; McLean Hospital; Department of Psychiatry and Behavioral Sciences, Emory University); John P. Rice (Department of Psychiatry, Washington University in Saint Louis School of Medicine); Victoria B. Risbrough (Department of Psychiatry, University of California, San Diego; Center of Excellence for Stress and Mental Health and Research Service, VA San Diego Healthcare System); Andrea L. Roberts (Department of Environmental Health, Harvard T.H. Chan School of Public Health, Harvard University); Alex O. Rothbaum (Department of Psychological Sciences, Case Western Reserve University); Barbara O. Rothbaum (Department of Psychiatry and Behavioral Sciences, Emory University); Peter Roy-Byrne (Department of Psychology, University of Washington); Ken Ruggiero (Department of Nursing and Department of Psychiatry, Medical University of South Carolina, Charleston); Ariane Rung (School of Public Health and Department of Epidemiology, Louisiana State University Health Sciences Center); Bart P. F. Rutten (School for Mental Health and Neuroscience and Department of Psychiatry and Neuropsychology, Maastricht Universitair Medisch Centrum, School for Mental Health and Neuroscience, Maastricht, the Netherlands); Nancy L. Saccone (Department of Psychiatry, Washington University in Saint Louis School of Medicine); Sixto E. Sanchez (Department of Medicine, Universidad Peruana de Ciencias Aplicadas Facultad de Ciencias de la Salud, Lima, Peru); Dick Schijven (Department of Translational Neuroscience and Department of Psychiatry, University Medical Center Utrecht Brain Center Rudolf Magnus); Soraya Seedat (Department of Psychiatry, Stellenbosch University Faculty of Medicine and Health Sciences); Antonia V. Seligowski (McLean Hospital; Department of Psychiatry, Harvard Medical School); Julia S. Seng (School of Nursing, University of Michigan, Ann Arbor); Christina M. Sheerin (Department of Psychiatry, Virginia Institute for Psychiatric and Behavioral Genetics); Derrick Silove (Department of Psychiatry, University of New South Wales); Alicia K. Smith (Department of Psychiatry and Behavioral Sciences and Department of Gynecology and Obstetrics, Emory University); Jordan W. Smoller (Analytic and Translational Genetics Unit and Department of Psychiatry, Massachusetts General Hospital; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard); Scott R. Sponheim (Mental Health Service Line, Minneapolis VA Health Care System; Department of Psychiatry, University of Minnesota); Dan J. Stein (South African Medical Research Council Unit on Risk and Resilience in Mental Disorders, Department of Psychiatry, University of Cape Town); Murray B. Stein (Department of Psychiatry, University of California, San Diego; Million Veteran Program and Psychiatry Service, VA San Diego Healthcare System); Jennifer S. Stevens (Department of Psychiatry and Behavioral Sciences, Emory University); Martin H. Teicher (Department of Psychiatry, Harvard Medical School; McLean Hospital); Wesley K. Thompson (Institute of Biological Psychiatry, Mental Health Centre, Sankt Hans, Roskilde, Denmark; KG Jebsen Centre for Psychosis Research, Norway Division of Mental Health and Addiction, Oslo, Norway; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH; Department of Psychiatry, University of California San Diego); Katy Torres (Department of Psychiatry, University of California, San Diego; Center of Excellence for Stress and Mental Health, Research Service, Veteran Affairs San Diego Healthcare System); Edward Trapido (School of Public Health and Department of Epidemiology, Louisiana State University Health Sciences Center); Monica Uddin (Genomics Program, University of South Florida College of Public Health, Tampa); Robert J. Ursano (Department of Psychiatry, Uniformed Services University, Bethesda, Maryland); Leigh Luella van den Heuvel (Department of Psychiatry, Stellenbosch University Faculty of Medicine and Health Sciences); Miranda van Hooff (Department of Psychiatry, University of Adelaide); Eric Vermetten (Psychotrauma Research Expert Group, Arq, Diemen, the Netherlands; Department of Psychiatry, Leiden University Medical Center, Leiden, the Netherlands; Research Center, Netherlands Defense Department, Utrecht; Department of Psychiatry, New York University School of Medicine); Christiaan H. Vinkers (Department of Anatomy and Neurosciences and Department of Psychiatry, Amsterdam University Medical Center [location VUmc], Amsterdam, the Netherlands); Joanne Voisey (Institute of Health and Behavioral Innovation, School of Biomedical Sciences, Queensland University of Technology); Yunpeng Wang (Institute of Biological Psychiatry, Mental Health Centre Sankt Hans; KG Jebsen Centre for Psychosis Research, Norway Division of Mental Health and Addiction, Oslo; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH); Zhewu Wang (Department of Psychiatry and Behavioral Sciences, Medical University of South Carolina; Department of Mental Health, Ralph H Johnson VA Medical Center, Charleston, South Carolina); Thomas Werge (Institute of Biological Psychiatry, Mental Health Centre Sankt Hans; The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH; Department of Medicine, University of Copenhagen); Michelle A. Williams (Department of Epidemiology, Harvard T.H. Chan School of Public Health, Harvard University); Douglas E. Williamson (Department of Psychiatry and Behavioral Sciences, Duke University; Research, Durham VA Medical Center); Sherry Winternitz (Department of Psychiatry, Harvard Medical School; McLean Hospital); Christiane Wolf (Psychiatry, Psychosomatics and Psychotherapy, University Hospital of Würzburg, Center of Mental Health); Erika J. Wolf (National Center for PTSD, VA Boston Healthcare System); Jonathan D. Wolff (McLean Hospital); Rachel Yehuda (Department of Psychiatry, Icahn School of Medicine at Mount Sinai; Department of Mental Health, James J Peters VA Medical Center, Bronx, New York); Keith A. Young (Department of Psychiatry, Baylor Scott and White Central Texas, Temple; COE for Research on Returning War Veterans, CTVHCS, Waco, Texas); Ross McD Young (Institute of Health and Biomedical Innovation and School of Psychology and Counseling, Faculty of Health, Queensland University of Technology); Hongyu Zhao (Department of Biostatistics, Yale University, New Haven, Connecticut); Lori A. Zoellner (Department of Psychiatry and Behavioral Sciences, University of Washington).
Contributor Information
Psychiatric Genomics Consortium Posttraumatic Stress Disorder:
Allison E Aiello, Lynn M Almli, Ananda B Amstadter, Søren B Andersen, Ole A Andreassen, Paul A Arbisi, Allison E Ashley-Koch, Elizabeth G Atkinson, S Bryn Austin, Esmina Avdibegovic, Dragan Babić, Marie Bækvad-Hansen, Dewleen G Baker, Jean C Beckham, Laura J Bierut, Jonathan I Bisson, Marco P Boks, Elizabeth A Bolger, Anders D Børglum, Bekh Bradley, Megan Brashear, Gerome Breen, Richard A Bryant, Angela C Bustamante, Jonas Bybjerg-Grauholm, Joseph R Calabrese, José M Caldas-de-Almeida, Chia-Yen Chen, Jonathan R I Coleman, Anders M Dale, Shareefa Dalvie, Mark J Daly, Nikolaos P Daskalakis, Jürgen Deckert, Douglas L Delahanty, Michelle F Dennis, Seth G Disner, Katharina Domschke, Laramie E Duncan, Alma Dzubur-Kulenovic, Christopher R Erbes, Alexandra Evans, Lindsay A Farrer, Norah C Feeny, Janine D Flory, David Forbes, Carol E Franz, Sandro Galea, Melanie E Garrett, Bizu Gelaye, Joel Gelernter, Elbert Geuze, Charles Gillespie, Aferdita Goci Uka, Scott D Gordon, Guia Guffanti, Magali Haas, Rasha Hammamieh, Michael A Hauser, Andrew C Heath, Sian M J Hemmings, David Michael Hougaard, Miro Jakovljevic, Marti Jett, Eric Otto Johnson, Ian Jones, Tanja Jovanovic, Angela G Junglen, Karen-Inge Karstoft, Milissa L Kaufman, Ronald C Kessler, Alaptagin Khan, Nathan A Kimbrel, Anthony P King, Nastassja Koen, Karestan C Koenen, Henry R Kranzler, William S Kremen, Bruce R Lawford, Lauren A M Lebois, Catrin E Lewis, Israel Liberzon, Sarah D Linnstaedt, Mark W Logue, Adriana Lori, Bozo Lugonja, Jurjen J Luykx, Michael J Lyons, Adam X Maihofer, Jessica Maples-Keller, Charles Marmar, Nicholas G Martin, Douglas Maurer, Matig R Mavissakalian, Alexander McFarlane, Regina E McGlinchey, Katie A McLaughlin, Samuel A McLean, Sarah McLeay, Divya Mehta, William P Milberg, Mark W Miller, Rajendra A Morey, Charles Phillip Morris, Ole Mors, Preben B Mortensen, Elliot C Nelson, Caroline M Nievergelt, Merete Nordentoft, Sonya B Norman, Meaghan O’Donnell, Holly K Orcutt, Matthew S Panizzon, Edward S Peters, Alan L Peterson, Matthew Peverill, Robert H Pietrzak, Melissa A Polusny, Xue-Jun Qin, Andrew Ratanatharathorn, Kerry J Ressler, John P Rice, Victoria B Risbrough, Andrea L Roberts, Alex O Rothbaum, Barbara O Rothbaum, Peter Roy-Byrne, Ken Ruggiero, Ariane Rung, Bart P F Rutten, Nancy L Saccone, Sixto E Sanchez, Dick Schijven, Soraya Seedat, Antonia V Seligowski, Julia S Seng, Christina M Sheerin, Derrick Silove, Alicia K Smith, Jordan W Smoller, Scott R Sponheim, Dan J Stein, Murray B Stein, Jennifer S Stevens, Martin H Teicher, Wesley K Thompson, Katy Torres, Edward Trapido, Monica Uddin, Robert J Ursano, Leigh Luella van den Heuvel, Miranda van Hooff, Eric Vermetten, Christiaan H Vinkers, Joanne Voisey, Yunpeng Wang, Zhewu Wang, Thomas Werge, Michelle A Williams, Douglas E Williamson, Sherry Winternitz, Christiane Wolf, Erika J Wolf, Jonathan D Wolff, Rachel Yehuda, Keith A Young, Ross McD Young, Hongyu Zhao, and Lori A Zoellner
Funding
This work was supported in part by the National Institutes of Health [grants T32MH020030 to M.J.L.; R01MH106595 to L.E.D., A.X.M., C.M.N.; R01HD059835 to B.G.; R01MH105379 to N.R.N.; R01MH108641 to N.R.N., and K02AA023239 to A.B.A.]. This work was funded by NIMH/U.S. Army Medical Research and Material Command [R01MH106595] and NIH 5U01MH109539. This work would have not been possible without the financial support provided by Stanley Center for Psychiatric Genetics at the Broad Institute, One Mind, and Cohen Veterans Bioscience.
Conflict of interest statement. M.B.S. has in the past 3 years received payments for editorial work from UpToDate, Biological Psychiatry, and Depression and Anxiety. He has also in the past 3 years been paid as a consultant for Aptinyx Pharmaceuticals, Bionomics, Janssen, Neurocrine, and Pfizer. He also owns founders shares and stock options in Resilience Pharmaceuticals and stock options in Oxeia Biopharmaceuticals. The remaining authors have nothing to disclose.
References
- 1. Benjet C, et al. The epidemiology of traumatic event exposure worldwide: results from the World Mental Health Survey Consortium. Psychol Med. 2016;46(2):327–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu H, et al. Association of DSM-IV posttraumatic stress disorder with traumatic experience type and history in the World Health Organization World Mental Health Surveys. JAMA Psychiatry. 2017;74(3):270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5th ed. Arlington, VA: American Psychiatric Association; 2013. [Google Scholar]
- 4. Ohayon MM, et al. Sleep disturbances and psychiatric disorders associated with posttraumatic stress disorder in the general population. Compr Psychiatry. 2000;41(6):469–478. [DOI] [PubMed] [Google Scholar]
- 5. Morin CM, et al. Epidemiology of insomnia. Sleep Med Clin. 2013;8(3):281–297. [Google Scholar]
- 6. Krystal AD. Psychiatric disorders and sleep. Neurol Clin. 2012;30(4):1389–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gehrman P, et al. Predeployment sleep duration and insomnia symptoms as risk factors for new-onset mental health disorders following military deployment. Sleep. 2013;36(7):1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pigeon WR, et al. Longitudinal relationships of insomnia, nightmares, and PTSD severity in recent combat veterans. J Psychosom Res. 2013;75(6):546–550. [DOI] [PubMed] [Google Scholar]
- 9. Mendlewicz J. Sleep disturbances: core symptoms of major depressive disorder rather than associated or comorbid disorders. World J Biol Psychiatry. 2009;10(4):269–275. [DOI] [PubMed] [Google Scholar]
- 10. O’Brien EM, et al. Severe insomnia is associated with more severe presentation and greater functional deficits in depression. J Psychiatr Res. 2011;45(8):1101–1105. [DOI] [PubMed] [Google Scholar]
- 11. Park SC, et al. Prevalence and clinical correlates of insomnia in depressive disorders: the CRESCEND study. Psychiatry Investig. 2013;10(4):373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kallestad H, et al. Impact of sleep disturbance on patients in treatment for mental disorders. BMC Psychiatry. 2012;12:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lind MJ, et al. Genetic pathways to insomnia. Brain Sci. 2016;6(4):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheerin CM, et al. The genetics and epigenetics of PTSD: overview, recent advances, and future directions. Curr Opin Psychol. 2017;14:5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lind MJ, et al. A longitudinal twin study of insomnia symptoms in adults. Sleep. 2015;38(9):1423–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan LE, et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 2018;23(3):666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nievergelt CM, et al. International meta-analysis of PTSD genome-wide association studies identifies sex-specific and ancestry-specific genetic risk loci. Nat Commun. 2019;10:4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Logue MW, et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2013;18(8):937–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lane JM, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones SE, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12(8):e1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lane JM, et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolf EJ, et al. Posttraumatic stress disorder and the genetic structure of comorbidity. J Abnorm Psychol. 2010;119(2):320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lind MJ, et al. An examination of the etiologic overlap between the genetic and environmental influences on insomnia and common psychopathology. Depress Anxiety. 2017;34(5):453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gregory AM, et al. A longitudinal twin and sibling study of associations between insomnia and depression symptoms in young adults. Sleep. 2016;39(11):1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sartor CE, et al. Common heritable contributions to low-risk trauma, high-risk trauma, posttraumatic stress disorder, and major depression. Arch Gen Psychiatry. 2012;69(3): 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koenen KC, et al. Common genetic liability to major depression and posttraumatic stress disorder in men. J Affect Disord. 2008;105(1–3):109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hammerschlag AR, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49(11):1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lane JM, et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. 2019;51(3): 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jansen PR, et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 2019;51(3):394–403. [DOI] [PubMed] [Google Scholar]
- 31. Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anttila V, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360(6395):eaap8757. doi:10.1126/science.aap8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi:10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davies NM, et al. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fry A, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank Participants with those of the general population. Am J Epidemiol. 2017;186(9): 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qaseem A, et al. Appropriate use of therapeutic interventions to foster high-value care. Ann Intern Med. 2016;165(11):831–832. [DOI] [PubMed] [Google Scholar]
- 38. Hemani G, et al. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195–R208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stein MB, et al. Genome-wide analysis of insomnia disorder. Mol Psychiatry. 2018;23(11):2238–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flory JD, et al. Comorbidity between post-traumatic stress disorder and major depressive disorder: alternative explanations and treatment considerations. Dialogues Clin Neurosci. 2015;17(2):141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swinkels CM, et al. The association of sleep duration, mental health, and health risk behaviors among U.S. Afghanistan/Iraq Era Veterans. Sleep. 2013;36(7):1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dashti HS, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10(1):1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watson NF, et al. Sleep duration and depressive symptoms: a gene–environment interaction. Sleep. 2014;37(2): 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCarthy MJ, et al. A survey of genomic studies supports association of circadian clock genes with bipolar disorder spectrum illnesses and lithium response. PLoS One. 2012;7(2):e32091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schuch JB, et al. The role of CLOCK gene in psychiatric disorders: evidence from human and animal research. Am J Med Genet B Neuropsychiatr Genet. 2018;177(2):181–198. [DOI] [PubMed] [Google Scholar]
- 46. Lind MJ, et al. Association of posttraumatic stress disorder with rs2267735 in the ADCYAP1R1 gene: a meta-analysis. J Trauma Stress. 2017;30(4):389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kawaguchi C, et al. Changes in light-induced phase shift of circadian rhythm in mice lacking PACAP. Biochem Biophys Res Commun. 2003;310(1):169–175. [DOI] [PubMed] [Google Scholar]
- 48. Hannibal J, et al. PACAP and glutamate are co-stored in the retinohypothalamic tract. J Comp Neurol. 2000;418(2): 147–155. [PubMed] [Google Scholar]
- 49. Duncan LE, et al. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacology. 2019;44(9):1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Border R, et al. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am J Psychiatry. 2019;176(5):376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jones SE, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10(1):343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Dalfsen JH, et al. The influence of sleep on human hypothalamic-pituitary-adrenal (HPA) axis reactivity: a systematic review. Sleep Med Rev. 2018;39:187–194. [DOI] [PubMed] [Google Scholar]
- 54. Colvonen PJ, et al. Recent advancements in treating sleep disorders in co-occurring PTSD. Curr Psychiatry Rep. 2018;20(7):48. [DOI] [PMC free article] [PubMed] [Google Scholar]