Abstract
Multidrug resistant (MDR) Salmonella are a leading cause of foodborne diseases and serious human health concerns worldwide. In this study we detected MDR Salmonella in broiler chicken along with the resistance genes and class 1 integron gene intl1. A total of 100 samples were collected from broiler farms comprising 50 cloacal swabs, 35 litter and 15 feed samples. Overall prevalence of Salmonella was 35% with the highest detected in cloacal swabs. Among the Salmonella, 30 isolates were confirmed as S. enterica serovar Typhimurium using molecular methods of PCR. Disk diffusion susceptibility test revealed that all the Salmonella were classified as MDR with the highest resistance to tetracycline (97.14%), chloramphenicol (94.28%), ampicillin (82.85%) and streptomycin (77.14%). The most prevalent resistance genotypes were tetA (97.14%), floR (94.28%), blaTEM-1 (82.85%) and aadA1 (77.14%). In addition, among the MDR Salmonella, 20% were positive for class 1 integron gene (intl1). As far as we know, this is the first study describing the molecular basis of antibiotic resistance in MDR Salmonella from broiler farms in Bangladesh. In addition to tetA, floR, blaTEM-1, aadA1 and intl1 were also detected in the isolated MDR Salmonella. The detection of MDR Salmonella in broiler chicken carrying intl1 is of serious public health concern because of their zoonotic nature and possibilities to enter into the food chain.
Keywords: multidrug resistant, Salmonella, poultry, litter, feed, integron, one health, Bangladesh
1. Introduction
Poultry farming is a profitable agricultural business globally, in Bangladesh; livestock and poultry contribute approximately 1.47% of the total GDP of the country [1]. In the last decade, tremendous development has been achieved in this sector in Bangladesh [2]. Poultry products including eggs and meat are excellent sources for humans’ daily protein requirement. The advancement of poultry production is being seriously hampered by management factors, such as biosecurity measures and infectious diseases [3]. Among the infectious diseases, salmonellosis caused by MDR Salmonella is a major constrain for poultry industries in Bangladesh [4] and is a worldwide epidemic causing millions of illness and deaths annually [5].
The chicken-adapted Salmonella serovar Gallinarum biovar Pullorum and Gallinarum are responsible for avian salmonellosis, e.g., pullorum disease and fowl typhoid, respectively [6]. In poultry, Salmonellosis is often associated with high mortality up to 90%, thus causing severe economic losses [7]. S. enterica serovar Typhimurium normally colonizes the cecum and is commonly found asymptomatic in cloaca in older chicks [7]. Day old chicks infected with Salmonella may show intestinal pathology and inflammatory responses [8]. Although birds have no obvious disease symptoms, S. enterica serovar Typhimurium can cause systemic metabolic changes in birds affecting production [9]. Human salmonellosis outbreaks are mostly associated with the consumption of poultry products contaminated with S. enterica serovar Typhimurium and S. enteritidis [10]. Therefore, the occurrence of Salmonella, particularly S. enterica serovar Typhimurium, in broiler farms is a public health concern as they can transmit to the food chain.
Modern food animal production, specifically broiler chicken production, is directly dependent on the use of antibiotics as growth promoters [11]. Antibiotics are also used in broiler chicken for treatment purposes. The escalated use of antibiotics causes the development of resistance through selection pressure [12]. Antimicrobial resistance (AMR) is an increasingly serious public health challenge [13]. It is speculated that if measures are not taken properly, by 2050 AMR will cause 300 million human deaths, financial losses will be equivalent to 100 trillion USD and an 11% fall in animal production [14]. By far, most of the affected countries will be the low and middle-income countries (LMICs) in Africa and Asia [15]. Bangladesh is located in the World Health Organization’s Southeast Asia region which is assessed as having a high risk of AMR [16].
Bacteria can acquire resistance genes through mobile genetic elements such as plasmids, transposons, integron, and insertion sequences (IS) elements [17]. Due to their mobile nature, these genetic elements can easily spread horizontally across many species of bacterial populations [18]. Humans can get exposed to antibiotic resistant bacteria through consumption of contaminated meat and eggs, or through direct transmission from colonized animals or manure and litter [19]. Because of their zoonotic nature, there are possibilities for the transmission of MDR Salmonella from poultry to humans throughout the food chain [20,21,22]. In recent years, the development of MDR among foodborne pathogens, such as Salmonella spp., have been associated with an increase in human mortality, and longtime hospitalization due to therapy failure [22].
The isolation of Salmonella from poultry was previously reported in Bangladesh, however, none of these studies focused on the molecular detection of resistance genes [23,24,25,26]. However, resistance genes were detected in Escherichia coli isolated from cloacal swabs of boiler chicks in Bangladesh [27,28]. Moreover, a study in Bangladesh, detected resistance genes in E. coli and Salmonella from dairy farms [29]. Another study detected integrons in Salmonella from chicken feces [30]. Therefore, we investigated the prevalence of resistance genes including integrons responsible for multidrug resistance in S. enterica serovar Typhimurium in broiler farms using different sample types (cloacal swabs, litter and feed).
2. Results
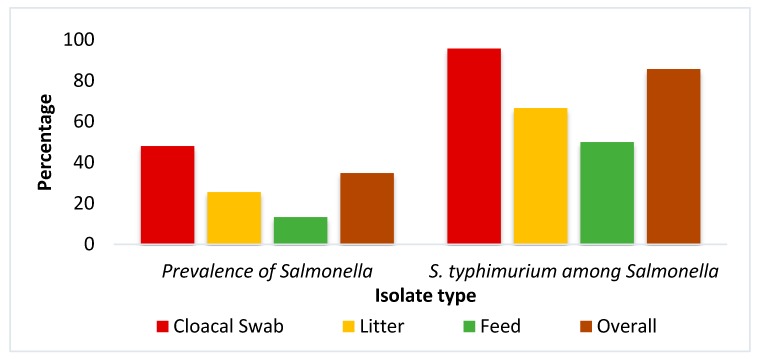
Among the 100 samples analyzed, 35 (35%) were found to be positive for the presence of Salmonella as evidenced from the isolation on selective media, followed by Gram staining, biochemical tests, and invA gene-targeted PCR (Figure 1). The prevalence of Salmonella was significantly (Chi-square test, p = 0.017) higher in cloacal samples (48%, n = 24) compared to all types of samples. The confirmed 35 isolates were inoculated into motility indole urea medium and 30 samples were found to be motile, suggesting that these motile Salmonella would be S. enterica serovar Typhimurium. Motile and non-motile Salmonella were also differentiated by PCR amplification of fliC gene specific for motile S. enterica serovar Typhimurium. All 30 motile isolates were also found to be positive for the fliC gene, confirming them as S. enterica serovar Typhimurium. The detection of S. enterica serovar Typhimurium was found to be the highest significantly in cloacal sample type (95.8%, n = 23) as compared to litter and feed samples (Chi-square test, p = 0.034).
Figure 1.
Prevalence of Salmonella spp. and S. enterica serovar Typhimurium in broiler farms.
From the antibiogram study, it was found that the isolated Salmonella were highly resistant to tetracycline (97.1%) followed by chloramphenicol (94.3%), ampicillin (82.9%), and streptomycin (77.1%), while the highest susceptibility rate was observed against cefixime (100%), followed by ertapenem (94.3%) and ciprofloxacin (85.7%) (Table 1). Salmonella from cloaca always showed a peak of resistance against all antibiotics and a statistically significant relationship was found for tetracycline, chloramphenicol and ampicillin. All 35 identified Salmonella were MDR with the highest 80% (n = 28) against three antibiotics classes, and only two isolates showed MDR phenotype against five antibiotic classes (Table 2).
Table 1.
Antibiotic resistance patterns of Salmonella in broiler farm in this study.
| Sample (n) | Antibiotic Resistance Pattern (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TE | p Value | C | p Value | AMP | p Value | S | p Value | CIP | p Value | ETP | p Value | CFM | p Value | |
| Cloacal Swab (24) | 24 (100) | <0.001 | 24 (100) |
0.010 | 23 (95.8) |
0.011 | 21 (87.5) |
0.012 | 4 (16.7) |
0.772 | 2 (8.3) |
0.615 | 0 (0) |
nc |
| Litter (9) | 9 (100) | 8 (88.9) |
5 (55.6) |
6 (66.7) |
1 (11.1) |
0 (0) |
0 (0) |
|||||||
| Feed (2) | 1 (50) | 1 (50) |
1 (50) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
|||||||
| Total (35) | 34 (97.1) | 33 (94.3) |
29 (82.9) |
27 (77.1) |
5 (14.3) |
2 (5.7) |
0 (0) |
|||||||
Note: TE, tetracycline; C, chloramphenicol; AMP, ampicillin; S, streptomycin; CIP, ciprofloxacin; ETP, ertapenem; CFM, cefixime; nc, not computed; n, no. of positive Salmonella.
Table 2.
Multidrug resistance profile of Salmonella used in this study.
| Isolates | No. of Antibiotic (Class) | Multidrug Profile | No. of Isolates (%) | Prevalence of MDR % |
|---|---|---|---|---|
|
Salmonella (n = 35) |
1 (1) | Any one of the tested antibiotics | 0 | 100 |
| 2 (2) | TE-AMP or other combination of two antibiotic classes | 0 | ||
| 3 (3) | TE-AMP-C | 28 (80) | ||
| TE-S-C | 26 (74.28) | |||
| 4 (4) | C-AMP-S-TE | 24 (68.57) | ||
| CIP-AMP-TE-S | 5 (14.28) | |||
| 5 (5) | C-AMP-S-TE-CIP | 1 (2.87) | ||
| C-AMP-S-TE-ETP | 1 (2.87) |
Note: TE, tetracycline; AMP, ampicillin; C, chloramphenicol; S, streptomycin; CIP, ciprofloxacin; ETP, ertapenem; MDR, multidrug resistance.
It was found that 34 (97.1%) out of 35 isolates were positive for tetracycline resistance gene tetA. The prevalence of floR, blaTEM-1, and aadA1 in Salmonella were 94.3%, 82.9%, and 77.1%, respectively, from different sources (Table 3). Salmonella isolated from cloacal swabs showed a statistically significant peak prevalence for all the resistance genes similar to their phenotype. Among the 35 isolates, 7 (20%) were found positive for class 1 integron gene intl1. Although cloacal isolates showed the highest prevalence of intl1, no statistically significant relationship was found when compared with isolates originated from litter and feed. These seven isolates carried multiple resistance genes (Table 4).
Table 3.
Prevalence of antibiotic resistance genes in Salmonella in broiler farm in the present study.
| Sample (n) | Antibiotic Resistance Gene (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| terA(TE) | p Value | floR (C) | p Value | blaTEM-1 (AMP) | p Value | aadA1 (S) | p Value | Class 1 Integron intl1 | p Value | |
| Cloacal Swab (24) | 24 (100) | <0.001 | 24 (100) |
0.010 | 23 (95.8) |
0.011 | 21 (87.5) |
0.012 | 6 (25) |
0.517 |
| Litter (9) | 9 (100) | 8 (88.9) |
5 (55.6) |
6 (66.7) |
1 (11.1) |
|||||
| Feed (2) | 1 (50) | 1 (50) |
1 (50) |
0 (0) |
0 (0) |
|||||
| Total (35) | 34 (97.1) | 33 (94.3) |
29 (82.9) |
27 (77.1) |
7 (20) |
|||||
Note: TE, tetracycline; AMP, ampicillin; C, chloramphenicol; S, streptomycin; n, no. of positive Salmonella.
Table 4.
Profiles of class 1 integron gene intl1 carrying Salmonella in the present study.
| Isolate number | Resistance Genes | MDR Profile |
|---|---|---|
| CS3 | terA, floR, blaTEM-1, aadA1 | C-AMP-S-TE |
| CS11 | terA, floR, blaTEM-1, aadA1 | C-AMP-S-TE-ETP |
| CS19 | terA, floR, blaTEM-1 | C-AMP-S-TE |
| CS22 | terA, floR, blaTEM-1, aadA1 | C-AMP-S-TE |
| CS41 | terA, floR, blaTEM-1, aadA1 | C-AMP-S-TE-CIP |
| CS43 | terA, floR, blaTEM-1, aadA1 | C-AMP-S-TE |
| L8 | terA, floR, aadA1 | C-AMP-S-TE |
Note: CS, cloacal swab; L, litter, MDR, multidrug resistance; C, chloramphenicol; TE, tetracycline; AMP, ampicillin; S, streptomycin; CIP, ciprofloxacin; ETP, ertapenem.
3. Discussion
Avian salmonellosis is a major threat for poultry industries since it is capable of causing heavy economic losses through mortality and reduced production. Salmonella are also important foodborne zoonotic pathogens. In this study, among the samples analyzed, 35% (35/100) were found to be positive for the presence of Salmonella as confirmed by PCR. Earlier, Al Mamun et al. [26] reported the prevalence of Salmonellosis in poultry as 23.53%, lower than the present findings, and Mahmud et al. [23] reported a prevalence of 37.9% in Bangladesh, which is quite similar to the current findings. The prevalence of Salmonella was significantly the highest in cloacal swabs (48%). Previously a study in Bangladesh, showed the highest rate of Salmonella occurrence in cloacal swabs (32%) among different samples of poultry [31] and another study found 48% Salmonella, similar to our result [25]. Litter (25.7%) was also found to be contaminated with Salmonella in the present study, whereas Islam et al. [32] reported 66.6% Salmonella in litter from broiler farms. Salmonella could be either motile or non-motile. The motile Salmonella are mostly associated with food products and are the major causes of salmonellosis in humans worldwide [33]. In the current study, 30 out of 35 Salmonella isolates were confirmed as motile Salmonella, i.e., S. enterica serovar Typhimurium by growing on motility indole urea agar and by fliC gene detection through PCR. Barua et al. [24] reported the presence of motile Salmonella in commercial broiler chicken farms as 11%. Another study in Bangladesh revealed 15.91% S. enterica serovar Typhimurium among the identified Salmonella [25]. Detection of S. enterica serovar Typhimurium in poultry implies the possibility of S. enterica serovar Typhimurium transmission via poultry originated food and may lead to foodborne illness in human [22].
These observed variations in the prevalence of Salmonella may be linked with various management factors such as biosecurity, hygiene, and sanitation of the farms. The presence of Salmonella spp. in cloacal swabs of healthy broiler chicken provides the evidence of persistent intestinal colonization of Salmonella spp. of the individual bird [34]. Moreover, their presence in cloacal swabs and poultry litter indicates that poultry droppings may act as vehicles for shedding Salmonella spp. to other birds [34]. In addition, poultry litter aeration can be a vital risk factor for spreading pathogens like Salmonella and can contaminate farm environments, causing birds to be at risk [35]. It is interesting to note that in this study, 13.3% of feed supplied to the broiler chicken were also found to be contaminated with Salmonella spp. Detection of Salmonella in poultry feed in Bangladesh is not uncommon since earlier Malorny et al. [36]; Islam et al. [32] and Al Mamun et al. [26] also reported presence of Salmonella in poultry feed. Our findings suggest that contaminated feed could be a potential source of Salmonella in poultry.
AMR is an escalating global health problem [37]. Treatment of infections caused by MDR bacteria are expensive and may be fatal. AMR can affect sustainable development goals (SDGs), especially those targeting hunger, health and economic growth [38]. MDR Salmonella has emerged as a major public health issue worldwide [39]. One of the classical examples of such public health issue is MDR S. enterica serovar Typhimurium phage type DT104, which was found to be resistant to several antibiotics, namely ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline [40]. The present study revealed that 97.1% to 77.1% of the isolated Salmonella showed resistance against commonly used antibiotics, namely tetracycline, ampicillin, streptomycin, and chloramphenicol. An alarming result was that all the tested isolates in the present study were found to be MDR. Development of antibiotic resistance in these bacteria could be a result of several factors including strong selective pressure resulting from the indiscriminate use of antibiotics [41]. Moreover, unpublished data suggested that, in many cases, these antibiotics are used as a growth promoter and are added to the poultry feed and water, respectively, by feed companies and the farmers. There is evidence of antibiotic residues in commercial feed in Bangladesh, acts as subnormal dose which also accelerates the emergence of antibiotic resistance [42]. Cloacal isolates always showed a higher resistance against different antibiotics than litter and feed isolates, which indicates that feces may play a vital role in the spread of resistant bacteria within the farm environment, including feed stored in the poultry shed.
The present study reported the detection of tetA (97.1%), floR (94.3), blaTEM-1 (82.9%) and aadA1 (77.1%) responsible for specific antibiotic resistance. The presence of several resistance genes in different samples may be due to integron that was detected in this study. Integrons are mobile genetic elements. They play important role in the transfer of clusters of genes including antibiotic resistance. Integrons could also be transferred horizontally among Salmonella serotypes [43,44]. Salmonella carrying class one integron int1 were found positive for multiple resistance genes. In this study, 20% isolates were found positive for the presence of the int1 gene, revealing the molecular basis of observed multidrug resistance [45]. These factors may facilitate the emergence of MDR pathogens. In many cases, resistance patterns are observed against those antimicrobials that are frequently used in veterinary practices [45,46].
MDR Salmonella are important to the etiology of bacterial foodborne associated deaths, particularly in the LMICs [47]. The WHO recognized Salmonella as one of the pathogens having severe impacts on human health [48]. Poultry and poultry products have been frequently reported to be involved in the outbreaks of salmonellosis [49]. Alarmingly, all the Salmonella isolated in the present study were found to be MDR including fluroquinolone-resistant Salmonella. The detection of MDR Salmonella in broiler chicken as evident in this study is of great public health concern. It is possible that these MDR Salmonella may transmit into the food chain in broiler meat and lead to serious illness in humans. Detection of mobile genetic elements, such as integron in these isolates make the situation more aggravated. Integrons are directly associated with resistance to antibiotics [50] and there are possibilities of their transmission to other bacterial species horizontally inside the broiler chicken gut. The present findings highlight the importance of practicing strict biosecurity, hygienic measures, proper litter management and safe storage of feed to reduce the load and spread of MDR Salmonella in broiler chicken and to ensure consumers’ health and safety. In addition, the implementation of one-health measure in Bangladesh is critical to early monitor and detect AMR in pathogens of zoonotic potential and to reduce the imprudent use of antibiotics in poultry farms.
4. Materials and Methods
4.1. Ethics Statement
The experimental procedures and protocols used in this study were approved by the Animal Welfare and Experimentation Ethics Committee of Bangladesh Agricultural University (approval number AWEEC/BAU/2018(20)
4.2. Collection of Samples and Isolation of Salmonella
A total of 100 samples (50 cloacal swabs, 35 litter samples, 15 feed samples) from five broiler (Gallus gallus domesticus) farms located in Mymensingh, Bangladesh were randomly collected in July 2017 and samples were transported immediately to the Department of Microbiology and Hygiene, Bangladesh Agricultural University, while maintaining sterile and cold chain conditions. From each farm, 10 cloacal swabs, 7 litter samples and 3 feed samples were collected. Isolation and identification of Salmonella from the collected samples were based on culture on selective media (XLD) (Hi Media, India), Gram staining and biochemical test as was previously described [29].
4.3. Extraction of Bacterial Genomic DNA
For the PCR, the genomic DNA was extracted using the boiling method [51]. In brief, initially a pure bacterial colony was mixed with 100 µL of distilled water in an Eppendorf tube followed by boiling for 10 min. After boiling the Eppendorf tube, it was immediately kept on ice to have cold shock followed by 10 min of centrifugation at 10,000 rpm. Finally, the supernatant was collected and used as a DNA template for PCR.
4.4. Molecular Detection of Salmonella
Salmonella was detected by PCR targeting the invA gene as previously described [52]. Motile Salmonella were detected by PCR targeting the fliC gene specific for S. enterica serovar Typhimurium [53]. Primers specific for invA and fliC genes are listed in Table 5. PCR was done in the final 25 µL containing nucleus free water (5.5 µL), master mixture (Promega) (12.5 µL), forward and reverse primer (1 µL each) and DNA template (5 µL). The PCR thermal profile consisted of 95 °C and 5 min for initial denaturation, followed by 29 cycles of denaturation at 95 °C for one-minute, variable annealing temperature (Table 5) for one-minute, elongation at 72 °C for one- minute and a final extension at 72 °C for 10 min. After completion, the PCR products were analyzed by running in 1.5% agarose gel electrophoresis.
Table 5.
PCR primers with sequence used in this study.
| Gene | Primer Sequence | Amplicon Size (bp) | Annealing Temperature | Reference |
|---|---|---|---|---|
| invA-F | CGGTGGTTTTAAGCGTACTCT T | 796 | 58 | [52] |
| invA-R | CGAATATGCTCCACAAGGTTA | |||
| Flic-C-F | CCCGCTTACAGGTGGACTAC | 433 | 58 | [53] |
| Flic-C-R | AGCGGGTTT TCGGTGGTTGT | |||
| aadA1-F | TATCAGAGGTAGTTG GCGTCAT | 484 | 55 | [57] |
| aadA1-R | GTTCCATAGCGTTAAGGTTTCATT | |||
| blaTEM-1-F | CATTTCCGTGTCGCCCTTAT | 793 | 56 | [58] |
| blaTEM-1-R | TCCATAGTTGCCTGACTCCC | |||
| floR-F | AACCCGCCCTCTGGATCAAGTCAA | 548 | 62 | [57] |
| floR-R | CAAATCACGGGCCACGCTGTATC | |||
| tetA-F | GGTTCACTCGAACGACGTCA | 577 | 57 | [59] |
| tetA-R | CTGTCCGACAAGTTGCATGA | |||
| intl1 A-F | GGCATCCAAGCAGCAAGC | 2000 | 55 | [60] |
| intl1 A-R: | AAG CAG ACT TGA CCT GAT |
4.5. Antimicrobial Susceptibility Testing
The antibiogram phenotype were determined by the Kirby-Bauer disk diffusion method [54] against seven commonly used antibiotics classes, namely penicillin (ampicillin-25 µg), cephalosporin (cefixime-5 µg), amphenicol (chloramphenicol-30 µg), fluoroquinolones (ciprofloxacin-5 µg), carbapenem (ertapenem-10 µg), aminoglycoside (streptomycin-10 µg) and tetracycline (tetracycline-30 µg). The antibiogram test was performed by disk diffusion method on Mueller-Hinton agar (Hi Media, India) plates with a concentration of bacteria equivalent to 0.5 McFarland standard, incubated for 18–24 h aerobically at 37 °C. The results of the antibiogram test were recorded as sensitive, intermediately sensitive, or resistant and the diameters of the zones of inhibition were compared with the diameters of interpretative tables provided by the Clinical and Laboratory Standards Institute (CLSI) [55]. Salmonella isolates that were found to be resistant to multiple antimicrobials (at least 3 classes of antibiotics) were considered as MDR [56].
4.6. Detection of Antibiotic Resistance and Class 1 Integron Gene
All the MDR phenotypes were subjected to PCR for the detection of aadA1, blaTEM-1, floR, and tetA genes responsible for resistance against streptomycin, beta lactam antibiotic, amphenicol (chloramphenicol), and tetracycline respectively, and class 1 integron gene intl1 [57,58,59,60]. The list of primers used to detect these genes are presented in Table 5.
4.7. Statistical Analysis
All the data was incorporated in Excel sheets (MS-2010) and analyzed by SPSS software (SPSS-24.0). Descriptive analysis was performed to calculate prevalence and the Chi-square test was performed to determine the level of significance. A p-value less than 0.05 (p-value < 0.05) was considered as statistically significant.
Acknowledgments
We are very much grateful to the farmers for giving access to the samples. The authors would also like to thank Department of Microbiology and Hygiene, Faculty of Veterinary Science Bangladesh Agricultural University, Mymensingh for the support during the research.
Author Contributions
Conceptualization, M.R., K.N.H.N. and T.R.; methodology, S.B.A., M.M. and M.R.; software, R.A., M.H. and A.S.; validation, M.R., A.S. and M.E.E.Z.; formal analysis, A.S. and T.R.; investigation, S.B.A., M.M., R.A. and M.H., resources, S.B.A., M.M. and M.R.; data curation, R.A., M.H. and A.S.; writing—original draft preparation, S.B.A., M.R., A.S. and K.N.H.N. writing—review and editing, M.R., M.E.E.Z., T.R., A.N. and A.S.; visualization, S.B.A. and A.S. supervision, M.R., K.N.H.N. and T.R.; project administration, M.R.; funding acquisition, M.R.; critical revisions and writing, M.R., M.E.E.Z., A.N., and T.R. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported in part by research fund from the Bangladesh Agricultural University Research System (BAURES) (Grant number: 2018/581/BAU).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.MOFL . Annual Report 2018–2019. Ministry of Fisheries and Livestock; Dhaka, Bangladesh: 2019. [(accessed on 2 November 2019)]. Available online: http://bbs.gov.bd/site/page/dc2bc6ce-7080-48b3-9a04-73cec782d0df/- [Google Scholar]
- 2.Rahman M. Growth of poultry industry in Bangladesh poverty alleviation and employment opportunity; Proceedings of the 3rd International Poultry Show and Seminar; Dhaka, Bangladesh. 28 February–2 March 2003; pp. 1–7. [Google Scholar]
- 3.Gonder E., Barnes H.J. Focal Ulcerative Dermatitis (“Breast Buttons”) in Marketed Turkeys. Avian Dis. 1987;31:52–58. doi: 10.2307/1590772. [DOI] [PubMed] [Google Scholar]
- 4.Haider M.G., Chowdhury E.H., Khan M.A.H.N.A., Hossain M.T., Rahman M.S., Song H.J., Hossain M.M. Experimental pathogenesis of pullorum disease with local isolate of Salmonella enterica serovar. Enterica subspecies Pullorum in pullets in Bangladesh. Korean J. Poultr. Sci. 2009;35:341–350. doi: 10.5536/KJPS.2009.35.4.341. [DOI] [Google Scholar]
- 5.Kariuki S., Gordon M.A., Feasey N., Parry C.M. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2015;33:C21–C29. doi: 10.1016/j.vaccine.2015.03.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrow P.A., Neto O.F. Pullorum disease and fowl typhoid—New thoughts on old diseases: A review. Avian Pathol. 2011;40:11–13. doi: 10.1080/03079457.2010.542575. [DOI] [PubMed] [Google Scholar]
- 7.Barrow P.A., Huggins M.B., Lovell M.A., Simpson J.M. Observations on the pathogenesis of experimental Salmonella typhimurium infection in chickens. Res. Vet. Sci. 1987;42:194–199. doi: 10.1016/S0034-5288(18)30685-4. [DOI] [PubMed] [Google Scholar]
- 8.Withanage G.S.K., Kaiser P., Wigley P., Powers C., Mastroeni P., Brooks H., Barrow P., Smith A., Maskell D., McConnell I. Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar typhimurium. Infect. Immun. 2004;72:2152–2159. doi: 10.1128/IAI.72.4.2152-2159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arsenault R.J., Napper S., Kogut M.H. Salmonella enterica Typhimurium infection causes metabolic changes in chicken muscle involving AMPK, fatty acid and insulin/mTOR signaling. Vet. Res. 2013;44:35. doi: 10.1186/1297-9716-44-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vose D., Koupeev T., Mintiens K. A quantitative microbiological risk assessment of Salmonella spp. in broiler (Gallus gallus) meat production. EFSA Support. Publ. 2011;8:183E. doi: 10.2903/sp.efsa.2011.EN-183. [DOI] [Google Scholar]
- 11.Costa M.C., Bessegatto J.A., Alfieri A.A., Weese J.S., JoãoFilho A.B., Oba A. Different antibiotic growth promoters induce specific changes in the cecal microbiota membership of broiler chicken. PLoS ONE. 2017;12:e0171642. doi: 10.1371/journal.pone.0171642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadde U.D., Oh S., Lillehoj H.S., Lillehoj E.P. Antibiotic growth promoters virginiamycin in and bacitracin methylene disalicylate alter the chicken intestinal metabolome. Sci. Rep. 2018;8:3592. doi: 10.1038/s41598-018-22004-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Davison H.C., Woolhouse M.E., Low J.C. What is antibiotic resistance and how can we measure it? Trends Microb. 2000;8:554–559. doi: 10.1016/S0966-842X(00)01873-4. [DOI] [PubMed] [Google Scholar]
- 14.Orubu E.S.F., Zaman M.H., Rahman M.T., Wirtz V.J. Veterinary Anti-Microbial Resistance Containment in Bangladesh: Evaluating the National Action Plan and scoping the evidence on implementation. J. Glob. Antimicrob. Resist. 2019 doi: 10.1016/j.jgar.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Adeyi O.O., Baris E., Jonas O.B., Irwin A., Berthe F.C.J., Le Gall F.G., Marquez P.V., Nikolic I.A., Plante C.A., Schneidman M., et al. Drug-Resistant Infections: A Threat to Our Economic Future. World Bank Group; Washington, DC, USA: 2017. [Google Scholar]
- 16.Chereau F., Opatowski L., Tourdjman M., Vong S. Risk assessment for antibiotic resistance in South East Asia. BMJ. 2017;358:j3393. doi: 10.1136/bmj.j3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair J.M., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbial. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 19.Paphitou N.I. Antimicrobial resistance: Action to combat the rising microbial challenges. Int. J. Antimicrob. Agents. 2013;42:S25–S28. doi: 10.1016/j.ijantimicag.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Mthembu T.P., Zishiri O.T., El Zowalaty M.E. Molecular Detection of Multidrug-Resistant Salmonella Isolated From Livestock Production Systems In South Africa. Infect. Drug Resist. 2019;12:3537. doi: 10.2147/IDR.S211618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mthembu T.P., Zishiri O.T., El Zowalaty M.E. Detection and Molecular Identification of Salmonella Virulence Genes in Livestock Production Systems in South Africa. Pathogens. 2019;8:124. doi: 10.3390/pathogens8030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jajere S.M. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World. 2019;12:504–521. doi: 10.14202/vetworld.2019.504-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahmud M.S., Bari M.L., Hossain M.A. Prevalence of Salmonella serovars and antimicrobial resistance profiles in poultry of Savar area, Bangladesh. Foodborne Pathog. Dis. 2011;8:1111–1118. doi: 10.1089/fpd.2011.0917. [DOI] [PubMed] [Google Scholar]
- 24.Barua H., Biswas P.K., Olsen K.P., Shil S.K., Christensen J.P. Molecular characterization of motile serovars of Salmonella enterica from breeder and commercial broiler poultry farms in Bangladesh. PLoS ONE. 2013;8:e57811. doi: 10.1371/journal.pone.0057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Islam M.J., Mahbub-E-Elahi A.T.M., Ahmed T., Hasan M.K. Isolation and identification of Salmonella spp. from broiler and their antibiogram study in Sylhet, Bangladesh. J. Appl. Biol. Biotechnol. 2016;4:046–051. [Google Scholar]
- 26.Al Mamun M.A., Kabir S.L., Islam M.M., Lubna M., Islam S.S., Akhter A.T., Hossain M.M. Molecular identification and characterization of Salmonella species isolated from poultry value chains of Gazipur and Tangail districts of Bangladesh. Afr. J. Microbiol. Res. 2017;11:474–481. [Google Scholar]
- 27.Al Azad M., Rahman A., Rahman M., Amin R., Begum M., Ara I., Fries R., Husna A., Khairalla A.S., Badruzzaman A.T.M., et al. Susceptibility and multidrug resistance patterns of Escherichia coli isolated from cloacal swabs of live broiler chickens in Bangladesh. Pathogens. 2019;8:118. doi: 10.3390/pathogens8030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobur M.A., Ievy S., Haque Z.F., Nahar A., Zaman S.B., Rahman M.T. Emergence of colistin-resistant Escherichia coli in poultry, house flies, and pond water in Mymensingh, Bangladesh. J. Adv. Vet. Anim. Res. 2019;6:50–53. doi: 10.5455/javar.2019.f311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobur M.A., Sabuj A.A.M., Sarker R., Rahman A.M.M.T., Kabir S.M.L., Rahman M.T. Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance. Vet. World. 2019;12:984–993. doi: 10.14202/vetworld.2019.984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begum K., Mannan S.J., Ahmed A. Antibiotic resistance, plasmids and integron profile of Salmonella species isolated from poultry farm and patients. Dhaka Univ. J. Pharm. Sci. 2016;15:209–214. doi: 10.3329/dujps.v15i2.30939. [DOI] [Google Scholar]
- 31.Karim M.R., Giasuddin M., Samad M.A., Mahmud M.S., Islam M.R., Rahman M.H., Yousuf M.A. Prevalence of Salmonella spp. in poultry and poultry products in Dhaka, Bangladesh. Int. J. Anim. Biol. 2017;3:18–22. [Google Scholar]
- 32.Islam M.M., Islam M.N., Sharifuzzaman F.M., Rahman M.A., Sharifuzzaman J.U., Sarker E.H., Shahiduzzaman M., Mostofa M., Sharifuzzaman M.M. Isolation and identification of Escherichia coli and Salmonella from poultry litter and feed. Int. J. Nat. Soc. Sci. 2014;1:1–7. [Google Scholar]
- 33.Whiley H., Ross K. Salmonella and eggs: From production to plate. Int. J. Environ. Res. Public Health. 2015;12:2543–2556. doi: 10.3390/ijerph120302543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saelinger C.A., Lewbart G.A., Christian L.S., Lemons C. Prevalence of Salmonella spp. in cloacal, fecal, and gastrointestinal mucosal samples from wild North American turtles. J. Am. Vet. Med. Assoc. 2006;229:266–268. doi: 10.2460/javma.229.2.266. [DOI] [PubMed] [Google Scholar]
- 35.Bodí S.G., Garcia A.V., García S.V., Orenga C.M. Litter aeration and spread of Salmonella in broilers. Poultr. Sci. 2013;92:2005–2011. doi: 10.3382/ps.2013-03078. [DOI] [PubMed] [Google Scholar]
- 36.Malorny B., Löfström C., Wagner M., Krämer N., Hoorfar J. Enumeration of Salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment. Appl. Environ. Microbiol. 2008;74:1299–1304. doi: 10.1128/AEM.02489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlén G., Blomqvist S., Almståhl A., Carlén A. Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections. J. Oral. Microbiol. 2012;4:10855. doi: 10.3402/jom.v4i0.10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clifford K., Darash D., da Costa C.P., Meyer H., Islam M.T., Meyer H., Klohe K., Winklerc A., Rahman M.T., Islam M.T., et al. The threat of antimicrobial resistance opportunities for a technology-integrated One Health approach. Bull. World Health Organ. 2018;96:662–664. doi: 10.2471/BLT.18.209585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall B.M., Levy S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011;24:718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulvey M.R., Boyd D.A., Olson A.B., Doublet B., Cloeckaert A. The genetics of Salmonella genomic island 1. Microbes Infect. 2006;8:1915–1922. doi: 10.1016/j.micinf.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 41.Wright G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 42.Molla M.A.M. Poultry Feed Laced with Antibiotics. [(accessed on 10 November 2019)];The Daily Star. 2019 Apr 13; Available online: https://www.thedailystar.net/frontpage/news/poultry-feed-laced-antibiotics-1729231.
- 43.White D.G., Zhao S., Sudler R., Ayer S., Friedman S., Chen S., McDermott P.F., McDermott S., Wagner D.D., Meng J. The isolation of antibiotic-resistant Salmonella from retail ground meats. N. Engl. J. Med. 2001;345:1147–1154. doi: 10.1056/NEJMoa010315. [DOI] [PubMed] [Google Scholar]
- 44.Hall R.M., Collis C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 45.Meng X., Zhang Z., Li K., Wang Y., Xia X., Wang X., Xi M., Meng J., Cui S., Yang B. Antibiotic Susceptibility and Molecular Screening of Class I Integron in Salmonella isolates recovered from retail raw chicken carcasses in China. Microbial. Drug Resist. 2017;23:230–235. doi: 10.1089/mdr.2015.0359. [DOI] [PubMed] [Google Scholar]
- 46.Ogunleye A.O., Carlson S. Characterization of a multidrug resistant Salmonella enterica give isolated from a lizard captured in a poultry house in Nigeria. Afr. J. Biomed. Res. 2017;20:53–58. [Google Scholar]
- 47.Smith S.I., Seriki A., Ajayi A. Typhoidal and non-typhoidal Salmonella infections in Africa. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:1913–1922. doi: 10.1007/s10096-016-2760-3. [DOI] [PubMed] [Google Scholar]
- 48.FAO-WHO . Microbiological Risk Assessment Series 19. World Health Organization; Geneva, Switzerland: Food and Agriculture Organization of the United Nations; Rome, Italy: 2009. [(accessed on 20 February 2020)]. Salmonella and Campylobacter in chicken meat. Available online: https://www.who.int/foodsafety/publications/mra19/en/ [Google Scholar]
- 49.Saravanan S., Purushothaman V., Murthy T.R.G.K., Sukumar K., Srinivasan P., Gowthaman V., Balusamy M., Atterbury R., Kuchipudi S.V. Molecular epidemiology of Nontyphoidal Salmonella in poultry and poultry products in India: Implications for human health. Indian J. Microbiol. 2015;55:319–326. doi: 10.1007/s12088-015-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jahan M.I., Rahaman M.M., Hossain M.A., Sultana M. Occurrence of intI1-associated VIM-5 carbapenemase and co-existence of all four classes of β-lactamase in carbapenem-resistant clinical Pseudomonas aeruginosa DMC-27b. J. Antimicrob. Chemother. 2019;75:86–91. doi: 10.1093/jac/dkz426. [DOI] [PubMed] [Google Scholar]
- 51.Sobur A., Haque Z.F., Sabuj A.A., Ievy S., Rahman A.T., El Zowalaty M.E., Rahman T. Molecular detection of multidrug and colistin-resistant Escherichia coli isolated from house flies in various environmental settings. Future Microb. 2019;14:847–858. doi: 10.2217/fmb-2019-0053. [DOI] [PubMed] [Google Scholar]
- 52.Fratamico P.M. Comparison of culture, polymerase chain reaction (PCR), TaqMan Salmonella, and Transia Card Salmonella assays for detection of Salmonella spp. in naturally-contaminated ground chicken, ground turkey, and ground beef. Mol. Cell. Probes. 2003;1:7215–7221. doi: 10.1016/S0890-8508(03)00056-2. [DOI] [PubMed] [Google Scholar]
- 53.O’Regan E., McCabe E., Burgess C., McGuinness S., Barry T., Duffy G. Development of a real‑time multiplex PCR assay for the detection of multiple Salmonella serotypes in chicken samples. BMC Microbiol. 2008;8:156. doi: 10.1186/1471-2180-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol-2009. ASM Microbe Library, American Society for Microbiology; New York, NY, USA: 2016. [(accessed on 29 August 2019)]. pp. 1–23. Available online: https://www.asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-DiffusionSusceptibility-Test-Protocol-pdf.pdf. [Google Scholar]
- 55.CLSI . Performance Standards for Antimicrobial Susceptibility Testing: 17th Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2016. [Google Scholar]
- 56.Sweeney M.T., Lubbers B.V., Schwarz S., Watts J.L. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018;73:1460–1463. doi: 10.1093/jac/dky043. [DOI] [PubMed] [Google Scholar]
- 57.Randall L.P., Cooles S.W., Osborn M.K., Piddock L.J.V., Woodward M.J. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004;53:208–216. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- 58.Walker R.A., Lindsay E., Woodward M.J., Ward L.R., Threlfall E.J. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb. Drug Resist. 2001;7:13–21. doi: 10.1089/107662901750152701. [DOI] [PubMed] [Google Scholar]
- 59.Sobur A., Hasan M., Haque E., Mridul A.I., Noreddin A., El Zowalaty M.E., Rahman T. Molecular Detection and Antibiotyping of Multidrug-Resistant Salmonella Isolated from Houseflies in a Fish Market. Pathogens. 2019;8:191. doi: 10.3390/pathogens8040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Navia M.M., Ruiz J., Vila J. Molecular characterization of the integrons in Shigella strains isolated from patients with traveler’s diarrhea. Diagn. Microbiol. Infect. Dis. 2004;48:175–179. doi: 10.1016/j.diagmicrobio.2003.10.007. [DOI] [PubMed] [Google Scholar]